Abstract
Background
Clinical detection of SARS-CoV-2 RNA in stools supports the idea of wastewater-based epidemiology (WBE) as a precious tool for COVID-19 environmental surveillance. Successful detection of SARS-CoV-2 RNA in untreated wastewaters has been reported in several countries. This study investigated the presence and persistence of viral RNA in treated and untreated wastewaters in Padua, Italy. An urban experimental network of sampling sites was tested for prospective surveillance activities.
Methods
Seven sampling sites (i.e. wastewater pumping stations, plant inlets and outlets) were selected from the two main municipal wastewater treatment plant systems. Eleven grab samples (9 untreated, 2 treated wastewaters) were collected on 2 dates. All samples were tested at t0 for SARS-CoV-2 RNA and t1 = 24 h to investigate its persistence, at room temperature and under refrigerated conditions. Overall, 33 sub-samples were concentrated by ultrafiltration and tested for molecular detection of viral RNA with two RT-qPCR assays.
Results
At t0, positivity for at least one RT-qPCR assay was achieved by 4/9 untreated wastewater samples and 2/2 tertiary treated samples. A minimum SARS-CoV-2 titer of 4.8–4.9 log10 gc/L was estimated. At t1, three refrigerated subsamples were positive as well. The two RT-qPCR assays showed differential sensitivity, with the N assay detecting 90% of successful amplifications.
Conclusions
SARS-CoV-2 RNA was detected in untreated and treated wastewaters. Its persistence after 24 h was demonstrated in subsamples kept at 4 °C. Hospitalization data suggested an approximate WBE detection power of 1 COVID-19 case per 531 inhabitants. The possible role of WBE in COVID-19 environmental surveillance is strongly supported by our findings. WBE can also provide precious support in the decision-making process of restriction policies during the epidemic remission phase. Optimization and standardization of laboratory methods should be sought in the short term, so that results from different studies can be compared with reliability.
Keywords: WBE, Environmental surveillance, SARS-CoV-2, COVID-19, Sewage
Graphical abstract
1. Introduction
Current evidence suggests that the novel Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic virus primarily spreads via droplet and airborne transmission (e.g. Li et al., 2020; Morawska and Cao, 2020; Zhang et al., 2020). Nevertheless, as previously described for SARS and MERS viruses (Chen et al., 2006), several authors are reporting how SARS-CoV-2 RNA can be found in stool samples of symptomatic, asymptomatic and pre-symptomatic COVID-19 patients (Jiang et al., 2020; Pan et al., 2020; Wölfel et al., 2020). Actually, detection of viral RNA in stools occurs in 16.5% to 100% of COVID-19 patients (La Rosa et al., 2020a), with estimated shedding of 102 to 108 RNA copies per gram (Pan et al., 2020; Randazzo et al., 2020; Wölfel et al., 2020) and observed duration of positivity ranging from 1 to >30 days after the onset of symptoms (Amirian, 2020). These clinical findings promptly lead to the idea that environmental surveillance and especially wastewater-based epidemiology (WBE) could serve as precious tools to monitor COVID-19 clusters (Carducci et al., 2020; Randazzo et al., 2020). In the past, WBE already proved to be a successful strategy for the surveillance and early detection of epidemic enteric virus outbreaks, e.g. poliovirus, norovirus and hepatitis A virus, as well as for tuning public health interventions (Ahmed et al., 2020; Hart and Halden, 2020; Hata and Honda, 2020; Randazzo et al., 2020; Sims and Kasprzyk-Hordern, 2020). Detection of SARS-CoV-2 RNA in untreated wastewater samples has been reported in Australia (Ahmed et al., 2020), USA (Wu et al., 2020), China (Wang et al., 2020), Japan (Haramoto et al., 2020), the Netherlands (Lodder and de Roda Husman, 2020; Medema et al., 2020), Spain (Randazzo et al., 2020) and Italy (La Rosa et al., 2020b; Rimoldi et al., 2020). Carducci et al. (2020) provided an interesting review that summarizes the main findings of some of the above studies. Laboratory methods mostly consist in wastewater sample pre-treatment (e.g. centrifugation, ultrafiltration), extraction of nucleic acids and molecular detection of viral RNA targets (e.g. Vogels et al., 2020), although rapid paper-based devices have recently been described as promising tools for SARS-CoV-2 in wastewater and other environmental samples (Mao et al., 2020). In addition, preliminary computational models have been proposed to estimate the prevalence of infection from WBE data along with the economic advantages deriving from environmental surveillance approaches (Ahmed et al., 2020; Hart and Halden, 2020).
