Abstract
Acetylcholine (ACh) regulates inflammation via α7 nicotinic acetylcholine receptor (α7 nAChR). Acetylcholinesterase (AChE), an enzyme hydrolyzing ACh, is expressed in immune cells suggesting non-classical function in inflammatory responses. Here, the expression of PRiMA-linked G4 AChE was identified on the surface of macrophages. In lipopolysaccharide-induced inflammatory processes, AChE was upregulated by the binding of NF-κB onto the ACHE promotor. Conversely, the overexpression of G4 AChE inhibited ACh-suppressed cytokine release and cell migration, which was in contrast to that of applied AChE inhibitors. AChEmt, a DNA construct without enzymatic activity, was adopted to identify the protein role of AChE in immune system. Overexpression of G4 AChEmt induced cell migration and inhibited ACh-suppressed cell migration. The co-localization of α7 nAChR and AChE was found in macrophages, suggesting the potential interaction of α7 nAChR and AChE. Besides, immunoprecipitation showed a close association of α7 nAChR and AChE protein in cell membrane. Hence, the novel function of AChE in macrophage by interacting with α7 nAChR was determined. Together with hydrolysis of ACh, AChE plays a direct role in the regulation of inflammatory response. As such, AChE could serve as a novel target to treat age-related diseases by anti-inflammatory responses.
Key words: Macrophage, AChE, Cholinergic anti-inflammatory pathway, α7 nAChR
Abbreviations: AChE, acetylcholinesterase; ACh, acetylcholine; BChE, butyrylcholinesterase; CDC42, cell division cycle; CAP pathway, cholinergic anti-inflammatory pathway; ChAT, choline acetyltransferase; DPZ, donepezil; GAL, galantamine hydrobromide; IL, interleukin; LPS, lipopolysaccharides; MLA, methyllycaconitine citrate salt; MMP, matrix metalloproteinase; nAChR, nicotinic AChR; NF-κB, nuclear factor-κB; PHA, PHA-543613; PRiMA, proline-rich membrane anchor; TNF-α, tumor necrosis factor α
Graphical abstract
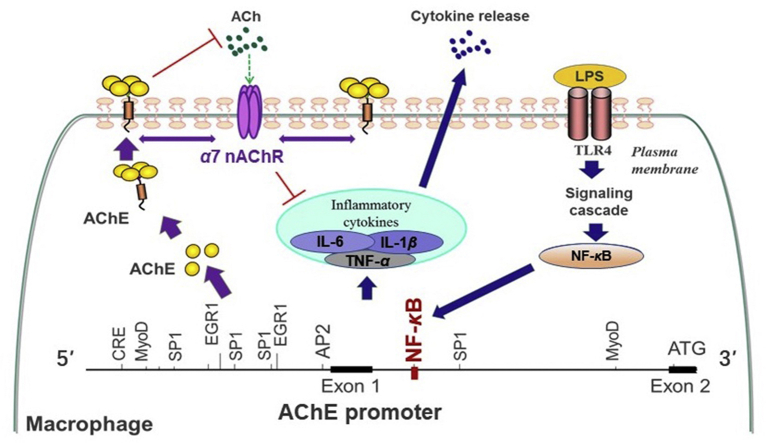
AChE regulates cholinergic anti-inflammatory pathway (CAP) in dual processes. First, AChE directly hydrolyzes ACh to promote inflammation via the inhibition of CAP. In parallel, NF-κB bound to ACHE promotor, which triggered the transcription of AChE during inflammation. Second, AChE enhanced the inflammatory responses via the interference with α7 nAChR.
1. Introduction
Acetylcholine (ACh) is the first identified neurotransmitter that defines the chemical nature of neurotransmission both in the central nervous system and the periphery. Besides neurotransmission, ACh is able to play anti-inflammatory roles in immune responses1 and synthesized by immune cells, e.g., T/B cell, macrophage, dendritic cell, and neutrophil. The ACh-activated responses could attenuate the release of interleukin (IL)-1β, IL-6 and tumor necrosis factor α (TNF-α) in human macrophages, suggesting its direct inhibitory effect in pro-inflammation1,2. In parallel, choline acetyltransferase (ChAT), the enzyme that produces ACh, is expressed in B cells, dendritic cells and macrophages3. Indeed, the efferent vagus nerve is being suggested to interplay between nervous and immune systems, which is known as cholinergic anti-inflammatory pathway (CAP). Today, CAP is the most notably for cholinergic signaling in regulating immune responses.
Nicotinic acetylcholine receptors (nAChRs) are a family of ligand-gated, pentameric ion channels. The α7 subunit of the nAChR was reported to be expressed in macrophages2,4. Indeed, CAP plays a critical role in regulating inflammatory response via interacting with peripheral α7 subunit-containing nAChR (α7 nAChR). During inflammation, the activation of α7 nAChR by ACh was associated with Ca2+ influx and inhibited production of pro-inflammatory cytokines by modulating nuclear factor κB (NF-κB) stimulation2. Besides, the activation of α7 nAChR inhibited lipopolysaccharides (LPS)-induced pro-migratory gene production, e.g., matrix metalloproteinase (MMP) 9, MMP2, cell division cycle 42 (CDC42), and macrophage migration via the JAK2/STAT3 signaling pathway5,6.
Acetylcholinesterase (AChE) is primarily responsible for its cholinergic function, i.e., the rapid hydrolysis of ACh into choline and acetate7, 8, 9. Alternative mRNA processing of the ACHE gene yields four transcripts encoding peptides, which differ only at their carboxyl termini but with same catalytic domain. This leads to different post-translational modifications, cellular localization and anchoring10,11. The four different types of AChE transcripts are AChER, AChEH, AChES and AChET8,12,13. In the brain and muscle, AChET was reported as predominant subunit10,14. In the brain, AChET localized and oligomerized with an anchoring protein, proline-rich membrane anchor (PRiMA), as PRiMA-linked tetrameric globular form (G4) AChE, which is the major and functional form14.
The existence of AChE has been widely reported in non-neural areas, where no cholinergic function is known to take place, suggesting the non-classical enzymatic functions of AChE in various systems9,15,16. High levels of AChE expression were found in activated B- and T-lymphocytes and thymocytes17, suggesting the possible function of AChE in inflammatory responses. In line to this notion, the binding site of NF-κB was revealed in mammalian ACHE gene18, which therefore suggested the potential role of NF-κB transcriptional factor in directing AChE expression during immune responses. Here, murine macrophage-like RAW 264.7 cells, peritoneal macrophages and bone marrow-derived macrophages were adopted to investigate the role of AChE in inflammatory responses.
2. Materials and methods
2.1. Chemicals
Acetylcholine chloride, PHA-543613 (PHA), lipopolysaccharides (LPS), methyllycaconitine citrate salt (MLA), donepezil hydrochloride (DPZ) and galantamine hydrobromide (GAL) were purchased from Sigma (St. Louis, MO, USA). Proper amount of chemical was dissolved in Mili Q water (Merck, Darmstadt, Germany) and stored at −20 °C.
2.2. Cell culture
Murine monocytes cell line (RAW 264.7) was obtained from American Type Culture Collection (Manassas, VA, USA). Cultured the cells in Dulbecco's modified Eagle's medium with 10% (v/v) FBS, 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C in a humidified atmosphere with 5% CO2 at 37 °C. Peritoneal macrophages were isolated from C57BL/6 J mice according to previously reported with some modifications19. Briefly, harvested the cells by washing the peritoneal cavity with ice-cold PBS at 5 mL per mouse followed centrifuged at 1000 rpm (Sorvall ST 8, Thermo Fisher Scientific, Suzhou, China) for 10 min. Then, the cells were cultured in RPMI-1640 medium supplemented with 10% (v/v) fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin. These macrophages were seeded on 6-well plates and maintained in a water-saturated atmosphere 5% CO2 at 37 °C. Non adherent cells were removed at 4 h after seeding.
2.3. AChE/BChE assay
Ellman method20, modified by adding an inhibitor of BChE activity (tetra-iso-propylpyrophosphoramide, 0.1 mmol/L) or an inhibitor of AChE activity (BW284c51, 20 μmol/L), was employed to determine the activities of AChE and butyrylcholinesterase (BChE). Samples were added to the reaction mixture and make sure the final concentrations of acetylthiocholine iodide at 0.625 mmol/L (ATCh; Sigma) and 5,5-dithiobis-2-nitrobenzoic acid (Sigma) 0.5 mmol/L in 80 mmol/L Na2HPO4. Then, the absorbance was measured at 412 nm. Specific enzyme activity was expressed as absorbance units/min/g of protein.
2.4. PCR and real-time quantitative PCR
RNAzol reagent (Molecular Research Center, Cincinnati, OH, USA) was used to extract the total RNA from cell cultures. Equal amounts of RNAs were reverse transcribed to cDNAs (Thermo Fisher Scientific; Waltham, MA, USA). PCR was performed to amplify targets in a 20 μL reaction mixture containing 1/50 reverse transcribed product, 0.4 U KAPA Taq DNA polymerase and 0.4 μmol/L of 5′–3′ primers. Samples were run in triplicate. Amplification was performed in a GeneAmp® 9700 PCR System for 35 cycles. Ten μL of the PCR products were separated by electrophoresis on 1% agarose gel. DNA sequencing was performed to confirm the identity of PCR products. Real-time quantitative PCR was performed by using Roche SYBR Green Master mix (Mannheim, Germany) with Rox reference dye for the target genes21. Transcript expression of genes was calculated by 18 S rRNA as the house keeping gene. The primers sequences were previously reported16,22 as follows. AChE: 5′-AAT CGA GTT CAT CTT TGG GCT CCC CC-3′ and 5′-CCA GTG CAC CAT GTA GGA GCT CCA-3′; AChER: 5′-CAG GGG ACC CCA ATG ACC CTC G-3′ and 5′-CCC ACT CCA TGC GCC TAC CGG T-3′; AChEH: 5′-CCG CGC AGC AAT ATG TGA GCC T-3′ and 5′-GCA GGT GCA AGG AGC CTC CGT-3′; AChET: 5′-TAG AGG TGC GGC GGG GAC TG-3′ and 5′-TGA GCA GCG CTC CTG CTT GC-3′; PRiMA: 5′-TCT GAC TGT CCT GGT CAT TTG CTA C-3′ and 5′-TCA CAC CGC AGC GTT CAC-3′; TNF-α: 5′-AGT GAC AAG CCT GTA GCC-3′ and 5′-AGG TTG ACT TTC TCC TGG-3′; MMP2: 5′-AGT GAC GGC TTC CTC TGG TGT T-3′ and 5′-CAG GGC TGT CCA TCT CCA TTG C-3′; CDC42: 5′-AGT GGC TCA GGG CAT GGC TTT C-3′ and 5′-ACT TTA CAG GCA GGC GGG CAT T-3′; 18 S rRNA: 5′-GAC TGT TAT GGT CAA GGT GAA-3′ and 5′-GAT AGT CAA GTT CGA CCG TC-3′.
2.5. ELISA analysis
A commercial ELISA kit (R&D System, Minneapolis, MN, USA) was employed to measure the protein release of TNF-α. Cell cultured medium was added to a 96-well plate with coating of anti-mouse TNF-α antibody, incubated at 37 °C for 2 h. Then, wells were washed by 400 μL wash buffer 4 times. Added biotinylated anti-mouse TNF-α antibody and incubated at 37 °C for 2 h. Then, washed 3 times by washing buffer. Followed by added 100 μL of substrate solution to each well and incubated for 30 min at room temperature in dark. Applied the stop solution and immediately measured the absorbance at 450 nm.
2.6. Sucrose density gradient analysis
Different molecular isoforms of AChE were separated by sucrose density gradient analysis22,23. Briefly, two hundred of cell extracts containing 200 mg protein were mixed with ALP and β-galactosidase standard, which the sedimentation coefficient is 6.1 and 16 S, respectively. Prepared continuous sucrose gradients from 5% to 20% with low salt lysis buffer (1 mmol/L EDTA, 1 mmol/L EGTA, 10 mmol/L HEPES, 150 mmol/L NaCl, and 0.2% Triton X-100, pH 7.5) in 12 mL centrifuge tubes. The mixtures were gently layered onto the top of prepared tubes. Centrifuged at 38,000 rpm (CP80WX, Hitachi, Hitachinaka, Japan) 4 °C for 16 h. Then, about 45 fractions (each ∼200 μL) were collected from the bottom of the sucrose gradients and tested the activity of AChE, β-galactosidase and ALP. Based on the known sedimentation coefficient of ALP and β-galactosidase, the sedimentation coefficients of each fractions could be calculated. And, the sedimentation coefficients of G1 and G4 isoforms has already known, which were approximately 4.3 S and 10.2 S, respectively. The molecular isoforms of AChE could be determined.
