Abstract
Obesity is a major risk factor for SARS-CoV-2 infection and COVID-19 severity. The underlying basis of this association is likely complex in nature. The host-cell receptor angiotensin converting enzyme 2 (ACE2) and the type II transmembrane serine protease (TMPRSS2) are important for viral cell entry. It is unclear whether obesity alters expression of Ace2 and Tmprss2 in the lower respiratory tract. Here, we show that: 1) Ace2 expression is elevated in the lung and trachea of diet-induced obese male mice and reduced in the esophagus of obese female mice relative to lean controls; 2) Tmprss2 expression is increased in the trachea of obese male mice but reduced in the lung and elevated in the trachea of obese female mice relative to lean controls; 3) in chow-fed lean mice, females have higher expression of Ace2 in the lung and esophagus as well as higher Tmprss2 expression in the lung but lower expression in the trachea compared to males; and 4) in diet-induced obese mice, males have higher expression of Ace2 in the trachea and higher expression of Tmprss2 in the lung compared to females, whereas females have higher expression of Tmprss2 in the trachea relative to males. Our data indicate diet- and sex-dependent modulation of Ace2 and Tmprss2 expression in the lower respiratory tract and esophagus. Given the high prevalence of obesity worldwide and a sex-biased mortality rate, we discuss the implications and relevance of our results for COVID-19.
Abbreviations: ACE2, angiotensin converting enzyme 2; COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TMPRSS2, transmembrane serine protease 2
1. Introduction
Since the worldwide spread of COVID-19 caused by SARS-CoV-2 [[1], [2], [3], [4]], epidemiological studies have highlighted obesity as a major risk factor for development of COVID-19, disease severity, and death [[5], [6], [7], [8], [9], [10]]. This association is not merely due to the high prevalence of obesity among world populations. A meta-analysis of 75 published studies from more than 10 countries in Asia, Europe, and North and South America confirmed a link between obesity and COVID-19 [9]. However, how obesity influences and contributes to the pathophysiology of COVID-19 is hotly debated—multiple mechanisms have been suggested, ranging from obesity-linked alteration in lung function, vascular health, systemic low-grade inflammation, altered immune response, and viral-bacterial interactions [[11], [12], [13], [14], [15], [16], [17]].
SARS-CoV-2 uses the host cell transmembrane carboxypeptidase ACE2 as a receptor to enter cells [[18], [19], [20]]. This process relies on the type II transmembrane serine protease TMPRSS2 to cleave and prime the viral spike protein for cell entry [18]. Expression of both ACE2 and TMPRSS2 in target cells is thought to be important for viral infection. Data from single-cell RNA sequencing reveal expression of ACE2 and TMPRSS2 in multiple cell types within the heart, lung, trachea, esophagus, kidney, gastrointestinal tract, and liver [[21], [22], [23], [24]]. Since SARS-CoV-2 is a respiratory virus that causes significant lung pathology [25] and is spread through person-to-person contact [26] or aerosolized nasal droplets [27,28], viral entry through the respiratory tract is thought to be the major route of infection.
In the present study, we hypothesized that obesity alters expression of ACE2 and TMPRSS2 in the lower respiratory tract and that these changes could potentially contribute to greater risk of developing COVID-19. To test this, we evaluated expression of Ace2 and Tmprss2 in the trachea, lung, and esophagus of chow-fed lean mice and obese mice of similar age chronically fed a high-fat diet (HFD). We observed both diet- and sex-dependent modulation of Ace2 and Tmprss2 expression in the lower respiratory tract (trachea and lung) and esophagus. Given the high prevalence of obesity worldwide [29] and sex biases in COVID-19–related death [30], this study has relevance and implications for COVID-19.