Italy has been the first out-of-Asia epicenter of the ongoing COVID-19 pandemic. On 30/01/2020 two imported cases, i.e. Chinese travelers, were confirmed in Rome, the capital city. The third case, an Italian citizen repatriated from China, was confirmed on 06/02/2020. The first autochthonous cases occurred on 21/02/2020, with the almost synchronous detection of two distinct outbreaks, in Lombardy and Veneto respectively (ISS, 2020). Both Regions are located in the NE of the Country. As of 22/09/2020, Italy reported 300,897 confirmed COVID-19 cases. Veneto scores as the fourth Italian Region for number of cases (26,004), of whom 5091 (19.6%) were confirmed for Padua Province (Protezione Civile, 2020). To the authors' knowledge, only two studies assessed and confirmed the presence of SARS-CoV-2 RNA in wastewaters in Italy (La Rosa et al., 2020b; Rimoldi et al., 2020). The first study tested 12 samples of untreated wastewater (i.e. plant influent), reporting positivity for 6 out of 12 samples. The second Italian study tested 8 untreated wastewater samples (4 positive samples) and 4 treated wastewater samples (i.e. plant effluent, no positive samples).
The present study reports the first detection of SARS-CoV-2 RNA in both treated and untreated wastewaters of Padua city, in the Veneto Region (NE Italy). Padua has 211,316 inhabitants (Padua Municipality, 2019) and it hosts one of the major COVID-19 hospitals of the Region. This pilot investigation also assesses the persistence of viral RNA in samples after 24 h from their collection, both at room temperature and under refrigerated conditions (4 °C). Moreover, in addition to wastewater treatment plant (WWTP) inlet and outlet, sampling sites at urban district level, i.e. minor wastewater pumping stations (WPSs) were also considered, in order to test a possible sampling site network for future COVID-19 environmental surveillance and WBE purposes.
2. Materials and methods
2.1. Choice of sampling sites
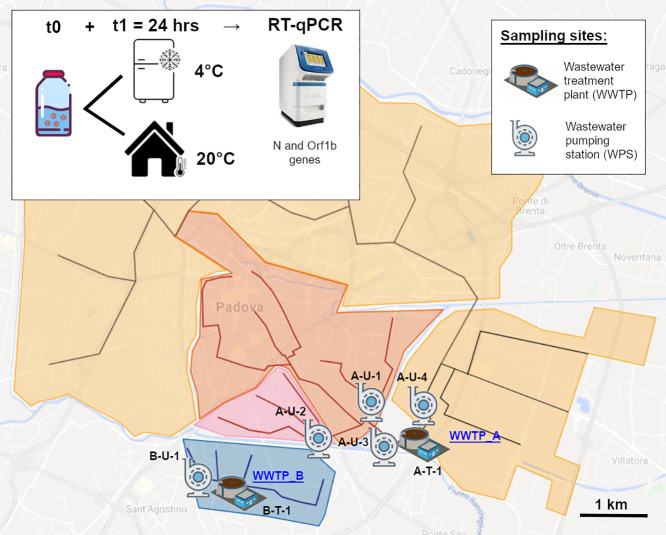
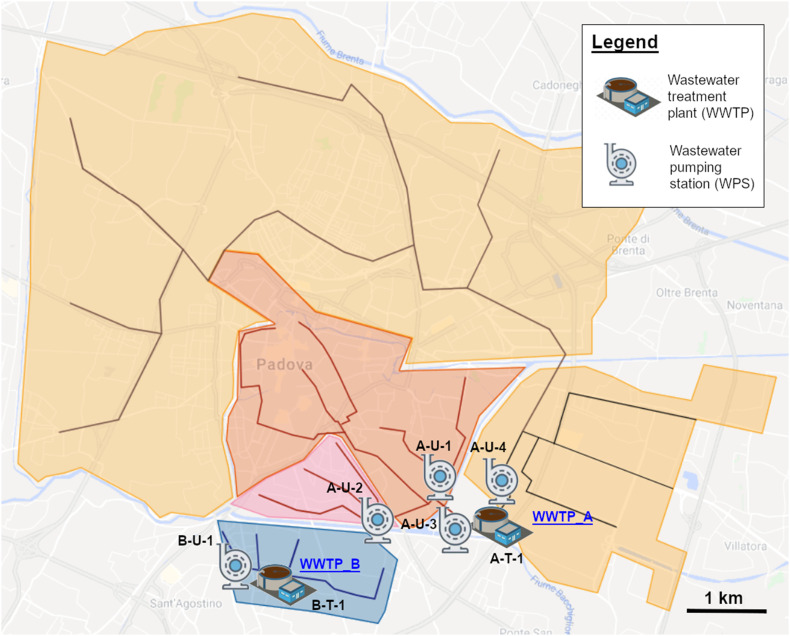
Padua's sewerage system, as for many historical Italian cities, is quite complex and encompasses separate sewer lines built over many decades (i.e. 1938 to recent years). Therefore, prior to field activities, an updated plan of the whole sewerage system (Acegas-Aps-Amga, personal communication, 20 April 2020) was thoroughly analyzed to select a network of the most appropriate and representative sampling sites (Fig. 1 ) for future WBE monitoring activities. In the end, 4 WPSs were selected, each representative of a precise urban district sewer line. Three of the selected WPSs flow into Padua's main wastewater treatment plant (WWTP_A) and one acts as inlet of the WWTP serving the Southern area of the city (WWTP_B). WWTP_A (197,000 population equivalent) and WWTP_B (13,000 PE) together count for roughly 80% of municipal users (Acegas-Aps-Amga, personal communication, 20 April 2020). Both WWTPs are activated sludge plants, performing disinfection with peracetic acid and terminal UV lamps as tertiary treatment. Effluents from WWTP_A and WWTP_B were also sampled (Table 1 ; Fig. 2 ).