2.7. Immunofluorescent staining
Cells were seeded on glass coverslip for 48 h and washed by PBS. Then, after fixed with 4% paraformaldehyde for 15 min, cells were blocked by 5% FBS with or without 0.2% Triton X-100 for 1 h. Stained with anti-AChE antibody at 1:100 for 16 h at 4 °C. After intensive washing, cells were labeled by secondary antibody (Alexa Fluor 488-conjugated anti-goat antibody) together with 4′,6-diamidino-2-phenylindole (DAPI) for 2 h. Cells were also stained with anti-NF-κB antibody, anti-α7 nAChR antibody, or anti-TLR4 antibody, followed with Alexa Fluor 555-conjugated anti-rabbit antibody (Sigma) or Alexa Fluor 647-conjugated anti-mouse antibody (Sigma). Glass coverslip were examined by a confocal fluorescence microscope (LSM710; Carl Zeiss MicroImaging, Jena, Germany).
2.8. Transwell® motility assay
RAW 264.7 cells were seeded on the top side of polycarbonate Transwell® filter of upper chamber in serum free medium with or without testing chemicals. Meanwhile, the culture medium (with 10% FBS) with or without testing chemical was added in the lower chamber. After 24 h, the cells on the top side of the chamber were gently scraped by cotton swabs. Cells migrated to the opposite side were fixed and stained by DAPI. The fluorescent (Ex/Em 358/461 nm) images were randomly captured for each well under Leica confocal software by 10× objective.
2.9. SDS-PAGE and western blot assay
Total protein of cell cultures was collected with the low salt lysis buffer added with 10 μmol/L aprotinin, 5 mmol/L benzamidine HCl, and 10 μmol/L leupeptin. Then, centrifuged at 13,200 rpm (5415R, Eppendorf, Hamburg, Germany) for 15 min at 4 °C. The supernatant was total protein from cell cultures. Homogenized samples to the same concretizations of protein. Lysed the homogenates by 2× lysis buffer and boiled for 10 min before performing on the gel electrophoresis14. Then, electrophoresis separated proteins in SDS-PAGE were transferred to nitrocellulose membrane by 1× transfer buffer, containing 0.192 mol/L glycine, 24 mmol/L Tris, and 15% ethanol. Stained the membrane to confirm the transfer and blocked with 5% dry milk in TBS-T for 1 h at room temperature. After washed by TBS-T, the membrane was incubated in primary anti-AChE antibody (1:500, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-flotinlin-2 antibody (1:1,000, Cell Signaling Technology; Danvers, MA, USA), anti-α7 nAChR antibody (1:1000, Cell Signaling Technology), anti-Toll-like receptor 4 (TLR 4) antibody (1:1000, Abcam, Cambridge, UK), anti-transferrin receptor antibody (1:500, Santa Cruz Biotechnology), anti-IKKα antibody (1:1000, Cell Signaling Technology), anti-IKKβ antibody (1:1,000, Cell Signaling Technology), anti-P-IKK α/β antibody (1:1000, Cell Signaling Technology), anti-NF-κB antibody (1:1000, Abcam), anti-IκBα antibody (1:1,000, BD Biosciences, San Jose, CA), anti-P-IκBα antibody (1:1,000, BD Biosciences), and anti-α-tubulin antibody (1:10,000, Sigma) for overnight at 4 °C. Washed three times by TBS-T, the membrane was incubated with 1:5000 diluted HRP-conjugated anti-goat, anti-mouse or anti-rabbit secondary antibody (Thermo Fisher Scientific) for 2 h at room temperature. Visualized the membrane by ECL method.
2.10. DNA construction and transfection
The cDNA encoding AChET or PRiMA was described previously14. Human NF-κB P65 was constructed in pCMV4 (Addgene, Watertown, MA, USA; ID: 21,966). The DNAs encompassing human AChE promoter with or without the NF-κB-binding site were sub-cloned into pGL3 vector (BD Biosciences) upstream of a luciferase reporter gene, described as pAChE-Luc and pAChEΔNF-κB-Luc. AChEmt (AChEWT residues S234, E365 and H478 were mutated to alanine) was described previously24. Transient transfection of RAW 264.6 cells or peritoneal macrophages with DNA constructs was performed by jetPRIMEr reagent. A β-galactosidase gene under a cytomegalovirus enhancer promoter was used to determine transfection efficiency. A commercial kit (Tropix, Bedford, MA, USA) was employed in luciferase assay. Briefly, washed cell cultures with PBS and resuspended in 100 mmol/L KH2PO4 (pH 7.8) supplemented with 1 mmol/L dithiothreitol and 0.2% Triton X-100. Tropix TR717 microplate luminometer was used to quantify the luminescent reaction. Luciferase activity was expressed as absorbance (up to 560 nm)/mg of protein.
2.11. Determination of ACh
ACh in culture medium were determined by a choline/acetylcholine assay kit (Abcam, ab65345). This choline/acetylcholine assay kit quantified ACh by a fluorometric or colorimetric method. Fluorometric method is higher sensitivity than colorimetric method, which was employed here. The free choline could turn to products, which reacted with the choline probe to generate fluorescence (Ex/Em 535/587 nm). To detect ACh in samples, AChE was added to the reaction, which converted ACh to choline. Then, the total choline can be measured. The concentration of ACh should be: total choline–free choline.
2.12. Preparation of lipid raft
Cells were collected for membrane raft preparation23. The preparation was carried at 4 °C. Briefly, washed cells with PBS and resuspended in buffer A (150 mmol/L NaCl, 50 mmol/L Tris-HCl, 10 μmol/L aprotinin, and 10 μmol/L leupeptin, pH 8.0). Then, sonicated the samples at low intensity 0.5 s three times. Between each sonication, the samples were stand on ice for 3 s to avoid heating. To get rid of the debris and nucleus, the samples were centrifuged at 500 × g, 5 min. The supernatant was collected and placed on ice for at least 1 h. One mL of the preparation was gradually mixed with 1 mL 80% sucrose in buffer A. The resulted 40% sucrose sample was loaded at the bottom of 12-mL centrifugation tube. Subsequently, 30% sucrose (4 mL) and 5% sucrose (5 mL) in buffer A were loaded in order. The gradient was centrifuged at 39,000 rpm (CP80WX, Hitachi) for 16 h. Collected 1 mL of each fraction from top to bottom. The activity of ALP and AChE were detected. Precipitated the proteins from fractions by 20% (final) trichloroacetic acid. Placed the samples on ice for 30 min and centrifuged at 13,200 rpm (5415 R, Eppendorf, Hamburg, Germany), 15 min. Saved the pallet and washed twice by cold acetone air dried to make sure no acetone was left. Then, dissolved and boiled the protein pallet in 1× lysis buffer, which was ready for Western blot assay.
2.13. Chromatin immunoprecipitation (ChIP) assay
ChIP kit (Abcam, ab500) was employed here. Gently fixed the cells with 4% formaldehyde and sonicated by using Q125 Sonicator (Qsonica, Melville, NY, USA). The specific NF-κB–DNA complex was immune-precipitated using anti-NF-κB antibody (Abcam). Primers flanking the NF-κB binding site of AChE promoter was used as follows: sense: 5′-CAA AAC TGC ACA CTT CCC ACA -3′; anti-sense: 5′-ATG CTA CAA TGC ACT CTG C-3′. The products were run at 1% agarose gel electrophoresis.
2.14. Co-immunoprecipitation
Anti-mouse AChE antibody E-19 (1:50; Santa Cruz), anti-α7 nAChR antibody (1:50, Cell Signaling Technology) or IgG (1:50; Santa Cruz) covalently coupled to 100 μL protein G-agarose beads (Thermo Fisher Scientific) overnight at 4 °C. Washed the beads 3 times with ice-cold extraction buffer. One hundred μL of antibody conjugated beads were incubated together with 200 μL (50 μg) of cell lysates extracted from AChET and PRiMA co-transfected RAW 264.7 cells. After the intensively washing with cold extraction buffer, the activity of AChE from the supernatant with protein G beads were determined. Then, centrifuged and discarded the supernatant. The beads were re-suspended and boiled in 100 μL 1× lysis buffer before performing on Western blot analysis.
2.15. Laser confocal fluorescent microscopy
Fluorometric measurements were performed by using a Leica Sp8 Confocal Microscope (Wetzlar, Germany), equipped with a 63 × objective. To measure the ion influx, Ca2+ concentration was monitored by using the fluorescent calcium indicator Fluo-4 AM (Invitrogen). RAW 264.7 cells were seeded on the SPL confocal dish (100350, Gyeonggi-Do, Korea). After 48 h, cells were incubated in a normal physiological solution (NPSS, 10 mmol/L glucose, 5 mmol/L KCl, 140 mmol/L NaCl, 1 mmol/L MgCl2, 1 mmol/L CaCl2, and 5 mmol/L HEPES, pH 7.4) containing 5 μmol/L Fluo-4 AM, 30 min, 37 °C. A23187 (Sigma; purity >98%), a Ca2+ ionophore, was used as a positive control. The amount of Ca2+ was evaluated by measuring the fluorescence intensity at Ex/Em 488/525 nm. The Ca2+ influx was displayed as a ratio of fluorescence relative to the intensity (Fn/F0).
2.16. Statistical analyses
All the result was represented as the mean ± standard error of mean (SEM) of at least three independent experiments. Statistical significance was determined by Student's t-test, one-way or two-way ANOVA with subsequent application of different multiple comparisons methods. Significant values were indicated by ∗P < 0.05; ∗∗P < 0.01; and ∗∗∗P < 0.001.
3. Results
3.1. AChE is expressed on the surface of macrophages
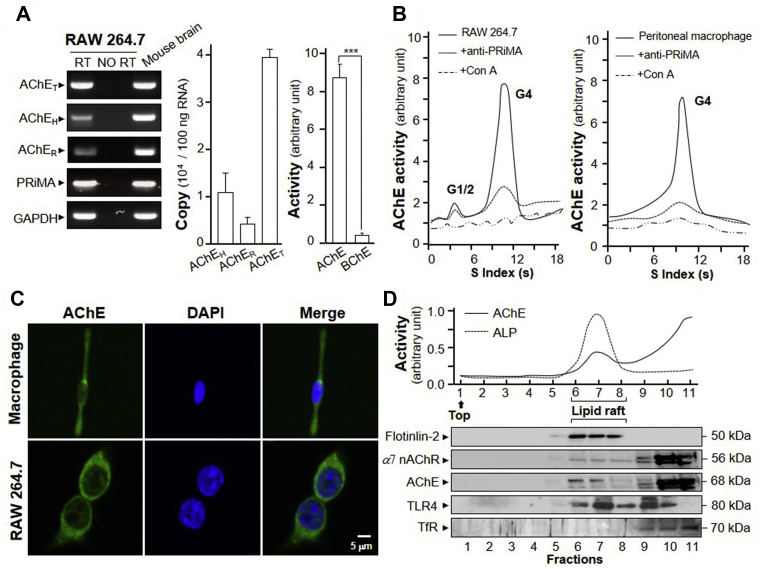
The splicing variants of AChE in macrophage was determined. Here, we quantified the AChE variants using real-time PCR coupled with standard curves, as described previously16. The amplicon of each AChE isoform was sub-cloned into pcDNA4; the generated plasmids were used for construction of standard curve, in which the Ct values were plotted against the corresponding copy number with good linearity. The splicing variants, AChEH, AChER and AChET, as well as PRiMA, were revealed in RAW 264.7 cells (Fig. 1A). The expression of AChET subunit in RAW 264.7 cell was higher than that of AChEH or AChER by 4- to 10-fold (Fig. 1A). AChE enzymatic activity in RAW 264.7 cell was robustly higher than butyrylcholinesterase (BChE) activity, suggesting an insignificant role of BChE here (Fig. 1A). In sucrose density gradient analysis, the G4 form of AChE was confirmed as the predominant form in cultured RAW 264.7 cell and peritoneal macrophage (Fig. 1B). To demonstrate the glycan composition of AChE in RAW 264.7 cell and peritoneal macrophages, Con A was employed here, which could bind to high-mannose glycan chains. In precursor and mature AChE, the glycan chains were abundant. Immobilized Con A precipitated most of the AChE activity in RAW 264.7 cell and peritoneal macrophage, which suggested the full glycosylation of AChE in both cells (Fig. 1B), as in other cells9. In addition, PRiMA antibody precipitated most of the AChE activity, suggesting the identity of PRiMA-linked G4 AChE (Fig. 1B). The surface expression of AChE in peritoneal macrophage and RAW 264.7 cell were recognized by anti-AChE antibody in absent of detergent (Fig. 1C). Probing the location of AChE, α7 nAChR and TLR4, the isolation of lipid raft was performed. The raft-enriched fractions (fractions 6–8) contained 60%–70% of total membrane-bound alkaline phosphatase (ALP), serving as a membrane raft marker (Fig. 1D). Western blot of the raft-enriched fractions showed the abundance of raft-associated protein, flotillin-225. The transferrin receptor, a non-lipid raft marker26, could not be detected in raft-enriched fractions. The expressions of AChE, α7 nAChR and TLR4 could be detected in the raft-enriched fractions, suggesting part of these cholinergic molecules was located in the raft fractions (Fig. 1D).
