Abstract
The E6 protein of both mucosal high-risk human papillomaviruses (HPVs) such as HPV-16, which have been causally associated with malignant tumors, and low-risk HPVs such as HPV-11, which cause the development of benign tumors, interacts with the cellular E3 ubiquitin ligase E6-associated protein (E6AP). This indicates that both HPV types employ E6AP to organize the cellular proteome to viral needs. However, whereas several substrate proteins of the high-risk E6-E6AP complex are known, e.g. the tumor suppressor p53, potential substrates of the low-risk E6-E6AP complex remain largely elusive. Here, we report on an affinity-based enrichment approach that enables the targeted identification of potential substrate proteins of the different E6-E6AP complexes by a combination of E3-selective ubiquitination in whole-cell extracts and high-resolution MS. The basis for the selectivity of this approach is the use of a ubiquitin variant that is efficiently used by the E6-E6AP complexes for ubiquitination but not by E6AP alone. By this approach, we identified ∼190 potential substrate proteins for low-risk HPV-11 E6 and high-risk HPV-16 E6. Moreover, subsequent validation experiments in vitro and within cells with selected substrate proteins demonstrate the potential of our approach. In conclusion, our data represent a reliable repository for potential substrates of the HPV-16 and HPV-11 E6 proteins in complex with E6AP.
Keywords: E3 ubiquitin ligase, E6AP/UBE3A, E6 oncoprotein, human papillomavirus, tumor virus, oncogene, ubiquitin, protein degradation
Because the coding capacity of viral genomes is rather limited, it is essential for viruses to reprogram the host cell metabolism according to viral need. This type of adaption can be achieved by various means, including modulation of host cell gene expression and exploitation of host cell regulatory proteins. Thus, in-depth analysis of viral regulatory proteins and their effect on host cell metabolism provides an attractive opportunity to obtain insight into mechanisms and processes governing cellular proteostasis. A prominent example is provided by papillomaviruses that contain a genome of ∼8 kb harboring only ∼10 genes (1–4). Thus, for viral propagation papillomaviruses largely depend on the host cell machinery.
Human papillomaviruses (HPVs) have been classified into different genera, with α-papillomaviruses comprising HPV types that infect mucosal and cutaneous epithelia (1–4). Based on molecular biological and clinical data, α-papillomaviruses with a tropism for mucosal epithelia can be roughly subdivided into “low-risk” and “high-risk” types. Whereas infection with low-risk HPVs (e.g. HPV-6 and HPV-11) induces the formation of benign lesions with no or only little risk to progress to cancer, women infected with high-risk HPVs (e.g. HPV-16 and HPV-18) have a significantly increased risk to develop malignant lesions, most notably cervical cancer (5, 6). This differential association with clinical lesions is also reflected in different biochemical properties of the major HPV oncoproteins, E6 and E7. For example, the high-risk E6 proteins utilize the cellular E3 ubiquitin ligase E6-associated protein (E6AP) to target the tumor suppressor p53 for ubiquitination and degradation (summarized in (7–9)), whereas low-risk E6 proteins bind to E6AP but do not target p53 (10, 11).
Because of their carcinogenic potential, many studies have focused on the characterization of the high-risk E6 proteins. Accordingly, a still-increasing number of potential interaction partners of high-risk E6 proteins has been reported, though the physiological relevance of many of these interactions remains unclear, with the exception of p53 and some PDZ domain-containing proteins (for reviews, see (12, 13)). Furthermore, most of the proteins reported to bind to high-risk E6 proteins do not appear to interact with low-risk E6s (12, 13). A remarkable exception is E6AP. E6AP is encoded by the UBE3A gene (14, 15) and has been causally implicated in the development of three human disorders. Loss of a functional E6AP protein and amplification of the UBE3A allele (“gain of function”) result in the neurodevelopmental disorders Angelman syndrome and Dup15q syndrome, respectively (16–22). Redirecting of E6AP by high-risk E6 proteins to substrates that in the absence of E6 are not targeted by E6AP, such as p53, is assumed to contribute to HPV-induced cervical carcinogenesis (8, 9). Indeed, the presence of E6AP is essential for HPV-induced cervical carcinogenesis in a transgenic mouse model (23).
High-risk and low-risk E6 proteins share significant similarity at the amino acid sequence level (∼50%) and, thus, are likely to play similar roles during the viral life cycle (3, 24). Because both interact with E6AP (10, 11), it is tempting to speculate that E6AP is a critical interaction partner of low-risk and high-risk E6 proteins and is utilized by these to target an overlapping spectrum of host proteins for ubiquitination. Yet, in particular, the potential targets of the low-risk E6-E6AP ubiquitin ligase complex remain largely elusive (13). In principle, there are at least two main possibilities to identify substrate proteins of ubiquitin ligases at the proteome level (summarized in (25)). Substrates can be identified by manipulating the expression levels of the respective ubiquitin ligase or of a subunit of a ubiquitin ligase complex, in this case E6 proteins, in cells (e.g. (26–28)). Although subsequent affinity-based purification procedures allow the identification of proteins whose ubiquitination status is affected by the presence or absence of the E6 proteins, it remains unclear whether the proteins identified represent direct substrates of the E6 proteins or whether their ubiquitination status is indirectly affected. Alternatively, potential substrate proteins can be identified in vitro (25), for instance by using whole-cell extracts. The advantage of a whole-cell extract system is that the ubiquitin machinery can be manipulated such that ubiquitination mainly depends on the activity of the ubiquitin ligase of interest, thereby minimizing the possibility that the ubiquitination status of a protein is indirectly affected. In any case, results obtained in any system have to be validated in vitro using recombinant proteins and in cell culture experiments.
To identify potential substrate proteins of both low-risk and high-risk E6 proteins, we established an affinity-based enrichment approach using whole-cell extracts and a dedicated biotin-tagged ubiquitin variant that in the absence of the E6 proteins is only poorly used by E6AP for ubiquitination. Enriched proteins were subsequently identified by high-resolution MS (LC–MS/MS) and label-free quantification. We further validated E6-mediated ubiquitination/degradation of some of the proteins identified both in vitro by using recombinant proteins and within cells by transient transfection experiments, demonstrating the potential of this approach.
Results
Establishment of an in vitro ubiquitin variant–based affinity enrichment approach
To enable purification and identification of potential substrates of the E6 proteins with high specificity (for a scheme of the general workflow, see Fig. 1A), we envisioned that a ubiquitin-based affinity approach should fulfill the following criteria. First, ubiquitin needs to be equipped with an affinity tag that does not interfere with E6-E6AP–mediated ubiquitination and allows enrichment of ubiquitinated proteins under harsh conditions to minimize the possibility of (co)purifying nonubiquitinated proteins. Second, E6AP was reported to catalyze mainly the formation of Lys-48–linked ubiquitin chains (poly-ubiquitination) (29, 30). Furthermore, poly-ubiquitination itself can render the identification of the respective protein by LC–MS/MS difficult, because the high number of ubiquitin-derived peptides poses a challenge to detect less-prominent peptides. Thus, a ubiquitin variant that is not or only poorly used by E6AP for poly-ubiquitination should prove helpful in circumventing this drawback. Third, the approach should depend on the presence of E6 proteins to increase the likelihood that the proteins identified are indeed substrates of the E6-E6AP complex rather than E6AP alone. A possibility to achieve this is the use of ubiquitin variants that are efficiently used by the E6-E6AP complex but not by E6AP alone, such as defined hydrophobic patch mutants (31).
Figure 1.
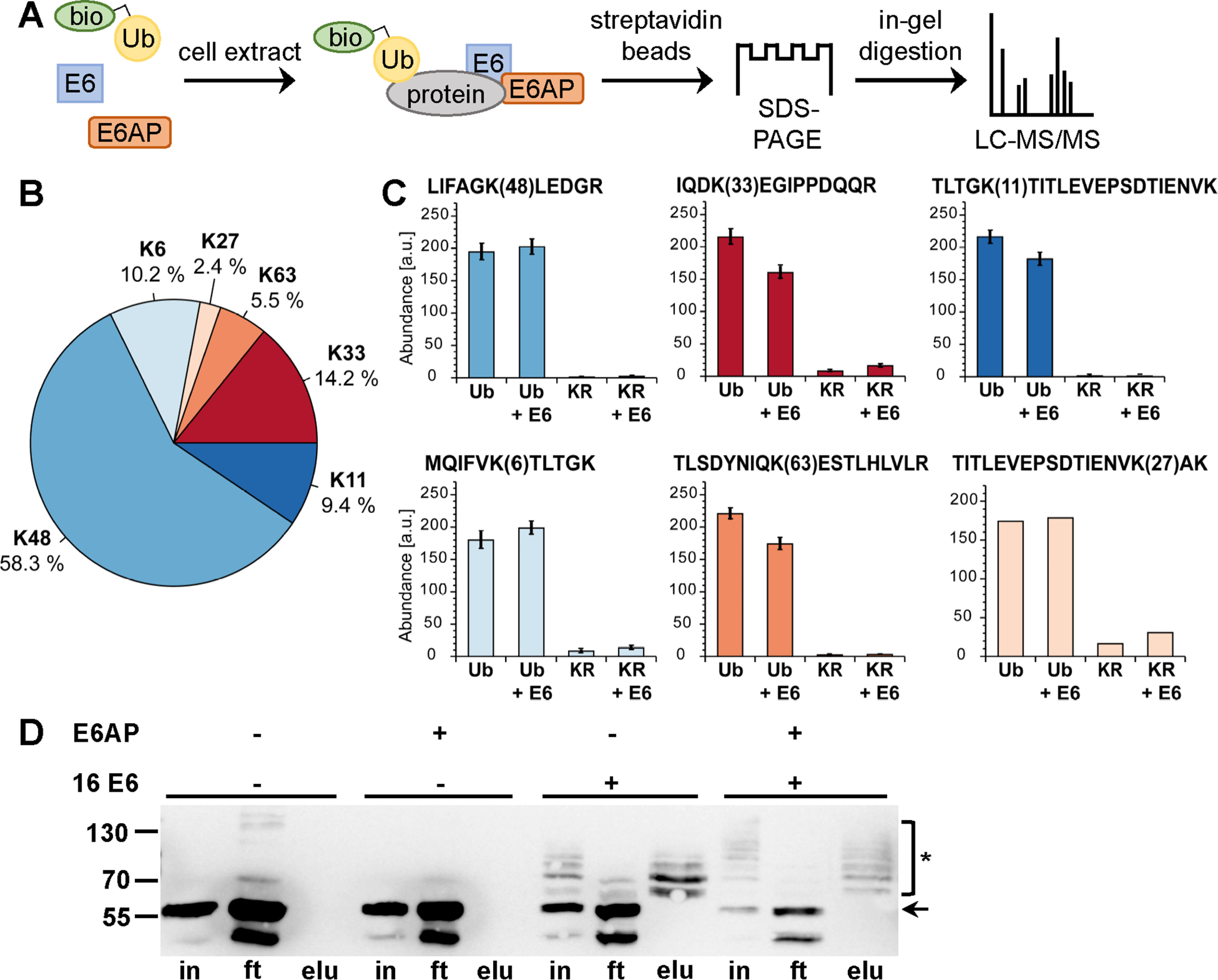
Biotinylated ubiquitin as a tool to identify substrates of the E6-E6AP ubiquitin ligase complex using cell extracts. A, schematic of the procedure. bio-Ub, biotinylated ubiquitin. B and C, determination of the lysine residues used for ubiquitin chain formation by E6AP and the E6-E6AP complex by TMT-based quantification of diGly-modified ubiquitin-derived peptides. B, relative distribution of all identified lysine residues harboring diGly modifications. C, bar charts showing the TMT-based quantification of all peptides identified with diGly modifications. Abundances are shown as grouped abundance over all peptide-spectrum matches for a respective peptide. Standard errors could not be calculated for the peptide modified at Lys-27 by diGly because of the low number of identified peptide-spectrum matches. Ub, WT ubiquitin; KR, Ub-K48/63R. D, HEK293T cell extract was incubated with a biotinylated ubiquitin variant (Ub-K48/63R; for details, see text) in the presence and absence of recombinant E6AP and a GST fusion protein of HPV-16 E6 as indicated. Ubiquitinated proteins were enriched via streptavidin-affinity purification and analyzed by Western blotting analysis using an anti-p53 antibody. Note that HEK293T cells express endogenous E6AP at levels that are sufficient to facilitate E6-mediated ubiquitination of p53; however, the efficiency is improved by addition of recombinant E6AP, as witnessed by the decrease in levels of nonmodified p53. The running positions of molecular mass markers, nonmodified p53 (arrow), and ubiquitinated forms of p53 (asterisk) are indicated. in, cell extract used for affinity purification after ubiquitination reaction; ft, proteins not bound to streptavidin beads; elu, proteins bound to streptavidin beads. Note that in the presence of both E6 and E6AP, p53 is almost quantitatively ubiquitinated, resulting in various ubiquitinated forms of p53 that differ in their migration behavior; i.e. the different p53 forms appear as a ladder. Because Ub-K48/63R is only poorly used by the E6-E6AP complex for poly-ubiquitination, each band of the ladder is likely to represent p53 forms that carry single ubiquitin moieties at a distinct number of lysine residues.
