Abstract
Purpose
Three genetic conditions—hereditary breast and ovarian cancer syndrome, Lynch syndrome, and familial hypercholesterolemia—have tier 1 evidence for interventions that reduce morbidity and mortality, prompting proposals to screen unselected populations for these conditions. We examined the impact of genomic screening on risk management and early detection in an unselected population.
Methods
Observational study of electronic health records (EHR) among individuals in whom a pathogenic/likely pathogenic variant in a tier 1 gene was discovered through Geisinger’s MyCode project. EHR of all eligible participants was evaluated for a prior genetic diagnosis and, among participants without such a diagnosis, relevant personal/family history, postdisclosure clinical diagnoses, and postdisclosure risk management.
Results
Eighty-seven percent of participants (305/351) did not have a prior genetic diagnosis of their tier 1 result. Of these, 65% had EHR evidence of relevant personal and/or family history of disease. Of 255 individuals eligible to have risk management, 70% (n = 179) had a recommended risk management procedure after results disclosure. Thirteen percent of participants (41/305) received a relevant clinical diagnosis after results disclosure.
Conclusion
Genomic screening programs can identify previously unrecognized individuals at increased risk of cancer and heart disease and facilitate risk management and early cancer detection.
Key words: genomic screening, BRCA1, BRCA2, Lynch syndrome, familial hypercholesterolemia
INTRODUCTION
Population screening for disease-causing genetic variants can identify individuals at risk for preventable cancers, heart disease, and other diseases who otherwise would not come to clinical attention. For example, our group and others have found that screening unselected cohorts for pathogenic/likely pathogenic BRCA1/2 variants can lead to improved ascertainment of individuals with these variants, 50% of whom did not have a personal or family history indicating their increased cancer risk.1–3 Further, genomic screening programs can identify individuals with early-onset cancers4 and improve medication management in individuals with high cholesterol levels.5
Three genetic conditions—hereditary breast and ovarian cancer (HBOC) syndrome (BRCA1 and BRCA2 genes), Lynch syndrome (MLH1, MSH2, MSH6, and PMS2 genes), and familial hypercholesterolemia (FH) (LDLR, APOB, and PCSK9 genes)—are recognized by the Centers for Disease Control and Prevention as having tier 1 evidence for interventions that reduce morbidity and mortality in individuals with certain personal and family history indications.6 At least 1% of the US population is thought to have genetic risk for these conditions,1,7–9 yet only a small minority of these individuals have been ascertained.10–12 Experts in clinical genetics, public health, and bioethics have highlighted these conditions as meeting criteria for a genomics-based screening program and noted the importance of gathering health outcomes data within such programs.13 Such outcomes include the rate of risk management among individuals with genetic results identified through a screening program and the degree to which genetic risk facilitates disease prevention and early detection.13
We sought to address these questions in an observational study that reviewed clinical data in a genomic screening program involving participants in Geisinger’s MyCode Community Health Initiative.14 We report outcomes of adult participants who received a report of a pathogenic/likely pathogenic variant in a gene that can cause one of the tier 1 genetic conditions. We focus on the degree to which the program identified individuals whose genetic status was previously unknown and on what care these participants had after receiving their genetic result. Findings from this study provide early evidence on the clinical utility of population screening for tier 1 genetic conditions and highlight opportunities to translate genetic information into clinical care.
MATERIALS AND METHODS
Setting and sample population
As detailed elsewhere,4,14 MyCode participants were enrolled throughout the Geisinger clinical enterprise without selection for disease risk (see Supplementary Fig. 1 for MyCode recruitment brochure). A subset of MyCode participants have undergone exome sequencing through the DiscovEHR collaboration with Regeneron Genetics Center15 and had sequence data analyzed for pathogenic/likely pathogenic variants in genes meeting the definition of actionability proposed by the American College of Medical Genetics and Genomics.16 Decisions on which genes to analyze were informed by interviews with patient and clinician stakeholders;17 review of genes’ actionability occurs annually using input from the Clinical Genome Resource (clinicalgenome.org), Geisinger clinicians, and genomic screening program leaders. All sequenced individuals had the same genes reviewed absent any knowledge of prior clinical genetic testing or diagnoses.
As a screening program that seeks to limit participant anxiety and health-care overutilization,18 we do not report variants of uncertain significance. We use a bioinformatic filtering process to select variants in genes of interest with the highest likelihood to be pathogenic/likely pathogenic. A variant scientist reviews all filtered variants to exclude variants of uncertain significance and likely benign/benign variants. To increase specificity and decrease the likelihood of reporting a variant with a classification discrepancy among clinical laboratories, we only clinically confirm variants classified in ClinVar as pathogenic/likely pathogenic by multiple laboratories without disagreements (2* status) or by an expert panel (3* status) or predicted loss-of-function variants in genes in which haploinsufficiency is an established mechanism of disease.
