Abstract
The current COVID-19 pandemic caused by a novel coronavirus SARS-CoV-2 urgently calls for a working therapeutic. Here we report a computation-based workflow for efficiently selecting a subset of FDA-approved drugs that can potentially bind to the SARS-CoV-2 main protease MPRO. The workflow started with docking (using Autodock Vina) each of 1615 FDA-approved drugs to the MPRO active site. This step selected 62 candidates with docking energies lower than −8.5 kcal/mol. Then, the 62 docked protein-drug complexes were subjected to 100 ns of molecular dynamics (MD) simulations in a molecular mechanics (MM) force field (CHARMM36). This step reduced the candidate pool to 26, based on the root-mean-square-deviations (RMSDs) of the drug molecules in the trajectories. Finally, we modeled the 26 molecules by a pseudo-quantum mechanical (ANI) force field, and ran 5-ns hybrid ANI/MM MD simulations of the 26 protein-drug complexes. ANI was trained by neural network models on quantum mechanical density functional theory (wB97X/6-31G(d)) data points. An RMSD cutoff winnowed down the pool to 12, and free energy analysis (MM/PBSA) produced the final selection of 9 drugs: dihydroergotamine, midostaurin, ziprasidone, etoposide, apixaban, fluorescein, tadalafil, rolapitant, and palbociclib. Of these, three are found to be active in literature reports of experimental studies. To provide physical insight into their mechanism of action, the interactions of the drug molecules with the protein are presented as 2D-interaction maps. These findings and mappings of drug-protein interactions may be potentially used to guide rational drug discovery against COVID-19.
Keywords: COVID-19, molecular dynamics, neural network, wB97X/6-31G(d), ANI/MM
Graphical Abstract
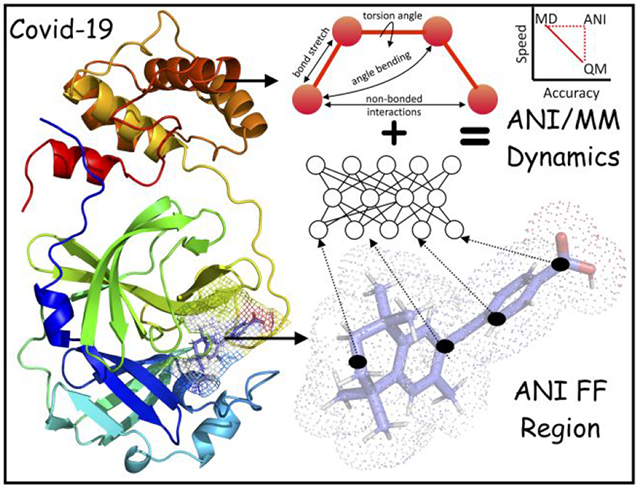
Introduction
The COVID-19 pandemic, caused by the novel coronavirus SARS-CoV-2 with crown-like spikes on the surface (Figure 1), is wreaking havoc on the whole world1. Since the outbreak of COVID-19 in late 2019, more than 36 million cases have been reported with over 1 million fatalities (source: Worldometer, Oct 8th, 2020). Coronaviruses can infect mammals and can then easily mutate to enable transfer from animals to humans2. SARS-CoV-2 spreads mainly from human to human and is rapidly becoming the world’s leading cause of death. Currently, no targeted vaccines or treatments are as yet available for SARS-CoV-2, and there is an urgent need to develop them. The aim of the present study is to use computational approaches to explore protein-drug interactions that can be useful in the fight against COVID-19.
Figure 1. Replication cycle of SARS-CoV-2.
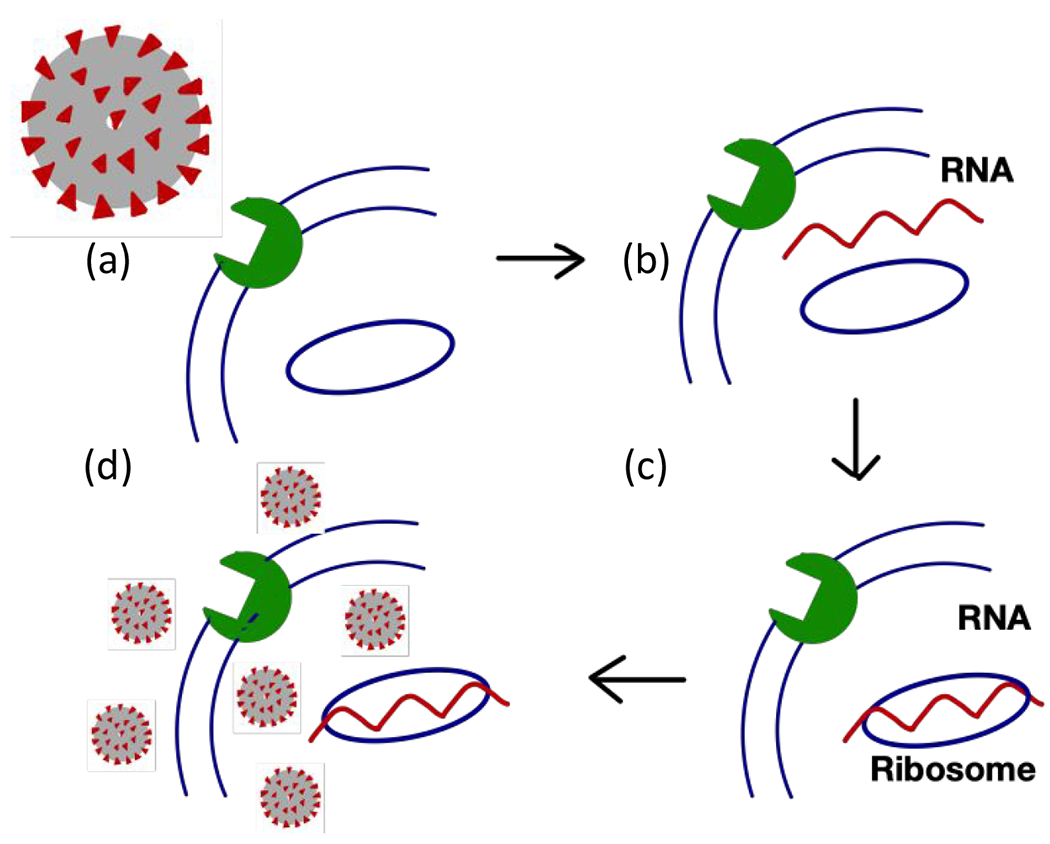
The virus invades a human cell by attaching its spike protein to a cell surface receptor (a). Upon entering the cell, the virus breaks up to release its genetic material (b). The viral RNA hijacks the ribosome of the host cell to produce viral proteins (c). Viral proteins and RNA are assembled into new viral particles, which are eventually released from the host cell to infect other cells (d). The vial main protease (MPRO) is essential for cleaving the viral polypeptide chain into functional proteins needed to assemble new viruses.
