Abstract
Genome-wide association studies (GWAS) of neurological diseases have identified thousands of variants associated with disease phenotypes. However, the majority of these variants do not alter coding sequences, making it difficult to assign their function. Here, we present a multi-omic epigenetic atlas of the adult human brain through profiling of single-cell chromatin accessibility landscapes and three-dimensional (3D) chromatin interactions of diverse adult brain regions across a cohort of cognitively healthy individuals. We developed a machine-learning classifier to integrate this multi-omic framework and predict dozens of functional single-nucleotide polymorphisms (SNPs) for Alzheimer’s disease (AD) and Parkinson’s disease (PD), nominating target genes and cell types for previously orphaned GWAS loci. Moreover, we dissected the complex inverted haplotype of the MAPT (encoding tau) PD risk locus, identifying putative ectopic regulatory interactions in neurons that may mediate this disease association. This work expands our understanding of inherited variation and provides a roadmap for the epigenomic dissection of causal regulatory variation in disease.
INTRODUCTION
AD and PD affect ~50 and ~10 million individuals world-wide, as two of the most common neurodegenerative disorders. Several large consortia have assembled GWAS that associate genetic loci with clinical diagnoses of probable AD dementia1–4 or probable PD5–7, or with their characteristic pathologic features. These efforts have led to the identification of dozens of potential risk loci for these diseases. However, most risk loci reside in noncoding regions and so it remains unclear if the nominated (often nearest) gene is functionally relevant for the disease, or if another gene is involved8.
Most functional noncoding SNPs would be predicted to exert their effects through alteration of gene expression via perturbation of transcription factor (TF) binding and regulatory element function8. Such regulatory elements are highly cell type-specific9, suggesting that the resultant effects of noncoding SNPs would be equally cell type-specific. Thus, comprehensive nomination of putative functional noncoding SNPs in the brain requires cataloging the regulatory elements that are active in every brain cell type in the correct organismal and regional context. These critical data hold the promise to illuminate the functional significance of genetic risk loci in the molecular pathogenesis of common neurodegenerative diseases.
Previous work has carefully mapped such cell type-specific gene regulatory landscapes in human brain, predominantly during early developmental time points10, in organoid culture systems11–13, or in induced pluripotent stem cell-derived cellular models14,15. Additional studies have profiled chromatin accessibility in macrodissected post-mortem adult human brain16–19. Such data sets have provided a rich resource for the nomination of putative functional SNPs in neurologic disease using multi-omic approaches10,14,17,20. Moreover, recent work has profiled chromatin accessibility and 3D chromatin conformation in primary brain cell types from resected pediatric brain tissue to explore the roles of noncoding SNPs in AD9. Lastly, innovative analytical approaches, for example leveraging machine learning (ML), have greatly expanded our ability to predict the functional effects of noncoding SNPs21–25. Cumulatively, this work has provided important advances in our understanding of the role of noncoding SNPs in disease predisposition, particularly in neurological disease.
Here, we build on the current understanding of inherited variation in neurodegenerative disease through implementation of a multi-omic framework that enables accurate prediction of functional noncoding SNPs. This framework layers bulk Assay for Transposase-accessible chromatin using sequencing (ATAC-seq)26, single-cell ATAC-seq (scATAC-seq)27, and HiChIP enhancer connectome28,29 data over a ML classifier to predict putative functional SNPs driving association with neurodegenerative diseases. Through these efforts, we pinpoint putative target genes and cell types of several noncoding GWAS loci in AD and PD, providing a roadmap for application of these data and technology to other neurological disorders and enabling a more comprehensive understanding of the role of inherited noncoding variation in disease.
RESULTS
Bulk chromatin accessibility landscapes in macrodissected tissue identify brain regional epigenomic heterogeneity
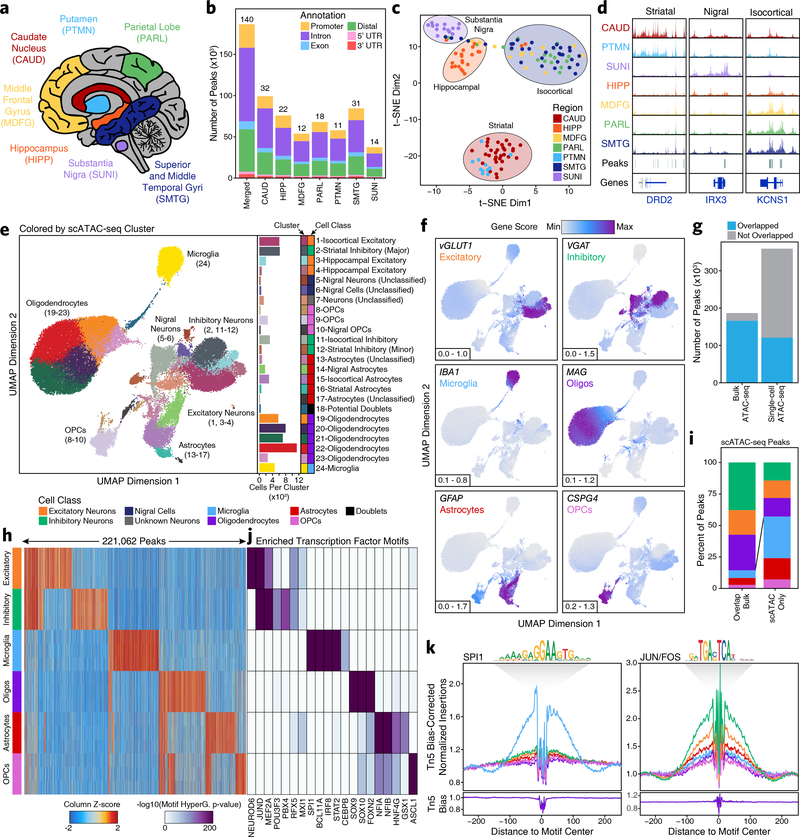
We profiled the bulk chromatin accessibility landscapes of 7 macrodissected brain regions across 39 cognitively healthy individuals to characterize the role of the noncoding genome in neurodegenerative diseases (Supplementary Table 1). These brain regions include distinct isocortical regions [superior and middle temporal gyri (SMTG), parietal lobe (PARL), and middle frontal gyrus (MDFG)], striatal regions [caudate nucleus (CAUD) and putamen (PTMN)], the hippocampus (HIPP), and the substantia nigra (SUNI) (Fig. 1a; see Methods). From these bulk ATAC-seq libraries, we compiled a merged set of 186,559 reproducible peaks (Fig. 1b and Supplementary Data Set 1). Here, a reproducible peak is defined as any peak that is called in at least 30% of the bulk ATAC-seq samples from any given brain region (Supplementary Fig. 1a; see Methods). Dimensionality reduction via t-distributed stochastic neighbor embedding (t-SNE) identified 4 distinct clusters of samples, grouped roughly by major brain region (Fig. 1c). While many region-specific peaks in chromatin accessibility could be identified from these bulk ATAC-seq data, most of these peaks corresponded to cell types predominantly present in a single region (Fig. 1d). A detailed analysis of these bulk ATAC-seq data primarily revealed region-specific differences in chromatin accessibility (Supplementary Fig. 1b–h & Supplementary Note 1).
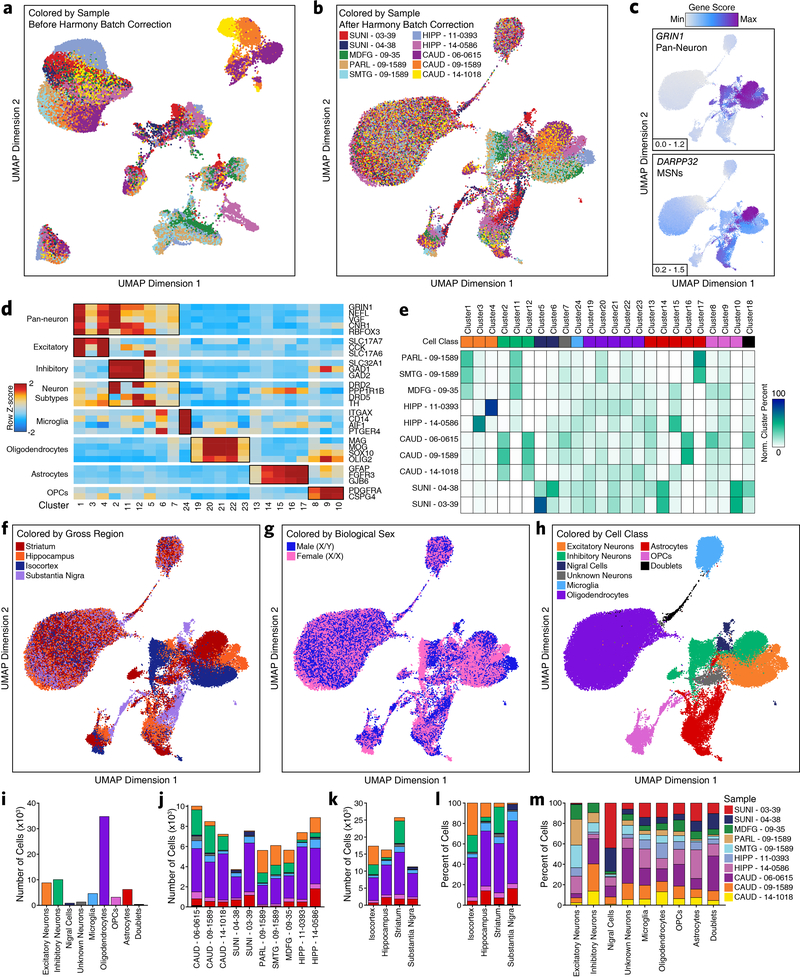
Fig. 1 – Single-cell ATAC-seq identifies cell type-specific chromatin accessibility in the adult brain.
a, Brain regions profiled in this study. b, Bar plot showing the number of reproducible peaks identified from samples in each brain region. The “Merged” bar represents the final merged peak set. The numbers above each bar represent the total number of biological samples profiled for each brain region. c, t-SNE dimensionality reduction of bulk ATAC-seq data. Each dot represents a single piece of tissue with technical replicates merged where applicable. d, Sequencing tracks of region-specific ATAC-seq peaks. From left to right, DRD2 (striatum-specific; chr11:113367951–113538919), IRX3 (substantia nigra-specific; chr16:54276577–54291319), and KCNS1 (isocortex-specific; chr20:45086706–45107665). Tracks have been normalized to the total number of reads in TSS regions. e, Left; UMAP dimensionality reduction after iterative LSI of scATAC-seq data from 10 different samples. Each dot represents a single cell (N = 70,631), colored by its corresponding cluster. Right; Bar plot showing the number of cells per cluster. f, Same as Figure 1e but each cell is colored by its gene activity score for the annotated lineage-defining gene. The minimum and maximum gene activity scores are shown in the bottom left of each panel. g, Bar plot showing the overlap of bulk ATAC-seq and scATAC-seq peak calls. “Bulk ATAC-seq” represents the number of peaks from the bulk ATAC-seq merged peak set that are overlapped by a peak called in our scATAC-seq merged peak set. “Single-cell ATAC-seq” represents the number of peaks from our scATAC-seq merged peak set that are overlapped by a peak called in our bulk ATAC-seq merged peak set. Overlap is considered as any overlapping bases. h, Heatmap representation of chromatin accessibility in binarized peaks (N = 221,062) from the scATAC-seq peak set. Each row represents an individual pseudo-bulk replicate (3 per cell type) and each column represents a peak. i, Bar plot of the percent of peaks from the scATAC-seq binarized peak set that overlap peaks identified by bulk ATAC-seq (“Overlap Bulk”) or are uniquely identified by scATAC-seq (“scATAC Only”). Only peaks found to be unique to a single cell type (N = 172,111) were used in this analysis. Bars are colored according to the legend above Fig. 1h. j, Motif enrichments of binarized peaks identified in Figure 1h. Due to redundancy in motifs, TF drivers were predicted using the average gene expression in GTEx brain samples and accessibility at TF promoters in cell class-grouped scATAC-seq profiles. k, Footprinting analysis of the SPI1 (left; CIS-BP M6484_1.02) and JUN/FOS (right; CIS-BP M4625_1.02) TFs across the 6 major cell classes.
Single-cell ATAC-seq captures regional and cell type-specific heterogeneity
To better understand brain-regional cell type-specific chromatin accessibility landscapes, we performed single-cell chromatin accessibility profiling in 10 samples spanning the isocortex (N = 3), striatum (N = 3), hippocampus (N = 2), and substantia nigra (N = 2) (Supplementary Table 1). In total, we profiled chromatin accessibility in 70,631 individual cells (Fig. 1e) after stringent quality control filtration (Supplementary Fig. 2a and Supplementary Data Set 2). Unbiased iterative clustering27,30 and Harmony-based batch correction of these single cells identified 24 distinct clusters (Fig. 1e and Extended Data Fig 1a–b), which were assigned to known brain cell types based on gene activity scores compiled from chromatin accessibility signal in the vicinity of key lineage-defining genes30,31 (Fig. 1f and Extended Data Fig. 1c–d; see Methods). Additionally, 13 of the 24 clusters showed regional specificity with some clusters composed almost entirely from a single brain region (Extended Data Fig. 1e–f and Supplementary Data Set 2). We did not identify any clusters that were clearly segregated by gender but the sample size used in this study was not powered to make such a determination (Extended Data Fig. 1g). Cumulatively, we defined 8 distinct cell classes, including the 6 main brain cell types (excitatory neurons, inhibitory neurons, microglia, oligodendrocytes, astrocytes, and OPCs), and identified one cluster (Cluster 18) as putative doublets that we excluded from downstream analyses (Fig. 1e and Extended Data Fig. 1h). These cell groupings varied largely in the total number of cells per grouping (Extended Data Fig. 1i) and showed distinct donor and regional compositions (Extended Data Fig. 1j–m).