Variants selected by this process were independently classified in a CLIA-certified clinical laboratory19 and, if confirmed as pathogenic/likely pathogenic, disclosed to participants and their primary care clinicians.4,20 Program staff called each participant to discuss the medical and familial implications of their result and coordinate genetic counseling, medical evaluation, and risk management in accord with published guidelines.21–24 The same risk management guidelines were discussed with all participants, including those with a personal or family history of cancer or hypercholesterolemia, and were personalized as indicated according to current disease status (e.g., active treatment for cancer or hypercholesterolemia). For individuals already undergoing some risk management due to average-risk screening guidelines or personal or family history of disease, differences in surveillance or treatment informed by the presence of their genetic variant were highlighted as part of the disclosure and consultation (e.g., even in those with known hypercholesterolemia, the genetic diagnosis of FH would trigger a recommendation to achieve a lower LDL-C goal).23
Participants
This paper focuses on all individuals with a tier 1 genetic result disclosed between May 2015 and February 2018, thereby allowing participants sufficient time to have the recommended disease risk management.
Ethics statement
The Geisinger Institutional Review Board approved the protocols for MyCode and for the research described below. Informed consent was obtained from all participants.
Outcomes
We assessed the percentage of all MyCode participants with a tier 1 genetic result disclosed during the study period who were not aware of their pathogenic/likely pathogenic variant via previous clinical genetic testing. To assess the impact of the genomic screening program in a group naïve to the medical implications of a genetic result, we also assessed the percentage of participants without previous clinical tier 1 genetic testing who had the following:
EHR evidence of a personal or family history of relevant disease prior to results disclosure.
A guideline-based risk management procedure21–24 postdisclosure (among those eligible by age, sex, and previous treatment to have risk management).
A relevant diagnosis postdisclosure.
We also assessed the association between participants’ characteristics and postdisclosure risk management.
Data sources and measurement
We reviewed the Geisinger electronic health record (EHR) in June–July 2018 for data on personal and family history, diagnoses post–results disclosure, and guidelines-based risk management procedures for each genetic condition.21–24 The Geisinger EHR includes records back to 1997. Review of predisclosure risk management procedures focused on the procedure closest to the disclosure date. Study clinicians logged data in a spreadsheet with defined fields. A second study clinician independently reviewed each chart in October–December 2018. All discrepancies between reviewers were resolved through joint review and consensus.
Supplementary Table 1 lists diagnoses, risk management procedures, and family history considered to be relevant for HBOC syndrome,25 Lynch syndrome,26 and FH.27 The EHR was reviewed for a clinical diagnosis of FH or Lynch syndrome (there are no clinical diagnostic criteria for HBOC syndrome). To be considered as having a prior genetic diagnosis of the relevant genetic condition, an individual’s record needed to have a genetic testing report showing the same pathogenic/likely pathogenic variant that was detected via MyCode genomic screening or a chart note that listed the same variant. Individuals with a HBOC-associated or Lynch-associated result were considered to have related family history if there was documentation of one first-degree relative or two second-degree relatives with relevant cancer, elements of cancer genetic counseling referral guidelines that are easily assessed in most EHRs. Participants with a FH-associated result were considered to have related family history if they had one first- or two second-degree relatives with high cholesterol or coronary artery disease. Family history analysis only included history recorded in the EHR prior to results disclosure. To determine whether postdisclosure diagnoses could be attributed to results disclosure, we reviewed documentation for the procedures that facilitated diagnoses.
Statistical methods
All data were summarized to assess for outliers and missing data. Data were described for all participants and stratified by tier 1 condition and by whether participants had a risk management behavior postdisclosure. Categorical variables were described using frequency and percentage; continuous variables were reported as median and interquartile ranges. To determine how representative study participants were of MyCode participants overall and of Geisinger patients overall, we used Pearson’s chi-square and Wilcoxon rank-sum tests to compare study participants’ characteristics to those of adult MyCode participants and to adult, active Geisinger patients (i.e., those with one or more clinical encounters in the last 24 months). Bivariate comparisons between those who had versus did not have any recommended risk management postdisclosure were calculated using Fisher’s exact and Wilcoxon rank-sum tests. These comparisons were performed among those who were eligible for risk management based on applicable guidelines.21–24 Participants whose EHR noted a predisclosure procedure (e.g., mammogram) but did not list the date of the procedure were counted as eligible to have that procedure postdisclosure. We fitted a multivariable logistic regression model that included variables with significant bivariate associations with postdisclosure risk management. Model results are reported as odds ratios (OR) and 95% confidence intervals (CI). All analyses were conducted using SAS v9.4 (SAS Institute Inc., Cary, NC). Computer code used to generate results can be obtained from the corresponding author.
RESULTS
Participant characteristics
Summary information is in Table 1. A pathogenic/likely pathogenic variant in a tier 1 gene was discovered in 349 participants during the analysis period (Supplementary Fig. 2). Two of these individuals had a variant in two tier 1 genes; all analyses below were performed separately for each tier 1 condition (i.e., resulting in N = 351). Participants were 54% female, 99% self-reported European ancestry, 99% self-reported non-Hispanic ethnicity, and had a median age of 63 years. Compared with adult MyCode participants and with active Geisinger patients, study participants had significantly higher rates of European ancestry and non-Hispanic ethnicity, older median age, and higher comorbidity index. Median time from results disclosure to EHR review was 21.8 months (range 0.6–43.4 months).
Table 1.
