Abstract
Purpose
We studied the penetrance of pathogenically classified variants in an elderly Dutch population from the Rotterdam Study, for which deep phenotyping is available. We screened the 59 actionable genes for which reporting of known pathogenic variants was recommended by the American College of Medical Genetics and Genomics (ACMG), and demonstrate that determining what constitutes a known pathogenic variant can be quite challenging.
Methods
We defined “known pathogenic” as classified pathogenic by both ClinVar and the Human Gene Mutation Database (HGMD). In 2628 individuals, we performed exome sequencing and identified known pathogenic variants. We investigated the clinical records of carriers and evaluated clinical events during 25 years of follow-up for evidence of variant pathogenicity.
Results
Of 3815 variants detected in the 59 ACMG genes, 17 variants were considered known pathogenic. For 14/17 variants the ClinVar classification had changed over time. Of 24 confirmed carriers of these variants, we observed at least one clinical event possibly caused by the variant in only three participants (13%).
Conclusion
We show that the definition of “known pathogenic” is often unclear and should be approached carefully. Additionally variants marked as known pathogenic do not always have clinical impact on their carriers. Definition and classification of true (individual) expected pathogenic impact should be defined carefully.
Keywords: ACMG genes, clinical interpretation, pathogenic variants, exome sequencing, penetrance
INTRODUCTION
Exome sequencing (ES) is of great value to detect rare, disease-causing genetic variants in affected individuals, and is applied in both diagnostic as well as research settings. However, evaluating whether a variant causes the disease can be challenging, even when this variant is predicted as potentially pathogenic by bioinformatic tools and classified as such in databases as the Human Gene Mutation Database (HGMD) and/or ClinVar. Increasingly, ES is being applied to large population-based settings with the potential to detect incidental or secondary findings.
Given these developments, the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG-AMP) has released a set of guidelines on interpretation of genetic variants for clinical interpretation.1
These guidelines include evidence like variant segregation through the affected individuals’ family, previously described presence of other disease-causing variants in the same gene, and knowledge of the functional mechanism of this gene in relation to the disease. Variants are classified in five classes based on clinical relevance: (1) benign, (2) likely benign, (3) uncertain significance, (4) likely pathogenic, and (5) pathogenic.1 Some databases, like ClinVar, directly follow this classification system.2 Other databases use their own adaptation of such a classification, such as HGMD.3
In 2013, Green et al. published a list of 56 genes involving rare monogenetic disorders for which preventive measures and/or treatments were available and recommended reporting to carriers of “incidental or secondary” findings, in clinical exome and genome sequencing data, regardless the diagnostic implication for which the sequencing was ordered.4 This list was updated by Kalia et al. in 2016, removing one gene and adding four others to a total of 59 genes.5 However, insufficient knowledge on penetrance of many variants, also in the categories of known pathogenic (KP) or expected pathogenic (EP) variants, makes interpretation challenging. Since then various studies have looked into the carrier status of pathogenic gene variants in larger and healthy populations and how pathogenicity scores are defined by different databases.6–10
Comparing interpretations of 99 variants of different classifications based on the ACMG-AMP guidelines of genetic variants in a Mendelian disease family setting showed a 71% to 92% agreement between 9 clinical laboratories.7 This indicates that clinical interpretation of genetic variants for the primary outcome (the Mendelian disease segregating in these families) yields similar conclusions for most patients in these diagnostic laboratories. In regard to secondary findings in sequencing data sets from non-family-based sources, investigations of several large population studies show that between 0.7% and 3.4% of their study population participants carry a KP or EP variant.6,8–10 Several of these studies used the list of 56 genes initially reported by Green et al.9,10 Other studies add additional genes considered to have a clear phenotype–genotype relation by clinical genetic specialists, like the 112–114 genes used by Dorschner et al. and Amendola et al.6,8 Most studies reported KP and EP carriers, although Amendola et al. and Jurgens et al. report respectively 0.7% and 0.9% carriers of only KP variants, suggesting almost 1% of the population carries a KP variant in the 56 ACMG genes.6,9 Yet, these studies lack an extensive clinical follow-up with information on health and disease status of the participants. And so, how many of these carriers of KP or EP variants actually have experienced clinically relevant phenotypes due to these variants is not yet clear.
