Abstract
Microcephaly is a rare, yet devastating, neurodevelopmental condition caused by genetic or environmental insults, such as the Zika virus infection. Microcephaly manifests with a severely reduced head circumference. Among the known heritable microcephaly genes, a significant proportion are annotated with centrosome-related ontologies. Centrosomes are microtubule-organizing centers, and they play fundamental roles in the proliferation of the neuronal progenitors, the neural stem cells (NSCs), which undergo repeated rounds of asymmetric cell division to drive neurogenesis and brain development. Many of the genes, pathways, and developmental paradigms that dictate NSC development in humans are conserved in Drosophila melanogaster. As such, studies of Drosophila NSCs lend invaluable insights into centrosome function within NSCs and help inform the pathophysiology of human microcephaly. This mini-review will briefly survey causative links between deregulated centrosome functions and microcephaly with particular emphasis on insights learned from Drosophila NSCs.
Introduction
Microcephaly is a neurological condition characterized by an abnormally small cerebral cortex and a head circumference that is more than two standard deviations below the population mean [1]. The characteristic small head of microcephalic individuals may manifest as the sole developmental phenotype, as in primary or non-syndromic microcephaly. Alternatively, microcephaly may present in conjunction with other comorbidities, also known as syndromic microcephaly. Those comorbidities include but are not limited to intellectual disability, epilepsy, eye abnormalities, short stature, etc. as observed in diverse human syndromes, such as primary recessive autosomal microcephaly, microcephalic osteodysplastic primordial dwarfism type II (MOPDII), Seckel syndrome, etc. (clinical manifestations of microcephaly reviewed in [2,3]).
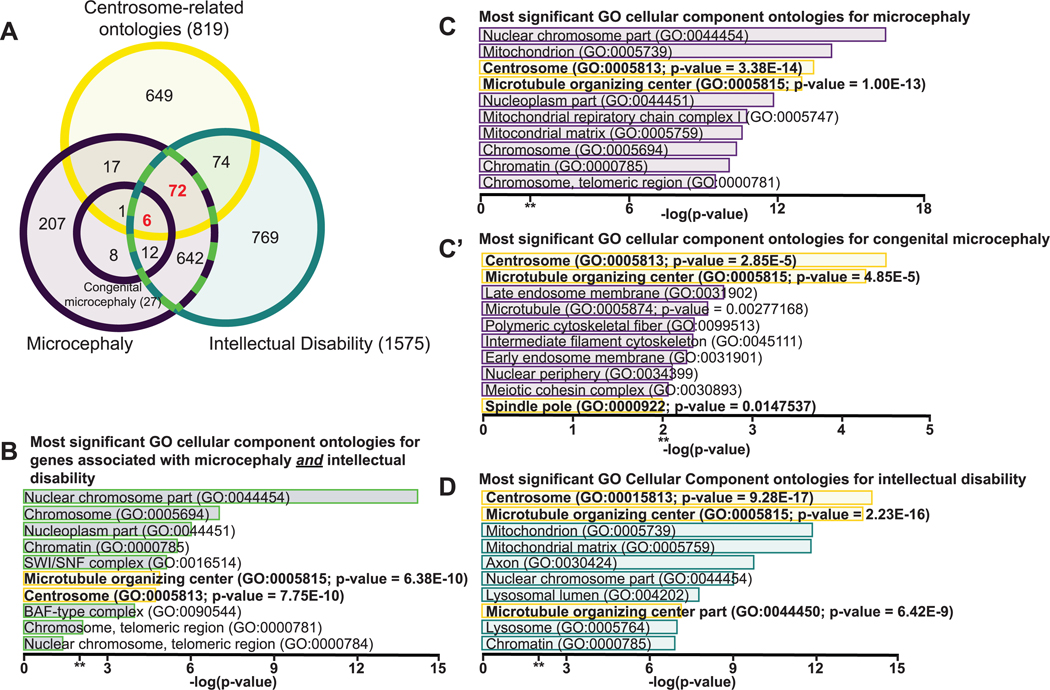
Not surprisingly, microcephaly is extremely genetically heterogenous. The human phenotype ontology (HPO) shows an association of the microcephaly phenotype (term HP:0000252) with more than 1400 diseases and 960 genes, with new causative genes routinely being discovered [4,5]. Intellectual disability, which is defined as an individual with an IQ score below 70 [6], is observed in ~50% of microcephaly cases and represents the most frequent microcephaly comorbidity [3,7,8]. HPO shows an association of the intellectual disability phenotype (term HP:0001249) with more than 2700 diseases and 1560 genes [4,5]. The American Psychiatric Association defines intellectual disability as a neurodevelopmental disorder characterized by significantly limited intellectual functioning that begins in childhood [9]. As both microcephaly and intellectual disability arise directly from aberrant neurodevelopment, it is not surprising that the two gene data sets have ~45–75% overlap (Figure 1A; Supplemental Tables S1 and S2). Using Enrichr gene analysis to focus on gene ontology (GO)-cellular components reveals that both the microcephaly gene data set and intellectual disability gene data set are significantly enriched for genes annotated with the centrosome (Figure 1B–D; yellow bars [10,11]). In fact, the number of genes annotated with at least one of the significantly enriched centrosome-related ontologies (centrosome (GO:0005815), microtubule-organizing center (GO:0005813), and spindle pole (GO:0000922)) represent ∼10% of each phenotype data set (Figure 1A; Supplemental Table S1).
Figure 1. Microcephaly associated genes are significantly enriched with centrosome genes.
(A) Venn-diagram depicts curated gene lists and overlap indicates the number of common genes present within each data set. A gene list curated by HPO indicates 1575 human genes are associated with intellectual disability (term HP:0001249; blue circle) and 965 human genes are associated with microcephaly (term HP:0000252; purple circle). Of these genes, 27 are also associated with congenital microcephaly (term: HP:0011451; purple circle inset). A gene list generated by combining all genes annotated with the following centrosome-related cell component ontology IDs curated by the Gene Ontology Resource: centrosomes (GO: 0005813), microtubule-organizing centers (GO: 0005815), and spindle pole (GO:0000922) contains 819 unique genes (yellow circle). The microcephaly phenotype and intellectual disability phenotype share 732 genes (dotted green outline). This overlap accounts for ∼75% of the microcephaly data set and ∼45% of the intellectual disability data set, indicating neurodevelopmental convergence between the neuroanatomical and behavioral phenotypes. Of the genes associated with microcephaly, 96 overlap with genes annotated with centrosome-related ontologies. Of the genes associated with intellectual disability, 152 overlap genes annotated with centrosome-related ontologies. The microcephaly and intellectual disability data sets share 78 common centrosome-related genes (red), representing ∼10% of the shared disease genes, indicating enrichment of the centrosome and centrosome-related cell components with both diseases. (B–D) Bar graphs show the most significant cellular components enriched in each data set as determined by Enrichr. P-values are displayed for centrosomes and centrosome-related cellular components (bolded text). (B) GO-cellular component analysis reveals that the centrosome and microtubule-organizing center are enriched among genes overlapping with both the microcephaly and intellectual disability phenotype data sets. (C) GO-cellular component analysis reveals that the centrosome and microtubule-organizing center are among the top five significantly enriched cellular components in the microcephaly gene data set. (C′) GO-cellular component analysis reveals that the centrosome and microtubule-organizing center are the most significantly enriched cellular components in the congenital microcephaly gene data set. (D) GO-cellular component analysis reveals that the centrosome is the most significantly enriched cellular component in the intellectual disability gene data set; **, P = 0.01.
Enrichments in centrosome genes are also noted in other microcephaly comorbidities. For example, genes associated with epilepsy (term HP:0001250) and eye abnormalities (term HP:0000478) are significantly enriched for centrosome annotation (P-value = 2.13 × 10−07 and P-value = 3.17 × 10−15, respectively). These observations highlight the prevalence of centrosome genes in microcephaly and some of its most frequent comorbidities.
Taken one step further, 30% of the genes associated with congenital microcephaly, defined as overt microcephaly at or before birth (term HP:0011451), are ontologically linked to the centrosome. Indeed, the centrosome and related microtubule-organizing center terms represent the two cellular components with the highest significant ontological enrichment for congenital microcephaly associated genes (Figure 1C′; yellow bars, P-value = 2.85 × 10−5 and Supplemental Table S2 [10–14]). This ontological analysis highlights the importance of centrosome regulation for normal brain development, morphology, and function. Consequently, centrosome-related microcephaly genes have been studied in depth (recently reviewed in [15–20]). Not only genetic deficiencies point to the centrality of centrosome-dependent mechanisms to microcephaly but also infectious agents like Zika, whose cardinal pathology is microcephaly, interfere with centrosome-related mechanisms [21–23].
