Summary
Serine, glycine, and other non-essential amino acids are critical for tumor progression, and strategies to limit their availability are emerging as potential cancer therapies1–3. However, the molecular mechanisms driving this response remain unclear, and the impact on lipid metabolism is relatively unexplored. Serine palmitoyltransferase (SPT) catalyzes the de novo biosynthesis of sphingolipids but also produces non-canonical 1-deoxysphingolipids (doxSLs) when using alanine as a substrate4,5. DoxSLs accumulate in the context of SPTLC1 or SPTLC2 mutations6,7 or low serine availability8,9 to drive neuropathy, and deoxysphinganine (doxSA) has been investigated as an anti-cancer agent10. Here we exploit amino acid metabolism and SPT promiscuity to modulate the endogenous synthesis of toxic doxSLs and slow tumor progression. Anchorage-independent growth reprograms a metabolic network involving serine, alanine, and pyruvate resulting in increased susceptibility to endogenous doxSL synthesis. Targeting the mitochondrial pyruvate carrier (MPC) promotes alanine oxidation to mitigate doxSL synthesis and improves spheroid growth, while direct inhibition of doxSL synthesis drives similar phenotypes. Restriction of dietary serine/glycine potently induces accumulation of doxSLs in xenografts while decreasing tumor growth. Pharmacological modulation of SPT rescues xenograft growth on serine/glycine-restricted diets, while reduction of circulating serine by inhibition of phosphoglycerate dehydrogenase (PHGDH) leads to doxSL accumulation and mitigates tumor growth. SPT promiscuity therefore links serine and mitochondrial alanine metabolism to membrane lipid diversity, which sensitizes tumors to metabolic stress.
Keywords: Serine, alanine, serine palmitoyltransferase, sphingolipids, 1-deoxysphinganine, mitochondrial pyruvate carrier, anchorage-independent growth, PHGDH, myriocin, ceramide
Alanine influences non-adherent cell growth
Anchorage-independent cell growth is a hallmark of aggressive tumors that reprograms metabolism to influence growth and survival11. We quantified metabolites in cancer cell lines grown as adherent or spheroid cultures and observed a pronounced increase in alanine secretion and intracellular abundance (Fig. 1a, Extended Data Fig. 1a–c). Consistent with prior studies12,13, spheroid culture decreased pyruvate dehydrogenase (PDH) flux and promoted reductive carboxylation by inducing PDH phosphorylation (Extended Data Fig. 1d–f), suggesting that alterations in mitochondrial metabolism influence alanine levels.
Figure 1. Alanine influences anchorage-independent cell growth.
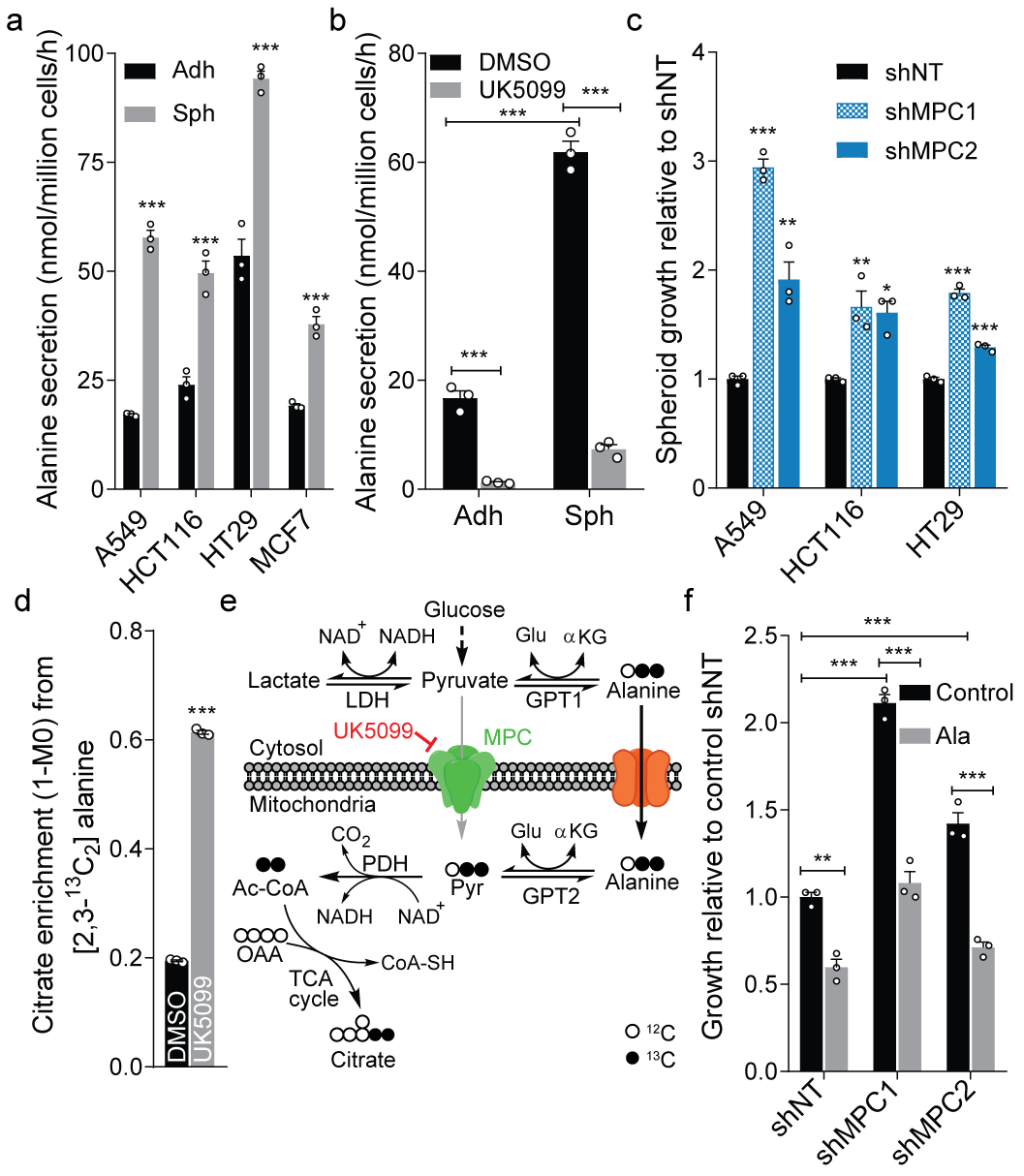
a. Alanine secretion by adherent (Adh) and spheroid (Sph) cultures (n=3 culture wells each).
b. Alanine secretion by HCT116 cells administered 5 μM UK5099 or DMSO control (n=3 culture wells each).
c. Spheroid growth of cells upon knockdown of MPC1 or MPC2 compared to shNT (n=3 culture wells each).
d. Citrate enrichment in HCT116 cells cultured with [2,3-13C2]alanine for 2 hours (n=3 culture wells each).
e. Schematic depicting alanine metabolism in response to MPC inhibition and tracing with [2,3-13C2]alanine. Cytosolic alanine bypasses MPC via GPT2.
f. Spheroid growth of HCT116 cells upon knockdown of MPC1, MPC2 (n=3 culture wells each).
Two-sided Student’s t-test (a, d), one-way ANOVA (c) or two-way ANOVA (b, f) with no adjustment for multiple comparisons. Similar results obtained in 2 (a, d) or 3 (b, c, f) independent experiments. Data represented as mean ± s.e.m. *P<0.05, **P<0.01, ***P<0.0001.
We previously observed that inhibition or knockdown of MPC reduces alanine levels in adherent cells14. The MPC inhibitor UK5099 profoundly decreased alanine secretion and abundance in anchorage-independent cultures (Fig 1b, Extended Data Fig. 1g–h). MPC expression is decreased in colon cancer, while its overexpression impairs spheroid formation and tumor growth15. Both MPC knockdown and UK5099 treatment significantly enhanced spheroid growth (Fig. 1c, Extended Data Fig. 1i), while proliferation under adherent conditions was unaffected (Extended Data Fig. 1j–m). Metabolic tracing studies using [U-13C6]glucose indicated that UK5099-treated HCT116 spheroids increased serine synthesis and decreased glucose oxidation, dramatically increasing the ratio of serine/alanine (Extended Data Fig. 1n–p). We therefore hypothesized that MPC inhibition promotes alanine oxidation and quantified citrate enrichment from [2,3-13C2]alanine in HCT116 cells, observing a significant increase in alanine flux into TCA metabolism with UK5099 treatment (Fig. 1d). Tumor cells effectively bypass MPC by synthesizing, transporting, and catabolizing alanine within mitochondria (Fig. 1e), which prevents its accumulation and enhances growth under anchorage-independent conditions.
These results suggest that alanine may be deleterious for spheroid growth. While alanine is abundant in plasma, it is absent from most tissue culture media. Intriguingly, alanine supplementation reduced spheroid biomass and mitigated the beneficial effects of MPC inhibition or knockdown on spheroid growth, while adherent growth was unaffected by alanine (Fig. 1f, Extended Data Fig. 1q–s). These observations highlight a potential mechanism through which MPC inhibition enhances anchorage-independent growth–decreased alanine levels. To better understand how alanine compares to other non-essential amino acids, we manipulated the availability of serine and glycine in spheroid cultures and quantified biomass, as these amino acids are highly exchanged in the cytosol and mitochondria16. Notably, removal of serine reduced spheroid growth to a similar or greater extent as alanine supplementation, while glycine in the absence of serine or formate also reduced growth (Extended Data Fig. 1t–v), suggesting these phenomena are metabolically linked.
Amino acid metabolism and doxSL synthesis
Serine fuels numerous biosynthetic pathways in tumors. To gauge the impact of serine restriction on different pathways, we compared KM values for different serine-utilizing enzymes (Fig. 2a)17. Several enzymes have KM values at or higher than our measured, intra-tumoral serine concentrations, indicating their affinity for serine could be limiting upon serine restriction. SPT is the only ubiquitously expressed enzyme in this group. Furthermore, SPT is promiscuous and can use alanine rather than serine as a substrate to produce toxic doxSLs4,5 (Fig. 2b). We therefore hypothesized that SPT function and lipid metabolism are altered under anchorage-independent or serine/glycine-free growth conditions and quantified sphingoid base diversity in these cultures.
Figure 2. DoxSL synthesis mitigates anchorage-independent cell growth.