Using these clusters, we then called peaks from scATAC-seq pseudo-bulk chromatin accessibility to create a union set of 359,022 reproducible peaks (Supplementary Data Set 3). Overall, 89% of bulk ATAC-seq peaks were overlapped by a peak called in the scATAC-seq data (Fig. 1g). Conversely, only 34% of scATAC-seq peaks were overlapped by a peak from the bulk ATAC-seq peak set (Fig. 1g). Consistent with a role for distal regulatory elements in cell type-specific gene regulation32, we found an enrichment in distal/intronic peaks and a depletion in promoter peaks in the peak set specifically identified via scATAC-seq (Extended Data Fig. 2a). To better understand the cell type specificity of the scATAC-seq peaks, we identified cell type-specific peaks through “feature binarization”, which identifies peaks that are uniquely accessible in a single cell type or subset of cell types33. This analysis identified 221,062 highly cell type-specific peaks within the 6 primary brain cell types, comprising > 60% of all peaks identified from our scATAC-seq data (Fig. 1h and Supplementary Data Set 4). These cell type-specific peaks were also enriched for distal/intronic peaks and depleted for promoter peaks (Extended Data Fig. 2b). Some of these peaks were shared across the different neuronal cell types while others were shared across astrocytes, OPCs, and oligodendrocytes (Fig. 1h, Extended Data Fig. 2c, and Supplementary Data Set 4). However, 48% of peaks called in our single-cell ATAC-seq data were specific to a single cell type (N = 172,111 peaks; Fig. 1h and Supplementary Data Set 4) with the vast majority of these cell type-specific peaks remaining undetected in our bulk ATAC-seq analyses. Consistent with previous work34, we found an enrichment of peaks from less abundant cell types (less than 20% of cells; i.e. microglia, astrocytes, and OPCs) within the set of peaks identified via scATAC-seq but not bulk ATAC-seq (Fig. 1i and Extended Data Fig. 1l). Similarly, examining per-cell accessibility at the peaks specifically identified via scATAC-seq, we found significantly fewer cells supporting these peaks (Extended Data Fig. 2d). These results highlight the utility of single-cell methods when cell type-specific peaks are difficult to identify from bulk tissues containing multiple distinct cell types at varying frequencies.
To predict which TFs may be responsible for establishing and maintaining these cell type-specific regulatory programs, we performed motif enrichment analyses of peaks specific to each cell type (Fig. 1j). We identified many known drivers of cell type identity, such as motifs specific to SOX9 and SOX10 in oligodendrocytes35,36, or to ASCL1 in OPCs37,38. Lastly, TF footprinting from our scATAC-seq-derived cell type-specific chromatin accessibility data showed enrichment of binding of key lineage defining TFs such as SPI1 in microglia39 and JUN/FOS in neurons40 (Fig. 1k). Notably, the three isocortical samples, derived from distinct brain regions, showed high similarity based on Pearson correlation, supporting their use as biological replicates (Extended Data Fig. 2e). These data provide reference cell profiles for cell type-specific deconvolution of bulk ATAC-seq data (Supplementary Fig. 3, Supplementary Data Set 5, and Supplementary Note 2) and identify brain regional heterogeneity in glial cells, such as astrocytes and OPCs (Supplementary Fig. 4, Supplementary Data Set 6, and Supplementary Note 3).
scATAC-seq identifies diverse neuronal subpopulations
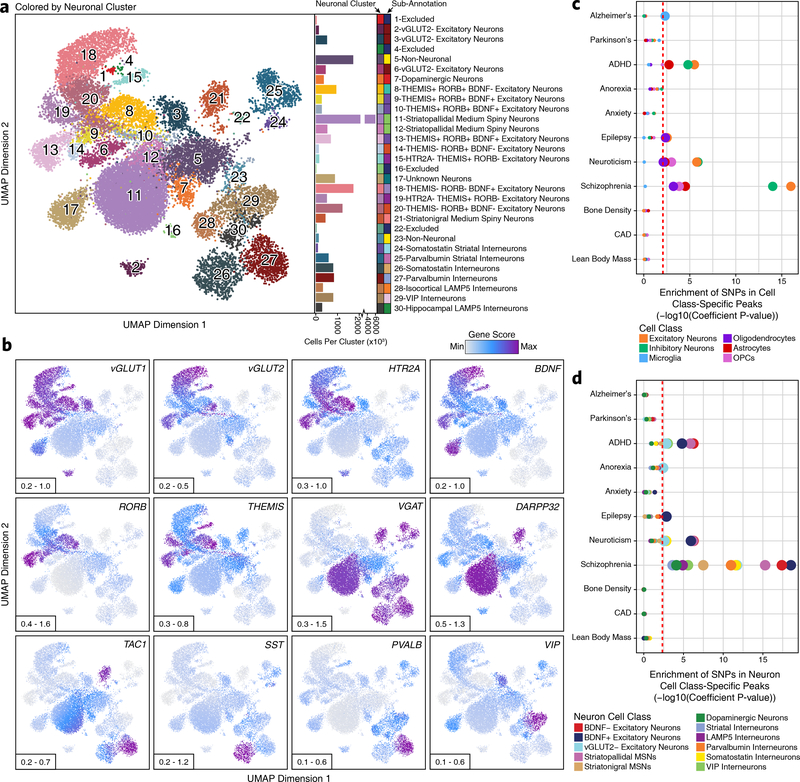
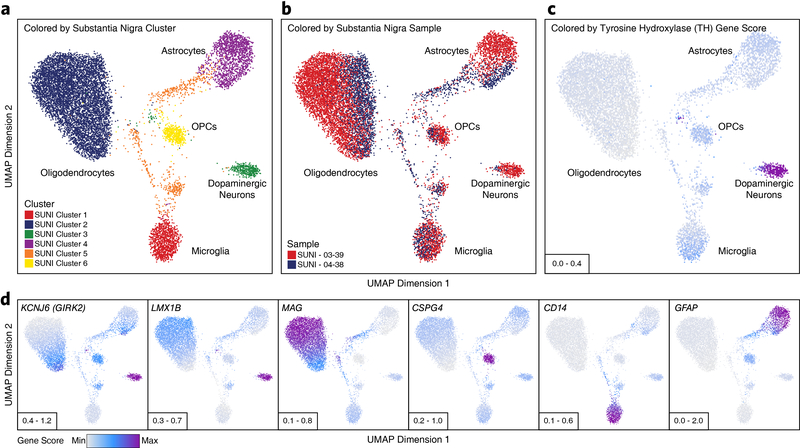
Given the well-understood diversity of neuronal types and functions, we sought to further subdivide our scATAC-seq data based on neuronal subtypes. Extracting all cells previously labeled as neurons (Clusters 1–7, 11, and 12; N = 21,116 cells), we performed unbiased iterative clustering followed by Harmony-based batch correction (Extended Data Fig. 3a–b), identifying 30 discrete neuronal clusters (Fig. 2a, Extended Data Fig. 3c, and Supplementary Data Set 2). For clarity, these are referred to as “neuronal clusters” to avoid confusion with the 24 clusters identified in our broad analysis above. Each neuronal cluster was interpreted to represent a unique neuronal cell type or cell state and annotated using gene activity scores for key lineage-defining genes (Fig. 2b and Extended Data Fig. 3d–e). This identified both broad neuronal classes (Extended Data Fig. 3f) and very granular neuronal subdivisions, even discriminating between striatopallidal (Neuronal Clusters 11–12) and striatonigral (Neuronal Cluster 21) medium spiny neurons, which both reside within the striatum but project to different brain areas (Fig. 2a and Extended Data Fig. 3g–h). These data identified neuronal cell class-specific peaks, genes, and TF activity (Supplementary Fig. 5, Supplementary Data Set 7, and Supplementary Note 4). While this analysis did identify a neuronal cluster corresponding predominantly to substantia nigra dopaminergic neurons (Neuronal Cluster 7), a key cell type lost in PD, we derived a more refined subset of tyrosine hydroxylase (TH)-positive dopaminergic neurons by sub-clustering only cells from the two substantia nigra samples (N = 403 dopaminergic neurons; Extended Data Fig. 4a–d).
Fig. 2 – Sub-clustering identifies diverse biologically relevant neuronal cell types in the adult brain.
a, Left; UMAP dimensionality reduction after iterative LSI of scATAC-seq data from neuronal cells from 10 different samples. Each dot represents a single cell (N = 21,116). Dots are colored by their corresponding neuronal sub-cluster. Neuronal cluster numbers are overlaid on the UMAP above each neuronal cluster centroid. Right; Bar plot showing the number of cells per cluster. Each neuronal cluster sub-annotation is labeled to the right of the bar plot and indicated by color. b, The same UMAP dimensionality reduction shown in Figure 2a but each cell is colored by its gene activity score for the annotated lineage-defining gene. The minimum and maximum gene activity scores are shown in the bottom left of each panel. c-d, LD score regression identifying the enrichment of GWAS SNPs from various brain-related and non-brain-related conditions in the peak regions of various (c) cell classes from the broad scATAC-seq clustering or (d) neuronal cell classes identified from the neuronal sub-clustering analysis. The dotted line represents the Bonferroni-corrected significance threshold for the LDSC coefficient P value (see Methods), adjusted for the number of cell classes tested. The size of the point for each cell class indicates whether this cell class passes the Bonferroni-corrected significance threshold (larger) or not (smaller).
Single-cell ATAC-seq pinpoints the cellular targets of GWAS polymorphisms
To understand if any particular cell type-specific regions of chromatin accessibility were enriched for neurodegenerative disease-associated SNPs, we performed LD score regression41 using a collection of relevant GWAS studies (Supplementary Table 2). Within the peak regions of our broad cell classes, cell type-specific LD score regression revealed a significant increase in per-SNP heritability for AD in the microglia peak set, reinforcing previous studies2,42,43 (Fig. 2c and Supplementary Data Set 8). Similar analyses in PD showed no significant enrichment in SNP heritability in any particular cell type, perhaps because the cellular bases of PD are more heterogeneous than AD (Fig. 2c). Though not a focus of the current study, we note that the data generated here can be used to inform the cellular ontogeny of any brain-related GWAS (Fig. 2c). We also confirmed that the heritability of GWAS SNPs from traits not directly related to brain cell types, such as lean body mass and coronary artery disease, was not significantly enriched in any of the tested brain cell types. To ensure that the lack of significance in cell class-specific peaks was not due to obfuscation of neuronal sub-types, we performed the same LD score regression analyses within the peak regions for the neuronal cell classes identified through sub-clustering (Fig. 2d and Extended Data Fig. 3h). This analysis confirmed our previous findings and showed no significant enrichment for AD or PD SNPs within the peak regions of any neuronal sub-classes (Fig. 2d).
Identification of putative enhancer-promoter interactions through chromatin conformation and cell type-specific co-accessibility
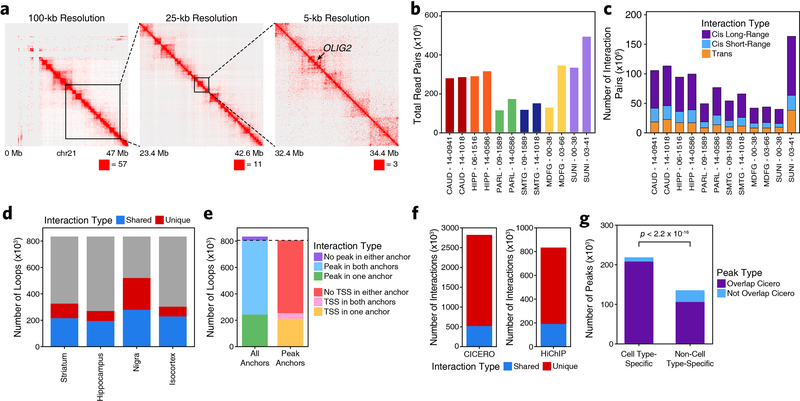
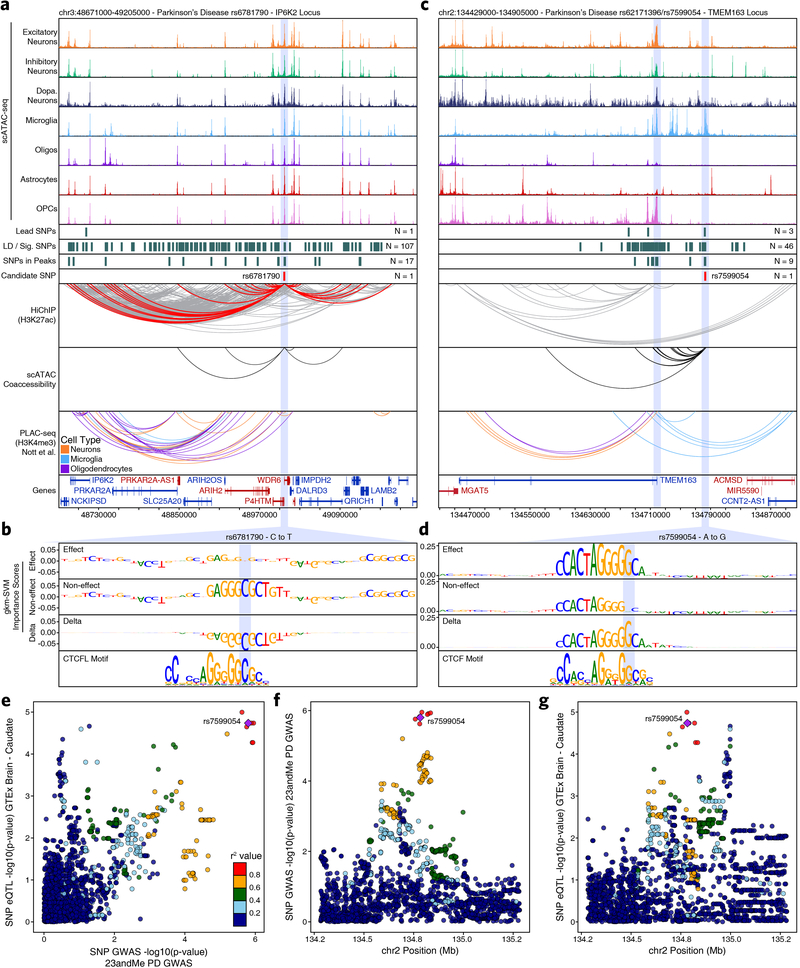
While our scATAC-seq data would enable us to identify the target cell types of functional noncoding SNPs, we sought to additionally identify the target genes of each GWAS locus. To do this, we mapped the enhancer-centric 3D chromatin architecture in multiple brain regions using HiChIP28 for histone H3 lysine 27 acetylation (H3K27ac), which marks active enhancers and promoters (Fig. 3a and Extended Data Fig. 5a). In total, we generated 3D interaction maps for 6 of the 7 regions profiled by ATAC-seq (putamen was excluded given the high overlap with the caudate nucleus), averaging 158 million valid interaction pairs identified per region (Extended Data Fig. 5b–c). We identified 833,975 predicted 3D interactions across all brain regions profiled, of which 331,730 (40%) were reproducible in at least two brain regions (Extended Data Fig. 5d and Supplementary Data Set 9). Of these loops, 67.4% had an ATAC-seq peak present in both anchors, 29.2% had an ATAC-seq peak present in one anchor, and 3.4% did not overlap any ATAC-seq peaks identified in either the bulk or scATAC-seq datasets (Extended Data Fig. 5e).
