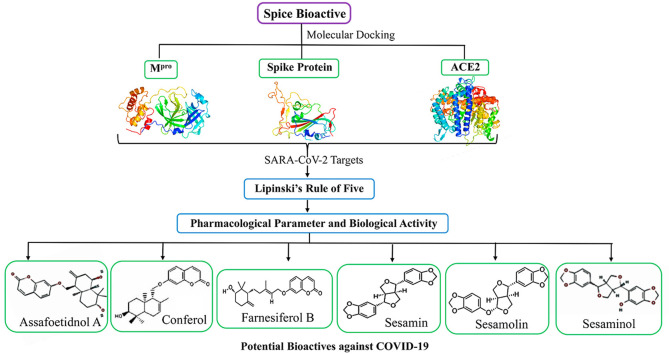
Abstract
Background
Coronavirus disease-2019 (COVID-19) is an infectious pandemic caused by SARS-CoV-2. SARS-CoV-2 main protease (Mpro) and spike protein are crucial for viral replication and transmission. Spike protein recognizes the human ACE2 receptor and transmits SARS-CoV-2 into the human body. Thus, Mpro, spike protein, and ACE2 receptor act as appropriate targets for the development of therapeutics against SARS-CoV-2. Spices are traditionally known to have anti-viral and immune-boosting activities. Therefore, we investigated the possible use of selected spice bioactives against the potential targets of SARS-CoV-2 using computational analysis.
Methods
Molecular docking analysis was performed to analyze the binding efficiency of spice bioactives against SARS-CoV-2 target proteins along with the standard drugs. Drug-likeness properties of selected spice bioactives were investigated using Lipinski's rule of five and the SWISSADME database. Pharmacological properties such as ADME/T, biological functions, and toxicity were analyzed using ADMETlab, PASS-prediction, and ProTox-II servers, respectively.
Results
Out of forty-six spice bioactives screened, six bioactives have shown relatively better binding energies than the standard drugs and have a higher binding affinity with at least more than two targets of SARS-CoV-2. The selected bioactives were analyzed for their binding similarities with the standard drug, remdesivir, towards the targets of SARS-CoV-2. Selected spice bioactives have shown potential drug-likeness properties, with higher GI absorption rate, lower toxicity with pleiotropic biological roles.
Conclusions
Spice bioactives have the potential to bind with the specific targets involved in SARS-CoV-2 infection and transmission. Therefore, spice-based nutraceuticals can be developed for the prevention and treatment of COVID-19.
Keywords: COVID-19, Drug-likeness, Molecular docking, Nutraceuticals, SARS-CoV-2, Spices
Graphical abstract
Highlights
-
•
The SARS-CoV-2 Mpro, spike protein, and human ACE2 are critical targets for COVID-19.
-
•
Spice bioactives from F. asafoetida and S. indicum potentially inhibit the SARS-CoV-2 targets.
-
•
Selected bioactives have drug-likeness properties along with optimal pharmacological and biological activities.
-
•
The development of spice-based nutraceuticals can be used for the prevention and treatment of COVID-19.
1. Introduction
Coronavirus disease-2019 (COVID-19) is causing severe infection and mortality around the world for the past ten months. According to WHO, more than 36 million are infected globally by this pandemic with casualties of more than one million lives by the mid of October 2020. Clinical characteristics of COVID-19 include asymptomatic to severe pneumonia and causing vital organ damages. The common COVID-19 symptoms are fever, cough, headache, fatigue, shortness of breath, muscle pain and sputum production, etc. [1]. The other characteristics of COVID-19 include hyperinflammatory response mediated by cytokine storm, intense lymphopenia, as well as considerable mononuclear cell infiltration into the various organs [2]. Currently, no specific medications are available to treat SARS-CoV-2-mediated disease [3]. Repurposing of anti-viral, anti-malarial, and angiotensin-converting enzyme 2 (ACE2) inhibitors are in progress. Recently, remdesivir, an anti-viral drug, has shown a glimpse of promise by shortening the recovery time of COVID-19 patients and evidenced to lower respiratory tract infection [4]. However, around 24.6% of patients who received remdesivir were reported with serious adverse events, including acute respiratory failure. In contrast, the most anticipated drug, hydroxychloroquine, has not lowered the mortality of COVID-19 patients who received it than the patients who received normal care [5]. Besides, many drugs are under clinical trials and unfortunately, none of the drugs has passed through the clinical trials, and the battle for vaccine development is in progress.
It is critical to understand the receptor-recognition mechanism and the viral biogenesis of COVID-19 to tackle its infection and pathogenesis. The primary infection of SARS-CoV-2 is facilitated by its binding with host cell receptors through an envelope-anchored spike protein [6]. The spike protein of SARS-CoV-2 comprises S1 and S2 domains; S1 is the receptor-binding domain, and S2 is the membrane fusion protein [7,8]. About 73% of spike protein has conserved to SARS-CoV; however, the SARS-CoV-2 spike protein has ten times higher affinity with the host cell binding receptor [7,9]. ACE2 is the receptor, wherein spike protein binds and infects the host cell. Molecular analysis of spike protein receptor binding motif (RBM)-human ACE2 complex determines various key residues of spike protein and hotspots residues of the ACE2 receptors involved in viral-host receptor interaction. The ACE2 receptor contains two viral-binding hotspots: Lys31 (hotspot 31) and Lys353 (hotspot 353). Hotspot 31 composed of a salt bridge between Lys31 and Glu35, and hotspot 353 consisting of a salt bridge between Lys353 and Asp38 concealed in a hydrophobic environment [[6], [7], [8]]. The critical amino acid residues of spike protein include Leu455, Phe486, Gln493, and Ser494 bind to ACE2.
Additionally, SARS-CoV-2 main protease (Mpro) is an essential protein involved in proteolytic maturation of pp1a and pp1ab polyproteins into non-structural protein (NSPs), which are involved in the replication and transcription of COVID-19. Mpro is a cysteine protease with Cys-His catalytic dyad situated in the cleft between domains I and II [10]. The functional role of Mpro and the attenuation of interaction between spike protein and ACE2 make these proteins attractive targets for drug development against COVID-19.
Natural compounds have served as a pioneer in drug development from ancient times. Many drugs and nutraceuticals have originated from natural sources such as herbs, medicinal plants, and spices. Recently, many computational studies have reported the potential bioactives from these natural sources against the targets of SARS-CoV-2 [[11], [12], [13], [14]]. Spices are not only liked for the taste and aroma but also used to treat various diseases [15]. Spice bioactives are known for their health benefits such as antioxidant, anti-inflammatory, and anti-microbial activities [[16], [17], [18]]. Curcumin, one of the most explored bioactive from turmeric, has shown anti-viral activity against many viruses, including hepatitis, and influenza viruses [19]. Furthermore, Wen et al., 2007 have reported curcumin inhibits the SARS-CoV replication and 3CL protease activity in vitro [20]. Recently, Rout et al., 2020 found that spice bioactive piperine has shown a higher binding affinity with the SARS-CoV-2 spike protein receptor-binding domain (RBD) and Mpro than few of the currently used drugs [21].
More recently, Elsayed and Khan, 2020 have correlated the prevalence of COVID-19 and the consumption of spices in 163 countries. Intriguingly, the nations with lower consumptions of spices per capita showed a higher number of COVID-19 cases as well as mortality per million populations. Further, the recovery rate was also higher in nations with higher consumption of spices [22]. The beneficial effect of spice bioactives might be due to their immunomodulatory and anti-inflammatory potential observed against various diseases [23]. The severity of COVID-19 escalates mostly because of hyperinflammatory response mediated by cytokine storm, leading to acute respiratory distress syndrome [2,24]. Spice bioactives are known to abrogate pro-inflammatory cytokines and have the potential to reduce respiratory distress [25,26]. Therefore, in our present study, we have investigated the potential therapeutic ability of spice bioactives against SARS-CoV-2 targets of Mpro, spike protein, and human ACE2 receptor by molecular modeling analysis. We also analyzed the drug-likeness and pharmacological characteristics of the potential spice bioactives for the development of therapeutics against SARS-CoV-2.
2. Materials and methods
2.1. Ligand preparation
The structures of standard drugs and spice ligand molecules were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Approved medicines for the treatment of HIV (darunavir, remdesivir, and ritonavir), ACE2 (losartan, valsartan) inhibitors were selected as standards for docking analysis against SARS-CoV-2 target proteins (Supplementary Table S1). The standard drugs were selected based on the existing anti-viral activity and ongoing trials against SARS-CoV-2. Based on the literature evidence, forty-six bioactives from twenty-eight spices were selected for docking analysis (Supplementary Table S2) [17,18,25]. Ligand molecules were converted into Protein Data Bank (PDB) format using Open Babel software (The Open Babel Package, version 2.3.1; http://openbabel.org) [27]. Further, ligand molecules were prepared for docking by converting it into PDBQT format using AutoDock Tools (ADT).