The main protease, or MPRO, of SARS-CoV-2 was identified shortly after the outbreak and its crystal structure was solved (Protein Data Bank (PDB) entry: 6LU7)3. There are now significant efforts aimed at developing drugs that can inhibit MPRO. However, no inhibitors against MPRO or other targets are available to treat COVID-19, as drug discovery is an expansive and time-consuming process4. When a new target protein is identified, a potential shortcut is to test, or repurpose, drugs that are FDA-approved. The concept of drug repurposing has proven to be successful in the past and is the most convenient method for screening drugs for novel diseases. Computations based on drug repurposing have identified the HIV antivirals Lopinavir and Ritonavir as potentially effective against COVID-19 5–6. The latter group and others have conducted drug repurposing computational analyses specifically targeting MPRO 6–9.
With the current advances in computational techniques in combination with physical chemistry methods that utilize machine learning algorithms, we are witnessing numerous impressive predictions in the field of drug discovery. These computations are used for screening and prediction of binding affinities and to generate fingerprint interactions with target proteins10. Protein-drug docking and molecular dynamics (MD) simulations can reveal interaction fingerprints that potentially hold key to design other potent drugs11. The accuracy of MD simulations relies on the parameterization of the forcefields12. Classical molecular mechanics (MM) forcefields, such as CHARMM13, AMBER14, and OPLS15, can probably model 99% of the properties of biomolecular systems by solving Newton’s equation of motion. However, the crucial 1% involves quantum chemistry (i.e., electronic and nuclear interactions) and is beyond the realm of classical forcefields16. Quantum chemistry methods, in particular density functional theory (DFT) and highly accurate coupled cluster (CCSD(T)/CBS), can provide accurate solutions to Schrödinger’s equation, but are too expensive both for large systems and for large-scale uses on even relatively small systems. To bridge the gap, machine learning methods, especially those based on neural networks, with augmentations in data, have become powerful to improve scalability without sacrificing accuracy17–18. Recently, the Roitberg group developed a suite of ANI forcefields, including ANI-2x and ANI-1ccx, that uses neural network-based training19–20. ANI-2x was trained on millions of small molecules, covering C-H-N-O-S-F-Cl atoms, against their DFT energies, whereas ANI-1ccx was trained on 500 thousand CCSD(T)/CBS data points but limited to C-H-N-O atoms. ANI-2x has similar accuracy to DFT but is 106 times faster, a speed that matches classical force fields. While DFT is limited approximately to 500 atoms and CCSD(T)/CBS to 10 atoms, ANI can be used on systems with ~10,000 atoms19–20. Moreover, with a speed comparable to classical forcefields, ANI is suitable for large-scale uses such as in drug screening or refinement.
Here we report computational drug repurposing against the MPRO protein using a workflow that encompasses several levels of sophistication, from docking all the way to MD simulations with the ANI-2x force field. Starting with 1615 FDA-approved drugs, docking selected the 62 most promising candidates. MD simulations with the CHARMM36 MM force field trimmed this list down to 26. Hybrid ANI/MM MD simulations produced a final list of 9 drugs, of which 3 are found to be active according to literature reports. Free energy analysis (based on MM/PBSA) and interaction mapping provided additional insight into the mechanism of target inhibition and guidance for rational drug discovery against COVID-19.
Computational Methodologies
Molecular Docking.
The crystal structure of MPRO (PDB entry 6LU7 chain A)3 was downloaded from the RCSB Protein Data Bank. To prepare MPRO for docking, we used AutoDock Tools (ADT)21 to assign charges and atom/bond types. For drug repurposing, we chose a database of 1615 drugs that are FDA-approved and readily available in the market. We obtained docking ready drugs from ZINC1522 and used open babel codes23 to perform file format conversion from SDF (structure data file from ZINC15) to PDBQT (used by Autodock Vina24). Screening against MPRO was performed using Autodock Vina, based on a 28 × 28 × 28 Å3 grid box centered at the active site, i.e., the pocket where the N3 inhibitor was bound in the crystal structure. Docking of each drug produced a score for filtering as well as a pose for further validation by MD simulations.
Classical Molecular Dynamics Simulations.
All MD simulations were conducted using NAMD25. We used the CHARMM-GUI Webserver26 to generate the CHARMM36m parameters and topology files for the protein, and the SwissParam server27 to generate topology and parameters for the drugs. Each protein-drug complex (produced by Autodock Vina) was solvated in a triclinic box using the TIP3P water model28. 0.15 M ions (Na+ and Cl−) were added to provide charge neutralization and electrostatic screening. The systems were subjected to 5000 steps of steepest descent energy minimization and equilibration under constant NVT (1 ns ) and constant NPT (2 ns). During the equilibration, position restraints were applied to both protein and drug molecules. The temperature (303 K) and pressure (1 atm) were controlled by the Langevin and Langevin piston methods29. The particle mesh Ewald method was used to treat long-range electrostatic interactions30. A 100-ns production run was then carried out for each equilibrated system at constant NPT without restraints. Snapshots were evenly sampled from 20 to 100 ns of the production run to (1) calculate average lig-RMSD, i.e., average (over 8000 snapshots) of the root-mean-square-deviations of the drug, after aligning the protein secondary structural elements to the snapshot at 20 ns; and (2) carry out MM/PBSA analysis (over 800 snapshots; see below).
Hybrid ANI/MM Molecular Dynamics Simulations.