Centrosomes are membrane-less organelles composed of two cylinder-shaped centrioles surrounded by a rich protein-matrix of pericentriolar material (PCM) and function as the microtubule-organizing centers of most animal cells. Centrosomes are responsible for preserving the genome during cell division and templating the primary cilia in quiescent cells [24]. The levels, composition, and organization of PCM oscillate in conjunction with the cell cycle, and these oscillatory behaviors dictate the microtubule-nucleating activity of centrosomes [25,26]. As cells enter mitosis, centrosomes duplicate and recruit PCM, a process called centrosome maturation. Following mitotic exit, centrosomes shed PCM and each daughter cell inherits a single centrosome. The processes of centrosome duplication and maturation are tightly regulated (as reviewed in [27–29]). Deregulation of centrosome number or activity manifests in developmental diseases, including congenital heart disease, ciliopathies (e.g. Bardet–Biedl syndrome), and microcephaly — the focus of this review [30–32]. These links to human disease underscore the importance of understanding centrosome function and regulation.
A significant cause of microcephaly is the depletion of the neural stem cells (NSCs) required for neurogenesis [33]. NSCs are the progenitor cells of the nervous system, and they undergo asymmetric cell division to yield one self-renewing stem cell and a daughter cell fated to differentiate into neurons or glia [34]. Centrosomes are critically important for NSC divisions. Centrosomes contribute to NSC polarity, engineer the bipolar mitotic spindle, and establish the invariant apical–basal cell division axis [35].
This mini-review will briefly survey causative links between deregulated centrosome functions and microcephaly with particular emphasis on insights learned from Drosophila NSCs. We will outline two crucial centrosome-dependent functions that are disrupted by different human microcephaly genes in asymmetrically dividing Drosophila NSCs. First, we will provide an overview of the intimate connection between centrosomes and polarity. We will review seminal studies outlining the importance of centrosome activity both in polarity establishment and asymmetric cell division. We will focus on the bidirectional communication of centrosomes and polarity factors and discuss the consequences following the disruption of this communication. Second, we will examine centrosomes at the spindle poles. We will focus on how disruption of centrosome number and activity affects spindle morphogenesis and NSC division. Finally, in addition to these established centrosome functions, we will also speculate on putative contributions of post-transcriptional mechanisms to centrosome regulation, as the ability of centrosomes to execute rapid transitions in composition and function remains incompletely understood.
Drosophila as a model to uncover cellular mechanisms of NSC divisions
Drosophila NSCs offer valuable insights into the fundamental cell biological mechanisms underlying microcephaly. Many of the genes implicated in human microcephaly are conserved in Drosophila (Supplemental Tables S1 and S2), and the loss of the some of these homologous genes can result in similar microcephaly phenotypes [36–39]. Indeed, several human microcephaly genes were originally identified in Drosophila from centrosome studies [40]. Notable similarities in human and Drosophila neurodevelopment further strengthen the utility of Drosophila to study neurodevelopmental disorders, such as microcephaly. For example, mammals and Drosophila share common progenitor lineages, their neuronal progeny undergo a regulated progression of fate determination, and many of the transcription factors that coordinate neuronal specification are conserved [20]. Finally, both mammalian and Drosophila NSCs share conserved polarity determinants and exhibit biased centrosome inheritance during asymmetric cell division through similar intrinsic mechanisms using conserved molecules [41–43].
In mammals and Drosophila, centrosomes are critical for normal neurodevelopment by supporting NSC proliferation and orienting the direction of asymmetric cell division [20]. Drosophila larval NSCs are a powerful model system to study paradigms of centrosome regulation in the context of neurodevelopment. In the developing Drosophila central brain, the NSCs are numerous, relatively large, close to the surface, rapidly dividing, amenable to a variety of imaging platforms, and genetically tractable. These unique features allow for the discovery of mechanisms underlying NSC centrosome regulation, many of which are conserved in mammals.
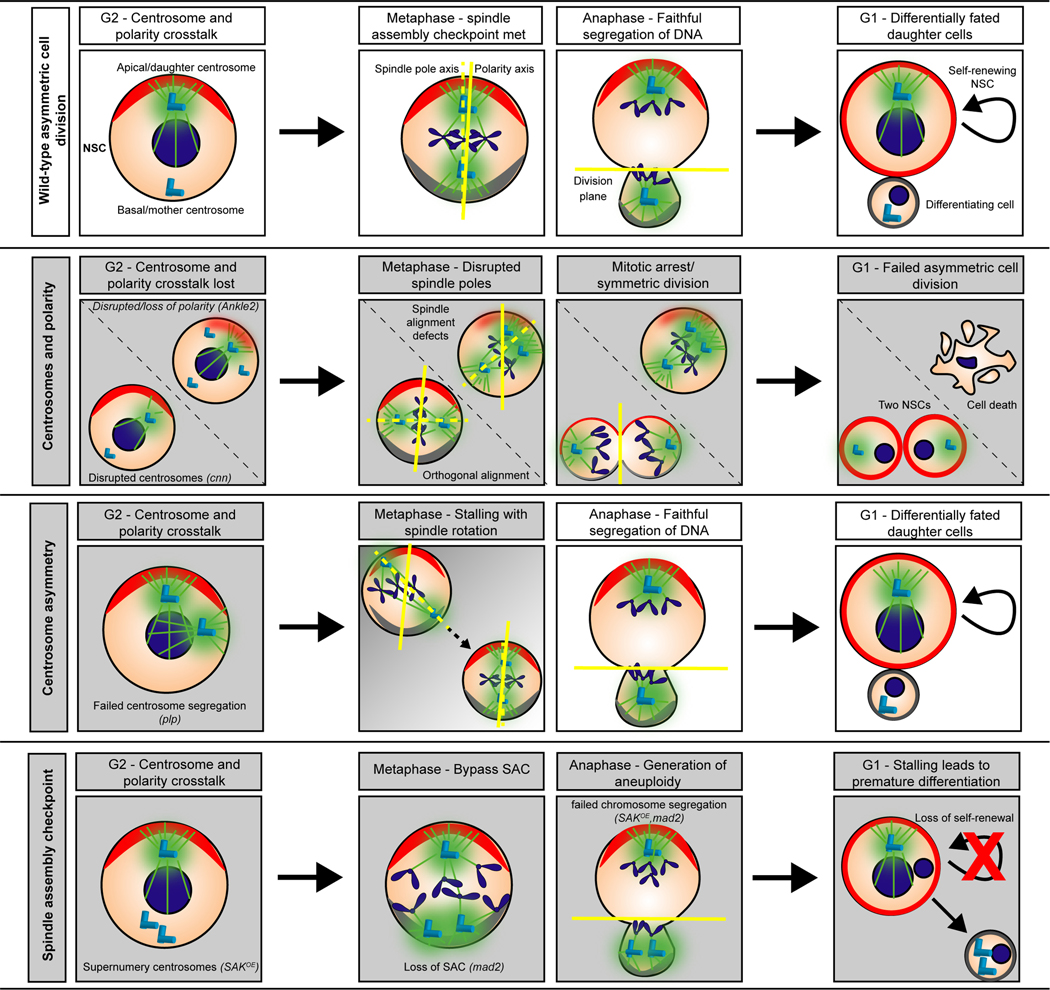
Live imaging studies revealed the two centrosomes within Drosophila NSCs do not recruit PCM synchronously; they undergo asymmetric centrosome maturation (Figure 2; Wild-type asymmetric cell division; [44,45]). Centrosomes are inherently asymmetric due to the varying age of their centrioles; an older (mother) centriole serves as the template for the formation of the younger (daughter) centriole. The daughter centrosome remains active, that is, recruits PCM and forms a microtubule aster, throughout the cell cycle and anchors to the apical cortex. In contrast, the mother centrosome is transiently inactivated during interphase and migrates throughout the cell until mitotic onset, at which point it anchors to the basal cortex and both centrosomes undergo mitotic maturation and rapidly assemble the bipolar spindle (Figure 3, Wild-type interphase) [46]. While centrosome asymmetry is not necessary for asymmetric cell division [47,48], it is required for the non-random segregation of the daughter centrosome to the stem cell and the mother centrosome to the differentiating cell [49,50]. Moreover, loss of centrosome asymmetry can compromise centrosome segregation, leading to centrosome numeracy anomalies (too many or too few inherited by the NSC) and resulting in spindle morphology defects, such as multipolar or monopolar spindles [48,51]. By informing the basic cell biology of asymmetric cell division in Drosophila NSCs, these studies have revealed insights into the pathophysiology of microcephaly (Figure 2).
