Abstract
CD4+Foxp3+ regulatory T (Treg) cells are key players in keeping excessive inflammation in check. Mounting evidence has shown that Treg cells exert much more diverse functions in both immunological and non-immunological processes. The development, maintenance and functional specification of Treg cells are regulated by multilayered factors, including antigens and TCR signaling, cytokines, epigenetic modifiers and transcription factors (TFs). In the review, we will focus on TFs by summarizing their unique and redundant roles in Treg cells under physiological and pathophysiological conditions. We will also discuss the recent advances of Treg trajectories between lymphoid organs and non-lymphoid tissues. This review will provide an updated view of the newly identified TFs and new functions of known TFs in Treg biology.
Keywords: Treg, Homeostasis, TCF1, LEF1, Precursor, Cofactor
Introduction
The activation of immune system is crucial for host defense against pathogens and cancer. However, excessive degree of immune activation causes deleterious consequences, evidenced by the development of autoimmune and inflammatory disorders [1, 2]. Negative feedback is crucial for keeping immune activation at an appropriate level. Multiple mechanisms have evolved as negative regulators of excessive immune responses [3]. Among them, CD4+ Foxp3+ regulatory (Treg) cells are the most pivotal players to keep autoimmunity and inflammation in check and maintain tissue homeostasis [4–8].
The concept of immunosuppression has been coined decades ago (reviewed in [9–12]). It has been reported in late 1960s that thymectomy 3 days after birth rendered the mice to develop autoimmune destruction of self-tissues such as ovaries and other organs [13]. Later studies have shown that CD4+ T cells were important players in inhibiting thymectomy-provoked autoimmune diseases [14]. Following studies had attempted to identify which subset of CD4+ T cells possessed this suppressive function [15–17]. One of the landmark studies was the identification of CD25 as a cell-surface marker and the validation of CD25+ CD4+ T cells as the bona fide suppressors [17].
In the early 2000s, several groups have reported the link between mutations of Foxp3 gene and the phenotypes in Scurfy mice or human IPEX patients (immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome [18–21]. Shortly after that, several independent studies demonstrated that Foxp3 was the TF that governed the generation of Treg lineage cells [22–24]. Ectopic expression of Foxp3 in CD4+ CD25− T cells converted these cells into CD4+ CD25+ “regulatory” cells [22]. The necessity of Foxp3 in Treg development was further supported by both loss- and gain-of-function studies, in which more Treg cells were found in Foxp3 over-expressing mice [23], and conversely, significantly fewer Treg cells were detected in Foxp3 knockout mice [24]. These findings clearly supported the lineage identity of Treg cells.
Foxp3 is still the most reliable marker for Treg lineage identity. Numerous studies have focused on investigating the expression and function of Foxp3 in mouse and human [25–29]. However, mounting evidence also show that Foxp3 alone is not sufficient to induce a full range of Treg transcriptional program [30–33]. Increasing number of other TFs and epigenetic modifiers have been reported to participate in almost every aspect of Treg biology (reviewed in [5, 34, 35]). Rapid advances have been made in identifying new TFs and new functions of known Treg-related TFs. This review will provide an updated view of how these TFs play their unique or redundant roles in Treg cells. We will first review the role of Foxp3 in Treg cells, and then summarize the effects of other TFs on regulating Foxp3 gene expression or collaborating with Foxp3 on protein level to control Treg gene expression. We will then discuss the increasingly emerging data on tissue Treg precursors and provide an integrated view of Treg trajectory. Recent updates on human Treg cells and their therapeutic potential will also be discussed.
Foxp3 and its cofactors
Foxp3 is a member of the forkhead family TFs [36]. It contains a C-terminal FKH domain, a proline-rich N-terminal (PRR) domain, a C2H2 zinc finger (ZF), and a central leucine zipper (LZ) domain [25]. Foxp3 binds to many genes related to the activation and function of conventional T (Tconv) cells or Treg cells [37, 38]. Further studies showed that Foxp3 is bound to the enhancers [39, 40] that already become accessible even before Foxp3 is expressed. These enhancers are also occupied by Foxp3 cofactors (e.g., Foxo proteins) in the precursor cells [39].
Foxp3 acts as both an activator and a repressor [32, 41–43]. Ectopic expression of Foxp3 in naive mouse CD4+ T cells induces the expression of a panel of canonical Treg signature genes, including Il2ra, Ctla4, Tnfrsf18, Itgae, Gpr83, and Nrp1 [22, 32], supporting an activator role of Foxp3. Conversely, Foxp3 is also bound to a large number of genes that are downregulated in Treg cells compared to Tconv cells [38]. In this case, Foxp3 acts as a repressor [44]. In activated Treg cells, Foxp3-binding sites show diminished accessibility of chromatin and selective deposition of histone H3 trimethylated at Lys27, which is associated with the recruitment of the histone methyltransferase EZH2 and downregulation of the expression of nearby genes [43]. Whether Foxp3 functions as an activator or a repressor is influenced by the configuration of the protein complex formed by Foxp3 and other TFs or histone modifiers [40]. For instance, Foxp3 acts primarily as an activator when forming a complex with RelA, Helios and Kat5 [40].
Foxp3 is pivotal for Treg lineage identity. Sustained expression of Foxp3 is a prerequisite of Treg lineage stability. Conversely, loss-of-expression of Foxp3 is an indication of compromised Treg lineage identity [45, 46]. Indeed, partially attenuated Foxp3 expression causes Treg to lose their identity and acquire effector T (Teff) cell characteristics [47]. Foxp3 has been considered crucial for Treg’s suppressive function. However, recent work has suggested that the suppressive function can be developed in Foxp3-deficient “Treg” cells by targeting their metabolic pathways [48]. Furthermore, the definition of Treg function has been much more broadened beyond the “suppression” of Teff cell responses [6, 49]. As a lineage-determinative TF, Foxp3 is the master regulator for Treg’s core functional modules, because unbiased high-throughput analyses have revealed that all kinds of Treg cells express a small number of Foxp3-dependent transcripts, onto which additional programs are added less uniformly [50]. However, additional factors are required for Treg’s functional adaptation and specification (discussed in detail below).
Cis-regulatory elements in the Foxp3 genomic locus
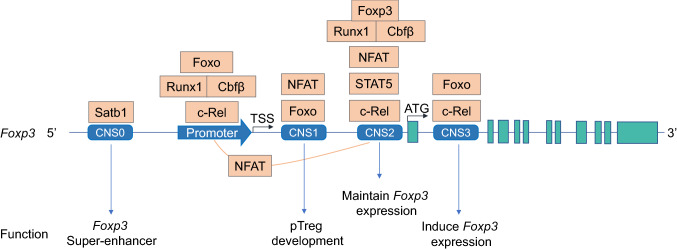
Several regulatory regions within Foxp3 genomic locus have been revealed (Fig. 1). The promoter locates about 6 kilobases upstream of the first exon of Foxp3. It can be activated by the TCR signaling, in cooperation with NFAT and AP1 [51]. Three conserved NFAT binding sites within 500 bases upstream from the transcriptional start site (TSS) have been identified in Foxp3 promoter region [51]. In Treg cells, this promoter region is highly demethylated, compared to that in Tconv cells [52]. Signaling mediated by tumor necrosis factor receptor 2 (TNFR2) can prevent CpG methylation in the Foxp3 promoter [53]. Nr4a nuclear receptors can activate Foxp3 promoter in that forced activation of Nr4a receptors bypasses low-strength TCR signaling to drive the Treg lineage development [54].
Fig. 1.
TFs regulate Foxp3 expression through binding to Foxp3 cis-regulatory regions. Several conserved noncoding sequences (CNSs) have been identified in the Foxp3 locus. Each of them has different function (detailed in the text). Multiple TFs are bound to different CNS regions and regulate Foxp3 super-enhancer formation, induction or stability in either a redundant or unique manner. In addition, CNS2 can form a physical loop with the Foxp3 promoter in an NFAT-dependent manner. TSS, transcription start site. ATG depicts the start site of translation
Cis-regulatory elements play an essential role in Treg lineage formation [55, 56]. Several conserved noncoding sequences (CNS) have been identified in Foxp3 regulatory regions [55, 56]. Each of these CNS regions exerts different functions in regulating Foxp3 induction, stability and Treg fate [55, 57–59]. CNS1 is preferentially needed for the generation of peripherally induced Treg cells (pTreg) [55], whereas CNS2 is required to maintain Foxp3 expression in committed Treg cells. CNS2 can form a physical loop with Foxp3 promoter [58]. NFAT activation can strengthen this promoter–enhancer loop interaction. The biological significance of this loop is to stabilize Foxp3 expression [58]. CNS3 is essential for the induction of Foxp3 via recruiting c-Rel to the Foxp3 locus [55]. In addition, recent studies have shown that long noncoding RNA (lncRNA) Flicr can modify chromatin accessibility in CNS3 [60].
CNS0 is a recently identified conserved regulatory region in Foxp3 genomic locus [57]. CNS0 locates approximately 8 kilobases upstream of the TSS of Foxp3. CNS0 is permissive for the binding of Satb1 and involved in chromatin modification and super-enhancer formation [57]. The role of Satb1 is to modify the epigenetic status of Foxp3 locus, making them permissive for the binding of other Treg-development related TFs [57]. Interestingly, in committed Treg cells, the expression Satb1 needs to be downregulated to a lower level, compared to their precursors or Tconvs [33, 42, 57]. Foxp3 represses Satb1 either directly, or indirectly via microRNA binding to the 3′ untranslated region of Satb1 [42]. Unwanted high level of Satb1 comprises Treg function and induces Teff cell cytokine expression in Treg cells [42]. While the expression of Satb1 is not as abundant as that in Tconv cells, the remaining Satb1 can form physical interaction with Foxp3 on the protein level [33]. In this case, Satb1 acts as a regulator for the auto-assembly of a core Treg TF circuit, including the upregulation of IRF4, GATA1 and Eos, and downregulation of endogenous LEF1 and Satb1 [33].
Transcriptional regulation of Foxp3 expression
TCR signaling
Natural Treg cells arise in the thymus. The specificity and affinity of TCR in thymocytes are the primary determinants for Treg lineage formation (reviewed by Hsieh et al. [61]). Earlier studies have found that high-affinity autoreactive TCR expression in thymocytes favors the generation of CD4+ CD25+ Treg cells [62, 63]. Along with TCR activation, costimulatory (CD28-CD80/CD86) and cytokine signaling (primarily IL-2) are required for thymic Treg generation. With these findings, a two-step model has been proposed to explain how Treg cells are generated in the thymus [61, 64, 65]. First, high-affinity TCR signaling instructs thymocytes to develop into the Treg precursors, characterized by the upregulation of CD25. At this stage, Foxp3 genomic locus is remodeled. Subsequently in step two, IL-2 (to a less extent IL-15) induces the expression of Foxp3 [64–67]. This model favors the perception that CD4 single positive Foxp3− CD25+ cells are the precursors of Treg cells in the thymus. However, a distinct population of the Treg progenitors in the thymus, characterized by low level of Foxp3 expression and lacking CD25 (Foxp3lo CD25−), has also been described [68–71]. In one recent study, Owen et al. have demonstrated that both types of precursors (namely Foxp3− CD25+ and Foxp3lo CD25−) can differentiate into bona fide Treg cells. Foxp3− CD25+ precursors exhibit a relatively higher level of the TCR affinity. In contrast, CD4+ thymocytes bearing relatively lower TCR affinity are more likely to differentiate into the Foxp3lo CD25− precursors. Further analysis revealed that Foxp3− CD25+ and Foxp3lo CD25− precursors have distinct TCR repertoires [71]. More interestingly, Treg cells derived from these two precursors have different functional properties in maintaining immune tolerance. For instance, Foxp3− CD25+ precursor-derived Treg cells have a superior capability to prevent experimental autoimmune encephalitis [71]. Together, these findings provide important insights into thymic Treg development.
