Abstract
Variations in MYT1L, a gene encoding a transcription factor expressed in the brain, have been associated with autism, intellectual disability, and schizophrenia. Here we provide an updated review of published reports of neuropsychiatric correlates of loss of function and duplication of MYT1L. Of 27 duplications all were partial; 33% were associated exclusively with schizophrenia, and the chromosomal locations of schizophrenia-associated duplications exhibited a distinct difference in pattern-of-location from those associated with autism and/or intellectual disability. Of 51 published heterozygous loss of function variants, all but one were associated with intellectual disability, autism, or both, and one resulted in no neuropsychiatric diagnosis. There were no reports of schizophrenia associated with loss of function variants of MYT1L (Fisher's exact p < .00001, for contrast with all reported duplications). Although the precise function of the various mutations remains unspecified, these data collectively establish the candidacy of MYT1L as a reciprocal mutation, in which schizophrenia may be engendered by partial duplications, typically involving the 3′ end of the gene, while developmental disability–notably autism–is associated with both loss of function and partial duplication. Future research on the specific effects of contrasting mutations in MYT1L may provide insight into the causal origins of autism and schizophrenia.
Keywords: autism, MYT1L, reciprocal mutation, schizophrenia
1 ∣. INTRODUCTION
MYT1L (myelin transcription factor 1-like gene), a zinc finger transcription factor found on the short arm of chromosome 2 and expressed in neuronal tissue, functions in genetic regulation, and its disruption is commonly associated with manifestations of neuropsychiatric disability. Previous reports of this association include publication of a series of MYT1L duplication cases with schizophrenia (Lee et al., 2012) and a number of reports of deletions associated with neurodevelopmental disorders, including intellectual disability (De Rocker et al., 2015; Tuwaijri & Alfadhel, 2019). The possibility that deletion versus duplication in a single gene might result in highly contrasting neuropsychiatric phenotypes motivates this updated appraisal of the published literature, because clarification of reciprocal effects of gene dosage could yield profound biological insights into distinctions between the associated neuropsychiatric disorders, in this case including contrasts between schizophrenia and autism. In this comprehensive appraisal, we consider associations of MYT1L with three neuropsychiatric phenotypes (autism, schizophrenia, and intellectual disability), how each relates to either loss of function or duplication in previously published reports, and how position of a partial duplication on the chromosome may be associated with phenotype.
In relation to contrasts between autism and schizophrenia, at least 10 other genes have been implicated in observations of pleiotropic effects of deletions resulting variously in one or the other phenotype (Vissers, Gilissen, & Veltman, 2016). Human loss of function mutations associated with these respective disparate phenotypes in one of the genes, SHANK3, were engineered in a mouse model, and revealed both overlapping and nonoverlapping synaptic, behavioral, and molecular signatures of disruption (Zhou et al., 2016). To our knowledge, MYT1L is the first gene associated with both autism and schizophrenia that exhibits a pattern of association suggesting reciprocal effects of deletion and partial duplication mutations resulting in these disparate disorders; these conditions have both contrasting and overlapping behavioral features and marked differences in developmental timing of symptom onset. Patterns of contrasting or reciprocal behavioral effects have been observed for a number of cytogenetic regions that harbor multiple genes, including 22q13.3 and 16p11.2 (Crespi, Stead, & Elliot, 2010).
Recently, reciprocal rearrangements in the Williams Beuren region of chromosome 7 (7q11.23) have been traced to the contrasting phenotypes of autism (in a number of duplication cases) and Williams Syndrome (most reported deletions), which is particularly intriguing because these syndromes can be construed as manifestations of hypo- and hyper-sociality (Mulle et al., 2014). More generally, reciprocal mutations have been categorized according to three models of gene expression: the additive model, the dominant model, and the U-shaped model. In the additive model, opposite directions of gene expression (increase and decrease) are associated with opposing phenotypes. An example of this additive class of variants is the historical association of 22q11.2 duplications with schizophrenia, and 22q11.2 deletions with possible protection from schizophrenia. In the U-shaped model, opposing directions of gene expression (increase and decrease) are associated with identical phenotypes. Finally, in the dominant model, increase or decrease of gene expression is associated with one phenotype while the converse level of gene expression (increase or decrease) has no effect on that specific trait (Deshpande & Weiss, 2018).