We combined the accurate ANI-2x forcefield for drugs with the CHARMM36m/TIP3P forcefields for proteins and solvent to run hybrid ANI/MM MD simulations31 of the MPRO-drug complexes (Figure 2), as implemented in the NAMD package32. In these hybrid ANI/MM simulations, the total potential energy (U) of the system was defined as the sum of the energies of the ANI region (i.e., the drug molecule) and the MM region (protein and solvent) and the interaction energy between the drug and the MM region31:
| (1) |
Figure 2. Structure of MPRO and the system for ANI/MM MD simulations.
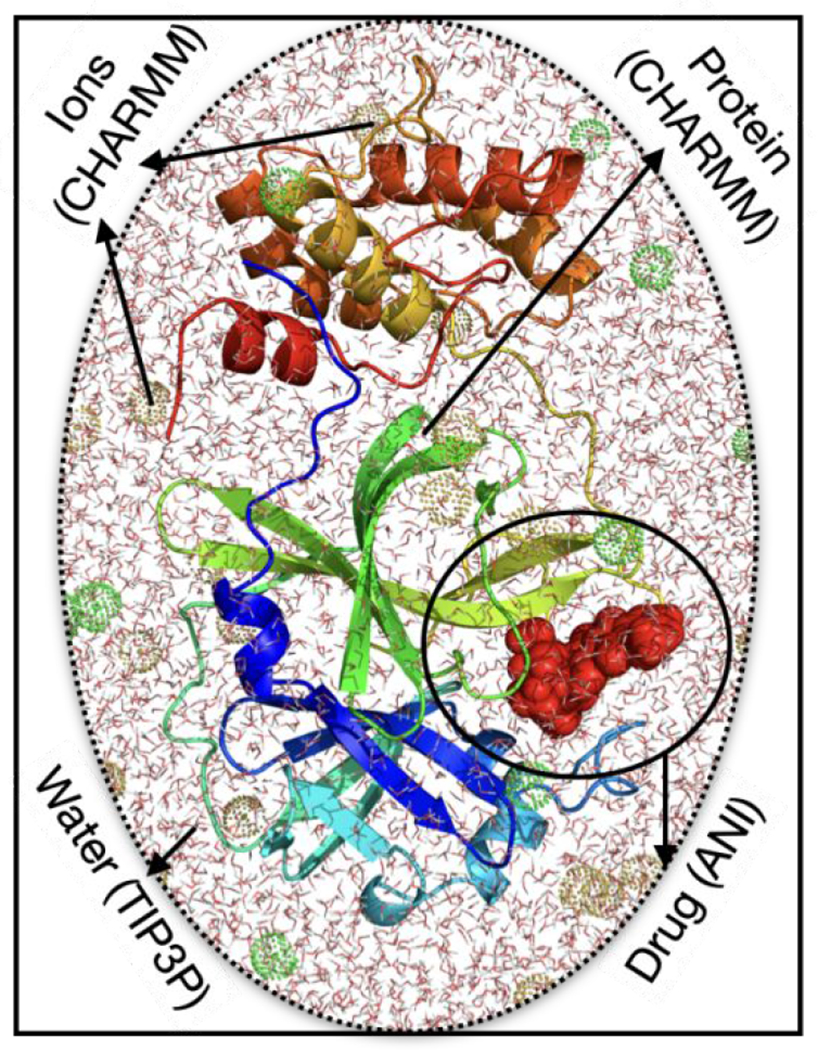
The system consists of a drug (red surface) bound to MPRO (cartoon representation), solvated with TIP3P water molecules and Na+ and Cl− ions (green and brown mesh bubbles) in a box shown with line representation. For the hybrid ANI/MM MD simulations, the protein and ions are modeled by the CHARMM forcefield, water is modeled as TIP3P, and ligand molecule is modeled by the ANI-2x force field.
The term comprised MM nonbonded interactions between the MM region and the drug, i.e., Coulombic and Lennard-Jones interactions between the ANI atoms and MM atoms31:
| (2) |
Starting from the last snapshot of the classical MD simulations, we ran 5 ns of ANI/MM MD simulations for each selected protein-drug complex. The NAMD input script for the ANI/MM simulations is listed in Supporting Information. From the 5 ns simulations, we sampled 2500 snapshots to calculate the average lig-RMSD (with the final snapshot of the classical MD simulations as the reference).
MM/PBSA Free Energy Calculations.
To compute the MM/PBSA free energy of protein-drug binding, we used CaFe (open source code for calculation of free energy) developed by Liu et al.33. MM/PBSA is an endpoint method for estimating binding free energies, by combining the molecular mechanics term for the gas-phase energy and Poisson-Boltzmann and surface area terms for polar and nonpolar solvation energies, respectively. Specifically, the MM term was similar to in equation (2) but the interaction was limited to between drug and protein atoms. The PB term, , was obtained by the APBS program34 (interfaced to CaFe), where the boundary conditions were set to Debye-Hückel values and charges were mapped to grids using cubic B spline. The SA term, , was calculated with the surface tension set to 0.00542 kcal/mol/Å2 and an offset of 0.92 kcal/mol. Finally, the binding free energy was summed and averaged over saved snapshots:
| (3) |
The MM/PBSA calculations were done on simulations of the complex only. Because of inaccuracy in conformational entropy calculations, we did not include such entropic contributions. Neglect of entropy tends to make larger ligands overly favorable. The same energy function was used whether the snapshots were from the classical MD simulations or from hybrid ANI/MM simulations. For the latter, we sampled 500 snapshots from the 5 ns trajectories.
Results
Our computational drug repurposing workflow against MPRO, the main protease of SARS-CoV-2, started with docking 1615 FDA-approved drugs (downloaded in dock ready form from ZINC15) to the active site of the MPRO crystal structure, using AutoDock Vina. Docking for each drug produced a score, representing the binding energy, and a pose for the protein-drug complex. After ranking the docking scores, we selected 62 candidates with scores equal to or less than −8.5 kcal/mol for further evaluations.