Figure 1.
AChE molecular isoforms in RAW 264.7 cells and peritoneal macrophages. (A) Total RNAs were extracted from RAW 264.7 cells and mouse brain for PCR to determine the presence of AChET, AChEH, AChER, PRiMA and GAPDH. PCR products were resolved on a 1% SYBR safe-stained agarose gel and visualized under UV light (Left panel). The identity of PCR product was confirmed by DNA sequencing. Mouse brain RNA served as a positive control. Expressions of AChE splicing variants of RAW 264.7 cell were measured by qPCR, coupled with standard curves. Data are expressed as numbers of copies (middle panel). Protein lysates of the RAW 264.7 cells were collected for AChE and BChE activity assay in arbitrary unit (right panel). Statistical significance was analyzed by Student's t-test. ∗∗∗P < 0.001. All values are the mean ± SEM, n = 4, each with triplicate samples. (B) Equal amount of proteins from RAW 264.7 cells (Left panel) or peritoneal macrophages (right panel) were incubated with Con A or agarose or PRiMA antibody. Supernatant was subjected to sucrose density gradient analysis. AChE activities are expressed in arbitrary unit. (C) Peritoneal macrophages and RAW 264.7 cells were cultured. Cells were fixed with 4% PFA for 15 min and stained with anti-AChE antibody, followed with goat Alexa Fluor 488-conjugated antibody. Nucleus was stained with DAPI. (D) AChE activity from RAW 264.7 cells in detergent-resistant (raft; fractions 6–8) and detergent-soluble (non-raft) fractions was determined after flotation in discontinuous sucrose gradients with 0.5% cold Triton X-100. Aliquots of each even fraction were analyzed by 8% SDS-PAGE. The expressions of flotillin-2 and transferrin receptor (TfR) are shown in Western blots as controls. Enzymatic activities of AChE and ALP are expressed in arbitrary units. ALP serves as control to identify raft-enriched fractions.
3.2. LPS positively regulates AChE transcription via NF-κB
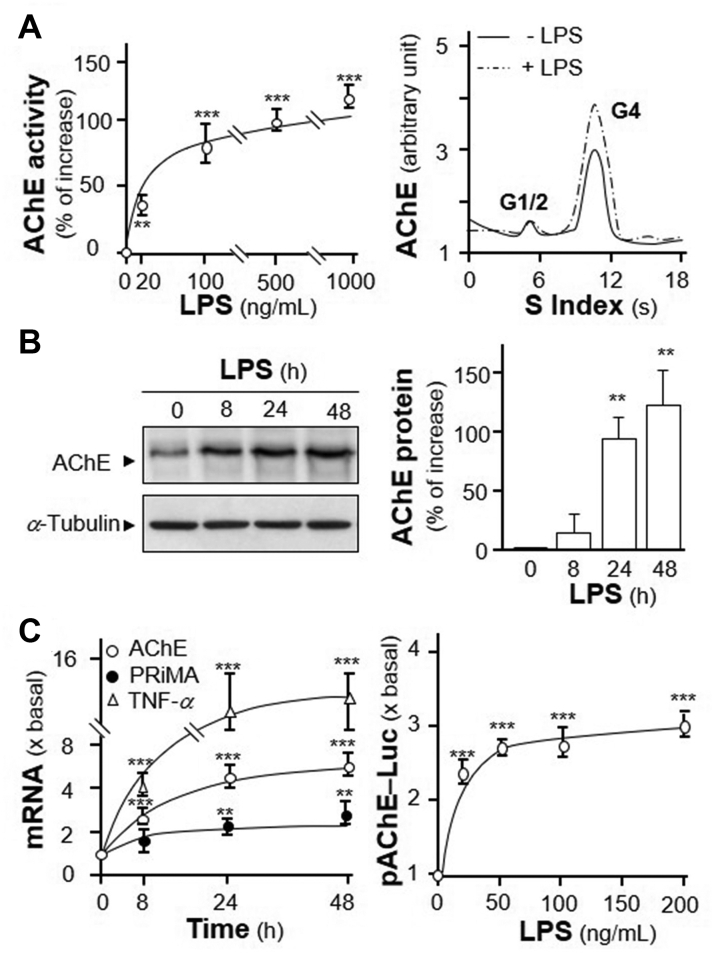
LPS acts as the prototypical endotoxin, promoting the secretion of pro-inflammatory cytokines, especially in monocytes, dendritic cells, macrophages and B cells27, including the secretion of TNF-α28. In cultured macrophage, LPS induced AChE activity in a dose-dependent manner (Fig. 2A). At the same time, G4 AChE was the major isoform being induced (Fig. 2A). AChE protein was induced by over 100% at 20 ng/mL of LPS treatment, which was in a time-dependent manner (Fig. 2B). The mRNA expression levels of AChE and PRiMA showed the upregulation of LPS on their transcriptions in a time-dependent manner. It reached the maximal induction at ∼4.2- and ∼2-fold at LPS treatment for 24 h, respectively (Fig. 2C). The mRNA expression of TNF-α was a positive control in inflammatory response. To further determine the regulation of LPS on ACHE gene transcription, the AChE promoter tagged with a luciferase gene, i.e. pAChE-Luc, was employed here. The promoter construct was transfected to RAW 264.7 cells, and then which were treated with LPS at various concentrations. The luciferase activities, driven by the promoter (pAChE-Luc), were increased by the challenge of LPS in a dose-dependent manner, reaching to a maximum of ∼3-fold at 50 ng/mL (Fig. 2C). Thus, the expression of AChE was upregulated during inflammatory process.
Figure 2.
LPS induces expression of AChE. (A) Cultured RAW 264.7 cells were challenged by LPS at various concentrations for 3 days. Cell lysates were collected for AChE activity assay (left panel) and sucrose density gradient analysis (right panel). (B) Cultured RAW 264.7 cells were induced by LPS at 20 ng/mL for different hours. Protein lysates of the cultures were performed Western blot assay (left panel). Quantification plot was shown in histograms (right panel). (C) Total RNAs was extracted from cultures as in (B) to perform quantitative PCR. The mRNA expressions of AChE, PRiMA and TNF-α were normalized with 18 S rRNA (left panel). Cultured cells were transiently transfected with promoter construct, pAChE-Luc, for 24 h. Then, cells were induced by LPS as in (A). Cell lysates were collected for luciferase assay. Statistical significance was analyzed by one-way ANOVA with subsequent application of Dunnett's multiple comparisons test. ∗∗P < 0.01, ∗∗∗P < 0.001 vs. control. Values are expressed as the fold, or percentage, of increase to basal reading, and are in means ± SEM, n = 4, each with triplicate samples.
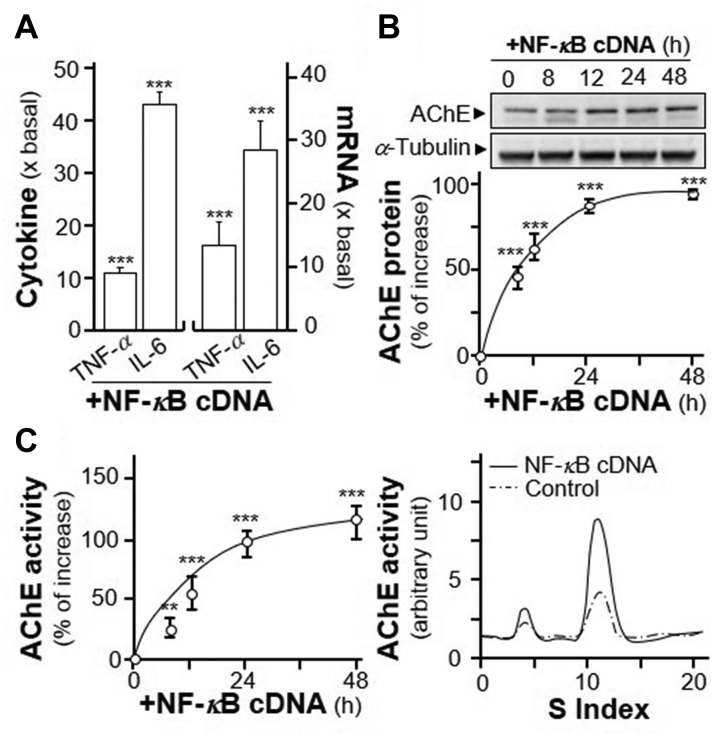
To further demonstrate the inductive effect of NF-κB on AChE expression, an overexpression system was employed. In NF-κB cDNA-transfected RAW 264.7 cells, the amounts of two cytokines, TNF-α and IL-6, were significantly induced at ∼10- and ∼40-fold, respectively (Fig. 3A). In parallel, the induction of TNF-α and IL-6 mRNA were ∼15- and ∼30-fold, as compared to control, respectively (Fig. 3A). Under the overexpression of NF-κB, AChE protein was induced in a time-dependent manner with the maximal induction at 100%, as compared to control (Fig. 3B). In parallel, the enzymatic activity of AChE was induced in a time-dependent manner in the NF-κB cDNA-transfected RAW 264.7 cells (Fig. 3C). Sucrose density gradient assay showed the induced AChE was G4 isoform (Fig. 3C).
Figure 3.
Over expression of NF-κB induces expression of G4 AChE. (A) RAW 264.7 cells were transiently transfected with pcDNA3 (as control) or NF-κB cDNA for 24 h. The cultures medium was collected for ELISA assay (left panel). Total RNA was extracted from cultures to perform quantitative PCR (right panel). Values were normalized with 18 S rRNA. Statistical significance was analyzed by Student's t-test. ∗∗∗P < 0.001. (B) RAW 264.7 cells were transiently transfected with NF-κB cDNA. The cultures were collected for Western blot assay (upper panel). The expression level was calculated using α-tubulin as an internal control. Quantification plot was shown in Lower panel. Statistical significance was analyzed by one-way ANOVA with subsequent application of Dunnett's multiple comparisons test. ∗∗∗P < 0.001. (C) RAW 264.7 cells were transiently transfected with NF-κB cDNA as in (B). The cultures were collected for AChE activity assay (left panel) and sucrose density gradient assay (right panel). Statistical significance was analyzed by one-way ANOVA with subsequent application of Dunnett's multiple comparisons test. ∗∗P < 0.01, ∗∗∗P < 0.001 vs. control. Data are expressed as the fold of basal value, or percentage of increase to basal reading, in mean ± SEM, n = 4, each with triplicate samples.