The crucial role of TCR signaling in thymic Treg cell development has also been demonstrated in other studies [62, 63, 72]. Downstream of TCR signaling, a number of TFs (including NF-κB, NFAT, AP1, CREB and ATF) have been reported to regulate Foxp3 expression [51, 52]. Foxp3 expression in Treg cells is controlled by both sequence-specific binding of CREB/ATF and DNA methylation of the CpG islands [52]. Recently, it has been reported that CARD11-BCL10-MALT1 (CBM) signaling mediates TCR-induced NF-κB activation in Treg cells [73] and controls the conversion of Treg cells from resting to effector stage under homeostatic conditions. MALT1 seems to play a central role in this signaling cascade [73, 74]. Deletion of Malt1 in Treg cells leads to scurfy-like lethal autoimmune diseases, which is caused by the deficit of the function not the number of Treg cells [75]. One study has reported that MALT1 supports Treg development in the thymus but suppresses Treg generation in the periphery during inflammation [76]. However, in established Treg cells, TCR signaling is dispensable for maintaining Foxp3 expression [77]. Nr4a family factors have been found to play important roles in guarding the completion of thymic Treg development [78]. Interestingly, these Nr4a factors are also involved in eliminating the Treg precursors that fail to develop into mature Treg cells [78].
NF-κB family TFs
In T cells, TCR signaling activates NF-κB [79]. The above mentioned CBM complex is a critical component in TCR-induced NF-κB activation. In mice deficient for Card11 (encoding Carma1), Treg development is halted due to the lack of thymic CD25+ GITR+ Foxp3− Treg precursors [80]. Constitutively active IKKβ can rescue the induction of Foxp3 in Card11-deficient Treg precursor cells [81].
The NF-κB family is composed of c-Rel, RelA (p65) and NF-κB1 (a.k.a., p105/50), belonging to the canonical NF-κB pathway, and NF-κB2 (p100/52) and RelB subunits of the non-canonical pathway [82]. NF-κB family members participate in multiple aspects of Treg development, stability and function (Table 1).
Table 1.
NF-kB family TFs in Treg biology
| Name | Key role | Mechanism | References |
|---|---|---|---|
| c-Rel | Thymic Treg development | [84] | |
| Maintain Foxp3 expression | Binding to Foxp3 CNS2, or promoter | [81, 83] | |
| Induce Foxp3 expression | Binding to Foxp3 CNS3 | [81] | |
| RelA | Promote eTreg differentiation | Form protein–protein complex with NF-κB1 (Ref. [89]) | [87, 89, 90] |
| Treg stability | [87, 88, 90] | ||
| NF-κB1 | Dispensable for Treg cell development | [91] |
Among the family members of NF-κB, c-Rel has been extensively studied in Treg biology. c-Rel-deficient mice show a severely reduced number of Treg cells compared to wild-type mice [83]. c-Rel exerts multiple functions in Treg cells. First, c-Rel is required for the development of thymic Treg cells [84]. Second, c-Rel ablation specifically impairs the generation and maintenance of activated Treg (aTreg) subset [85]. Third, c-Rel deficient T cells lose their competence to be converted into Treg cells by TGF-β signaling [83]. However, the role of c-Rel in pTreg induction remains controversial. Some studies have reported that TGF-β induced pTreg generation is severely impaired in c-Rel deficient naïve T cells [86], whereas others have shown that c-Rel-deficient naïve CD4+ T cells normally upregulates Foxp3 when stimulated by TGF-β [84]. c-Rel is dispensable for Treg suppressive function, since c-Rel-deficient Treg cells are fully suppressive [84]. Mechanistically, c-Rel binds to multiple sites of Foxp3 regulatory regions. By binding to Foxp3 CNS2, c-Rel demethylates the CpG sites in that region [81]. In addition, c-Rel can also bind to Foxp3 promoter and form a large enhanceosome, containing c-Rel, RelA/p65, NFAT, SMAD and CREB [83]. Interestingly, SMAD and CREB are first bound to Foxp3 enhancers, but later they move to the promoter to participate in the formation of Foxp3 enhanceosome [83]. In addition, c-Rel can also bind to Foxp3 CNS3 and regulate the induction of Foxp3 [81].
RelA/p65 is another canonical NF-κB family protein. Conditional inactivation of RelA in Treg cells induces autoimmune diseases [87]. RelA and c-Rel have different and partially redundant roles in developing and mature Treg cells [88]. RelA is constitutively active in naïve and effector Treg cells [87]. Several studies have shown that RelA promotes effector Treg (eTreg) generation [87, 89, 90] and Treg lineage stability [87, 88, 90]. The role of RelA in maintaining eTreg pool is through binding to NF-κB1 [89].
The role of other NF-κB family proteins remains to be investigated. It has been reported that NF-κB1 is dispensable for Treg cell development [91]. NF-κB1-deficient mice have relatively normal numbers of Treg cells [83, 84, 91]. Moreover, the deficiency of IκB(NS), a member of Bcl3 family atypical IκB proteins, does not lead to the reduction of Treg precursor cells [92].
Other TFs
In addition to the aforementioned NF-κB family TFs, increasing number of other TFs have been demonstrated to regulate Foxp3 expression. Through binding to the Foxp3 regulatory regions, these TFs exert either redundant or unique roles (Fig. 1). Among them, STAT5 mainly binds to Foxp3 CNS2 and acts downstream of IL-2 and other common gamma chain cytokines [93]. This STAT5-CNS2 axis is crucial for maintaining stable expression of Foxp3 and Treg lineage identity [59]. Optimal activation (phosphorylation) of STAT5 is critical for Treg competitive fitness [94, 95]. Additional factors and signals have been found to regulate the expression and activation of STAT5. For instance, TCF1 can bind to the promotor of Stat5b (to a less extend to Stat5a) and regulate its expression [95]. Moreover, in the absence of Helios, STAT5 activation is diminished with a concomitant reduction of Foxp3 expression [96]. Agonists of TNFRSF members (namely GITR, OX40 and TNFR2) enhance the responsiveness of STAT5 in Treg precursors [97]. Furthermore, serine–threonine kinase Mst1 has been shown as an amplifier for STAT5 activity in Treg cells [98]. Dedicator of cytokinesis 8 (DOCK8) is another protein required for optimal STAT5 activation and Treg fitness [99, 100]. On the other hand, PD-1/PD-L1 signaling inhibits STAT5 phosphorylation in Treg cells. PD-L1 blockade upregulates STAT5 phosphorylation in Treg cells ex vivo [101]. In addition, microRNA155 confers competitive fitness to regulatory T cells via repressing SOCS1, which acts as a negative regulator for STAT5 [94]. Taken together, multiple factors and mechanisms are evolved to ensure an optimal STAT5 activation, which is crucial for Treg development in the thymus [65, 102] and competitive fitness in the periphery [94, 95].
Foxo1 and Foxo3, two forkhead family members, can regulate Foxp3 promoter activity [103–105]. T cell-specific depletion of Foxo1 results in multiorgan immune infiltration, and augmented germinal center (GC) responses [104, 105]. Foxo proteins bind to the promoter, CNS1 and CNS3 of Foxp3. Mutation of the proximal Foxo-binding site results in repressed CNS1 transactivation [103, 104]. There is functional redundancy between Foxo1 and Foxo3 [103, 104]. Through binding to both CNS1 and CNS3, Foxo proteins participate in the regulations of both thymic and TGF-β-induced Treg development [103–105]. Foxo proteins are also critical for Treg function [106]. This is mainly through the axis of Foxo–Akt in that Treg cells express high amounts of Foxo1 and suppress Akt activation [106]. In the absence of Foxo, IFNγ production is elevated in Treg cells, which contributes to the loss of function of Treg cells [106]. While these findings support a critical role of Foxo proteins in Treg differentiation and function, later studies have revealed that Foxo1 is actually repressed in aTreg cells. Downregulation of Foxo is required for Treg homing to non-lymphoid organs [107]. Interestingly, Treg cells at the tumor sites exhibit a profound downregulation of Foxo signaling. Based on this, expression of an Akt-insensitive Foxo1 mutant at a low dose can deplete tumor-associated Treg cells [107].
NFAT and SMAD are also involved in regulating Foxp3 expression [52, 64, 108]. NFAT binds to Foxp3 CNS1, together with SMAD3, to facilitate TGF-β-induced Foxp3 expression [108]. In addition, NFAT also binds to Foxp3 CNS2 upon TCR activation [58]. Interestingly, CNS2 can form a loop interaction with Foxp3 promoter in an NFAT-dependent manner to stabilize Foxp3 transcription [58].
Runx1 can regulate Foxp3 expression [109, 110]. Treg cell-specific deficiency of Runx1, or Cbfb, a cofactor for all Runx proteins, induces lymphoproliferation, autoimmune diseases and hyperproduction of IgE [110]. Runx1 and its cofactor Cbfβ form a heterodimer which binds to Foxp3 CNS2 [109, 110] and promoter [109, 111]. Later studies found that the Cbfβ–Runx1 complex is also required for Foxp3 protein binds to its own CNS2, in a CpG demethylation-dependent manner [55].
Protein–protein complexes formed by Foxp3 and its cofactors
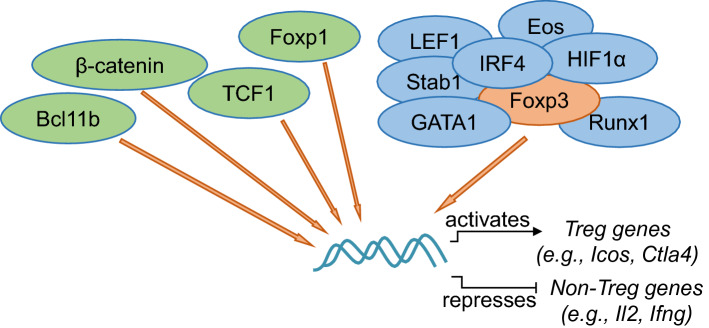
Foxp3 forms large protein complexes with its binding partners [112]. A few TFs have been demonstrated to cooperate with Foxp3 for binding to the same set of target genes (Fig. 2). Using a combination of systems biology and functional validation approaches, five TFs (namely, GATA1, Eos, Satb1, IRF4 and LEF1) have been found to act redundantly as Foxp3 cofactors [33]. A common feature of these Foxp3 cofactors is to enhance the transcriptional activity of Foxp3. This is due to that the cofactors (e.g., GATA1) can robustly enhance the occupancy of Foxp3 to its target genes, such as Icos [33]. This study provides experimental evidence to support a model, whereby multiple TFs act as Foxp3 cofactors to enhance the functionality of Foxp3, thus “locking in” a stable Treg transcriptional program. Further studies have found that Foxp3 is bound to pre-accessible enhancers occupied by its cofactors in Treg precursors [39]. Interestingly, a recent study has reported that the binding sites of TCF1, a TCF/LEF family TF, substantially overlapped with that by Foxp3 in Treg cells [113]. A considerable proportion of Foxp3-bound genes are also bound by β-catenin, an upstream regulator for TCF1/LEF1 [114]. However, the co-occupancy by β-catenin negatively impacts Foxp3′s transcriptional activity [114], suggesting a multifaceted regulation by the co-binding of Foxp3 and cofactors to target genes. A large number of Foxp3-bound genomic sites in Treg cells are occupied by Foxp1. As a consequence, Foxp1 markedly enhances the binding of Foxp3 to these sites [115], an effect similar to that by GATA1 [33]. As such, Foxp1 and Foxp3 coordinate the expression of Ctla4, a key gene for Treg function [116]. Bcl11b binds to the genomic loci of Treg genes in both human and mouse Treg cells, overlapping with Foxp3 binding [117, 118]. The absence of Bcl11b leads to reduced chromatin accessibility in these bound genes [117]. As a functional consequence, Treg-specific ablation of Bcl11b induces the onset of autoimmune diseases in mice, due to the compromised Foxp3 activity [117, 118]. Thus, multiple TFs (such as GATA1, Foxp1 and Bcl11b) collaborate with Foxp3 to enhance the binding of Foxp3 to its target genes. Whether each of these cofactors participate in the regulation of different functional properties of Treg cells remains to be determined.
