ABSTRACT
Candida glabrata is a prominent pathogenic yeast which exhibits a unique ability to survive the harsh environment of host immune cells. In this study, we describe the role of the transcription factor encoded by the gene CAGL0F09229g, here named CgTog1 after its Saccharomyces cerevisiae ortholog, as a new determinant of C. glabrata virulence. Interestingly, Tog1 is absent in the other clinically relevant Candida species (C. albicans, C. parapsilosis, C. tropicalis, C. auris), being exclusive to C. glabrata. CgTog1 was found to be required for oxidative stress resistance and for the modulation of reactive oxygen species inside C. glabrata cells. Also, CgTog1 was observed to be a nuclear protein, whose activity up-regulates the expression of 147 genes and represses 112 genes in C. glabrata cells exposed to H2O2, as revealed through RNA-seq-based transcriptomics analysis. Given the importance of oxidative stress response in the resistance to host immune cells, the effect of CgTOG1 expression in yeast survival upon phagocytosis by Galleria mellonella hemocytes was evaluated, leading to the identification of CgTog1 as a determinant of yeast survival upon phagocytosis. Interestingly, CgTog1 targets include many whose expression changes in C. glabrata cells after engulfment by macrophages, including those involved in reprogrammed carbon metabolism, glyoxylate cycle and fatty acid degradation. In summary, CgTog1 is a new and specific regulator of virulence in C. glabrata, contributing to oxidative stress resistance and survival upon phagocytosis by host immune cells.
KEYWORDS: Candida glabrata, virulence, phagocytosis, CgTog1, oxidative stress, RNA-seq based transcriptomics
Introduction
Candida glabrata is a prominent cause of invasive candidiasis, best known for its ability to resist azole antifungal therapy [1–5]. In contrast, the mechanisms allowing it to interact with the host and cause disease are more elusive, as typical virulence features displayed by the main pathogen Candida albicans are absent in C. glabrata [6].
Due to the absence of hyphae formation, secretion of hydrolases and host tissue damage, C. glabrata virulence has been associated with its ability to survive immune attack, rather than active immune escape or host tissue invasion (reviewed in [6,7]). Indeed, C. glabrata was found to be able to proliferate inside phagocytes, eventually leading to cell lysis due to intracellular fungal growth [8,9]. Genome-wide studies have granted a sneak peek into possible pathways that mediate C. glabrata immune survival [10]. The Oxidative Stress Response (OSR) pathway is regarded as a possible pathway for phagocyte survival, since oxidative stress caused by the production of Reactive Oxygen Species (ROS) is considered one of the main antifungal mechanisms applied by phagocytes [11,12].
The OSR in yeasts is centered on the activation of antioxidant systems based on the activity of catalase (CTA1), superoxide dismutases (SOD1/2), thioredoxin and glutathione pathways [13–16]. In yeasts, the transcription factors (TFs) Yeast-AP1 and Skn7 are the most well studied oxidative stress regulators [13,17]. Despite their well-defined role in mediating OSR, the actual role of this network (and the participation of each player) during phagocyte attack and virulence needs further clarification.
In this study, we identified the transcription factor CgTog1 as a regulator of oxidative stress resistance and phagocyte survival in C. glabrata. In a previous screening for zinc cluster transcription factor functions, CgTog1 had been shown to confer a slight increase in ketoconazole resistance, but no effect on hydrogen peroxide (H2O2) tolerance was detected under the chosen experimental conditions [18]. In previous studies, CgYap1 alone was not found to be required for C. glabrata macrophage survival nor virulence [13,19]. On the other hand, CgSkn7 was described to be involved in virulence, but not macrophage survival [13,20]. This study provides evidence for the new TF CgTog1 as a regulator required for OSR and phagocyte survival. In Saccharomyces cerevisiae, the Tog1 homolog has been implicated in oxidative stress tolerance, but it is best known for its role in regulation of fatty acid utilization genes and utilization of alternative carbon sources [21,22], a phenotype not observed in CgTog1.
Given the identification of CgTOG1 as a new determinant of OSR in C. glabrata, its role in the transcriptome-wide response to H2O2 was assessed. The determination of CgTog1 targets enabled the identification of target genes sharing differential expression upon phagocytosis, which can be the basis for its role in hemocyte survival in a Galleria mellonella infection model. Additionally, this TF is absent in other Candida species (C. albicans, C. parapsilosis, C. tropicalis, C. auris), indicating a possible virulence regulatory network which is unique to C. glabrata.
Results
CgTog1 confers resistance to oxidative stress, but is not involved in the utilization of alternative carbon sources in C. glabrata
Upon phagocytosis, pathogens are faced with a multitude of antimicrobial stresses in the phagosome environment, including carbon limitation, which is often translated by the activation of pathways for the utilization of alternative carbon sources [6,10,23–26]. The TF Tog1 in the nonpathogenic yeast S. cerevisiae is required for the utilization of alternative carbon sources and resistance to oxidative stress [21]. Given the close phylogenetic relationship between S. cerevisiae and C. glabrata, we searched for a possible ortholog in the pathogenic yeast and assessed its functional conservation. Syntenic context visualized at YGOB [27] and protein homology searches show the TF encoded by C. glabrata ORF (open reading frame) CAGL0F09229g to be homologous to the S. cerevisiae Tog1 TF (26.46% identity). As such, this gene is henceforth designated CgTOG1.
A possible role for CgTOG1 in the utilization of alternative carbon sources was investigated by comparing the growth of the wild type and the Δcgtog1 mutant in the presence of lactate, glycerol or oleate. The deletion of CgTOG1 does not result in growth impairment in the presence of non-fermentative carbon sources, as observed by spot assays (Figure 1a). Accordingly, CgTOG1 overexpression via an expression plasmid did not provide any additional fitness for growth in the same conditions (Figure 1a). The attained results indicate that CgTOG1 is not required for C. glabrata growth using alternative carbon sources, indicating that this TF does not share the same function as its S. cerevisiae counterpart. Another relevant antimicrobial mechanism applied in the phagosome is the production of ROS [6,10,14,28]. C. glabrata is notoriously more resistant to oxidative stress than other yeasts [15], which can be related with its ability to survive the phagosome environment. The participation of CgTOG1 in oxidative stress resistance was investigated by spot assays, comparing the growth of the wild type and the Δcgtog1 mutant in the presence of the oxidative stress inducers H2O2 and menadione. The Δcgtog1 strain is more susceptible to H2O2 stress than the wild type strain (Figure 1b), indicating that it is required for C. glabrata tolerance to oxidative stress. Accordingly, its overexpression contributed to increased resistance against the same oxidative stress agent (Figure 1b). The overexpression of CgTOG1 from the expression plasmid was validated by RT-PCR. Quantification of CgTOG1 overexpression was found to be between 6 and 7-fold higher relative to the native CgTOG1 expression (Supplementary Figure S1).
Figure 1.
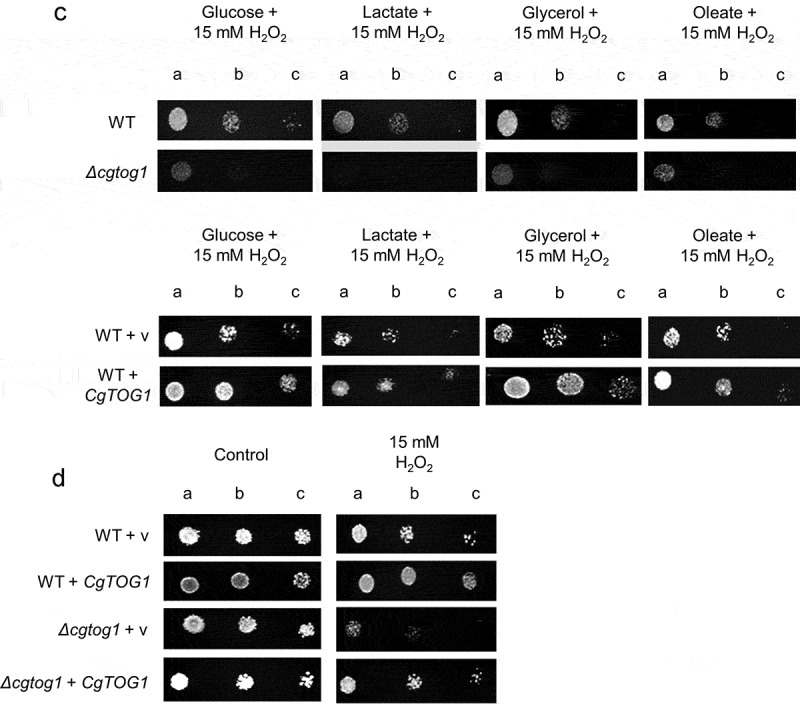
(Continued)
Figure 1.