Fig. 1.
Map of sampling sites. The figure reports a simplified scheme of Padua sewerage system. The city main wastewater treatment plant (WWTP-A) has two separate inlets. The red and pink areas (city center) are served by two district wastewater pumping stations (WPSs), A-U-1 and A-U-2. Especially, A-U-1 drains sewage from the city hospital district. Wastewater from A-U-1 and A-U-2 then merge and reach WWTP-A as a unique influent (A-U-3). The second WWTP_A influent originates from the orange area (site A-U-4). The separate Southern blue area is served by WWTP_B. The plant influent and effluent were sampled (B-U-1 and B-T-1). (Figure adapted from Acegas-Aps-Amga technical sewerage plan, personal communication, 20 April 2020). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
List of sampling sites. IDs intuitively report the draining WWTP (either A or B), wastewater typology (U – untreated or T – treated) and a progressive number. A brief description of each site is also provided. Sites 3, 1 and 8 were only sampled on 05/05/2020. The total population served by WWTP A and B is of 162,460 citizens.
| Wastewater treatment plant | ID | Site brief description | Wastewater | Population (2019) | Pop. coverage | 23/04/20 sampling | 05/05/20 sampling |
|---|---|---|---|---|---|---|---|
| WWTP-A | A-U-1 | WPS “Forcellini” | Untreated | 36,042 | 17% | Yes | Yes |
| A-U-2 | WPS “Crescini” | Untreated | 21,778 | 10% | Yes | Yes | |
| A-U-3 | Influent 1 “City Center” (A-U-1+2) | Untreated | 57,830 | 27% | No | Yes | |
| A-U-4 | Influent 2 “Fossetta” | Untreated | 91,860 | 44% | Yes | Yes | |
| A-T-1 | Effluent | Tertiary treated | 149,690 | 71% | No | Yes | |
| WWTP-B | B-U-1 | WPS & influent “Guizza” | Untreated | 12,770 | 6% | Yes | Yes |
| B-T-1 | Effluent | Tertiary treated | No | Yes |
Fig. 2.
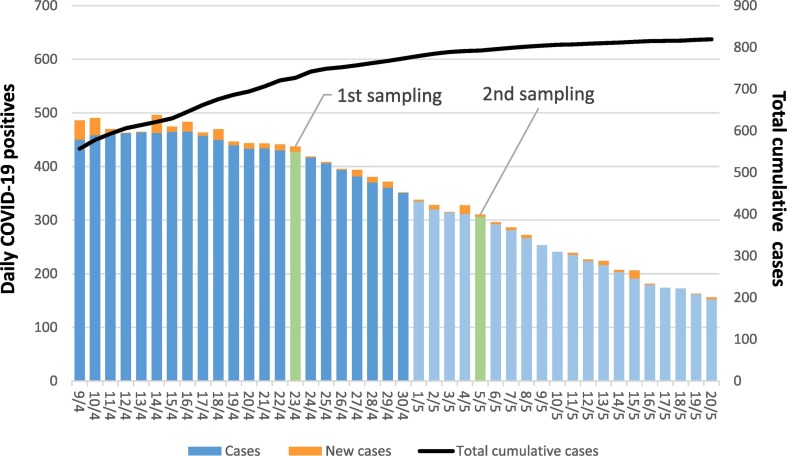
Graphical summary of COVID-19 epidemiological data for Padua Municipality. The figure shows a time frame of ±15 days from sampling dates (i.e. April 23rd and May 5th), reporting daily COVID-19 cases and the total cumulative case number, as reference for the local epidemic trend. Point prevalence was of 202 and 145 cases per 100,000 inhabitants on the first and on the second sampling date, that is 427 and 306 COVID-19 cases in the municipality, respectively (clinical surveillance data derived from: Protezione Civile, 2020).
