Abstract
To better understand cell-to-cell heterogeneity, advanced analytical tools are in a growing demand for elucidating chemical compositions of each cell within a population. However, the progress of single-cell chemical analysis has been restrained by the limitations of small cell volumes and minute cellular concentrations. Here, we present a rapid and sensitive method for investigating the lipid profiles of isolated single cells using infrared matrix-assisted laser desorption electrospray ionization mass spectrometry (IR-MALDESI-MS). In this work, HeLa cells were dispersed onto a glass slide, and the cellular contents were ionized by IR-MALDESI and measured using a Q-Exactive HF-X mass spectrometer. Importantly, this approach does not require extraction and/or enrichment of analytes prior to MS analysis. Using this approach, 45 distinct lipid species, predominantly phospholipids, were detected and putatively annotated from the single HeLa cells. The proof-of-concept study demonstrates the feasibility and efficacy of IR-MALDESI-MS for rapid lipidomic profiling of single cells, which provides an important basis for future work on differentiation between normal and diseased cells at various developmental states, which can offer new insights into cellular metabolic pathways and pathological processes. Although not yet accomplished, we believe this approach can be readily used as an assessment tool to compare the number of identified species during source evolution and method optimization (intra-laboratory), and also disclose the complementary nature of different direct analytical approaches for the coverage of different types of endogenous analytes (inter-laboratory).
Keywords: Single cell analysis, Lipidome, Mass spectrometry, IR-MALDESI, Orbitrap
Graphical Abstract
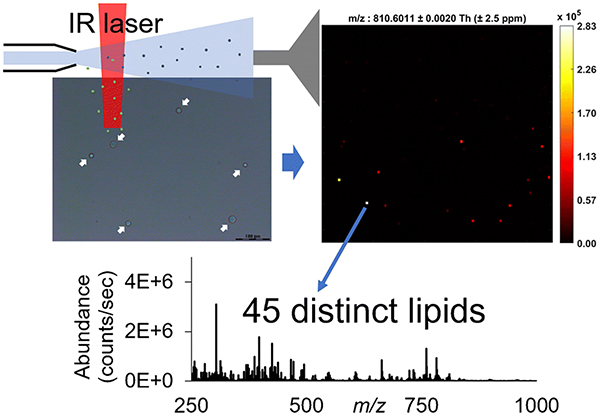
INTRODUCTION
Heterogeneity is an intrinsic attribute, existing everywhere in biological systems. It is reported that even cells originating from the same genome have diverse chemical and physiological features because of their different microenvironments and stochastic processes [1–3]. These unicellular properties carry critical biological information regarding cell activities and molecular functions [4], for disease diagnoses [5, 6] and drug treatments [7]. However, currently biological research focuses on bulk analyses of complex tissues or large cell populations, which yields an averaged overview but lacks the unique insight into individual cells. Owing to the limited sample volume and thus low amounts of constituents in individual cells, probing chemical compositions within a single cell remains challenging, which largely hinders the advancement of this field.
Mass spectrometry (MS) has been regarded as a promising technique to explore chemical contents in cell analyses [1, 8], greatly benefitting from its typically high sensitivity, label-free nature, and relatively high throughput. Over the past few decades, breakthroughs in MS instrumentation and methodology have revolutionized the field of single-cell characterization. A great deal of single-cell work has been carried out with liquid-based methods such as capillary electrophoresis electrospray ionization MS (CE-ESI-MS) which is preceded by using an offline organic extraction [9–12]. However, these approaches usually demand strict sample clean-up and efficient extraction of target analytes for the following MS measurements, which not only are time-consuming [1, 12] but also result in the loss of metabolites from cells [9, 11]. Some in-situ methods, such as secondary ion mass spectrometry (SIMS) and matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS), allow for direct sampling of the cellular contents; thus, have been emerging as powerful and high-throughput alternatives for single-cell studies. Although SIMS and MALDI have achieved remarkable results [13, 14], the regular requirements of operation under vacuum conditions lead to tedious sample preparation and loss of volatile molecules. Moreover, the need for an organic matrix in MALDI increases background signals in the low m/z region, which interfere with the detection of low-molecular-weight analytes [15]. To overcome these obstacles, ambient ionization methods have attracted considerable interest, of which one widely-performed strategy is desorption electrospray ionization (DESI), which has revealed its ability of molecular profiling of single cells like human cheek cells [16], bovine [17], and porcine oocytes [18]. Along with DESI, other commonly used ambient methods promoting in-situ single-cell studies include probe electrospray ionization (PESI) [19–21], laser ablation electrospray ionization (LAESI) [22], and laser desorption/ionization droplet delivery (LDIDD) [23].
Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) is another innovative in-situ ionization method operated under ambient conditions. In IR-MALDESI analyses, a mid-IR laser is utilized to resonantly excite the O−H symmetric and asymmetric stretching bands of endogenous water and/or exogenously deposited ice matrix, inducing ablation of biological samples with few or no sample treatments [24, 25]. The desorbed neutrals are subsequently ionized when they encounter an orthogonal beam of electrospray droplets [26–28]. Due to its matrix- and label-free feature, high salt tolerance [29], and simple preparation procedure, IR-MALDESI-MS has been extensively used to measure lipids and metabolites in a variety of native biological samples, ranging from soft tissues (e.g., hen ovaries [30]), to hard tissues (e.g., mouse bones [31]), to whole-body animals (e.g., zebrafish [32]). Moreover, the capability of IR-MALDESI-MS to detect tissue-specific species, especially cholesterol, at the cellular level was illustrated in our previous work by incorporating the oversampling method [33].
Herein, we report a rapid mass spectrometric method of profiling the lipidome of single HeLa cells dispersed on a glass slide by using IR-MALDESI-MS without extra sample preparative steps. IR-MALDESI was coupled to a Q-Exactive HF-X mass spectrometer. The high capacity transfer tube (HCTT) and electrodynamic ion funnel in HF-X reduce ion losses and boost sensitivity through effective ion transfer [34], enhancing molecular coverage in isolated single cells.
EXPERIMENTAL
Materials
Phosphate buffered saline (PBS) buffer was purchased from Gibco (Grand Island, NY, USA). Acetonitrile, methanol, formic acid, and water in Optima grade were purchased from Fisher Chemical (Fair Lawn, NJ, USA). Nitrogen gas used to purge the sample stage enclosure was purchased from Arc3 Gases (Raleigh, NC, USA).
Cell Culture
HeLa cells (human cervix adenocarcinoma cell line, CCL-2TM) were bought from ATCC (Manassas, VA, USA) and cultured in the Pierce lab (NCSU, Raleigh, USA). Briefly, cells were grown in DMEM with 10% FBS (Thermo Fisher Scientific) and 1× Pen/Strep and incubated at 37 °C in an atmosphere with 95% air and 5% CO2 for 3 days. The medium was renewed once after 12 h incubation. Cells were collected when the flask was ~80% confluent. Cell stocks were kept in growth medium supplemented with 5% (v/v) DMSO at −80 °C for ~12 months until used for the time of analysis.
Single-Cell Sample Preparation
The workflow of sample preparation for the single-cell IR-MALDESI-MS analysis is summarized and depicted in Figure 1. Briefly, the frozen cell stock was thawed in a 37 °C water bath within 1 min and then centrifuged for 5 min at 1000 rpm (Sorvall ST16R centrifuge, Thermo Fisher Scientific, Waltham, MA, USA) to produce a cell pellet. The original cell medium was discarded using a pipette tip. Then the cell pellet was rinsed twice in 1 mL 1× PBS buffer, centrifuged, and resuspended in 330 μL 1× PBS buffer to keep the cells intact, yielding the original cell suspension. In order to establish the optimal cell density, a 10-fold serial dilution in PBS was performed and a set of working cell suspensions (i.e., 10-fold dilution, 100-fold dilution, etc.) was obtained until we observed that the HeLa cells were sufficiently isolated (i.e., the cell-to-cell distance should be larger than the laser spot diameter which is 100 μm on target). 5 μL of each suspension was deposited onto a glass slide for microscope inspection under the view of a LMD7000 microscope (Leica Microsystems, Buffalo Grove, IL, USA). The glass slide deposited with the optimal cell suspension was chosen for subsequent IR-MALDESI-MS single-cell analysis.
Figure 1.