Recent studies have shown that the occurrence of KP variants is higher in the healthy normal population than expected based on the frequency in the Mendelian disease patient cohorts in which these variants have been originally identified. For example, Minikel et al. showed that the prevalence of missense variants in the dominant prion disease gene PRNP was 30-fold higher in the general population than expected based on prion disease prevalence.11 A similar observation was made for ASXL1 and other intellectual disability genes by Ropers et al.12 On a larger scale, Saleheen et al. showed that 1317 genes were predicted to be completely knocked out in at least 1 of 10,503 adult Pakistani individuals, caused by the large rate of consanguinity in this population, but in many cases without obvious phenotype.13 Similarly, Lek et al. showed that 3230 genes in their Exome Aggregation Consortium database of 60,706 individuals harbored damaging variants without a currently established disease phenotype.14 They also showed that each participant carried on average 54 variants that might be considered pathogenic by ClinVar or HGMD, often at higher than expected frequencies, even for homozygous variants in genes for recessive inheritance. Finally, Chen et al. identified 13 carriers of severe Mendelian pathogenic variants in a large cohort of nearly 600,000 participants,15 who did not show the expected phenotypes and were considered nonpenetrant or resilient to these variants. Results like these show that many potentially pathogenic variants have a lower than expected penetrance in healthy populations and thus should be interpreted with caution.
In our study, we combined ES data with clinical information of 2628 participants of the longitudinal Rotterdam Study. This is a prospective, population-based cohort study of elderly subjects 45 years and older, living in a suburb of Rotterdam since 1990, and of whom we have almost 30 years of follow-up information from clinical records and detailed physical examination every 4–5 years.16 In the ES data we evaluated different variant classifications for the 59 ACMG genes, using and comparing ClinVar and HGMD to ascertain known pathogenic variants, and then retrospectively look into the clinical history of carriers to evaluate possible variant pathogenicity and penetrance. Additionally, we analyzed overall changes of variant classification over time in the different database versions of ClinVar, in particular for the identified known pathogenic variants observed in our study population.
MATERIALS AND METHODS
Details on collection and processing of exome sequencing data from the Rotterdam Study have been described previously.17 In short, DNA of 2628 participants was sequenced to an average depth of 56× using NimbleGen SeqCap v2 capture and Illumina’s Hiseq2000. Data was processed using BWA, picard, samtools and GATK. Variants were called using GATKs HaplotypeCaller. Variants with a variant quality over sequencing depth (QD) < 5 were filtered out. Variants in the 59 ACMG genes were extracted and annotated using Annovar, including minor allele frequencies (MAFs) from the Genome Aggregation Database (gnomAD, Karczewski et al., 2019, unpublished data), Combined Annotation Dependent Depletion (CADD) scores, and multiple versions of the ClinVar database, including the most recently available version (2018-03-06).2,18 Variants were annotated to HGMD (v17.3) by batch filtering in the HGMD professional database.3 No additional filtering was performed based on CADD score or population MAF.
Identifying known pathogenic variants
To identify KP variants in our data set we utilized the largest and most commonly used databases of clinical interpretation of genetic variants: the National Center for Biotechnology Information (NCBI) ClinVar database and the Human Gene Mutation Database (HGMD). We categorized the classifications from both databases for all variants detected in the 59 ACMG genes according to the five major classifications outlined in the ACMG-AMP guidelines, to be able to compare classifications in both databases.1 Specific additional evidence criteria from ClinVar were not assessed at this point.
We added the category for absence from databases with a zero as follows: 0: absent from database; 1: benign; 2: likely/probable benign or likely/probably nonpathogenic; 3: unknown, untested, or uncertain; 4: likely/probably pathogenic; and 5: pathogenic. When multiple classifications for the same variant were available in ClinVar, they were averaged (e.g., a 4–4–5 variant is classified as class 4, while a 4–5–5 variant is classified as 5). HGMD classifications were coded in a similar manner: 0: absent from database; 3: no clinical interpretation available (NA) or functional polymorphism (FP); 4: disease polymorphism (DP), disease functional polymorphism (DFP), or possible disease mutation (DM?); and 5: disease mutation (DM). Classes 1 and 2 are not present in HGMD. Variants classified as class 5 in both ClinVar and HGMD were considered KP variants. All KP variants were checked in the latest online ClinVar database (date: April 2020) to confirm the pathogenic classification for the phenotype of which the gene was included in the ACMG recommendations. From this time point, the ClinVar star rating score was extracted for each variant, as well as the number of submissions, as indicated in Table 1.
Table 1.
Annotation of 17 known pathogenic variants.