2. Materials and methods
2.1. Mice
C57BL/6J male and female mice were housed in polycarbonate cages on a 12-h light-dark photocycle with ad libitum access to water and food, with no more than five adult mice per cage. Mice were fed either standard laboratory chow (Tekland 2018SX, Envigo, Indianapolis, IN) or a HFD (60% kcal derived from fat; D12492, Research Diets, New Brunswick, NJ) beginning at 6 weeks of age. At termination of the study, food was removed for 2 h before euthanasia. All animal protocols were approved by the Institutional Animal Care and Use Committee of The Johns Hopkins University School of Medicine (Protocol # MO16M431).
2.2. Body composition analysis
Body composition analyses for total fat and lean mass were determined using a quantitative magnetic resonance instrument (Echo-MRI-100, Echo Medical Systems, Waco, TX) in the Mouse Phenotyping Core facility at Johns Hopkins University School of Medicine.
2.3. Tissue collection
Whole lung, trachea, and esophagus were immediately harvested from euthanized mice, flash-frozen in liquid nitrogen, and stored at −80 °C until analysis.
2.4. Quantitative real-time PCR
Total RNA was isolated from lung, trachea, and esophagus using Trizol reagent (Life Technologies, Frederick, MD), reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA), and subjected to quantitative real-time PCR analyses using SsOAdvanced Universal SYBR Green Supermix (Bio-Rad) on a CFX Connect Real-Time System (Bio-Rad). A total of 50 ng (lung samples) or 100 ng (trachea and esophagus samples) of reverse-transcribed cDNA were used in each real-time PCR reaction. Data were normalized to the average of two house-keeping genes (β-actin and cyclophilin A) and expressed as relative mRNA levels using the ΔCt method [31]. Real-time PCR primers used in this study were: Ace2 forward, 5′- TCCAGACTCCGATCATCAAGC-3′ and reverse, 5′-GCTCATGGTGTTCAGAATTGTGT-3’; Tmprss2 forward, 5′-CA GTCTGAGCACATCTGTCCT-3′ and reverse, 5′-CTCGGAGCATACTGAGGCA-3’; β-actin forward, 5′-AGTGTGACGTTGACATCCGTA-3′ and reverse, 5′-GCCAGAGCAGTAATCTCCTTCT-3’; cyclophilin A forward, 5′-GAGCTGTTTGCAGACAAAGTTC-3′ and reverse, 5′-CCC TGGCACATGAATCCTGG-3’.
2.5. Statistical analysis
All data are presented as mean ± SEM. Single-variable comparisons between two groups of data were performed using two-tailed Student’s t-test with 95% confidence intervals (i.e. male chow vs male HFD, in which gene expression is measured and diet is the variable; and male chow vs female chow, in which gene expression is measured and sex is the variable). Comparisons were not made across two variables, for example, male chow vs female HFD. Prism 8 (GraphPad Software, La Jolla, CA, USA) was used for statistical analyses, and differences were considered to be statistically significant at P < 0.05.
3. Results
Age, body weight, total fat and lean mass, and lung weight of male and female mice fed control chow or a HFD are indicated in Table 1 . As expected, five months of chronic high-fat feeding resulted in significant weight gains and fat mass accrual in both male and female mice relative to control chow-fed mice.
Table 1.
Age, sex, weight, total fat and lean mass, and lung weight of lean and obese mice.