Fig. 2.
TFs cooperate with Foxp3 to activate Treg genes or repress non-Treg genes. Increasing number of TFs have been reported to cooperate with Foxp3 to regulate target gene expression. Some of them (e.g., TCF1, Foxp1 and Bcl11b) have been found to co-occupy with Foxp3 onto the target genes. Others (e.g., GATA1, Runx1) are reported to promote Foxp3’s transcriptional activity to either induce the expression of Treg genes (such as Icos, Ctla4) or repress non-Treg genes (such as Il2, Ifng). Changes of Foxp3 domains may alter the interactions between Foxp3 and other TFs (such as IRF4, Eos and HIF1α). TFs that have been reported to have physical interactions with Foxp3 on the protein level are marked in blue
These findings together suggest that many TFs act as Foxp3 cofactors to regulate its transcriptional activity. Many of these TFs are physiologically associated with Foxp3 (Fig. 2). For instance, the physical interaction between Foxp3 and Runx1 leads to the suppression of Il2, Ifng, and induction of Treg genes [119]. Studies have also suggested that the configuration of the Foxp3–cofactor complex impacts the function of Treg cells [112, 120, 121]. For instance, the N-terminus of Foxp3 protein is essential for it to interact with a number of TFs and regulators, including IRF4, HIF1α and Eos [120, 121]. Disrupted N-terminus of Foxp3 protein alters the binding pattern to its cofactors. In Foxp3tm2Ayr mice, a fusion of EGFP to the N-terminal of Foxp3 enhances the binding of IRF4, whereas weakens that of HIF1α. One biological consequence of this shifted binding pattern is that autoimmune diabetes is accelerated, whereas rheumatoid arthritis is suppressed when crossed to the genetically susceptible mouse strains. However, it remains to be determined whether multiple cofactors coexist in one protein complex, or each of them independently cooperate with Foxp3 to form separate complexes in individual Treg cells.
Treg homeostasis
Homeostatic regulation of the differentiation and function of Treg cells is essential for these cells to exert their physiological roles [122–124]. The pool of Treg cells in peripheral lymphoid organs is maintained at an appropriate size and diversity. The latter is influenced by the activation status, TCR repertoires, and specialized functional subsets. Multiple mechanisms have been evolved to maintain Treg homeostasis, including the balance between proliferation and apoptosis [125], the dependence and negative feedback of Treg on paracrine IL-2 [122], and metabolic regulations [126, 127]. These mechanisms have been discussed elsewhere [122, 128]. We here focus on summarizing recent advances of the classification of Treg subsets and highlighting newly identified TFs in Treg homeostatic differentiation (Fig. 3).
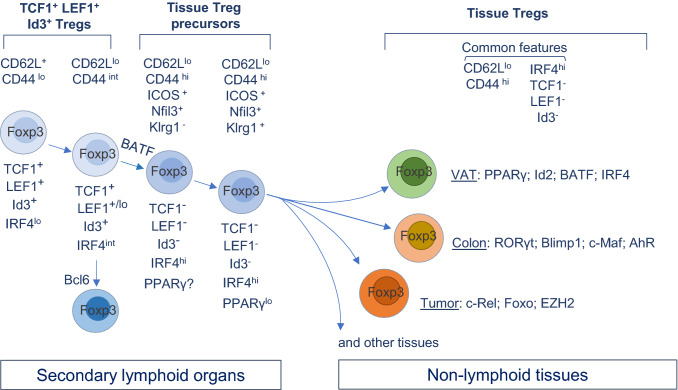
Fig. 3.
Transcriptional regulations of Treg homeostasis, tissue Treg precursors and tissue-specific Treg phenotype. Treg cells undergo a process of activation and differentiation in the SLOs. This is correlated with the upregulation (e.g., IRF4) and downregulation (e.g., TCF1, LEF1 and Id3) of different TFs. Recent studies have identified tissue Treg precursors in the SLOs, marked by either low level of PPARγ, the expression of Nfil3, or the “turning-off” of TCF1, LEF1 and Id3. Nfil3+ Klrg1+ subset is considered a more mature stage of tissue Treg precursors in the SLOs. Upon arriving at each tissue, Treg cells undertake a further differentiation process mediated by tissue local factors. For instance, PPARγ drives phenotypic and functional maturation of adipose tissue Treg cells, and Blipm1 drives intestinal Treg cells to produce IL-10. Of note, the development of Tfr cells occurs at a relatively earlier stage, prior to the “turning-off” of TCF1, LEF1, or Id3
The compositions of the Treg pool
Based on the expression of two cell-surface markers CD62L and CD44, Treg cells in peripheral lymphoid organs can be roughly separated into resting (or naïve) versus activated (or effector) pools [122, 124]. CD62L+ CD44lo resting Treg (rTreg) cells also express higher levels of CCR7 [124], which guides rTreg cells to gain more access to the paracrine IL-2 in the T cell zones of the secondary lymphoid organs (SLOs) [124]. In contrast, CD62L− CD44hi Treg cells rely relatively less on IL-2 for survival. Instead, they can respond to the ICOS/ICOSL signaling for their maintenance [124]. Recently, several studies have revealed more details of the heterogeneity of the Treg pool in the SLOs. For instance, based on the expression of TCF1 (and LEF1 in a similar pattern), peripheral Treg cells can be clearly separated into three subpopulations: CD62L+ CD44lo TCF1+; CD62L− CD44int/hi TCF1+ and CD62L− CD44hi TCF1−. Transcriptome analysis has revealed clear distinctions among them, with the CD62L− CD44hi TCF1− subset showing a full-spectrum of the mature Treg phenotype [95]. In another study, based on the expression of Id3, a very similar profile of Treg subpopulations has been revealed [129], suggesting that Id3 and TCF1 have a synchronized expression pattern in Treg cells. Furthermore, using a combination of CD62L, ICOS and TIGIT, Dias et al. have separated Treg cells into naive (CD62L+ICOS−TIGIT−), activated (CD62L−ICOS−TIGIT−) and effector (CD62L−ICOS+TIGIT+) subsets [130]. To what extent the “three-subset” classification of Treg cells in these studies is interchangeable remains to be determined. However, transcriptome profiling analyses performed by all three studies have suggested a high likelihood of that the “turning-on” of ICOS and TIGIT is correlated with the simultaneous “turning-off” of TCF1, LEF1 and Id3. Thus, bulk Treg pool in the peripheral organs can be further divided into phenotypically distinct subpopulations (Fig. 3). How the transition and equilibrium among these subpopulations are regulated remains to be investigated.
TCR, TFs and Treg homeostatic differentiation
TCR signaling is essential for the differentiation of Treg cells and the acquisition of the activated phenotype [77]. TCR sequencing analysis reveals that although TCR signal intensity does not affect the ratio between resting and activated Treg cells, it shapes the composition of the aTreg pool [50]. Following TCR activation, the expression of IRF4 is upregulated. IRF4 plays a key role in mobilizing Treg cells from resting to activated status [131]. Depletion of IRF4 restrains Treg cells at the resting stage. IRF4-deficient Treg cells fail to acquire the effector phenotype even in the presence of activation signals. These cells not only lack ICOS expression and IL-10 production, but also are impaired in the expression of activation markers and molecules required for homing, such as CD62L, CD103 and CCR6, and suppressor function, such as CTLA4 [131]. In line with this, IRF4 expression downstream of TCR signaling in Treg cells contributes to the optimal suppressive function for Treg cells [77]. The AP-1 transcription factor, JunB, promotes an IRF4-dependent transcription program in eTreg cells [132]. Mechanistically, JunB facilitates the accumulation of IRF4 at its target genes, including Icos and Ctla4. In addition, mTOR functions downstream of antigenic signals to drive IRF4 expression and mitochondrial metabolism, and accordingly, deletion of mitochondrial transcription factor A (Tfam) severely impairs Treg suppressive function and eTreg generation [133]. Interestingly, IRF4 expression in thymic epithelial cells is required to prime these stromal cells for Treg development in the thymus [134]. In that setting, IRF4 regulates the expression of chemokines and costimulatory molecules in thymic epithelial cells to favor a microenvironment for Treg development.
In contrast to the expression pattern of IRF4, the expression of TCF1 and LEF1 exhibits a stepwise downregulation in Treg cells during the differentiation from resting, activated to effector stage [95]. Treg-specific ablation of Tcf7 (encoding TCF1) and Lef1 doesn’t affect bulk Treg homeostasis. However, it alters the composition of the Treg pool by increasing the fraction of eTreg cells, and a simultaneous decrease of rTreg and aTreg proportions. These data suggest that TCF1 and LEF1 act as a gatekeeper to restrain the differentiation of Treg cells from resting and activated stage to effector stage. Therefore, in contrast to the differentiation-promoting effect elicited by IRF4, TCF1 and LEF1 act in a redundant manner to maintain resting and activated Treg cells. It is likely that TCF1 and LEF1 can antagonize the effects of IRF4 in Treg homeostatic differentiation.
The expression of Bach2 also shows a stepwise downregulation in Treg cells following the path from resting to activated/effector stage [95]. Functionally, Bach2 prevents premature differentiation of eTreg cells and limits IL-10 production [135]. This effect is likely due to that Bach2 can counteract the DNA-binding activity of IRF4 and limit chromatin accessibility, thereby attenuating IRF4-dependent regulation. At the same time, Bach2 is required for the development of pTreg cells in the gastrointestinal tract, an effect can be recapitulated by the depletion of IRF4. Therefore, Bach2 counterbalances the effects of IRF4 to achieve an equilibrium between thymus-derived and peripherally induced Treg cells and also influence their activation/differentiation status. One interesting (and surprising) finding is that IRF4 plays a dampening role in pTreg generation [135]. IRF4 has been shown to promote Treg (mainly tTreg) differentiation [131]. How IRF4 differentially influences tTreg and pTreg development remains to be defined. In addition to IRF4, Dias et al. have found that Myb can also promote tTreg differentiation, but has no effect on pTreg cells [130].
E-proteins and inhibitor of DNA-binding (Id) proteins have been shown to play important roles in TCR-mediated Treg differentiation. Following TCR activation, E-proteins are downregulated. Decreased expression or activity of E-proteins is a prerequisite for TCR-induced CD25 expression, NF‐κB activation [136] and eTreg differentiation [137]. The role of Id proteins in Treg cells is more complicated. TCR signaling represses Id3 expression in Treg cells. Lowered level of Id3 is associated with an activation of follicular regulatory T (Tfr) phenotype [138]. However, sustained lower abundance of both Id2 and Id3 interferes with normal development of Tfr cells. Depletion of both Id2 and Id3 in Treg cells impairs Treg maintenance and anatomical distribution [138]. While Id3 is downregulated following TCR activation, the expression of Id2 is increased [129, 139]. The opposite directions of Id2 and Id3 expression suggest that they may control different modules of Treg biology. Indeed, Id2 is critically needed for certain types of tissue Treg cells. Treg-specific deletion of Id2 alone preferentially reduces Treg cells in adipose tissue, not in the SLOs. Moreover, Id2 deficiency leads to decreased expression of adipose tissue Treg-associated markers (e.g., ST2, CCR2, KLRG1 and GATA3) [139]. In contrast, Id3 has different functions. Treg cells residing in non-lymphoid tissues do not express Id3 [129]. Adoptive transfer experiments and transcriptome analyses show a reduction of Id3 in Treg cells between lymphoid organs and those residing at tissue sites [129]. Based on these data, a stepwise differentiation process marked by downregulation of Id3 has been proposed for tissue resident Treg cells. In addition, Id3 is involved in induced Treg (iTreg) generation, because ablation of Id3 in T cells leads to compromised generation of iTreg cells and biased differentiation towards the Th17 fate [140]. Id3 also helps maintain Foxp3 stability via a TF circuit involving E47, Spi-B, and SOCS3 [141]. However, how essential Id3 is needed for stable Foxp3 expression remains to be determined, especially given that Id3 is absent in most if not all, tissue Treg cells.