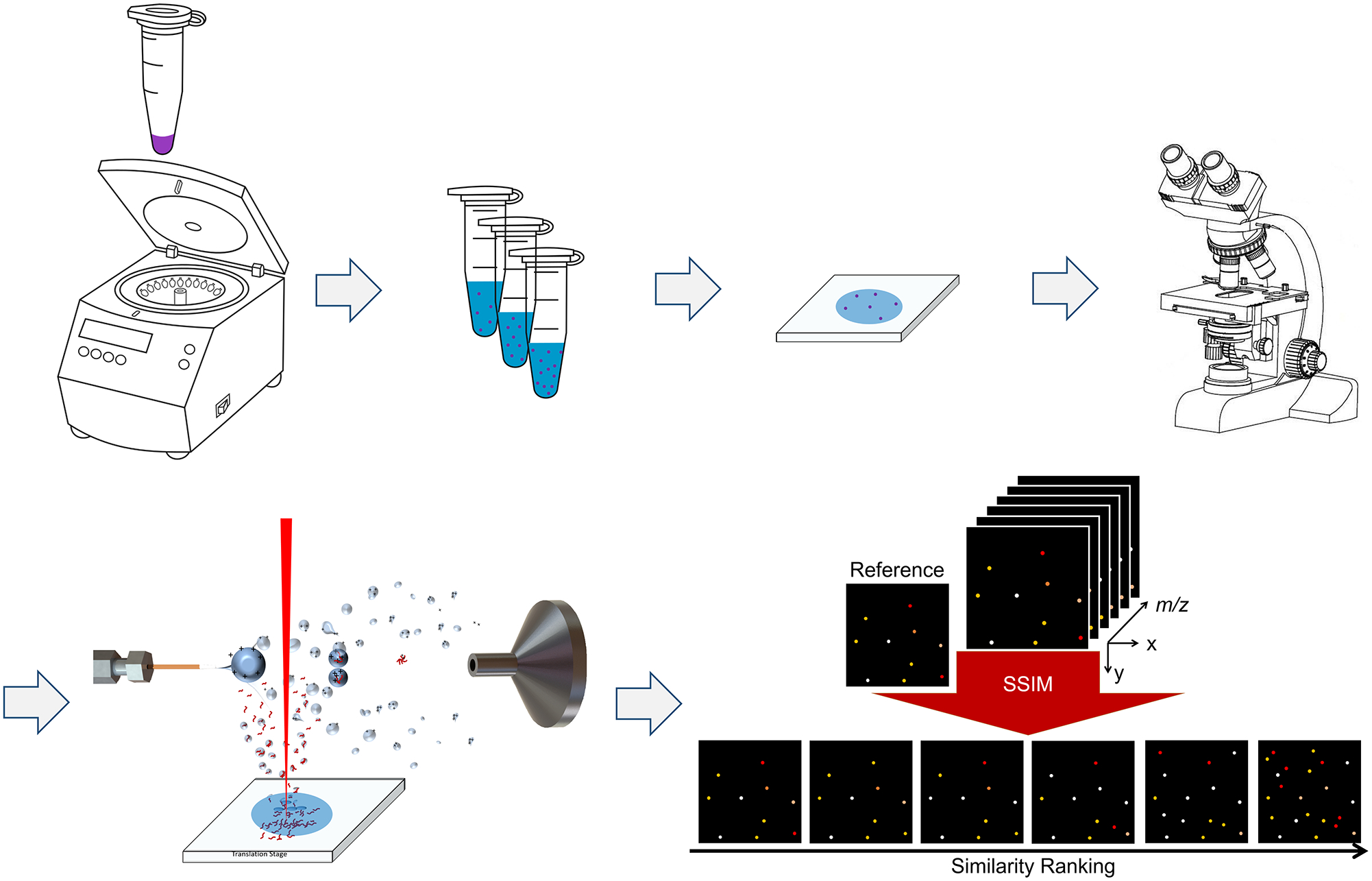
Schematic workflow of single-cell IR-MALDESI-MS analysis. Ion maps in the bottom-right corner were manually drawn to illustrate the SSIM analysis and the peak picking process.
IR-MALDESI-MS Analysis
The experiment was performed on a home-built IR-MALDESI system described thoroughly in previous studies [25, 35]. A “burst-mode” 2970-nm laser (JGM Associates Inc, MA, USA) system [36] was used in this experiment. The laser was set to generate 5 pulses per burst with a total energy of ~ 0.5 mJ/burst at a maximum pulse rate of 10 kHz on the target. An ice matrix was formed on the sample surface after the stage was cooled to −10 °C and was maintained by purging the enclosure with dry nitrogen to a relative humidity of approximately 10% during the entire experiment. The ablated materials were further ionized with an orthogonal electrospray plume. The electrospray solvent was composed of 60% (v/v) aqueous acetonitrile modified with 0.2% formic acid, delivered by a syringe pump (Fusion 101, Thermo Fisher Scientific, Bremen, Germany) at a flow rate of 1.0 μL/min and applied with a voltage of 3.20 kV. A Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) was coupled to the IR-MALDESI source for mass detection. The automatic gain control function (AGC) was turned off, and the injection time (IT) was fixed to 25 ms to keep the laser desorption and ion acquisition events coordinated and to capture a maximum number of endogenous ions. All the MS data was acquired in the positive mode at m/z 250–1000 with a mass resolving power of 120,000 (FHWM at 200 m/z). Parts per million (ppm) mass measurement accuracy can be achieved by using lock mass calibrants [37], which were polysiloxane at m/z 371.1012 [M+H+]+ and diisooctyl phthalate at m/z 391.2843 [M+H+]+. A region-of-interest (ROI) of 9.0 mm-by-8.5 mm on the glass slide was sampled with a Rastir step size of 90 μm-by-100 μm, resulting in a total of 8,500 pixels.
Data Analysis
The raw mass spectra files were converted into mzML format by the msconvert from the ProteoWizard software package [38], and then converted to imzML [39] format by the imzML converter [40]. The imzML files were then loaded into MSiReader [41, 42] (version 1.02, available at https://msireader.ncsu.edu/) built in the MatLab (R2019b; MathWorks, Natick, MA, USA) environment for data analysis. All ion heatmaps were generated in “hot” colormap with ± 2.5 ppm m/z tolerance. To locate the pixels corresponding to cells instead of the background, the ion image of [phosphatidylcholine PC (38:4)+H+]+ at m/z 810.6011 was mapped since it was highly abundant in our previous rat liver data acquired by IR-MALDESI-MS. By comparing one scan containing m/z 810.6011 (single-cell pixel) to the blank scans using the MSiPeakfinder tool in MSiReader, a list of 747 centroid m/z values was pre-generated and considered as cell-specific ion candidates, showing 2× or higher abundance ratios in the single-cell pixel. The MSiCorrelation [43] tool in MSiReader was then utilized to assign a structural similarity (SSIM) index score to each ion image from the list based on the spatial similarity to the reference ion image at m/z 810.6011. Ions with similar spatial distributions to the reference one were highly ranked and manually picked out, and their accurate monoisotopic masses (< ± 2.5 ppm mass error) were putatively annotated via database searches in METLIN [44]. The duplicate ions with the same chemical formula but different adducts, e.g., Na+, were manually filtered out, resulting in the final list of 45 distinct lipid ions.
RESULTS AND DISCUSSION
Cell Dispersion at Different Cell Densities
As one of the most widely investigated human cell lines in the biomedical field [45], HeLa cells were chosen as a well-established model to analyze in this work. Additionally, HeLa cells have a relatively large cell diameter ~40 μm [46], making them easy to visualize under the microscope. After simple pretreatment of the cell stock, a serial 10-fold cell dilution was conducted to fully isolate cells for the following single-cell MS measurement. 5 μL of each cell suspension was directly deposited onto a microscope slide, covering an area of ~1 cm2. The extent of cell dispersion from each suspension was confirmed via a LMD7000 microscope with 20× magnification before the droplet was totally dried out and formed salt crystals, which could mask the presence of the cells. One important observation was that multiple cells clustered together in the droplet from the original cell suspension, revealing that this cell density was too high (Figure 2a and 2b). It can be seen that the cells from the 10-fold dilution were well spread and separated with the distances between the cells more than 100 μm (Figure 2c and 2d). Given the laser spot was measured to be 90 × 100 μm on a 10 μm-thick rat liver section, this cell density was desirable for single-cell IR-MALDESI-MS experiments, enabling each laser shot to target no more than one HeLa cell, hence avoiding obscuring chemical information from other neighboring cells.
Figure 2.