2.2. Protein preparation
The main therapeutic targets of COVID-19 subjected to the investigation were SARS-CoV-2 Mpro, spike protein, and human ACE2 receptor. The three-dimensional coordinates of the target molecules were retrieved from PDB with PDB IDs 6LU7 (Mpro), 6W41 (spike protein) and, 1R42 (ACE2) [10,[28], [29], [30]]. The water molecules available in the PDB were deleted, and polar hydrogens were added, followed by stabilizing the charges [9]. Finally, the 3D coordinates were converted into PDBQT format using ADT [31].
2.3. Molecular docking
SARS-CoV-2 Mpro, spike protein, and human ACE2 were used as the target molecules for docking with the spice bioactive ligands, and the molecular docking study was performed using AutoDock Vina [32]. Grid boxes were generated covering the catalytic site of SARS-CoV-2 Mpro and the interacting critical residues of the SARS-CoV-2 spike protein-human ACE2 complex using ADT with dimensions relative to the ligands (XYZ) with a resolution of 1 Å. Each docking calculations were repeated three times using different seeds and retaining the remaining values as default. The binding energy for each ligand-receptor docked complexes was obtained from the AutoDock Vina. Final protein-ligand interactive models were chosen based on the binding energy as well as the potential hydrogen bonds (H-bonds) and hydrophobic contacts. Protein-ligand interaction profiler (PLIP) (https://projects.biotec.tu-dresden.de/plip-web/plip/) was used to calculate H-bonds and non-bonded interactions. 3D stereo figures of protein-ligand interactions were rendered using PyMOL (https://pymol.org/2).
2.4. Drug-likeness properties
Lipinski's rule of five (RO5) is one of the significant parameters in drug discovery. The best interacting ligand molecules were subjected to Lipinski's rule (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp) [33]. Drug-likeness, lipophilicity, and solubility characteristics of spice bioactives were analyzed using OSIRIS Property Explorer (https://www.organic-chemistry.org/prog/peo/). SWISS-ADME server (http://www.swissadme.ch/) was used to evaluate the Ghose, Veber, Egan, and Muegge rules.
2.5. ADME/toxicity prediction
The ADME/T provides absorption, distribution, metabolism, excretion, and toxicity of the given compounds. The pharmacokinetics and pharmacodynamics properties of the selected spice bioactives were evaluated by the ADMETlab server (http://admet.scbdd.com). The acquired categorical and numerical values were transformed into qualitative units based on ADMETlab server explanation and interpretation.
2.6. Evaluation of biological activity
The biological activities of the selected spice bioactives were evaluated through Prediction of Activity Spectra for Substances (PASS)-Way2Drug server (http://www.pharmaexpert.ru/passonline/). The potentially active or inactive ligands were predicted through Pa and Pi values. The potential active compound should have Pa values close to one and Pi values close to zero.
2.7. Prediction of toxicity
The toxicity parameters of selected spice bioactives were predicted through the ProTox-II server (http://tox.charite.de/protox_II/) and OSIRIS Property Explorer (https://www.organic-chemistry.org/prog/peo) [34].
3. Results and discussion
3.1. Molecular docking analysis
Molecular docking is a versatile tool to study the interaction between a small molecule and a protein at the atomic level, predict the binding efficiency, types of interactions between the ligand and receptors [35,36]. Interventions of COVID-19 require a suitable target to develop drug molecules. The biological functions of Mpro, spike protein, and ACE2 receptors have qualified to be the appropriate targets to develop drugs against COVID-19. Spice bioactives are well known for their anti-viral, anti-inflammatory, and immune-boosting activities [16]. Here, we have investigated the binding efficiency of spice bioactives with SARS-CoV-2 Mpro, spike protein, and human ACE2 receptors using computational analysis. Initially, we have screened the binding affinity of various standard drugs of HIV and ACE2 inhibitors with target proteins (Supplementary Table S1). Among the standard drugs, remdesivir, an anti-retroviral and protease inhibitor, has shown a higher affinity towards SARS-CoV-2 Mpro and human ACE2 receptor with the binding energies (BEs) of −8.3 kcal/mol and −6.4 kcal/mol, respectively. Whereas, another anti-retroviral drug ritonavir has shown a strong binding affinity towards SARS-CoV-2 spike protein with a BE of −7.4 kcal/mol. Similar to standard drugs, several spice bioactives have demonstrated a higher binding affinity with SARS-CoV-2 target proteins (Supplementary Table S2). Sesamin, a bioactive from sesame plant, has shown the higher BE of −8.2 kcal/mol with Mpro, while conferol from asafoetida has shown the higher BEs with spike protein (−7.4 kcal/mol) and ACE2 receptor (−7.1 kcal/mol). Based on the average BEs of the standard drugs, BEs of −7.5 kcal/mol for Mpro, −7.0 kcal/mol for spike protein, and −6.5 kcal/mol for ACE2 are set as an optimal threshold to narrow down the potential spice bioactives. The potential bioactives are selected for further analysis that satisfy (i) the threshold BE, and (ii) having a superior binding affinity against at least two targets of SARS-CoV-2.
Based on the above two conditions, six spice bioactives were selected out of forty-six bioactives. The selected bioactives (assafoetidnol A, conferol, farnesiferol B, sesamin, sesaminol, and sesamolin) are studied for their chemical interactions with the binding sites and pharmacological characteristics (Table 1 ). Interestingly, all these bioactives are predominantly from asafoetida (Ferula asafoetida) and sesame seed (Sesamum indicum). Traditionally, F. asafoetida and S. indicum have anti-microbial, especially anti-viral activities [37,38]. F. asafoetida is known to use for the treatment of respiratory problems, including bronchitis, asthma, and whooping coughs [39]. Additionally, these spice bioactives have exhibited several beneficial activities, including antifungal, antioxidant, hepatoprotective, anti-inflammatory, and have immune-boosting properties [16,[38], [39], [40], [41], [42]].
Table 1.
Binding energies (BEs) (kcal/mol) of selected bioactives from the spices with of SARS-CoV-2 Mpro, spike protein, and human ACE2 and their molecular interactions. The name of the bioactives are highlighted in boldface, and the first three bioactives are belonging to asafoetida, and the last three bioactives are belonging to sesame seed. * represents standard drug.