Treg functional diversity
“Division-of-labor” of Treg subsets
The primary role of Treg cells is to restrain excessive inflammation. However, each inflammation is different in nature. For instance, CD4+ Teff responses can be classified into different types (including Th1, Th2, Th17, Tfh, etc.). How do Treg cells effectively keep each of these Teff responses in check? Treg cells have evolved various mechanisms to keep various kinds of inflammation in check. TFs play a key role in driving the functional specification of Treg cells. Several studies have provided clear evidence showing that Treg cells can acquire the expression of Teff cell-associated TFs such as T-bet, IRF4 and STAT3. By doing so, Treg cells can more effectively suppress the corresponding Th responses, namely, Th1, Th2 and Th17, respectively [142–144]. Later studies have confirmed this notion. For instance, T-bet+ CXCR3+ Treg cells are preferentially enriched in the pancreatic islets of animal models of type 1 diabetes (T1D), in which Th1 responses are a key component of disease pathogenesis. Indeed, ablation of T-bet in Treg cells unleashes islet inflammation and exacerbates diabetes [145]. Tfr cells, a Treg functional subset, have a unique role in regulating follicular helper T (Tfh) and germinal center (GC) B cell responses [146]. A key feature of Tfr cells is the expression of CXCR5 and Bcl6. Bcl6 is a TF associated with Tfh development [147–149]. Treg-specific ablation of Bcl6 leads to a reduced Tfr pool and the onset of autoimmune diseases [150]. TCF1 has been found to act upstream of Bcl6 [95, 113]. Indeed, ablation of TCF1 (and LEF1 due to redundancy) almost completely eliminates Tfr generation [95]. Thus, Treg cells co-opt a TF axis of TCF1/LEF1-Bcl6 to differentiate into Tfr cells, which control Tfh responses. Later studies have found more TFs and transcriptional regulators to guide Treg cells to gain specialized functions. For instance, transcriptional regulator Rbpj guides Treg cells to specifically restrain Th2 responses, including their own excessive Th2-like differentiation potential [151].
These models explain how Treg cells work. Following this rule, it can be speculated that Treg cells in an analogous manner may op-opt the corresponding TFs (such as GATA3 and RORγt) to suppress Th2 and Th17 responses, respectively. GATA3, a master TF for Th2 cell differentiation [152] is abundantly expressed in Treg cells, at least in certain subsets [153–155]. However, GATA3 does not seem to restrict Treg cells to control Th2 responses. Instead, studies have found that GATA3 is critical for maintaining Foxp3 expression [153] and lineage stability especially under inflammatory conditions [154]. GATA3-deficient Treg cells have reduced expression of Foxp3 and other Treg suppressive genes and exhibit a broader defect in control of Th1, Th2, and Th17 responses. In parallel, RORγt, a TF for Th17 differentiation [156], is expressed by a subset of intestinal Treg cells [157, 158]. Gut microbiota and retinoid acid are involved in the development of RORγt+ Treg cells [157, 158]. In terms of their function, one study showed that RORγt+ Treg cells are needed to restrain intestinal Th2 responses [157], whereas another study found that these Treg cells can control both Th1 and Th17 responses [158]. These discrepancies may be due to the differences of colonized microbiota, disease model, or cytokine milieu.
This functional adaption model to explain each of these TFs in Treg functional heterogeneity may be oversimplified. The role of aforementioned TFs (such as IRF4, T-bet, STAT3) is not merely restricted to the adaptation mechanisms by Treg cells to restrain the corresponding CD4+ Teff responses. For example, IRF4 has been found to play a more general role in promoting the transition of Treg cells from resting to activated phenotype following TCR activation [77, 131]. A recent study shows that T-bet+ Treg cells can also dampen CD8+ T cell activation [159]. Last, STAT3 can act upstream of TCF1 to regulate Tfr development [160]. Furthermore, while TCF1 and LEF1 drive Tfr development, they are also required for the optimal response to IL-2 in Treg cells [95] and the repression of non-Treg lineage genes [113].
Treg functional adaptation to environmental cues
Mounting evidence has suggested that Treg cells can adapt to local cues and acquire special functional properties. This is best exemplified by the reported Treg phenotypes and specific functions in non-lymphoid tissues [161–164]. This topic has been comprehensively reviewed elsewhere [6, 7]. Here, we will provide an update of recent advances of Treg functional adaptation and highlight a few examples to show how TFs act as important regulators linking local environmental cues and Treg adaptation.
A recent study using an integrated approach of microarray, scRNA-seq and ATAC-seq analyses has revealed new features of how Treg’s tissue tropism is established. One of the key findings from this study is that chromatin structures of tissue Treg genes have been primed in the SLOs, such as spleen, through the establishment of the pan-Treg open chromatin regions [165]. Local tissue-derived factors help remodel additional regulatory elements, forming pan-tissue or tissue-specific chromatin modifications. On top of these epigenomic preparations, the effects of TFs are unfolded in two layers. First, all tissue Treg cells show repeated enrichments of motifs for bZIP [165, 166] and GATA family [166] TFs, suggesting that certain members of these families are the main drivers of the expression of the shared tissue Treg gene signature [165, 166]. Within each tissue, distinct TFs (such as Ets, nuclear receptor, or Runx families) are induced. Together with the primary pan-tissue Treg TFs, a feed-forward loop is formed for the induction of tissue-specific Treg gene signature [165]. Below we will use adipose tissue, colon and tumor as examples to discuss how Treg functional adaptation in each condition is regulated by different TFs.
Transcriptional regulation of adipose tissue Treg phenotype and function
The identification and functional characterization of Treg cells in adipose tissue has opened a new arena of understanding the Treg’s role in tissue homeostasis (reviewed in [6, 167]). The abundance of Treg cells in abdominal fat is anti-correlated with the degree of insulin resistance [161]. In humans, Treg frequencies negatively correlate with body mass index but are comparable between type 2 diabetes (T2D) and non-T2D individuals [168]. These Treg cells express high level of PPARG, CCR4, PRDM1 and CXCL2, but not ST2 [168].
Peroxisome proliferator-activated receptor (PPAR)γ is a key driver for visceral adipose tissue (VAT) Treg accumulation, phenotype and function [155]. PPARγ interacts with Foxp3 on the protein level to induce VAT Treg-specific gene expression. A recent study has found that that a small population of PPARγlo Treg cells are present in lymphoid organs (e.g., spleen), and already gain part of the VAT Treg gene signature. However, the majority of the VAT Treg unique genes are only “turned on” locally in the adipose tissue [169]. Thus, tissue-derived focal factors play an important role in programming Treg phenotype and function.
Additional TFs have been found to participate in VAT Treg phenotype and function. Id2 is found to promote VAT Treg survival [139]. Loss of Id2 increases the death of VAT Treg cells due to increased FAS expression. BATF and IRF4 have been reported to bind to regulatory regions of Pparg and Il1rl1 (encoding ST2, the receptor for IL-33) and regulate their expression [170]. IL-33 is a critical factor in inducing VAT Treg phenotype and function [170, 171]. ST2 is abundantly expressed in VAT Treg cells, in a Myd88-dependent manner [170, 171]. However, the enrichment for ST2+ Tregs in VAT only partially depends PPARγ. Instead, MHCII and IL-33 play a dominant role in driving the accumulation of VAT Treg cells [170, 171].
Transcriptional regulation of intestinal Treg phenotype and function
Blimp1 plays a more specific role in Treg activation and differentiation. Blimp1+ Treg cells are enriched at mucosal sites [131]. IL-10 is a key mediator of Treg’s function in mucosal barriers [172, 173]. Blimp1 is the key TF driving IL-10 production in Treg cells [131, 174]. Moreover, Blimp1 has also been found to prevent the expression of Th17 cytokines, in particular in the intestinal Foxp3+ RORγt+ Treg subset [175]. In the absence of Blimp1, the Il17 locus becomes activated. Blimp1-deficient RORγt+ Treg cells lose suppressor function and instead provoke intestinal inflammation. Thus, through promoting IL-10 production and suppressing the alternative fate, Blimp1 ensures the function of Treg cells at mucosal barriers. Recent studies have revealed more functions of Blimp1 in Treg cells. For instance, genetic ablation of Blimp1 leads to hypermethylation of Foxp3 CNS2 and loss of Foxp3 expression [176].
In addition to Blimp1, three recent studies have found that c-Maf plays a critical role in driving intestinal RORγt+ Treg generation and host-microbe symbiosis [177–179]. Two of them have shown that c-Maf is needed for IL-10 production in Treg cells [177, 179], whereas the third one has argued that IL-10 production in Treg cells is c-Maf independent [178]. Aryl hydrocarbon receptor (AhR) is an environmental sensor. AhR is important for in vivo Treg function, but not Foxp3 expression. AhR expression regulates Treg homing to the gut. In the gut, depending on the types of ligands, AhR regulates the balance between Treg and Th17 differentiation, thus the outcome of local immune responses [180, 181]. A recent study has found that AhR is more abundantly expressed in pTreg cells in the gut [182]. Therefore, multiple TFs are involved in regulating Treg phenotypes and functions at mucosal barriers. However, it remains to be determined how these TFs cooperate with other each or simply each of them controls different subsets of intestinal Treg cells.
Treg cells in tumors
Tumor-associated Treg cells have become another important frontier for basic and translational immunology [183–187]. In general, Treg cells at tumor sites exhibit an activated phenotype, expressing high levels of Ctla4, Tigit and Tnfrsf9 in multiple mouse tumors (including MC38, B16 and CT26) and TNFRSF4, TNFRSF9, and TNFRSF18 in human colorectal carcinomas [188]. In fact, there is a significant overlap of the tumor Treg signature between the mouse and human datasets [188]. Interestingly, the expression of IL10 is downregulated in tumor Treg cells, suggesting IL-10 is unlikely the effector cytokine for Treg’s function in tumor microenvironment. Studies from individual cases of tumors have revealed more features of Treg cells. For instance, CCR8+ Treg cells densely populate human beast carcinomas and correlate with the grade and type of breast cancers [189]. In colorectal cancer (CRC), two populations of FOXP3+ T cells are detected: FOXP3hi cells have a typical eTreg phenotype and are immunosuppressive. However, a population of non-immunosuppressive FOXP3lo T cells are also enriched in the CRC. The abundance of the latter is correlated with a better cancer prognosis [190].
It can be speculated that the deletion of Treg cells could augment anti-tumor immunity. Indeed, an early animal study has shown that the depletion Treg cells using anti-CD25 mAb prior to the tumor inoculation, increases the infiltration of CD8+ T cells and IL-2 production [191]. However, a key challenge for targeting tumor Treg cells is how to specifically deplete Treg cells infiltrating into tumor tissues without dampening anti-tumor Teff cells and provoking systemic autoimmunity. Following studies have reported various methods to selectively remove tumor-associated Treg cells. For instance, CCR4+ Treg cells are the dominant population of Treg cells in human melanoma tissue. Ex vivo depletion of CCR4+ Treg cells and subsequent in vitro stimulation of the depleted cell population with tumor antigen (NY-ESO1 in this case) efficiently induces antigen-specific CD4 and CD8 T cell responses. In vivo administration of an anti-CCR4 mAb reduces the number of eTreg cells and augments tumor antigen-specific CD8+ T cell responses in adult T-cell leukemia–lymphoma patients [192].
