Abstract
The finding that transformed mouse B-1 and B-2 progenitors give rise to B-cell acute lymphoblastic leukemias (B-ALLs) with varied aggressiveness suggests that B cell lineage might also be a factor in the initiation and progression of pediatric B-ALLs in humans. If this is the case, we hypothesized that human pediatric B-ALLs would share gene expression patterns with mouse B-1 or B-2 progenitors. We tested this premise by deriving a distinct 30-gene B-1 and B-2 progenitor signature that was applied to a microarray dataset of human pediatric ALLs. Cluster analysis revealed that CRLF2, E2A-PBX1, ERG, and ETV6-RUNX1 leukemias were B-1-like, whereas BCR-ABL1, hyperdiploid, and MLL leukemias were B-2-like. Examination of the 30-gene signature in two independent datasets of pediatric ALLs supported this result. Our data suggest that common genetic subtypes of human ALL have their origin in the B-1 or B-2 lineage.
Keywords: Precursor B-Cell Lymphoblastic Leukemia-Lymphoma, Gene Expression, Transcriptome, Gene Expression Profiles, B-Lymphocyte Subsets, Precursor Cells, B-Lymphoid
Introduction
B-acute lymphoblastic leukemia (B-ALL), the most common pediatric malignancy, is associated with chromosomal rearrangements and mutations that play a role in disease initiation and progression1–3. These genetic events have been used to stratify patients into risk groups and likelihood of therapeutic failure. However, the possibility that B-cell lineage also influences the initiation and/or progression of disease has generally not been considered. In this regard, while B-cell development is traditionally viewed as a linear process, it is now appreciated that distinct types of B-lymphocytes are produced in separable fetal and adult waves of development4.
The first B-cells to arise in the fetus are an innate-like lymphocyte population referred to as B-1 B-cells. These cells preferentially localize to serous cavities where they spontaneously secrete immunoglobulins of limited diversity5. Candidate human B-1 B-cells that share functional properties with their mouse counterparts have been described6. B-1 development wanes by late gestation coincident with the emergence of B-2 cells7. B-2 cells are the predominant B-cell population in the spleen and lymph nodes and include a major subpopulation of follicular B-cells that, in response to T-cell help, undergo class switching and somatic hypermutation. Since many translocations associated with leukemia occur in utero8 when B-1 lymphopoiesis peaks and B-2 development initiates, a logical hypothesis is B-ALL can be associated with either lineage.
The ability to resolve phenotypically distinct B-1 and B-2 progenitors in the mouse7 made it possible to test if both types of B-cell progenitor could initiate ALL. To do so, we transduced B-1 and B-2 progenitors with the BCR-ABL1 oncogene and injected the cells into syngeneic recipients. We found that, whereas both populations initiated B-ALL, B-1 progenitors did so with a more rapid kinetics and nearly two-fold higher tumor burden9. These observations provided evidence that, in addition to the particular genetic mutation, B-cell lineage significantly influences ALL development.
These pre-clinical data raise the question as to whether human B-ALLs can be classified as B-1-like or B-2-like. Comparing gene expression data across species has been used to establish associations with disease phenotypes, and this raised the possibility that the recently established whole transcriptome database for murine B-1 and B-2 progenitors10 could be used to probe the extensive gene expression data available for human B-ALL subtypes. One study successfully used this approach and reported that human pediatric ALLs with CRLF2 rearrangements may have a B-1 origin11. We now report the results of a more global comparison of the murine B-1 and B-2 transcriptomes to the different human B-ALL subtypes and demonstrate that a module of only 30 genes can be used to associate different forms of human B-ALL with the B-1 or B-2 lineages.