Figure 2. Multiple centrosome-dependent cellular mechanisms are disrupted by homologous human microcephaly genes.
Cartoons depict the process of NSC proliferation in control (wild-type; top row) versus various mutant conditions. Asymmetric cell division defects are highlighted with gray-filled boxes. NSCs (peach circles) are oriented along the apical–basal axis with the apical polarity markers (red arc) and basal polarity determinants (gray arc) shown. Top row: During wild-type asymmetric cell division, two centrosomes (light blue cylinders) are present in late interphase. The apical centrosome is an active microtubule-organizing center with rich levels of PCM (green), while the basal centrosome is inactive (no PCM). Just prior to the onset of mitosis, cortical basal polarity (gray arc) is established. During metaphase, the spindle pole axis (dotted yellow line) aligns along the polarity axis (solid yellow line); both centrosomes are fully mature/active by this point. During anaphase, the chromosomes and polarity markers are segregated, and the cell divides along the division plane (yellow line). This asymmetric cell division generates one larger self-renewing stem cell (red outline) and one smaller differentiating cell (gray outline). 2nd row: Centrosomes and polarity. In either centrosome (e.g. cnn) or polarity (e.g. Ankle2) mutants, resultant defects include centrosome amplification with spindle morphogenesis defects or randomized spindle pole alignment, leading to failed asymmetric cell division. These errant divisions lead to cell death or symmetric cell divisions (two NSCs). 3rd row: Centrosome asymmetry. Although centrosome phenotypes are observed in interphase (note the two active centrosomes), NSCs mutant for centrosome asymmetry genes rotate misaligned spindle poles before the onset of anaphase (gray → white gradient) and then resume normal asymmetric cell division (white boxes). Not shown, some stem cells missegregate their centrosomes, resulting in too many or too few centrosomes, which may compromise NSC survival. Bottom row: Spindle assembly checkpoint. NSCs mutant for both centrosome genes and components of the SAC generate aneuploid NSCs, which undergo premature differentiation, essentially depleting the NSC pool.
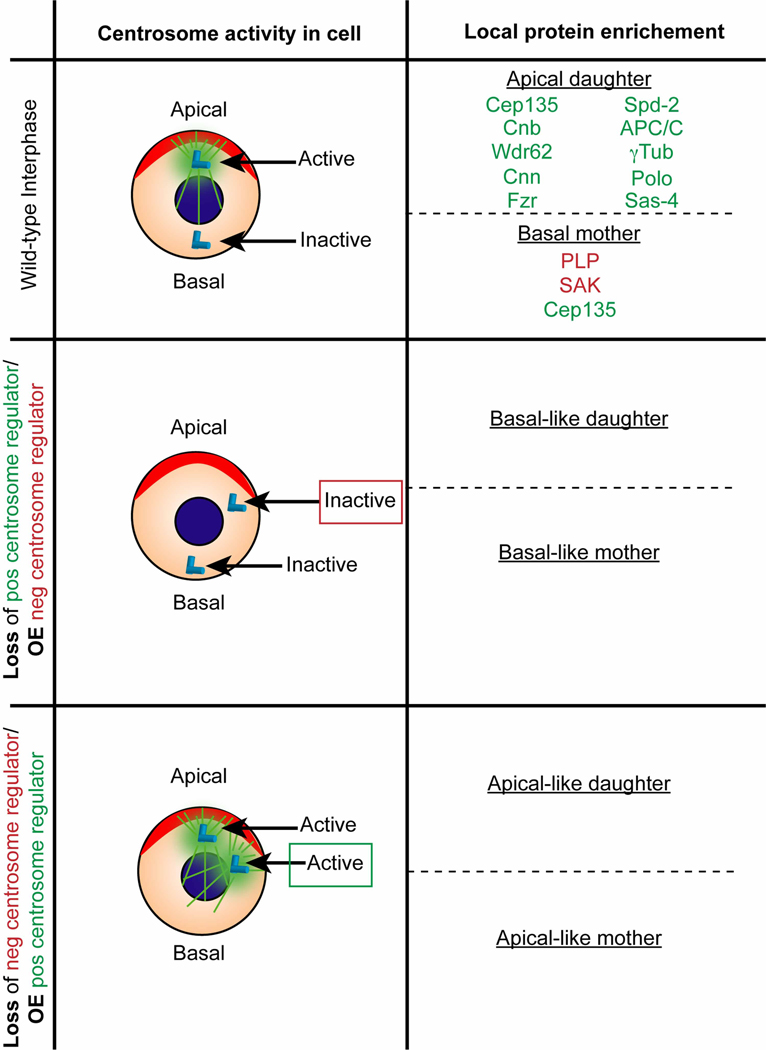
Figure 3. Asymmetric protein localization directs different centrosome activity levels in interphase NSCs.
Normally, wild-type interphase NSCs exhibit asymmetric centrosome activity levels. Top row: In normal cells, the apical, daughter centrosome has high levels of PCM (green cloud) surrounding the centrioles (light blue cylinders), and it nucleates microtubules (green lines). Proteins enriched on the apical centrosome include those that promote microtubule nucleation (local protein enrichment; apical-like daughter, green font). Conversely, the mother, basal centrosome has little to no PCM. Proteins localized on the basal centrosome frequently have negative centrosome-regulating activities (basal-like mother, red font). Centrosome activity level becomes symmetrical when centrosome regulator genes are lost or overexpressed. Middle row: Loss of a positive regulator of centrosome activity (e.g. cnb) or overexpression of a negative centrosome regulator of centrosome activity (e.g. SAK) leads to two inactive, basal-like centrosomes during interphase. Bottom row: Conversely, Loss of a negative regulator of centrosome activity (e.g. plp) or overexpression of a positive regulator of centrosome activity (e.g. cnb) results in two active, apical-like centrosomes during interphase.
Despite the intriguing observation of biased centrosome inheritance, the functional consequences of these inheritance patterns have yet to be identified in Drosophila. In contrast, randomization of centrosome inheritance in the mouse neocortex led to neural progenitor depletion and premature differentiation, suggesting that the biased inheritance of the mother centrosome by the progenitor cells helps maintain their position and stem-ness [43]. These findings are linked to the biased inheritance of the ciliary remnant, which remains attached to the mother centriole and promotes efficient cilia formation upon mitotic exit [52]. Functions of biased centrosome inheritance in Drosophila await discovery.
Centrosomes and polarity
The asymmetric division of the NSCs achieves the segregation of the apical versus basal-localized cell fate determinants, a process coupled to NSC polarization [53–56]. Apical cortical polarity is established during late interphase/prophase and is distinguished by the localization of the Par-complex, defined by Bazooka (Baz)/Par-3, Par-6, and atypical protein kinase C (αPKC), which then recruits the adapter protein Inscuteable (Insc) [57]. Insc interacts with and recruits Partner of Inscuteable (Pins), which contains GoLoco motifs required to associate with the heterotrimeric G-protein subunit Gαi [58,59]. The primary function of Pins/Gαi is to align the bipolar mitotic spindle along the apical–basal polarity axis via interactions with Mushroom body defective (Mud), the Drosophila ortholog of NuMA [60–62].
Conversely, basal polarity is established after apical polarity. Localization of the cell fate determinants Numb, Prospero (Pros), Brain tumor (Brat), and Staufen (Stau) to the basal cortex is mediated by the adapter proteins Miranda (Mira) and Partner of Numb (Pon) and the tumor suppressors Lethal giant larvae (L(2)gl), Discs large (Dlg1), and Scribble (Scrib) [63–68]. Restriction of the apical and basal domains is achieved largely through inhibitory phosphorylation events by αPKC [69,70].