To address the first criterion, we adapted a published procedure that results in site-specific modification of ubiquitin at Lys-6, presumably because of the catalytic microenvironment of the ε-amino group of K6 (32, 33). To biotinylate ubiquitin, we employed sulfo-N-hydroxysuccinimide long-chain biotin (Fig. S1B), and MS analysis of the resulting product revealed that ubiquitin was indeed primarily biotinylated at Lys-6 (Fig. S1, C and D). Because E6AP mainly catalyzes the formation of Lys-48–linked ubiquitin chains (29, 30), we expected that biotinylation of ubiquitin at Lys-6 should not, or only moderately, interfere with E6AP activity. To determine whether this is the case, we performed E6AP auto-ubiquitination assays (34) and ubiquitination assays with whole-cell extracts derived from HEK293T cells (35) that express high levels of WT p53. Indeed, E6AP was efficiently auto-ubiquitinated in the presence of biotinylated ubiquitin (Fig. S2A). Similarly, addition of HPV-16 E6 and E6AP to HEK293T cell extracts resulted in quantitative poly-ubiquitination of p53 under the conditions used. The ubiquitinated forms of p53 were enriched by incubation with streptavidin beads and subjected to Western blotting analysis using a p53-specific antibody (Fig. S2B). However, LC–MS/MS analysis of the eluates did not result in the identification of a significant number of p53-derived peptides (not shown).
To address the second criterion, we resorted to a ubiquitin variant in which lysine residues 48 and 63 are replaced by arginine (Ub-K48/63R), because this variant should only be poorly used by E6AP for poly-ubiquitination. Surprisingly, it turned out that Ub-K48/63R is not only poorly used by E6AP for poly-ubiquitination (Fig. 1B) but for ubiquitination in general (Fig. S3, A–C). Moreover, Ub-K48/63R is efficiently used by the E6-E6AP complex (Fig. 1D and Fig. S3). In a simplified view, E6AP-mediated ubiquitination is a two-step process. In the first step, E6AP forms a thioester complex with ubiquitin, and in the second step, E6AP catalyzes the covalent attachment of ubiquitin to substrate proteins by isopeptide bond formation. Thioester complex formation assays with WT ubiquitin and with Ub-K48/63R clearly showed that E6AP can readily form thioester complexes with either of these and that the efficiency is not affected by E6 (Fig. S3D). This strongly indicates that the E6 proteins rescue the inability of E6AP to transfer Ub-K48/63R to substrate proteins, which is reminiscent of the results we previously obtained with hydrophobic patch mutants of ubiquitin (31). Because E6 also does not affect the lysine residues of ubiquitin used by E6AP for poly-ubiquitination (Fig. 1C), Ub-K48/63R fulfills not only the second but also the third criterion. Thus, Lys-6–biotinylated Ub-K48/63R is ideally suited to identify proteins that are targeted by the E6-E6AP complex, but not by E6AP alone.
Identification of potential substrate proteins of HPV E6 proteins
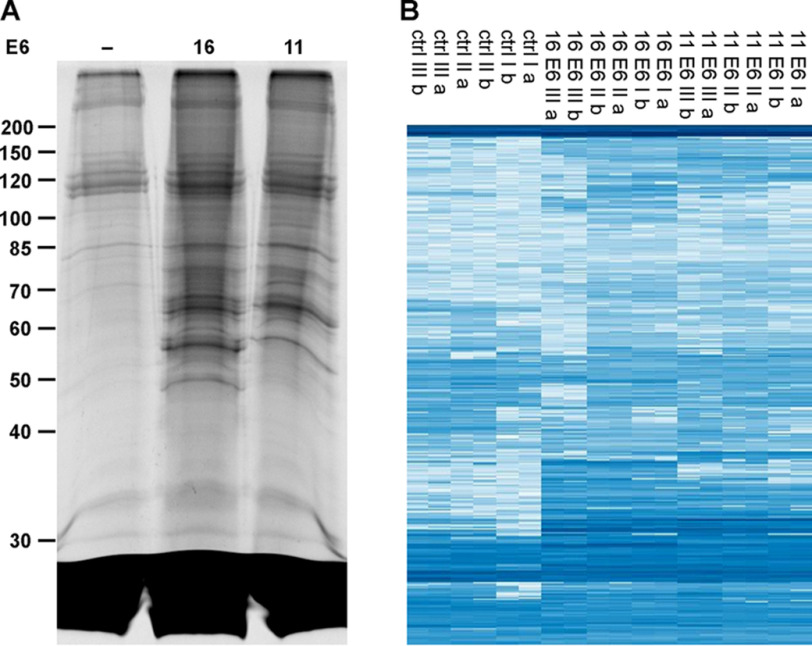
To identify substrate proteins of HPV-16 E6 (high risk) and HPV-11 E6 (low risk), we employed whole-cell extracts derived from HaCaT cells (36). HaCaT cells are spontaneously immortalized keratinocytes, and because keratinocytes constitute the host cells of HPVs, they represent an appropriate means to obtain first insights into the substrate pattern of HPV E6 proteins. HaCaT cell extracts were incubated in the presence of recombinant ubiquitin-like modifier–activating enzyme 1 (UBA1), ubiquitin-conjugating enzyme UbcH7, biotinylated Ub-K48/63R, and E6AP in the absence and presence of GST fusion proteins of HPV-16 E6 or HPV-11 E6. Ubiquitinated proteins were enriched by affinity chromatography using streptavidin beads and upon elution separated by SDS-PAGE (Fig. 2A). After cutting the gel in slices and in-gel tryptic digestion, the samples were analyzed by LC–MS/MS followed by label-free quantification. In total, 1509 proteins were identified in at least two of three biological replicates (HPV-16 E6, HPV-11 E6, and control reaction in the absence of any E6), and the corresponding intensities are shown as a heat map in Fig. 2B. Of these, 199 and 179 proteins were significantly enriched by (false discovery rate (FDR) = 0.001, s0 = 2, n = 3, two-sample t test) and, thus, identified as potential substrate proteins of HPV-16 E6 and HPV-11 E6, respectively, with 109 of these shared by both E6 proteins (Fig. 3C and Supporting data 1; a gene ontology annotation analysis is shown in Fig. S4). The corresponding volcano plots are shown in Fig. 3, A and B.
Figure 2.
Identification of potential substrate proteins of HPV-16 E6 and HPV-11 E6. A, HaCaT cell extract was incubated with recombinant E1, UbcH7, E6AP, and biotinylated Ub-K48/63R in the absence (–) and presence of HPV-16 (16) or HPV-11 (11) E6. Ubiquitinated proteins were enriched by streptavidin-affinity chromatography, eluted under denaturing conditions, separated by SDS-PAGE, and visualized by Coomassie Blue staining. The gel shown is representative of three independent biological replicate experiments. Running positions of molecular mass markers are indicated. B, the individual lanes of the gel were cut into slices. Note that the region of the gel corresponding to proteins with an apparent molecular mass below 30 kDa was not analyzed, because it mainly contains free Ub-K48/63R and streptavidin. Upon tryptic digestion, proteins were identified and quantified by LC–MS/MS. In total, 1509 proteins were identified and are shown with their corresponding intensities in a heat map. Columns show the three biological replicates (I, II, III), with each replicate being measured as a technical duplicate (a, b). Each row corresponds to one protein, with its abundance increasing from light blue to dark blue.
Figure 3.
