Abstract
Thiol-based redox regulation is a post-translational protein modification for controlling enzyme activity by switching oxidation/reduction states of Cys residues. In plant cells, numerous proteins involved in a wide range of biological systems have been suggested as the target of redox regulation; however, our knowledge on this issue is still incomplete. Here we report that 3-phosphoglycerate dehydrogenase (PGDH) is a novel redox-regulated protein. PGDH catalyzes the first committed step of Ser biosynthetic pathway in plastids. Using an affinity chromatography-based method, we found that PGDH physically interacts with thioredoxin (Trx), a key factor of redox regulation. The in vitro studies using recombinant proteins from Arabidopsis thaliana showed that a specific PGDH isoform, PGDH1, forms the intramolecular disulfide bond under nonreducing conditions, which lowers PGDH enzyme activity. MS and site-directed mutagenesis analyses allowed us to identify the redox-active Cys pair that is mainly involved in disulfide bond formation in PGDH1; this Cys pair is uniquely found in land plant PGDH. Furthermore, we revealed that some plastidial Trx subtypes support the reductive activation of PGDH1. The present data show previously uncharacterized regulatory mechanisms of PGDH and expand our understanding of the Trx-mediated redox-regulatory network in plants.
Keywords: Arabidopsis thaliana, 3-phosphoglycerate dehydrogenase, redox regulation, thiol, thioredoxin, plant biochemistry
To tune cellular physiology, a number of proteins in the cell undergo several post-translational modifications. Thiol-based redox regulation is one of such mechanisms; it controls enzyme activity by switching the oxidation/reduction states of Cys residues (e.g. formation/cleavage of disulfide bonds). A small ubiquitous protein thioredoxin (Trx) is largely responsible for the redox regulation. Trx contains the highly conserved amino acid sequence WCGPC at the active site. By using two Cys residues in this motif, Trx catalyzes a dithiol-disulfide exchange reaction with its target proteins, allowing modulation of their enzyme activities. Trx is thus critical for transmitting reducing power to redox-regulated proteins and adjusting cellular functions in response to changes in local redox environments (1, 2).
Trx-mediated redox-regulatory system is ubiquitously found in all kingdoms of life. Among them, the system in plant chloroplasts has attracted much attention due to its unique mode of action related to photosynthesis. Upon illumination, photochemical reactions are triggered in the thylakoid membrane, generating the reducing power. Trx receives a part of reducing power from ferredoxin (Fd) via Fd-Trx reductase (FTR), and in turn transfers it to several redox-regulated proteins. This redox cascade ensures light-responsive coordination of chloroplast functions, which has long been recognized as the molecular basis of the redox-regulatory system in chloroplasts (3, 4).
Another characteristic of the chloroplast system is the emergence of multiple Trx subtypes, categorized into f-, m-, x-, y-, and z-types (5, 6). They have different midpoint redox potentials and protein surface charges, conferring functional diversity to each of the Trx subtypes (e.g. distinct target selectivity) (7–11). In addition, other proteins serving as the mediator of reducing power have been reported to reside in chloroplasts. A most well-known example is the NADPH-Trx reductase C (NTRC); this protein contains both an NADPH-Trx reductase (NTR) domain and a Trx domain in a single polypeptide, and thus works in the redox regulation relying on NADPH (12). Recent studies have proposed specific and important roles of NTRC in redox regulation (13–15). All these data raise a novel hypothesis that the redox-regulatory system in chloroplasts constitutes a complex network, allowing flexible control of chloroplast functions. Uncovering its whole organization is currently a major challenge in the field of redox study (16–22).
To achieve this goal, it is also important to grasp what chloroplast proteins are redox-regulated. Classically, only a limited number of chloroplast proteins have been known as the target of redox regulation (e.g. several Calvin-Benson cycle enzymes and ATP synthase CF1-γ subunit) (3, 4, 23, 24). In the 2000s, several proteomics-based methods for comprehensively identifying Trx-interacting proteins have been developed (25–29), opening the possibility of unprecedented diversity of redox-regulated proteins in chloroplasts. Furthermore, these integral analyses have been applied to other cellular compartments, including other types of plastids (30), mitochondria (31, 32), and cytosol (33). It is now speculated that a broad spectrum of proteins may be under redox regulation in plant cells (34, 35); however, our consistent understanding of this issue is still weak. To overcome this limitation, we need to verify the possibility of redox regulation while paying attention to each of potential target proteins individually. A plastidial enzyme, 3-phosphoglycerate dehydrogenase (PGDH), is one of such targets that remains to be studied at a biochemical level (27, 30, 36).