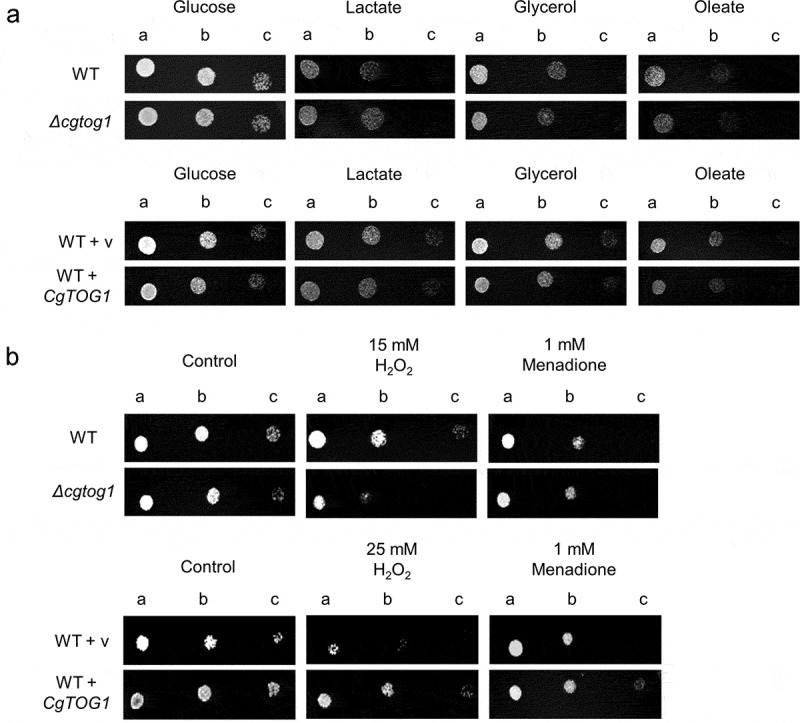
CgTOG1 confers resistance to oxidative stress inducers but is not required for the utilization of alternative carbon sources. (a) Comparison of spot growth assays of the KUE100 C. glabrata wild type and derived Δcgtog1 deletion mutant, as well as the L5U1 C. glabrata wild type strain, harboring the pGREG576 cloning vector, or the pGREG576_MTI_CgTOG1 expression plasmid, in the presence of glucose, lactate, glycerol or oleate as carbon sources. (b) Comparison of spot growth assays of the KUE100 C. glabrata wild type and derived Δcgtog1 deletion mutant, as well as the L5U1 C. glabrata wild type strain, harboring the pGREG576 cloning vector, or the pGREG576_MTI_CgTOG1 expression plasmid, in the presence of oxidative stress inducers H2O2 and menadione. (c) Comparison of spot growth assays of the KUE100 C. glabrata wild type and derived Δcgtog1 deletion mutant, as well as the L5U1 C. glabrata wild type strain, harboring the pGREG576 cloning vector, or the pGREG576_MTI_CgTOG1 expression plasmid, in the presence of distinct carbon sources and the oxidative stress inducer H2O2. (d) Comparison of spot growth assays of the KUE100::URA- C. glabrata wild type strain and the derived KUE100_Δcgtog1:URA- deletion mutant, harboring the pGREG576 cloning vector, or the pGREG576_MTI_CgTOG1 expression plasmid, in the presence of the oxidative stress inducer H2O2. The inocula were prepared as described in the materials and methods section. Cell suspensions used to prepare the spots were 1:5 (b) and 1:25 (c) dilutions of the cell suspension used in (a). The displayed images are representative of at least three independent experiments
Since the use of alternative carbon sources and oxidative stress represent concurrent environmental stresses felt by microbial pathogens in the phagosome, we investigated a possible role of CgTog1 in these conditions. The Δcgtog1 strain displays increased susceptibility when compared to the wild type strain, whereas CgTOG1 overexpression confers a fitness advantage (Figure 1c). We could not clearly see an effect of CgTOG1 deletion in the susceptibility to oxidative stress in cells growing in alternative carbon sources, when compared to oxidative stress alone, except eventually for lactate. These results indicate that CgTog1 may play a role in mediating C. glabrata response to phagosome combined antimicrobial stresses. Considering the absence of phenotypic differences when probing growth defects using alternative carbon sources alone, these results seem to indicate that resistance to H2O2 may be the most direct impact of CgTog1 expression. To further assess this, we investigated the ability of CgTog1 to rescue the susceptibility phenotype of the Δcgtog1 mutant. Accordingly, the expression of CgTOG1 was able to rescue the observed susceptibility phenotype to H2O2 (Figure 1d), indicating a role for this TF in oxidative stress resistance.
Altogether, this data indicates that CgTog1 mediates oxidative stress resistance in C. glabrata. Moreover, it suggests that the role of Tog1 has partially diverged in C. glabrata relative to the model yeast S. cerevisiae, which may be the result of co-evolution with the human host.
CgTog1 contributes to the reduce the intracellular concentration of ROS during oxidative stress
The susceptibility phenotype of the Δcgtog1 mutant to the oxidative stress inducer H2O2 led us to investigate if this TF contributes to control the concentration of ROS in C. glabrata cells as part of its mode of action. The presence of intracellular ROS during H2O2 exposure was assessed using the fluorescent probe H2DCFDA. A significant increase in the intracellular concentration of ROS inside C. glabrata cells was observed after 1 h of H2O2 stress (Figure 2). An even stronger increase in intracellular ROS concentration was observed in Δcgtog1 cells (Figure 2), indicating that CgTOG1 is required to reduce the formation of intracellular ROS in C. glabrata during oxidative stress cause by H2O2.
Figure 2.
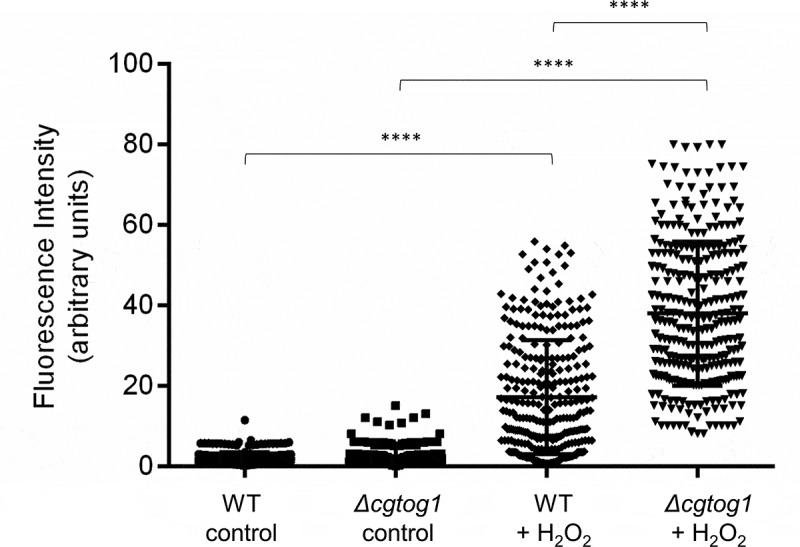
CgTOG1 contributes to reduce the intracellular accumulation of ROS. Comparison of intracellular ROS concentration of the KUE100 C. glabrata wild type and derived Δcgtog1 deletion mutant in control conditions or upon 1 h of H2O2 stress. The estimation of intracellular ROS is based on the fluorescence intensity values exhibited by yeast cells upon incubation with the cell-permeant 2ʹ,7ʹ-dichlorodihydrofluorescein diacetate indicator H2DCFDA. The displayed values correspond to at least three independent experiments. Error bars represent the corresponding standard deviation
CgTog1 is constitutively localized in the cell nucleus
To screen for conditions that might lead to CgTog1 activation, the subcellular localization of the CgTog1_GFP fusion protein was inspected by fluorescence microscopy in medium containing oleate as an alternative carbon source and during H2O2 stress. However, the subcellular localization of CgTog1_GFP was unchanged during growth in glucose or oleate as carbon source, being always localized to the nucleus, as verified by co-localization with DAPI nuclear staining (Figure 3a,b). The same pattern was observed before and after H2O2 exposure (Figure 3c,d). Therefore, neither carbon source nor H2O2 treatment change the relative distribution of CgTog1 in C. glabrata cells, indicating that activation of its target genes is not based on translocation of the transcription factor to the nucleus. This observation uncovers a distinct activation mechanism from other ORS regulators, such as Yap1, which is imported to the nucleus upon oxidative stress [29].
Figure 3.