| Male | P | Female | P | |||
|---|---|---|---|---|---|---|
| N | 9 | 8 | 10 | 10 | ||
| Diet | Chow | High-fat diet | Chow | High-fat diet | ||
| Age (months) | 6 | 6.5 | 6 | 6.5 | ||
| Weight (g), mean ± SEM | 29.0 ± 0.45 | 45.2 ± 1.49 | <0.0001 | 21.9 ± 0.76 | 36.5 ± 1.50 | <0.0001 |
| Total fat mass (g), mean ± SEM | 4.80 ± 0.46 | 18.6 ± 0.94 | <0.0001 | 2.60 ± 0.23 | 12.6 ± 0.98 | <0.0001 |
| Total lean mass (g), mean ± SEM | 21.1 ± 0.41 | 20.8 ± 0.37 | 0.600 | 17.2 ± 0.44 | 17.1 ± 0.34 | 0.907 |
| Lung weight (g), mean ± SEM | 0.148 ± 0.002 | 0.159 ± 0.004 | 0.017 | 0.140 ± 0.005 | 0.156 ± 0.003 | 0.008 |
3.1. Obesity alters expression of Ace2 and Tmprss2 in lung, trachea, and esophagus
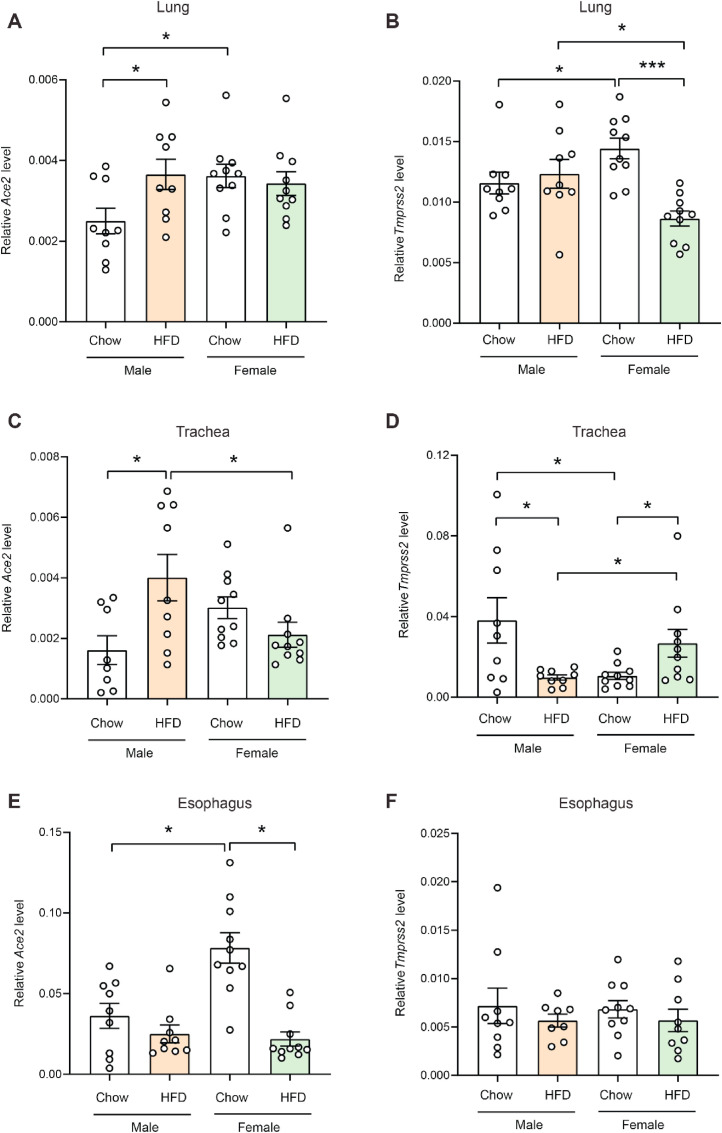
Quantitative real-time PCR showed that in obese male mice fed an HFD, expression of Ace2 was significantly elevated in the lung and trachea relative to chow-fed lean male mice (Fig. 1 A,C). In esophagus, Ace2 expression did not significantly differ between obese and lean male mice (Fig. 1E). In obese female mice, however, expression of Ace2 in the esophagus was significantly reduced relative to lean female animals (Fig. 1E). In contrast to Ace2 expression, Tmprss2 expression was significantly reduced in the trachea of obese male mice relative to lean controls (Fig. 1D). In obese female mice, Tmprss2 expression was significantly reduced in the lung but elevated in the trachea compared to female lean controls (Fig. 1B,D).
Fig. 1.