2.2. Wastewater sampling
A total of 11 wastewater samples were collected on two dates, 23 April and 5 May 2020 (Table 1). No rainfall occurred at least in the 12 h that preceded the sampling. For each sampling site, 2 sterile 1000 mL HDPE bottles (Corning® Gosseling™, France) were filled with wastewater, for a total volume of 2000 mL. Field personnel wore standard personal protective equipment (PPE) for wastewater sampling (i.e. safety footwear, gloves, FFP3 face mask and face shield). Grab sampling was carried out in the morning, between 9:00 and 10:30 am, so as to detect the “morning flush”, as supported both by literature (Wang et al., 2005; Randazzo et al., 2020) and by local WWTPs technical records (Acegas-Aps-Amga, personal communication, April 2020). Wastewater pH and temperature were determined with a portable pH tester (Hanna Instruments, Italy). Samples were carried to the laboratory on ice (4 °C), where 100 mL of each sample was immediately processed. The two collected bottles were then kept one at room temperature (r.t. 20 °C) and one at 4 °C. SARS-CoV-2 RNA molecular detection was also performed after 24 h (t1), for subsamples stored in both temperature conditions.
2.3. Concentration method
Concentration method was chosen after briefly reviewing some recently published protocols (Ye et al., 2016; Blanco et al., 2019; Ahmed et al., 2020). Among these, the detailed experimental method comparison carried out by Ye et al. (2016) made us lean towards an ultrafiltration protocol over ultracentrifugation or PEG precipitation methods. An optimized ultrafiltration procedure is described as the one with best recovery of enveloped virus from wastewater samples (average recovery = 22%) and, in our opinion, it also grants fast processing coupled to little sample manipulation. According to the procedure, 100 mL of each sample was pre-filtered on 0.22 μm polyether sulfone (PES) (Stericup-GP 250 mL Express Plus, Merck, Germany). A 6 mL volume of pre-filtered sample was then concentrated with centrifugal 10 kDa filter units (Amicon Ultra, Merck, Germany) in a swinging bucket rotor at 4000 ×g for 10 min, with a final recovery of 150–200 μL of concentrated solute.
2.4. RNA extraction and molecular detection
An internal RNA positive control (Seegene RP Allplex kit component, Korea) was added to concentrated samples before extraction and it also served to check for inhibitors of quantitative reverse transcription polymerase chain reaction (RT-qPCR). Viral RNA extraction was performed on 150 μL of concentrated sample with a commercial kit (QIAamp viral RNA mini kit, Qiagen, Germany), following the manufacturer's instructions. Molecular detection of SARS-Cov-2 RNA was performed with two WHO-shared RT-qPCR assays targeting genes ORF1b-nsp14 and N (Chu et al., 2020). Primers and dual-labeled probes (Table 2 ) were provided from Thermo Fisher Scientific (US). Synthetic dsDNA fragments were used as positive controls and were also purchased by Thermo Fisher. For each PCR run, 2 positive and 2 negative controls were also included. All assays were performed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems, USA). Positivity was attributed only to reactions with cycle threshold (Ct) <40 (Medema et al., 2020; Randazzo et al., 2020).
Table 2.
Sequences of primers and probes. Sequence of forward (F) and reverse (R) primers and of dual-labeled probes are reported. Expected amplicon size and cycling parameter are given, as per Chu et al. (2020).