| Spices and its bioactives | SARS-CoV-2 Mpro |
SARS-CoV-2 spike protein |
Human ACE2 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| BE | H-Bond | Hydrophobic interactions | BE | H-Bond | Hydrophobic interactions | BE | H-Bond | Hydrophobic interactions | |
| *Remdesivir | −8.3 | Leu141, Gly143, His163, Glu166 | Thr25, Thr26, Leu27, His41, Met 49, Met165, Glu166, Gln189 | −6.6 | Arg403, Tyr453, Gln493, Gly 496, Asn501 | Leu455, Phe456, Tyr489 | −6.4 | His34 | Asn33, His34, Glu37, Asp38, Tyr41, Lys353 |
|
F.asafoetida Assafoetidnol A |
−7.4 | Tyr54 | Thr25, Leu27, His41, Asn142, Cys145, Met165, Gln189 | −7.0 | Phe490, Ser494 | Phe456, Tyr489, Gln493 | −6.9 | Asp38 | Asp30, His34, Tyr41, Lys353 |
|
F. asafoetida Conferol |
−7.6 | His163 | Thr25, Leu27, His41, Cys145, Met165, Glu166, Gln189 | −7.4 | – | Leu452, Leu455, Phe456, Tyr489, Phe490, Ser494 | −7.1 | Asp38 | His34, Glu37, Lys353 |
|
F. asafoetida Farnesiferol B |
−7.2 | Thr26, Asp 187 | Thr25, His41, Tyr54, Asn142, Cys145, Met165, Gln189 | −7.0 | Arg403, Gln493 | Lys417, Tyr453, Leu 455, Phe456, Tyr489 | −6.4 | Arg 393 | Asn33, His34, Glu37, Tyr41, Lys353 |
|
S. indicum Sesamin |
−8.2 | Gly143, His163 |
Thr25, His41, Phe140, Cys145, Glu166 | −7.2 | Gln493 | Leu455, Phe456, Tyr489, Phe490 | −6.4 | – | His34, Glu37, Asp38, Tyr41, Lys353 |
|
S. indicum Sesaminol |
−7.8 | Thr26, His41, Gly143, His163, Met165 |
Thr25, Phe140, Cys145, Glu166 | −7.0 | Tyr489, Gln493, Ser494 | Phe490 | −6.5 | His34, Asp38 | Glu37, Tyr41, Leu45, Lys353 |
|
S. indicum Sesamolin |
−7.7 | Thr45, Ser 46, Gly143, His163 | Thr25, His41, Cys145, Glu166 | −7.0 | Ser494 | Leu452, Leu455, Phe456, Tyr489, Phe490, Gln493 | −6.5 | Glu37 | His34, Asp38, Tyr41, Lys353 |
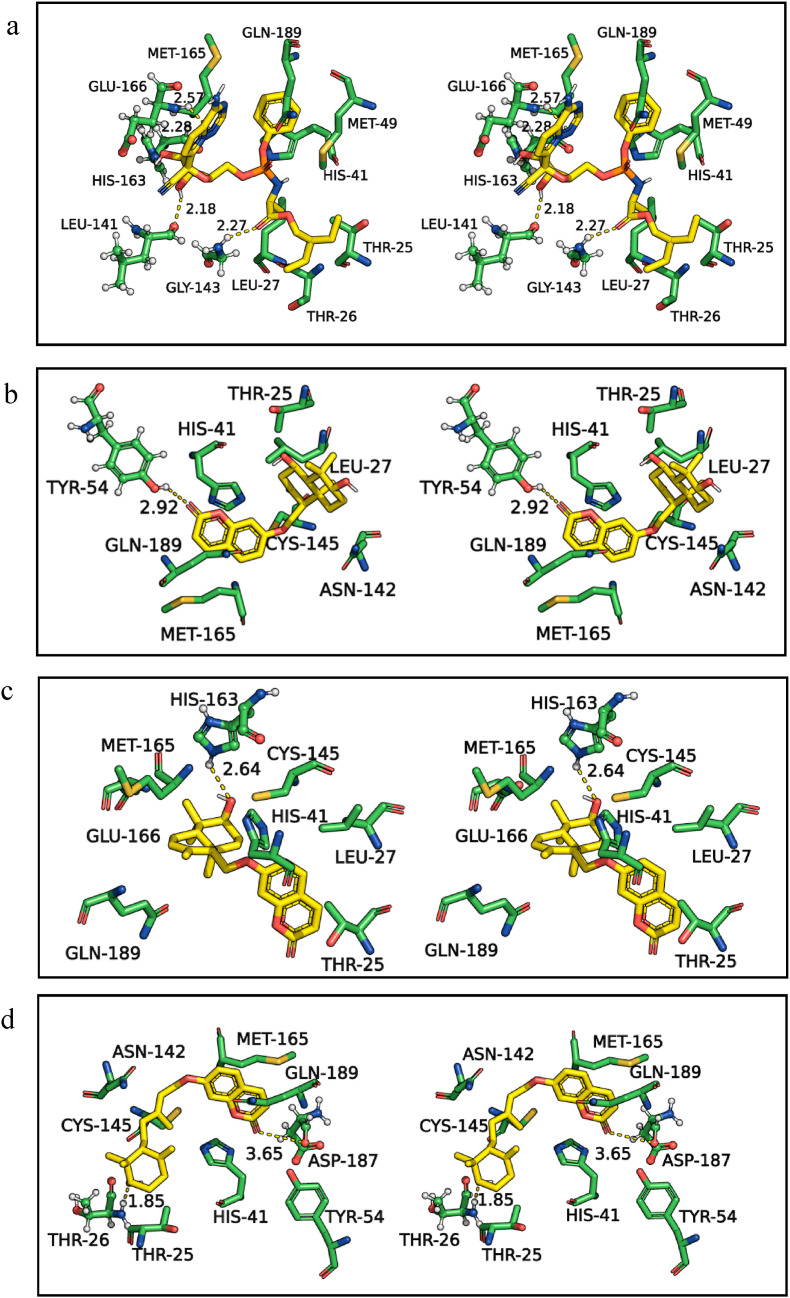
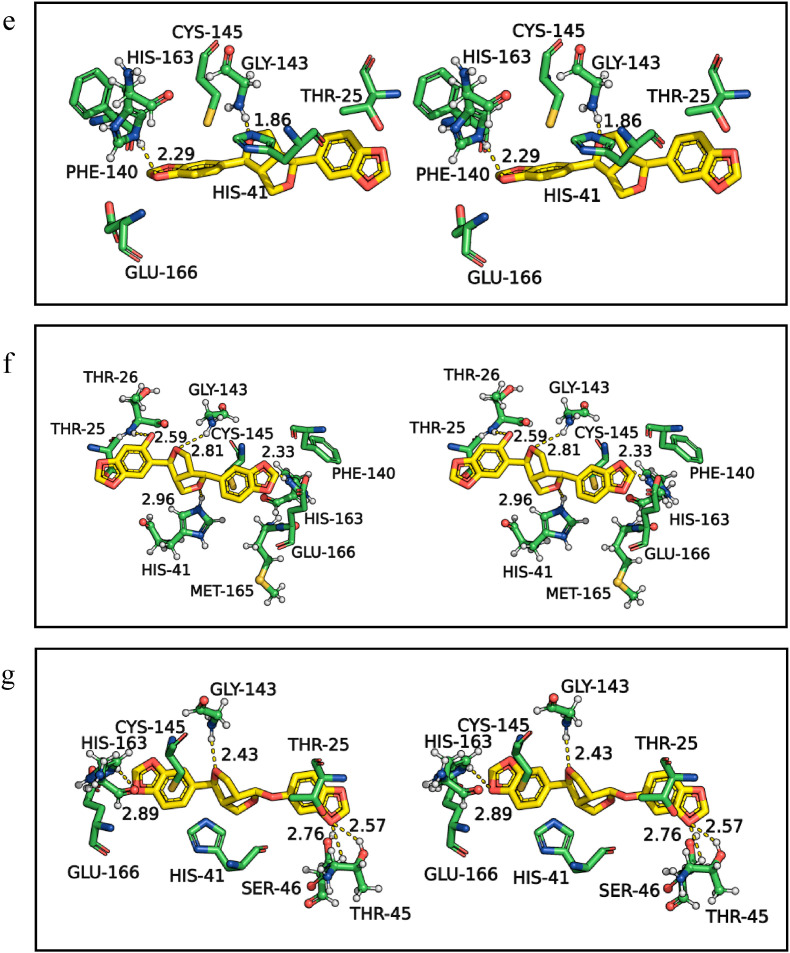
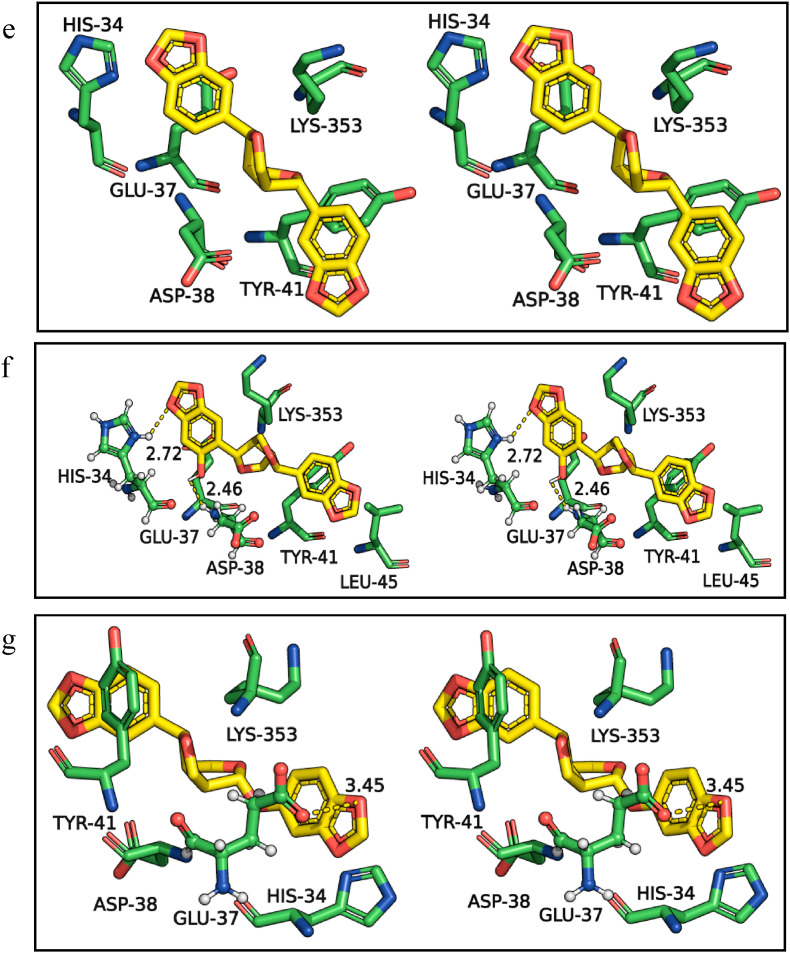
Further, the similarity in the binding efficiency of selected spice bioactives is compared with the standard drug remdesivir, which showed higher BEs with the targets of SARS-CoV-2 (Table 1 and Fig. 1 ). The H-bonds and hydrophobic interactions of remdesivir (Fig. 1a) with Mpro are compared with the spice bioactives assafoetidnol A (Fig. 1b), conferol (Fig. 1c), farnesiferol B (Fig. 1d), sesamin (Fig. 1e), sesaminol (Fig. 1f), and sesamolin (Fig. 1g). The remdesivir formed four H-bonds with Mpro residues Leu141, Gly143, His163, and Glu166, in addition to several hydrophobic interactions at the catalytic site (Fig. 1a, and Table 1). Among the selected spice bioactives, sesamin has shown the highest binding affinity with Mpro. The sesamin formed two H-bonds with Gly143 and His163, as well as several hydrophobic interactions at the catalytic site (Fig. 1e). Overall the amino acid residues of Mpro Thr25, His41, Gly143, Cys145, His163, Glu166, and Gln189 showed frequent interactions with the standard drugs as well as with spice bioactives (Table 1, Supplementary Table S1). The His41 of Mpro is also involved in salt bridge and π interaction with assafoetidnol A, and farnesiferol B. Thus, the results suggest that the spice bioactives interact with the key residues of Mpro as similar to the standard drugs (Fig. 1, Table 1, and Supplementary Table S1).