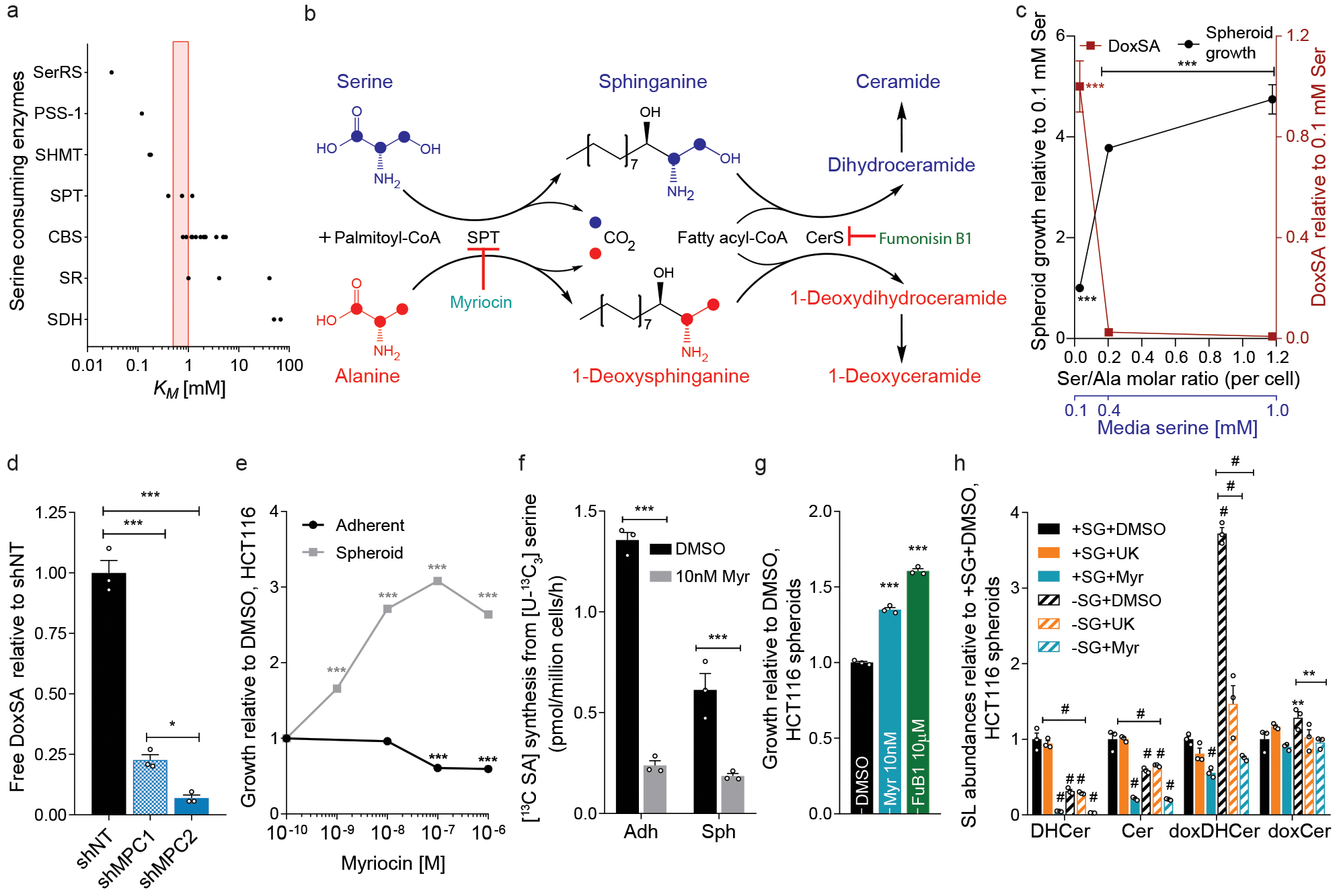
a. KM values for serine-utilizing enzymes. Red bar indicates tumor serine concentrations in mice fed control or –SG diets.
b. Schematic depicting synthesis of canonical sphingolipids from serine (blue) and doxSLs from alanine (red) via promiscuous SPT activity and ceramide synthase (CerS).
c. Free doxSA (red) and spheroid growth (black) of HCT116 after 5 days of growth in media with altered serine (n=3 culture wells each).
d. DoxSA levels in spheroid cultures of HCT116 cells upon MPC1 or MPC2 knockdown (n=3 culture wells each).
e. Growth of HCT116 adherent cultures (n=12 culture wells) and spheroids (n=3 culture wells) in the presence of myriocin.
f. [13C]sphinganine synthesis from [U-13C3]serine tracer in HCT116 cells treated with 10 nM myriocin (n=3 culture wells each).
g. HCT116 spheroid growth in the presence of myriocin (Myr) and fumonisin B1 (FuB1) (n=3 culture wells each).
h. Relative abundances of summed ceramide species in HCT116 spheroids cultured in –SG, UK5099, or 10 nM myriocin conditions (n=3 culture wells each).
One-way ANOVA (d, g) or two-way ANOVA (c, e, f, h) was performed for each comparison with no adjustment for multiple comparisons. Similar results obtained in 2 (c-h) independent experiments. Data represented as mean ± s.e.m. *P<0.05, **P<0.01, *** or # P<0.0001.
Restriction of serine from culture medium promoted doxSA accumulation and reduced HCT116 spheroid growth (Fig. 2c). Free doxSA levels and the doxSA/SA ratio were increased in spheroid cultures and further elevated by removal of serine/glycine and supplementation of alanine (Extended Data Fig. 2a–c). Consistent with our results linking MPC flux to alanine and spheroid growth, MPC knockdown significantly decreased doxSA levels in spheroids (Fig. 2d). These results suggest that aberrant doxSL production due to serine/glycine deprivation and/or alanine supplementation constrains anchorage-independent growth. Administration of exogenous sphingoid bases further highlighted the toxicity of doxSLs over canonical sphingolipids in spheroid cultures. Exogenous doxSA blocked the ability of UK5099 to enhance spheroid growth (Extended Data Fig. 2d). While the canonical sphingoid bases sphinganine (SA) and sphingosine (SO) had minimal impact, both doxSA and deoxysphingosine (doxSO) reduced spheroid growth while causing accumulation of 1-deoxydihydroceramides (doxDHCer) and/or 1-deoxyceramides (doxCer) species (Extended Data Fig. 2e–f).
SPT flux, doxSLs, and spheroid growth
To orthogonally test whether amino acid metabolism drives changes in spheroid growth by modulating doxSLs, we directly targeted SPT using myriocin and quantified its impact on HCT116 cell growth (Fig. 2b). Intriguingly, myriocin increased the growth of spheroids in a dose-dependent manner, while adherent growth was minimally impacted at low doses (Fig. 2e). We confirmed that 10 nM myriocin inhibited SPT flux by quantifying incorporation of [U-13C3]serine into SA (Fig. 2f). Myriocin reduced SA, doxSA, the doxSA/SA ratio as well as alanine-induced doxSA accumulation in spheroids (Extended Data Fig. 2g–i). SPT inhibition rescued anchorage-independent growth in the presence of alanine when tested in various colon cancer cell lines (Extended Data Fig. 2j), further linking modulation of alanine levels to potential doxSA toxicity. DoxSA becomes toxic upon N-acylation and formation of doxDHCer (Fig. 2b); inhibition of ceramide synthases using fumonisin B1 (FuB1) enhanced spheroid growth like myriocin (Fig. 2g, Extended Data Fig. 2k), highlighting the toxicity of doxDHCer. In contrast to canonical sphingolipids that are catabolized via sphingosine-1-phosphate (S1P), doxSLs are metabolized via an alternate pathway induced by fenofibrate (FeF)18. FeF treatment decreased doxSA levels upon modulation of serine/glycine or alanine while improving spheroid growth in alanine-supplemented cultures (Extended Data Fig. 2l–m). These results provide evidence that altering the synthesis or degradation of doxSLs improves anchorage-independent growth.
We next examined how different ceramide species are altered upon modulation of serine, alanine, and SPT flux in HCT116 spheroids (Fig. 2h). Serine/glycine-free media potently increased doxDHCer levels, and both SPT and MPC inhibition blocked this accumulation, while doxCer species were not dramatically altered in these cells. As expected19, serine/glycine-restriction and myriocin both reduced canonical DHCer and Cer species. Importantly, while myriocin reduced levels of all ceramides, UK5099 treatment only reduced doxDHCer species. These results provide evidence that doxDHCer accumulation (rather than reduced ceramide synthesis) sensitizes cancer cells to stresses associated with anchorage-independent culture to reduce growth.
–SG diet drives doxSL synthesis in tumors
In contrast to in vitro systems, serine, alanine, and sphingolipid metabolism are highly buffered in the context of in vivo physiology. We next fed mice bearing HCT116 xenograft tumors either Control diet or an isonitrogenous diet lacking serine and glycine (–SG), as both must be removed since they buffer one another via one-carbon metabolism. Nitrogen balance was maintained by increasing other non-essential amino acids, including alanine (Supplementary Table 1). Mice administered –SG diet bore smaller tumors that had altered levels of serine, glycine, and alanine, with tumor size correlating negatively with the duration of –SG diet feeding (Fig. 3a–c, Extended Data Fig. 3a–b). Importantly, doxSLs were the most significantly altered lipids in tumors isolated from mice fed –SG diet, exhibiting 5–8-fold increased doxSA, doxDHCers, and doxCers (Fig. 3d–e; Extended Data Fig. 3c–d). Thus, dietary removal of serine and glycine results in toxic doxSL accumulation within tumors. Liver doxCer species showed a more modest increase compared to tumors in mice fed –SG diet, suggesting that tumor cells have a greater propensity to synthesize/accumulate doxSLs than other tissues (Extended Data Fig. 3e). In contrast to doxSLs, tumors maintained similar abundances of canonical sphingolipids such as DHCer, Cer, sphingomyelin, and S1P in mice fed Control versus –SG diets (Fig. 3e; Extended Data Fig. 3f–i), highlighting key differences in restricting serine/glycine in vivo versus cell culture (Fig. 2h). Finally, we detected a significant decrease in lactosylceramides (Fig. 3f), which are essential precursors for complex membrane lipids.
Figure 3. Serine restriction drives 1-deoxysphingolipid synthesis in vivo.