Expression of Ace2 and Tmprss2 in lean and obese mice. Real-time PCR analysis of Ace2 and Tmprss2 transcript levels in lung (A, B), trachea (C, D), and esophagus (E, F) of lean male (n = 9), obese male (n = 8), lean female (n = 10), and obese female (n = 10) mice. All expression data were normalized to the average of two house-keeping genes (β-actin and cyclophilin A). All data are expressed as mean ± SEM. ∗P < 0.05; ∗∗∗P < 0.001.
3.2. Sex differences in Ace2 and Tmprss2 expression in chow-fed lean mice
In chow-fed lean mice, females had significantly higher expression of Ace2 in the lung and esophagus than males (Fig. 1A,E). Although expression of Ace2 in trachea also trended higher in lean females compared to lean males, the differences were not significant (Fig. 1C). Interestingly, chow-fed female mice had significantly higher Tmprss2 expression in the lung but significantly lower expression in the trachea compared to chow-fed male mice (Fig. 1B,D). Unlike in the lower respiratory tract, no sex differences were noted in expression of Tmprss2 in the esophagus of chow-fed animals (Fig. 1F).
3.3. Sex differences in Ace2 and Tmprss2 expression in HFD-fed obese mice
When comparing obese male and female mice, males had significantly higher expression of Ace2 in the trachea and Tmprss2 in the lung than females (Fig. 1B and C). Obese female mice, however, had significantly higher expression of Tmprss2 in the trachea relative to obese male mice (Fig. 1D).
4. Discussion
Our data indicate sexually dimorphic expression patterns of Ace2 and Tmprss2 in both lean and diet-induced obese mice. Although it is not known at present whether expression of ACE2 and TMPRSS2 in the lung, trachea, and esophagus of humans also exhibit similar patterns of sexual dimorphism, our data have relevance and implications for COVID-19.
Initial viral loads are thought to impact the severity of COVID-19. Indeed, patients with more severe COVID-19 have higher viral loads in the respiratory tract (throat, bronchoalveolar lavage fluid, or sputum) and longer viral persistence than those who experience milder disease [[32], [33], [34]]. Not surprisingly, expression levels of the viral host receptor ACE2 and cell entry-associated molecules (e.g., TMPRSS2) are thought to be important and relevant factors influencing viral loads and infection. Although elevated expression of ACE2 in the upper airway (nasal and olfactory epithelium) may be linked to anosmia in patients with COVID-19 [35,36], its expression in the lung appears to plays a crucial role in SARS-CoV-induced lung injury in animal models [37].
Of the comorbidities associated with COVID-19, obesity consistently ranks among the highest risk factors influencing COVID-19 diagnosis (>46% higher), hospitalization (113% higher), ICU admission (74% higher), and mortality (48% higher) based on a meta-analysis of 109,367 individuals from 75 published studies from more than 10 countries [9]. Obesity also appears to shift COVID-19 severity to younger ages [8]. Epidemiological data further suggest that men have ∼2.5% greater mortality due to COVID-19 than women [30].
How do sex and obesity influence the pathobiology of COVID-19? Many host factors, both genetic and behavioral, are likely at play, with different degrees of importance and contribution to overall disease severity and viral persistence [[11], [12], [13], [14], [15], [16], [17]]. The present study sought to address one basic question—does obesity alter expression of Ace2 and Tmprss2 in the lower respiratory tract and esophagus of male and female mice? Our data show that obese male mice indeed had significantly higher Ace2 expression in the lung than lean controls. Although obese female mice had comparable Ace2 levels in the lung as female lean controls or obese male mice, Tmprss2 transcript levels in the lung were significantly reduced relative to lean controls and obese male mice.
The trachea connects the upper airway (nose and throat) to the lung and thus represents an additional high virus-to-tissue contact zone. In this organ, Ace2 expression was also significantly higher in obese male mice relative to lean male controls or to obese female mice. Together, these observations may potentially account, at least in part, for the association of obesity with SARS-CoV-2 infection and COVID-19 severity, as well as the male-biased mortality rate. Interestingly, expression of Tmprss2 in trachea was significantly lower in obese male mice relative to lean male controls and obese female mice. It has been suggested that other cellular proteases (e.g., furin and cathepsin B/L) may potentially be used by SARS-CoV-2 to enter Tmprss2-negative cells [18,38]. Thus, elevated Ace2 viral receptor expression in the trachea of obese male mice, despite lower expression of Tmprss2, may still favor viral entry if other cellular proteases can substitute for Tmprss2.