While localization of the Par-complex to the apical cortex represents the upstream step in NSC polarization, there is also a partially redundant microtubule-dependent pathway that contributes to polarity [71,72]. Therefore, centrosomes functioning as microtubule-organizing centers contribute to the cell-intrinsic functions that ensure polarity establishment [73,74]. A requirement for centrosomes in the establishment of basal cortical polarity, for example, was demonstrated by genetically removing centrioles. sas-4, the Drosophila ortholog of the human microcephaly gene CENPJ, is essential for centriole assembly. Removal of sas-4 results in a depletion of centrosomes over time, permitting the examination of centrosome requirements in various tissues. Homozygous sas-4 adults are morphologically normal, yet partially inviable due to ciliary defects that impair locomotion and feeding. In larval NSCs, loss of sas-4 did not alter apical polarity, as Insc localization was unaffected. However, in a subset of NSCs, the basal adapter protein Mira failed to localize, consistent with the ideas that apical polarity can proceed normally through the centrosome-independent Par/Insc pathway and that centrosomes contribute to aspects of basal polarization. It is interesting to note that while acentrosomal microtubule spindles permit bipolar spindle formation and chromosome segregation in sas-4 mutants, ∼50% of sas-4 NSCs show spindle alignment errors and some NSCs divide symmetrically, supporting a role for centrosomes in efficient asymmetric cell division [75].
The microtubule-dependent pathway requires astral microtubules, the plus-end-directed microtubule motor kinesin heavy chain 73 (Khc-73), and Dlg1, a membrane-associated guanylate kinase (MAGUK) protein, to recruit the Pins/Gαi complex to the apical cortex. NSCs lacking insc fail to localize the Par-complex to the apical cortex, yet they retain the ability to recruit Pins, Gαi, and Dlg1. Microtubule depolymerization results in a dose-dependent decrease in Pins/Gαi apical cortical localization in insc mutants, demonstrating a role for microtubules to polarize the NSC cortex. Genetic ablation of astral microtubules validated these findings [72]. To ensure mitotic spindle orientation, the Pins/Gαi complex interacts with the NuMA-related Mud protein [60–62]. Taken together, these data highlight the importance of centrosome-nucleated microtubules for NSC polarization and invariant spindle orientation (Figure 2; Centrosomes and polarity).
Indeed, there is significant cross-talk between centrosomes and NSC polarity. When apical polarity is disrupted in pins mutants, the apical centrosome is initially competent to nucleate astral microtubules, but is unable to maintain apical centrosome identity throughout interphase [45]. Likewise, when polarity is blocked, as in Ankle2 mutants or NSCs exposed to the Zika virus protein NS4A, which interacts with ANKLE2 protein, numerous centrosome phenotypes are observed, including centrosome amplification and misaligned spindle poles (Figure 2; Centrosomes and polarity) [21,22]. Disruption of the Ankle2 pathway generates microcephalic Drosophila larvae due to impaired polarization, reduced NSC divisions, excessive apoptosis, and a reduction in NSCs [76]. We speculate that the centrosome phenotypes also contribute to increased apoptosis (Figure 2; Centrosomes and polarity). In some cases, errant spindle morphogenesis leads to a failure to satisfy the spindle assembly checkpoint (SAC) and results in p53-mediated cell death [77]. Live imaging mitotic progression in Ankle2 vs control NSCs may further inform mechanisms of Ankle2-dependent microcephaly. Nonetheless, this work highlights the interplay between cortical polarity establishment and centrosome function, and loss of either axis can have devasting consequences on Drosophila and/or human brain development.
Genetic mutants and pharmacological experiments reveal that disruption of the cross-talk between centrosome activity and polarity cues results in deleterious consequences to asymmetric cell division. While loss of neither the microcephaly gene CDK5RAP2/centrosomin (cnn) nor the NuMA-related mud gene impairs polarization, mitotic spindle orientation becomes randomized (Figure 2; Centrosomes and polarity) [56,60]. Similarly, when astral microtubules are lost in asterless (asl) or anastral spindle 2 (ana2) mutants, or by treatment with microtubule antagonists, the polarity axis is no longer invariant [45,78,79]. When microtubules are destabilized using colchicine, centrosomes shed their PCM, migrate freely through the cell, and polarity is lost. Restoration of microtubule nucleation through UV-inactivation of colchicine, however, permits reactivation of the centrosome and the formation of a new polarity axis along a random axis dependent upon centrosome position, suggesting that the centrosome is responsible for the maintenance of the invariant orientation of the polarity axis [78]. In summary, centrosome microtubule-nucleating activity and cortical polarity are intimately linked through multiple, nonlinear pathways throughout interphase. Disruption of this centrosome-polarity cross-talk impairs asymmetric cell division. That being said, not all centrosome genes affect polarity but are still implicated in human microcephaly through other cellular processes.
Is centrosome asymmetry dispensable in Drosophila NSCs?
While loss of some microcephaly associated genes results in a similar phenotype in Drosophila, others do not. For example, loss of Drosophila spindle defective 2 (spd-2) [80,81], cnn [82], Cep135/bld-10 [83,84], or pericentrin-like protein (plp) [48,85] severely impairs centrosome function and NSC divisions, yet does not yield a microcephalic phenotype. Nevertheless, these genes, as well as polo kinase (polo) and centrobin (cnb), are critical for centrosome asymmetry in interphase NSCs [37,38,47–49,86,87]. WD repeat domain 62 (Wdr62) is also required for centrosome asymmetry; however, wdr62 mutant flies are microcephalic. Nonetheless, the microcephaly phenotype associated with wdr62 loss is likely due to prolonged cell divisions, not centrosome asymmetry [37,88].
Centrosome asymmetry is established through both positive and negative interactions. To generate spatial and temporal asymmetries, proteins that promote the recruitment/stability of PCM are enriched on the apical centrosome, such as Polo, Cnn, Spd-2, and Cnb (Figure 3, Wild-type interphase). Loss of one of these centrosome activators results in a stem cell with symmetrical centrosomes that act in a basal-like centrosome manner (Figure 3, middle row) [37,38,47,49,50,86]. The microcephaly gene Ninein (Nin)/Bsg25D is also asymmetrically localized to centrosomes when overexpressed, but appears dispensable for normal centrosome function [89], suggesting that not all asymmetrically localized proteins act directly on centrosome regulation. Conversely, PLP and Plk4/SAK, which promote PCM shedding, are enriched on the basal centrosome. When either of these proteins are lost, the resulting stem cell has two symmetrical centrosomes that act in an apical-like centrosome manner (Figure 3, bottom row) [48,86]. Conversely, overexpression of a centrosome activator such as Cnb, which is normally only enriched on the apical centrosome, generates symmetrically apical-like centrosomes [47]. Additionally, overexpression of SAK also generates symmetrical centrosomes, however, these centrosomes are inactive [86]. Intriguingly, Cep135, which is also required to promote the down-regulation of the basal centrosome, is uniformly distributed on apical and basal centrosomes. However, loss of Cep135 also up-regulates the activity of the basal centrosome, suggesting that Cep135 likely interacts with an asymmetrically regulated protein in order to generate these spatial asymmetries [38]. Although microcephaly is not observed in Drosophila mutants lacking most of these centrosome asymmetry genes, they do present with many mitotic defects. For example, defects in centrosome segregation, spindle orientation, and centrosome number are consistently observed when two symmetrical centrosomes are present (Figure 2; Centrosome asymmetry).
In humans, heritable microcephaly is most commonly associated with mutations in ASPM [90]. Loss of the Drosophila homolog abnormal spindle (asp) also results in microcephaly, as well as centrosome segregation and spindle orientation defects similar to the defects observed in centrosome asymmetry mutants [91]. Expression of a full-length asp transgene recues brain size and microtubule defects in asp mutants. In contrast, expression of an N-terminal asp fragment or a full-length transgene lacking a domain required for interaction with Calmodulin (aspΔIQ) rescues the microcephaly phenotype without rescuing the spindle morphology defects, suggesting the bent, unfocused spindles typical of asp mutants are insufficient to cause microcephaly — other mechanisms are at play [91]. Given that both centrosome asymmetry mutants and animals rescued of asp-dependent microcephaly have morphologically wild-type brains, it appears that Drosophila neurogenesis is resistant to certain perturbations of centrosome activity to which human neurogenesis may be more sensitive. Increased sensitization to microcephaly may arise in mammals, for example, because of additional microtubule-dependent functions, such as neuronal migration, required for cell positioning in the developing, stratified neocortex [92].
