Abstract
Background
m6A modification affects the pathological progress of many diseases by affecting RNA stability and translocation. YTHDC2, a m6A reader, is associated with multiple cancers; however, little is known of its role in non‐small cell lung cancer (NSCLC).
Methods
The GEPIA, Oncomine and GEO databases were analyzed to assess expression of YTHDC2 in NSCLC patients. Quantitative polymerase chain reaction, western blot and immunohistochemistry were used to detect YTHDC2 expression in different NSCLC cell lines (H1299, H460, H292 and A549) and patients. The effects of YTHDC2 on NSCLC cell lines (A549 and H1299) proliferation and migration were employed using CCK8 and transwell assays. The potential target RNAs of YTHDC2 were obtained from the POSTAR database. Functional enrichment analysis of YTHDC2 targeted RNAs was performed using the Metascape database.
Results
GEPIA, Oncomine and GEO databases showed low expression of YTHDC2 in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) patients. YTHDC2 expression was significantly decreased in different NSCLC cell lines and our clinical samples. Moreover, low expression of YTHDC2 was significantly associated with poor differentiation, lymph node metastasis, tumor size and stage. In addition, YTHDC2 could suppress the proliferation and migration ability of A549 and H1299 cell lines. Kaplan‐Meier Plotter database analysis revealed that patients with low level of YTHDC2 had a significantly poor prognosis. Finally, functional enrichment analysis of YTHDC2 targeted RNAs indicated several enriched pathways related to cancer.
Conclusions
These findings elucidate that YTHDC2 suppresses tumorigenesis in NSCLC, indicating that YTHDC2 may be a promising therapeutic target for NSCLC.
Key points
Significant findings of the study
This study demonstrated that the downregulation of YTHDC2 promotes tumor progression and predicts poor prognosis in non‐small cell lung cancer (NSCLC).
What this study adds
YTHDC2 might be a promising therapeutic target for non‐small cell lung cancer (NSCLC).
Keywords: Epigenetics, N6‐methyladenosine, non‐small cell lung cancer, prognosis, YTHDC2
We revealed that the expression of YTHDC2 was decreased in NSCLC tissues and cell lines. Gain and loss of function assays demonstrated that YTHDC2 could suppress proliferation and migration ability of NSCLC cells. Moreover, YTHDC2 might be a remarkable prognostic marker for NSCLC.
Introduction
Lung cancer, one of the most prevalent malignancies in adults, ranks first among the leading cause of cancer‐related mortality worldwide. 1 , 2 Non‐small cell lung cancer (NSCLC) accounts for the majority of newly diagnosed lung cancer cases. Although surgery combined with chemotherapy, as well as immunotherapy, have improved prognosis, the five‐year survival rate of NSCLC patients is still poor. 3 , 4 Therefore, identifying novel biomarkers and therapeutic targets for NSCLC is essential.
N6‐methyladenosine (m6A) modification is the most prevalent internal modification in eukaryotic mRNA. 5 m6A modification regulates a wide spectrum of biological processes such as the regulation of mRNA splicing, stability and translocation. 6 , 7 , 8 m6A modification is installed by methyltransferases (termed as “m6A writers”), removed by demethylases (termed as “m6A erasers”) and recognized by binding proteins (termed as “m6A readers”). 9 , 10 One class of direct and robust m6A readers are proteins containing the YTH domain, including YTHDF1‐3 and YTHDC1‐2. 11 , 12
YTHDC2, a m6A reader, has been previously explored in different diseases, including cancer. For example, Bailey et al. demonstrated that YTHDC2 regulates the transition from proliferation to differentiation in the germline. 13 Another study found that Ythdc2 suppresses liver steatosis via regulation of mRNA stability of lipogenic genes. 14 Fanale et al. reported that the YTHDC2 gene could be a potential candidate for pancreatic cancer susceptibility and a useful marker of early detection. 15 In addition, loss of YTHDC2 promoted the proliferation rate of esophageal squamous‐cell carcinoma (ESCC) by affecting several cancer‐related signaling pathways. 16 However, whether YTHDC2 is associated with NSCLC remains unknown and needs to be explored further.
In the present study, we aimed to analyze the expression of YTHDC2 in NSCLC cell lines and patients, and evaluate the correlation of YTHDC2 expression with clinicopathological variables of NSCLC patients. The potential biological functions of YTHDC2 in NSCLC cell lines were also investigated. Bioinformatic analysis were then performed to assess whether expression of YTHDC2 is related to prognostic value in NSCLC patients. Finally, the potential target RNAs of YTHDC2 were searched and functional enrichment analysis performed.
Methods
Cell lines and cell culture
The human bronchial epithelial (HBE) cell line was obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS). Human NSCLC cell lines (H1299, H460, H292 and A549) were obtained from the Shanghai Cell Bank (Shanghai, China) and cultured in RPMI1640 medium supplemented with 10% FBS (Gibco, NY, USA). All cell lines were cultured in 5% CO2 at 37°C in a humidified incubator.
Clinical specimen collection
A total of 96 NSCLC tissues and 31 adjacent normal tissues were obtained from 96 NSCLC patients who had undergone surgery at the First Affiliated Hospital of China Medical University from 2017 to 2018. This study was approved by Ethics Committee of the hospital. Written informed consent was obtained from all patients.