This review and meta-analysis was motivated by a steady accumulation of cases representing a range of neuropsychiatric abnormality associated with MYT1L, and the potential biological significance of understanding discrepancies between loss of function and partial duplication mutations. Specifying the patterns of association between disruptions in individual genes and syndromes of developmental and behavioral abnormality can offer critical clues to understanding the effects of genetic variation on causation of neuropsychiatric disorders.
2 ∣. METHODS
We conducted a search of the Medline database and Database of Genomic Variants for reports of human subjects with variations in the gene MYT1L and identified 24 published reports encompassing 78 presumably nonoverlapping cases. We use the label “loss of function” to refer to the following classes of variants: deletions, frameshift, nonsense, and splicing variants, because these are likely to result in lack of production of mRNA from these alleles. We also label missense variants that are predicted to be damaging as loss of function, although it is possible that such variants may have other effects at the protein level. We define “duplications” as copy number gains that consist of at least 100,000 nucleotides and which incorporate at least a portion of the coding region of the MYT1L gene. Partial duplications are those variants which only include some, but not all, of the coding region of the gene. Depending on the orientation and location of the duplicated region, possible consequences of such variants are difficult to predict, especially for those that only include the 3′ portion of the gene. It is possible that these variants may cause abnormal splicing, incorrect RNA processing via disruption of the 3′UTR, and/or disruption of transcriptional regulatory elements. There were 51 MYT1L loss of function cases within 14 of the papers (Blanchet et al., 2017; Bonaglia, Giorda, & Zanini, 2014; D'Angelo et al., 2018; De Rocker et al., 2015; De Rubeis et al., 2014; Doco-Fenzy et al., 2014; Loid et al., 2018; Mayo et al., 2015; Rio et al., 2013; Stevens et al., 2011; Tu et al., 2014; Tuwaijri & Alfadhel, 2019; Vlaskamp et al., 2017; Wang et al., 2016). We also identified 27 partial MYT1L duplication cases in 11 papers in PubMed (Braddock, Del Campo, Reiff, & Stein, 2018; Coe et al., 2014; Cooper et al., 2011; De Rocker et al., 2015; ISC, 2008; Jakobsson et al., 2008; Meyer, Axelsen, Sheffield, Patil, & Wassink, 2012; Suktitipat et al., 2014; Van den Bossche et al., 2013; Vrijenhoek et al., 2008; Walsh et al., 2008).
When collecting and analyzing data from these reports, it is important to recognize that different papers placed emphasis on different phenotypes based on the specific motivation or hypothesis of the manuscript. Some papers resulted from analyses of cohorts of patients known to have specific phenotypes, among whom copy number variants (CNVs) were analyzed. These phenotypes included schizophrenia (ISC, 2008; Van den Bossche et al., 2013; Vrijenhoek et al., 2008; Walsh et al., 2008), autism spectrum disorder (Braddock et al., 2018; De Rubeis et al., 2014; Wang et al., 2016), syndromic obesity (D'Angelo et al., 2018), co-occurring intellectual disability and obesity (Loid et al., 2018), and epilepsy (Vlaskamp et al., 2017), and these reports found mutations of MYT1L to be commonly associated with these conditions. Most of the cases reported, however, were derived from reports of patients known to have mutations of MYT1L whose phenotypic data was secondarily retrieved and analyzed (Blanchet et al., 2017; Bonaglia et al., 2014; De Rocker et al., 2015; Doco-Fenzy et al., 2014; Mayo et al., 2015; Meyer et al., 2012; Stevens et al., 2011; Tu et al., 2014; Tuwaijri & Alfadhel, 2019). One example of this type of report is a study of a pair of monozygotic twins with mutations in MYT1L, one twin having a deletion and the other twin having a mosaic mutation (1/3 deletion, 1/3 duplication, 1/3 normal) (Rio et al., 2013); the latter twin (with mosaicism) was excluded from this updated review. Lee et al. (2012) previously summarized information from nine reported cases of MYT1L duplications, two of which were excluded from this review (Ikeda et al., 2010; Kirov et al., 2009), because it could not be clearly established that MYT1L duplications were operative in the respective patients. We also reviewed the Database of Genomic Variants (MacDonald, Ziman, Yuen, Feuk, & Scherer, 2013) in order to examine the frequency of partial duplications of MYT1L in control populations. We used a threshold of 50 kb for inclusion of a duplication case in this review. We found six additional MYT1L duplication cases in four separate reports presenting no neuropsychiatric diagnoses (Coe et al., 2014; Cooper et al., 2011; Jakobsson et al., 2008; Suktitipat et al., 2014).
