Abstract
Recent advances in methods for enrichment and mass spectrometric analysis of intact glycopeptides have produced large-scale glycoproteomics datasets, but interpreting this data remains challenging. We present MSFragger-Glyco, a glycoproteomics mode of the MSFragger search engine, for fast and sensitive identification of N- and O-linked glycopeptides and open glycan searches. Reanalysis of recent N-glycoproteomics data resulted in annotation of 80% more glycopeptide-spectrum matches (glycoPSMs) than previously reported. In published O-glycoproteomics data, our method more than doubled the number of glycoPSMs annotated when searching the same glycans as the original search and yielded 4–6-fold increases when expanding searches to include additional glycan compositions and other modifications. Expanded searches also revealed many sulfated and complex glycans that remained hidden to the original search. With greatly improved spectral annotation, coupled with the speed of index-based scoring, MSFragger-Glyco makes it possible to comprehensively interrogate glycoproteomics data and illuminate the many roles of glycosylation.
Introduction
Glycosylation is a ubiquitous and heterogeneous post-translational modification (PTM) of proteins used by cells to accomplish a wide variety of critical tasks and provide a flexible response to a changing environment1,2. Altered glycosylation profiles have been detected or implicated in numerous cancers and other diseases, making the comprehensive characterization of protein glycosylation critical to improving our understanding of health and disease3–5. Analysis of intact glycopeptides by tandem mass spectrometry (MS) has the potential to simultaneously determine the sites and compositions of glycans on a proteome-wide scale but presents several challenges due to the unique characteristics of glycosylation. Enrichment of glycopeptides is required to overcome low ionization efficiencies in positive ion mode6, and the heterogeneity of glycans, both at a given site and in the occupancy of possible sites in a protein, presents significant challenges to interpretation of intact glycopeptide MS data7,8. Recent advances in enrichment and mass spectrometric analysis of intact glycopeptides have begun to produce large-scale, high-quality datasets from a range of organisms and sample types9–13. The ability to produce glycoproteomic data at this scale has the potential to generate a paradigm shift in understanding the role of glycosylation in health and disease.
Interpretation of proteome-scale intact glycopeptide mass spectrometry data remains challenging, however. The most common method for interpreting glycopeptide mass spectra is similar to the treatment of other PTMs in proteomics database searches, i.e., to search all or a subset of potential glycans as variable modifications on all possible glycosylation sites. Several existing proteomics search engines have been adapted to search glycosylation as a variable modification14–16, and several glycopeptide-specific tools tailored to particular MS acquisition methods also use variable glycan modifications for small-scale searches17–20. The variable modification approach has two major limitations: first, the large number of possible glycans can result in a combinatorial explosion of possible configurations for peptides that contain multiple potential glycosylation sites, which is particularly problematic for the analysis of peptides with densely clustered O-linked glycans. The second is the highly labile nature of glycosylation during vibrational activation, including the collisional activation used solo or in a hybrid mode in the vast majority of glycopeptide analyses. The variable modification approach employed by many proteomic search tools, e.g. SEQUEST16, looks for fragment ions containing the intact glycan, even though glycan fragmentation during collisional activation makes b- and y-type ions very unlikely to retain intact glycan(s). Some search engines, e.g. Comet21, allow neutral losses to be specified for labile modifications, but the diversity of possible glycans results in a large number of neutral loss masses, which presents challenges when searching many possible glycan compositions.
An alternative approach can be found in the open search method22–29. In open searches, the peptide mass is determined by matching fragment ions without knowledge of the precursor, and the difference between the matched sequence mass and the observed precursor mass, called the “delta mass” or “mass offset,” is the mass of any unspecified modifications to the sequence. Crucially, for modifications that are labile, this strategy captures their presence on the precursor via the delta mass without requiring their presence on fragment ions, enabling a larger proportion of the observed fragment ions to be matched. A subset of open search, called a mass offset30 or multinotch23 search, uses this strategy to look only for a known set of delta masses of interest, such as those of known or potential glycans. These strategies have previously been employed for searching glycoproteomics data in iterative searches where an initial search pass is used to reduce computational complexity prior to the main search31. An iterative open search method has also been described using Protein Prospector32 in full proteome searches33,34. This approach was designed for electron transfer dissociation (ETD) data, in which glycans are not fragmented, limiting its potential for use with collisional or hybrid activation methods. Another iterative approach is that of pGlyco 28, which performs an initial search for glycan mass offsets from a large glycan database and scores those spectra on the presence of Y-type ions, before sending high-scoring candidates to a second peptide search with a full proteome database. The iterative strategy reduces computational complexity associated with the large glycan database, but requires that glycans generate abundant Y ion fragments, limiting its applicability to N-glycans fragmented by collision-induced dissociation (CID) or higher energy collisional dissociation (HCD).
Here we present MSFragger-Glyco, a glycoproteomics mode for the MSFragger search engine that applies the concept of open and mass offset search strategies to searching glycoproteomics data in a single pass, made computationally practicable by the fragment ion indexing approach of MSFragger22. Importantly, spectra are searched for all fragment ion series of interest simultaneously, including any of Y, b/y with no glycan or (optionally) with a single HexNAc remaining, and c/z ions containing the intact glycan, depending on the activation method(s) employed. This ensures that the score associated with any spectrum is generated from all fragment ions that can be reasonably expected, without noise from highly unlikely fragments, resulting in greatly improved confidence for the identification of labile glycans. Taking advantage of the ultrafast indexed-based searching, complex searches including hundreds of possible glycans and open searches can be accomplished in a matter of seconds to minutes per raw file. We applied MSFragger-Glyco to several state-of-the-art glycoproteomics datasets of N- and O-linked glycopeptides, comparing against published glycoPSM identifications from Byonic15, pGlyco 28, and SEQUEST16. In all cases, the glycan mass offset strategy of MSFragger-Glyco provided a substantial increase in the number of spectra that could be successfully annotated, and corresponding increases in the numbers of glycopeptides, proteins, and sites identified that could be identified from the data. For O-glycoproteomics in particular, this approach offered 2–6-fold improvements in glycoPSMs identified over recently published results from the same raw data, indicating the potential of MSFragger-Glyco for widespread improvement in the analysis of glycoproteomic data.
Results
Development of MSFragger-Glyco
MSFragger-Glyco takes advantage of the localization-aware (i.e. including shifted fragment ions) open search strategy35, with several modifications specific to glycopeptides (Fig. 1). Fragmentation of glycopeptides results in a complex milieu of products, especially in hybrid activation techniques such as EThcD and AI-ETD10,11,36–39. Because fragmentation of glycans is typically lower energy than that of the peptide backbone during CID/HCD, it is unusual to observe the intact mass of the glycan on b- or y-type fragment ions. This presents a challenge to typical (variable modification) searches, in that the observed mass of the precursor no longer matches that of the sequence that can be detected from the fragment ions.
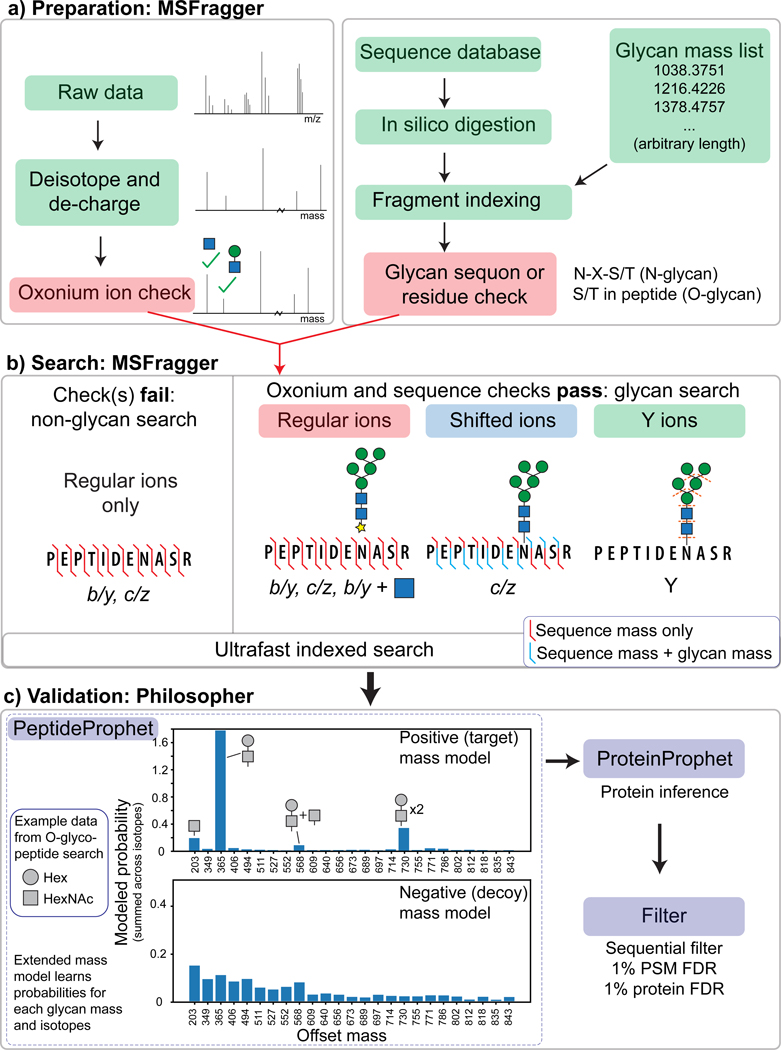
Figure 1.