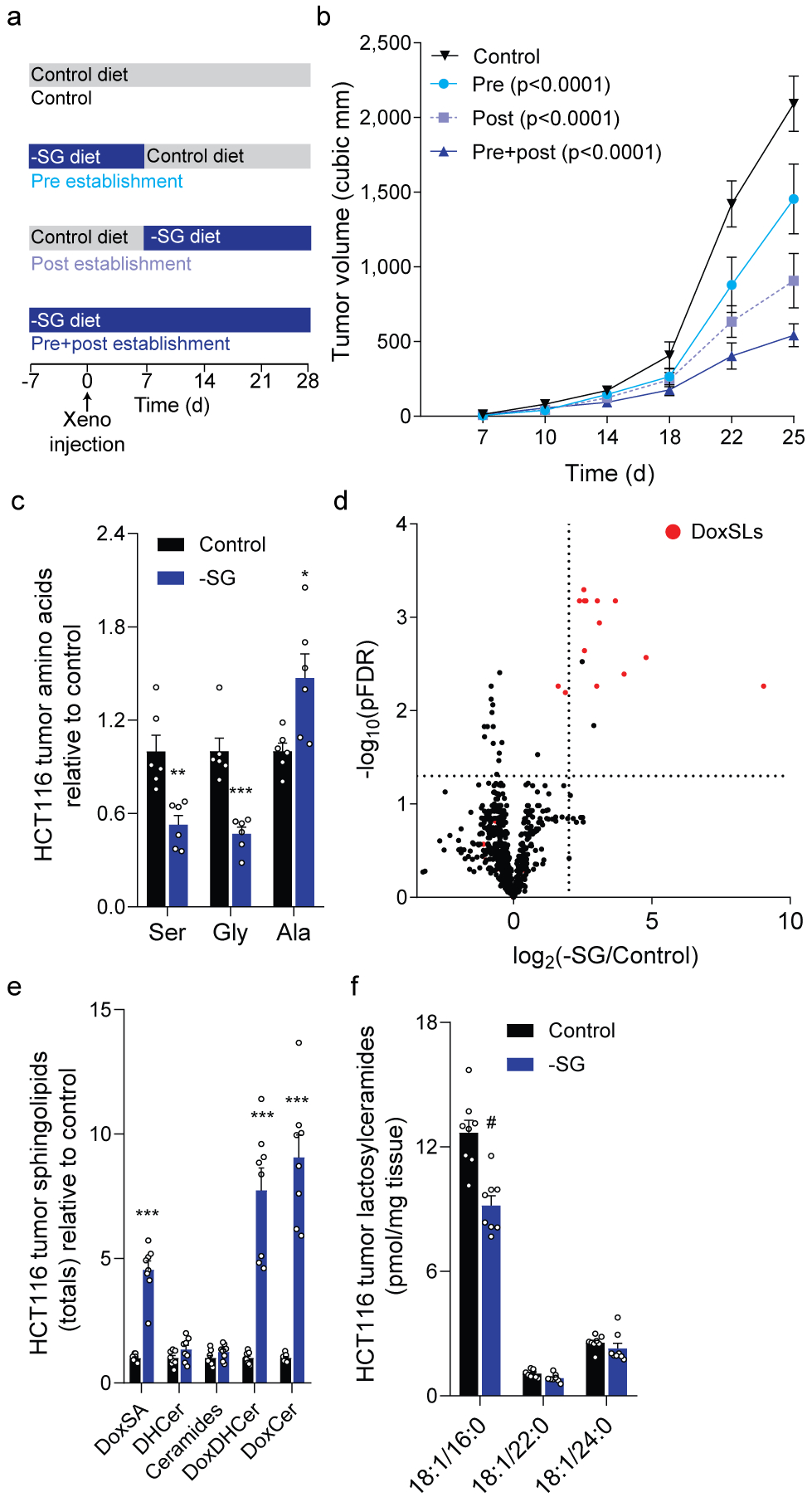
a. Schematic depicting feeding strategies in mice bearing HCT116 xenografts.
b. HCT116 xenograft size over time (n=16 tumors for each).
c. Serine, glycine, and alanine abundance in HCT116 xenografts (n=6 tumors each diet).
d. Volcano plot depicting lipid alterations in HCT116 xenografts (n=4 tumors each diet).
e. f. Abundances of (e) summed ceramides and (f) lactosylceramides in HCT116 xenografts (n=8 tumors each diet).
Two-way ANOVA (b) or two-sided Student’s t-test (c, e, f) was performed with no adjustment for multiple comparisons. Data represented as mean ± s.e.m. *P<0.05, **P<0.01, *** or # P<0.0001.
Serine/glycine-restricted diets also reduced growth of HT29 xenograft tumors while driving doxSL accumulation (Extended Data Fig. 3j–m), and these tumors also had reduced levels of lactosylceramide (Extended Data Fig. 3n). We also attempted to enhance the potency of isonitrogenous –SG diets by further increasing alanine (–SG+A), but this diet had no additional impacts on tumor growth, alanine abundance, or sphingolipid levels (Extended Data Fig. 3j–m). Expression of glutamic-pyruvic transaminases, Gpt1 and Gpt2, were significantly increased in liver (Extended Fig. 3o), suggesting the liver compensates to dispose of excess alanine and serine/glycine-restriction is the primary driver of doxSL accumulation.
Consistent with prior reports3, nucleotide monophosphates were significantly decreased within tumors fed –SG diet (Extended Data Fig. 3p). Reduced (GSH) and oxidized (GSSG) glutathione abundances were not considerably impacted by –SG diet alone (Extended Data Fig. 3q). Collectively, these data indicate that dietary serine/glycine restriction drives tumor doxSL accumulation in addition to other stresses, and the combination is deleterious for tumor progression.
SPT and PHGDH inhibition modulate doxSLs
We next attempted to rescue HCT116 xenograft growth on –SG diets by inhibiting SPT and doxSL synthesis, administering vehicle or myriocin at 0.03 or 0.3 mg/kg intraperitoneally. Interestingly, tumor growth was significantly increased when mice were treated with 0.03 mg/kg but not 0.3 mg/kg myriocin (Fig. 4a). Both doses significantly decreased doxDHCer and ceramides within tumors (Fig. 4b, Extended Data Fig. 4a–d); however, only the high-dose myriocin reduced liver ceramides (Fig. 4c, Extended Data Fig. 4e–h). Indeed, tumor-bearing mice fed –SG diet and administered high-dose myriocin showed more weight loss compared to other groups (Extended Data Fig. 4i), indicating that endogenous lipid metabolism was more drastically altered at this dosage. These data suggest that low dose myriocin rescued tumor growth by 1) mitigating doxSL synthesis in the –SG diet and 2) better maintaining hepatic lipid physiology to support provision of canonical sphingolipids (Extended Data Fig. 4j).
Figure 4. Pharmacological inhibition of SPT or PHGDH modulates tumor growth in vivo.
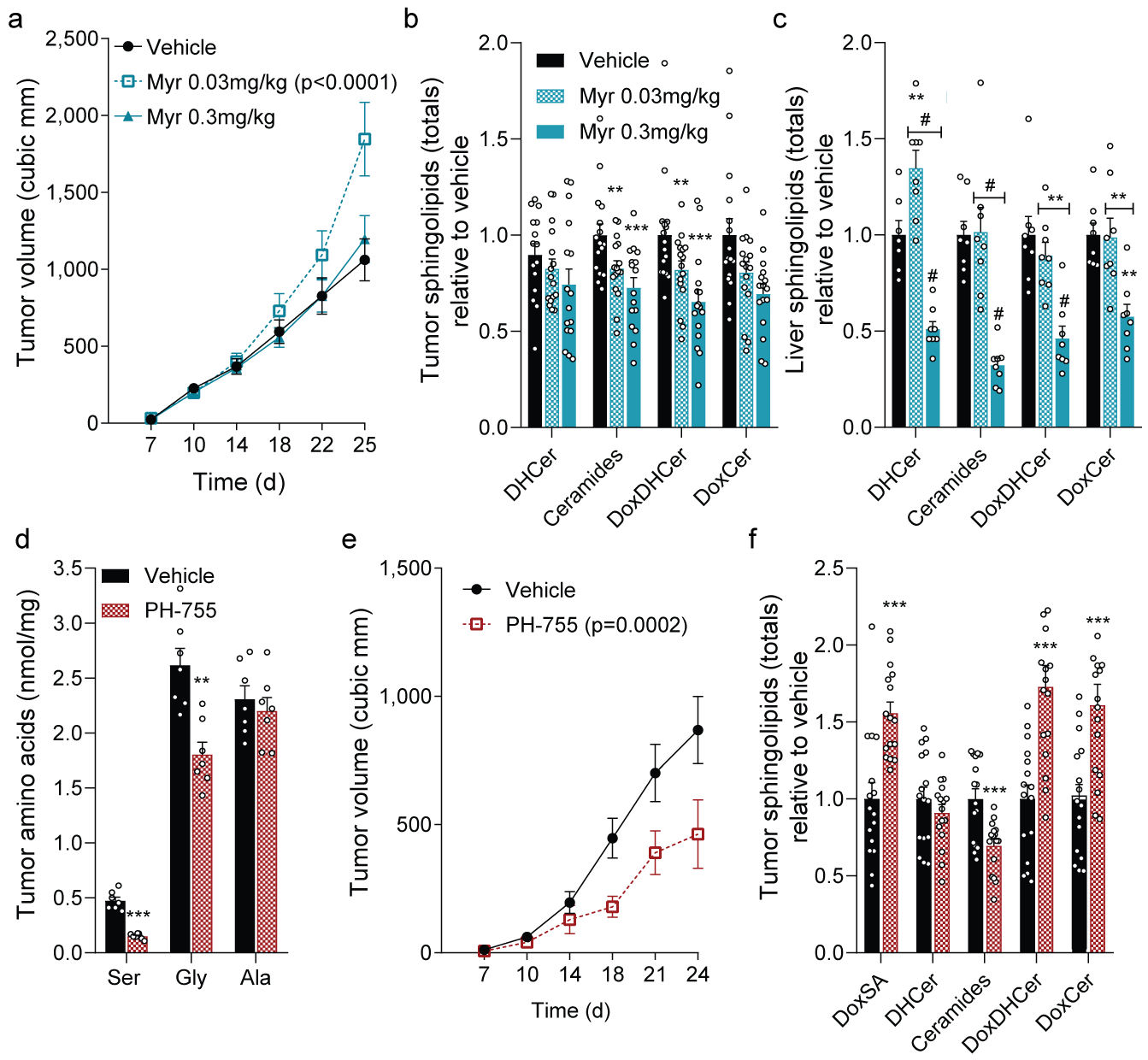
a. HCT116 xenograft size in mice fed -SG diet and administered vehicle, low-dose or high-dose myriocin (n=16 tumors each treatment).
Relative abundances of summed ceramides in (b) xenograft tumors (n=16 each treatment) and (c) liver (n=8 each treatment) from mice in (a).
d. Amino acid levels in HCT116 xenograft tumors from mice administered vehicle or PH-755 (n=7 tumors each treatment).
e. HCT116 xenograft size in mice administered vehicle (n=16 tumors) or PH-755 (n=14 tumors).
f. Relative abundances of summed ceramides from HCT116 xenografts from mice administered vehicle or PH-755 (n=16 tumors each treatment).
Two-way ANOVA (a-c, e) or two-sided Student’s t-test (d, f) were performed with no adjustment for multiple comparisons. Data represented as mean ± s.e.m. **P<0.01, *** or # P<0.0001.
Finally, we attempted to reduce serine levels, drive doxSL synthesis, and manipulate tumor growth in serine/glycine-replete diets via pharmacological inhibition of PHGDH. PH-755 decreases serine synthesis and reduces circulating serine/glycine when administered to mice20, and this compound potently decreased serine synthesis in HCT116 spheroid cells (Extended Data Fig. 4k). PHGDH inhibition significantly reduced growth at 0.4 mM (regular DMEM concentrations) while increasing doxSA abundance (Extended Data Fig. 4l–m). Notably, serine supplementation rescued spheroid growth and reduced doxSA levels in the presence and absence of PH-755 (Extended Data Fig. 4l–m). PHGDH inhibition also promoted doxSL accumulation in A549 and MCF7 cells, with spheroid cultures showing more responsiveness than adherent cells (Extended Data Fig. 4n). Finally, we administered PH-755 to mice fed a standard chow diet and initiated xenograft tumors one week later. Levels of serine/glycine in tumors and plasma were significantly reduced along with tumor size in treated animals (Fig 4d–e, Extended Data Fig. 4o). PHGDH inhibition caused tumor doxSLs to accumulate, while ceramides were decreased (Fig. 4f). Thus, manipulation of PHGDH or SPT flux in tumor-bearing mice alters the balance of canonical sphingolipids and doxSLs to influence tumor progression.