Study participants’ demographic and health characteristics, stratified by tier 1 condition (n = 351a).
| Characteristic | FH (n = 93) | HBOC (n = 202) | Lynch (n = 56) | All study participants (n = 351a) | MyCode participants (n = 202,024) | P valuec | Active Geisinger patientsd (n = 917,910) | P valuec |
|---|---|---|---|---|---|---|---|---|
| Female sex | 56 (60.2%) | 103 (51.0%) | 32 (57.1%) | 191 (54.4%) | 123,244 (61.0%) | 0.041 | 507,599 (55.3%) | 0.94 |
| Race | 0.005 | <0.001 | ||||||
| White | 92 (98.9%) | 201 (99.5%) | 56 (100%) | 349 (99.4%) | 194,226 (96.1%) | 855,152 (93.2%) | ||
| African American | 1 (1.1%) | 1 (0.5%) | – | 2 (0.6%) | 5009 (2.5%) | 38,705 (4.2%) | ||
| Other | – | – | – | – | 2789 (1.4%) | 24,053 (2.6%) | ||
| Ethnicity | 0.026 | <0.001 | ||||||
| Non-Hispanic/Non-Latino | 92 (98.9%) | 199 (98.5%) | 56 (100%) | 348 (99.2%) | 196,922 (97.5%) | 880,702 (96.0%) | ||
| Other/Hispanic/Latino | 1 (1.1%) | 3 (1.5%) | – | 3 (0.8%) | 5102 (2.5%) | 37,208 (4.0%) | ||
| Current smoker | 13 (14.0%) | 43 (21.3%) | 11 (19.6%) | 67 (19.1%) | 37,920 (18.8%) | 0.87 | 177,877 (19.4%) | 0.89 |
| Alive at initial data pull | 93 (100%) | 194 (96.0%) | 56 (100%) | 343 (97.7%) | N/A | – | N/A | – |
| Median age in years (IQR) | 62.7 (51.2, 72.0) | 62.6 (50.6–72.1) | 62.8 (53.8–73.8) | 62.7 (51.0–72.2) | 55.0 (40.0, 67.0) | <0.001 | 51.0 (34.0, 66.0) | <0.001 |
| Median Charlson comorbidity index (IQR) | 5 (1–7) | 4 (2– 6) | 4 (2–6.5) | 4 (2–6) | 2 (0, 4) | <0.001 | 1 (0, 3) | <0.001 |
| Median follow-up in months (IQR, range)b | 14.4 (12.8–30.5, 7.4–43.3) | 24.2 (21.1–32.8, 0.6–43.3) | 14.7 (12.6–28.9, 8.0–36.3) | 21.8 (14.5–30.6, 0.6–43.3) | N/A | – | N/A | – |
| Prior genetic diagnosis | 0/93 (0%) | 39/202 (19.3%) | 7/56 (12.5%) | 46/351 (13.1%) | Not assessed | – | Not assessed | – |
FH familial hypercholesterolemia, HBOC hereditary breast and ovarian cancer syndrome, IQR interquartile range.
aTwo participants had a variant in two genes; analyses were performed separately for each tier 1 condition.
bNumber of months from date genetic testing report was uploaded to electronic health record to date of chart review completion.
cCompared with study participants.
dClinical encounter within last 24 months.
Eighty-seven percent of participants (305/351) were unaware of their genetic result via clinical genetic testing that occurred prior to MyCode results disclosure. None of the participants with a FH-associated variant identified by MyCode were already aware of it. Individuals with a prior genetic diagnosis (n = 46) were excluded from further analyses.
Clinical outcomes
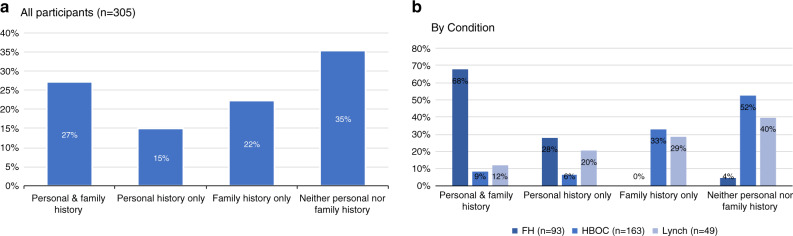
Among the 305 participants without prior knowledge of their genetic result, 42% had EHR evidence of a personal history of relevant disease prior to results disclosure and 50% had EHR evidence of a family history of relevant disease, for a total of 65% who had EHR evidence of a personal and/or family history of relevant disease (Fig. 1). None of the participants with a FH or Lynch syndrome variant had a prior clinical diagnosis of FH or Lynch syndrome. Relevant personal and family history were significantly more common among those with a FH result than among those with a HBOC or Lynch syndrome result (p < 0.001 for personal and family history of FH vs. HBOC and FH vs. Lynch).
Fig. 1. Relevant personal and family history among participants without prior genetic diagnosis.
a Relevant personal and family history in cohort. b Relevant personal and family history by genetic condition. FH familial hypercholesterolemia, HBOC hereditary breast and ovarian cancer syndrome, Hx history.
