Abstract
Enzymes that cleave ATP to activate carboxylic acids play essential roles in primary and secondary metabolism in all domains of life. Class I adenylate-forming enzymes share a conserved structural fold but act on a wide range of substrates to catalyze reactions involved in bioluminescence, nonribosomal peptide biosynthesis, fatty acid activation, and β-lactone formation. Despite their metabolic importance, the substrates and functions of the vast majority of adenylate-forming enzymes are unknown without tools available to accurately predict them. Given the crucial roles of adenylate-forming enzymes in biosynthesis, this also severely limits our ability to predict natural product structures from biosynthetic gene clusters. Here we used machine learning to predict adenylate-forming enzyme function and substrate specificity from protein sequences. We built a web-based predictive tool and used it to comprehensively map the biochemical diversity of adenylate-forming enzymes across >50,000 candidate biosynthetic gene clusters in bacterial, fungal, and plant genomes. Ancestral phylogenetic reconstruction and sequence similarity networking of enzymes from these clusters suggested divergent evolution of the adenylate-forming superfamily from a core enzyme scaffold most related to contemporary CoA ligases toward more specialized functions including β-lactone synthetases. Our classifier predicted β-lactone synthetases in uncharacterized biosynthetic gene clusters conserved in >90 different strains of Nocardia. To test our prediction, we purified a candidate β-lactone synthetase from Nocardia brasiliensis and reconstituted the biosynthetic pathway in vitro to link the gene cluster to the β-lactone natural product, nocardiolactone. We anticipate that our machine learning approach will aid in functional classification of enzymes and advance natural product discovery.
Keywords: natural product biosynthesis, acetyl-CoA synthetase, enzyme catalysis, bioinformatics, coenzyme A (CoA), β-lactone synthetases, adenylate-forming enzymes, machine learning, Nocardia, substrate specificity
Adenylation is a widespread and essential reaction in nature to transform inert carboxylic acid groups into high energy acyl-AMP intermediates. Class I adenylate-forming enzymes catalyze reactions for natural product biosynthesis, firefly bioluminescence, and the activation of fatty acids with CoA (1). The conversion of acetate to acetyl-CoA by a partially purified acetyl-CoA ligase was first described by Lipmann in 1944 (2). Since then, enzymes with this conserved structural fold have been found to activate over 200 different substrates including aromatic, aliphatic, and amino acids. To encompass the major functional enzyme classes, the term “ANL superfamily” was proposed based on the acyl-CoA ligases, nonribosomal peptide synthetases (NRPSs), and luciferases (3).
Most ANL superfamily enzymes have two-step reaction mechanisms: adenylation followed by thioesterification. During the thioesterification step, ANL enzymes undergo a dramatic conformational change involving a 140° domain rotation of the C-terminal domain (3). Thioester bond formation results from nucleophilic attack, typically by a phosphopantetheine thiol group. A notable exception to phosphopantetheine is the use of molecular oxygen by firefly luciferase to convert d-luciferin to a light-emitting oxidized intermediate (3). Other interesting exceptions include functionally divergent ANL enzymes within the same pathway that catalyze the adenylation and thioesterification reactions separately (4, 5). The ANL superfamily has recently expanded to include several new classes of enzymes including the fatty acyl-AMP ligases (6), aryl polyene adenylation enzymes (7), and β-lactone synthetases (8). Strikingly, an ANL enzyme involved in cremeomycin biosynthesis was shown to use nitrite to catalyze late stage nitrogen–nitrogen bond formation in diazo-containing natural products (9). The discovery of new reactions in the ANL superfamily over 75 years after Lipmann's initial report suggests that these enzymes still have unexplored biocatalytic potential, particularly in specialized metabolic pathways.
No computational tools currently exist for prediction and functional classification of ANL enzymes at the superfamily level. Still, the development of one previous platform, which is no longer supported or available, showed that this class of enzymes is amenable to computational predictions of substrate and function (10). Bioinformatics tools have also developed to predict substrates for NRPS adenylation (A) domains (11, 12). Genome mining approaches using NRPS A domain prediction tools have proved useful to access the biosynthetic potential of unculturable organisms and link “orphan” natural products with their biosynthetic gene clusters. For example, NRPS A domain predictions guided the discovery of the biosynthetic machinery for the leinamycin family of natural products (13) and enabled sequence-based structure prediction of several novel lipopeptides by Zhao et al. (14). However, Zhao et al. reported limitations in existing tools in that they could not predict the chain length of lipid tails incorporated into lipopeptides. Lipid tails are prevalent in natural products and are incorporated by fatty acyl-AMP or acyl-CoA ligase enzymes, both in the ANL superfamily. These enzymes are among the most well-studied subclasses of ANL enzymes. For less-studied subclasses, enzyme functions and substrates are even more challenging to predict. Hence, a computational tool encompassing all classes of ANL enzymes would constitute a major step toward more accurate structural prediction of natural product scaffolds.
Here, we used machine learning to develop a predictive platform for ANL superfamily enzymes and map their substrate-and-function landscape across 50,064 candidate biosynthetic gene clusters in bacterial, fungal, and plant genomes. We detected candidate β-lactone synthetases in uncharacterized biosynthetic gene clusters from pathogenic Nocardia spp. and experimentally validated the gene cluster in vitro to link it to the orphan β-lactone compound, nocardiolactone. Overall, this research provides a proof of principle toward the use of machine learning for classification of enzyme substrates to guide natural product discovery.
Results
Machine learning accurately predicts ANL enzyme function and substrate specificity
A global analysis of protein family domains revealed that ANL superfamily enzymes (PF00501; AMP-binding domains) are the third most abundant domain in known natural product biosynthetic pathways (Table S1). Despite the essential and varied roles of ANL enzymes, there is no single database that catalogs their biosynthetic diversity. Therefore, we mined the literature, MIBiG (15), and UniProtKB (16) for ANL enzymes with known substrate specificities. We then constructed a training set of >1,700 ANL protein sequences paired with their functional class, substrate(s), kinetic data, and crystal structures if solved. As reported previously by Gulick (3) and others, ANL superfamily enzymes in our training set were divergent at the sequence level but shared a common structural fold and core motifs including (Y/F)(G/W)X(A/T)E and (S/T)GD critical for ATP binding and catalysis.
We defined nine major functional classes on the basis of having enough experimentally characterized enzymes to enable classification by machine learning: short-chain acyl-CoA synthetases (C2–C5, SACS), medium-chain acyl-CoA synthetases (C6–C12, MACS), long-chain acyl-CoA synthetases (C13–C17, LACS), very-long-chain acyl-CoA synthetases (C18+), fatty acyl-AMP ligases (FAAL), luciferases (LUC), β-lactone synthetases (BLS), aryl-CoA ligases (ARYL), and NRPS A domains. In addition, we trained a separate model to predict enzyme substrate specificity. Although prediction at the level of an individual substrate is desirable, the broad substrate specificity of some classes of ANL enzymes limited the resolution of our predictions to groups of chemically similar compounds. For example, one class of LACS has demonstrated activity with fatty acids with chain lengths ranging from C8 to C20 (17). There were over 200 chemically distinct substrates in our training set, many with just one experimental example. Therefore, we clustered substrates based on chemical similarity (Tanimoto coefficient) to identify 15 groups for broad level substrate classification (Table S2).
Previously, 34 amino acids within 8 Å of the gramicidin synthetase active site were shown to be critical for accurate prediction of NRPS A domain substrate specificity (11). Because of the high level of structural conservation between NRPS A domains and other proteins in the superfamily, we hypothesized that these 34 active site residues would also be important features for ANL substrate prediction. Using an AMP-binding profile Hidden Markov Model (pHMM), we extracted 34 active site residues from our >1,700 training set sequences. Further inspection of the superfamily-wide pHMM alignment revealed the presence of a fatty acyl-AMP ligase-specific insertion (FSI) of 20 amino acids not present in other superfamily members (Fig. S1). The FSI has been suggested to inhibit the 140° domain rotation of the C-terminal domain and is critical for the rejection of CoA-SH as an acceptor molecule (6). Because the FSI was shown experimentally to be an important feature to distinguish FAAL from LACS enzymes, we extracted 20 FSI residues from each sequence and appended them to our feature vector for a total of 54 residues. Each amino acid was further encoded as 15 normalized real numbers corresponding to different physicochemical properties including hydrophobicity, volume, secondary structure, and electronic properties (11). The physicochemical properties were then used to train machine learning models to predict enzyme function and substrate (Fig. 1A).
Figure 1.