Ser is an indispensable amino acid involved in many physiological events in all organisms. In plant cells, there are three different pathways for Ser biosynthesis, including the photorespiratory glycolate pathway (in mitochondria), the glycerate pathway (in the cytosol and peroxisomes), and the phosphorylated pathway (in plastids; Fig. 1) (37, 38). The glycolate pathway has been considered to be of major importance especially under photorespiratory conditions. However, recent reverse-genetic studies indicate that the phosphorylated pathway plays a critical role in supporting plant metabolism and development (39–41). The first step of this pathway, that is the oxidation of 3-phosphoglycerate (3-PGA), is catalyzed by PGDH (Fig. 1). PGDH can be classified into three types based on domain structure; plant PGDH belongs to the type I enzyme composed of four domains, the substrate-binding domain, the nucleotide-binding domain, the allosteric substrate-binding domain, and the Asp kinase-chorismate mutase-TyrA domain (42). Despite its postulated importance in ensuring Ser homeostasis in plants, the regulatory mechanisms of PGDH are largely unclear especially at a post-translational level.
Figure 1.
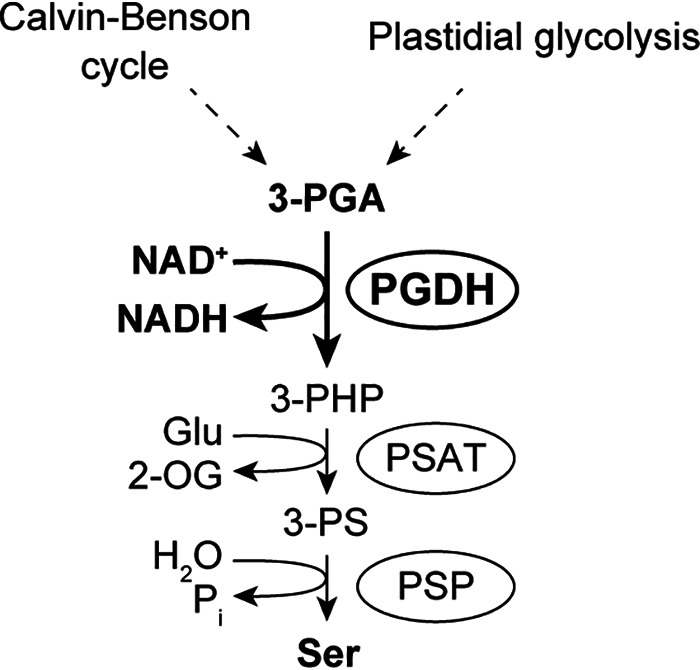
Plastidial Ser biosynthesis mediated by the phosphorylated pathway. The following abbreviations were used: 2-OG, 2-oxoglutarate; 3-PHP, 3-phosphohydroxypyruvate; 3-PS, 3-phosphoserine; PSAT, 3-phosphoserine aminotransferase; PSP, 3-phosphoserine phosphatase.
In this study, we have addressed the possibility of PGDH redox regulation using a biochemical procedure. Our data indicate that (i) a specific isoform of Arabidopsis PGDH, PGDH1, forms the intramolecular disulfide bond, lowering PGDH enzyme activity; (ii) its disulfide bond is formed between Cys residues uniquely conserved in land plant PGDH; and (iii) some plastid-localized Trx subtypes can reduce and activate PGDH1.
