Abstract
Copy number variations (CNVs) of the human 16p11.2 genetic locus are associated with a range of neurodevelopmental disorders, including autism spectrum disorder, intellectual disability and epilepsy. In this review, we delineate genetic information and diverse phenotypes in individuals with 16p11.2 CNVs, and synthesize preclinical findings from transgenic mouse models of 16p11.2 CNVs. Mice with 16p11.2 deletions or duplications recapitulate many core behavioral phenotypes, including social and cognitive deficits, and exhibit altered synaptic function across various brain areas. Mechanisms of transcriptional dysregulation and cortical maldevelopment are reviewed, along with potential therapeutic intervention strategies.
Keywords: 16p11.2 deletion and duplication, autism spectrum disorders, clinical phenotypes, mouse models, prefrontal cortex
The Link between 16p11.2 CNVs and Neurodevelopmental Disorders
Genetic factors comprise a large proportion of the risk for neurodevelopmental disorders (NDDs) such as autism spectrum disorder (ASD), schizophrenia, and intellectual disability (ID) [1]. Copy number variations (CNVs, i.e. deletion or duplication) of various susceptible genetic loci predispose individuals to these NDDs and other developmental abnormalities [2, 3]. The human 16p11.2 gene locus (chromosome 16, position 11.2) is a ~500–600 kb region containing 27–29 genes [4–7] located on the proximal short arm of chromosome 16 [8]. Deletions and duplications of 16p11.2 have highly pleiotropic phenotypic effects, with strong links to ASD [8–19], ID [8, 10, 12, 14–16, 19–21], motor/developmental delay [8–10, 12, 13, 15–19, 22], dysmorphic features [8, 12, 16, 17, 22], and epilepsy/seizures [10, 12, 17]. Deletions of 16p11.2 are associated with increased head circumference (macrocephaly) [10, 12, 17, 23] and obesity [10, 12, 16, 20], whereas duplications often result in below average head size (microcephaly) [10, 17, 23, 24] and low body weight/BMI [10, 24]. Schizophrenia (SZ) appears to be associated more strongly with 16p11.2 duplications [9, 25–27].
The literature on 16p11.2 CNVs is large and growing. This review provides a synthesis of the most common neurodevelopmental phenotypes associated with 16p11.2 deletions/duplications (Figure 1). Studies performed in animal models of 16p11.2 CNVs have also begun highlighting core neurobiological mechanisms. Here we discuss key pathways and mechanisms of dysfunction identified through these studies (Figure 2 and Figure 3), which may provide explanations for some of the phenomena observed in human 16p11.2 CNVs.
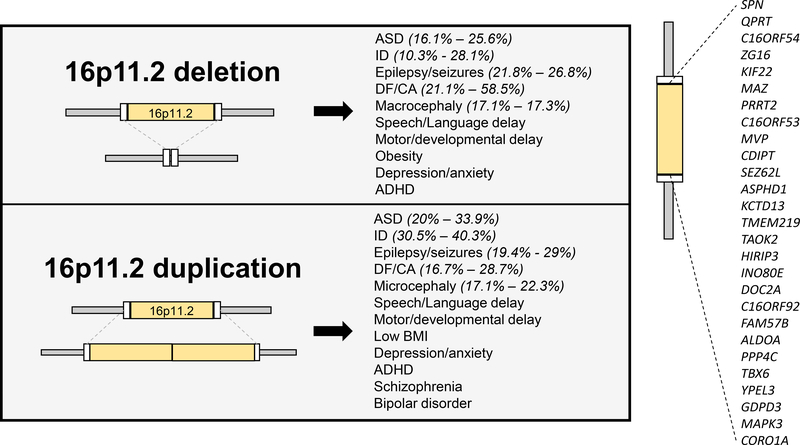
Figure 1. Common phenotypes in carriers of 16p11.2 CNVs.
Both 16p11.2 deletions and duplications predispose individuals to ASD, ID, epilepsy/seizures, and DF/CA with high penetrance, in addition to several other common phenotypes. Numbers inside parentheses indicate ranges of reported penetrance within cohorts of 16p11.2 CNV patients. Inset: Genes in the human 16p11.2 region. ASD: autism spectrum disorder; ID: intellectual disability; DF/CA: dysmorphic features/congenital anomalies.
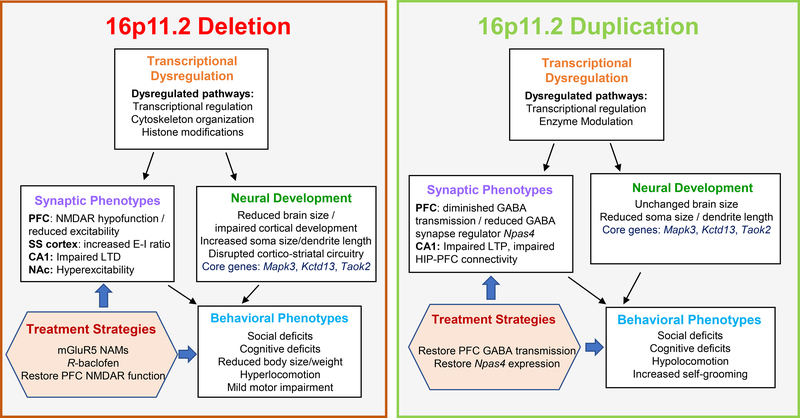
Figure 2. Summary of main preclinical findings from mouse models of 16p11.2 deletion and duplication.
Studies in 16p11.2 CNV transgenic mice have identified several core behavioral abnormalities, along with associated neurobiological disruptions. The deletion or duplication of genes within the 16p11.2 region drives transcriptional dysregulation of various downstream pathways. These transcriptional changes in turn lead to the altered developmental trajectories and synaptic changes across distributed brain regions. A range of behavioral phenotypes are reported in both 16p11.2 deletion and duplication models, likely driven by structural/functional neurological changes. Preclinical studies have identified several therapeutic approaches targeting selected disrupted systems. *KEY FIGURE
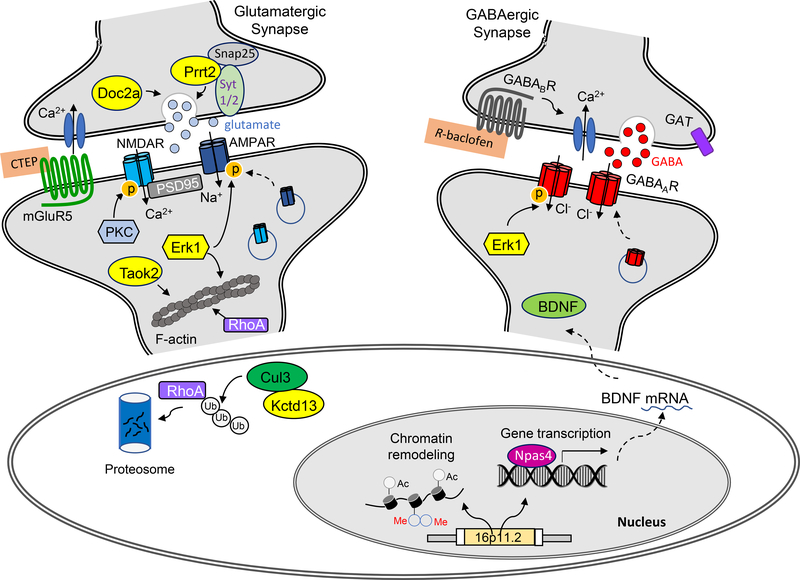
Figure 3. Schematic showing molecular and cellular mechanisms of 16p11.2 genes and interacting partners in nucleus, soma and synapses.
Proteins encoded by genes in 16p11.2 region are labeled with yellow. At glutamatergic synapses, Prrt2 and Doc2a are involved in regulating presynaptic transmitter release. TaoK2 and Erk1 act on postsynaptic actin cytoskeleton via RhoA signaling, which contributes to the deficits of spine structure and synaptic excitation associated with 16p11.2 deletions. Kctd13 is an adaptor of the E3 ligase Cul3, controlling the degradation of RhoA and other protein substrates. The therapeutic agent CTEP acts on mGluR5 to modify glutamatergic responses. At GABAergic synapses, the therapeutic agent R-baclofen acts on presynaptic GABAB receptors to regulate GABA release. Erk1 regulates the phosphorylation of GABAA receptor channels. In the nucleus, 16p11.2 CNVs could induce the alterations of chromatin remodeling and gene transcription. The expression of Npas4, a key regulator of GABA synapses via BDNF, is downregulated in 16p11.2 duplication mice, which contributes to the deficits of synaptic inhibition.