Fig. 1.
Three-dimensional stereo figures representing molecular interactions of SARS-CoV-2 Mpro (PDB ID: 6LU7) with standard drug remdesivir (a) and spice bioactives assafoetidnol A (b), conferol (c), farnesiferol B (d), sesamin (e), sesaminol (f), sesamolin (g). The amino acids of the Mpro within 4 Å proximity are shown. The H-bonds are represented in dotted lines and distances (Å) are marked. Mpro amino acids and spice bioactives are shown as sticks. The carbon atoms are colored in green for Mpro and the spice bioactives carbons are colored in yellow. The nitrogen and oxygen atoms are colored in blue and red, respectively, for all molecules.
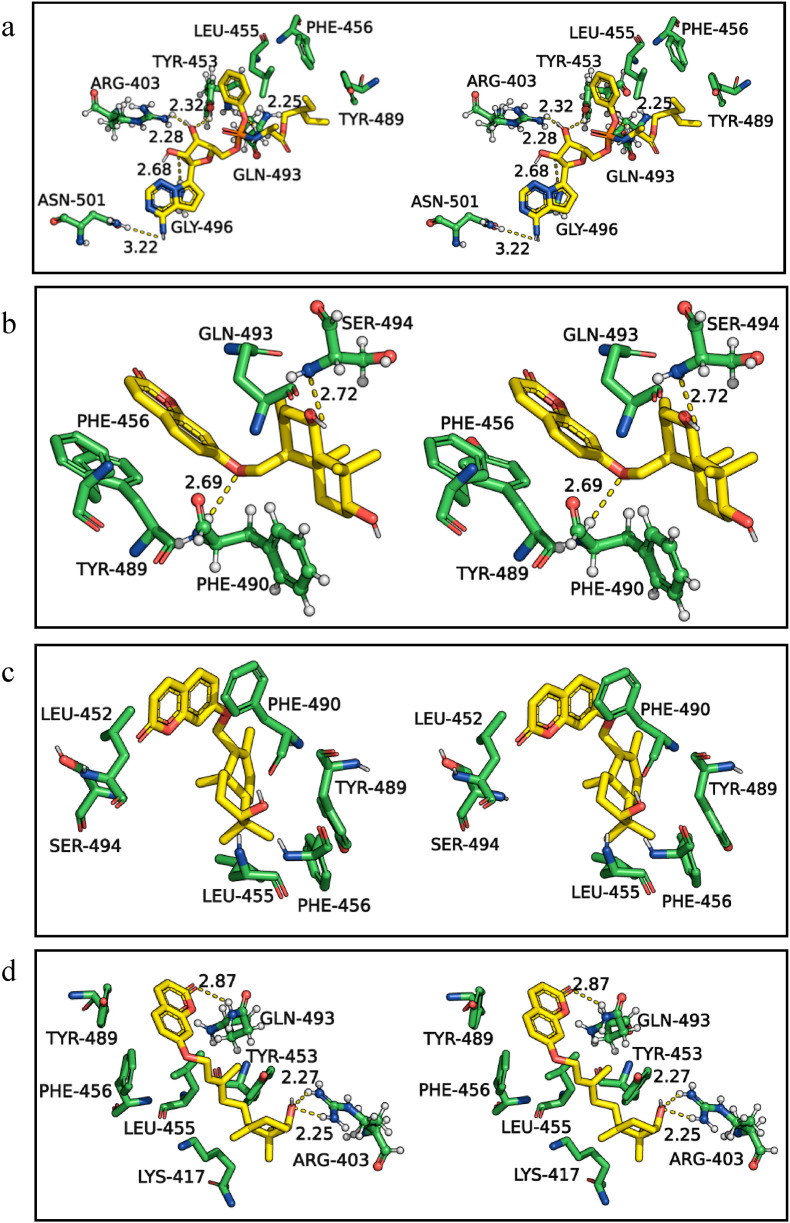
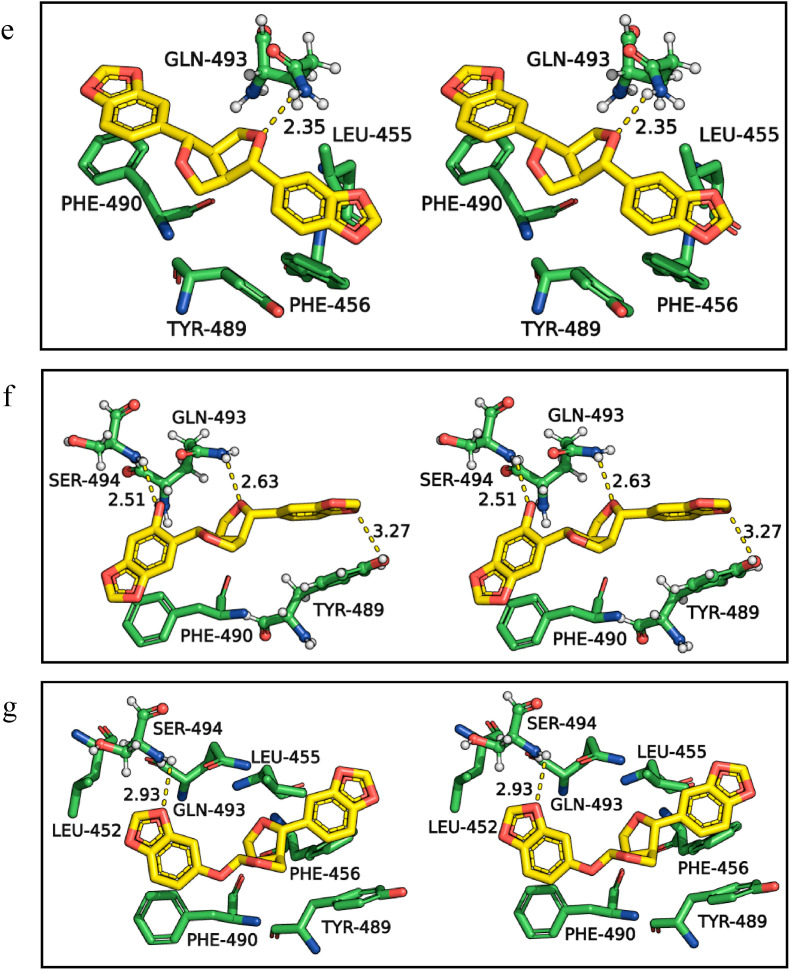
During viral transmission, the residues of the receptor-binding motif of spike protein bind with ACE2 receptor hotspots. The receptor-binding motif of spike protein consists of Leu 455, Phe486, Gln493, and Ser494 residues, which play a crucial role in interacting with the ACE2 receptors [8,28,43,44]. The molecular interactions of SARS-CoV-2 spike protein obtained from the docking studies with remdesivir (Fig. 2 a), and spice bioactives assafoetidnol A (Fig. 2b), conferol (Fig. 2c), farnesiferol B (Fig. 2d), sesamin (Fig. 2e), sesaminol (Fig. 2f), and sesamolin (Fig. 2g) are shown in Fig. 2. Remdesivir formed five H-bonds with spike protein residues Arg403, Tyr453, Gln493, Gln498, and Asn501 and several hydrophobic interactions with the spike protein receptor-binding motif. The amino acid residues Gln493 and Ser494 of spike protein frequently interact with all the spice bioactives through H-bonds except the conferol. However, among the spice bioactives, conferol and sesamolin have hydrophobic interactions with key residues of spike protein such as Leu452, Leu455, Phe456, Tyr489, Phe490, and Gln493 (Fig. 2c and 2g and Table 1). In general, the Leu455, Phe456, Tyr489, and Phe490 of spike protein are the key residues majorly involved in hydrophobic interactions with spice bioactives. Interestingly, Phe490 is also involved in π interactions with assafoetidnol A and farnesiferol B.