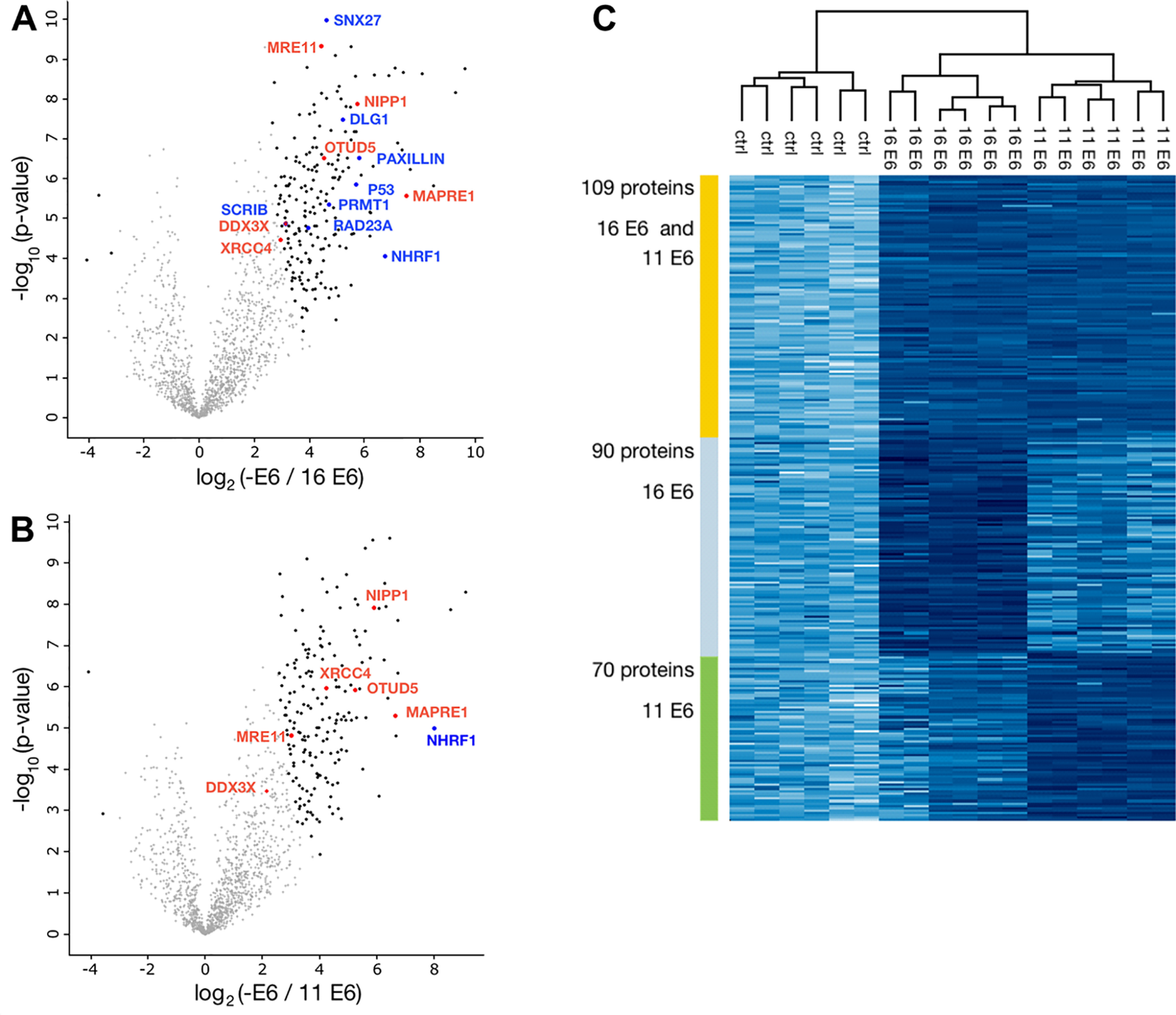
Analysis of potential substrate proteins of HPV-16 E6 and HPV-11 E6. A and B, volcano plots of proteins that were significantly enriched in the presence of the E6 proteins and thus represent potential substrate proteins of HPV-16 E6 and HPV-11 E6, respectively. Significant enrichment was determined relative to proteins identified in the reactions in the absence of E6 proteins (Supporting data 1). Plotted is the log2 fold change versus the negative logarithm of the p-values. Black dots indicate significant enrichment (FDR = 0.001, S0 = 2, n = 3, two-sample t test). Blue dots/protein names indicate significant enrichment of previously reported substrate proteins or interaction partners of the E6 proteins or of E6AP (i.e. RAD23A). Red dots/protein names indicate proteins that were not reported previously to represent substrates of the E6 proteins and were selected for validation. C, heat map of proteins significantly enriched for both HPV-16 and HPV-11 E6 and HPV-16 E6 or HPV-11 E6.
Closer inspection of the results obtained by LC–MS/MS showed that for 67 (HPV-16 E6) and 44 (HPV-11 E6) of the enriched proteins at least one peptide harboring a diGly motif was identified (Supporting data 1), strongly indicating that the lysine residue within the respective peptide was ubiquitinated (37). Furthermore, streptavidin beads were stringently washed upon incubation with the ubiquitination reaction mixtures under conditions (6 m urea and 6 m guanidinium chloride) that in general result in protein denaturation but do not interfere with the biotin-streptavidin interaction (38). Taken together, our data strongly indicate that the vast majority of the enriched proteins identified were indeed ubiquitinated. Notably, several proteins, including p53, zonula occludens-1 protein Zo-1 (DLG1), SCRIBBLE, Paxillin, protein arginine N-methyltransferase 1 (PRMT1), sorting nexin-27 (SNX27), and Na+/H+ exchange regulatory cofactor NHE-RF1 (NHERF1), that were previously reported to interact with or represent substrate proteins of, HPV-16 E6 or for both E6 proteins (39–46) were identified (Fig. 3, A and B and Supporting data 1). In conclusion, the results obtained indicate that our approach enables the identification of substrates of the ubiquitin system in an E6-specific manner.
Validation of substrate proteins
To obtain further evidence for the robustness and “E6-specificity” of the ubiquitin-based affinity enrichment approach, we selected a few proteins for which a cDNA was readily available (XRCC4, MRE11, OTU deubiquitinase 5 (OTUD5), nuclear inhibitor of protein phosphatase 1 (NIPP1), DDX3X, and microtubule-associated protein RP/EB 1 (MAPRE1)) to determine their ability to serve as substrates for the E6-E6AP complex in vitro and in transient transfection experiments in cells. To do so, the respective cDNAs were cloned into an expression vector that allows the expression of the respective proteins both in a combined in vitro transcription/translation system and upon transfection within cells. Furthermore, the expressed proteins harbor an HA-tag at their N terminus, enabling their detection by Western blotting analysis. With the exception of the microtubule-associated protein MAPRE1 (alias EB1; data not shown) (47), the results obtained by the affinity-enrichment approach could be validated in vitro and within cells. The results for XRCC4 and OTUD5 are presented in the following paragraphs; the results obtained for MRE11, a component of DNA damage repair pathways (48); NIPP1, an inhibitor of protein phosphatase PP1 (49); and DDX3X, a member of the DEAD box family of RNA helicases (50), are provided in Fig. S7, Fig. S8, and Fig. S9, respectively.
XRCC4 is an intrinsic component of the nonhomologous end-joining pathway for the repair of DNA double-strand breaks (DSBs) (51). To confirm that XRCC4 represents a potential substrate of the E6 proteins, we first performed in vitro ubiquitination assays. In vitro translated radiolabeled XRCC4 was incubated in the absence or presence of various combinations of E6AP and the E6 proteins under standard ubiquitination conditions (“Experimental procedures”). As shown in Fig. 4A, XRCC4 is efficiently ubiquitinated by HPV-16 E6 and somewhat less efficiently by HPV-11 E6 in the presence of E6AP. Notably, XRCC4 was also ubiquitinated by E6AP alone. However, subsequent time course experiments revealed that E6AP alone is significantly less active in XRCC4 ubiquitination than the E6-E6AP complex (Fig. S5A). We also performed GST pulldown experiments to determine whether a stable complex is formed between the E6 proteins and XRCC4. However, the data obtained suggest that the interaction is rather weak (i.e. of low affinity) and is not, or barely, detectable under the conditions used (Fig. S5B). Finally, the effect of the E6 proteins on XRCC4 levels in cells was determined in transient transfection experiments. For this, we used H1299-K3 cells in which endogenous E6AP expression is stably down-regulated by RNAi to ∼10–15% of the levels in parental H1299 cells (11). This showed that in the presence of HPV-16 E6 or HPV-11 E6, levels of ectopically expressed XRCC4 are decreased (Fig. 4B). Importantly, this decrease was significantly enhanced in the presence of ectopically expressed WT E6AP, whereas in the presence of a catalytically inactive E6AP mutant (E6AP-C820A), levels of XRCC4 were similar to those in the absence of E6 proteins. Although we were not able to detect ubiquitinated forms of XRCC4 (see “Discussion”), the finding that the presence of catalytically active E6AP is required to reduce XRCC4 levels strongly indicates that the E6 proteins target XRCC4 for ubiquitination and subsequent degradation.
Figure 4.
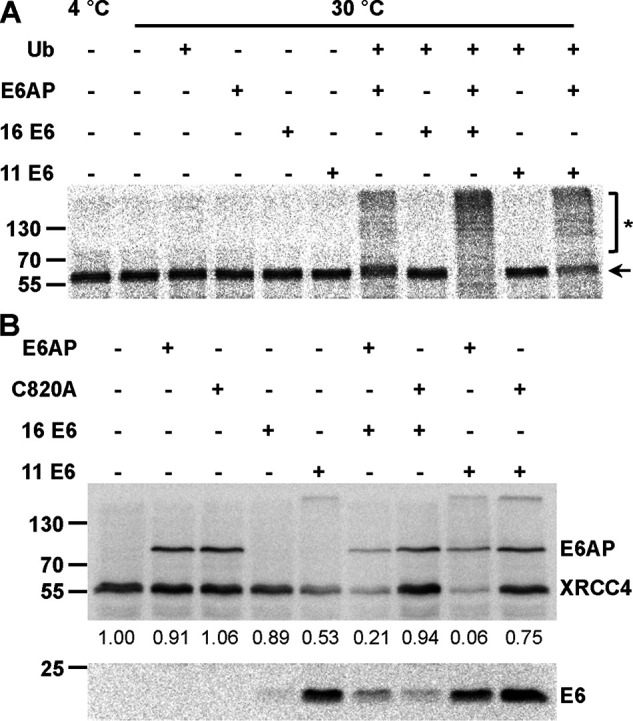
Validation of XRCC4 as a potential substrate protein of the HPV E6 proteins. A, in vitro translated, radiolabeled XRCC4 was incubated with E1 and UbcH7 in the absence and presence of ubiquitin (Ub), baculovirus-expressed E6AP, and GST fusion proteins of HPV-16 E6 or HPV-11 E6 as indicated. Upon 90 min at 30 °C, reactions were stopped and analyzed by SDS-PAGE followed by fluorography. Running positions of the nonmodified form and ubiquitinated forms of XRCC4 are indicated by an arrow and an asterisk, respectively. B, H1299 cells, in which endogenous E6AP expression is stably down-regulated by RNAi, were transfected with expression constructs encoding HA-tagged forms of XRCC4, E6AP, a catalytically inactive E6AP mutant (C820A), HPV-16 E6, and HPV-11 E6 as indicated. 24 h after transfection, protein extracts were prepared and the levels of the various proteins were determined by Western blotting analysis using an anti-HA antibody. Note that prior to SDS-PAGE, the relative transfection efficiency of each transfection was determined (see “Experimental procedures”) to adjust the amounts of the extracts used for analysis. Relative levels of HA-XRCC4 are indicated. The analysis shown is representative of three independent experiments. Running positions of molecular mass markers are indicated. Of note, XRCC4 levels are significantly decreased in the presence of HPV-11 E6 alone, which is likely explained by the notion that H1299-K3 express residual levels of endogenous E6AP. Importantly, XRCC4 levels are further decreased in the presence of ectopically expressed E6AP and increased in the presence of a catalytically inactive mutant (C820A), strongly indicating that HPV-11 E6 targets XRCC4 for degradation in an E6AP-dependent manner.