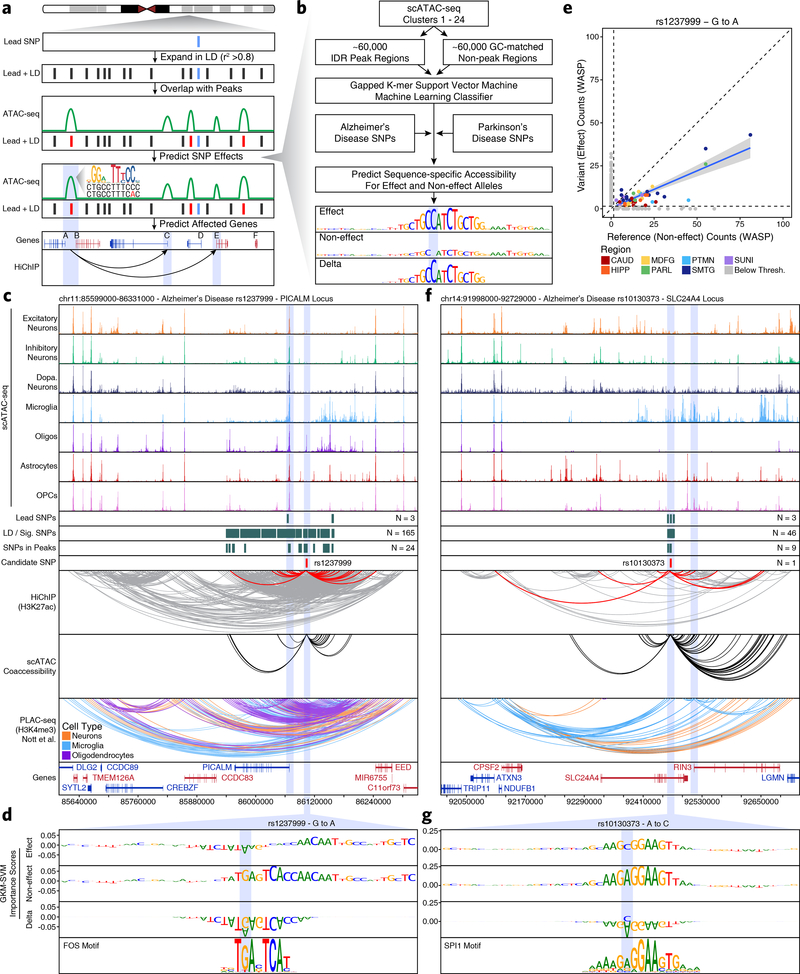
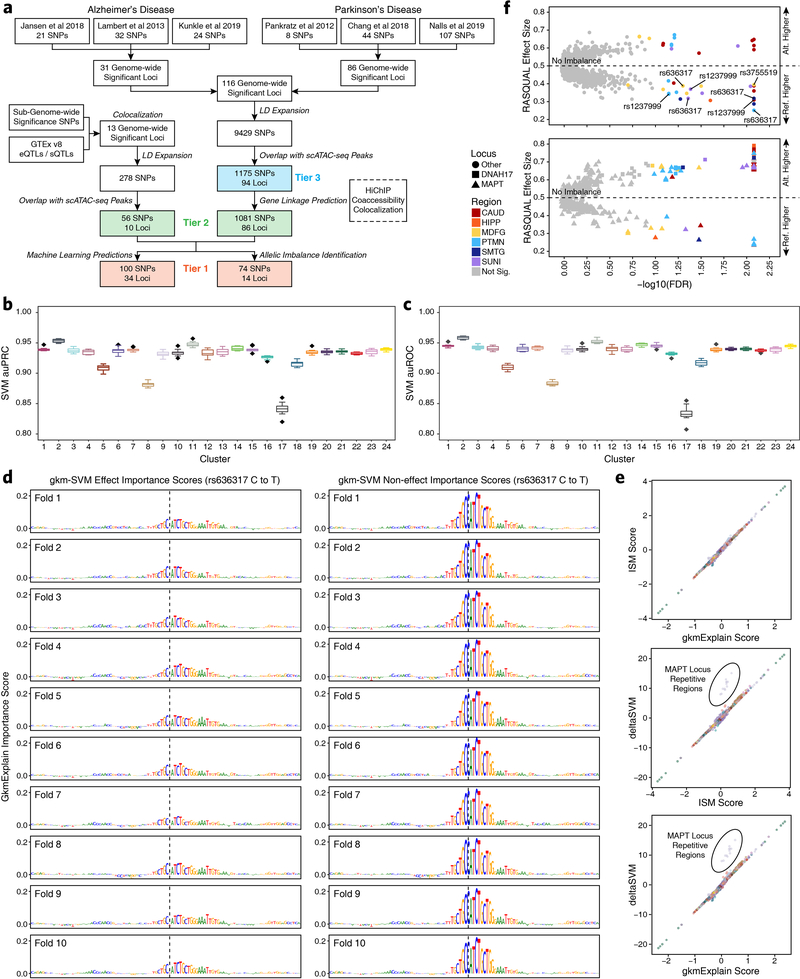
Fig. 3 – Machine learning predicts functional polymorphisms in AD and PD.
a, Schematic of the overall strategy for tiered identification of putative functional SNPs and their corresponding gene targets. b, Schematic of the gkm-SVM machine learning approach used to predict which noncoding SNPs alter TF binding and chromatin accessibility. c,f, Normalized scATAC-seq-derived pseudo-bulk tracks, H3K27ac HiChIP loop calls, co-accessibility correlations, and publicly available H3K4me3 PLAC-seq loop calls (Nott et al. 2019) in the (c) PICALM gene locus (chr11:85599000–86331000) and (f) SLC24A4 locus (chr14:91998000–92729000). scATAC-seq tracks represent the aggregate signal of all cells from the given cell type and have been normalized to the total number of reads in TSS regions. For HiChIP, each line represents a FitHiChIP loop call connecting the points on each end. Red lines contain one anchor overlapping the SNP of interest. For co-accessibility, only interactions involving the accessible chromatin region of interest are shown. For PLAC-seq, MAPS loop calls from microglia (blue), neurons (orange), and oligodendrocytes (purple) are shown. d,g, GkmExplain importance scores for each base in the 50-bp region surrounding (d) rs1237999 and (g) rs10130373 for the effect and non-effect alleles from the gkm-SVM model corresponding to (d) oligodendrocytes (Cluster 21) and (g) microglia (Cluster 24). The predicted motif affected by the SNP is shown at the bottom and the SNP of interest is highlighted in blue. e, Dot plot showing allelic imbalance at rs1237999. The bulk ATAC-seq counts for the reference/non-effect (G) allele and variant/effect (A) allele are plotted. Each dot represents an individual bulk ATAC-seq sample (N = 140) colored by brain region. Samples where fewer than 3 reads were present to support both the reference and variant allele (i.e. presumed homozygotes or samples with insufficient sequencing depth) are shown in grey. The blue line represents a linear regression of the non-grey points and the grey box represents the 95% confidence interval of that regression.
Additionally, correlated variation of chromatin accessibility in peaks across single cells has been shown to predict functional interactions between regulatory elements31,44. Using this co-accessibility framework, we predicted regulatory interactions from our scATAC-seq data from the variation across all cells (Extended Data Fig. 5f), identifying 2,822,924 putative pairwise interactions between regions of chromatin accessibility (Supplementary Data Set 9). This set of interactions showed only moderate overlap (~20%) with our HiChIP data, consistent with the ability of this technique to identify cell type-specific regulatory interactions, whereas HiChIP of bulk brain tissue is better suited for identification of more shared regulatory interactions (Extended Data Fig. 5f–g). Together, these two techniques define a compendium of putative regulatory interactions in the various brain regions studied here, thus enabling downstream linkage of GWAS SNPs to putative target genes.
A tiered multi-omic approach to predicting functional noncoding SNPs
To annotate functional effects of GWAS polymorphisms, we first compiled a comprehensive set of putative disease-relevant SNPs in AD and PD, taking into account the propensity of nearby SNPs to be co-inherited based on linkage disequilibrium (LD). We identified (i) any SNPs passing genome-wide significance (P < 5 × 10−8) in recent GWAS1–3,5–7, (ii) any SNPs exhibiting colocalization of GWAS and eQTL signal (FINEMAP/eCAVIAR colocalization posterior probability > 0.01), and (iii) any SNPs in linkage disequilibrium with a SNP in the previous two categories based off of an LD R2 value greater than or equal to 0.8 calculated from Phase 1 genotypes of individuals of European ancestry in the 1000 Genomes dataset (Supplementary Table 2, see Methods). In total, this identified 9,707 SNPs including 3,245 unique SNPs across 44 loci associated with AD and 6,496 across 86 loci associated with PD, with a single locus containing 34 SNPs appearing in both diseases.
Using this catalog of putative disease-relevant noncoding polymorphisms, we developed a tiered multi-omic approach to predict functional noncoding GWAS polymorphisms by (i) overlapping these SNPs with peaks of chromatin accessibility in our bulk or scATAC-seq data (Tier 3), (ii) identifying the subset of Tier 3 SNPs that may also affect predicted regulatory interactions (Tier 2), and (iii) predicting which Tier 2 SNPs might directly affect TF binding (Tier 1) (Fig. 3a and Extended Data Fig. 6a).
To predict these Tier 1 SNPs that might directly affect TF binding, we implemented a ML framework to score the allelic effect of a SNP on chromatin accessibility. Using the gapped k-mer support vector machine (gkm-SVM) framework45, we trained predictive regulatory sequence models of chromatin accessibility from each of the 24 broad clusters derived from our scATAC-seq data (Fig. 3b and Supplementary Table 2; see Methods). The gkm-SVM models for all 24 scATAC-seq clusters exhibited high prediction performance on held-out test sequences (Extended Data Fig. 6b–c) and across a 10-fold validation scheme (Extended Data Fig. 6d). We used three complementary approaches, GkmExplain22, in silico mutagenesis46, and deltaSVM21 to predict the allelic impact of candidate SNPs on chromatin accessibility in each cluster by providing the sequences corresponding to both alleles of each SNP to the models for each of the 24 clusters. All three approaches showed high concordance of predicted allelic effects across all candidate SNPs (Extended Data Fig. 6e).
As an orthogonal metric for Tier 1 SNPs, we performed allelic imbalance analyses with our bulk ATAC-seq data using the robust allele-specific quantification and quality control (RASQUAL) statistical framework23 (Extended Data Fig. 6f and Supplementary Data Set 10; see Methods). Allelic imbalance refers to the differential chromatin accessibility observed between two alleles when one allele is more readily bound by a TF.
Using this tiered approach, we identified genes and molecular processes that could be implicated in AD and PD (Supplementary Fig. 6a–d & Supplementary Note 5). To avoid overinterpretation, we focused our downstream analyses on the subset of GWAS loci that were most likely to involve noncoding regulation based on absence of any LD SNPs in coding regions (Supplementary Fig. 6e and Supplementary Table 2).
Machine learning predicts putative functional SNPs and identifies the molecular ontogeny of disease associations
This multi-omic approach identified two main categories of novel associations within our Tier 1 SNPs: established disease-related genes where the precise causative SNP remains unknown, and genes previously not implicated in disease etiology. Many studies have investigated the role of genes such as PICALM47, SLC24A448, BIN19,49, and MS4A6A50 in AD since their implication in the disease by GWAS. However, it remains unclear which polymorphisms drive these associations. In the case of PICALM, our models predicted a potential functional variant (rs1237999) disrupting a putative FOS/AP1 factor binding site within an oligodendrocyte-specific regulatory element 35 kb upstream of PICALM (Fig. 3c–d). Moreover, rs1237999 showed significant allelic imbalance with the variant (effect) allele showing diminished accessibility in bulk ATAC-seq data from heterozygotes across multiple brain regions (Fig. 3e and Supplementary Data Set 10). Lastly, rs1237999 showed 3D interaction with both PICALM and the EED gene, a polycomb-group family member involved in maintaining a repressive transcriptional state. This expands the potential functional role of this association to a novel gene and specifically points to a role for oligodendrocytes which were not previously implicated in this phenotypic association47.