Fig. 2.
Three-dimensional stereo figures representing molecular interactions of SARS-CoV-2 spike protein (PDB ID: 6W41) with standard drug remdesivir (a) and spice bioactives assafoetidnol A (b), conferol (c), farnesiferol B (d), sesamin (e), sesaminol (f), sesamolin (g).
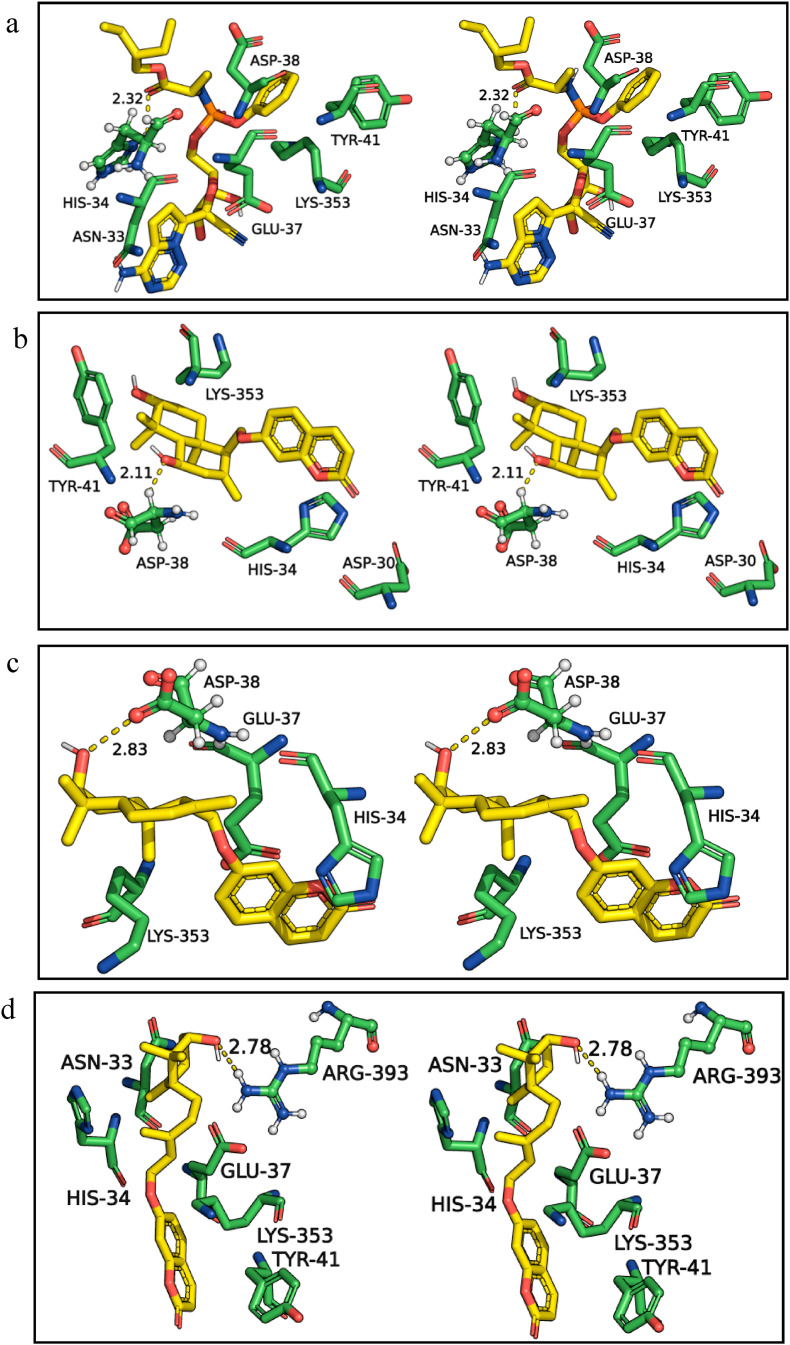
Based on the recent structural and molecular dynamics studies, many important amino acid residues of the ACE2 receptor are recognized in interacting with its partner proteins [8,45]. The human ACE2 receptor contains hotspot Lys31 and hotspot Lys353. The spike protein identifies these hotspots, and their interactions are essential for transmitting viral infection. Our docking results also showed the selected spice bioactives, and the standard drugs have strong interactions with hotspot residues of the ACE2 receptor (Table 1). Fig. 3 summarizes the molecular interactions of human ACE2 receptor with standard drug remdesivir (Fig. 3a) and spice bioactives assafoetidnol A (Fig. 3b), conferol (Fig. 3c), farnesiferol B (Fig. 3d), sesamin (Fig. 3e), sesaminol (Fig. 3f), sesamolin (Fig. 3g). While the standard drugs, darunavir, remdesivir, and ritonavir formed H-bonds with His34, Glu37, the spice bioactives prefer the adjacent residue Asp38 for the H-bond. In terms of hydrophobic interactions both the standard drugs as well as the spice bioactives interact with His34, Glu37, Asp38, Tyr41, and the hotspot residue Lys353 (Fig. 3, and Table 1, Supplementary Table S1). The amino acids His34 (assafoetidnol A, and conferol) and Lys353 (farnesiferol B) also interact with spice bioactives through salt bridges.
Fig. 3.
Three-dimensional stereo figures representing molecular interactions of human ACE2 receptor (PDB ID:1R42 11R42) with standard drug remdesivir (a) and selected spice bioactives assafoetidnol A (b), conferol (c), farnesiferol B (d), sesamin (e), sesaminol (f), sesamolin (g).
Overall, the molecular docking results suggest that the chosen spice bioactives have a strong affinity towards the Mpro and may hinder the interaction between spike protein and ACE2.
