Abstract
Mutations in the molecular co-chaperone Bcl2-associated athanogene 3 (BAG3) are found to cause dilated cardiomyopathy (DCM), resulting in systolic dysfunction and heart failure, as well as myofibrillar myopathy (MFM), which is characterized by protein aggregation and myofibrillar disintegration in skeletal muscle cells. Here, we generated a CRISPR/Cas9-induced Bag3 knockout zebrafish line and found the complete preservation of heart and skeletal muscle structure and function during embryonic development, in contrast to morpholino-mediated knockdown of Bag3. Intriguingly, genetic compensation, a process of transcriptional adaptation which acts independent of protein feedback loops, was found to prevent heart and skeletal muscle damage in our Bag3 knockout model. Proteomic profiling and quantitative real-time PCR analyses identified Bag2, another member of the Bag protein family, significantly upregulated on a transcript and protein level in bag3-/- mutants. This implied that the decay of bag3 mutant mRNA in homozygous bag3-/- embryos caused the transcriptional upregulation of bag2 expression. We further demonstrated that morpholino-mediated knockdown of Bag2 in bag3-/- embryos evoked severe functional and structural heart and skeletal muscle defects, which are similar to Bag3 morphants. However, Bag2 knockdown in bag3+/+ or bag3+/- embryos did not result in (cardio-)myopathy. Finally, we found that inhibition of the nonsense-mediated mRNA decay (NMD) machinery by knockdown of upf1, an essential NMD factor, caused severe heart and skeletal muscle defects in bag3-/- mutants due to the blockade of transcriptional adaptation of bag2 expression. Our findings provide evidence that genetic compensation might vitally influence the penetrance of disease-causing bag3 mutations in vivo.
Author summary
One form of genetic compensation is described as transcriptional adaptation of gene expression triggered by deleterious gene mutations. Although the precise molecular mechanism that induces genetic compensation needs to be defined, it represents a powerful biological phenomenon that warrants genetic robustness. We find that antisense-mediated knockdown of Bag3 in zebrafish embryos causes heart failure and myopathy. By contrast, CRISPR/Cas9-induced depletion of Bag3 does not result in the abrogation of heart and skeletal muscle function in zebrafish embryos. We find here that transcriptional activation of the Bag family member bag2 is capable of restoring heart and skeletal muscle function in bag3 mutant embryos, whereas this compensatory mechanism is not present in the bag3 morphants. Furthermore, we show that nonsense-mediated decay of bag3 mRNA is the molecular trigger for the compensatory upregulation of bag2. Our study provides evidence that genetic compensation via transcriptional adaptation is a vital modulator of disease peculiarity and penetrance in bag3 mutant zebrafish and that this biological phenomenon might also be active in certain human BAG3 mutation carriers.
Introduction
Compensatory transcriptional adaptation of gene expression in response to malignant and harmful gene mutations is a powerful mechanism to warrant genetic robustness. Interestingly, this genetic compensation was recently demonstrated to be triggered by a decay of the mutated mRNA which results in the transcriptional upregulation of one or more related and compensation-competent genes [1–3]. Recently, Rossi and coworkers found that CRISPR/Cas9-induced mutation of egfl7, an endothelial ECM gene, did not result in obvious defects in zebrafish. Whereas morpholino-modified antisense oligonucleotide (morpholino, MO)-mediated egfl7 depletion led to severe vascular malformations [3]. Interestingly, Emilins, also important ECM genes, were found to be transcriptionally upregulated in egfl7 mutant zebrafish, but not in morphant embryos, suggesting their compensatory potential to buffer the organism to egfl7 loss. Additionally, Sztal et al. recently found genetic compensation in a zebrafish model of Actin deficiency [4]. Genetic actc1b mutant zebrafish embryos only show very mild muscle defects due to transcriptional upregulation and functional compensation by an α-Actin paralogue. Interestingly, morpholino-mediated actc1b depletion did not result in compensatory upregulation of other Actin family members thereby leading to severe myopathy in vivo [4].
B-cell lymphoma 2 (Bcl2)-associated athanogene 3 (BAG3) is a member of the BAG protein family which acts as a molecular co-chaperone by physically interacting with chaperone molecules such as 14-3-3 proteins, Hsp70, and small heat shock proteins (HSPBs) [5]. One of the main biological functions of BAG3 is the regulation of autophagy and the degradation of misfolded proteins to warrant orchestrated protein homeostasis [5, 6]. Mammalian BAG3 is strongly expressed in the heart and skeletal muscle, where it mainly localizes to sarcomeric Z-disks, but is also expressed in the brain and peripheral nervous system [7]. Mutations in the Bag3 gene are described to cause a diverse spectrum of disease phenotypes including striated muscle diseases such as dilated cardiomyopathy (DCM) or myofibrillar myopathy (MFM). Furthermore, decreased BAG3 protein levels were found in failing human hearts [8].
In this study, we uncovered and explored an example of functional genetic compensation in a CRISPR/Cas9-induced zebrafish model of Bag3-deficiency. We and others found that targeted morpholino-mediated knockdown of Bag3 in zebrafish embryos resulted in severe structural and functional heart and skeletal muscle defects [9–11]. Interestingly and in contrast to these studies, we found here that the CRISPR/Cas9-induced deletion of 19 nucleotides in exon 2 of the bag3 gene leading to a frame shift and thereby introducing a premature stop codon did not evoke heart and skeletal muscle defects in homozygous mutant embryos. We determined that the degradation of mutated bag3 mRNA triggered the compensatory transcriptional upregulation of the BAG protein family member bag2, which was able to functionally compensate for the loss of Bag3.
Our findings present another example of genetic compensation in vivo, underlining the importance of this exciting biological mechanism in buffering the malignancy of specific gene mutations and, thereby, mediating genetic robustness.
Results
Targeted knockout of zebrafish bag3 by CRISPR/Cas9 does not provoke heart and skeletal muscle dysfunction
In humans, mutations in BAG3 (Bcl2-associated athanogene 3) were found to cause dilated cardiomyopathy (DCM) and myofibrillar myopathies (MFM) [9, 12]; however, the molecular BAG3-associated underpinnings are poorly understood. To study the in vivo roles of BAG3 in more detail, we generated a Bag3-deficient zebrafish line by using the CRISPR/Cas9 genome-editing technology.
Although teleosts went through whole genome duplication during evolution [13, 14], only one single bag3 ortholog of the human BAG3 gene can be found on zebrafish chromosome 13. Zebrafish Bag3 shows 41% overall amino acid homology to human BAG3, with up to 77% sequence identity in distinct functional domains such as the WW domain (S1 Fig). As in humans, zebrafish bag3 transcripts are enriched in the heart and skeletal muscle during embryogenesis and are more predominantly expressed in the adult cardiac muscle [10, 15].
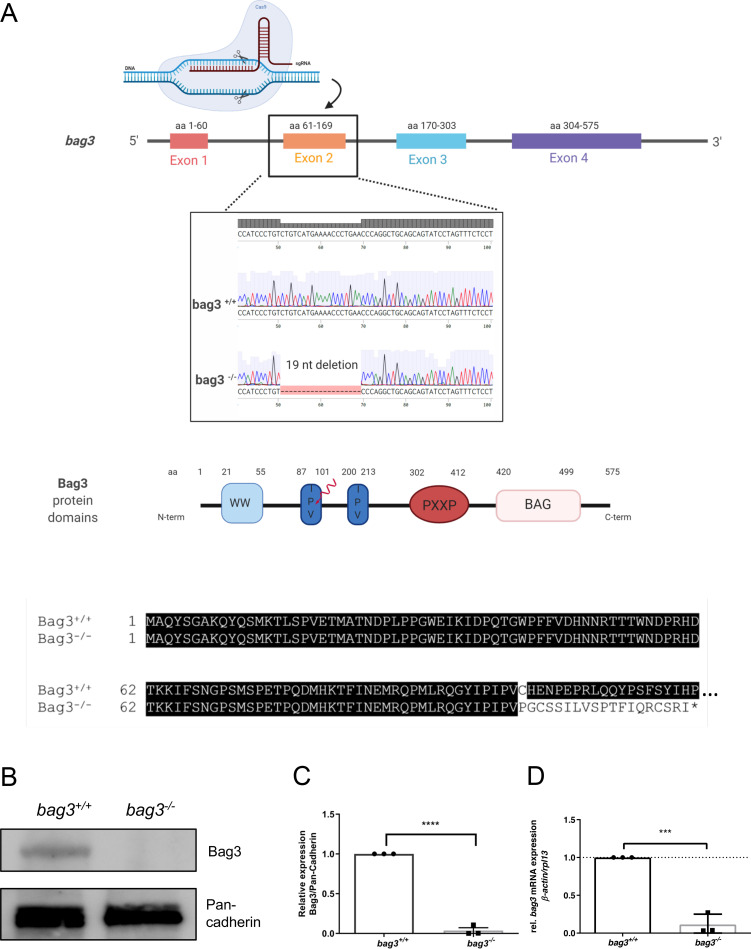
To generate a loss-of-function zebrafish model by introducing CRISPR/Cas9-induced mutations in the bag3 gene, we first designed a CRISPR-RNA (crRNA) targeting exon 2 of the bag3 genomic sequence. By co-injecting synthetic tracrRNA, bag3-specific crRNA, and Cas9 protein, a frameshift mutation in the bag3 gene was generated, leading to an alternative stop codon and premature termination of protein translation. Specifically, a mutant allele with a 19-nucleotide deletion was identified among F1 fish and the mutant line was propagated (Fig 1A). The putative mutation leads to the loss of the crucial Bag3 protein domains IPV and PXXP as well as the BAG domain (Fig 1A). To confirm Bag3 deficiency, we performed immunoblot assays and found the complete loss of wild-type Bag3 proteins in homozygous mutant bag3-/- embryos (N = 3, P<0.0001) (Fig 1B and 1C). We also performed quantitative RT-PCR analysis using homozygous mutant bag3-/- embryos and found bag3 mRNA levels significantly downregulated (N = 3, P<0.0001) (Fig 1D). This suggested that nonsense-mediated mRNA decay (NMD) was triggered by the CRISPR/Cas9-induced gene mutation.
Fig 1. Generation of zebrafish bag3 knockout by CRISPR/Cas9 gene editing.