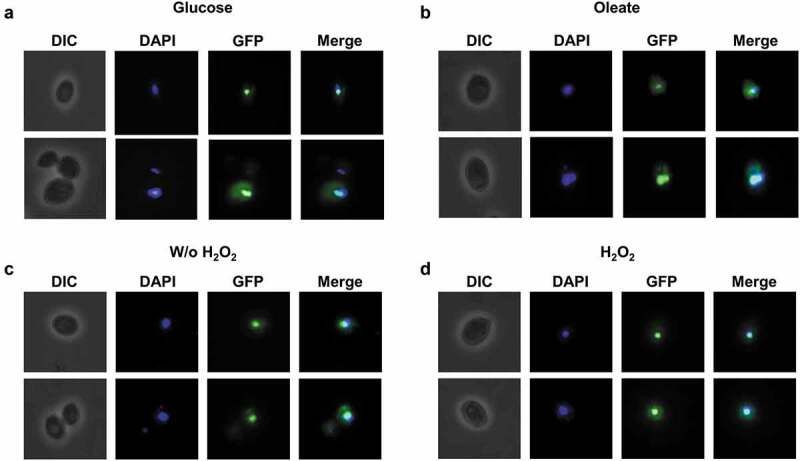
CgTOG1 is localized to the nucleus. Fluorescence of L5U1 C. glabrata cells harboring the pGREG576_MTI_CgTOG1 plasmid after 5 h of copper-induced recombinant protein production in BM-U medium. (a,b) Cells were washed with PBS buffer and transferred to YP medium with glucose or oleate as carbon source for 1 h. (c,d) Cells were washed with PBS buffer and transferred to fresh medium with or without 15 mM H2O2 for 1 h. Results indicate that CgTog1_GFP fusion protein localized to the nucleus under all conditions tested. The displayed images are representative of at least three independent experiments
Oxidative stress induces CgCTA1 expression in a CgTOG1-independent manner
The finding that CgTOG1 is required for C. glabrata oxidative stress resistance exerted by H2O2 motivated us to inquire if the catalase gene CgCTA1 is regulated by this TF. The transcript levels of the CgCTA1 gene were quantified by RT-PCR in the presence or absence of H2O2 and its expression in the Δcgtog1 mutant was compared to that of the wild type parental strain. The basal expression of the catalase gene is higher in Δcgtog1 cells when compared to wild type cells. A strong upregulation of CgCTA1 expression (~20-fold) was observed after 1 h of H2O2 stress (Figure 4). Interestingly, the induction of catalase gene expression is affected by CgTOG1 deletion, showing a 4-fold decrease compared to wild type induction. However, given the different basal expression levels, both wild type and Δcgtog1 mutant strains attain similar CgCTA1 expression levels during H2O2 stress (Figure 4).
Figure 4.
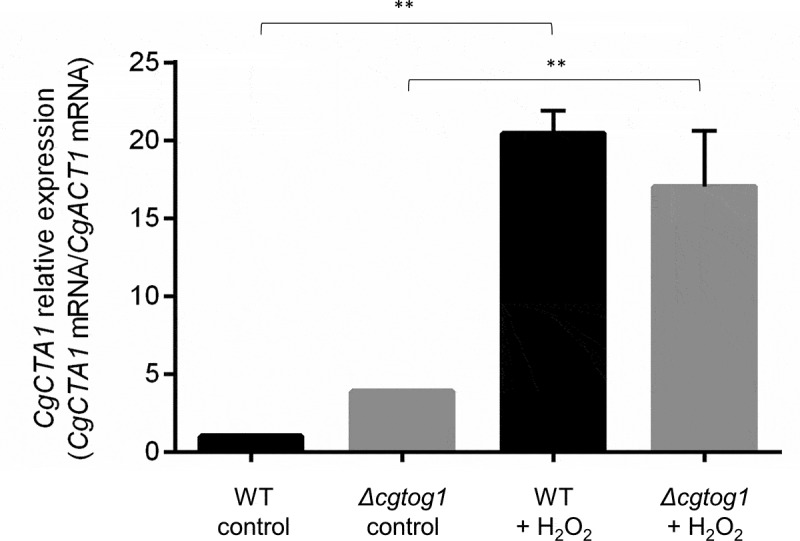
CgCTA1 expression is independent of CgTOG1. Comparison of the variation of CgCTA1 transcript levels determined by RT-PCR in the KUE100 C. glabrata wild type and derived Δcgtog1 deletion mutant in control conditions or upon 1 h of 15 mM H2O2 stress. Transcript levels of CgACT1 were used for normalization. Expression values are the average of at least three independent experiments. Error bars represent the corresponding standard deviation
Role of CgTOG1 in the transcriptome-wide changes occurring in response to H2O2 in C. glabrata
The Δcgtog1 susceptibility phenotype to H2O2 independent of catalase expression warranted further investigation into the role of this transcription factor in mediating oxidative stress resistance. To clarify the role of CgTog1 in C. glabrata response to H2O2, transcriptome-wide expression changes occurring upon H2O2 exposure were assessed and the participation of CgTOG1 in the observed expression program was evaluated.
The expression of 2558 genes was altered in C. glabrata cells following exposure of 1 h to 15 mM H2O2 (log2 fold change > 0.5 or < −0.5, adjusted p-value < 0.05) (Supplementary Table S1). These conditions appear to elicit a significant stress, translated by the pronounced transcriptional response: the upregulation of 1168 (46%) and the downregulation of 1390 (54%) genes (Figure 5a). As expected, this dataset includes the upregulation of genes involved in the core oxidative stress response: the hallmark catalase gene (CgCTA1), the superoxide dismutase gene (CgSOD1), oxidative stress resistance regulators (CgYAP1, CgSKN7) and several peroxidases and reductases (CgTSA1, CgTRR1, CgTRX2, CgTRX3, CgMXR1, CgSRX1, CgGRX8, CgGRX4, CgGPX2).
Figure 5.
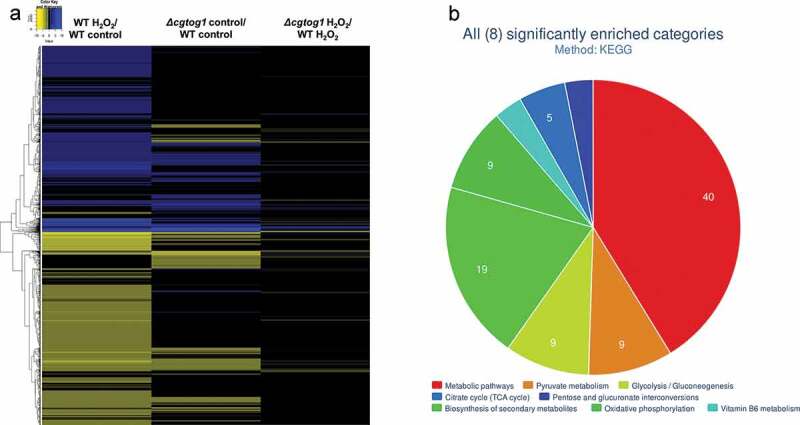
C. glabrata gene expression during oxidative stress and its regulation by CgTOG1. (a) Heatmap depicting the global transcriptomics response to H2O2 in KUE100 C. glabrata wild type and the derived Δcgtog1. (b) Systematic analysis of differentially expressed genes regulated by CgTOG1 with the help of enriched KEGG pathways calculated with FungiFun2. The significantly enriched pathways are shown
To understand the role of CgTog1 in the oxidative stress response of C. glabrata, the differential basal gene expression of Δcgtog1 vs wild type cells and the differential gene expression in the presence of H2O2 was further evaluated. Deletion of CgTog1 was found to deregulate the basal expression of 607 genes, resulting in increased expression of 233 genes and reduced expression of 374 genes. CgTog1 was found to control the basal expression of antioxidant genes and redox processes (CgGPX2, CgIDP1, CgTSA1, CgERV1, CgPAN5). Interestingly, CgTog1 was found to significantly contribute to the basal expression of protein folding/proteasome homeostasis genes (CgPRE1/3/4/5/7, CgRPN1/5/7/11, CgPUP3).
While assessing the impact of CgTog1 deletion during oxidative stress, 112 genes display increased expression, while 147 genes show decreased expression (Figure 5a; Supplementary Table S1). To determine the possible CgTog1-regulated genes that may mediate the oxidative stress resistance phenotype, those found to display altered expression in Δcgtog1 cells in the presence of H2O2 but not in control conditions were analyzed. In this dataset, the expression of 115 genes was found to be activated by CgTog1 specifically under H2O2 stress, while 89 genes were found to be repressed. In accordance with our previous RT-PCR data, the catalase gene CgCTA1 is upregulated during oxidative stress, but its final expression level is not affected by CgTOG1 deletion. Among the activated genes, 13 were also found overexpressed in the wild type response to H2O2, which could represent the core gene set activated by CgTog1 to mediate the oxidative stress response (Table 1). This subset includes the ORF CAGL0F00649g, encoding a putative kinase with similarity to ScRck2, known to be involved in the oxidative stress response in S. cerevisiae [30]. Other than this target, the transcriptome profile of Δcgtog1 cells did not reveal deregulation over core OSR genes, but rather mitochondrial function, redox and energy pathways (CgADH6, CgPDC6, CgALD6, CgMRP10, CgCYB2, CgDLD1, CgCYT1), as analyzed using FungiFun2 [31] (Figure 5b).
Table 1.
List of CgTog1 activated genes which are also upregulated in the wild type oxidative stress response caused by H2O2. Expression values represent log2 fold changes
| ORF/Gene Name | S. cerevisisae homolog | Function | WT H2O2/WT control | Δcgtog1 H2O2/WT H2O2 |
|---|---|---|---|---|
| MIG1 | MIG1 | Transcription factor involved in glucose repression | 2,58 | −1,79 |
| CAGL0F05709g | ATC1 | Cation stress response | 2,56 | −1,24 |
| CAGL0H02491g | COX7 | Subunit VII of cytochrome c oxidase | 2,48 | −0,68 |
| CAGL0F00649g | Response to oxidative and osmotic stress | 2,47 | −3,63 | |
| CAGL0I00116g | Unknown function | 2,43 | −1,06 | |
| CAGL0B02079g | AZR1 | Plasma membrane transporter | 2,30 | −1,56 |
| NCE103 | NCE103 | Beta carbonic anhydrase | 2,24 | −1,45 |
| CAGL0A01606g | HOP2 | Meiosis-specific protein | 1,93 | −0,85 |
| CAGL0E00649g | PTC6 | Mitophagy | 1,67 | −0,88 |
| CAGL0D02640g | Unknown function | 1,62 | −3,64 | |
| CAGL0M12001g | Unknown function | 1,07 | −1,76 | |
| CAGL0G07931g | MRPS12 | Mitochondrial translation | 0,86 | −0,70 |
| CAGL0K07942g | Unknown function | 0,61 | −0,66 |
The transcription factor CgTOG1 is required for Candida glabrata survival to phagocytosis
The ability of pathogens to cope with oxidative stress in the phagosome environment is a key virulence feature that mediates phagocyte survival. The role of CgTOG1 in conferring resistance to oxidative stress prompted us to investigate its impact in C. glabrata survival upon phagocytosis using the Galleria mellonella infection model hemocytes, the insect equivalent to mammalian lymphocytes.