Similarly, the SLC24A4 locus harbors a small LD block with 46 SNPs that all reside within an intron of SLC24A4. Previous work has implicated both SLC24A4 and the nearby RIN3 gene in this association but the true mediator remains unclear51,52. Our multi-omic approach identifies a single SNP, rs10130373, which occurs within a microglia-specific peak, disrupts an SPI1 motif, and communicates specifically with the promoter of the RIN3 gene (Fig. 3f–g). This is consistent with the role of RIN3 in the early endocytic pathway which is crucial for microglial function and of particular disease relevance in AD53. We identify similar examples in the BIN1 and MS4A6A loci (Extended Data Fig. 7 & Supplementary Note 6).
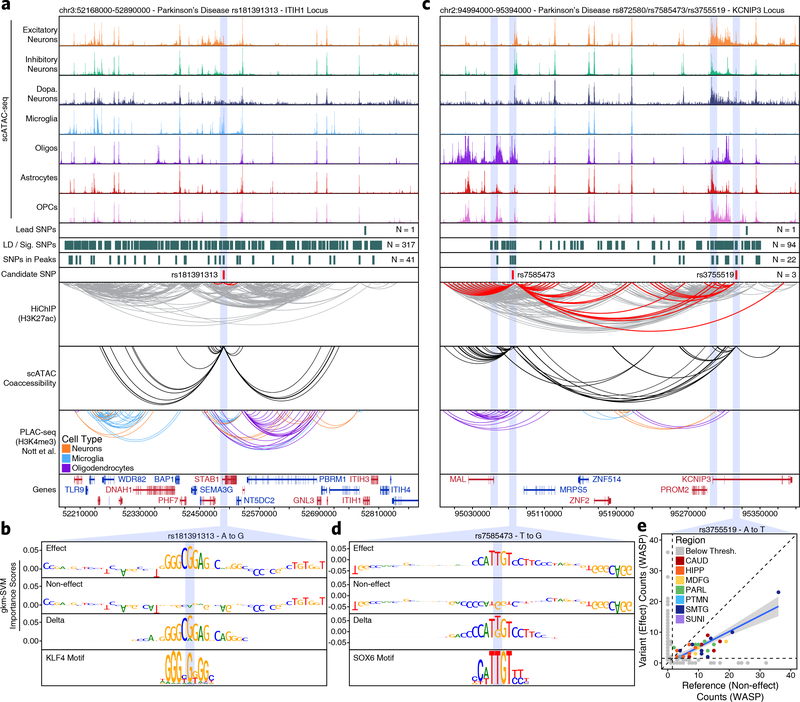
Moreover, the true promise in studying these noncoding polymorphisms is the identification of novel genes affected by disease-associated variation. The ITIH1 GWAS locus occurs within a 600-kb LD block harboring 317 SNPs and no plausible gene association has been made to date. We nominate rs181391313, a SNP occurring within a putative microglia-specific intronic regulatory element of the STAB1 gene (Fig. 4a). STAB1 is a large transmembrane receptor protein that functions in lymphocyte homing and endocytosis of ligands such as low density lipoprotein, two functions consistent with a role for microglia in PD54. This SNP is predicted to disrupt a KLF4 binding site, consistent with the role of KLF4 in regulation of microglial gene expression55 (Fig. 4b). Similarly, the KCNIP3 GWAS locus resides in a 300-kb LD block harboring 94 SNPs. Our results identify two putative mediators of this phenotypic association with different functional interpretations (Fig. 4c). First, rs7585473 occurs > 250 kb upstream of the lead SNP and disrupts an oligodendrocyte-specific SOX6 motif in a peak found to interact with the MAL gene, implicated in myelin biogenesis and function (Fig. 4d). Alternatively, we find rs3755519 in a neuronal-specific intronic peak within the KCNIP3 gene with clear interaction with the KCNIP3 gene promoter. While this SNP does not show a robust ML prediction, nor reside within a known motif, significant allelic imbalance supports its predicted functional alteration of TF binding (Fig. 4e and Supplementary Data Set 10). Furthermore, this SNP is associated with KCNIP3 expression in three bulk brain regions from the GTEx database (frontal cortex, P = 4.04 × 10−7; hippocampus, P = 1.45 × 10−7; cerebellum, P = 3.47 × 10−8) and fine-mapping analysis places rs3755519 within the 95% credible set of causal SNPs in all three brain regions. Together, these SNPs provide competing interpretations of this locus, implicating oligodendrocyte- and neuron-specific functions, and demonstrating the complexities of interpretation of functional noncoding SNPs. We additionally noted that many SNPs appear to disrupt binding sites related to CTCF (Extended Data Fig. 8 & Supplementary Note 6).
Fig. 4 – Vertical integration of multi-omic data and machine learning nominates gene targets in AD and PD.
a,c, Normalized scATAC-seq-derived pseudo-bulk tracks, H3K27ac HiChIP loop calls, co-accessibility correlations, and publically available H3K4me3 PLAC-seq loop calls (Nott et al. 2019) in (a) the ITIH1 gene locus (chr3:52168000–52890000) or (c) the KCNIP3 locus (chr2:94994000–95394000). scATAC-seq tracks represent the aggregate signal of all cells from the given cell type and have been normalized to the total number of reads in TSS regions. For HiChIP, each line represents a FitHiChIP loop call connecting the points on each end. Red lines contain one anchor overlapping the SNP of interest. For co-accessibility, only interactions involving the accessible chromatin region of interest are shown. For PLAC-seq, MAPS loop calls from microglia (blue), neurons (orange), and oligodendrocytes (purple) are shown. b,d, GkmExplain importance scores for each base in the 50-bp region surrounding (b) rs181391313 or (d) rs7585473 for the effect and non-effect alleles from the gkm-SVM model corresponding to (b) microglia (Cluster 24) or (d) oligodendrocytes (Cluster 21). The predicted motif affected by the SNP is shown at the bottom and the SNP of interest is highlighted in blue. e, Dot plot showing allelic imbalance at rs3755519. The bulk ATAC-seq counts for the reference/non-effect (A) allele and variant/effect (T) allele are plotted. Each dot represents an individual bulk ATAC-seq sample (N = 140) colored by brain region. Samples where fewer than 3 reads were present to support both the reference and variant allele (i.e. presumed homozygotes or samples with insufficient sequencing depth) are shown in grey. The blue line represents a linear regression of the non-grey points and the grey box represents the 95% confidence interval of that regression.
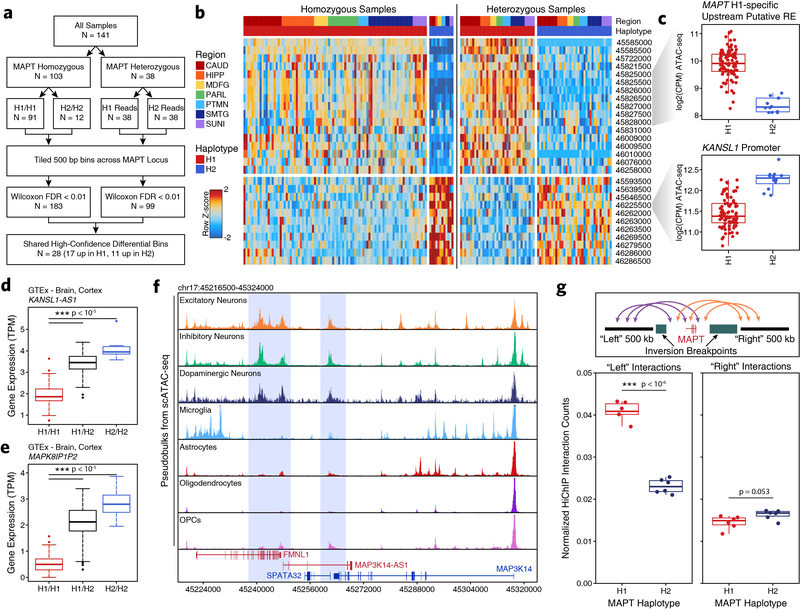
Epigenomic dissection of the MAPT locus explains haplotype-specific changes in local gene expression
One of the strongest PD-associated risk loci is the MAPT gene, which encodes tau proteins whose pathological, hyperphosphorylated aggregates form neurofibrillary tangles in AD56. However, despite this long-known genetic association, it remains unclear how the MAPT locus may play a role in PD. The MAPT locus is present within a large 1.8-Mb LD block and manifests as two distinct haplotypes, H1 and H2, which differ by (i) > 2,000 SNPs across the two haplotypes and (ii) an ~1-Mb inversion that includes the MAPT gene57,58 (Fig. 5a). Previous reports have nominated multiple explanations for how these alterations are associated with PD, including increased MAPT expression in the H1 haplotype59,60 (Fig. 5b), different ratios of splice isoforms61–63, and the use of alternative promoters64. We created a haplotype-specific map of chromatin accessibility and 3D chromatin interactions at the MAPT locus (Fig. 5c). Using data from heterozygote H1/H2 individuals, we split reads into H1 and H2 haplotypes based on the presence of one of the 2,366 haplotype divergent SNPs (Supplementary Table 2; see methods). We tiled the region into non-overlapping 500-bp bins (to avoid biases in peak calling) and performed a Wilcoxon rank sum test to identify regions differentially accessible both between H1/H1 and H2/H2 homozygotes and between split reads from H1/H2 heterozygotes (Extended Data Fig. 9a–b). This identified 28 differentially accessible bins including an H1-specific putative regulatory element located 68 kb upstream of the MAPT promoter and the promoter of the KANSL1 gene located 330 kb downstream of MAPT (Fig. 5d (asterisks) and Extended Data Fig. 9c). Using our HiChIP data, we performed haplotype-specific virtual 4C to determine if any changes in chromatin accessibility were accompanied by changes in 3D chromatin interaction frequency. We identified H2-specific 3D interactions between a putative domain boundary upstream of MAPT (labeled “A”) and the region surrounding the KANSL1 promoter (labeled “B”) spanning a distance of > 600 kb inside the inversion breakpoints (Fig. 5d). Additionally, the H1-specific putative regulatory element upstream of MAPT showed increased interaction with a second putative regulatory element intronic to MAPT as well as with the MAPT promoter (Fig. 5d).
Fig. 5 – Epigenetic deconvolution of the MAPT locus explains haplotype-associated transcriptional changes.
a, The MAPT locus (chr17:44905000–46895000) showing all genes, the predicted locations of the inversion breakpoints, and the 2,366 haplotype-divergent SNPs used for haplotype-specific analyses. b, Gene expression of the MAPT gene from GTEx cortex brain samples subdivided based on MAPT haplotype (N = 117 H1/H1, 78 H1/H2, 10 H2/H2). The lower and upper ends of the box represent the 25th and 75th percentiles and the internal line represents the median. The whiskers represent 1.5 multiplied by the inter-quartile range. Outliers are shown as individual dots. Significance determined by Wilcoxon rank sum test. c, Schematic for the allelic analysis of the MAPT region. d, HiChIP (top) and bulk ATAC-seq (middle) sequencing tracks of the region representing the MAPT locus inside of the predicted inversion breakpoints (chr17:45510000–46580000; bottom). Each track represents the merge of all available H1 or H2 reads from all heterozygotes. HiChIP and ATAC-seq tracks represent unnormalized data from heterozygotes where reads were split based on haplotype. HiChIP is shown as a virtual 4C plot where the anchor is indicated by a dotted line and the signal represents paired-end tag counts overlapping a 10-kb bin. Regions showing significant haplotype bias in ATAC-seq are marked by an asterisk (Wilcoxon rank sum test). e, GTEx cortex gene expression of genes in the MAPT locus comparing H1 homozygotes (N = 117) to H1/H2 (N = 78). Regions A and B are shown as in Figure 5d. * P < 0.05 by Wilcoxon rank sum test after multiple hypothesis correction. f, HiChIP (top) and cell type-specific scATAC-seq (middle) sequencing tracks of the region representing the MAPT locus outside of the predicted inversion breakpoints (bottom). HiChIP tracks for bulk homozygote H1 or H2 samples (normalized based on reads-in-loops) are shown at the top while haplotype-specific tracks from heterozygotes (unnormalized) are shown below. In each HiChIP plot, the anchor represents the MAPT promoter. scATAC-seq tracks represent the aggregate signal of all cells from the given cell type and have been normalized to the total number of reads in TSS regions. g, Schematic illustrating the predicted haplotype-specific change in long-distance interaction between the MAPT promoter and the predicted distal regulatory element identified in Figure 5d. Regions marked A and B represent the same regions marked in Figure 5d-e.
To better understand how these epigenetic changes impact haplotype-specific gene expression, we used RNA-seq data from the GTEx database. In addition to the previously mentioned haplotype-specific differences in MAPT expression (Fig. 5b), we also identified significant changes in gene expression near the largest changes in chromatin accessibility and 3D interaction (“A” and “B”; Fig. 5e and Extended Data Fig. 9d–e). These increases in gene expression could play a functional role in MAPT haplotype-mediated pathologic changes or, more likely, be a non-functional byproduct of the genomic inversion.
These analyses illuminate how the genomic region inside the MAPT inversion breakpoints differs between the H1 and H2 haplotypes; alternatively, the inversion could alter MAPT gene expression by changing the relative orientation of the MAPT gene to enhancers and promoters outside of the breakpoints. In support of this, we identified a long-distance putative regulatory element located 650 kb upstream of the MAPT gene that showed elevated interaction with the MAPT promoter specifically in the H1 haplotype (Fig. 5f). Indeed, we found multiple neuron-specific putative regulatory elements in this upstream region, consistent with the known neuron-specific expression of MAPT (Extended Data Fig. 9f), and an increase in overall 3D interaction between this upstream region and the region surrounding MAPT inside of the inversion breakpoints (Extended Data Fig. 9g). Additional studies will be necessary to demonstrate functional effects of these predicted regulatory interactions (Fig. 5g).