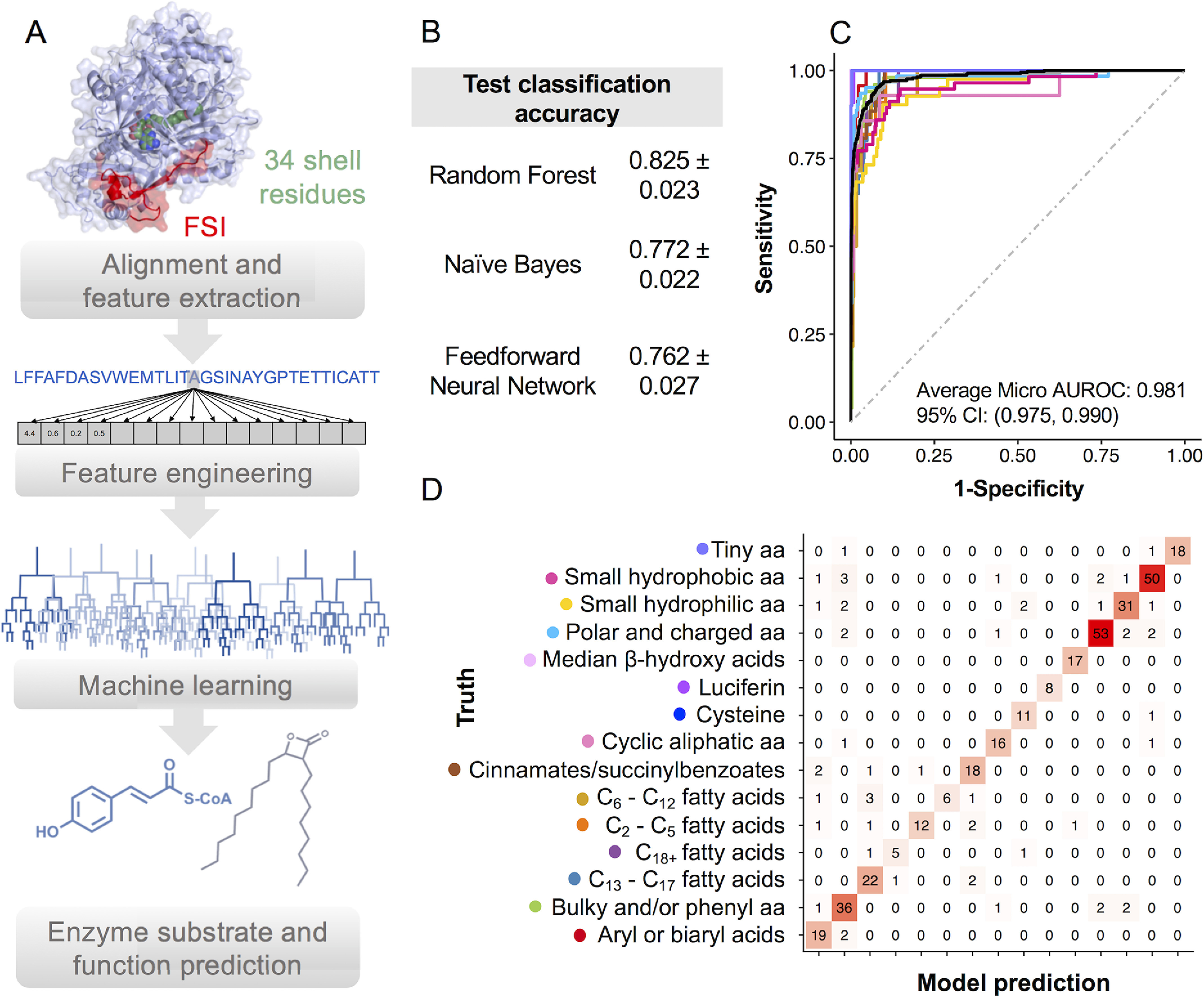
Machine learning to predict substrate and function of adenylate-forming enzymes from protein sequences. A, the machine learning workflow includes extracting 34 active site residues (green) and FAAL-specific loop (red) residues and encoding them as a vector of physiochemical properties. Separate classifiers are trained to predict substrate specificity and enzyme function. B, hold-out test set accuracy scores for three different classification methods evaluated in this study. C, AUROC for substrate specificity predictions. Colors correspond to different substrates and macro (gray) and micro (black) AUROC averages. Red, aryl/biaryl acids; green, bulky/phenyl aa; blue, C13–C17 fatty acids; purple, C18+ fatty acids; orange, C2–C5 acids; tan, C6–C12 fatty acids; brown, succinylbenzoic acids; hot pink, cyclic aliphatic aa; dark blue, cysteine; fuchsia, luciferin; light pink, β-hydroxy acids; turquoise, polar and charged aa; goldenrod, small hydrophilic aa; deep pink, small hydrophobic aa; lavender, tiny aa. D, confusion matrix of predicted versus truth for ANL substrate specificity on hold-out test set. Predictions for functional class are presented in Fig. S2.
Three different machine learning algorithms were evaluated for our classification problem: feedforward neural networks, naïve Bayes, and random forest. Random forest performed slightly better than its counterparts for both function and substrate classification problems (Fig. 1B and Fig. S2). Naïve Bayes also performed well but was significantly slower than random forest in run time. The feedforward neural network performed the worst, likely because of a relatively small number of training samples, and was also the slowest model to train. On the basis of speed and accuracy, we chose to proceed with the random forest algorithm. Our best performing model achieved 82.5 ± 2.3% test set classification accuracy for substrate specificity and 83.3 ± 3.0% for functional class prediction (Fig. 1B and Fig. S2). The average area under the receiver operating characteristic curve (micro-average AUROC) was 0.981 for substrate group prediction and 0.978 for functional class (Fig. 1C and Fig. S2). Within-class accuracy was highest for the FAAL and NRPS A domains, most likely because of a larger amount of experimental data for these protein families (Fig. S2). Both of our classifiers also performed well with more specialized enzymes that accept single substrates such as the BLS and LUC classes (Fig. S2). We speculate that enzymes with specialized functions might have distinct “active-site patterns” that could be learned by our algorithms because of the preference of these enzymes for single substrates (e.g. d-luciferin). Our machine learning model consistently performed the worst on substrate classification for enzymes with broad substrate specificity such as the aryl/biaryl acids and C6–C11 chain length fatty acids and aryl acids (Fig. 1D).
We developed a web application, AdenylPred (z.umn.edu/AdenylPred), for adenylation prediction, to make our machine learning models publicly available. Users can upload their sequences of adenylate-forming enzymes in multi-FASTA or GenBank format either as nucleotide or as protein sequences. Predictions and probability scores between 0 and 1 (with scores >0.6 designated as confident predictions) are reported for both functional classification and substrate specificity. The entire ANL enzyme training set is also available in a searchable database format. Overall, the web app provides an interactive interface for users with little computational experience. A pared-down command-line version of the tool is also available for download for the analysis of large data sets.
AdenylPred validation with widely distributed ANL superfamily sequences
To evaluate AdenylPred performance with an entirely separate set of ANL sequences, we mined the literature for newly characterized adenylate-forming enzymes that had not been included in the initial training and testing sets. We assembled a new benchmark set of 40 protein sequences from insects, fungi, cyanobacteria, and other prokaryotes (Table S3). Of these, 27 of 40 had been directly verified through protein purification and in vitro biochemical assays or through MS-based analysis of cell lysates. The substrates and functions of remaining 13 of 40 enzymes could still be inferred with confidence using biosynthetic logic on the basis of experimental work such as natural product structure elucidation or deletions of other biosynthetic genes in the cluster (Table S3). Sequences in the benchmark set were also divergent from the training set with as low as 27.4% amino acid sequence identity to the top BLAST hit in the training set (mean 52.5%; Table S3). Substrate preferences of enzymes in the benchmark set included >30 compounds such as benzoxazolinate, 3-cyanobutanoate, and 3-(methylthio)propanoate (Fig. S3). The benchmark set also included a number of unusual or multifunctional adenylate-forming enzymes involved in diazo-group formation (9), formylation (18), and amide-bond formation (19, 20).
The overall accuracy of AdenylPred for all 40 sequences independent of probability score was 83% for functional class prediction and 73% for substrate specificity prediction (Table S3). Additionally, AdenylPred had 100% functional class and substrate specificity prediction accuracy for all sequences with probability scores >0.6. There were 25 of 40 sequences that had functional group probability scores >0.6 and 11 of 40 that had substrate scores >0.6. For these “high-confidence” sequences, the average percentage of aa identity was 58.9% to the training set, whereas for substrate specificity it was 71%. The overall classification accuracy scores indicate that AdenylPred has utility in predicting functions and substrates for a range of different enzymes in the ANL superfamily. As expected, prediction accuracy is positively associated with both the AdenylPred probability score and similarity of the query sequence to the training set. In particular, ANL sequences with low AdenylPred probability scores are promising candidates for experimental investigation because they may highlight ANL enzymes with as-yet undiscovered functions and substrates.
Specialized ANL enzymes may have evolved from an ancestral scaffold utilizing CoA-SH
We next used a phylogenetic approach to investigate the functional divergence of ANL superfamily enzymes (Fig. 2). Maximum-likelihood phylogenetic analysis revealed that some functional classes, including the BLS, LUC, and NRPS enzymes, formed tight monophyletic clades, whereas others, including enzymes in the ARYL, SACS, and MACS classes, were dispersed throughout the tree. The wide phylogenetic distribution of the ARYL sequences in particular indicated many ARYL enzymes were more closely related to different protein subfamilies than to each other. This observation could likely be explained by two evolutionary scenarios: 1) CoA ligase activity arose independently several times throughout ANL superfamily evolution or 2) radial divergence of the superfamily occurred from an ancestral scaffold similar to contemporary CoA-ligase-like enzymes (21). To investigate these scenarios further, we used a maximum-likelihood approach for ancestral sequence reconstruction to estimate the 10 most likely sequences for the predicted ancestral protein at the root of the ANL phylogeny (22). We used AdenylPred to extract sequence features and predict function and substrate for our reconstructed ancestral proteins. AdenylPred classified 10 of 10 of the most likely ancestral ANL proteins as the aryl-CoA ligases most likely to activate aryl and biaryl derivatives as substrates (probability score = 0.6). We also tested a maximum-likelihood ancestral reconstruction using only 34 active-site residues as the seed sequences rather than full-length sequences and obtained similar ARYL predictions (probability score = 0.7). These results suggest that the active sites of our reconstructed ancestral ANL proteins were most similar to contemporary CoA-ligase enzymes in the ANL superfamily.
Figure 2.