Database search
The expression of YTHDC2 in multiple types of cancer was analyzed using the GEPIA (http://gepia.cancer-pku.cn) and Oncomine (https://www.oncomine.org/resource/login.html) online databases. For the GEPIA database, we defined expression value as log2 (TPM + 1), while for the Oncomine database, expression value was defined as log2 (median‐centered ratio). In addition, three sets of microarray data of NSCLC tissues were downloaded from Gene Expression Omnibus (GEO) database (accession numbers: GSE63459, GSE74706 and GSE32665). For the GEO database, we defined expression value as log2 (expression). We used Kaplan‐Meier Plotter (http://kmplot.com/private) to assess the prognostic value of YTHDC2 among the cancer patients with different cancer types. The potential target RNAs of YTHDC2 were obtained from the POSTAR database (http://lulab.life.tsinghua.edu.cn/postar/rbp2.php). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of YTHDC2 targeted RNAs were performed in the online tool Metascape (https://metascape.org/gp/index.html#/main/step1). Methylation prediction of YTHDC2 promoter was carried out using online MethPrimer software (http://www.urogene.org/methprimer2/). JASPAR (http://jaspar.genereg.net/) and UCSC (http://genome.ucsc.edu/) databases were used to predict potential transcription factors in YTHDC2 promoter.
Vector, small interfering RNA and transfection
The cDNA sequences of YTHDC2 were subcloned into the pcDNA3.1 vector (Invitrogen, CA, USA). Short interfering RNAs (siRNAs) were synthesized by GenePharma (Shanghai, China). The siRNA sequences used here were as follows: siYTHDC2#1, 5′‐GCCUUGGAUGUAAAUCUCUTT‐3′; siYTHDC2#2, 5′‐CGGAAGCUAAAUCGAGCCUTT‐3′. The plasmid or siRNA were transfected into NSCLC cell lines using Lipofectamine 3000 (Invitrogen, CA, USA) according to the manufacturer's instructions.
Immunohistochemistry (IHC)
The NSCLC tissues were collected, paraffin‐embedded, and cut into 4 μm serial sections. Sections were incubated with anti‐YTHDC2 (1:500 dilution, Abcam, ab220160) at 4°C overnight, followed by incubation with secondary antibodies. The sections were then visualized under a microscope. YTHDC2 staining intensity was scored as follows: 0 (no staining); 1 (weak); 2 (moderate) and 3 (high). Percentage scores were assigned as follows: 1 (1%–25%); 2 (26%–50%); 3 (51%–75%) and 4 (76%–100%). The final IHC score was calculated by multiplying the intensity score with the percentage of positive cells. YTHDC2 status was regarded as low YTHDC2 expression (score < 4) or high expression (score ≥ 4).
RNA extraction and quantitative polymerase chain reaction (qPCR)
TRIzol reagent (Takara, Dalian, China) was used to separate total RNA from NSCLC cell lines. The mRNA was then reverse transcribed into cDNA using the cDNA Reverse Transcription Kit (Takara). For YTHDC2, the forward primer was 5′‐CAAAACATGCTGTTAGGAGCCT‐3′ and the reverse primer was 5′‐CCACTTGTCTTGCTCATTTCCC‐3′, the amplicon size was 136 base pairs. For GAPDH, the forward primer was 5′‐ATCATCCCTGCCTCTACTG‐3′ and the reverse primer was 5′‐TGCTTCACCACCTTCTTG‐3′, the amplicon size was 176 base pairs. qPCR was performed in duplicate with SYBR green reagent (Takara). GAPDH was used for normalization.
Western blot analysis
Serum samples were resolved on SDS‐PAGE gels followed by standard western blot. The following primary antibodies were used: YTHDC2 (1:1000 dilution, Abcam, ab220160), GAPDH (1:1000 dilution, Santa Cruz Biotechnology, sc‐365 062).
Cell counting Kit‐8 (CCK8) and transwell assay
For CCK8 assay, NSCLC cell lines (A549 and H1299) were seeded into 96‐well plates (5 × 103 cells per well). The plate was incubated for the indicated time periods (24, 48, 72, 96 and 120 hours) in an incubator and 10 μL CCK8 solution added to each well, and cultured for a further 2 hours. The 450 nm light absorption value was read with a microplate reader.
For transwell assay, a 24 well transwell system (Corning, NY, USA) was used. 1 × 105 cells in 100 μL of the culture medium were added into the upper chamber, while 600 μL medium supplemented with 10% FBS were added into the lower chamber. After 24 hours of incubation, cells on the lower surfaces of the membrane were fixed, stained, photographed and counted.
Statistical analysis
Statistical analysis was conducted using SPSS 20.0 software. A chi‐square test was performed to calculate the relationship between YTHDC2 expression and clinicopathological features. The differences in data between groups were performed by using unpaired or paired Student's t‐test. A two‐tailed P < 0.05 was considered statistically significant.
Results
YTHDC2 expression downregulated in NSCLC tissues and cell lines
To explore the expression of YTHDC2 in NSCLC, we first examined two NSCLC databases from GEPIA and Oncomine and observed significantly low expression of YTHDC2 in LUAD and LUSC patients (Fig 1a,b). We then also evaluated the expression of YTHDC2 by searching three GEO datasets (GSE63459, GSE74706 and GSE32665). The results demonstrated a lower expression of YTHDC2 in NSCLC tumors than normal tissues (Fig 1c–e). To verify the results of bioinformatic analysis, we analyzed YTHDC2 expression in H1299, H460, H292, A549 and HBE cell lines. Lower levels of YTHDC2 were detected in NSCLC cell lines compared with HBE, both in mRNA and protein levels (Fig 1f,g).
