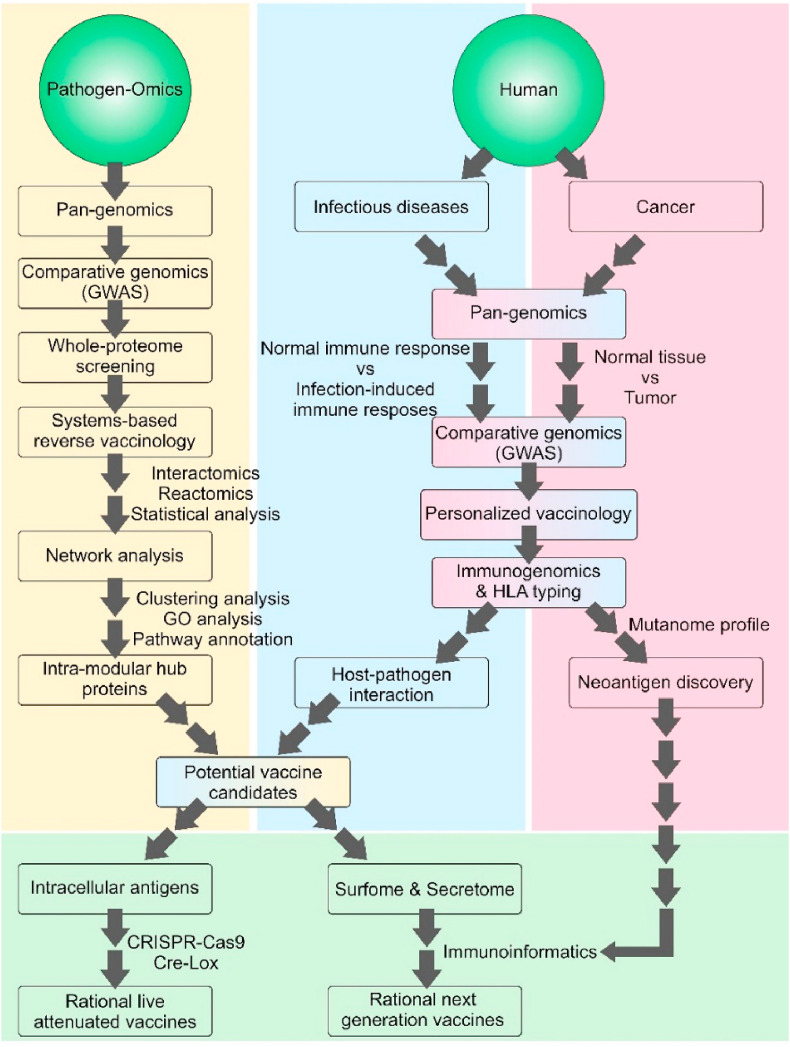
During the last 30 years, alongside the increasing progress in the genome sequencing technologies and development of biomedical computing sciences (e.g., bioinformatics and systems biology), several vaccinology-related software, computational/statistical methods, and databases have been established to analyze, predict, and annotate different features in vaccinology [1]. Data obtained from the whole-genome or pan-genome sequencing analyses provide in-depth information about the target pathogen's antigenic repertoire, in which high-throughput data analysis results in the identification of candidate antigens (Ag). Now, the dreadful experience of the COVID-19 pandemic emphasizes the urgent need of mankind to revisit the scientific approaches towards fast and cost-effective development of vaccines and immunomediciens with high precision and effectiveness such as self-amplifying mRNA, and DNA vaccines (ClinicalTrials.gov IDs: NCT04283461 and NCT04336410, respectively) [2,3]. The vaccine production against SARS-CoV-2 began soon after the first full-genome sequence of the virus (NCBI accession no. NC_045512.2) and its subsequent bioinformatics analysis [4]. Of note, the evolutionary process in microorganisms over time is one of their main immune escape mechanisms. Therefore, full genome profiling and subsequent high-throughput omics data analyses appear to be paramount in the vaccinology against the emerging and re-emerging infections (EREIs) and rapid mutation developing pathogens especially RNA viruses (e.g., different types of influenza virus) [5][6]. Moreover, the gene expression profiling technologies (e.g., RNA-sequencing and microarray) provide key insights into the differentially expressed protein-encoding genes, which can be analyzed through the regulatory and protein-protein interaction (PPI) networks, systems-based gene annotation, and gene ontology enrichment analysis (e.g., biological pathways, molecular functions, and cellular components) [7,8]. The genomic and transcriptomic patterns can be followed by the interactomics inferences as an integrated gene regulatory and/or PPI network analysis, which can be used in precision vaccinology. In precision vaccinology, both the individual's immunogenetic and immunogenomics profile and the vaccine-induced protective immunity can be predicted through the systems biology-based meta-analysis [9]. Currently, it is known that immune cell networks are controlled by many different high polymorphic immune system-related genes. Further, many entities such as butyrophilin (e.g., BTN1A/2A/3A), antiviral effectors, major histocompatibility complex (MHC) proteins such as class I polypeptide-related sequence A, killer cell immunoglobulin-like receptors, toll-like receptors (e.g., TLR1,2,3,4,6,7,8, and 9), and their related signaling molecules, as well as human leukocyte antigen (HLA) are the key players in the vaccine-induced immune responses. Multiple vaccine-induced immune responses might be mainly due to the inter-individual genetic variations that are measurable by the meta-analysis methods like genome-wide association study [9]. These bioinformatics approaches can be used in the personalized screening strategies to identify (i) the personal mutations in the immune-related genes, (ii) single nucleotide polymorphisms especially during the HLA class I and II loci typing, and (iii) person-specific