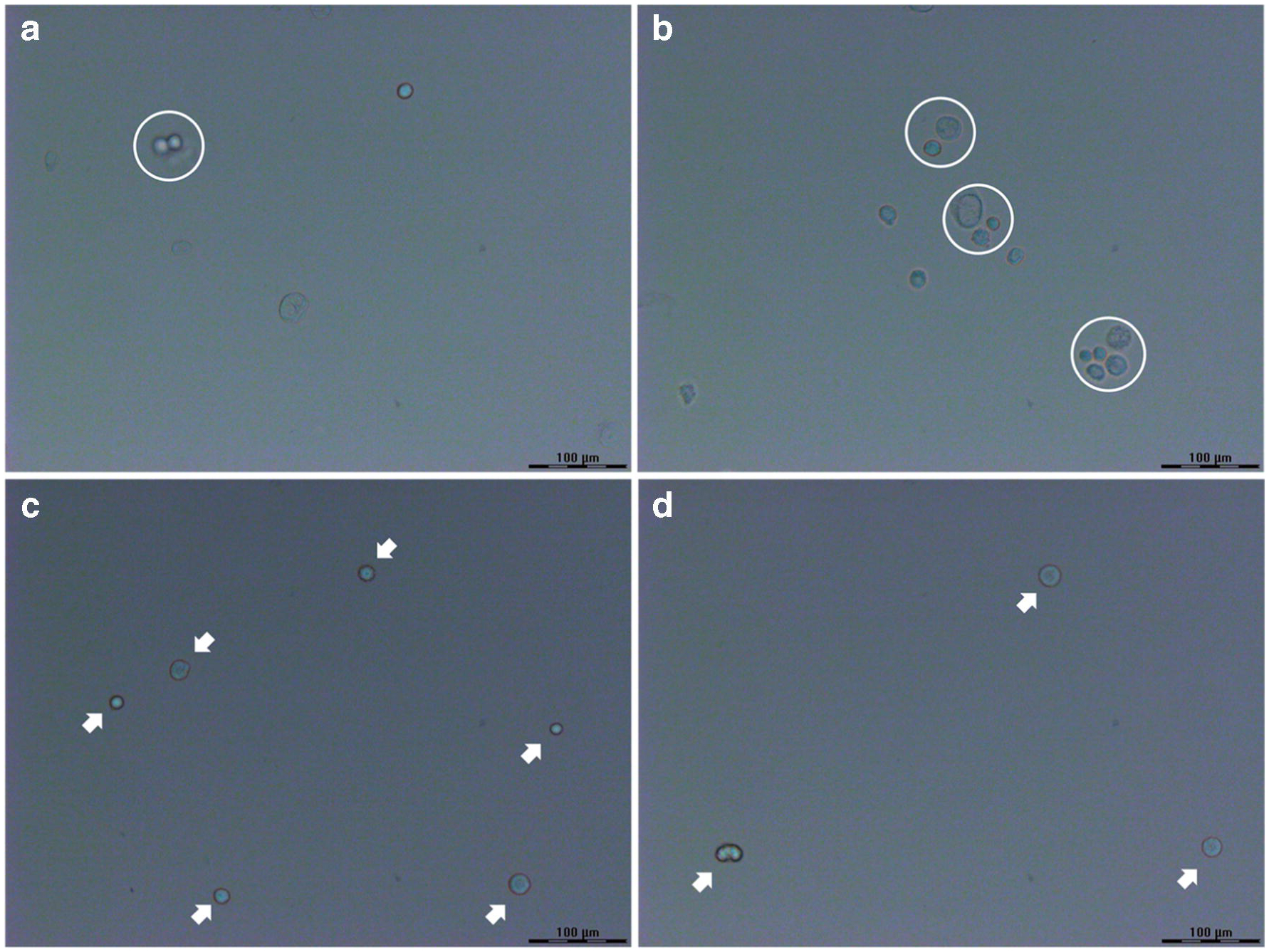
Optical images of HeLa cells dispersed on the glass slides from (a, b) the original cell suspension and (c, d) the 10-fold dilution. The cell clusters in (a, b) are indicated by white circles, and the isolated single cells in (c, d) are pointed by white arrows. Scale bar = 100 μm.
Single Cell Analysis
To determine the locations of the single cells and to generate the cell-specific peak list, the lipid ion [PC (38:4) + H]+ at m/z 810.6011 was selected as the reference due to its significant abundance in the rat liver while absent in the background using IR-MALDESI-MS (data not shown) and existence in HeLa cells reported in the literature [47]. The mass spectra acquired from a single cell pixel containing [PC (38:4)+H]+ at m/z 810.6011 and a background pixel are compared and shown in Figure 3, where 747 mass peaks with >2× higher abundances in the single cell pixels were picked out preliminarily, most of which fell into the mass region of 650–1000 (Figure 3b). Ion images of each m/z value from the list were generated with a mass tolerance of ± 2.5 ppm, then evaluated against the reference image and sorted by their distribution similarity. Among the ions in the pre-generated list, 98 of them exhibited the same spatial distribution as that of m/z 810.6011 and hence were considered as cell-specific ions, and were putatively annotated by matching their experimental m/z values to known metabolites in the METLIN database with a mass tolerance of ± 2.5 ppm. The majority of the ions were tentatively identified as PEs or PCs, which is not surprising because PEs and PCs are one of the primary components of cellular membranes for key cellular functions, e.g., energy storage and cellular signaling [48], and hence have been proposed as valuable biomarkers with great specificity for various diseases such as cancer [49, 50]. Since odd-numbered fatty acids are rare in mammalian cells, the PE and PC species were matched with even-numbered carbon chains. Additionally, several PC ions show a characteristic neutral loss of 59.0735 corresponding to the head group of PCs, i.e., (CH3)3N. Although on the basis of mass measurement accuracy both [LysoPC (15:0)+H+-H2O]+ and [LysoPE (18:0)+H+-H2O]+ are possible identifications for m/z 464.3137, this ion is labeled as [LysoPC (15:0)+H+-H2O]+ in that in the positive mode LysoPCs are much more likely to lose water than LysoPEs according to the literature [51]. Some compounds of different monoisotopic masses were assigned with the same elemental compositions but with different cation adducts (e.g., Na+), which suggests that adding different kinds of dopants such as silver dopants [52] into electrospray solvents might increase the lipidomic coverage in future studies. The duplicate species were manually removed from the list in the following step, reducing the number of lipid ions from 98 detected ions to 45 unique lipid species. The detailed information of all the distinct ions was provided in Table 1, all of which were shown with mass measurement accuracy within ± 2.5 ppm. This number of identifications outperforms some vacuum methods [14, 53, 54] as well as some ambient approaches [16, 19, 55–57]. However, it should be aware that in regards to the diverse ionization mechanisms, mass spectrometer techniques as well as sample subjects, it is hard to directly compare our results with previous reports solely based on the number of identifications; other indicators such as molecular complementarity are also in the primary consideration.
Figure 3.