The SAC as a microcephaly fail-safe
Another intriguing hypothesis that we favor as to how Drosophila NSCs are able to resist failed asymmetric cell division involves the SAC. The SAC prevents misaligned or errant spindle poles (e.g. bent, monopolar, or multipolar microtubule spindles) from continuing through mitosis. This checkpoint is active in the presence of unattached kinetochores. Once all kinetochores are stably attached to microtubules, the cell cycle stall is lifted and the cell can proceed into anaphase [93]. We favor a mechanism in which the spindle orientation defects resulting from centrosome asymmetry loss are corrected prior to anaphase due to the ‘fail-safe’ action of the SAC. In many of these mutants, spindle orientation is defective and slight mitotic stalling is observed [37,56,94]. Is this due to a delay in satisfying the SAC? Through live imaging, the disorientated spindles can be seen to correctly orient themselves prior to anaphase [38,47,48,60], strongly suggesting a connection to the SAC. A pressing question in centrosome-regulated neurodevelopment is, therefore, what happens to centrosome mutants without this likely fail-safe?
Mitotic slippage occurs when components of the SAC are compromised, thereby allowing abnormal mitoses to proceed, typically resulting in chromosomal missegregation and genome instability [95]. Likewise, the requirement for proper centrosome regulation and activity to maintain genomic stability has been previously reviewed [96]. Although sas-4 mutants lack centrosomes, they proceed through larval neurogenesis and develop an average-sized brain [75]. However, if the SAC is bypassed through loss of mad2, the resulting sas-4,mad2 double-mutant is microcephalic [97]. It is important to note that mad2 mutant NSCs divide normally [98], highlighting that the microcephaly phenotype is due to a combination of the loss of centrosomes as well as loss of the SAC.
Centrosome amplification coupled with loss of the SAC also results in microcephaly. Centrosome amplification can arise from repeated rounds of centrosome duplication, failed centrosome segregation during cytokinesis, or failed cytokinesis [99,100]. Overexpression of the master kinase regulating centriole duplication, SAK, results in centrosome amplification [101–103]. When coupled with loss of the SAC through depletion of mad2, the resulting NSC divisions are significantly error-prone and genetically unstable (Figure 2; Spindle assembly checkpoint). Loss of mad2 paired with overexpression of SAK (mad2;SAKOE) causes aneuploidy as a consequence of lagging chromosomes/failed DNA segregation and cytokinesis failure. Brains that develop from mad2;SAKOE larva have fewer NSCs and are microcephalic, highlighting the critical role of centrosomes in maintaining genome integrity during cell division [94]. It is important to note that these NSCs still stall in mitosis, perhaps due to redundancy within the SAC. The loss of NSCs in these aneuploid models is not due to an increase in apoptosis or necropsy, but rather premature differentiation [94]. Overexpression of cell differentiation factors can also induce premature differentiation [56]; therefore, the extra chromosomes resulting from failed chromosome segregation may contribute to premature differentiation. Although only a few microcephaly genes have been tested in the mad2 background, others, such as cnn,mad2 double-mutants, do show aneuploidy [98], suggesting that the SAC is a fail-safe that prevents microcephaly in many of these models (Figure 2; Spindle assembly checkpoint).
Emerging roles of post-transcriptional control in preventing microcephaly
In the mammalian brain, defects in NSC proliferation, differentiation, and neuronal migration contribute to microcephaly and other neurodevelopmental disorders. Essential to these processes is the precise control of gene expression. While understanding the contributions of post-transcriptional regulation in brain development is an emergent field, many RNA-binding proteins implicated in diverse processes, including RNA editing, splicing, export, localization, translation, and turnover, are associated with microcephaly [104]. Likewise, recent work in Drosophila highlights post-transcriptional regulation of deadpan, pros, and Myc mRNAs is important for neurodevelopment [105–107]. In mammalian models, haploinsufficiency of three core exon-junction components (EJC; Magoh, Rbm8a, and Eif4a) results in microcephaly associated with prolonged progenitor cell cycles leading to progenitor loss, neural depletion, and increased rates of apoptosis [108–111]. Intriguingly, pharmacologically stalling NSC mitotic progression is sufficient to phenocopy these responses [108,112]. Although centrosomes are unaffected in EJC mutants [108], these studies raise the possibility that other mutations that alter mitotic progression, perhaps by altering the post-transcriptional regulation of centrosome genes, could similarly impair neurodevelopment.
The idea that post-transcriptional control of centrosome genes may influence neurodevelopment is supported by recent work highlighting the alternative splicing of Nin. Gene expression profiling uncovered alternatively spliced variants of the microcephaly gene Nin differentially expressed in mammalian progenitors versus neurons [113]. Nin localizes to the mother centriole and promotes its maturation and is conserved in mammals and Drosophila [89,114]. Zhang et al. found the Nin protein product encoded by the progenitor-enriched isoform localized to centrioles, whereas the neuronal variant remained cytoplasmic. Ectopic expression of the neuronal Nin variant led to premature differentiation and depletion of the neuronal progenitors [113]. These data reveal that alternative splicing generates variants of a centrosome gene that are differentially localized (centrosome versus cytoplasm) and expressed (progenitor versus neuron). Moreover, these findings provide a link between post-transcriptional regulation via alternative splicing to centrosome asymmetry within neural progenitors, as Nin localizes to the mother centriole. Interestingly, differential expression of Nin-orthologous Bsg25D isoforms was also noted in Drosophila NSCs versus neurons [115], although these variants await functional characterization. Alternative splicing coupled with differential expression may contribute to the regulation of other centrosome genes and influence neurodevelopment.
The mechanisms that regulate the spatial and temporal regulation of centrosome asymmetry throughout NSC asymmetric cell division remain incompletely understood. One intriguing hypothesis is that these rapid transitions in composition and organization are mediated, in part, by post-transcriptional mechanisms, which may include RNA localization and/or local RNA translation. For example, mRNAs of several centrosome genes, including Bsg25D mRNA, localize near centrosomes within syncytial Drosophila embryos [116]. For a comprehensive review on the relationship between RNA localization and centrosomes, we refer the reader to [117]. We speculate that mRNAs encoding positive or negative regulators of centrosome maturation may be preferentially enriched, locally translated, or stabilized at the apical versus basal NSC centrosome. Supporting this possibility, local translation of centrosome genes was recently reported in non-neuronal contexts [118,119]. We surmise that differential localization, translation, and/or stability of centrosome genes within NSCs would profoundly affect neurodevelopment and that dysregulation of these processes would likely contribute to pathogenic phenotypes, including microcephaly.
Mutations in several RNA-binding proteins, which often bind the 3′-untranslated regions (UTRs) of their target RNAs, are associated with human microcephaly [104]. Some of these microcephaly associated RNA-binding proteins are ontologically associated with centrosomes (Supplemental Tables S1 and S2). Likewise, a mutation in the 3′UTR of the human microcephaly gene MECP2 has also been identified in a patient with microcephaly [120]. Expanded use of whole-genome sequencing (as opposed to exome sequencing) of microcephaly patients may uncover additional causative mutations within UTRs. Moreover, these studies strongly suggest that mutations in RNA-binding proteins that impinge on centrosome gene regulation, or mutations within centrosome gene regulatory motifs (e.g. UTRs), likely also contribute to microcephaly.
While hundreds of RNA-binding proteins are expressed in the mammalian neonatal brain, only a handful are functionally characterized and most RNA targets await discovery [104]. As centrosome dynamics throughout the cell cycle clearly play a fundamental role in brain development, and RNA-binding proteins also contribute to the dynamic processes regulating neurodevelopment, whether disruption of RNA-binding proteins leads to dysregulation of centrosome activity represents a key unexplored mechanism of microcephaly. We predict that Drosophila models will continue to serve as valuable tools to address some of these critical questions. We are only just beginning to understand the mechanisms that govern centrosome regulation, and regulation by RNA-binding proteins is an intriguing paradigm to explore.
Summary
NSCs are neural progenitors required for neurogenesis that undergo asymmetric cell division along an invariant apical–basal polarity axis. Centrosomes are microtubule-organizing centers that orient and engineer the mitotic spindle required for NSC divisions. Deregulation of centrosome activity impairs multiple aspects of NSC divisions, including polarization, spindle orientation, spindle morphogenesis, and faithful segregation of the genome. Consequently, genetic lesions in centrosome genes represent the astounding majority of causative mutations associated with congenital human microcephaly. Studies in Drosophila NSC models have proved invaluable for the discovery of microcephaly genes and their pathophysiology, particularly with respect to centrosome function and regulation.
Supplementary Material
Perspectives.
Importance to field: NSCs are neural progenitors required for normal brain development whose stereotypical self-renewing divisions are regulated by centrosomes. Centrosome dysfunction depletes the NSC pool and is the leading cause of microcephaly, a neurodevelopmental disorder defined by a characteristically small brain size.