DISCUSSION
Here, we provide a high-resolution epigenetic characterization of the role of inherited noncoding variation in AD and PD. Our integrative multi-omic framework and ML classifier predicted dozens of functional SNPs, nominating gene and cellular targets for each noncoding GWAS locus. These predictions both inform well-studied disease-relevant genes, such as BIN1 in AD, and suggest novel gene-disease associations, such as STAB1 in PD. This expands our understanding of inherited variation in AD and PD and provides a roadmap for epigenomic dissection of noncoding variation in neurodegenerative and other complex genetic diseases.
Together, this multi-omic resource captures the regional and cellular gene regulatory machinery that governs phenotypic expression of noncoding variation, thus allowing to the identification of the majority of polymorphisms that could putatively affect gene expression through overlap with peaks of chromatin accessibility (Tier 3). To further refine these putative functional variants, we identified the subset of polymorphisms that could be mapped to gene targets through 3D chromatin interactions or co-accessibility networks (Tier 2). Finally, we employed a ML approach to predict the subset of polymorphisms likely to perturb TF binding and validated these predictions with measurements of allelic imbalance (Tier 1). In total we implicate ~5 times as many genes in the phenotypic association of AD and PD and nominate functional noncoding variants for dozens of previously orphaned GWAS loci. Additionally, through our integrative analysis, we provide a comprehensive epigenetic characterization of the MAPT gene locus (discussed in detail in the Supplementary Note 7). The functional predictions made through our ML classifier and integrative analytical approach greatly expand our understanding of noncoding contributions to AD and PD. More broadly, this work represents a systematic approach to understanding inherited variation in disease and provides an avenue towards the nomination of novel therapeutic targets that previously remained obscured by the complexity of the regulatory machinery of the noncoding genome.
METHODS
Code Availability
All custom code used in this work is available in the following GitHub repository: https://github.com/kundajelab/alzheimers_parkinsons.
Publicly Available Data Used In This Work
All QTL analysis was performed using GTEx v8. Additionally, we downloaded full-genome summary statistics of GWAS associations for three Alzheimer’s cohorts1–3 and two Parkinson’s cohorts6,65; however, it should be noted that these cohorts are not all mutually exclusive. The Parkinson’s disease full GWAS summary statistics from Chang et al. were obtained through a research agreement with 23andMe. These summary statistics included those generated by 23andMe (N = 6,476 PD-affected individuals and 302,042 disease-free controls) but not summary statistics from individuals incorporated into meta-analysis from the original publication. All GWAS data used in this study (except the data protected through our research agreement with 23andMe) have been compiled for ease of reproducibility and is available under doi 10.1101/2020.01.06.896159 here: https://zenodo.org/record/3817811. Additionally, we obtained MAPS-based loop calls directly from published PLAC-seq data from microglia, neurons, and oligodendrocytes9.
Genome Annotations
All data are aligned and annotated to the hg38 reference genome.
Sequencing
Bulk ATAC-seq, and HiChIP were sequenced using an Illumina HiSeq 4000 with paired-end 75-bp reads. Single-cell ATAC-seq was sequenced using an Illumina NovaSeq 6000 with an S4 flow cell with paired-end 99 bp reads.
Sample acquisition and patient consent
Primary brain samples were acquired post-mortem with IRB-approved informed consent from Stanford University, the University of Washington, or Banner Health. Human donor sample sizes were chosen to provide sufficient confidence to validate methodological conclusions. Human brain samples were collected with an average post-mortem interval of 3.9 hours (range 2.0 – 6.9 hours). These brain regions include distinct isocortical regions [superior and middle temporal gyri (SMTG, Brodmann areas 21 and 22), parietal lobe (PARL, Brodmann area 39), and middle frontal gyrus (MDFG, Brodmann area 9)], striatum at the level of the anterior commissure [caudate nucleus (CAUD) and putamen (PTMN)], hippocampus (HIPP) at the level of the lateral geniculate nucleus, and the substantia nigra (SUNI) at the level of the red nucleus. Macrodissected brain regions were flash frozen in liquid nitrogen. Some samples were embedded in Optimal Cutting Temperature (OCT) compound. All samples were stored at −80°C until use. Due to the limiting nature of these primary samples, this unique biological material is not available upon request.
Isolation of nuclei from frozen tissue chunks and bulk ATAC-seq data generation
Nuclei were isolated from frozen tissue as described previously19,33. This protocol, including the transposition reaction, is now available on protocols.io (dx.doi.org/10.17504/protocols.io.6t8herw). Briefly, frozen tissue fragments were Dounce homogenized to create a suspension of nuclei. Nuclei were purified using an iodixanol gradient and washed in resuspension buffer (RSB). Nuclei were counted and, for each replicate, 50,000 nuclei were aliquoted into a separate tube containing RSB with 0.1% Tween-20. Nuclei were pelleted and transposed as described in the protocol linked above according to the Omni-ATAC transposition conditions19. Transposed fragments were purified and amplified as described previously26 with slight modification. Briefly, transposed fragments were pre-amplified for 3 cycles. The concentration of pre-amplified fragments was determined by qPCR and this concentration was used to estimate the total number of cycles required to obtain 160 fmol of fragments. A second PCR was performed to amplify the pre-amplified fragments for the desired number of cycles. Final libraries were again purified. Prior to sequencing, libraries were pooled and run on a 6% PAGE gel and excess primers and primer dimers below 125 bp were removed. Libraries were sequenced on an Illumina HiSeq4000 instrument as described above. After isolation and bulk ATAC-seq, remaining nuclei were cryopreserved in BAM Banker (Wako Chemicals) and stored at −80°C for use in other assays such as scATAC-seq and HiChIP.
Statistics
All statistical tests performed are included in the figure legends or methods where relevant.
ATAC-seq Data Processing
The ENCODE DCC ATAC-seq pipeline (doi:10.5281/zenodo.211733) (V1.1.7) was used to process bulk ATAC-seq samples, starting from fastq files. The pipeline was executed with IDR enabled and the IDR threshold set to 0.05. The GRCh38 reference genome assembly was used, keeping only the primary chromosomes chr1 - chr22, chrX, chrY, chrM. The pipeline was executed with ATAQC enabled, using GENCODE version 29 TSS annotations. Biological replicates were analyzed individually, with the two technical replicates for each bio-rep provided as inputs to the “atac.bams” argument of the pipeline. Other arguments to the pipeline were kept at their defaults.
ATAC-seq Peak Calling
Pipeline peak calls underwent several levels of filtering to identify credible peak sets. The IDR optimal peak set from the DCC pipeline for each biological replicate was determined. It was observed that although the IDR peaks for individual biological replicates were corrected for multiple testing, the high number of biological samples in the dataset served as another source of multiple testing error. To address this source of error, tagAlign files for all biological replicates for a given brain region/ condition were concatenated. The DCC pipeline (v1.1.7) was subsequently executed on the merged tagAlign files as single-replicate inputs. The pipeline generated pseudo-replicates from the input tagAlign files for each brain region/condition. Optimal IDR peaks were called from the pseudo-replicates. This set of IDR peaks was filtered to keep peaks supported by 30% or more of IDR peaks from the pipeline runs on individual biological replicates.
Sample-by-peak count matrices were then generated from the resulting set of filtered peaks. Filtered peaks from the pooled tagAlign files were concatenated and truncated to within 200 bp of the summit (100 bp flank kept upstream and downstream of the peak summit). These 200-bp regions were merged with the bedtools66 merge command to avoid merging peaks with low levels of overlap. The bedtools coverage -counts was used to compute the number of tagAlign reads that overlapped each peak region in the pseudo-replicates in the merged tagAlign dataset. This analysis yielded a total of n = 186,559 peaks combined across the brain regions.
Motif enrichment
Motif enrichment was performed using the hypergeometric test as described previously33,67.
Feature Binarization
Identification of “unique” peaks from ATAC-seq data was performed as described previously33. Briefly, for each of the cell classes (termed “groups” here), we created 3 pseudo-bulk replicates which were used to create a counts matrix of insertion counts within each peak of the scATAC-seq peak set. This counts matrix was then log-normalized using ‘edgeR::cpm(mat,log = TRUE,prior.count = 3)’. We then calculated the intra-group mean and intra-group standard deviation across every peak in the scATAC-seq peak set. Then, for each peak, we rank the groups by their intra-group mean. Then, we iterate from the second lowest group asking whether the mean of that group is greater than the maximum intra-group mean plus the intra-group standard deviation of the next-lowest sample. This iterative process proceeds until a group is identified that meets this criterion. This point is defined as the break point and all groups with a higher intra-group mean are classified as positive for this peak and given a value of “1”. All groups below the break point are given a value of “0”. If a peak does not have a break point it is discarded. This peak “binarization” procedure classifies all “1s” as being higher than every individual “0”. This also captures the peaks that are unique to multiple groups. We kept all combinations that were unique to 3 or fewer groups. To facilitate multiple hypothesis testing, we computed a contrast matrix for all observed combinations and ran limma’s eBayes test on the log-normalized counts matrix. We then extracted all of the FDR-adjusted P values from differential testing keeping those peaks that were below an FDR of 0.001. This resulted in the classification of 221,062 peaks.
Sequencing Tracks
Sequencing tracks were created using the WashU Epigenome Browser. All sequencing tracks of a given locus have the same y-axis. All tracks show data that have been normalized by “reads-in-peaks” (for ATAC-seq) or “reads-in-loops” for HiChIP to account for differences in signal-to-background ratios across multiple samples, unless otherwise stated. For all sequencing tracks, genes that are on the plus strand (i.e. 5’ to 3’ in the left to right direction) are shown in red and genes that are on the minus strand (i.e. 5’ to 3’ in the right to left direction) are shown in blue to enable identification of the TSS.
LD score regression
We apply stratified LD score regression, a method for partitioning heritability from GWAS summary statistics, to sets of cell type-specific ATAC-seq peaks to identify disease-relevant cell types for Alzheimer’s and Parkinson’s diseases along with other brain-related GWAS traits. Using our single-cell ATAC-seq data, peak coordinates were first converted from hg38 to hg19 for analysis with GWAS data. We followed the LD score regression tutorial (https://github.com/bulik/ldsc/wiki) as used previously41 for single-cell specific analysis68. We used brain related GWAS summary statistics such as Alzheimer’s1, Parkinson’s6, Schizophrenia69, Anorexia Nervosa70, Attention Deficit Hyperactivity Disorder (ADHD)71, Anxiety72, Neuroticism73 and Epilepsy74 (Supplementary Table 2 and https://zenodo.org/record/3817811). To serve as controls, we also used summary statistics for GWAS of traits not obviously linked to brain tissues such as Lean Body Mass75, Bone Mineral Density76 and Coronary Artery Disease77. In particular, we looked at the regression coefficient P value, indicative of the contribution of this annotation to trait heritability, conditional on the baseline model described previously41.
Allele counts from ATAC-seq data
The WASP mapping pipeline (https://github.com/bmvdgeijn/WASP/tree/master/mapping) was used to reduce biases in mapping and in filtering duplicate reads. Reads were mapped using bowtie2 to the UCSC hg38 reference genome. Variants were called on the resulting bam files using bcftools mpileup (v1.9) to produce VCF files. These VCF files and the WASP-corrected bam files were used as input for the GATK ASEReadCounter tool to obtain allele counts and their mapping quality. These allele counts were used to visualize significant allelic imbalance as determined by RASQUAL (see below). For plotting, samples that lacked at least 3 read counts for both the reference and alternate alleles were inferred to be either homozygous or too low coverage to presume heterozygosity. However, we note that these allele counts were only used for display purposes and did not contribute to any determination of significance for allelic imbalance.
Allelic imbalance from ATAC-seq data using RASQUAL
We intersected the coordinates of all LD-expanded candidate AD and PD GWAS and colocalization SNPs with peaks from our ATAC-seq data to obtain the candidate SNPs that we tested for allele-specific effects on chromatin accessibility. We used the createASVCF.sh script from the RASQUAL23 GitHub repository (https://github.com/natsuhiko/rasqual) to obtain the allele-specific counts at each candidate SNP for all samples. We used the fitAseNullMulti function from the QuASAR78 GitHub repository to calculate for each donor the posterior probability of the three possible genotypes at all of the candidate SNP positions using all available brain region samples from that donor and assigned the genotype at each position to be the one with the highest posterior probability. Next, using these allele-specific counts and genotypes and the allele frequencies from the 1000 Genomes Project79 for each candidate SNP, we created a VCF file for each brain region, which included the allele-specific counts and genotypes from only the samples that originated from those respective regions. Similarly, we created region-specific counts matrices, which contain columns of ATAC-seq read counts for each feature only from the samples that originated from the respective regions. We also ran the makeOffset.R script from the RASQUAL repository with a list of GC contents, corresponding to the GC content of each feature in the counts matrix, as an argument to generate the sample specific offset terms file for each brain region. Since RASQUAL is run on each feature from the counts matrix independently of other features, we further split the region-specific input VCF files, counts matrices, and offset files by chromosome and used the text2bin.R script from the RASQUAL repository to convert the region and chromosome-specific input counts matrices and offset files into the binary format required by RASQUAL.
Finally, we ran RASQUAL using the input VCF file, counts matrix, and offset file from each of the 22 chromosomes (chromosomes 1 – 22; chromosome X and chromosome Y did not have any candidate SNPs) from each of the brain regions and tested each candidate SNP present in each feature in the counts matrix. To test for genome-wide significance of each putative chromatin accessibility QTL (caQTL), we ran RASQUAL with the --random-permutation option along with the same inputs 10 times to generate a background set of null q-values. For each brain region, we used the empirical distribution of null q-values to identify those SNPs that have a q-value lower than the 10% False Discovery Rate (FDR) threshold as significant caQTLs as recommended by the authors (https://github.com/natsuhiko/rasqual/issues/21).