| Organism | Target gene | Assay name | Sequence (5′-3′) | Amplicon size | Cycling parameters | Reference |
|---|---|---|---|---|---|---|
| SARS-CoV-2 | Orf1b | HKU-ORF1b-nsp14F | F-TGGGGYTTTACRGGTAACCT | 132 bp | Chu et al., 2020 | |
| HKU-ORF1b-nsp14R | R-AACRCGCTTAACAAAGCACTC | 50 °C for 5 min; | ||||
| HKU-ORF1b-nsp141P | P-FAM-TAGTTGTGATGCWATCATGACTAG-QSY | 95 °C for 20 s; | ||||
| N | HKU-NF | F-TAATCAGACAAGGAACTGATTA | 110 bp | 40 cycles of 95 °C for 5 s; | ||
| HKU-NR | R-CGAAGGTGTGACTTCCATG | 60 °C for 30 s. | ||||
| HKU-NP | P-FAM-GCAAATTGTGCAATTTGCGG-QSY |
3. Results
SARS-CoV-2 RNA presence was assessed for all collected samples. Moreover, persistence after 24 h from sampling was evaluated in subsamples stored at r.t. and 4 °C. Overall, molecular analysis was carried out on 33 different subsamples. Among these, 9 subsamples from 4 different sampling sites resulted positive for at least one RT-qPCR assay (Table 3 ). In detail, sites A-U-3 and B-U-1 (WWTP_A and WWTP_B influents) never reported any positivity for SARS-CoV-2 RNA. Site A-U-1 (WWTP_A WPS) and site A-U-4 (WWTP_A influent) were positive at t0 in both sampling dates. Moreover, on 05/05/2020 sample A-U-1 positivity was detected also at t1 (4 °C). Samples A-T-1 and B-T-1 (WWTP_A and WWTP_B effluents) were collected only on 05/05/2020 and reported positivity at t0. A-T-1 also resulted positive after 24 h at 4 °C. A-U-2 showed an inconsistent pattern on the second sampling date, since SARS-CoV-RNA was not detected at t0 but only at t1 in the refrigerated subsample. Actually, the N and Orf1b RT-qPCR assays gave inconsistent results. The majority of positive amplifications were indeed obtained with the N assay. In detail, 9 were positive for N, whereas the Orf1b assay didn't succeed in detecting SARS-CoV-2 RNA but in one sample (A-U-1, at t0 on 23/04/2020). Direct quantitation of positive samples was not performed. Nevertheless, since the reported limit of detection (LOD) of the implemented RT-qPCR assay is 2.5 genome copies (gc) per μL (Chu et al., 2020), a minimum SARS-CoV-2 titer of 4.8–4.9 log10 gc/L can be estimated.
Table 3.
Molecular detection of SARS-CoV-2 RNA in wastewater samples. Collected samples were tested at t0 and at t1 = 24 h, with subsamples being kept at room temperature and at 4 °C. Two RT-qPCR assays were performed on all extracts, targeting genes N and Orf1b, with an estimated limit of detection (LOD) of 2.5 gc/L (Chu et al., 2020). Average recovery for enveloped viruses from wastewater with the adopted ultrafiltration protocol is estimated at 22% (Ye et al., 2016).
| Sampling date: 23/04/2020 | |||||||
|---|---|---|---|---|---|---|---|
| Plant | t0 |
t1 (r.t.) |
t1 (4 °C) |
||||
| ID | N | Orf1b | N | Orf1b | N | Orf1b | |
| WWTP_A | A-U-1 | + | + | − | − | − | − |
| A-U-2 | − | − | − | − | − | − | |
| A-U-4 | + | − | − | − | − | − | |
| WWTP_B | B-U-1 | − | − | − | − | − | − |
| Sampling date: 05/05/2020 | |||||||
|---|---|---|---|---|---|---|---|
| Plant | t0 |
t1 (r.t.) |
t1 (4 °C) |
||||
| ID | N | Orf1b | N | Orf1b | N | Orf1b | |
| WWTP_A | A-U-1 | + | − | − | − | + | − |
| A-U-2 | − | − | − | − | + | − | |
| A-U-3 | − | − | − | − | − | − | |
| A-U-4 | + | − | − | − | − | − | |
| B-T-1 | + | − | − | − | + | − | |
| WWTP_B | B-U-1 | − | − | − | − | − | − |
| B-T-1 | + | − | − | − | − | − | |
4. Discussion
The present study investigated the presence of SARS-CoV-2 RNA in treated and untreated wastewater samples and also evaluated the virus persistence at 24 h, under two different temperature conditions. Moreover, an experimental network of sampling sites (i.e. district WPS, WWTP inlets and outlets) was tested for future COVID-19 environmental surveillance activities. From the analytic perspective, the first issue to be addressed was that of choosing a suitable recovery and concentration method. Recent studies already outlined the lack of a standard concentration method (Carducci et al., 2020; La Rosa et al., 2020a). A recent ultrafiltration protocol was adapted, since it had been especially optimized for the recovery of enveloped viruses from wastewater (Ye et al., 2016). A volume of about 100 mL was pre-filtered on 0.22 μm PES membranes (Stericup®, Merck-Millipore, Germany). Six milliliter of pre-filtered samples were then concentrated using centrifugal filter units Amicon® Ultra with a 10 kDa cutoff (Merck-Millipore, Germany). A minor flaw of this method is that – if using the suggested centrifugal filter units - it only allows to process small volumes of wastewater. Some authors (Ahmed et al., 2020; Medema et al., 2020) overcame this problem using bigger centrifugal devices, i.e. Centricon® Plus units, but unfortunately, we could not purchase such items due to the strict lockdown measures that were enforced in Italy starting from 08/03/2020.