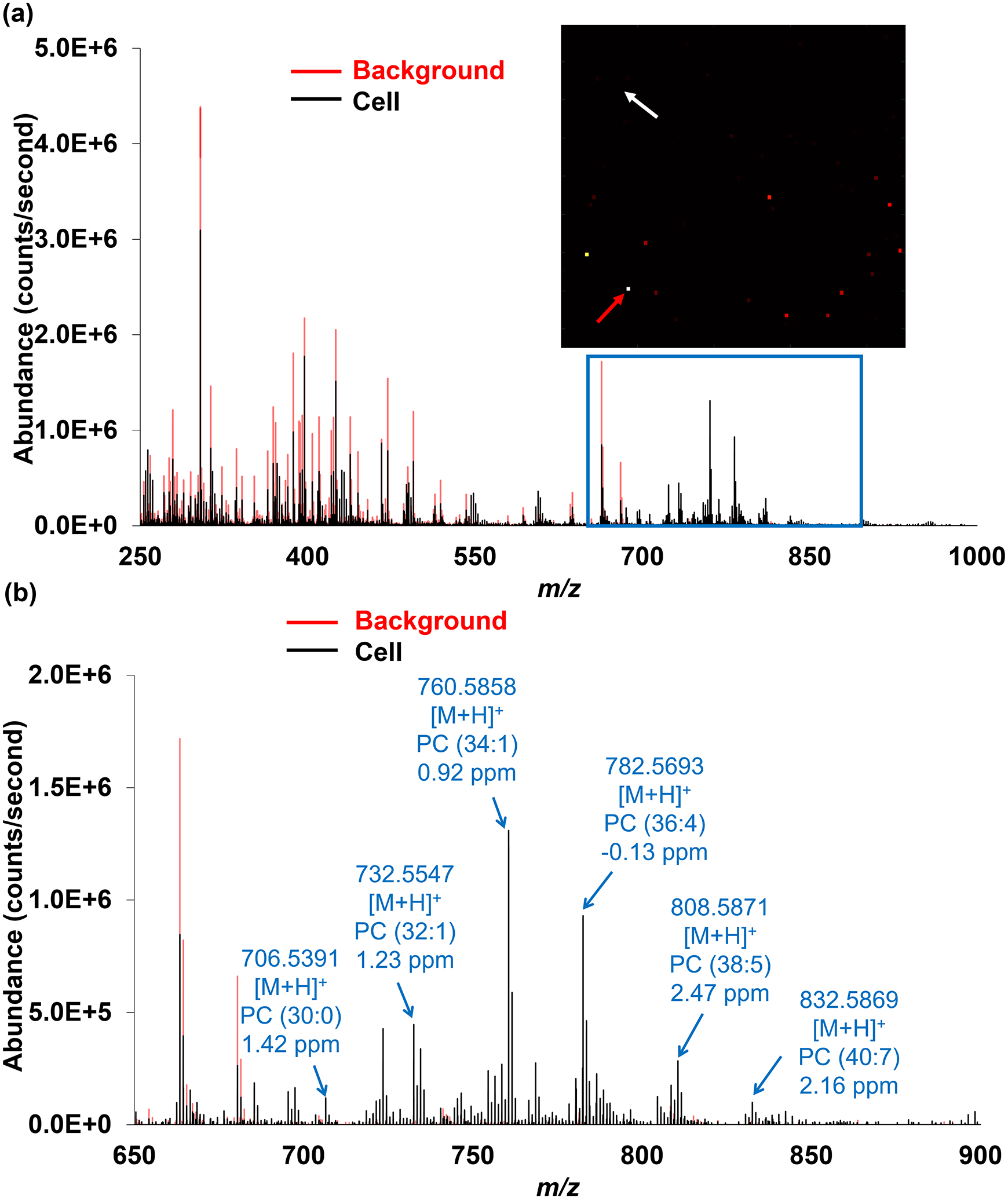
(a) Positive ion mode mass spectrum (m/z 250–1000) from a single cell (pointed by the red arrow) and a background pixel (pointed by the white arrow). (b) Expanded spectrum in the range of m/z 650–900 where most cell-specific ions were detected. Representative peaks are labeled and putatively annotated based on the accurate mass measurements. See Table 1 for a complete list of the detected distinct lipid ions.
Table 1.
List of detected ions with putative identifications. Elemental compositions were determined based on mass measurement accuracy (MMA) and spectral accuracy using MS1 data.
| Experimental m/z | Theoretical m/z | Adduct | Formula | Potential ID | MMA [ppm] |
|---|---|---|---|---|---|
| 369.3515 | 369.3516 | H+-H2O | C27H46O | Cholesterol | −0.27 |
| 464.3137 | 464.3136 | H+-H2O | C23H48NO7P | LysoPC (15:0) | 0.22 |
| 478.3289 | 478.3292 | H+-H2O | C24H50NO7P | LysoPC (16:0) | −0.63 |
| 617.5148 | 617.5140 | H+ | C39H68O5 | DG (36:4) | 1.30 |
| 667.5299 | 667.5296 | H+ | C43H70O5 | DG (40:7) | 0.45 |
| 683.4650 | 683.4646 | Na+ | C43H64O5 | DG (40:10) | 0.59 |
| 697.4797 | 697.4803 | H+- N(CH3)3 | C42H78NO8P | PC (34:3) | −0.86 |
| 703.5735 | 703.5749 | H+ | C39H79N2O6P | SM (d34:1) | −1.99 |
| 706.5391 | 706.5381 | H+ | C38H76NO8P | PC (30:0) | 1.42 |
| 718.5375 | 718.5381 | H+ | C39H76NO8P | PE (34:1) | −0.84 |
| 720.5542 | 720.5538 | H+ | C39H78NO8P | PE (34:0) | 0.56 |
| 724.5261 | 724.5252 | Na+ | C39H76NO7P | PE (P-34:1); PE (O-34:2) | 1.24 |
| 730.5397 | 730.5381 | H+ | C40H76NO8P | PC (32:2) | 2.19 |
| 732.5547 | 732.5538 | H+ | C40H78NO8P | PC (32:1) | 1.23 |
| 734.5716 | 734.5723 | NH4+ | C47H72O5 | DG (44:10) | −0.95 |
| 740.5230 | 740.5225 | H+ | C41H74NO8P | PE (36:4) | 0.68 |
| 744.5547 | 744.5538 | H+ | C41H78NO8P | PE (36:2) | 1.21 |
| 746.5703 | 746.5694 | H+ | C41H80NO8P | PE (36:1) | 1.21 |
| 746.6053 | 746.6053 | H+ | C42H84NO7P | PC (P-34:0); PC (O-34:1) | 0.00 |
| 750.5413 | 750.5408 | Na+ | C41H78NO7P | PE (P-36:2); PE (O-36:3) | 0.67 |
| 752.5579 | 752.5589 | H+ | C43H78NO7P | PE (P-38:4); PE (O-38:5) | −1.33 |
| 756.5527 | 756.5514 756.5538 |
Na+ H+ |
C40H80NO8P C42H78NO8P |
PC (32:0) PC (34:3) |
1.72 −1.45 |
| 758.5689 | 758.5694 | H+ | C42H80NO8P | PC (34:2) | −0.66 |
| 760.5858 | 760.5851 | H+ | C42H82NO8P | PC (34:1) | 0.92 |
| 762.5080 | 762.5068 | H+ | C43H72NO8P | PE (38:7) | 1.57 |
| 764.5224 | 764.5225 | H+ | C43H74NO8P | PE (38:6) | −0.13 |
| 772.5856 | 772.5851 | H+ | C43H82NO8P | PE (38:2) | 0.65 |
| 774.6011 | 774.6007 | H+ | C43H84NO8P | PE (38:1) | 0.52 |
| 780.5526 | 780.5538 | H+ | C44H78NO8P | PC (36:5) | −1.54 |
| 782.5693 | 782.5694 | H+ | C44H80NO8P | PC (36:4) | −0.13 |
| 786.5994 | 786.6007 | H+ | C44H84NO8P | PC (36:2) | −1.65 |
| 788.5191 | 788.5201 | Na+ | C43H76NO8P | PE (38:5) | −1.27 |
| 788.6175 | 788.6164 | H+ | C44H86NO8P | PC (36:1) | 1.39 |
| 790.5350 | 790.5357 | Na+ | C43H78NO8P | PE (38:4) | −0.89 |
| 794.5695 | 794.5694 | H+ | C45H80NO8P | PE (40:5) | 0.13 |
| 794.6073 | 794.6058 | H+ | C46H84NO7P | PC (P-38:4); PC (O-38:5) | 1.89 |
| 796.5869 | 796.5851 | H+ | C45H82NO8P | PE (40:4) | 2.26 |
| 804.5550 | 804.5538 | H+ | C46H78NO8P | PC (38:7) | 1.49 |
| 806.5711 | 806.5694 | H+ | C46H80NO8P | PC (38:6) | 2.11 |
| 808.5871 | 808.5851 | H+ | C46H82NO8P | PC (38:5) | 2.47 |
| 810.6011 | 810.6007 | H+ | C46H84NO8P | PC (38:4) | 0.49 |
| 830.5681 | 830.5694 | H+ | C48H80NO8P | PC (40:8) | −1.57 |
| 832.5869 | 832.5851 | H+ | C48H82NO8P | PC (40:7) | 2.16 |
| 834.6024 | 834.6007 | H+ | C48H84NO8P | PC (40:6) | 2.04 |
| 858.6001 | 858.6007 | H+ | C50H84NO8P | PC (42:8) | −0.70 |
PC = Phosphatidylcholine, DG = Diglyceride, SM = Sphingomyelin, PE = Phosphatidylethanolamine.
The co-localization of the cellular components aided in verifying the presence of the single cells but did not offer sufficient information to make a definitive identification of each detected compound. To further validate the accuracy of the identifications, the theoretical carbon isotopic distribution of each lipid ion was determined and compared against the experimental value. Examples shown in Figure 4 indicate good agreement between the experimental and theoretical relative abundances for four representative lipids at m/z 760.5854, 782.5693, 808.5871, and 369.3516.
Figure 4.