Discussion
Using combinatorial interventions that influence spheroid growth by targeting distinct nodes of serine, alanine, and sphingolipid metabolism, we identify SPT promiscuity and the aberrant production of doxSLs as a molecular driver of serine-restriction phenotypes in cancer. Our in vivo studies demonstrate that doxSL synthesis occurs in tumors in the context of whole-body physiology. Furthermore, our lipidomic results suggest that doxDHCer may compromise the metabolism of glycosphingolipids, which are important components of cell membranes critical for tumor growth21. SPT therefore integrates distinct metabolic signals from glycolysis (serine) and mitochondria (alanine) to influence sphingolipid diversity and tumor progression. This link may provide additional insights into why specific tumors are most sensitive to serine deprivation strategies22 or amplify PHGDH23,24.
DoxSLs accumulate in various disease states, including genetically-driven neuropathy, metabolic syndrome, and macular telangiectasia6,7,9,25. These lipids drive neurotoxicity, though the cell-type specific mechanisms are not yet clear26,27. Myriocin at low doses influenced SLs and doxSLs in a tissue-specific manner, suggesting that toxic doxSLs could be selectively reduced by such treatments, similar to serine supplementation28. On the other hand, inhibition of PHGDH lowered tumor serine levels and promoted doxSL accumulation even on a serine-replete diet. These in vivo results highlight the complex metabolic interactions between tumors and tissues which contribute to cancer progression.
Finally, our studies emphasize the functional impact of enzyme promiscuity in biological systems. KM values of SPT for serine and alanine are tuned to physiologically relevant concentrations of amino acids. SPT and fatty acid synthase (FASN) both exhibit catalytic activity with diverse acyl-CoA species29,30, and their promiscuity may allow discrete metabolic signals to regulate lipid diversity, localization, or function. Thus, doxSLs may serve as stress signals to influence membrane composition or cell fate. While additional studies are needed to understand why these molecules are toxic to cancer cells and neurons, our findings highlight SPT as a metabolically responsive “switch” in the lipidome that can be co-opted to decrease tumor growth.
Materials and Methods
Reagents
Media and sera were purchased from Life Technologies. Glucose and amino acid isotope tracers were purchased from Cambridge Isotopes Inc. Sphingolipid standards were purchased from Avanti polar lipids. All other reagents were purchased from Sigma-Aldrich unless otherwise noted.
Cell lines
All cell lines were purchased from ATCC and authenticated. For cell line authentication, short tandem repeat (STR) analysis was performed on isolated genomic DNA with the GenePrint® 10 System from Promega, and peaks were analyzed using GeneMarker HID from Softgenetics (v2.9.5). Allele calls were searched against STR databases as previously described31. MCF7 cells were used due to their demonstrated regulation of PDH in spheroid culture12 and routine use in sphingolipid biosynthesis studies. All cell lines were grown in regular growth medium (DMEM) supplemented with 10% FBS, 100 units/mL of penicillin and 100 μg/mL of streptomycin, in a humidified cell culture incubator at 37°C unless otherwise stated.
Spheroid formation and growth measurements
For spheroid growth analyses, a semi confluent adherent culture of cells was detached with 0.05% Trypsin at room temperature. After the cells have detached, the trypsin is neutralized with 10 times excess of growth media and centrifuged. The cells are then singularized by trituration and counted with a hemocytometer. 2×104 singularized cells were plated on each well of a 6-well ultra-low attachment dish (Corning, Cat.no. 3471) in appropriate media. The media was changed on day 3, day 5. For experiments comparing spheroid and adherent cultures, adherent cells were also given media changes on day 3 and day 5 for consistency. The media changes were accomplished by spinning the spheroids at 50 × g for 3 minutes. For amino acid-restricted growth, cells were cultured in media reconstituted from glucose, glutamine, and amino acid free media (Hyclone, custom formulation) supplemented with amino acids at appropriate concentrations from a 100x stock solution of amino acids without serine and glycine (detailed in Supplementary Table 2). Serine, glycine and alanine were supplemented from a 1000x stock solutions made separately according to the experiment and the media was supplemented with 10% dialyzed FBS (Thermo Fischer Scientific, Cat. No. 26400044). For sphingolipid analyses, 2 ×105 cells were plated on 10 cm ultra-low attachment plates (Corning, Cat.no. 3262) in appropriate culture media. For counting spheroid numbers, spheroids were transferred onto a 24- well tissue culture plate and the entire area of each well were imaged using EVOS FL auto microscope (Life Technologies, software v31201) with a 2x objective. The images were then used to count the number of spheroids with the software image J (v1.52a). For counting cell numbers from spheroids, the spheroids were trypsinized for 5 minutes at 37°C, with intermittent mixing, the cells were then counted using a hemocytometer. For protein estimation, spheroids were transferred to 1.5 mL Eppendorf tubes, spun at 50 × g for 3 minutes and the media was aspirated. The spheroids were then lysed using M-PER mammalian protein extraction reagent (Thermo Fischer Scientific, Cat. No. 78501). Proteins were quantified using BCA Assay kit according to manufacturer’s instructions (Lambda biotech, Cat.no. G1002).
Stable isotope tracing
The cells were cultured in media reconstituted from glucose, glutamine and amino acid-free media (Sigma Aldrich Cat. No. D5030) supplemented with 13C labeled tracer of interest, unlabeled components and 10% dialyzed FBS. Cells were given a media change 1 hour before the addition of tracer with growth media. Cells were rinsed with PBS before the addition of tracing media. The time of addition of tracer media was designated time 0. Metabolites were extracted at the indicated time periods as indicated in text. For spheroid cultures, the media changes were accomplished by spinning the spheroids in 1.5 mL Eppendorf tubes at 50 × g for 3 minutes.
Aqueous metabolite extraction and gas chromatograph – mass spectrometry (GC-MS) analysis
Cells were washed once with saline (0.9% w/v NaCl). 500 μL of methanol at – 80 °C was added to quench metabolic reactions. 200 μL of 5 μg/mL ice-cold norvaline in water was added to each well and the cells were collected by scraping. The lysate was then transferred to a fresh 1.5 mL Eppendorf tube containing 500 μL of chloroform. The tubes were vortexed for 10 minutes and centrifuged at 16,000 × g for 10 minutes to allow phase separation. The upper aqueous layer was collected and dried under vacuum. Metabolite derivatization and GC-MS analysis (MassHunter GC/MS Acquisition (vB.07.04.2260)) was performed as described in detail elsewhere1. Briefly, metabolite derivatization was performed with a GerstelMultiPurpose Sampler (MPS 2XL) using methoxylamine hydrochloride (MP Biomedicals, Cat. No. 0215540525) in pyridine and N-tert-butyldimethylsily-N-methyltrifluoroacetamide (MTBSTFA) with 1% tert-butyldimethylchlorosilane (tBDMS) (Regis Technologies). The derivatized metabolites were analyzed by an Agilent 7890B gas chromatograph (GC) equipped with a DB-35MS column [30 m (length) × 0.25mm (i.d.) Agilent J&W Scientific] and connected with an Agilent 5977C mass spectrometer (MS). For metabolite separation, the GC oven was held at 100°C for 2 min followed by increasing the temperature to 300°C at a ramp rate of 10°C/min, and held for 3 min. The eluates from the GC were subjected to electron impact ionization. MS scanning was performed over M/Z range of 100–650 m/z. The quadrupole was held at 150 °C.
Isotopologue distributions and total metabolite abundances were computed by the integration of mass fragments using a MATLAB (R2014b) based algorithm INCA with corrections for natural isotope abundances32. For absolute quantification of amino acids, a 13C,15N-labeled amino-acid mixture (Cambridge Isotope Laboratories, Cat. No MSK-A2–1.2) was used as an internal standard. Details on specific fragments are provided elsewhere33.
Sphingolipid extraction for liquid chromatograph- mass/mass spectrometry (LC-MS/MS) analysis
Cell lines
Adherent cells were washed and scraped into 500μl of methanol, 400μl saline and 100μl of water spiked with deuterated internal standards ((100 picomoles of D7- Sphingosine (Avanti polar lipids, Cat. No. 860657), 20 picomoles of D7-Sphinganine (Avanti polar lipids, Cat. No. 860658), 2 picomoles of D3-Deoxysphinganine (Avanti polar lipids, Cat. No. 860474), 200 picomoles of C15 Ceramide-d7 (d18:1-d7/15:0) (Avanti polar lipids, Cat. No. 860681), 100 picomoles of C13-dihydroceramide-d7 (d18:0-d7/13:0), 10 picomoles of Glucosylsphingosine (d18:1-d7), (Avanti polar Lipids Cat. No. 860695), 100 picomoles of Glucosylceramide (d18:1-d7/15:0), (Avanti polar Lipids Cat. No. 330729), 100 picomoles of Lactosylceramide (d18:1-d7/15:0), (Avanti polar Lipids Cat. No. 330727), 200 picomoles of Sphingomyelin (d18:1/18:1-d9), (Avanti polar Lipids Cat. No. 791649)). Spheroids were washed with saline by pelleting at 50 × g for 3 minutes, removing supernatant, and the pellet was extracted with 500 μl methanol, 400 μl saline, and 100 μl of water spiked with deuterated internal standards as described earlier. The cells were homogenized with a ball mill (Retsch Mixer Mill MM 400) at 30 Hz for 3 minutes. One hundred μL of the homogenate was dried under airflow and redissolved in 20 μL of M-PER buffer (Thermo Fisher Scientific Cat. No. 78501) for protein estimation with BCA assay. One mL of chloroform was added to the remaining methanol extract and the tubes were vortexed and spun at 16000 × g for 10 minutes to allow phase separation. The organic phase was separated and saved. Sphingolipids were re-extracted from the polar phase by adding 1 mL of chloroform, followed by 3 μL of formic acid, and the tubes were vortexed and centrifuged, and the organic phase was collected and combined with the organic phase saved earlier and dried under air-flow. The dried organic extracts were resuspended in 100 μL of 0.2% formic acid and 1 mM ammonium formate in methanol. Next, the tubes were sonicated in a bath sonicator for 10 minutes and spun at 16,000 × g for 10 minutes at 4 °C. The supernatant was then analyzed with LC-MS/MS. The ion transitions, collision energies and fragmentor voltages for LC-MS/MS are provided in Supplementary Table 3.
Tissues
Fifteen to twenty-five mg of frozen tissue were homogenized with a ball mill (Retsch Mixer Mill MM 400) at 30 Hz for 3 minutes in 500 μL −20°C methanol, 400 μL of ice-cold saline and 100 μL of ice-cold MiliQ water, spiked with deuterated internal standards as described earlier. The mixture was then transferred into a 2 mL Eppendorf tube containing 1 mL of chloroform. The samples were vortexed for 5 min followed by centrifugation at 4°C for 5 min with 15 000 × g. The organic phase was collected and 2 μL of formic acid was added to the remaining polar phase which was re-extracted with 1 mL of chloroform. Combined organic phases were dried and the pellet was resuspended in 100 μL of buffer containing 100% methanol, 1 mM ammonium formate and 0.2% formic acid.