(A) Structure of the zebrafish bag3 gene and protein. Exon 2 is the target for the CRISPR/Cas9 gene editing in zebrafish bag3. The CRISPR/Cas9-induced mutation (19 bp deletion) in bag3 is shown in bag3 mutant DNA sequencing chromatogram. The 19 nucleotides deletion in bag3-/- leads to a frame shift, the introduction of a premature stop codon and thereby the premature termination of Bag3 translation, as demonstrated by the alignment of the Bag3-/- and Bag3+/+ aminoacid sequences (only partial aminoacid sequence shown) (B-C) Immunoblot analysis of 72 hpf bag3+/+ embryo protein lysates compared to lysates obtained from bag3-/- clutchmates with antibody against zebrafish Bag3. The figure shows one representative immunoblot from three independent experiments (N = 3, mean ± SD, P<0.0001 determined using two-tailed t-test). (D) Quantitative real-time PCR of bag3+/+ and bag3-/- embryos at 72 hpf shows significant downregulation of bag3 mRNA levels in bag3-/- embryos (N = 3, mean ± SD, P = 0.0004 determined using two-tailed t-test).
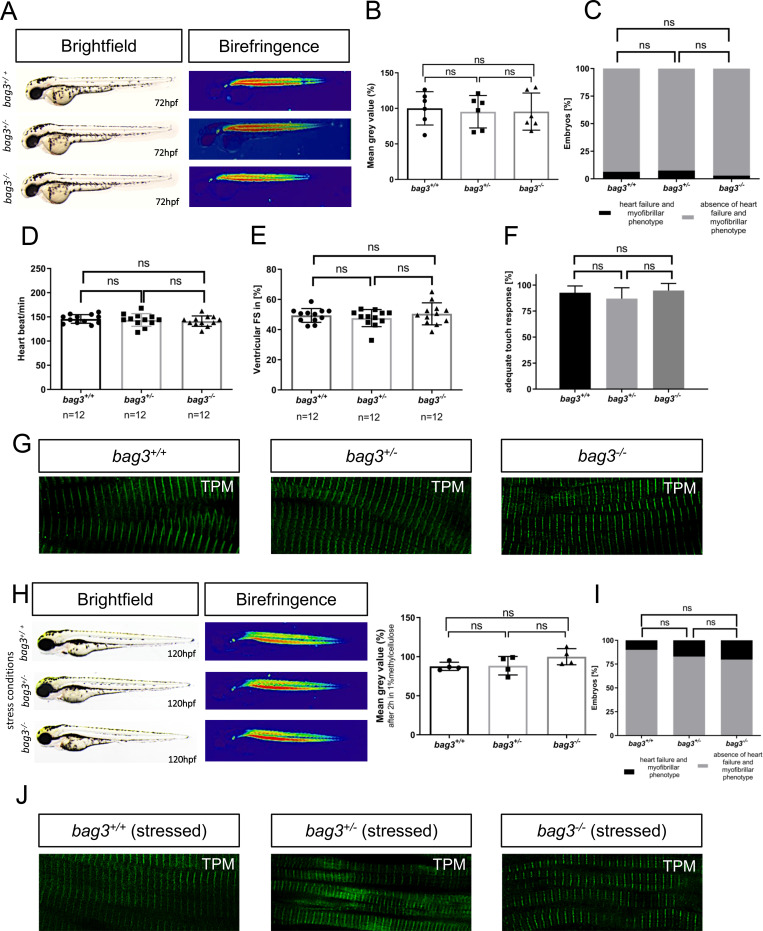
In-crosses of heterozygous carriers yielded offspring demonstrating the regular mendelian genotypic ratio of 25% homozygous wild-type (bag3+/+), 50% heterozygous (bag3+/-) and 25% homozygous mutant (bag3-/-) embryos (N = 3, n = 100) (Fig 2A). Interestingly, none of the zebrafish embryos displayed morphological malformations of the heart or skeletal muscle [16] (Fig 2A–2C). Since the high organization of striated muscle tissue is able to polarize light, the myofibrillar disorganization within muscle tissue can therefore be easily visualized by a reduction in the birefringence signal intensity. In bag3-/- embryos, birefringence signal intensity from the skeletal muscle was similar to the intensity in bag3+/- and bag3+/+, demonstrating preserved muscle structure in Bag3-deficient zebrafish embryos (Fig 2B). Additionally, bag3-/- embryos did not develop functional heart or skeletal muscle defects under normal conditions as demonstrated by the assessment of the heart rate (N = 3, n = 12, P = 0.6026. HR bag3+/+: 146±9.03 heart beat/min; HR bag3+/-: 144±13.12 heart beat/min; HR bag3-/-: 141±10.71 heart beat/min, at 72 hpf), contractile force by fractional shortening (FS) measurements (N = 3, n = 12, P = 0.5061. FS bag3+/+: 49.41±4.57%; FS bag3+/-: 47.64±5.67%; FS bag3-/-: 50.47±7.291%, at 72 hpf), and touch evoked assays (N = 3, n = 90, P = 0.7851. Mot bag3+/+: 92.67±3.48%; Mot bag3+/-: 86.99±4.54%; Mot bag3-/-: 94.7±3.34%, at 72 hpf) (Fig 2D–2F; S1–S8 Movies). Muscle structure in bag3+/- and bag3-/- embryos was indistinguishable from that of bag3+/+ embryos as demonstrated by Tropomyosin immunostainings (Fig 2G). Additionally, transmission electron microscopy (TEM) analyses of heart and skeletal muscle ultrastructure revealed no alterations between wild-type and bag3-/- sarcomeric organization (S3A Fig). We found highly organized myofilaments in bag3-/- mutant embryos with thin and thick filaments in well-aligned bundles and discernible A-, I-, M-bands and Z-disks. Moreover, as shown by immunostainings and confocal imaging, atrial and ventricular cardiomyocytes of bag3-/- mutant embryos express myosin heavy chains (MHC) in a proper cardiac chamber specific manner, suggesting regular heart chamber specification (S3B Fig).
Fig 2. Genetic loss of bag3 does not interfere with heart and skeletal muscle function.
(A) Lateral view of brightfield and birefringence images for bag3+/+, bag3+/- and bag3-/- embryos at 72 hpf exhibiting homogenous birefringence signals. (B) Densitometric analysis of birefringence does not report muscle disorganization in bag3-/- embryos. Representative samples are shown (n = 6, One-way ANOVA followed by tukey's multiple comparison analysis, P = 0.9688). (C) Embryos at 72 hpf reveal no myopathic phenotype (N = 3, n = 100, two-tailed value for Fisher´s exact test. bag3+/+ and bag3+/- P = 0.8096; bag3+/+ and bag3-/- P = 0.4949; bag3+/- and bag3-/- P = 0.1925). (D) Heart rate quantification at 72 hpf reveals no impairments under physiological conditions (N = 3, n = 12, mean ± S.D. One-way ANOVA followed by tukey's multiple comparison analysis, P = 0.6026). (E) Quantification of ventricular FS at 72 hpf does not reveal contractile dysfunction (N = 3, n = 12, mean ± S.D. One-way ANOVA followed by tukey's multiple comparison analysis, P = 0.5061). (F) Quantification of the touch evoked assay. bag3-/- embryos reveal no significant difference in responsiveness upon mechanical stimulus (N = 3, n = 90, mean ± S.D. One-way ANOVA followed by tukey's multiple comparison analysis, P = 0.7851). (G) Immunostaining of bag3+/+, bag3+/- and bag3-/- embryos at 72 hpf, with sarcomeric Tropomyosin, showing no muscle fiber disruptions. (H) Brightfield and birefringence images of mutant embryos at 120 hpf under stressed conditions reveal no myopathic phenotype, which was confirmed by densitometric analysis of birefringence signals. Representative samples are shown (n = 4, One-way ANOVA followed by tukey's multiple comparison analysis, P = 0.1824). (I) Embryos, under stressed conditions, do not develop myopathic phenotype (N = 4, n = 50, two-tailed value for Fisher´s exact test. bag3+/+ and bag3+/- P = 0.6050; bag3+/+ and bag3-/- P = 0.8320; bag3+/- and bag3-/- P = 0.8582). (J) Immunostaining of bag3+/+, bag3+/- and bag3-/- embryos at 120 hpf under stressed conditions (2 hours in 1% methylcellulose), with sarcomeric Tropomyosin, showing no muscle fiber disruptions.
Since Bag3 loss did not result in phenotypic alterations during development under normal physiological conditions, we aimed to mechanically overload the skeletal muscle in Bag3-deficient zebrafish embryos. Mutant bag3-/- embryos were incubated in 1% methylcellulose, leading to increased viscosity of the media, which is known to significantly enhance workload on skeletal muscle [10]. Interestingly, increased muscle workload resulted in no significant elevation of myopathic phenotypes in bag3-/- embryos compared to bag3+/- and bag3+/+ clutchmates (N = 4, n = 50, bag3+/+ and bag3+/- P = 0.6050; bag3+/+ and bag3-/- P = 0.8320; bag3+/- and bag3-/- P = 0.8582). (Fig 2H). Similar to our observations on unstressed bag3 mutant embryos, methylcellulose-incubated bag3+/+, bag3+/-, and bag3-/- embryos did not display structural failure of myofibers following treatment (Fig 2J). This demonstrates that loss of Bag3 in our CRISPR/Cas9-induced zebrafish line does not cause cardiomyopathy and skeletal muscle dysfunction under physiological or stressed conditions. Additionally, we found that homozygous mutant bag3-/- fish can be raised to adulthood and they are fertile. However, we found a significant increase in the mortality rate of bag3-/- zebrafish starting from 9 month of age as depicted in a Kaplan-Meier survival curve (S2 Fig), which is consistent with the recently published findings by Ding et al[17].