Selection of candidate SNPs for ATAC-seq overlap analysis, HiChIP interaction tests, and gkm-SVM model-based allelic effect scores
Our goal was to identify SNPs with a causal effect on any of the selected GWAS traits. To minimize the chances of excluding causal GWAS SNPs, we selected the set of all variants achieving a genome-wide significant P value < 5 × 10−8 for any GWAS trait. We then added in any lead SNPs from the colocalization analysis that achieved CLPP score of > 0.01, even those that did not pass the genome-wide significance value of P < 5 × 10−8. We also included all trait-associated SNPs curated from two other Parkinson’s studies6,7. In these studies, full summary statistics were not publicly available for the entire genome because meta-analysis was applied only to the subset of SNPs reaching genome-wide significance in a previous Parkinson’s GWAS. We then computed the full set of SNPs that had LD R2 ≥ 0.8 with at least one of the SNPs in the set selected above. These LD calculations were performed on Phase 1 genotypes of individuals of European ancestry in the 1000 Genomes dataset, provided in full here (https://zenodo.org/record/3404275#.Xlw62XVKhhE). Pairwise LD values of all variants in the above subset were calculated via plink (v.1.90). These pairwise LD values were used to identify 1000 Genomes SNPs with R2 ≥ 0.8 with the SNPs in our dataset. Together, these LD buddies plus the original set of trait-relevant SNPs comprised the set of SNPs tested in our subsequent functional analyses.
Testing GWAS loci for overlap with ATAC-seq peaks
We tested all SNPs in the above set for overlap with ATAC-seq peaks from two different annotation formats. The first annotation consisted of bulk ATAC-seq peaks identified in one of 7 brain regions. The second annotation consisted of cluster-specific peaks from single-cell ATAC-seq data. For each variant selected for functional analysis, we determined all cellular contexts in which an ATAC-seq peak contained this variant, as well as the nearest peak if no peak contained the variant.
Single-cell ATAC-seq library generation
Cryopreserved nuclei were thawed on ice and 65,000 nuclei were transferred to a tube containing 1 ml of RSB-T [10 mM Tris-HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween]. Nuclei were pelleted at 500 RCF for 5 minutes at 4°C in a fixed angle rotor. The supernatant was fully removed using two pipetting steps (p1000 to remove down to the last 100 μl, then p200 to remove all remaining supernatant). This pellet was then gently resuspended in 12 μl of 1× Nuclei Buffer (10x Genomics). To transpose, 5 μl of this nuclei suspension (containing 27,000 nuclei) was transferred to a tube containing 10 μl of transposition mix (10x Genomics). This reaction mixture was incubated at 37°C for 1 hour to transpose. The remainder of library generation was completed as described in the 10x Genomics Single Cell ATAC Regent Kits User Guide (v1 Chemistry).
Single-cell ATAC-seq LSI clustering and visualization
Single-cell ATAC-seq clustering analysis was performed using an alpha version of the ArchR software80. To cluster our scATAC-seq data (for both broad clustering and neuronal sub-clustering), we first identified a robust set of peak regions followed by iterative LSI clustering27,30. Briefly, we created 1-kb windows tiled across the genome and determined whether each cell was accessible within each window (binary). Next, we identified the top 50,000 accessible windows across all samples (accounting for GC bias) and performed an LSI dimensionality reduction (TF-IDF transformation followed by Singular Value Decomposition SVD) on these windows followed by Harmony batch correction81. We then performed Seurat82 clustering (FindClusters v2.3) on the harmonized LSI dimensions at a resolution of 0.8, 0.4 and 0.2, keeping the clustering for which the minimum cluster size was greater than 100 cells (0.2 if this condition is not met). For each cluster, we called peaks on the Tn5-corrected insertions (each end of the Tn5-corrected fragments) using the MACS2 callpeak command with parameters ‘--shift −75 --extsize 150 --nomodel --call-summits --nolambda --keep-dup all -q 0.05’. The peak summits were then extended by 250 bp on either side to a final width of 501 bp, filtered by the ENCODE hg38 blacklist (https://www.encodeproject.org/ annotations/ENCSR636HFF/), and filtered to remove peaks that extend beyond the ends of chromosomes. We then created a non-overlapping set of extended summits across all of these peaks as described previously27,30.
We then counted the accessibility for each cell in these peak regions to create an accessibility matrix. We then adopted the iterative LSI clustering approach27,30 to unbiasedly identify clusters that are due to biological vs. technical variation. Briefly, we computed the TF-IDF transformation as described by Cusanovich et al.83. To do this, we divided each index by the colSums of the matrix to compute the cell “term frequency”. Next, we multiplied these values by log(1 + ncol(matrix)/rowSums(matrix)), which represents the “inverse document frequency”. This yields a TF-IDF matrix that can be used as input to irlba’s SVD implementation in R. We then used Harmony to batch correct the LSI dimensions in R. Using the first 25 reduced dimensions as input into a Seurat object, crude clusters were identified using Seurat’s (v2.3) SNN graph clustering FindClusters function with a resolution of 0.2. We then calculated the cluster sums from the binarized accessibility matrix and then log-normalized using edgeR’s ‘cpm(matrix, log = TRUE, prior.count = 3)’ in R. Next, we identified the top 25,000 varying peaks across all clusters using ‘rowVars’ in R. This was done on the cluster log-normalized matrix rather than the sparse binary matrix because: (1) it reduced biases due to cluster cell sizes, and (2) it attenuated the mean-variability relationship by converting to log space with a scaled prior count. The 25,000 variable peaks were then used to subset the sparse binarized accessibility matrix and recompute the TF-IDF transform. We used SVD on the TF-IDF matrix to generate a lower dimensional representation of the data by retaining the first 25 dimensions. We then used Harmony to batch correct the LSI dimensions in R. We then used these reduced dimensions as input into a Seurat object and crude clusters were identified using Seurat’s (v.2.3) SNN graph clustering FindClusters function with a resolution of 0.6. This process was repeated a third time with a resolution of 1.0. Then, these same reduced dimensions were used as input to Seurat’s ‘RunUMAP’ with default parameters and plotted in ggplot2 using R.
Single-cell ATAC-seq gene activity scores
Gene activity scores are based on the observation that chromatin accessibility within the gene body, at the promoter, and at distal regulatory elements is correlated with gene expression30,31,80,84. Gene scores were calculated using ArchR v0.9.480 with default parameters. Briefly, ArchR infers gene activity scores using a distance-weighted accessibility model that aggregates accessibility signal inside the gene body and in the local genomic region. The resulting gene activity scores were additionally imputed using MAGIC85 to reduce noise due to scATAC-seq data sparsity.
Identification of clusters and cell types from scATAC-seq data
Different clusters and cell types were manually identified using promoter accessibility and gene activity scores for various lineage-defining genes. Microglia (Cluster 24) were identified based on accessibility near the IBA1, CD14, CD11C, PTGS1, and PTGS2 genes. Astrocytes (Clusters 13–17) were identified based on accessibility near the GFAP and FGFR3 genes. Excitatory neurons (Clusters 1, 3, and 4 were identified based on accessibility near the SLC17A6 and SLC17A7 genes. Inhibitory neurons (Cluster 2, 11, and 12) were identified based on accessibility near the GAD2 and SLC32A1 genes. Medium spiny neurons (most of Cluster 2) were identified based on accessibility near the DARPP32 gene. Oligodendrocytes (Clusters 19–23) were identified based on accessibility near the MAG and SOX10 genes. OPCs (Clusters 8–10) were identified based on accessibility near the PDGFRA gene. All neuronal subsets were identified primarily as neurons based on accessibility near the NEFL, RBFOX3, VGF, and GRIN1 genes and then subdivided based on the region of origin and the accessibility near other genes mentioned above.
Single-cell ATAC-seq peak calling
For scATAC-seq peak calling from clusters or manually defined cell types, all single cells belonging to the given group were pooled together. These pooled fragment files were converted to the paired-end tagAlign format and processed with version 1.4.2 of the ENCODE DCC ATAC-seq pipeline. The conversion to tagAlign was performed as follows. For fragments on the positive strand, the read start coordinate was the fragment start coordinate, zero-indexed. The read end coordinate was the fragment start coordinate plus the read length (99 bp). For fragments on the negative strand, the read start coordinate was the fragment end coordinate, zero-indexed. The read start coordinate was the fragment end coordinate minus the read length (99 bp). Then, these tagAlign files were used as input to the DCC ATAC-seq pipeline. IDR optimal peak sets with an IDR threshold of 0.05 were determined for each cluster by the pipeline, using pseudo-bulk replicate tagAligns for the cluster. Other pipeline parameters were the same as for bulk ATAC-seq data (see above).
Single-cell ATAC-seq pseudo-bulk replicate generation and differential accessibility comparisons
For differential comparisons of clusters or cell types, including Pearson correlation determination, non-overlapping pseudo-bulk replicates were generated from groups of cells. For each cell grouping (i.e a cluster or a cell type), a minimum of 300 cells was required in order to make at least two non-overlapping pseudo-bulk replicates of 150 cells each. A maximum of 3 pseudo-bulk replicates was made per group if the total number of cells per group was greater than 450 cells. Cells were randomly deposited into one of the pseudo-bulk replicates and all available cells were used. In this way, the non-overlapping pseudo-bulk replicates are agnostic to which donor the cell came from but aware of individual cells (i.e. all reads from a given cell are deposited into the same pseudo-bulk replicate). These pseudo-bulk replicates were then used for differential comparisons using DESeq286.
Identification of neuronal cell class-specific peaks, TF motifs, and genes
ArchR (version 0.9.4) was used to call peaks (using “addReproduciblePeakSet) and identify cell class-specific peaks and genes (using “getMarkerFeatures”). The cell class-specific peaks were tested from motif enrichment (using “peakAnnoEnrichment”).
Transcription factor footprinting
TF footprinting was performed as described previously33.
HiChIP library generation
HiChIP library generation was performed as described previously28. One million cryopreserved nuclei were used per experiment. Enzyme MboI was used for restriction digest. Sonication was performed on a Covaris E220 instrument using the following settings: duty cycle 5, peak incident power 140, cycles per burst 200, time 4 minutes. All HiChIP was performed using H3K27ac as the target (Abcam ab4729).
HiChIP data analysis
HiChIP paired-end sequencing data were processed using HiC-Pro87 version 2.11.0 with a minimum mapping quality of 10. FitHiChIP88 was used to identify “peak-to-all” interactions using peaks called from the one-dimensional HiChIP data. A lower distance threshold of 20 kb and an upper distance threshold of 2 Mb were used. Bias correction was performed using coverage-specific bias.
HiChIP linkage of SNPs to genes
To link SNPs to genes, we identified FitHiChIP loops that contained a SNP in one anchor and a TSS in the other anchor. This was performed for all LD-expanded SNPs to identify the full complement of genes that could be putatively implicated in AD and PD.
gkm-SVM machine learning classifier training and testing
Identification of MAPT haplotypes
The MAPT haplotype block is part of one of the largest LD blocks in the human genome. To identify SNPs that belong exclusively to either the H1 or H2 haplotype, we used minor allele frequencies from dbSNP version 151. SNPs were required to be within the coordinates of the MAPT inversion breakpoints (hg38 chr17:45551578–46494237) and to have a minor allele frequency between 8.4% and 9%. While there are undoubtedly haplotype specific SNPs outside this frequency range, we chose this range to be as conservative as possible and to pick SNPs that showed minimal haplotype switching. Each SNP was verified to track with the predicted haplotype using LDLink89. This resulted in 2,366 SNPs that could be confidently called as haplotype divergent.
MAPT locus differential expression analysis
A 900-kb block of variants in strong LD at the MAPT locus hampered the resolution of colocalization methods for identifying causal variants and/or genes at this locus. To probe this locus more deeply, we assembled a list of 2,366 variants uniquely found in either the H1 or the H2 haplotype of the MAPT locus (described above). For each of the 838 individuals genotyped in GTEx v8, we counted the number of variants in support of either haplotype. We designated individuals as homozygous if they possessed less than 1% of variants favoring the opposite haplotype and heterozygous if 45% to 55% of variants supported either haplotype. This determined the individual’s haplotype in all but six cases, which were excluded from the remainder of the MAPT analysis. In total, we identified 539 individuals with the H1/H1 haplotype, 260 with H2/H1, and 33 with H2/H2. Our a priori gene of interest was MAPT, whose expression had previously been demonstrated to be higher in H1 than H2 haplotypes. At a nominal cutoff of P < 0.05, we confirmed this expected direction of differential MAPT expression (higher in H1 haplotypes) in multiple tissues, with the strongest contrasts in “Brain - Cortex”.
We then extended our analysis to include all genes expressed in any of the brain tissues from GTEx v8. We compared the log2-fold change of gene expression (TPM) between H1/H1 and H1/H2 individuals, given that these subgroups had the largest sample size. A change was considered statistically significant if a Wilcoxon rank-sum test between the two groups produced a P value of < 0.05 / (total # genes) / (total # tissues). We also performed pairwise Wilcoxon rank-sum test comparisons for each gene in each brain tissue between all 3 pairings of haplotypes.
MAPT haplotype-specific ATAC-seq and HiChIP analysis
For both ATAC-seq and HiChIP, reads from heterozygote donors were re-mapped to an N-masked genome (using bowtie2 or HiCPro, respectively) where all dbSNP v151 positions were masked to “N”. After alignment, SNPsplit90 was used to divide reads mapping to either the H1 or H2 haplotypes based on the presence of one of the 2,366 haplotype-divergent SNPs identified above. In this way, reads mapping to regions that lack a haplotype-divergent SNP could not be assigned in an allelic fashion to either the H1 or H2 haplotypes and were ignored. For track-based visualizations of haplotype-specific data, all available data from a given haplotype were merged agnostically to what brain region the data were derived from. For visualtization of ATAC-seq and HiChIP data from H1/H2 heterozygotes, no normalization was performed because each sample is internally controlled for allelic depth. To identify regions with haplotype-specific chromatin accessibility in the MAPT locus, the entire locus was tiled into non-overlapping 500 bp bins and the number of Tn5 transposase insertions were counted for each haplotype in each bin for each sample. A Wilcoxon signed-rank test was used to determine if the difference between H1 and H2 for each bin was significant after multiple hypothesis correction (FDR < 0.01).