The amount of viable C. glabrata cells found within hemocytes was assessed up to 48 h of co-culture. After 1 h, no differences can be observed in the survival of internalized C. glabrata between the wild type and the Δcgtog1 mutant strains. However, after only 8 h the population of wild type cells found within hemocytes was found to be 2.5-fold higher than that of the Δcgtog1 mutant (Figure 6a). Consistent with the previous results, the overexpression of CgTOG1 was found to lead to a 35% increase in cell proliferation within hemocytes at 48 h of internalization (Figure 6b). Overall, these results show that the TF CgTog1 is involved in C. glabrata virulence in the G. mellonella infection model by enabling yeast survival inside phagocytic cells.
Figure 6.
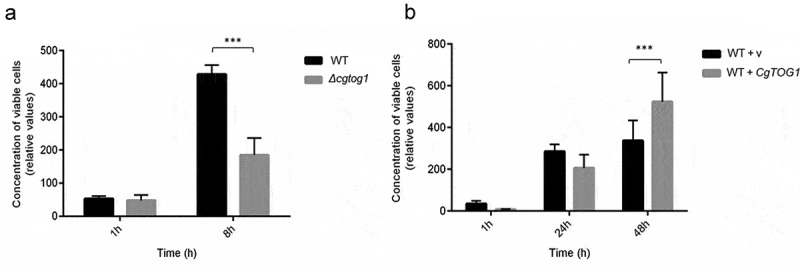
CgTOG1 is required for survival upon phagocytosis. (a) The concentration of viable KUE100 C. glabrata wild type (black bars) or derived Δcgtog1 deletion mutant (gray bars) after co-culture with G. mellonella hemocytes using a MOI of 1:5. (b) The concentration of viable L5U1 C. glabrata wild type strain, harboring the pGREG576 cloning vector (black bars), or the pGREG576_MTI_CgTOG1 expression plasmid (gray bars) after co-culture with G. mellonella hemocytes using a MOI of 1:5. The displayed results are relative to the concentration of viable cells inoculated at time zero and are the average of at least six independent experiments. Error bars represent the corresponding standard deviation
The role of CgTog1 regulon in C. glabrata virulence
Upon phagocytosis pathogens activate alternative carbon source pathways due to glucose starvation (e.g., glyoxylate cycle) and oxidative stress response genes [10,24,25]. Moreover, both glucose starvation and oxidative stress have been described to elicit a somewhat similar transcriptional response in C. glabrata [10,13,25].
The role of CgTog1 in mediating hemocyte survival and oxidative stress resistance, together with its regulation of energy/metabolism genes, including players in the TCA and glyoxylate cycles (Table 1; Supplementary Table S1), led us to compare the genes activated by CgTog1 in control conditions or H2O2 stress with the described transcriptome-wide response induced in murine and human macrophage-engulfed C. glabrata cells [8,24], as to ascertain its relevance in this context. An overlap of 93 genes was revealed (Figure 7a, Supplementary Table S1), with the most enriched biological functions featuring: amino acids metabolism, carbon/energy, and lipid and fatty acid metabolism and transport, as analyzed by FungiFun2 [31] (Figure 7b). From the 22 genes upregulated in macrophages and activated by CgTog1 in response to H2O2, 4 were also upregulated in the wild type transcriptional response to oxidative stress: CgMIG1, CgNCE103, CAGL0F00649g, and CAGL0F05709g (CgATC1).
Figure 7.
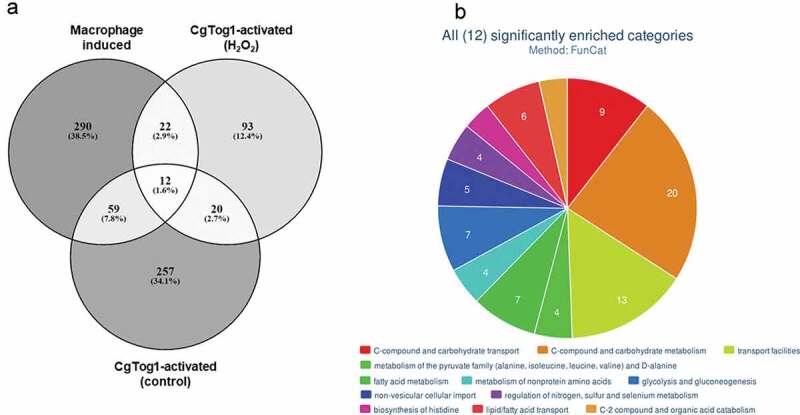
The CgTOG1 regulon in the transcriptomics context of macrophage engulfed cells. (a) Number of differentially expressed genes that constitute the CgTOG1 activated genes in either control conditions or H2O2 stress, compared to the activated transcriptional response of C. glabrata upon macrophage engulfment. Overlapping genes were identified based on their systematic ORF designations. (b) Systematic analysis of genes commonly activated by CgTOG1 and upon macrophage engulfment, with the help of enriched FunCat functional categories calculated with FungiFun2. The significantly enriched pathways are shown
Interestingly, a search for gene syntenic context using CGOB [32] and ScTOG1 as query revealed no homologs in Candida spp. from the CTG clade (C. albicans, C. parapsilosis, C. tropicalis) or C. auris. Likewise, Blastp search for amino acid similarity to the CgTog1 protein did not reveal any unequivocal homolog (best match = E-value 10−16) in any of the aforementioned species. This indicates that the Tog1 TF is present in the two phylogenetically close yeasts C. glabrata and S. cerevisiae (albeit with distinct functions) but is absent in the remaining (distantly related) Candida species. These facts appear to support a unique function for CgTOG1 as a virulence determinant in which is specific of C. glabrata.
Discussion
In this study, the TF CgTog1 was found to be a determinant of virulence in C. glabrata. Unlike its S. cerevisiae counterpart, it is not required for growth in alternative carbon sources, but rather for oxidative stress resistance. The limited homology between the two Tog1 proteins can be a signature of its evolution toward a distinct function. The two proteins share 26% identity, which is mainly localized in the zinc-finger DNA binding domain. Both proteins harbor a fungal transcription factor regulatory middle homology region (MHR), typically present in TFs with a N-terminal GAL4-like zinc finger DNA-binding domain and has a regulatory role. Interestingly, the MHR is significantly shorter in CgTog1 (80–92 amino acids long) when compared to ScTog1 (223–249 amino acids longs). This could represent an important feature underlying the different phenotypes regulated by the two Tog1 proteins.
The apparent loss of CgTog1 function in alternative carbon source utilization and focus on oxidative stress response may possibly correlate with the evolution of C. glabrata in the human host, where the production of ROS is a major antimicrobial strategy inside phagocytic cells [11,12]. Indeed, CgTOG1 is shown in this study to modulate the concentration of intracellular ROS, indicating a role either in ROS detoxification or intracellular redox balance, and to mediate C. glabrata survival upon phagocytosis.
Transcriptomics profiling during H2O2 exposure revealed that the response of C. glabrata is quite extensive, including the activation of core OSR genes with additional concurrent transcriptional patterns. Previous transcriptomics studies investigating the response of C. glabrata to oxidative stress have probed a lower concentration of H2O2 and a shorter exposure time [13,33]. However, while this concentration is stressful for S. cerevisiae, it is only mildly so for C. glabrata [33]. The distinct experimental conditions may justify why our study observed a larger transcriptional response (>1000 activated genes in this study versus >200 activated genes in previous studies). As expected, both approaches allowed to capture the activation of core OSR genes (CgCTA1, CgTRR1, CgTRX2, CgTSA1, CgGPX2), while our study also identified additional and possibly partial indirect responses. Considering the higher resistance of C. glabrata to oxidative stress, the use of more stressful conditions in this study presumably allowed to capture a more complete picture of its transcriptional response. Enriched groups include regulation of transcription, denoting the activation of multiple regulatory pathways (CgHOG1, CgCAD1, CgYAP5, CgYAP7, CgHAP1). Interestingly, response to high levels of H2O2 also encompasses increased expression of the OSR regulators CgYAP1 and CgSKN7, and not only their target genes. Moreover, the comparison between the transcriptional response between H2O2 stress and phagocytosis reveals a common activation of oxidative stress genes (CgCTA1, CgTRR1, CgTRX2, CgGRE2(B)), which conforms to the notion that yeast cells suffer from oxidative stress upon phagocytosis. Additionally, common transcriptional responses include activation of utilization of alternative carbon sources (CgCAT8, CgMIG1, CgPOX1) and energy pathways such as the glyoxylate (CgICL1) and TCA cycles (CgKGD1, CgSDH2, CgFUM1, CgACO1, CgSDH2). These findings highlight an overlap in the transcriptional response to high oxidative stress and phagocytosis, mostly translated in the activation of not only OSR genes, but also key energetic pathways. This may underlie the success of C. glabrata as an oxidative and phagocytosis resistant pathogen.