| ClinVar annotations | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Variant | Identifier | HGVS_transcript | HGVS_Predicted_Protein | Carrier | Sanger | MAF gnomAD | CADD | 2014–2018 | 2020 stars | Submissions |
| RET | chr10:43609097_G>A | rs79781594 | NM_020630.5:c.1853G>A | NP_065681.1(LRG_518p2):p.(Cys618Tyr) | A1 | + | . | 19 | 5,5,5,5,5 | 2/4 | 2 |
| PTEN | chr10:89685307_T>C | rs398123317 | NM_000314.5:c.202T>C | NP_000305.3(LRG_311p1):p.(Tyr68His) | B1 | + | . | 23 | 0,0,5,3,5 | 2/4 | 1 |
| KCNQ1 | chr11:2591882_G>A | rs179489 | NM_000218.2:c.502G>A | NP_000209.2(LRG_287p1):p.(Gly168Arg) | C1 | + | 0.000012 | 30 | 3,4,4,4,5 | 2/4 | 3 |
| KCNQ1 | chr11:2591949_G>A | rs120074178 | NM_000218.2:c.569G>A | NP_000209.2(LRG_287p1):p.(Arg190Gln) | D1 | + | 0.000004 | 35 | 4,4,5,4,5 | 2/4 | 4 |
| MYBPC3 | chr11:47356671_G>A | rs387907267 | NM_000256.3:c.2827C>T | NP_000247.2(LRG_386p1):p.(Arg943Ter) | E1 | + | 0.000012 | 39 | 5,5,5,5,5 | 2/4 | 9 |
| MYL2 | chr12:111348980_C>G | rs199474813 | NM_000432.3:c.403–1G>C | NP_000423.2(LRG_393p1):p.? | F1 | + | 2/4 | 2 | |||
| F2 | + | ||||||||||
| F3 | + | ||||||||||
| F4 | + | 0.000045 | 16 | 3,4,4,4,5 | |||||||
| F5 | + | ||||||||||
| F6 | + | ||||||||||
| F7 | + | ||||||||||
| MYL2 | chr12:111356937_C>T | rs104894368 | NM_000432.3:c.64G>A | NP_000423.2(LRG_393p1):p.(Glu22Lys) | G1 | + | 0.000020 | 33 | 5,3,4,4,5 | 2/4 | 9 |
| BRCA2 | chr13:32900281_AA>- | rs397507739 | NM_000059.3:c.469_470del | NP_000050.2(LRG_293p1):p.(Lys157ValfsTer25) | H1 | + | . | . | 4,4,4,4,5 | 3/4 | 6 |
| BRCA2 | chr13:32930747_G>T | rs397507922 | NM_000059.3:c.7617+1G>T | NP_000050.2(LRG_293p1):p.? | I1 | + | . | 21 | 0,5,3,5,5 | 2/4 | 4 |
| BRCA2 | chr13:32936831_G>A | rs81002873 | NM_000059.3:c.7976+1G>A | NP_000050.2(LRG_293p1):p.? | J1 | - | . | 29 | 4,4,4,4,5 | 3/4 | 4 |
| J2 | - | ||||||||||
| BRCA1 | chr17:41209068_C>T | rs80358150 | NM_007300.3:c.5340+1G>A | NP_009231.2:p.? | K1 | + | . | 18 | 0,3,5,4,5 | 3/4 | 13 |
| DSC2 | chr18:28667778_T>C | rs397514042 | NM_024422.3:c.631–2A>G | NP_077740.1:p.? | L1 | + | 0.000016 | 9 | 5,3,4,5,5 | 1/4 | 2 |
| DSG2 | chr18:29116261_G>A | rs121913009 | NM_001943.3:c.1520G>A | NP_001934.2(LRG_397p1):p.(Cys507Tyr) | M1 | + | . | 15 | 5,5,5,5,5 | 0/4 | 1 |
| LDLR | chr19:11210962_G>A | rs267607213 | NM_000527.4:c.131G>A | NP_000518.1(LRG_274p1):p.(Trp44Ter) | N1 | + | . | 25 | 5,5,3,3,5 | 2/4 | 13 |
| RYR1 | chr19:38948185_C>T | rs118192172 | NM_000540.2:c.1840C>T | NP_000531.2(LRG_766p1):p.(Arg614Cys) | O1 | + | 16 | 3,3,3,4,5 | 3/4 | 5 | |
| O2 | + | 0.000097 | |||||||||
| O3 | + | ||||||||||
| RYR1 | chr19:38986923_C>T | rs118192177 | NM_000540.2:c.6617C>T | NP_000531.2(LRG_766p1):p.(Thr2206Met) | P1 | + | 0.000012 | 17 | 0,3,4,3,5 | 2/4 | 2 |
| RYR1 | chr19:39071043_G>A | rs118192168 | NM_000540.2:c.14545G>A | NP_000531.2(LRG_766p1):p.(Val4849Ile) | Q1 | + | 0.000016 | 17 | 5,4,4,4,5 | 3/4 | 2 |
For each variant the genomic location (build hg37), single-nucleotide polymorphism (SNP) identifier, Human Genome Variation Society (HGVS) coding, minor allele frequency (MAF) in the gnomAD exome database, CADD score, and ClinVar classification class for the five tested version time points (2014–2018) are indicated. All variants had classification 5 according to the Human Gene Mutation Database (HGMD) and the 2018 version of ClinVar. In addition, the pathogenic classification of each variant was confirmed in the most recent online ClinVar database (09-04-2020, 9 April 2020). For each variant the 2020 ClinVar star rating and number of submission are shown. In ClinVar, the following definition is giving for these star classifications: 0: no assertion criteria provided; 1: criteria provided, conflicting interpretation; 2: criteria provided, multiple submitters; 3: reviewed by expert panel. The column “Sanger” denotes confirmed (+) (24 samples) or unconfirmed (-) (2 samples) by Sanger sequencing.