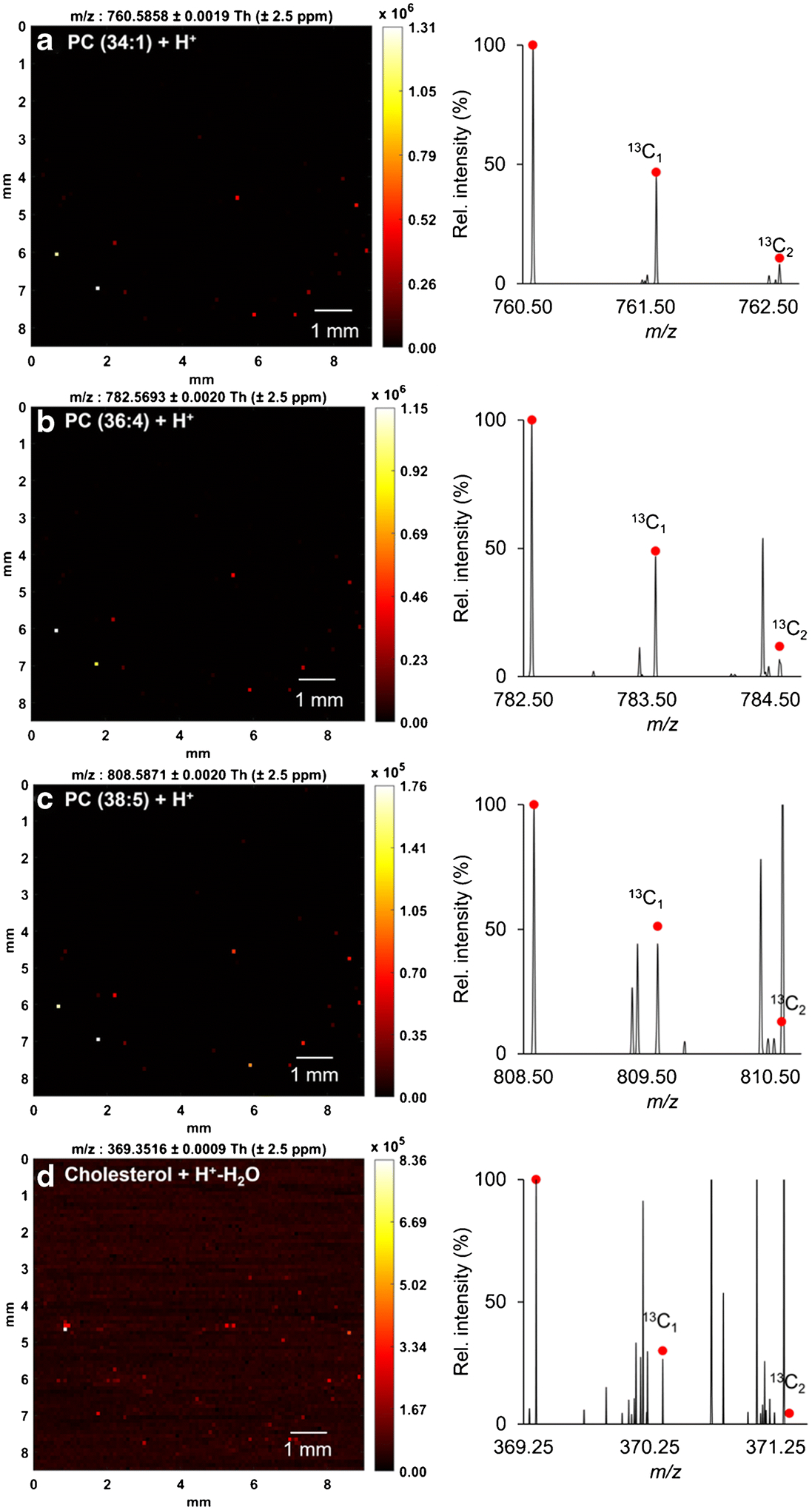
Ion heatmaps (left) and mass spectra (right) of four representative lipids from a single HeLa cell. (a) m/z 760.5854 ([M+H+]+), (b) m/z 782.5693 ([M+H+]+), (c) m/z 808.5871 ([M+H+]+), (d) m/z 369.3516 ([M+H+-H2O]+). Experimental data are overlaid with the theoretical isotopic distribution in red dots.
The data collection of 8,500 (100 × 85) scans was completed in 42 min. It can be seen from the extracted ion chronograms (Figure 5) that 34 scans out of 8,500 showing two major cell-specific MS features at m/z 760.5858 and 782.5693 were indicative of the presence of 34 single cells. In this context, the automatic scanning process was performed at a rate of ~1.2 min/cell, calculated by dividing 42 min by 34 cells. It is noteworthy that the latest IR-MALDESI control software, RastirX, allows arbitrarily shaped ROIs on a sample rather than a rectangular ROI around the entire sample [58]. This will help substantially shorten the duration of single-cell data acquisition by accurately recording the spatial coordinates of single cells when implementing a microscope-linked camera and then automatically scanning the small subregions where cells are located [8] meanwhile minimizing the unwanted nearby areas. Under the same parameters, the RastirX will theoretically accelerate the acquisition rate up to 0.33 second/cell and 0.55 second/cell at a resolving power of 120,000 and 240,000, respectively.
Figure 5.
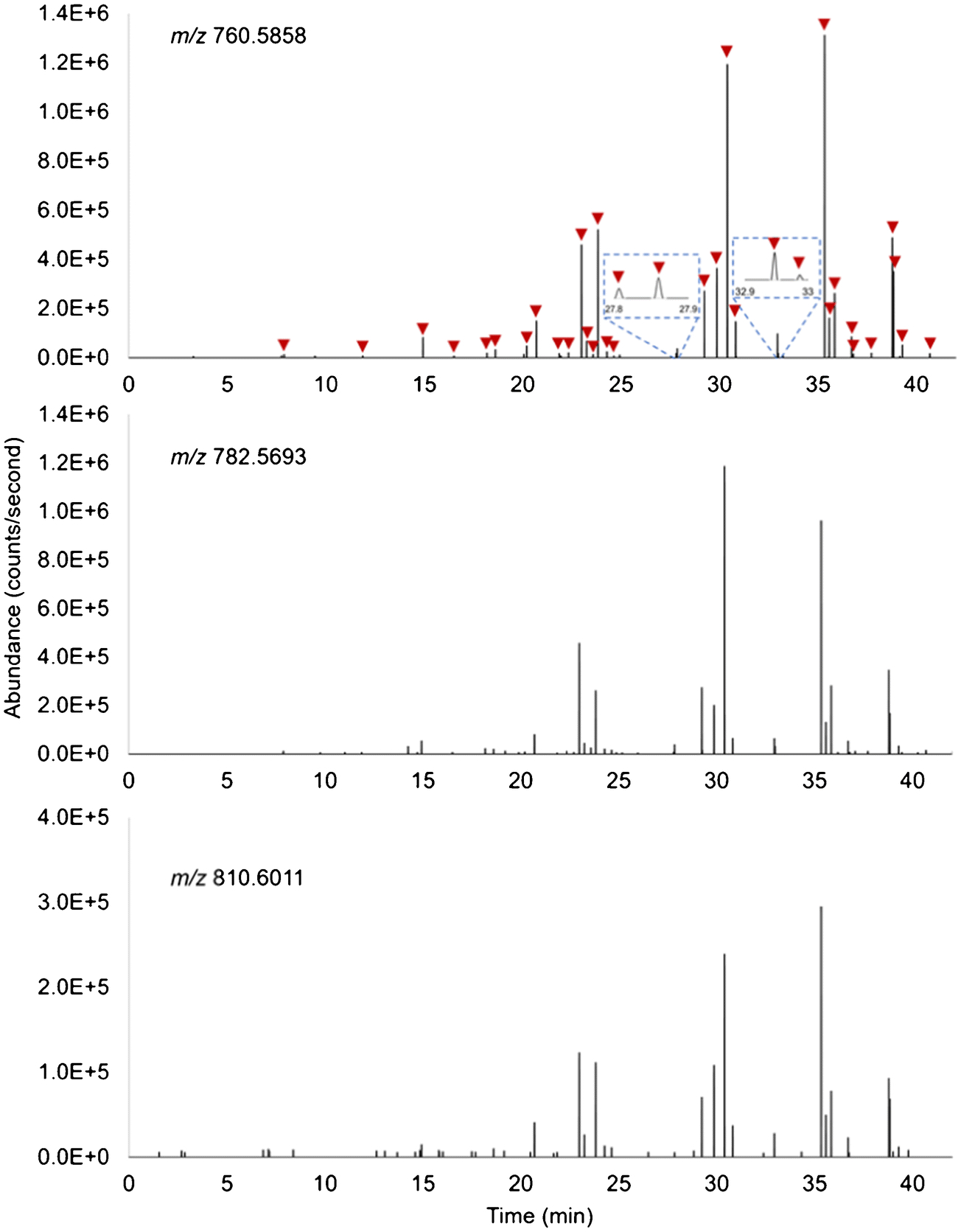
Extracted ion chronograms of lipids at m/z 760.5858 (top), 782.5693 (middle) and 810.6011 (bottom). Each time point recorded in the chronograms correlates to a specific sampling position (pixel) due to stage movement. The sharp signal rise and drop reflect the cell locations. 34 pixels exhibiting both ions at m/z 760.5858 and 782.5693 were considered as cell-located which are marked by red arrows.