OTUD5 is a member of the OTU family of deubiquitinating enzymes and has been implicated in the regulation of p53 stability and, like XRCC4, in DSB repair (52, 53). Similar to XRCC4, E6AP alone was able to ubiquitinate OTUD5 in vitro to some extent, and this ubiquitination was stimulated by the E6 proteins (Fig. 5A and Fig. S6). Furthermore, in transient transfection experiments, coexpression of the E6 proteins resulted in a significant decrease in OTUD5 levels in the presence of WT E6AP but not in the presence of E6AP-C820A (Fig. 5B). Unlike XRCC4, an interaction of OTUD5 with the E6 proteins, in particular with HPV-16 E6, was readily observed in GST pulldown experiments, indicating that the E6 proteins have a higher affinity for OTUD5 than for XRCC4. In any case, the data obtained in in vitro ubiquitination experiments and in the cotransfection experiments clearly indicate that at least under the conditions used, both XRCC4 and OTUD5 represent substrate proteins for the HPV E6 proteins.
Figure 5.
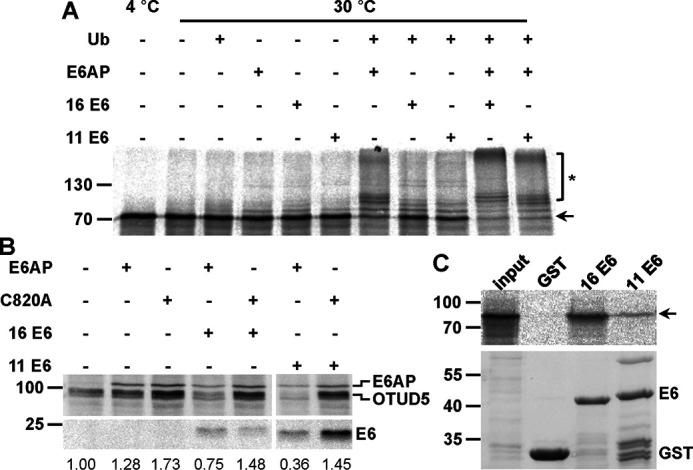
Validation of OTUD5 as a potential substrate protein of the HPV E6 proteins. A, in vitro translated, radiolabeled OTUD5 was incubated with E1 and UbcH7 in the absence and presence of ubiquitin (Ub), baculovirus-expressed E6AP, and GST fusion proteins of HPV-16 or HPV-11 E6 as indicated. Upon 90 min at 30 °C, reactions were stopped and analyzed by SDS-PAGE, followed by fluorography. Running positions of the nonmodified form and ubiquitinated forms of OTUD5 are indicated by an arrow and an asterisk, respectively. B, H1299 cells, in which endogenous E6AP expression is stably down-regulated by RNAi, were transfected with expression constructs encoding HA-tagged forms of OTUD5, E6AP, a catalytically inactive E6AP mutant (C820A), HPV-16, and HPV-11 E6 as indicated. 24 h after transfection, protein extracts were prepared and the levels of the various proteins were determined by Western blotting analysis using an anti-HA antibody. Note that prior to SDS-PAGE, the relative transfection efficiency of each transfection was determined (see “Experimental procedures”) to adjust the amounts of the extracts used for analysis. Relative levels of HA-OTUD5 are indicated. The analysis shown is representative of three independent experiments. Running positions of molecular mass markers are indicated. C, GST fusion proteins of HPV-16 E6 and HPV-11 E6 or GST alone bound to GSH Sepharose beads were mixed with in vitro translated, radiolabeled OTUD5. Upper panel, upon incubation for 90 min at 4 °C, beads were washed and eluates were analyzed by SDS-PAGE, followed by fluorography. input, 10% of OTUD5 used in the binding reactions. Lower panel, amount of GST proteins used in the binding reactions as determined by SDS-PAGE and Coomassie Blue staining.
In conclusion, our validation experiments in vitro and in cell culture confirm both the selectivity and the robustness of our cell extract-based affinity enrichment approach in detecting bona fide substrates of the HPV-11 E6 and HPV E6 proteins in complex with E6AP.
Discussion
Because E3 ubiquitin ligases mediate the recognition of substrate proteins and thereby the specificity of the ubiquitin-conjugation system (54), there is considerable interest to identify the substrate spectrum of ubiquitin ligases, in particular of those involved in human disease. Over the years, a number of approaches for the identification of substrates of ubiquitin ligases have been reported, ranging from in vitro assays with phage display libraries, protein microarrays, or cell extracts to modulation of expression/activity of the ubiquitin ligase of interest within cells with subsequent analysis of changes in protein expression levels in general or in the pattern of ubiquitinated proteins at the proteomic scale (summarized in (25)). Each of these approaches has its advantages but also its pitfalls and limitations (25). Here, we employed a biotin-tagged ubiquitin variant in combination with cell extracts to identify potential substrate proteins of the ubiquitin ligase E6AP in complex with the E6 protein of HPV-16 or HPV-11. Compared with protein microarrays or phage display libraries, the use of cell extracts has the advantages that potential substrate proteins are posttranslationally modified, which may be required for specific recognition by the E3 ligase of interest, and that ubiquitination is performed in the context of the entire proteome, reducing the risk that proteins are ubiquitinated in a nonspecific manner. Because cell extracts can be spiked with high amounts of the ubiquitin ligase of interest, it is likely that the proteins identified as ubiquitination substrates indeed represent direct targets of the ubiquitin ligase and that their ubiquitination status is not indirectly affected, as can happen by modulating the activity of ubiquitin ligases within cells.
As shown in Fig. S3, Ub-K48/63R is not, or only poorly, used for ubiquitination by E6AP alone, whereas it is efficiently used by E6AP in the presence of the HPV E6 proteins. This unexpected observation warrants a note of caution when lysine-deficient ubiquitin mutants are employed to determine the actual lysine residues that are used by a given E3 ubiquitin ligase for ubiquitin chain formation. To come to a sound conclusion, it needs to be shown that the E3 ubiquitin ligase in question can use the respective ubiquitin mutant for ubiquitination at all. Regardless, with respect to the identification of substrates of the E6-E6AP ubiquitin ligase complex rather than substrates of E6AP alone, the use of Ub-K48/63R represents the main advantage of our approach, because it enables the selective identification of substrates of the E6-E6AP complex. Indeed, we identified several proteins that were previously reported to be substrates of the E6-E6AP complex, including p53 and several PDZ domain–containing proteins that are not recognized by E6AP in the absence of E6 (39–46). The selectivity and robustness of our cell extract–based affinity enrichment approach is further demonstrated by the results obtained in validation experiments for previously unknown substrate proteins of the E6 proteins. In five of six cases tested (XRCC4, OTUD5, MRE11, NIPP1, and DDX3X), E6AP-mediated ubiquitination in vitro was either dependent on or significantly stimulated by the presence of the E6 proteins. Moreover, in transient transfection experiments, a decrease in levels of the respective proteins was observed only in the presence of the E6 proteins and catalytically active E6AP. However, efforts to detect ubiquitinated forms of the substrate proteins in cotransfection experiments in cells, for example by coexpression of His-tagged ubiquitin, failed. However, we were also not able to detect ubiquitinated forms of p53 in such assays (55). Although the reason for this remains to be determined, the finding that the decrease was not observed in the presence of a catalytically inactive E6AP mutant (E6AP-C820A) strongly indicates that the E6-E6AP complex targets the respective proteins for ubiquitination and subsequent degradation.
In contrast to previously reported substrates of the HPV-16 E6 protein (e.g. p53 and PDZ domain-containing proteins) (39–46), we were not able to detect an interaction of the E6 protein with in vitro translated XRCC4 and NIPP1 under the conditions of a coprecipitation experiment. This may indicate that the interactions are mediated by additional proteins not present in the rabbit reticulocyte extract used for in vitro translation. This appears to be unlikely, however, because an interaction was also not observed when extracts derived from cells ectopically expressing XRCC4 or NIPP1 were employed (data not shown). Alternatively and more likely, E6 or the E6-E6AP complex binds to these proteins with an affinity too low to be detectable under the conditions of a coprecipitation experiment (note that because XRCC4 and NIPP1 are ubiquitinated in vitro by the E6-E6AP complex, they have to bind to this enzymatic complex). Such low-affinity enzyme-substrate interactions can also be observed by proximity-labeling approaches, which have been employed to identify substrates of E3 ubiquitin ligases in cells (e.g. (27, 56, 57)). In this context, it should be noted that not all proteins identified by our approach are necessarily physiologically relevant substrates of the E6-E6AP complex. For instance, in some cases proteins that interact with but are not ubiquitinated by the E6-E6AP complex within cells may also be ubiquitinated in vitro because of the conditions used.
Because the E6 proteins of both low-risk and high-risk HPVs interact with E6AP and because little is known about potential substrates of the low-risk E6-E6AP complex, we originally set out to identify proteins that serve as potential substrates for HPV-16 E6 and/or HPV-11 E6. We speculate that proteins such as XRCC4, MRE11, OTUD5, and NIPP1 that are recognized by both E6 proteins are important for common aspects of the viral life cycles, whereas those that are recognized by either HPV-16 E6 or HPV-11 E6 have type-specific functions. These functions are likely related to peculiarities of the respective life cycle, which in the case of HPV-16 E6 may also be important for HPV-induced carcinogenesis. p53 represents a prime example for the latter (5, 8, 9, 23); however, its role for the life cycle of high-risk HPVs and the reason that it is not targeted by low-risk HPVs remain enigmatic. Intriguingly, OTUD5 has been reported to be involved in p53 stability regulation (52). Thus, one could envision that HPV-11 E6 indirectly affects p53 stability and function by targeting OTUD5. Unfortunately, it will be difficult to prove this possibility because HPVs go through a peculiar life cycle. In a simplified view, initial infection occurs in primary keratinocytes of a stratified epithelium, whereas viral replication and production take place in the differentiated layers (3, 4). In consequence, it is still difficult to propagate HPVs under cell culture conditions and, moreover, we do not know at which stage of the viral life cycle interference with p53 function may be of importance. Similarly, we identified XRCC4 and MRE11 as potential substrate proteins of HPV-11 E6 and HPV-16 E6. Both XRCC4 and MRE11 are involved in DSB repair, though in different pathways (48, 51). Whereas degradation of these proteins may contribute to the mutagenic, and thus oncogenic, potential of HPV-16 E6, the potential significance of these interactions for the viral life cycle is more difficult to explain because viral replication appears to depend on functional DSB repair pathways (58, 59). Again, however, E6-mediated degradation of XRCC4 and/or MRE11 may, for instance, play a role in the early stages of viral infection but not in later ones. In conclusion, our data represent a reliable repository for potential substrates of the HPV-16 and HPV-11 E6 proteins, but additional studies will be required to eventually obtain insight into the physiological functions of E6-mediated ubiquitination/degradation of the respective proteins.