From the original reports, we extracted demographic data (sex and age), genetic data (mutation type, genomic coordinates, and inheritance pattern), and also noted presence or absence of a diagnosis of intellectual disability, autism, schizophrenia, epilepsy, and obesity while noting any reported dysmorphisms. In addition, we collected occipital frontal circumference (OFC) data from each report, and characterized each patient as macrocephalic, microcephalic, or neither; we standardized our measures by defining macrocephaly as having an OFC measurement greater than the 97th percentile or greater than 2 SDs away from the mean, and microcephaly as having an OFC measurement less than the third percentile or <−2 SDs away from the mean.
PMIDs and references for each case are also included, and cases that were presented in the original reports as DECIPHER cases were notated with their DECIPHER IDs (Supporting Information Data S1). After synthesizing all published clinical reports of MYT1L mutations, we then analyzed the data with respect to mutation type and corresponding neuropsychiatric disorder category/ies (schizophrenia, autism, and intellectual disability). Translocations, inherited single nucleotide variants (SNVs), and ring chromosomes in the aforementioned case reports were excluded from data collection.
In the process of data collection, it was noted that the genetic nomenclature was inconsistent among the papers. In order to create a data set with consistent and comparable mutation locations, Mutalyzer (Wildeman, van Ophuizen, den Dunnen, & Taschner, 2008) was used to convert all CNV mutations to “arr2p25.3(1,896,431-2,062,854)x3” format and all SNV mutations to “c.1579G>A; p.(Gly527Arg)” format, according to Ensemble transcript NM_001303052.2.
In addition to examining previously published data regarding mutations of MYT1L specifically, we also created a review table organizing several previously reported reciprocal relationships for neuropsychiatric phenotypes associated with specific CNVs (Supporting Information Data S2).
3 ∣. DATA ANALYSIS
We first categorized all cases on the basis of whether or not they had schizophrenia, autism, intellectual disability, some combination of these, or no neuropsychiatric diagnosis. Cases presenting features of autism were included in the autism category irrespective of documentation of full diagnostic confirmation. For each primary diagnosis (schizophrenia, intellectual disability, and autism), we constructed 2 × 2 contingency tables (Table 1) depicting association of respective partial duplication and loss of function mutations with presence or absence of each condition, considered one at a time, and without regard to their overlap. Fischer's exact test was used to examine the strength of associations within the context of the published literature.
TABLE 1.
Contingency tables and Fischer's exact test statistic values for neuropsychiatric phenotypes versus MYT1L variants
| Neuropsychiatric phenotype |
|||||||
|---|---|---|---|---|---|---|---|
| ASDa |
SCHZb |
IDc |
|||||
| + | − | + | − | + | − | ||
| MYT1L variants | Duplication | 6 | 21 | 10 | 17 | 6 | 21 |
| Loss of function | 22 | 29 | 0 | 51 | 46 | 5 | |
Abbreviations: ASD, autism spectrum disorder; dup, duplication; ID, intellectual disability; lof, loss of function; SCHZ, schizophrenia.
ASD+/ASD-Fischer's exact test statistic value, versus dup/lof = .0849, NOT significant. (This pertains to all reported variants and should not infer that specific sequence variants would not reach the statistical threshold individually for an effect on these phenotypes.)
SCHZ+/SCHZ-Fischer's exact test statistic value, versus dup/lof < .0001, Significant.
ID+/ID-Fischer's exact test statistic value, versus dup/lof < .0001, Significant.
We also constructed contingency tables for our macrocephaly data and used Fischer's exact tests to examine the strength of the associations between macrocephaly versus deletion or partial duplication, and macrocephaly versus presence or absence of an autism diagnosis.
We then examined the locations of the duplications to determine whether specific exon intervals and/or adjacent chromosomal regions might account for the diversity of neuropsychiatric manifestations observed in duplication patients. Two particular MYT1L duplication cases (Braddock et al., 2018; Vrijenhoek et al., 2008) did not report coordinates for the specific mutations, therefore the mutation coordinates were notated as “2p25.3” in Supporting Information Data S1, and these cases were included in the counts for statistical comparison with neuropsychiatric phenotype but were excluded from the gene map.
Finally, we reviewed the Exome Aggregation Consortium (ExAC) (Lek et al., 2016), and the Genome Aggregation Database (gnomAD) (Karczewski et al., 2019), in order to contextualize the loss of function variants in relation to frequencies of the observed sequence variants in the general population. In addition, we reviewed the Database of Genomic Variants in order to examine the frequency of partial duplications of MYT1L in control populations (MacDonald et al., 2013).