The molecular detection of SARS-CoV-2 RNA was performed adopting two assays, based on the N and Orf1b genes (Chu et al., 2020). Actually, several custom RT-qPCR assays have been designed for the clinical diagnosis of SARS-CoV-2 and, so far, some of them (e.g. Corman et al., 2020) have been used also on wastewater samples. To our knowledge, this is the first study to detect SARS-CoV-2 RNA in environmental samples using the N and Orf1b-nsp14 clinical assays originally designed by Chu et al. (2020). However, as mentioned, RT-qPCR amplifications produced incongruous results between the two assays. Among positive amplifications, 9 out of 10 (90%) were given by the N gene assay. Actually, the Orf1b assay successfully detected viral RNA in a single sample (i.e. A-U-1, t0, 23/04/2020), matching the N assay result. To this point, Chu and colleagues eventually discussed some preliminary results that suggested a 10× analytical sensitivity of the N assay over the Orf1b one in detecting positive samples (Chu et al., 2020). Accordingly, authors using other RT-qPCR assays for the detection of SARS-CoV-2 in wastewaters also reported non-coherent responses between assays targeting different genes (Ahmed et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Rimoldi et al., 2020). Especially, one study highlighted that when viral titer is <10 gc/μL, the power of some RT-qPCR assays in differentiating true from false negatives can vary (Vogels et al., 2020). All positive amplifications obtained in the present investigation were characterized by Ct > 37, so that very low viral titers in samples could similarly explain the discordant results between N and Orf1b assays. Moreover, non-homogenous subsampling can also supervene whenever the target gene is present in a few copies (Taylor et al., 2019; Ahmed et al., 2020). The SARS-CoV-2 RNA titers estimated by the present study (4.8–4.9 log 10 gc/L) are in range with those reported by other studies worldwide, e.g. 4 to 6 log 10 gc/L (Randazzo et al., 2020; Wurtzer et al., 2020; Wu et al., 2020).
Similar data have recently been discussed in terms of potential effectiveness of SARS-CoV-2 environmental surveillance and some computational models proposed a theoretical power of detecting 1 COVID-19 case/100 to 2,000,000 non-infected people (e.g. Ahmed et al., 2020; Hart and Halden, 2020; Hata and Honda, 2020). However, we did not attempt any prevalence estimate due to the considerable uncertainty related to a multiplicity of variables that need further investigation. Some variables are laboratory-related (e.g. lack of accurate quantification of viral RNA in wastewater samples; variable recovery efficiency). Other variables depend on the host-pathogen interaction (e.g. range of virions shed per g of feces; geographic variation in the proportion of infected people with positive stool samples) (Ahmed et al., 2020; Pan et al., 2020; Randazzo et al., 2020; Wölfel et al., 2020).
Wastewater samples tested in the present study outline an interesting scenario for Padua's experimental surveillance network. Positive samples were found in both WWTP_A and WWTP_B systems. At t0, positivity was detected for 4 out of 9 untreated wastewater samples and 2 out of 2 tertiary treated samples. In detail, with regard to WWTP_A, WPS A-U-1 resulted positive on both sampling dates. This WPS collects wastewater drained from the historical center of the city and its hospital district (with a dedicated COVID-19 hospital), that encompass a population of 36,052 inhabitants (Table 1). On 23/04/2020 and on 05/05/2020 the number of COVID-19 patients hospitalized in non-intensive care unit was of 64 and 49, respectively (Azienda Zero, 2020). On the same dates, there were a total of 427 and 306 COVID-19 cases in the Municipality (Protezione Civile, 2020), that is 363 and 257 cases, without considering the hospitalized ones. Assuming that COVID-19 cases had a homogeneous distribution in the Municipal territory (i.e. no major clusters were registered on these dates), the proportion of COVID-19 cases for WPS A-U-1 at-home population would be 62 and 44, respectively. If we add the hospitalized cases the WPS A-U-1 positives, we can estimated that on the two sampling dates there were 126 and 93 infected people possibly spreading SARS-CoV-2 virions with their stools. In the end, A-U-1 positivity can be read as the successful detection of 93 cases per 36,052 inhabitants, i.e. 1 / 388. Although obtained with a rough calculation, this value takes into account precise small-scale numbers and provides a quite reliable estimate of WBE true potentiality. Should an urban surveillance network be implemented, A-U-1 shall be carefully considered, as it could serve as a reference sampling point. WPS A-U-2 was negative, as well as A-U-3 (WWTP_A first influent). Actually, A-U-1 and A-U-2 wastewaters merge before reaching WWTP_A and flow into the plant as A-U-3. Therefore, it could be hypothesized that negative A-U-2 wastewater could have diluted positive A-U-1 below the LOD, so that A-U-3 results negative as well. A-U-4 (WWTP_A second influent) was also positive on both sampling dates. It is representative of a quite broad urban area embracing the city Western, Northern and Eastern neighborhoods. Ideally, a sampling point should be