Figure 1.
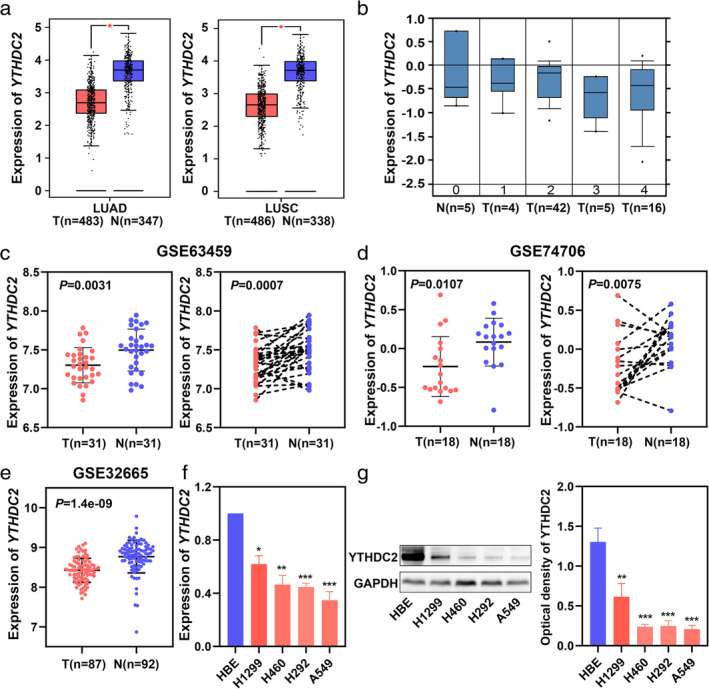
YTHDC2 is downregulated in NSCLC tissues and cell lines. (a) YTHDC2 gene expression analysis based on GEPIA database. (b) YTHDC2 gene expression analysis based on Oncomine database: 0 (no value), 1 (large cell lung carcinoma), 2 (lung adenocarcinoma), 3 (small cell lung carcinoma), and 4 (squamous cell lung carcinoma). (c, d, e) Expression of YTHDC2 in NSCLC tissues and normal tissues from three GEO datasets (GSE63459, GSE74706 and GSE32665). P‐values were calculated using a two‐sided unpaired or paired Student's t‐test. (f) The mRNA expression of YTHDC2 in HBE and different NSCLC cell lines were determined by qPCR. P‐values were calculated using a two‐sided unpaired Student's t‐test. (g) The protein expression of YTHDC2 in HBE and different NSCLC cell lines were determined by western blot. P‐values were calculated using a two‐sided unpaired Student's t‐test.*, P < 0.05; **, P < 0.01; ***, P < 0.001. T, tumor tissues; N, normal tissues.
YTHDC2 expression and clinicopathological variables in NSCLC patients
To further investigate the YTHDC2 protein expression in NSCLC patients, IHC was performed in 96 NSCLC tissues and 31 adjacent normal tissues. YTHDC2 was found to be mainly expressed in the cytoplasm (Fig 2a–f). Compared to adjacent normal tissues, THDC2 expression was decreased in NSCLC tissues (Fig 2a–f). In addition, we investigated the relationship between YTHDC2 expression and clinicopathological variables in NSCLC patients. Low expression of YTHDC2 was found to be significantly associated with poor differentiation, lymph node metastasis, tumor size and stage (Table 1), but was not significantly interrelated with other clinicopathological variables including age, gender and histological type (Table 1).
Figure 2.
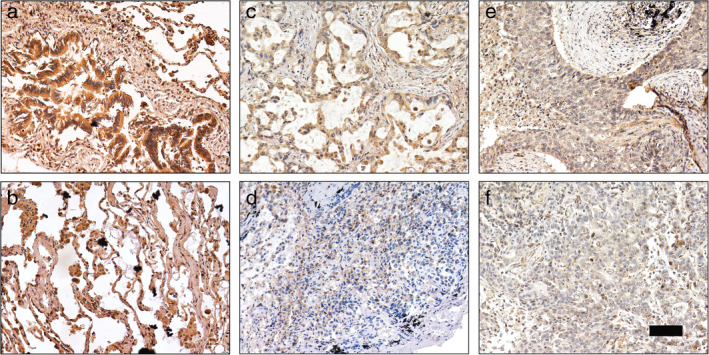
IHC staining of YTHDC2 in NSCLC tissues and adjacent normal lung tissues. (a) Strong staining of YTHDC2 in normal bronchial epithelial cells. (b) Strong staining of YTHDC2 in normal alveolar cells. (c) Moderate staining of YTHDC2 in well‐differentiated lung adenocarcinoma. (d) Weak staining of YTHDC2 in poorly differentiated lung adenocarcinoma. (e) Moderate staining of YTHDC2 in well‐differentiated lung squamous cell carcinoma. (f) Weak staining of YTHDC2 in poorly differentiated lung squamous cell carcinoma. Scale bars, 100 μm.
Table 1.