Analysis of Clinical Data from Humans with 16p11.2 CNVs
Prevalence of 16p11.2 CNVs
Genetics Home Reference estimates that 16p11.2 deletions and duplications each affect about 3 in every 10,000 individuals [28, 29]. A predictive algorithm estimated 16p11.2 deletions to affect 1 in every 3,021 live births, and duplications 1 in every 4,216 [30]. These estimates are supported by large genetic screenings [20, 24, 31]. 16p11.2 deletions have been reported at rates of 0.028% - 0.043% in the general population, while duplications have been reported between 0.035% - 0.053% (Table 1). Triplications of 16p11.2 have also been reported [32], though the frequency of this presumably rarer CNV is unknown.
Table 1.
Prevalence of 16p11.2 CNVs in the general population and within groups of clinical cohorts with various neurodevelopmental disorders.
| Ref. | 16p11.2 deletion prevalence (%) | 16p11.2 duplication prevalence (%) | Sample Group Description |
|---|---|---|---|
| General population data | |||
| [31] | 110/396,725 (0.028) | 138/396,725 (0.035) | General population (UK Biobank) |
| [24] | 25/58,635 (0.043) | 31/58,635 (0.053) | General European population |
| [20] | 4/11,856 (0.034) | - | General Swiss, Finnish, and Estonian population |
| Clinical cohorts | |||
| [17] | 27/7,400 (0.36) | 18/7,400 (0.24) | DD/MR, DF, seizures, CA, ASD, ADHD, or failure to thrive. |
| [26] | 98/38,779 (0.25) | 59/38,779 (0.15) | Unexplained physical and/or intellectual disabilities, with or without DF. |
| [20] | 9/312 (2.9) | - | CA and/or DD with obesity |
| [20] | 22/3,947 (0.56) | - | CA and/or DD without obesity |
| [20] | 11/2,772 (0.40) | - | Childhood/adult obesity (combined datasets) |
| [24] | 119/31,424 (0.38) | 73/31,424 (0.23) | DD/ID |
| [15] | 20/3,450 (0.58) | - | DD, ID, DF or MCA |
| [33] | 14/4,284 (0.33) | - | MR or multiple congenital anomalies |
| [16] | 45/9,773 (0.46) | 32/9,773 (0.33) | ASD, DD, DF, CA or seizures |
| [13] | 5/512 (0.98%) | 4/512 (0.78%) | DD, MR, or suspected ASD |
DD: developmental delay; MR: mental retardation; DF: dysmorphic features; (M)CA:(multiple) congenital anomalies; ASD: autism spectrum disorder; ID: intellectual disability
Genetic screenings of individuals with NDDs find starkly higher rates of 16p11.2 CNVs. Data compiled from eight studies [13, 15–17, 20, 24, 26, 33] screening individuals with either ASD, ID, developmental delay (DD), dysmorphic features (DF), multiple congenital anomalies (MCA), obesity, or seizures find 16p11.2 deletions at rates of 0.25–2.9%, and duplications at rates between 0.15–0.78% (Table 1).
Inheritance Patterns of 16p11.2 CNVs
We compiled data from 13 studies in which the rates of de novo vs. inherited cases of 16p11.2 deletions and duplications were reported [10, 12, 13, 15–17, 19, 20, 26, 33–36]. In studies with at least 50 subjects, between 60–76% of 16p11.2 deletions were reported as de novo events [10, 12, 34, 36], while only 16–29% of 16p11.2 duplications occurred de novo [10, 19, 34, 36]. Both the deletion and duplication appear to be preferentially maternally inherited.
Differing pathological severity of 16p11.2 deletions/duplications may underlie these dissimilar inheritance patterns. One study estimates that the penetrance of any pediatric phenotype in deletion carriers is 62.4%, relative to only 11.2% in duplication carriers [37]. The more severe outcomes associated with 16p11.2 deletions preclude these patients from having children, resulting in lower rates of inherited deletions [34]. However, de novo and inherited deletion carriers do not perform differently on several cognitive tasks [35]. De novo and inherited duplication carriers also show no differences in most clinical outcomes [19], and on cognitive tests [10]. Altogether, these results suggest that 16p11.2 deletions are more likely to be de novo events than duplications.
Neurodevelopmental Phenotypes Associated with 16p11.2 CNVs
Autism spectrum disorder (ASD)
Genetic screenings of ASD patients repeatedly identify 16p11.2 deletions & duplications, placing them among the strongest genetic risk factors for ASD [11, 13, 16, 38]. Data accumulated from 14 studies on the penetrance of ASD in 16p11.2 deletion and duplication carriers are shown in Table 2 [9, 10, 12, 14–17, 19–21, 23, 33–35]. In multiple studies of subjects with 16p11.2 deletion or duplication, autistic features or a formal ASD diagnosis were reported in 16.1% - 25.6% of deletion carriers, and 20% - 33.9% of duplication carriers. Overall, ASD appears to be a highly penetrant phenotype in both 16p11.2 duplications and deletions.
Table 2.
Penetrance of neurodevelopmental disorders and physical abnormalities among 16p11.2 deletion and 16p11.2 duplication carriers.
| # 16p11.2 deletion carriers |
# 16p11.2 duplication carriers |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ref. | ASD | ID | E/S | Mac. | DF/CA | ASD | ID | E/S | Mic. | DF/CA |
| [34] | 41/217 | 61/217 | - | - | - | 26/114 | 36/114 | - | - | - |
| [35] | 3/62 | - | - | - | - | 5/44 | - | - | - | - |
| [23] | 4/25 | 5/25 | - | 6/25 | - | 1/17 | 1/17 | - | 4/17 | - |
| [17] | 3/11 | - | 5/16 | 11/16 | 5/16 | 0/10 | - | 3/10 | 6/10 | 5/10 |
| [14] | 0/14 | 4/14 | - | - | - | 2/17 | 3/17 | - | - | - |
| [10] | 51/317 | - | 69/317 | - | 67/317 | 36/180 | 47/154 | 35/180 | 48/215 | 30/180 |
| [15] | 6/21 | - | - | - | - | - | - | - | - | - |
| [21] | 20/78 | 8/78 | - | - | - | - | - | - | - | - |
| [33] | 0/14 | - | 3/14 | - | 9/14 | - | - | - | - | - |
| [16] | 9/16 | - | 2/18 | - | - | - | - | 1/10 | - | - |
| [20] | 4/24 | - | - | - | - | - | - | - | - | - |
| [12] | 8/55 | - | 47/195 | 29/170 | 76/130 | - | - | - | - | - |
| [9] | - | - | - | - | - | 4/21 | - | - | - | - |
| [19] | - | - | - | - | - | 21/62 | 25/62 | - | - | - |
| [36] | - | - | 22/82 | 14/81 | - | - | - | 22/76 | 13/76 | - |
| [24] | - | - | - | - | - | - | - | - | - | 29/101 |
ASD: autism spectrum disorder; ID: intellectual disability; E/S: epilepsy/seizures; Mac: macrocephaly; Mic: microcephaly; DF/CA: dysmorphic features/congenital anomalies.
Intellectual Disability/Cognitive Impairment
Table 2 summarizes findings from 6 studies on the penetrance of ID in 16p11.2 deletion and duplication carriers [10, 14, 19, 21, 23, 34]. In studies of at least 50 subjects, ID was reported in 10.3% - 28.1% of 16p11.2 deletion carriers, and 30.5% - 40.3% of duplication carriers. Note that several of these studies were conducted in clinically ascertained populations of 16p11.2 CNV carriers, so the estimate of ID penetrance may be exaggerated.