Materials and methods
Generation of 30-gene B-1 and B-2 progenitor signature
A previously published RNA-seq data set derived from highly purified B-1 and B-2 progenitors derived from mice of varied ages (GSE81411)10 was used to develop a B-1/B-2 progenitor signature. We pooled data from fetal (embryonic day 15) and neonatal (post-natal day 2) mice together so that B-1 and B-2 progenitors each had a combined dataset. From these data we selected human orthologs that were present in St. Jude pediatric ALL microarray data (GSE26281). When more than one mouse probe could be mapped to a human gene, the most variable probe was selected. We identified the most differentially expressed genes between B-1 and B-2 progenitors (p <0.001, log2 fold change>2). The 574 differentially expressed genes were filtered to remove genes that were expressed at very low levels (at least one RNA-seq read count of 0). The Immgen microarray gene expression database was then used to identify genes that are expressed in B cells2. To this end, genes were ranked by log2(maximum expression value across all B cell populations), or Bmax value. 153 genes were selected for further analysis, as this threshold yielded a balanced number of B-1 (n=76) and B-2 (n=77) progenitor genes.
To refine this list of 153 progenitor genes to those relevant in human pediatric B-ALL, we selected the 25% most variable B-1 and B-2 genes12 in GSE26281 and used them to calculate z-scores. We defined sample-specific B-1 z-scores by summing the z-scores of genes up-regulated in B-1 progenitors and dividing it by the square-root of the number of those genes. Similarly, we calculated sample-specific B-2 z-scores using the genes up-regulated in B-2 progenitors. We then calculated the relative B-1 vs. B-2 signal in each B-ALL sample by subtracting the B-2 from the B-1 sample-specific z-scores. B-ALL samples were classified as B-1-like if the resulting signal was > 0.3 or as B-2-like if the signal was < −0.3. Based on these criteria, 6 samples were considered B-1-like and 12 samples were considered B-2-like. We then compared gene expression between the B-1-like and B-2-like samples. We identified 30 genes (20 B-1, 10 B-2) that were significantly increased in either B-1-like or B-2-like B-ALL samples using a moderated t-test13. Our cutoff for significance was p-value < 0.05 and log2 fold change > 2. Supplementary Table 1 summarizes the expression levels of the 30-gene set in B-1 and B-2 progenitors from GSE81411.
Determination of the relative strength of the B-1 and B-2 progenitor signal of a sample
The 30 genes identified above were the basis for determining a B-1 or B-2 association in the ALL genetic databases. We utilized a weighted sum of those 30 genes to derive a sample score for each B-ALL sample. We performed principal component analysis on the 30 genes and considered the absolute rotations as the weights of the B-1 and B-2 up-regulated genes, assigning a negative weight to B-1 up-regulated genes and a positive weight to B-2 up-regulated genes. For each sample, we calculated the sample score as a weighted sum of the gene expression and scaled the values to range between −100 and 100 based on all the samples in a dataset. We used a Kruskal Wallis test to determine whether subtypes within a dataset could be distinguished as B-1-like or B-2-like.
Validation of subtype associations using random gene sets
In the Dutch Childhood Oncology Group dataset (GSE13351) we evaluated whether subtypes were more B1-like or B2-like by utilizing the scoring system described above. We used a resampling-based approach to evaluate whether the assignment of subtypes as relatively B-1-like or B-2-like could have been expected by chance. We calculated the sample scores for 30 random genes in GSE13351 using the approach mentioned in the previous paragraph, and then repeated this calculation for 10,000 permutations of 30 random genes. To determine how B-1-like (or B-2-like) a subtype was, we calculated the proportion of times the mean resampling-based sample score was less (or greater) than the mean observed score obtained from our 30-gene B-1 and B-2 progenitor signature. We applied this approach to the ETV6-RUNX1 and hyperdiploid samples.