Clinicopathological variables and YTHDC2 expression in 96 NSCLC patients
| Characteristics | Number | High | Low | χ2 | P‐value |
|---|---|---|---|---|---|
| Age (years) | |||||
| <60 | 39 | 15 | 24 | 0.026 | 0.872 |
| ≧60 | 57 | 21 | 36 | ||
| Gender | |||||
| Male | 63 | 20 | 43 | 2.589 | 0.108 |
| Female | 33 | 16 | 17 | ||
| Differentiation | |||||
| Well | 45 | 25 | 20 | 11.782 | 0.001 |
| Moderate and poor | 51 | 11 | 40 | ||
| Histological type | |||||
| Adenocarcinoma | 55 | 22 | 33 | 0.343 | 0.558 |
| Squamous cell carcinoma | 41 | 14 | 27 | ||
| Tumor size | |||||
| ≤3 cm | 45 | 23 | 22 | 6.696 | 0.010 |
| >3 cm | 51 | 13 | 38 | ||
| Lymph node metastasis | |||||
| Positive | 54 | 15 | 39 | 4.978 | 0.026 |
| Negative | 42 | 21 | 21 | ||
| Tumor stage | |||||
| I + II | 60 | 29 | 31 | 8.012 | 0.005 |
| III | 36 | 7 | 29 | ||
P‐values were calculated using chi‐square test.
Overexpression of YTHDC2 inhibits NSCLC cell proliferation and migration
Because YTHDC2 had been determined to be significantly associated with poor differentiation, lymph node metastasis, tumor size and stage, we next evaluated whether YTHDC2 might be related to tumorigenesis in NSCLC. We selected A549 cells with low YTHDC2 expression for plasmid transfection. We then inhibited YTHDC2 expression with siRNAs (YTHDC2#1, YTHDC2#2) in H1299 cells. As expected, significantly increased or decreased mRNA and protein expression of YTHDC2 was detected by qPCR and western blot analysis (Fig 3a,b). We then performed CCK8 and transwell assays to investigate whether YTHDC2 might be involved in NSCLC cell proliferation and migration. The proliferation and migration ability of A549 cells were significantly decreased after YTHDC2 overexpression. (Fig 3c,e). Conversely, the proliferation and migration rate of H1299 cells were drastically elevated after YTHDC2 knockdown (Fig 3d,e).
Figure 3.
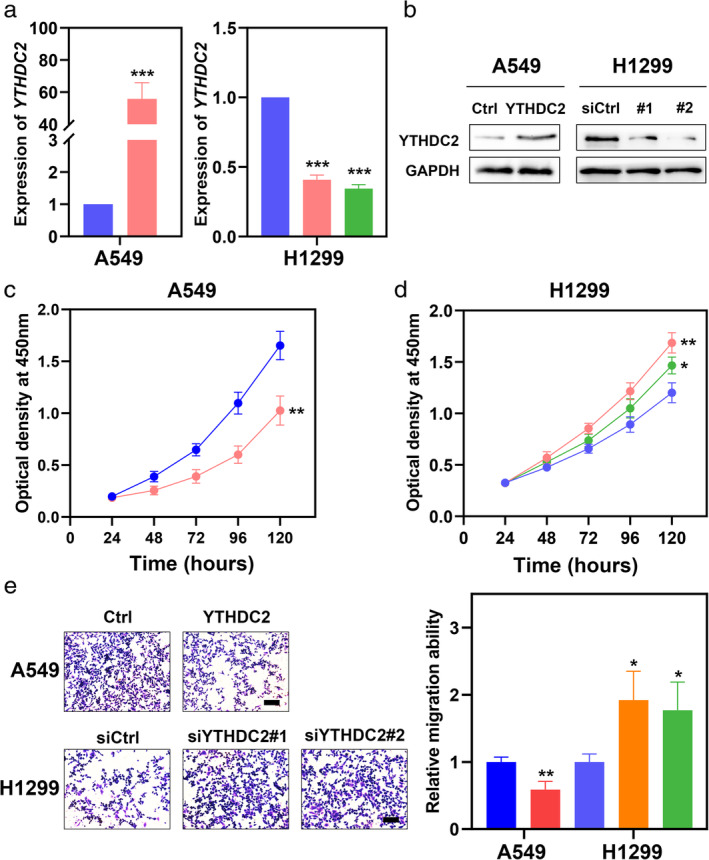
YTHDC2 inhibits proliferation and migration of NSCLC cell lines. (a, b) Empty or YTHDC2 expressing plasmids were transfected into A549 cells ( ) Ctrl (
) Ctrl ( ) YTHDC2 (
) YTHDC2 ( ) siCtrl (
) siCtrl ( ) siYTHDC2#1 (
) siYTHDC2#1 ( ) siYTHDC2#2. Negative control or YTHDC2‐siRNAs (siYTHDC2 #1, #2) were transfected into H1299 cells. The mRNA and protein expression of YTHDC2 was verified by qPCR and western blot. P‐values were calculated using a two‐sided unpaired Student's t‐test. (c, d) CCK8 assay was performed in A549 and H1299 cell lines transfected with YTHDC2 expressing plasmids or YTHDC2‐siRNAs (
) siYTHDC2#2. Negative control or YTHDC2‐siRNAs (siYTHDC2 #1, #2) were transfected into H1299 cells. The mRNA and protein expression of YTHDC2 was verified by qPCR and western blot. P‐values were calculated using a two‐sided unpaired Student's t‐test. (c, d) CCK8 assay was performed in A549 and H1299 cell lines transfected with YTHDC2 expressing plasmids or YTHDC2‐siRNAs ( ) Ctrl (
) Ctrl ( ) YTHDC2 (
) YTHDC2 ( ) siCtrl (
) siCtrl ( ) siYTHDC2#1 (
) siYTHDC2#1 ( ) siYTHDC2#2. P‐values were calculated using a two‐sided unpaired Student's t‐test. (e) Transwell assay was performed in A549 and H1299 cell lines transfected with YTHDC2 expressing plasmids or YTHDC2‐siRNAs (
) siYTHDC2#2. P‐values were calculated using a two‐sided unpaired Student's t‐test. (e) Transwell assay was performed in A549 and H1299 cell lines transfected with YTHDC2 expressing plasmids or YTHDC2‐siRNAs ( ) Ctrl (
) Ctrl ( ) YTHDC2 (
) YTHDC2 ( ) siCtrl (
) siCtrl ( ) siYTHDC2#1 (
) siYTHDC2#1 ( ) siYTHDC2#2. P‐values were calculated using a two‐sided unpaired Student's t‐test. Scale bars, 100 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
) siYTHDC2#2. P‐values were calculated using a two‐sided unpaired Student's t‐test. Scale bars, 100 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Prognostic value of YTHDC2 in lung cancer patients
To determine whether YTHDC2 expression might be associated with overall survival (OS) and post progression survival (PPS) in lung cancer patients, we used bioinformatic methods to analyze the Kaplan‐Meier Plotter database. The results revealed that patients with a low level of YTHDC2 had significantly shorter OS and PPS than YTHDC2 high expression patients (Fig 4a,b). Moreover, low expression of YTHDC2 was correlated with worse OS in LUAD patients, but not in LUSC patients (Fig 4c,d).