We previously reported that the E6 proteins do not only affect the substrate spectrum of E6AP but also act as potent activators of E6AP (31, 60). This notion is further supported by the finding that E6AP can employ Ub-K48/63R for ubiquitination in the presence of E6 but not, or only poorly, in its absence. Intriguingly, we identified RAD23 in our study, which was previously reported to be an E6-independent substrate of E6AP (61). Thus, it seems possible, if not likely, that in the list of identified substrates, additional proteins are present that also serve as substrates for E6AP in the absence of E6 proteins. Notably, the substrate(s) of E6AP, whose deregulation contributes to the development of the Angelman syndrome and/or the Dup15q syndrome, are still not known. We therefore propose that the E6 proteins can serve as potent tools to identify such proteins, in particular by designing and employing E6 variants that can still activate E6AP but have lost the ability to interact with their own substrates such as p53 and PDZ domain-containing proteins. Eventually, identification of the “normal” (i.e. E6-independent) substrate spectrum of E6AP may not only provide insight into the normal and pathophysiological cellular functions of E6AP but also into why the HPV E6 proteins utilize E6AP and not another E3 ubiquitin ligase for viral purposes.
Experimental procedures
Plasmids and antibodies
Bacterial expression constructs for the ubiquitin-conjugating enzyme UbcH7, ubiquitin, and GST fusion proteins of HPV-16 E6 (GST-16 E6) and HPV-11 E6 (GST-11 E6), and expression constructs (in vitro translation, transient transfection experiments) encoding HA-tagged WT E6AP (isoform 1), the HA-tagged catalytically inactive mutant E6AP-C820A (substitution of Cys-820 by Ala; numbering according to isoform 1 (15)), and HA-tagged HPV-16 E6 and HPV-11 E6, were described previously (11, 31, 62). cDNAs encoding the ubiquitin mutant Ub-K48/63R (substitution of Lys-48 and Lys-63 by Arg) and HA-tagged forms of XRCC4, OTUD5, MRE11, NIPP1, and DDX3X were generated by PCR-based approaches and cloned into pET3a and pcDNA3, respectively.
Where indicated, HA-tagged proteins were detected by the mouse mAb HA.11 (Covance) and p53 was detected with the mouse mAb DO-1 (Calbiochem).
Protein expression and purification
UBA1 and E6AP were expressed in the baculovirus system. High Five insect cells were plated on a 10-cm dish to 60% confluency. After 90 min, the medium was exchanged, and the cells were infected with the respective baculovirus. After 44 h incubation at 27 °C, the cells were harvested, washed with PBS, and resuspended in lysis buffer (100 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1% Nonidet-P40, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 100 μm Pefabloc, and 1 mm DTT). Upon incubation on ice for 90 min, the lysate was cleared by centrifugation and the supernatant was loaded onto Q-Sepharose beads packed in a gravity-flow column (GE Healthcare). For lysate derived from one 10-cm plate, 100-μl beads pre-equilibrated with 20 ml of wash buffer (25 mm Tris-HCl, pH 7.4, and 125 mm NaCl) were used. Upon loading of the lysate, the beads were washed with 25 ml of wash buffer (25 mm Tris-HCl, pH 7.4, and 200 mm NaCl). E1 and E6AP were eluted in 1-ml fractions with 300 mm NaCl and 350 mm NaCl, respectively, in elution buffer (25 mm Tris-HCl, pH 7.4, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 100 μm Pefabloc, and 1 mm DTT). Fractions containing the respective protein were pooled.
C-terminally His-tagged UbcH7 was expressed in E. coli BL21 (DE3) cells. Cells from 500 ml of bacterial culture were pelleted by centrifugation and resuspended in 10 ml of lysis buffer (PBS, 1% Triton X-100, 10 mm imidazole, 1 mm DTT, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 100 μm Pefabloc). After 10 min of incubation on ice, the cells were sonicated and centrifuged. The supernatant was incubated with 1 ml of nickel-nitrilotriacetic acid beads pre-equilibrated in lysis buffer. After 1 h of incubation at 4 °C, the beads were spun down, washed with 15 ml of wash buffer 1 (PBS and 0.1% Triton X-100), then washed with 20 ml of wash buffer 2 (PBS, 0.1% Triton X-100, 1 mm DTT, and 50 mm imidazole), then washed with 10 ml of wash buffer 3 (25 mm Tris-HCl, 300 mm NaCl, 0.1% Triton X-100, 1 mm DTT, and 50 mm imidazole, pH 7.5), and finally loaded onto a gravity-flow column. The beads were washed with 10 ml of wash buffer 3 and UbcH7 was eluted with 6× 1 ml of elution buffer (25 mm Tris-HCl, pH 7.5, 300 mm NaCl, 1 mm DTT, 250 mm imidazole, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 100 μm Pefabloc). UbcH7-containing fractions were pooled and dialyzed against 2 liters of dialysis buffer (25 mm Tris-HCl, pH 7.5, 300 mm NaCl, 0.2 mm DTT, and 5% glycerol).
GST fusion proteins of HPV-16 and HPV-11 E6 were expressed in E. coli BL21 (DE3) cells. Cell pellets derived from 1 liter of bacterial culture were resuspended in 30 ml of lysis buffer (PBS, 1% Triton X-100, 1 mm DTT, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 100 μm Pefabloc), sonicated, and centrifuged. The supernatant was added to 150 μl of GSH Sepharose beads (GE Healthcare) pre-equilibrated in lysis buffer. Upon incubation for 90 min at 4 °C, the beads were spun down and washed with 3× 1 ml of washing buffer (PBS, 0.1% Triton X-100, and 1 mm DTT), and the GST fusion proteins were eluted with 5× 150 μl of elution buffer (25 mm Tris-HCl, pH 8.0, 25 mm GSH, and 1 mm DTT).
Ubiquitin and the ubiquitin mutant Ub-K48/63R were expressed in E. coli BL21 (DE3) cells. Upon lysis in 25 mm sodium acetate (NaOAc), pH 4.0, and 1 mm DTT, extracts were heated to 70 °C for 20 min, centrifuged, and loaded onto a 1-ml HiTrap SP Sepharose High Performance column (GE Healthcare). The proteins were eluted by a linear gradient of NaCl from 25 mm to 500 mm in 25 mm NaOAc, pH 4.0, and 1 mm DTT. Ubiquitin-containing fractions were pooled and applied to size exclusion chromatography (Superdex 75 Hi Load 20/60; GE Healthcare). Ubiquitin-containing fractions were pooled and dialyzed against H2O (Milli-Q). Purified ubiquitin was lyophilized and stored at 4 °C.
Generation of biotinylated ubiquitin and biotinylated Ub-K48/63R
For biotinylation of ubiquitin, sulfo-N-hydrosuccinimide long-chain biotin (Fig. S1) (sulfo-NHS-LC-biotin; Thermo Fisher Scientific) was used. 3 mg of ubiquitin or Ub-K48/63R were mixed with sulfo-NHS-LC-biotin in a 1:3 molar ratio in 1 ml of 50 mm NaAc (pH 6.5). Upon overnight incubation at 4 °C, the reaction was stopped by addition of 40 mm glycine and incubation for 40 min at 30 °C. The mixture was passed through a Minsart RC4 filter (Sartorius) and subjected to size exclusion chromatography (Superdex 75 10/300GL; GE Healthcare). Ubiquitin was eluted with 25 mm Tris-HCl, pH 7.5, 50 mm NaCl, and 1 mm DTT, and fractions were analyzed by SDS-PAGE followed by Coomassie Blue staining. Ubiquitin-containing fractions were pooled and analyzed by MS (for further details, see Fig. S1).
Generation of TAMRA-labeled ubiquitin and TAMRA-labeled Ub-K48/63R
To attach the fluorescent dye 5(6)-carboxytetramethylrhodamine (TAMRA), ubiquitin or Ub-K48/63R was dissolved in 50 mm NaOAc (pH 7.2) and incubated for 30 min at 4 °C. TAMRA-NHS (Invitrogen) was freshly dissolved in DMSO and added to ubiquitin or Ub-K48/63R in a molar ratio of 1:3. The final reaction concentrations were 3 mg/ml ubiquitin or Ub-K48/63R, 0.55 mg/ml TAMRA-NHS, and 10% DMSO (v/v). Upon overnight incubation at 4 °C, the reaction was quenched with 50 mm Tris-HCl, pH 8.0, for 30 min at 30 °C. 30 ml of 25 mm NaOAc, pH 4.0, were added, pH was adjusted to 4.0, and the solution was passed through a 0.22-μm syringe filter. Ub-T (ubiquitin labeled with TAMRA) was further purified via ion exchange chromatography using a 1-ml HiTrap SP HP column (GE Healthcare) with a gradient of 20 column volumes from 0% buffer B to 100% buffer B (buffer A: 25 mm NaOAc, pH 4.0; buffer B: 25 mm NaOAc and 1 m NaCl, pH 4.0). 4-ml Amicon filter devices with a cutoff of 10 kDa (Millipore) were used for further purification and buffer exchange to storage buffer (25 mm Tris-HCl, 50 mm NaCl, and 1 mm DTT, pH 7.5).
Ubiquitin thioester complex formation assay
To determine the ability of E6AP to form thioester complexes with Ub-K48/63R compared with WT ubiquitin, 0.4 μm UBA1, 2 μm UbcH7, and 0.5 μm E6AP were incubated with 0.3 μm ubiquitin and 0.3 μm Ub-T or 0.3 μm Ub-K48/63 and 0.3 μm Ub-K48/63-T for various times at 30 °C in 20-μl volumes. In addition, reactions contained 25 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.1 mm DTT, 2 mm ATP, and 10 mm MgCl2. Reactions were stopped by incubating the mixtures for 15 min at 30 °C in urea loading buffer (2× urea loading buffer: 8 m urea, 0.1 m Tris-HCl, pH 6.8, 10% glycerol, 4% SDS, and 0.001% bromphenol blue) or urea loading buffer containing 25 mm DTT (reducing conditions). The samples were separated on 7.5–15% SDS-PAGE gradient gels at 4 °C and analyzed by fluorescence scan at 532 nm.
Tandem mass tag–based quantification of diGly-modified ubiquitin-derived peptides
E6AP auto-ubiquitination assays (34) were performed with either WT ubiquitin or Ub-K48/63R in the presence or absence of GST-16 E6 in biological triplicates and analyzed via SDS-PAGE, followed by staining with colloidal Coomassie Blue. The part of the gel containing E6AP and the ubiquitinated forms thereof (≥100 kDa) were excised and processed essentially as described (63). In short, Coomassie Blue–stained bands were destained with 100 mm ammonium bicarbonate/acetonitrile (1:1, v/v), reduced with 10 mm DTT in 100 mm ammonium bicarbonate for 30 min at 56 °C, and alkylated with 55 mm iodoacetamide in 100 mm ammonium bicarbonate for 30 min at room temperature in the dark. After buffer exchange to 100 mm HEPES, pH 7.8, the peptides were digested with trypsin (13 ng/μl, Promega, V5111; cleavage after Lys and Arg but not before Pro) at 37 °C overnight. The digested peptides were extracted twice with acetonitrile/5% formic acid (2:1, v/v) and vacuum concentrated. Peptides were resuspended in 50 μl 100 mm HEPES, pH 8, before being labeled with tandem mass tag (TMT) 6-plex according to the manufacturer's protocol (Thermo Fisher Scientific). TMT labels (126, 127, 129, and 130) were thoroughly dissolved in 41 μl of anhydrous acetonitrile before 20 μl were added to each sample which contained ∼12 μg peptides. Labeling was performed for 60 min at 28 °C before the reaction was quenched with 4 μl of 5% hydroxylamine for 15 min. Then, all four conditions were pooled for each of the three replicates and vacuum concentrated. Prior to MS analysis, the peptides were desalted via C18 spin tips (Thermo Fisher Scientific), dried, and resuspended in 50 μl of 5% acetonitrile/0.1% formic acid for MS analysis.