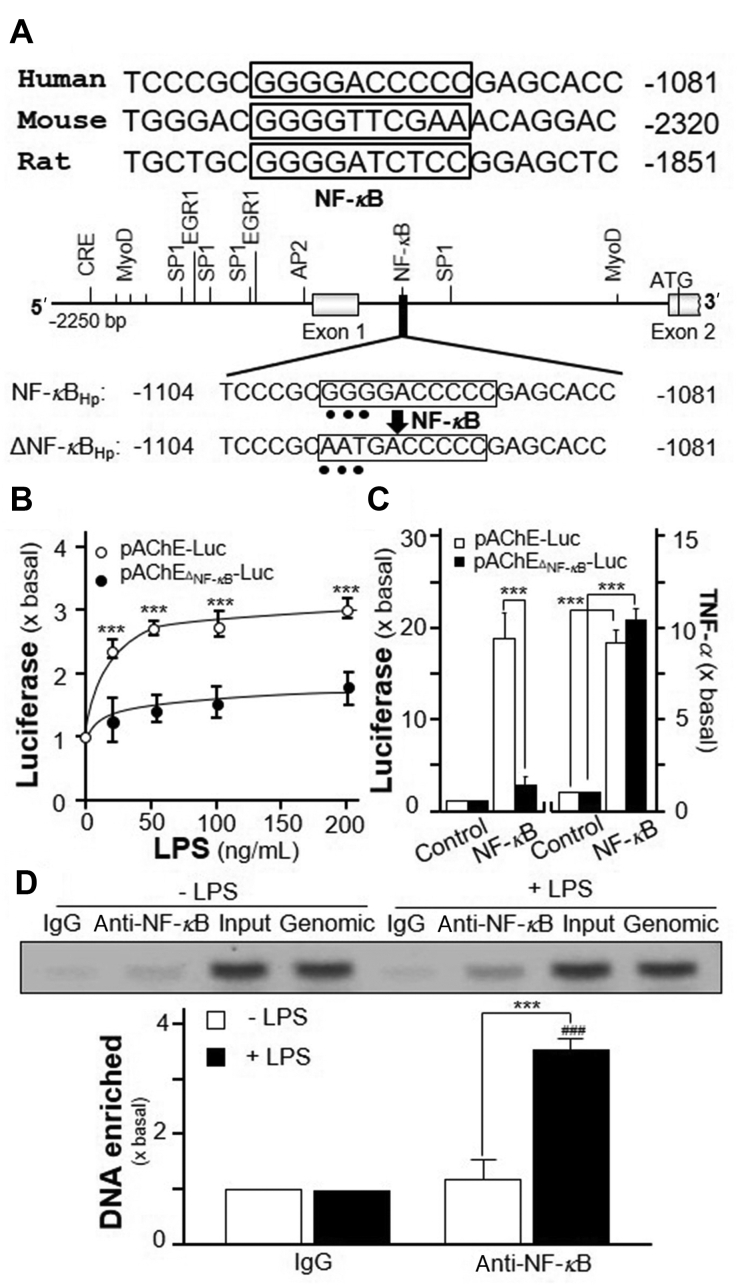
The NF-κB binding sequences in promoters of various mammalian ACHE genes are conserved in human, mouse and rat (Fig. 4A). To determine the function of NF-κB on ACHE gene, pAChEΔNF-κB-Luc, having mutation on NF-κB-binding site of human ACHE promoter, was used to transfect into RAW 264.7 cells (Fig. 4A). The NF-κB-binding site sequence (GGG GAC CCC C) was mutated into AAT GAC CCC C on human AChE promoter. The application of LPS, even in high dose, in transfected RAW 264.7 cells showed no induction on pAChEΔNF-κB-Luc signal (Fig. 4B), as well as no induction by NF-κB over expression (Fig. 4C). As a control, pAChE-Luc activity was activated by the challenge of LPS. In parallel, the co-transfection of NF-κB cDNA with pAChE-Luc, or pAChEΔNF-κB-Luc, robustly induced the release of TNF-α, as a control for over expression of NF-κB in DNA-transfected cells (Fig. 4C). Moreover, the binding of NF-κB with human ACHE gene promotor was determined by chromatin immunoprecipitation (ChIP). In the input control group, the NF-κB occupancy at human ACHE promoter was barely detected (Fig. 4D). However, NF-κB occupancy was induced to ∼4-fold of enrichment after the application of LPS, as compared to the control (Fig. 4D). The primers flanking NF-κB-binding site of the ACHE promoter were used for DNA amplification. The upregulation of AChE transcription via NF-κB binding site on the promotor during inflammatory process therefore was proposed here.
Figure 4.
LPS regulates AChE transcription via NF-κB binding site. (A) A schematic diagram of human ACHE promoter with key transcription elements was shown. The NF-κB-binding site sequence (GGG GAC CCC C) was mutated into AAT GAC CCC C on the human promoter. (B) Cultured RAW 264.7 cells were transiently transfected with pAChE-Luc and its mutant pAChEΔNF-κB-Luc for 24 h before application of LPS for 2 days. The cultures were collected for luciferase assay. Statistical significance was analyzed by two-way ANOVA with subsequent application of Bonferroni's multiple comparisons test. ∗∗∗P < 0.001 vs. pAChEΔNF-κB-Luc at different LPS concentrations. (C) Cultured RAW 264.7 cells were transiently co-transfected pAChE-Luc or pAChEΔNF-κB-Luc with or without cDNA encoding NF-κB. The cultures were collected 2 days later for luciferase assay (Left panel), as well as for ELISA assay of TNF-α (Right panel). Statistical significance was analyzed by two-way ANOVA with subsequent application of Bonferroni's multiple comparisons test. ∗∗∗P < 0.001. (D) RAW 264.7 cells were treated with or without LPS at 20 ng/mL for 3 days. The ChIP assay was performed. The primers flanking the NF-κB-binding site of the ACHE promoter were used: sense: ATG CTA CAA TGC ACT CTG C; antisense: CAA AAC TGC ACA CTT CCC ACA. Results were normalized with each pull-down. Statistical significance was analyzed by two-way ANOVA with subsequent application of Bonferroni's multiple comparisons test. ∗∗∗P < 0.001. ###P < 0.001 vs. LPS IgG group. All the values are expressed as the fold of basal value, mean ± SEM, n = 4, each with triplicate samples.
3.3. AChE enhances LPS-induced inflammation
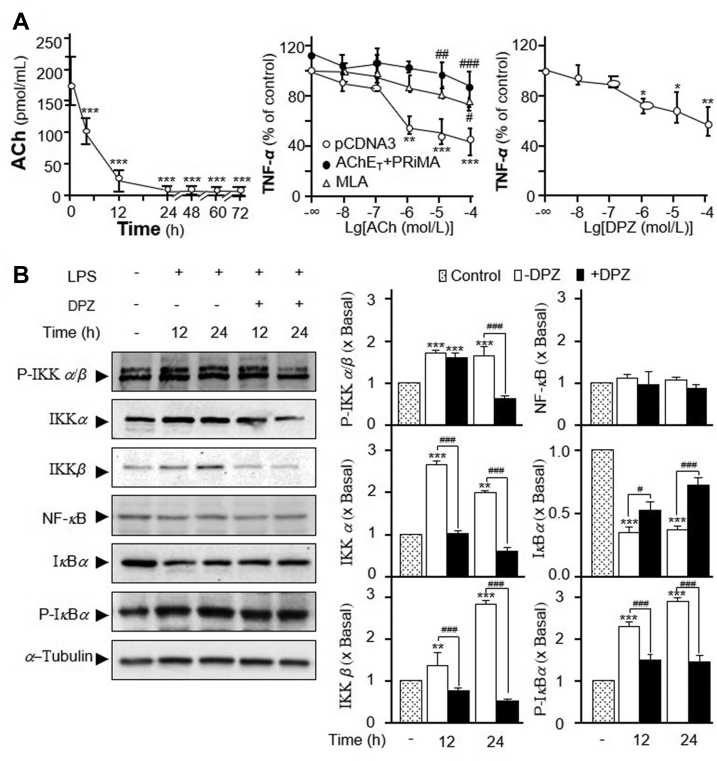
In order to investigate the immunological role of ACh and/or AChE in cultured macrophage, the existence of ACh in culture medium is of critical importance. The content of ACh in culture medium was dramatically decreased after the incubation with RAW 264.7 cells. After 4 h of incubation, ACh declined to only half of the initial culture medium, at ∼100 pmol/mL (Fig. 5A, left panel). Until to 12 h, ACh decreased to ∼30 pmol/mL, close to the detection limit of this assay. Beyond 24 h, ACh was negligible. In order to ignore the impact of ACh deriving from the medium, the experiment was performed after 16 h of seeding. Besides, the culture medium did not show AChE/BChE activity. Here, the release of TNF-α was measured in LPS-treated macrophage. ACh is known to inhibit the LPS-induced TNF-α release in macrophage2. In cultured macrophages, the suppression of LPS-induced the release of TNF-α reached to a maximum at 1 μmol/L of applied ACh, ∼60% of reduction (Fig. 5A, middle panel). This outcome was similar as previous reports2. In addition, the pre-treatment of methyllycaconitine citrate salt (MLA), an α7 nAChR antagonist, blocked the anti-inflammation effect triggered by ACh (Fig. 5A, middle panel). Overexpression of PRiMA-linked G4 AChE in the cultures reduced ACh level in medium, which thereafter blocked the anti-inflammation, as mediated by ACh. The suppression of TNF-α release by ACh was almost fully recovered by overexpression of PRiMA-linked G4 AChE (Fig. 5A, middle panel). In parallel, the treatment of donepezil, an AChE inhibitor, suppressed LPS-induced TNF-α release in a dose-dependent manner: the maximal suppression of TNF-α release by donepezil was at ∼40% (Fig. 5A, right panel). The aforementioned results therefore could be fully accounted by the amount of ACh in the medium.
Figure 5.
The effects of AChE on TNF-α release and NF-κB translocation in RAW 264.7 cells. (A) Culture medium of RAW 264.7 cells were collected at different hours. Dried by nitrogen and resolved in 100 μL choline assay buffer for ACh assay. Statistical significance was analyzed by one-way ANOVA with subsequent application of Dunnett's multiple comparisons test. ∗∗∗P < 0.001 vs. control (left panel). RAW 264.7 cells were transiently transfected with pcDNA3, or AChET with PRiMA cDNA, for 24 h before the pre-treatment with or without MLA (5 μmol/L). Then, application of LPS (20 ng/mL) with ACh at different concentrations onto transfected RAW 264.7 cells. The culture medium was performed ELISA assay to determine the release of TNF-α. Statistical significance was analyzed by two-way ANOVA with subsequent application of Tukey's multiple comparisons test. ∗∗P < 0.01, ∗∗∗P < 0.001 vs. control; #P < 0.05,##P < 0.01,###P < 0.001 vs. pcDNA3 group (middle panel). RAW 264.7 cells were co-treated with LPS 20 ng/mL, ACh at 1 μmol/L and donepezil (DPZ) at various concentrations. Statistical significance was analyzed by one-way ANOVA with subsequent application of Dunnett's multiple comparisons test. ∗P < 0.05, ∗∗P < 0.01 vs. control (right panel). (B) Cultured RAW 264.7 cells were treated with LPS 20 ng/mL, ACh at 1 μmol/L together with or without DPZ at 100 μmol/L for various hours. The amounts of p-IKKβ (∼87 kDa), IKKβ (∼87 kDa), IKKα (∼85 kDa), IκBα (∼39 kDa), p-IκBα (∼40 kDa), and NF-κB P65 subunit (65 kDa) were determined by using specific antibodies. α-Tubulin served as a loading control (Left panel). The band intensity was calibrated (Right panel). Statistical significance was analyzed by two-way ANOVA with subsequent application of Tukey's multiple comparisons test. ∗∗P < 0.01, ∗∗∗P < 0.001 vs. control; #P < 0.05, ###P < 0.001 vs. LPS without DPZ group. Data are percentage of control or in fold of change (X basal) to control). Values are shown mean ± SEM, n = 4, each with triplicate samples.