CTLA4 is highly expressed in Treg cells at tumor sites [188, 193]. Treg-specific depletion of CTLA4 more effectively eradicates inoculated tumors in mice [193, 194]. An interesting finding is that ipilimumab, a human anti-CTLA4 mAb, can engage FcγRIIIA-expressing monocytes, resulting in the lysis of Treg cells in a manner of antibody-dependent cell-mediated cytotoxicity [195]. PD-1 is also expressed at a relatively higher level in tumor Treg cells than the circulating counterparts [196]. However, surprisingly, anti-PD-1 treatment increases the proliferation of Treg cells in some gastric cancer patients. In fact, PD-1 blockade has been found to enhance Treg cell suppressive capacity in vitro [196]. Similar phenomenon has been reported in animal studies [196]. Thus, blocking CTLA4 or PD-1 induces different outcomes in tumor Treg cells. Moreover, in human melanoma and mouse MC38 tumor models, anti-CTLA4 induces the expansion of the ICOS+ Th1-like effector T cells, whereas anti-PD-1 predominantly induces the expansion of specific tumor-infiltrating exhausted-like CD8 T cell subsets. Combinational approach of Treg depletion and immune checkpoint blockade has also been tested in various cancers. For example, an Fc-optimized anti-CD25 mAb synergizes with anti-PD1 mAb to more effectively eradicate established tumors in mice [197].
IL-33 plays an important role in the accumulation and function of Treg cells at tumor sites [198–200]. Epidermis-derived IL-33 escalates a tumor-promoting immune environment in chronic allergic contact dermatitis (ACD). IL-33 promotes Treg accumulation in the ACD. Mice lacking IL-33 are protected from chronic ACD and skin cancer compared to wild-type controls [198]. In the case of CRC, tumor-infiltrating Treg cells express high level of ST2 [199]. Genetic ablation of Il1rl1 (encoding ST2) reduces Treg infiltration and enhances the frequencies of CD8+ T cells, together decreasing tumor burden [199, 201]. The number of activated ST2 expressing Treg cells is also increased in blood and tumor lesions of the CRC patients [199]. Another study has reported a Treg-intrinsic role of IL-33 in regulating the expansion and function of these cells at tumor sites [200]. They found that Treg cells can also produce IL-33. In a melanoma model, Foxp3Cre Il33fl/fl mice exhibited delayed tumor growth compared to control Foxp3Cre mice [200]. Together, these studies have revealed important role of IL-33 in regulating Treg accumulation and function at tumor sites in either Treg cell-intrinsic or extrinsic manner.
TFs and histone modifiers have been reported to regulate Treg phenotype and function in tumor. For instance, Foxo1, while promoting Treg cell suppression of lymphoproliferative diseases [105, 106], is downregulated in activated Treg cells, concomitant with the repression of Foxo1-target genes [107]. This change becomes more profound in tumor-infiltrating Treg cells. A mutant form of Foxo1 refractory to the inhibition by Akt is sufficient to deplete tumor-associated Treg cells, activate effector CD8+ T cells, and inhibit tumor growth without causing autoimmunity. These data suggest that Foxo pathway can be harnessed in a dose-dependent manner to selectively remove Treg cells in tumor with modest effect on systemic Treg distribution and function.
In addition to depletion of Treg cells in tumors, modulation of Treg stability has also been examined to boost anti-tumor immunity. For instance, NF-κB family member c-Rel regulates the expression of activated Treg genes (such as Tnfrsf8, Klrg1, Il1r2, Tigit, Ccr8). Importantly, c-Rel controls a specific genetic program in Treg cells that is required for inhibition of the anti-melanoma protective immune response mediated by CD8+ T cells [85]. As such, pharmacological inhibition of c-Rel impairs Treg stability by reducing the expression of Treg core genes including Foxp3, Il2ra (CD25), and Ikzf2 (Helios). With this, tumor growth is halted by the c-Rel inhibitor [85]. EZH2 is upregulated in tumor-infiltrating Treg cells in melanoma [202]. Pharmacological inhibition of EZH2 destabilizes Foxp3 expression and slows tumor growth. Disruption of EZH2 activity in Treg cells, either pharmacologically or genetically, induces the expression of pro-inflammatory genes in Treg cells. Of note, inhibition of EZH2 activity selectively reprograms the function of tumor-infiltrating Treg cells without systemically altering Treg function, thus reducing the risk of systemic autoimmunity [202]. These findings suggest that Treg stability at tumor sites can be targeted to convert Treg cells into pro-inflammatory Teff cells. As a consequence, it remodels the tumor microenvironment and enhances the recruitment and function of CD8+ and CD4+ Teff cells, together eradicating tumor.
Tissue Treg precursors
The accumulated knowledge about tissue Treg cells raises important questions: where do Treg cells in non-lymphoid tissues come from? What is the relationship between tissue Treg cells and their counterparts in lymphoid organs? Several recent studies have provided important clues about the connections of Treg cells between SLOs and non-lymphoid tissues (Fig. 3). First, using the PPARγ reporter mice, Li et al. have proposed a two-step model of VAT Treg differentiation. As a first step, part of the VAT Treg transcriptome component, in particular T cell activation related genes, has already been induced in a subset of PPARγlo Treg cells in the spleen, suggesting that a priming has occurred in these cells prior to their migration to the tissue sites. The second step occurs at local tissues and is driven by tissue-derived factors [169]. VAT Treg cells proliferate locally with little contribution of conversion from Tconv cells. Factors such as IL-33 and antigens not only promote their expansion, also shape their TCR repertoire [171].
The identification of splenic PPARγlo Treg cells suggests the existence of tissue Treg precursors in the SLOs. These cells have already gained partial VAT Treg transcriptional program, mainly related to T cell activation [169]. In another study, Nfil3+ Treg cells have been identified as the pan-tissue Treg precursors [166]. Further analysis has divided these lymphoid organ Nfil3+ Treg cells into two subsets, based on the expression of Klrg1 [166]. Klrg1− Nfil3+ Treg cells constitute about 10% in spleen and lymph nodes (LNs) whereas the Klrg1+ Nfil3+ subset is about 4% in spleen and even lower in LNs (1–2%). Both subsets possess the common accessible chromatin regions shared between all examined tissue Treg types. RNA-velocity analysis and adoptive transfer assay show that Klrg1+ Nfil3+ subset is at a further developmental stage towards the tissue Treg phenotype. Of note, the TCR repertoire in Klrg1− Nfil3+ Treg cells becomes much more restricted (even more in Klrg1+ Nfil3+ subset) compared to that in the Klrg1− Nfil3− Treg cells. What drives the selection of TCRs remains poorly understood.
Treg trajectory and the priming of the precursors in lymphoid organs have also been reported in another study focusing on investigating the ontogeny of colon and skin Treg cells [203]. Using scRNA-seq and pseudo-time ordering algorithm, Miragaia et al. have attempted to define the trajectories among Treg cell populations in skin, colon and the respective draining LNs. They have found that at steady state, a core gene signature is shared by the LN–skin and LN–colon trajectories, suggesting a common mechanism is evolved to regulate the migration of Treg cells from the draining LNs to the tissue sites. Their analyses have also suggested that a further adaptation process would happen in a tissue-specific manner [203]. Interestingly, a similar trajectory was recapitulated in a melanoma model [203]. An earlier study has reported a bidirectional trafficking of Treg cells between skin and skin draining LNs in mice [204]. However, to what extent the homing of Treg cells from tissue to the LNs remains to be determined as later studies have counterargued this possibility [166, 171].
On transcriptional level, BATF is critical for the development of Treg precursors in lymphoid organs [166]. In the absence of BATF, both Klrg1− and Klrg1+ precursors are markedly reduced with a concomitant reduction of Treg numbers in all examined tissues namely colon, skin and VAT [166]. In line with this, an earlier study focusing on VAT Treg has shown that BATF is required for VAT Treg differentiation through regulating ST2 and PPARγ expression [170]. BATF has also been reported to play a critical role in human tissue Treg cells. Patients with FOXP3A384T mutations develop tissue-restricted autoimmunity. The A384T mutation in FOXP3 results in the repressed expression of BATF, which is responsible for impaired tissue Treg fitness [205].
Treg differentiation is associated with a progressive loss of Id3, TCF1 or LEF1 expression [95, 129]. Of note, the percentage of Id3− or TCF1− Treg cells in the SLOs is quite similar to that of the reported Nfil3+ Treg cells by Delacher et al. Furthermore, in both studies [95, 129], the expression of Klrg1 is restricted to Id3− or TCF1− Treg subset. In fact, flow cytometric analysis has revealed that only a subset of TCF1− Treg cells express Klrg1, reminiscent of the expression pattern of Klrg1 in Nfil3+ cells [166]. Based on these data, it is very likely that the TCF1− (Id3−) Treg subset overlaps with the Nfil3+ Treg cells, with the Klrg1+ subset (also Nfil3+ TCF1− Id3−) being the more mature stage of tissue Treg precursors in the SLOs. Therefore, a reoriented TF network with the upregulation of PPARγ and BATF, and downregulation of TCF1 and Id3 is permissive for the transitional stage of Treg, prior to populating non-lymphoid tissues.
Closing remarks and future perspectives
There is little doubt that Treg cells play a pivotal role in keeping autoimmune responses in check. Treg cells can exert their “suppressive” capacity in multiple ways (reviewed in [206–209]). Indeed, numerous studies have proved that an impairment of Treg’s suppressive capacity leads to severe autoimmune or inflammatory consequences. However, mounting evidence has also shown that Treg cells can regulate a broad range of immunological and non-immunological processes [6]. In many cases, the function of Treg cells is not merely explained by their “suppressive” capability [95, 210]. For instance, the ablation of both Tcf7 and Lef1 does not alter Treg’s “classical” function to suppress Teff cell proliferation and dendritic cell maturation, but does cause severe pathological consequences in vivo evidenced by the early and spontaneous onset of systemic autoimmune diseases [95]. Emerging evidence shows that Treg cells also participate in tissue repair and regeneration [49, 162, 210]. This has been evidenced in various models of tissue injury. For instance, Treg cells rapidly accumulate in the acutely injured skeletal muscle of mice. The IL-33/ST2 axis plays a key role in driving the accumulation of Treg cells in injured muscle [211]. Muscle Treg cells acquire special phenotypic and functional properties, presumably making these cells more effective in tissue repair [162, 171, 211]. On the other hand, punctual depletion of Treg cells during the repair process prolongs pro-inflammatory infiltrate and impairs muscle repair [162]. In animal models of infectious lung injury, Treg cells play a major role in tissue repair, distinct from that in suppression of immune responses [210]. Amphiregulin is a key molecule produced by lung Treg cells to mediate tissue protection and maintain barrier integrity. Of note, amphiregulin deficiency does not alter Treg suppressive function [210]. Thus, different factors and mechanisms are used by Treg cells to partake in the repair and regeneration processes in a tissue-specific manner. Identifying these molecular mediators and pathways will have both theoretical and applicable impacts.