Workflow of MSFragger-Glyco. a) Data and database preparation. Raw MS/MS spectra are processed (left) and searched against a protein database (right). Peptides containing a possible glycosite have additional glyco-specific fragments added to the index. b) Spectra are searched against indexed peptides. If the spectrum contains oxonium ion(s) and the peptide being considered contains a possible glycosite, a glycan search is performed (right); if either check fails, a regular search is performed. Shifted ions (blue) contain the intact mass of the glycan on the peptide while regular ions (red) contain only the masses of the amino acid residues. c) FDR filtering is performed using Philosopher. Plot of probabilities learned by PeptideProphet for a subset of mass offsets searched (mass 0 to 850 Da, masses with target probability >1% only) from O-glycopeptide data shows high probability for common compositions (e.g. HexNAc-Hex at 365) in the positive model, whereas the negative model shows similar probability for across mass shifts. Probabilities displayed are summed across isotope errors (0/1/2), resulting in probabilities than can exceed a value of 1.
MSFragger-Glyco can perform a fully open search (i.e. allow any mass offset) for exploratory interrogation of the data, and improved sensitivity can be achieved by restricting allowed mass offsets a set of user-provided glycan masses. An arbitrary number of these glycan masses can be supplied to MSFragger-Glyco and set to correspond to either N- or O-glycans. To improve glycopeptide searches, MSFragger-Glyco considers additional ion types, including Y ions (user-specified) and b/y ions with a single HexNAc residue remaining. In addition, it performs sequence motif checks for peptides and an oxonium ion check for spectra. For each peptide that contains a potential glycosite (N-X-S/T for N-glycans or S/T for O-glycans by default), Y and (optionally) b/y + HexNAc ions are added to the fragment index (Fig 1a, right). The glycan mass offsets are only searched for peptides that contain a potential glycosite and for spectra that contain sufficiently abundant oxonium ions (above a user-defined threshold, 10% relative intensity by default). For all other spectra, a regular search is performed with no mass offsets or glycan masses allowed (Fig. 1b, left).
Potential peptide-spectrum matches (PSMs) are processed using PeptideProphet40 and ProteinProphet41 using the Philosopher42 toolkit, and filtered to 1% PSM and protein-level FDR. In doing so, we utilize the extended mass model of PeptideProphet to independently model peptides with different mass offsets22, corresponding to different glycan masses in this case. As a result, PSMs with similar database search scores may have very different modeled probabilities if, for example, one has a mass offset corresponding to a commonly observed glycan and the other the mass offset of a rare glycan (Fig. 1c, top) (see Methods for details).
MSFragger-Glyco Greatly Improves Identification of Labile Glycan Spectra
We evaluated the performance of the MSFragger-Glyco mass offset method for N-glycoproteomic data using publicly available mouse brain tissue N-glycosylation data from Riley et al.10. This dataset, generated using HCD and hybrid activation method AI-ETD, represents the largest number of glycosylation sites found in such tissue to date. An example MS/MS spectrum of a glycopeptide selected for HCD fragmentation, shown in Fig. 2a, illustrates why the MSFragger-Glyco mass offset search strategy offers substantial benefits over the typical variable modification search. The spectrum is dominated by Y and B (oxonium) ions resulting from fragmentation of the glycan, while only a small fraction of the ion current comes from fragmentation of the peptide backbone, typically following extensive or complete fragmentation of the glycan. Peaks that would be considered in a variable modification search, i.e. peaks explained by the peptide sequence with an intact glycan present on Asn-9, are shown in red, and peaks matched by the mass offset strategy are shown in blue. By matching glycans as mass offsets between observed sequence and precursor masses instead of direct modifications, peptide fragments can be matched successfully following loss of the glycan (light blue), unlike in the variable modification search. In addition, MSFragger-Glyco adds the Y (dark blue) and b/y + HexNAc (medium blue) ion series to the mass offset search for glycopeptides, which represent the majority of matched ions in this spectrum. As a result, the conventional variable modification search matches 8 ions and <5% of the total ion current, while with the MSFragger-Glyco’s mass offset search with glycan-specific ion types matches 21 ions and >50% of the total ion current, generating a far more confident PSM. For AI-ETD spectra, glycan fragmentation from the laser irradiation results in a similar effect, although typically less so than for HCD since the degree of glycan fragmentation is lower, and some c/z-type ions can be observed with the glycan intact10. Importantly, the MSFragger-Glyco glycan offset search can include shifted ions35, which contain the intact glycan for c/z-type fragments, allowing for matching of all ion types observed in AI-ETD and other hybrid methods, such as EThcD.
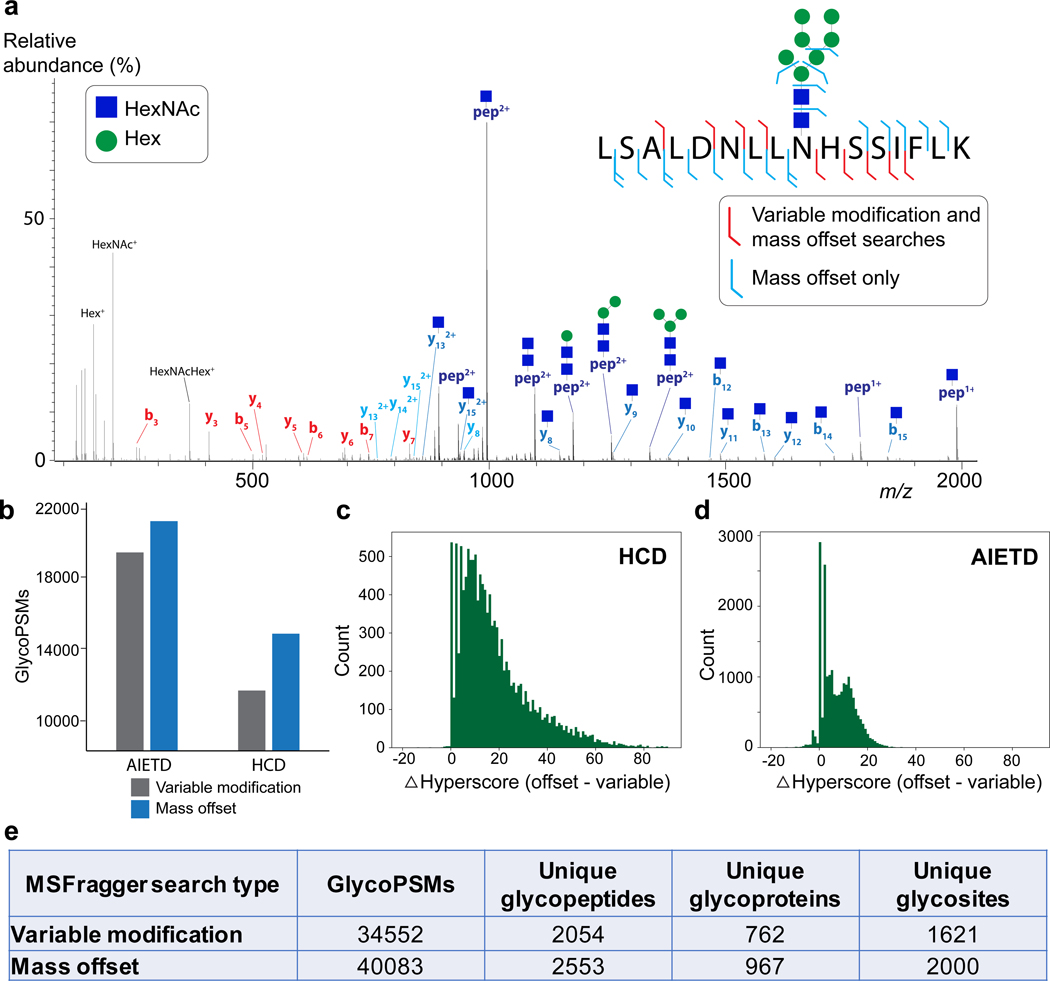
Figure 2.