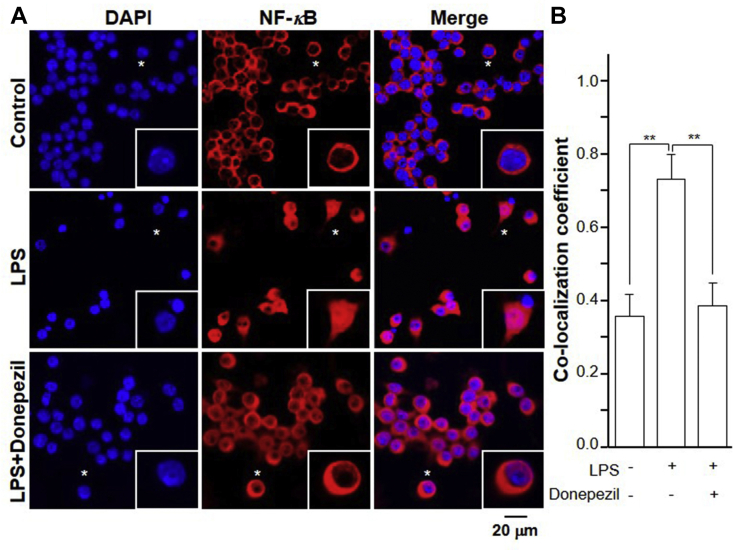
ACh binds and activates α7 nAChR, and thereafter inhibits degradation of IκBα and NF-κB nuclear translocation29. NF-κB, a protein complex consisting of P65 protein subunit30, regulates the transcription of DNA, cytokine production and cell survival. The dynamic translocation of NF-κB into the nucleus could contribute to the inflammatory gene expression. LPS application induced the expressions of IKKα, IKKβ, and the phosphorylation of IKKβ, in time-dependent manners. These inductions could be completely blocked by co-treatment of donepezil (Fig. 5B). Donepezil indirectly increased the concentration of ACh by inhibiting its hydrolysis. The expression of total NF-κB was not altered in the treatment of LPS or donepezil (Fig. 5B). Moreover, LPS suppressed expression of IκBα and induced expression of phosphorylated IκBα in time-dependent manners. These effects were reversed by co-treatment with donepezil (Fig. 5B). Using immunofluorescence staining, NF-κB and nucleus were identified by red and blue color, respectively (Fig. 6). LPS application in the cultures significantly induced the nuclear accumulation of NF-κB by ∼2-fold (Fig. 6). Co-treatment with donepezil significantly reduced the LPS-induced NF-κB nuclear accumulation.
Figure 6.
Immunofluorescent staining of NF-κB translocation. (A) Cultured RAW 264.7 cells were treated with LPS 20 ng/mL, ACh at 1 μmol/L together with or without donepezil at 100 μmol/L for 24 h. Immunofluorescence staining localization of NF-κB P65 by antibody, and nuclei staining by DAPI in RAW 264.7 cells. Asterisks indicate nuclei with NF-κB staining, also the enlarged cells in bottom right corner. (B) The co-localizing pixel for NF-κB P65 in channel 1 (Ch1) was calculated relative to the total number of pixels for the nuclei (T1) by using Zeiss co-localization coefficient function software. Statistical significance was analyzed by one-way ANOVA with subsequent application of Tukey's multiple comparisons test. ∗∗P < 0.01. All values are expressed as the mean ± SEM. n = 4, each with triplicate samples.
3.4. AChE enhances the migration of RAW 264.7 cells
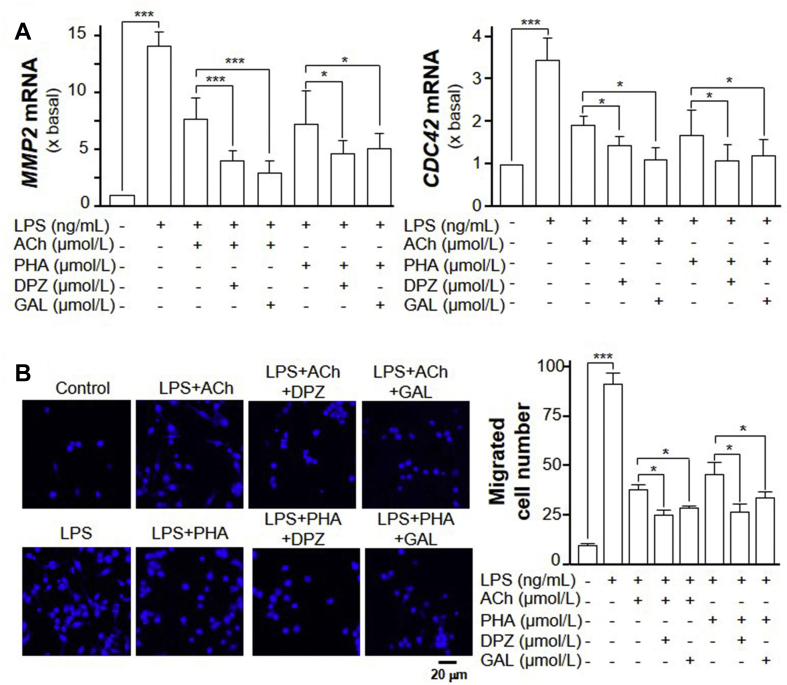
Activation of macrophage induces inflammatory reactions, including the migration of macrophages31. The migration of macrophage within the brain is of critical importance in causing neuro-inflammatory diseases. CDC42 and MMP2 are two pro-migratory genes, which can be elevated by LPS application in RAW 264.7 cells5,32. Application of LPS significantly induced the number of migrated RAW 264.7 cells, as well as the mRNA expressions of MMP2 and CDC42, in dose-dependent manner (Supporting Information Fig. S1). Here, the applied LPS at 100 ng/mL in cultured macrophages significantly increase the mRNA expressions of CDC42 and MMP2, which were ∼4- and ∼13-fold higher, respectively, as compared to control (Fig. 7A). ACh has been shown to inhibit LPS-induced MMP-9 production and cell migration via α7 nAChR in RAW 264.7 cells6. In line to this observation, applied ACh markedly suppressed the LPS-induced mRNA upregulation of CDC42 and MMP2 in the cultures. The suppression was ∼30% and ∼50%, respectively (Fig. 7A). PHA-543613 (PHA), an α7 nAChR agonist, was found to markedly suppress the LPS-induced mRNA upregulation of CDC42 and MMP2 in RAW 264.7 cells, similar as that of ACh (Fig. 7A). Donepezil and galantamine were shown to enhance the effect of ACh, or PHA, in suppressing the LPS-induced mRNA expressions of CDC42 and MMP2 (Fig. 7A).
Figure 7.
AChE activity regulates the migration of RAW 264.7 cells. (A) RAW 264.7 cells were treated with LPS together with or without ACh (1 μmol/L), donepezil (DPZ; 100 μmol/L), galantamine (GAL, 10 μmol/L) and PHA-543613 (PHA, 10 μmol/L) for 24 h. The expression of MMP2 (left panel) and CDC42 (right panel) mRNAs were determined by quantitative PCR analysis. (B) Cells were treated as in (A), Transwell® motility assay was performed. The migrated RAW 264.7 cells were counted. A representative image of the migrated cells is shown. The quantification data is shown in right panel. Statistical significance was analyzed by one-way ANOVA with subsequent application of Tukey's multiple comparisons test. ∗P < 0.05, ∗∗∗P < 0.001. Data are expressed as the migration cell number or the fold of basal value where control value is set as 1, mean ± SEM, n = 4, each with triplicate samples.
The Transwell® motility assay was performed to quantify the motility of cell migrating vertically from one side of membrane to the opposite side. LPS treatment significantly promoted the migration of RAW 264.7 cells (Fig. 7B). After 24 h of treatment with LPS in the culture, the migrated cells were ∼8-fold higher than that of control (Fig. 7B). After LPS and ACh co-treatment, the number of migrated cells was significant less than the LPS-treated group. At the same time, the application of PHA decreased the number of migrated RAW 264.7 cells. In addition, the treatment of donepezil, or galantamine, further enhanced the effect of ACh and PHA in inhibiting cell migration (Fig. 7B).
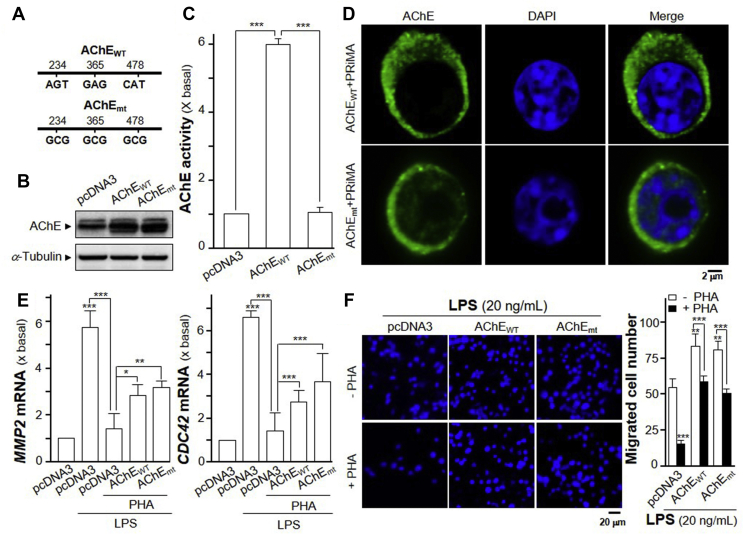
To investigate whether AChE enzymatic activity playing role in inflammatory responses, a DNA mutant of AChET subunit without enzymatic activity was constructed. AChEWT residues S234, E365 and H478 of the catalytic triad contributing to its enzymatic activity were all mutated to alanine (A) to generate AChEmt (S234A, E365A, H478A) cDNA construct (Fig. 8A). This mutated AChE protein contained same protein size as the wild type AChE but showed no enzymatic activity (Fig. 8B and C). Recognized by immuno-staining, the PRiMA-linked AChEmt protein was being expressed on the surface of cultured RAW 264.7 cells, similar as that of AChEWT (Fig. 8D). LPS at 20 ng/mL, a sub-maximal concentration, was used in the following experiments. In LPS-induced RAW 264.7 cells, the induced MMP2 and CDC42 expressions were markedly abolished by the application of PHA (Fig. 8E). The overexpression of AChEWT or AChEmt in cultured macrophages reversed, almost by half, the suppression of PHA in the mRNA expressions of MMP2 and CDC42 (Fig. 8E). The constructs of AChEWT and AChEmt showed similar magnitude of suppression, suggesting the enzymatic activity might not be relevant in this scenario. In Transwell® motility assay, the overexpression of AChEWT and AChEmt further induced the LPS-induced cell migration by over 30% (Fig. 8F). The application of PHA on LPS-induced RAW 264.7 cells significantly suppressed the migration of RAW 264.7 cells, as compared to control. Overexpression of AChEWT and AChEmt reversed, almost to complete, the suppression of PHA in migration of RAW 264.7 cells (Fig. 8F).
Figure 8.
Overexpression of AChEWT or AChEmt affects migration of RAW 264.7 cells. (A) Diagram shows wild type AChE (AChEWT) and AChE activity-deleted mutant (AChEmt). AChEWT S234 (AGT), E365 (GAG), H478 (CAT) were mutated to A (GCG). (B) RAW 264.7 cells were transiently transfected with pcDNA3 or AChEWT or AChEmt with PRiMA cDNA for 24 h. Protein of the cultures were collected for Western blot assay. α-Tubulin served as loading control. (C) AChE activity of the culture from (B) was measured. Statistical significance was analyzed by one-way ANOVA with subsequent application of Tukey's multiple comparisons test. ∗∗∗P < 0.001. (D) Peritoneal macrophages and RAW 264.7 cells were transiently transfected with pcDNA3 or AChEWT and AChEmt with PRiMA cDNA for 24 h. Cells were fixed with 4% PFA for 15 min and stained with anti-AChE antibody without Triton X-100, followed with the goat Alexa Fluor 488-conjugated antibody. Nucleus was stained with DAPI. Representative images were shown. (E) RAW 264.7 cells were transiently transfected with cDNAs as in (C). Transfected cells were treated with LPS (20 ng/mL) together with or without PHA (10 μmol/L) for 24 h. The expressions of MMP2 (left panel) and CDC42 (right panel) mRNAs were determined by quantitative PCR analysis. Statistical significance was analyzed by one-way ANOVA with subsequent application of Tukey's multiple comparisons test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (F) Cells were treated as in (E). The migrated RAW 264.7 cells were counted. Representative images of the migrated cells are shown. The quantification data is shown in right panel. Statistical significance was analyzed by two-way ANOVA with subsequent application of Tukey's multiple comparisons test. ∗∗P < 0.01, ∗∗∗P < 0.001. Data are expressed as the migration cell number or the fold of basal value where control value is set as 1, mean ± SEM, n = 4, each with triplicate samples.