4 ∣. RESULTS
Our Medline search yielded 78 patient cases within 24 published reports. A schematic summarizing the phenotypic correlates of deletion and duplication is presented in Figure 1.
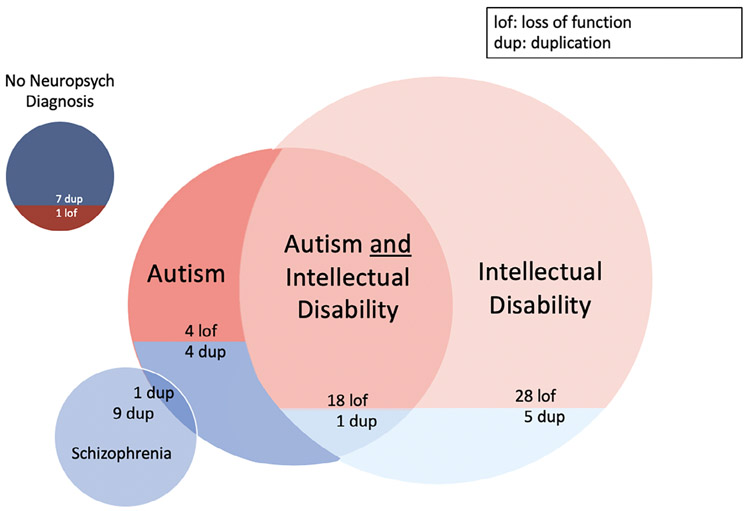
FIGURE 1.
Counts of phenotypic manifestations for loss of function versus duplication. Venn diagram depicting the number of MYT1L loss of function (lof, red) and duplication (dup, blue) cases presenting with each individual phenotype of interest, including their overlap; color coding matches that of Supporting Information Data S1, where comprehensive patient and reference data can be found
4.1 ∣. Duplications
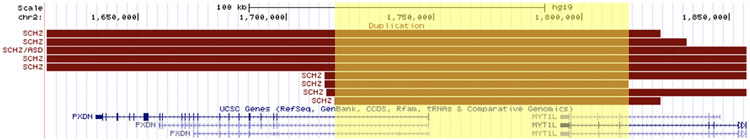
Of the 27 published cases of partial MYT1L duplications, 9 were reported to have presented with schizophrenia only, 5 with intellectual disability only, 4 with autism only, one with autism and intellectual disability, one with co-occurring diagnoses of schizophrenia and autism, and 7 patients with no appreciable neuropsychiatric disorder. When considering the genomic coordinates for duplications of MYT1L associated with schizophrenia, all but one encompassed part or all of the neighboring gene, PXDN. All cases associated with schizophrenia (for which we were able to extract genomic coordinate data) overlapped with respect to a large interval from the first intron of PXDN through Intron 22 of MYT1L (chr2: 1,716,437-1,815,909, hg19), as shown in Figure 2. Several cases associated with intellectual disability had duplications outside of that interval that overlapped with one another; three of the four cases, however, were members of a single biological family (De Rocker et al., 2015).
FIGURE 2.
Gene map of duplication cases of MYT1L presenting schizophrenia. Gene map depicting mutation coordinates for all MYT1L duplication cases presenting schizophrenia (including those with co-occurring neuropsychiatric disorders), with critical region (the first intron of PXDN through Intron 22 of MYT1L (chr2: 1,716,437-1,815,909, hg19)) highlighted in yellow
4.2 ∣. Loss of function variants
Of the 51 published MYT1L loss of function cases (predominantly point mutations), 28 presented with intellectual disability only; 18 presented with both intellectual disability and autism; 4 presented diagnoses and/or characteristics of autism only; and 1 exhibited none of the aforementioned neuropsychiatric disorders. There were no reports of schizophrenia in any of the loss of function cases.
Next, we considered the likelihood that the absence of schizophrenia among loss of function cases could have occurred by chance. Fischer's exact p for presence/absence of schizophrenia in relation to published reports of duplication versus deletion (from all reports in the literature) was p < .0001. Fischer's exact p for presence/absence of intellectual disability in relation to published duplication versus deletion (from all reports in the literature) was also p < .0001, indicating a statistically significant relationship between deletions of MYT1L and diagnoses of intellectual disability. Autism did not reach statistical significance for a specific association with loss of function or partial duplication (Table 1).