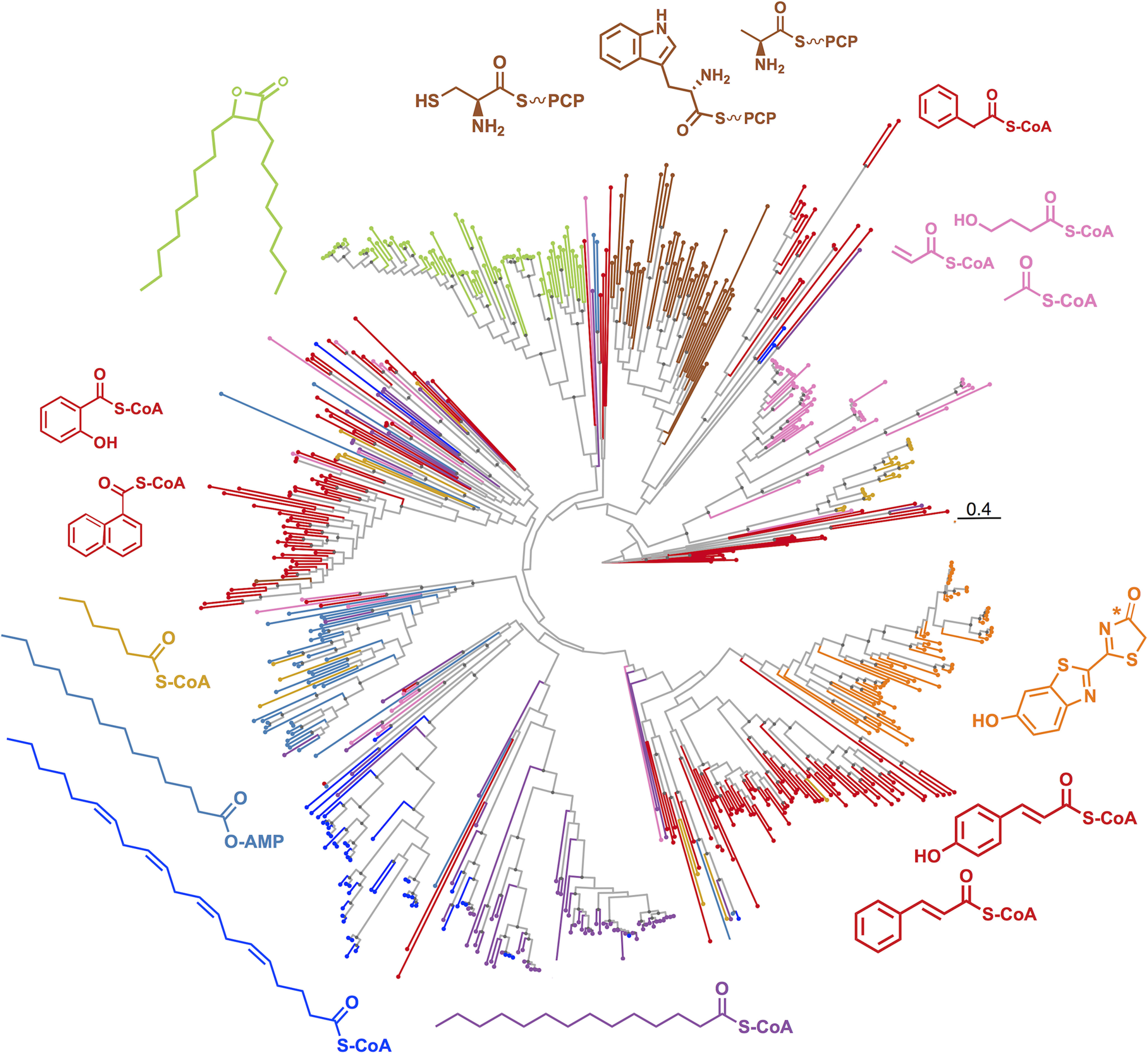
Maximum-likelihood phylogenetic tree of characterized protein sequences in the ANL superfamily. Tree was computed using the Jones–Taylor–Thornton matrix–based model of amino acid substitution and colored by functional enzyme class. Some enzyme classes such as BLS, NRPS, and LUC form monophyletic clades, whereas other sequences, i.e. the ARYL class are dispersed throughout the tree, suggest evolutionary divergence. Red, ARYL; green, BLS; dark blue, very-long-chain acyl-CoA synthetase; orange, LUC; light blue, FAAL; brown, NRPS; purple, LACS; pink, SACS; gold, MACS. Gray node circles represent bootstrap support >75% at branch points. Bar, 0.4 aa substitutions per site.
Sequence similarity networking suggests radial divergence of the ANL superfamily
To map the functional distribution of adenylation enzymes encoded in 50,064 candidate biosynthetic gene clusters, we applied AdenylPred to a taxonomically diverse and representative collection of bacterial, fungal, and plant genomes. We extracted all standalone adenylate-forming enzyme sequences from 50,064 candidate biosynthetic gene clusters detected using antiSMASH (23), fungiSMASH (24), and plantiSMASH (25). To visualize results, we constructed a sequence similarity network of adenylate-forming enzymes in which each node represents a group of proteins that share >40% amino acid sequence identity (Fig. 3A). Nodes were colored by their functional class predicted using AdenylPred. The sequence similarity network displayed a topology in which most functional protein subfamilies showed higher sequence similarity with “core” sequences than with any other subfamily (Fig. 3A). For all nodes in the core of the clustering diagram with AdenylPred probability scores >0.6, 80% were predicted to be aryl-CoA synthetases. To test the robustness of the network topology with a different sequence set, we constructed a sequence similarity network from a smaller set of all AMP-binding domains from the manually curated MIBiG database (Fig. S4). Again, we recovered the same topology with the core containing mostly CoA-utilizing enzymes. Of the 48,250 full-length AMP-binding hits that we analyzed with AdenylPred, 79% of the high-confidence hits were predicted to be in either the ARYL or NRPS classes (Fig. 3A). There were no predicted luciferases in the biosynthetic gene clusters, which was expected because insect genomes including fireflies were not included in the genome set. Notably, the number of predicted FAALs in our data set outnumbered LACS more than 10-fold (3,673 to 322). Because both FAALs and LACSs both accept long-chain fatty acids as substrates, these findings support previous reports that the majority of lipid tails in lipopeptides, and other natural products may be incorporated through FAAL-mediated activity rather than CoA activation (26). β-Lactone formation and CoA activation of long-chain fatty acids were the least common functions catalyzed by ANL enzymes in candidate biosynthetic gene clusters (Fig. 3B).
Figure 3.
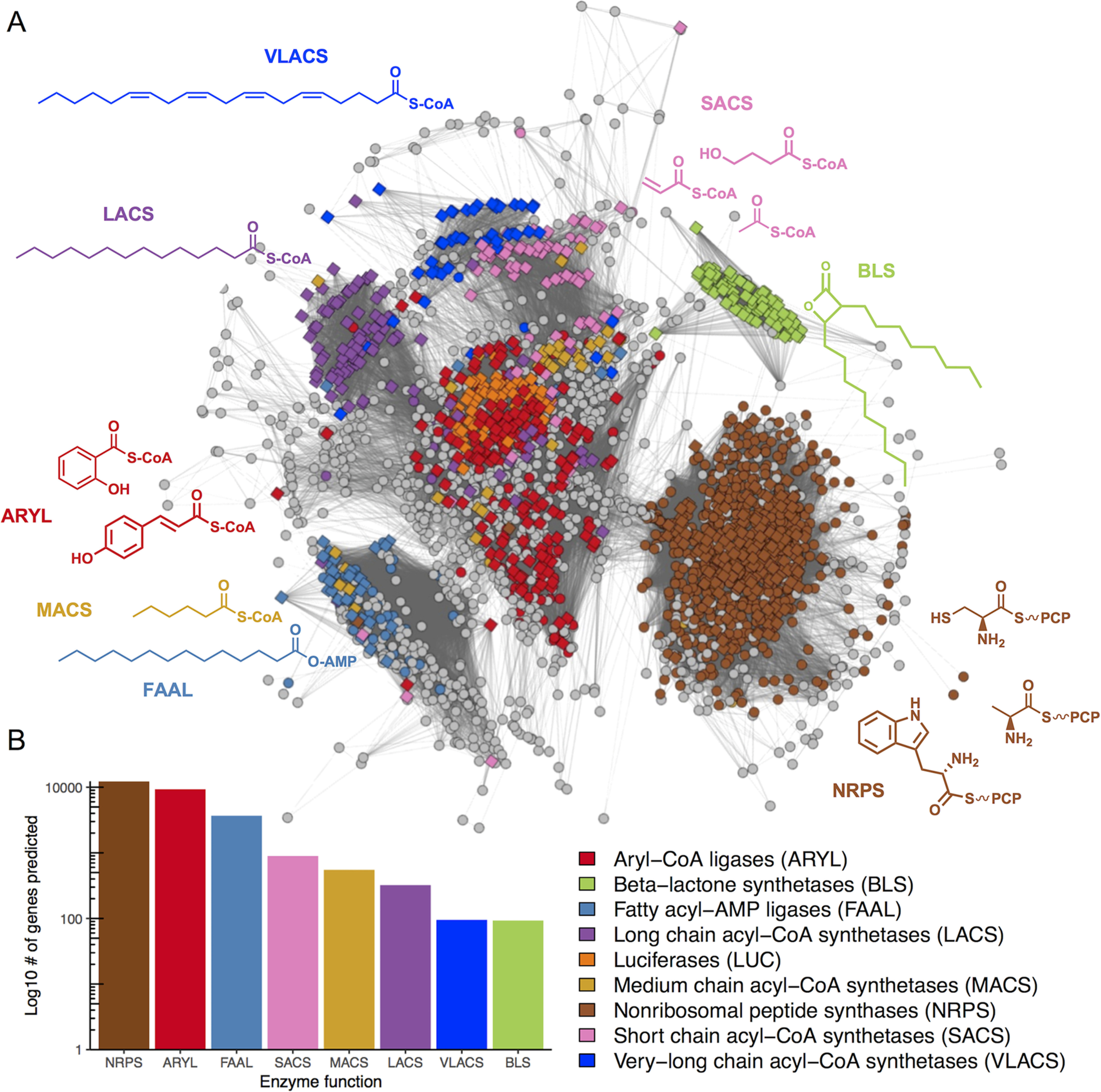
Predicted functional distribution of adenylation enzymes encoded in 50,064 candidate biosynthetic gene clusters. A, sequence similarity network of all standalone AMP-binding pHMM hits extracted from candidate biosynthetic gene clusters identified in >24,000 bacterial, fungal, and plant genomes. Diamonds correspond to training set sequences, and circles represent AMP-binding hits extracted from biosynthetic gene clusters. The network was trimmed to a BLAST e-value threshold of 1 × 10−36. Circles with a probability >0.6 are colored by their prediction, whereas sequences colored gray had “no confident prediction.” B, bar plot of relative counts for different functional classes of ANL enzymes within biosynthetic gene clusters (AdenylPred prediction probability > 0.6). VLACS, very-long-chain acyl-CoA synthetase.