3.5. Protein interference of AChE with α7 nAChR
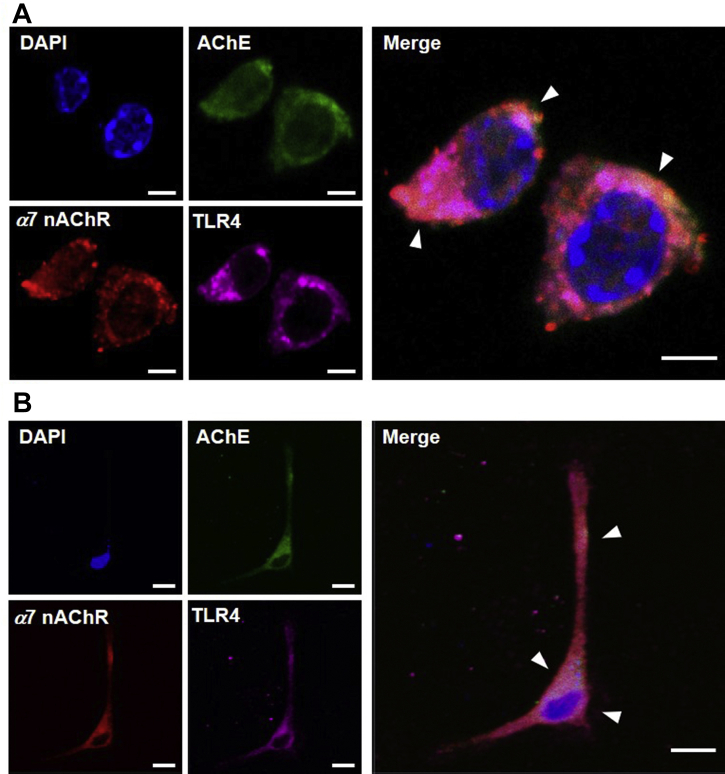
The results of the aforementioned experiments showed AChE, α7 nAChR and TLR4 proteins located, at least partially, in lipid raft of RAW 264.7 cells. Together with the result of AChE protein enhancing migration of RAW 264.7 cells, the potential interactions of AChE with α7 nAChR, or TLR4, was proposed. We investigated the co-localization of AChE with α7 nAChR and TLR4 in cultured RAW 264.7 cells (Fig. 9A), peritoneal macrophages (Fig. 9B) and bone marrow-derived macrophages (Supporting Information Fig. S2B). Immunofluorescence staining of AChE, α7 nAChR and TLR4 by their primary antibody was performed, which was shown as green, red and magenta, respectively. Nuclei were stained by DAPI, shown as blue color. AChE, α7 nAChR and TLR4 were located at surface of these cells. In merged images, as shown by arrows, the co-localization of AChE, α7 nAChR and TLR4 was identified (Fig. 9).
Figure 9.
AChE, α7 nAChR and TLR4 are co-localized on surface of RAW 264.7 cells and peritoneal macrophages. Immunofluorescence staining of AChE, α7 nAChR and TLR4 by their primary antibody, and nuclei staining by DAPI (blue), was performed. AChE was followed with the Alexa Fluor 488-conjugated anti-goat antibody (green). α7 nAChR was followed with the Alexa Fluor 555-conjugated anti-rabbit antibody (red). TLR4 was followed with the Alexa Fluor 647-conjugated anti-mouse antibody (Magenta). (A) Representative images of RAW 264.7 cells were shown. Bar = 5 μm. (B) Representative images of peritoneal macrophages were shown. Bar = 10 μm, n = 4.
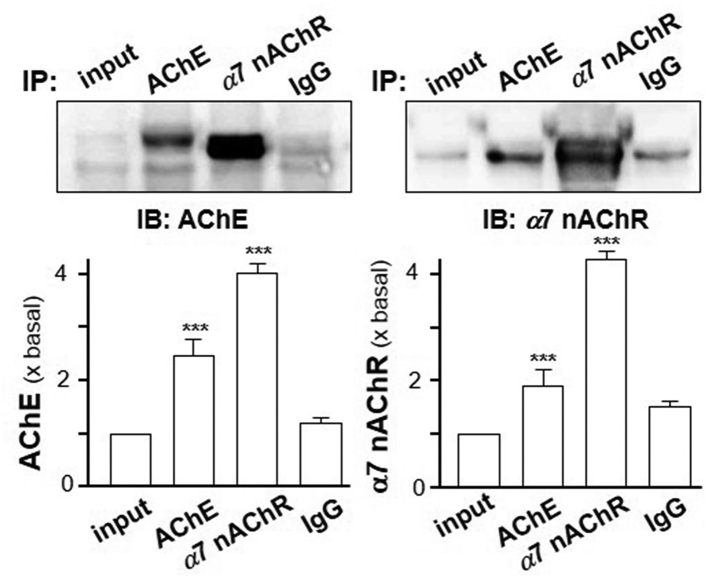
Since the co-localization of AChE and α7 nAChR was determined, the possible association of the two proteins aroused our curiosity. Cultured RAW 264.7 cells were transiently transfected with AChEWT and PRiMA cDNA for 24 h. The cell lysate of transfected RAW 264.7 cells were undergone Co-IP assay. Western blot analysis showed a clear background in the bead of IgG control group. In anti-α7 nAChR antibody group, the band of AChE was detected from the beads (Fig. 10). Vice versa, α7 nAChR (∼42 kDa) was also detected from the beads in anti-AChE antibody group (Fig. 10). Thus, it suggested that AChE and α7 nAChR could be Co-IP together.
Figure 10.
Association of α7 nAChR with AChE in RAW 264.7 cells. RAW 264.7 cells were transiently transfected with AChEWT with PRiMA cDNA for 24 h. The cell lysates were collected for co-immunoprecipitation by either anti-AChE (1:20) or anti-α7 nAChR antibody (1:50). The precipitated complexes on beads were subjected to Western blot by anti-AChE or anti-α7 nAChR antibody. AChE (∼76 kDa) and α7 nAChR (∼42 kDa) were identified. Representative gels are shown. Statistical significance was analyzed by one-way ANOVA with subsequent application of Dunnett's multiple comparisons test. ∗∗∗P < 0.001. Values are in mean ± SEM, n = 4.
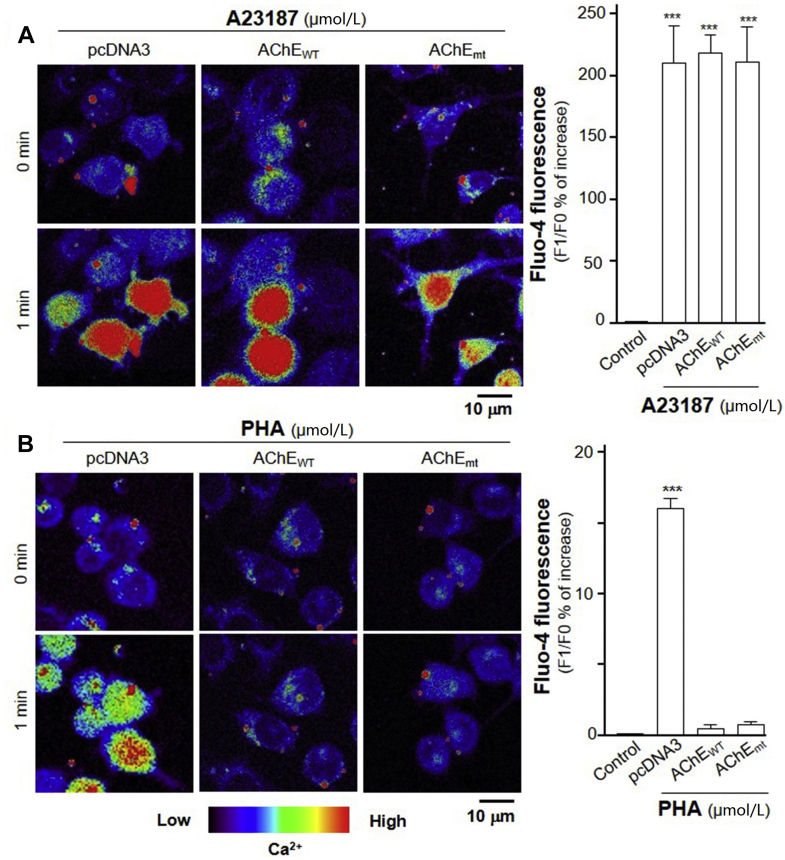
The nAChR has been known to be differentially permeable to Ca2+ influx, and indeed α7 nAChR is a very efficient way to raise cytoplasmic Ca2+ level33. By having an association of AChE with α7 nAChR, we hypothesized the influx of Ca2+ could be altered by this association. In order to eliminate the role of enzymatic activity in this process, we employed wild type AChEWT and mutated AChEmt cDNAs in the transfected RAW 264.7 cells. The Ca2+ influx was investigated in the cultures by using Fluo-4 AM, a Ca2+ indicator. Serving as a positive control, A23187, a calcium ionophore, was applied onto the transfected cells. The applied A23187 induced Ca2+ increase in pcDNA3, AChEWT and AChEmt + PRiMA transfected cells, suggesting overexpression of G4 AChE with or without enzymatic activity did not affect the ability of calcium change (Fig. 11A). The Ca2+ influx was investigated after treatment of PHA in transfected RAW 264.7 cells. The increase of Ca2+ influx was found after 1-min of PHA treatment. The maximal induction was over 15% (Fig. 11B). Overexpression of G4 AChE with or without enzymatic activity completely inhibited the PHA-induced Ca2+ influx (Fig. 11B). As to eliminate the error in DNA transfection efficiency, low magnification was used to determine the Ca2+ change, and which showed similar result as that in high magnification (Supporting Information Fig. S3). The above-mentioned results supported the interaction of AChE with α7 nAChR on the surface of cells, and AChE protein could inhibit the downstream Ca2+ influx of activated α7 nAChR.
Figure 11.
Overexpression of AChEWT or AChEmt inhibits PHA-induced Ca2+ mobilization. (A) RAW 264.7 cells were transiently transfected AChEWT or AChEmt with PRiMA cDNA for 24 h. Cultured RAW 264.7 cells were labeled with fluorescent Ca2+ indicator Fluo-4 AM for 30 min. Fluorimetric measurement was performed after the treatment of A23187, a Ca2+ ionophore at 1 μmol/L. (B) Transfected RAW 264.7 cells, as in (A), were labeled with fluorescent Ca2+ indicator Fluo-4 AM for 30 min. PHA at 10 μmol/L was applied. The amounts of Ca2+ were evaluated by measuring the fluorescence intensity. Micrographs were taken by a confocal microscope (Right panel). Quantification of Ca2+ was displayed as a ratio of fluorescence intensity at any time (Fn) to the control at time 0 (F0) in the cultures (left panel). Statistical significance was analyzed by one-way ANOVA with subsequent application of Dunnett's multiple comparisons test. ∗∗∗P < 0.001. Values are in mean ± SEM, n = 4.
4. Discussion
Vigilant defense is necessary for survival during injury and attack. In the presence of invaders, immune system continuously surveys the body. CAP is a complicated interplay between immune and nervous systems, which dominantly happens in blood and mucosa. This signaling pathway is acting on α7 nAChR locating on the surface of blood's macrophages34, 35, 36. In view of possible clinical application of CAP, the potential usage of neural input to control immune responses attracted more and more attention today.
Exposure of human macrophages to nicotine or ACh effectively inhibited the release of TNF-α, IL-1, IL-6 and IL-18 in response to LPS2. Tissue macrophages produce most of TNF-α that appears systemically during an excessive inflammatory response. The binding of cholinergic receptor with its ligand in macrophage decreased the synthesis of proinflammatory cytokines (TNF-α, IL-1 and IL-18) but not for anti-inflammatory cytokines, IL-102: this cholinergic effect was similar to our findings that application of ACh decreased markedly the LPS-induced TNF-α release in RAW 264.7 cells. However, the mRNA level of TNF-α was not inhibited by ACh, which suggested the inhibited cytokine synthesis should be at post-transcriptional level. Contrary to the robust sensitivity of macrophage to ACh, the cytokine-inhibiting action of ACh in monocytes was less effective than macrophages; because the synthesis of cytokines in monocytes could only by triggered by very high concentration of the agonists2. Hence, macrophage should be a good model in investigating the role of AChE in immune system.