Summary of current thinking: Although well known for their roles in spindle orientation and organization, centrosomes impinge upon most aspects of NSC division.
Future directions: Dynamic centrosome regulation is still poorly understood in the context of the asymmetrically dividing NSC. Unknowns include how centrosomes are asymmetrically regulated in time and space, how Drosophila NSCs can overcome centrosome asymmetry defects that impair mammalian neurodevelopment, and whether RNA-binding proteins participate in the regulation of NSC centrosomes.
Acknowledgements
We thank members of the Lerit laboratory for insightful comments and discussions.
Funding
This work was supported by the Orphan Disease Center grant CDKL5–19-102–01 to V.F. and NIH grants T32GM008490 (B.V.R.), 1RF1AG060285 to V.F., and 5K22HL126922 to D.A.L.
Abbreviations
- EJC
exon-junction complex
- GO
gene ontology (http://geneontology.org/)
- HPO
human phenotype ontology (https://hpo.jax.org/)
- MTOC
microtubule-organizing center
- NSC
neural stem cell
- OMIN
online Mendelian inheritance in man (https://www.ncbi.nlm.nih.gov/omim)
- ORPHA
rare disease code from orpha.net (https://www.orpha.net/consor/cgi-bin/index.php)
- PCM
pericentriolar material
- SAC
spindle assembly checkpoint
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Allanson JE, Cunniff C, Hoyme HE, McGaughran J, Muenke M and Neri G. (2009) Elements of morphology: standard terminology for the head and face. Am. J. Med. Genet. A 149A, 6–28 10.1002/ajmg.a.32612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pirozzi F, Nelson B and Mirzaa G. (2018) From microcephaly to megalencephaly: determinants of brain size. Dialogues Clin. Neurosci. 20, 267–282 10.31887/DCNS.2018.20.4/gmirzaa [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Passemard S, Kaindl AM and Verloes A. (2013) Microcephaly. Handb. Clin. Neurol. 111, 129–141 10.1016/B978-0-444-52891-9.00013-0 [DOI] [PubMed] [Google Scholar]
- 4.Kohler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, Gourdine JP et al. (2019) Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 47, D1018–D1D27 10.1093/nar/gky1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CE, Singleton KS, Wallin M and Faundez V. (2020) Rare genetic diseases: nature’s experiments on human development. iScience 23, 101123 10.1016/j.isci.2020.101123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semichov SB (1984). [Theoretical basis of the newest American classification of mental disorders (diagnostic and statistical manual of mental disorders, 3d edition–DSM-III, Washington, D.C., APA, 1980, 494 p.)]. Zh. Nevropatol. Psikhiatr. Im. S. S. Korsakova 84, 457–461 PMID:6720187 [PubMed] [Google Scholar]
- 7.Dolk H. (1991) The predictive value of microcephaly during the first year of life for mental retardation at seven years. Dev. Med. Child Neurol. 33, 974–983 10.1111/j.1469-8749.1991.tb14813.x [DOI] [PubMed] [Google Scholar]
- 8.Watemberg N, Silver S, Harel S and Lerman-Sagie T. (2002) Significance of microcephaly among children with developmental disabilities. J. Child Neurol. 17, 117–122 10.1177/088307380201700205 [DOI] [PubMed] [Google Scholar]
- 9.Neurodevelopmental Disorders in Diagnostic and Statistical Manual of Mental Disorders, 5th ed. American Psychiatric Association, Arlington [Google Scholar]
- 10.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV et al. (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 14, 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z. et al. (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al. (2000) Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi H, Muruganujan A, Ebert D, Huang X and Thomas PD (2019) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–DD26 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Gene Ontology C. (2019) The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47(D1), D330–D3D8 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J. et al. (2005) A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37, 353–355 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- 16.Degrassi F, Damizia M and Lavia P. (2019) The mitotic apparatus and kinetochores in microcephaly and neurodevelopmental diseases. Cells 9, 49 10.3390/cells9010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marthiens V and Basto R. (2020) Centrosomes: the good and the bad for brain development. Biol. Cell 112, 153–172 10.1111/boc.201900090 [DOI] [PubMed] [Google Scholar]
- 18.Mochida GH (2009) Genetics and biology of microcephaly and lissencephaly. Semin. Pediatr. Neurol. 16, 120–126 10.1016/j.spen.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill RS, Schoborg TA and Rusan NM (2018) Same but different: pleiotropy in centrosome-related microcephaly. Mol. Biol. Cell 29, 241–246 10.1091/mbc.E17-03-0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homem CC, Repic M and Knoblich JA (2015) Proliferation control in neural stem and progenitor cells. Nat. Rev. Neurosci. 16, 647–659 10.1038/nrn4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah PS, Link N, Jang GM, Sharp PP, Zhu T, Swaney DL et al. (2018) Comparative flavivirus-host protein interaction mapping reveals mechanisms of Dengue and Zika virus pathogenesis. Cell 175, 1931–45.e18 10.1016/j.cell.2018.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Link N, Chung H, Jolly A, Withers M, Tepe B, Arenkiel BR et al. (2019) Mutations in ANKLE2, a ZIKA virus target, disrupt an asymmetric cell division pathway in Drosophila neuroblasts to cause microcephaly. Dev. Cell 51, 713–29.e6 10.1016/j.devcel.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devakumar D, Bamford A, Ferreira MU, Broad J, Rosch RE, Groce N. et al. (2018) Infectious causes of microcephaly: epidemiology, pathogenesis, diagnosis, and management. Lancet Infect. Dis. 18, e1–e13 10.1016/S1473-3099(17)30398-5 [DOI] [PubMed] [Google Scholar]
- 24.Nigg EA and Raff JW (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- 25.Palazzo RE, Vogel JM, Schnackenberg BJ, Hull DR and Wu X. (2000) Centrosome maturation. Curr. Top. Dev. Biol. 49, 449–470 10.1016/S0070-2153(99)49021-0 [DOI] [PubMed] [Google Scholar]
- 26.Khodjakov A and Rieder CL (1999) The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146, 585–596 10.1083/jcb.146.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nigg EA and Holland AJ (2018) Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19, 297–312 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nigg EA and Stearns T. (2011) The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13, 1154–1160 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu J, Hagan IM and Glover DM (2015) The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 7, a015800 10.1101/cshperspect.a015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dieks JK, Baumer A, Wilichowski E, Rauch A and Sigler M. (2014) Microcephalic osteodysplastic primordial dwarfism type II (MOPD II) with multiple vascular complications misdiagnosed as Dubowitz syndrome. Eur. J. Pediatr. 173, 1253–1256 10.1007/s00431-014-2368-5 [DOI] [PubMed] [Google Scholar]
- 31.Lorenzo-Betancor O, Blackburn PR, Edwards E, Vazquez-do-Campo R, Klee EW, Labbe C. et al. (2018) PCNT point mutations and familial intracranial aneurysms. Neurology 91, e2170–e2e81 10.1212/WNL.0000000000006614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blacque OE and Leroux MR (2006) Bardet–Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell Mol. Life Sci. 63, 2145–2161 10.1007/s00018-006-6180-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barkovich AJ, Guerrini R, Kuzniecky RI, Jackson GD and Dobyns WB (2012) A developmental and genetic classification for malformations of cortical development: update 2012. Brain 135(Pt 5), 1348–69 10.1093/brain/aws019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao MS, Carpenter M and Vemuri MC (2012) Neural Development and Stem Cells, Springer, New York [Google Scholar]
- 35.Lesage B, Gutierrez I, Marti E and Gonzalez C. (2010) Neural stem cells: the need for a proper orientation. Curr. Opin. Genet. Dev. 20, 438–442 10.1016/j.gde.2010.04.013 [DOI] [PubMed] [Google Scholar]
- 36.Jana SC, Bettencourt-Dias M, Durand B and Megraw TL (2016) Drosophila melanogaster as a model for basal body research. Cilia 5, 22 10.1186/s13630-016-0041-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair A R, Singh P, Salvador Garcia D, Rodriguez-Crespo D, Egger B and Cabernard C. (2016) The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in Drosophila neuroblasts. Cell Rep. 14, 1100–1113 10.1016/j.celrep.2015.12.097 [DOI] [PubMed] [Google Scholar]
- 38.Singh P, Ramdas Nair A and Cabernard C. (2014) The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr. Biol. 24, 1548–1555 10.1016/j.cub.2014.05.050 [DOI] [PubMed] [Google Scholar]