A study of 270 duplication carriers and 442 deletion carriers found that FSIQ was significantly lower than intrafamilial controls in both 16p11.2 duplication carriers (18.0 points) and deletion carriers (22.1 points) [10]. Several supporting studies also reported lower FSIQ scores in 16p11.2 deletion [12, 35] and duplication [19, 35] patients than intrafamilial controls. Higher variability in FSIQ scores is observed among duplication carriers than deletion carriers [10, 19], suggesting a broader range of cognitive impairments in duplication patients. In one group of 16p11.2 duplication carriers, FSIQ scores range from 33–129, with 17% displaying severe impairment (<55), while 33% fall within the average range or better [19]. Interestingly, 16p11.2 duplication patients with ASD display more severe cognitive impairments than those without ASD [10], indicating that the severity of cognitive phenotypes may correlate with the pathogenesis of other neurodevelopmental disorders, particularly ASD.
Epilepsy
Epilepsy/seizures ranks among the most common phenotypes observed in both 16p11.2 deletions and duplications [10, 12, 16, 17, 33, 36] (Table 2). In studies of at least 50 subjects, epilepsy/seizures has been reported in 21.8% - 26.8% of deletion carriers, and 19.4% - 29% of duplication carriers. Epilepsy is thus a highly penetrant phenotype in 16p11.2 deletions and duplications.
Developmental Delay/Language Impairment
Motor delays are reported at rates of 57.1% [15] and 50% [17] in deletion carriers, and in 60% of duplication carriers [17]. A very delayed (>24 mo.) age of first walking is observed in 6.1% of deletion carriers and 15.9% of duplication carriers [10]. Balance impairment and gait abnormalities are reported in both 16p11.2 CNVs [39]. Developmental coordination disorder is reported at rates of 32% [23] and 57.7% [21] in deletion carriers, and 46.8% in duplication carriers [19]. Speech articulation abnormalities [36], phonological processing disorder [21, 23], language or communication disorder [14, 21, 23], speech or language deficits [15, 16], and speech delay [17, 33] are present at high rates in deletion carriers. Duplications are similarly associated with speech articulation abnormalities [36], speech delay [17, 19], and language deficits [16]. One study performed deep phenotyping of speech and language skills in 16p11.2 deletion carriers, providing further insight into the specific impairments present [40]. Interestingly, motor control of speech is impaired in 16p11.2 deletion carriers [41], indicating that the motor and language deficits may be fundamentally linked.
Schizophrenia/Psychosis
In a 2009 study, 21 duplications and 1 deletion were detected among 5,877 schizophrenia (SZ) patients, indicating a 14.5-fold increased SZ risk in 16p11.2 duplication patients [9]. One follow-up study reported 6 duplications in 659 SZ patients [42]. An analysis of 22 screenings in various populations found 98 duplications in 36,676 SZ patients (0.26%), with only 12 duplications in 48,331 controls (0.025%), indicating a 10.79-fold increase in risk for developing SZ [43]. Psychotic symptoms have also been reported in duplication (7/114 [6.1%]) and deletion (5/217 [2.3%]) carriers [34]. In one study, both 16p11.2 deletion and duplication carriers exhibited psychotic symptoms, though only the duplication was a significant predictor of psychotic symptoms [44]. Thus, 16p11.2 duplications predispose robustly to SZ and associated psychotic symptoms, with greater penetrance than 16p11.2 deletions. The link between 16p11.2 duplications and SZ is particularly interesting, considering the growing perspective of SZ as a neurodevelopmental disorder [45]. The overlapping genetic predisposition by 16p11.2 CNVs to both SZ and more typical NDDs like ASD and ID appears to support a neurodevelopmental origin of SZ [46].
Other psychiatric phenotypes in 16p11.2 CNVs
16p11.2 deletions and duplications are linked to several other psychiatric conditions, including depression [14, 47] and anxiety [12, 14, 18, 19, 23, 34]. Attention-deficit/hyperactivity disorder (ADHD) is also observed in 16p11.2 CNV carriers [12, 15–17, 19, 23, 48], at rates as high as 29% (63/217) and 42% (48/114) in deletion and duplication carriers, respectively [34]. Bipolar disorder is also reported in 16p11.2 duplication carriers [9, 14, 24].
Dysmorphic features/congenital anomalies
Summarized data from published reports on dysmorphic features/congenital anomalies (DF/CA) in 16p11.2 CNVs are shown in Table 2 [10, 12, 17, 24, 33]. In studies of at least 50 subjects, DF/CA were reported in in 21.1% - 58.5% of deletion carriers, and 16.7% - 28.7% of duplication carriers, indicating high penetrance in both CNVs.
Obesity/Body Mass Index (BMI)
Several reports indicate higher BMI in 16p11.2 deletion patients [10, 12, 23]. 16p11.2 deletions are present at higher-than-expected rates in obese individuals [31], and are more prevalent in developmental delay (DD) patients with obesity than those without obesity [20]. Conversely, BMI of 16p11.2 duplication patients is significantly lower than controls [10, 24]. Birth parameters for 16p11.2 duplication carriers are generally normal [24], indicating a postnatal effect.
Cephalic phenotypes & neurostructural changes
Certain physical phenotypes, such as head size, display dosage-dependent effects in 16p11.2 CNVs. Deletions of 16p11.2 are associated with macrocephaly [12, 17, 23, 36], while duplications are associated with microcephaly [10, 17, 23]. Summarized data on penetrance of microcephaly/macrocephaly in 16p11.2 CNVs are shown in Table 2 [10, 12, 17, 23, 36]. In studies of at least 50 subjects, macrocephaly was reported in 17.1% - 17.3% of deletion carriers, while microcephaly was reported in 17.1% - 22.3% of duplication carriers. Several studies also report increased or decreased head circumference in deletion [10, 12, 24, 36] or duplication carriers [10, 24], respectively.
MRI studies show reciprocal brain volume changes, with increased gray/white matter in 16p11.2 deletions, and decreased gray/white matter in 16p11.2 duplications [14, 23, 49, 50]. The increased or decreased axial diffusivity of white matter and the thickening or thinning of the corpus callosum have also been found in deletion or duplication patients, respectively [51–53]. Regional volumetric differences are also found in 16p11.2 CNV humans. Several brain areas exhibit the increased volume in deletion carriers and reduced volume in duplications, including insula, calcarine cortex [49, 50], accumbens, pallidum [54], transverse temporal gyrus [49, 50], caudate, putamen [49, 54], and thalamus [23]. Increased or decreased cortical surface area has been reported in 16p11.2 deletions or duplications, respectively [14, 23, 49]. 16p11.2 duplications also have the reduced cortical thickness [14, 23], reduced hippocampal volume [49] and enlarged ventricles [50, 53, 55].
Sex differences in 16p11.2 CNVs
Several neurodevelopmental disorders display a sex bias where males are at a higher risk [56]. In agreement with this, the ratio of males to females with either ASD or ID shows a male predominance in both 16p11.2 CNVs [57], suggesting that being female is a protective factor when predicting overall ASD severity in either 16p11.2 CNV [58]. However, another study found that sex was not a significant predictor of psychosis in 16p11.2 CNV carriers [44]. Thus, the association between 16p11.2 CNVs and ASD/ID, but not psychosis, may involve a sex bias.
Insights from Preclinical Studies in 16p11.2 CNV Mouse Models
Here we describe behavioral phenotypes in 16p11.2 CNV mouse models, and synthesize the major biological takeaways and implications (Figure 2 and Figure 3). Three lines of 16p11.2 CNV mice have been generated: 16p11.2 mice (Mills) carrying deletion (16p11.2+/−) or duplication (16p11.2dp/+) of 7F4 region (Slx1b-Sept1) syntenic to human 16p11.2 [5]; 16p11.2 mice (Dolmetsch) with deletion of the Coro1a-Spn interval [4]; 16p11.2 mice (Herault) with deletion/duplication of the Sult1a1-Spn genetic interval [59]. Behavioral phenotypes in 16p11.2 deletion or duplication mice are summarized in Table 3.
Table 3.