In primitive plants and unicellular organisms, the widely distribution of ACh was reported. It suggested the ACh expressed its biological function in non-nervous system37,38. Not surprisingly, the detection of ACh in blood has been reported; although ACh was unlikely to be present in blood due to high activity of cholinesterase, i.e., AChE and BChE. The quantification of ACh in blood was determined in various species, including dog, horse, rabbit and human39: the concentration was ranged from 10 pmol/mL–150 nmol/mL according to different species and measurements. The measurement of ACh in human blood was performed by using different methods, such as polarography, pyrolysis-gas chromatography–mass spectrometry39. Thus, the existence of ACh in blood suggests a possible role of which in regulating immune system40.
The presence of ACh in immune cells was proposed to contribute to the blood circulating ACh41. Firstly, the fraction from human peripheral blood mononuclear leukocyte, consisting mainly of lymphocyte and a small fraction of monocyte, was confirmed having ACh39. Subsequently, the existence of ACh in immune cells have been detected in various human leukemic cell lines, also rat lymphocytes, including T and B cells41. ACh was identified in spleen, though it should not be cholinergically innervated42. These findings provided a proper explanation for ACh function in immune responses. Moreover, human T cell line has higher ACh concentration than pre-lymphoma, monocytic and B cells41. Especially, the content of ACh in CD4+ T cells was much higher than that in CD8+ T cells41 (Fujii et al., 2017). Due to the robust sensitivity of macrophage to ACh, these non-neuronal cells, producing ACh, might participate in modulating the function of adjacent macrophages38.
The intracellular ACh content in macrophage and dendritic cell has not been fully understood. Only one study showed low level of ACh was released and could be measured from lung macrophages; but these levels were very close to the detection limit of assay43. In accordance with higher ACh content, higher activity of ChAT was observed in rat T cell, as compared to that in B cell44. ChAT mRNA of human lung and alveolar macrophages was analyzed using RT-PCR of a rather long cycles (45 cycles), suggesting a marginal expression of ChAT mRNA43. In addition, splenic macrophage also expressed ChAT, which is conformed in transgenic mice3. Upon activation with LPS, macrophage has the possibility for ChAT-catalyzed ACh synthesis in a cell-intrinsic manner3,45. In macrophage, application of LPS and Toll-like receptor agonists induced expressions of ChAT mRNA and enzyme protein3. Hence, the release of ACh from macrophage upon activation with LPS needs to be further confirmed. Thus, ACh synthesized in macrophages might regulate innate immune responses by modulating cytokine production, including TNF-α.
The expression of AChE has been reported in various immune cells and organs, e.g., macrophage, T cell, B cell and spleen11,46. Interestingly, the expression and activity of AChE have been shown to be modulated during inflammation in animal models11,46. The activity of AChE was increased in LPS-administrated mice47. So far, the anti-inflammatory effect of AChE inhibitors has been widely investigated, experimentally and clinically, in animal models and human. In LPS-administrated animal model, donepezil, tacrine, rivastigmine and galantamine significantly attenuated the neuroinflammation by decreasing the LPS-induced levels of IL-2 and TNF-α36,47. In murine endotoxemia, peripheral administration of galantamine significantly reduced serum TNF-α level through α7 nAChR-dependent CAP signaling48; the pre-treatment and post-treatment of physostigmine significantly reduced the capillary leakage and the leukocyte–endothelial interaction by activating CAP49; Eserine and JWH-133 inhibited central and peripheral inflammation by reducing IL-6, E-selectin and vascular cell adhesion molecule 150. In rats suffering from rheumatoid arthritis, galantamine significantly reduced all biomarkers of inflammation, showing a novel therapeutic function51. AChE inhibitors are currently the most established medication worldwide for the treatment of Alzheimer's disease (AD). In Alzheimer's patients, donepezil down-regulated expression and production of inflammatory cytokines (IL-1, IL-6 and TNF-α), and increased anti-inflammatory cytokine (IL-4) in peripheral blood mononuclear cells52. Thus, AChE inhibitor has been proposed to be therapeutic agent in various inflammatory diseases.
Cell specific regulation of AChE expression under inflammatory conditions have been demonstrated in monocyte-derived cultures of human macrophages, in which the mRNA of AChE, especially AChET isoform, was generally upregulated in inflammatory situations46. The upregulation of AChE in macrophage could affect ACh level46. Hence, Shapira et al.53 indicated that ACHE gene promoter contains DNA binding sites that can potentially recruit transcription factors involving in inflammatory response, i.e. NF-κB factor. Another study supported the intracellular AChE interacting with receptor for activated C kinase 1, which thereafter could meditate the activation of NF-κB54. Our current results support the notion that AChE expression was tightly regulated by inflammatory action, triggered by endotoxin. The transcription of AChE was significantly induced during inflammation in RAW 264.7 cells, which was upregulated by the binding of NF-κB with AChE promotor. Such regulation will surely be relevant for the action of ACh within the context of CAP. Moreover, AChE, even in the absent of enzymatic activity, affected the cell migration suggesting possible adhesion properties of the enzyme. Indeed, AChE possesses similar sequence with a protein family known as cholinesterase-like adhesion molecules, e.g. neurotactin, gliotactin, glutactin and neuroligins55. The putative sites of AChE responsible for AChE adhesive properties have been proposed55, and the specific inhibitors for these sites have been reported to decrease adhesion of osteoblastic cells56.
For over a decade, α7 nAChR and AChE has been known physically interacted during development57. In neuronal proliferation and differentiation, the two proteins played an integral role58. Fossier59 envisaged the non-hydrolytic action of AChE caused up-regulation of the AChR. Then, Bond60 demonstrated that peptides derived from the C-terminus of AChE selectively bond with α7 nAChR and upregulated its expression. Meanwhile, activation of α7 nAChR reciprocally upregulated the expression of AChE. Here, we indicated for the first time that the spatial co-localization and interaction of α7 nAChR and AChE was identified in immune cells. The intracellular Ca2+ increase, triggered by α7 nAChR activation, could be inhibited by overexpression of AChE. Hypothetically, the tertiary structure of AChE might be too bulky, which could interfere the binding sites of α7 nAChR with its agonists. Alternatively, the structure of AChE may constrain α7 nAChR into a conformation, which occludes the residues necessary for its agonist's binding. All the mentioned evidences supported our findings on the association of AChE with α7 nAChR.
Inflammation clearly occurs as a major risk factor underlying aging and age-related diseases, such as AD, cancer and arthritis61. It had been reported that AChE was upregulated in pathologically vulnerable regions of AD. But, the involvement of AChE in inflammation in this area has not been seriously considered. In immune cells, the role of AChE in CAP was complex and involved in dual processes. On one hand, AChE directly hydrolyzes ACh to promote inflammation via inhibition of CAP. In parallel, NF-κB transcription factor bound to ACHE gene promotor, which triggered the transcription of AChE during inflammation. On the other hand, AChE protein enhanced the inflammatory responses via interacting with α7 nAChR, which firstly indicated the direct role of AChE in CAP, independent of hydrolyzing ACh. As such, AChE could serve as a novel target to treat age-related diseases by anti-inflammatory responses.
Acknowledgments
This study was supported by Shenzhen Science and Technology Committee Research Grant (JCYJ20170413173747440, ZDSYS201707281432317, JCYJ20180306174903174, CKFW2016082916015476, China), China Post-doctoral Science Foundation (2019M653087), Zhongshan Municipal Bureau of Science and Technology (ZSST20SC03, China), Guangzhou Science and Technology Committee Research Grant (GZSTI16SC02 and GZSTI17SC02, China), Hong Kong RGC Theme-based Research Scheme (T13-607/12R, China); Hong Kong Innovation Technology Fund (UIM/340, UIM/385, ITS/500/18FP, TCPD/17–9, PD18SC01 and HMRF18SC06, China).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.05.005.
Author contributions
Etta Y.L. Liu designed and performed most of the experiments and wrote the paper. Yingjie Xia and Miranda L. Xu cultured the mouse peritoneal macrophages and prepared the slides for immunofluorescent staining. Xiangpeng Kong performed the TNF-α ELISA assay. Maggie S.S. Guo prepared the SDS-PAGE gel for Western blot assay. Anna X.D. Yu amplified parts of the construct used in the paper. Brody Z.Y. Zheng helped to culture RAW 264.7 cells. Shinghung Mak revised and gave suggestions on the revision of the paper. Karl W.K. Tsim proposed the project, analyzed data and wrote the paper with Etta Y.L. Liu.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Fernández-Cabezudo M.J., George J.A., Bashir G., Mohamed Y.A., Al-Mansori A., Qureshi M.M. Involvement of acetylcholine receptors in cholinergic pathway-mediated protection against autoimmune diabetes. Front Immunol. 2019;10:1038. doi: 10.3389/fimmu.2019.01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borovikova L.V., Ivanova S., Zhang M., Yang H., Botchkina G.I., Watkins L.R. Vagus nerve stimulation attenuates the systemic inflammatory response to endotoxin. Nature. 2000;405:458–462. doi: 10.1038/35013070. [DOI] [PubMed] [Google Scholar]
- 3.Reardon C., Duncan G.S., Brüstle A., Brenner D., Tusche M.W., Olofsson P.S. Lymphocyte-derived ACh regulates local innate but not adaptive immunity. Proc Natl Acad Sci U S A. 2013;110:1410. doi: 10.1073/pnas.1221655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiguchi N., Kobayashi D., Saika F., Matsuzaki S., Kishioka S. Inhibition of peripheral macrophages by nicotinic acetylcholine receptor agonists suppresses spinal microglial activation and neuropathic pain in mice with peripheral nerve injury. J. Neuroinflammation. 2018;15:96. doi: 10.1186/s12974-018-1133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lone B.A. MMP-9, MMP-2 profile in different mice tissues and LPS induced MMP-9 and MMP-2 expression in RAW 264.7 cells, down regulated by emodin treatment. IRJIMS. 2014;1:1–7. [Google Scholar]
- 6.Yang Y.H., Li D.L., Bi X.Y., Sun L., Yu X.J., Fang H.L. Acetylcholine inhibits LPS-induced MMP-9 production and cell migration via the α7 nAChR-JAK2/STAT3 pathway in RAW264.7 cells. Cell Physiol Biochem. 2015;36:2025–2038. doi: 10.1159/000430170. [DOI] [PubMed] [Google Scholar]
- 7.Massoulié J., Bon S. The molecular forms of cholinesterase and acetylcholinesterase in vertebrates. Annu Rev Neurosci. 1982;5:57–106. doi: 10.1146/annurev.ne.05.030182.000421. [DOI] [PubMed] [Google Scholar]
- 8.Massoulié J., Pezzementi L., Bon S., Krejci E., Vallette F.M. Molecular and cellular biology of cholinesterases. Prog Neurobiol. 1993;41:31–91. doi: 10.1016/0301-0082(93)90040-y. [DOI] [PubMed] [Google Scholar]
- 9.Xu M.L., Luk W.K.W., Bi C.W.C., Liu E.Y.L., Wu K.Q.Y., Yao P. Erythropoietin regulates the expression of dimeric form of acetylcholinesterase during differentiation of erythroblast. J Neurochem. 2018;146:390–402. doi: 10.1111/jnc.14448. [DOI] [PubMed] [Google Scholar]
- 10.Bon S., Massoulié J. Quaternary associations of acetylcholinesterase. I. Oligomeric associations of T subunits with and without the amino-terminal domain of the collagen tail. J Biol Chem. 1997;272:3007–3015. doi: 10.1074/jbc.272.5.3007. [DOI] [PubMed] [Google Scholar]