Comparison of mass offset and variable modification-type searches for N-linked glycopeptides. a) Example HCD tandem mass spectrum of peptide LSALDNLLNHSSIFLK with glycan HexNAc2Hex7. Fragment ions that match the identification assuming the intact glycan is present at N-9 (variable modification-type) are colored red. Note that none of the fragments annotated in red contain the glycosite. Fragments in blue correspond to the mass offset search, including Y ions, b/y ions without the glycan or with a single HexNAc (blue square) remaining. Oxonium ions are shown in black (not all are labeled). b) Number of glycoPSMs obtained for MSFragger-Glyco mass offset search (orange) or variable modification search (blue) from AIETD and HCD spectra. c) Score difference (mass offset hyperscore – variable modification hyperscore) for spectra that were annotated in both search types for HCD spectra and (d) for AI-ETD spectra, showing a larger improvement in scores for HCD data. e) Table of results for MSFragger-Glyco mass offset and variable modification searches of 16 glycans.
The comparison between the MSFragger-Glyco mass offset search and a variable modification search on the complete dataset, using the same set of 16 glycans, is shown in Fig 2b (see Methods for details). For both HCD and AI-ETD activation methods, the mass offset search annotated many more glycoPSMs than the variable modification search and, as expected, the degree of improvement was larger in HCD spectra (24% increase) than AI-ETD (8% increase) (Fig. 2b). In many cases, the both searches successfully identified a glycoPSM, but with very different levels of confidence. For HCD spectra, nearly all glycoPSMs (>95%) had a higher score in the mass offset search, with over 60% having a substantial increase of more than 10 (Fig. 2c). As expected, the effect was less pronounced in AI-ETD spectra due to the lower degree of glycan fragmentation, but 81% of all glycoPSMs still scored higher in the mass offset search, with 33% having an increase of more than 10 (Fig. 2d). Overall, the increased scores and confidence of the mass offset search resulted in nearly 5,000 more glycoPSMs than in the variable modification search, translating to 20–25% increases in the number of unique glycopeptides, glycoproteins, and glycosites observed in the data (Fig. 2e). The ability of the mass offset search to capture fragment ions after glycan fragmentation thus gives it a unique advantage over traditional variable modification searches, resulting in increased annotation of spectra and ultimately of identified glycopeptides and glycoproteins.
Large-scale N-glycoproteomics with MSFragger-Glyco
To demonstrate the utility of MSFragger-Glyco for large-scale glycoproteomics analyses, we searched the HCD-pd-AI-ETD data from Riley et al. using the same 182 possible glycan compositions and protein database used in their search, as well as the same digestion and non-glycan modification parameters, and compared to the results reported by Riley et al. In their original publication, Riley et al. used Byonic15, a commercial platform that uses a variable modification-type search with support for glycoproteomics, to analyze the data. As with other variable modification-type searches, Byonic places potential N-glycans on peptides containing possible glycosylation sites and looks for fragments of those peptides that contain the intact glycan, b/y ions that have lost the glycan and Y ions.
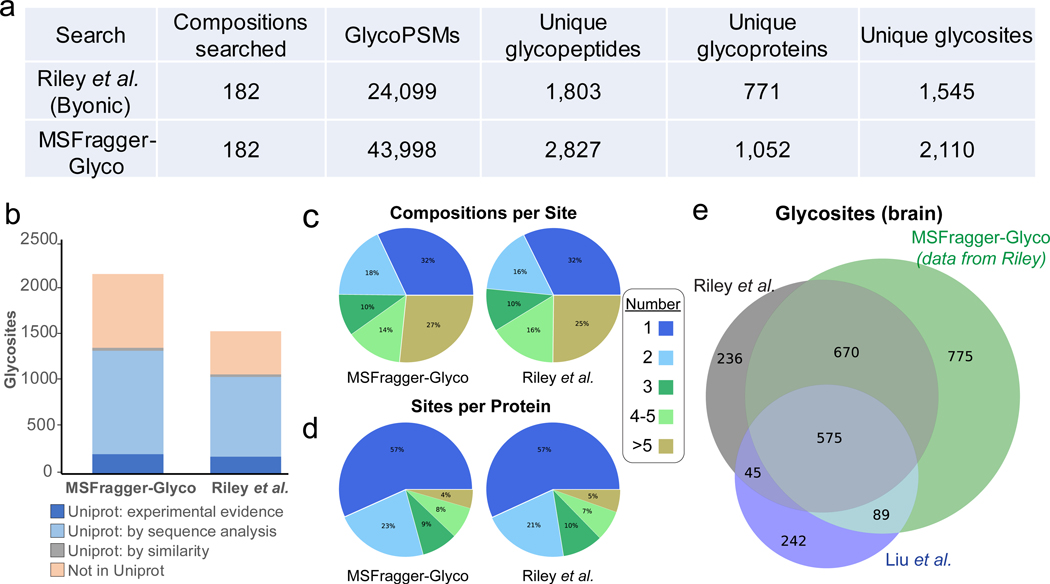
MSFragger-Glyco obtained a dramatic increase in the number of spectra that can be successfully matched to glycopeptides, with 43,998 glycoPSMs to the 24,099 reported in Riley et al (Fig. 3a). This increase in identified spectra translated to a 56% increase in the total number of unique glycopeptide sequences detected, and a 36% increase in unique glycoproteins and glycosylation sites identified across the entire dataset (Fig. 3a). The main MSFragger-Glyco search took ~1.5 minutes per raw file on a desktop computer (6 cores, 32 GB RAM), which, to our knowledge, is substantially faster than many existing tools (Supplementary Table 1). Both our MSFragger-Glyco search and Riley et al. report glycosites with a mixture of UniProt annotation levels, with similar proportions of sites at each level (Fig. 3b). The additional glycosites detected by MSFragger-Glyco are split roughly evenly between previously annotated in Uniprot and not. Despite the large increases in detected glycosites, the distribution of glycosites observed per glycoprotein and in glycan compositions observed per site remain very similar to those reported in Riley et al. (Fig. 3c, d). Compared to Riley et al. and Liu et al.7, another study with N-glycosylation data from mouse brain tissue, our analysis of the Riley et al. data shows excellent overlap with the previously detected glycosites while adding nearly 800 new glycosites not detected in either previous analysis (Fig. 3e). As Liu et al. used pGlyco 2.0 to annotate their N-glycoproteomics data, we re-analyzed this dataset with MSFragger-Glyco to obtain a comparison with this software package as well. MSFragger-Glyco obtained more than twice as many glycoPSMs as reported in Liu et al. across all tissue types (Supplementary Figure 1a). Glycosites detected by MSFragger-Glyco showed excellent overlap with those found in Liu et al., as well as recovering a large number of sites not annotated by Liu et al. that were identified in the analysis of Riley et al. (Supplementary Figure 1b, c).
Figure 3.
Comparison of MSFragger-Glyco and original analysis for N-glycan datasets. a) Direct comparison with identical protein databases and possible glycan compositions between MSFragger-Glyco and previously published results from Riley et al. b) Comparison of found glycosites to UniProt, color coded by evidence type. c) Number of glycan compositions per glycosite (all compositions, not separated by composition type) and d) Number of glycosites per protein found in MSFragger-Glyco and Riley et al.10, showing very high similarity in both cases. e) Observed glycosites from MSFragger-Glyco and original analysis of Riley et al. compared with mouse brain glycosites from Liu et al.
Overall, these results indicate that MSFragger-Glyco mass offset search performs exceptionally well for analyzing large-scale N-glycoproteomics data in HCD and hybrid activation modes. The 80% increase in glycoPSMs annotated from the same raw data with identical glycan and protein databases resulted in notable increases in glycoprotein and glycosite annotation, including confirmation of predicted glycosites and annotation of novel ones.