Behavioral phenotypes present in various mouse models carrying 16p11.2 deletion (16p11.2+/−) or duplication (16p11.2dp/+).
| Ref. | Mouse origin/genetic interval | Behavioral Phenotypes |
|---|---|---|
| 16p11.2+/− mice | ||
| [5] | Mills lab Slx1b-Sept1 |
• Reduced body size/body weight • Hyperlocomotion |
| [62] | Mills lab Slx1b-Sept1 |
• Barnes maze deficits • Reduced social approach |
| [61] | Mills lab Slx1b-Sept1 |
• Reduced body weight • More complex locomotor trajectory/shorter latency to approach stimulus • Deficits in righting from upside-down position • Normal 3 chamber social preference/social novelty recognition • Normal startle/pre-pulse inhibition (PPI) |
| [67] | Mills lab Slx1b-Sept1 |
• Delayed learning in FR1 continuous reinforcement nose-poke task (males only) • Earlier response termination in a progressive ratio nose-poke task (males only) • Deficits in the five-choice serial reaction time test (5-CSRTT) • No difference in sucrose preference test |
| [70] | Mills lab Slx1b-Sept1 |
• Hyperlocomotion over 24-hour period (both sexes). • Reduced sleep time (males only) and less time in NREM sleep (REM normal) |
| [66] | Mills lab Slx1b-Sept1 |
• Reduced open arm time in the elevated plus maze • Novel object recognition deficits |
| [65] | Mills lab Slx1b-Sept1 |
• Deficits in contextual fear conditioning (reduced freezing). • Impaired memory acquisition and extinction in inhibitory avoidance task |
| [64] | Mills lab Slx1b-Sept1 |
• Impaired object recognition memory • Reduced freezing in context-dependent aversive learning task • Open field hyperlocomotion |
| [64] | Dolmetsch lab Coro1a-Spn |
• Deficient object location memory • Reduced male-to-female nose-to-nose/nose-to-anogenital sniffing, following time, following bouts, and ultrasonic vocalizations. • Normal open field locomotion |
| [4] | Dolmetsch lab Coro1a-Spn |
• Reduced body length/body weight • Severely impaired startle response • Lack of gait fluidity; frequent tremor • Hyperlocomotion in home cage and in an activity chamber • Increased hanging; reduced grooming; reduced resting; increased circling behavior • Impaired novel object recognition |
| [63] | Dolmetsch lab Coro1a-Spn |
• Reduced body weight • Reduced male-to-female anogenital sniff, follow time, and ultrasonic vocalizations (mixed-genotype housed only) • Increased open arm time in elevated plus maze (mixed-genotype housed only) • Reduced immobility time in tail suspension test (mixed-genotype housed only) • Impaired novel object recognition (mixed-genotype housed only) • Impaired object location memory (mixed-genotype housed only) • Increased arena exploration • Normal 3 chamber social preference |
| [60] | Dolmetsch lab Coro1a-Spn |
• Reduced body weight • Reduced male-female ultrasonic vocalizations, anogenital sniff, and follow • Severe deficits in startle response and pre-pulse inhibition • Deafness, confirmed via auditory brainstem response test • Delayed hot plate response (reduced pain sensitivity) • Normal olfaction |
| [68] | Dolmetsch lab Coro1a-Spn |
• Impaired novel object recognition • Impaired object location memory • Mild social novelty recognition deficits • Delayed acquisition & reversal in a touch screen discrimination task • Inability to swim • Normal contextual and cued fear conditioning |
| [59] | Herault lab Sult1a1-Spn |
C57BL/6N inbred background • Increased dark cycle vertical activity • Increased time in center of open field • Increased jumping and rearing • Impaired object recognition memory • Increased hindlimb errors in notched bar test. • Normal social interaction |
| C57BL/6N-C3B hybrid background • Increased dark cycle ambulatory/vertical activity and light cycle vertical activity • Increased climbing behavior • Reduced social interaction • Impaired object recognition memory • Normal rotarod motor coordination • Normal open field locomotion and time in center |
||
| 16p11.2dp/+ mice | ||
| [5] | Mills lab Slx1b-Sept1 |
• Hypolocomotion • Increased grooming |
| [72] | Mills lab Slx1b-Sept1 |
• 3 chamber social preference deficits (normal social novelty recognition) • Social approach deficits • Temporal order recognition memory (TORM) deficits • Increased self-grooming • Hypolocomotion. • Normal novel object recognition (NOR) • Normal startle response/PPI • Lack of MK-801-induced hyperlocomotion • Normal elevated plus maze and rotarod |
| [73] | Mills lab Slx1b-Sept1 |
• Hypolocomotion (males only) • Reduced time in center of open field (males only) • Reduced ratio of open:closed arm time in elevated plus maze (males only) • Normal startle response • Reduced PPI (females only) • Increased distance between cagemates • Reduced time spent in close proximity to cagemates • Impaired performance in “N-back” WM task • Impaired performance in continuous performance task |
| [59] | Herault lab Sult1a1-Spn |
C57BL/6N inbred background • Reduced light cycle ambulatory/vertical activity and dark cycle vertical activity • Open field hypolocomotion and reduced time in center • Enhanced object recognition memory • Normal social interaction • Normal motor coordination in notched bar test |
| C57BL/6N-C3B hybrid background • Reduced dark cycle vertical activity • Reduced social interaction • Decreased climbing behavior • Enhanced object recognition memory • Normal open field locomotion/time in center • Normal rotarod motor coordination |
||
Behavioral Phenotypes in 16p11.2 Deletion or Duplication Mice Recapitulate Neurodevelopmental Deficits of Human Carriers
All 3 lines of 16p11.2+/− mice display social deficits in various testing paradigms [59–64], and an array of cognitive deficits [4, 59, 62–68]. Sleep abnormalities [69, 70] and anxiety [66] have been reported in 16p11.2+/− mice (Mills). Startle response [4, 60] and pre-pulse inhibition (PPI) [60] are severely impaired in the 16p11.2+/− mice (Dolmetsch). However, these mice are deaf [60], which likely underlies startle/PPI deficits and may be related to altered ultrasonic vocalizations. Mild motor deficits are also present in these models [4, 61, 68], consistent with human phenotypes. However, 16p11.2+/− models show reduced body size/weight [4, 5, 60, 61, 63], in contrast to the human obesity phenotype. Hyperlocomotion is also broadly reported in 16p11.2+/− mice [4, 5, 59, 61, 63, 64, 70], another behavioral feature that does not coincide directly with human symptomology.
16p11.2+/− mice display impairments in cognition, sociability and motor function, coinciding with phenotypes present in human 16p11.2 deletion carriers. However, several phenotypes, such as body weight, are discordant between mice and humans. Another limitation is that many deficits present in deletion carriers, such as apraxia of speech, cannot be tested or modeled in animals [71]. Thus, 16p11.2+/− mice may represent a suitable preclinical model system for evaluating mechanisms of certain social and cognitive dysfunctions, but several of the deficits associated with 16p11.2 deletions cannot be modeled in these animals.
The 16p11.2dp/+ mice (Mills) exhibit social and cognitive deficits [72, 73] along with increased repetitive self-grooming [5, 72], with the absence of motor coordination deficits [72]. Two behavioral phenotypes associated with SZ, PPI of startle responses and MK-801-induced hyperlocomotion, were not observed in 16p11.2dp/+ mice [72], despite a report on female-specific PPI deficits [73]. Based on these results, the SZ-linked changes should be further investigated in 16p11.2dp/+ mice. 16p11.2dp/+ mice also exhibit hypolocomotion [5, 72, 73], in direct contrast to 16p11.2+/− animals. Hypolocomotion and enhanced object recognition memory are reported in 16p11.2dp/+ mice (Herault) on a C57BL/6N inbred genetic background, whereas on a F1 C57BL/6N-C3B hybrid background, mice additionally exhibit social approach deficits [59]. These studies indicate that 16p11.2dp/+ mice recapitulate several, but not all, human 16p11.2 duplication phenotypes, including social deficits, repetitive behaviors, and cognitive impairment.
Sex-Specific Behavioral Phenotypes in Mouse Models of 16p11.2 CNVs
Male, but not female, 16p11.2+/− mice (Mills) display reduced and altered pup isolation calls, indicating sex-specific communication impairments in perinatal development [74]. Additionally, male 16p11.2+/− mice (Mills) exhibit sleep deficits [70], and impairments in a reward-directed learning task [67], while females do not. In 16p11.2dp/+ mice (Mills), male-specific reductions in locomotion, time in the center of an open field, and ratio of open arm/closed arm time in the elevated plus maze have also been reported [73].