Results
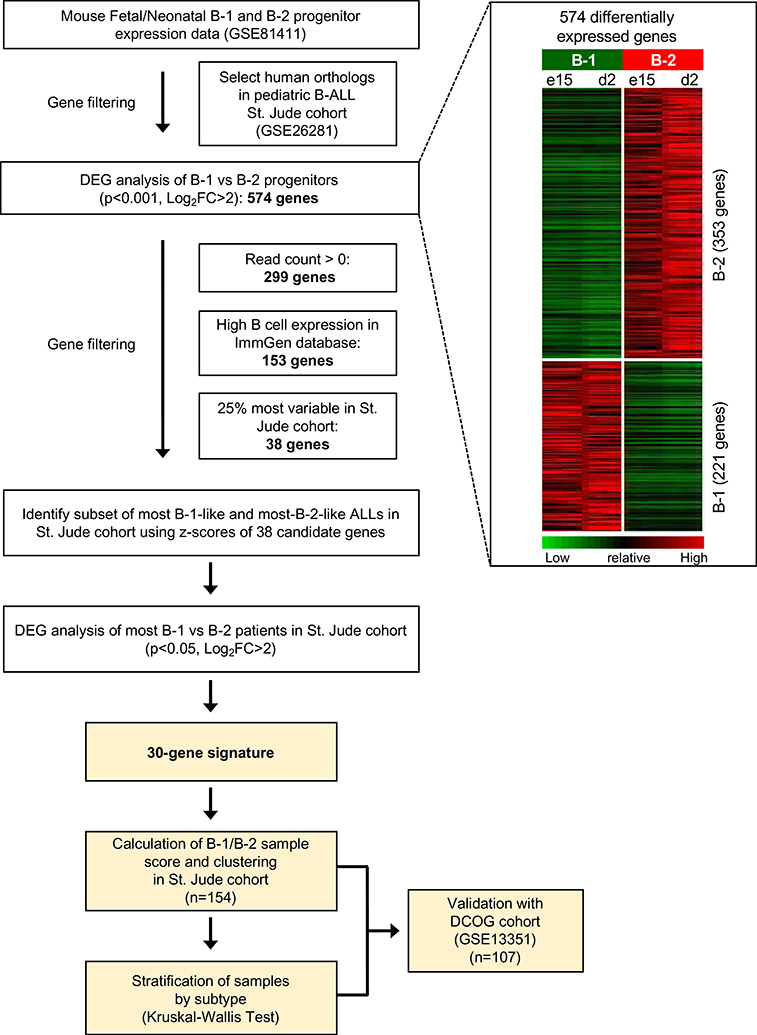
We recently reported the whole transcriptome RNA-seq profiles of B-1 and B-2 progenitors isolated from fetal and neonatal mice (GSE81411)10. We analyzed this database with the goal of identifying B-1 and B-2 progenitor signatures in different subtypes of human B-ALL. First, we selected mouse B-1 and B-2 progenitor genes in GSE81411 that could be mapped to human orthologs in the Affymetrix HG-U133A microarray platform. This platform had been used for transcriptome profiling of our discovery cohort containing 127 human pediatric B-ALLs from St. Jude Children’s Research Hospital (GSE26281). Differential gene expression analysis identified 574 genes with human orthologs that were differentially expressed in mouse B-1 and B-2 progenitor cells. We then selected genes with a read count greater than zero, as zero read count genes were either absent from or expressed at low levels in B cell progenitors (Supplementary Figure 1). Of the 153 genes that were highly expressed in B cells, we selected the 25% most variable genes in GSE26281. The resultant set of 38 genes was refined to include genes that could distinguish between B-1-like and B-2-like pediatric B-ALL samples. To this end, we defined B-1-like and B-2-like pediatric B-ALL samples in GSE26281 by using the 38 gene set. 30 genes were differentially expressed between B-1-like and B-2-like pediatric B-ALLs in GSE26281 and included in the final B-1/B-2 progenitor signature.
The identification of a limited number of genes that could classify ALLs as B-1-like or B-2-like could have diagnostic relevance. We therefore dissected our 153-gene set to determine if a minimal set could be identified. To this end, we mapped the 153-gene signature onto 127 human pediatric B-ALLs from the St. Jude Children’s Research Hospital discovery cohort (GSE26281)14. Z-scores for the 25% most variable genes were calculated and used to classify B-ALLs as B-1-like or B-2-like (detailed description in Methods). Within this subset of highly variable genes, we identified 30 genes that particularly distinguished B-1-like and B-2-like B-ALLs (p < 0.05, fold change > 2) (Table 1). This 30-gene signature was applied to the St Jude as well as Dutch Childhood Oncology Group (DCOG, GSE13351) human ALL gene expression datasets14,15.