Morpholino-mediated ablation of Bag3 specifically results in (cardio)-myopathy
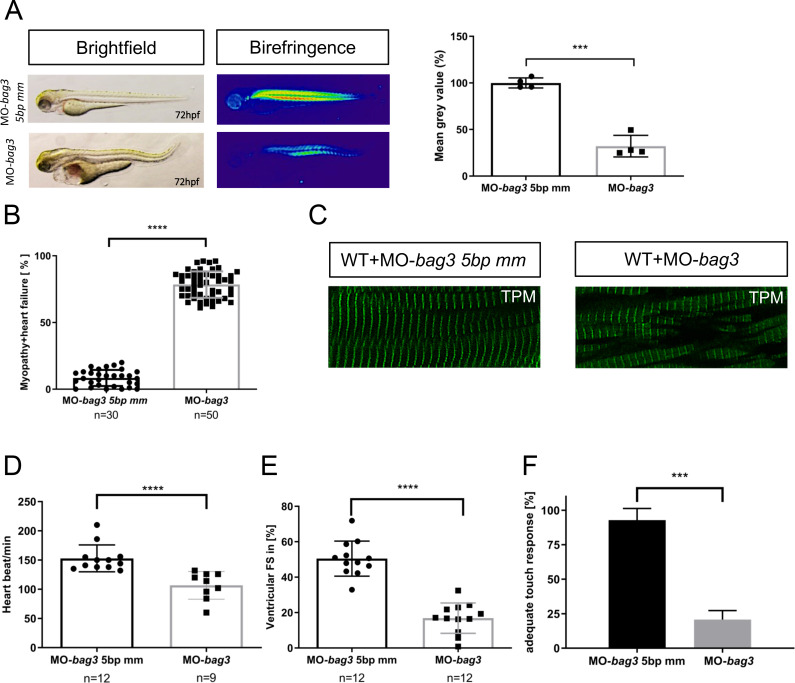
Recently, different morpholino-mediated knockdown studies in zebrafish showed that loss of Bag3 led to severe heart and skeletal muscle defects and (cardio-)myopathy [9, 10, 18]. Similarly, we found severe cardiac and skeletal muscle defects (Fig 3A and 3B) associated with myofibrillar disorganization (Fig 3A–3C), diminished cardiac contractile function (Fig 3D and 3E; S9 Movie), and significantly reduced motility (Fig 3F) in zebrafish embryos injected with a morpholino targeting the splice donor site of exon 2 in bag3 [9]. By contrast, injection of a specific 5bp mismatch control morpholino (MO-bag3 5bp mm) unable to target bag3 did not evoke any cardiac and skeletal muscle defects (Fig 3A–3F).
Fig 3.
Targeted knock-down of Bag3 leads to (cardio-)myopathy in zebrafish (A) Brightfield and birefringence images of MO-bag3 and MO-bag3 5bp mm injected embryos developed (cardio-)myopathy phenotype at 72 hpf. The densitometric analysis of birefringence supports the presence of myopathy phenotype only in bag3 morphants. Representative samples are shown (n = 4, P = 0.0003 determined using two tailed t-test). (B) bag3 splice MO injected embryos develop (cardio-)myopathy phenotype (78.42±9.88%) whereas control-injected embryos developed no pathological phenotype (78.42±9.89%) (N = 3, n = 30/50 mean± SD P<0.0001 determined using two-tailed t-tests). (C) Tropomyosin immunostaining of MO-bag3 and MO-bag3 5bp mm embryos at 72 hpf shows that embryos injected with Bag3 splice MO develop muscle fiber disruptions. (D) Heart rate quantification of bag3 morphants reveals impairments at 72 hpf (N = 3, n = 9/12, P = 0.6026. HR 5bp-mismatch-MO injected embryos: 153±22.98 heart beat/min; HR bag3 morphants: 106±23.58 heart beat/min; mean ± S.D. P<0.0001 determined using two-tailed t-tests). (E) FS of bag3 morphant ventricles at 72 hpf is significantly reduced (16,88±8.56%), compared to MO-bag3 5bp mm injected embryos (FS: 50.48±9.90%) (N = 3, n = 12; Mean± SD P<0.0001 determined using two-tailed t-tests). (F) Touch evoked assay reveals significant reduction in responsiveness upon mechanical stimulus for bag3 morphants (20.78±4.46%) and not for 5bp-mismatch-MO injected embryos (92.78±4.40%) (N = 3, n = 40, mean ± S.D. P = 0.0005, determined using two-tailed t-tests).
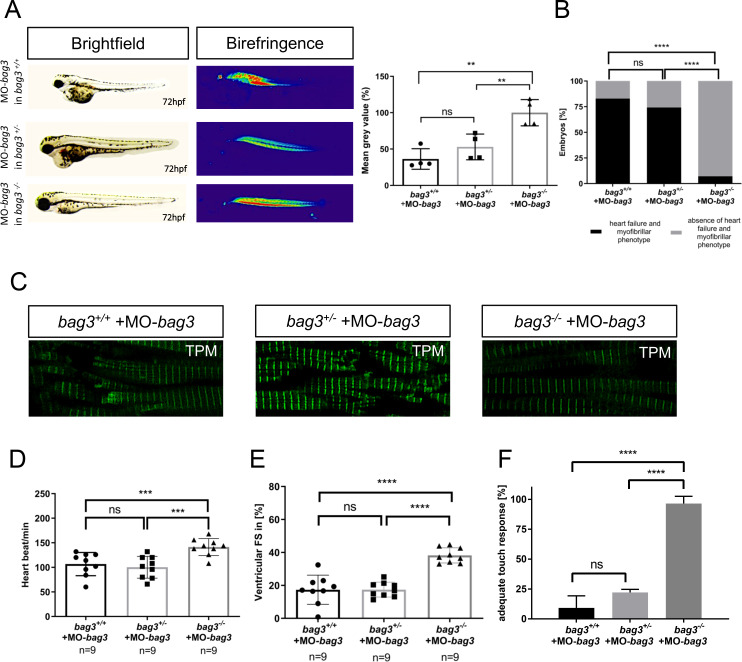
To validate whether these findings were specifically caused by the loss of Bag3 or were due to off-target effects of the morpholino, we injected the same concentration of the bag3-specific morpholino (MO-bag3) into mutant bag3-/- embryos as well as clutchmate controls (bag3+/- and bag3+/+ embryos). Next, we screened these embryos for specific bag3-associated phenotypic alterations. Similar to MO-bag3-injected wild-type embryos (N = 3, n = 140/160 P<0.0001) (Fig 3A), bag3+/- and bag3+/+ embryos injected with MO-bag3 showed severe pericardial edema and muscle fiber disorganization as well as heart and skeletal muscle dysfunction (N = 3, n = 40 P<0.0001) (Fig 4A–4F). By contrast, injection of MO-bag3 into homozygous mutant bag3-/- embryos did not cause cardiomyopathy and skeletal muscle dysfunction (Fig 4A–4F). Specifically, muscle structure (evaluated by Tropomyosin immunostainings) in MO-bag3-injected bag3+/- and bag3+/+ embryos was indistinguishable from wild-type embryos injected with MO-bag3 (Fig 4A and 4C). By contrast, muscle structure in bag3-/- embryos injected with MO-bag3 is well preserved and indistinguishable from wild-type muscle structure (Fig 4A–4C). Moreover, bag3-/- embryos injected with MO-bag3 did not develop functional heart and skeletal muscle defects compared to MO-bag3-injected bag3+/+ and bag3+/- embryos as indicated by the assessment of the heart rate (N = 3, n = 9, P = 0.0008. HR bag3+/+: 106±23.58 heart beat/min; HR bag3+/-: 100 ±22.21 heart beat/min; HR bag3-/-: 141±17.26 heart beat/min, at 72 hpf), contractile force by FS measurements (N = 3, n = 9, P<0.0001. FS bag3+/+: 17.38±8.84%; FS bag3+/-: 17.44±4.54%; FS bag3-/-: 38.23±4.59%, at 72 hpf), and touch evoked assays (N = 3, n = 60, P<0.0001. Mot bag3+/+: 9.23±5.69%; Mot bag3+/-: 22.14±2.25%; Mot bag3-/-: 96.49±3.18%, at 72 hpf) (Fig 4D–4F; S2, S4, S9 and S10 Movies).
Fig 4.
Knockdown of Bag3 leads to heart and skeletal muscle dysfunctions only in bag+/+ and bag3+/- embryos. (A) Brightfield and birefringence images of bag3+/+, bag3+/- and bag3-/- embryos at 72 hpf injected with MO-bag3. Densitometric analysis of birefringence (n = 4). Representative samples are shown (One-way ANOVA followed by tukey's multiple comparison analysis, P = 0.0012). (B) Only bag3-/- embryos injected with MO-bag3 do not show (cardio-)myopathy, whereas bag3+/+and bag3+/- develop the characteristic bag3 morphant phenotype (N = 3, n = 40, mean ± S.D., P<0.0001, two-tailed value for Fisher´s exact test. bag3+/+ and bag3+/- P = 0.5346; bag3+/+ and bag3-/- P<0.0001; bag3+/- and bag3-/- P<0.0001). (C) Tropomyosin immunostainings of bag3+/+, bag3+/-, and bag3-/- embryos injected with MO-bag3 at 72 hpf reveal that only bag3-/- mutant embryos injected with MO-bag3 does not develop muscle fiber disruptions. (D) Heart rate quantification at 72 hpf reveals impairments only in bag3+/+and bag3+/- embryos injected with MO-bag3 (N = 3, n = 9, mean ± S.D. One-way ANOVA followed by tukey's multiple comparison analysis, P = 0.0008). (E) Quantification of ventricular FS at 72 hpf reveals contractile dysfunctions only in bag3+/+and bag3+/- embryos injected with MO-bag3 (N = 3, n = 9, mean ± S.D. One-way ANOVA followed by tukey's multiple comparison analysis, P<0.0001). (F) bag3-/- embryos injected with MO-bag3 show proper flight response upon mechanical stimulus, whereas bag3+/+and bag3+/- embryos injected with MO-bag3 do not show an adequate touch response (N = 3, n = 60, mean ± S.D. One-way ANOVA followed by tukey's multiple comparison analysis, P<0.0001).
These findings clearly demonstrate the specificity of the morpholino-mediated Bag3 knockdown and indicate that CRISPR/Cas9-induced Bag3 ablation might trigger genetic compensation to prevent (cardio-)myopathy.