Deciphering complex, large-scale O-glycoproteomics data with MSFragger-Glyco
The several types of O-linked glycosylation are known to play important biological roles but have not been studied as extensively as N-linked glycosylation, due in part to additional challenges in enriching and analyzing O-linked glycans and glycopeptides. As in the case of N-glycosylation, O-linked glycans are highly labile during vibrational activation and occur in a wide variety of compositions. Unlike N-linked glycans, however, there is no consensus sequon for O-glycosylation, and some types of O-glycans are densely clustered in regions of protein sequence43,44. These factors make analysis of O-glycoproteomic MS/MS data particularly challenging, as many compositions potentially occurring on multiple sites within the same peptide results in a massive search space for any comprehensive O-glycan search by traditional methods. To date, large-scale O-glycoproteomics has proven challenging, with the most successful studies relying on simplifying the complexity of the O-glycoproteome by only generating a subset of O-glycans types, e.g. with SimpleCells45, by enzymatic reduction of glycan complexity13, or by searching for a small subset of highly abundant glycans in data that potentially contains many more compositions.
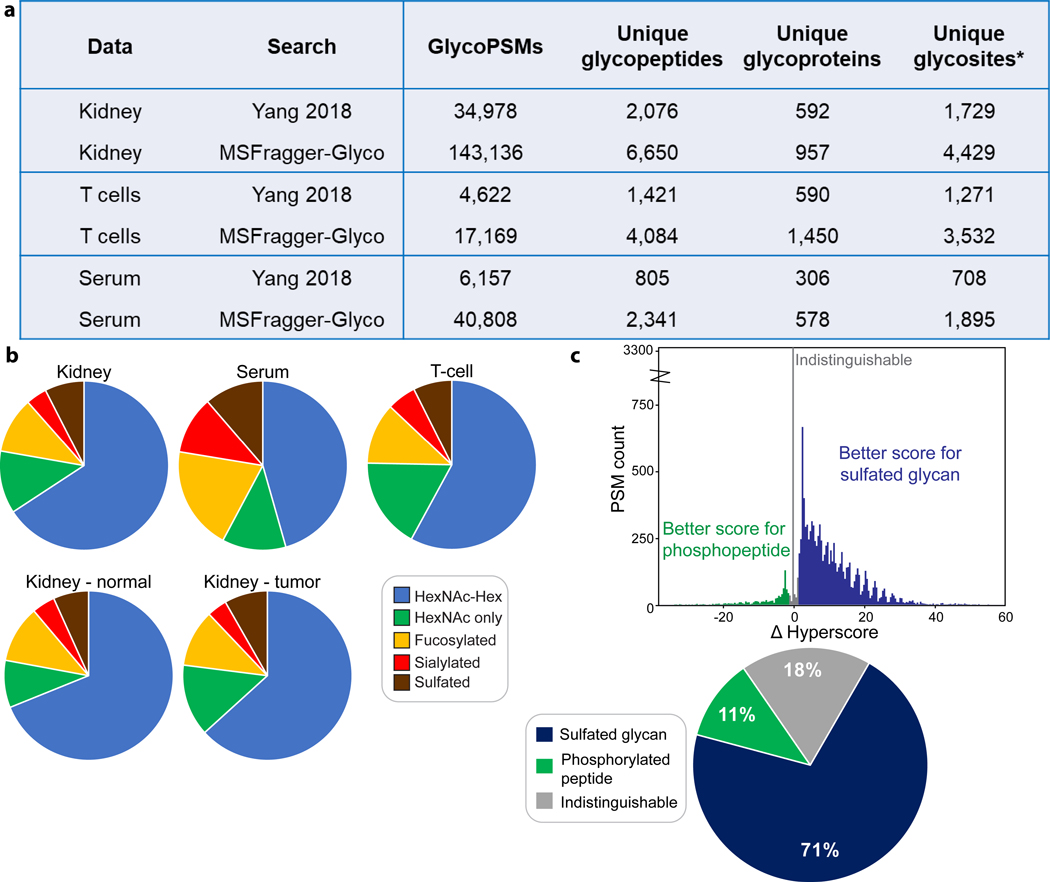
To evaluate the use of MSFragger-Glyco for large-scale O-glycoproteomics, we analyzed data from a recent study by Yang et al.12, which used a bacterial enzyme dubbed “OpeRATOR” that cleaves N-terminal to O-glycosylated Ser and Thr residues. The authors developed a protocol using this enzyme to enrich and analyze glycopeptides from human kidney tissue, serum, and T-cells with HCD MS/MS. The data was searched using SEQUEST in a variable modification mode with two possible glycans on Ser/Thr residues, resulting in the identification of nearly 35,000 glycoPSMs in the kidney tissue data (Table 1). This resulted in 12 total glycan compositions observed at the peptide level due to co-occurrence of glycans at multiple residues within some peptides. This is an important distinction when comparing with the mass offset search of MSFragger-Glyco, as the mass offset is computed at the peptide level, including all glycan modifications present on separate residues as a single mass. To compare search strategies, we performed both variable modification and mass offset searches in MSFragger-Glyco with the same two glycan types and search parameters. The variable modification search gave results similar to the SEQUEST search, finding 38,632 glycoPSMs. To perform the equivalent comparison with the mass offset method, we searched the 12 peptide-level compositions (corresponding to 2 glycan types allowed on multiple sites) searched by Yang et al. as mass offsets with MSFragger-Glyco, obtaining over 77,000 glycoPSMs, or more than double those found in the original search (Table 1). Even searching just two mass offsets (i.e. disallowing any peptides with multiple glycosites in the mass offset search) still resulted in a large increase, with over 50,000 glycoPSMs annotated. Because O-glycans readily dissociate in the HCD fragmentation used in acquiring these data, the vast majority of fragment ions lack glycans or glycan fragments entirely. The mass offset search is able to match these unmodified ions and use the offset between sequence and precursor masses to determine the glycan mass, enabling confident annotation of otherwise challenging spectra.
Table 1.
Comparison of MSFragger-Glyco and Yang et al.12 O-glycoproteomics search results in variable modification and mass offset modes. Note that variable modification searches with 2 glycan masses can identify multiple glycosites per peptide, resulting a total of 12 compositions at the peptide level found in Yang et al. The mass offset search used these same 12 glycan masses for comparison.
| Search | Search type | Glycan compositions searched | GlycoPSMs | Unique glycopeptides | Unique glycoproteins |
|---|---|---|---|---|---|
| Yang 2018 (SEQUEST) | Variable Modification | 12 | 34,978 | 2,076 | 592 |
| MSFragger-Glyco | Variable Modification | 12 | 38,632 | 2,171 | 508 |
| MSFragger-Glyco | Mass Offset | 12 | 77,236 | 4,121 | 709 |
| MSFragger-Glyco | Mass Offset | 300 | 143,136 | 6,650 | 957 |
While the mass offset strategy considering equivalent modifications resulted in vastly more glycoPSMs than the previous searches, we sought to use the speed of MSFragger-Glyco to comprehensively analyze all glycopeptides present in the data. To do so, we first performed an exploratory, fully open search with MSFragger-Glyco to generate a list of abundant glycan compositions, then performed a mass offset search using this list. The open search revealed a large number of glycan compositions present in the data that were missed in the original searches, including fucosylated, sialylated, and sulfated glycans as well as masses corresponding to multiple glycans present on the same peptide. A total of 300 glycan compositions identified using open search were searched with the mass offset method, resulting in annotation of 143,136 glycoPSMs (Table 1), or 4 times as many as in the original search and nearly double the number from the 12-composition mass offset search. We found many additional glycopeptides, resulting in the identification of 365 more glycoproteins and many more potential glycosites than the original search of Yang et al.
Several factors contributed to this massive increase in annotated spectra, including our expansion of the peptide search space as well as searching for many more types of glycans. Because OpeRATOR cleaves at glycosylated Ser/Thr, but the sites of glycosylation are not known in advance, Yang et al. digested their protein database by cleaving at all Ser/Thr residues but allowing up to 5 missed cleavages per peptide to allow for residues that may not be glycosylated. Taking advantage of ultrafast indexed searching, we were able to allow up to 10 missed cleavages by OpeRATOR, and variable modifications including oxidation (M), guanidinylation and carbamidomethylation (K), and deamidation (N, Q), after the exploratory open search revealed substantial amounts of these modifications in the data. Guanidinylation is also very close in mass to the difference between a Hex residue and a HexNAc (with +1 isotope error), but these could be clearly resolved as guanidinylation is not labile (Supplementary Figure 2). These searches with very large peptide digestion and variable modification spaces, plus 300 potential glycan compositions, were still completed in a matter of minutes per raw file, despite complexity that would be prohibitive for many search tools.
