Abstract
Purpose
Structural variants (SVs) may be an underestimated cause of hereditary cancer syndromes given the current limitations of short-read next-generation sequencing. Here we investigated the utility of long-read sequencing in resolving germline SVs in cancer susceptibility genes detected through short-read genome sequencing.
Methods
Known or suspected deleterious germline SVs were identified using Illumina genome sequencing across a cohort of 669 advanced cancer patients with paired tumor genome and transcriptome sequencing. Candidate SVs were subsequently assessed by Oxford Nanopore long-read sequencing.
Results
Nanopore sequencing confirmed eight simple pathogenic or likely pathogenic SVs, resolving three additional variants whose impact could not be fully elucidated through short-read sequencing. A recurrent sequencing artifact on chromosome 16p13 and one complex rearrangement on chromosome 5q35 were subsequently classified as likely benign, obviating the need for further clinical assessment. Variant configuration was further resolved in one case with a complex pathogenic rearrangement affecting TSC2.
Conclusion
Our findings demonstrate that long-read sequencing can improve the validation, resolution, and classification of germline SVs. This has important implications for return of results, cascade carrier testing, cancer screening, and prophylactic interventions.
Keywords: hereditary cancer, variant interpretation, structural variants, genome sequencing, long-read sequencing
INTRODUCTION
A significant amount of genetic variation in the human genome is due to structural variants (SVs), such as deletions, duplications, inversions, and translocations.1,2 Genome sequencing allows high-resolution hypothesis-free analysis of variants in known and novel disease genes, and thus may improve rates of molecular diagnosis by overcoming some of the limitations of targeted clinical assays. Next-generation sequencing (NGS) is the most widely used sequencing technology, and is based on the generation of short (50–300 bp) reads that are aligned to a reference genome or assembled into longer contiguous sequences (contigs) prior to alignment. Accurate alignment and variant calling in NGS is challenging due to regions of low sequence complexity, repetitive elements, and strong GC bias in the human genome, reducing the sensitivity and specificity for novel variant discovery. This indicates a need for improved approaches to characterize genetic variation, particularly for large or complex variants.
Moderate- to high-penetrance germline variants in cancer predisposition genes underlie 5–10% of all cancers. However, the prevalence of SVs in clinical and research cancer cohorts is likely underestimated due to the technical and computational limitations of multigene panel sequencing, exome sequencing, and genome sequencing.3 Recently, long-read sequencing (LRS) has been used to characterize complex genetic variation in human genomes and aid in the diagnosis of rare disorders.4,5 To investigate the contribution of germline SVs to cancer susceptibility, we used short- and long-read genome sequencing to elucidate pathogenic germline SVs in advanced cancer patients enrolled in a genomics-based precision medicine program. Here, we describe the application of nanopore sequencing to correctly interpret and classify SVs that could not be resolved through short-read genome sequencing.
MATERIALS AND METHODS
Ethics statement
This study was approved by the University of British Columbia Research Ethics Committee, and written informed consent was obtained for all participants (REB H12–00137, H14–00681, H16–00291).
Short-read sequencing
Short-read genome sequencing was previously performed on Illumina HiSeq platforms in normal tissue samples for 669 advanced cancer patients enrolled in the BC Cancer Personalized OncoGenomics (POG) program (NCT02155621).6 Putative SVs were identified in genomes aligned to the human reference genome version hg19 using multiple copy-number and SV calling tools (Supplementary Materials and Methods). Fourteen SVs that were predicted to have a deleterious impact on gene expression or function in at least 1 of 98 cancer predisposition genes were subsequently identified through manual review in the Integrated Genomics Viewer (IGV) (Table S1).7 Variants in five known carriers previously identified by clinical guideline-based testing were used to evaluate the sensitivity of SV calling through Illumina genome sequencing.