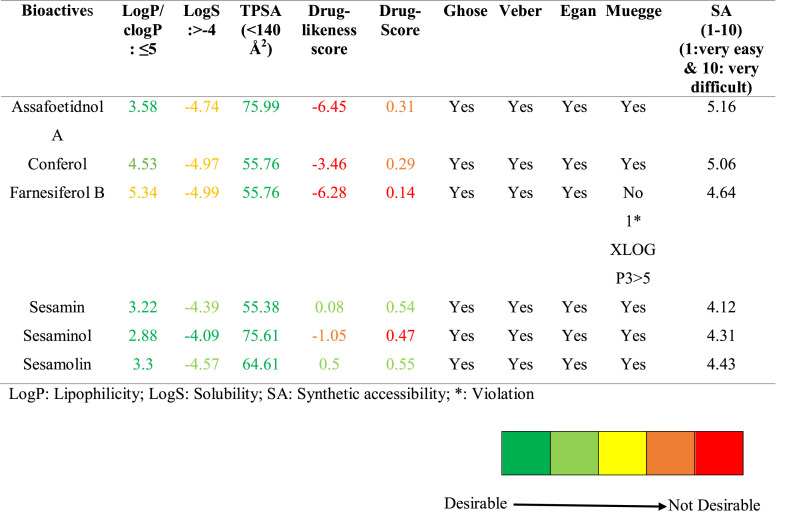
3.2. Drug-likeness properties
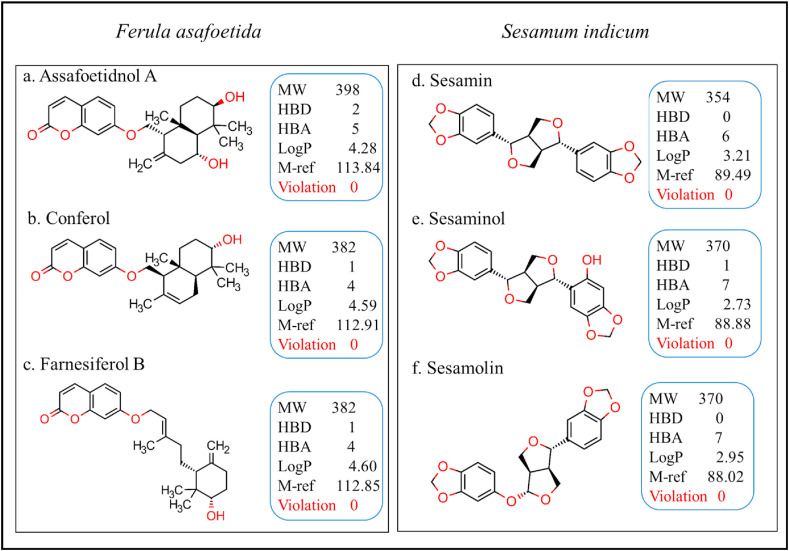
The investigation of the drug-likeness properties accelerates drug discovery and development procedures. Lipinski's rule of five (RO5) is one of the key factors to qualify to become a drug [33]. Six selected bioactives from the spices of F. asafoetida and S. indicum have passed on the ‘RO5’ without any single violations (Fig. 4 ). Analysis of Ghose filter rule, Veber rule, Egan rule, and Muegge rule related to drug-likeness properties were also performed (Table 2 ). These parameters have their principles to identify a bioactive function as an effective drug. The rule of Ghose filter is logP value: -0.4 to 5.6, molecular weight: 160–480 g/mol, molar refractivity: 40–130, and the total number of atoms range: 20–70 [46]. Whereas in Veber rule, ligand should contain ten or fewer numbers of rotatable bonds with ≤140 Å2 of the polar surface area [47]. In Egan rule, the absorption of a drug molecule depends on two factors: the polar surface area (PSA) and AlogP98 (the logarithm of the partition coefficient between n-octanol and water). The Muegge rule follows an effective drug that has to pass a pharmacophore point filter [48,49]. All the bioactives selected for our study were perfectly fit into the Ghose filter rule, Veber rule, and Egan rule without any violation. Except for farnesiferol B, all other bioactives have qualified the Muegge rule.
Fig. 4.
Lipinski's Rule of Five (RO5) for selected bioactives from the spices. MW: Molecular weight (<500 g/mol), HBD: H-bond donor (<5), H-bond acceptor (<10), LogP (<5), M-ref: Molar Refractivity (40–130).
Table 2.
The drug-likeness properties of bioactives from spices.
Likewise, except farnesiferol B, all other bioactives were found to be less lipophilic and having more absorption, as shown in Table 2. In the case of solubility, sesamin, sesaminol, and sesamolin have better solubility compared to other compounds. All bioactives have shown optimal cell permeability and good oral bioavailability due to the less topological polar surface area (TPSA). Synthetic accessibility (SA) is used to analyze the possibility and feasibility of the synthesis of target compounds [50]. Chemical synthesis of these compounds is feasible as they belong in the range of about 4–5 in the 1–10 scale of easy to difficult, as shown in Table 2.
3.3. ADME/toxicity prediction
ADME/T is an important parameter to understand the fate of a compound when administered orally. There is a link between oral administration of a drug and drug level present in the blood-stream through human intestinal absorption (HIA) [51]. Commonly, the in vitro permeability study is performed in the Caco-2 cell line, and it mimics the intestinal absorption of the compound. Therefore, we analyzed the ADME/T properties of selected spice bioactives and presented in Table 3.1 .
Table 3.1 ADME/T test of selected bioactives from spices.
|
Absorption Properties |
Assafoetidnol A | Conferol | Farnesiferol B | Sesamin | Sesaminol | Sesamolin |
|---|---|---|---|---|---|---|
| Caco-2 Permeability (Optimal: higher than −5.15 Log unit or −4.70 or −4.80) | Optimal −5.022 cm/s |
Optimal −4.941 cm/s |
Optimal −4.949 cm/s |
Optimal −4.853 cm/s |
Optimal −5.04 cm/s |
Optimal −4.887 cm/s |
| Human Intestinal Absorption (HIA)≥30%: HIA+; <30%: HIA- | + (0.558) | + (0.605) | + (0.604) | + (0.613) | + (0.571) | + (0.568) |
| P-glycoprotein Substrate | ---(0.016) | ---(0.016) | ---(0.009) | ---(0.058) | ---(0.063) | ---(0.022) |
| P-glycoprotein Inhibitor | ++ (0.858) | ++ (0.868) | +++ (0.949) | + (0.663) | + (0.543) | ++ (0.821) |
| Bioavailability Score | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 |
| GI absorption | High | High | High | High | High | High |
|
Distribution Properties |
Assafoetidnol A | Conferol | Farnesiferol B | Sesamin | Sesaminol | Sesamolin |
| PPB (Plasma Protein Binding): 90% | High (95.70%) | High (96.23%) | High (96.65%) | Littte less (79.80%) | Little less (79.42%) | Little less (80.09%) |
| Blood-Brain Barrier (BBB) BB ratio≥0.1: BBB+; BB ratio <0.1: BBB- | + (0.623) | -(0.486) | + (0.617) | +++ (0.991) | ++ (0.861) | +++ (0.98) |
| VD (Volume Distribution) 0.04–20 L/kg | 0.132 L/kg | 0.104 L/kg | −0.056 L/kg | 0.243 L/kg | −0.094 L/kg | −0.067 L/kg |
|
Metabolism Properties |
Assafoetidnol A | Conferol | Farnesiferol B | Sesamin | Sesaminol | Sesamolin |
| P450 CYP1A2 inhibitor | ---(0.191) | ---(0.154) | ---(0.231) | ++ (0.765) | + (0.541) | + (0.518) |
| P450 CYP1A2 Substrate | -(0.452) | -(0.41) | + (0.514) | + (0.615) | + (0.516) | + (0.569) |
| P450 CYP3A4 inhibitor | ---(0.286) | ---(0.297) | -(0.401) | + (0.598) | + (0.648) | ++ (0.705) |
| P450 CYP3A4 substrate | ++ (0.723) | + (0.615) | + (0.622) | + (0.518) | + (0.62) | + (0.534) |
| P450 CYP2C9 inhibitor | ---(0.16) | ---(0.148) | ---(0.254) | ---(0.205) | -(0.345) | + (0.508) |
| P450 CYP2C9 substrate | + (0.532) | + (0.637) | + (0.559) | -(0.351) | -(0.458) | -(0.377) |
| P450 CYP2C19 inhibitor | ---(0.227) | -(0.349) | -(0.359) | ++ (0.767) | ++ (0.723) | ++ (0.856) |
| P450 CYP2C19 substrate | + (0.598) | + (0.596) | + (0.58) | + (0.627) | + (0.665) | + (0.672) |
| P450 CYP2D6 inhibitor | -(0.402) | -(0.377) | -(0.45) | +++ (0.9) | ++ (0.851) | ++ (0.85) |
| P450 CYP2D6 substrate | + (0.543) | + (0.591) | -(0.469) | + (0.569) | + (0.559) | + (0.61) |
|
Excretion Properties |
Assafoetidnol A | Conferol | Farnesiferol B | Sesamin | Sesaminol | Sesamolin |
| T 1/2 (Half Life Time) (>8 h: high; 3 h < Cl < 8 h: moderate; <3 h: low) |
Low (1.48 h) |
Low (1.89 h) |
Low (1.84 h) |
Low (1.99 h) |
Low (1.74 h) |
Low (1.58 h) |
|
Toxicity Properties |
Assafoetidnol A | Conferol | Farnesiferol B | Sesamin | Sesaminol | Sesamolin |
| hERG (hERG Blockers) | + (0.508) | + (0.614) | ++ (0.705) | - (0.492) | -(0.394) | -(0.488) |
| AMES (Ames Mutagenicity) | -(0.366) | -(0.348) | -(0.336) | -(0.354) | ---(0.276) | -(0.342) |
| DILI (Drug Induced Liver Injury) | + (0.514) | + (0.502) | + (0.522) | ++ (0.724) | ++ (0.718) | ++ (0.728) |
All the selected bioactives have shown satisfactory Caco-2 permeability and human intestinal absorption (HIA). Interestingly, none of the selected bioactive functions as P-glycoprotein (P-gp) substrate, in contrast, they function as P-gp inhibitors. The inhibition of P-gp enhances the intracellular concentration of the compound by inhibiting the drug efflux proteins [52]. After absorption, drugs flow to the other parts of the body through the blood and then metabolized in the liver. In the liver, a group of enzymes from the Cytochrome P450 family can breakdown the absorbed drug molecules and eliminate from the human body as bile and urine [53]. Specific bioactives act as a substrate of these enzymes; thus, they will be metabolized through that corresponding CYP450 enzyme.