Lipidomic Variability among Cells
Investigation of the cell-to-cell heterogeneity among HeLa cells is another focus of this work. An overview in Figure 6a shows the number of lipid ions detected varies from cell to cell. 11 cells show more than 30 out of the 45 lipid species whereas 12 cells contain fewer than 10. There seems to be a positive correlation between the abundances of lipid ions and the frequency of them being detected in these 34 cells (Figure 6b), i.e., lipids detected with higher abundances were observed in more cells. This could be associated with the limit of detection of our method. To give a deeper insight into the cellular heterogeneity, the distributions of the abundances of representative ions are visualized in box plots (Figure 7), showing great divergences among individual cells where ion abundances of lipids can differ over 2 orders of magnitude. The percentage relative standard deviations (%RSDs) in ion abundances of all 45 lipids calculated from the 34 cells range from 61 to 294 (summarized in the Electronic Supplementary Material, ESM). All these results presented above may suggest the intrinsic lipidome differences within a cell population. Nevertheless, it should be pointed out that the evidence here can only be used to present a preliminary picture of lipidomic variability among cells; the validity needs to be confirmed in the future work by applying an internal standard for relative quantification to account for the analytical variability.
Figure 6.
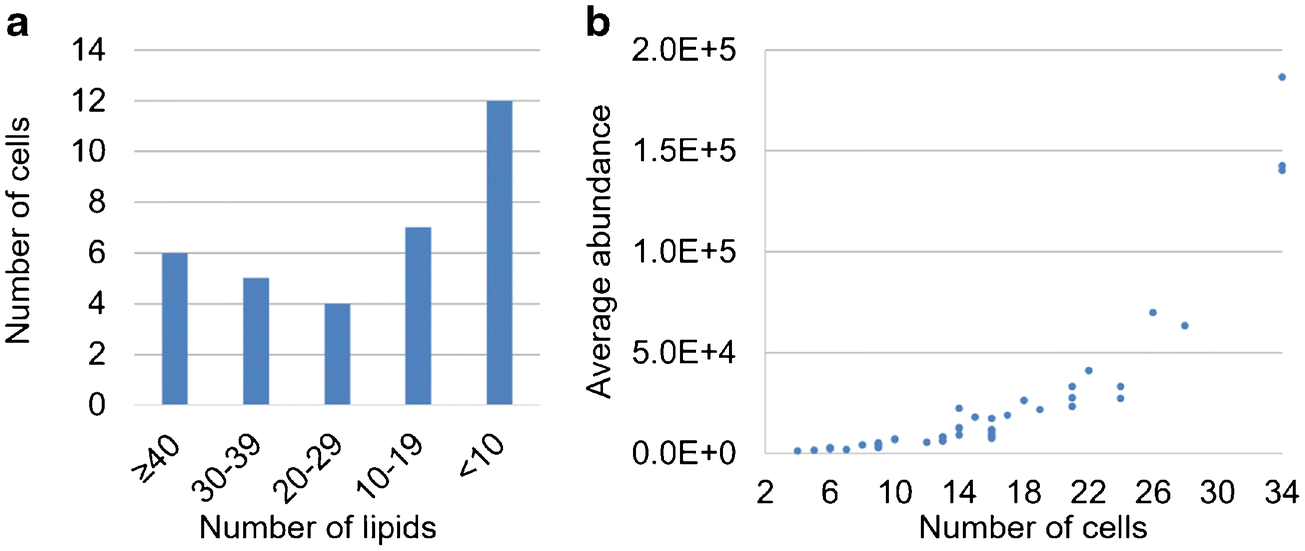
(a) Bar plot summarizes the frequencies of detecting different numbers of lipid species in the 34 cells. (b) Scatter plot exhibits a positive correlation between the ion abundance and the detection frequency.
Figure 7.
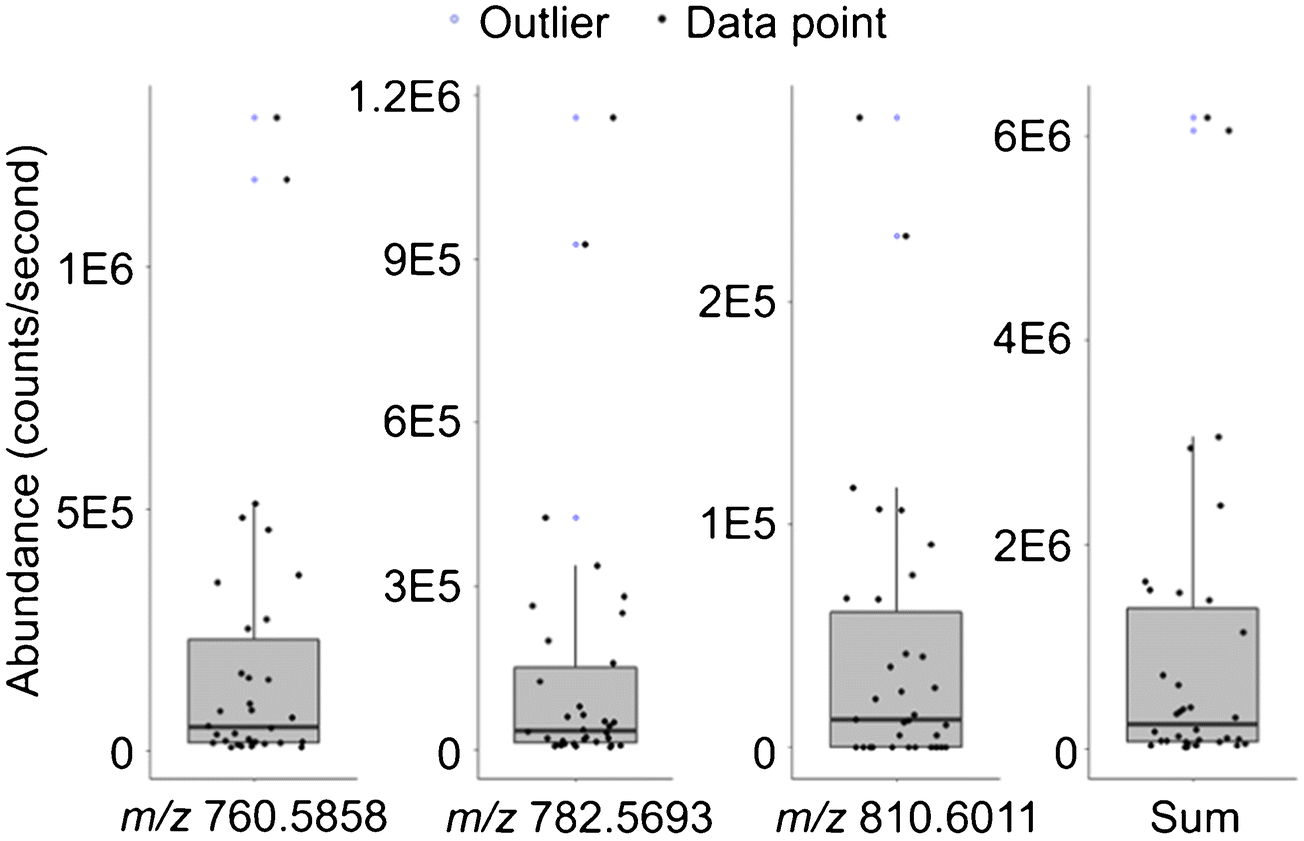
Box plots display distributions of abundances of representative lipid ions (corresponding chronograms shown in Figure 5) acquired from 34 single cells. Sum (far right) represents the summed abundances of all putatively identified lipid ions provided in Table 1 except cholesterol due to the high ion abundance at m/z 369.3515 in the background. The %RSD of the ion abundance (n=34) is 164, 181, 158 and 163 from left to right. Individual data points used to build the box plots are plotted with black filled circles for better visualization. Outliers located outside the whiskers of the box plots (1.5× interquartile range above the third quartile) are represented by blue open circles.