Oxford nanopore long-read sequencing
LRS was performed in 13 cases where archived normal DNA was available (Table S2). Genome libraries were constructed for high molecular weight DNA purified from peripheral blood, and sequenced on the Oxford Nanopore Technology MinION or PromethION. Base calling and read alignment were performed using Guppy version 3 and Minimap2, respectively, and alignments were visualized in IGV.8,9 Variant calling was performed for samples sequenced on the PromethION using Sniffles v1.0.11.10 Paired tumor genome sequencing and transcriptome sequencing (RNA-seq) were assessed for somatic variants, loss of heterozygosity (LOH), somatic single-nucleotide variant (SNV) signatures, alternative splicing, and fusion transcript expression as previously described (Supplementary Materials and Methods).6,11
RESULTS
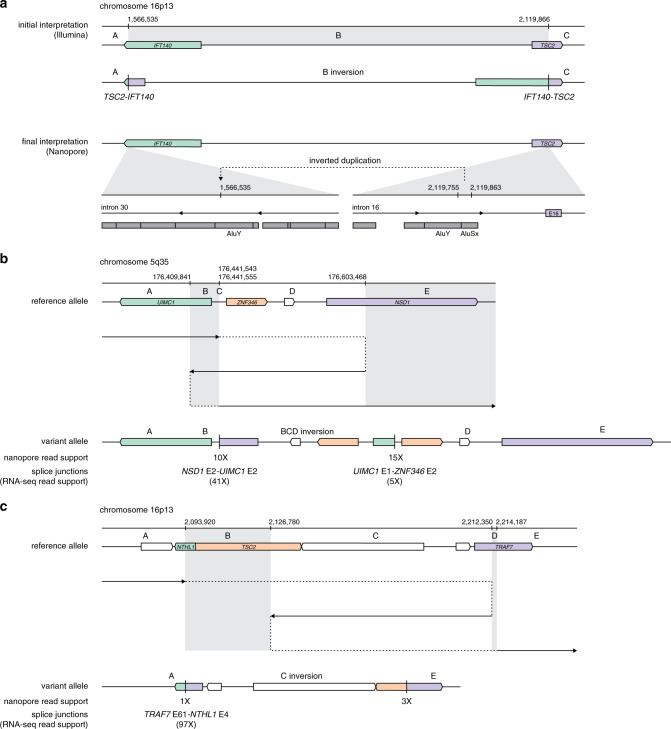
Twelve candidate germline SVs were identified in 14 individuals by short-read genome sequencing, of whom 5 were known carriers (Table 1). Eight deletions, two inversions, and two complex rearrangements were predicted to disrupt the coding sequence of at least one known cancer predisposition gene. Although most variants were detected by multiple short-read SV calling tools and inferred through contig-level read support, three variants were identified by only one tool, including one with prior clinical validation (Table S2). Surprisingly, three unrelated individuals without medical histories suggestive of tuberous sclerosis complex (TSC) were found to carry a recurrent and predicted pathogenic event on chromosome 16p13 identified through short-read genome sequencing (Fig. 1a). LRS performed in cases 1–3 revealed that an inverted duplication of an Alu element from TSC2 intron 16 into IFT140 intron 30 was miscalled by both DELLY and Manta and could not be resolved through manual review, consistent with ambiguous alignment of short reads at these loci (Table S2 and Figs. S1–S3). This finding, in addition to the lack of clinical phenotype in any of the carriers, led to the classification of this variant as likely benign.
Table 1.
Variant information and patient characteristics for known or suspected deleterious germline structural variants detected through short-read genome sequencing.
| Case ID | Resolved variant | SRS evidence | Descriptive utility of LRS | Coding sequence impact | ACMG/AMP classification (criteria) | Indication for clinical genetics assessment |
|---|---|---|---|---|---|---|
| Cases 1–3 | NC_000016.9:g.1566535_1566536ins2119755_2119863inv | Variant miscalleda | Variant reinterpretation and confirmation of false-positive finding | None | Likely benign (BS2) | No referral |
| Case 4 | NC_000005.9:g.176441544_176441555delins176409841_176603468inv | PR, SR, contig | Resolution of variant configuration | NSD1 5’UTR-exon 2 duplication | Likely benign (BS2) | No referral |
| Case 5 | NC_000016.9:g.2093921_2214187delins2126780_2212350inv | PR, SR, contig | Resolution of variant configuration | TSC2 5’UTR-exon 25 deletion | Pathogenic (PVS1, PM2, PP4) | Tuberous sclerosis complex |
| NTHL1 5’UTR-exon 3 deletion | Pathogenic (PVS1, PM2) | Autosomal recessive NTHL1-associated polyposisc | ||||
| Case 6 | NM_000051.3(ATM):c.2467–527_8851–2114del | Read depth | Resolution of breakpoints near flanking repetitive elements | ATM exons 17–61 deletion | Pathogenic (PVS1, PM2) | ATM-associated cancer susceptibility |
| Case 7 | NM_058216.2(RAD51C):c.706–1013_837+296delins706–469_837+296inv | SRb | Resolution of 5’ breakpoint and flanking deletion | RAD51C exon 5 deletion | Likely pathogenic (PVS1 [strong], PM2) | Moderate-penetrance ovarian cancer susceptibility |
| Case 8 | NM_000051.3(ATM):c.1065+647_1236–369del | Contig | Confirmation | ATM exon 9 deletion | Pathogenic (PVS1, PM2) | ATM-associated cancer susceptibility |
| Case 9 | NC_000017.10:g.41217614_41295110del | PR, SR, contig, read depth | Confirmation | BRCA1 5’ UTR-exon 17 deletion | Pathogenic (PVS1, PM2) | HBOC |
| Case 10 | NM_007294.3(BRCA1):c.547+946_4186–1194del | Read depth | Confirmation | BRCA1 exons 9–12 deletion | Pathogenic (PVS1, PM2) | HBOC |
| Case 11 | NC_000002.11:g.47545553_47674137del | PR, SR, contig, read depth | Confirmation | EPCAM deletion MSH2 5’UTR-exon 7 deletion | Pathogenic (PVS1, PM2) | Lynch syndrome |
| Case 12 | NM_000135.2(FANCA):c.792+452_1826+222del | PR, SR, contig, read depth | Confirmation | FANCA exons 9–20 deletion | Pathogenic (PVS1, PM2) | Autosomal recessive Fanconi anemiac |