Results
PGDH physically interacts with Trx
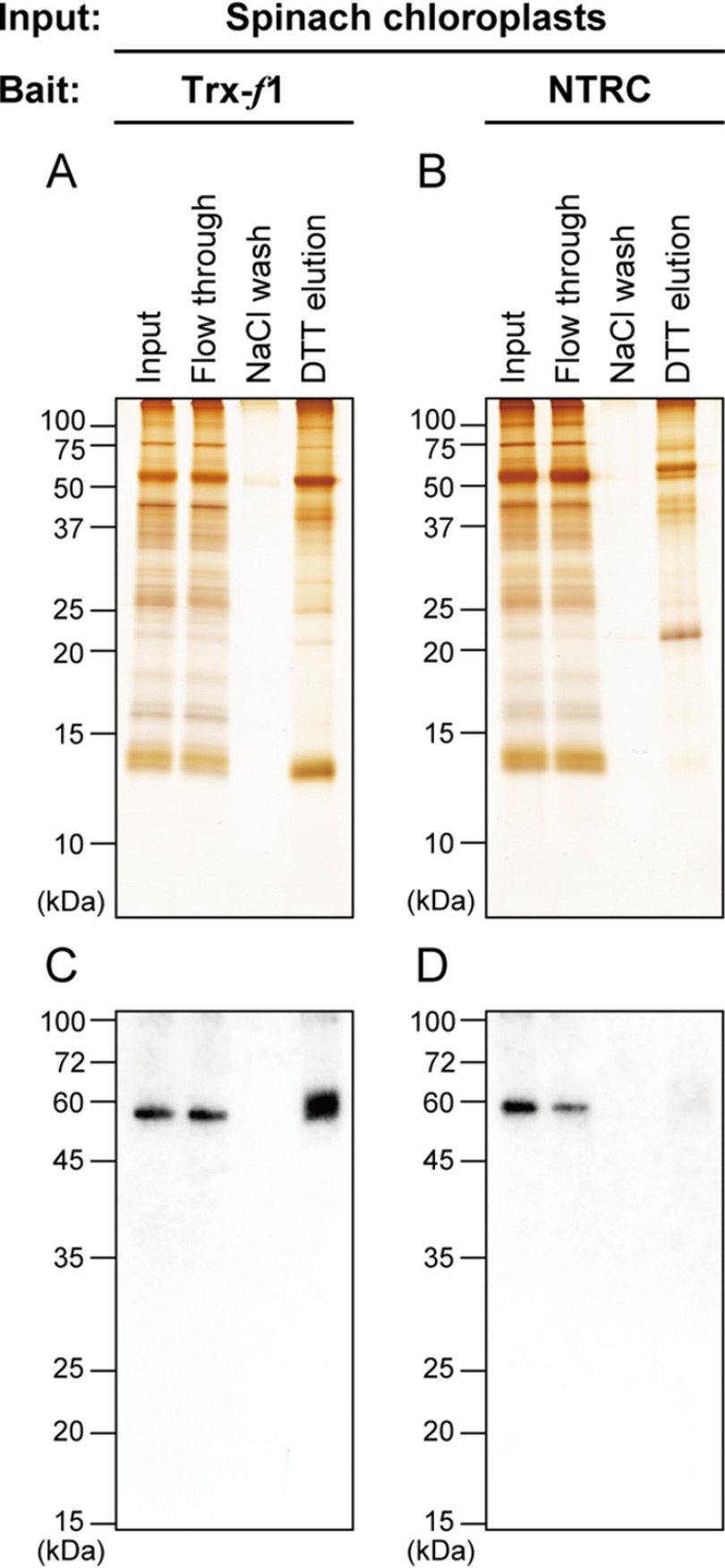
We first investigated whether PGDH has a cross-talk with redox-regulatory factors, such as Trx. For this purpose, we applied the affinity chromatography-based screening method (25). One of Arabidopsis plastidial Trx isoforms, Trx-f1, or Arabidopsis NTRC, each of which was prepared in the form of monocysteinic variant (13), was used as bait in this experiment. Chloroplast soluble proteins extracted from spinach leaves were loaded onto a Trx-f1- or NTRC-immobilized affinity chromatography column. The proteins associated with Trx-f1 or NTRC via the mixed-disulfide bond were eluted by a reducing agent DTT. As shown in Fig. 2A (for Trx-f1) and Fig. 2B (for NTRC), different SDS-PAGE profiles of DTT-eluted proteins were evident between assays using Trx-f1 and NTRC, because of their distinct target selectivity (13). Immunoblotting analyses indicated that PGDH was bound to Trx-f1, but not to NTRC (Fig. 2, C and D). After eluted by DTT, the molecular weight of PGDH seems to be slightly varied (Fig. 2C), which may reflect the change in PGDH redox state. These results suggest that PGDH may be redox-regulated in a Trx-dependent manner.