Interestingly, CgTOG1 regulates oxidation-reduction processes, but it does not control the expression of typical OSR genes, including the catalase gene CgCTA1. Validation of CgCTA1 expression by RT-PCR was performed and revealed an apparent upregulation of the catalase gene when CgTOG1 is deleted. This may indicate a role in CgCTA1 basal expression regulation. However, no regulation of catalase gene expression by CgTog1 under H2O2 stress was found in the transcriptomics results. This data was validated by RT-PCR, which shows a reduction in CgCTA1 induction in the absence of the TF, but to levels similar to those attained in the wild type strain.
By analyzing the regulation of CgCTA1 using the PathoYeastract database [34,35], its expression is activated by four TFs, including the OSR regulators CgYAP1 and CgSKN7. CgTOG1 is not known to be part of the regulons of either CgYAP1 or CgSKN7, nor it regulates either of them as determined by our data, indicating that its mode of action is outside of the traditional OSR network. In fact, it exerts transcriptional regulation that mimics the hallmark of macrophage-internalized pathogens: reprogrammed carbon metabolism based on increased nucleogenesis, glyoxylate cycle and fatty acid degradation pathways [23]. It is striking that a determinant of OSR presents such a regulatory portrait, even during H2O2 stress. These observations appear to be consistent with studies in S. cerevisiae that show respiration disruption is correlated with H2O2 sensitivity [36,37], while reduced cofactors (e.g., NADPH) are especially relevant in oxidative stress conditions, as NADPH is a final electron donor in glutathione and thioredoxin systems [38]. This data is further corroborated by other C. glabrata studies reporting mitochondrial processes and activity of dehydrogenases as crucial under oxidative stress [13].
Upon phagocytosis, reprogramming of nutrient metabolic pathways, particularly carbon and nitrogen biosynthesis, takes place due to nutrient limitation [8,14,24]. Our transcriptomics analysis identified CgTOG1 as a regulator of these central responses to phagosome stress, which is concordant with its G. mellonella hemocyte survival phenotype and the overlap between activated genes in both glucose starvation and OSR in C. glabrata [13]. Previous studies reported the upregulation of gluconeogenesis, glyoxylate and methylcitrate cycles and oxidation of fatty acids upon C. glabrata phagocytosis by murine or human macrophages [8,24]. In this study, CgTOG1 was found to contribute to the activation of these pathways, including previously identified genes in such responses, such as CgMLS1 (glyoxylate cycle), CgICL2 and CgPDH1 (methylcitrate cyle), CgFBP1 and CgPCK1 (gluconeogenesis). Interestingly, CgMDH2, encoding an enzyme involved in the glyoxylate cycle and gluconeogenesis that interacts with Pck1 and Fbp1 in S. cerevisiae [39], is also regulated by CgTog1. The genes CgIDP2, CgACS1 and CgICL2, which are activated upon non-fermentable carbon source growth in S. cerevisiae [40–42], are also regulated by CgTog1. Furthermore, the glucose limitation response TF CgMIG1 was found to be induced by CgTOG1 and in the wild type response to H2O2, being induced in macrophages as well, which hints for another regulatory network involved in C. glabrata virulence. Other than regulation over central carbon and energy pathways, CgTog1 also shares regulation over β-oxidation of fatty acids and fatty acid metabolism genes that are activated during phagocytosis, including CgYAT1, CgYAT2, CgCAT2, CgCRC1, CgYMC2, CgFAA2 and CgECI1. Fatty acid β-oxidation is recognized as a possible major energy generation pathway in phagocytosed Candida cells [24,25,43]. The regulation of fatty acid metabolism by CgTog1 may therefore represent an additional basis for the hemocyte survival phenotype mediated by CgTOG1.
Several of the genes concurrently found to be activated during phagocytosis and by CgTog1 are involved in mitochondrial functions and peroxisomal activity, with potential impact on the redox state of the cells (e.g., fatty acid oxidation). This indicates a possible role for CgTog1 in the key regulation of central carbon metabolism and energy pathways, although not required for growth on alternative carbon sources. This would be consistent with the role of CgTog1 in mediating hemocyte survival: specific pathways would be activated for the uptake of alternative carbon sources with a concurrent role of CgTog1 in prompting central carbon metabolism reshuffling through, for instance, the glyoxylate cycle. Such a role would correlate well with control over the cells redox state via regulation of oxidative phosphorylation and fatty acid oxidation, which in turn can impact resistance to oxidative stress. In conjunction, adaptive energy metabolism and oxidative state are the main drivers of pathogen survival upon phagocytosis and the CgTog1 is found as a central piece in this response.
Differential regulation over nitrogen metabolism pathways is also a feature of CgTOG1. Previous studies have reported the upregulation of arginine and lysine biosynthetic pathways [24], while our data also indicates an enrichment in the histidine pathway. Consistently, our results also highlight an enrichment in amino acid transport activity, including the previously observed general amino acid permease CgGAP1 [24]. The conclusions drawn from this study reinforce the importance of multi stress resistance for prevalence within the host. Importantly, this TF is not only functionally distinct from its counterpart in the nonpathogenic S. cerevisiae, but it is absent from other clinically relevant Candida spp., including C. albicans, C. parapsilosis, C. tropicalis and C. auris. It is plausible that distinct evolutionary paths have occurred for C. glabrata to retain this TF, while its evolution with the human host may have contributed for CgTog1 to have become distinct from its S. cerevisiae counterpart.
Overall, this study describes a new regulator of virulence in C. glabrata. Our results show that the CgTog1 TF mediates phagocyte survival by limiting the accumulation of intracellular ROS and a concerted regulation over OSR, nutrient limitation response and energy metabolism. This transcriptional pattern constitutes the hallmark of pathogen survival upon phagocytosis and the prevalent role of CgTOG1 in this mechanism has been confirmed by the activation of common genes during oxidative stress and upon C. glabrata phagocytosis. Together with the transcriptomics data showing a role for CgTOG1 in the transcriptional response to phagocytosis, a prominent function in C. glabrata virulence is proposed. We present evidence for a specific regulator of C. glabrata survival in immune cells, which could add another piece to the puzzle on how this species is capable of not only survive but replicate inside phagocytes.
Materials and methods
Strains, plasmids and growth media
Candida glabrata parental strain KUE100 [44] and derived single deletion mutants were batch-cultured at 30ºC, with orbital agitation (250 rpm) in basal medium (BM) or Yeast extract-Peptone-Dextrose (YPD) medium. BM has the following composition (per liter): 1.7 g yeast nitrogen base without amino acids or NH4+ (Difco), 20 g glucose (Merck) and 2.65 g (NH4)2SO4 (Merck). YPD has the following composition (per liter): 20 g glucose (Merck), 20 g Peptone (Merck) and 10 g Yeast extract (Merck). C. glabrata strain L5U1 (cgura3Δ0, cgleu2Δ0) [19], kindly provided by John Bennett from the National Institute of Allergy and Infectious Diseases, NIH, Bethesda, USA, was grown in BM supplemented with 20 mg/L uracil and 60 mg/L leucine. L5U1 strains harboring pGREG576 derived plasmids were grown in the same medium lacking uracil. KUE100::URA- and the derived KUE100_Δcgtog1:URA- strains, constructed in this study, harboring pGREG576 derived plasmids were grown in BM. Solid media contained, besides the above-indicated ingredients, 20 g/L agar (Iberagar). The plasmid pGREG576 was obtained from the Drag&Drop collection [45]. The plasmid pV1382 [46] was obtained from Addgene (Addgene plasmid# 111436).
Disruption of C. glabrata CgTOG1 (ORF CAGL0F09229g)
The deletion of the C. glabrata gene encoded by ORF CAGL0F09229g, addressed in this study, was carried out in the parental strain KUE100, using the method described by Ueno et al. [47]. Genes of interest were replaced by a DNA cassette including the CgHIS3 gene, through homologous recombination. The PCR primers used to generate the replacement cassette for each gene and the primers used for PCR confirmation of gene deletion are present in Supplementary Table S2. The pHIS906 plasmid including CgHIS3 was used as a template and transformation was performed as described previously [44].