Ceramide species were quantified via liquid chromatography mass-spectrometry (Agilent 6460 QQQ, MassHunter LC/MS Acquisition (vB.08.02)). Corresponding transitions for different species from each classes of sphingolipids such as Ceramides, DHceramides, doxCeramides, doxDHceramides, sphingomyelins, lactosylceramide were identified using MassHunter optimizer software (MassHunter Workstation Software for Triple Quadrupole (Optimizer vB.08.02, Quantitative Analysis vB08.00, Agilent Technologies)) utilizing commercially available standards and their described exact masses. Unlabeled standards were spiked into tissue extracts to confirm the identity of representative lipid species. The commercially available deoxyceramide standards (Avanti) have been shown to differ in their double bond positions18,34 from native deoxy ceramides produced in cells and tissues; hence we identified and validated the native deoxyceramide species empirically by using nutritional and pharmacological interventions in xenograft models of colon cancer. Ceramides were separated on C8 column (Spectra 3 μm C8SR 150 × 3mm ID, Peeke Scientific, CA) as previously described35. Mobile phase A was composed of 100% HPLC grade water containing 2 mM ammonium formate and 0.2% formic acid and mobile phase B consisted of 100% methanol containing 0.2% formic acid and 1 mM ammonium formate. The gradient elution program consisted of the following profile: 0 min, 82% B; 3 min, 82% B; 4 min, 90% B, 18 min, 99% B; 25 min, 99%, 27 min, 82% B, 30 min, 82 %B. Column re-equilibration followed each sample and lasted 10 min. The capillary voltage was set to 3.5 kV, the drying gas temperature was 350 °C, the drying gas flow rate was 10 L/min, and the nebulizer pressure was 60 psi. Ceramide species were analyzed by SRM of the transition from precursor to product ions at associated optimized collision energies and fragmentor voltages (Supplemental Table 3). Ceramide and dihydroceramide species were then quantified from spiked internal standards.
Nucleotide quantification
Nucleotides from tumor samples were extracted with 500 μL of 80 % methanol (cooled to −80° C) containing internal standard (ATP-γ-S). Samples were sonicated, vortexed and incubated for 4h at −80° C. Samples were centrifuged (14,000 × g, 4° C, 10 min), supernatants were transferred to new tubes and dried under vacuum. Samples were reconstituted in 45 μL of solution A (see below) prior to injection. Nucleotides were analyzed according to Wu et al.36. A Dionex Ultimate 3000 LC system (Thermo) coupled to a TSQ Quantiva mass spectrometer (Thermo) fitted with an Atlantis T3 reversed phase column (3 μm, 100 × 2.1 mm i.d., Waters) was used. The following LC solvents were used: solution A, 100 mM HFIP and 8.6 mM TEA in water solution B, 10 % acetonitrile in mobile phase A. At a flow rate of 0.5 mL/min, the following gradient was utilized: 0 to 10 % B from 0 to 6 min, 10 to 0 % B from 6 to 6.1 min, maintain 0 % B from 6.1 to 10 min. The injection volume was 10 μL, the column oven temperature was set to 45° C and the autosampler kept at 4° C. MS analyses were performed using electrospray ionization in positive and negative ion modes, with spay voltages of 3.5 and −2.5 kV, respectively, ion transfer tube temperature of 325° C, and vaporizer temperature of 310° C. Multiple reaction monitoring (MRM) was performed by using mass transitions between specific parent ions into corresponding fragment ions for each analyte.
Glutathione quantification
Polar metabolites were extracted from samples according to Yuan et al., 20128. Briefly, polar metabolites from tumor samples were extracted with 500 μL of 80 % methanol (cooled to −80° C) containing heavy internal standards (Metabolomics Amino Acids Standard, MSK-A2, Cambridge Isotope Laboratories). Samples were sonicated, vortexed and incubated for 4h at −80° C. Samples were centrifuged (14,000 × g, 4° C, 10 min), supernatants were transferred to new tubes and dried under vacuum in a speedvac. Samples were reconstituted in 45 μL water prior to injection. Polar metabolites were analyzed on a Dionex Ultimate 3000 LC system (Thermo) coupled to a TSQ Quantiva mass spectrometer (Thermo) fitted with a XBridge Amide HILIC column (3.5 μm, 100 × 4.6 mm i.d., Waters). The following LC solvents were used: solution A, 95:5 water:acetonitrile, 20 mM ammonium hydroxide, 20 mM ammonium acetate, solution B, 100 % acetonitrile. The following gradient was utilized: at a flow rate of 0.4 mL/min with a gradient consisting of 85–60 % B in 2 min, 60–50 % B in 8 min, 50–2 % B in 2 min, 2 % B for 4 min, up to 85 % B in 1 min and reequilibrate at 85 % B for 7 min, for a total run time of 24 min. The injection volume was 10 μL, the column oven temperature was set to 25° C and the autosampler kept at 4° C. MS analyses were performed using electrospray ionization in positive and negative ion modes, with spay voltages of 3.5 and −3.5 kV, respectively, ion transfer tube temperature of 325° C, and vaporizer temperature of 275° C. Multiple reaction monitoring (MRM) was performed by using mass transitions between specific parent ions into corresponding fragment ions for each analyte. Reduced glutathione (GSH) parent mass was 308.1, transitions were 179.0, 162.0, 76.1 and 233.1. Oxidized glutathione (GSSH) parent mass was 613.2, transitions were 355.1, 231.0, 484.2 and 409.1. As internal standard, heavy proline parent mass was 122.1, transitions were 75.1 and 46.2.
RNA isolation and quantitative RT-PCR
Total RNA was purified from frozen tissues using Trizol Reagent (Life Technologies) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg of total RNA using iScript Reverse Transcription Supermix for RT-PCR (Bio-Rad Laboratories) according to the manufacturer’s instructions. Individual 10 μl SYBR Green real-time PCR reactions consisted of 2 μl of diluted cDNA, 5 μl of SYBR Green Supermix (Bio-Rad), and 1 μl of each 5 μM forward and reverse primers. For standardization of quantification, 18 S was amplified simultaneously. The PCR was carried out on 96-well plates on a CFX Connect Real time System (Bio-Rad CFX Manager v3.1) using a three-stage program provided by the manufacturer: 95 °C for 3 min, 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Gene-specific primers used are listed in Supplementary Table 4. PCR data for intergene comparison were corrected for primer efficiency.
Small molecule stock solutions
UK5099: Sigma, Cat. No. PZ0160. The stock solution was made at a concentration of 50 mM in DMSO and the aliquots were stored in −20°C until use. The cells were treated at a concentration of 5 μM.
Myriocin: Sigma, Cat.No. M1177. The stock solution was made at a concentration of 1 mM in DMSO and the aliquots were stored in −20°C until use. The cells were treated at a concentration of 10 nM.
Fumonisin B1: Sigma, Cat.No. F147. The stock solution was made at a concentration of 10 mM in DMSO and the aliquots were stored in −20°C until use. The cells were treated at a concentration of 10 μM.
Fenofibrate: Sigma, Cat.No. PHR1246. The stock solution was made at a concentration of 20 mM in DMSO and the aliquots were stored in −20°C until use. The cells were treated at a concentration of 20 μM.
PH-755: The compound was provided by Raze Therapeutics. The stock solution was made at a concentration of 5 mM in DMSO and the aliquots were stored in −20°C until use. The cells were treated at a concentration of 5 μM.
shRNA mediated knockdown of mitochondrial pyruvate carrier (MPC)
MPC knock down was carried out as previously described14
Lentiviral shRNA targeting human
Mpc1 (NM_016098.1– 19s1c1): CCGGGCTGCCTTACAAGTATTAAATCTCGAGATTTAATACTTGTAAGGCAGCTTTTT,
Mpc2 (NM_015415.2–400s21c1): CCGGGTCAAGATACTCACTTGTAATCTCGAGATTACAAGTGAGTATCTTGACTTTTTG,
or a non-targeting control construct was packaged in HEK293T cells with pLKO vector with packaging genes Vsv-G, Gag-Pol, and Rev using Fugene 6 as a transfection reagent. The 293T culture medium containing the lentiviral particles was collected and filtered with 0.45-μm syringe filter. For transduction, cells cultured in 6-well plate were incubated with 0.5 mL of the viral suspension for 6 hours before adding 2 mL growth medium. Transduced cells were then selected by growing with 2 μg/mL puromycin.
In vivo Xenograft assay
Animal handling and care followed the NIH Guide for Care and Use of Laboratory Animals. The experimental protocols were approved by the UCSD Institutional Animal Care and Use Committee. 5 week old outbred female athymic nude mice (RRID:IMSR_JAX:007850) were purchased from JAX mice. Mice were housed in groups of four and food and water were provided ad libitum. HCT116 or HT29 cells cultured in 15 cm dishes were trypsinized and singularized. The trypsin was washed with 5 times excess growth medium and the cells were counted. The cells were then resuspended in PBS at a density of 15 × 106 cells/mL. 200 μL aliquots of the cell suspension (3 million cells) were made and kept on ice until injection. The cells were injected subcutaneously onto the hind flanks of 7-week-old athymic nude mice by gently lifting the skin. For dietary amino acid modulation, mice were pre-fed with a control diet or serine, glycine- free diet (-SG) or serine, glycine-free, extra alanine diet (-SG +A) custom formulation from Envigo, (Supplementary Table 1) for a week prior to xenograft injection. Tumor growth was measured twice, weekly with calipers. The tumor volumes were calculated by using the formula V= 0.5(L × B2) where V is the volume, L is the major diameter, and W is the minor diameter. The major diameter was not allowed to exceed 20mm, in accordance with UCSD IACUC guidelines.
Myriocin was formulated at 2 mg/ml stock in methanol and stored at −20°C until use. On the day of injection, 120 μl stock solution was sonicated for 5 minutes in a bath sonicator, and solutions of 60 μg/ml and 6 μg/ml myriocin were made in PBS. Appropriate volumes were injected intraperitoneally to attain 0.3 mg/kg or 0.03 mg/kg every other day. PH-755 suspension was made by dispersing 2g of PH-755 in 40 ml PBS containing 0.5% methyl cellulose and 0.5% tween 80 in a bath sonicator for 20 minutes. The suspension was then stored as 1 mL aliquots at −20°C until use. On the day of drug administration, the aliquots were sonicated for 5 minutes in a bath sonicator and the drug suspension or vehicle was administered via oral gavage at a dose of 300 mg/kg, twice a day, using straight, stainless-steel oral gavage needles (22g × 1.5”, 1.25mm, Cadence scientific, Cat. No. 7920).
Immunoblotting
Adherent cells were washed with cold PBS and harvested by scraping on ice with M-PER mammalian protein extraction reagent (Thermo Fischer scientific, Cat. No. 78501) freshly supplemented with 1x HALT protease inhibitor cocktail (Thermo Fisher Scientific, Cat. No. 78430). Spheroids were washed by spinning at 50 × g for 3 minutes and resuspending in M-PER. BCA assay was used to determine protein concentrations. An equal amount of proteins was loaded onto 10% SDS-PAGE gels. The proteins were transferred onto a nitrocellulose membrane. The membrane was blocked using 1 % BSA in tris buffered saline with 0.1% Tween 20 (TBST) for 1 h and incubated with primary antibody at 4°C overnight. Anti-PDH antibody (rabbit polyclonal, Cell Signaling Technologies, Cat. No. 2784S, Lot 2, 1:1000), Anti-PDH-E1α (pSer293) (Rabbit polyclonal Millipore Sigma, Cat. No. AP1062, Lot 2972838 1:1000), or anti β-Actin (rabbit monoclonal, Cell signaling technology, Cat. No 4970, Lot 15, 1:1000). The immunoblots were then incubated with secondary antibody for 1 h at room temperature, Monoclonal anti-rabbit antibody HRP-conjugate (Cell Signaling Technology, Cat. No 7074, Lot 27, 1:1000). To image, blots were incubated with ECL substrate (BioRad Cat. No. 1705061) and imaged with a BioRad ChemiDoc XRS+ imaging station equipped with Image Lab (v5.2.1).