To assess the reliability of the docking step, we exhaustively searched the literature for experimental information on the inhibitory activities of the 62 candidates. We found 10 of the 62 candidates with reported IC50 or KD data against MPRO, and divided the 10 into three categories according to efficacy: active (A) with IC50 < 10 μM; or KD < 100 μM; moderately active (MA) with 10 μM < IC50 < 20 μM or 100 μM < KD < 200 μM; and inactive (I) with IC50 > 20 μM or KD > 200 μM3, 35–36. Among the docking-selected candidates, 3, 3, and 4 are in the A, MA, I categories, respectively. The A-category drugs are atovaquone (IC50 = 1.5 μM)37, midostaurin (KD = 43.5 μM)35, and tadalafil (KD = 52.2 μM)35. The MA-category drugs are dihydroergotamine (KD = 107.6)35, simeprevir (IC50 = 13.74 μM)36, and mefloquine (IC50 = 14.1 μM)38. The I-category drugs are pimozide (IC50 = 42 μM)39, itraconazole (IC50 = 111 μM)39, amphotericin B (reported as “did not inhibit SARS-CoV-2 infection”)40, and azelastine (IC50 = 20-100 μM)41.
As negative control, we took a random sample of 62 drugs that were filtered by the docking step (i.e., with score > −8.5 kcal/mol; Supporting Information Table S1), and searched for experimental information on them. Only two of these drugs were found in experimental studies. Elbasvir “did not inhibit SARS-CoV-2 infection” 40, which suggests that it is inactive against MPRO. On the other hand, quinidine showed some activity in a SARS-CoV-2 replication inhibition assay 42, but the possible target proteins were unknown. Taken together, we conclude that the docking step is successful in selecting candidates that are likely to be effective in inhibiting MPRO, albeit with a tendency to also predict false positives.
In order to reduce the pool of drug candidates and hopefully filter out the false positives from the docking step, we turned to MD simulations with a classical forcefield (CHARMM36m), starting with the docking-generated pose for each drug. We were able to obtain parameters for 58 of the 62 drug candidates from the SwissParam server 27. For each of the 58 protein-drug complex, we carried out 100 ns classical MD simulations. From the last 80 ns of the simulations, we calculated the average lig-RMSD and MM/PBSA binding free energies for each of the 58 drug candidates (Fig. 3). We then used lig-RMSD as a filter: drug with lig-RMSD > 4 Å, indicating unstable binding, were filtered, while drugs with lig-RMSD < 4 Å, of which there were 26, were selected for further evaluation in the next step. Comparing the 26 selected candidates against the 10 drugs with experimental information for MPRO binding, one (pimozide) of the 4 drugs in the I category was correctly filtered, but we also lost one (atovaquone) in the A category and one (simeprevir) in the MA category. So the retained drugs in the A, MA, and IA categories were 2, 2, and 3, respectively. The docking scores, lig-RMSDs, and 2D structures of the selected 26 drugs are shown in Supporting Information Table S2.
Figure 3. Average lig-RMSDs and MM/PBSA binding free energies for 58 drugs in classical MD simulations.
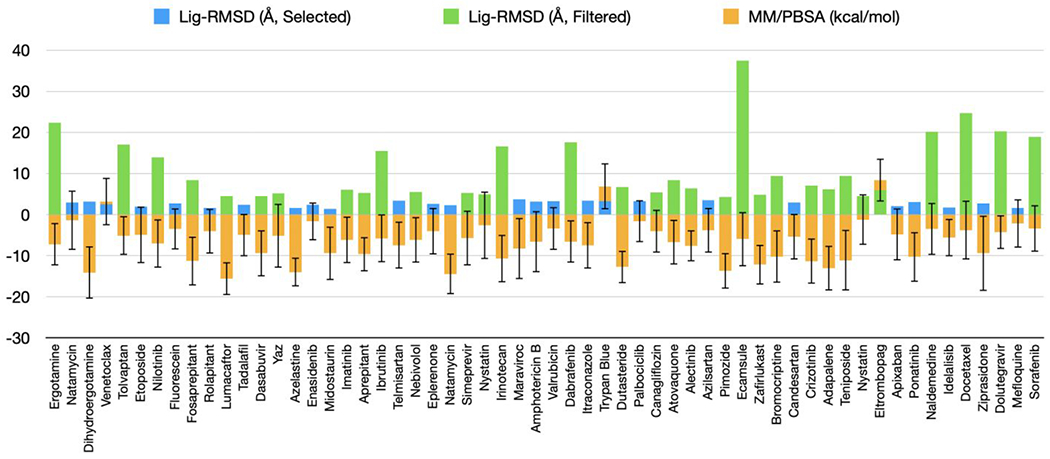
The blue bars represent drugs with an average lig-RMSD below 4 Å and green bars represent drugs with an average lig-RMSD above 4 Å. The orange bars represent average MMPBSA binding free energies (in kcal/mol) with standard deviations. The drugs are ordered according to docking scores. Not included are 4 drugs with no available force-field parameters.
To further winnow down the list of candidate drugs and potentially refine the protein-drug poses, we ran 5 ns hybrid ANI/MM MD simulations. Filtering first by the average lig-RMSD, at a 5 Å cutoff, selected 12 drugs (Figure 4). All the three drugs in the I category were now correctly removed, along with one in the MA category. So now two active drugs and one moderately active drug, but no inactive drugs, were in the selection. We also added a second filter, by MM/PBSA binding free energy. Three of the 12 drugs with MM/PBSA binding free energy > 0 kcal/mol were further removed. The final set of 9 drugs still contain the experimentally validated two active ones (midostaurin and tadalafil) and one moderately active one (dihydroergotamine). Moreover, two of the active drugs, dihydroergotamine and midostaurin, have the lowest MM/PBSA binding free energies, −17.9 and −16.2 kcal/mol, respectively, among the final set of 9 drugs. The 3D structures of dihydroergotamine and midostaurin bound to MPRO are shown in Figure 5.
Figure 4. Average lig-RMSD and MM/PBSA binding free energies in ANI/MM MD simulations.