Table 1.
List of top 30 B-1 and B-2 progenitor genes from fetal (embryonic day 15) and neonatal (post-natal day 2) mice with differential expression between B-1-like and B-2-like human pediatric B-ALL
| Rank (most B2-like to most B1-like) Gene Symbol Log2 Fold Change | ||
|---|---|---|
| 1 | MN1 | 5.1274 |
| 2 | FLT3 | 4.9679 |
| 3 | TYROBP | 4.7341 |
| 4 | AHNAK | 4.6172 |
| 5 | BST2 | 4.1191 |
| 6 | ID2 | 3.8286 |
| 7 | S100A4 | 3.643 |
| 8 | IFITM1 | 3.5862 |
| 9 | IRF8 | 3.1419 |
| 10 | ANXA2 | 3.0173 |
| 11 | CD72 | −2.5982 |
| 12 | NEIL1 | −2.8807 |
| 13 | CERK | −2.8863 |
| 14 | TRAF5 | −2.9106 |
| 15 | CD79B | −2.957 |
| 16 | BCL2L1 | −3.048 |
| 17 | BCL7A | −3.3489 |
| 18 | BACH2 | −3.3864 |
| 19 | VPREB3 | −3.5482 |
| 20 | IRF4 | −3.5803 |
| 21 | AKAP12 | −3.6366 |
| 22 | POU2AF1 | −3.8021 |
| 23 | LIG4 | −4.1534 |
| 24 | CDC25B | −4.4413 |
| 25 | IGLL1 | −4.4786 |
| 26 | LGR5 | −5.3029 |
| 27 | POLM | −5.7146 |
| 28 | CD19 | −5.9779 |
| 29 | LEF1 | −6.2369 |
| 30 | RASGRP1 | −6.2671 |
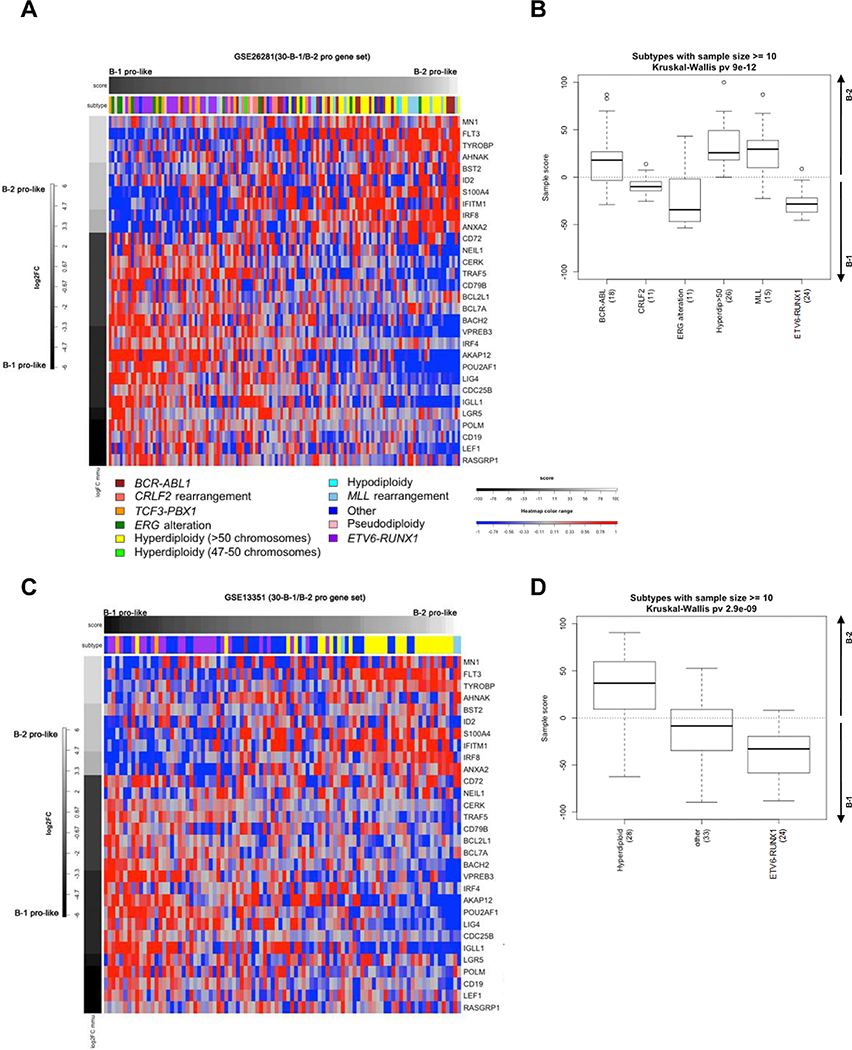
In the St. Jude cohort, principal component analysis was used to weigh our signature of 30 genes for pediatric ALLs that span 12 genetic subtypes. The resulting sample scores were scaled to range from −100 to 100, correlating with most B-1-like to most B-2-like. We ranked pediatric ALLs by sample score and used a heatmap to display the Log2 fold change (Log2FC) for each of the 30 genes (Figure 2A). We observed that genetic subtypes with n≥10 were distinguished as B-1 or B-2-like (Figure 2B, p = 9 × 10−13, Kruskal Wallis). Our data demonstrate that