Bag2 mediated genetic compensation triggered by CRISPR/Cas9-induced Bag3 deletion prevents heart and skeletal muscle dysfunction
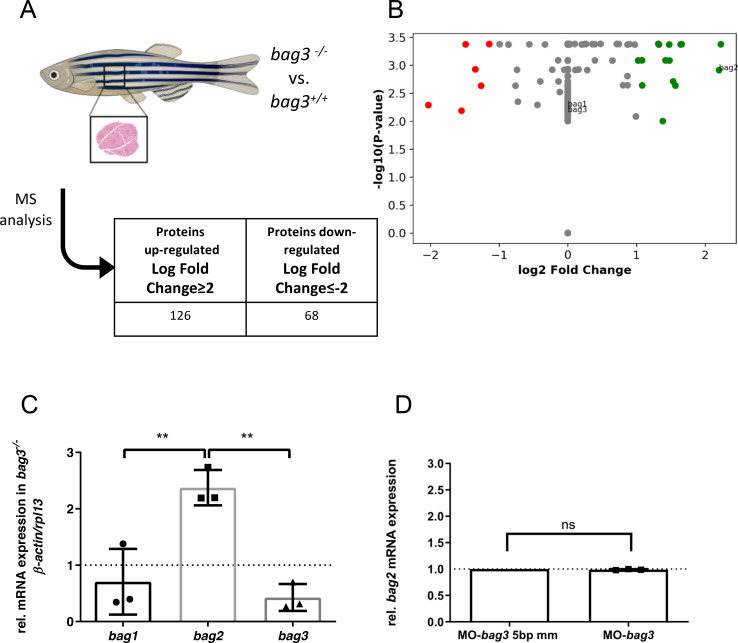
Next, to assess whether genetic compensation is activated in our CRISPR/Cas9-induced bag3 mutants, we performed a mass spectrometry (MS)-based proteomic analysis on muscle tissue derived from adult homozygous bag3-/- mutants compared to homozygous wild-type bag3+/+ clutchmates (Fig 5A). The obtained proteomics profiles revealed 68 down- and 126 up-regulated proteins (log fold change >2 and <-2) (Fig 5B; S2 Table). As expected, Bag3 was not detected in homozygous bag3-/- mutant muscle tissue (Fig 5B), confirming Bag3 deficiency in muscle tissue of our CRISPR/Cas9-induced mutant line. Interestingly, among the most up-regulated proteins, we found levels of Bag2, a member of the Bag protein family, significantly elevated (Log Fold Change = 2.2). This suggested Bag2 could be a potential functional compensator for the loss of Bag3.
Fig 5. Bag2 mediates genetic compensation in bag3 knockout zebrafish.
(A) Schematic description of proteomic workflow. Acquired spectra were analyzed against the Uniprot zebrafish database using MaxQuant. (B) Detection levels of label free quantification (LFQ) intensity ratio between bag3-/- and bag3+/+ zebrafish samples of three orthologs of the Bag family protein, Bag1, Bag2 and Bag3. The values are shown as Log2 fold change ≥2 and ≤2. (C) Quantitative real-time PCR of bag3-/- and bag3+/+ zebrafish embryos at 72 hpf shows significant upregulation of bag2 mRNA levels, whereas bag1 transcript levels are not increased (N = 3, mean±S.D, One-way ANOVA followed by tukey's multiple comparison analysis **bag1 vs bag2 P = 0.0057, ** bag2 vs bag3 P = 0.0026). (D) Quantitative real-time PCR of MO-bag3- or MO-bag3 5bp mm-injected embryos shows no upregulation of bag2 mRNA levels in bag3 morphants at 72 hpf. (n = 3, mean±S.D, P = 0.1080 determined using two-tailed t-test).
Next, to validate transcriptional adaptation/upregulation of Bag2 in bag3-/- mutants, we performed quantitative real-time PCR analyses of bag1, bag2, and bag3. Initially, we confirmed that the three bag family members, bag1-3, are expressed in wild-type zebrafish during development and in the adult skeletal muscle (S4A–S4D Fig). In bag3-/- mutant embryos and adult skeletal muscle, bag1 mRNA levels were transcriptionally unaltered compared to wild-types, whereas bag3 mRNA levels were significantly reduced (N = 3, **P = 0.0071, ***P = 0.0007) (Fig 5C; S4E Fig), confirming bag3 mutant mRNA decay. In accordance with the proteomic analyses, bag2 transcript levels were found to be significantly upregulated in bag3-/- mutant embryos and adult zebrafish skeletal muscle (Fig 5C; S4E Fig). By contrast, bag2 transcript levels were not found to be upregulated in zebrafish embryos injected with bag3-specific morpholinos (MO-bag3) (Fig 5D).
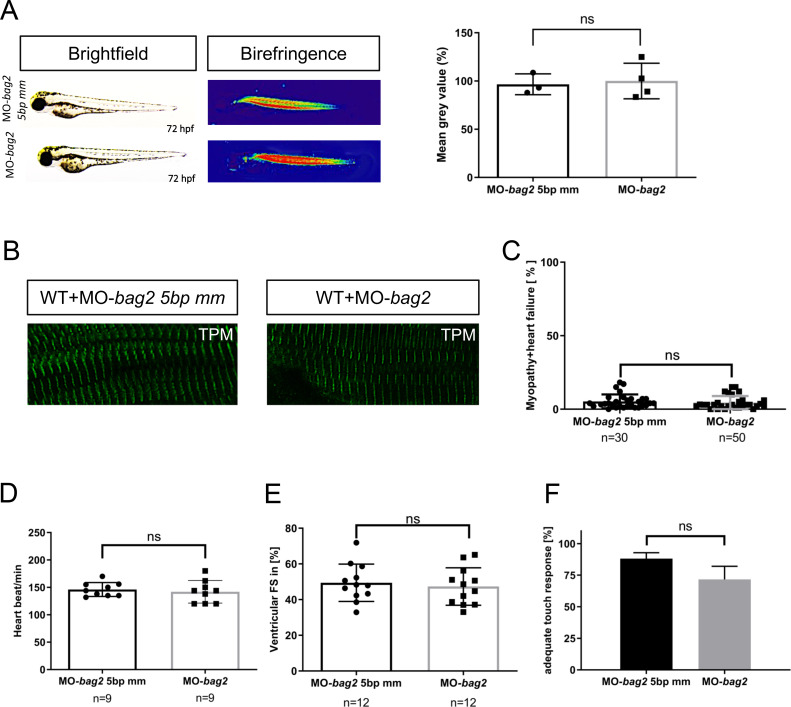
Bag2 (Bcl2-associated athanogene 2) is a molecular co-chaperone and a member of the Bag protein family that shows high structural similarity to Bag3. In zebrafish, Bag2 was found to be expressed in the heart and skeletal muscle [19]. To first investigate the role of Bag2 in vivo, we inactivated zebrafish bag2 by injecting a morpholino against the splice donor site of exon 2 (MO-bag2) into one-cell stage zebrafish embryos. When injected with either MO-bag2 or the 5bp mismatch control morpholino (MO-bag2 5bp mm), injected embryos (N = 3, n = 30/50, P<0.5010) did not develop (cardio-)myopathy (Fig 6A–6F)). We found normal cardiac and skeletal muscle structure and function (Fig 6A and 6B), unconfined cardiac contractile function (Fig 6D and 6E), and undisturbed motility (Fig 6F) in bag2 morphant embryos. This demonstrates that Bag2 is dispensable for early heart and skeletal muscle development and function.
Fig 6. Targeted knockdown of bag2 does not lead to heart and muscles dysfunction in zebrafish embryos.
(A) Brightfield and birefringence images and the densitometric analysis of birefringence (n = 4) of MO-bag2 and MO-bag2 5bp mm injected embryos reveal that bag2 morphants do not develop a (cardio)-myopathy phenotype by 72 hpf. Representative samples are shown (P = 0.7891 determined using two tailed t-test). (B) Tropomyosin immunostainings of MO-bag2- and MO-bag2 5bp mm-injected embryos at 72 hpf demonstrate that bag2 morphants do not develop fiber disruptions. (C) bag2 morphants do not develop (cardio-)myopathy (only 4.50±4.43% of embryos exhibit a phenotype) (N = 3, n = 30/50, Mean± SD P = 0.5010 determined using two-tailed t-tests). (D) Heart rate quantification of bag2 morphants (146±12.64 heart beat/min), at 72 hpf does not show any impairment compared to bag2 5bp-mismatch morphants (142±20.57 heart beat/min) (N = 3, n = 9, mean ± S.D. P = 0.6490 determined using two-tailed t-tests). (E) FS of bag2 morphant ventricles at 72 hpf is not reduced (47.34±10.49%) compared to 5bp-mismatch-MO injected embryos (FS: 49.44±10.48%) (N = 3, n = 12, Mean± SD P = 0.6279 determined using two-tailed t-tests). (F) Touch evoked assay for bag2 morphants does not reveal a significant reduction in responsiveness to mechanical stimulus (71.67±4.22%) (N = 3, n = 50/25, mean ± S.D. P = 0.0941, determined using two-tailed t-tests).
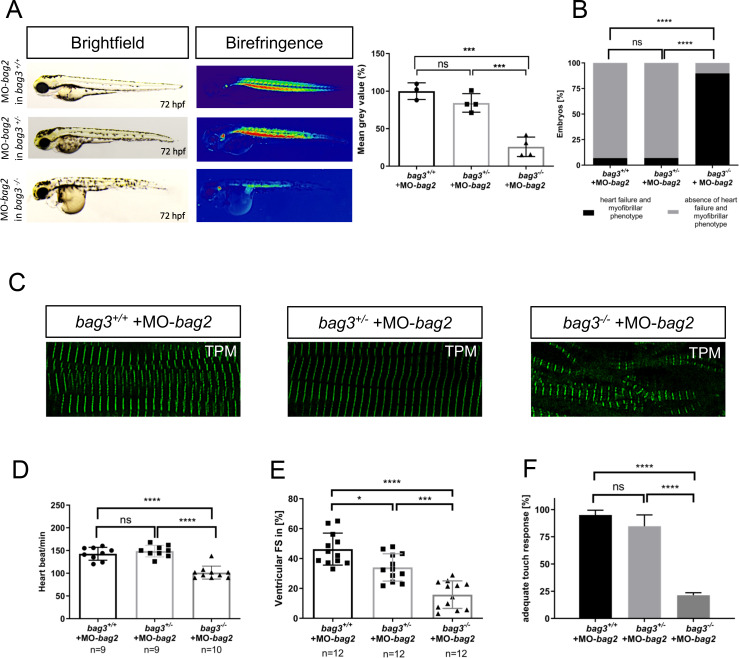
Next, to investigate the role of Bag2 as putative compensatory factor in the CRISPR/Cas9-induced Bag3 knockout line, we inactivated bag2 by injecting MO-bag2 into bag3+/+, bag3+/-, and bag3-/- embryos. While Bag2 knockdown in bag3+/+ and bag3+/- embryos did not result in heart or skeletal muscle malformation and dysfunction (Fig 7A–7F), 89.76 ± 4.37% of bag3-/- mutant embryos injected with MO-bag2 developed severe heart and skeletal muscle defects comparable to the phenotypic characteristics observed in bag3 morphants (N = 3, n = 50, P<0.0001) (Fig 7A–7F). In detail, we found that MO-bag2 injection into bag3-/- embryos caused severe disruption of muscle structure and ultrastructure (Fig 7C; S3A Fig), whereas sarcomeric organization in MO-bag2-injected bag3+/+ and bag3+/- embryos was completely unaffected (Fig 7C). Furthermore, bag3-/- embryos injected with MO-bag2 showed severe functional heart and skeletal muscle defects compared to MO-bag2-injected bag3+/+ and bag3+/- embryos as assessed by the measurement of the heart rate (N = 3, n = 9/10, P<0.0001. HR bag3+/+: 143±14.18 heart beat/min; HR bag3+/-: 148±12.65 heart beat/min; HR bag3-/-: 101±14.27 heart beat/min, at 72 hpf), contractile force (N = 3, n = 12, P<0.0001. FS bag3+/+: 46.33±10.72%; FS bag3+/-: 34.07±9.17%; FS bag3-/-: 15.83±9.19%, at 72 hpf), and touch evoked assays (N = 3, n = 80/50, P<0.0001. Mot bag3+/+: 95±4.41%; Mot bag3+/-: 84.68±10.38%; Mot bag3-/-: 21.33±2.31%, at 72 hpf) (Fig 7D–7F; S3, S5 and S11 Movies). By contrast, specification of atrial and ventricular chambers was not affected by the knockdown of bag2 in bag3-/- embryos (S3B Fig). Efficiency of the bag2 knockdown was demonstrated by RT-PCR-based mRNA splicing assays (S6B Fig).