Antibodies for western blotting were validated for human reactivity by the manufacturer and used per their instructions. Additional information on validation (publication, certificate, manufacturer western blot) can be found on the manufacturers’ websites listed below:
Anti-PDH: (rabbit polyclonal, Cell Signaling Technologies, Cat. No. 2784S, Lot 2, 1:1000): validation by manufacturer (correct size, https://www.cellsignal.com/products/primary-antibodies/pyruvate-dehydrogenase-antibody/2784) and publication PMID: 25910802, PMID: 31231686
Anti-PDH-E1α (pSer293): (Rabbit polyclonal Millipore Sigma Cat. No. AP1062, Lot 2972838, 1:1000): validation by manufacturer (correct size, https://www.emdmillipore.com/US/en/product/PhosphoDetect-Anti-PDH-E1-pSer293-Rabbit-pAb,EMD_BIO-AP1062) and publication PMID: 17474719, PMID: 17208939, PMID: 11486000
Anti-β-Actin: (rabbit monoclonal, Cell signaling technology, Cat. No 4970, Lot 15, 1:1000): validation by manufacturer (correct size, https://www.cellsignal.com/products/primary-antibodies/b-actin-13e5-rabbit-mab/4970) and publication PMID: 31516608, PMID: 31367191, PMID: 30881499
Anti-rabbit antibody HRP-conjugate (Cell Signaling Technology, Cat. No 7074, Lot 27, 1:1000): validation by manufacturer (correct size, https://www.cellsignal.com/products/secondary-antibodies/anti-rabbit-igg-hrp-linked-antibody/7074)
Quantification and statistical analysis
All results were depicted as mean ± s.e.m. of at least three biological replicates as indicated in figure legends using GraphPad Prism (v8.0.1). P values were calculated using a Student’s two-tailed t-test, ordinary one way, or two-way ANOVA where, * P value < 0.05; **P < 0.01; *** or # P value <0.001.
Extended Data
Extended Data Figure 1. Mitochondrial pyruvate transport and amino acid metabolism influence spheroid growth.
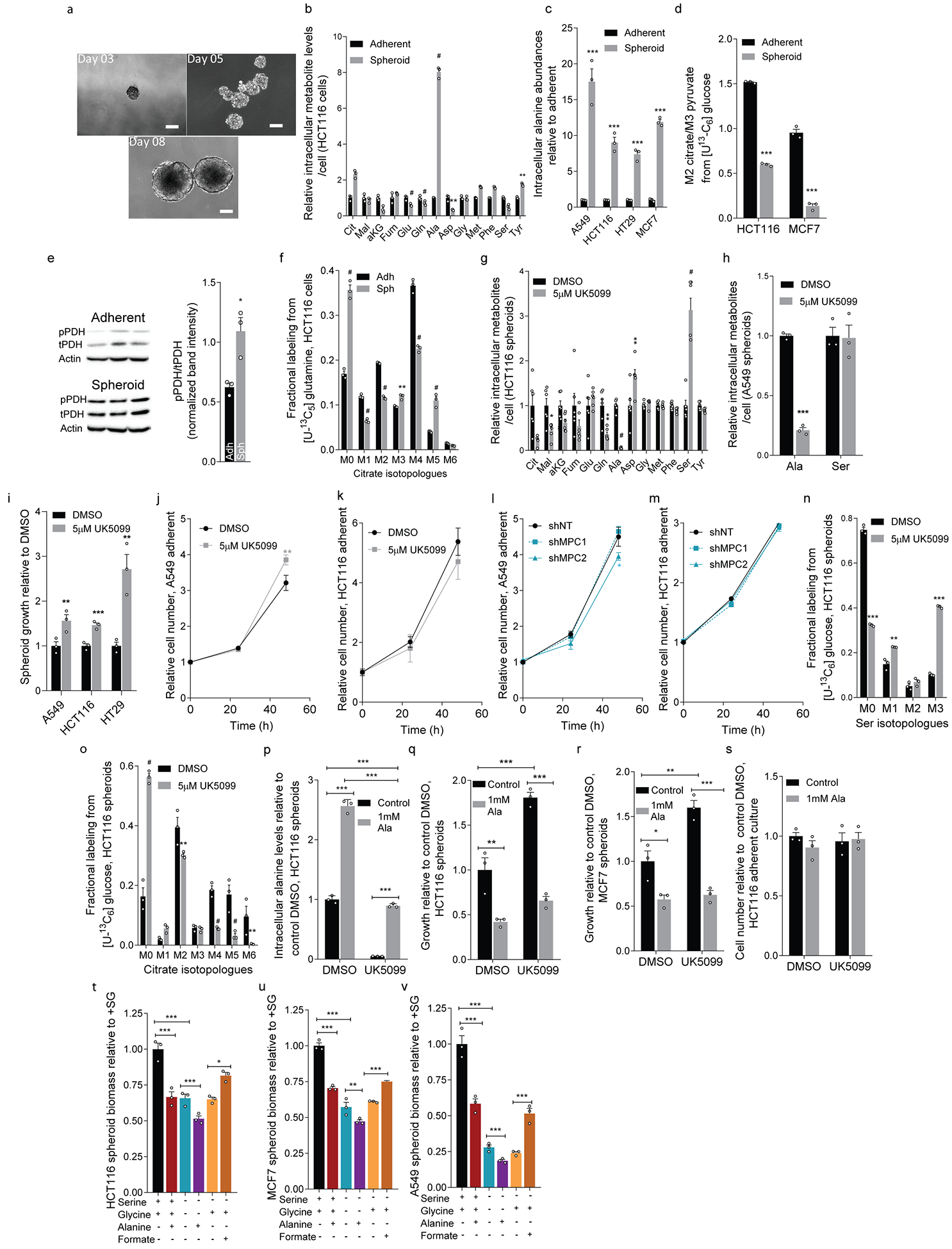
a. HCT116 spheroid growth from 3-, 5-, and 8-day cultures. Scale bars indicate 100 μm.
b. Metabolite levels in HCT116 adherent and spheroid cultures (n=3 culture wells each).
c.Alanine levels in adherent and spheroid cultures (n=3 culture wells each condition and cell line).
d. Isotopic labeling (M2 citrate/M3 pyruvate) in HCT116 and MCF7 cells cultured with [U-13C6]glucose for 24 hours (n=3 culture wells each).
e. Protein expression of phosphorylated PDH (pPDH), total PDH (tPDH), and β-actin in HCT116 cells. Each lane derived from a single culture well, processed in parallel, and used for quantitation. For gel source data, see Supporting files.
f. Citrate labeling in HCT116 cells cultured with [U-13C5]glutamine in HCT116 (n=3 culture wells each condition).
g. Metabolite levels upon UK5099 treatment in HCT116 spheroid cultures (n=6 culture wells each).
h. Abundances of alanine and serine in A549 spheroid cultures upon treatment with UK5099 (n=3 culture wells each).
i. Spheroid growth in cells upon UK5099 treatment (n=3 culture wells each).
j, k. Adherent growth of (j) A549 and (k) HCT116 cells upon treatment with UK5099 (n=3 culture wells each).
l, m. Adherent growth of (l) A549 and (m) HCT116 cells upon MPC1 or MPC2 knockdown compared to control (shNT) (n=3 culture wells each)
n, o. Isotopologue distributions of (n) serine and (o) citrate in HCT116 spheroid cultures traced with [U-13C6]glucose for 24 hours (n=3 culture wells each)
p. Alanine abundances in HCT116 spheroids in the presence of 1 mM alanine and UK5099 (n=3 culture wells each).
q, r. Spheroid growth of (q) HCT116 and (r) MCF7 cells grown in the presence of UK5099 and alanine (n=3 culture wells each condition).
s. Cell number of adherent HCT116 cells in the presence of UK5099 and alanine (n=3 culture wells each).
t-v. Spheroid biomass in (t) HCT116, (u) MCF7, and (v) A549 cells grown in the presence or absence of 0.4 mM serine, 0.4 mM glycine, 1 mM alanine, and 1 mM formate (n=3 culture wells for each cell line and condition).
Two-sided Student’s t-test (b-i, n, o), one-way ANOVA (t-v) or two-way ANOVA (j-m, p-s) was performed for each comparison with no adjustment for multiple comparison. Similar results obtained in 2 (d, e, h, p, r), 3 (c, f-g, i, n-o, q), or 4 (b) independent experiments. Data represented as mean ± s.e.m. *P<0.05, **P<0.01, ***P<0.0001.
Extended Data Figure 2. 1-deoxysphingolipid synthesis and degradation influence spheroid growth in vitro.
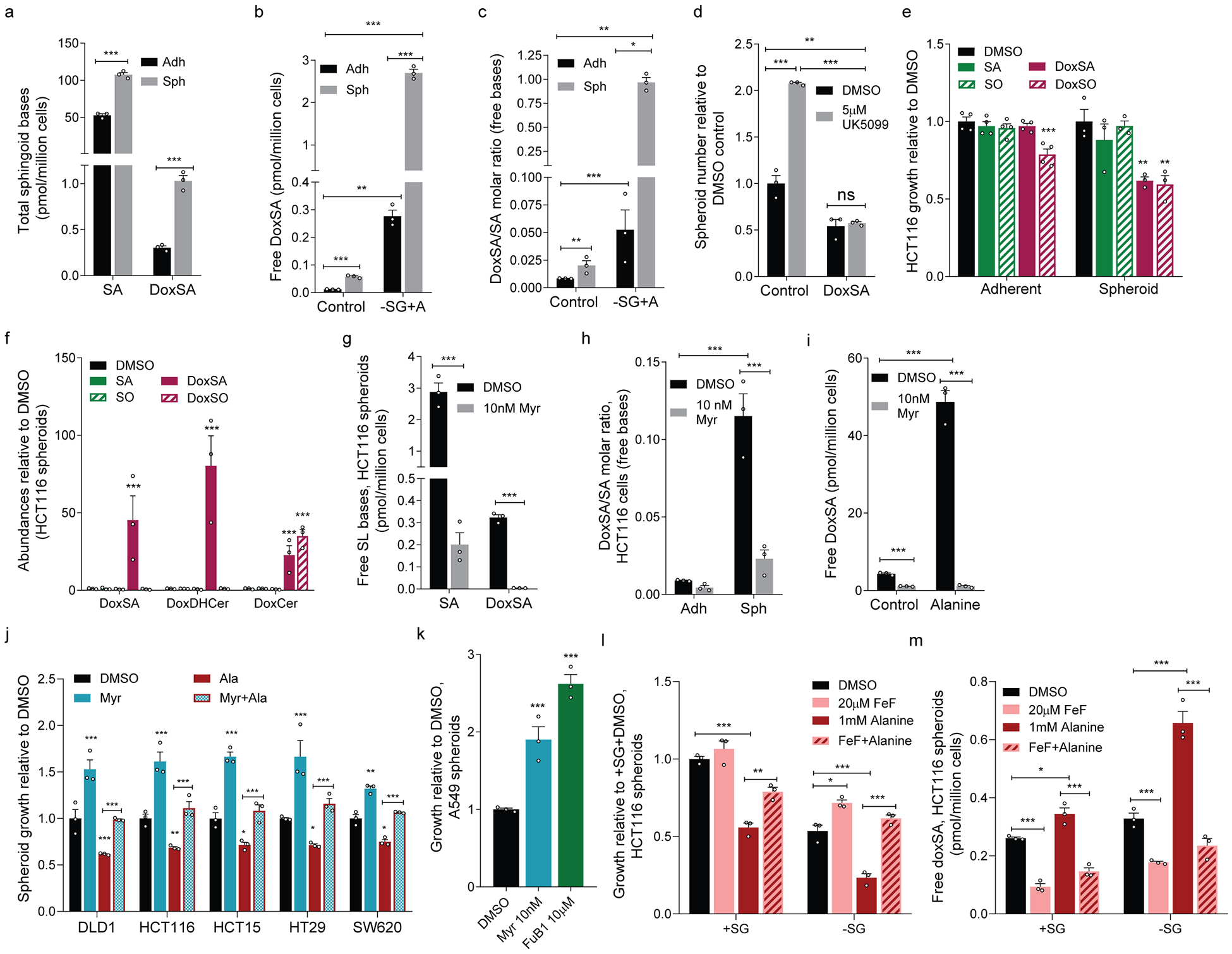
a. Total (hydrolyzed) SA and doxSA abundances in adherent and spheroid cultures of HCT116 cells (n=3 culture wells each).