With regard to our statistical analyses for macrocephaly, 9% of our cases, all with loss of function variants, presented with macrocephaly; no significant associations were found for macrocephaly comparing subjects with loss of function to those with partial duplication, or comparing subjects manifesting the presence or absence of an autism diagnosis. We also note that 9% of our cases (three duplications, four loss of function) presented with microcephaly.
Because the data presented here consists solely of clinically ascertained data, we reviewed the ExAC, which includes 60,706 unrelated individuals, and the gnomAD, which includes 125,748 exome sequences and 15,708 whole-genome sequences from unrelated individuals (many of which overlap with the ExAC data), and identified only one loss of function SNV in each database, demonstrating the extreme rarity of MYT1L loss of function in a healthy population. This observation is consistent with a reported a pLI score of 1.00 and an o/e ratio of 0.02 (0.01–0.09) (Karczewski et al., 2019; Lek et al., 2016) for MYT1L. In addition, upon reviewing the Database of Genomic Variants, we found only six cases with partial MYT1L duplication in a group of 54,000 control individuals (frequency of 0.0001), demonstrating the extreme rarity of these variants (MacDonald et al., 2013).
5 ∣. DISCUSSION
By assimilating all published data on MYT1L and its reported neuropsychiatric manifestations, marked association between MYT1L duplications and schizophrenia was found: 10 of 27 reported duplications resulted in schizophrenia, while none of the 51 deletion cases did. Although evidence of this specific association has been previously documented (Lee et al., 2012), this review expands the number of MYT1L duplication cases reported and considers all published reports of deletion cases. Moreover, the chromosomal locations of the MYT1L partial duplication cases presenting schizophrenia exhibited a pattern that was distinct from those with reported associations to other developmental disorders. Based on the accumulated data, a critical region on the p-arm of chromosome 2 from the first intron of PXDN through Intron 22 of MYT1L (chr2: 1,716,437-1,815,909, hg19), is associated with schizophrenia, as shown in Figure 2. This review also confirms a significant association between intellectual disability and under-expression of MYT1L (De Rocker et al., 2015; Tuwaijri & Alfadhel, 2019). 98% of the 51 deletion cases reported were characterized by a diagnosis of intellectual disability, autism, or both; in total, 43% of the deletion cases resulted in features or a diagnosis of autism.
A number of studies have highlighted the role of MYT1L as a laboratory tool for neuronal cell differentiation and maturation, in the context of which the function of MYT1L and clues to its potential role in neuropsychiatric syndromes have been identified. Along with ASCL1 and BRN2, MYT1L is a transcription factor that efficiently converts mouse fibroblasts (embryonic and postnatal) into functional neurons in vitro (Vierbuchen et al., 2010). Pang et al. demonstrated the ability of these transcription factors to convert human fibroblasts into functional iN cells (Pang et al., 2011), and it was later shown that MYT1L functions in the neuronal maturation aspect of the reprogramming process (Chanda et al., 2014). More recent work has shown that MYT1L works to actively repress nonneuronal differentiation of cells, maintaining the identity of neurons in their development and maturation (Mall et al., 2017).
In general, these data support Deshpande and Weiss' U-shaped model of gene dosage for reciprocal mutations in MYT1L including intellectual disability and autism, and their dominant model with regard to mutations in MYT1L presenting schizophrenia. The possibility that variation in dosage of a single gene could alternately engender schizophrenia (a majority of partial duplications) and autism (a near-majority of deletions) warrants continued investigation of MYT1L, given its remarkable potential to illuminate contrasts between these often catastrophic neuropsychiatric conditions which share many behavioral features but differ profoundly in timing of onset and the presence/absence of overt psychosis. Pleiotropic effects of highly disruptive mutations to at least 10 genes have variously resulted in autism and schizophrenia in different individuals (Vissers et al., 2016), but none have previously exhibited diagnostic contrasts on the basis of partial duplication versus deletion. Recently, transgenic mice with autism- and schizophrenia-linked mutations in SHANK3 revealed both overlapping and nonoverlapping synaptic, behavioral, and molecular effects of the mutations (Zhou et al., 2016), and similar studies are warranted for deletion and duplication in MYT1L; however, we know of no previous publication involving a transgenic mouse model of deletion or duplication in this gene.