Fig 7. Knockdown of bag2 in bag3-/- mutants causes heart and skeletal muscle disruptions.
(A) Brightfield and birefringence images and densitometric analysis of birefringence (n = 4) of bag3+/+, bag3+/- and bag3-/- embryos at 72 hpf injected with 200μM of MO-bag2. Representative samples are shown (One-way ANOVA followed by tukey's multiple comparison analysis, P<0.0004). (B) bag3-/- embryos injected with MO-bag2 develop (cardio-)myopathy (89.76±4.37%), whereas bag3+/+ (93.10±0.33%) and bag3+/- (94.77±2.35%) embryos are unaffected by MO-bag2 injection (N = 3, n = 50, mean ± S.D., P<0.0001, two-tailed value for Fisher´s exact test). (C) Tropomyosin immunostainings of bag3+/+, bag3+/- and bag3-/- embryos injected with MO-bag2 at 72 hpf. Only MO-bag2-injected bag3-/- embryos show muscle fiber disruptions. (D) Heart rate quantification at 72 hpf reveals impairments only in bag3-/- embryos injected with MO-bag2 (N = 3, n = 9/10, mean ± S.D. One-way ANOVA followed by tukey's multiple comparison analysis, P<0.0001). (E) FS of ventricles of bag2 splice MO injected in bag3-/- embryos at 72 hpf (FS: 15.83±9.19%) is significantly reduced compared to the FS measured in bag3+/+ (FS: 46.33±10.72%) and bag3+/- (FS: 34.07±9.17%) injected embryos (N = 3, n = 12; mean± SD One-way ANOVA followed by tukey's multiple comparison analysis, *P = 0.0110,***P = 0.0002, ****P< 0.0001). (F) bag3-/- embryos injected with MO-bag2 reveal significantly impaired responsiveness upon mechanical stimulus compared to bag3+/+and bag3+/- embryos injected with MO-bag2 (N = 3, n = 80/50, mean ± S.D. One-way ANOVA followed by tukey's multiple comparison analysis, P<0.0001).
Similar experiments as described for bag2 were conducted for bag1, but no phenotypic alterations could be recognized neither after injection of bag1-specific morpholinos into wild-type embryos nor bag3-/- embryos. This further supports the hypothesis that bag2 and not bag1 effectively compensates for the loss of Bag3 in bag3-/- embryos (S5A–S5E Fig, S6B and S6C Fig and S13 and S14 Movies).
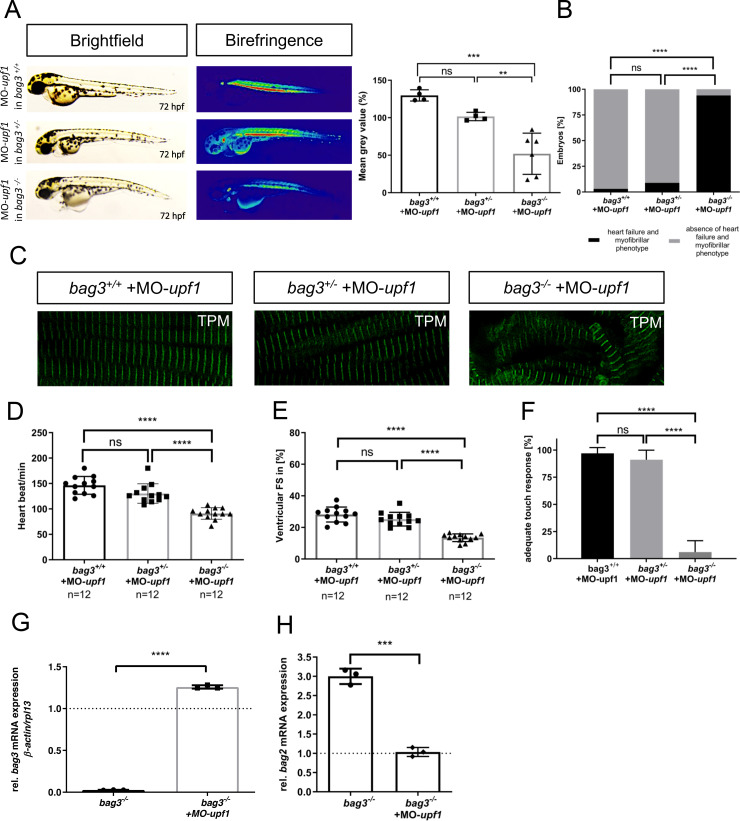
Blockade of bag3 mRNA decay inhibits transcriptional adaptation and induces cardiomyopathy and skeletal muscle dysfunction in Bag3-deficient zebrafish
To investigate whether the inhibition of nonsense-mediated decay (NMD) of mRNA and the subsequent blockade of the transcriptional adaptation machinery [1–3] can induce heart and skeletal muscle defects in bag3-/- embryos, we inactivated upf1, a factor known to be crucial for regular NMD function[20, 21]. We found that morpholino-mediated knockdown of upf1 (MO-upf1) in bag3+/+ and bag3+/- embryos did not result in heart and skeletal muscle defects, whereas knockdown of upf1 in bag3-/- embryos led to severe structural and functional heart and skeletal muscle defects (Fig 8A–8F). Muscle structure was disrupted in bag3-/- embryos injected with MO-upf1 (Fig 8C), while sarcomeric organization in MO-upf1-injected bag3+/+ and bag3+/- embryos was preserved (Fig 8C). Furthermore, MO-upf1 injection into bag3-/- embryos caused severe functional heart and skeletal muscle defects, however, MO-upf1-injected bag3+/+ and bag3+/- embryos showed no phenotypic alterations (Fig 8D–8F; S6, S7 and S12 Movies). High efficiency of the morpholino-mediated upf1 knockdown was confirmed by RT-PCR-based mRNA splicing assays (S6A Fig).
Fig 8. Knockdown of upf1 blocks mutant mRNA decay required for transcriptional adaptation in bag3-/- mutants.
(A) Brightfield and birefringence images and densitometric analysis of birefringence (n = 4/6) of bag3+/+, bag3+/- and bag3-/- embryos at 72 hpf injected with 50μM MO-upf1. Individual samples are shown (One-way ANOVA followed by tukey's multiple comparison analysis, **P=0.0053, ***P=0.0002). (B) bag3-/- embryos injected with MO-upf1 develop (cardio-)myopathy (93.94±10.50%), whereas bag3+/+ (96.97±5.25%) and bag3+/- (91.09±8.82%) embryos are devoid of any phenotype (N = 3, n = 40, mean ± S.D., P<0.0001, two-tailed value for Fisher´s exact test). (C) Tropomyosin immunostainings of bag3+/+, bag3+/- and bag3-/- embryos injected with MO-upf1 at 72 hpf reveal muscle fiber disruptions due to the blockage of the transcriptional adaptation only in bag3-/- injected embryos. (D) Heart rate quantification at 72 hpf reveals impairments only in bag3-/- embryos injected with MO-upf1 (N = 3, n = 12, HR bag3+/+: 146±17.49 heart beat/min; HR bag3+/-: 130±19.18 heart beat/min; HR bag3-/-: 91±11.39 heart beat/min, mean ± S.D. One-way ANOVA followed by tukey's multiple comparison analysis, P<0.0001). (E) FS of ventricles of bag3-/- embryos injected with MO-upf1 is significantly reduced at 72 hpf (FS: 13.48±2.438%) compared to the FS measured in bag3+/+ (FS: 28.17±4.753%) and bag3+/- (FS: 25.23±4.328%) morphants (N = 3, n = 12; mean± SD One-way ANOVA followed by tukey's multiple comparison analysis, P<0.0001). (F) bag3-/- embryos injected with MO-upf1 (6.06±10.5%) reveal significant difference in responsiveness upon mechanical stimulus compared to bag3+/+(96.97±5.25%) and bag3+/- (91.09±8.82%) injected embryos (N = 3, n = 35, mean ± S.D. One-way ANOVA followed by tukey's multiple comparison analysis, P<0.0001). (G) Quantitative real-time PCR of bag3-/- +MO-upf1 and bag3-/- zebrafish embryos at 72 hpf shows restored bag3 mRNA levels only in bag3-/- embryos injected with MO-upf1 (N = 3, mean ± SD, P<0.0001 determined using two-tailed t-test). (H) Quantitative real-time PCR of bag3-/- embryos injected with MO-upf1and uninjected bag3-/- control embryos at 72 hpf reveals the downregulation of bag2 mRNA levels only in bag3-/- + MO-upf1 embryos (N = 3, mean ± SD, P = 0.0004 determined using two-tailed t-test).
Finally, to assess whether inhibiting NMD in bag3-/- embryos indeed resulted in the preservation of bag3 mRNA levels and reduction of bag2 mRNA levels, we performed qRT-PCR analyses in MO-upf1-injected bag3-/- embryos. Indeed, we found the preservation of mutant bag3 mRNA levels as well as the diminution of bag2 mRNA levels to physiological levels in bag3-/- embryos injected with MO-upf1, (Fig 8G and 8H), indicating that blocking the NMD of bag3 mRNA effectively inhibits the transcriptional adaptation of bag2 expression.
In summary, our findings demonstrate that transcriptional adaptation of bag2 expression in a model of CRISPR/Cas9-induced Bag3 ablation is involved in the process of genetic compensation to preserve heart and skeletal muscle structure and function in vivo.