Extended Data
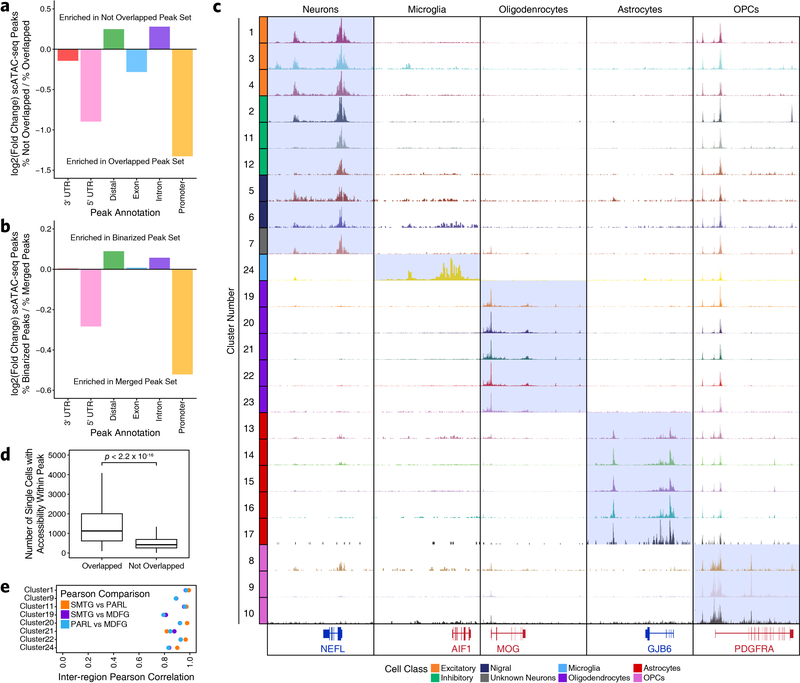
Extended Data Fig. 1. Region-centric scATAC-seq identifies cellular and regional heterogeneity in chromatin accessibility in adult brain.
a-b, UMAP dimensionality reduction (a) prior to and (b) after batch correction with Harmony of scATAC-seq data from 10 different samples. Each dot represents a single cell (N = 70,631). Dots are colored by the sample of origin. Color labels are shown in Extended Data Figure 1b. c, The same UMAP dimensionality reduction shown in Extended Data Figure 1b but each cell is colored by its gene activity score for the annotated lineage-defining gene. Gene activity scores were imputed using MAGIC. Grey represents the minimum gene activity score while purple represents the maximum gene activity score for the given gene. The minimum and maximum scores are shown in the bottom left of each panel. The gene of interest and the cell type that it identified are shown in the upper left of each panel. MSNs – medium spiny neurons. d, Heatmap of cell type-specific markers used to define the cell type corresponding to each cluster. Color represents the row-wise Z-score of chromatin accessibility in the vicinity of each gene for each cluster. e, Cluster residence heatmap showing the percent of each cluster that is composed of cells from each sample. Cell numbers were normalized across samples prior to calculating cluster residence percentages to account for differences in total pass filter cells per sample. f-h, UMAP dimensionality reduction as shown in Extended Data Figure 1b but colored by (f) the gross brain region from which each cell was obtained, (g) the biological sex of the donor for each cell, or (h) the predicted cell class for each cell. i-k, Bar plot showing the number of cells identified in our scATAC-seq data from (i) each of the annotated cell classes, (j) each of the annotated donors/samples, or (k) each of the gross brain regions subdivided based on cell class. Color represents the predicted cell class as shown in the legend of Extended Data Figure 1h. l-m, Bar plot showing the percentage of cells in our scATAC-seq data from (l) each of the gross brain regions subdivided based cell class or (m) each of the annotated cell classes subdivided based on donor/sample of origin. Color represents (l) the predicted cell class as shown in the Extended Data Figure 1h or (m) the biological sample from which the cells were obtained.
Extended Data Fig. 2. Cellular heterogeneity in brain tissue necessitates single-cell approaches to capture biological complexity.
a-b, Bar plot of the log2(Fold Change) in the percent of peaks mapping to various genomic annotations comparing peaks from (a) the scATAC-seq peak set that are not overlapped by a peak from the bulk ATAC-seq peak set to peaks that are overlapped by a peak from the bulk ATAC-seq peak set or (b) the scATAC-seq peak set that are were identified as cell type-unique through feature binarization to all peaks from the scATAC-seq peak set. c, Sequencing tracks of lineage-defining factors shown across all 24 scATAC-seq clusters (except Cluster 18 – putative doublets). From left to right, NEFL (neurons; chr8:24933431–24966791), AIF1 (aka IBA1, microglia; chr6:31607841–31617906), MOG (oligodendrocytes; chr6:29652183–29699713), GJB6 (astrocytes; chr13:20200243–20239571), and PDGFRA (OPCs; chr4:54209541–54303643). d, Box and whiskers plots showing the distribution of the number of single cells from our scATAC-seq data showing accessibility within (left) each peak from the set of peaks from the scATAC-seq peak set that overlap a peak from the bulk ATAC-seq peak set (N = 120,941 peaks) and (right) each peak from the set of peaks from the scATAC-seq peak set that do not overlap a peak from the bulk ATAC-seq peak set (N = 238,081 peaks). The lower and upper ends of the box represent the 25th and 75th percentiles and the internal line represents the median. The whiskers represent 1.5 multiplied by the inter-quartile range. P-value determined by Kolmogorov–Smirnov test. e, Dot plot showing the inter-region Pearson correlation of pseudo-bulk replicates comprised of all cells from either SMTG, PARL, or MDFG within each of the clusters shown. The clusters shown were selected based on biological relevance (i.e. clusters annotated as “substantia nigra astrocytes” should not be compared across isocortical regions) and on cluster size (i.e. clusters with small numbers of isocortical cells would not provide robust comparisons).
Extended Data Fig. 3. Neuronal sub-clustering identifies diverse biologically relevant populations of neurons.
a-d, UMAP dimensionality reduction of neuronal cells (identified as Clusters 1, 2, 3, 4, 5, 6, 7, 11, and 12 from Figure 1e) (a) prior to or (b-d) after batch correction with Harmony of scATAC-seq data from 10 different samples. Each dot represents a single cell (N = 21,116). Dots are colored by (a-b) the sample of origin, (c) the neuronal sub-cluster (repeated from Figure 2a), or (d) its gene activity score for the annotated lineage-defining gene. In (d), gene activity scores were imputed using MAGIC. Grey represents the minimum gene activity score while purple represents the maximum gene activity score for the given gene. The minimum and maximum scores are shown in the bottom left of each panel. The gene of interest is shown in the upper right of each panel. e, Heatmap of gene activity scores for all neuronal markers used in identifying relevant cell types for neuronal sub-clusters. Color represents the column-wise z-scores for each gene across all neuronal sub-clusters with values thresholded at −2 and +2. Neuronal cluster “major annotation” is shown by color along with a cluster description to the right of the plot. f-h, The same UMAP dimensionality reduction shown in Extended Data Figure 3c but cells are colored by (f) the major cell class annotation, (g) a more granular neuronal sub-annotation, or (h) the neuronal cell class annotation. Assignment was made based on gene activity scores of lineage-defining genes. The cell class annotation shown in (h) was used to perform LD score regression analysis.
Extended Data Fig. 4. Sub-clustering of cells from the substantia nigra identifies TH-positive dopaminergic neurons.
a-d, UMAP dimensionality reduction after iterative LSI of scATAC-seq data from substantia nigra cells from 2 different samples. Each dot represents a single cell (N = 11,199). Dots are colored by (a) their corresponding substantia nigra sub-cluster, (b) the sample of origin, or (c-d) its gene activity score for (c) the tyrosine hydoxylase (TH) gene, a specific marker of dopaminergic neurons or (d) other lineage-defining genes. In (c-d), gene activity scores were imputed using MAGIC. Grey represents the minimum gene activity score while purple represents the maximum gene activity score for the TH gene. In (a-c), the minimum and maximum scores are shown in the bottom left of the figure. Predicted cluster cell type identities are overlaid on the UMAPs.
Extended Data Fig. 5. HiChIP and co-accessibilty predict enhancer-promoter interactions in primary adult human brain.
a, Heatmap representation of HiChIP interaction signal at 100-kb, 25-kb, and 5-kb resolution at the OLIG2 locus. Sample shown represents the substantia nigra from donor 03–41. Signal is normalized to the square root of the coverage. The maximum value of the color range and the coordinates along chromosome 21 are shown below each panel. b, Bar plots showing the total number of paired-end reads sequenced for each HiChIP library generated in this study. Color represents the brain region from which the data was generated. c, Bar plots showing the number of valid interaction pairs identified in HiChIP data from all samples profiled in this study. Color represents the type of interaction identified. d, Bar plot showing the overlap of FitHiChIP loop calls from the 4 gross brain regions profiled. Color indicates whether the loop was identified in a single region (unique) or more than one region (shared). e, Bar plot showing the classification of FitHiChIP loop calls based on whether the loop call contained an ATAC-seq peak (from either the bulk ATAC-seq peak set or the scATAC-seq peak set) or TSS in one, both, or no anchor. f, Bar plots showing the number of Cicero-predicted co-accessibility-based peak links that are observed in HiChIP (left) or the number of HiChIP-based FitHiChIP loop calls that are predicted as peak links by Cicero. g, Bar plot showing the number of cell type-specific peaks (defined as peaks identified through feature binarization; N = 221,062) or non-cell type-specific peaks (defined as scATAC-seq peaks that were not identified through feature binarization; N = 137,960) that overlap or do not overlap a Cicero-predicated co-accessibility linkage. Significance determined by Kolmogorov-Smirnov test.
Extended Data Fig. 6. A multi-omic tiered approach leverages machine learning to predict functional noncoding SNPs in AD and PD.
a, Flow chart of the analytical framework used to prioritize noncoding SNPs and predict functionality. The highest confidence SNPs (Tier 1) are supported by either machine learning predictions, allelic imbalance, or both. Moderate confidence SNPs (Tier 2) are supported by the presence of the SNP within a peak and a HiChIP loop or co-accessibility peak link that connects the SNP to a gene. Lower confidence SNPs (Tier 3) are only supported by the presence of the SNP in a peak. b-c, Box plot showing the area under (b) the precision-recall curve or (c) the receiver-operating characteristics curve for the gkm-SVM machine learning classifier. Performance for each of the 24 broad clusters is shown with dots representing outliers. The lower and upper ends of the box represent the 25th and 75th percentiles. The whiskers represent 1.5 multiplied by the inter-quartile range. The center line represents the median. d, GkmExplain importance scores shown across all 10 folds for each base across a 100-bp window surrounding rs636317 for the effect (left) and noneffect (right) bases. e, Dot plots showing comparison of the GkmExplain score, ISM score, and deltaSVM score. Each dot represents an individual SNP test in a given fold. Dot color represents the GWAS locus number. The only off-diagonal dots (circled) correspond to repetitive regions within the MAPT locus where the deltaSVM score appears to be particularly sensitive. f, Dot plot showing allelic imbalance assessed by RASQUAL across all bulk ATAC-seq data used in this study from a region-specific analysis. Significance is assessed by RASQUAL (see Methods). Dot color indicates the brain region found to have significant allelic imbalance. Grey dots do not pass significance testing based on an empircal distribution of permuted null q-values and a 10% false discovery rate. A RASQUAL effect size greater than 0.5 indicates that the alternate allele is enriched while less than 0.5 indicates that the reference allele is enriched. The plot is divided to show SNPs within the MAPT and DNAH17 loci (bottom) and SNPs in all other loci (top). SNPs mentioned in downstream analyses are highlighted by red text.
Extended Data Fig. 7. Multi-omic characterization of well-studied AD-related GWAS loci pinpoints putative functional noncoding SNPs.
a,c, Normalized scATAC-seq-derived pseudo-bulk tracks, H3K27ac HiChIP loop calls, co-accessibility correlations, and publically available H3K4me3 PLAC-seq loop calls (Nott. et al. 2019) in (a) the BIN1 gene locus (chr2:127045000–127182000) and (c) the MS4A gene locus (chr11:60023000–60554000). scATAC-seq tracks represent the aggregate signal of all cells from the given cell type and have been normalized to the total number of reads in TSS regions, enabling direct comparison of tracks across cell types. For HiChIP, each line represents a FitHiChIP loop call connecting the points on each end. Red lines contain one anchor overlapping the SNP of interest while grey lines do not. For co-accessibility, only interactions involving the accessible chromatin region of interest are shown. For PLAC-seq, MAPS loop calls from microglia (blue), neurons (orange), and oligodendrocytes (purple) are shown. b,d, GkmExplain importance scores for each base in the 50-bp region surrounding (b) rs13025717 or (d) rs636317 for the effect and non-effect alleles from the gkm-SVM model for microglia (Cluster 24). The predicted motif affected by the SNP is shown at the bottom and the SNP of interest is highlighted in blue. e, Dot plot showing allelic imbalance at rs636317. Significance of allelic imbalance was determined by RASQUAL. The bulk ATAC-seq counts determined by WASP and ASEReadCounter for the reference/non-effect (A) allele and variant/effect (T) allele are plotted. Each dot represents an individual bulk ATAC-seq sample (N = 140) colored by the brain region from which the sample was collected. Samples where fewer than 3 reads were present to support both the reference and variant allele (i.e. presumed homozygotes or samples with insufficient sequencing depth) are shown in grey. The blue line represents a linear regression of the non-grey points and the grey box represents the 95% confidence interval of that regression.