Eighty-four percent (255/305) of participants were eligible by age, sex, and prior treatment to have recommended disease risk management during the postdisclosure analysis period (Table 2). Seventy percent of these (179/255) had at least one recommended risk management procedure (e.g., breast magnetic resonance imaging [MRI], colonoscopy, lipid panel) after results disclosure (Tables 2 and 3). Of those who had a risk management procedure, 59% (105/179) had at least one such procedure within the syndrome-recommended interval prior to results disclosure (e.g., breast MRI within past year).21–24 Among the 132 eligible participants who had not had any recommended risk management in the risk-based interval prior to disclosure, 56% (n = 75) had a recommended risk management procedure postdisclosure.
Table 2.
Risk management performance and postdisclosure diagnoses among eligible participants without prior genetic diagnosis.
| FH (n = 93) | HBOC (n = 163) | Lynch (n = 49) | All (n = 305) | |
|---|---|---|---|---|
| Risk management eligiblea | 93/93 (100%) | 114/163 (69.9%) | 48/49 (98.0%) | 255/305 (83.6%) |
| Risk management predisclosureb | 69/93 (74.2%) | 43/114 (37.7%) | 11/48 (22.9%) | 123/255 (48.2%) |
| Risk management postdisclosurec | 78/93 (83.9%) | 82/114 (71.9%) | 19/48 (39.5%) | 179/255 (70.2%) |
| New diagnosis postdisclosured | 26/93 (28.0%) | 10/163 (6.1%) | 5/49 (10.2%) | 41/305 (13.4%) |
FH familial hypercholesterolemia, HBOC hereditary breast and ovarian cancer syndrome.
aEligible by age, sex, and prior treatment to have risk management per published guidelines during postdisclosure analysis period.
bHad at least one risk management procedure within recommended time period prior to genetic results disclosure.
cHad at least one risk management procedure during postdisclosure follow-up period.
dDiagnosis of relevant disease or clinical finding during postdisclosure follow-up period.
Table 3.
Type of postdisclosure risk management among eligible participants, by genetic condition.
| Condition (participants who performed risk management) | Risk management procedure | Number of participants per procedure | |
|---|---|---|---|
| Familial hypercholesterolemia (78) | LDL-C measurement (via lipid panel or LDL-direct) | 78 | |
| Cardiology appointment | 37 | ||
| Changed existing lipid-lowering therapy | 22 | ||
| Stress echocardiogram | 12 | ||
| Carotid ultrasound | 11 | ||
| Initiated lipid-lowering therapy | 7 | ||
| Pharmacy visita | 3 | ||
| Coronary artery bypass graft (CABG) | 2 | ||
| Carotid endarterectomy | 2 | ||
| Percutaneous coronary intervention (PCI) | 2 | ||
| Lipoprotein (a) measurement | 2 | ||
| Cardiac magnetic resonance imaging (MRI) | 0 | ||
| Calcium scoring | 0 | ||
| Hereditary breast and ovarian cancer syndrome (82) | Females (n = 52) | Males (n = 30) | |
| Mammogram | 48 | 3 | |
| Breast magnetic resonance imaging (MRI) | 28 | N/A | |
| Risk-reducing salpingo-oophorectomy | 16 | N/A | |
| Inherited risk breast clinic appointment | 23 | 1 | |
| CA-125 testing | 13 | N/A | |
| Transvaginal ultrasound | 10 | N/A | |
| Risk-reducing mastectomy | 9 | N/A | |
| Chemoprevention (tamoxifen or raloxifene) | 5 | N/A | |
| Prostate specific antigen (PSA) testing | N/A | 29 | |
| Lynch syndrome (19) | Females (n = 11) | Males (n = 8) | |
| Colonoscopy | 11 | 8 | |
| Inherited risk gastrointestinal clinic appointment | 7 | 3 | |
| Upper endoscopy | 7 | 5 | |
| Risk-reducing salpingo-oophorectomy | 2 | N/A | |
| Prophylactic hysterectomy | 1 | N/A | |
aFor familial hypercholesterolemia medication management.
Thirteen percent of participants (41/305) without prior knowledge of their genetic result had a relevant clinical diagnosis added to their EHR after results disclosure (Table 2). Nine of the ten participants diagnosed with a HBOC-associated lesion postdisclosure had stage 0, I, or II disease (Supplementary Table 2); the other had a serous tubal intraepithelial carcinoma, a precursor lesion for HBOC-associated ovarian cancer.28 All five participants diagnosed with Lynch-associated findings postdisclosure had adenomas (four with colon adenomas and one with sebaceous adenomas). Eight of these 15 participants with a HBOC- or Lynch-associated variant (53%) had EHR documentation of the diagnosis being attributed to results disclosure. The others were diagnosed due to symptoms (e.g., a participant with an ampulla of Vater cancer) or did not have documentation that attributed the diagnosis to results disclosure.
Of the 26 participants with a FH variant who had a postdisclosure diagnosis, 20 were diagnosed with a LDL-C >190 mg/dL, 5 were diagnosed with atherosclerosis, 4 with claudication and/or peripheral vascular disease, 2 with a cerebrovascular accident, and 7 with physical features of FH (3 with xanthoma/xanthelasma; 4 with corneal arcus). Seventeen of these individuals (65%) had EHR documentation of the diagnosis being attributed to results disclosure.