Phenotypic validation of carriers
Phenotypic events of all study participants are collected weekly by automated linking of the general practitioners' records and diagnoses made by medical specialists, as detailed in the Supplemental methods. These events are compared with all medical records, letters from medical specialists, and discharge reports. All events were confirmed by trained research assistants. Participants are interviewed about all events at their next study visit.19
For each KP variant carrier, the events and respective age at event were extracted. For each carrier of a KP variant with an event of interest, four clinicians evaluated the potential causal relationship between the variant and the event, giving consideration to the age at which the event occurred. Ties were broken by the first author. For events marked by a majority all occurrences of this event in the data set were collected. For each event, the average age at event and the standard deviation were determined. The age at event of the KP carrier was expressed as a z-score, by calculating the number of standard deviations from the average event age across the 2628 participants with ES data available.
Confirmation by Sanger sequencing
All carriers of KP variants classified as class 5 by both ClinVar and HGMD were validated using Sanger sequencing. Primers were designed and produced by Baseclear B.V. (Leiden, The Netherlands). Optimal primer annealing temperature was determined using gradient polymerase chain reaction (PCR) on control DNA samples. Sanger sequencing of variants in BRCA1/2 was performed at our department of clinical genetics, where this is routinely performed for diagnostic purposes. Sanger sequencing for the other variants was performed by Baseclear B.V. Results were checked manually to verify the variants. Primer sequences and Sanger results are available in Supplemental results 1. Variants not confirmed by Sanger sequencing were retained as to not bias further interpretation (two variants in BRCA2), as is addressed in the discussion.
Ethics statement
The Rotterdam Study has been approved by the Medical Ethics Committee of Erasmus MC (registration number MEC 02.1015) and by the Dutch Ministry of Health, Welfare and Sport (Population Screening Act WBO, license number 1071272–159521-PG). This study has been entered into the Netherlands National Trial Register (www.trialregister.nl) and into the World Health Organization (WHO) International Clinical Trials Registry Platform (www.who.int/ictrp/network/primary/en/) under shared catalog number NTR6831. All participants provided written informed consent to participate in the study and to have their information obtained from treating physicians.
RESULTS
Identification of known pathogenic variant carriers
Exome sequencing was performed on 2628 Rotterdam Study (RS) participants and after filtering and quality control (QC) resulted in a total of 703,990 genomic variants, as was previously described.17 Of these, 3815 variants were located in one of the 59 ACMG genes.5 All these 3815 variants were classified using both the HGMD and ClinVar databases, resulting in six classes—0 (absent from database), 1 (benign), 2 (likely benign), 3 (uncertain), 4 (likely pathogenic), or 5 (pathogenic)—per database.
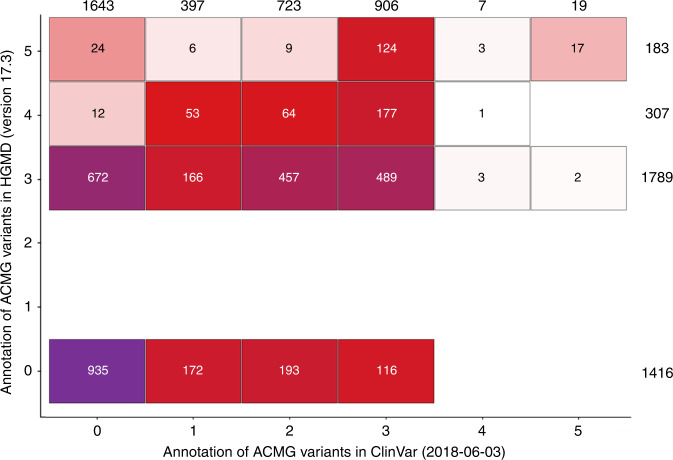
The 3815 variants were classified and grouped according to this system as indicated in Fig. 1, comparing their classification in both databases. The 119 variants in autosomal recessive genes MUTYH or ATP7B were excluded from this figure and analyzed separately. Of the resulting 3696 variants, 935 variants (25%) were absent from both databases. An additional 708 variants (19%) were present in HGMD but not in ClinVar and another 481 variants (13%) were present in ClinVar but not in HGMD. Thus, the remaining 1691 variants (43%) were classified by both databases. Furthermore, HGMD classifies 183 of these variants (5%) as pathogenic (class 5) versus only 19 by ClinVar (0.5%). In total 17 variants are classified as pathogenic by both of the databases (0.5% of all variants), and are here defined as known pathogenic (KP) variants. In total, 24 participants were confirmed by Sanger validation to carry one of these 17 KP variants (0.9% of all participants). An additional two carriers of a single variant in BRCA2 were identified, but were found to be false positives by Sanger validation. These variants were retained as not to bias further interpretation, but are carefully marked in subsequent tables.