TMT-labeled peptides were analyzed on an Orbitrap Fusion Tribrid mass spectrometer coupled to an EASY-nLC 1200 system (Thermo Fisher Scientific). The peptides were separated on an Acclaim PepMap C18 column (50 μm × 150 mm, 2 μm, 100 Å) at a flow rate of 300 nl/min using a 90-min gradient (4 min 0.1% formic acid, 75 min linear gradient from 0–35% acetonitrile/0.1% formic acid, 1 min linear gradient from 35–80% acetonitrile/0.1% formic acid, 10 min 80% acetonitrile/0.1% formic acid). TMT-labeled peptides were measured using a synchronous precursor selection-MS3 method (64) with a full cycle time of 3 s. Full scan mass spectrum was acquired at a resolution of 120K (at 200 m/z) with a maximum injection time of 50 ms, an automatic gain control (AGC) target of 4e5 and a scan range 375-1500 m/z. For MS2 scans, only ions with an intensity threshold of 5e3 were isolated, fragmented with 35% collision-induced dissociation and measured in the Ion Trap in rapid scan mode with an isolation window of 0.7 m/z, a maximum injection time of 50 ms and an AGC target of 1e4. For the MS3 scan precursor ion exclusion (low 18, high 5), TMT as isobaric tag loss exclusion and synchronous precursor selection set to 10 precursor ions were selected. The MS3 scan was acquired at a resolution of 60K with an MS isolation window of 2 m/z, higher-energy collisional dissociation fragmentation with normalized collision energy of 65%, a scan range of 100–500 m/z, a maximum injection time of 118 ms, and an AGC target of 1e5.
Raw data files were processed using the Proteome Discoverer v 2.2.0.388 (Thermo Fisher Scientific). MS/MS data were searched via the SEQUEST search node with the following parameter settings: trypsin digestion allowing for up to 2 missed cleavages, minimum peptide length of 5 amino acids, 10 ppm precursor mass tolerance, and 0.6 ppm fragment mass tolerance. Fixed modifications were set to carbamidomethylation at cysteine residues and the TMT label at peptide N termini, and dynamic modifications were set to oxidation of methionine residues, diGly modification at lysine residues and TMT label at lysine residues. Reporter ions were quantified via the Reporter Ions Quantifier node from MS3 spectra based on their S/N ratio with a co-isolation threshold ≤ 30 and average reporter S/N threshold ≥ 50. Quantification results were visualized as grouped abundances of the summed abundances of all replicates for the respective channel.
In vitro ubiquitination with whole-cell extracts or in vitro translated proteins
For ubiquitination of in vitro translated proteins, 1 μl of rabbit reticulocyte lysate-translated 35S-labeled substrate (XRCC4, OTUD5, MRE11, NIPP1, and DDX3X) was incubated with 100 ng of UBA1, 50 ng of UbcH7, varying amounts of baculovirus-expressed E6AP, and 10 μg of ubiquitin in the absence or presence of GST-16 E6 or GST-11 E6 (200 ng) in 20-μl volumes. In addition, the reactions contained 25 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm DTT, 2 mm ATP, and 4 mm MgCl2. After incubation at 30 °C for 90 min, total reaction mixtures were electrophoresed in 10% or 12.5% SDS-polyacrylamide gels, and 35S-labeled proteins were detected by fluorography.
For ubiquitination assays with cell extracts, HEK293T or HaCaT cells were lysed in 0.1 m Tris-HCl, pH 8.0, 0.1 m NaCl, 1% IGEPAL (MP Biomedicals), 1 mm Pefabloc, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mm DTT for 30 min at 4 °C. Upon clearance by centrifugation, protein concentration was determined by the BCA protein assay kit according to the manufacturer's instructions (Thermo Fisher Scientific). In vitro ubiquitination was performed with 30 μg of cell extract, 75 ng of E1, 75 ng of UbcH7, indicated amounts of baculovirus-expressed E6AP, 200 ng of GST-16 E6 or GST-11 E6, and 6 μg of biotinylated ubiquitin or Ub-K48/63R in 30-μl volumes. In addition, the reaction mixtures contained 25 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm DTT, 2 mm ATP, and 4 mm MgCl2. For determination of potential substrates of the E6 proteins, 5 mg of HaCaT cell extract were employed and the reaction mixture was up-scaled accordingly. Reactions were incubated at 30 °C for 90 min.
Affinity purification of ubiquitinated proteins
To isolate proteins modified by biotinylated ubiquitin or biotinylated Ub-K48/63R, total reaction mixtures were mixed with High Capacity Streptavidin beads (Thermo Fisher Scientific) in a ratio of 1.3 μg of biotinylated ubiquitin to 2 μl of beads pre-equilibrated in 25 mm Tris-HCl, pH 7.5, and 50 mm NaCl. Upon incubation for 60 min at room temperature, the mixtures were transferred into columns (empty laboratory columns, 5-ml column volume, 35-μm filter pore size; Mobitec). The beads were washed in a sequential manner with 5 column volumes each of 1) 6 m urea, 25 mm Tris-HCl, pH 7.5, and 0.5% SDS; 2) 6 m guanidinium chloride and 25 mm Tris-HCl, pH 8.5; 3) 6 m urea, 1 m NaCl, and 25 mm Tris-HCl, pH 7.5; 4) 25 mm Tris-HCl, pH 7.5, and 500 mm NaCl; 5) 25 mm Tris-HCl, pH 7.5, and 250 mm NaCl; 6) 5 mm NH4OAc, pH 5; and 7) 25 mm Tris-HCl, pH 7.5. In addition, washing buffers contained 1 mm DTT, and the beads were washed after each washing step with 5 column volumes of H2O (Milli-Q) supplemented with 1 mm DTT. Finally, proteins bound to streptavidin beads were eluted by boiling in 125 mm Tris-HCl, pH 6.8, 20% (v/v) glycerol, 4% (w/v) SDS, 0.001% (w/v) bromphenol blue, and 200 mm DTT at 95 °C for 5 min. Eluates were separated by SDS-PAGE followed by Western blotting analysis using an anti-p53 antibody (experiments with HEK293T cell extracts) or by staining with colloidal Coomassie Blue (HaCaT cell extracts). To identify potential substrates of the E6 proteins, ubiquitination reactions with HaCaT cell extracts were performed in three independent experiments (“biological replicates”).
Protein identification by LC–MS/MS
In-gel digestion was performed essentially as described (63). The individual lanes (see Fig. 2) were sliced into seven pieces and washed at 37 °C with 50 mm ammonium bicarbonate/acetonitrile (1:1, v/v) until gel pieces appeared colorless. After washing with 50 mm ammonium bicarbonate, proteins were reduced by addition of 10 mm DTT in 50 mm ammonium bicarbonate for 60 min at 56 °C. Then, proteins were alkylated with 50 mm iodoacetamide in 50 mm ammonium bicarbonate. After 60 min at room temperature, gel pieces were washed with 50 mm ammonium bicarbonate/acetonitrile (1:1, v/v) and dried in 100% acetonitrile. For digestion, gel pieces were fully covered with trypsin solution (13 ng/μl; Promega, V5111; cleavage after Lys and Arg but not before Pro) and incubated overnight at 37 °C. Finally, peptides were extracted twice from the gel pieces in 5% formic acid/acetonitrile (1:2, v/v), freeze-dried, and desalted using U-C18 ZipTips (Millipore). Peptides were eluted in 2 μl of 0.1% TFA/75% acetonitrile and diluted with 21 μl of 0.1% formic acid.
Tryptic peptides were separated on an EASY-nLC 1200 system (Thermo Fisher Scientific) at a flow rate of 300 nl/min using a 45-min gradient from 5% acetonitrile, 0.1% formic acid to 35% acetonitrile, 0.1% formic acid. This was followed by a 6-min gradient from 35% acetonitrile, 0.1% formic acid to 45% acetonitrile, 0.1% formic acid and subsequently by a 1-min gradient from 45% acetonitrile, 0.1% formic acid to 80% acetonitrile and a washing step with 80% acetonitrile for 8 min. Mass spectra were collected on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) operated in data-dependent mode with dynamic exclusion set at 45 s and a total cycle time of 3 s. A resolution of 120,000 (at m/z 200) with an automatic gain control ion target value of 4e5 and a maximum injection time of 50 ms was chosen to acquire full scan MS spectra in the Orbitrap. Most intense precursors with charge states of 2–7 and intensities greater than 5e3 were selected for MS/MS experiments using collision-induced dissociation with 35% collision energy. In the quadrupole, isolation was carried out with an isolation window of 1.6 m/z. The automatic gain control ion target was set to 2e3 with a maximum injection time of 300 ms. MS/MS spectra were acquired in the ion trap at a rapid scan rate. For MS/MS selection, monoisotopic peak determination was set to peptide. The biological replicates of each affinity purification were measured as technical duplicates.
Raw files from LC–MS/MS measurements were evaluated using MaxQuant (version 1.5.4.1) using the andromeda search engine (65, 66) with default settings and match between runs and label-free quantification (LFQ) (minimum ratio count 2) enabled. For the analysis, the digestion mode was set to “specific” using “Trypsin/P” as protease, which also cleaves after the amino acids lysine and arginine if a proline follows in the amino acid sequence, and a maximum of two missed cleavage sites were allowed. Carbamidomethyl was considered as a fixed modification, and oxidation of methionine and acetylation of protein N termini were considered as variable modifications. Mass tolerances were set to 4.5 ppm for precursor ions measured in the Orbitrap and to 0.5 Da for fragment ions measured in the ion trap. The complete search parameters are deposited with the raw files at PRIDE PXD017335 (see below). The human reference proteome downloaded from the UniProt database (download date 2018-02-06; 71,804 entries) and an integrated database of common contaminants (containing 245 protein entries) were used to identify protein identities.