Given the proposed specificity of the OpeRATOR enzyme, Yang et al. assumed that all glycopeptides would contain a single glycosylation site at their N-terminus but noted the possibility that additional glycosites could be present if enzymatic cleavage at glycosylated Ser/Thr was imperfect. Glycosites reported in Fig. 4a are computed as in Yang et al., assuming the peptide N-terminal Ser or Thr is the only glycosite in the peptide, for purposes of comparison, though this likely underestimates the true number of glycosites in all searches. Our results show abundant evidence of these missed cleavages, particularly in cases where several glycosylated residues occur in series (Supplementary Figure 3). The mass offset search annotates multiply glycosylated peptides as containing a single composite mass offset, which works well when the glycans have largely dissociated from peptide fragment ions. This approach is not ideal for determining the exact location of each glycosite, but the HCD fragmentation used in this study resulted in the majority of glycopeptide spectra lacking any fragment ions retaining intact glycan(s) or glycan fragments, precluding data-driven localization in any case.
Figure 4.
Expanded O-glycan searches across tissues. a) Table of glycoPSMs, glycopeptides, glycoproteins, and glycosites from Yang et al.12 and MSFragger-Glyco reanalysis. *Glycosites are computed as in Yang et al. b) Glycan composition types detected in expanded searches by tissue type (Kidney (all), Serum, and T-cell (top), normal vs tumor Kidney tissue (bottom). Pie charts are simplified such that any glycan containing fucose is counted as fucosylated (for example), resulting in glycans that contain multiple composition types being counted in multiple categories. Overall composition trends are thus approximate. c) Results of competitive phospho-vs-sulfo search for Kidney data. Delta (hyper)score was computed by subtracting the score of the phosphorylated top hit from the score of the sulfated-glycan top hit of the same peptide. Histogram of delta scores (top) and proportion of delta scores indicating sulfated glycan, phosphopeptide, or indistinguishable (−2 < delta score < 2) (bottom).
Applying the 300-composition search to the serum and T-cell datasets presented in Yang et al. yielded several interesting observations. In each case, the number of glycoPSMs annotated by our glycan mass offset search was dramatically increased compared to those reported by Yang et al., with 3.7 times as many glycoPSMs for the T-cell data and 6.6 times as many glycoPSMs for the serum data (Fig. 4a). The larger increase in PSMs in the serum samples can be attributed to the much greater proportion of fucosylated, sialylated, and sulfated glycans detected in serum (together comprising nearly half of all glycoPSMs) as compared to kidney or T-cell samples (20–25% of glycoPSMs), as the original search by Yang et al. did not consider these glycan types (Fig. 4b). Yang et al. also highlight differences in glycosylation sites and occupancy between tumor and normal kidney tissue samples. We find many additional glycoPSMs that broadly support the conclusion that glycosylation is increased in the tumor data, though observed only minor differences in glycan compositions between the normal and tumor kidney tissues (Fig. 4b).
The initial open search also revealed a large number of PSMs containing phosphorylation or sulfation, which are challenging to distinguish due to a mass difference of only 9.5 mDa. Peptide backbone fragmentation is often, though not always, a lower energy pathway than phosphate loss, particularly under conditions of high charge mobility46. In contrast, the O-glycans in this dataset were observed to nearly always dissociate from the peptide, raising the possibility of distinguishing between phosphorylation and sulfation based on whether the modification mass is retained on fragment ions or not. We performed a competitive search that allowed both phosphorylation as a variable modification (fragments retain the additional mass) and sulfated glycans as mass offsets (fragments do not retain the additional mass) and computed a delta score comparing sulfated and phosphorylated possibilities. We observed (Fig. 4c, top) strong evidence for the presence of many sulfated glycans, as over 70% of all possibilities had a higher score for the sulfated glycan and exhibited large delta scores, with just 11% obtaining a clearly higher phosphopeptide score, and 18% indistinguishable. Analysis of sulfated glycopeptides is extremely challenging, particularly in the possible presence of phosphopeptides. Further validation of these results would be required to draw biological conclusions, but the clear distinction between scores obtained illustrates the capabilities of MSFragger-Glyco to interrogate highly complex and challenging data, allowing us to identify large numbers of sulfated O-glycopeptides that would otherwise go unannotated in this dataset.
Discussion
Here, we demonstrate that MSFragger-Glyco provides superior performance for labile modifications, which we use to dramatically improve annotation of glycopeptide spectra. In reanalyzing several large-scale glycoproteomics datasets, we provide increases of 80–560% in the total number of glycoPSMs annotated, ultimately identifying many more glycoproteins and glycosylation sites from the same raw data. MSFragger-Glyco’s ultrafast index-based scoring enables complex searches for hundreds of glycan compositions and several variable modifications, and even fully open searches, in reasonable time. We have demonstrated the potential of these capabilities for both N- and O-linked glycoproteomics data, revealing hundreds of additional glycosites in the N-linked data of Riley et al. and delving into the complexity of O-glycans found in several human samples from Yang et al., uncovering trends in composition that were invisible to the original analysis and distinguishing between phosphorylation and glycan sulfation.
The open and mass offset-style searches employed in MSFragger-Glyco offer dramatic improvements in annotating peptides that have lost partial or complete glycans during fragmentation. Identification of peptides containing multiple glycans is also enhanced with these searches, particularly when glycans have been entirely lost during fragmentation, as in the case of the O-glycopeptide data from Yang et al. These searches offer mixed performance in localizing multiple glycans in a single peptide, as the addition of b/y + HexNAc fragments provides additional information for localization as compared to a variable modification search. However, multiple glycosylation sites on a single peptide are treated as a single mass offset, potentially resulting in poor localization if glycans are only partially fragmented. Identifying the peptide and combined mass of the glycans present recovers many glycopeptides that would not otherwise be annotated, even if exact site localization remains challenging. If there are sufficient glycan-containing fragment ions present, a post-search analysis could utilize the known peptide and glycans to perform a multi-site assignment.
As improvements to glycopeptide enrichment and analysis by MS continue, the quality and complexity of available data will continue to grow. MSFragger-Glyco provides a powerful platform to enable the next generation of glycoproteomics, combining an improved search method for labile glycans with the speed to perform very complex analyses. This method can be extended to glycoproteomic searches in other organisms and systems using by changing the glycan masses, Y and oxonium ions, and/or glycosylation sites in the search parameters. Similarly, MSFragger-Glyco can be used to search other labile modifications that are challenging to assess with conventional search strategies. These capabilities offer the potential to elucidate many areas of biology and disease that have proven challenging or even intractable, including the intricacies of O-glycosylation and large-scale analysis of sulfated and other complex glycans. Further development of post-search analysis tools to take advantage of these capabilities is ongoing, including conversion of matched masses to specific glycans and quantitative analysis of glycoproteomics data.
Online Methods
Raw Data Preparation
In PXD01153310, N-glycopeptides from mouse brain tissue were lectin enriched (Concanavalin A) and analyzed by HCD-pd-AI-ETD LC-MS/MS on an Orbitrap Fusion Lumos mass spectrometer. Downloaded raw data files were centroided and converted to the mzML spectral format using MSConvert47, with HCD and AI-ETD scans filtered to separate mzML files. PXD009476 contains O-glycopeptides enriched from human kidney tissue, CEM T-cells, and human serum using an extraction procedure dubbed ExOO12. Enriched O-glycopeptides were analyzed by HCD LC-MS/MS on Q-Exactive HF Orbitrap mass spectrometer. Downloaded raw data files were centroided and converted to the mzML spectral format using MSConvert version 3.0.19296-ebe17a86f. Liu et al.8 enriched N-glycopeptides from five mouse tissues with ZIC-HILIC and acquired stepped-energy HCD spectra on an Orbitrap Fusion instrument. Downloaded raw data files were centroided and converted to the mzML spectral format using MSConvert. Further details regarding sample preparation and MS analysis can be found in the corresponding publications.