Some limitations of the present study are noted, however. (1) Only Ace2 and Tmprss2 transcript levels were assessed, and we presumed that ACE2 and TMPRSS2 protein abundance correlated with their mRNA levels. (2) Quantifications of Ace2 and Tmprss2 transcript levels were performed on bulk RNA isolated from whole tissue; as such, we could not ascertain the cell source responsible for altered expression of Ace2 and Tmprss2 in the lung and trachea of obese mice. However, recent single-cell RNA sequencing analyses identified type II pneumocytes and ciliated cells as the two major cell types in the human lung that express ACE2 and TMPRSS2 [[21], [22], [23], [24]]. (3) Ace2 and Tmprss2 expression levels were only assessed in ∼6-month-old mice, corresponding to young human adults. COVID-19 mortality is significantly higher in people >70 years old [39]. Thus, further assessment of Ace2 and Tmprss2 expression in midlife (∼1-year-old) and geriatric (∼1.5-year-old) mice is warranted.
In summary, our study provides valuable insights into dynamic expression of the SARS-CoV-2 cell entry receptor and an important associated protease in the context of obesity and sex. This information will help inform our ongoing understanding of COVID-19 pathophysiology.
Author contributions
DCS and GWW contributed to the experimental design, performed the experiments, and analyzed and interpreted the data; GWW wrote the manuscript.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Please note that all Biochemical and Biophysical Research Communications authors are required to report the following potential conflicts of interest with each submission. If applicable to your manuscript, please provide the necessary declaration in the box above.
All third-party financial support for the work in the submitted manuscript.
All financial relationships with any entities that could be viewed as relevant to the general area of the submitted manuscript.
All sources of revenue with relevance to the submitted work who made payments to you, or to your institution on your behalf, in the 36 months prior to submission.
Any other interactions with the sponsor of outside of the submitted work should also be reported. (5) Any relevant patents or copyrights (planned, pending, or issued).
Any other relationships or affiliations that may be perceived by readers to have influenced, or give the appearance of potentially influencing, what you wrote in the submitted work. As a general guideline, it is usually better to disclose a relationship than not.
Acknowledgements
This work was partly supported by grants from the National Institutes of Health (DK084171 to GWW).
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caussy C., Pattou F., Wallet F., Simon C., Chalopin S., Telliam C., Mathieu D., Subtil F., Frobert E., Alligier M., Delaunay D., Vanhems P., Laville M., Jourdain M., Disse E., Consortium C.O.H., Lille C.-O.S.G. Prevalence of obesity among adult inpatients with COVID-19 in France. Lancet Diabetes Endocrinol. 2020;8:562–564. doi: 10.1016/S2213-8587(20)30160-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caussy C., Wallet F., Laville M., Disse E. Obesity is associated with severe forms of COVID-19. Obesity. 2020;28:1175. doi: 10.1002/oby.22842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dugail I., Amri E.Z., Vitale N. Biochimie; 2020. High Prevalence for Obesity in Severe COVID-19: Possible Links and Perspectives towards Patient Stratification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kass D.A., Duggal P., Cingolani O. Obesity could shift severe COVID-19 disease to younger ages. Lancet. 2020;395:1544–1545. doi: 10.1016/S0140-6736(20)31024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popkin B.M., Du S., Green W.D., Beck M.A., Algaith T., Herbst C.H., Alsukait R.F., Alluhidan M., Alazemi N., Shekar M. Individuals with obesity and COVID-19: a global perspective on the epidemiology and biological relationships. Obes. Rev. 2020;21(11) doi: 10.1111/obr.13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalligeros M., Shehadeh F., Mylona E.K., Benitez G., Beckwith C.G., Chan P.A., Mylonakis E. Association of obesity with disease severity among patients with coronavirus disease 2019. Obesity. 