Cloning of the C. glabrata CgTOG1 (ORF CAGL0F09229g)
The pGREG576 plasmid from the Drag&Drop collection was used as described before to clone and express the C. glabrata CgTOG1 gene [48–53]. pGREG576 was acquired from Euroscarf and contains a galactose inducible promoter (GAL1), the yeast selectable marker URA3 and the GFP gene, encoding a Green Fluorescent Protein (GFPS65T), which allows monitoring of the expression and subcellular localization of the cloned fusion protein. CgTOG1 DNA was generated by PCR, using genomic DNA extracted from the sequenced CBS138 C. glabrata strain. The designed primers contain, besides a region with homology to the first and last 22 nucleotides of the CgTOG1 coding region (italic), nucleotide sequences with homology to the cloning site flanking regions of the pGREG576 vector (underlined). The amplified fragments were co-transformed into the parental S. cerevisiae strain BY4741 with the pGREG576 vector, previously cut with the restriction enzyme SalI (NZYTech), to obtain the pGREG576_CgTOG1 plasmid. Since the GAL1 promoter only allows a slight expression of downstream genes in C. glabrata, to visualize by fluorescence microscopy the subcellular localization of the CgTog1 protein in C. glabrata, new constructs were obtained. The GAL1 promoter present in the pGREG576_CgTOG1 plasmid was replaced by the copper-inducible MTI C. glabrata promoter [54], giving rise to the pGREG576_MTI_CgTOG1 plasmid. The MTI promoter DNA was generated by PCR, using genomic DNA extracted from the sequenced CBS138 C. glabrata strain. The designed primers contain, besides a region with homology to 27 nucleotides in the beginning and the last 27 nucleotides of the 1000 bp upstream region of the MTI coding sequence (italic), nucleotide sequences with homology to the cloning site flanking regions of the pGREG576 vector (underlined). The amplified fragment was co-transformed into the parental strain BY4741 with the pGREG576_CgTOG1 plasmid, previously cut with SacI (NZYTech) and NotI (NZYTech) restriction enzymes to remove the GAL1 promoter, to generate the pGREG576_MTI_CgTOG1 plasmid. The recombinant plasmids pGREG576_CgTOG1 and pGREG576_MTI_CgTOG1 were obtained through homologous recombination in S. cerevisiae and verified by DNA sequencing. All primers used are present in Supplementary Table S2.
Disruption of C. glabrata CgURA3 (ORF CAGL0I03080g)
The disruption of the C. glabrata URA3 gene encoded by ORF CAGL0I03080g, was carried out in the parental strain KUE100 and the derived Δcgtog1 mutant, using the CRISPR-Cas9 system from Vyas et al. [46]. Briefly, a CgURA3 gRNA sequence selected from the resources made available by Vyas et al. [46] was cloned in the pV1382 plasmid, previously linearized with the restriction enzyme BsmBI (NEB). The CgURA3 gRNA was obtained by oligonucleotide annealing and the product ligated into the previously linearized pV1382 plasmid to obtain the pV1382_CgURA3 vector. The construct was verified by DNA sequencing. The plasmid was transformed into the parental strain KUE100 or the derived Δcgtog1 mutant and cells were then directly plated on 5-Fluoroorotic acid (5-FOA) to select for URA- cells. Sequential passages in nonselective medium (YPD) were performed to avoid detrimental effects of further Cas9 expression and CgURA3 loss of function was further confirmed by the inability to grow in medium without uracil. The introduction of pGREG576 derived plasmids in the edited strains was able to rescue the growth impairment in the absence of uracil. Sequencing of the CgURA3 gene from the selected candidates revealed the existence of frameshifts within the ORF, thus resulting in premature stop codons as it is expected from Non-Homologous End Joining (NHEJ) correction of double-strand breaks. All primers used are present in Supplementary Table S2.
Alternative carbon source utilization assays
The ability to use alternative carbon sources of the parental strain KUE100 was compared to that of the deletion mutant KUE100_Δcgtog1 by spot assays. The ability of CgTOG1 gene expression to increase wild type utilization of alternative carbon sources was also examined in the URA3- L5U1 C. glabrata strain, using the pGREG576_MTI_CgTOG1 centromeric plasmid. The same plasmid was used to assess the ability of CgTOG1 to recue susceptibility to oxidative stress, examined in the KUE100::URA- and derived deletion mutant KUE100_Δcgtog1::URA- strains.
KUE100 C. glabrata and derived deletion mutant KUE100_Δcgtog1 cell suspensions used to inoculate agar plates were mid-exponential cells grown in YPD, until culture OD600nm = 0.5 ± 0.05 was reached. Cells were washed and diluted in sterile water to obtain suspensions with OD600nm = 0.05 ± 0.005. These cell suspensions and subsequent dilutions (1:5; 1:25) were applied as 4 µL spots onto the surface of solid YP plates, using distinct carbon sources: 2% glucose, 0.75% lactate, 1% glycerol or 0.1% oleic acid + 0.5% Tween80. L5U1 harboring the pGREG576_MTI_CgTOG1 C. glabrata cell suspensions used to inoculate the agar plates were mid-exponential cells grown in BM-U supplemented with leucine and 100 µM CuSO4, until culture OD600nm = 0.5 ± 0.05 was reached. The cells were washed and diluted in sterile water to obtain suspensions with OD600nm = 0.05 ± 0.005. KUE100::URA- and the derived deletion mutant KUE100 Δcgtog1::URA- cell suspensions, harboring the pGREG576_MTI_CgTOG1 plasmid, used to inoculate the agar plates, were mid-exponential cells grown in BM supplemented with 100 µM CuSO4, until culture OD600nm = 0.5 ± 0.05 was reached. The cells were washed and diluted in sterile water to obtain suspensions with OD600nm = 0.05 ± 0.005
These cell suspensions and subsequent dilutions (1:5; 1:25) were applied as 4 µL spots onto the surface of solid YP plates supplemented with 100 µM CuSO4 and the concentrations of alternative carbon sources previously referred.
Oxidative stress susceptibility assays
The susceptibility of the parental strain KUE100 toward toxic concentrations of oxidative stress inducers was compared to that of the deletion mutant KUE100_Δcgtog1 by spot assays. The ability of CgTOG1 gene expression to increase wild type resistance to the referred conditions was also examined in the URA3- L5U1 C. glabrata strain, using the pGREG576_MTI_CgTOG1 centromeric plasmid.
KUE100 C. glabrata and derived deletion mutant KUE100_Δcgtog1 cell suspensions used to inoculate agar plates were mid-exponential cells grown in BM, until culture OD600nm = 0.5 ± 0.05 was reached and then diluted in sterile water to obtain suspensions with OD600nm = 0.05 ± 0.005. These cell suspensions and subsequent dilutions (1:5; 1:25) were applied as 4 µL spots onto the surface of solid BM, supplemented with adequate chemical stress concentrations. L5U1 harboring the pGREG576_MTI_CgTOG1 C. glabrata cell suspensions used to inoculate the agar plates were mid-exponential cells grown in BM-U supplemented with leucine and 100 µM CuSO4, until culture OD600nm = 0.5 ± 0.05 was reached and then diluted in sterile water to obtain suspensions with OD600nm = 0.05 ± 0.005. These cell suspensions and subsequent dilutions (1:5; 1:25) were applied as 4 µL spots onto the surface of solid BM-U supplemented with leucine, 100 µM CuSO4 and the concentrations of oxidative stress inducers previously referred.
Intracellular ROS quantification
The presence of intracellular ROS was assessed by the cell-permeant 2ʹ,7ʹ-dichlorodihydrofluorescein diacetate indicator (H2DCFDA; 25 mg/mL in DMSO, Santa Cruz Biotechnology, Inc.). C. glabrata cell suspensions from strains KUE100 and KUE100_∆cgtog1 were prepared in BM until a standard culture OD600nm = 0.5 ± 0.05 was reached and transferred to the same medium with or without 15 mM H2O2. After 1 h of incubation, cells were centrifuged (17,500 × g for 5 minutes), washed twice and resuspended in PBS buffer to a final 107 cells/mL aliquots. H2DCFDA was added to 1 mL of 4 × 107 cells/mL to a final concentration of 0.2 µL/mL and cell suspensions were incubated in the dark with orbital agitation (20 minutes, 250 rpm). Cells exposed to H2DCFDA were centrifuged (17,500 × g for 5 minutes), washed twice and resuspended in 1 mL PBS buffer. H2DCFDA-fluorescence was detected by fluorescence microscopy in a Zeiss Axioplan microscope (Carl Zeiss MicroImaging), using excitation and emission wavelength of 395 and 509 nm, respectively. Fluorescence images were captured using a cooled Zeiss Axiocam 503 color (Carl Zeiss Microscopy) and the images were analyzed with the ZEN lite software from ZEISS microscopy. Cell-to-cell fluorescence intensity was defined as the average of pixel by pixel intensity in the selected region of interest and a minimum of 100 cells/experiment were used.
CgTog1 subcellular localization assessment
The subcellular localization of the CgTog1 protein was determined based on the observation of L5U1 C. glabrata cells transformed with the pGREG576_MTI_CgTOG1 plasmid. These cells express the CgTog1_GFP fusion protein, whose localization may be determined using fluorescence microscopy. For experiments with distinct carbon sources, C. glabrata cell suspensions were prepared in BM-U supplemented with leucine and 100 µM CuSO4, until a standard culture OD600nm = 0.5 ± 0.05 was reached and transferred to YPD medium or YP medium with 0.1% oleic acid + 0.5% Tween80. For experiments with H2O2 stress, cell suspensions were prepared in BM-U supplemented with leucine and 100 µM CuSO4, until a standard culture OD600nm = 0.5 ± 0.05 was reached and transferred to fresh medium with or without 15 mM H2O2. After 1 h of incubation cells were washed twice and resuspended in 500 µL PBS buffer to which 2 µL of a DAPI solution (1 mg/mL in DMSO) was added. Cell suspensions were incubated in the dark with orbital agitation (30 minutes, 250 rpm) and centrifuged (17,500xg for 5 minutes), washed twice and resuspended in PBS buffer. The distribution of CgTog1_GFP fusion protein in C. glabrata living cells was detected by fluorescence microscopy in a Zeiss Axioplan microscope (Carl Zeiss MicroImaging), using excitation and emission wavelength of 395 and 509 nm (GFP) or 358 and 461 nm (DAPI), respectively. Fluorescence images were captured using a cooled Zeiss Axiocam 503 color (Carl Zeiss Microscopy).