Open and mass offset searches with MSFragger-Glyco
The extension of open searching to include shifted ions for improved scoring with simultaneous localization of the mass shift (termed localization-aware open search) has been described elsewhere22,35. Briefly, searching “shifted ions,” or those resulting from the addition of a known (mass offset search) or unknown (open search) delta mass to a peptide sequence, as well as “regular ions,” or those resulting only from fragmentation of a database peptide, improves the sensitivity and quality of open (and mass offset) search results. Shifted ions can be indexed for rapid search by subtracting the delta mass from the observed precursor mass, enabling MSFragger to search shifted and regular fragment ions from a peptide simultaneously. Glycopeptide spectra are searched by providing potential glycan compositions as a list of allowed mass offsets. Raw spectra were deisotoped and de-charged in MSFragger-Glyco prior to analysis, which proved particularly helpful for high-mass glycopeptides. Spectra were searched with various ion types, depending on the activation method (Fig. 1b). For CID/HCD (vibrational activation only), only regular ions are searched as the glycan is assumed to have dissociated from peptide (either entirely, or partially to form Y-ions or b/y ions with a single HexNAc remaining) (see Supplementary Table 2 for Y ions considered). For ETD/ECD (electronic activation only), regular and shifted ions are matched (as in a typical MSFragger open/mass offset search) assuming the glycan remains intact on the peptide. For hybrid activation (both vibrational and electronic), only regular b/y-type ions (no shifted b/y) are searched along with regular and shifted c/z-type ions to match all possible peptide and glycan fragmentation products simultaneously. A sequon check was added to ensure that only peptides with a potential glycosite are allowed to have Y, b~, or y~ fragment ions and be matched to a mass offset (in mass offset mode). An oxonium ion check was added to ensure that only spectra with evidence of glycan fragmentation can be matched to a glycopeptide. The oxonium ion check searches for a user-provided list of diagnostic fragment ions (the default list is provided in Supplementary Table 3) using the same fragment mass tolerance used in search. Intensities of all diagnostic ions found are summed and compared to the intensity of the base peak in the spectrum. If the summed intensity is greater than the user-provided threshold (default is 10% of the base peak), the spectrum is considered a potential glyco-spectrum. Mass offsets (glycan modifications) are only considered for spectra that pass this oxonium check and matches to glycan-specific fragment ions (Y, b~, and y~) are discarded if the spectrum does not pass the oxonium check.
FDR control for glycoPSMs
Our FDR approach is designed for large-scale glycoproteomics, in which sufficiently many glycopeptide spectra are available for the target-decoy approach to FDR to be used, and is essentially the same as the procedure for FDR control of open search results. Filtering was performed with Philosopher (v.3.2.5) (https://philosopher.nesvilab.org/)42, including PeptideProphet (v5.2.1) modeling of peptide probabilities, ProteinProphet (v5.2.1) protein inference, and Philosopher’s internal filter for FDR control. A combined target and decoy (reversed) protein database is supplied to MSFragger-Glyco. Reversed N-glycan sequons are checked in reversed (decoy) peptides to ensure the same number of potential glycopeptides are searched in both target and decoy databases. The extended mass model of PeptideProphet is used as described in Kong et al.22 to model probabilities for each mass offset (glycan mass) independently to account for the differing probabilities of rare and common glycans. For example, probability distributions for a subset of O-glycan masses (Fig. 1c, top) show a high modeled probability for a delta mass of 365 Da, corresponding to the very common HexNAc-Hex glycan, while a delta mass of 349 Da, corresponding to the much less common HexNAc-Fuc glycan, has nearly zero probability. Delta mass values are binned at a width of 1 Da in PeptideProphet, and precursor isotope error peaks are summed in this example, which is why probabilities can exceed a value of 1. The distribution of delta mass probabilities for decoys (Fig. 1c, “negative”) shows roughly even probabilities for all glycan delta mass values, as hits to decoy peptides are expected to occur randomly without enrichment for specific glycan compositions. Following PeptideProphet, protein inference is performed using standard open search settings in ProteinProphet and filtering is performed in Philosopher to 1% PSM and protein levels. A sequential filtering step is then applied to remove any PSMs matched to proteins that did not pass 1% protein-level FDR.
MSFragger-Glyco computes a hyperscore based on the number of matched fragments and their intensity, which is used to generate an expectation value that is passed to PeptideProphet for modeling. All ion types are considered equally when scoring, including Y ions. As Y ions include the complete mass of the peptide (and have no sequence position dependence), it is possible to obtain high-scoring false positives matching the full Y ion series from a peptide with sufficiently similar mass to the true peptide. As these random events are equally likely for target and decoy peptides, this increases the score require to pass FDR filtering, potentially reducing sensitivity if many Y ions masses are allowed. PeptideProphet parameters were as follows: extended mass model (4000 Da), glycan flag to separately model peptides containing the N-glycan sequon (for N-glycan data only), semi-parametric modeling using expectation scores only, cLevel −2. ProteinProphet was used with default settings except ‘maxppmdiff,’ which was set to 20,000,000 to ensure peptides containing glycan mass offsets were not filtered out. Philosopher filtering was performed at 1% PSM, peptide, and protein levels, followed by sequential filtering of PSMs from the final protein list.
The extended mass model of PeptideProphet functions as the primary method of controlling FDR in glycopeptide-spectrum matching, though glycan FDR is not explicitly controlled. FDR control in glycoproteomics remains challenging48,49, and it is critical to ensure that FDR control and validation is used appropriately to rule out incorporation of low-confidence identifications into reported results. To validate that our approach appropriately controlled FDR for glycoPSMs, we performed several checks. First, there was not a significant difference between the FDR rates for glycoPSMs and non-glycoPSMs, and FDRs for PSMs containing each type of glycan all individually remain near 1% in all analyses performed (Supplementary Tables 8, 9). Second, searches were performed with an equal number of target and decoy glycans provided to MSFragger-Glyco as mass offsets to search to confirm that glycans not present in the data are not found at rates exceeding the expected FDR. Decoy glycans were generated by shifting common glycan masses by +20 Da (N-glycan) or +10 Da (O-glycan), after confirming there was no overlap with other commonly occurring glycans. In total, 0.6% (N-glycan) and 1.3% (O-glycan) of glycoPSMs were matched to decoy glycans (Supplementary Tables 4, 5), which broadly agree with the expected PSM FDR of 1%.
Variable modification searches
A modified version of MSFragger-Glyco was used to perform the comparative variable modification search for N-glycan data. 16 glycan masses were specified as variable modifications on N-X-S/T. The MSFragger-Glyco code was modified to allow specification of the full sequon for a variable modification (the standard version of MSFragger allows specification of single residues only). Oxidized Met was allowed (up to 2 per peptide) but no other variable modifications were allowed. Only 1 glycan was allowed per peptide. All other parameters were as in the N-glycan mass offset searches. For variable modification searches in O-glycan data, standard MSFragger-Glyco was used with HexNAc-Hex (365.1322) and HexNAc (203.0897) specified on Ser/Thr residues (up to 3 each per peptide). Oxidation of Met (up to 2 per peptide) and guanidinylation of Lys (up to 2 per peptide) were allowed as well, and the maximum total number of variable modifications per peptide was set to 4. To match the search used in Yang et al., peptides of length 7 to 46 residues were considered, allowing up to 5 missed cleavages by OpeRATOR. All other parameters were identical to those used in the O-glycan mass offset searches.
N-glycan mass offset search
MzML files were searched with MSFragger-Glyco using 182 mass offsets (Supplementary Table 6), identical to those used by Riley et al.10, against the glycoprotein database used by Riley et al. containing 3,574 entries with decoys added using Philosopher. Trypsin digestion with up to 3 missed cleavages was specified with variable modifications of oxidized Met, protein N-terminal acetylation, and peptide N-terminal pyroglutamate. Peptides containing the consensus sequon (N-X-S/T) and decoy (reversed) peptides containing the reversed sequon were considered as potential glycopeptides. Only spectra containing oxonium ion peaks with summed intensity at least 10% of the base peak were considered for glycan searches. Data was deisotoped and de-charged in MSFragger-Glyco, calibrated, and searched with mass tolerances for precursors and products of 20 and 10 ppm, respectively. Errors in monoisotopic peak detection by the instrument were allowed (+1 and +2 Da). Precursor and electron transfer-no dissociation peaks were removed, and data was square root-transformed prior to analysis. For AI-ETD data, b,y,c,z, and Y ions were considered in searching; for HCD data, b,y,Y and b,y + HexNAc ions were considered. No b or y ions containing the intact glycan were considered in either mode. Spectra were visualized using Byonic viewer and PDV50 to determine appropriate ion types during development. Search results from all raw files and both activation modes were processed together using Philosopher. PSMs and glycopeptides/proteins/sites were compared to those reported in the supporting information of Riley et al. and Liu et al.8 using custom Python 3.7 scripts, pyOpenMS (v2.4)51, and Biopython (v1.74)52. Unique glycopeptides refer to unique peptide sequences present in at least one glycoPSM. Unique glycopeptides were defined as different peptide sequences only, i.e. different glycans and/or other modifications to an existing sequence did not count as additional unique glycopeptides. Glycosites were assigned to the position of the glycan sequon (N-X-S/T) in each glycopeptide, with peptides containing multiple sites excluded from site-specific analyses.