On bivariate analyses, personal and family history of relevant disease, postdisclosure genetic counseling, genetic condition (higher performance among those with a FH variant and lower performance among those with a Lynch variant, compared with performance among those with a HBOC variant), and having a recommended risk management procedure within the recommended time interval prior to disclosure were each significantly associated with having one or more recommended risk management procedures postdisclosure (Table 4). On logistic regression analysis including each of these variables plus age and female sex, postdisclosure genetic counseling (OR = 2.90 [95% CI = 1.52, 5.56], p = 0.001), genetic condition (HBOC set as reference, FH OR = 1.68 [95% CI = 0.59, 4.80], p = 0.33; Lynch OR = 0.25 [95% CI = 0.12, 0.56], p < 0.001), and prior risk management (OR = 3.36 [95% CI = 1.64, 6.87], p < 0.001) remained significantly associated with having a recommended risk management procedure after results disclosure.
Table 4.
Association between participants’ characteristics and postdisclosure performance of risk management among eligible participants.
| All participants (n = 255) | Postdisclosure risk management (n = 179) | No postdisclosure risk management (n = 76) | OR (95% CI), p value | |
|---|---|---|---|---|
| Female sex | 149 (58.4%) | 109 (60.9%) | 40 (52.6%) | 1.40 (0.82, 2.41), p = 0.22 |
| Race | N/A | |||
| White | 253 (99.2%) | 178 (99.4%) | 75 (98.7%) | |
| Black | 2 (0.8%) | 1 (0.6%) | 1 (1.3%) | |
| Ethnicity | N/A | |||
| Non-Hispanic/Non-Latino | 252 (98.2%) | 176 (98.3%) | 76 (100%) | |
| Other/Hispanic/Latino | 3 (1.2%) | 3 (1.7%) | 0 (0%) | |
| Current smoker | 51 (20.0%) | 31 (17.3%) | 20 (26.3%) | 0.58 (0.31, 1.11), p = 0.10 |
| Alive at initial data pull | 251 (98.4%) | 178 (99.4%) | 73 (96.0%) | N/A |
| Median age in years (IQR) | 62.4 (52.1, 70.8) | 62.7 (53.0, 70.7) | 61.2 (50.5, 71.7) | 1.01 (0.99, 1.03), p = 0.36 |
| Median Charlson comorbidity index (IQR) | 3.0 (1.0, 6.0) | 4.0 (2.0, 6.0) | 3.0 (1.0, 6.0) | 1.05 (0.96, 1.15), p = 0.32 |
| Relevant personal history | 121 (47.4%) | 95 (53.1%) | 25 (34.2%) | 2.18 (1.25, 3.80), p = 0.006 |
| Relevant family history | 137 (53.7%) | 108 (60.3%) | 29 (38.2%) | 2.46 (1.42, 4.28), p = 0.001 |
| Postdisclosure diagnosis | 41 (16.1%) | 41 (22.9%) | 0 (0%) | N/A |
| Genetic counseling visit postdisclosure | 141 (55.3%) | 107 (59.8%) | 34 (44.7%) | 1.84 (1.07, 3.16), p = 0.028 |
| Tier 1 genetic condition | ||||
| HBOC | 114 (44.7%) | 82 (45.8%) | 32 (42.1%) | Ref |
| FH | 93 (36.5%) | 78 (43.6%) | 15 (19.7%) | 2.03 (1.02, 4.03), p = 0.044 |
| Lynch | 48 (18.8%) | 19 (10.6%) | 29 (38.2%) | 0.26 (0.13, 0.52), p < 0.001 |
| Predisclosure risk management | 123 (48.2%) | 105 (58.7%) | 18 (23.7%) | 4.57 (2.49, 8.39), p < 0.001 |
CI confidence interval, FH familial hypercholesterolemia, HBOC hereditary breast and ovarian cancer syndrome, IQR interquartile range.
DISCUSSION
This observational study of clinical outcomes of individuals with a pathogenic/likely pathogenic variant in a tier 1 gene disclosed via a population genomic screening program highlights the program’s ability to identify at-risk individuals previously unaware of their variant and facilitate disease risk management. The study demonstrates the promise of more comprehensive ascertainment of individuals with genetic risk for actionable conditions,12 and serves as a foundation for further research that is necessary prior to routine implementation of genomic screening in clinical care.
While nearly 90% of participants were not ascertained through prior clinical genetic testing, lack of previous ascertainment did not mean that they were not at increased disease risk. Indeed, two-thirds of participants without prior ascertainment had EHR evidence of a personal and/or family history of relevant disease, a rate similar to that in a smaller study that performed more comprehensive phenotyping.29 Importantly, more than 10% had a relevant clinical diagnosis added to their EHR between results disclosure and chart review, highlighting the potential of genomic screening programs to reduce morbidity and mortality through early cancer detection or initiation of lipid-lowering therapy.