Gene expression measurement by RT-PCR
The transcript levels of the CgCTA1 or CgTOG1 genes were determined by quantitative real-time PCR (RT-PCR). For CgCTA1 expression, KUE100 and KUE100_Δcgtog1 strains were grown in BM; for CgTOG1 expression, L5U1 strains harboring the pGREG576 cloning vector or the pGREG576_MTI_CgTOG1 expression plasmid were grown in BM-U supplemented with leucine and 100 µM CuSO4. Cells were grown until mid-exponential phase and transferred to fresh medium (control) or fresh medium containing 15 mM H2O2 and harvested after 1 h incubation. Samples were immediately frozen at −80ºC until RNA extraction. RT-PCR was performed as described elsewhere [51–53]. Synthesis of cDNA for real time RT-PCR experiments, from total RNA samples, was performed using the MultiscribeTM reverse transcriptase kit (Applied Biosystems) and the 7500 RT-PCR Thermal Cycler Block (Applied Biosystems), following the manufacturer’s instructions. The quantity of cDNA for the following reactions was kept around 10 ng. The subsequent RT-PCR step was carried out using SYBR® Green (NZYTech) reagents with default parameters established by the manufacturer and the primers in Supplementary Table S2. The CgACT1 gene transcript levels were used as an internal reference.
Total RNA extraction
C. glabrata strains KUE100 and KUE100_∆cgtog1 were grown in BM until mid-exponential phase. Subsequently, cells were transferred to fresh medium (control) or fresh medium containing 15 mM H2O2 and were harvested after 1 h of incubation. Total RNA was isolated using an Ambion Ribopure-Yeast RNA kit, according to manufacturer’s instructions.
Library preparation
Prior to RNA-seq analysis quality control measures were implemented. Concentration of RNA was ascertained via fluorometric analysis on a Thermo Fisher Qubit fluorometer. Overall quality of RNA was verified using an Agilent Tapestation instrument. Following initial QC steps sequencing libraries were generated using the Illumina Truseq Stranded Total RNA library prep kit with ribosomal depletion via RiboZero Gold according to the manufacturer’s protocol. Briefly, ribosomal RNA was depleted via pull down with bead-bound ribosomal-RNA complementary oligomers. The RNA molecules were then chemically fragmented and the first strand of cDNA was generated using random primers. Following RNase digestion the second strand of cDNA was generated replacing dTTP in the reaction mix with dUTP. Double stranded cDNA then underwent adenylation of 3ʹ ends following ligation of Illumina-specific adapter sequences. Subsequent PCR enrichment of ligated products further selected for those strands not incorporating dUTP, leading to strand-specific sequencing libraries. Final libraries for each sample were assayed on the Agilent Tapestation for appropriate size and quantity. These libraries were then pooled in equimolar amounts as ascertained via fluorometric analyses. Final pools were absolutely quantified using qPCR on a Roche LightCycler 480 instrument with Kapa Biosystems Illumina Library Quantification reagents.
Gene expression analysis
Strand specific RNA-seq library preparation and sequencing was carried out as a paid service by the NGS core from Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA. Paired-end reads (Illumina HiSeq 3000 PE150, 2 × 150 bp, 2 Gb clean data) were obtained from KUE100 and KUE100_∆cgtog1. Two replicates of each sample were obtained from three independent RNA isolations, subsequently pooled together. Raw data is available at Gene Expression Omnibus (GEO) under the accession number GSE148288. Reads were trimmed using Skewer (v0.2.2) [55] and aligned to the C. glabrata CBS138 reference genome, obtained from the Candida Genome Database (CGD) (http://www.candidagenome.org/), using TopHat (v2.1.1) [56] with parameters -p 12 (number of threads), -g 1 (maximum amount of times a read can be mapped to the genome), – b2-very-sensitive (preset option) and – library-type fr-firststrand (account for strand specificity). HTSeq (v0.7.1) [57] was used to count mapped reads per ORF Differentially expressed genes were identified using DESeq2 [58] with an adjusted p-value threshold of 0.05 and a log2 fold change threshold of −0.5 and 0.5. Default parameters in DESeq2 were used.
C. glabrata phagocytosis assays in G. mellonella hemocytes
G. mellonella hemocytes were isolated as described before [59]. Briefly, hemolymph was extracted from last instar larvae and mixed in with anticoagulant buffer (98 mM NaOH, 145 mM NaCl, 17 mM EDTA and 41 mM citric acid; pH 4.5) in a 1:1 proportion. After centrifugation at 250 g for 10 min and 4ºC, hemocytes were recovered and wash twice with PBS. Hemocytes pellet was suspended in Grace Insect Medium (GIM) (Thermo Fisher Scientific) supplemented with 10% (v/v) fetal bovine serum, 1% (w/v) glutamine, and 1% (w/v) antibiotic/antimycotic solution (10,000 units penicillin G, 10 mg Streptomycin, 25 mg/l amphotericin B), viable cells were counted with a hemocytometer and incubated at 25ºC in 24-well plates with a concentration of 2 × 105 cell/ml, one day prior to be used.
Cultures of C. glabrata cells were grown until mid-exponential phase (OD600nm = 0.4–0.6) and the appropriate volume was collected to have 7 × 102 cells/ml in PBS. Galleria hemocyte monolayer medium was replaced with GIM without antimycotics, and then cells were infected with the yeast suspensions with a multiplicity of infection (MOI) of 1:5. After 1 h of infection at 37°C, the hemocytes were carefully washed twice with cell culture medium and fresh GIM medium was added. Viable intracellular yeast cells were measured after 1, 8, 24 or 48 h of infection, upon hemocyte lysis with 0.5%(v/v) Triton X-100 for 20 min and CFUs were counted by performing serial dilutions and plating in YPD agar plates.
Statistical analysis
All experiments represent the average of three or more independent experiments. Error bars represent the standard deviation. Statistical analyses were performed using one-way ANOVA with Tukey’s correction. Significance levels are attributed as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
Acknowledgments
We acknowledge John Bennett from the National Institute of Allergy and Infectious Diseases, NIH, Bethesda, USA, for kindly providing the L5U1 strain.