Figure 4.
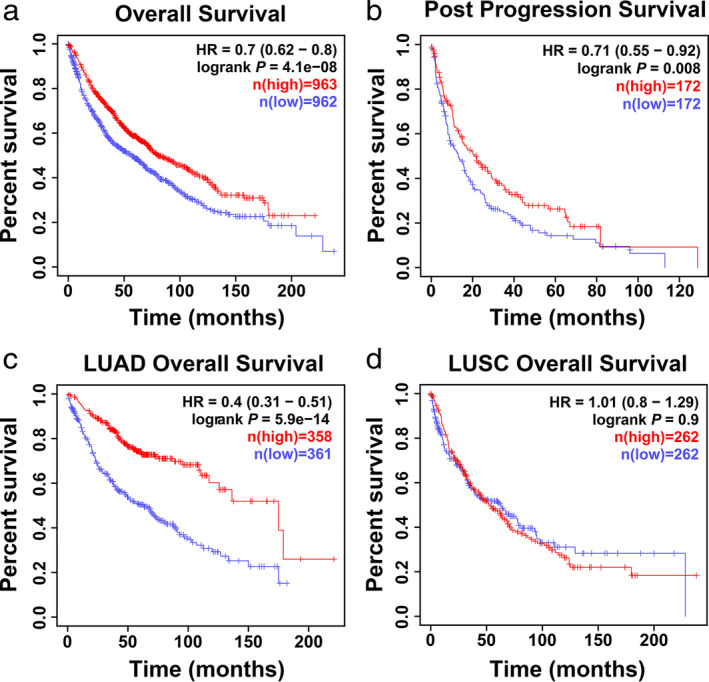
YTHDC2 predicts poor prognosis in lung cancer patients. (a, b) Overall survival and post progression survival analysis of all lung cancer patients from the Kaplan‐Meier Plotter database ( ) low (
) low ( ) high (
) high ( ) low (
) low ( ) high. (c, d) Overall survival analysis of LUAD and LUSC patients from the Kaplan‐Meier Plotter database (
) high. (c, d) Overall survival analysis of LUAD and LUSC patients from the Kaplan‐Meier Plotter database ( ) low (
) low ( ) high (
) high ( ) low (
) low ( ) high.
) high.
Expression and prognostic value of YTHDC2 in multiple types of cancer
The expression and prognostic value of YTHDC2 in other cancer types were also conducted in our study. The GEPIA results revealed that YTHDC2 expression was downregulated in multiple types of cancer, including bladder carcinoma (BLCA), rectal adenocarcinoma (READ), uterine corpus endometrial carcinoma (UCEC) and ovarian cancer (OV) (Fig 5a–d). Moreover, low YTHDC2 expression was significantly associated with worse overall survival among the cancer patients with BLCA, READ, UCEC and OV (Fig 6a–d).
Figure 5.
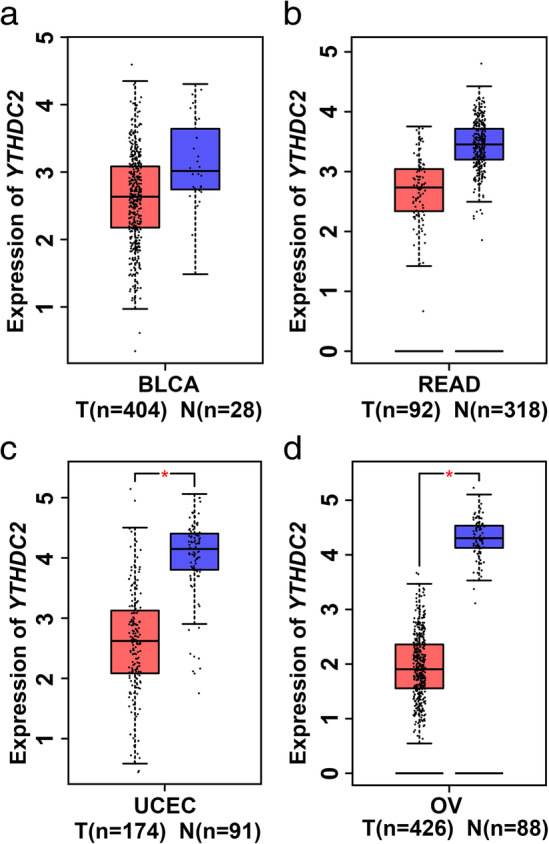
YTHDC2 is downregulated in many cancers. YTHDC2 gene expression analysis based on the GEPIA database. (a) Bladder carcinoma (BLCA). (b) Rectal adenocarcinoma (READ). (c) Uterine corpus endometrial carcinoma (UCEC). (d) Ovarian cancer (OV). T, tumor tissues; N, normal tissues.