| Case 13 | NM_024675.3(PALB2):c.2835–282_3113+1377del | PRb | Confirmation | PALB2 exons 9–10 deletion | Pathogenic (PVS1, PM2) | Moderate-penetrance breast cancer susceptibility |
| Case 14d | NM_000546.5(TP53):c.−28–252_920–15del | PR, SR, contig | NA | TP53 exons 2–9 deletion | Pathogenic (PVS1, PM2, PP4) | Li–Fraumeni syndrome |
ACMG/AMP American College of Medical Genetics and Genomics/Association for Molecular Pathology, FHx family history, HBOC hereditary breast and ovarian cancer, LRS long-read genome sequencing, NA not applicable, PR paired reads, SR split reads, SRS short-read genome sequencing.
aThe predicted variant, NC_000016.9:g.1566535_2119866inv, was miscalled by short-read genome sequencing based on paired reads, split reads, and contigs in three unrelated cases. This variant was subsequently found by nanopore sequencing to reflect an inverted duplication of an Alu element from TSC2 intron 16 into intron 30 of IFT140.
bGermline variants in cases 7 and 13 were additionally supported by multiple lines of read evidence in matched tumor tissue.
cClinical referral on the basis of carrier status for recessive syndromes should be considered in the context of family structure and medical history.
dCase 14 was not assessed by Oxford Nanopore sequencing.
Fig. 1. Schematic representations of candidate structural variants resolved using long-read sequencing.
(a) A recurrent event identified in cases 1, 2, and 3 and predicted to be pathogenic was reinterpreted as a likely benign intronic variant based on Oxford nanopore sequencing. Illumina short-read genome sequencing data supported a long-range inversion on chromosome 16p13 with breakpoints in IFT140 and TSC2 (upper), while nanopore sequencing data showed an insertion in intron 30 of IFT140 likely arising from an Alu element in intron 16 of TSC2 (lower). (b, c) A pathogenic complex variant in case 4 (b) and a likely benign variant in case 5 (c) characterized by nanopore sequencing. The path of long-read alignments to the reference genome is denoted by a solid black arrow, indicating the putative direction of DNA replication on the variant allele with dashed lines indicating positions of template switching and reinitiation of replication.
A novel complex rearrangement was identified on chromosome 5q35 in case 4, who was shown to carry a 194-kb inverted duplication flanked by a small indel at the breakpoint junction (Figs. 1b and S4). Two fusion transcripts, NSD1-UIMC1 and UIMC1-ZNF346, were identified by RNA-seq, but configuration of the variant determined from LRS indicated that undisrupted copies of both NSD1 and UIMC1 were maintained on the variant allele. Given the individual’s unremarkable medical history, with no known diagnosis of Sotos syndrome, this variant was classified as likely benign. In contrast, LRS in case 5 indicated that a complex variant identified on chromosome 16p13.3 involved an 85-kb inversion with breakpoints in TSC2 and TRAF7 flanked by two deletions, resulting in partial loss of NTHL1 and TSC2 (Figs. 1c and S5). Furthermore, LOH at the locus in the individual’s tumor indicated that the complex germline rearrangement involved only one allele (Table S3). This case had a prior history of TSC and has been previously described.12
Nanopore sequencing further informed SV breakpoints in two cases and confirmed simple deletions in six additional cases (Figs. S6–S13). Sequence analysis at the breakpoint junctions found that repetitive elements were present at most breakpoints, suggesting that they contributed to both the formation of large SVs and miscalling of a recurrent variant (Table S4). Long tracks of homology in two cases indicated that variant formation may have been a consequence of break-induced replication. Notably, the breakpoints of a partial ATM deletion in case 6 were predicted to occur near two long interspersed nuclear elements (LINEs), of which a single copy could be mapped to the PromethION reads (Fig. S6). Many SV breakpoints had simple blunt ends or small indels in the absence of microhomology, short regions of shared nucleotide identity, characteristic of products of nonhomologous end joining.13 Microhomology near the breakpoints in cases 4, 7, and 11 suggested that these events may have arisen through microhomology-mediated end joining or microhomology-mediated break-induced replication. Likely as a consequence of breakpoint sequence homology, a 544-bp deletion at the 5’ breakpoint of a RAD51C exon 5 inversion in case 7 was not confidently captured by Illumina sequencing (Fig. S7).