AdenylPred-guided discovery of β-lactone synthetases in biosynthetic gene clusters
β-Lactone synthetases are among the most recently discovered members of the ANL superfamily (8), and a comprehensive analysis of their prevalence in natural product biosynthetic gene clusters had never been conducted. We examined AdenylPred hits for β-lactone synthetases from our collection of 50,064 candidate biosynthetic gene clusters. As expected, we detected candidate β-lactone synthetases in gene clusters responsible for the biosynthesis of known β-lactone natural products including ebelactone and lipstatin (27, 28). Predicted candidate β-lactone synthetases were also detected in 48 distinct bacterial and fungal genera, with the highest-confidence hits in the Planctomycetes, Deltaproteobacteria, and Actinobacteria (Fig. S5). To experimentally characterize one of these predictions, we selected a candidate β-lactone synthetase sequence that was highly conserved within the bacterial genus Nocardia (Fig. S5). Previously, a β-lactone natural product, nocardiolactone, had been isolated from pathogenic strains of Nocardia spp. (29). However, no follow-up studies on nocardiolactone were published, and the biosynthetic gene cluster was never reported, resulting in nocardiolactone being labeled an orphan natural product. Based on this, we hypothesized that the β-lactone synthetases detected by AdenylPred in Nocardia genomes were involved in β-lactone biosynthesis in the natural product nocardiolactone.
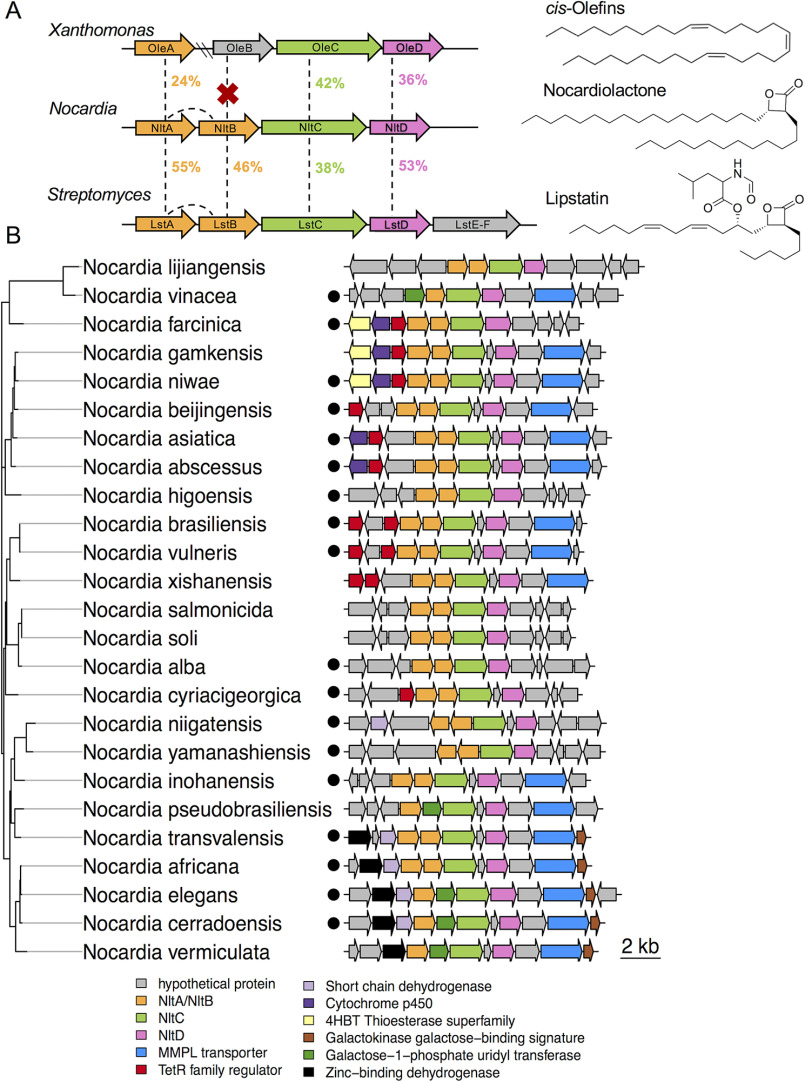
We identified three flanking genes around the predicted β-lactone synthetase genes in Nocardia spp. that shared synteny with biosynthetic genes for the β-lactone natural product, lipstatin (Fig. 4A). The lipstatin cluster also encodes NRPS and formyltransferase enzymes that attach an N-formyl leucine to the di-alkyl backbone (27). However, NRPS and formyltransferase genes conserved in the lipstatin cluster were absent from all candidate nocardiolactone clusters in Nocardia (Fig. 4B). We hypothesized that this cluster of four biosynthetic genes, termed nltABCD, might encode all the necessary enzymes for Nocardia spp. to produce a di-alkyl β-lactone product similar to lipstatin but lacking an amino acid side chain. Predicted functions of genes in the nltABCD biosynthetic cluster correspond to the reactions required to produce the orphan structure of the β-lactone product, nocardiolactone (Fig. 4A), prompting us to characterize the biosynthetic enzymes and pathway experimentally.
Figure 4.
Proposed nocardiolactone biosynthetic gene cluster. A, synteny between published bacterial cis-olefin and lipstatin gene clusters with the proposed nocardiolactone biosynthetic gene cluster. Percentages correspond to amino acid identity. B, representatives of the proposed nocardiolactone biosynthetic cluster in Nocardia. Maximum-likelihood phylogenetic tree is based on NltC amino acid sequence distance estimated using the Jones–Taylor–Thornton model of amino acid substitution. Sequences corresponding to Nocardia isolated from humans are designated by black circles.
In vitro reconstitution of the nocardiolactone pathway links the biosynthetic gene cluster to its orphan natural product
Nocardiolactone was originally isolated in 1999 from a pathogenic strain of Nocardia brasiliensis and other unidentified strains of Nocardia spp. that are not available in public culture collections (29). Genetic manipulation in Nocardia remains challenging; therefore we opted instead to reconstitute the complete biosynthetic pathway in vitro by heterologously expressing and purifying individual nltABCD pathway enzymes. This approach gave us full control to determine the function of each pathway enzyme through biochemical analysis of intermediates and comparison with synthetic standards.
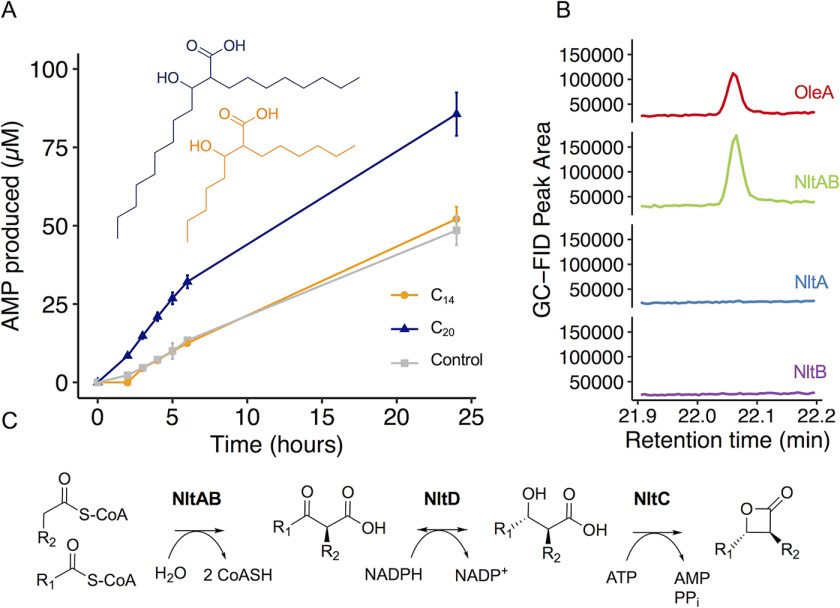
We cloned and expressed the gene encoding a candidate β-lactone synthetase, termed NltC, from a publicly available N. brasiliensis genome. NltC purified as a 60-kDa monomer. To test NltC for β-lactone synthetase activity, we synthesized syn- and anti-2-octyl-3-hydroxydodecanoic acid diastereomers as substrate analogs and added purified NltC, ATP, and MgCl2 (Fig. 5A). Compared with no enzyme controls, we observed NltC-catalyzed formation of cis-and trans-β-lactones by 1H NMR after overnight reactions (Fig. S6A). The coupling constants were consistent with synthetic standards for cis- and trans-β-lactones of comparable chain length (8). AMP release is also commonly used as a readout for ANL enzyme activity. In a time-course analysis of AMP release by NltC, we observed activity with C20 chain length β-hydroxy acids but no activity with C14 length analogs above the level of the no-substrate control (Fig. 5A). NltC also showed weak activity with 2-hexyldecanoic acid as a substrate mimic, but not with 10-nonadecanol, suggesting adenylation of the carboxylic acid rather than hydroxyl group (Fig. S6B). These results are consistent with the reported adenylation activity for the β-lactone synthetase from Xanthomonas campestris and with most enzymes in the ANL superfamily (30). Overall, these results support AdenylPred-guided predictions that NltC in N. brasiliensis is a functional β-lactone synthetase.
Figure 5.
Biochemical characterization of nocardiolactone biosynthetic enzymes. A, time-course analysis of NltC activity with di-alkyl β-hydroxy acids with carbon backbones of length C20 (blue) and C14 (orange) compared with a no-substrate control (gray). NltC prefers longer chain β-hydroxy acids (C20) and shows no discernable activity with C14 β-hydroxy acids above the level of the no-substrate control. B, co-expressed NltA and NltB enzymes condense 2 myristoyl-CoAs to form 2-myristoyl-3-ketomyristic acid. The resulting ketone (14-heptacosanone) from the breakdown of 2-myristoyl-3-ketomyristic acid was observable by GC-MS. The enzymatic product of NltAB was identical to a 14-heptacosanone control produced by WT X. campestris OleA but was not observed to be catalyzed by NltA or NltB enzymes purified individually. C, proposed biosynthetic pathway for nocardiolactone. R1, C18H37; R2, C13H27.