This work was supported by “Fundação para a Ciência e a Tecnologia” (FCT) [Contract PTDC/BII-BIO/28216/2017 and PhD grants to PP and MG] as well as by the Programa Operacional Regional de Lisboa 2020 (LISBOA-01-0145-FEDER-022231, the BioData.pt Research Infrastructure). Funding received by iBB-Institute for Bioengineering and Biosciences from FCT-Portuguese Foundation for Science and Technology (UIDB/04565/2020) is also acknowledged. This study was partly supported by a Joint Usage/Research Program of the Medical Mycology Research Center, Chiba University.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Tsai HF, Krol AA, Sarti KE, et al. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants, antimicrob. Agents Chemother. 2006;50(4):1384–1392. doi: 10.1128/AAC.50.4.1384-1392.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vermitsky JP, Earhart KD, Smith WL, et al. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Molecular Microbiology. 2006;61(3):704–722. [DOI] [PubMed] [Google Scholar]
- [3].Caudle KE, Barker KS, Wiederhold NP, et al. Genomewide expression profile analysis of the candida glabrata pdr1 regulon. Eukaryot Cell. 2011. doi: 10.1128/EC.00073-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Whaley SG, Rogers PD.. Azole Resistance in Candida glabrata. Curr Infect Dis Rep. 2016;18:41. [DOI] [PubMed] [Google Scholar]
- [5].Cavalheiro M, Costa C, Pais P, et al. Acquition of resistance to azole antifungal drugs in candida species: imidazoles versus triazoles. Adv Med Biol. 2017;108:183–210. [Google Scholar]
- [6].Galocha M, Pais P, Cavalheiro M, et al. Divergent approaches to virulence in C. albicans and C. glabrata: two sides of the same coin. Int J Mol Sci. 2019;20. doi: 10.3390/ijms20092345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pais P, Galocha M, Viana R, et al. Microevolution of the pathogenic yeasts Candida albicans and Candida glabrata during antifungal therapy and host infection. Microb Cell. 2019;6:142–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Seider K, Brunke S, Schild L, et al. The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J Immunol. 2011;187:3072–3086. [DOI] [PubMed] [Google Scholar]
- [9].Dementhon K, El-Kirat-Chatel S, Noël T. Development of an in vitro model for the Multi-Parametric quantification of the cellular interactions between Candida yeasts and phagocytes. PLoS One. 2012. doi: 10.1371/journal.pone.0032621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pais P, Galocha M, Teixeira MC. Genome-wide response to drugs and stress in the pathogenic yeast Candida glabrata. Prog Mol Subcell Biol. 2019;155–193. doi: 10.1007/978-3-030-13035-0_7 [DOI] [PubMed] [Google Scholar]
- [11].Vieira OV, Botelho RJ, Grinstein S. Phagosome maturation: aging gracefully. Biochem J. 2002;366:689–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Haas A. The phagosome: compartment with a license to kill.. Traffic. 2007;8:311–330. [DOI] [PubMed] [Google Scholar]
- [13].Roetzer A, Klopf E, Gratz N, et al. Regulation of Candida glabrata oxidative stress resistance is adapted to host environment. FEBS Lett. 2011. doi: 10.1016/j.febslet.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fukuda Y, Tsai HF, Myers TG, et al. Transcriptional profiling of Candida glabrata during phagocytosis by neutrophils and in the infected mouse spleen. Infect Immun. 2013;81:1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cuéllar-Cruz M, Briones-Martin-del-Campo M, Cañas-Villamar I, et al. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p, Eukaryot. Cell. 2008. doi: 10.1128/EC.00011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Briones-Martin-Del-Campo M, Orta-Zavalza E, Juarez-Cepeda J, et al. The oxidative stress response of the opportunistic fungal pathogen Candida glabrata. Rev Iberoam Micol. 2014;31:67–71. [DOI] [PubMed] [Google Scholar]
- [17].Ikner A, Shiozaki K. Yeast signaling pathways in the oxidative stress response. Mutat Res Mol Mech Mutagen. 2005;569:13–27. [DOI] [PubMed] [Google Scholar]
- [18].Klimova N, Yeung R, Kachurina N, et al. Phenotypic analysis of a family of transcriptional regulators, the zinc cluster proteins, in the human fungal pathogen Candida glabrata. G3 (Bethesda). 2014. doi: 10.1534/g3.113.010199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen KH, Miyazaki T, Tsai H-F, et al. The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene. 2007;386:63–72. [DOI] [PubMed] [Google Scholar]
- [20].Saijo T, Miyazaki T, Izumikawa K, et al. Skn7p is involved in oxidative stress response and virulence of candida glabrata. Mycopathologia. 2010. doi: 10.1007/s11046-009-9233-5 [DOI] [PubMed] [Google Scholar]
- [21].Thepnok P, Ratanakhanokchai K, Soontorngun N. The novel zinc cluster regulator Tog1 plays important roles in oleate utilization and oxidative stress response in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2014;450:1276–1282. [DOI] [PubMed] [Google Scholar]
- [22].Akache B, Wu K, Turcotte B. Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res. 2001;29:2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001. doi: 10.1038/35083594 [DOI] [PubMed] [Google Scholar]
- [24].Kaur R, Ma B, Cormack BP. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci U S A. 2007;104:7628–7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rai MN, Balusu S, Gorityala N, et al. Functional genomic analysis of Candida glabrata-macrophage interaction: role of chromatin remodeling in virulence. PLoS Pathog. 2012. doi: 10.1371/journal.ppat.1002863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chew SY, Ho KL, Cheah YK, et al. Glyoxylate cycle gene ICL1 is essential for the metabolic flexibility and virulence of Candida glabrata. Sci Rep. 2019;9:2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Byrne KP. Visualizing syntenic relationships among the hemiascomycetes with the yeast gene order browser. Nucleic Acids Res. 2006. doi: 10.1093/nar/gkj041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li L, Naseem S, Sharma S, et al. Flavodoxin-like proteins protect Candida albicans from oxidative stress and promote virulence. PLoS Pathog. 2015;11:e1005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. Embo J. 1997. doi: 10.1093/emboj/16.7.1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bilsland E, Molin C, Swaminathan S, et al. Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol Microbiol. 2004. doi: 10.1111/j.1365-2958.2004.04238.x [DOI] [PubMed] [Google Scholar]
- [31].Priebe S, Kreisel C, Horn F, et al. FungiFun2: A comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics. 2015. doi: 10.1093/bioinformatics/btu627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Maguire SL, ÓhÉigeartaigh SS, Byrne KP, et al. Comparative genome analysis and gene finding in Candida species using CGOB. Mol Biol Evol. 2013. doi: 10.1093/molbev/mst042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Roetzer A, Gregori C, Jennings AM, et al. Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors. Mol Microbiol. 2008;69:603–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Monteiro PT, Oliveira J, Pais P, et al. YEASTRACT+: a portal for cross-species comparative genomics of transcription regulation in yeasts. Nucleic Acids Res. 2019. doi: 10.1093/nar/gkz859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Monteiro PT, Pais P, Costa C, et al. The PathoYeastract database: an information system for the analysis of gene and genomic transcription regulation in pathogenic yeasts. Nucleic Acids Res. 2017;45:D597–D603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Grant CM, MacIver FH, Dawes IW. Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. FEBS Lett. 1997. doi: 10.1016/S0014-5793(97)00592-9 [DOI] [PubMed] [Google Scholar]
- [37].Thorpe GW, Fong CS, Alic N, et al. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci U S A. 2004. doi: 10.1073/pnas.0305888101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gutiérrez-Escobedo G, Orta-Zavalza E, Castaño I, et al. Role of glutathione in the oxidative stress response in the fungal pathogen Candida glabrata. Curr Genet. 2013. doi: 10.1007/s00294-013-0390-1 [DOI] [PubMed] [Google Scholar]
- [39].Gibson N, McAlister-Henn L. Physical and genetic interactions of cytosolic malate dehydrogenase with other gluconeogenic enzymes. J Biol Chem. 2003;278:25628–25636. [DOI] [PubMed] [Google Scholar]
- [40].Haselbeck RJ, McAlister-Henn L. Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J Biol Chem. 1993;268:12166. [PubMed] [Google Scholar]
- [41].Kratzer S, Schüller HJ. Carbon source-dependent regulation of the acetyl-coenzyme A synthetase-encoding gene ACSI from saccharomyces cerevisiae. Gene. 1995;161:75–79. [DOI] [PubMed] [Google Scholar]
- [42].Luttik MAH, Kötter P, Salomons FA, et al. The Saccharomyces cerevisiae ICL2 gene encodes a mitochondrial 2-methylisocitrate lyase involved in propionyl-coenzyme a metabolism. J Bacteriol. 2000;182:7007–7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lorenz MC, Bender JA, Fink GR. Transcriptional Response of Candida albicans upon Internalization by Macrophages. Eukaryot Cell. 2004;3:1076–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ueno K, Uno J, Nakayama H, et al. Development of a highly efficient gene targeting system induced by transient repression of YKU80 expression in Candida glabrata. Eukaryot Cell. 2007;6:1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jansen G, Wu C, Schade B, et al. Drag&Drop cloning in yeast. Gene. 2005;344:43–51. [DOI] [PubMed] [Google Scholar]
- [46].Vyas VK, Bushkin GG, Bernstein DA, et al. New CRISPR mutagenesis strategies reveal variation in repair mechanisms among fungi. MSphere. 2018. doi: 10.1128/msphere.00154-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ueno K, Matsumoto Y, Uno J, et al. Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS One. 2011;6:e24759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Costa C, Pires C, Cabrito TR, et al. Candida glabrata drug: H+antiporter CgQdr2 confers imidazole drug resistance, being activated by transcription factor CgPdr1, Antimicrob. Agents Chemother. 2013. doi: 10.1128/AAC.00811-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Costa C, Henriques A, Pires C, et al. The dual role of candida glabrata drug: H+antiporter CgAqr1 (ORF CAGL0J09944g) in antifungal drug and acetic acid resistance. Front Microbiol. 2013;4. doi: 10.3389/fmicb.2013.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Costa C, Nunes J, Henriques A, et al. Candida glabrata drug: H+antiporterCgTpo3 (ORF CAGL0I10384G): role in azole drug resistance and polyamine homeostasis. J Antimicrob Chemother. 2014;69:1767–1776. [DOI] [PubMed] [Google Scholar]
- [51].Pais P, Pires C, Costa C, et al. Membrane proteomics analysis of the Candida glabrata response to 5-flucytosine: unveiling the role and regulation of the drug efflux transporters CgFlr1 and CgFlr2. Front Microbiol. 2016;7. doi: 10.3389/fmicb.2016.02045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pais P, Costa C, Pires C, et al. Membrane proteome-wide response to the antifungal drug clotrimazole in Candida glabrata : role of the transcription factor CgPdr1 and the Drug: H+antiporters CgTpo1_1 and CgTpo1_2. Mol Cell Proteomics. 2016. doi: 10.1074/mcp.M114.045344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Romão D, Cavalheiro M, Mil-Homens D, et al. Determinant of Candida glabrata virulence: the acetate exporter CgDtr1. Front Cell Infect Microbiol. 2017. DOI: 10.3389/fcimb.2017.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zordan RE, Ren Y, Pan S-J, et al. Expression Plasmids for Use in Candida glabrata. G3: Genes | Genomes | Genetics. 2013;3:1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Jiang H, Lei R, Ding S-W, et al. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Mil-Homens D, Barahona S, Moreira RN, et al. Stress response protein BolA influences fitness and promotes Salmonella enterica serovar Typhimurium virulence. Appl Environ Microbiol. 2018. doi: 10.1128/AEM.02850-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