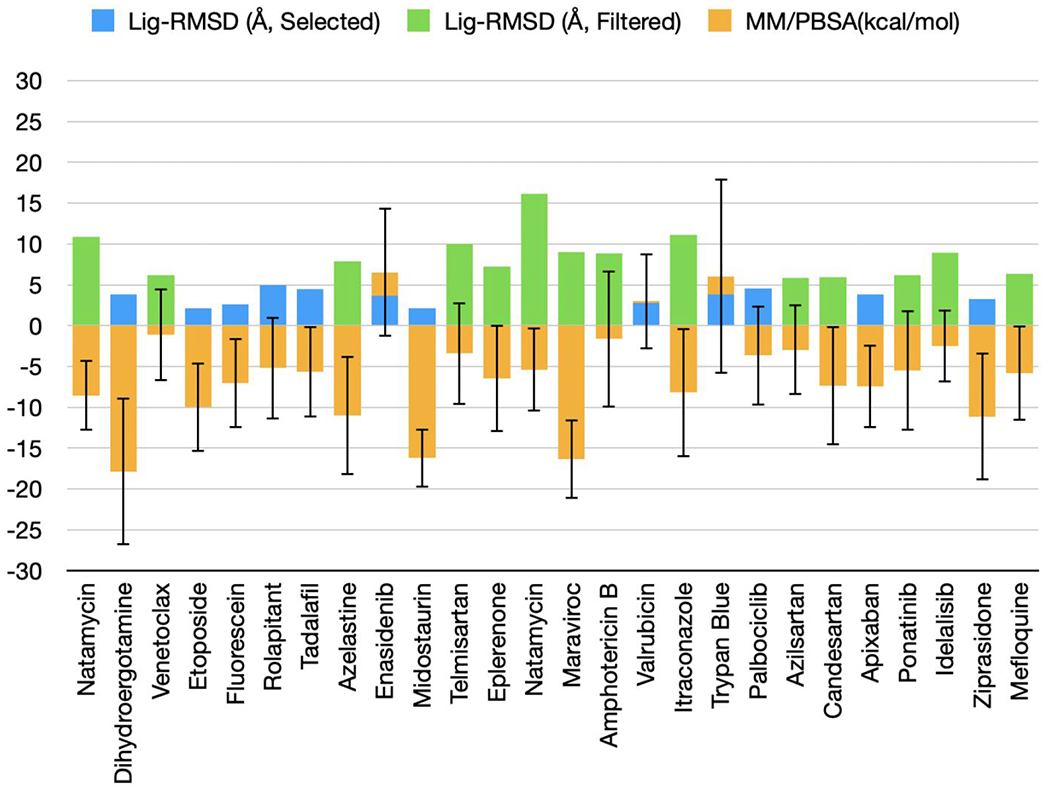
The blue bars represent selected drugs with average lig-RMSD below 5 Å and green bars represent filtered drugs with lig-RMSD above 5 Å. The orange bars represent average MM/PBSA binding free energies with standard deviations. The drugs are ordered according to average lig-RMSD.
Figure 5. 3D structures of dihydroergotamine and midostaurin in complex with MPRO.
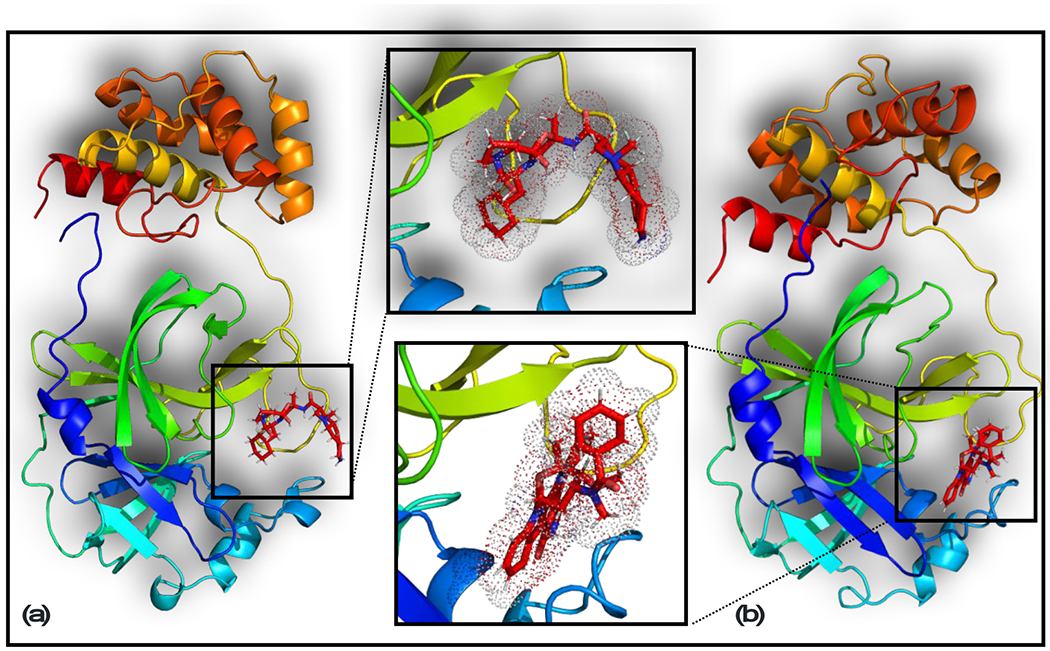
The last snapshots of these complexes in ANI/MM MD simulations are shown. (a) Dihydroergotamine; (b) Midostaurin.
The MM/PBSA binding free energies and their decompositions for the 26 candidates evaluated by ANI/MM MD simulations are listed in Supporting Information Table S3. We also compared these results with the counterparts calculated from the classical MD simulations (Figure 3). For all the 9 drugs in the final selection, the MM/PBSA binding free energies improved on going from the classical MD simulations to ANI/MM MD simulations, with an average decrease of −3.0 kcal/mol. (Supporting Information Table S4). In comparison, among the 17 filtered candidates, 9 had increases in MM/PBSA binding free energies on going from the classical MD simulations to ANI/MM MD simulations. So the ANI/MM MD simulations clearly improved both the reliabilities of the drug selection and the interactions of the selected drugs with the target proteins. This is especially notable since the MM/PBSA energy function was the same and it was the refined protein-drug configurations that were responsible for the enhanced protein-drug binding stability in the ANI/MM MD simulations.