Fig. 1. Classification of clinically relevant variants in 2628 Rotterdam Study participants in the 59 American College of Medical Genetics and Genomics–Association for Molecular Pathology (ACMG-AMP) genes according to ClinVar version 2018 and the Human Gene Mutation Database (HGMD).
Classes are defined per the ACMG-AMP guidelines: (1) benign, (2) likely benign, (3) uncertain, (4) likely pathogenic, (5) pathogenic. Variants absent from the database are coded as 0. The classifications for HGMD were converted to class 3 (No interpretation available (NA), functional polymorphism (FP) and disease polymorphism (DP)), class 4 (disease functional polymorphism (DFP), possible disease mutations (DM?)) and class 5 (disease mutation (DM). For visualization purposes, the variants observed in autosomal recessive genes ATP7B and MUTYH are not shown. The numbers at the sides are sums for that respective classification.
Additionally, 8 of the 119 variants in MUTHY and ATP7B were classified as pathogenic by both HGMD and ClinVar (not shown), but only as autosomal recessive inheritance, thus in homozygous state. In total, 50 carriers were observed for any of these 8 variants, all in a heterozygous state. No compound heterozygosity was detected. Heterozygous variants in these genes were not considered as KP and thus they were not followed up further.
Variation in ClinVar clinical classification over time
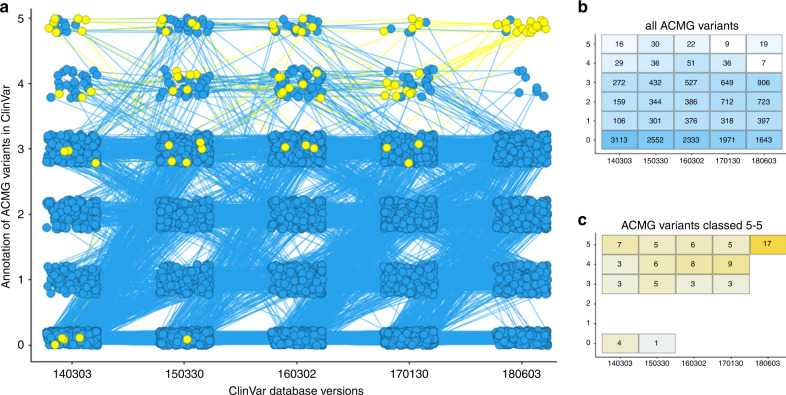
We have downloaded ClinVar database versions from the years 2014 until 2018. For HGMD the most recent online version was used (v17.3). Comparing the clinical classification for the 3815 ACMG variants identified in our study population between ClinVar database versions shows that classification largely changes over time, as shown in Fig. 2. First, in 2014 only 582 variants were present in ClinVar (16%), versus 2052 in 2018 (56%), a 3.5-fold increase. This increase was most notable for variants of class 1: benign (3.7-fold increased), class 2: likely benign (4.5-fold increased), and class 3: uncertain significance (3.3-fold increased). Whereas class 5: pathogenic remained almost unchanged (1.2-fold increase) and class 4: likely pathogenic decreased 4.1-fold decrease). The migration of classification for the 17 known pathogenic variants (as classified in version 2018) is marked separately in Fig. 2. As shown, only between 5 and 7 of these 17 KP variants were classified as pathogenic at the same time at any given ClinVar version in the previous years. In fact, only 3 of the 17 KP variants remained at class 5 in all tested previous versions of ClinVar. The classification per variant per ClinVar version is indicated in Table 1. All variants were confirmed pathogenic at the online version of ClinVar (dated April 2020). Five of the 17 variants received a three star score in ClinVar (reviewed by expert panel), and 10 received a two star score (multiple submitters, no conflicting interpretation). A single variant received a one star score (multiple submitters, conflicting interpretation), and one variant received a zero star score (no assertion criteria provided).
Fig. 2. Classification changes of all variants detected in the 59 ACMG genes by ClinVar over time.
(a) Classification of all variants detected in one of the 59 American College of Medical Genetics and Genomics (ACMG) genes in 2628 participants of the Rotterdam Study population according to ClinVar at different time points: March 2014 (date 140303), March 2015 (date 150330), March 2016 (date 160302), January 2017 (date 170130), and June 2018 (date 180603). Each variant is connected by a line between all five versions. Marked in yellow are the 17 known pathogenic variants classified as category 5 by the most recent versions of ClinVar (version 180603) and the Human Gene Mutation Database (HGMD) (version 17.3). (b) The number of variants in each class of each ClinVar database version. (c) The class at each database version for the 17 variants that were classified as 5 in ClinVar in 2018 and by HGMD 17.3 (marked yellow in a). For visualization purposes, the variants observed in autosomal recessive genes ATP7B and MUTYH are not shown.