Treg-based therapies are currently undergoing active pre-clinical and clinical investigations, aiming for the treatment of autoimmune diseases (such as type 1 diabetes, T1D), organ transplantation and graft-versus-host diseases [8, 212, 213]. Several strategies have been tested and optimized to generate therapeutic-grade Treg cells. For instance, IL-2 and rapamycin have been shown to expand Treg cells. Expansion of Treg cells following low-dose IL-2 treatment has also been reported in T1D patients [214, 215]. However, major challenges remain. Some of the recent advances tackling them are discussed below. First, the purity of isolated human Treg cells remains to be improved [216]. It is known that FOXP3 can be transiently expressed in activated human T cells [217]. Indeed, human FOXP3+ CD4+ T cells are a mixture of three phenotypically and functionally distinct subpopulations [218]. Both FOXP3hi CD45RA− and FOXP3lo CD45RA+ subsets are immunosuppressive in vitro. However, the FOXP3lo CD45RA− subset does not possess a suppressive capacity. Instead, they produce inflammatory cytokines, indicative of a promiscuous expression of FOXP3 in activated Teff cells. The gating strategy (CD25+CD127lo) for sorting human Treg cells [219, 220] does not exclude the possible contamination of activated Teff cells. More studies are needed to best identify the “genuine” human Treg cells [221, 222]. Second, selective expansion of human Treg in vivo is another task that needs be prioritized for developing Treg-based therapy. Engineering tools to modify IL-2 or IL-2R, or anti-IL-2 antibodies have demonstrated a targeted expansion of Treg cells [223, 224]. These effects could pave the road for next-generation Treg therapy [225]. Last, learned from the studies in animal models, it is persuasive that different populations of Treg cells that have been programed with specific functional properties are needed to best treat the corresponding human diseases. One of the strategies to produce functionally tailored Treg subsets is to induce the expression of certain TFs that can drive Treg cells to acquire the needed functional specificity. Conversely, knockdown of Treg “locking” TFs may have therapeutic implications in anti-tumor immunity [226].
Acknowledgements
We thank the Fu lab members for insightful discussions. This work was funded by the US National Institute of Health (AI139753 to W.F.). We apologize to those individuals whose work could not be cited here due to space limitation.
Abbreviations
- AhR
Aryl hydrocarbon receptor
- Akt
AKT serine/threonine kinase 1
- AP1
Activator protein 1
- ATF
Activating transcription factor
- Bach2
BTB domain and CNC homolog 2
- BATF
Basic leucine zipper ATF-like transcription factor
- Bcl10
B-cell lymphoma/leukemia 10
- Bcl11b
B-cell lymphoma/leukemia 11b
- Bcl6
B-cell lymphoma/leukemia 6
- Blimp1
B-lymphocyte-induced maturation protein 1
- CARD11
Caspase recruitment domain family member 11
- Cbfβ
Core-binding factor subunit beta
- c-Maf
V-maf musculoaponeurotic fibrosarcoma oncogene homolog
- CREB
CAMP responsive element binding protein 1
- c-Rel
REL proto-oncogene, NF-κB subunit
- CTLA4
Cytotoxic T-lymphocyte associated protein 4
- DOCK8
Dedicator of cytokinesis 8
- E47
Transcription factor 3
- Eos
IKAROS family zinc finger 4
- EZH2
Enhancer of Zeste homolog 2
- Foxo1
Forkhead box O1
- Foxo3
Forkhead box O3
- Foxp1
Forkhead box P1
- Foxp3
Forkhead box P3
- GATA1
GATA binding protein 1
- GATA3
GATA binding protein 3
- Helios
IKAROS family zinc finger 2
- HIF1α
Hypoxia inducible factor 1 subunit alpha
- Id3
Inhibitor of DNA binding 3, HLH protein
- IKKβ
Inhibitor of nuclear factor kappa B kinase subunit beta
- IRF4
Interferon regulatory factor 4
- IκB
Inhibitor of nuclear factor kappa B
- JunB
JunB proto-oncogene, AP-1 transcription factor subunit
- Klrg1
Killer cell lectin like receptor G1
- LEF1
Lymphoid enhancer binding factor 1
- MALT1
Mucosa-associated lymphoid tissue lymphoma translocation protein 1
- NFAT
Nuclear factor of activated T cells
- Nfil3
Nuclear factor, interleukin 3 regulated
- NF-κB
Nuclear factor kappa B subunit 1
- Nr4a
Nuclear receptor subfamily 4 group A
- PPARγ
Peroxisome proliferator-activated receptor gamma
- Rbpj
Recombination signal binding protein for immunoglobulin kappa J region
- RelA
RELA proto-oncogene, NF-κB subunit
- RORγt
RAR-related orphan receptor gamma
- Runx1
RUNX family transcription factor 1
- Satb1
SATB homeobox 1
- SMAD
Sma- and mad-related protein
- SOCS1
Suppressor of cytokine signaling 1
- SOCS3
Suppressor of cytokine signaling 3
- Spi-B
Spi-B transcription factor
- STAT5
Signal transducer and activator of transcription 5
- T-bet
T-box expressed in T cells
- TCF1
T-cell factor 1
Author contributions
Conceptualization: KW and WF. Writing and editing: KW and WF
Compliance with ethical standards
Conflict of Interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lee YS, Wollam J, Olefsky JM. An integrated view of immunometabolism. Cell. 2018;172:22–40. doi: 10.1016/j.cell.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125:3347–3355. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pentcheva-Hoang T, Corse E, Allison JP. Negative regulators of T-cell activation: potential targets for therapeutic intervention in cancer, autoimmune disease, and persistent infections. Immunol Rev. 2009;229:67–87. doi: 10.1111/j.1600-065X.2009.00763.x. [DOI] [PubMed] [Google Scholar]
- 4.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panduro M, Benoist C, Mathis D. Tissue tregs. Annu Rev Immunol. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whibley N, Tucci A, Powrie F. Regulatory T cell adaptation in the intestine and skin. Nat Immunol. 2019;20:386–396. doi: 10.1038/s41590-019-0351-z. [DOI] [PubMed] [Google Scholar]
- 8.Raffin C, Vo LT, Bluestone JA. Treg cell-based therapies: challenges and perspectives. Nat Rev Immunol. 2020;20:158–172. doi: 10.1038/s41577-019-0232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tada T, Taniguchi M, Takemori T. Properties of primed suppressor T cells and their products. Transplant Rev. 1975;26:106–129. doi: 10.1111/j.1600-065x.1975.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 10.Green DR, Flood PM, Gershon RK. Immunoregulatory T-cell pathways. Annu Rev Immunol. 1983;1:439–463. doi: 10.1146/annurev.iy.01.040183.002255. [DOI] [PubMed] [Google Scholar]
- 11.Dorf ME, Benacerraf B. Suppressor cells and immunoregulation. Annu Rev Immunol. 1984;2:127–157. doi: 10.1146/annurev.iy.02.040184.001015. [DOI] [PubMed] [Google Scholar]
- 12.Asherson GL, Colizzi V, Zembala M. An overview of T-suppressor cell circuits. Annu Rev Immunol. 1986;4:37–68. doi: 10.1146/annurev.iy.04.040186.000345. [DOI] [PubMed] [Google Scholar]
- 13.Nishizuka Y, Sakakura T. Thymus and reproduction: sex-linked dysgenesia of the gonad after neonatal thymectomy in mice. Science. 1969;166:753–755. doi: 10.1126/science.166.3906.753. [DOI] [PubMed] [Google Scholar]
- 14.Sakaguchi S, Takahashi T, Nishizuka Y. Study on cellular events in post-thymectomy autoimmune oophoritis in mice. II. Requirement of Lyt-1 cells in normal female mice for the prevention of oophoritis. J Exp Med. 1982;156:1577–1586. doi: 10.1084/jem.156.6.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powrie F, Mason D. OX-22high CD4+ T cells induce wasting disease with multiple organ pathology: prevention by the OX-22 low subset. J Exp Med. 1990;172:1701–1708. doi: 10.1084/jem.172.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKeever U, et al. Adoptive transfer of autoimmune diabetes and thyroiditis to athymic rats. Proc Natl Acad Sci USA. 1990;87:7618–7622. doi: 10.1073/pnas.87.19.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 18.Chatila TA, et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106:R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wildin RS, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 20.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 21.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 22.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 23.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 24.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 25.Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 26.Kwon HK, Chen HM, Mathis D, Benoist C. FoxP3 scanning mutagenesis reveals functional variegation and mild mutations with atypical autoimmune phenotypes. Proc Natl Acad Sci USA. 2018;115:E253–E262. doi: 10.1073/pnas.1718599115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgiev P, Charbonnier LM, Chatila TA. Regulatory T cells: the many faces of Foxp3. J Clin Immunol. 2019;39:623–640. doi: 10.1007/s10875-019-00684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, et al. Biochemistry and therapeutic implications of mechanisms involved in FOXP3 activity in immune suppression. Curr Opin Immunol. 2007;19:583–588. doi: 10.1016/j.coi.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Ramsdell F, Ziegler SF. FOXP3 and scurfy: how it all began. Nat Rev Immunol. 2014;14:343–349. doi: 10.1038/nri3650. [DOI] [PubMed] [Google Scholar]
- 30.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 31.Lin W, et al. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 32.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Fu W, et al. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nat Immunol. 2012;13:972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cretney E, Kallies A, Nutt SL. Differentiation and function of Foxp3(+) effector regulatory T cells. Trends Immunol. 2013;34:74–80. doi: 10.1016/j.it.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Kitagawa Y, Wing JB, Sakaguchi S. Transcriptional and epigenetic control of regulatory T cell development. Prog Mol Biol Transl Sci. 2015;136:1–33. doi: 10.1016/bs.pmbts.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Zaiss DMW, Coffer PJ. Forkhead box transcription factors as context-dependent regulators of lymphocyte homeostasis. Nat Rev Immunol. 2018;18:703–715. doi: 10.1038/s41577-018-0048-9. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Y, et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 38.Marson A, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon HK, Chen HM, Mathis D, Benoist C. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat Immunol. 2017;18:1238–1248. doi: 10.1038/ni.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Beyer M, et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol. 2011;12:898–907. doi: 10.1038/ni.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arvey A, et al. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2014;15:580–587. doi: 10.1038/ni.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopes JE, et al. Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J Immunol. 2006;177:3133–3142. doi: 10.4049/jimmunol.177.5.3133. [DOI] [PubMed] [Google Scholar]
- 45.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawant DV, Vignali DA. Once a Treg, always a Treg? Immunol Rev. 2014;259:173–191. doi: 10.1111/imr.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 48.Charbonnier LM, et al. Functional reprogramming of regulatory T cells in the absence of Foxp3. Nat Immunol. 2019;20:1208–1219. doi: 10.1038/s41590-019-0442-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell C, Rudensky A. Roles of regulatory t cells in tissue pathophysiology and metabolism. Cell Metab. 2020;31:18–25. doi: 10.1016/j.cmet.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zemmour D, et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol. 2018;19:291–301. doi: 10.1038/s41590-018-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mantel PY, et al. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176:3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- 52.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tseng WY, et al. TNF receptor 2 signaling prevents DNA methylation at the Foxp3 promoter and prevents pathogenic conversion of regulatory T cells. Proc Natl Acad Sci USA. 2019;116:21666–21672. doi: 10.1073/pnas.1909687116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sekiya T, et al. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat Immunol. 2013;14:230–237. doi: 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- 55.Zheng Y, et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 57.Kitagawa Y, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol. 2017;18:173–183. doi: 10.1038/ni.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Liang Y, LeBlanc M, Benner C, Zheng Y. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell. 2014;158:734–748. doi: 10.1016/j.cell.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng Y, et al. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 2014;158:749–763. doi: 10.1016/j.cell.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zemmour D, Pratama A, Loughhead SM, Mathis D, Benoist C. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc Natl Acad Sci USA. 2017;114:E3472–E3480. doi: 10.1073/pnas.1700946114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsieh CS, Lee HM, Lio CW. Selection of regulatory T cells in the thymus. Nat Rev Immunol. 2012;12:157–167. doi: 10.1038/nri3155. [DOI] [PubMed] [Google Scholar]
- 62.Jordan MS, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:283–284. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 63.Kawahata K, et al. Generation of CD4(+)CD25(+) regulatory T cells from autoreactive T cells simultaneously with their negative selection in the thymus and from nonautoreactive T cells by endogenous TCR expression. J Immunol. 2002;168:4399–4405. doi: 10.4049/jimmunol.168.9.4399. [DOI] [PubMed] [Google Scholar]
- 64.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 65.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice Implications for the nonredundant function of IL-2. Immunity. 2012;17:167–178. doi: 10.1016/s1074-7613(02)00367-9. [DOI] [PubMed] [Google Scholar]