human B-ALLs with CRLF2, ERG, and ETV6-RUNX1 mutations have a more B-1-like signature. Although only a small number of cases were present in the dataset, E2A-PBX1 cases also have a more B1-like signature. In contrast, BCR-ABL1, hyperdiploid, and MLL subtypes have a more B-2-like signature.
Figure 2.
A 30-gene cluster can identify ALLs with B-1 and B-2 signatures. (A) Heatmap of supervised clustering of 127 pediatric ALLs (St. Jude Children’s Research Hospital, GSE26281) using 30 B-1 and B-2 progenitor genes. Columns represent individual samples, ranked by sample score from most B-1-like to most B-2-like, left to right. Rows represent each gene, ranked from most B-2-like to most B-1-like, top to bottom. The blue/red color scale indicates Z scores. (B) Sample scores for ALL subtypes (n≥10) in the St. Jude dataset that are more B-1-like (negative score) or more B-2-like (positive score). (C) Heatmap of supervised clustering of 107 pediatric ALLs (Dutch Childhood Oncology Group, GSE13351) using 30 B-1 and B-2 progenitor genes. Data presented as in Figure 2A. (D) Sample scores for ALL subtypes (n≥10) in the DCOG dataset. Data presented as in Figure 2B.
We applied our mapping strategy to an independent human B-ALL dataset (DCOG, GSE13351; Figure 2C) in order to provide further support for our conclusions. We found that the 30-gene signature again, distinguished ETV6-RUNX1 ALLs as B-1-like and hyperdiploid ALLs as B-2-like (Figure 2D, p = 2.9 × 10−9, Kruskal Wallis). These results demonstrate that the gene signature of B-1 and B-2 progenitors was reflected in common B-ALL subtypes.