b. Free doxSA abundances in HCT116 cultures in normal medium or medium without serine/glycine but containing 1 mM alanine (–SG+A) (n=3 culture wells each).
c. DoxSA/SA molar ratio in HCT116 cells in cultures from (b) (n=3 culture wells each).
d. HCT116 spheroid growth in the presence or absence of 10 nM doxSA and treated with vehicle (DMSO) or UK5099 (n=3 culture wells each).
e. Adherent (n=4 culture wells each) and spheroid (n=3 culture wells each) biomass of HCT116 cells treated with vehicle or 50 nM sphingoid bases.
f. Free doxSA, summed doxDHCer and summed doxCer in spheroid cultures from (e) (n=3 culture wells each).
g. SA and doxSA abundances in HCT116 spheroid cultures treated with vehicle or 10 nM myriocin (Myr; n=3 culture wells each).
h. DoxSA/SA molar ratio in HCT116 adherent and spheroid cultures treated with vehicle or 10 nM Myr (n=3 culture wells each).
i. Free DoxSA abundances in HCT116 spheroid cells in the presence or absence of 1 mM alanine and treated with vehicle or 10 nM Myr (n=3 culture wells each).
j. Spheroid growth of cell lines cultured in the presence or absence of 1 mM alanine (red), 10 nM Myr (blue), or both (red outline with checkered blue fill) (n=3 culture wells each condition).
k. A549 spheroid growth under 5 days of culture in Myr or 10 μM fumonisin B1 (FuB1) (n=3 culture wells each).
l, m. HCT116 (l) spheroid growth and (m) free doxSA in the presence (+SG) or absence (–SG) of 0.4 mM serine/serine and treated with DMSO, FeF, 1 mM alanine, or both (n=3 culture wells for each condition).
Two-sided Student’s t-test (a, g), two-way ANOVA (b, c, d, h, i, l, m) or one-way ANOVA (e, f, j, k) was performed for each comparison with no adjustment for multiple comparison. Similar results obtained in 2 (b-e) or 3 (a, g-i) independent experiments. Data represented as mean ± s.e.m. *P<0.05, **P<0.01, *** or # P<0.0001.
Extended Data Figure 3. Dietary serine/glycine restriction alters tumor growth and ceramide metabolism.
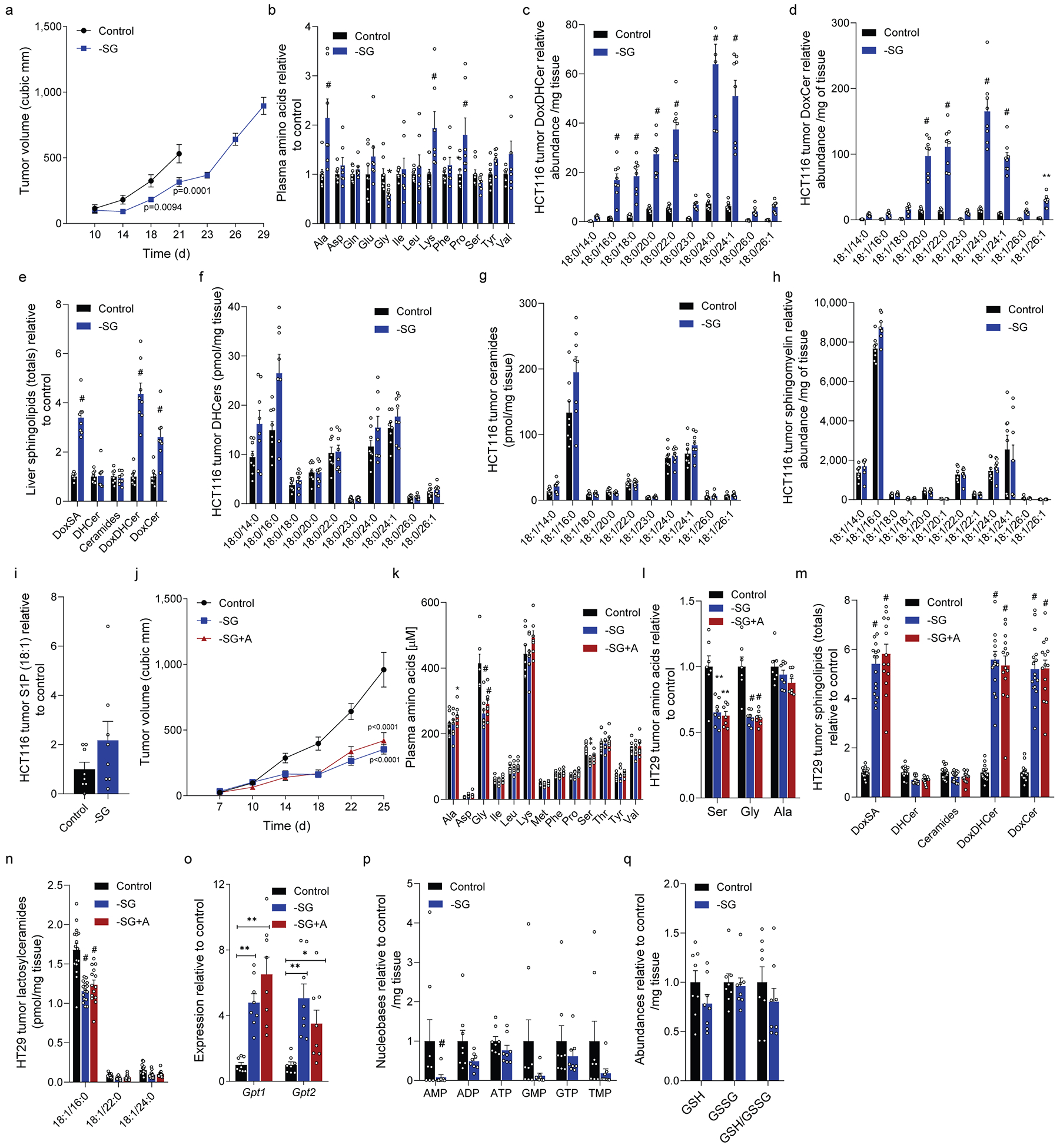
a. HCT116 xenograft size in mice fed control (n=16) or –SG (n=15 day 21) diet.
b. Plasma amino acids from mice fed control (n=8) or –SG (n=7) diet.
c, d. Abundances of (c) doxDHCer and (d) doxCer in HCT116 xenograft tumors (n=8 each diet).
e. Abundances of total sphingolipid species in the livers from HCT116 tumor-bearing mice fed control or –SG diet (n=8 each diet).
f-i. Abundances of (f) DHCer, (g) Ceramides, (h) sphingomyelins, and (i) S1P in HCT116 xenografts from mice fed control or –SG diet (n=8 each diet).
j. HT29 xenograft size in mice fed control (n=16), –SG (n=16), or –SG+A (n=14) diets.
k. Plasma amino acids in mice in (j) fed control (n=8), –SG (n=8), or –SG+A (n=7) diets.
l. Tumor amino acids from mice in (j) fed control (n=7), –SG (n=8), or –SG+A (n=7).
m. DoxSA and summed ceramide species in HT29 xenograft tumors from mice fed control (n=16), –SG (n=16), or –SG+A diets (n=14).
n. Abundances of lactosylceramides in HT29 xenograft tumors from mice fed control (n=16), –SG (n=16), or –SG+A diets (n=14).
o. Gpt1 and Gpt2 expression in liver tissue of mice fed control, –SG, or –SG+A diets (n=8 for each).
p. Nucleotide phosphate abundances in HCT116 xenograft tumors from mice fed control, –SG, or –SG+A diets (n=8 for each).
q. Reduced glutathione (GSH), oxidized glutathione (GSSG), and GSH/GSSG measurements of HCT116 xenograft tumors from mice fed control, –SG, or –SG+A diets (n=8 each).
Two-sided Student’s t-test (b-i, p, q) or two-way ANOVA (a, j, k, l, m, n, o), was performed with no adjustment for multiple comparison. Data are represented as mean ± s.e.m. *P<0.05, **P<0.01, *** or # P<0.0001.
Extended Data Figure 4. Influence of SPT and PHGDH inhibition on sphingolipid metabolism.