To gain further insight into the enhanced protein-drug interactions by ANI, we compared the last snapshots from the classical and ANI/MM MD simulations of the final 9 drugs. The results are presented as 2D interaction maps in Figure 6. ANI/MM produced additional interactions (hydrogen bonding and nonbonded interactions) not sampled in classical MD simulations. For example, dihydroergotamine formed additional hydrogen bonds, whereas midostaurin formed additional nonbonded interactions in the ANI/MM snapshots. Thus ANI was indeed able to refine protein-drug poses.
Figure 6. 2D interaction maps of the final selection of 9 drugs with MPRO.
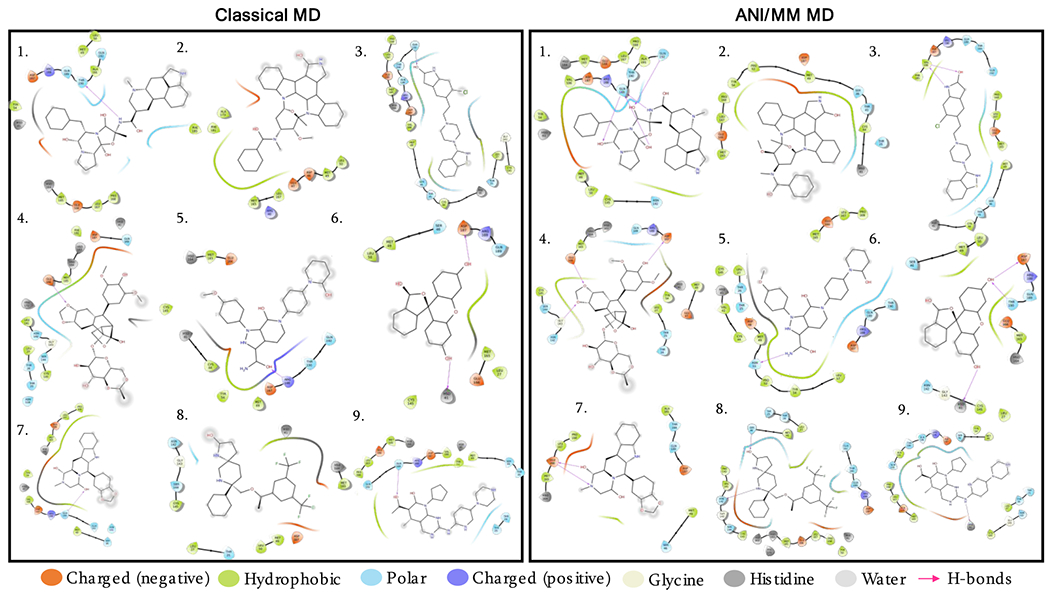
The last snapshots of classical and ANI/MM MD trajectories are used to generate the 2D interaction maps. Colored drops represent different properties of interacting residues of the protein. 1. Dihydroergotamine; 2. Midostaurin; 3. Ziprasidone; 4. Etoposide; 5. Apixaban; 6. Fluorescein; 7. Tadalafil; 8. Rolapitant; 9. Palbociclib. Drugs are in ascending orders of their ANI/MM MM/PBSA binding free energies.
Discussion and Conclusion
We have presented an investigation on the development of potential inhibitors against the main protease of SARS-CoV-2 using a computational drug repurposing approach. The scientific community is devoting a tremendous amount of effort to characterizing potential drugs to inhibit this virus, yet much more information and effort is required before a unique treatment can be approved43. Systematic studies on the interactions of viral proteins with FDA-approved drugs are crucial for understanding the binding behaviors of these proteins and can aid in accelerating the development of biochemical assays44. MD simulations with classical forcefields can describe many important drug-protein interactions, but they tend to miss crucial details at the electronic and nuclear levels16. These missed details can be recovered when classical forcefields are combined with quantum calculations such as DFT and CCSD(T)/CBS that can provide the most accurate descriptions of electronic and nuclear effects for small drug molecules. In this work, a neural network-trained force field was used to study interactions of repurposed drug molecules with the MPRO protein. The workflow developed in this study (Figure 7), the interaction maps, and the structures of selected protein-drug complexes may be useful for designing novel drugs that can be used against COVID-19.
Figure 7. Computational drug repurposing workflow.
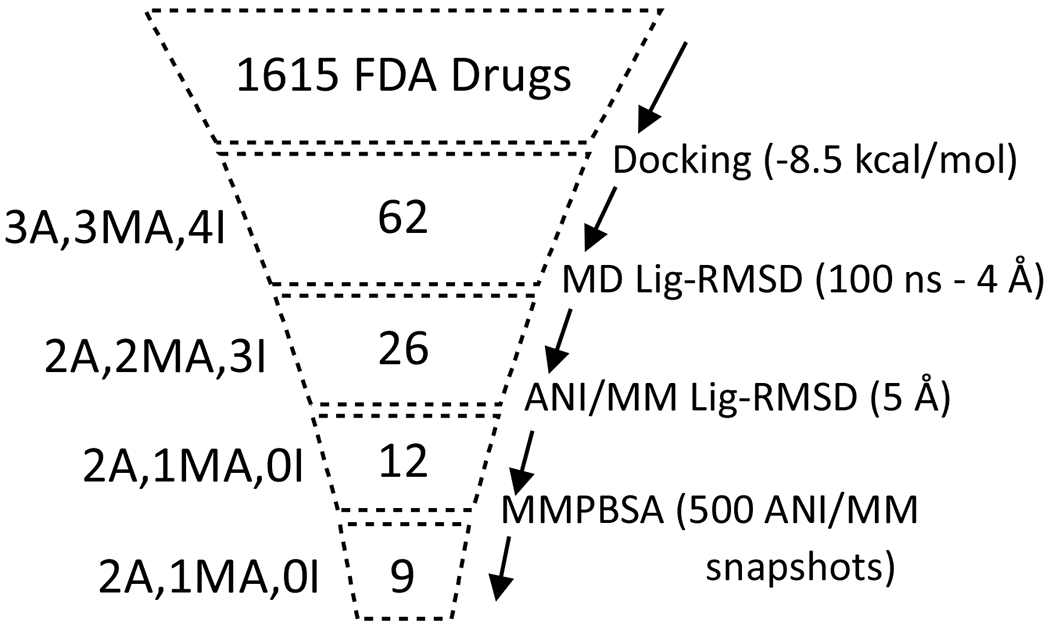
The number of selected compounds at each step is shown, with the corresponding method of selection indicated on the right. Shown the left are the numbers of experimentally studied compounds in three categories: A (active), MA (moderately active), and I (inactive), that are retained in each step.