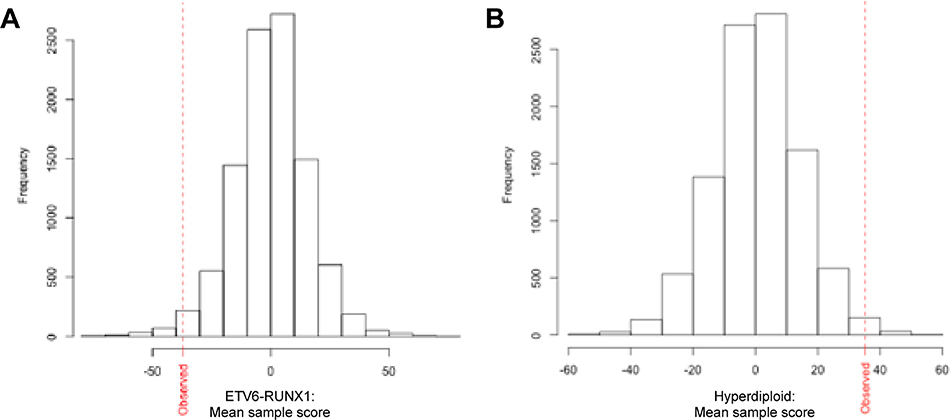
We further evaluated the robustness of our 30-gene signature by comparing its performance to randomly generated signatures. A resampling based approach was used to generate thousands of 30-gene signatures with randomly assigned B-1 and B-2 progenitor genes. We then used each random signature to calculate the B-1/B-2 sample score for ETV6-RUNX1 and hyperdiploidy subtypes in the DCOG validation cohort. We found that our signature was more robust at classifying ETV6-RUNX1 ALLs as B-1-like (p = 0.016; Figure 3A) and hyperdiploid ALLs as B-2-like (p = 0.0093; Figure 3B) than random signatures. These results highlighted the strength of the derived 30-gene signature in classifying pediatric B-ALL subtypes as resembling the B-1 or B-2 progenitor lineage.
Figure 3.
Set of 30 B-1 and B-2 progenitor genes clusters ETV6-RUNX1 ALLs with B-1 genes and hyperdiploid ALLs with B-2 genes more robustly than do random gene sets. Histogram of mean sample scores of (A) ETV6-RUNX1 and (B) hyperdiploid B-ALLs in DCOG cohort calculated for 10,000 random sets of 30 genes compared to mean sample score observed with our set of 30 selected genes (red dotted line). Sample scores are ranked left to right from most B-1-like to most B-2-like. For ETV6-RUNX1 B-ALLs, the proportion of times the permuted gene sets were less than the observed value was 0.016. For hyperdiploid B-ALLs, the proportion of times the permuted gene sets were greater than the observed value was 0.0093.
Discussion
Our previous study in which mouse B-1 and B-2 progenitors were transduced with BCR-ABL1 revealed that B-1 ALL was rapidly progressive while B-2 disease was more indolent9. Thus, our initial expectation was that all B-1-like human B-ALLs should be more aggressive and respond less well to treatment. The fact that ERG-mutated and CRLF2 leukemias have these characteristics is consistent with this prediction. However, ETV6-RUNX1 B-ALL, which was classified as B-1-like, is low risk and has a good prognosis. Conversely, our prediction that B-2-like ALLs should be indolent was clearly not the case as B-2-like MLL leukemias are considered higher risk. These observations indicate that B-1 or B-2 character alone is not a predictor of aggressive disease progression. Instead, how the particular genetic lesion functions in the distinct landscape of the B-1 or B-2 progenitor transcriptomes must be considered.
Previous results have shown that human pediatric CRLF2 B-ALLs have a similar gene expression profile to B-1 lineage ALLs in NUP98-PH23 (NP23) transgenic mice11. With a completely independent method, we also identified an association between the CRLF2 genetic subtype and mouse B-1 progenitor cells, providing further evidence for the classification of CRLF2 ALLs as B-1-like. In addition, we were able to classify five additional genetic subtypes of B-ALL with gene expression profiles that correlate with B cell lineage. A distinction from prior work is that the current study assesses the presence of both B-1 and B-2 progenitor profiles in human B-ALL subtypes, rather than focusing on the B-1 lineage. In broadening our reference gene expression profiles to include both lineages, this study is the first to classify human pediatric ALLs as B-1-like or B-2-like.
A key question is why human leukemias have a B-1 or B-2 signature. One possibility is that the B-1 and B-2 profile indicates that disease initiated in a B-1 or B-2 specified precursor. This possibility is consistent with our data showing that BCR-ABL1 transduced B-1 progenitors can initiate B-ALL9 and another study showing that B-cell leukemia in NP23 transgenic mice is associated with a B-1 progenitor phenotype11. However, we cannot exclude the possibility that a particular translocation or other genetic event activates B-1 or B-2 transcriptional programs in B-cell progenitors. These are not mutually exclusive possibilities, and distinguishing between them will ultimately be dependent upon the identification and manipulation of candidate human B-1 and B-2 lineage cells.