Discussion
Genetic compensation by transcriptional adaptation was recently described to contribute to genetic robustness that guarantees viability and fitness of an organism in the presence of malignant and harmful gene variations and mutations [1, 2]. Here, we describe compensatory transcriptional adaptation in a CRISPR/Cas9-induced zebrafish model of Bag3-deficiency. Whereas morpholino-mediated ablation of Bag3 in zebrafish embryos led to severe heart failure and myopathic phenotypes, the knockout of Bag3 by CRISPR/Cas9 technology was not accompanied by phenotypic alterations in the developing embryo. We found that in our CRISPR/Cas9-induced bag3 mutant embryos, the expression of the Bag protein family member Bag2 was significantly upregulated.
El-Brolosy et al. proposed an explanatory model for the discrepancy in transcriptional adaptation in mutants and morphants. It was found that degradation of the mutated mRNA by mRNA degradation pathways is the molecular trigger for the upregulation of genes that exhibit sequence similarity thereby enabling functional compensation [1, 22, 23]. Pharmacological but also genetic inhibition of NMD (e.g. by the inactivation of Upf1, a factor pivotal for NMD) led to a significant reduction of mutated mRNA and the loss compensatory transcriptional adaptation [2, 23, 24].
BAG3 is a molecular co-chaperone that interacts with heat shock proteins to facilitate important functions in protein homeostasis such as autophagy [5, 6]. Interestingly, BAG3 is predominantly expressed in the heart and skeletal muscle cells and mutations in the BAG3 gene are associated with different variants of cardiomyopathies such as hypertrophic cardiomyopathy (HCM) and DCM or protein aggregation diseases such as MFM [8–10, 15]. In 2011, two independent genome-wide association studies (GWAS) linked mutations in the human BAG3 gene to DCM [9, 12]. Based on these initial findings, human genetic studies confirmed these results and introduced BAG3 as a major DCM causing gene with an incidence of up to 6.7% of all DCM cases [17, 25, 26]. To date, several different human BAG3 mutations are described to be associated with DCM. Remarkably, DCM-associated BAG3 mutations are frequently found to result in the truncation of the BAG3 protein and are assumed to cause the complete loss of BAG3 function [25, 27]. These findings are supported by analyses in conditional, cardiomyocyte-specific Bag3 knockout mice that also develop DCM shortly after birth [28]. To the best of our knowledge, DCM-associated truncating human BAG3 mutations, predominately frame shift and nonsense mutations [25], were never analyzed for the induction of the NMD of the mutated bag3 mRNA and thereby the activation of putative compensatory mechanisms.
Global disruption of the Bag3 gene in homozygous mice led to lethality after 4 weeks of age. These BAG3 knockout mice were characterized by severe myopathic phenotypes including disruption of Z-disk architecture and myofibrillar degeneration [15]. Cardiac-specific knockout of BAG3 by the use of cardiomyocyte-specific α-myosin heavy chain Cre-transgenic mice (αMHC-Cre mice) significantly impaired structure and function of the contractile apparatus in cardiomyocytes resulting in DCM and premature death of these conditional Bag3 knockout mice during their first year of life [28]. Fang and co-workers did not observe increased expression of other BAG family members such as Bag 1 or Bag2 in their conditional Bag3 knockout model [28], implying that their knockout strategy might not activate NMD-related genetic compensation. Knockdown of Bag3 in the zebrafish model using morpholino-modified antisense oligonucleotides resulted in the development of cardiac contractile dysfunction and cardiomyopathy [9] as well as a contraction-dependent myofibrillar disintegration and skeletal muscle dysfunction, two of the characteristics of BAG3-related myofibrillar myopathies (MFM) [10, 11]. Very recently, Ding et al. described that TALEN-mediated ablation of Bag3 led to cardiomyopathy in adult zebrafish most likely due to interference with protein degradation and homeostasis [17].
Here, we describe a CRISPR/Cas9-induced zebrafish model of Bag3 deficiency and we show that bag3 mutant zebrafish embryos, in contrast to bag3 morphants, do not develop heart and skeletal muscle defects. Expression of Rpl10, the 60S ribosomal protein L10 and Bag2 were found to be strongly induced in bag3-/- embryos. Loss of Rpl10 was shown to cause neurodevelopmental defects in zebrafish [29], but does not show any sequence or domain similarities to Bag3 making Rpl10 unlikely to functionally compensate for the loss of Bag3. By contrast, Bag2, a member of the Bag protein family, shows high sequence and domain similarities to Bag3 turning Bag2 into a perfect candidate to compensate for the loss of Bag3 function. In humans, BAG2 is expressed in several tissues, including brown adipose and the lung but also the heart [30]. Unfortunately, not much is known about the in vivo function of Bag2. To date, no bag2-null animal models are described. Here, we knocked down Bag2 by morpholinos in the zebrafish embryo and found no phenotypic alterations in the morphants. By contrast, injection of MO-bag2 into homozygous mutant bag3-/- zebrafish embryos led to severe heart and skeletal muscle defects, resembling the phenotypic alterations observed in bag3 morphants. This suggests that Bag2 functionally compensates for the loss of Bag3 in our genetic model of Bag3 deficiency. El-Brolosy and colleagues found that blocking NMD in hbegfaΔ7, vegfaa and vclaΔ13 zebrafish mutants by genetically inactivating Upf1 led to the preservation of the mutated mRNA levels and, subsequently, to the loss of transcriptional adaptation [1]. Similar to these studies, we found that NMD of the mutated bag3 mRNA triggers the transcriptional adaptation of bag2 expression since blocking NMD by the injection of upf1-specific morpholinos was able to preserve mutant bag3 mRNA levels and to reduce the bag2 mRNA levels to normal levels in homozygous mutant bag3-/- embryos. These findings imply that bag3 mRNA degradation is the main trigger of transcriptional adaptation in bag3-/- zebrafish embryos. Interestingly, bag3-/- zebrafish display increased mortality starting from 9 month of age. We found that bag2 mRNA expression was still increased in the adult skeletal muscle in bag3-/- zebrafish, suggesting that transcriptional adaptation of bag2 expression is able to compensate for the loss of bag3 during embryogenesis but not in the adult zebrafish. Whether this is due to insufficient compensatory bag2 levels or specific Bag2-independent molecular pathomechanisms in the adult zebrafish is unknown but should be investigated in future studies.
Recent findings from zebrafish and mouse studies suggest that genetic compensation is a widespread phenomenon which plays a vital role in the phenotypic peculiarity in response to malignant and harmful gene mutations. Human genetics studies also suggest that genetic compensation might be an active mechanism to influence clinical disease manifestation in response to severely mutated alleles [31–33]. Further elucidation of the molecular mechanisms that control genetic compensation might pave the way to develop therapeutic interventions to foster genetic robustness to a malignant mutation.
Materials and methods
Zebrafish strains and injection procedures
The present study was performed after appropriate institutional approvals (Tierforschungszentrum (TFZ) Ulm University, No. 0183), which conform to EU Directive 2010/63/EU. Care and breeding of zebrafish, Danio rerio, was conducted as described previously [34, 35]
Morpholino-modified antisense oligonucleotides (MOs; Gene Tools, LLC, Oregon, USA) were directed against the splice-acceptor/donor site of zebrafish bag3, bag2, bag1 and upf1. As negative controls 5 bp mismatch MOs were injected at the same concentration as the respective MO. All MOs were resolved in 0.2 M KCl to a final amount of 200 μM MO-bag3 and MO-bag3 5bp mismatch; 200μM MO-bag2 and MO-bag2 5bp mismatch; 50μM MO-upf1 and MO-upf1 5bp mismatch; 200μM MO-bag1 and MO-bag1 5bp mismatch. Injections were performed into one-cell stage zebrafish embryos.
For CRISPR/Cas9 injections, 400 ng/μl recombinant Cas9 protein (Eupheria GmbH, Germany) was mixed with synthetic tracrRNA (100 ng/μl) and a gene-specific crRNA (Eurofins Genomics, Germany) against bag3 (50 ng/μl) in 200 mM KCl. Sequences of CRISPR RNA oligonucleotides and Morpholinos are summarized in S1 Table.
RNA extraction and quantitative real-time PCR
For RNA extraction, 30 wild-type embryos and 30 bag3-/- embryos were collected at 72 hpf. To extract the RNA the RNeasy® Mini Kit was used (Qiagen) according to the manufacturer's instructions. Reverse transcription was performed by using SuperScript® III Reverse Transcriptase (Life Technologies), 1μg total RNA and oligo(dT) primer. Quantitative real-time PCR was carried out according to standard protocols using SYBR-Green master mix (Roche) and a Roche LightCycler 480 II. cDNA was generated as described above from 72 hpf embryos. To correct for sample to sample variation, housekeeping genes β-actin and rpl13 were used for normalization [36]. Sequences of primers used are summarized in S1 Table.
Immunoblotting
For immunoblot analysis, from 72 hpf wild-type and bag3-/- embryos, proteins lysates were prepared. 30μg of protein lysate was boiled in 5x Laemmli Buffer, separated on an SDS-PAGE (8–16%), and blotted onto polyvinylidene difluoride (PVDF) membranes. Then, the membranes were blocked using 5% milk powder in TBST for 1h at RT and incubated with primary antibodies overnight at 4°C. The membranes were washed and incubated with polyclonal anti-rabbit-IgG antibody conjugated to horseradish peroxidase. All membranes were developed using the Pierce ECL Western Blotting Substrate (Thermo Scientific) and a luminescent image analyzer (Image Quant Las4000 mini) [36].
The Bag3 antibody (1:1000 in 5% BSA) was designed against the c-terminus of zebrafish Bag3 (epitope position 213–226: LSQSSHPTREKIYR) and purchased from BioGenes GmbH, Berlin. Pan-cadherin (1:4000 in 5% milk) was purchased from Abcam (ab16505).
Immunostaining
WT, bag+/+, bag3+/-, and bag3-/- injected (MO-bag3, MO-bag2 and MO-upf1) and non-injected embryos were euthanized with tricaine at 72 hpf. Embryos were fixed in 4% paraformaldehyde overnight at 4°C and embedded in 4% low melting agarose (melting temperature ≤65°C; Sigma) dissolved in distilled water. Longitudinal sections were cut with a Leica VT1200S vibratome to a thickness of 100μm and incubated in CAS-block solution (008120, Thermo Fisher) for 30 minutes (diluted 1:10 in distilled water). Staining was conducted in CAS-block (diluted 1:10 in distilled water). Tropomyosin staining was performed with 1:50 CH1 antibody (Hybridoma Bank) overnight at 4°C followed by 1:100 Alexa-Fluor 488 (Invitrogen, A-21121) for 2 h at RT.