- 11.Shaked I., Meerson A., Wolf Y., Avni R., Greenberg D., Gilboa-Geffen A. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Massoulié J. The origin of the molecular diversity and functional anchoring of cholinesterases. Neurosignals. 2002;11:130–143. doi: 10.1159/000065054. [DOI] [PubMed] [Google Scholar]
- 13.Mishra N., Friedson L., Hanin G., Bekenstein U., Volovich M., Bennett E.R. Antisense miR-132 blockade via the AChE-R splice variant mitigates cortical inflammation. Sci Rep. 2017;7:42755. doi: 10.1038/srep42755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen V.P., Choi R.C., Chan W.K., Leung K.W., Guo A.J., Chan G.K. The assembly of proline-rich membrane anchor (PRiMA)-linked acetylcholinesterase enzyme: glycosylation is required for enzymatic activity but not for oligomerization. J Biol Chem. 2011;286:32948–32961. doi: 10.1074/jbc.M111.261248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mor I., Sklan E.H., Podoly E., Pick M., Kirschner M., Yogev L. Acetylcholinesterase-R increases germ cell apoptosis but enhances sperm motility. J Cell Mol Med. 2008;12:479–495. doi: 10.1111/j.1582-4934.2008.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q., Fung A.H.Y., Xu M.L., Poon K., Liu E.Y.L., Kong X.P. Microphthalmia-associated transcription factor up-regulates acetylcholinesterase expression during melanogenesis of murine melanoma cells. J Biol Chem. 2018;293:14417–14428. doi: 10.1074/jbc.RA118.003729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richier P., Arpagaus M., Toutant J.P. Glycolipid-anchored acetylcholinesterases from rabbit lymphocytes and erythrocytes differ in their sensitivity to phosphatidylinositol-specific phospholipase C. BBA Biomembranes. 1992;1112:83–88. doi: 10.1016/0005-2736(92)90257-m. [DOI] [PubMed] [Google Scholar]
- 18.Getman D.K., Mutero A., Inoue K., Taylor P. Transcription factor repression and activation of the human acetylcholinesterase gene. J Biol Chem. 1995;270:23511–23519. doi: 10.1074/jbc.270.40.23511. [DOI] [PubMed] [Google Scholar]
- 19.He G., Zhang X., Chen Y., Chen J., Li L., Xie Y. Isoalantolactone inhibits LPS-induced inflammation via NF-κB inactivation in peritoneal macrophages and improves survival in sepsis. Biomed Pharmacother. 2017;90:598–607. doi: 10.1016/j.biopha.2017.03.095. [DOI] [PubMed] [Google Scholar]
- 20.Ellman G.L., Courtney K.D., Andres V., Featherstone R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 21.Winer J., Jung C.K.S., Shackel I., Williams P.M. Development and validation of real-time quantitative reverse transcriptase–polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 22.Liu E.Y.L., Xu M.L., Jin Yan, Wu Q.Y., Dong T.T.X., Tsim K.W.K. Genistein, a phytoestrogen in soybean, induces the expression of acetylcholinesterase via G protein-coupled receptor 30 in PC12 Cells. Front Mol Neurosci. 2018;11:59. doi: 10.3389/fnmol.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie H.Q., Liang D., Leung K.W., Chen V.P., Zhu K.Y., Chan W.K. Targeting acetylcholinesterase to membrane rafts: a function mediated by the proline-rich membrane anchor (PRiMA) in neurons. J Biol Chem. 2010;285:11537–11546. doi: 10.1074/jbc.M109.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du A., Xie J., Guo K., Yang L., Wan Y., OuYang Q. A novel role for synaptic acetylcholinesterase as an apoptotic deoxyribonuclease. Cell Discov. 2015;1:15002. doi: 10.1038/celldisc.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neumann-Giesen C., Falkenbach B., Beicht P., Claasen S., LÜErs G., Stuermer C.A. Membrane and raft association of reggie-1/flotillin-2: role of myristoylation, palmitoylation and oligomerization and induction of filopodia by overexpression. Biochem J. 2004;378:509. doi: 10.1042/BJ20031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harder T., Simons K. Clusters of glycolipid and glycosylphosphatidylinositol-anchored proteins in lymphoid cells: accumulation of actin regulated by local tyrosine phosphorylation. Eur J Immunol. 1999;29:556–562. doi: 10.1002/(SICI)1521-4141(199902)29:02<556::AID-IMMU556>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Sweet M.J., Hume D.A. Endotoxin signal transduction in macrophages. J Leukoc Biol. 1996;60:8–26. doi: 10.1002/jlb.60.1.8. [DOI] [PubMed] [Google Scholar]
- 28.Beutler B., Krochin N., Milsark I.W., Luedke C., Cerami A. Control of cachectin (tumor necrosis factor) synthesis: mechanisms of endotoxin resistance. Science. 1986;232:977. doi: 10.1126/science.3754653. [DOI] [PubMed] [Google Scholar]
- 29.Treinin M., Papke R.L., Nizri E., Ben-David, Mizrachi T., Brenner T. Role of the α7 nicotinic acetylcholine receptor and RIC-3 in the cholinergic anti-inflammatory pathway. Cent Nerv Syst Agents Med Chem. 2017;17:90–99. doi: 10.2174/1871524916666160829114533. [DOI] [PubMed] [Google Scholar]
- 30.Siebenlist U., Franzoso G., Brown K. Structure, regulation and function of NF-kappa B. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 31.Akira S., Takeda K., Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 32.Nobes C.D., Hall A. Rho, Rac, and Cdc 42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 33.Shen J.X., Yakel J.L. Nicotinic acetylcholine receptor-mediated calcium signaling in the nervous system. Acta Pharmacol Sin. 2009;30:673–680. doi: 10.1038/aps.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wessler I., Kirkpatrick C.J. Acetylcholine beyond neurons: the non-neuronal cholinergic system in humans. Br J Pharmacol. 2008;154:1558–1571. doi: 10.1038/bjp.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellier J.P., Kimura H. Peripheral type of choline acetyltransferase: biological and evolutionary implications for novel mechanisms in cholinergic system. J Chem Neuroanat. 2011;42:225–235. doi: 10.1016/j.jchemneu.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Pohanka M. Inhibitors of acetylcholinesterase and butyrylcholinesterase meet immunity. Int J Mol Sci. 2014;15:9809–9825. doi: 10.3390/ijms15069809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burgen A.S.V. The background of the muscarinic system. Life Sci. 1995;56:801–806. doi: 10.1016/0024-3205(95)00013-v. [DOI] [PubMed] [Google Scholar]
- 38.Wessler I., Kirkpatrick C.J., Racké K. Non-neuronal acetylcholine, a locally acting molecule, widely distributed in biological systems: expression and function in humans. Pharmacol Therapeut. 1998;77:59–79. doi: 10.1016/s0163-7258(97)00085-5. [DOI] [PubMed] [Google Scholar]
- 39.Kawashima K., Fujii T. Extraneuronal cholinergic system in lymphocytes. Pharmacol Ther. 2000;86:29–48. doi: 10.1016/s0163-7258(99)00071-6. [DOI] [PubMed] [Google Scholar]
- 40.Wessler I., Kirkpatrick C., Racké K. The cholinergic ‘pitfall’: acetylcholine, a universal cell molecule in biological systems, including humans. Clin Exp Pharmacol Physiol. 1999;26:198–205. doi: 10.1046/j.1440-1681.1999.03016.x. [DOI] [PubMed] [Google Scholar]
- 41.Fujii T., Mashimo M., Moriwaki Y., Misawa H., Ono S., Horiguchi K. Expression and function of the cholinergic system in immune cells. Front Immunol. 2017;8:1085. doi: 10.3389/fimmu.2017.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dale H.H., Dudley H.W. The presence of histamine and acetylcholine in the spleen of the ox and the horse. J Physiol. 1929;68:97–123. doi: 10.1113/jphysiol.1929.sp002598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koarai A., Traves S.L., Fenwick P.S., Brown S.M., Chana K.K., Russell R.E.K. Expression of muscarinic receptors by human macrophages. Eur Respir J. 2012;39:698. doi: 10.1183/09031936.00136710. [DOI] [PubMed] [Google Scholar]
- 44.Rinner I., Schauenstein K. Detection of choline-acetyltransferase activity in lymphocytes. J Neurosci Res. 1993;35:188–191. doi: 10.1002/jnr.490350209. [DOI] [PubMed] [Google Scholar]
- 45.Kawashima K., Yoshikawa K., Fujii Y.X., Moriwaki Y., Misawa H. Expression and function of genes encoding cholinergic components in murine immune cells. Life Sci. 2007;80:2314–2319. doi: 10.1016/j.lfs.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 46.de Oliveira P., Gomes A.Q., Pacheco T.R., Vitorino de Almeida V., Saldanha C., Calado A. Cell-specific regulation of acetylcholinesterase expression under inflammatory conditions. Clin Hemorheol Microcirc. 2012;51:129–137. doi: 10.3233/CH-2011-1520. [DOI] [PubMed] [Google Scholar]
- 47.Tyagi E., Agrawal R., Nath C., Shukla R. Effect of anti-dementia drugs on LPS induced neuroinflammation in mice. Life Sci. 2007;80:1977–1983. doi: 10.1016/j.lfs.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 48.Pavlov V.A., Parrish W.R., Rosas-Ballina M., Ochani M., Puerta M., Ochani K. Brain acetylcholinesterase activity controls systemic cytokine levels through the cholinergic anti-inflammatory pathway. Brain Behav Immun. 2009;23:41–45. doi: 10.1016/j.bbi.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peter C., Schmidt K., Hofer S., Stephan M., Martin E., Weigand M.A. Effects of physostigmine on microcirculatory alterations during experimental endotoxemia. Shock. 2010;33:405–411. doi: 10.1097/SHK.0b013e3181b77e82. [DOI] [PubMed] [Google Scholar]
- 50.Gamal M., Moawad J., Rashed L., El-Eraky W., Saleh D., Lehmann C. Evaluation of the effects of Eserine and JWH-133 on brain dysfunction associated with experimental endotoxemia. J Neuroimmunol. 2015;281:9–16. doi: 10.1016/j.jneuroim.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Gowayed M.A., Refaat R., Ahmed W.M. El-Abhar HS Effect of galantamine on adjuvant-induced arthritis in rats. Eur J Pharmacol. 2015;764:547–553. doi: 10.1016/j.ejphar.2015.07.038. [DOI] [PubMed] [Google Scholar]
- 52.Reale M., Iarlori C., Gambi F., Feliciani C., Salone A., Toma L. Treatment with an acetylcholinesterase inhibitor in Alzheimer patients modulates the expression and production of the pro-inflammatory and anti-inflammatory cytokines. J Neuroimmunol. 2004;148:162–171. doi: 10.1016/j.jneuroim.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Shapira M., Tur-Kaspa I., Bosgraaf L., Livni N., Grant A.D., Grisaru D. A transcription-activating polymorphism in the ACHE promoter associated with acute sensitivity to anti-acetylcholinesterases. Hum Mol Genet. 2000;9:1273–1281. doi: 10.1093/hmg/9.9.1273. [DOI] [PubMed] [Google Scholar]
- 54.Waiskopf N., Ofek K., Gilboa-Geffen A., Bekenstein U., Bahat A., Bennett E.R. AChE and RACK1 promote the anti-inflammatory properties of fluoxetine. J Mol Neurosci. 2014;53:306–315. doi: 10.1007/s12031-013-0174-6. [DOI] [PubMed] [Google Scholar]
- 55.Gilbert M.M., Auld V.J. Evolution of CLAMS (cholinesterase-like adhesion molecules): structure and function during development. Front Biosci. 2005;10:2177–2192. doi: 10.2741/1689. [DOI] [PubMed] [Google Scholar]
- 56.Inkson C.A., Brabbs A.C., Grewal T.S., Skerry T.M., Genever P.G. Characterization of acetylcholinesterase expression and secretion during osteoblast differentiation. Bone. 2004;35:819–827. doi: 10.1016/j.bone.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 57.Broide R.S., Robertson R.T., Leslie F.M. Regulation of alpha 7 nicotinic acetylcholine receptors in the developing rat somatosensory cortex by thalamocortical afferents. J Neurosci. 1996;16:2956–2971. doi: 10.1523/JNEUROSCI.16-09-02956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Z., Zhang J., Berg D.K. Role of endogenous nicotinic signaling in guiding neuronal development. Biochem Pharmacol. 2007;74:1112–1119. doi: 10.1016/j.bcp.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fossier P., Baux G., Tauc L. Possible role of acetylcholinesterase in regulation of postsynaptic receptor efficacy at a central inhibitory synapse of Aplysia. Nature. 1983;301:710–712. doi: 10.1038/301710a0. [DOI] [PubMed] [Google Scholar]
- 60.Bond C.E., Zimmermann M., Greenfield S.A. Upregulation of alpha 7 nicotinic receptors by acetylcholinesterase C-terminal peptides. PLoS One. 2009;4:e4846. doi: 10.1371/journal.pone.0004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung H.Y., Sung B., Jung K.J., Zou Y., Yu B.P. The molecular inflammatory process in aging. Antioxidants Redox Signal. 2006;8:572–581. doi: 10.1089/ars.2006.8.572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.