2020;28:1200–1204. doi: 10.1002/oby.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bansal R., Gubbi S., Muniyappa R. Metabolic syndrome and COVID 19: endocrine-immune-vascular interactions shapes clinical course. Endocrinology. 2020;161 doi: 10.1210/endocr/bqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mauvais-Jarvis F. Aging, male sex, obesity, and metabolic inflammation create the perfect storm for COVID-19. Diabetes. 2020;69:1857–1863. doi: 10.2337/dbi19-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchis-Gomar F., Lavie C.J., Mehra M.R., Henry B.M., Lippi G. Obesity and outcomes in COVID-19: when an epidemic and pandemic collide. Mayo Clin. Proc. 2020;95:1445–1453. doi: 10.1016/j.mayocp.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefan N., Birkenfeld A.L., Schulze M.B., Ludwig D.S. Obesity and impaired metabolic health in patients with COVID-19. Nat. Rev. Endocrinol. 2020;16:341–342. doi: 10.1038/s41574-020-0364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaughan C.J., Cronin H., Ryan P.M., Caplice N.M. Thromb Haemost; 2020. Obesity and COVID-19: A Virchow’s Triad for the 21st Century. [DOI] [PubMed] [Google Scholar]
- 16.Bornstein S.R., Dalan R., Hopkins D., Mingrone G., Boehm B.O. Endocrine and metabolic link to coronavirus infection. Nat. Rev. Endocrinol. 2020;16:297–298. doi: 10.1038/s41574-020-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruglikov I.L., Shah M., Scherer P.E. Obesity and diabetes as comorbidities for COVID-19: underlying mechanisms and the role of viral-bacterial interactions. Elife. 2020;9 doi: 10.7554/eLife.61330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sungnak W., Huang N., Becavin C., Berg M., Queen R., Litvinukova M., Talavera-Lopez C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L., Network H.C.A.L.B. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., 2nd, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.E., Barbry P., Leslie A., Kiem H.P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J., H.C.A.L.B.N.E.a. lung-network@humancellatlasorg. H.C.A.L.B. Network SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035 e1019. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meselson M. Droplets and aerosols in the transmission of SARS-CoV-2. N. Engl. J. Med. 2020;382:2063. doi: 10.1056/NEJMc2009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collaboration N.C.D.R.F. Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19.2 million participants. Lancet. 2016;387:1377–1396. doi: 10.1016/S0140-6736(16)30054-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chanana N., Palmo T., Sharma K., Kumar R., Graham B.B., Pasha Q. Sex-derived attributes contributing to SARS-CoV-2 mortality. Am. J. Physiol. Endocrinol. Metab. 2020;319:E562–E567. doi: 10.1152/ajpendo.00295.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., Wang Z., Li J., Li J., Feng C., Zhang Z., Wang L., Peng L., Chen L., Qin Y., Zhao D., Tan S., Yin L., Xu J., Zhou C., Jiang C., Liu L. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Yan L.M., Wan L., Xiang T.X., Le A., Liu J.M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M., Shen W., Rowan N.R., Kulaga H., Hillel A., Ramanathan M., Jr., Lane A.P. Eur Respir J; 2020. Elevated ACE2 expression in the olfactory neuroepithelium: implications for anosmia and upper respiratory SARS-CoV-2 entry and replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bilinska K., Jakubowska P., Von Bartheld C.S., Butowt R. Expression of the SARS-CoV-2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem. Neurosci. 2020;11:1555–1562. doi: 10.1021/acschemneuro.0c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldstein J.R., Lee R.D. Demographic perspectives on the mortality of COVID-19 and other epidemics. Proc. Natl. Acad. Sci. U. S. A. 2020;117:22035–22041. doi: 10.1073/pnas.2006392117. [DOI] [PMC free article] [PubMed] [Google Scholar]