The Perseus software (version 1.5.4.1) (67) was used to further process acquired data. Identified proteins were filtered for reverse hits, for proteins that were only identified by site, and for common contaminants. LFQ intensities were log2 transformed. Proteins detected in at least four replicates (of six replicate experiments per experimental group) were selected, and missing values were imputed for the total matrix from a normal distribution (width = 0.3 and shift = 1.8), based on the assumption that these proteins were below the detection limit. The LFQ values of the 1509 identified proteins were visualized as a heat map (Fig. 2B and Supporting data 1). A two-sample t test (FDR = 0.001, S0 = 2) in combination with a permutation-based FDR estimation was used to identify significantly enriched proteins, comparing LFQ intensities of each experiment group (HPV-16 E6 and HPV-11 E6) with the respective control group without E6. The data were visualized in volcano plots showing the log2 fold change versus the negative logarithmized p-values (Fig. 3, A and B). The log2-transformed LFQ values of the 269 proteins that were significantly enriched with HPV-16 E6, HPV-11 E6, or both are listed in Supporting data 1; the Z-score normalized LFQ values (per row) are visualized as a heat map with hierarchical cluster analysis (Euclidian distance) of the columns in Fig. 3C. To determine potential ubiquitination sites, significant hits were further analyzed with Proteome Discoverer (version 2.2.0.3.88), allowing diGly as dynamic modification on lysine residues and N termini (Supporting data 1).
Gene Ontology annotation analysis
To find classes of molecular functions that are overrepresented among the 199 proteins enriched with HPV-16 E6 or the 179 proteins enriched with HPV-11 E6 relative to the human reference genome, the Gene Ontology (GO) annotations for “molecular function complete” (released 2019-12-09) were mapped and an overrepresentation test (released 2019-07-11) was applied using PANTHER (68). Of the 199 proteins enriched with HPV-16 E6 and the 179 proteins enriched with HPV-11 E6, 193 and 176 proteins, respectively, could be mapped to GO annotations. The Homo sapiens genome with 20,851 IDs was used as reference. For the overrepresentation test, a Fisher's exact test with an FDR correction (<0.05) was applied. The fold enrichment of molecular functions classes that were over- or under-represented for HPV-16 E6 and HPV-11 E6 were plotted in % as stacked bar charts (69) with python (version 3.7.3; RRID:SCR_008394) (Fig. S4). Note that most of the proteins mapped to more than one GO annotation term.
GST pulldown experiments
For pulldown experiments, 0.5 μg GST-16 E6, GST-11 E6, or GST bound to 5 μl GSH Sepharose beads were incubated with 10 μl of in vitro translated radiolabeled protein of interest in 0.1 m Tris-HCl, pH 8.0, 0.1 m NaCl, 1% IGEPAL (MP Biomedicals), 1 mm Pefabloc, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mm DTT in a total volume of 200 μl for 90 min at 4 °C. Then, the beads were washed three times with 1 ml of 0.1 m Tris-HCl, pH 8.0, 0.1 m NaCl, 1% IGEPAL (MP Biomedicals), 1 mm Pefabloc, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mm DTT. Finally, bound proteins were eluted by boiling in 125 mm Tris-HCL, pH 6.8, 20% (v/v) glycerol, 4% (w/v) SDS, 0.001% (w/v) bromphenol blue, and 200 mm DTT at 95 °C for 5 min. The eluates were subjected to SDS-PAGE and the proteins were visualized by fluorography.
Transient transfection experiments
H1299-K3 cells (stable knockdown of E6AP expression) (11) were grown in DMEM supplemented with 10% FBS. For degradation assays, one well (80–90% confluency) of a 6-well culture plate (Sarstedt) was transfected with expression constructs encoding HA-tagged E6AP or the catalytically inactive mutant E6AP-C820A (1.5 μg), HA-tagged HPV-16 E6 (2.5 μg) or HPV-11 E6 (0.4 μg), and HA-tagged versions of the protein of interest (XRCC4, OTUD5, MRE11, NIPP1, and DDX3X) by lipofection (Lipofectamine 2000; Invitrogen). In addition, a β-galactosidase expression construct was co-transfected to determine relative transfection efficiencies, and total DNA amounts in all transfections were equalized by addition of empty vector (pcDNA3). 24 h after transfection, the cells were lysed in 0.1 ml of 0.1 m Tris-HCl, pH 8.0, 0.1 m NaCl, 1% IGEPAL (MP Biomedicals), 1 mm Pefabloc, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mm DTT. Volumes of cell lysates used for SDS-PAGE and Western blotting analysis were adjusted based on transfection efficiencies, as determined by measuring β-galactosidase activity in duplicates. Levels of E6AP, the E6 proteins, and the proteins of interest were monitored by an anti-HA antibody followed by quantification of the intensity of the signals with the Aida 4.08 software package (Raytest).
Experimental design and statistical rationale
In total, 126 samples were analyzed by LC–MS/MS. For each condition (i.e. HPV-16 E6 and HPV-11 E6) and for the control (reaction in the absence of E6 proteins), seven gel pieces were analyzed from biological triplicates (i.e. separately expressed and purified batches of protein). Each of these 63 samples was measured in technical duplicates. For the two-sample t test (S0 = 2) that was used to identify significantly enriched proteins, a permutation-based FDR estimation was applied (FDR ≤ 0.001). To find classes of molecular functions that are over- or under-represented among the proteins significantly enriched with HPV-16 E6 or HPV-11 E6, a Fisher's exact test with an FDR correction (<0.05) was applied. No data were excluded.
Data availability
All data generated or analyzed during this study are included in this article and in the supporting information. The MS raw files of the affinity-based enrichment of interacting proteins have been deposited to the Proteome Xchange Consortium via the PRIDE partner repository (70) with the project accession number PXD017335. The MS data for the TMT-based quantification are available via the Proteome Xchange Consortium with the project accession number PXD020660.
Supplementary Material
Acknowledgments
We are grateful to Silke Büstorf, Lea Güntner, Franziska Guba, Matthias Heiser, and Nicole Richter-Müller for technical assistance.
This article contains supporting information.
Author contributions—F. A. E., F. S., and M. S. conceptualization; F. A. E., C. S., and D. E. validation; F. A. E., C. S., D. E., J. J., A. S.-M., and F. S. investigation; F. A. E., C. S., D. E., J. J., A. S.-M., and F. S. methodology; F. A. E., C. S., D. E., J. J., A. S.-M., F. S., and M. S. writing-review and editing; F. S. and M. S. supervision; F. S. and M. S. funding acquisition; M. S. writing-original draft; M. S. project administration.
Funding and additional information—This work was supported by the German Research Foundation Grant SFB969, project B2 (to M. S.), and the Emmy Noether-Program of the German Research Foundation Grant STE 2517/1-1 (to F. S.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- HPV
- human papillomavirus
- E6AP
- E6-associated protein
- OTU
- ovarian tumor
- OTUD
- OTU deubiquitinase
- DSB
- double-strand break
- TMT
- tandem mass tag
- Ub
- ubiquitin
- UBA
- ubiquitin-like modifier–activating enzyme
- Ubc
- ubiquitin-conjugating enzyme
- NaOAc
- sodium acetate
- TAMRA
- 5(6)-carboxytetramethylrhodamine
- LFQ
- label-free quantification.
References
- 1. de Villiers E. M., Fauquet C., Broker T. R., Bernard H. U., and Zur Hausen H. (2004) Classification of papillomaviruses. Virology 324, 17–27 10.1016/j.virol.2004.03.033 [DOI] [PubMed] [Google Scholar]
- 2. de Villiers E. M. (2013) Cross-roads in the classification of papillomaviruses. Virology 445, 2–10 10.1016/j.virol.2013.04.023 [DOI] [PubMed] [Google Scholar]
- 3. Doorbar J., Quint W., Banks L., Bravo I. G., Stoler M., Broker T. R., and Stanley M. A. (2012) The biology and life-cycle of human papillomaviruses. Vaccine 30 Suppl 5, F55–F70 10.1016/j.vaccine.2012.06.083 [DOI] [PubMed] [Google Scholar]
- 4. Harden M. E., and Munger K. (2017) Human papillomavirus molecular biology. Mutat. Res. Rev. Mutat. Res. 772, 3–12 10.1016/j.mrrev.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zur Hausen H. (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2, 342–350 10.1038/nrc798 [DOI] [PubMed] [Google Scholar]
- 6. Schiffman M., Castle P. E., Jeronimo J., Rodriguez A. C., and Wacholder S. (2007) Human papillomavirus and cervical cancer. Lancet 370, 890–907 10.1016/S0140-6736(07)61416-0 [DOI] [PubMed] [Google Scholar]
- 7. Mantovani F., and Banks L. (2001) The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 20, 7874–7887 10.1038/sj.onc.1204869 [DOI] [PubMed] [Google Scholar]
- 8. Scheffner M., and Whitaker N. J. (2003) Human papillomavirus-induced carcinogenesis and the ubiquitin-proteasome system. Semin. Cancer Biol. 13, 59–67 10.1016/S1044-579X(02)00100-1 [DOI] [PubMed] [Google Scholar]
- 9. Beaudenon S., and Huibregtse J. M. (2008) HPV E6, E6AP and cervical cancer. BMC Biochem. 9, S4 10.1186/1471-2091-9-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brimer N., Lyons C., and Vande Pol S. B. (2007) Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology 358, 303–310 10.1016/j.virol.2006.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuballa P., Matentzoglu K., and Scheffner M. (2007) The role of the ubiquitin ligase E6-AP in human papillomavirus E6-mediated degradation of PDZ domain-containing proteins. J. Biol. Chem. 282, 65–71 10.1074/jbc.M605117200 [DOI] [PubMed] [Google Scholar]
- 12. White E. A., and Howley P. M. (2013) Proteomic approaches to the study of papillomavirus-host interactions. Virology 435, 57–69 10.1016/j.virol.2012.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wallace N. A., and Galloway D. A. (2015) Novel functions of the human papillomavirus E6 oncoproteins. Annu. Rev. Virol. 2, 403–423 10.1146/annurev-virology-100114-055021 [DOI] [PubMed] [Google Scholar]
- 14. Sutcliffe J. S., Jiang Y. H., Galijaard R. J., Matsuura T., Fang P., Kubota T., Christian S. L., Bressler J., Cattanach B., Ledbetter D. H., and Beaudet A. L. (1997) The E6-AP ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region. Genome Res. 7, 368–377 10.1101/gr.7.4.368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamamoto Y., Huibregtse J. M., and Howley P. M. (1997) The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics 41, 263–266 10.1006/geno.1997.4617 [DOI] [PubMed] [Google Scholar]
- 16. Kishino T., Lalande M., and Wagstaff J. (1997) UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 15, 70–73 10.1038/ng0197-70 [DOI] [PubMed] [Google Scholar]
- 17. Matsuura T., Sutcliffe J. S., Fang P., Galjaard R. J., Jiang Y. H., Benton C. S., Rommens J. M., and Beaudet A. L. (1997) De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat. Genet. 15, 74–77 10.1038/ng0197-74 [DOI] [PubMed] [Google Scholar]