Our workflow encompasses computations at several levels of sophistication. Our starting point is a database of 1615 FDA-approved drugs. Using molecular docking, the number of candidate drugs was reduced to 62 with the best docking scores. To sample the conformational space of drug-protein complexes, MD simulations were conducted first using a classical force field alone, which were then combined with the neural network-trained force field ANI to provide an accurate description of the interaction profiles of the drugs with the protein. Whereas docking and classical MD simulations are routinely used in drug discovery, here we used hybrid ANI/MM MD simulations to investigate interactions that may be ignored by classical MD simulations but could prove useful for guiding experimental drug design. Additionally, combining ANI/MM MD trajectories with endpoint MM/PBSA free-energy calculations assists in obtaining physically important information related to drug binding. The MM/PBSA calculations can be further improved in the future to handle the ANI/MM interactions.
We have used experimental information in the literature to assess each step of our workflow. Among the 62 docking-selected candidates, 10 compounds had experimental data on their MPRO binding affinities, with 6 active or moderately active and 4 inactive. We tracked whether these compounds were filtered or selected in each step of our workflow. Our final selection of 9 drugs contained three of the experimentally validated active or moderately active compounds and none of the inactive compounds. The workflow thus appears to be very effective according to this measure. Importantly, our ANI/MM MD simulations improved the binding stability of the 9 selected drugs.
Supplementary Material
Acknowledgement
This work was supported by National Institutes of Health Grant GM118091.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Supporting Information
The Supporting Information is available at.
References
- 1.Xu Z; Shi L; Wang Y; Zhang J; Huang L; Zhang C; Liu S; Zhao P; Liu H; Zhu L; Tai Y; Bai C; Gao T; Song J; Xia P; Dong J; Zhao J; Wang FS, Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020, 8 (4), 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J; Li F; Shi ZL, Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019, 17 (3), 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Z; Du X; Xu Y; Deng Y; Liu M; Zhao Y; Zhang B; Li X; Zhang L; Peng C; Duan Y; Yu J; Wang L; Yang K; Liu F; Jiang R; Yang X; You T; Liu X; Yang X; Bai F; Liu H; Liu X; Guddat LW; Xu W; Xiao G; Qin C; Shi Z; Jiang H; Rao Z; Yang H, Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582 (7811), 289–293. [DOI] [PubMed] [Google Scholar]
- 4.Hughes JP; Rees S; Kalindjian SB; Philpott KL, Principles of early drug discovery. Br J Pharmacol 2011, 162 (6), 1239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nutho B; Mahalapbutr P; Hengphasatporn K; Pattaranggoon NC; Simanon N; Shigeta Y; Hannongbua S; Rungrotmongkol T, Why Are Lopinavir and Ritonavir Effective against the Newly Emerged Coronavirus 2019? Atomistic Insights into the Inhibitory Mechanisms. Biochemistry 2020, 59 (18), 1769–1779. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Fast Identification of Possible Drug Treatment of Coronavirus Disease-19 (COVID-19) through Computational Drug Repurposing Study. J Chem Inf Model 2020, 60 (6), 3277–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elmezayen AD; Al-Obaidi A; Sahin AT; Yelekci K, Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J Biomol Struct Dyn 2020, DOI: 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huynh T; Wang H; Luan B, In Silico Exploration of the Molecular Mechanism of Clinically Oriented Drugs for Possibly Inhibiting SARS-CoV-2’s Main Protease. J Phys Chem Lett 2020, 11 (11), 4413–4420. [DOI] [PubMed] [Google Scholar]
- 9.Muralidharan N; Sakthivel R; Velmurugan D; Gromiha MM, Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J Biomol Struct Dyn 2020, DOI: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 10.Vamathevan J; Clark D; Czodrowski P; Dunham I; Ferran E; Lee G; Li B; Madabhushi A; Shah P; Spitzer M; Zhao S, Applications of machine learning in drug discovery and development. Nat Rev Drug Discov 2019, 18 (6), 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sliwoski G; Kothiwale S; Meiler J; Lowe EW Jr., Computational methods in drug discovery. Pharmacol Rev 2014, 66 (1), 334–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang LP; Martinez TJ; Pande VS, Building Force Fields: An Automatic, Systematic, and Reproducible Approach. J Phys Chem Lett 2014, 5 (11), 1885–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanommeslaeghe K; Hatcher E; Acharya C; Kundu S; Zhong S; Shim J; Darian E; Guvench O; Lopes P; Vorobyov I; Mackerell AD Jr., CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 2010, 31 (4), 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J; Wolf RM; Caldwell JW; Kollman PA; Case DA, Development and testing of a general amber force field. J Comput Chem 2004, 25 (9), 1157–74. [DOI] [PubMed] [Google Scholar]
- 15.Harder E; Damm W; Maple J; Wu C; Reboul M; Xiang JY; Wang L; Lupyan D; Dahlgren MK; Knight JL; Kaus JW; Cerutti DS; Krilov G; Jorgensen WL; Abel R; Friesner RA, OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J Chem Theory Comput 2016, 12 (1), 281–296. [DOI] [PubMed] [Google Scholar]
- 16.Eichinger M; Tavan P; Hutter J; Parrinello M, A hybrid method for solutes in complex solvents: Density functional theory combined with empirical force fields. J Chem Phys 1999, 110 (21), 10452–10467. [Google Scholar]
- 17.Balabin RM; Lomakina EI, Neural network approach to quantum-chemistry data: accurate prediction of density functional theory energies. J Chem Phys 2009, 131 (7), 074104. [DOI] [PubMed] [Google Scholar]