Figure 6.
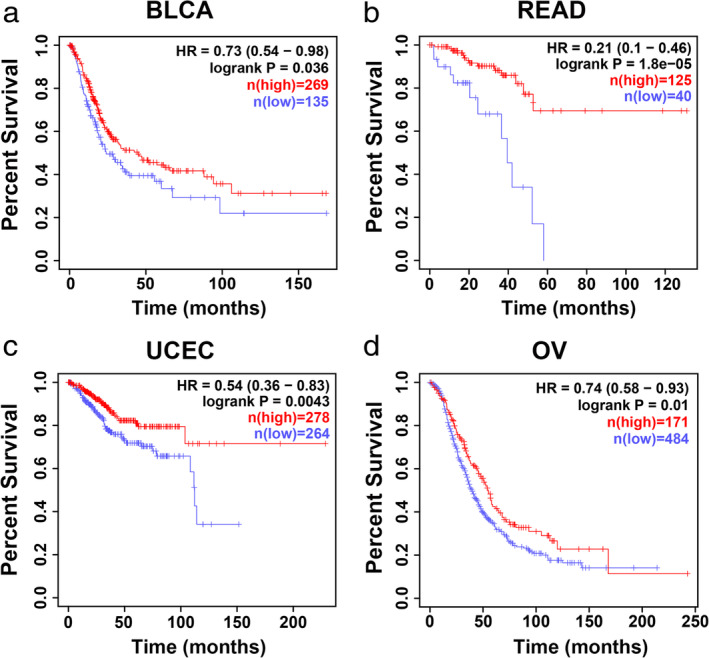
YTHDC2 predicts poor prognosis in multiple types of cancer. Overall survival analysis of cancer patients from the Kaplan‐Meier Plotter database. (a) Bladder carcinoma (BLCA) ( ) low (
) low ( ) high. (b) Rectal adenocarcinoma (READ) (
) high. (b) Rectal adenocarcinoma (READ) ( ) low (
) low ( ) high. (c) Uterine corpus endometrial carcinoma (UCEC) (
) high. (c) Uterine corpus endometrial carcinoma (UCEC) ( ) low (
) low ( ) high. (d) Ovarian cancer (OV) (
) high. (d) Ovarian cancer (OV) ( ) low (
) low ( ) high.
) high.
Functional and pathway enrichment analysis of YTHDC2 targeted RNAs
As an RNA binding protein, YTHDC2 can affect different biological functions by directly binding to different RNAs. We explored the potential target RNAs of YTHDC2 using the POSTAR database. Subsequently, a total of 640 YTHDC2 targeted RNAs were identified (Table S1).GO and KEGG enrichment analysis were then conducted for YTHDC2 targeted RNAs by using the Metascape database. In GO analysis, the target RNAs of YTHDC2 were enriched in nuclear‐transcribed mRNA catabolic process, cadherin binding and translation (Fig 7a and Table S2). KEGG analysis suggested that the target RNAs of YTHDC2 were mostly enriched in ribosome, microRNAs in cancer and transcriptional misregulation in cancer biological pathways (Fig 7b and Table S3).
Figure 7.
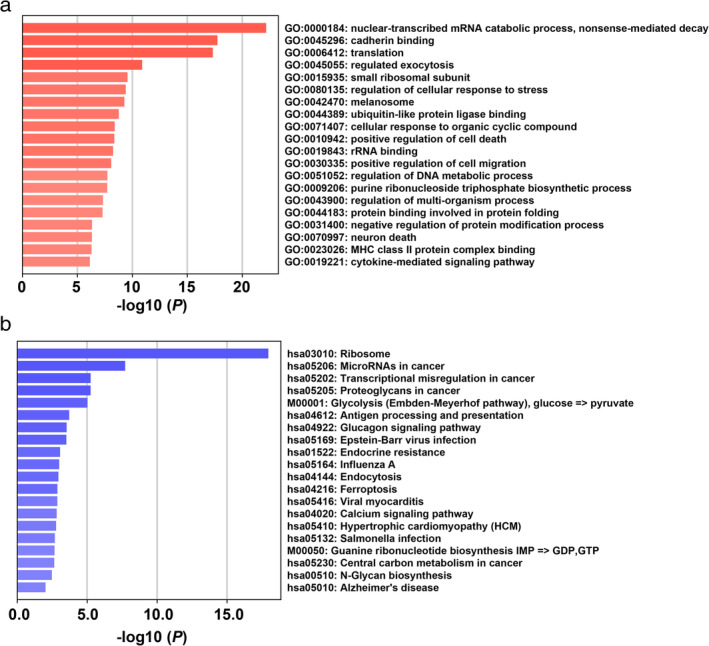
Functional enrichment analysis of YTHDC2 targeted RNAs. (a) GO enrichment results. (b) KEGG pathway enrichment results.