- 18. Clayton-Smith J., and Laan L. (2003) Angelman syndrome: a review of the clinical and genetic aspects. J. Med. Genet. 40, 87–95 10.1136/jmg.40.2.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dagli A., Buiting K., and Williams C. A. (2012) Molecular and clinical aspects of Angelman syndrome. Mol. Syndromol. 2, 100–112 10.1159/000328837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glessner J. T., Wang K., Cai G., Korvatska O., Kim C. E., Wood S., Zhang H., Estes A., Brune C. W., Bradfield J. P., Imielinski M., Frackelton E. C., Reichert J., Crawford E. L., Munson J., et al. (2009) Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459, 569–573 10.1038/nature07953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hogart A., Wu D., LaSalle J. M., and Schanen N. C. (2010) The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol. Dis. 38, 181–191 10.1016/j.nbd.2008.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith S. E., Zhou Y. D., Zhang G., Jin Z., and Stoppel D. C., and Anderson M. P. (2011) Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci. Transl. Med. 3, 103–197 10.1126/scitranslmed.3002627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shai A., Pitot H. C., and Lambert P. F. (2010) E6-associated protein is required for human papillomavirus type 16 E6 to cause cervical cancer in mice. Cancer Res. 70, 5064–5073 10.1158/0008-5472.CAN-09-3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Egawa N., and Doorbar J. (2017) The low-risk papillomaviruses. Virus Res. 231, 119–127 10.1016/j.virusres.2016.12.017 [DOI] [PubMed] [Google Scholar]
- 25. Iconomou M., and Saunders D. N. (2016) Systematic approaches to identify E3 ligase substrates. Biochem. J. 473, 4083–4101 10.1042/BCJ20160719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burande C. F., Heuzé M. L., Lamsoul I., Monsarrat B., Uttenweiler-Joseph S., and Lutz P. G. (2009) A label-free quantitative proteomics strategy to identify E3 ubiquitin ligase substrates targeted to proteasome degradation. Mol. Cell. Proteomics 8, 1719–1727 10.1074/mcp.M800410-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sarraf S. A., Raman M., Guarani-Pereira V., Sowa M. E., Huttlin E. L., Gygi S. P., and Harper J. W. (2013) Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thompson J. W., Nagel J., Hoving S., Gerrits B., Bauer A., Thomas J. R., Kirschner M. W., Schirle M., and Luchansky S. J. (2014) Quantitative Lys–Gly-Gly (diGly) proteomics coupled with inducible RNAi reveals ubiquitin-mediated proteolysis of DNA damage-inducible transcript 4 (DDIT4) by the E3 ligase HUWE1. J. Biol. Chem. 289, 28942–28955 10.1074/jbc.M114.573352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang M., and Pickart C. M. (2005) Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J. 24, 4324–4333 10.1038/sj.emboj.7600895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim H. C., and Huibregtse J. M. (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell Biol. 29, 3307–3318 10.1128/MCB.00240-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mortensen F., Schneider D., Barbic T., Sladewska-Marquardt A., Kühnle S., Marx A., and Scheffner M. (2015) Role of ubiquitin and the HPV E6 oncoprotein in E6AP-mediated ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 112, 9872–9877 10.1073/pnas.1505923112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Macdonald J. M., Haas A. L., and London R. E. (2000) Novel mechanism of surface catalysis of protein adduct formation. NMR studies of the acetylation of ubiquitin. J. Biol. Chem. 275, 31908–31913 10.1074/jbc.M000684200 [DOI] [PubMed] [Google Scholar]
- 33. Shang F., Deng G., Liu Q., Guo W., Haas A. L., Crosas B., Finley D., and Taylor A. (2005) Lys6-modified ubiquitin inhibits ubiquitin-dependent protein degradation. J. Biol. Chem. 280, 20365–20374 10.1074/jbc.M414356200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nuber U., Schwarz S. E., and Scheffner M. (1998) The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur. J. Biochem. 254, 643–649 10.1046/j.1432-1327.1998.2540643.x [DOI] [PubMed] [Google Scholar]
- 35. DuBridge R. B., Tang P., Hsia H. C., Leong P. M., Miller J. H., and Calos M. P. (1987) Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell Biol. 7, 379–387 10.1128/mcb.7.1.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., and Fusenig N. E. (1988) Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106, 761–771 10.1083/jcb.106.3.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., and Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- 38. González M., Argaraña C. E., and Fidelio G. D. (1999) Extremely high thermal stability of streptavidin and avidin upon biotin binding. Biomol. Eng. 16, 67–72 10.1016/S1050-3862(99)00041-8 [DOI] [PubMed] [Google Scholar]
- 39. Werness B. A., Levine A. J., and Howley P. M. (1990) Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248, 76–79 10.1126/science.2157286 [DOI] [PubMed] [Google Scholar]
- 40. Scheffner M., Werness B. A., Huibregtse J. M., Levine A. J., and Howley P. M. (1990) The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63, 1129–1136 10.1016/0092-8674(90)90409-8 [DOI] [PubMed] [Google Scholar]
- 41. Kiyono T., Hiraiwa A., Fujita M., Hayashi Y., Akiyama T., and Ishibashi M. (1997) Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. U. S. A. 94, 11612–11616 10.1073/pnas.94.21.11612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakagawa S., and Huibregtse J. M. (2000) Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell Biol. 20, 8244–8253 10.1128/.20.21.8244-8253.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vande Pol S. B., Brown M. C., and Turner C. E. (1998) Association of bovine papillomavirus type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif. Oncogene 16, 43–52 10.1038/sj.onc.1201504 [DOI] [PubMed] [Google Scholar]
- 44. Hsu C. H., Peng K. L., Jhang H. C., Lin C. H., Wu S. Y., Chiang C. M., Lee S. C., Yu W. C., and Juan L. J. (2012) The HPV E6 oncoprotein targets histone methyltransferases for modulating specific gene transcription. Oncogene 31, 2335–2349 10.1038/onc.2011.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ganti K., Massimi P., Manzo-Merino J., Tomaić V., Pim D., Playford M. P., Lizano M., Roberts S., Kranjec C., Doorbar J., and Banks L. (2016) Interaction of the human papillomavirus E6 oncoprotein with sorting nexin 27 modulates endocytic cargo transport pathways. PLoS Pathog. 12, e1005854 10.1371/journal.ppat.1005854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Drews C. M., Case S., and Vande Pol S. B. (2019) E6 proteins from high-risk HPV, low-risk HPV, and animal papillomaviruses activate the Wnt/β-catenin pathway through E6AP-dependent degradation of NHERF1. PLoS Pathog. 15, e1007575 10.1371/journal.ppat.1007575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Akhmanova A., and Steinmetz M. O. (2015) Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16, 711–726 10.1038/nrm4084 [DOI] [PubMed] [Google Scholar]
- 48. Paull T. T. (2018) 20 Years of Mre11 biology: no end in sight. Mol. Cell 71, 419–427 10.1016/j.molcel.2018.06.033 [DOI] [PubMed] [Google Scholar]
- 49. Trinkle-Mulcahy L., Ajuh P., Prescott A., Claverie-Martin F., Cohen S., Lamond A. I., and Cohen P. (1999) Nuclear organisation of NIPP1, a regulatory subunit of protein phosphatase 1 that associates with pre-mRNA splicing factors. J. Cell Sci. 112, 157–168 [DOI] [PubMed] [Google Scholar]
- 50. Valiente-Echeverría F., Hermoso M. A., and Soto-Rifo R. (2015) RNA helicase DDX3: at the crossroad of viral replication and antiviral immunity. Rev. Med. Virol. 25, 286–299 10.1002/rmv.1845 [DOI] [PubMed] [Google Scholar]
- 51. Pannunzio N. R., Watanabe G., and Lieber M. R. (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 293, 10512–10523 10.1074/jbc.TM117.000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luo J., Lu Z., Lu X., Chen L., Cao J., Zhang S., Ling Y., and Zhou X. (2013) OTUD5 regulates p53 stability by deubiquitinating p53. PLoS ONE 8, e77682 10.1371/journal.pone.0077682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li F., Sun Q., Liu K., Han H., Lin N., Cheng Z., Cai Y., Tian F., Mao Z., Tong T., and Zhao W. (2019) The deubiquitinase OTUD5 regulates Ku80 stability and non-homologous end joining. Cell. Mol. Life Sci. 76, 3861–3873 10.1007/s00018-019-03094-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zheng N., and Shabek N. (2017) Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 10.1146/annurev-biochem-060815-014922 [DOI] [PubMed] [Google Scholar]
- 55. Glockzin S., Ogi F. X., Hengstermann A., Scheffner M., and Blattner C. (2003) Involvement of the DNA repair protein hHR23 in p53 degradation. Mol. Cell Biol. 23, 8960–8969 10.1128/mcb.23.24.8960-8969.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Coyaud E., Mis M., Laurent E. M., Dunham W. H., Couzens A. L., Robitaille M., Gingras A. C., Angers S., and Raught B. (2015) BioID-based identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 ligase substrates. Mol. Cell. Proteomics 14, 1781–1795 10.1074/mcp.M114.045658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. O'Connor H. F., Swaim C. D., Canadeo L. A., and Huibregtse J. M. (2018) Ubiquitin-activated interaction traps (UBAITs): tools for capturing protein-protein interactions. Methods Mol. Biol. 1844, 85–100 10.1007/978-1-4939-8706-1_7 [DOI] [PubMed] [Google Scholar]
- 58. Bristol M. L., Das D., and Morgan I. M. (2017) Why human papillomaviruses activate the DNA damage response (DDR) and how cellular and viral replication persists in the presence of DDR signaling. Viruses 9, 268 10.3390/v9100268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Prati B., Marangoni B., and Boccardo E. (2018) Human papillomavirus and genome instability: from productive infection to cancer. Clinics (Sao Paulo) 73, e539s 10.6061/clinics/2018/e539s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sailer C., Offensperger F., Julier A., Kammer K. M., Walker-Gray R., Gold M. G., Scheffner M., and Stengel F. (2018) Structural dynamics of the E6AP/UBE3A-E6-p53 enzyme-substrate complex. Nat. Commun. 9, 4441 10.1038/s41467-018-06953-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kumar S., Talis A. L., and Howley P. M. (1999) Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J. Biol. Chem. 274, 18785–18792 10.1074/jbc.274.26.18785 [DOI] [PubMed] [Google Scholar]
- 62. Kühnle S., Mothes B., Matentzoglu K., and Scheffner M. (2013) Role of the ubiquitin ligase E6AP/UBE3A in controlling levels of the synaptic protein Arc. Proc. Natl. Acad. Sci. U. S. A. 110, 8888–8893 10.1073/pnas.1302792110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- 64. McAlister G. C., Nusinow D. P., Jedrychowski M. P., Wühr M., Huttlin E. L., Erickson B. K., Rad R., Haas W., and Gygi S. P. (2014) MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 10.1021/ac502040v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tyanova S., Temu T., and Cox J. (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- 67. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 68. Mi H., Muruganujan A., Ebert D., Huang X., and Thomas P. D. (2019) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hunter J. D. (2007) Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9, 90–95 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- 70. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and in the supporting information. The MS raw files of the affinity-based enrichment of interacting proteins have been deposited to the Proteome Xchange Consortium via the PRIDE partner repository (70) with the project accession number PXD017335. The MS data for the TMT-based quantification are available via the Proteome Xchange Consortium with the project accession number PXD020660.