- 67.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 68.Tai X, et al. Foxp3 transcription factor is proapoptotic and lethal to developing regulatory T cells unless counterbalanced by cytokine survival signals. Immunity. 2013;38:1116–1128. doi: 10.1016/j.immuni.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marshall D, et al. Differential requirement for IL-2 and IL-15 during bifurcated development of thymic regulatory T cells. J Immunol. 2014;193:5523–5533. doi: 10.4049/jimmunol.1402144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Łyszkiewicz M, et al. miR-181a/b-1 controls thymic selection of Treg cells and tunes their suppressive capacity. PLoS Biol. 2019;17:e2006716. doi: 10.1371/journal.pbio.2006716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Owen D, et al. Thymic regulatory T cells arise via two distinct developmental programs. Nat Immunol. 2019;20:195–205. doi: 10.1038/s41590-018-0289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002;3:756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 73.Rosenbaum M, et al. Bcl10-controlled Malt1 paracaspase activity is key for the immune suppressive function of regulatory T cells. Nat Commun. 2019;10:2352. doi: 10.1038/s41467-019-10203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gewies A, et al. Uncoupling Malt1 threshold function from paracaspase activity results in destructive autoimmune inflammation. Cell Rep. 2014;9:1292–1305. doi: 10.1016/j.celrep.2014.10.044. [DOI] [PubMed] [Google Scholar]
- 75.Cheng L, Deng N, Yang N, Zhao X, Lin X. Malt1 protease is critical in maintaining function of regulatory T cells and may be a therapeutic target for antitumor immunity. J Immunol. 2019;202:3008–3019. doi: 10.4049/jimmunol.1801614. [DOI] [PubMed] [Google Scholar]
- 76.Brustle A, et al. MALT1 is an intrinsic regulator of regulatory T cells. Cell Death Differ. 2017;24:1214–1223. doi: 10.1038/cdd.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nat Immunol. 2014;15:1070–1078. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sekiya T, et al. Nr4a receptors regulate development and death of labile Treg precursors to prevent generation of pathogenic self-reactive cells. Cell Rep. 2018;24:1627–1638. doi: 10.1016/j.celrep.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 79.Thome M, Tschopp J. TCR-induced NF-kappaB activation: a crucial role for Carma1, Bcl10 and MALT1. Trends Immunol. 2003;24:419–424. doi: 10.1016/s1471-4906(03)00177-7. [DOI] [PubMed] [Google Scholar]
- 80.Molinero LL, et al. CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J. Immunol. 2009;182:6736–6743. doi: 10.4049/jimmunol.0900498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Long M, Park SG, Strickland I, Hayden MS, Ghosh S. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921–931. doi: 10.1016/j.immuni.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 82.Hayden MS, Ghosh S. Regulation of NF-kappaB by TNF family cytokines. Semin Immunol. 2014;26:253–266. doi: 10.1016/j.smim.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruan Q, et al. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Isomura I, et al. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J Exp Med. 2009;206:3001–3014. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grinberg-Bleyer Y, et al. NF-kappaB c-Rel is crucial for the regulatory T cell immune checkpoint in cancer. Cell. 2017;170:1096–1108. doi: 10.1016/j.cell.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Visekruna A, et al. c-Rel is crucial for the induction of Foxp3(+) regulatory CD4(+) T cells but not T(H)17 cells. Eur J Immunol. 2010;40:671–676. doi: 10.1002/eji.200940260. [DOI] [PubMed] [Google Scholar]
- 87.Messina N, et al. The NF-kappaB transcription factor RelA is required for the tolerogenic function of Foxp3(+) regulatory T cells. J Autoimmun. 2016;70:52–62. doi: 10.1016/j.jaut.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 88.Oh H, et al. An NF-kappaB transcription-factor-dependent lineage-specific transcriptional program promotes regulatory T cell identity and function. Immunity. 2017;47:450–465. doi: 10.1016/j.immuni.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vasanthakumar A, et al. The TNF receptor superfamily-NF-kappaB axis is critical to maintain effector regulatory T cells in lymphoid and non-lymphoid tissues. Cell Rep. 2017;20:2906–2920. doi: 10.1016/j.celrep.2017.08.068. [DOI] [PubMed] [Google Scholar]
- 90.Ronin E, et al. The NF-kappaB RelA transcription factor is critical for regulatory T cell activation and stability. Front Immunol. 2019;10:2487. doi: 10.3389/fimmu.2019.02487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Deenick EK, et al. c-Rel but not NF-kappaB1 is important for T regulatory cell development. Eur J Immunol. 2010;40:677–681. doi: 10.1002/eji.201040298. [DOI] [PubMed] [Google Scholar]
- 92.Schuster M, et al. IkappaB(NS) protein mediates regulatory T cell development via induction of the Foxp3 transcription factor. Immunity. 2012;37:998–1008. doi: 10.1016/j.immuni.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 93.Yao Z, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu LF, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang BH, et al. TCF1 and LEF1 control Treg competitive survival and Tfr development to prevent autoimmune diseases. Cell Rep. 2019;27:3629–3645. doi: 10.1016/j.celrep.2019.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim HJ, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350:334–339. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mahmud SA, et al. Costimulation via the tumor-necrosis factor receptor superfamily couples TCR signal strength to the thymic differentiation of regulatory T cells. Nat Immunol. 2014;15:473–481. doi: 10.1038/ni.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shi H, et al. Hippo kinases Mst1 and Mst2 sense and amplify IL-2R-STAT5 signaling in regulatory T cells to establish stable regulatory activity. Immunity. 2018;49:899–914. doi: 10.1016/j.immuni.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Janssen E, et al. DOCK8 enforces immunological tolerance by promoting IL-2 signaling and immune synapse formation in Tregs. JCI Insight. 2017;2(19):e94298. doi: 10.1172/jci.insight.94298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Singh AK, et al. DOCK8 regulates fitness and function of regulatory T cells through modulation of IL-2 signaling. JCI Insight. 2017;2(19):e94275. doi: 10.1172/jci.insight.94275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Franceschini D, et al. PD-L1 negatively regulates CD4+ CD25+Foxp3+ Tregs by limiting STAT-5 phosphorylation in patients chronically infected with HCV. J Clin Invest. 2009;119:551–564. doi: 10.1172/JCI36604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burchill MA, et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harada Y, et al. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J Exp Med. 2010;207:1381–1391. doi: 10.1084/jem.20100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ouyang W, et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 105.Kerdiles YM, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ouyang W, et al. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 2012;491:554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Luo CT, Liao W, Dadi S, Toure A, Li MO. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature. 2016;529:532–536. doi: 10.1038/nature16486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tone Y, et al. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 109.Rudra D, et al. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kitoh A, et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–620. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 111.Klunker S, et al. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med. 2009;206:2701–2715. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rudra D, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol. 2012;13:1010–1019. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xing S, et al. Tcf1 and Lef1 are required for the immunosuppressive function of regulatory T cells. J Exp Med. 2019;216:847–866. doi: 10.1084/jem.20182010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van Loosdregt J, et al. Canonical Wnt signaling negatively modulates regulatory T cell function. Immunity. 2013;39:298–310. doi: 10.1016/j.immuni.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 115.Konopacki C, Pritykin Y, Rubtsov Y, Leslie CS, Rudensky AY. Transcription factor Foxp1 regulates Foxp3 chromatin binding and coordinates regulatory T cell function. Nat Immunol. 2019;20:232–242. doi: 10.1038/s41590-018-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ren J, et al. Foxp1 is critical for the maintenance of regulatory T-cell homeostasis and suppressive function. PLoS Biol. 2019;17:e3000270. doi: 10.1371/journal.pbio.3000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Drashansky TT, et al. Bcl11b prevents fatal autoimmunity by promoting Treg cell program and constraining innate lineages in Treg cells. Sci Adv. 2019;5:eaaw0480. doi: 10.1126/sciadv.aaw0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hasan SN, et al. Bcl11b prevents catastrophic autoimmunity by controlling multiple aspects of a regulatory T cell gene expression program. Sci Adv. 2019;5:0706. doi: 10.1126/sciadv.aaw0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ono M, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 120.Darce J, et al. An N-terminal mutation of the Foxp3 transcription factor alleviates arthritis but exacerbates diabetes. Immunity. 2012;36:731–741. doi: 10.1016/j.immuni.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bettini ML, et al. Loss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity. 2012;36:717–730. doi: 10.1016/j.immuni.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liston A, Gray DH. Homeostatic control of regulatory T cell diversity. Nat Rev Immunol. 2014;14:154–165. doi: 10.1038/nri3605. [DOI] [PubMed] [Google Scholar]
- 123.Campbell DJ. Control of regulatory T cell migration, function, and homeostasis. J Immunol. 2015;195:2507–2513. doi: 10.4049/jimmunol.1500801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smigiel KS, Srivastava S, Stolley JM, Campbell DJ. Regulatory T-cell homeostasis: steady-state maintenance and modulation during inflammation. Immunol Rev. 2014;259:40–59. doi: 10.1111/imr.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pierson W, et al. Antiapoptotic Mcl-1 is critical for the survival and niche-filling capacity of Foxp3(+) regulatory T cells. Nat Immunol. 2013;14:959–965. doi: 10.1038/ni.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang K, et al. Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature. 2017;548:602–606. doi: 10.1038/nature23665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.He N, et al. Metabolic control of regulatory T cell (Treg) survival and function by Lkb1. Proc Natl Acad Sci USA. 2017;114:12542–12547. doi: 10.1073/pnas.1715363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shi H, Chi H. Metabolic control of Treg cell stability, plasticity, and tissue-specific heterogeneity. Front Immunol. 2019;10:2716. doi: 10.3389/fimmu.2019.02716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sullivan JM, Hollbacher B, Campbell DJ. Cutting edge: dynamic expression of Id3 defines the stepwise differentiation of tissue-resident regulatory T cells. J Immunol. 2019;202:31–36. doi: 10.4049/jimmunol.1800917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dias S, et al. Effector regulatory T cell differentiation and immune homeostasis depend on the transcription factor Myb. Immunity. 2017;46:78–91. doi: 10.1016/j.immuni.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 131.Cretney E, et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 132.Koizumi SI, et al. JunB regulates homeostasis and suppressive functions of effector regulatory T cells. Nat Commun. 2018;9:5344. doi: 10.1038/s41467-018-07735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chapman NM, et al. mTOR coordinates transcriptional programs and mitochondrial metabolism of activated Treg subsets to protect tissue homeostasis. Nat Commun. 2018;9:2095. doi: 10.1038/s41467-018-04392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Haljasorg U, et al. Irf4 expression in thymic epithelium is critical for thymic regulatory T cell homeostasis. J Immunol. 2017;198:1952–1960. doi: 10.4049/jimmunol.1601698. [DOI] [PubMed] [Google Scholar]
- 135.Sidwell T, et al. Attenuation of TCR-induced transcription by Bach2 controls regulatory T cell differentiation and homeostasis. Nat Commun. 2020;11:252. doi: 10.1038/s41467-019-14112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gao P, et al. Dynamic changes in E-protein activity regulate T reg cell development. J Exp Med. 2014;211:2651–2668. doi: 10.1084/jem.20132681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Han X, et al. E-protein regulatory network links TCR signaling to effector Treg cell differentiation. Proc Natl Acad Sci USA. 2019;116:4471–4480. doi: 10.1073/pnas.1800494116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miyazaki M, et al. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat Immunol. 2014;15:767–776. doi: 10.1038/ni.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frias AB, Jr, et al. The transcriptional regulator Id2 is critical for adipose-resident regulatory T cell differentiation, survival, and function. J Immunol. 2019;203:658–664. doi: 10.4049/jimmunol.1900358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Maruyama T, et al. Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nat Immunol. 2011;12:86–95. doi: 10.1038/ni.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rauch KS, et al. Id3 maintains Foxp3 expression in regulatory T cells by controlling a transcriptional network of E47, Spi-B, and SOCS3. Cell Rep. 2016;17:2827–2836. doi: 10.1016/j.celrep.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 142.Koch MA, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tan TG, Mathis D, Benoist C. Singular role for T-BET + CXCR3 + regulatory T cells in protection from autoimmune diabetes. Proc Natl Acad Sci USA. 2016;113:14103–14108. doi: 10.1073/pnas.1616710113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sage PT, Sharpe AH. T follicular regulatory cells. Immunol Rev. 2016;271:246–259. doi: 10.1111/imr.12411. [DOI] [PubMed] [Google Scholar]