CONCLUSIONS
In this work, we demonstrate the viability of the high-performance IR-MALDESI-Q Exactive HF-X-MS to profile endogenous lipid species from single mammalian cells without additional sample pretreatment. A total of 45 unique cellular lipids from single Hela cells were measured and putatively identified based on their exact m/z’s and carbon isotopic distributions. The number and coverage of identified compounds can be further increased by optimizing the MS parameters by means of exploring different polarity/ m/z ranges and adding dopants into the ESI solvent. With the profiling technique presented here, each sample slide with a deposited area around 1 cm2 can be processed within ~42 min of acquisition time (~1.2 min/cell), allowing for MS analysis in a rapid manner. A considerable reduction in the duration of single-cell data acquisition (<1 second per cell) can be expected with the implementations of RastirX and a microscope-linked camera in the future work. Cell-to-cell heterogeneity was eventually examined in terms of the detected lipids and their abundances, showing noticeable differences across cells with the %RSDs ranging from 61 to 294. Follow-on studies will focus on the development of a relative quantification strategy to reveal genuine biological variability among individual cells. Furthermore, we envision the single-cell sample serves to quantitatively benchmark the attributes of different MS approaches in terms of the number and diversity of detected biomolecules and their ion abundances; this approach might form the basis for conducting an inter-laboratory study.
Supplementary Material
ACKNOWLEDGEMENTS
This work was performed in part by the Molecular Education, Technology and Research Innovation Center (METRIC) at NC State University, which is supported by the State of North Carolina. The authors gratefully acknowledge the financial support received from the National Institutes of Health (R01GM087964). The authors gratefully thank Dr. Crissi Martinez and Professor. Josh Pierce at NCSU for providing the cultured HeLa cells.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
DISCLOSURES
The authors declare that we have no competing financial or non-financial conflicts of interests.
REFERENCES
- 1.Comi TJ, Do TD, Rubakhin SS, Sweedler JV. (2017) Categorizing Cells on the Basis of their Chemical Profiles: Progress in Single-Cell Mass Spectrometry. J Am Chem Soc 139:3920–3929. 10.1021/jacs.6b12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE (2005) Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 122:289–301. 10.1016/j.cell.2005.05.010 [DOI] [PubMed] [Google Scholar]
- 3.Meacham CE, Morrison SJ (2013) Tumour heterogeneity and cancer cell plasticity. Nature 501:328–337. 10.1038/nature12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackermann M (2015) A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 13:497–508. 10.1038/nrmicro3491 [DOI] [PubMed] [Google Scholar]
- 5.Candia J, Maunu R, Driscoll M, Biancotto A, Dagur P, McCoy JP, Sen HN, Wei L, Maritan A, Cao K, Nussenblatt RB, Banavar JR, Losert W (2013) From Cellular Characteristics to Disease Diagnosis: Uncovering Phenotypes with Supercells. PLoS Comput Biol 9:1–10. 10.1371/journal.pcbi.1003215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D, Bodovitz S (2010) Single cell analysis: The new frontier in “omics”. Trends Biotechnol 28:281–290. 10.1016/j.tibtech.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heath JR, Ribas A, Mischel PS (2016) Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov 15:204–216. 10.1038/nrd.2015.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ong TH, Kissick DJ, Jansson ET, Comi TJ, Romanova EV, Rubakhin SS, Sweedler JV. (2015) Classification of Large Cellular Populations and Discovery of Rare Cells Using Single Cell Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Anal Chem 87:7036–7042. 10.1021/acs.analchem.5b01557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lapainis T, Rubakhin SS, Sweedler JV. (2009) Capillary electrophoresis with electrospray ionization mass spectrometric detection for single-cell metabolomics. Anal Chem 81:5858–5864. 10.1021/ac900936g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV. (2011) Metabolic differentiation of neuronal phenotypes by single-cell capillary electrophoresis-electrospray ionization-mass spectrometry. Anal Chem 83:6810–6817. 10.1021/ac2015855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu JX, Aerts JT, Rubakhin SS, Zhang XX, Sweedler JV. (2014) Analysis of endogenous nucleotides by single cell capillary electrophoresis-mass spectrometry. Analyst 139:5835–5842. 10.1039/c4an01133c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemes P, Rubakhin SS, Aerts JT, Sweedler JV. (2013) Qualitative and quantitative metabolomic investigation of single neurons by capillary electrophoresis electrospray ionization mass spectrometry. Nat Protoc 8:783–799. 10.1038/nprot.2013.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Passarelli MK, Ewing AG, Winograd N (2013) Single-cell lipidomics: Characterizing and imaging lipids on the surface of individual Aplysia californica neurons with cluster secondary ion mass spectrometry. Anal Chem 85:2231–2238. 10.1021/ac303038j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schober Y, Guenther S, Spengler B, Römpp A (2012) Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal Chem 84:6293–6297. 10.1021/ac301337h [DOI] [PubMed] [Google Scholar]
- 15.Calvano CD, Monopoli A, Cataldi TRI, Palmisano F (2018) MALDI matrices for low molecular weight compounds: an endless story? Anal Bioanal Chem 410:4015–4038. 10.1007/s00216-018-1014-x [DOI] [PubMed] [Google Scholar]
- 16.Bergman HM, Lanekoff I (2017) Profiling and quantifying endogenous molecules in single cells using nano-DESI MS. Analyst 142:3639–3647. 10.1039/c7an00885f [DOI] [PubMed] [Google Scholar]
- 17.González-Serrano AF, Pirro V, Ferreira CR, Oliveri P, Eberlin LS, Heinzmann J, Lucas-Hahn A, Niemann H, Cooks RG (2013) Desorption Electrospray Ionization Mass Spectrometry Reveals Lipid Metabolism of Individual Oocytes and Embryos. PLoS One 8:1–11. 10.1371/journal.pone.0074981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirro V, Oliveri P, Ferreira CR, González-Serrano AF, Machaty Z, Cooks RG (2014) Lipid characterization of individual porcine oocytes by dual mode DESI-MS and data fusion. Anal Chim Acta 848:51–60. 10.1016/j.aca.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan N, Rao W, Kothapalli NR, Liu R, Burgett AWG, Yang Z (2014) The Single-probe: A miniaturized multifunctional device for single cell mass spectrometry analysis. Anal Chem 86:9376–9380. 10.1021/ac5029038 [DOI] [PubMed] [Google Scholar]
- 20.Liu R, Pan N, Zhu Y, Yang Z (2018) T-Probe: An Integrated Microscale Device for Online in Situ Single Cell Analysis and Metabolic Profiling Using Mass Spectrometry. Anal Chem 90:11078–11085. 10.1021/acs.analchem.8b02927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong X, Zhao Y, Cai S, Fu S, Yang C, Zhang S, Zhang X (2014) Single cell analysis with probe ESI-mass spectrometry: Detection of metabolites at cellular and subcellular levels. Anal Chem 86:3809–3816. 10.1021/ac500882e [DOI] [PubMed] [Google Scholar]
- 22.Samarah LZ, Khattar R, Tran TH, Stopka SA, Brantner CA, Parlanti P, Veličković D, Shaw JB, Agtuca BJ, Stacey G, Paša-Tolić L, Tolić N, Anderton CR, Vertes A (2020) Single-Cell Metabolic Profiling: Metabolite Formulas from Isotopic Fine Structures in Heterogeneous Plant Cell Populations. Anal Chem 92:7289–7298. 10.1021/acs.analchem.0c00936 [DOI] [PubMed] [Google Scholar]