Among the ten pathogenic and likely pathogenic SVs identified in this cohort, seven were associated with LOH and four tumors showed significant contributions from somatic SNV signatures with characterized genetic etiologies: signature 30 was associated with homozygous loss of NTHL1 in case 5; signature 3 suggested homologous recombination deficiency caused by loss of BRCA1 and PALB2 in cases 9 and 13, respectively; and signature 6 supported mismatch repair deficiency in case 11 (Fig. S14).14 Tumor RNA-seq demonstrated aberrant splicing in several cases with intragenic SVs and sufficient read coverage at the splice junction, thus providing additional support for variant pathogenicity in these cases (Table S3).
DISCUSSION
The average human genome contains approximately 2500 SVs, including balanced rearrangements such as inversions and translocations, and unbalanced rearrangements such as large deletions, duplications, and insertions.15 Microscopic SVs, those typically larger than ~3 Mb, are found at a high frequency in certain disorders and have historically been assessed using karyotyping or microarrays. However, submicroscopic SVs require molecular approaches with a higher resolution to determine variant configuration and to allow for accurate clinical interpretation. Our findings suggest that SVs are a rare cause of cancer susceptibility, underlying 1.5% of cases in an advanced adult cancer cohort (n = 669). Short-read genome sequencing demonstrated 100% sensitivity in variant detection for five known carriers, and identified pathogenic and likely pathogenic variants in five additional cases without prior genetic diagnoses. However, Illumina genome sequencing was insufficient to accurately and fully resolve 5 of 12 unique SVs, including two likely benign variants.
Recently, LRS has allowed the molecular diagnosis of SVs causing Mendelian disease in cases where clinical assays or short-read genome sequencing have been unsuccessful.4,5 Insertions, balanced SVs, and complex rearrangements that consist of three or more breakpoints are particularly difficult to characterize using NGS given the inferential nature of SV detection through contig-, split read-, flanking read-, or depth of coverage–based approaches. Although breakpoints of LINE-mediated variants remain difficult to assess by both sequencing technologies, long reads may span the entirety of homologous sequences or capture multiple breakpoints to inform haplotype configuration. For example, LRS helped resolve a single-exon inversion in RAD51C that would have been missed through targeted NGS and whose 5’ breakpoint was incompletely determined by short-read genome sequencing. As demonstrated by Rhees et al., the precise characterization of SV breakpoints is critical to guide the development of targeted clinical assays for familial, recurrent, or founder variants that may be undetectable through standard clinical assays in known or suspected hereditary cancer families.16
Although many carriers in our cohort had a personal and/or family history suggestive of moderate- to high-penetrance cancer susceptibility, 4 carriers (40%) did not have a previous personal or family history indicating referral for genetic counseling and testing (Table S5). This finding is consistent with previous reports suggesting that less than half of carriers identified through population genetic testing meet current clinical testing criteria.17 The significance of accurate variant interpretation, particularly in individuals who do not meet phenotype-based criteria, was highlighted by case 3 who was referred for clinical testing on the basis of the miscalled inversion in TSC2 and LOH in their tumor. At the time of referral, polymerase chain reaction (PCR)–based validations of the predicted breakpoint junctions were unsuccessful; however, LRS later characterized the true variant as a small inverted duplication in a deep intronic region of IFT140. On the basis of accurate variant resolution, classifications for this variant and a complex rearrangement at the locus of NSD1 were downgraded to likely benign. This ultimately prevented clinical referral for two cases without suspicious personal or family medical history.