In contrast, some bioactives act as inhibitors of these enzymes, which disturb the biodegradation process [54,55]. Here, sesamin, sesaminol, and sesamolin from the spices function as P450 CYP1A2 inhibitors. Sesamolin also functions as a P450 CYP2C9 inhibitor. Most of the selected bioactives have shown shorter half-life due to the secondary metabolism of spices [56].
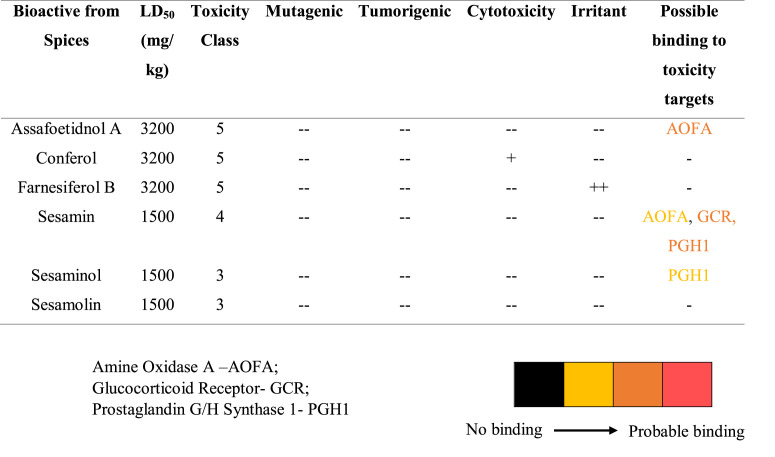
3.4. Prediction of adverse and toxic effect
The ProTox-II server categorizes the molecules according to the Globally Harmonized System of Classification and Labelling of Chemicals (GHS). According to the GHS, the toxicity of any compound taken orally is classified into five categories based on the LD50 ranges. Class 1- fatal (LD50 ≤ 5), class 2- fatal (5 < LD50 ≤ 50), class 3- toxic (50 < LD50 ≤ 300), class 4- harmful (300 < LD50 ≤ 2000), class 5- may be harmful (2000 < LD50 ≤ 5000), and if LD50 > 5000 is considered as non-toxic [34]. The prophesied LD50 value for assafoetidnol A, conferol, and farnesiferol B was 3200 mg/kg, and all of them belonged to class 5, whereas LD50 of sesamin, sesaminol, and sesamolin was 1500 mg/kg and belonged to class 4 (Table 3.2 ). Although most of the selected bioactives fall under the toxicity class of 4–5, their toxicity will be further reduced when consumed as the whole spice. Fortunately, none of the spice bioactive predicted to have hepatotoxicity, cytotoxicity, mutagenic, and tumorigenicity.
Table 3.2 Adverse and toxic effects of selected bioactives from spices.
3.5. Estimation of biological activities
Prediction of activity spectra for substances (PASS) was performed with selected bioactives from the spices and presented in Supplementary Table S3. Two factors are involved in PASS prediction: Pa and Pi. Pa represents the probability “to be active” of a ligand, whereas Pi represents the probability “to be inactive” of a ligand, and their values are ranging from 0 to 1. Most of the selected spice bioactives were probably effective against the SARS, except farnesiferol B. The selected spice bioactives are probable to have anti-inflammatory, respiratory analeptics, and moderately anti-viral properties.
These selected spices are already known to have several promising activities, mainly antioxidant, anti-inflammatory, immunomodulatory, and anti-viral [38,57]. Primarily, F. asafoetida can function as an anti-viral agent against influenza A (H1N1) and herpes virus type 1 (HSV-1) [42,58]. According to Chinese researchers, F. asafoetida is effective against influenza A (H1N1) viruses [58]. Similarly, the roots and leaves of S. indicum can be effective as anti-viral agents against chickenpox and measles, as well as this plant also functions as an immunomodulatory agent [37,59]. Further, sesame essential oil and sesamol regulate the pro-inflammatory function of macrophages and dendritic cells as well as promote Th2 response [59]. Therefore, the inclusion of these two spices in the daily diet may enhance immunity and fight against SARS-CoV-2 infection.
4. Conclusion
Currently, humankind is facing a dreadful situation due to the emergence of the COVID-19 pandemic. Various natural dietary phytochemicals, including spice bioactives, are known to possess plenty of health benefits, including antioxidant, anti-inflammatory, and anti-viral activities. In our study, we have convincingly demonstrated the potentiality of spice bioactives in targeting SARS-CoV-2 Mpro, spike protein, and human ACE2 receptors. The inhibition of these target proteins may lead to either attenuation of viral replication or reduce the infectivity of this virus. Furthermore, it is required to investigate the in vitro and in vivo effect of these selected spice bioactives in inhibiting the activity of these targets. Fortunately, most of the potential bioactives can be obtained by consuming asafoetida and sesame spices. The selected spice bioactives have also displayed various druggable properties with desirable biological aspects. Thus, incorporation of asafoetida and sesame in the diet might intervene in the COVID-19 infection as a preventive and prophylactic approach. Further, the development of potential bioactive-based nutraceuticals might be effective against COVID-19, as a quick relief before discovering any specific drugs.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
JN acknowledges senior research fellowships from DBT, Government of India, and PM and DJ from CSIR-UGC, Government of India. AAAS acknowledges the research grant provided by a MAHE intramural grant (MAHE/DREG/PhD/IMF/2019). We also acknowledge CSIR-CFTRI for the support and infrastructure facility.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2020.104102.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Ge H., Wang X., Yuan X., Xiao G., Wang C., Deng T., Yuan Q., Xiao X. The epidemiology and clinical information about COVID-19. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39(6):1011–1019. doi: 10.1007/s10096-020-03874-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20(6):355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H., Zhou Y., Zhang M., Wang H., Zhao Q., Liu J. Updated approaches against SARS-CoV-2, antimicrob. Agents Chemother. 2020;64(6) doi: 10.1128/AAC.00483-20. e00483-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of covid-19 — preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/nejmoa2007764. NEJMoa2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Recovery Collaborative Group, Horby P., Mafham M., Linsell L., Bell J.L., Staplin N., Emberson J.R., Wiselka M., Ustianowski A., Elmahi E., Prudon B., Whitehouse T., Felton T., Williams J., Faccenda J., Underwood J., Baillie J.K., Chappell L.C., Faust S.N., Jaki T., Jeffery K., Lim W.S., Montgomery A., Rowan K., Tarning J., Watson J.A., White N.J., Juszczak E., Haynes R., Landray M.J. Effect of hydroxychloroquine in hospitalized patients with covid-19. N. Engl. J. Med. 2020:1–11. doi: 10.1056/NEJMoa2022926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/jvi.00127-20. e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 9.Choudhary S., Malik Y.S., Tomar S. Identification of SARS-CoV-2 cell entry inhibitors by drug repurposing using in silico structure-based virtual screening approach. Front. Immunol. 2020 doi: 10.3389/fimmu.2020.01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 11.Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., Chugh A., Giri A.P., Kulkarni M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Khafaji K., Al-DuhaidahawiL D., Taskin Tok T. Using integrated computational approaches to identify safe and rapid treatment for SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1764392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar D., Kumari K., Jayaraj A., Kumar V., Kumar R.V., Dass S.K., Chandra R., Singh P. Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1752310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J. Biomol. Struct. Dyn. 2020:1–6. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 15.Gunasekar M., Geemon K., Mariwala S.J. Health benefits of bioactive molecules from spices and aromatic plants. Indian Soc. Spices. 2012;21(2):87–101. [Google Scholar]
- 16.Vázquez-Fresno R., Rosana A.R.R., Sajed T., Onookome-Okome T., Wishart N.A., Wishart D.S. Herbs and spices- biomarkers of intake based on human intervention studies - a systematic review. Genes Nutr. 2019;14(1):18. doi: 10.1186/s12263-019-0636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubió L., Motilva M.J., Romero M.P. Recent advances in biologically active compounds in herbs and spices: a review of the most effective antioxidant and anti-inflammatory active principles. Crit. Rev. Food Sci. Nutr. 2013;53(9):943–953. doi: 10.1080/10408398.2011.574802. [DOI] [PubMed] [Google Scholar]
- 18.Shahidi F., Ambigaipalan P. Phenolics and polyphenolics in foods, beverages and spices: antioxidant activity and health effects - a review. J. Funct. Foods. 2015;18:820–897. doi: 10.1016/j.jff.2015.06.018. [DOI] [Google Scholar]