- 18.Morawietz T; Behler J, A density-functional theory-based neural network potential for water clusters including van der Waals corrections. J Phys Chem A 2013, 117 (32), 7356–66. [DOI] [PubMed] [Google Scholar]
- 19.Devereux C; Smith JS; Davis KK; Barros K; Zubatyuk R; Isayev O; Roitberg AE, Extending the Applicability of the ANI Deep Learning Molecular Potential to Sulfur and Halogens. J Chem Theory Comput 2020, 16 (7), 4192–4202. [DOI] [PubMed] [Google Scholar]
- 20.Smith JS; Nebgen BT; Zubatyuk R; Lubbers N; Devereux C; Barros K; Tretiak S; Isayev O; Roitberg AE, Approaching coupled cluster accuracy with a general-purpose neural network potential through transfer learning. Nat Commun 2019, 10 (1), 2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forli S; Huey R; Pique ME; Sanner MF; Goodsell DS; Olson AJ, Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 2016, 11 (5), 905–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sterling T; Irwin JJ, ZINC 15--Ligand Discovery for Everyone. J Chem Inf Model 2015, 55 (11), 2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Boyle NM; Banck M; James CA; Morley C; Vandermeersch T; Hutchison GR, Open Babel: An open chemical toolbox. J Cheminform 2011, 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trott O; Olson AJ, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010, 31 (2), 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kale L; Schulten K, Scalable molecular dynamics with NAMD. J Comput Chem 2005, 26 (16), 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jo S; Kim T; Iyer VG; Im W, CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 2008, 29 (11), 1859–1865. [DOI] [PubMed] [Google Scholar]
- 27.Zoete V; Cuendet MA; Grosdidier A; Michielin O, SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem 2011, 32 (11), 2359–2368. [DOI] [PubMed] [Google Scholar]
- 28.Price DJ; Brooks CL 3rd, A modified TIP3P water potential for simulation with Ewald summation. J Chem Phys 2004, 121 (20), 10096–10103. [DOI] [PubMed] [Google Scholar]
- 29.Feller SE; Zhang YH; Pastor RW; Brooks BR, Constant pressure molecular dynamics simulation: The Langevin piston method. J Chem Phys 1995, 103 (11), 4613–4621. [Google Scholar]
- 30.Essmann U; Perera L; Berkowitz ML; Darden T; Lee H; Pedersen LG, A smooth particle mesh Ewald method. J Chem Phys 1995, 103 (19), 8577–8593. [Google Scholar]
- 31.Lahey S-LJ; Rowley CN, Simulating protein-ligand binding with neural network potentials. Chem Sci 2020, 11 (9), 2362–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melo MCR; Bernardi RC; Rudack T; Scheurer M; Riplinger C; Phillips JC; Maia JDC; Rocha GB; Ribeiro JV; Stone JE; Neese F; Schulten K; Luthey-Schulten Z, NAMD goes quantum: an integrative suite for hybrid simulations. Nat Methods 2018, 15 (5), 351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H; Hou T, CaFE: a tool for binding affinity prediction using end-point free energy methods. Bioinformatics 2016, 32 (14), 2216–2218. [DOI] [PubMed] [Google Scholar]
- 34.Baker NA; Sept D; Joseph S; Holst MJ; McCammon JA, Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 2001, 98 (18), 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y; Zhang J; Wang N; Zhang Y; Yang Y; Yuan Y; Jing H; Liu X; Wu S; Luo P; Zhang W; Lu D; Zeng H; Zou Q, High-throughput Screening and Experimental Identication of Potent Drugs Targeting SARS-CoV-2 Main Protease. 2020, DOI: 10.21203/rs.3.rs-40014/v1. [DOI]
- 36.Ma C; Sacco MD; Hurst B; Townsend JA; Hu Y; Szeto T; Zhang X; Tarbet B; Marty MT; Chen Y; Wang J, Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res 2020, 30 (8), 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farag A; Wang P; Boys I; L. Eitson J; Ohlson M; Fan W; McDougal M; Ahmed M; W. Schoggins J; Sadek H, Identification of Atovaquone, Ouabain and Mebendazole as FDA Approved Drugs Tar-geting SARS-CoV-2 (Version 4). 2020, DOI: 10.26434/chemrxiv.12003930. [DOI]
- 38.Ellinger B; Bojkova D; Zaliani A; Cinatl J; Claussen C; Westhaus S; Reinshagen J; Kuzikov M; Wolf M; Geisslinger G; Gribbon P; Ciesek S, Identification of inhibitors of SARS-CoV-2 in-vitro cellular toxicity in human (Caco-2) cells using a large scale drug repurposing collection. 2020, DOI: 10.21203/rs.3.rs-23951/v1. [DOI]
- 39.Vatansever EC; Yang K; Kratch KC; Drelich A; Cho CC; Mellot DM; Xu S; Tseng CK; Liu WR, Targeting the SARS-CoV-2 Main Protease to Repurpose Drugs for COVID-19. bioRxiv 2020, DOI: 10.1101/2020.05.23.112235. [DOI] [Google Scholar]
- 40.Drayman N; Jones KA; Azizi SA; Froggatt HM; Tan K; Maltseva NI; Chen S; Nicolaescu V; Dvorkin S; Furlong K; Kathayat RS; Firpo MR; Mastrodomenico V; Bruce EA; Schmidt MM; Jedrzejczak R; Munoz-Alia MA; Schuster B; Nair V; Botten JW; Brooke CB; Baker SC; Mounce BC; Heaton NS; Dickinson BC; Jaochimiak A; Randall G; Tay S, Drug repurposing screen identifies masitinib as a 3CLpro inhibitor that blocks replication of SARS-CoV-2 in vitro. bioRxiv 2020, DOI: 10.1101/2020.08.31.274639. [DOI] [Google Scholar]
- 41.Ghahremanpour MM; Tirado-Rives J; Deshmukh M; Ippolito JA; Zhang CH; de Vaca IC; Liosi ME; Anderson KS; Jorgensen WL, Identification of 14 Known Drugs as Inhibitors of the Main Protease of SARS-CoV-2. bioRxiv 2020, DOI: 10.1101/2020.08.28.271957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Touret F; Gilles M; Barral K; Nougairede A; van Helden J; Decroly E; de Lamballerie X; Coutard B, In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci Rep 2020, 10 (1), 13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kupferschmidt K; Cohen J, Race to find COVID-19 treatments accelerates. Science 2020, 367, 1412–1413. [DOI] [PubMed] [Google Scholar]
- 44.Rismanbaf A, Potential Treatments for COVID-19; a Narrative Literature Review. Arch Acad Emerg Med 2020, 8 (1), e29. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