Genome sequencing allows unbiased characterization of SV breakpoints, unlike relative and targeted approaches such as MLPA and NGS panels, that are influenced by variable efficiency in primer binding, probe hybridization, and target amplification. Despite the current limitations of LRS, including the necessity for high molecular weight DNA, higher error rate, and increased cost, this technology is particularly beneficial in the genetic diagnosis of monogenic disorders where NGS has failed to identify a causal variant. Many nonrecurrent SVs result from template switching between homologous repetitive elements, which are inherently difficult to map with short reads. As both we and others have shown, such variants are inaccurately or incompletely captured by NGS.13 This was exemplified by two complex rearrangements that could only be resolved through LRS, and one false-positive inversion that was refractory to accurate interpretation based on short-read sequencing. Recent studies have further shown the potential of amplification-free target enrichment for the sensitive detection of small variants and SVs at increased coverage and reduced costs.18
As clinical genome sequencing becomes more widely used for molecular diagnoses in a variety of genetic syndromes, there is a need for standardized guidelines for the identification and validation of SVs using high-throughput sequencing technology. Considering the limitations of NGS, LRS offers a complementary approach in the diagnostic odyssey of patients and families where standard clinical testing is uninformative.
Supplementary information
Acknowledgements
We wish to extend our deepest gratitude to the individuals who participated in this research. We would also like to thank Jessica Nelson, Katherine Mui, Lindsay Zibrik, Kevin Sauve, Young Cheng, Correy Lim, and Kirstin Brown for program support, data collection, data deposition, and editing assistance. The POG program is supported by Genome Canada and Genome BC (202SEQ, 212SEQ, 12002), the Canada Foundation for Innovation (20070, 30198, 30981, 33408), and the BC Knowledge Development Fund. We gratefully acknowledge the generous support of the BC Cancer Foundation and Genome British Columbia (B20POG). M.L.T. is supported by the University of British Columbia Clinician Investigator Program. K.O. is supported by a Canadian Institutes for Health Research Postdoctoral Fellowship award. K.A.S. is supported by the Canadian Institutes of Health Research and the Michael Smith Foundation for Health Research.
Disclosure
The authors declare no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: My Linh Thibodeau, Kieran O’Neill, Katherine Dixon.
Contributor Information
Kasmintan A. Schrader, Email: ischrader@bccancer.bc.ca.
Steven J. M. Jones, Email: sjones@bcgsc.ca.
Supplementary information
The online version of this article (10.1038/s41436-020-0880-8) contains supplementary material, which is available to authorized users.
References
- 1.Gibbs RA, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain M, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng DT, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics. 2017;10:33. doi: 10.1186/s12920-017-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchis-Juan A, et al. Complex structural variants in Mendelian disorders: identification and breakpoint resolution using short- and long-read genome sequencing. Genome Med. 2018;10:95. doi: 10.1186/s13073-018-0606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merker JD, et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet Med. 2018;20:159–163. doi: 10.1038/gim.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones MR, et al. Successful targeting of the NRG1 pathway indicates novel treatment strategy for metastatic cancer. Ann Oncol. 2017;28:3092–3097. doi: 10.1093/annonc/mdx523. [DOI] [PubMed] [Google Scholar]
- 7.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wick RR, Judd LM, Holt KE. Performance of neural network basecalling tools for Oxford nanopore sequencing. Genome Biol. 2019;20:129. doi: 10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sedlazeck FJ, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao EY, et al. Homologous recombination deficiency and platinum-based therapy outcomes in advanced breast cancer. Clin Cancer Res. 2017;23:7521–7530. doi: 10.1158/1078-0432.CCR-17-1941. [DOI] [PubMed] [Google Scholar]
- 12.Wong H-L, et al. Molecular characterization of metastatic pancreatic neuroendocrine tumors (PNETs) using whole-genome and transcriptome sequencing. Cold Spring Harb Mol Case Stud. 2018;4:a002329.. doi: 10.1101/mcs.a002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho CMB, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhees J, Arnold M, Boland CR. Inversion of exons 1-7 of the MSH2 gene is a frequent cause of unexplained Lynch syndrome in one local population. Fam Cancer. 2014;13:219–225. doi: 10.1007/s10689-013-9688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metcalfe KA, et al. Screening for founder mutations in BRCA1 and BRCA2 in unselected Jewish women. J Clin Oncol. 2010;28:387–391. doi: 10.1200/JCO.2009.25.0712. [DOI] [PubMed] [Google Scholar]
- 18.Gilpatrick T, et al. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat Biotechnol. 2020;38:433–438. doi: 10.1038/s41587-020-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.