To further characterize enzymes involved in nocardiolactone biosynthesis, we purified two upstream thiolase family proteins, NltA and NltB. Recently, homologous enzymes in the lipstatin biosynthetic pathway, LstA and LstB were shown to form a functional heterodimer to catalyze “head-to-head” Claisen condensation of two acyl-CoAs (31). We hypothesized NltA and NltB might catalyze a similar reaction to form the nocardiolactone backbone (Fig. 5B). When heterologously expressed, NltA was mostly insoluble and formed inclusion bodies even when co-expressed with chaperones. We also attempted to purify a NltA homolog from a closely related organism, Nocardia yamanashiensis, and again observed inclusion body formation. In contrast, NltB could be purified with a moderate yield (∼28 mg/liters of culture). When tested individually, NltA and NltB protein preparations did not display activity with any chain length (C8–C16) acyl-CoA substrate tested. However, when nltA and nltB were co-expressed on the same plasmid, we obtained soluble protein that actively catalyzed the Claisen condensation of long-chain acyl-CoAs to β-keto acids (Fig. 5B). Size-exclusion chromatography indicated that NltA and NltB formed a heterodimer that eluted at 72 kDa.
From homology modeling and sequence analysis, we observed that the NltB sequence was 45 amino acids shorter than NltA and lacked a Cys-His-Asn catalytic triad, similar to reports for LstB relative to LstA (31). Although no crystal structure of LstAB is available, site-directed mutagenesis revealed that a conserved glutamate in LstB (Glu60) was required for condensation activity (31). In the crystal structure of the physiological homodimer of OleA from X. campestris, a similarly positioned glutamate from the β-chain was shown to enter the active site of the α-chain in the OleA homodimer to deprotonate and activate the α-carbon of the substrate (32). Based on homology modeling and structural alignments with OleA, Glu57 from NltB may be similarly poised for α-carbon deprotonation in the NltA active site (Fig. S7A). We used site-directed mutagenesis to mutate the NltB Glu57 to either Ala or Gln. Claisen condensation activity was abolished in NltABE57A and NltABE57Q mutants compared with WT NltAB (Fig. S7B). Taken together, these results suggest NltA and NltB may form a functional heterodimer with NltBE57 required to catalyze the Claisen condensation of two fatty acyl-CoA substrates.
The final enzyme in the nocardiolactone cluster, NltD, belongs to the short-chain reductase superfamily. NltD purified as a 78-kDa fusion protein with a maltose-binding protein tag. NltD has an N-terminal conserved nucleotide binding motif (Rossman fold) and a SXnYXXXK catalytic triad characteristic of short-chain reductase superfamily members (33). NltD shares 53% amino acid identity with the lipstatin reductase (LstD) and 36% identity with X. campestris OleD, a 2-alkyl-3-ketoalkanoic acid reductase involved in olefin biosynthesis (Fig. 4A). Because studies on OleD demonstrated 2-alkyl-3-ketoalkanoic acids are unstable, we monitored the reaction in reverse with 2-alkyl-3-hydroxyalkanoic acid substrates using a spectrophotometric assay for NADPH formation (33). Purified NltD catalyzed NADP+-dependent conversion of 2-alkyl-3-hydroxyalkanoic acid to 2-alkyl-3-ketoalkanoic acid with both C20 and C14 di-alkyl β-hydroxy acid substrates at a rate similar to a X. campestris OleD control (Fig. S8).
We next reconstituted the entire pathway to produce nocardiolactone-like analogs by combining purified pathway enzymes with decanoyl-CoA, NADPH, ATP, and MgCl2 (Fig. 5C and Fig. S9). Because of instability and loss of activity of purified NltAB over time, we substituted the functionally equivalent and stable homodimer, OleA, from X. campestris (34). After overnight incubation, we observed formation of a di-alkyl β-lactone natural product (Fig. S9). Based on in vitro evidence, we propose nocardiolactone biosynthesis is initiated via “head-to-head” Claisen condensation of two fatty acyl-CoA substrates catalyzed by a heterodimeric interaction between NltA and NltB. NltD then reduces the di-alkyl β-keto acid to a di-alkyl β-hydroxy acid in an NADPH-dependent manner. Finally, intramolecular ring closure of a β-hydroxy acid to a β-lactone is catalyzed by the ATP-dependent β-lactone synthetase NltC (Fig. 5C). Overall, these results link the nltABCD biosynthetic gene cluster in N. brasiliensis to the orphan natural product nocardiolactone.
The nocardiolactone gene cluster is enriched in human pathogens
With the biosynthetic gene cluster identified, we next probed the taxonomic distribution and abundance of the nocardiolactone pathway in Nocardia genomes. We used AdenylPred to identify putative β-lactone synthetases and detected nltC homologs with flanking nltABD genes in 94 of 159 complete Nocardia genomes in the PATRIC database (35). Notably, the strict nltABCD cluster was not detected in any closely related genera such as Rhodococcus, Streptomyces, or Mycobacterium, suggesting that nocardiolactone biosynthesis may be specific to the genus Nocardia. We observed the nocardiolactone gene cluster was more prevalent among strains of pathogenic clinical isolates from human patients than in strains isolated from other sources (Fig. 4B). The complete nltABCD cluster was detected in 68% of genomes of distinct species of human pathogenic Nocardia relative to 27% isolated from nonhuman sources (p = 0.002, Fisher's exact test). We also queried a separate data set comprised of 169 clinical isolates of Nocardia from human patients6 and recovered a similar proportion of complete nocardiolactone cluster hits among clinical isolates (112 of 169, ∼66%). Although only correlative, the enrichment warrants further research and suggests an association between the presence of the nocardiolactone cluster and Nocardia pathogenicity.
Discussion
Based on our ancestral reconstruction, we propose that ancient ANL enzymes had an active site most similar to contemporary enzymes that use CoA-SH as an acceptor molecule. This evolutionary scenario is supported by phylogenetic analysis and the composition of the “core” of the ANL sequence similarity network (Fig. 3A). Many other enzyme superfamilies do not have this radial topology and instead tend to show patterns of sequential functional divergence (36, 37). However, Babbitt and co-workers (21) detected a radial topology in the nitroreductase superfamily and provided multiple lines of evidence supporting the possibility that this network topology indicated divergent evolution of the superfamily from a minimal flavin-binding scaffold. Similarly, our results suggest that ANL sequences may have undergone divergent evolution from ancestral enzymes with CoA-ligase–like scaffolds toward more specialized functions.
The proposed evolutionary trajectory of the ANL superfamily is supported by experimental evidence for low-level CoA-ligase activity in many extant ANL enzymes that primarily perform other functions. For example, Linne et al. (38) tested the CoA-ligase activity of five different NRPS A domains. Surprisingly, all of the NRPS A domains tested were also able to synthesize acyl-CoAs in vitro. Enzymatic CoA-ligase activity of the NRPS A domains varied proportionally to their evolutionary similarity with a native acyl-CoA synthetase, suggesting that greater sequence divergence resulted in more specialized NRPS A domain activity and reduced bifunctionality (38). Firefly luciferases were also demonstrated to be bifunctional as CoA-ligases (39), and luciferase activity was conferred to an acyl-CoA ligase from a nonluminescent click beetle by just a single point mutation (40). Arora et al. (6) showed that FAAL enzymes likely lost their CoA-ligase activity because of the FSI. Indeed, deletion of the FSI conferred acyl-CoA ligase activity in FAAL28 from Mycobacterium tuberculosis. The fact that single mutations can revert many enzymes back to a CoA-ligase state supports the hypothesis that the ANL superfamily arose from an ancestral enzyme using CoA-SH as an acceptor molecule. Nevertheless it is challenging to conclusively rule out the alternative hypothesis that CoA-ligase activity arose independently several times in the evolution of the ANL superfamily. Overall, phylogenetic ancestral reconstruction coupled with AdenylPred analysis yielded new insights into the evolutionary structure–function relationships among adenylate-forming enzymes.
Within the ANL superfamily, the β-lactone synthetases were the most recently discovered family, and the extent of their role in natural product biosynthesis remains poorly understood (8, 30). We used AdenylPred to identify >90 β-lactone synthetases in uncharacterized biosynthetic gene clusters from 48 different genera (Fig. S5), which is significantly more than the eight known biosynthetic gene clusters for β-lactone natural products reported to date (41). The disparity between the number of predicted β-lactone biosynthetic gene clusters and known β-lactone natural products has several possible explanations. One reason may be limited discovery because of the reactivity and thermal instability of β-lactones. β-Lactones are strained rings that can rapidly hydrolyze in aqueous solutions (42) or thermally decarboxylate, thus hampering their detection by common analytical methods such as GC-MS (8). It is also plausible many biosynthetic gene clusters with β-lactone synthetases are not expressed under normal laboratory conditions. Another explanation is the undetected role of β-lactones as intermediates in the biosynthesis of other chemical moieties (8). Chemists have long referred to β-lactones as “privileged structures” for the total synthesis of compounds with a variety of functional groups including β-lactams, γ-lactones, and alkenes (41, 42). Our findings suggest microbes might also use β-lactones as intermediates because we detected a number of β-lactone synthetase hits in gene clusters known to make natural products without final β-lactone moieties such as polyunsaturated fatty acids. Indeed, β-lactone synthetases in oleABCD gene clusters were recently linked to production of the final alkene moiety in the biosynthesis of a C31 polyunsaturated hydrocarbon product (43). Such cases, if more widespread, may have also escaped detection because most of the predicted gene clusters with β-lactone synthetases were not detected by tools like antiSMASH until recently (23), prohibiting the discovery of such biosynthetic pathways through genome mining approaches. The recent feature implementation in antiSMASH to detect likely β-lactone moieties now enables further research into the role of β-lactones as intermediates in the biosynthesis of hydrocarbons and other natural products (23).
Among the predicted gene clusters containing β-lactone synthetase homologs, we detected a conserved four-gene cluster in Nocardia that we linked to the orphan β-lactone natural product, nocardiolactone. Numerous biosynthetic gene clusters have been detected in Nocardia spp., and Nocardia genomes were shown to have as many type I polyketide synthases and NRPS gene clusters as Streptomyces genomes (44). Despite this, only a small number of biosynthetic gene clusters in Nocardia have been experimentally verified. Recently, the biosynthetic gene cluster for the nargenicin family of macrolide antibiotics was discovered in human pathogenic strains of Nocardia arthriditis (45). Khosla and co-workers (46) also reported on a unique class of orphan polyketide synthases in the genomes of Nocardia isolates from human patients with nocardiosis. Based on the conservation of this gene cluster in clinical isolates from nocardiosis patients, Khosla proposed the product might play a role in Nocardia pathogenicity in human hosts. Similarly, we found that the nocardiolactone gene cluster was significantly more abundant in the genomes of human pathogenic strains compared with isolates from nonhuman sources. The enrichment supports the hypothesis that nocardiolactone could play a role in the pathogenicity of Nocardia; however, in vivo studies are required.