resistance/susceptibility to diseases. All these approaches are critical in terms of the prediction of the mechanisms underlying the heterogeneity of human immune responses to the vaccination. In the case of cancer vaccinology, through the pan-genomic analysis, a detailed cancer mutanome profile can be achieved. Such data, as an irreplaceable comparative tumor-normal map, are used to discover the whole-genome or whole-exome in a patient- and tumor-specific approaches [10,11]. The raw pan-genomic data can be analyzed using comparative statistical and bioinformatics methods to map the possible tumor-relevant single nucleotide variations, and even insertion/deletion and translocations. Next, the identified cancer mutanome profile can be interpreted by different databases (e.g., COSMIC, IntOGen, TARGET, TCGA, etc.) to massively annotate the genomic and molecular features of the target cancer [12]. Following such analyses, possible explicable neoantigens (or neoepitopes) are explored. However, to predict the immunogenic behavior of the identified neoantigens, the exome- or whole-genome sequencing results should be paired with the cancer proteomics data via predictor algorithms. Likewise, patient-specific HLA typing and their capability in eliciting the T-cell responses is a key step to predict which neoepitopes are successfully presented by the MHC molecules. Besides, omics data enable vaccinologists to depict a high-resolution blueprint of the target pathogen. Such data can be used to identify (i) targets for the vaccine design and gene-editing strategies, (ii) molecular mechanisms of the host-pathogen interactions, and (iii) functional modules involved in the pathogen's survival and invasion, infection-associated proteome changes, gene regulatory pathways, co-expressed gene modules, functional hub protein(s), and so forth [13]. The network-based whole proteome analysis of an infectious agent provides two groups of intra-modular hub (iMhub) proteins, including (i) surfome/secretome/immunome that offer the potential vaccine Ags for the gene- or peptide-based recombinant vaccine development, and (ii) intracellular iMhubs with minimal off-target potential that can be used for the pathogen's genome editing. In the latter approach, technologies such as CRISPR-Cas9 and Cre-Lox systems can be used to produce “rational live-attenuated vaccines” by executing the targeted genetic change(s) in one or more identified iMhub. The target antigen should have (i) a critical role in the pathogen virulence (not in the cell division system), (ii) a differential expression pattern during the invasion, and (iii) no homologous with the host's body. As a future direction, the next generation vaccines can be associated with three main interlinked approaches, including (i) pan-omics systems-based data analysis for developing personalized therapeutic or prophylactic vaccines, (ii) manipulation of the target pathogen's genome using the gene-editing technologies to produce non-invasive "live attenuated-like vaccines", and (iii) rapid developing and high-efficient vaccine platforms (e.g., DNA and mRNA vaccines) for combat against multidrug-resistant superbugs, healthcare-associated germs and infections, and EREIs. As illustrated in Fig. 1 , the vaccine design approach needs to be revisited and the related computational/in-silico technologies should be taken into account towards the development of next-generation vaccines.