Discussion
Several studies have revealed that m6A modification is involved in different diseases, including pulmonary hypertension, rheumatoid arthritis, type 2 diabetes and acute myeloid leukemia. 17 , 18 , 19 , 20 Moreover, further studies on the biological functions of m6A modification related protein in lung cancer have been reported in recent years. For example, m6A demethylase ALKBH5 (m6A eraser) has been shown to inhibit tumor growth and metastasis by reducing YTHDFs‐mediated YAP expression and inhibit miR‐107/LATS2‐mediated YAP activity in NSCLC. 21 Another m6A methyladenosine METTL3 (m6A writer) induced miR‐143‐3p has been reported to promote the brain metastasis of lung cancer via regulation of VASH1. 22 In addition, Liu et al. demonstrated that m6A demethylase FTO (m6A eraser) promotes cell proliferation and invasion in lung squamous cell carcinoma by regulating MZF1 expression. 23 However, m6A binding proteins (m6A readers) remain unexplored in NSCLC.
In the present study, we demonstrated that YTHDC2 expression at both mRNA and protein levels were decreased in NSCLC cell lines and tissues, in agreement with the data from multiple databases (GEPIA, Oncomine and GEO). Similarly, Yang et al. reported that YTHDC2 is downregulated in ESCC tumor tissues. 16 Several studies using multiple databases have revealed low expression of YTHDC2 in LUA. 24 , 25 Compared with published papers, we explored the expression of YTHDC2 in NSCLC tissues and cell lines. In contrast, another study reported that YTHDC2 was highly expressed in human colon cancer. 26 Therefore, the expression of YTHDC2 in various types of cancer is different. We intend to explore this mechanism in future studies. Our functional experiments suggested that YTHDC2 could inhibit NSCLC proliferation and migration, which is different to the data reported by Tanabe et al. 27 However, our results are consistent with the findings in ESCC. 16 Finally, the prognostic value of YTHDC2 in NSCLC was conducted via online database. Consistent with the conclusions in rectal cancer and head and neck squamous cell carcinoma, the Kaplan‐Meier Plotter database validated that low expression of YTHDC2 indicates a poor prognosis in multiple types of cancer, including NSCLC. 28 , 29 However, YTHDC2 is associated with OS in LUAD rather than in LUSC. The reason for this could be that the underlying signaling regulation for YTHDC2‐mediated tumor progression in LUAD and LUSC may be different, and this needs to be explored further.
YTHDC2 expression has been found to be low in lung cancer and different cancer types. Methylation of gene promoters and transcription factor regulation can lead to decreased gene expression. Here, we explored the underlying mechanisms of low expression of YTHDC2 in NSCLC. First, we obtained the 2 Kb sequence of YTHDC2 promoter through the UCSC database (Homo sapiens: chr5:113511694–113 513 693, the human reference version: GRCh38/hg38). By searching online databases, we found one CpG island in the YTHDC2 promoter (Fig S1). In addition, we analyzed the potential transcription factors in YTHDC2 promoters using the JASPAR and UCSC databases. The results showed that there are many potential transcription factors in YTHDC2 promoters, including TBX and Fox family members (Fig S2).
In addition to YTHDC2, other members of m6A readers also play various roles in tumorigenesis. YTHDF1 has been reported to participate in the development of human colorectal cancer. 30 Additionally, YTHDF2 has been found to promote liver cancer stem cell phenotypes and cancer metastasis by modulating the m6A methylation of OCT4 mRNA. 31 Aberrant overexpression of YTHDC1 in colon adenocarcinoma has also been previously reported. 32 Our findings and these results suggest that m6A readers may be a potential therapeutic target.
There are several deficiencies in the present study. First, survival analysis was not performed due to the short follow‐up time of patients included in the study. Second, the detailed mechanism of how YTHDC2 participates in the biological function of NSCLC remain unknown, and further research is needed to explore its mechanism. RNA binding proteins (RBPs) bind their target RNAs through one or more RNA‐binding domains and change the function of the bound RNAs. 33 We intend to explore the potential RNAs which may interact with YTHDC2 in future studies.
In summary, this study revealed that YTHDC2 expression was decreased in NSCLC tissues and cell lines. Gain and loss of function assays demonstrated that YTHDC2 could suppress the proliferation and migration ability of NSCLC cells. Moreover, YTHDC2 might be a remarkable prognostic marker for NSCLC. Finally, functional enrichment analysis of YTHDC2 targeted RNAs indicated several enriched pathways related to cancer. This study suggests that YTHDC2 might be a potential therapeutic target for NSCLC.
Disclosure
The authors declare no competing interests.
Supporting information
Figure S1 The CpG island in promoter of YTHDC2.
Figure S2 The potential transcription factors in promoter of YTHDC2.
Table S1 The detailed list of YTHDC2 targeted RNAs.
Table S2 Gene ontology (GO) enrichment analysis of 640 YTHDC2 targeted RNAs.
Table S3 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis of 640 YTHDC2 targeted RNAs.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81970084 and 81902986) and the General Hospital of Tianjin Medical University Youth Incubation Foundation (Grant No. ZYYFY2018030).
Contributor Information
Shulei Sun, Email: slsun@tmu.edu.cn.
Jie Cao, Email: tmucaojie@163.com.