- 19.Praditya D., Kirchhoff L., Brüning J., Rachmawati H., Steinmann J., Steinmann E. Anti-infective properties of the golden spice curcumin. Front. Microbiol. 2019;10:912. doi: 10.3389/fmicb.2019.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G., Lee C.K., Chang S.T., Kuo C.J., Lee S.S., Hou C.C., Hsiao P.W., Chien S.C., Shyur L.F., Yang N.S. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007;50(17):4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 21.Rout J., Swain B.C., Tripathy U. In silico investigation of spice molecules as potent inhibitor of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1819879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elsayed Y., Khan N.A. Immunity-boosting spices and the novel coronavirus. ACS Chem. Neurosci. 2020;11(12):1696–1698. doi: 10.1021/acschemneuro.0c00239. [DOI] [PubMed] [Google Scholar]
- 23.Kunnumakkara A.B., Sailo B.L., Banik K., Harsha C., Prasad S., Gupta S.C., Bharti A.C., Aggarwal B.B. Chronic diseases, inflammation, and spices: how are they linked? J. Transl. Med. 2018 doi: 10.1186/s12967-018-1381-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guldiken B., Ozkan G., Catalkaya G., Ceylan F.D., Ekin Yalcinkaya I., Capanoglu E. Phytochemicals of herbs and spices: health versus toxicological effects. Food Chem. Toxicol. 2018;119:37–49. doi: 10.1016/j.fct.2018.05.050. [DOI] [PubMed] [Google Scholar]
- 26.Patel V.J., Biswas Roy S., Mehta H.J., Joo M., Sadikot R.T. Alternative and natural therapies for acute lung injury and acute respiratory distress syndrome. BioMed Res. Int. 2018;2018:2476824. doi: 10.1155/2018/2476824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3(1):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan M., Wu N.C., Zhu X., Lee C.C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., Patane M.A., Pantoliano M.W. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279(17) doi: 10.1074/jbc.M311191200. 17996–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N. The protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. www.rcsb.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris G.M., Goodsell D.S., Pique M.E., Lindstrom W. “Lindy”, Huey R., Forli S., Hart W.E., Halliday S., Belew R., Olson A.J. Autodock 4 and AutoDockTools 4: automated docking with selective receptor flexiblity. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Drwal M.N., Banerjee P., Dunkel M., Wettig M.R., Preissner R. ProTox: a web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014;42(W1) doi: 10.1093/nar/gku401. W53–W58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salam A.A.A., Nayek U., Sunil D. Homology modeling and docking studies of bcl-2 and bcl-xL with small molecule inhibitors: identification and functional studies. Curr. Top. Med. Chem. 2019;18(31):2633–2663. doi: 10.2174/1568026619666190119144819. [DOI] [PubMed] [Google Scholar]
- 36.Kamath P.R., Sunil D., Ajees A.A. Synthesis of indole–quinoline–oxadiazoles: their anticancer potential and computational tubulin binding studies. Res. Chem. Intermed. 2016;42(6):5899–5914. doi: 10.1007/s11164-015-2412-8. [DOI] [Google Scholar]
- 37.Nagpurkar M., Patil N.M. A review on sesame-an ethno medicinally significant oil crop. Int. J. Life Sci. Pharma Res. 2017;7(2):L58–L63. [Google Scholar]
- 38.Amalraj A., Gopi S. Biological activities and medicinal properties of Asafoetida: a review. J. Tradit. Complement. Med. 2017;7(3):347–359. doi: 10.1016/j.jtcme.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahendra P., Bisht S. Ferula asafoetida : traditional uses and pharmacological activity. Pharm. Rev. 2012;6(12):141. doi: 10.4103/0973-7847.99948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu M.S., Aquino L.B.B., Barbaza M.Y.U., Hsieh C.L., De Castro-Cruz K.A., Yang L.L., Tsai P.W. Anti-inflammatory and anticancer properties of bioactive compounds from sesamum indicum L. - a review. Molecules. 2019;24:1–28. doi: 10.3390/molecules24244426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afroz M., Zihad S.M.N.K., Uddin S.J., Rouf R., Rahman M.S., Islam M.T., Khan I.N., Ali E.S., Aziz S., Shilpi J.A., Nahar L., Sarker S.D. A systematic review on antioxidant and antiinflammatory activity of Sesame (Sesamum indicum L.) oil and further confirmation of antiinflammatory activity by chemical profiling and molecular docking. Phyther. Res. 2019;33(10):2585–2608. doi: 10.1002/ptr.6428. [DOI] [PubMed] [Google Scholar]
- 42.Ghannadi A., Fattahian K., Shokoohinia Y., Behbahani M., Shahnoush A. Anti-viral evaluation of sesquiterpene coumarins from ferula assa-foetida against HSV-1, Iran. J. Pharm. Res. 2014;13:523–530. doi: 10.22037/ijpr.2014.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi C., Sun X., Ye J., Ding L., Liu M., Yang Z., Lu X., Zhang Y., Ma L., Gu W., Qu A., Xu J., Shi Z., Ling Z., Sun B. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020;17(6):621–630. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veeramachaneni G.K., Thunuguntla V.B.S.C., Bobbillapati J., Bondili J.S. Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1773318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghose A.K., Viswanadhan V.N., Wendoloski J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999;1(1):55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 47.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 48.Egan W.J., Merz K.M., Baldwin J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000;43(21):3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 49.Muegge I., Heald S.L., Brittelli D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001;44(12):1841–1846. doi: 10.1021/jm015507e. [DOI] [PubMed] [Google Scholar]
- 50.Ertl P., Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminf. 2009;1(1):8. doi: 10.1186/1758-2946-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radchenko E.V., Dyabina A.S., Palyulin V.A., Zefirov N.S. Prediction of human intestinal absorption of drug compounds. Russ. Chem. Bull. 2016;65:576–580. doi: 10.1007/s11172-016-1340-0. [DOI] [Google Scholar]
- 52.Abdallah H.M., Al-Abd A.M., El-Dine R.S., El-Halawany A.M. P-glycoprotein inhibitors of natural origin as potential tumor chemo-sensitizers: a review. J. Adv. Res. 2015;6(1):45–62. doi: 10.1016/j.jare.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glue P., Clement R.P. Cytochrome P450 enzymes and drug metabolism - basic concepts and methods of assessment, Cell. Mol. Neurobiol. 1999;19(3):309–323. doi: 10.1023/A:1006993631057. [DOI] [PubMed] [Google Scholar]
- 54.Li A.P. Screening for human ADME/Tox drug properties in drug discovery, Drug Discov. Today Off. 2001;6:357–366. doi: 10.1016/S1359-6446(01)01712-3. [DOI] [PubMed] [Google Scholar]
- 55.Guengerich F.P. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 1999;39(1):1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 56.Hodges R.E., Minich D.M. Modulation of metabolic detoxification pathways using foods and food-derived components: a scientific review with clinical application. J. Nutr. Metab. 2015;2015:760689. doi: 10.1155/2015/760689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dravie E.E., Kortei N.K., Essuman E.K., Tettey C.O., Boakye A.A., Hunkpe G. Antioxidant, phytochemical and physicochemical properties of sesame seed (Sesamum indicum L) Sci. African. 2020 doi: 10.1016/j.sciaf.2020.e00349. [DOI] [Google Scholar]
- 58.Lee C.L., Chiang L.C., Cheng L.H., Liaw C.C., Abd El-Razek M.H., Chang F.R., Wu Y.C. Influenza A (H1N1) antiviral and cytotoxic agents from Ferula assa-foetida. J. Nat. Prod. 2009;72(9):1568–1572. doi: 10.1021/np900158f. [DOI] [PubMed] [Google Scholar]
- 59.Khorrami S., Daneshmandi S., Mosayebi G. Sesame seeds essential oil and Sesamol modulate the pro-inflammatory function of macrophages and dendritic cells and promote Th2 response. Med. J. Islam. Repub. Iran. 2018;32:98. doi: 10.14196/mjiri.32.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.