Fig. 1.
Schematic illustration of next-generation vaccines from pan-genomics to personalized vaccinology. GWAS: Genome-wide association study. HLA: Human leukocyte antigen. GO: Gene ontology.
References
- 1.Diehl A.D., Lee J.A., Scheuermann R.H., Blake J.A. Ontology development for biological systems: immunology. Bioinformatics. 2007;23:913–915. doi: 10.1093/bioinformatics/btm029. [DOI] [PubMed] [Google Scholar]
- 2.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N. An mRNA vaccine against SARS-CoV-2 - preliminary report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiyotani K., Toyoshima Y., Nemoto K., Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J Hum Genet. 2020;65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parvizpour S., Pourseif M.M., Razmara J., Rafi M.A., Omidi Y. Epitope-based vaccine design: a comprehensive overview of bioinformatics approaches. Drug Discov Today. 2020;25(6):1034–1042. doi: 10.1016/j.drudis.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Frost S.D.W., Magalis B.R., Kosakovsky Pond S.L. Neutral theory and rapidly evolving viral pathogens. Mol Biol Evol. 2018;35:1348–1354. doi: 10.1093/molbev/msy088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahbar Saadat Y., Pourseif M.M., Zununi Vahed S., Barzegari A., Omidi Y., Barar J. Modulatory role of vaginal-isolated lactococcus lactis on the expression of miR-21, miR-200b, and TLR-4 in CAOV-4 cells and in silico revalidation. Probiotics Antimicrob Proteins. 2020;12(3):1083–1096. doi: 10.1007/s12602-019-09596-9. [DOI] [PubMed] [Google Scholar]
- 8.Rogers L.R.K., de Los Campos G., Mias G.I. Microarray gene expression dataset Re-analysis reveals variability in influenza infection and vaccination. Front Immunol. 2019;10:2616. doi: 10.3389/fimmu.2019.02616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poland G.A., Ovsyannikova I.G., Kennedy R.B. Personalized vaccinology: a review. Vaccine. 2018;36:5350–5357. doi: 10.1016/j.vaccine.2017.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moradi A., Pourseif M.M., Jafari B., Parvizpour S., Omidi Y. Nanobody-based therapeutics against colorectal cancer: precision therapies based on the personal mutanome profile and tumor neoantigens. Pharmacol Res. 2020;156:104790. doi: 10.1016/j.phrs.2020.104790. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X., Sharma P.K., Peter Goedegebuure S., Gillanders W.E. Personalized cancer vaccines: targeting the cancer mutanome. Vaccine. 2017;35:1094–1100. doi: 10.1016/j.vaccine.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagan J., Van Allen E.M. Next-generation sequencing to guide cancer therapy. Genome Med. 2015;7:80. doi: 10.1186/s13073-015-0203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q., Ma C., Gearing M., Wang P.G., Chin L.S., Li L. Integrated proteomics and network analysis identifies protein hubs and network alterations in Alzheimer's disease. Acta Neuropathol Commun. 2018;6:19. doi: 10.1186/s40478-018-0524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]