Nocardiolactone was first isolated from the mycelia of Nocardia spp. rather than the fermentation broth, suggesting the compound is cell-associated (29). The long, waxy di-alkyl tails of nocardiolactone resemble mycolic acids and would likely embed in the cell membrane. The cell wall composition varies between different species of Nocardia but is known to consist primarily of trehalose dimycolate and several other unidentified hydrophobic compounds (47). Studies on Nocardia virulence found that cell-surface composition was a critical determinant for attachment and penetration of host cells (48). Cell wall–associated lipids in N. brasiliensis were also shown to induce a strong inflammatory response (47). The role of hydrophobic natural products in Nocardia pathogenicity and infection remains a rich and untapped direction for future research.
In summary, we conducted the first global analysis of adenylate-forming enzymes in >50,000 candidate biosynthetic gene clusters from all domains of life. Our machine learning approach yielded evolutionary insights into ANL superfamily divergence from a core scaffold similar to contemporary CoA-ligases toward enzymes with more specialized functions such as β-lactone formation. AdenylPred analysis also detected >90 β-lactone synthetases in gene clusters in Nocardia spp. that were enriched in the genomes of human pathogens. Through in vitro pathway reconstitution, we were able to link this gene cluster family to the orphan natural product nocardiolactone. These findings demonstrate how machine learning methods can be used to pair gene clusters with orphan secondary metabolites and advance understanding of natural product biosynthesis.
Experimental procedures
Sequence similarity network
Candidate biosynthetic gene clusters from >24,000 bacterial genomes in the antiSMASH database version 2 (49) were combined with precalculated plantiSMASH output (25) and results from fungiSMASH analysis of 1,100 fungal genomes (accession numbers available at https://github.com/serina-robinson/adenylpred_analysis/). The AMP-binding pHMM (PF00501) was used to query all genes in the data set with default parameters, returning 213,993 significant hits. Because the distribution and identity of NRPS adenylation domains have already been analyzed in detail by Chevrette et al. (12), we opted to analyze only standalone AMP-binding pHMM hits. All sequences with a condensation domain (PF00668) in the same coding sequence were filtered out, leaving 71,331 “standalone” AMP-binding HMM hits. Of these, partial sequences (less than 150 amino acids in length) were removed for a total of 63,395 sequences. Because of computational limitations of visualizing large networks, the sequences were clustered using CD-HIT (50) with a word size of 2 and 40% sequence similarity cutoff to yield 2,344 cluster representatives. Cluster representatives were combined with the AdenylPred training set sequences to observe their relation to sequences with known specificity. A sequence similarity network was constructed by calculating pairwise BLAST (51) similarities between all sequences. The network was examined over a comprehensive range of e-value cutoffs and visualized using the igraph package (52).
Training set construction
The AdenylPred training set was pulled and manually curated from three databases: UniProtKB, MIBiG, and the most up-to-date NRPS A domain training set from SANDPUMA (12). AMP-binding enzymes (PF00501) in the UniProtKB database that had experimental evidence at the protein level were extracted and linked to their substrate through literature mining and manual verification. The MIBiG database was queried with the AMP-binding HMM (PF00501). MIBiG sequences were extracted and linked to their product and substrate when reported in the literature. The SANDPUMA NRPS A domain data set (12) was randomly down-sampled with stratification by substrate class to balance the training set classes for substrate and enzyme function prediction. A dictionary of databases and other bioinformatics resources used in analysis here are defined in Table S4. Training set sequences were grouped into 15 substrate groups and 9 enzyme functional classes.
Machine learning methods
Protein sequences in the training set were aligned with the AMP-binding HMM using HMMAlign (53). Residues within 8 Å of the active site were extracted and encoded as tuples of physicochemical properties as described by Röttig et al. (11). The FSI was extracted, one-hot encoded, and concatenated to the vector of active site vector properties for a total of 585 sequence features. Three different machine learning algorithms were trained for multiclass classification: random forest, naïve Bayes, and feedforward neural networks. Descriptions of these algorithms and other machine learning terms commonly used throughout the manuscript are provided in Table S5. The data were split with stratified sampling into 75% training and 25% test sets. Tuning parameters for all models were adjusted by grid search using 10-fold cross-validation repeated with five iterations. The confidence of random forest predictions can be assessed using a nonparametric probability estimation for class membership calculated as a value between 0 and 1 (54). Based on the distribution of prediction probabilities, we set an empirical threshold for prediction confidence of 0.6 (60%), below which all substrates are listed as “no confident result,” although the best prediction is still provided to the user.
AdenylPred availability
The web application is available at z.umn.edu/adenylpred (shortened URL) or https://srobinson.shinyapps.io/AdenylPred/. The command-line version of the tool is available at https://github.com/serina-robinson/adenylpred.
Phylogenetic analysis and ancestral reconstruction
Training set sequences were aligned were aligned using HMMAlign (53) and terminal ends of the alignment were trimmed. The phylogeny of the entire training set was estimated using RAxML version 8.2.9 (55) with the Jones–Taylor–Thornton matrix–based model of amino acid substitution and a discrete gamma model with 20 rate categories. For ancestral sequence reconstruction, training set sequences were redundancy filtered to 40% amino acid identity with a word size of 2 with CD-HIT (50). FastML was used to reconstruct the most likely ancestral sequences for internal nodes of the tree (22). AdenylPred was then used to predict the functional class and substrate of the root ancestral sequence in the ANL superfamily.
HPLC analysis of NltC activity
HPLC analysis of ATP, ADP, and AMP was conducted using an Agilent 1100 series instrument with a C18 eclipse plus (Agilent) column mounted with a C18 guard column. The reactions were carried out in glass HPLC vials with a total volume of 500 μl in assay buffer (50 mm Tris base adjusted to pH 8.0 with HCl). The reactions were initiated with the addition of 0.5 μm enzyme (15 µg) to 100 μm of ATP, 100 μm of substrate, and 2% EtOH originating from the substrate stocks. Separation of ATP, ADP, and AMP was observed after 9 min under isocratic conditions with 95% 100 mm H2KPO4 (pH 6.0 with KOH) and 5% methanol while monitoring at 259 nm.
Cloning, expression, and purification of nocardiolactone biosynthetic enzymes
Genes encoding NltA (WP_042260942.1), NltB (WP_042260944.1), NltC (WP_042260945.1), and NltD (WP_042260949.1) from N. brasiliensis, and NltA from N. yamanashiensis (WP_067710538.1) were codon-optimized for Escherichia coli and synthesized by Integrated DNA Technologies. The synthetic gene encoding NltC was cloned into a pET30b+ vector with NdeI and HindIII restriction sites with a C-terminal His6 tag. The gene encoding NltD was cloned into a modified pMAL-c5x vector with a tobacco etch virus protease cut site added at NdeI and HindIII restriction sites and a N-terminal His6 tag and expressed as a fusion with maltose-binding protein. The gene encoding NltA from N. yamanashiensis was cloned into a pET28b+ at NdeI and XhoI restriction sites. The genes encoding NltA and NltB from N. brasiliensis were cloned individually as in the case of N. yamanashiensis and combined with a ribosome-binding site-like sequence (tttgtttaactttaagaaggaga) inserted into a single pET28b+ vector with a N-terminal His6 tag. Accession numbers and codon-optimized plasmid sequences are available in the supporting information. Constructs were cloned by Gibson assembly into DH5α cells and verified by Sanger sequencing. Sequence-verified plasmids were transformed into BL21 (DE3) cells (NEB). Starter cultures (5 ml) were grown in Terrific Broth overnight at 37 °C with kanamycin selection. Cultures of 1 liter with 75 µg/ml kanamycin were grown to an optical density of 0.5 at 37 °C, induced by the addition of 1 ml of isopropyl β-d-1-thiogalactopyranoside (1 m stock) and further incubated for 19 h at 15 °C. Induced cells were harvested at 4000 × g with a Beckman centrifuge and frozen at −80 °C. The cell pellets were thawed on ice and resuspended in 10 ml of buffer containing 500 mm NaCl, 20 mm Tris base, and 10% glycerol at pH 7.4. Elution buffer for the nickel column was the same but with the addition of 400 mm imidazole. NltD purification buffer required the addition of 0.025% Tween 20. Cell pellets were lysed with two or three cycles in a French pressure cell (1500 p.s.i.) and centrifuged for 60 min at 17,000 × g. Supernatants were filtered through a 0.45-μm low protein binding filter (Corning), loaded into a GE Life Sciences ÄKTA fast liquid protein chromatography system, and injected onto a GE Life Sciences HisTrap HP 5-ml column. After washes to remove nonspecifically bound proteins, His-tagged proteins were eluted with a stepwise gradient of 5, 10, 15, and 80% elution buffer over 2.5 column volumes at 1 ml/min and collected in 2-ml fractions. Protein concentration was determined by the method of Bradford (56) using the Bio-Rad protein assay dye reagent concentrate and a standard curve prepared from a 2 mg/ml bovine albumin standard (Thermo Scientific). The desired protein fractions were pooled, analyzed by SDS-PAGE, flash-frozen in liquid N2, and stored at −80 °C.
GC-MS
Separation and identification of metabolites was accomplished by GC-MS (Agilent 7890a and 5975c) equipped with a 30-m × 0.25-mm inner diameter × 0.25-μm DB-1ms capillary column with outflow split to flame ionization and MS detectors. The formation of the unstable β-keto acid catalyzed by OleA and NltAB could be observed as its ketone breakdown product by GC-MS as published previously (33). Olefins from the complete thermal decarboxylation of β-lactone products were detected without derivatization by comparison to synthetic β-lactone standards described previously (8, 30). The β-hydroxy acids required methylation of the carboxylic acid group by diazomethane for detection by GC-MS. Ethereal alcoholic solutions of diazomethane were prepared from N-methyl-N-nitroso-p-toluenesulfonamide (Sigma–Aldrich). All samples were extracted with tert-methyl butyl ether and mixed with 50 μl of diazomethane solution. One microliter of each sample was injected into the injection port (230 °C). The 25-min program was as follows: hold 80 °C for 2 min; ramp linearly to 320 °C for 20 min; and hold 320 °C for 3 min.