The preponderance of personal history and postdisclosure diagnoses in participants with a FH result compared with those with a hereditary cancer result suggests that genomic screening programs may best achieve clinical benefits if they screen for different conditions at different ages.13 While none of the participants with a FH result were previously aware of the result or had a clinical diagnosis of FH, nearly all had a personal history of relevant diagnoses (e.g., hypercholesterolemia, coronary artery disease, physical exam findings of cholesterol accumulation), and more than a quarter had a relevant diagnosis postdisclosure (e.g., new notation of LDL-C >190 mg/dL, new diagnosis of atherosclerosis). These missed opportunities to prevent atherosclerotic disease highlight the importance of beginning genomic screening for FH in childhood23,24,30 so that parents and children can adopt healthy lifestyles and initiate statin therapy when indicated. In contrast, those with a HBOC or Lynch result had a significantly lower rate of relevant personal history, yet still had several early-stage cancers and preinvasive lesions diagnosed postdisclosure, suggesting that genomic screening for these conditions can begin at a later age.
The finding that more than two-thirds of eligible participants across all three conditions chose to participate in some postdisclosure risk management suggests that genetic information can guide patient care in an unselected population. However, it is challenging to unambiguously attribute a specific health behavior to the disclosure of the genetic result in this unselected population in which some participants have already had risk management procedures due to average-risk screening guidelines or personal or family history of disease. This raises the possibility of misattribution of a health behavior (i.e., attributing a health behavior to the genetic result when it occurred for other reasons or attributing a health behavior to a previously established care plan when the behavior occurred due to the genetic result).
In spite of this uncertainty, several factors support an association between results disclosure and undergoing recommended risk management. A conservative approach to assignment of attribution based on explicit EHR documentation found that 61% (25/41) of postdisclosure diagnoses of cancer, adenomas, and FH features could be attributed to the genetic result. Additionally, more than half of those without prior risk-based management before disclosure had a recommended risk management procedure after disclosure. Further, postdisclosure risk management was not limited to individuals with personal or family history of relevant disease (which were not significantly associated with postdisclosure risk management in the logistic regression model). Finally, participants underwent several risk management procedures that would be uncommon among average-risk individuals but are recommended for those with a genetic diagnosis (see Table 3 for rates of participants with an FH variant who changed lipid-lowering therapy; females with an HBOC variant who had a breast MRI, risk-reducing salpingo-oophorectomy, or risk-reducing mastectomy; and participants with a Lynch syndrome variant who had an upper endoscopy or inherited risk clinic appointment).
The rates of postdisclosure risk management and diagnoses we observed highlight opportunities to increase the population health impact of a genomic screening program. Interventions that improve the rate of recommended risk management could lead to increased diagnoses of early-stage disease, particularly if the lower rate of risk management we observed among individuals with a Lynch syndrome variant persists in future studies. Our findings suggest that embedding genomic screening programs in health-care systems with ready access to genetic counselors can be associated with adherence. We found that having genetic counseling postdisclosure was significantly associated with undergoing recommended risk management. This finding, which is consistent with previous research on the role of genetics professionals in motivating cancer risk management,31 occurred even in a logistic regression model that included prior recommended risk management.
Our study’s limitations indicate that much research remains to be done before genomic screening can be deemed to have clinical utility. Although diagnosing cancer at earlier stages is associated with higher 5-year survival than at later stages,32 diagnosing a disease post–results disclosure will not necessarily lead to improved health outcomes in that individual and can introduce confounders such as lead time bias that inflate estimates of benefit. Longer-term studies of health outcomes in individuals with risk identified via genomic screening are required for a more thorough assessment of clinical utility. Additionally, our study was conducted within a racially and ethnically homogeneous population in a single health-care system. This may limit the generalizability of our findings and calls for evaluation of outcomes of genomic screening programs, like the one in All of Us,33 that occur under different conditions than ours, including those in more racially and ethnically diverse cohorts, younger cohorts with fewer comorbidities, and that occur outside of a health-care system (e.g., via a direct-to-consumer genomic screening test). Similarly, we acknowledge that by enrolling in MyCode, participants may have been inclined to value the utility of genomic screening results.
Although our process of double-coding charts and resolving discrepancies via consensus decreased the likelihood of missing or miscoding relevant data, the EHR has limitations as a data source. It fails to capture diagnoses and behaviors that occur outside our health-care system and can have incomplete reporting of relevant family history.1 Such incompleteness could lead to underestimates of the rates of individuals with prior genetic diagnoses, relevant clinical diagnoses before or after disclosure, and relevant risk management before or after disclosure. Further, the indication for a particular procedure is not always clearly documented, obscuring the clinical impact of the genetic result. Thus, it may be most useful to consider our reported rates of diagnoses and risk management as a floor on which future studies that include multiple data sources, including patient-reported outcomes, will build.
Finally, a comprehensive assessment of disease risk in participants was beyond the scope of this study. It is possible, given the higher comorbidity index in study participants compared with MyCode participants overall and with the overall Geisinger patient population (Table 1), that fewer postdisclosure diagnoses would occur in a healthier population. Future studies that include a comparison group and more thoroughly assess participants’ nongenetic disease risk may better address this question.
The classic World Health Organization principles for screening for disease,34 since updated for genomic technologies,35 can inform research that ensures the benefits of risk reduction outweigh the risks of health-care overutilization and the attendant financial and psychosocial costs. Although early evidence suggests that these costs will be modest,36,37 further research is required.38 Additionally, the prevalence and penetrance of pathogenic/likely pathogenic variants in clinically actionable genes in diverse, unselected populations are still being defined.1,29,36,39,40
Population genomic screening programs can identify previously unrecognized individuals at increased genetic risk of cancer and heart disease and facilitate risk management and early cancer detection, as evidenced by our study of clinical outcomes in individuals with a tier 1 genetic result. This study also highlights the need to determine optimal statistical methods for addressing the underlying uncertainties in EHR data analysis. Future longitudinal, pragmatic trials that include patient-reported outcomes and health-care utilization in diverse cohorts will further elucidate the clinical utility of genomic screening in unselected populations.