In mice, B-1 and B-2 progenitors have been shown to have differences in developmental and survival pathways9,10. It may be possible to build upon these finding to develop new avenues for therapeutics that are targeted to B-1 and B-2 proliferation and survival pathways. Although many patients with pediatric B-ALL are cured, there may be opportunities to develop less toxic therapeutic approaches and better outcomes for cases that remain high risk.
Supplementary Material
Figure 1.
Strategy for identification and validation of a 30-gene B-1/B-2 progenitor signature for pediatric B-ALL. Flowchart describing the experimental design used to generate a B-1/B-2 progenitor signature. Mouse B-1 and B-2 progenitor RNA-seq data were selected from GSE81411. Genes were filtered for human orthologs in the St. Jude discovery cohort (GSE26281), then used for differentially expressed gene (DEG) analysis (p<0.001, Log2 fold change (FC) >2). The 574 resulting genes were filtered by removing genes with a read count > 0 in all GSE81411 samples and selecting B cell genes from ImmGen. Of the 153 remaining genes, the 25% most variable genes were applied to human pediatric B-ALL samples in the St. Jude cohort to identify B-1-like and B-2-like samples. 30 of the differentially expressed genes between mouse B-1 and B-2 progenitors reached statistical power for distinguishing B-1-like and B-2-like ALL samples (p<0.05, Log2FC>2). The 30-gene signature was applied to the St. Jude cohort and the DCOG validation cohort (GSE13351).
Acknowledgements
This work was supported by NIH grants R21-CA173028 (K.D. and S.C.K.) and P30-CA082103 (R.R., H.B., and A.B.O).
Footnotes
Disclosure of Conflicts of Interest
No relevant conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Iacobucci I, Mullighan CG. Genetic Basis of Acute Lymphoblastic Leukemia. J Clin Oncol. 2017;35(9):975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts KG. Genetics and prognosis of ALL in children vs adults. Hematology Am Soc Hematol Educ Program. 2018;2018(1):137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teitell MA, Pandolfi PP. Molecular genetics of acute lymphoblastic leukemia. Annu Rev Pathol. 2009;4:175–198. [DOI] [PubMed] [Google Scholar]
- 4.Montecino-Rodriguez E, Dorshkind K. B-1 B Cell Development in the Fetus and Adult. Immunity. 2012;36(1):13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantor AB, Herzenberg LA. Origin of murine B cell lineages. Annu. Rev. Immunol. 1993;11:501–538. [DOI] [PubMed] [Google Scholar]
- 6.Griffin DO, Holodick NE, Rothstein TL. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70−. J. Exp. Med. 2011;208(1):67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell–specified progenitor. Nature Immunology. 2006;7(3):293–301. [DOI] [PubMed] [Google Scholar]
- 8.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nature Reviews Cancer. 2003;3(9):639–649. [DOI] [PubMed] [Google Scholar]
- 9.Montecino-Rodriguez E, Li K, Fice M, Dorshkind K. Murine B-1 B Cell Progenitors Initiate B-Acute Lymphoblastic Leukemia with Features of High-Risk Disease. The Journal of Immunology. 2014;192(11):5171–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montecino-Rodriguez E, Fice M, Casero D, et al. Distinct Genetic Networks Orchestrate the Emergence of Specific Waves of Fetal and Adult B-1 and B-2 Development. Immunity. 2016;45(3):527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gough SM, Goldberg L, Pineda M, et al. Progenitor B-1 B-cell acute lymphoblastic leukemia is associated with collaborative mutations in 3 critical pathways. Blood Advances. 2017;1(20):1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Painter MW, Davis S, Hardy RR, et al. Transcriptomes of the B and T Lineages Compared by Multiplatform Microarray Profiling. The Journal of Immunology. 2011;186(5):3047–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueroa ME, Chen S-C, Andersson AK, et al. Integrated genetic and epigenetic analysis of childhood acute lymphoblastic leukemia. J Clin Invest. 2013;123(7):3099–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. The Lancet Oncology. 2009;10(2):125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.