With regard to the specific function of MYT1L, MYT1L is expressed in oligodendrocyte lineage cells during both myelination and remyelination, and, more specifically, overexpression of MYT1L promotes the differentiation of oligodendrocyte progenitor cells (OPCs), while under-expression inhibits this OPC differentiation process (Shi et al., 2018). MYT1L promotes OPC differentiation by initiating the transcription of Olig1, another transcription factor that initiates the transcription of more myelin-related genes (Shi et al., 2018). These data suggest that expression of MYT1L may have a broad effect on the myelination of the central nervous system. Schizophrenia is a neuropsychiatric disorder that features white matter abnormality, and we report here a statistically significant association of schizophrenia with partial duplication of MYT1L. It is our hope that this report and the accumulation of case reports surrounding MYT1L provide a foundation for further inquiry into the relationship of this gene to neuropsychiatric abnormalities.
Reciprocal relationships of CNV gains and losses have been previously observed, whose phenotypic contrasts we summarize in Supporting Information Data S2. We excluded sequence variants (as have been reported for SCN1A, SCN2A, RAI1, and CACNA1) from the table, since they are highly specific and difficult to compare to CNV's, but briefly summarize notable phenotypic consequences of gain and loss of copy number in these genes as follows: Duplication of SCN1A is associated with presence of familial hemiplegic migraines, while loss of function variants are associated with Dravet syndrome (Heyne et al., 2019). Similarly, duplication of CACNA1 is associated with familial hemiplegic migraines, and loss of function variants in mice demonstrate episodic ataxia and presence of a neurodevelopmental disorder presenting with epilepsy (Heyne et al., 2019). Gain of function of SCN2A is associated with early onset epilepsy, while loss of function of SCN2A is associated with autism or later onset epilepsy (Heyne et al., 2019). Finally, duplication of RAI1 in mice has been associated with hyperactivity while loss of function in mice has been associated with obesity and abnormal EEG patterns (Neira-Fresneda & Potocki, 2015). Each of these associations, individually considered with respect to deletion or duplication, can be categorized as following Deshpande and Weiss' dominant model of gene expression in which gain or loss of copy number is associated with one phenotype, while the opposite direction of gene expression has no effect on that phenotype, but rather influences an alternate phenotype (Deshpande & Weiss, 2018).
In conclusion, these data strongly suggest a role of MYT1L in diverse neuropsychiatric outcomes that at least partially diverge on the basis of specificity of disruption to the gene. Future research on the specific effects of contrasting mutations in MYT1L may provide insight into the causal origins of autism and schizophrenia.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development, the National Heart, Lung, and Blood Institute, and the National Human Genome Research Institute of the National Institutes of Health under Award Numbers U54HD087011 (the Intellectual and Developmental Disabilities Research Center at Washington University), K12HL120002, and K08HG010154. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding information
Eunice Kennedy Shriver National Institute of Child Health and Human Development, Grant/ Award Number: U54HD087011; National Heart and Lung Institute, Grant/Award Number: K12HL120002; National Human Genome Research Institute, Grant/Award Number: K08HG010154
Footnotes
CONFLICT OF INTEREST
Dr Constantino receives royalties from Western Psychological Services for the commercial distribution of the Social Responsiveness Scale. No royalties were generated by the use of the instrument for this research study. For the remaining authors none were declared.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- Blanchet P, Bebin M, Bruet S, Cooper GM, Thompson ML, Duban-Bedu B McNeill A (2017). MYT1L mutations cause intellectual disability and variable obesity by dysregulating gene expression and development of the neuroendocrine hypothalamus. PLoS Gene, 13, e1006957 10.1371/journal.pgen.1006957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, & Zanini S (2014). A new patient with a terminal de novo 2p25.3 deletion of 1.9 Mb associated with early-onset of obesity, intellectual disabilities and hyperkinetic disorder. Molecular Cytogenetics, 7, 53 10.1186/1755-8166-7-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock A, Del Campo M, Reiff MI, & Stein MT (2018). Disruptive behavior, global developmental delay, and obesity in a 5-year-old boy with a chromosome microduplication. Journal of Developmental and Behavioral Pediatrics, 39, 81–84. 10.1097/DBP.0000000000000528 [DOI] [PubMed] [Google Scholar]