16p11.2 CNV Mouse Models Exhibit Synaptic Dysfunction in Distributed Brain Regions
The 16p11.2+/− mice (Mills) display deficient NMDA-receptor-mediated glutamatergic transmission and reduced frequency of action potential firing in medial prefrontal cortex (mPFC) pyramidal neurons [62], along with increased excitation-inhibition (E-I) ratio in somatosensory cortex [75]. Additionally, GABAergic neurons in the ventral medulla display hyperpolarized resting membrane potential and increased membrane resistance [69]. Long term potentiation and depression (LTP/LTD) appear to be intact in hippocampal CA1 of 16p11.2+/− mice (Mills), though protein-synthesis-dependent mGluR5 LTD is impaired [65]. Recordings from CA1 of 16p11.2+/− mice (Herault) reveal intact synaptic transmission, along with modest but not statistically significant reductions in LTP [59]. Nucleus accumbens (NAc) medium spiny neurons (MSNs) in 16p11.2+/− mice (Dolmetsch) display increased AMPAR/NMDAR ratio and decreased paired pulse ratios (PPR), along with increased miniature EPSC frequency [4]. The authors hypothesize that these results are due to increased release probability of excitatory synapses on NAc MSNs [4]. Synaptic phenotypes in 16p11.2+/− mice evidently vary across brain areas, suggesting that 16p11.2 deletion may cause region- or cell-type-specific impairments.
The 16p11.2dp/+ mice (Herault) show impaired LTP in CA1 [59]. However, these findings have not yet been confirmed in other 16p11.2dp/+ models, and may benefit from behavioral validation with corresponding memory tasks such as contextual fear conditioning [65]. The 16p11.2dp/+ mice (Mills) display GABAergic synaptic deficits in mPFC pyramidal neurons and increased action potential firing rates [72]. Restoring mPFC GABAergic synaptic activity is sufficient to reverse the social and cognitive deficits [72], implicating prefrontal cortical synaptic dysfunction in 16p11.2 duplication pathology. In addition, disrupted connectivity between hippocampal-orbitofrontal and hippocampal-amygdala circuits has been found in 16p11.2dp/+ mice (Mills), as well as reduced expression of several GABAergic markers, including parvalbumin, calbindin and somatostatin in frontal cortex [73].
16p11.2 CNV Induce Broad Transcriptional Disruptions in Mice and Human Cells
Sequencing studies have revealed broad transcriptional disruptions in 16p11.2 CNVs. RNA-sequencing identifies 2,344 and 1,504 significant differentially expressed genes (DEGs) in the cortex of 16p11.2+/− and 16p11.2dp/+ mice (Mills), respectively, as well as 908 and 1,290 nominally significant DEGs in lymphoblastoid cell lines (LCLs) from human 16p11.2 deletion or duplication carriers, respectively [76]. Gene ontology (GO) analysis of DEGs in mouse cortex indicates the strongest disruption of genes related to “regulation of transcription”. The top pathways implicated in human LCLs are “microtubule cytoskeleton organization” and “cell surface receptor-linked signal transduction”.
A separate RNA-seq experiment in mPFC of 16p11.2dp/+ mice (Mills) identifies 388 DEGs (277 downregulated, 111 upregulated), confirming broad transcriptional disruption extending far beyond the genes located within the 16p11.2 region [72]. GO analysis indicates that the largest portion of downregulated genes are classified as “Transcription Factors” (17.7%), consistent with prior RNA-seq data [76]. The largest portion of upregulated DEGs (21.6%) are identified as “Enzyme Modulators”. These findings highlight the wide transcriptional impacts of 16p11.2 CNVs, but further studies should clarify the specific contributions of 16p11.2 genes to these far-reaching downstream transcriptional disruptions and their links to the heterogeneous associated behavioral phenotypes.
Abnormal Cortical Development Driven by 16p11.2 CNVs Involves Several Genes in 16p11.2 Region
Abnormal cortical development has been proposed as a core mechanism in 16p11.2 CNVs. As reviewed earlier, 16p11.2 deletions are associated with macrocephaly, whereas duplications are linked to microcephaly. Human induced pluripotent stem cell (iPSC)-derived neurons from 16p11.2 deletion/duplication carriers display corresponding features, with increased soma size/dendrite length in 16p11.2 deletion neurons, and reduced size/dendrite length in duplication neurons [77]. In contrast, 16p11.2+/− mice display reduced brain size and reductions in upper layer cortical projection neurons, driven by increased progenitor proliferation and premature cell cycle exit, resulting in depleted progenitor pools and maldeveloped cortical structures [66]. The precise reasons for the reduced brain size remain unclear, although the reduced brain size is associated with reduced body weight phenotype in 16p11.2+/− mice, suggesting that at least in part, a broader developmental physiological impairment might be involved.
Cortico-striatal circuits have been implicated in the pathophysiology of 16p11.2 deletion. Various structural abnormalities are reported in the striatum and cortex of 16p11.2+/− mice (Dolmetsch), along with an increased population of Drd2+ MSNs in the striatum, and reduced Drd1+ neurons in the cortex [4]. An elevated number of MSNs and the increased expression of Drd2 in striatum are found in male 16p11.2+/− mice (Mills) [4, 67]. Disrupted synaptic function in striatal MSNs is thought to underlie locomotion-related behavioral phenotypes in 16p11.2+/− mice [4].
The MAPK3 gene located within the 16p11.2 region encodes the signaling molecule ERK1. Human genetic screenings link MAPK3 signaling to ASD [78], and ERK dysregulation is associated with ASD-related behavioral phenotypes in mice [79]. This pathway is also linked to ID [80]. Cultured pyramidal neurons from 16p11.2dp/+ mice exhibit increased ERK1 phosphorylation, and greater dendritic arborization, which can be reversed by ERK inhibition [81]. These findings support a role for Mapk3 in cortical development abnormalities associated with 16p11.2 CNVs.
The 16p11.2 gene KCTD13 is also associated with cranial size phenotypes. In zebrafish, out of all 16p11.2 gene transcripts, only KCTD13 overexpression drives a microcephalic phenotype with reduced brain cell counts, whereas its suppression produces a macrocephalic phenotype with increased brain cell counts [6]. However, these findings do not generalize to mice: two follow-up studies reported normal brain size in Kctd13-deficient mice [82, 83]. Kctd13-deficient mice display reduced dendritic length/complexity and spine density, which is linked to downstream RhoA overexpression [82]. Kctd13-deficient mice also display cognitive deficits in several memory tasks [83], raising the possibility that similar mechanisms may contribute to ID pathogenesis in 16p11.2 CNVs.
Kctd13 is one of the adaptors of Cullin 3 (Cul3), a core component of the E3 ubiquitin ligase complex mediating protein ubiquitination and degradation [84]. The Cul3 gene is one of the top-ranking high-risk factors for autism and related neurodevelopmental disorders [85, 86]. In mice, Cul3 deficiency in the forebrain or PFC induces social interaction impairment and sensory-gating deficiency, as well as NMDA receptor hypofunction, whereas Cul3 loss in the striatum causes stereotypic behaviors, as well as cell type-specific changes in neuronal excitability [87]. Abnormality in gene transcription or protein translation may underlie the involvement of Cul3 in neurodevelopmental disorders [87, 88]. It has also been suggested that dysregulation of the KCTD13-Cul3-RhoA pathway during the critical period for establishing the connectivity of 16p11.2 proteins with their co-expressed partners determines the abnormal brain sizes associated with 16p11.2 CNVs [89].
Knockout of the 16p11.2 gene Taok2 produces cognitive and social deficits in mice, and also results in increased brain volume [90]. Taok2-deficient mice display reduced dendritic growth and deficient excitatory synaptic transmission in PFC through a mechanism involving the reduced RhoA expression [90], suggesting that the 16p11.2 genes Kctd13 and Taok2 may act upon convergent downstream molecules. Taok2 has also been implicated in synaptic stabilization and spine maturation [91].
In humans, mutations in MAPK3 [92] and TAOK2 [93, 94] but not in KCTD13 [85, 95] have been found in ASD cases. Missense variants in MAPK3 are identified in ASD probands [85, 92], but no loss of function (LoF) MAPK3 mutations have been reported in ASD, leaving the direct link to ASD uncertain. LoF variants in TAOK2 are identified in ASD probands [93, 94], however it is not a top-ranking ASD risk factor by large genetic screening [85]. Additionally, results from rodent studies of individual genes need to be considered with caution, as the phenotypes of 16p11.2 CNVs may depend on the collective effects of multiple genes in 16p11.2 locus.