Verification of NltC β-lactone synthetase activity by 1H NMR
The reaction mixtures were set up in separate funnels with 1 mg of NltC, 20 mg of ATP, and 30 mg of MgCl2·6H2O in 100 ml of 200 mm NaCl and 20 mm NaPO4 buffer (pH 7.4). The reactions were initiated with the addition of 1.5 ml of 2-octyl-3-hydroxydodecanoic acids dissolved in EtOH (1.0 mg/ml stock). As an internal standard, 10 μl of 1-bromo-naphthalene (0.5 mg/ml stock) was added to each reaction mixture. The reactions were allowed to run for 24 h before three successive extractions were performed with 10, 5, and 5 ml of dichloromethane. The samples were evaporated at room temperature before solvation in CDCl3 for 1H NMR (400 MHz). Fig. S6A shows the 1H NMR spectra of 2-octyl-3-hydroxydodecanoic acids allowed to react overnight with NltC compared with a no-enzyme control. Chemical shifts for synthetic 2-octyl-3-hydroxydodecanoic acid starting materials and cis- and trans-3-octyl-4-nonyloxetan-2-one products were reported by Christenson et al. (8).
NltD activity assay
The activity of NltD/OleD by NADPH-dependent consumption of 2-alkyl-3-ketoalkanoic acid was monitored using the method described by Bonnett et al. (33). The reaction was measured in reverse because the β-keto acid substrate for NltD/OleD homologs was previously shown to be unstable and undergoes rapid decarboxylation to a ketone (33). Briefly, progress was tracked by the change in absorbance at 340 nm by the formation or consumption of NADPH (ε340 = 6,220 m−1 cm−1) in UV-transparent 96-well plates (Greiner, Sigma–Aldrich) measured using a SpectraMax Plus microplate reader (Molecular Devices).
General synthetic procedures
Synthesis and purification of racemic 2-octyl-3-hydroxydodecanoic acid was performed as described previously (8, 57) via α-carbon deprotonation of decanoic acid by lithium diisopropylamide followed by the addition of decanal to form 2-octyl-3-hydroxydodecanoate. An identical synthetic method was used to synthesize a racemic diastereomeric mix of 2-hexyl-3-hydroxyoctanoic acid from hexanal and octanoic acid as described by Robinson et al. (30).
Data availability
Scripts and raw data for analyses and figures presented in this manuscript are available on GitHub: https://github.com/serina-robinson/adenylpred_analysis. A searchable database for the training set is also available at z.umn.edu/adenylpred.
Supplementary Material
Acknowledgments
We acknowledge Satria Kautsar for assistance in extracting candidate biosynthetic gene clusters. Aalt-Jan van Dijk is recognized for insightful discussions on machine learning. Kelly Aukema and Mike Freeman are acknowledged for manuscript edits and discussion. We are grateful to Jorge Navarro-Muñoz for providing the output from fungiSMASH analysis.
This article contains supporting information.
Author contributions—S. L. R., M. H. M., and L. P. W. conceptualization; S. L. R., S. J. P., T. P. S., and M. H. M. resources; S. L. R` B. R. T. data curation; S. L. R. and B. R. T. software; S. L. R. formal analysis; S. L. R., M. H. M., and L. P. W. funding acquisition; S. L. R., B. R. T., and S. J. P. validation; S. L. R., B. R. T., M. D. S., and S. J. P. investigation; S. L. R. visualization; S. L. R., B. R. T., and M. H. M. methodology; S. L. R. writing-original draft; S. L. R., T. P. S., M. H. M., and L. P. W. project administration; S. L. R., B. R. T., M. D. S., S. J. P., T. P. S., M. H. M., and L. P. W. writing-review and editing; M. H. M. and L. P. W. supervision.
Funding and additional information—S.L.R. is supported by the National Science Foundation Graduate Research Fellowship under NSF grant number 00039202 and a Graduate Research Opportunities Worldwide (GROW) fellowship to the Netherlands supported by the NSF and the Netherlands Organization for Scientific Research (NWO) grant number 040.15.054/6097 (to S. L. R. and M. H. M.).
Conflict of interest—M. H. M. is a co-founder of Design Pharmaceuticals and on the scientific advisory board of Hexagon Bio.
S. J. Pidot and T. P. Stinear, manuscript in preparation.
- NRPS
- nonribosomal peptide synthetase
- A domain
- adenylation domain
- SACS
- short-chain acyl-CoA synthetase
- MACS
- medium-chain acyl-CoA synthetase
- LACS
- long-chain acyl-CoA synthetase
- FAAL
- fatty acyl-AMP ligase
- LUC
- luciferase
- BLS
- β-lactone synthetase
- ARYL
- aryl-CoA ligase
- pHMM
- profile Hidden Markov Model
- FSI
- fatty acyl-AMP ligase-specific insertion
- AUROC
- area under the receiver operating characteristic curve
- aa
- amino acid(s).
References
- 1. D'Ambrosio H. K., and Derbyshire E. R. (2020) Investigating the role of class I adenylate-forming enzymes in natural product biosynthesis. ACS Chem. Biol. 15, 17–27 10.1021/acschembio.9b00865 [DOI] [PubMed] [Google Scholar]
- 2. Lipmann F. (1944) Enzymatic synthesis of acetyl phosphate. J. Biol. Chem. 155, 55–70 [Google Scholar]
- 3. Gulick A. M. (2009) Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827 10.1021/cb900156h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang N., Rudolf J. D., Dong L. B., Osipiuk J., Hatzos-Skintges C., Endres M., Chang C. Y., Babnigg G., Joachimiak A., Phillips G. N., and Shen B. (2018) Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme. Nat. Chem. Biol. 14, 730–737 10.1038/s41589-018-0061-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bera A. K., Atanasova V., Gamage S., Robinson H., and Parsons J. F. (2010) Structure of the d-alanylgriseoluteic acid biosynthetic protein EhpF, an atypical member of the ANL superfamily of adenylating enzymes. Acta Crystallogr. D Biol. Crystallogr. 66, 664–672 10.1107/S0907444910008425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arora P., Goyal A., Natarajan V. T., Rajakumara E., Verma P., Gupta R., Yousuf M., Trivedi O. A., Mohanty D., Tyagi A., Sankaranarayanan R., and Gokhale R. S. (2009) Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis. Nat. Chem. Biol. 5, 166–173 10.1038/nchembio.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cimermancic P., Medema M. H., Claesen J., Kurita K., Brown L. C., Mavrommatis K., Pati A., Godfrey P. A., Koehrsen M., Clardy J., Birren B. W., Takano E., Sali A., Linington R. G., and Fischbach M. A. (2014) Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 10.1016/j.cell.2014.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christenson J. K., Richman J. E., Jensen M. R., Neufeld J. Y., Wilmot C. M., and Wackett L. P. (2017) β-Lactone synthetase found in the olefin biosynthesis pathway. Biochemistry 56, 348–351 10.1021/acs.biochem.6b01199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Waldman A. J., and Balskus E. P. (2018) Discovery of a diazo-forming enzyme in cremeomycin biosynthesis. J. Org. Chem. 83, 7539–7546 10.1021/acs.joc.8b00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Khurana P., Gokhale R. S., and Mohanty D. (2010) Genome scale prediction of substrate specificity for acyl adenylate superfamily of enzymes based on active site residue profiles. BMC Bioinformatics 11, 57 10.1186/1471-2105-11-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Röttig M., Medema M. H., Blin K., Weber T., Rausch C., and Kohlbacher O. (2011) NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362–W367 10.1093/nar/gkr323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chevrette M. G., Aicheler F., Kohlbacher O., Currie C. R., and Medema M. H. (2017) SANDPUMA: ensemble predictions of nonribosomal peptide chemistry reveal biosynthetic diversity across Actinobacteria. Bioinformatics 33, 3202–3210 10.1093/bioinformatics/btx400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pan G., Xu Z., Guo Z., Ma M., Yang D., Zhou H., Gansemans Y., Zhu X., Huang Y., Zhao L. X., and Jiang Y. (2017) Discovery of the leinamycin family of natural products by mining actinobacterial genomes. Proc. Natl. Acad. Sci. U.S.A. 114, E11131–E11140 10.1073/pnas.1716245115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao H., Liu Y. P., and Zhang L. Q. (2019) In silico and genetic analyses of cyclic lipopeptide synthetic gene clusters in Pseudomonas sp. 11K1. Front. Microbiol. 10, 544 10.3389/fmicb.2019.00544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kautsar S. A., Blin K., Shaw S., Navarro-Muñoz J. C., Terlouw B. R., van der Hooft J. J. J., van Santen J. A., Tracanna V., Suarez Duran H. G., Pascal Andreu V., Selem-Mojica N., Alanjary M., Robinson S. L., Lund G., Epstein S. C., et al. (2020) MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 48, D454–D458 10.1093/nar/gkz882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. UniProt Consortium, (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujino T., Kang M. J., Suzuki H., Iijima H., and Yamamoto T. (1996) Molecular characterization and expression of rat acyl-CoA synthetase 3. J. Biol. Chem. 271, 16748–16752 10.1074/jbc.271.28.16748 [DOI] [PubMed] [Google Scholar]