Figure 2.
Identification of PGDH as a Trx-interacting protein. A and B, affinity chromatography-based screening of Arabidopsis Trx-f1 (A) or NTRC (B) target candidate proteins in chloroplasts. Protein elution profiles were analyzed by SDS-PAGE, followed by silver staining. C and D, validation of PGDH binding to Trx-f1 (C) or NTRC (D). PGDH was detected by immunoblotting analysis using a PGDH antibody.
Arabidopsis PGDH1, but not PGDH2 and PGDH3, is activated by cleavage of intramolecular disulfide bond
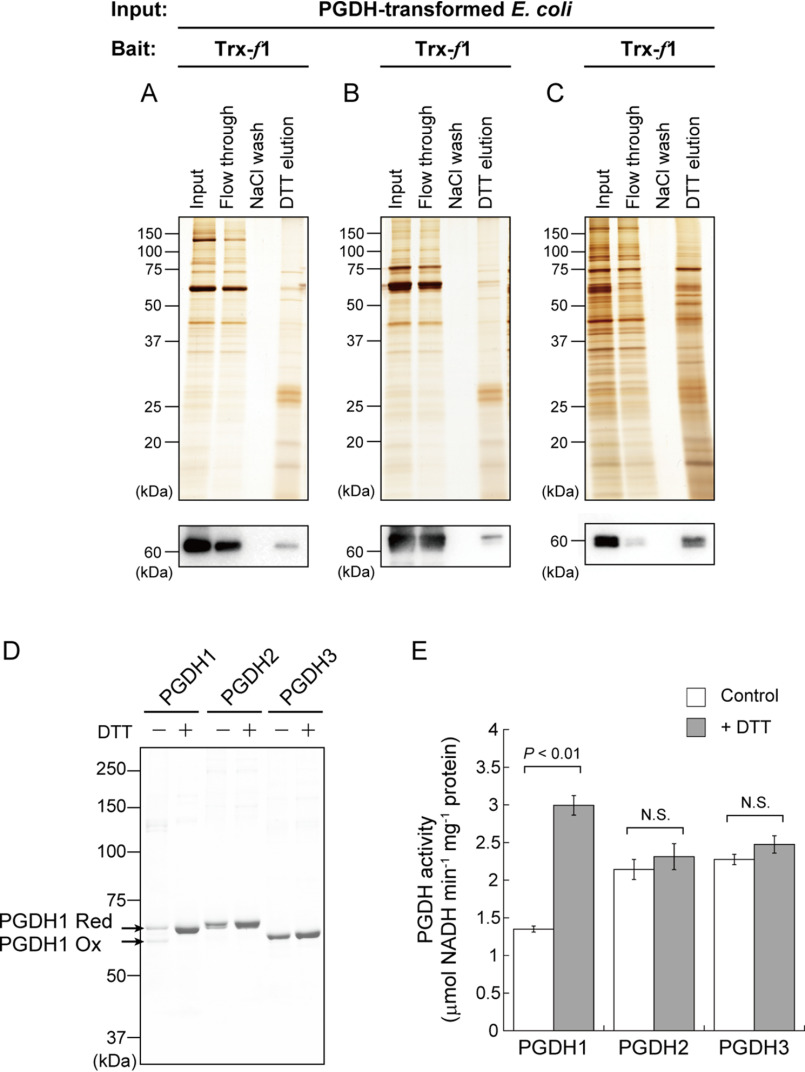
In Arabidopsis thaliana, three nuclear genes encoding PGDH (PGDH1, At4g34200; PGDH2, At1g17745; and PGDH3, At3g19480) are identified (38). All PGDH isoforms were confirmed to be targeted to plastids (39, 41). To test whether each PGDH isoform can bind to Trx, affinity chromatography experiments were conducted on Escherichia coli that expressed PGDH1–3 individually (Fig. 3, A–C). In all cases, PGDH was detected in DTT-eluted fractions. These results give an implication that all of PGDH isoforms in Arabidopsis are redox-regulated as the target of Trx. However, we cannot conclude the possibility for PGDH redox regulation at this stage, because it is known that affinity chromatography-based methods often traps pseudo-target proteins of Trx during the incubation process (43). To verify this issue, it is necessary to clarify (i) Trx-dependent reduction, (ii) redox-dependent change in the activity, and (iii) Cys residues responsible for the redox regulation. Following studies were designed to address these points.