References
- 1. Chen W, Zheng R, Baade PD et al Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115–32. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70: 7–30. [DOI] [PubMed] [Google Scholar]
- 3. Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019; 16: 341–55. [DOI] [PubMed] [Google Scholar]
- 4. Yuwen D, Ma Y, Wang D et al Prognostic role of circulating exosomal miR‐425‐3p for the response of NSCLC to platinum‐based chemotherapy. Cancer Epidemiol Biomarkers Prev 2019; 28: 163–73. [DOI] [PubMed] [Google Scholar]
- 5. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell 2017; 169: 1187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer 2019; 18: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6‐methyladenosine and gene expression control. Nat Rev Mol Cell Biol 2014; 15: 313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m6A methylation in cancer. Cancer Res 2019; 79: 1285–92. [DOI] [PubMed] [Google Scholar]
- 9. Zhao Y, Shi Y, Shen H, Xie W. m6A‐binding proteins: The emerging crucial performers in epigenetics. J Hematol Oncol 2020; 13: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen M, Wong CM. The emerging roles of N6‐methyladenosine (m6A) deregulation in liver carcinogenesis. Mol Cancer 2020; 19: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang X, Lu Z, Gomez A et al N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature 2014; 505: 117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi H, Wang X, Lu Z et al YTHDF3 facilitates translation and decay of N6‐methyladenosine‐modified RNA. Cell Res 2017; 27: 315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bailey AS, Batista PJ, Gold RS et al The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife 2017; 6: e26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou B, Liu C, Xu L et al N6‐methyladenosine reader protein Ythdc2 suppresses liver steatosis via regulation of mRNA stability of lipogenic genes. Hepatology 2020. 10.1002/hep.31220. [DOI] [PubMed] [Google Scholar]
- 15. Fanale D, Iovanna JL, Calvo EL et al Germline copy number variation in the YTHDC2 gene: Does it have a role in finding a novel potential molecular target involved in pancreatic adenocarcinoma susceptibility? Expert Opin Ther Targets 2014; 18: 841–50. [DOI] [PubMed] [Google Scholar]
- 16. Yang N, Ying P, Tian J et al Genetic variants in m6A modification genes are associated with esophageal squamous‐cell carcinoma in the Chinese population. Carcinogenesis 2020; 41: 761–8. [DOI] [PubMed] [Google Scholar]
- 17. Su H, Wang G, Wu L, Ma X, Ying K, Zhang R. Transcriptome‐wide map of m6A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genomics 2020; 21: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mo XB, Zhang YH, Lei SF. Genome‐wide identification of N6‐methyladenosine (m6A) SNPs associated with rheumatoid arthritis. Front Genet 2018; 9: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Jesus DF, Zhang Z, Kahraman S et al m6A mRNA methylation regulates human beta‐cell biology in physiological states and in type 2 diabetes. Nat Metab 2019; 1: 765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paris J, Morgan M, Campos J et al Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 2019; 25: 137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jin D, Guo J, Wu Y et al m6A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs‐mediated YAP expression and inhibiting miR‐107/LATS2‐mediated YAP activity in NSCLC. Mol Cancer 2020; 19: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang H, Deng Q, Lv Z et al N6‐methyladenosine induced miR‐143‐3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer 2019; 18: 181. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Liu J, Ren D, Du Z et al m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun 2018; 502: 456–64. [DOI] [PubMed] [Google Scholar]
- 24. Zhuang Z, Chen L, Mao Y et al Diagnostic, progressive and prognostic performance of m6A methylation RNA regulators in lung adenocarcinoma. Int J Biol Sci 2020; 16: 1785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu J, Wang M, Hu D. Deciphering N6‐methyladenosine‐related genes signature to predict survival in lung adenocarcinoma. Biomed Res Int 2020; 2020: 2514230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanabe A, Tanikawa K, Tsunetomi M et al RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF‐1 alpha mRNA is translated. Cancer Lett 2016; 376: 34–42. [DOI] [PubMed] [Google Scholar]
- 27. Tanabe A, Konno J, Tanikawa K, Sahara H. Transcriptional machinery of TNF‐alpha‐inducible YTH domain containing 2 (YTHDC2) gene. Gene 2014; 535: 24–32. [DOI] [PubMed] [Google Scholar]
- 28. Zhuang J, Lin C, Ye J. m6A RNA methylation regulators contribute to malignant progression in rectal cancer. J Cell Physiol 2020; 235: 6300–6. [DOI] [PubMed] [Google Scholar]
- 29. Zhao X, Cui L. Development and validation of a m6A RNA methylation regulators‐based signature for predicting the prognosis of head and neck squamous cell carcinoma. Am J Cancer Res 2019; 9: 2156–69. [PMC free article] [PubMed] [Google Scholar]
- 30. Bai Y, Yang C, Wu R et al YTHDF1 regulates tumorigenicity and cancer stem cell‐like activity in human colorectal carcinoma. Front Oncol 2019; 9: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang C, Huang S, Zhuang H et al YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 2020; 39: 4507–18. [DOI] [PubMed] [Google Scholar]
- 32. Liu X, Liu L, Dong Z et al Expression patterns and prognostic value of m6A‐related genes in colorectal cancer. Am J Transl Res 2019; 11: 3972–91. [PMC free article] [PubMed] [Google Scholar]
- 33. Lunde BM, Moore C, Varani G. RNA‐binding proteins: Modular design for efficient function. Nat Rev Mol Cell Biol 2007; 8: 479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The CpG island in promoter of YTHDC2.
Figure S2 The potential transcription factors in promoter of YTHDC2.
Table S1 The detailed list of YTHDC2 targeted RNAs.
Table S2 Gene ontology (GO) enrichment analysis of 640 YTHDC2 targeted RNAs.
Table S3 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis of 640 YTHDC2 targeted RNAs.