Immunostaining of heart muscle myosin heavy chains MF20 (1:10, mouse monoclonal IgG2b; Hybridomabank) in co-stain with S46 (1:50, mouse monoclonal IgG1, Hybridoma Bank) was performed on Dent's fixed embryos at 72 hpf. Secondary antibodies goat anti-mouse IgG1 Alexa Fluor 488 and Alexa 555 goat ant-mouse IgG2b (both Invitrogen) were applied in dilutions of 1:100. Images were acquired using the Leica SP8 microscopes (Leica Mikrosysteme Vertrieb GmbH, Wetzlar, Germany).
Mass spectrometry
Protein lysates derived from muscle tissue of adult homozygous bag3-/- mutant and homozygous wild-type (bag3+/+) controls (age: 3 month) were separated using SDS-PAGE and the entire lane was subsequently processed as described previously [37] for MS analysis.
Samples were analyzed as previously described [37] by employing an U3000 RSLCnano (Thermo Fisher Scientific, Idstein, Germany) for peptide separation online coupled to an LTQ Orbitrap Velos Pro (Thermo Fisher Scientific, Bremen, Germany) mass spectrometry.
A database search was performed using MaxQuant Ver. 1.6.3.4 (www.maxquant.org) [38]. Using the built-in Andromeda search engine [39], MS/MS spectra were correlated with the UniProt zebrafish database (www.uniprot.org) for peptide identification. Carbamidomethylated cysteine was considered as a fixed modification along with oxidation (M), and acetylated protein N-termini as variable modifications. False Discovery rates were set on both, peptide and protein levels, to 0.01. For quantitation, LFQ was enabled within MaxQuant using default parameters.
Birefringence analysis
Images were taken with an Olympus SZX 16 microscope and movies were recorded with a Leica DM IL LED microscope. The functional assessment of cardiac contractility was carried out as previously described [34, 40].
Regarding the birefringence analysis, 72 hpf zebrafish embryos were anesthetized with tricaine and embedded in 2.5% methylcellulose, in a glass petri dish, making sure that they were lying as flat as possible. Images were acquired between two polarizing filters on an Olympus SZX16 with a DP72 camera and the Olympus Stream software. After acquisition, the inverted images were analysed using the Fiji software and the mean grey value of the selected area of the skeletal muscles was measured. Data analysis was performed normalizing the mean intensity with the selected area as previously described [16, 41]. For the publication, the analysed images where optimized from black and white to colored heat map using the tool display “LUT” and selecting the “physics” option in Fiji software.
Functional assessment
For the touch evoked assay, zebrafish embryos at 72 hpf were touched by a needle tip and their flight response was analysed. An immediate and straightforward flight was regarded as “adequate” while no movement or a delayed/incomplete response were regarded as “inadequate”. Movies of 10–30 seconds were acquired for each sample to show the larval response to the needle touch. For each experiment the number of embryos with adequate response to the touch was converted to a percentage, analysed, and plotted in a bar graph using GraphPad Prism software[11, 42].
To increase the workload on the skeletal muscles, embryos at 120 hpf were incubated in 1% methylcellulose for 2 hours as previously described [10].
Statistical analysis
Statistical analyses were performed using GraphPad Prism software. All the experiments were conducted in triplicate using different biological samples.
All results are expressed as mean ± standard deviation (S.D.) and statistical analysis were performed as indicated in the figure legends. A p-value smaller than 0.05 was regarded as statistically significant.
Supporting information
Overall amino acid identity between human and zebrafish BAG3.
(PDF)
(PDF)
(a) Transmission electron microscopic (TEM) analysis of parasagittal and transversal sections through cardiac and skeletal muscle cells of WT, bag3-/- and bag3-/- + MO-bag2 embryos at 72 hpf. In contrast to the WT and bag3-/-, bag3-/- + MO-bag2 cardiomyocytes and fast-twitch skeletal muscle fibers show disrupted sarcomeric structures. (b) Hearts at 72 hpf stained with chamber-specific myosin antibodies, MF20 (red) and S46 (green) (MF20 marks the entire heart and S46 is atrium specific). Heart chamber specification appears to be normal in all analyzed embyros.
(PDF)
The graph shows a very similar expression pattern for bag1 and bag2. (d) qPCR showing bag1, bag2 and bag3 levels in adult zebrafish skeletal muscles (N = 3, mean±S.D, One-way ANOVA followed by tukey's multiple comparison analysis P = 0.8457). (e) bag2 transcript levels are significantly increased in skeletal muscle of adult bag3-/- zebrafish (N = 3, mean±S.D, One-way ANOVA followed by tukey's multiple comparison analysis ***P = 0.0003).
(PDF)
(a) Brightfield and birefringence images of bag1 splice MO injected WT and bag3-/- embryos at 72 hpf don´t show any (cardio)-myopathy phenotype. The densitometric analysis of birefringence signals supports the absence of sarcomeric disorganization. Individual samples are shown (n = 4, P>0.9999 determined using two tailed t-test). (b) MF20 and S46 immunostainings of WT and bag3-/- embryos at 72 hpf injected with MO-bag1 reveal regular specification of the cardiac chambers. (c) Heart rate quantification of bag1 morphants and bag3-/- + MO-bag1 at 72 hpf reveals no functional cardiac impairments (N = 3, n = 9/10. HR MO-bag1 injected WT embryos: 167±11.40 heart beat/min; HR bag3-/- +MO-bag1: 167±9.98 heart beat/min; mean ± S.D. P = 0.9894 determined using two-tailed t-tests). (d) Ventricular FS of bag1 morphants (20.59±8.47%) and bag3-/- + MO-bag1 embryos (FS: 22.12±4.20%) at 72 hpf is unaltered (N = 3, n = 9; Mean± SD P = 0.6373 determined using two-tailed t-tests).
(PDF)
(a) Injection of MO-upf1 (splice-blocking morpholino) results in the partial skipping of upf1 exon 1, leading to a frame shift, a premature stop codon and the premature termination of Upf1 translation [43, 44]. (b) Injection of MO-bag2 (splice-blocking morpholino) results in the integration of the intron 2, a frame shift, a premature stop codon and the premature termination of Bag2 translation. (c) Injection of splice MO-bag1 (splice-blocking morpholino) results in the skipping of the exon 2, a frame shift, a premature stop codon and the premature termination of Bag1 translation.
(PDF)
Additional transmission electron microscopic (TEM) pictures of parasagittal and transversal sections through cardiac muscle cells of WT, bag3-/- and bag3-/- + MO-bag2 embryos at 72 hpf.
(PDF)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(DOCX)
(XLSX)
Acknowledgments
We thank Regine Baur, Renate Durst, Karin Strele, Katrin Vogt, and Sabrina Diebold for their excellent technical assistance. We thank Dr. Sebastian Wiese and the Core Unit Mass Spectrometry and Proteomics (Ulm University) for the Mass-Spectrometry analysis.
The Fis 1a and 5a on this manuscript were done using biorender.com software.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) JU2859/2-1 and JU2859/7-1 (SJ) and the German Federal Ministry of Education and Research (BMBF) (e:Med-SYMBOL-HF grant #01ZX1407A; e:Med-coNfirm grant #01ZX1708C) (SJ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gunther S, Fukuda N, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568(7751):193–7. Epub 2019/04/05. 10.1038/s41586-019-1064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Brolosy MA, Stainier DYR. Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet. 2017;13(7):e1006780 Epub 2017/07/14. 10.1371/journal.pgen.1006780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524(7564):230–3. Epub 2015/07/13. 10.1038/nature14580 . [DOI] [PubMed] [Google Scholar]
- 4.Sztal TE, McKaige EA, Williams C, Ruparelia AA, Bryson-Richardson RJ. Genetic compensation triggered by actin mutation prevents the muscle damage caused by loss of actin protein. PLoS Genet. 2018;14(2):e1007212 Epub 2018/02/08. 10.1371/journal.pgen.1007212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sturner E, Behl C. The Role of the Multifunctional BAG3 Protein in Cellular Protein Quality Control and in Disease. Front Mol Neurosci. 2017;10:177 Epub 2017/07/07. 10.3389/fnmol.2017.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klimek C, Kathage B, Wordehoff J, Hohfeld J. BAG3-mediated proteostasis at a glance. J Cell Sci. 2017;130(17):2781–8. Epub 2017/08/16. 10.1242/jcs.203679 . [DOI] [PubMed] [Google Scholar]
- 7.Behl C. Breaking BAG: The Co-Chaperone BAG3 in Health and Disease. Trends Pharmacol Sci. 2016;37(8):672–88. Epub 2016/05/06. 10.1016/j.tips.2016.04.007 . [DOI] [PubMed] [Google Scholar]
- 8.Feldman AM, Begay RL, Knezevic T, Myers VD, Slavov DB, Zhu W, et al. Decreased levels of BAG3 in a family with a rare variant and in idiopathic dilated cardiomyopathy. J Cell Physiol. 2014;229(11):1697–702. Epub 2014/03/14. 10.1002/jcp.24615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Norton N, Li D, Rieder MJ, Siegfried JD, Rampersaud E, Züchner S, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88(3):273–82. Epub 2011/02/25. 10.1016/j.ajhg.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruparelia AA, Oorschot V, Vaz R, Ramm G, Bryson-Richardson RJ. Zebrafish models of BAG3 myofibrillar myopathy suggest a toxic gain of function leading to BAG3 insufficiency. Acta Neuropathol. 2014;128(6):821–33. Epub 2014/10/02. 10.1007/s00401-014-1344-5 . [DOI] [PubMed] [Google Scholar]
- 11.Buhrdel JB, Hirth S, Kessler M, Westphal S, Forster M, Manta L, et al. In vivo characterization of human myofibrillar myopathy genes in zebrafish. Biochemical and biophysical research communications. 2015;461(2):217–23. 10.1016/j.bbrc.2015.03.149 . [DOI] [PubMed] [Google Scholar]
- 12.Villard E, Perret C, Gary F, Proust C, Dilanian G, Hengstenberg C, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32(9):1065–76. Epub 2011/04/01. 10.1093/eurheartj/ehr105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. Epub 2013/04/17. 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kettleborough RN, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496(7446):494–7. Epub 2013/04/17. 10.1038/nature11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Homma S, Iwasaki M, Shelton GD, Engvall E, Reed JC, Takayama S. BAG3 deficiency results in fulminant myopathy and early lethality. Am J Pathol. 2006;169(3):761–73. 10.2353/ajpath.2006.060250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith LL, Beggs AH, Gupta VA. Analysis of skeletal muscle defects in larval zebrafish by birefringence and touch-evoke escape response assays. J Vis Exp. 2013;(82):e50925 Epub 2013/12/13. 10.3791/50925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding Y, Dvornikov AV, Ma X, Zhang H, Wang Y, Lowerison M, et al. Haploinsufficiency of mechanistic target of rapamycin ameliorates bag3 cardiomyopathy in adult zebrafish. Dis Model Mech. 2019. Epub 2019/09/08. 10.1242/dmm.040154 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruparelia AA, Oorschot V, Ramm G, Bryson-Richardson RJ. FLNC myofibrillar myopathy results from impaired autophagy and protein insufficiency. Hum Mol Genet. 2016;25(11):2131–42. Epub 2016/10/30. 10.1093/hmg/ddw080 . [DOI] [PubMed] [Google Scholar]
- 19.Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. (http://zfin.org).2004.