Phenotypic evaluation of known pathogenic carriers
We extracted 94 International Statistical Classification of Diseases and Related Health Problems, Tenth Revision (ICD10)–coded clinical events for the 26 KP carriers, from 9165 coded clinical events across our 2628 study participants, in addition to the age at each event, shown in Fig. 3. In total, 18 events (20%) in 10 different individuals were marked by at least one clinical referee as possibly related to the KP variant. Nine events (10%) in three carriers (indicated with an asterisk in Fig. 3) were marked by at least three referees.
Fig. 3. Twenty-six carriers of 17 known pathogenic (KP) variants, one shown on each line.
The column “Sanger” denotes confirmed (+) (24 samples) or unconfirmed (-) (2 samples) by Sanger sequencing. For each carrier, their recorded clinical events are displayed in 5-year intervals. The events are coded using the International Statistical Classification of Diseases and Related Health Problems, Tenth Revision (ICD10) classification system. The last column denotes the primary disease for which the gene was included in the American College of Medical Genetics and Genomics (ACMG) recommendations. Events marked with a “++” are evaluated by at least 3 of the 5 referees (3 of 4 clinicians or 2 clinicians and the first author) as possibly explained by the variant for which the patient was a carrier. Those carriers are marked by an asterisk and shown in bold. Events marked with a single “+” were marked by only 1 or 2 referees. ICD10 codes in alphabetical order: neoplasm of C18: colon, C19: rectosigmoid junction, C34: bronchus, C44: skin, C45: mesothelioma, C50: breast, C61: prostate, C66: ureter, C67: bladder. D47: other neoplasm of uncertain behavior. F00: Alzheimer disease, F01: vascular dementia, G20: Parkinson disease, G45: transient ischemic attack. H25: cataract, H35: retinopathy, H40: glaucoma. I20: angina pectoris, I21: myocardial infarction, I25: ischemic heart disease, I46: cardiac arrest, I48: atrial fibrillation, I50: heart failure, I61: intercerebral hemorrhage, I63: cerebral infarct, I64: stroke, I80: deep vein thrombosis. J15: pneumonia, J44: chronic obstructive pulmonary disease, J96: respiratory failure. M96: postprocedural skeletal disorder. R99: death of unknown cause. Fractures of S22: rib, S32: lumbar spine, S52: forearm, S62: wrist, S72: femur, S92: foot.
Frequency of ICD10 events in entire study population
Nine ICD10-coded clinical events in three carriers were considered linked to the detected variant. For each we calculated the prevalence and average age in the rest of the Rotterdam Study population for which we have ES data available (n = 2628).17 The results for these nine events are shown in Supplemental table 3. All events occurred commonly in this population: I20: angina pectoris (in 4.9% of the 2628 participants, average age of the event is 72 ± 8), I21: myocardial infarction (10.5%, average age 79 ± 8), I46: cardiac arrest (4.6%, average age 81 ± 8), I48: atrial fibrillation (19.8%, average age 77 ± 10), I50: heart failure (24.9%, average age 80 ± 8), and R99: death with cause unknown (6.3%, average age 87 ± 7). For all events selected by the referees the age at event was earlier than the average age at event across the 2628 participants for which ES data were available, although all events fell within 1.5 standard deviation.
DISCUSSION
From 3815 variants that we found in 59 reported ACMG genes in ES data of 2628 participants from the Rotterdam Study, we confirmed 24 participants to carry a total of 17 “known” pathogenic (KP) variants, comprising 0.9% of our study population. Two additional carriers of a single variant in BRCA2 were identified, but this variant proved false positive after Sanger validation, despite passing all exome sequencing QC and filtering criteria. Upon investigation, the variant was supported by a small number of reads and would have been filtered out in single-sample data processing (i.e., the fact of two putative carriers strengthened the variant quality in calling). Thus, this result indicates we should be careful in the way we handle and interpret this kind of data. Validation by Sanger sequencing in our case was required for a reliable result. This is in line with previous findings, where <2% of all variants identified through ES could not be confirmed, and variants of high clinical relevance should be confirmed beyond doubt.20,21
The proportion of 0.9% KP carriers is similar to what was found in previous studies.6,8–10 Upon investigation by four clinicians, 10 variant carriers (of 26) were observed with at least one ICD10-coded clinical event deemed possibly related to their KP variant, according to at least one of the referees. Only in three carriers (13%) was at least one clinical event considered to be related to the identified variant by a majority of the referees. In all of these carriers it was difficult to determine if the ICD10-based clinical events were caused by these variants, as these events occur frequently in the population. As a result, no information was reported back to any of the carriers or their relatives.