Data from Liu et al. was searched in N-glycan mode in MSFragger-Glyco with the same glycan database used in the original search of Liu et al. (7,884 entries, corresponding to 1,670 unique masses) against the Uniprot mouse proteome database (downloaded 9/24/2019, 17,019 entries) with decoys added in Philosopher. Tryptic digestion was performed with 2 missed cleavages allowed, precursor and fragment mass tolerances of 20 ppm and 10 ppm, respectively, and b, y, b~, y~, and Y ions were used. Y and oxonium ion masses were identical to those used in searching the data of Riley et al.
O-glycan mass offset search
Kidney, Serum, and T cell-derived samples were searched separately with MSFragger-Glyco using reviewed human sequences from UniProtKB (downloaded 08/22/2019, 20464 sequences in total) with decoys and common contaminants added using the Philosopher database command. The OpeRATOR enzyme used in Yang et al.12 cleaves N-terminal to O-glycosylated Ser and Thr. Protein sequences were pre-digested at S/T with up to 10 missed cleavages, as not all Ser and Thr residues are glycosylated and the sites of glycosylation are not known in advance, except for 12-composition searches comparing directly to published results, in which 5 missed cleavages at Ser and Thr were allowed. The resulting peptides were introduced to MSFragger-Glyco as protein sequences in a custom database and digested with Trypsin, allowing 1 missed cleavage. Variable modifications of oxidation (M), guanidinylation and carbamidomethylation (K), and deamidation (N, Q) were specified after initial searches revealed high levels of each in the data. A list of 300 O-glycans (Supplementary Table 7) was curated from open search results on the data and passed to MSFragger-Glyco as a mass offset list. Peptides were required to contain at least one S/T residue to be considered for glycan search, and spectra were required to have summed oxonium ion intensity at least 10% of the base peak. Data was deisotoped and de-charged in MSFragger-Glyco, calibrated with parameter optimization, and searched with mass tolerances for precursors and products of 20 ppm and 10 ppm, respectively. Errors in monoisotopic peak detection by the instrument were allowed (+1 and +2 Da). Precursor peaks were removed, and intensities were square root-transformed prior to analysis. Only unshifted (no glycan) b and y ions were considered in searching as very few spectra retained any glyco-related fragments following HCD. Filtering and validation were performed in Philosopher as for N-glycan AI-ETD data, with the exception of no glycan motif modeling in PeptideProphet.
For competitive search of phosphorylation vs glycosylation, searches were conducted as above except for the following changes. Variable phosphorylation (up to 2 per peptide) was allowed on S, T, Y, and glycan mass offsets included up to 2 sulfations (450 compositions total). The top 25 hits were reported to the pepXML output file to allow comparison of multiple possibilities per spectrum. Only spectra corresponding to PSMs containing either a sulfated glycan or a phosphorylation were considered for comparison. Delta scores were computed by subtracting the top phosphorylation hit from the top sulfated glycan hit (ensuring the same peptide was matched in each case). As shifted ions were disabled for all O-glycan searches, the mass offset search cannot match any ions retaining the modification mass on the fragments. Any case in which the delta score was less than 2 was considered to be indistinguishable. This delta score threshold for indistinguishable possibilities was chosen arbitrarily and is intended as a general illustration of the ability to distinguish these possibilities rather than a true identification cutoff or localization score.
Statistics
MSFragger-Glyco runtimes reported for analysis of Riley et al. N-glycoproteomics data are reported as the average of 3 repeated runs (details in Supplementary Table 1), with standard deviation reported. PSM modeling, validation, and FDR calculations were performed using existing tools as described above, and no additional statistical tests were performed on the output. A summary of statistical methods can be found in the Life Sciences Reporting Summary available with the online version of this paper.
Data Availability
N- and O-linked glycoproteomics raw data was downloaded from the PRIDE Archive53 and Proteome Xchange54, http://proteomecentral.proteomexchange.org, with accession numbers PXD011533 (Riley et al.10 N-glycan data), PXD009476 (Yang et al.12 O-glycan data), and PXD005411, PXD005412, PXD005413, PXD005553, and PXD005555 (Liu et al.8 N-glycan data). Processed search results (raw data, MSFragger output files, and processed peak tables) that support the findings of this study are available in PRIDE (accession number PXD021196).
Code Availability
The MSFragger-Glyco program was developed in the cross-platform Java language, and incorporated in the MSFragger search engine (https://msfragger.nesvilab.org/) starting with version 3.0, which can be accessed at www.nesvilab.org/software.
Supplementary Material
Acknowledgements
This work was funded in part by NIH grants R01-GM-094231 and U24-CA210967.
Footnotes
Ethics Declaration
The authors declare no competing financial or non-financial interests.
Editorial summary:
MSFragger-Glyco allows identification of N- and O-linked glycopeptides using the localization-aware open search strategy of the MSFragger search engine
References
- 1.Varki A. Biological roles of glycans. Glycobiology 27, 3–49, doi: 10.1093/glycob/cww086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thaysen-Andersen M, Packer NH & Schulz BL Maturing glycoproteomics technologies provide unique structural insights into the N-glycoproteome and its regulation in health and disease. Molecular and Cellular Proteomics 15, 1773–1790, doi: 10.1074/mcp.O115.057638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang D & Zaia J. Why glycosylation matters in building a better flu vaccine. Molecular & Cellular Proteomics, mcp.R119.001491-mcp.R001119.001491, doi: 10.1074/mcp.r119.001491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsico, G., Russo, L., Quondamatteo, F. & Pandit, A. Vol. 4 537–552 (Cell Press, 2018). [DOI] [PubMed]
- 5.Schedin-Weiss S, Winblad B & Tjernberg LO The role of protein glycosylation in Alzheimer disease. FEBS Journal 281, 46–62, doi: 10.1111/febs.12590 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Wohlgemuth J, Karas M, Eichhorn T, Hendriks R & Andrecht S. Quantitative site-specific analysis of protein glycosylation by LC-MS using different glycopeptide-enrichment strategies. Analytical Biochemistry 395, 178–188, doi: 10.1016/j.ab.2009.08.023 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Rudd PM & Dwek RA Glycosylation: Heterogeneity and the 3D structure of proteins. Critical Reviews in Biochemistry and Molecular Biology 32, 1–100, doi: 10.3109/10409239709085144 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Liu MQ et al. PGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nature Communications 8, doi: 10.1038/s41467-017-00535-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suttapitugsakul S, Sun F & Wu R. Recent Advances in Glycoproteomic Analysis by Mass Spectrometry. Analytical Chemistry, acs.analchem.9b04651-acs.analchem.04659b04651, doi: 10.1021/acs.analchem.9b04651 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riley NM, Hebert AS, Westphall MS & Coon JJ Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nature Communications 10, 1311–1311, doi: 10.1038/s41467-019-09222-w (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiding, K. R., Bondt, A., Franc, V. & Heck, A. J. R. Vol. 108 260–268 (Elsevier B.V., 2018).
- 12.Yang W, Ao M, Hu Y, Li QK & Zhang H. Mapping the O‐glycoproteome using site‐specific extraction of O‐linked glycopeptides (EXoO). Molecular Systems Biology 14, doi: 10.15252/msb.20188486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King SL et al. Characterizing the O -glycosylation landscape of human plasma, platelets, and endothelial cells. Blood Advances 1, 429–442, doi: 10.1182/bloodadvances.2016002121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bollineni RC, Koehler CJ, Gislefoss RE, Anonsen JH & Thiede B. Large-scale intact glycopeptide identification by Mascot database search. Scientific Reports 8, doi: 10.1038/s41598-018-20331-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bern, M., Kil, Y. J. & Becker, C. Vol. 40 13.20.11–13.20.14 (John Wiley & Sons, Inc., 2012).