- 20.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16(11):665–77. Epub 2015/09/23. 10.1038/nrm4063 . [DOI] [PubMed] [Google Scholar]
- 21.Akimitsu N. Messenger RNA surveillance systems monitoring proper translation termination. J Biochem. 2008;143(1):1–8. Epub 2007/11/01. 10.1093/jb/mvm204 . [DOI] [PubMed] [Google Scholar]
- 22.Ma Z, Zhu P, Shi H, Guo L, Zhang Q, Chen Y, et al. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature. 2019;568(7751):259–63. Epub 2019/04/03. 10.1038/s41586-019-1057-y . [DOI] [PubMed] [Google Scholar]
- 23.Peng J. Gene redundancy and gene compensation: An updated view. J Genet Genomics. 2019;46(7):329–33. Epub 2019/07/19. 10.1016/j.jgg.2019.07.001 . [DOI] [PubMed] [Google Scholar]
- 24.Kurosaki T, Popp MW, Maquat LE. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol. 2019;20(7):406–20. 10.1038/s41580-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domínguez F, Cuenca S, Bilińska Z, Toro R, Villard E, Barriales-Villa R, et al. Dilated Cardiomyopathy Due to BLC2-Associated Athanogene 3 (BAG3) Mutations. J Am Coll Cardiol. 2018;72(20):2471–81. 10.1016/j.jacc.2018.08.2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franaszczyk M, Bilinska ZT, Sobieszczańska-Małek M, Michalak E, Sleszycka J, Sioma A, et al. The BAG3 gene variants in Polish patients with dilated cardiomyopathy: four novel mutations and a genotype-phenotype correlation. J Transl Med. 2014;12:192 Epub 2014/07/09. 10.1186/1479-5876-12-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chami N, Tadros R, Lemarbre F, Lo KS, Beaudoin M, Robb L, et al. Nonsense mutations in BAG3 are associated with early-onset dilated cardiomyopathy in French Canadians. Can J Cardiol. 2014;30(12):1655–61. Epub 2014/10/02. 10.1016/j.cjca.2014.09.030 . [DOI] [PubMed] [Google Scholar]
- 28.Fang X, Bogomolovas J, Wu T, Zhang W, Liu C, Veevers J, et al. Loss-of-function mutations in co-chaperone BAG3 destabilize small HSPs and cause cardiomyopathy. J Clin Invest. 2017;127(8):3189–200. Epub 2017/07/24. 10.1172/JCI94310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks SS, Wall AL, Golzio C, Reid DW, Kondyles A, Willer JR, et al. A novel ribosomopathy caused by dysfunction of RPL10 disrupts neurodevelopment and causes X-linked microcephaly in humans. Genetics. 2014;198(2):723–33. Epub 2014/10/16. 10.1534/genetics.114.168211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin L, Guo J, Zheng Q, Zhang H. BAG2 structure, function and involvement in disease. Cell Mol Biol Lett. 2016;21:18 Epub 2016/09/20. 10.1186/s11658-016-0020-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34(5):531–8. Epub 2016/04/12. 10.1038/nbt.3514 . [DOI] [PubMed] [Google Scholar]
- 32.Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, Barnes MR, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352(6284):474–7. Epub 2016/03/05. 10.1126/science.aac8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sulem P, Helgason H, Oddson A, Stefansson H, Gudjonsson SA, Zink F, et al. Identification of a large set of rare complete human knockouts. Nat Genet. 2015;47(5):448–52. Epub 2015/03/26. 10.1038/ng.3243 . [DOI] [PubMed] [Google Scholar]
- 34.Rottbauer W, Just S, Wessels G, Trano N, Most P, Katus HA, et al. VEGF-PLCgamma1 pathway controls cardiac contractility in the embryonic heart. Genes & development. 2005;19(13):1624–34. 10.1101/gad.1319405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Just S, Hirth S, Berger IM, Fishman MC, Rottbauer W. The mediator complex subunit Med10 regulates heart valve formation in zebrafish by controlling Tbx2b-mediated Has2 expression and cardiac jelly formation. Biochemical and biophysical research communications. 2016;477(4):581–8. 10.1016/j.bbrc.2016.06.088 . [DOI] [PubMed] [Google Scholar]
- 36.Hirth S, Buhler A, Buhrdel JB, Rudeck S, Dahme T, Rottbauer W, et al. Paxillin and Focal Adhesion Kinase (FAK) Regulate Cardiac Contractility in the Zebrafish Heart. PloS one. 2016;11(3):e0150323 10.1371/journal.pone.0150323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gross A, Kracher B, Kraus JM, Kuhlwein SD, Pfister AS, Wiese S, et al. Representing dynamic biological networks with multi-scale probabilistic models. Commun Biol. 2019;2:21 Epub 2019/01/25. 10.1038/s42003-018-0268-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. Epub 2008/11/26. 10.1038/nbt.1511 . [DOI] [PubMed] [Google Scholar]
- 39.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–805. Epub 2011/01/25. 10.1021/pr101065j . [DOI] [PubMed] [Google Scholar]
- 40.Meder B, Just S, Vogel B, Rudloff J, Gartner L, Dahme T, et al. JunB-CBFbeta signaling is essential to maintain sarcomeric Z-disc structure and when defective leads to heart failure. J Cell Sci. 2010;123(Pt 15):2613–20. Epub 2010/07/08. 10.1242/jcs.067967 . [DOI] [PubMed] [Google Scholar]
- 41.Molt S, Buhrdel JB, Yakovlev S, Schein P, Orfanos Z, Kirfel G, et al. Aciculin interacts with filamin C and Xin and is essential for myofibril assembly, remodeling and maintenance. J Cell Sci. 2014;127(Pt 16):3578–92. 10.1242/jcs.152157 . [DOI] [PubMed] [Google Scholar]
- 42.Kustermann M, Manta L, Paone C, Kustermann J, Lausser L, Wiesner C, et al. Loss of the novel Vcp (valosin containing protein) interactor Washc4 interferes with autophagy-mediated proteostasis in striated muscle and leads to myopathy in vivo. Autophagy. 2018;14(11):1911–27. Epub 2018/07/17. 10.1080/15548627.2018.1491491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slijkerman R, Goloborodko A, Broekman S, de Vrieze E, Hetterschijt L, Peters T, et al. Poor Splice-Site Recognition in a Humanized Zebrafish Knockin Model for the Recurrent Deep-Intronic c.7595-2144A>G Mutation in USH2A. Zebrafish. 2018;15(6):597–609. Epub 2018/10/03. 10.1089/zeb.2018.1613 . [DOI] [PubMed] [Google Scholar]
- 44.Wittkopp N, Huntzinger E, Weiler C, Saulière J, Schmidt S, Sonawane M, et al. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 2009;29(13):3517–28. Epub 2009/05/04. 10.1128/MCB.00177-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overall amino acid identity between human and zebrafish BAG3.
(PDF)
(PDF)
(a) Transmission electron microscopic (TEM) analysis of parasagittal and transversal sections through cardiac and skeletal muscle cells of WT, bag3-/- and bag3-/- + MO-bag2 embryos at 72 hpf. In contrast to the WT and bag3-/-, bag3-/- + MO-bag2 cardiomyocytes and fast-twitch skeletal muscle fibers show disrupted sarcomeric structures. (b) Hearts at 72 hpf stained with chamber-specific myosin antibodies, MF20 (red) and S46 (green) (MF20 marks the entire heart and S46 is atrium specific). Heart chamber specification appears to be normal in all analyzed embyros.
(PDF)
The graph shows a very similar expression pattern for bag1 and bag2. (d) qPCR showing bag1, bag2 and bag3 levels in adult zebrafish skeletal muscles (N = 3, mean±S.D, One-way ANOVA followed by tukey's multiple comparison analysis P = 0.8457). (e) bag2 transcript levels are significantly increased in skeletal muscle of adult bag3-/- zebrafish (N = 3, mean±S.D, One-way ANOVA followed by tukey's multiple comparison analysis ***P = 0.0003).
(PDF)
(a) Brightfield and birefringence images of bag1 splice MO injected WT and bag3-/- embryos at 72 hpf don´t show any (cardio)-myopathy phenotype. The densitometric analysis of birefringence signals supports the absence of sarcomeric disorganization. Individual samples are shown (n = 4, P>0.9999 determined using two tailed t-test). (b) MF20 and S46 immunostainings of WT and bag3-/- embryos at 72 hpf injected with MO-bag1 reveal regular specification of the cardiac chambers. (c) Heart rate quantification of bag1 morphants and bag3-/- + MO-bag1 at 72 hpf reveals no functional cardiac impairments (N = 3, n = 9/10. HR MO-bag1 injected WT embryos: 167±11.40 heart beat/min; HR bag3-/- +MO-bag1: 167±9.98 heart beat/min; mean ± S.D. P = 0.9894 determined using two-tailed t-tests). (d) Ventricular FS of bag1 morphants (20.59±8.47%) and bag3-/- + MO-bag1 embryos (FS: 22.12±4.20%) at 72 hpf is unaltered (N = 3, n = 9; Mean± SD P = 0.6373 determined using two-tailed t-tests).
(PDF)
(a) Injection of MO-upf1 (splice-blocking morpholino) results in the partial skipping of upf1 exon 1, leading to a frame shift, a premature stop codon and the premature termination of Upf1 translation [43, 44]. (b) Injection of MO-bag2 (splice-blocking morpholino) results in the integration of the intron 2, a frame shift, a premature stop codon and the premature termination of Bag2 translation. (c) Injection of splice MO-bag1 (splice-blocking morpholino) results in the skipping of the exon 2, a frame shift, a premature stop codon and the premature termination of Bag1 translation.
(PDF)
Additional transmission electron microscopic (TEM) pictures of parasagittal and transversal sections through cardiac muscle cells of WT, bag3-/- and bag3-/- + MO-bag2 embryos at 72 hpf.
(PDF)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(AVI)
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.