sought for each district but access to suitable WPSs was not granted due to sewerage maintenance works. Since SARS-CoV-2 RNA was found upstream WWTP_A, positivity of plant effluent A-T-1 should not come unexpected. Smaller WWTP_B reported negative influent (B-U-1) but positive effluent (B-T-1). Implementing a proportion analogous to that of A-U-1, we can assume that at least 1 COVID-19 case out of 582 to 822 inhabitants was detected from A-U-4 and B-T-1 samples. Moreover, the B-U-1/B-T-1 discrepancy, besides advoking a “false negative”, could be technically explained if taking into account the WWTP retention time (16–18 h), so that sampled effluent is actually representative of the evening before, rather than paired to the co-sampled influent. A similar result (negative influent/positive effluent) was also reported in Japan and attributed by authors to possible higher analytical LOD for influent samples due to their turbidity and content of suspended solids (Haramoto et al., 2020). To our knowledge, only one study tested treated wastewaters for SARS-CoV-2 RNA in Italy, although without reporting any positive sample (Rimoldi et al., 2020). Of course it would have been be precious to investigate virus vitality in these samples, but it was precluded by the non-availability of a BSL 3 facility. Nevertheless, it should be reminded that the current evidence-based opinion supports the adequacy of routine wastewater treatments in efficiently abating enveloped viruses such as SARS-CoV-2 (Ahmed et al., 2020; Carducci et al., 2020; WHO, 2020). Moreover, besides the exploratory inclusion of WWTP effluents in the present investigation, in our opinion it is not advisable to collect for surveillance purposes a sample that bears an informative delay equal to the WWTP full retention time (i.e. 24 h). Overall, in the future choice of definitive WBE sampling sites, at least every WWTP influent should be sampled. However, as shown by the above discussed results, the sampling of upstream sites, such a minor WPSs, could be even more informative. A sampling site that is representative for a precise urban district, e.g. main sewer line, is probably the best choice for public health purposes and rapid implementation of preventive control measures. During a period of epidemic remission or whenever a second wave is expected to occur, positivity in a specific sampling site would allow prompt identification of connected households and backward tracing of possible asymptomatic and pre-symptomatic SARS-coV-2 carriers.
In addition, the present study also assessed the persistence of viral RNA in collected samples. After 24 h from sampling, three positive samples were found (A-U-1; A-U-2; A-T-1 sampled on 05/05/20). Precisely, only subsamples kept at +4 °C resulted positive, whilst those stored at r.t. did not report any successful amplification. Consistently, low temperature is known to slow viral HCoV RNA degradation (Carducci et al., 2020). Actually, A-U-2 showed the only incongruent result. SARS-CoV-2 RNA was found in its t1 refrigerated subsample, although it was negative at t0. A-U-2 could possibly show a t0 “false negative”, either due to very low viral titers or a subsampling error. PCR inhibitors could also be present in the sample, despite the presence of an internal positive control that validated the RT-qPCR reaction.
Developments of this pilot investigation are already in progress. Wastewater samples are currently being collected and stored at −20 °C. Although real-time surveillance is not feasible, samples will be tested whenever a COVID-19 “second wave” or a smaller local outbreak should occur. In the meanwhile, we are also willing to strengthen interdisciplinary relationships and integration with local public health stakeholders, so that a joint and complete surveillance strategy, i.e. clinical and environmental, can be pursued.
5. Conclusions
The present study tested an experimental network of 7 distinct sampling sites for hypothetical COVID-19 environmental surveillance in Padua city, Veneto Region, NE Italy. The two main wastewater treatment plant (WWTP) systems were considered, as they are representative of 94% of the total city users. In addition to the two WWTPs inlets and outlets, wastewater pumping stations were selected to provide detailed information at urban district level as well. SARS-CoV-2 RNA was successfully detected by RT-qPCR both in untreated and treated wastewaters, although the pathogen vitality could not be determined. Among positive sampling sites, the pumping station collecting wastewater from Padua city center and the hospital district (site A_U_1) resulted especially informative. Punctual hospitalization data for the two sampling dates suggested an approximate effectiveness in detecting about 1 COVID-19 case out of 500 inhabitants. The role of WBE as a powerful tool for COVID-19 environmental surveillance is supported by the findings of the present investigation. WBE could also help in the swift decision-making and implementation of local restriction policies. Laboratory methods, i.e. concentration method and RT-qPCR assays, still need to be optimized. Especially, standardization should be sought in the short term, so that results from different research groups can be compared with reliability.