- 16.Eng JK, McCormack AL & Yates JR An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry 5, 976–989, doi: 10.1016/1044-0305(94)80016-2 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Zhu Z, Hua D, Clark DF, Go EP & Desaire H. GlycoPep detector: A tool for assigning mass spectrometry data of N-linked glycopeptides on the basis of their electron transfer dissociation spectra. Analytical Chemistry 85, 5023–5032, doi: 10.1021/ac400287n (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu CY et al. Automated Glycan Sequencing from Tandem Mass Spectra of N-Linked Glycopeptides. Analytical Chemistry 88, 5725–5732, doi: 10.1021/acs.analchem.5b04858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, Xin L, Shan B, Lajoie GA & Ma B. GlycoMaster DB: Software to assist the automated identification of N-linked glycopeptides by tandem mass spectrometry. Journal of Proteome Research 13, 3881–3895, doi: 10.1021/pr401115y (2014). [DOI] [PubMed] [Google Scholar]
- 20.Mayampurath A et al. Computational framework for identification of intact glycopeptides in complex samples. Analytical Chemistry 86, 453–463, doi: 10.1021/ac402338u (2014). [DOI] [PubMed] [Google Scholar]
- 21.Eng JK, Jahan TA & Hoopmann MR Comet: An open-source MS/MS sequence database search tool. Proteomics 13, 22–24, doi: 10.1002/pmic.201200439 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D & Nesvizhskii AI MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nature Methods 14, 513–520, doi: 10.1038/nmeth.4256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solntsev SK, Shortreed MR, Frey BL & Smith LM Enhanced Global Post-translational Modification Discovery with MetaMorpheus. Journal of Proteome Research 17, 1844–1851, doi: 10.1021/acs.jproteome.7b00873 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Creasy DM & Cottrell JS Error tolerant searching of uninterpreted tandem mass spectrometry data. Proteomics 2, 1426–1434, doi: (2002). [DOI] [PubMed] [Google Scholar]
- 25.Ma CWM & Lam H. Hunting for unexpected post-translational modifications by spectral library searching with tier-wise scoring. Journal of Proteome Research 13, 2262–2271, doi: 10.1021/pr401006g (2014). [DOI] [PubMed] [Google Scholar]
- 26.Chick JM et al. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nature Biotechnology 33, 743–749, doi: 10.1038/nbt.3267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahrné E, Nikitin F, Lisacek F & Müller M. QuickMod: A tool for open modification spectrum library searches. Journal of Proteome Research 10, 2913–2921, doi: 10.1021/pr200152g (2011). [DOI] [PubMed] [Google Scholar]
- 28.Chi H et al. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nature Biotechnology 36, 1059–1066, doi: 10.1038/nbt.4236 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Na S, Bandeira N & Paek E. Fast multi-blind modification search through tandem mass spectrometry. Molecular and Cellular Proteomics 11, M111.010199-M010111.010199, doi: 10.1074/mcp.M111.010199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swearingen KE et al. A Tandem Mass Spectrometry Sequence Database Search Method for Identification of O-Fucosylated Proteins by Mass Spectrometry. Journal of Proteome Research 18, 652–663, doi: 10.1021/acs.jproteome.8b00638 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trinidad JC, Schoepfer R, Burlingame AL & Medzihradszky KF N- and O-Glycosylation in the murine synaptosome. Molecular and Cellular Proteomics 12, 3474–3488, doi: 10.1074/mcp.M113.030007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chalkley RJ, Baker PR, Medzihardszky KF, Lynn AJ & Burlingame AL In-depth analysis of tandem mass spectrometry data from disparate instrument types. Molecular and Cellular Proteomics 7, 2386–2398, doi: 10.1074/mcp.M800021-MCP200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chalkley RJ & Baker PR Use of a glycosylation site database to improve glycopeptide identification from complex mixtures. Analytical and Bioanalytical Chemistry 409, 571–577, doi: 10.1007/s00216-016-9981-2 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Medzihradszky KF, Kaasik K & Chalkley RJ Tissue-specific glycosylation at the glycopeptide level. Molecular and Cellular Proteomics 14, 2103–2110, doi: 10.1074/mcp.M115.050393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu F et al. Identification of Modified Peptides using Localization-aware Open Search. Nature Communications 11, 4065, doi: 10.1038/s41467-020-17921-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seipert RR et al. Factors that influence fragmentation behavior of N-linked glycopeptide ions. Analytical Chemistry 80, 3684–3692, doi: 10.1021/ac800067y (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wuhrer M, Deelder AM & Van Der Burgt YEM Mass spectrometric glycan rearrangements. Mass Spectrometry Reviews 30, 664–680, doi: 10.1002/mas.20337 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Ledvina AR et al. Infrared photoactivation reduces peptide folding and hydrogenatom migration following ETD tandem mass spectrometry. Angewandte Chemie - International Edition 48, 8526–8528, doi: 10.1002/anie.200903557 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vékey K et al. Fragmentation characteristics of glycopeptides. International Journal of Mass Spectrometry 345–347, 71–79, doi: 10.1016/j.ijms.2012.08.031 (2013). [DOI] [Google Scholar]
- 40.Keller A, Nesvizhskii AI, Kolker E & Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry 74, 5383–5392, doi: 10.1021/ac025747h (2002). [DOI] [PubMed] [Google Scholar]
- 41.Nesvizhskii AI, Keller A, Kolker E & Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry 75, 4646–4658, doi: 10.1021/ac0341261 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Leprevost FD et al. Philosopher: a versatile toolkit for shotgun proteomics data analysis. Nature Methods 17, 869–870, doi: 10.1038/s41592-020-0912-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hang, H. C. & Bertozzi, C. R. Vol. 13 5021–5034 (2005). [DOI] [PubMed]
- 44.Jensen PH, Kolarich D & Packer NH Mucin-type O-glycosylation - putting the pieces together. FEBS Journal 277, 81–94, doi: 10.1111/j.1742-4658.2009.07429.x (2010). [DOI] [PubMed] [Google Scholar]
- 45.Yang Z et al. The GalNAc-type O-glycoproteome of CHO cells characterized by the simplecell strategy. Molecular and Cellular Proteomics 13, 3224–3235, doi: 10.1074/mcp.M114.041541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potel, C. M., Lemeer, S. & Heck, A. J. R. Vol. 91 126–141 (American Chemical Society, 2019).
Methods-only References
- 47.Kessner D, Chambers M, Burke R, Agus D & Mallick P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536, doi: 10.1093/bioinformatics/btn323 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu H, Khatri K, Klein J, Leymarie N & Zaia J. A review of methods for interpretation of glycopeptide tandem mass spectral data. Glycoconjugate Journal 33, 285–296, doi: 10.1007/s10719-015-9633-3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu H, Khatri K & Zaia J. Algorithms and design strategies towards automated glycoproteomics analysis. Mass Spectrometry Reviews 36, 475–498, doi: 10.1002/mas.21487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li K, Vaudel M, Zhang B, Ren Y & Wen B. PDV: An integrative proteomics data viewer. Bioinformatics 35, 1249–1251, doi: 10.1093/bioinformatics/bty770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Röst HL, Schmitt U, Aebersold R & Malmström L. pyOpenMS: A Python-based interface to the OpenMS mass-spectrometry algorithm library. Proteomics 14, 74–77, doi: 10.1002/pmic.201300246 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Cock PJA et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423, doi: 10.1093/bioinformatics/btp163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Research 47, D442–D450, doi: 10.1093/nar/gky1106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deutsch EW et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Research 45, D1100–D1106, doi: 10.1093/nar/gkw936 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
N- and O-linked glycoproteomics raw data was downloaded from the PRIDE Archive53 and Proteome Xchange54, http://proteomecentral.proteomexchange.org, with accession numbers PXD011533 (Riley et al.10 N-glycan data), PXD009476 (Yang et al.12 O-glycan data), and PXD005411, PXD005412, PXD005413, PXD005553, and PXD005555 (Liu et al.8 N-glycan data). Processed search results (raw data, MSFragger output files, and processed peak tables) that support the findings of this study are available in PRIDE (accession number PXD021196).