- Chanda S, Ang CE, Davila J, Pak C, Mall M, Lee QY, … Wernig M (2014). Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Reports, 3, 282–296. 10.1016/j.stemcr.2014.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, Van Bon BW, Vulto-van Silfhout AT, Bosco P, … Schuurs-Hoeijmakers JH (2014). Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nature Genetics, 46, 1063–1071. 10.1038/ng.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, … Abdel-Hamid H (2011). A copy number variation morbidity map of developmental delay. Nature Genetics, 43, 838–846. 10.1038/ng.909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi B, Stead P, & Elliot M (2010). Comparative genomics of autism and schizophrenia. Proceedings of the National Academy of Sciences of the United States of America, 107, 1736–1741. 10.1073/pnas.0906080106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo CS, Varela MC, de Castro CIE, Otto PA, Perez ABA, Lourenço CM, … Koiffmann CP (2018). Chromosomal microarray analysis in the genetic evaluation of 279 patients with syndromic obesity. Molecular Cytogenetics, 11, 14 10.1186/s13039-018-0363-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rocker N, Vergult S, Koolen D, Jacobs E, Hoischen A, Zeesman S, … Menten B (2015). Refinement of the critical 2p25.3 deletion region: The role of MYT1L in intellectual disability and obesity. Genetics in Medicine, 17, 460–466. 10.1038/gim.2014.124 [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, … Buxbaum JD (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature, 515, 209–215. 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A, & Weiss LA (2018). Recurrent reciprocal copy number variants: Roles and rules in neurodevelopmental disorders. Developmental Neurobiology, 78, 519–530. 10.1002/dneu.22587 [DOI] [PubMed] [Google Scholar]
- Doco-Fenzy M, Leroy C, Schneider A, Petit F, Delrue MA, Andrieux J, … Geneviève D (2014). Early-onset obesity and paternal 2pter deletion encompassing the ACP1, TMEM18, and MYT1L genes. European Journal of Human Genetics, 22, 471–479. 10.1038/ejhg.2013.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyne HO, Palmer D, Iqbal S, Baez-Nieto D, Brunklaus A, Johannesen K, … Scholl U (2019). Predicting functional effects of missense variants in voltage-gated sodium and calcium channels. Bio-Rxiv, 671453 10.1101/671453 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Aleksic B, Kirov G, Kinoshita Y, Yamanouchi Y, Kitajuma T, … Nakao I (2010). Copy number variation in schizophrenia in the Japanese population. Biological Psychiatry, 67, 283–286. 10.1016/j.biopsych.2009.08.034 [DOI] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. (2008). Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature, 455, 237–241. 10.1038/nature07239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, … Bras JM (2008). Genotype, haplotype and copy-number variation in worldwide human populations. Nature, 451, 998–1003. 10.1038/nature06742 [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, … MacArthur DG (2019). Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv, 531210 10.1101/531210 [DOI] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, … O'Donovan MC (2009). Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Human Molecular Genetics, 18, 1497–1503. 10.1093/hmg/ddp043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Mattai A, Long R, Rapoport JL, Gogtay N, & Addington AM (2012). Microduplications disrupting the MYT1L gene (2p25.3) are associated with schizophrenia. Psychiatric Genetics, 22, 206–209. 10.1097/YPG.0b013e328353ae3d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, … Exome Aggregation Consortium. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536, 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loid P, Mäkitie R, Costantini A, Viljakainen H, Pekkinen M, & Mäkitie O (2018). A novel MYT1L mutation in a patient with severe early-onset obesity and intellectual disability. American Journal of Medical Genetics. Part A, 176, 1972–1975. 10.1002/ajmg.a.40370 [DOI] [PubMed] [Google Scholar]
- MacDonald JR, Ziman R, Yuen RK, Feuk L, & Scherer SW (2013). The database of genomic variants: A curated collection of structural variation in the human genome. Nucleic Acids Research, 42, D986–D992. 10.1093/nar/gkt958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall M, Kareta MS, Chanda S, Ahlenius H, Perotti N, Zhou B, … Wernig M (2017). Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature, 544, 245–249. 10.1038/nature21722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo S, Rosello M, Monfort S, Oltra S, Orellana C, & Martínez F (2015). Haploinsufficiency of the MYT1L gene causes intellectual disability frequently associated with behavioral disorder. Genetics in Medicine, 17, 683–684. 10.1038/gim.2015.86 [DOI] [PubMed] [Google Scholar]