Supplementary information
Acknowledgements
Regeneron Genetics Center funds the cost of MyCode recruitment and research exome sequencing. The genomic screening program and observational study described in this paper were funded by institutional support from Geisinger.
Disclosure
None of the authors has a financial relationship with Regeneron Genetics Center. A.H.B.’s work has been funded by the National Institutes of Health (NIH) and the Marcus Foundation. He has received compensation as a section editor for the Journal of Genetic Counseling. He holds an equity stake in MeTree and You, Inc. L.K.’s work has been funded by the NIH, Patient-Centered Outcomes Research Institute (PCORI), and the Geisinger Health Plan Quality Fund. The work of M.L.B.S., M.A.K., T.S., M.L.G.H., M.B., L.B., A.L.L., L.K.J., A.K.R., and A.C.S. has been funded by the NIH. J.H.’s work has been funded by the NIH, Geisinger Health Plan, and Progenity, Inc. H.F.W. receives compensation as a member of the scientific advisory boards of the Simons Foundation and Helix. He previously received compensation as a member of the scientific review board of the Howard Hughes Medical Institute. C.L.M.’s work has been funded by the NIH and Simons Foundation. Dr. Ledbetter’s work has been funded by the NIH and the Marcus Foundation. He has received stock options as a scientific consultant to Clear Genetics, Natera and X-Therma. M.S.W.’s work has been funded by the NIH. He receives royalties from Elsevier. L.R.F., R.S., and H.R. declare no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41436-020-0876-4) contains supplementary material, which is available to authorized users.
References
- 1.Manickam K, Buchanan AH, Schwartz MLB, et al. Exome sequencing-based screening for BRCA1/2 expected pathogenic variants among adult biobank participants. JAMA Netw Open. 2018;1:e182140. doi: 10.1001/jamanetworkopen.2018.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabai-Kapara E, Lahad A, Kaufman B, et al. Population-based screening for breast and ovarian cancer risk due to BRCA1 and BRCA2. Proc Natl Acad Sci U S A. 2014;111:14205–14210. doi: 10.1073/pnas.1415979111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abul-Husn NS, Soper ER, Odgis JA, et al. Exome sequencing reveals a high prevalence of BRCA1 and BRCA2 founder variants in a diverse population-based biobank. Genome Med. 2019;12:2. doi: 10.1186/s13073-019-0691-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchanan AH, Manickam K, Meyer MN, et al. Early cancer diagnoses through BRCA1/2 screening of unselected adult biobank participants. Genet Med. 2018;20:554–558. doi: 10.1038/gim.2017.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones LK, Kulchak Rahm A, Manickam K, et al. Healthcare utilization and patients’ perspectives after receiving a positive genetic test for familial hypercholesterolemia. Circ Genom Precis Med. 2018;11:e002146. doi: 10.1161/CIRCGEN.118.002146. [DOI] [PubMed] [Google Scholar]
- 6.CDC. Office of Public Health Genomics. Genomic tests and family history by levels of evidence. 2014. http://www.cdc.gov/genomics/gtesting/tier.htm. Accessed 28 April 2015.
- 7.Abul-Husn NS, Manickam K, Jones LK, et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science. 2016;354:aaf7000.. doi: 10.1126/science.aaf7000. [DOI] [PubMed] [Google Scholar]
- 8.Win AK, Jenkins MA, Dowty JG, et al. Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2017;26:404–412. doi: 10.1158/1055-9965.EPI-16-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Austin MA, Hutter CM, Zimmern RL, Humphries SE. Genetic causes of monogenic heterozygous familial hypercholesterolemia: a HuGE prevalence review. Am J Epidemiol. 2004;160:407–420. doi: 10.1093/aje/kwh236. [DOI] [PubMed] [Google Scholar]
- 10.Hampel H, de la Chapelle A. The search for unaffected individuals with Lynch syndrome: do the ends justify the means? Cancer Prev Res. 2011;4:1–5. doi: 10.1158/1940-6207.CAPR-10-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordestgaard BG, Chapman MJ, Humphries SE, et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 2013;34:3478–3490a. doi: 10.1093/eurheartj/eht273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King MC, Levy-Lahad E, Lahad A. Population-based screening for BRCA1 and BRCA2: 2014 Lasker Award. JAMA. 2014;312:1091–1092. doi: 10.1001/jama.2014.12483. [DOI] [PubMed] [Google Scholar]
- 13.Murray MF, Evans JP, Angrist M, et al. A proposed approach for implementing genomics-based screening programs for healthy adults: discussion paper. NAM Perspectives. Washington, DC: National Academy of Medicine; 2018. 10.31478/201812a.