- 39.Thornton GK and Woods CG (2009) Primary microcephaly: do all roads lead to Rome? Trends Genet. 25, 501–510 10.1016/j.tig.2009.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sunkel CE (1988) Glover DM. polo, a mitotic mutant of Drosophila displaying abnormal spindle Poles. J. Cell Sci. 89(Pt 1), 25–38 PMID: 3417791 [DOI] [PubMed] [Google Scholar]
- 41.Li S, Wang H and Groth C. (2014) Drosophila neuroblasts as a new model for the study of stem cell self-renewal and tumour formation. Biosci. Rep. 34, e00125 10.1042/BSR20140008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knoblich JA (2010) Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11, 849–860 10.1038/nrm3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB and Shi SH (2009) Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461, 947–955 10.1038/nature08435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rusan NM and Peifer M. (2007) A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177, 13–20 10.1083/jcb.200612140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H and Gonzalez C. (2007) Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell 12, 467–474 10.1016/j.devcel.2007.01.021 [DOI] [PubMed] [Google Scholar]
- 46.Reina J and Gonzalez C. (2014) When fate follows age: unequal centrosomes in asymmetric cell division. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130466 10.1098/rstb.2013.0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Januschke J, Reina J, Llamazares S, Bertran T, Rossi F, Roig J. et al. (2013) Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15, 241–248 10.1038/ncb2671 [DOI] [PubMed] [Google Scholar]
- 48.Lerit DA and Rusan NM (2013) PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202, 1013–1022 10.1083/jcb.201303141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conduit PT and Raff JW (2010) Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20, 2187–2192 10.1016/j.cub.2010.11.055 [DOI] [PubMed] [Google Scholar]
- 50.Januschke J, Llamazares S, Reina J and Gonzalez C. (2011) Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2, 243 10.1038/ncomms1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marthiens V, Piel M and Basto R. (2012) Never tear us apart–the importance of centrosome clustering. J. Cell Sci. 125(Pt 14), 3281–92 10.1242/jcs.094797 [DOI] [PubMed] [Google Scholar]
- 52.Paridaen JT, Wilsch-Brauninger M and Huttner WB (2013) Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell 155, 333–344 10.1016/j.cell.2013.08.060 [DOI] [PubMed] [Google Scholar]
- 53.Broadus J, Fuerstenberg S and Doe CQ (1998) Staufen-dependent localization of prospero mRNA contrtibutes to neuroblast daughter-cell fate. Nature 391, 792–795 10.1038/35861 [DOI] [PubMed] [Google Scholar]
- 54.Vessey JP, Amadei G, Burns SE, Kiebler MA, Kaplan DR and Miller FD (2012) An asymmetrically localized Staufen2-dependent RNA complex regulates maintenance of mammalian neural stem cells. Cell Stem Cell 11, 517–528 10.1016/j.stem.2012.06.010 [DOI] [PubMed] [Google Scholar]
- 55.Freeman MR and Doe CQ (2001) Asymmetric Prospero localization is required to generate mixed neuronal/glial lineages in the Drosophila CNS. Development 128, 4103–4112 PMID: 11641232 [DOI] [PubMed] [Google Scholar]
- 56.Cabernard C and Doe CQ (2009) Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell 17, 134–141 10.1016/j.devcel.2009.06.009 [DOI] [PubMed] [Google Scholar]
- 57.Loyer N and Januschke J. (2020) Where does asymmetry come from? Illustrating principles of polarity and asymmetry establishment in Drosophila neuroblasts. Curr. Opin. Cell Biol. 62, 70–77 10.1016/j.ceb.2019.07.018 [DOI] [PubMed] [Google Scholar]
- 58.Schaefer M, Petronczki M, Dorner D, Forte M and Knoblich JA (2001) Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107, 183–194 10.1016/S0092-8674(01)00521-9 [DOI] [PubMed] [Google Scholar]
- 59.Yu F, Morin X, Cai Y, Yang X and Chia W. (2000) Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100, 399–409 10.1016/S0092-8674(00)80676-5 [DOI] [PubMed] [Google Scholar]
- 60.Siller KH, Cabernard C and Doe CQ (2006) The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 8, 594–600 10.1038/ncb1412 [DOI] [PubMed] [Google Scholar]
- 61.Izumi Y, Ohta N, Hisata K, Raabe T and Matsuzaki F. (2006) Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 8, 586–593 10.1038/ncb1409 [DOI] [PubMed] [Google Scholar]
- 62.Bowman SK, Neumuller RA, Novatchkova M, Du Q and Knoblich JA (2006) The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 10, 731–742 10.1016/j.devcel.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 63.Caussinus E and Hirth F. (2007) Asymmetric stem cell division in development and cancer. Prog. Mol. Subcell. Biol. 45, 205–225 10.1007/978-3-540-69161-7_9 [DOI] [PubMed] [Google Scholar]
- 64.Gonzalez C. (2007) Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 8, 462–472 10.1038/nrg2103 [DOI] [PubMed] [Google Scholar]
- 65.Ohshiro T, Yagami T, Zhang C and Matsuzaki F. (2000) Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature 408, 593–596 10.1038/35046087 [DOI] [PubMed] [Google Scholar]
- 66.Peng CY, Manning L, Albertson R and Doe CQ (2000) The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature 408, 596–600 10.1038/35046094 [DOI] [PubMed] [Google Scholar]
- 67.Albertson R and Dlg DC (2003) Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat. Cell Biol. 5, 166–170 10.1038/ncb922 [DOI] [PubMed] [Google Scholar]
- 68.Shen CP, Knoblich JA, Chan YM, Jiang MM, Jan LY and Jan YN (1998) Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev. 12, 1837–1846 10.1101/gad.12.12.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Atwood SX and Prehoda KE (2009) aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol. 19, 723–729 10.1016/j.cub.2009.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Betschinger J, Mechtler K and Knoblich JA (2003) The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422, 326–330 10.1038/nature01486 [DOI] [PubMed] [Google Scholar]
- 71.Siegrist SE and Doe CQ (2007) Microtubule-induced cortical cell polarity. Genes Dev. 21, 483–496 10.1101/gad.1511207 [DOI] [PubMed] [Google Scholar]
- 72.Siegrist SE and Doe CQ (2005) Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell 123, 1323–1335 10.1016/j.cell.2005.09.043 [DOI] [PubMed] [Google Scholar]
- 73.Doe CQ (2008) Neural stem cells: balancing self-renewal with differentiation. Development 135, 1575–1587 10.1242/dev.014977 [DOI] [PubMed] [Google Scholar]
- 74.Betschinger J and Knoblich JA (2004) Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 14, R674–R685 10.1016/j.cub.2004.08.017 [DOI] [PubMed] [Google Scholar]
- 75.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A. et al. (2006) Flies without centrioles. Cell 125, 1375–1386 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- 76.Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G. et al. (2014) A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159, 200–214 10.1016/j.cell.2014.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fong CS, Mazo G, Das T, Goodman J, Kim M, O’Rourke BP et al. (2016) 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. eLife 5, e16270 10.7554/eLife.16270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Januschke J and Gonzalez C. (2010) The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J. Cell Biol. 188, 693–706 10.1083/jcb.200905024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang C, Li S, Januschke J, Rossi F, Izumi Y, Garcia-Alvarez G. et al. (2011) An ana2/ctp/mud complex regulates spindle orientation in Drosophila neuroblasts. Dev. Cell 21, 520–533 10.1016/j.devcel.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 80.Giansanti MG, Bucciarelli E, Bonaccorsi S and Gatti M. (2008) Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 18, 303–309 10.1016/j.cub.2008.01.058 [DOI] [PubMed] [Google Scholar]
- 81.Dix CI and Raff JW (2007) Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17, 1759–1764 10.1016/j.cub.2007.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vaizel-Ohayon D and Schejter ED (1999) Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr. Biol. 9, 889–898 10.1016/S0960-9822(99)80393-5 [DOI] [PubMed] [Google Scholar]