Neurostructural Changes in Mouse Models of 16p11.2 CNVs
In contrast to the human data [14, 23, 49, 50], 16p11.2+/− mice (Mills) do not exhibit changes in gray matter [7], but they show male-specific increases in medial and peristriatal fiber tracts [7], coinciding with the increased medial fiber tracts in human deletion carriers [14]. Increased volumes of several brain areas, including the hypothalamus, midbrain, cerebellar cortex, striatum, nucleus accumbens and globus pallidus, are observed in 16p11.2+/− mice (Mills, Dolmetsch) [4, 5, 96], concordant with the increased volume of these structures in human deletion patients [54]. No significant differences were found between 16p11.2dp/+ (Mills) and WT mice, though trends toward reduced volume were observed in several regions [5].
Therapeutic Interventions: Targeting Glutamatergic and GABAergic Systems
Many valuable findings have been drawn from attempted intervention strategies in 16p11.2 mouse models. Inhibitory avoidance (IA) deficits are observed in 16p11.2+/− mice (Mills) [65]. In a mouse model of Fragile X Syndrome, IA deficits are reversed via chronic post-adolescent administration of the mGluR5 negative allosteric modulator (NAM) CTEP [97]. This approach was thus tested in 16p11.2+/− mice and proved effective in ameliorating IA deficits [65].
The GABAB-receptor selective agonist R-baclofen was tested in 16p11.2+/− mice (Mills and Dolmetsch), as R-baclofen displayed therapeutic efficacy in Fmr1-deficient mice with a similar synaptic plasticity phenotype [64]. R-baclofen improves performance of 16p11.2+/− mice in several cognitive tasks, while hyperlocomotion and USVs are unaffected [64]. However, R-baclofen and several mGluR5 NAMs have produced negative results in clinical trials with Fragile X patients [98]. Thus, findings from preclinical studies in animal models should be interpreted with caution. It has also been demonstrated that GABAA receptor availability is unaffected in human 16p11.2 deletion carriers [99], though this does not preclude the possibility of presynaptic GABAergic alterations or effects on other GABA receptor subunits.
As reviewed earlier, 16p11.2+/− mice exhibit NMDAR deficits and hypoactivity in PFC pyramidal neurons [62]. Both humans and mice carrying 16p11.2 deletions display reduced prefrontal cortical connectivity [100], further suggesting a role for PFC disruption in 16p11.2 CNVs. Chemogenetic activation of PFC leads to increased NMDAR phosphorylation and function, resulting in amelioration of social and cognitive deficits [62]. This study suggests NMDAR hypofunction in PFC as a core mechanism in 16p11.2 deletion-linked phenotypes, consistent with the significant involvement of NMDARs in ASD [101–103].
As to the rescue of phenotypes in 16p11.2dp/+ mice (Mills), a recent study from our group [72] revealed that restoring expression of Npas4, a key regulator of GABA synapses [104, 105] reverses GABAergic deficits and ameliorates social and cognitive deficits in these mice, but not repetitive grooming behavior. The therapeutic potential of pharmaceutical interventions to boost GABAergic transmission by targeting Npas4 or related molecules for 16p11.2 duplication syndrome awaits to be further explored.
Concluding Remarks and Future Perspectives
Clinical and preclinical investigations illustrate the diverse neurobiological impact of 16p11.2 CNVs. A number of highly penetrant developmental phenotypes are linked to both 16p11.2 deletions and duplications (Figure 1). Given the effects of 16p11.2 CNVs on 27–29 genes, these mutations have the capacity to cause broad and severe downstream biochemical insults across various brain areas. The array of cellular changes in 16p11.2 models, including transcriptional and synaptic dysregulation, aberrant cell proliferation and cortical development, provides a window into the molecular pathologies underlying behavioral syndromes associated with 16p11.2 CNVs. Restoring E/I balance and synaptic plasticity by targeting glutamate and GABA systems is suggested as a core intervention strategy (Figure 2).
Characterizations of 16p11.2 CNV mouse models have illuminated several important pathophysiological clues, though more work must be done before one could come up with a clear disease mechanism (see Outstanding Questions). Future studies could expand electrophysiological, biochemical and genomic investigations into stem cell-derived neuronal models from human 16p11.2 CNV carriers, to test the existence of similar molecular and cellular aberrations. Additionally, optogenetic and chemogenetic approaches could be used to investigate long-range and local circuits in 16p11.2 CNV mice, which will help explain how the distributed physiological changes integrate to produce broad behavioral phenotypes. To identify the genes in the 16p11.2 region that drive morphological and functional alterations, future studies could use CRISPR-Cas9 technology to manipulate individual 16p11.2 genes or combinations of them in human iPSC-derived neurons or transgenic mice. Combined transcriptomic analyses of 16p11.2 CNV mice and human RNAseq datasets have identified overrepresentation of pathways related to histone methylation [76], thus future studies are encouraged to explore the role of epigenetic modifications of gene expression in 16p11.2 CNVs. The ultimate goal is to find mechanism-based treatment strategy for neurodevelopmental disorders related to 16p11.2 CNVs and beyond. Key molecular targets or biological pathways will guide translational research for therapeutic intervention.
OUTSTANDING QUESTIONS BOX.
What are the mechanistic links between the molecular, electrophysiological, and behavioral abnormalities driven by 16p11.2 CNVs? How do synaptic disruptions across different brain regions integrate to produce diverse behavioral phenotypes in 16p11.2 CNVs?
Are the structural and functional changes found in 16p11.2 deletion or duplication mice relevant to the pathophysiological mechanisms of human 16p11.2 syndromes? Do human stem cell-derived neurons from 16p11.2 CNV carriers display analogous electrophysiological, biochemical, and transcriptional phenotypes to those observed in corresponding mouse models?
Which genes in the chromosome 16p11.2 region are core drivers of the molecular, cellular and behavioral aberrations associated with 16p11.2 CNVs?
What is the role of transcriptomic change in the pathogenesis of 16p11.2 syndromes? Given the prominent role of chromatin remodelers and histone modifiers in neurodevelopmental disorders, including ASD and ID,is epigenetic dysregulation of gene expression also critically involved in 16p11.2 CNVs?
How might preclinical findings from 16p11.2 CNV mouse models translate to clinical treatment of humans with 16p11.2 CNVs or ASD in general? What is the therapeutic value of targeting the NMDAR or mGluR system in 16p11.2 deletions and GABA system in 16p11.2 duplications?
HIGHLIGHTS.
16p11.2 deletions or duplications predispose individuals to neurodevelopmental diseases, including autism spectrum disorder, intellectual disability, epilepsy/seizures, dysmorphic features, congenital anomalies, macrocephaly and microcephaly.
16p11.2 CNV mouse models recapitulate many of the human behavioral phenotypes, with distinct representation of social and cognitive deficits.
Various forms of synaptic dysfunction are observed across distributed brain areas in 16p11.2 CNV mouse models.
Broad transcriptional dysregulation is found in 16p11.2 CNV mouse models and human carriers, extending far beyond genes within the 16p11.2 region.
Several 16p11.2 genes, including Mapk3, Kctd13, Taok2, are highly involved in abnormal cortical development in 16p11.2 CNVs.
Possible therapeutic intervention strategies include the restoration of E/I balance and synaptic plasticity by targeting glutamate and GABA systems.