- Meyer KJ, Axelsen MS, Sheffield VC, Patil SR, & Wassink TH (2012). Germline mosaic transmission of a novel duplication of PXDN and MYT1L to two male half-siblings with autism. Psychiatric Genetics, 22,137–140. 10.1097/YPG.0b013e32834dc3f5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG, Pulver AE, McGrath JA, Wolyniec PS, Dodd AF, Cutler DJ, … Warren ST (2014). Reciprocal duplication of the Williams-Beuren syndrome deletion on chromosome 7q11. 23 is associated with schizophrenia. Biological Psychiatry, 75, 371–377. 10.1016/j.biopsych.2013.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neira-Fresneda J, & Potocki L (2015). Neurodevelopmental disorders associated with abnormal gene dosage: Smith-Magenis and Potocki-Lupski syndromes. Journal of Pediatric Genetics, 4, 159–167. 10.1055/s-0035-1564443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, … Wernig M (2011). Induction of human neuronal cells by defined transcription factors. Nature, 476, 220–223. 10.1038/nature10202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio M, Royer G, Gobin S, de Blois MC, Oziliou C, Bernheim A, … Malan V (2013). Monozygotic twins discordant for submicroscopic chromosomal anomalies in 2p25.3 region detected by array CGH. Clinical Genetics, 84, 31–36. 10.1111/cge.12036 [DOI] [PubMed] [Google Scholar]
- Shi Y, Shao Q, Li Z, Gonzalez GA, Lu F, Wang D, … Cao L (2018). Myt1L promotes differentiation of oligodendrocyte precursor cells and is necessary for remyelination after lysolecithin-induced demyelination. Neuroscience Bulletin, 34, 247–260. 10.1007/s12264-018-0207-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SJ, van Ravenswaaij-Arts CM, Janssen JW, Klein Wassink-Ruiter JS, van Essen AJ, Dijkhuizen T, … Engelen JJ (2011). MYT1L is a candidate gene for intellectual disability in patients with 2p25.3 (2pter) deletions. American Journal of Medical Genetics. Part A, 155A, 2739–2745. 10.1002/ajmg.a.34274 [DOI] [PubMed] [Google Scholar]
- Suktitipat B, Naktang C, Mhuantong W, Tularak T, Artiwet P, Pasomsap E, … Yimwadsana B (2014). Copy number variation in Thai population. PLoS One, 9, e104355 10.1371/journal.pone.0104355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, Zeng J, Cong X, Yan A, Lin Y, Zhang X, … Lan F (2014). Genetic diagnosis and analysis of related genes for a pedigree with 2p25 and 12p13 cryptic rearrangements. Zhonghua Yi Xue Yi Chuan Xue Za Zhi, 31, 444–448. 10.3760/cma.j.issn.1003-9406.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Tuwaijri A, & Alfadhel M (2019). MYT1L mutation in a patient causes intellectual disability and early onset obesity: A case report and review of the literature. Journal of Pediatric Endocrinology & Metabolism, 32, 409–413. 10.1515/jpem-2018-0505 [DOI] [PubMed] [Google Scholar]
- Van Den Bossche MJ, Strazisar M, Cammaerts S, Liekens AM, Vandeweyer G, Depreeuw V, … Del-Favero J (2013). Identification of rare copy number variants in high burden schizophrenia families. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 162B, 273–282. 10.1002/ajmg.b.32146 [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, & Wernig M (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature, 463, 1035–1041. 10.1038/nature08797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, Gilissen C, & Veltman JA (2016). Genetic studies in intellectual disability and related disorders. Nature Reviews. Genetics, 17, 9–18. 10.1038/nrg3999 [DOI] [PubMed] [Google Scholar]
- Vlaskamp DRM, Callenbach PMC, Rump P, Giannini LAA, Dijkhuizen T, Brouwer OF, & van Ravenswaaij-Arts CMA (2017). Copy number variation in a hospital-based cohort of children with epilepsy. Epilepsia Open, 2, 244–254. 10.1002/epi4.12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Genetic Risk Outcomes in Psychosis (GROUP) Consortium, Sabatti C, … Veltman JA (2008). Recurrent CNVs disrupt three candidate genes in schizophrenia patients. American Journal of Human Genetics, 83, 504–510. 10.1016/j.ajhg.2008.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, … Sebat J (2008). Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science, 320, 539–543. 10.1126/science.1155174 [DOI] [PubMed] [Google Scholar]
- Wang T, Guo H, Xiong B, Stessman HA, Wu H, Coe BP Eichler EE (2016). De novo genic mutations among a Chinese autism spectrum disorder cohort. Nature Communications, 7, 13316 10.1038/ncomms13316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildeman M, van Ophuizen E, den Dunnen JT, & Taschner PE (2008). Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Human Mutation, 29, 6–13. 10.1002/humu.20654 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Kaiser T, Monteiro P, Zhang X, Van der Goes MS, Wang D, … Feng G (2016). Mice with Shank3 mutations associated with ASD and schizophrenia display both shared and distinct defects. Neuron, 89, 147–162. 10.1016/j.neuron.2015.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.