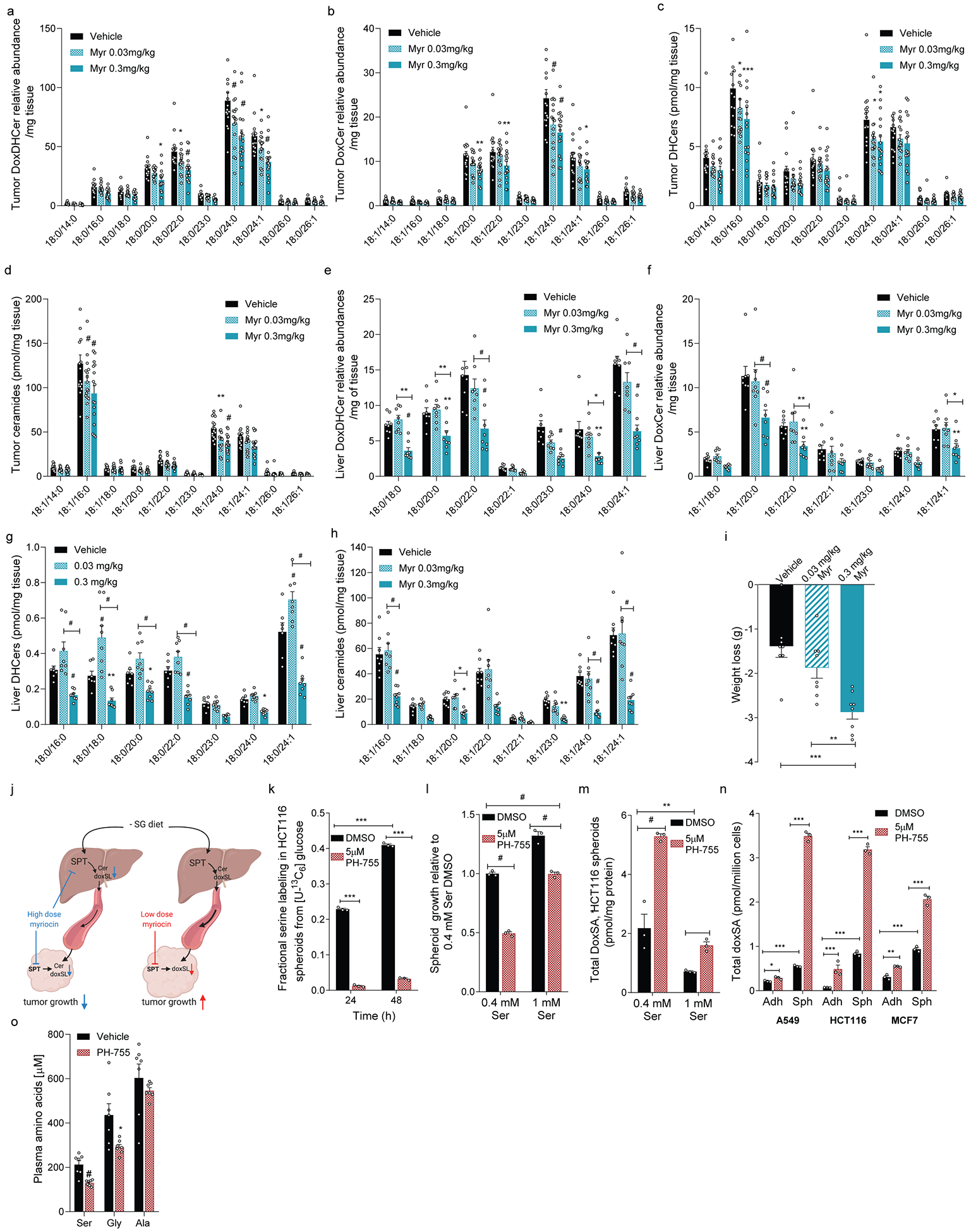
a-d.Abundances of (a) DoxDHCer, (b) DoxCer, (c) DHCer, and (d) Ceramides in HCT116 xenograft tumors from mice fed –SG diet and administered vehicle, 0.03 mg/kg myriocin, or 0.3 mg/kg myriocin (n=16 for each treatment).
e-h.Abundances of (e) DoxDHCer, (f) DoxCer, (g) DHCer, and (h) Ceramides in livers from HCT116 xenograft-bearing mice fed –SG diet and administered vehicle (n=8 for e, f, h and n=7 for g), 0.03 mg/kg myriocin (n=8), or 0.3 mg/kg myriocin (n=8).
i. Weight loss in mice after tumor inoculation in mice administered vehicle (n=7), 0.03 mg/kg myriocin (n=8), or 0.3 mg/kg myriocin (n=8).
j. Schematic describing in vivo sphingolipid physiology under high- and low-dose myriocin treatments generated using BioRender.
k. Fractional labeling of serine (1-M0) from HCT116 spheroids cultured with [U-13C6]glucose for 24 hours and treated with vehicle or 5μM PH-755 (n=3 culture wells for each).
l. HCT116 spheroid growth in media containing 0.4 mM or 1.0 mM serine and treated with vehicle or 5μM PH-755 (n=3 culture wells for each).
m. Total DoxSA abundances in HCT116 spheroids grown for 5 days in media with 0.4 mM or 1 mM serine and treated with vehicle or 5μM PH-755 (n=3 culture wells for each).
n. Total DoxSA abundances in A549, HCT116 and MCF7 spheroids treated with vehicle or 5μM PH-755 (n=3 culture wells each).
o. Plasma serine, glycine and alanine in mice treated with vehicle or PH-755 (n=7 each).
Two-way ANOVA (a-h, k-n), one-way ANOVA (i), or two-sided Student’s t-test (o) was performed with no adjustment for multiple comparison. Similar results were obtained in 2 independent experiments for k-n. Data represented as mean ± s.e.m. *P<0.05, **P<0.01, *** or # P<0.0001.
Supplementary Material
Acknowledgments
We would like to thank Marin Gantner (Lowy Medical Research Institute, La Jolla, CA), Martin Friedlander (The Scripps Research Institute, La Jolla, CA) and all members of the Metallo laboratory for support and helpful discussions. We are grateful to Nello Mainolfi, Vipin Suri, Adam Friedman, and Mark Manfredi of Raze Therapeutics (Cambridge, MA) for providing PH-755. This work was supported by the NIH (R01CA188652 and R01CA234245 to C.M.M.; U54CA132379), a Camille and Henry Dreyfus Teacher-Scholar Award (to C.M.M.), the National Science Foundation (NSF) Faculty Early Career Development (CAREER) Program (1454425 to C.M.M.), the Helmsley Center for Genomic Medicine (to A.F.M.P) and funding from Ferring Foundation (to A.S.). This work was also supported by NIH grants to the Salk Institute Mass Spectrometry Core (P30CA014195, S10OD021815).
Footnotes
Competing interests
The authors declare no competing interests.
Data availability
Source data for immunoblots are provided as Supplementary Information. Additional data that support findings are available from the corresponding author upon reasonable request.
References
- 1.Knott SRV et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature 554, 378–381, doi: 10.1038/nature25465 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LeBoeuf SE et al. Activation of Oxidative Stress Response in Cancer Generates a Druggable Dependency on Exogenous Non-essential Amino Acids. Cell Metabolism, doi: 10.1016/j.cmet.2019.11.012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maddocks ODK et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546, doi: 10.1038/nature11743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan J & Merrill AH 1-Deoxysphingolipids Encountered Exogenously and Made de Novo : Dangerous Mysteries inside an Enigma. Journal of Biological Chemistry 290, 15380–15389, doi: 10.1074/jbc.R115.658823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lone MA, Santos T, Alecu I, Silva LC & Hornemann T 1-Deoxysphingolipids. Biochim Biophys Acta Mol Cell Biol Lipids 1864, 512–521, doi: 10.1016/j.bbalip.2018.12.013 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Eichler FS et al. Overexpression of the wild-type SPT1 subunit lowers desoxysphingolipid levels and rescues the phenotype of HSAN1. The Journal of neuroscience : the official journal of the Society for Neuroscience 29, 14646–14651, doi: 10.1523/JNEUROSCI.2536-09.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penno A et al. Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J Biol Chem 285, 11178–11187, doi: 10.1074/jbc.M109.092973 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esaki K et al. L-serine deficiency elicits intracellular accumulation of cytotoxic deoxysphingolipids and lipid body formation. Journal of Biological Chemistry 290, doi: 10.1074/jbc.M114.603860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gantner ML et al. Serine and Lipid Metabolism in Macular Disease and Peripheral Neuropathy. New England Journal of Medicine, NEJMoa1815111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baird RD et al. Phase I safety, pharmacokinetic, and pharmacogenomic trial of ES-285, a novel marine cytotoxic agent, administered to adult patients with advanced solid tumors. Molecular Cancer Therapeutics 8, 1430–1437, doi: 10.1158/1535-7163.MCT-08-1167 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Schafer ZT et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–113, doi: 10.1038/nature08268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532, 255–258, doi: 10.1038/nature17393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker SJ et al. LKB1 promotes metabolic flexibility in response to energy stress. Metab Eng 43, 208–217, doi: 10.1016/j.ymben.2016.12.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vacanti NM et al. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol Cell 56, 425–435, doi: 10.1016/j.molcel.2014.09.024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schell JC et al. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Molecular cell 56, 400–413, doi: 10.1016/j.molcel.2014.09.026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis CA et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell 55, 253–263, doi: 10.1016/j.molcel.2014.05.008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeske L, Placzek S, Schomburg I, Chang A & Schomburg D BRENDA in 2019: a European ELIXIR core data resource. Nucleic Acids Research 47, D542–D549, doi: 10.1093/nar/gky1048 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alecu I et al. Cytotoxic 1-deoxysphingolipids are metabolized by a cytochrome P450-dependent pathway. Journal of Lipid Research 58, 60–71, doi: 10.1194/jlr.M072421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao X et al. Serine Availability Influences Mitochondrial Dynamics and Function through Lipid Metabolism. Cell Reports 22, 3507–3520, doi: 10.1016/j.celrep.2018.03.017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez AE et al. Serine Metabolism Supports Macrophage IL-1β Production. Cell Metabolism 29, 1003–1011.e1004, doi: 10.1016/j.cmet.2019.01.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guri Y et al. mTORC2 Promotes Tumorigenesis via Lipid Synthesis. Cancer Cell 32, 807–823 e812, doi: 10.1016/j.ccell.2017.11.011 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Maddocks ODKK et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature 544, 372–376, doi: 10.1038/nature22056 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Locasale JW et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature Genetics 43, 869–874, doi: 10.1038/ng.890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Possemato R et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350, doi: 10.1038/nature10350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Othman A et al. Lowering plasma 1-deoxysphingolipids improves neuropathy in diabetic rats. Diabetes 64, 1035–1045, doi: 10.2337/db14-1325 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Oswald MC, West RJ, Lloyd-Evans E & Sweeney ST Identification of dietary alanine toxicity and trafficking dysfunction in a Drosophila model of hereditary sensory and autonomic neuropathy type 1. Hum Mol Genet 24, 6899–6909, doi: 10.1093/hmg/ddv390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alecu I et al. Localization of 1-deoxysphingolipids to mitochondria induces mitochondrial dysfunction. J Lipid Res 58, 42–59, doi: 10.1194/jlr.M068676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garofalo K et al. Oral l-serine supplementation reduces production of neurotoxic deoxysphingolipids in mice and humans with hereditary sensory autonomic neuropathy type 1. Journal of Clinical Investigation 121, 4735–4745, doi: 10.1172/JCI57549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han G et al. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proceedings of the National Academy of Sciences of the United States of America 106, 8186–8191, doi: 10.1073/pnas.0811269106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace M et al. Enzyme promiscuity drives branched-chain fatty acid synthesis in adipose tissues. Nat Chem Biol 14, 1021–1031, doi: 10.1038/s41589-018-0132-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu M et al. A resource for cell line authentication, annotation and quality control. Nature 520, 307–311, doi: 10.1038/nature14397 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Young JD INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics 30, 1333–1335, doi: 10.1093/bioinformatics/btu015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cordes T & Metallo CM Quantifying Intermediary Metabolism and Lipogenesis in Cultured Mammalian Cells Using Stable Isotope Tracing and Mass Spectrometry. Methods Mol Biol 1978, 219–241, doi: 10.1007/978-1-4939-9236-2_14 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Steiner R et al. Elucidating the chemical structure of native 1-deoxysphingosine. J Lipid Res 57, 1194–1203, doi: 10.1194/jlr.M067033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bielawski J et al. Comprehensive quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods Mol Biol 579, 443–467, doi: 10.1007/978-1-60761-322-0_22 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Wu J et al. Quantitative analysis of intracellular nucleoside triphosphates and other polar metabolites using ion pair reversed-phase liquid chromatography coupled with tandem mass spectrometry. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences 1006, 167–178, doi: 10.1016/j.jchromb.2015.10.030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