- 14.Carey DJ, Fetterolf SN, Davis FD, et al. The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet Med. 2016;18:906–913. doi: 10.1038/gim.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewey FE, Murray MF, Overton JD, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:aaf6814. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 16.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 17.Faucett WA, Davis FD. How Geisinger made the case for an institutional duty to return genomic results to biobank participants. Appl Transl Genomics. 2016;8:33–35. doi: 10.1016/j.atg.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard S, Sun S, Regier DA. Balancing uncertainty with patient autonomy in precision medicine. Nat Rev Genet. 2019;20:251–252. doi: 10.1038/s41576-019-0111-9. [DOI] [PubMed] [Google Scholar]
- 19.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams MS, Buchanan AH, Davis FD, et al. Patient-centered precision health in a learning health care system: Geisinger’s genomic medicine experience. Health Aff. 2018;37:757–764. doi: 10.1377/hlthaff.2017.1557. [DOI] [PubMed] [Google Scholar]
- 21.NCCN. Clinical Practice Guidelines in Oncology. Genetic/familial high-risk assessment: breast and ovarian. Version 1.2018. 2017. https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf. Accessed 27 March 2019.
- 22.NCCN. Clinical Practice Guidelines in Oncology. Genetic/familial high-risk assessment: colorectal. Version 1.2018. 2018. https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf. Accessed 27 March 2019.
- 23.Gidding SS, Champagne MA, de Ferranti SD, et al. The agenda for familial hypercholesterolemia: a scientific statement from the American Heart Association. Circulation. 2015;132:2167–2192. doi: 10.1161/CIR.0000000000000297. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg AC, Hopkins PN, Toth PP, et al. Familial hypercholesterolemia: screening, diagnosis and management of pediatric and adult patients: clinical guidance from the National Lipid Association Expert Panel on Familial Hypercholesterolemia. J Clin Lipidol. 2011;5(3 (Suppl)):S1–S8. doi: 10.1016/j.jacl.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Petrucelli N, Daly MB, Pal T. BRCA1- and BRCA2-associated hereditary breast and ovarian cancer. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews. Seattle, WA: University of Washington; 1993.
- 26.Kohlmann W, Gruber SB Lynch syndrome. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews. Seattle, WA: University of Washington; 1993.
- 27.Youngblom E, Pariani M, Knowles JW Familial Hypercholesterolemia. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews. Seattle, WA: University of Washington; 1993.
- 28.Dietl J. Revisiting the pathogenesis of ovarian cancer: the central role of the fallopian tube. Arch Gynecol Obstet. 2014;289:241–246. doi: 10.1007/s00404-013-3041-3. [DOI] [PubMed] [Google Scholar]
- 29.Johnston JJ, Lewis KL, Ng D, et al. Individualized iterative phenotyping for genome-wide analysis of loss-of-function mutations. Am J Hum Genet. 2015;96:913–925. doi: 10.1016/j.ajhg.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daniels SD, Benuck I, Christakis DA, et al. Expert panel on integrated guidelines for cardiovascular health and risk reduction in children and adolescents. National Heart Lung and Blood Institute. 2012. https://www.nhlbi.nih.gov/files/docs/peds_guidelines_sum.pdf. Accessed 10 May 2019.
- 31.Pal T, Lee JH, Besharat A, et al. Modes of delivery of genetic testing services and the uptake of cancer risk management strategies in BRCA1 and BRCA2 carriers. Clin Genet. 2014;85:49–53. doi: 10.1111/cge.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howlader N, Noone AM, Krapcho M, et al. SEER cancer statistics review, 1975-2013. National Cancer Institute. 2016. http://seer.cancer.gov/csr/1975_2013/. Accessed 21 October 2016.
- 33.Denny JC, Rutter JL, Goldstein DB, et al. The “All of Us” research program. N Engl J Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson JMG, Jungner G. Principles and practice of screening for disease. World Health Organization. 1968. https://apps.who.int/iris/handle/10665/37650. Accessed 26 March 2019.
- 35.Andermann A, Blancquaert I, Beauchamp S, Dery V. Revisiting Wilson and Jungner in the genomic age: a review of screening criteria over the past 40 years. Bull World Health Organ. 2008;86:317–319. doi: 10.2471/BLT.07.050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hart MR, Biesecker BB, Blout CL, et al. Secondary findings from clinical genomic sequencing: prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study. Genet Med. 2019;21:1100–1110. doi: 10.1038/s41436-018-0308-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Bao Y, Riaz M, et al. Population genomic screening of all young adults in a health-care system: a cost-effectiveness analysis. Genet Med. 2019;21:1958–1968. doi: 10.1038/s41436-019-0457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson JF, Roden DM, Orlando LA, Ramirez AH, Mensah GA, Williams MS. Building evidence and measuring clinical outcomes for genomic medicine. Lancet. 2019;394:604–610. doi: 10.1016/S0140-6736(19)31278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haer-Wigman L, van der Schoot V, Feenstra I, et al. 1 in 38 individuals at risk of a dominant medically actionable disease. Eur J Hum Genet. 2019;27:325–330. doi: 10.1038/s41431-018-0284-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Natarajan P, Gold NB, Bick AG, et al. Aggregate penetrance of genomic variants for actionable disorders in European and African Americans. Sci Transl Med. 2016;8:364ra151. doi: 10.1126/scitranslmed.aag2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.