ACKNOWLEDGMENTS
This work was supported by grants from Nancy Lurie Marks Family Foundation and National Institutes of Health (MH112237) to Z.Y.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest related to this work (financial or else).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mitchell KJ, The genetics of neurodevelopmental disease. Curr Opin Neurobiol, 2011. 21(1):p. 197–203. [DOI] [PubMed] [Google Scholar]
- 2.Grayton HM, et al. , Copy number variations in neurodevelopmental disorders. Prog Neurobiol, 2012. 99(1): p. 81–91. [DOI] [PubMed] [Google Scholar]
- 3.Redon R, et al. , Global variation in copy number in the human genome. Nature, 2006. 444(7118): p. 444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Portmann T, et al. , Behavioral abnormalities and circuit defects in the basal ganglia of a mouse model of 16p11.2 deletion syndrome. Cell Reports, 2014. 7: p. 1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horev G, et al. , Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. PNAS, 2011. 108(41): p. 17076–17081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golzio C, et al. , KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature, 2012. 485(7398): p. 363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar VJ, et al. , Linking spatial gene expression patterns to sex-specific brain structural changes on a mouse model of 16p11.2 hemideletion. Transl Psychiatry, 2018. 8(1): p. 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barber JC, et al. , 16p11.2-p12.2 duplication syndrome; a genomic condition differentiated from euchromatic variation of 16p11.2. Eur J Hum Genet, 2013. 21(2): p. 182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy SE, et al. , Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet, 2009. 41(11): p. 1223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Angelo D, et al. , Defining the Effect of the 16p11.2 Duplication on Cognition, Behavior, and Medical Comorbidities. JAMA Psychiatry, 2016. 73(1): p. 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar RA, et al. , Recurrent 16p11.2 microdeletions in autism. Human Molecular Genetics, 2008. 17(4): p. 628–638. [DOI] [PubMed] [Google Scholar]
- 12.Zufferey F, et al. , A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. Journal of Medical Genetics, 2012. 49: p. 660–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss LA, et al. , Association between microdeletion and microduplication at 16p11.2 and autism. The New England Journal of Medicine, 2008. 358: p. 667–675. [DOI] [PubMed] [Google Scholar]
- 14.Maillard AM, et al. , The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Molecular Psychiatry, 2014. 20: p. 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson E, et al. , Cognitive and behavioral characterization of 16p11.2 deletion syndrome. Journal of Developmental & Behavioral Pediatrics, 2010. 31: p. 649–657. [DOI] [PubMed] [Google Scholar]
- 16.Rosenfeld JA, et al. , Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. Journal of Neurodevelopmental Disorders, 2010. 2: p. 26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinawi M, et al. , Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. Journal of Medical Genetics, 2010. 47(5): p. 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez BA, et al. , Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J Med Genet, 2010. 47(3): p. 195–203. [DOI] [PubMed] [Google Scholar]
- 19.Green Snyder L, et al. , Autism Spectrum Disorder, Developmental and Psychiatric Features in 16p11.2 Duplication. J Autism Dev Disord, 2016. 46(8): p. 2734–48. [DOI] [PubMed] [Google Scholar]
- 20.Walters RG, et al. , A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature, 2010. 463(7281): p. 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson E, et al. , The cognitive and behavioral phenotype of the 16p11.2 deletion in a clinically ascertained population. Biological Psychiatry, 2015. 77: p. 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballif BC, et al. , Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet, 2007. 39(9): p. 1071–3. [DOI] [PubMed] [Google Scholar]
- 23.Qureshi AY, et al. , Opposing brain differences in 16p11.2 deletion and duplication carriers. J Neurosci, 2014. 34(34): p. 11199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacquemont S, et al. , Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature, 2011. 478(7367): p. 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinberg S, et al. , Common variant at 16p11.2 conferring risk of psychosis. Mol Psychiatry, 2014. 19(1): p. 108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahoo T, et al. , Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med, 2011. 13(10): p. 868–80. [DOI] [PubMed] [Google Scholar]
- 27.Kirov G, et al. , De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry, 2012. 17(2): p. 142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genetics Home Reference 16p11.2 deletion Available from: https://ghr.nlm.nih.gov/condition/16p112-deletion-syndrome#statistics.
- 29.Genetics Home Reference 16p11.2 duplication Available from: https://ghr.nlm.nih.gov/condition/16p112-duplication#statistics.
- 30.Gillentine MA, et al. , An estimation of the prevalence of genomic disorders using chromosomal microarray data. J Hum Genet, 2018. 63(7): p. 795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford K, et al. , Medical consequences of pathogenic CNVs in adults: analysis of the UK Biobank. J Med Genet, 2019. 56(3): p. 131–138. [DOI] [PubMed] [Google Scholar]
- 32.Wallace AS, et al. , Longitudinal report of child with de novo 16p11.2 triplication. Clin Case Rep, 2018. 6(1): p. 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bijlsma EK, et al. , Extending the phenotype of current rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. European Journal of Medical Genetics, 2009. 52: p. 77–87. [DOI] [PubMed] [Google Scholar]
- 34.Niarchou M, et al. , Psychiatric disorders in children with 16p11.2 deletion and duplication. Transl Psychiatry, 2019. 9(1): p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hippolyte L, et al. , The Number of Genomic Copies at the 16p11.2 Locus Modulates Language, Verbal Memory, and Inhibition. Biol Psychiatry, 2016. 80(2): p. 129–139. [DOI] [PubMed] [Google Scholar]
- 36.Steinman KJ, et al. , 16p11.2 deletion and duplication: Characterizing neurologic phenotypes in a large clinically ascertained cohort. Am J Med Genet A, 2016. 170(11): p. 2943–2955. [DOI] [PubMed] [Google Scholar]
- 37.Rosenfeld JA, et al. , Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med, 2013. 15(6): p. 478–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall CR, et al. , Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet, 2008. 82(2): p. 477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldman S, et al. , Quantitative gait assessment in children with 16p11.2 syndrome. J Neurodev Disord, 2019. 11(1): p. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mei C, et al. , Deep phenotyping of speech and language skills in individuals with 16p11.2 deletion. Eur J Hum Genet, 2018. 26(5): p. 676–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demopoulos C, et al. , Abnormal Speech Motor Control in Individuals with 16p11.2 Deletions. Sci Rep, 2018. 8(1): p. 1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W, et al. , Study of the association between Schizophrenia and microduplication at the 16p11.2 locus in the Han Chinese population. Psychiatry Res, 2018. 265: p. 198–199. [DOI] [PubMed] [Google Scholar]
- 43.Chang H, et al. , Rare and common variants at 16p11.2 are associated with schizophrenia. Schizophr Res, 2017. 184: p. 105–108. [DOI] [PubMed] [Google Scholar]
- 44.Jutla A, et al. , Psychotic symptoms in 16p11.2 copy-number variant carriers. Autism Res, 2020. 13(2): p. 187–198. [DOI] [PubMed] [Google Scholar]
- 45.Insel TR, Rethinking schizophrenia. Nature, 2010. 468(7321): p. 187–93. [DOI] [PubMed] [Google Scholar]
- 46.Owen MJ, et al. , Neurodevelopmental hypothesis of schizophrenia. Br J Psychiatry, 2011. 198(3): p. 173–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kendall KM, et al. , Association of Rare Copy Number Variants With Risk of Depression. JAMA Psychiatry, 2019. 76(8): p. 818–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gudmundsson OO, et al. , Attention-deficit hyperactivity disorder shares copy number variant risk with schizophrenia and autism spectrum disorder. Transl Psychiatry, 2019. 9(1): p. 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin-Brevet S, et al. , Quantifying the Effects of 16p11.2 Copy Number Variants on Brain Structure: A Multisite Genetic-First Study. Biol Psychiatry, 2018. 84(4): p. 253–264. [DOI] [PubMed] [Google Scholar]
- 50.Cardenas-de-la-Parra A, et al. , Developmental trajectories of neuroanatomical alterations associated with the 16p11.2 Copy Number Variations. Neuroimage, 2019. 203: p. 116155. [DOI] [PubMed] [Google Scholar]
- 51.Owen JP, et al. , Aberrant white matter microstructure in children with 16p11.2 deletions. J Neurosci, 2014. 34(18): p. 6214–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang YS, et al. , Reciprocal white matter alterations due to 16p11.2 chromosomal deletions versus duplications. Hum Brain Mapp, 2016. 37(8): p. 2833–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Owen JP, et al. , Brain MR Imaging Findings and Associated Outcomes in Carriers of the Reciprocal Copy Number Variation at 16p11.2. Radiology, 2018. 286(1): p. 217–226. [DOI] [PubMed] [Google Scholar]
- 54.Sonderby IE, et al. , Dose response of the 16p11.2 distal copy number variant on intracranial volume and basal ganglia. Mol Psychiatry, 2020. 25(3): p. 584–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Filges I, et al. , Brain MRI abnormalities and spectrum of neurological and clinical findings in three patients with proximal 16p11.2 microduplication. Am J Med Genet A, 2014. 164A(8): p. 2003–12. [DOI] [PubMed] [Google Scholar]