- 147.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 150.Fu W, et al. Deficiency in T follicular regulatory cells promotes autoimmunity. J Exp Med. 2018;215:815–825. doi: 10.1084/jem.20170901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Delacher M, et al. Rbpj expression in regulatory T cells is critical for restraining TH2 responses. Nat Commun. 2019;10:1621. doi: 10.1038/s41467-019-09276-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 153.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wohlfert EA, et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121:4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cipolletta D, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17 + T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 157.Ohnmacht C, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 158.Sefik E, et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Levine AG, et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature. 2017;546:421–425. doi: 10.1038/nature22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xu L, et al. The kinase mTORC1 promotes the generation and suppressive function of follicular regulatory T cells. Immunity. 2017;47:538–551. doi: 10.1016/j.immuni.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 161.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, Rudensky AY. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. 2012;150:29–38. doi: 10.1016/j.cell.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Scharschmidt TC, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 2015;43:1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.DiSpirito JR et al. (2018) Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci Immunol 3 [DOI] [PMC free article] [PubMed]
- 166.Delacher M, et al. Precursors for nonlymphoid-tissue treg cells reside in secondary lymphoid organs and are programmed by the transcription factor BATF. Immunity. 2020;52:295–312. doi: 10.1016/j.immuni.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nat Immunol. 2013;14:1007–1013. doi: 10.1038/ni.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wu D, et al. Characterization of regulatory T cells in obese omental adipose tissue in humans. Eur J Immunol. 2019;49:336–347. doi: 10.1002/eji.201847570. [DOI] [PubMed] [Google Scholar]
- 169.Li C, et al. TCR transgenic mice reveal stepwise, multi-site acquisition of the distinctive fat-treg phenotype. Cell. 2018;174:285–299. doi: 10.1016/j.cell.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vasanthakumar A, et al. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat Immunol. 2015;16:276–285. doi: 10.1038/ni.3085. [DOI] [PubMed] [Google Scholar]
- 171.Kolodin D, et al. Antigen- and cytokine-driven accumulation of regulatory T cells in visceral adipose tissue of lean mice. Cell Metab. 2015;21:543–557. doi: 10.1016/j.cmet.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Murai M, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 174.Cretney E, et al. Characterization of Blimp-1 function in effector regulatory T cells. J Autoimmun. 2018;91:73–82. doi: 10.1016/j.jaut.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 175.Ogawa C, et al. Blimp-1 functions as a molecular switch to prevent inflammatory activity in Foxp3(+)RORgammat(+) regulatory T cells. Cell Rep. 2018;25:19–28. doi: 10.1016/j.celrep.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Garg G, et al. Blimp1 prevents methylation of Foxp3 and loss of regulatory T cell identity at sites of inflammation. Cell Rep. 2019;26:1854–1868. doi: 10.1016/j.celrep.2019.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xu M, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 2018;554:373–377. doi: 10.1038/nature25500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wheaton JD, Yeh CH, Ciofani M. Cutting Edge: c-Maf is required for regulatory T cells to adopt RORgammat(+) and follicular phenotypes. J Immunol. 2017;199:3931–3936. doi: 10.4049/jimmunol.1701134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Neumann C, et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat Immunol. 2019;20:471–481. doi: 10.1038/s41590-019-0316-2. [DOI] [PubMed] [Google Scholar]
- 180.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 181.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 182.Ye J, et al. The aryl hydrocarbon receptor preferentially marks and promotes gut regulatory T cells. Cell Rep. 2017;21:2277–2290. doi: 10.1016/j.celrep.2017.10.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tanaka A, Sakaguchi S. Targeting Treg cells in cancer immunotherapy. Eur J Immunol. 2019;49:1140–1146. doi: 10.1002/eji.201847659. [DOI] [PubMed] [Google Scholar]
- 184.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 185.Joshi NS, et al. Regulatory T cells in tumor-associated tertiary lymphoid structures suppress anti-tumor T cell responses. Immunity. 2015;43:579–590. doi: 10.1016/j.immuni.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Malchow S, et al. Aire-dependent thymic development of tumor-associated regulatory T cells. Science. 2013;339:1219–1224. doi: 10.1126/science.1233913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Maj T, et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat Immunol. 2017;18:1332–1341. doi: 10.1038/ni.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Magnuson AM, et al. Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types. Proc Natl Acad Sci USA. 2018;115:E10672–E10681. doi: 10.1073/pnas.1810580115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Plitas G, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Saito T, et al. Two FOXP3CD4 T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:674–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 191.Shimizu J, Yamazaki S, Sakaguchi S. Induction of tumor immunity by removing CD25 + CD4 + T cells: a common basis between tumor immunity and autoimmunity. J Immunol. 1999;163:5211–5218. [PubMed] [Google Scholar]
- 192.Sugiyama D, et al. Anti-CCR4 mAb selectively depletes effector-type FoxP3 + CD4 + regulatory T cells, evoking antitumor immune responses in humans. Proc Natl Acad Sci USA. 2013;110:17945–17950. doi: 10.1073/pnas.1316796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ha D, et al. Differential control of human Treg and effector T cells in tumor immunity by Fc-engineered anti-CTLA-4 antibody. Proc Natl Acad Sci USA. 2019;116:609–618. doi: 10.1073/pnas.1812186116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wing K, et al. CTLA-4 control over Foxp3 + regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 195.Romano E, et al. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proc Natl Acad Sci USA. 2015;112:6140–6145. doi: 10.1073/pnas.1417320112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kamada T, et al. PD-1(+) regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc Natl Acad Sci USA. 2019;116:9999–10008. doi: 10.1073/pnas.1822001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Arce Vargas F et al. (2017) Fc-Optimized Anti-CD25 depletes tumor-infiltrating regulatory t cells and synergizes with PD-1 blockade to eradicate established tumors. Immunity 46:577–586 [DOI] [PMC free article] [PubMed]
- 198.Ameri AH, et al. IL-33/regulatory T cell axis triggers the development of a tumor-promoting immune environment in chronic inflammation. Proc Natl Acad Sci USA. 2019;116:2646–2651. doi: 10.1073/pnas.1815016116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pastille E, et al. The IL-33/ST2 pathway shapes the regulatory T cell phenotype to promote intestinal cancer. Mucosal Immunol. 2019;12:990–1003. doi: 10.1038/s41385-019-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hatzioannou A, et al. An intrinsic role of IL-33 in Treg cell-mediated tumor immunoevasion. Nat Immunol. 2020;21:75–85. doi: 10.1038/s41590-019-0555-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Li A, et al. IL-33 signaling alters regulatory T cell diversity in support of tumor development. Cell Rep. 2019;29:2998–3008. doi: 10.1016/j.celrep.2019.10.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wang D, et al. Targeting EZH2 reprograms intratumoral regulatory T cells to enhance cancer immunity. Cell Rep. 2018;23:3262–3274. doi: 10.1016/j.celrep.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Miragaia RJ, et al. Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity. 2019;50:493–504. doi: 10.1016/j.immuni.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tomura M, et al. Activated regulatory T cells are the major T cell type emigrating from the skin during a cutaneous immune response in mice. J Clin Invest. 2010;120:883–893. doi: 10.1172/JCI40926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hayatsu N, et al. Analyses of a mutant Foxp3 allele reveal BATF as a critical transcription factor in the differentiation and accumulation of tissue regulatory T cells. Immunity. 2017;47:268–283. doi: 10.1016/j.immuni.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 206.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sakaguchi S, Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T. Regulatory T cells: how do they suppress immune responses? Int Immunol. 2009;21:1105–1111. doi: 10.1093/intimm/dxp095. [DOI] [PubMed] [Google Scholar]
- 208.Yamaguchi T, Wing JB, Sakaguchi S. Two modes of immune suppression by Foxp3(+) regulatory T cells under inflammatory or non-inflammatory conditions. Semin Immunol. 2011;23:424–430. doi: 10.1016/j.smim.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 209.Wing JB, Sakaguchi S. Multiple treg suppressive modules and their adaptability. Front Immunol. 2012;3:178. doi: 10.3389/fimmu.2012.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Arpaia N, et al. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kuswanto W, et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity. 2016;44:355–367. doi: 10.1016/j.immuni.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Tang Q, Bluestone JA. Regulatory T-cell therapy in transplantation: moving to the clinic. Cold Spring Harb Perspect Med. 2013;3(11):a015552. doi: 10.1101/cshperspect.a015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Perdigoto AL, Chatenoud L, Bluestone JA, Herold KC. Inducing and administering Tregs to treat human disease. Front Immunol. 2015;6:654. doi: 10.3389/fimmu.2015.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rosenzwajg M, et al. Low-dose interleukin-2 fosters a dose-dependent regulatory T cell tuned milieu in T1D patients. J Autoimmun. 2015;58:48–58. doi: 10.1016/j.jaut.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hartemann A, et al. Low-dose interleukin 2 in patients with type 1 diabetes: a phase 1/2 randomised, double-blind, placebo-controlled trial. Lancet Diabetes Endocrinol. 2013;1:295–305. doi: 10.1016/S2213-8587(13)70113-X. [DOI] [PubMed] [Google Scholar]
- 216.Hoffmann P, et al. Only the CD45RA + subpopulation of CD4 + CD25high T cells gives rise to homogeneous regulatory T-cell lines upon in vitro expansion. Blood. 2006;108:4260–4267. doi: 10.1182/blood-2006-06-027409. [DOI] [PubMed] [Google Scholar]
- 217.Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4 + T cells. Eur J Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- 218.Miyara M, et al. Functional delineation and differentiation dynamics of human CD4 + T cells expressing the FoxP3 transcription factor. Immunity. 2009;30:899–911. doi: 10.1016/j.immuni.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 219.Liu W, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4 + T reg cells. J Exp Med. 2006;203:1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Seddiki N, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med. 2006;203:1693–1700. doi: 10.1084/jem.20060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bin Dhuban K, et al. Coexpression of TIGIT and FCRL3 identifies Helios + human memory regulatory T cells. J Immunol. 2015;194:3687–3696. doi: 10.4049/jimmunol.1401803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Fuhrman CA, et al. Divergent phenotypes of human regulatory T cells expressing the receptors TIGIT and CD226. J Immunol. 2015;195:145–155. doi: 10.4049/jimmunol.1402381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Trotta E, et al. A human anti-IL-2 antibody that potentiates regulatory T cells by a structure-based mechanism. Nat Med. 2018;24:1005–1014. doi: 10.1038/s41591-018-0070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sockolosky JT, et al. Selective targeting of engineered T cells using orthogonal IL-2 cytokine-receptor complexes. Science. 2018;359:1037–1042. doi: 10.1126/science.aar3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ferreira LMR, Muller YD, Bluestone JA, Tang Q. Next-generation regulatory T cell therapy. Nat Rev Drug Discov. 2019;18:749–769. doi: 10.1038/s41573-019-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Akimova T, et al. Human lung tumor FOXP3+ Tregs upregulate four “Treg-locking” transcription factors. JCI Insight. 2017;22(16):e94075. doi: 10.1172/jci.insight.94075. [DOI] [PMC free article] [PubMed] [Google Scholar]