Extended Data Fig. 8. Multi-omic characterization of noncoding SNPs identifies novel genes implicated in PD.
a,c, Normalized scATAC-seq-derived pseudo-bulk tracks, H3K27ac HiChIP loop calls, co-accessibility correlations, and publically available H3K4me3 PLAC-seq loop calls (Nott. et al. 2019) in (a) the IP6K2 gene locus (chr3:48671000–49205000) or (c) the TMEM163 gene locus (chr2:134429000–134905000). scATAC-seq tracks represent the aggregate signal of all cells from the given cell type and have been normalized to the total number of reads in TSS regions, enabling direct comparison of tracks across cell types. For HiChIP, each line represents a FitHiChIP loop call connecting the points on each end. Red lines contain one anchor overlapping the SNP of interest while grey lines do not. For co-accessibility, only interactions involving the accessible chromatin region of interest are shown. For PLAC-seq, MAPS loop calls from microglia (blue), neurons (orange), and oligodendrocytes (purple) are shown. b,d, GkmExplain importance scores for each base in the 50-bp region surrounding (b) rs6781790 or (d) rs7599054 for the effect and non-effect alleles from the gkm-SVM model for (b) astrocytes (Cluster 15) or (d) microglia (Cluster 24). The predicted motif affected by the SNP is shown at the bottom and the SNP of interest is highlighted in blue. e, Dot plot comparing the –log10(p-value) from 23andMe PD GWAS data with the –log10(p-value) from GTEx Caudate eQTL data of SNPs in the TMEM163 locus. Each dot represents an individual SNP. Dot color represents the r2 value of LD with the lead SNP (rs7599054 – purple diamond) within a European reference population. f-g, Dot plots showing the genomic coordinates of each SNP and the –log10(p-value) from (f) 23andMe PD GWAS data or (g) GTEx Caudate eQTL data. Dots are colored as in Extended Data Figure 8e. In (e-g), p-values are based on genome-wide chi-squared statistics from the relevant GWAS and eQTL studies.
Extended Data Fig. 9. Epigenomic dissection of the MAPT locus.
a, Flowchart illustrating the analytical scheme used to identify bins with significant allelic imbalance across the H1 and H2 MAPT haplotypes. b, Heatmaps showing chromatin accessibility in 500-bp bins identified as having significantly different accessibility across MAPT haplotypes. Regions are shown for homozygous samples without allelic read splitting (left) and for heterozygous samples after allelic read splitting (right). Bin start coordinates are shown to the right. c, Box and whiskers plots for multiple regions which show differential chromatin accessibility across the H1 and H2 MAPT haplotypes. Each dot represents a single homozygous H1 (N = 91) or homozygous H2 (N = 12) sample. Heterozygotes are not shown. The lower and upper ends of the box represent the 25th and 75th percentiles. The whiskers represent 1.5 multiplied by the inter-quartile range. The center line represents the median. d-e, Gene expression of (d) the KANSL1-AS1 gene or (e) the MAPK8IP1P2 gene shown as a box plot from GTEx cortex brain samples subdivided based on MAPT haplotype. The lower and upper ends of the box represent the 25th and 75th percentiles. The whiskers represent 1.5 multiplied by the inter-quartile range. The center line represents the median. ***p < 10−5 based on Wilcoxon rank sum test. N = 117 H1/H1, 78 H1/H2, and 10 H2/H2. f, Sequencing tracks from pseudo-bulk data derived from predicted cell types in scATAC-seq data. This region represents a zoomed in view of the predicted distal regulatory region (chr17:45216500–45324000) that interacts with the MAPT promoter in the H1 haplotype. Putative neuron-specific regulatory elements are highlighted in blue. g, Box plots showing differential HiChIP interaction signal occurring between regions within the MAPT inversion and regions outside the inversion (“left” or “right”). The schematic at the top explains the analysis performed. The box plots show normalized HiChIP interaction counts for the H1 (N = 6) and H2 (N = 6) haplotypes for upstream/“left” interactions and downstream/“right” interactions. P-value determined by paired two-sided t-test.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH NS062684, AG057707, AG053959, AG047366 (to T.M.), HG007735 (to H.Y.C.), HG009431 (to S.B.M./A.K.), AG066490 (to S.B.M.), and AG059918 (to M.R.C.). Additional support for patient sample collection provided by NIH AG005136 and AG019610. Sequencing data for this project were generated on an Illumina HiSeq 4000 supported in part by NIH award S10OD018220. Additional resources at the Stanford Center for Genomics and Personalized Medicine Sequencing Center were supported by NIH S10OD025212. M.R.C. is supported by the American Society of Hematology Scholar Award. A.S. is supported by the Stanford BioX Bowes fellowship. M.J.G. and T.E. are supported by NLM training grant 5T15LM007033–36. M.J.G. is additionally supported by a Stanford Graduate Fellowship. H.Y.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
COMPETING INTERESTS STATEMENT
H.Y.C. is a co-founder of Accent Therapeutics, Boundless Bio, and an advisor to 10x Genomics, Arsenal Biosciences, Spring Discovery. S.B.M. is on the scientific advisory board of MyOme. A.K. is a consultant for Biogen Inc. A.S. is a consultant for MyoKardia. W.J.G. is a consultant for Guardant Health, 10x Genomics, and Protillion Biosciences.
DATA AVAILABILITY
All data generated in this work are available through GEO accession GSE147672. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147672
To facilitate broad access to our data, we have created a WashU Epigenome browser session (Session ID: drS3o1n4kJ) for our scATAC-seq data in the following track formats: (i) broad cell types (“Corces_scATAC_BroadCellTypes”), (ii) broad clusters (“Corces_scATAC_BroadClusters”), (iii) neuron subclusters (“Corces_scATAC_NeuronSubClusters”), and (iv) neuron subclustered cell types / LDSC groups (“Corces_scATAC_NeuronSubCellTypes”). These tracks are accessible via the following link - http://epigenomegateway.wustl.edu/legacy/?genome=hg38&session=drS3o1n4kJ.
REFERENCES (MAIN TEXT)
- 1.Kunkle BW et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019 513 51, 414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jansen I et al. Genetic meta-analysis identifies 10 novel loci and functional pathways for Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert J-C et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beecham GW et al. Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer’s Disease and Related Dementias. PLoS Genet 10, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pankratz N et al. Meta-analysis of Parkinson’s Disease: Identification of a novel locus, RIT2. Ann. Neurol. 71, 370–384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang D et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 49, 1511–1516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nalls MA et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallagher MD & Chen-Plotkin AS The Post-GWAS Era: From Association to Function. Am. J. Hum. Genet. 102, 717–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nott A et al. Brain cell type – specific enhancer – promoter interactome maps and disease-risk association. Science 1139, 1134–1139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amiri A et al. Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trevino AE et al. Chromatin accessibility dynamics in a model of human forebrain development. Science 367, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowakowski TJ et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song M et al. Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nat. Genet. 51, 1252–1262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajarajan P et al. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fullard JF et al. An atlas of chromatin accessibility in the adult human brain. Genome Res. 28, 1243–1252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fullard JF et al. Open chromatin profiling of human postmortem brain infers functional roles for non-coding schizophrenia loci. Hum. Mol. Genet. 26, 1942–1951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryois J et al. Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nat. Commun 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corces MR et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sey NYA et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat. Neurosci. 23, 583–593 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee D et al. A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet. 47, 955–961 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrikumar A, Prakash E & Kundaje A GkmExplain: Fast and accurate interpretation of nonlinear gapped k-mer SVMs. Bioinformatics 35, i173–i182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumasaka N, Knights AJ & Gaffney DJ High-resolution genetic mapping of putative causal interactions between regions of open chromatin. Nat. Genet. 51, 128–137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amlie-Wolf A et al. InferNo: Inferring the molecular mechanisms of noncoding genetic variants. Nucleic Acids Res. 46, 8740–8753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulirsch JC et al. Interrogation of human hematopoiesis at single-cell and single-variant resolution. Nat. Genet. 51, 683–693 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satpathy AT et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mumbach MR et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mumbach MR et al. Enhancer connectome in primary human cells reveals target genes of disease-associated DNA elements. Nat. Genet. 49, 1602–1612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granja JM et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 37, 1458–1465 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pliner HA et al. Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol. Cell 71, 858–871.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corces MR et al. Lineage-specific and single cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corces MR et al. The chromatin accessibility landscape of primary human cancers. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKeown MR et al. Superenhancer analysis defi nes novel epigenomic subtypes of non-APL AML, including an RARα dependency targetable by SY-1425, a potent and selective RARα agonist. Cancer Discov. 7, 1136–1153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stolt CC et al. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17, 1677–1689 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I & Wegner M Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 18, 237–250 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondo T & Raff M Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development 127, 2989–2998 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Nakatani H et al. Ascl1/Mash1 promotes brain oligodendrogenesis during myelination and remyelination. J. Neurosci. 33, 9752–9768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith AM et al. The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia 61, 929–942 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Schlingensiepen KH et al. The role of Jun transcription factor expression and phosphorylation in neuronal differentiation, neuronal cell death, and plastic adaptations in vivo. Cell. Mol. Neurobiol. 14, 487–505 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hemonnot AL, Hua J, Ulmann L & Hirbec H Microglia in Alzheimer disease: Well-known targets and new opportunities. Front. Cell. Infect. Microbiol. 9, 1–20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Efthymiou AG & Goate AM Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener. 12, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghandi M et al. GkmSVM: An R package for gapped-kmer SVM. Bioinformatics 32, 2205–2207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bromberg Y & Rost B Comprehensive in silico mutagenesis highlights functionally important residues in proteins. Bioinformatics 24, 207–212 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu W, Tan L & Yu JT The Role of PICALM in Alzheimer’s Disease. Mol. Neurobiol. 52, 399–413 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Stage E et al. The effect of the top 20 Alzheimer disease risk genes on gray-matter density and FDG PET brain metabolism. Alzheimer’s Dement. Diagnosis, Assess. Dis. Monit. 5, 53–66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrew RJ et al. Reduction of the expression of the late-onset Alzheimer’s disease (AD) risk-factor BIN1 does not affect amyloid pathology in an AD mouse model. J. Biol. Chem. 294, 4477–4487 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma J, Yu JT & Tan L MS4A Cluster in Alzheimer’s Disease. Mol. Neurobiol. 51, 1240–1248 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Rouka E et al. Differential recognition preferences of the three Src Homology 3 (SH3) domains from the adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3). J. Biol. Chem. 290, 25275–25292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larsson M et al. GWAS findings for human iris patterns: Associations with variants in genes that influence normal neuronal pattern development. Am. J. Hum. Genet. 89, 334–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kajiho H et al. RIN3: A novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J. Cell Sci. 116, 4159–4168 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Lecours C et al. Microglial implication in Parkinson’s disease: Loss of beneficial physiological roles or gain of inflammatory functions? Front. Cell. Neurosci. 12, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaushik DK, Gupta M, Das S & Basu A Krüppel-like factor 4, a novel transcription factor regulates microglial activation and subsequent neuroinflammation. J. Neuroinflammation 7, 1–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schellenberg GD & Montine TJ The genetics and neuropathology of Alzheimer’s disease. Acta Neuropathol. 124, 305–323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stefansson H et al. A common inversion under selection in Europeans. Nat. Genet. 37, 129–137 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Zody MC et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet. 40, 1076–1083 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valenca GT et al. The Role of MAPT Haplotype H2 and Isoform 1N/4R in Parkinsonism of Older Adults. PLoS One (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allen M et al. Association of MAPT haplotypes with Alzheimer’s disease risk and MAPT brain gene expression levels. Alzheimer’s Res. Ther. 6, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pascale E et al. Genetic architecture of MAPT gene region in parkinson disease subtypes. Front. Cell. Neurosci. 10, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beevers JE et al. MAPT Genetic Variation and Neuronal Maturity Alter Isoform Expression Affecting Axonal Transport in iPSC-Derived Dopamine Neurons. Stem Cell Reports 9, 587–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lai MC et al. Haplotype-specific MAPT exon 3 expression regulated by common intronic polymorphisms associated with Parkinsonian disorders. Mol. Neurodegener. 12, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huin V et al. Alternative promoter usage generates novel shorter MAPT mRNA transcripts in Alzheimer’s disease and progressive supranuclear palsy brains. Sci. Rep. 7, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 65.Pankratz N et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum. Genet. 124, 593–605 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan AR & Hall IM BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heinz S et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 49, 1576–1583 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Duncan L et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am. J. Psychiatry 174, 850–858 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Demontis D et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Otowa T et al. Meta-analysis of genome-wide association studies of anxiety disorders. Mol. Psychiatry 21, 1391–1399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okbay A et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anney RJL et al. Genetic determinants of common epilepsies: A meta-analysis of genome-wide association studies. Lancet Neurol. 13, 893–903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zillikens MC et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat. Commun 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kemp JP et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet. 49, 1468–1475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Howson JMM et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat. Genet. 49, 1113–1119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harvey CT et al. QuASAR: Quantitative allele-specific analysis of reads. Bioinformatics 31, 1235–1242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Auton A et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Granja JM et al. ArchR : An integrative and scalable software package for single-cell chromatin accessibility analysis. biorxiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Korsunsky I et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cusanovich DA et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fulco CP et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Dijk D et al. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 1–21 (2014) doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Servant N et al. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bhattacharyya S, Chandra V, Vijayanand P & Ay F Identification of significant chromatin contacts from HiChIP data by FitHiChIP. Nat. Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Machiela MJ & Chanock SJ LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krueger F & Andrews SR SNPsplit: Allele-specific splitting of alignments between genomes with known SNP genotypes [version 2; referees: 3 approved]. F1000Research 5, 1–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.