- 83.Blachon S, Cai X, Roberts KA, Yang K, Polyanovsky A, Church A. et al. (2009) A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics 182, 133–144 10.1534/genetics.109.101709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mottier-Pavie V and Megraw TL (2009) Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol. Biol. Cell 20, 2605–2614 10.1091/mbc.e08-11-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martinez-Campos M, Basto R, Baker J, Kernan M and Raff JW (2004) The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165, 673–683 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gambarotto D, Pennetier C, Ryniawec JM, Buster DW, Gogendeau D, Goupil A. et al. (2019) Plk4 regulates centriole asymmetry and spindle orientation in neural stem cells. Dev. Cell 50, 11–24.e10 10.1016/j.devcel.2019.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li K and Kaufman TC (1996) The homeotic target gene centrosomin encodes an essential centrosomal component. Cell 85, 585–596 10.1016/S0092-8674(00)81258-1 [DOI] [PubMed] [Google Scholar]
- 88.Chen JF, Zhang Y, Wilde J, Hansen KC, Lai F and Niswander L. (2014) Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat. Commun. 5, 3885 10.1038/ncomms4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng Y, Mennella V, Marks S, Wildonger J, Elnagdi E, Agard D. et al. (2016) The Seckel syndrome and centrosomal protein Ninein localizes asymmetrically to stem cell centrosomes but is not required for normal development, behavior, or DNA damage response in Drosophila. Mol. Biol. Cell 27, 1740–1752 10.1091/mbc.e15-09-0655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nicholas AK, Swanson EA, Cox JJ, Karbani G, Malik S, Springell K. et al. (2009) The molecular landscape of ASPM mutations in primary microcephaly. J. Med. Genet. 46, 249–253 10.1136/jmg.2008.062380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schoborg T, Zajac AL, Fagerstrom CJ, Guillen RX and Rusan NM (2015) An Asp-CaM complex is required for centrosome-pole cohesion and centrosome inheritance in neural stem cells. J. Cell Biol. 211, 987–998 10.1083/jcb.201509054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bond J and Woods CG (2006) Cytoskeletal genes regulating brain size. Curr. Opin. Cell Biol. 18, 95–101 10.1016/j.ceb.2005.11.004 [DOI] [PubMed] [Google Scholar]
- 93.Lara-Gonzalez P, Westhorpe FG and Taylor SS (2012) The spindle assembly checkpoint. Curr. Biol. 22, R966–R980 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 94.Gogendeau D, Siudeja K, Gambarotto D, Pennetier C, Bardin AJ and Basto R. (2015) Aneuploidy causes premature differentiation of neural and intestinal stem cells. Nat. Commun. 6, 8894 10.1038/ncomms9894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rieder CL and Maiato H. (2004) Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 7, 637–651 10.1016/j.devcel.2004.09.002 [DOI] [PubMed] [Google Scholar]
- 96.Lerit DA and Poulton JS (2016) Centrosomes are multifunctional regulators of genome stability. Chromosome Res. 24, 5–17 10.1007/s10577-015-9506-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Poulton JS, Cuningham JC and Peifer M. (2017) Centrosome and spindle assembly checkpoint loss leads to neural apoptosis and reduced brain size. J. Cell Biol. 216, 1255–1265 10.1083/jcb.201607022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buffin E, Emre D and Karess RE (2007) Flies without a spindle checkpoint. Nat. Cell Biol. 9, 565–572 10.1038/ncb1570 [DOI] [PubMed] [Google Scholar]
- 99.Godinho SA, Kwon M and Pellman D. (2009) Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 28, 85–98 10.1007/s10555-008-9163-6 [DOI] [PubMed] [Google Scholar]
- 100.Godinho SA and Pellman D. (2014) Causes and consequences of centrosome abnormalities in cancer. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 369, 20130467 10.1098/rstb.2013.0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G and Godinho SA (2011) Centrosomes and cilia in human disease. Trends Genet. 27, 307–315 10.1016/j.tig.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Habedanck R, Stierhof YD, Wilkinson CJ and Nigg EA (2005) The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7, 1140–1146 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- 103.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD and Nigg EA (2007) Plk4-induced centriole biogenesis in human cells. Dev. Cell 13, 190–202 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 104.Lennox AL, Mao H and Silver DL (2018) RNA on the brain: emerging layers of post-transcriptional regulation in cerebral cortex development. Wiley Interdiscip. Rev. Dev. Biol. 7, e290 10.1002/wdev.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Komori H, Golden KL, Kobayashi T, Kageyama R and Lee CY (2018) Multilayered gene control drives timely exit from the stem cell state in uncommitted progenitors during Drosophila asymmetric neural stem cell division. Genes Dev. 32, 1550–1561 10.1101/gad.320333.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Samuels TJ, Arava Y, Jarvelin AI, Robertson F, Lee JY, Yang L. et al. (2020) Neuronal upregulation of Prospero protein is driven by alternative mRNA polyadenylation and Syncrip-mediated mRNA stabilisation. Biol. Open 9, bio049684 10.1242/bio.049684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Samuels TJ, Jarvelin AI, Ish-Horowicz D and Davis I. (2020) Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. eLife 9, e51529 10.7554/eLife.51529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pilaz LJ, McMahon JJ, Miller EE, Lennox AL, Suzuki A, Salmon E. et al. (2016) Prolonged mitosis of neural progenitors alters cell fate in the developing brain. Neuron 89, 83–99 10.1016/j.neuron.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ et al. (2010) The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat. Neurosci. 13, 551–558 10.1038/nn.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mao H, Pilaz LJ, McMahon JJ, Golzio C, Wu D, Shi L. et al. (2015) Rbm8a haploinsufficiency disrupts embryonic cortical development resulting in microcephaly. J. Neurosci. 35, 7003–7018 10.1523/JNEUROSCI.0018-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mao H, McMahon JJ, Tsai YH, Wang Z and Silver DL (2016) Haploinsufficiency for core exon junction complex components disrupts embryonic neurogenesis and causes p53-mediated microcephaly. PLoS Genet. 12, e1006282 10.1371/journal.pgen.1006282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mitchell-Dick A, Chalem A, Pilaz LJ and Silver DL (2019) Acute lengthening of progenitor mitosis influences progeny fate during cortical development in vivo. Dev. Neurosci. 41, 300–317 10.1159/000507113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang X, Chen MH, Wu X, Kodani A, Fan J, Doan R. et al. (2016) Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell 166, 1147–62.e15 10.1016/j.cell.2016.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Delgehyr N, Sillibourne J and Bornens M. (2005) Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118(Pt 8), 1565–75 10.1242/jcs.02302 [DOI] [PubMed] [Google Scholar]
- 115.Berger C, Harzer H, Burkard TR, Steinmann J, van der Horst S, Laurenson AS et al. (2012) FACS purification and transcriptome analysis of Drosophila neural stem cells reveals a role for Klumpfuss in self-renewal. Cell Rep. 2, 407–418 10.1016/j.celrep.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T. et al. (2007) Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 117.Ryder PV and Lerit DA (2018) RNA localization regulates diverse and dynamic cellular processes. Traffic 19, 496–502 10.1111/tra.12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sepulveda G, Antkowiak M, Brust-Mascher I, Mahe K, Ou T, Castro NM et al. (2018) Co-translational protein targeting facilitates centrosomal recruitment of PCNT during centrosome maturation in vertebrates. eLife 7, e34959 10.7554/eLife.34959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bergalet J, Patel D, Legendre F, Lapointe C, Benoit Bouvrette LP, Chin A. et al. (2020) Inter-dependent centrosomal co-localization of the cen and ik2 cis-natural antisense mRNAs in Drosophila. Cell Rep. 30, 3339–52.e6 10.1016/j.celrep.2020.02.047 [DOI] [PubMed] [Google Scholar]
- 120.Coutinho AM, Oliveira G, Katz C, Feng J, Yan J, Yang C. et al. (2007) MECP2 coding sequence and 3’UTR variation in 172 unrelated autistic patients. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144B, 475–483 10.1002/ajmg.b.30490 [DOI] [PubMed] [Google Scholar]
- 121.Harripaul R, Vasli N, Mikhailov A, Rafiq MA, Mittal K, Windpassinger C. et al. (2018) Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol. Psychiatry 23, 973–984 10.1038/mp.2017.60 [DOI] [PubMed] [Google Scholar]
- 122.Hu H, Kahrizi K, Musante L, Fattahi Z, Herwig R, Hosseini M. et al. (2019) Genetics of intellectual disability in consanguineous families. Mol. Psychiatry 24, 1027–1039 10.1038/s41380-017-0012-2 [DOI] [PubMed] [Google Scholar]
- 123.Santos-Cortez RLP, Khan V, Khan FS, Mughal ZU, Chakchouk I, Lee K, et al. (2018) Novel candidate genes and variants underlying autosomal recessive neurodevelopmental disorders with intellectual disability. Hum. Genet. 137, 735–752 10.1007/s00439-018-1928-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.