We consulted two main databases for clinical interpretation: HGMD and ClinVar.2,3 Comparing their clinical classification for the ACMG variants identified in our study population we observed disagreement in which variants are classified as pathogenic. In total 17 variants were categorized as class 5 by both databases, 19 in total by ClinVar, and 183 in total by HGMD.
Of concern is a large portion of classifications that differ between both databases, such as the 59 variants classified as class 4 or 5 (likely pathogenic or pathogenic) in HGMD and class 1 (benign) in ClinVar. These most likely stem from overestimation of pathogenicity of HGMD, as has been described before.22,23 This disagreement illustrates the challenge of clinically interpreting genetic variants, especially in a research setting, and how different individuals, laboratories, or databases might reach different conclusions for the same variant. Even when restricting to variants classified as class 5 in both databases, it appears that such variants can be carried without obvious phenotypic consequence.
Additionally, we investigated the clinical classification within ClinVar in different releases over five years (from 2014 to 2018). We observe that the clinical interpretation of many variants has changed over time, where many variants moved toward class 1 (benign), 2 (likely benign), or 3 (uncertain significance). Over this period various genomic variant resources have surfaced and impacted variant interpretation, including the gnomAD database, which now contains data from 125,748 exomes and 15,708 whole genomes from population studies. Additionally the ACMG/AMP criteria were released during this time frame and influenced how consistently labs were applying evidence. One example of this is the reclassification for BRCA1 and BRCA2 variants over time, most often downgrading.24,25 Traditionally the classification of (pathogenic) variants was based on the ascertainment from the more severe Mendelian disorders. Now, with more data available from population studies, reduced penetrance of variants is becoming clearer as is demonstrated by these kind of variants found in individuals without a Mendelian phenotype.11–14,26 By including information about penetrance in healthy populations, the changes in variant classification may stabilize over time.
Although ClinVar contributes greatly to centralizing publicly available clinical genetic information, it does not contain local databases maintained by clinical genetic laboratories. This could result in classification differences of variants between laboratories, and may challenge research efforts to utilize clinical genetic classifications by the more conservative ACMG-AMP criteria. Thus, our definition of a KP variant may be less stringent than that used by a clinical genetic laboratory. Furthermore, several of the variants we indicated as KP have limited information available in ClinVar. In the most recently checked online version (April 2020), two variants had a star classification of less than 2. Five additional variants had only one or two submissions in ClinVar at this time. These results demonstrate the need for additional clinical genetic information to completely classify such variants. Nevertheless, we have attempted to retain the most likely true pathogenic variants as possible using publicly available information. We believe that most of these variants would retain their pathogenic classifications under ACMG-AMP evaluation in clinical genetic laboratories. However, it is possible that the percentage of carriers (0.9%) and fraction of expressivity in these carriers (13%) is lower than under complete clinical genetic evaluation.
For the clinical evaluation of our KP carriers we used the ICD10-coded records that report clinical events during standard clinical practice and during Rotterdam Study research participation. We collected 9165 ICD10-coded events for 2628 study participants, providing unique insight into the health of such a typical elderly population. In 0.9% of this population we observed a KP variant, but only 13% of these carriers (0.13% of the whole study population) presented an ICD10-coded event that could be related to the variant. For none of them was this effect obvious. Due to these results, no events were reported back to any of these carriers, and thus we were not able to collect additional, more detailed, phenotypic information.
Our study demonstrated that the definition of a KP variant is ambiguous between databases, but also within different versions of the same database. This might lead to differences in reporting depending on the used evidence for classification. Specifically, information on the occurrence of KP variants in healthy populations is needed to correctly estimate the penetrance of such variants, and this information should be considered in the recommendations. Currently, several studies have demonstrated that approximately 1% of the population carries a KP defined as such by different databases. Our results based on a thorough clinical follow-up evaluation in subjects 55 years and older linked only 0.13% of events to the presence of a KP variant. This suggest that KP variants are less likely to lead to a phenotype in their carriers, and that such reduced penetrance should be considered when reporting back results to carriers in population-based studies. Overall, our results indicate that reporting back of pathogenic ACMG variants should be approached carefully in these kind of studies.