CRediT authorship contribution statement
Tatjana Baldovin: Conceptualization, Methodology, Funding acquisition, Laboratory and field investigation, Writing - Original Draft. Irene Amoruso: Conceptualization, Methodology, Visualization, Laboratory and field Investigation, Writing - Original Draft. Marco Fonzo: Data Curation, Writing - Review & Editing. Alessandra Buja: Formal analysis, Writing - Review & Editing. Vincenzo Baldo: Project administration, Resources, Writing - Review & Editing. Silvia Cocchio: Formal analysis, Writing - Review & Editing. Chiara Bertoncello: Conceptualization, Writing - Review & Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We wish to thank the Direzione Acqua of AcegasApsAmga S.p.A. for their genuine interest in this research project and precious field support. We also wish to thank Yinyin Ye for her kind and prompt feedback when invoked on Researchgate.
Editor: Yolanda Picó
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirian E.S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azienda Zero SARS-CoV-2 in Veneto. Official daily reports. 2020. www.azero.veneto.it/-/emergenza-coronavirus Available from: [last access on: Nov 4th 2020]
- Blanco A., Abid I., Al-Otaibi N., Pérez-Rodríguez F.J., Fuentes C., Guix S., Pintó R.M., Bosch A. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food Environ. Virol. 2019;11:184–192. doi: 10.1007/s12560-019-09378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carducci A., Federigi I., Dasheng L., Thompson J.R., Marco V. Making waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020 doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Zhang X.J., Wang Y., Zhu L.X., Liu J. Waste water disinfection during SARS epidemic for microbiological and toxicological control. Biomed. Environ. Sci. 2006;19:173–178. [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;555:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;138875 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Honda R. Potential sensitivity of wastewater monitoring for SARS-CoV-2: comparison with Norovirus cases. Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c02271. (acs.est.0c02271) [DOI] [PubMed] [Google Scholar]
- ISS Istituto Superiore di Sanità – ISS per COVID-19 (institutional portal) 2020. www.iss.it/coronavirus (last accessed on: 10 Sep 2020)
- Jiang X., Luo M., Zou Z., Wang X., Chen C., Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J. Med. Virol. 2020:1807–1809. doi: 10.1002/jmv.25941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;115899 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. lancet. Gastroenterol. Hepatol. 2020 doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padua Municipality Annuario statistico Comunale – Sez. 2 – Popolazione. 2019. www.padovanet.it/informazione/padova-cifre (last accessed on: 15 Sep 2020)
- Pan Y., Zhang D., Yang P., Poon L., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protezione Civile COVID-19 – Situazione in Italia. Daily bulletin. 2020. www.salute.gov.it/portale/nuovocoronavirus (last accessed on: 15 Sep 2020)
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.C., Nadeau K., Abbasi M., Lachance C., Nguyen M., Fenrich J. The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 2019;37:761–774. doi: 10.1016/j.tibtech.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O. El, Wong P., Yang Y., Cheemarla N.R., White E.B., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., Dela Cruz C.S., Iwasaki A., Ko A.I., Landry M.L., Foxman E.F., Grubaugh N.D. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5 doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J., Guo T., Zhen B., Kong Q., Yi B., Li Z., Song N., Jin M., Xiao W., Zhu X., Gu C., Yin J., Wei W., Yao W., Liu C., Li J., Ou G., Wang M., Fang T., Wang G., Qiu Y., Wu H., Chao F., Li J. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan hospital and the 309th Hospital of the Chinese People’s Liberation Army. Water Sci. Technol. 2005;52:213–221. doi: 10.2166/wst.2005.0266. [DOI] [PubMed] [Google Scholar]
- Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y., Zhuo L., Zhong Z., Qu T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020;94:103–106. doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Water, sanitation, hygiene, and waste management for SARS-CoV-2, the virus that causes COVID-19 - Interim guidance. 2020. www.who.int/publications/i/item/water-sanitation-hygiene-and-waste-management-for-the-covid-19-virus-interim-guidance Available from: [last access on: Nov 4th 2020]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5:1–9. doi: 10.1128/msystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. 2020. Evaluation of Lockdown Impact on SARS-CoV-2 Dynamics Through Viral Genome Quantification in Paris Wastewaters. medRxiv 2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhang R., Li Y., Zhang A.L., Wang Y., Molina M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. 2020;202009637 doi: 10.1073/pnas.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]