- 56.Werling DM and Geschwind DH, Understanding sex bias in autism spectrum disorder. Proc Natl Acad Sci U S A, 2013. 110(13): p. 4868–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polyak A, Rosenfeld JA, and Girirajan S, An assessment of sex bias in neurodevelopmental disorders. Genome Med, 2015. 7: p. 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hudac CM, et al. , Evaluating heterogeneity in ASD symptomatology, cognitive ability, and adaptive functioning among 16p11.2 CNV carriers. Autism Res, 2020(Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 59.Arbogast T, et al. , Reciprocal Effects on Neurocognitive and Metabolic Phenotypes in Mouse Models of 16p11.2 Deletion and Duplication Syndromes. PLoS Genet, 2016. 12(2): p. e1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang M, et al. , 16p11.2 deletion syndrome mice display sensory and ultrasonic vocalization deficits during social interactions. Autism Research, 2015. 8(5): p. 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brunner D, et al. , Comprehensive Analysis of the 16p11.2 Deletion and Null Cntnap2 Mouse Models of Autism Spectrum Disorder. PLoS One, 2015. 10(8): p. e0134572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W, et al. , Chemogenetic Activation of Prefrontal Cortex Rescues Synaptic and Behavioral Deficits in a Mouse Model of 16p11.2 Deletion Syndrome. J Neurosci, 2018. 38(26): p. 5939–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang M, et al. , In tribute to Bob Blanchard: Divergent behavioral phenotypes of 16p11.2 deletion mice reared in same-genotype versus mixed-genotype cages. Physiol Behav, 2015. 146: p. 16–27. [DOI] [PubMed] [Google Scholar]
- 64.Stoppel LJ, et al. , R-Baclofen Reverses Cognitive Deficits and Improves Social Interactions in Two Lines of 16p11.2 Deletion Mice. Neuropsychopharmacology, 2018. 43(3): p. 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian D, et al. , Contribution of mGluR5 to hippocampal pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nature Neuroscience, 2015. 18(2): p. 182–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pucilowska J, et al. , The 16p11.2 deletion mouse model of autism exhibits altered cortical progenitor proliferation and brain cytoarchitecture linked to the ERK MAPK pathway. J Neurosci, 2015. 35(7): p. 3190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grissom NM, et al. , Male-specific deficits in natural reward learning in a mouse model of neurodevelopmental disorders. Mol Psychiatry, 2017. 23: p. 544–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang M, et al. , 16p11.2 deletion mice display cognitive deficits in touchscreen learning and novelty recognition tasks. Learning and Memory, 2015. 22(12): p. 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu HC, et al. , Altered sleep architecture, rapid eye movement sleep, and neural oscillation in a mouse model of human chromosome 16p11.2 microdeletion. Sleep, 2019. 42(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Angelakos CC, et al. , Hyperactivity and male-specific sleep deficits in the 16p11.2 deletion mouse model of autism. Autism Research, 2017. 10: p. 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fedorenko E, et al. , A highly penetrant form of childhood apraxia of speech due to deletion of 16p11.2. Eur J Hum Genet, 2016. 24(2): p. 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rein B, et al. , Reversal of synaptic and behavioral deficits in a 16p11.2 duplication mouse model via restoration of the GABA synapse regulator Npas4. Mol Psychiatry, 2020(Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bristow GC, et al. , 16p11 Duplication Disrupts Hippocampal-Orbitofrontal-Amygdala Connectivity, Revealing a Neural Circuit Endophenotype for Schizophrenia. Cell Rep, 2020. 31(3): p. 107536. [DOI] [PubMed] [Google Scholar]
- 74.Agarwalla S, et al. , Male-specific alterations in structure of isolation call sequences of mouse pups with 16p11.2 deletion. Genes Brain Behav, 2020: p. e12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antoine MW, et al. , Increased Excitation-Inhibition Ratio Stabilizes Synapse and Circuit Excitability in Four Autism Mouse Models. Neuron, 2019. 101(4): p. 648–661 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blumenthal I, et al. , Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am J Hum Genet, 2014. 94(6): p. 870–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deshpande A, et al. , Cellular Phenotypes in Human iPSC-Derived Neurons from a Genetic Model of Autism Spectrum Disorder. Cell Rep, 2017. 21(10): p. 2678–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pinto D, et al. , Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet, 2014. 94(5): p. 677–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yufune S, et al. , Transient Blockade of ERK Phosphorylation in the Critical Period Causes Autistic Phenotypes as an Adult in Mice. Sci Rep, 2015. 5: p. 10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borrie SC, et al. , Cognitive Dysfunctions in Intellectual Disabilities: The Contributions of the Ras-MAPK and PI3K-AKT-mTOR Pathways. Annu Rev Genomics Hum Genet, 2017. 18: p. 115–142. [DOI] [PubMed] [Google Scholar]
- 81.Blizinsky KD, et al. , Reversal of dendritic phenotypes in 16p11.2 microduplication mouse model neurons by pharmacological targeting of a network hub. Proc Natl Acad Sci U S A, 2016. 113(30): p. 8520–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Escamilla CO, et al. , Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature, 2017. 551(7679): p. 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arbogast T, et al. , Kctd13-deficient mice display short-term memory impairment and sex-dependent genetic interactions. Hum Mol Genet, 2019. 28(9): p. 1474–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Genschik P, Sumara I, and Lechner E, The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J, 2013. 32(17): p. 2307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DeRubeis S, et al. , Synaptic, transcriptional and chromatin genes disrupted in autism. Nature, 2014. 515(7526): p. 209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stessman HA, et al. , Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet, 2017. 49(4): p. 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rapanelli M, et al. , Behavioral, circuitry, and molecular aberrations by region-specific deficiency of the high-risk autism gene Cul3. Mol Psychiatry, 2019(Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 88.Dong Z, et al. , CUL3 Deficiency Causes Social Deficits and Anxiety-like Behaviors by Impairing Excitation-Inhibition Balance through the Promotion of Cap-Dependent Translation. Neuron, 2020. 105(3): p. 475–490 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin GN, et al. , Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron, 2015. 85(4): p. 742–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Richter M, et al. , Altered TAOK2 activity causes autism-related neurodevelopmental and cognitive abnormalities through RhoA signaling. Mol Psychiatry, 2019. 24(9): p. 1329–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yadav S, et al. , TAOK2 Kinase Mediates PSD95 Stability and Dendritic Spine Maturation through Septin7 Phosphorylation. Neuron, 2017. 93(2): p. 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schaaf CP, et al. , Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum Mol Genet, 2011. 20(17): p. 3366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lim ET, et al. , Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat Neurosci, 2017. 20(9): p. 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.RK CY, et al. , Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci, 2017. 20(4): p. 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Satterstrom FK, et al. , Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell, 2020. 180(3): p. 568–584 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ellegood J, et al. , Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Mol Psychiatry, 2015. 20(1): p. 118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Michalon A, et al. , Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron, 2012. 74(1): p. 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berry-Kravis EM, et al. , Drug development for neurodevelopmental disorders: lessons learned from fragile X syndrome. Nat Rev Drug Discov, 2018. 17(4): p. 280–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Horder J, et al. , GABAA receptor availability is not altered in adults with autism spectrum disorder or in mouse models. Sci Transl Med, 2018. 10(461). [DOI] [PubMed] [Google Scholar]
- 100.Bertero A, et al. , Autism-associated 16p11.2 microdeletion impairs prefrontal functional connectivity in mouse and human. Brain, 2018. 141(7): p. 2055–2065. [DOI] [PubMed] [Google Scholar]
- 101.Won H, et al. , Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature, 2012. 486(7402): p. 261–5. [DOI] [PubMed] [Google Scholar]
- 102.Qin L, et al. , Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition. Nat Neurosci, 2018. 21(4): p. 564–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee EJ, Choi SY, and Kim E, NMDA receptor dysfunction in autism spectrum disorders. Curr Opin Pharmacol, 2015. 20: p. 8–13. [DOI] [PubMed] [Google Scholar]
- 104.Lin Y, et al. , Activity-dependent regulation of inhibitory synapse development by Npas4. Nature, 2008. 455(7217): p. 1198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bloodgood BL, et al. , The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature, 2013. 503(7474): p. 121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]