- 18. Bauman K. D., Li J., Murata K., Mantovani S. M., Dahesh S., Nizet V., Luhavaya H., and Moore B. S. (2019) Refactoring the cryptic streptophenazine biosynthetic gene cluster unites phenazine, polyketide, and nonribosomal peptide biochemistry. Cell Chem. Biol. 26, 724–736 10.1016/j.chembiol.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petchey M., Cuetos A., Rowlinson B., Dannevald S., Frese A., Sutton P. W., Lovelock S., Lloyd R. C., Fairlamb I. J. S., and Grogan G. (2018) The broad aryl acid specificity of the amide bond synthetase McbA suggests potential for the biocatalytic synthesis of amides. Angew. Chem. Int. Ed. 57, 11584–11588 10.1002/anie.201804592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du Y. L., Alkhalaf L. M., and Ryan K. S. (2015) In vitro reconstitution of indolmycin biosynthesis reveals the molecular basis of oxazolinone assembly. Proc. Natl. Acad. Sci. U.S.A. 112, 2717–2722 10.1073/pnas.1419964112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akiva E., Copp J. N., Tokuriki N., and Babbitt P. C. (2017) Evolutionary and molecular foundations of multiple contemporary functions of the nitroreductase superfamily. Proc. Natl. Acad. Sci. U.S.A. 114, E9549–E9558 10.1073/pnas.1706849114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ashkenazy H., Penn O., Doron-Faigenboim A., Cohen O., Cannarozzi G., Zomer O., and Pupko T. (2012) FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 40, W580–W584 10.1093/nar/gks498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., Medema M. H., and Weber T. (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 10.1093/nar/gkz310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blin K., Wolf T., Chevrette M. G., Lu X., Schwalen C. J., Kautsar S. A., Suarez Duran H. G., de Los Santos E. L. C., Kim H. U., Nave M., Dickschat J. S., Mitchell D. A., Shelest E., Breitling R., Takano E., et al. (2017) antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36–W41 10.1093/nar/gkx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kautsar S. A., Suarez Duran H. G., Blin K., Osbourn A., and Medema M. H. (2017) plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 45, W55–W63 10.1093/nar/gkx305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galica T., Hrouzek P., and Mareš J. (2017) Genome mining reveals high incidence of putative lipopeptide biosynthesis NRPS/PKS clusters containing fatty acyl-AMP ligase genes in biofilm-forming cyanobacteria. J. Phycol. 53, 985–998 10.1111/jpy.12555 [DOI] [PubMed] [Google Scholar]
- 27. Bai T., Zhang D., Lin S., Long Q., Wang Y., Ou H., Kang Q., Deng Z., Liu W., and Tao M. (2014) Operon for biosynthesis of lipstatin, the β-lactone inhibitor of human pancreatic lipase. Appl. Environ. Microbiol. 80, 7473–7483 10.1128/AEM.01765-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wyatt M. A., Ahilan Y., Argyropoulos P., Boddy C. N., Magarvey N. A., and Harrison P. H. (2013) Biosynthesis of ebelactone A: isotopic tracer, advanced precursor and genetic studies reveal a thioesterase-independent cyclization to give a polyketide β-lactone. J. Antibiotics 66, 421–430 10.1038/ja.2013.48 [DOI] [PubMed] [Google Scholar]
- 29. Mikami Y., Yazawa Y., Tanaka Y., Ritzau M., and Gräfeb U. (1999) Isolation and structure of nocardiolactone, a new dialkyl-substituted β-lactone from pathogenic Nocardia strains. Nat. Prod. Lett. 13, 277–284 10.1080/10575639908048798 [DOI] [Google Scholar]
- 30. Robinson S. L., Christenson J. K., Richman J. E., Jenkins D. J., Neres J., Fonseca D. R., Aldrich C. C., and Wackett L. P. (2019) Mechanism of a standalone β-lactone synthetase: new continuous assay for a widespread ANL superfamily enzyme. ChemBioChem. 20, 1701–1711 10.1002/cbic.201800821 [DOI] [PubMed] [Google Scholar]
- 31. Zhang D., Zhang F., and Liu W. (2019) A KAS-III heterodimer in lipstatin biosynthesis nondecarboxylatively condenses C8 and C14 fatty acyl-CoA substrates by a variable mechanism during the establishment of a C22 aliphatic skeleton. J. Am. Chem. Soc. 141, 3993–4001 10.1021/jacs.8b12843 [DOI] [PubMed] [Google Scholar]
- 32. Goblirsch B. R., Jensen M. R., Mohamed F. A., Wackett L. P., and Wilmot C. M. (2016) Substrate trapping in crystals of the thiolase OleA identifies three channels that enable long chain olefin biosynthesis. J. Biol. Chem. 291, 26698–26706 10.1074/jbc.M116.760892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bonnett S. A., Papireddy K., Higgins S., del Cardayre S., and Reynolds K. A. (2011) Functional characterization of an NADPH dependent 2-alkyl-3-ketoalkanoic acid reductase involved in olefin biosynthesis in Stenotrophomonas maltophilia. Biochemistry 50, 9633–9640 10.1021/bi201096w [DOI] [PubMed] [Google Scholar]
- 34. Frias J. A., Richman J. E., Erickson J. S., and Wackett L. P. (2011) Purification and characterization of OleA from Xanthomonas campestris and demonstration of a non-decarboxylative Claisen condensation reaction. J. Biol. Chem. 286, 10930–10938 10.1074/jbc.M110.216127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wattam A. R., Abraham D., Dalay O., Disz T. L., Driscoll T., Gabbard J. L., Gillespie J. J., Gough R., Hix D., Kenyon R., Machi D., Mao C., Nordberg E. K., Olson R., Overbeek R., et al. (2014) PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42, D581–D591 10.1093/nar/gkt1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baier F., and Tokuriki N. (2014) Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 426, 2442–2456 10.1016/j.jmb.2014.04.013 [DOI] [PubMed] [Google Scholar]
- 37. Hicks M. A., Barber A. E., Giddings L. A., Caldwell J., O'Connor S. E., and Babbitt P. C. (2011) The evolution of function in strictosidine synthase-like proteins. Proteins 79, 3082–3098 10.1002/prot.23135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Linne U., Schäfer A., Stubbs M. T., and Marahiel M. A. (2007) Aminoacyl-coenzyme A synthesis catalyzed by adenylation domains. FEBS Lett. 581, 905–910 10.1016/j.febslet.2007.01.066 [DOI] [PubMed] [Google Scholar]
- 39. Oba Y., Ojika M., and Inouye S. (2003) Firefly luciferase is a bifunctional enzyme: ATP-dependent monooxygenase and a long chain fatty acyl-CoA synthetase. FEBS Lett. 540, 251–254 10.1016/S0014-5793(03)00272-2 [DOI] [PubMed] [Google Scholar]
- 40. Oba Y., Iida K., and Inouye S. (2009) Functional conversion of fatty acyl-CoA synthetase to firefly luciferase by site-directed mutagenesis: a key substitution responsible for luminescence activity. FEBS Lett. 583, 2004–2008 10.1016/j.febslet.2009.05.018 [DOI] [PubMed] [Google Scholar]
- 41. Robinson S. L., Christenson J. K., and Wackett L. P. (2019) Biosynthesis and chemical diversity of β-lactone natural products. Nat. Prod. Rep. 36, 458–475 10.1039/C8NP00052B [DOI] [PubMed] [Google Scholar]
- 42. Wang Y., Tennyson R. L., and Romo D. (2004) β-Lactones: intermediates for natural product total synthesis and new transformations. Heterocycles 64, 605–658 10.3987/REV-04-SR(P)3 [DOI] [Google Scholar]
- 43. Allemann M. N., Shulse C. N., and Allen E. E. (2019) Linkage of marine bacteria polyunsaturated fatty acid and long-chain hydrocarbon biosynthesis. Front. Microbiol. 10, 702 10.3389/fmicb.2019.00702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Komaki H., Ichikawa N., Hosoyama A., Takahashi-Nakaguchi A., Matsuzawa T., Suzuki K. I., Fujita N., and Gonoi T. (2014) Genome based analysis of type-I polyketide synthase and nonribosomal peptide synthetase gene clusters in seven strains of five representative Nocardia species. BMC Genomics 15, 323 10.1186/1471-2164-15-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pidot S. J., Herisse M., Sharkey L., Atkin L., Porter J. L., Seemann T., Howden B. P., Rizzacasa M. A., and Stinear T. P. (2019) Biosynthesis and ether-bridge formation in nargenicin macrolides. Angew. Chem. Int. Ed. 58, 3996–4001 10.1002/anie.201900290 [DOI] [PubMed] [Google Scholar]
- 46. Kuo J., Lynch S. R., Liu C. W., Xiao X., and Khosla C. (2016) Partial in vitro reconstitution of an orphan polyketide synthase associated with clinical cases of nocardiosis. ACS Chem. Biol. 11, 2636–2641 10.1021/acschembio.6b00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trevino-Villarreal J. H., Vera-Cabrera L., Valero-Guillén P. L., and Salinas-Carmona M. C. (2012) Nocardia brasiliensis cell wall lipids modulate macrophage and dendritic responses that favor development of experimental actinomycetoma in BALB/c mice. Infect. Immun. 80, 3587–3601 10.1128/IAI.00446-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Beaman B. L. (1996) Differential binding of Nocardia asteroides in the murine lung and brain suggests multiple ligands on the nocardial surface. Infect. Immun. 64, 4859–4862 10.1128/IAI.64.11.4859-4862.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blin K., Pascal Andreu V., de los Santos E. L. C., Del Carratore F., Lee S. Y., Medema M. H., and Weber T. (2019) The antiSMASH database version 2: a comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 47, D625–D630 10.1093/nar/gky1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fu L., Niu B., Zhu Z., Wu S., and Li W. (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Csardi G., and Nepusz T. (2006) The igraph software package for complex network research. InterJournal 1695, 1–9 [Google Scholar]
- 53. Eddy S. R. (2011) Accelerated profile HMM searches. PLoS Comp. Biol. 7, e1002195 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Malley J. D., Kruppa J., Dasgupta A., Malley K. G., and Ziegler A. (2012) Probability machines. Methods Inf. Med. 51, 74–81 10.3414/ME00-01-0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bradford M. M. (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 57. Mulzer J., Brüntrup G., Hartz G., Kühl U., Blaschek U., and Böhrer G. (1981) Additionen von Carbonsäure-Dianionen an α,β-ungesättigte Carbonylverbindungen-Steuerung der 1,2-/1, 4-Regioselektivität durch sterische Substituenteneffekte. Chemische Berichte 114, 3701–3372 10.1002/cber.19811141123 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Scripts and raw data for analyses and figures presented in this manuscript are available on GitHub: https://github.com/serina-robinson/adenylpred_analysis. A searchable database for the training set is also available at z.umn.edu/adenylpred.