Several causes for the reduced penetrance could play a role in our population. First, our study population is an elderly population, in which carriers reached late adulthood (55 years or older) despite carrying a potentially pathogenic variant.16 Therefore, our population contains survival bias and the penetrance of some of these variants might be higher in younger populations. Additionally, these participants were investigated in a research setting, and despite the rigorous phenotype collection in the Rotterdam Study they may have exhibited subtle clues missed during examination, such as subclinical deviations or specific relevant family history, which is often used in ACMG-AMP evaluation but could not be collected in this setting. Conversely, this data set is representative for many hospital populations in which (secondary) genetic testing is most likely to occur.16 Second, the expected penetrance is not standardly included in the classification of a pathogenic variants. Thus, variants in class 5 can have variable penetrance and those variants we observe in an elderly research population are likely those with lower penetrance. Considering penetrance on top of the five-class system might facilitate more accurate interpretation. Third, such severely reduced penetrance of KP variants in population-based settings could indicate a strong influence of the genomic context of the functional effects of KP variants in such normal healthy population-dwelling subjects. While in Mendelian disease families the penetrance is usually substantially higher, also here penetrance can be variable and the genomic context might play a role due to the complex way in which different inherited variants or modifiers can influence the phenotype.27
Conclusion
We show that the definition of “known pathogenic” is often not clear and should be approached carefully. Variants marked as KP may have (severely) reduced penetrance. Definition and classification of true (individual) expected pathogenic impact should include, for example, the use of multiple data sources, the pathogenicity prediction over time, and an assessment of the penetrance of the variant in healthy control populations.
Supplementary information
Acknowledgements
We thank the participants of the ERGO population study for their participation in this research; Emma van de Ende, Merel Mol, Eline van der Valk, and Anela Blazevic for interpretation of clinical events in variant carriers; and Mila Jhamai, Joost Verlouw, and Marijn Verkerk for their help in generating the exome sequencing data set. We thank Jolande Verkroost-van Heemst for coordinating clinical follow-up data collection and Joyce van Meurs for supporting the project. We thank Sergio Chavez, Wout Deelen, and Joan Kromosoeto for supporting and performing the Sanger sequencing experiments.
Disclosure
The authors declare no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41436-020-0900-8) contains supplementary material, which is available to authorized users.
References
- 1.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landrum MJ, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stenson PD, et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green RC, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalia SS, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 6.Amendola LM, et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res. 2015;25:305–315. doi: 10.1101/gr.183483.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amendola LM, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorschner MO, et al. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jurgens J, et al. Assessment of incidental findings in 232 whole-exome sequences from the Baylor-Hopkins Center for Mendelian Genomics. Genet Med. 2015;17:782–788. doi: 10.1038/gim.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olfson E, et al. Identification of medically actionable secondary findings in the 1000 Genomes. PLoS One. 2015;10:e0135193. doi: 10.1371/journal.pone.0135193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minikel EV, et al. Quantifying prion disease penetrance using large population control cohorts. Sci Transl Med. 2016;8:322ra9. doi: 10.1126/scitranslmed.aad5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ropers HH, Wienker T. Penetrance of pathogenic mutations in haploinsufficient genes for intellectual disability and related disorders. Eur J Med Genet. 2015;58:715–718. doi: 10.1016/j.ejmg.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Saleheen D, et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- 16.Ikram MA, et al. The Rotterdam Study: 2018 update on objectives, design and main results. Eur J Epidemiol. 2017;32:807–850. doi: 10.1007/s10654-017-0321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rooij JGJ, et al. Population-specific genetic variation in large sequencing data sets: why more data is still better. Eur J Hum Genet. 2017;25:1173–1175. doi: 10.1038/ejhg.2017.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rentzsch P, et al. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leening MJ, et al. Methods of data collection and definitions of cardiac outcomes in the Rotterdam Study. Eur J Epidemiol. 2012;27:173–185. doi: 10.1007/s10654-012-9668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck TF, et al. Systematic evaluation of Sanger validation of next-generation sequencing variants. Clin Chem. 2016;62:647–654. doi: 10.1373/clinchem.2015.249623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lincoln SE, et al. A rigorous interlaboratory examination of the need to confirm next-generation sequencing-detected variants with an orthogonal method in clinical genetic testing. J Mol Diagn. 2019;21:318–329. doi: 10.1016/j.jmoldx.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cassa CA, Tong MY, Jordan DM. Large numbers of genetic variants considered to be pathogenic are common in asymptomatic individuals. Hum Mutat. 2013;34:1216–1220. doi: 10.1002/humu.22375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kundu K, et al. Determination of disease phenotypes and pathogenic variants from exome sequence data in the CAGI 4 gene panel challenge. Hum Mutat. 2017;38:1201–1216. doi: 10.1002/humu.23249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mighton C, et al. Correction: Variant classification changes over time in BRCA1 and BRCA2. Genet Med. 2019;21:2406–2407. doi: 10.1038/s41436-019-0526-x. [DOI] [PubMed] [Google Scholar]
- 25.Mighton C, et al. Variant classification changes over time in BRCA1 and BRCA2. Genet Med. 2019;21:2248–2254. doi: 10.1038/s41436-019-0493-2. [DOI] [PubMed] [Google Scholar]
- 26.Narasimhan VM, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352:474–477. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deltas C. Digenic inheritance and genetic modifiers. Clin Genet. 2018;93:429–438. doi: 10.1111/cge.13150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.