- 23.Lee JK, Jansson ET, Nam HG, Zare RN (2016) High-Resolution Live-Cell Imaging and Analysis by Laser Desorption/Ionization Droplet Delivery Mass Spectrometry. Anal Chem 88:5453–5461. 10.1021/acs.analchem.6b00881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokhart MT, Rosen E, Thompson C, Sykes C, Kashuba ADM, Muddiman DC (2015) Quantitative mass spectrometry imaging of emtricitabine in cervical tissue model using infrared matrix-assisted laser desorption electrospray ionization. Anal Bioanal Chem 407:2073–2084. 10.1007/s00216-014-8220-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robichaud G, Barry JA, Muddiman DC (2014) IR-MALDESI mass spectrometry imaging of biological tissue sections using ice as a matrix. J Am Soc Mass Spectrom 25:319–328. 10.1007/s13361-013-0787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu A, Muddiman DC (2019) Internal Energy Deposition in Infrared Matrix-Assisted Laser Desorption Electrospray Ionization With and Without the Use of Ice as a Matrix. J Am Soc Mass Spectrom 30:2380–2391. 10.1007/s13361-019-02323-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampson JS, Hawkridge AM, Muddiman DC (2006) Generation and Detection of Multiply-Charged Peptides and Proteins by Matrix-Assisted Laser Desorption Electrospray Ionization (MALDESI) Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. J Am Soc Mass Spectrom 17:1712–1716. 10.1016/j.jasms.2006.08.003 [DOI] [PubMed] [Google Scholar]
- 28.Dixon RB, Muddiman DC (2010) Study of the ionization mechanism in hybrid laser based desorption techniques. Analyst 135:880–882. 10.1039/b926422a [DOI] [PubMed] [Google Scholar]
- 29.Fideler J, Johanningsmeier SD, Ekelöf M, Muddiman DC (2019) Discovery and quantification of bioactive peptides in fermented cucumber by direct analysis IR-MALDESI mass spectrometry and LC-QQQ-MS. Food Chem 271:715–723. 10.1016/j.foodchem.2018.07.187 [DOI] [PubMed] [Google Scholar]
- 30.Nazari M, Muddiman DC (2016) Polarity switching mass spectrometry imaging of healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Analyst 141:595–605. 10.1039/c5an01513h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khodjaniyazova S, Hanne NJ, Cole JH, Muddiman DC (2019) Mass spectrometry imaging (MSI) of fresh bones using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Anal Methods 11:5929–5938. 10.1039/c9ay01886g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stutts WL, Knuth MM, Ekelöf M, Mahapatra D, Kullman SW, Muddiman DC (2020) Methods for Cryosectioning and Mass Spectrometry Imaging of Whole-Body Zebrafish. J Am Soc Mass Spectrom 31:768–772. 10.1021/jasms.9b00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nazari M, Muddiman DC (2015) Cellular-level mass spectrometry imaging using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) by oversampling. Anal Bioanal Chem 407:2265–2271. 10.1007/s00216-014-8376-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scientific T (2017) Thermo Scientific Q Exactive HF-X Hybrid Quadrupole-Orbitrap MS System. thermofisher.com/QExactiveHFX
- 35.Robichaud G, Barry JA, Garrard KP, Muddiman DC (2013) Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J Am Soc Mass Spectrom 24:92–100. 10.1007/s13361-012-0505-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ekelöf M, Manni J, Nazari M, Bokhart M, Muddiman DC (2018) Characterization of a novel miniaturized burst-mode infrared laser system for IR-MALDESI mass spectrometry imaging. Anal Bioanal Chem 410:2395–2402. 10.1007/s00216-018-0918-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen JV, de Godoy LMF, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M (2005) Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 4:2010–2021. 10.1074/mcp.T500030-MCP200 [DOI] [PubMed] [Google Scholar]
- 38.Kessner D, Chambers M, Burke R, Agus D, Mallick P (2008) ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 24:2534–2536. 10.1093/bioinformatics/btn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schramm T, Hester A, Klinkert I, Both JP, Heeren RMA, Brunelle A, Laprévote O, Desbenoit N, Robbe MF, Stoeckli M, Spengler B, Römpp A (2012) ImzML - A common data format for the flexible exchange and processing of mass spectrometry imaging data. J Proteomics 75:5106–5110. 10.1016/j.jprot.2012.07.026 [DOI] [PubMed] [Google Scholar]
- 40.Race AM, Styles IB, Bunch J (2012) Inclusive sharing of mass spectrometry imaging data requires a converter for all. J Proteomics 75:5111–5112. 10.1016/j.jprot.2012.05.035 [DOI] [PubMed] [Google Scholar]
- 41.Robichaud G, Garrard KP, Barry JA, Muddiman DC (2013) MSiReader: An open-source interface to view and analyze high resolving power MS imaging files on matlab platform. J Am Soc Mass Spectrom 24:718–721. 10.1007/s13361-013-0607-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bokhart MT, Nazari M, Garrard KP, Muddiman DC (2018) MSiReader v1.0: Evolving Open-Source Mass Spectrometry Imaging Software for Targeted and Untargeted Analyses. J Am Soc Mass Spectrom 29:8–16. 10.1007/s13361-017-1809-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ekelöf M, Garrard KP, Judd R, Rosen EP, Xie DY, Kashuba ADM, Muddiman DC (2018) Evaluation of Digital Image Recognition Methods for Mass Spectrometry Imaging Data Analysis. J Am Soc Mass Spectrom 29:2467–2470. 10.1007/s13361-018-2073-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, Koellensperger G, Huan T, Uritboonthai W, Aisporna AE, Wolan DW, Spilker ME, Benton HP, Siuzdak G (2018) METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal Chem 90:3156–3164. 10.1021/acs.analchem.7b04424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cells HeLa (1951). In: Br. Soc. Immunol https://www.immunology.org/hela-cells-1951
- 46.Milo R, Phillips R (2015) Cell Biology by the Numbers. Garland Science, New York [Google Scholar]
- 47.Naguib A, Bencze G, Engle DD, Chio IIC, Herzka T, Watrud K, Bencze S, Tuveson DA, Pappin DJ, Trotman LC (2015) P53 mutations change phosphatidylinositol acyl chain composition. Cell Rep 10:8–19. 10.1016/j.celrep.2014.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brügger B (2014) Lipidomics: Analysis of the Lipid Composition of Cells and Subcellular Organelles by Electrospray Ionization Mass Spectrometry. Annu Rev Biochem 83:79–98. 10.1146/annurev-biochem-060713-035324 [DOI] [PubMed] [Google Scholar]
- 49.Yan F, Zhao H, Zeng Y (2018) Lipidomics: a promising cancer biomarker. Clin Transl Med 7:21–23. 10.1186/s40169-018-0199-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bandu R, Mok HJ, Kim KP (2018) PHOSPHOLIPIDS AS CANCER BIOMARKERS: MASS SPECTROMETRY-BASED ANALYSIS. Mass Spectrom Rev 37:107–138. 10.1002/mas.21510 [DOI] [PubMed] [Google Scholar]
- 51.Milne S, Ivanova P, Forrester J, Alex Brown H (2006) Lipidomics: An analysis of cellular lipids by ESI-MS. Methods 39:92–103. 10.1016/j.ymeth.2006.05.014 [DOI] [PubMed] [Google Scholar]
- 52.Meier F, Garrard KP, Muddiman DC (2014) Silver dopants for targeted and untargeted direct analysis of unsaturated lipids via infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Rapid Commun Mass Spectrom 28:2461–2470. 10.1002/rcm.7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dueñas ME, Essner JJ, Lee YJ (2017) 3D MALDI Mass Spectrometry Imaging of a Single Cell: Spatial Mapping of Lipids in the Embryonic Development of Zebrafish. Sci Rep 7:1–10. 10.1038/s41598-017-14949-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Do TD, Comi TJ, Dunham SJB, Rubakhin SS, Sweedler JV. (2017) Single Cell Profiling Using Ionic Liquid Matrix-Enhanced Secondary Ion Mass Spectrometry for Neuronal Cell Type Differentiation. Anal Chem 89:3078–3086. 10.1021/acs.analchem.6b04819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen F, Lin L, Zhang J, He Z, Uchiyama K, Lin JM (2016) Single-Cell Analysis Using Drop-on-Demand Inkjet Printing and Probe Electrospray Ionization Mass Spectrometry. Anal Chem 88:4354–4360. 10.1021/acs.analchem.5b04749 [DOI] [PubMed] [Google Scholar]
- 56.Kompauer M, Heiles S, Spengler B (2016) Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat Methods 14:90–96. 10.1038/nmeth.4071 [DOI] [PubMed] [Google Scholar]
- 57.Huang Q, Mao S, Khan M, Zhou L, Lin JM (2018) Dean flow assisted cell ordering system for lipid profiling in single-cells using mass spectrometry. Chem Commun 54:2595–2598. 10.1039/c7cc09608a [DOI] [PubMed] [Google Scholar]
- 58.Garrard KP, Ekelöf M, Khodjaniyazova S, Bagley MC, Muddiman DC (2020) A Versatile Platform for Mass Spectrometry Imaging of Arbitrary Spatial Patterns. J Am Soc Mass Spectrom. 10.1021/jasms.0c00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