Figure 3.
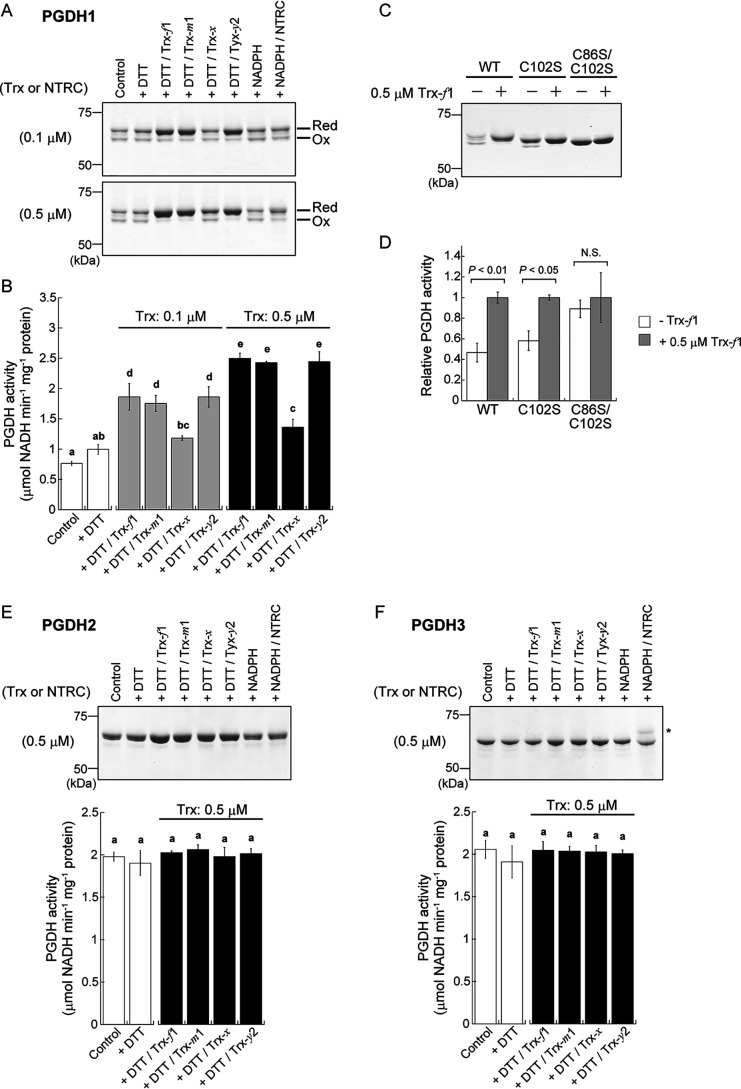
Redox sensitivity of Arabidopsis PGDH. A-C, affinity chromatography-based test for binding of PGDH1 (A), PGDH2 (B), or PGDH3 (C) to Arabidopsis Trx-f1. Proteins extracted from each PGDH-transformed E. coli cells were loaded as input. Protein elution profiles were analyzed by SDS-PAGE, followed by silver staining. PGDH was detected by immunoblotting analysis using a PGDH antibody. D, redox shift assay of PGDH using the thiol-modifying reagent. Each PGDH (1.2 μm) was incubated with or without 10 mm DTT for 15 min. PGDH was then labeled with maleimide-PEG11-biotin and loaded on nonreducing SDS-PAGE. Ox, oxidized form; Red, reduced form. E, enzyme activity measurement of PGDH. Each PGDH (1.2 μm) was incubated with or without 1 mm DTT for 15 min. PGDH activity was then monitored. Data are shown as the mean ± S.D. (n = 4). Statistical analyses were performed using the Student's t test. N.S., not significant.
We prepared PGDH1–3 from Arabidopsis as the purified recombinant proteins (Fig. S1). We examined whether the redox state of PGDH is variable or not (Fig. 3D). Redox shift assays using a thiol-modifying reagent indicated that PGDH1 mainly existed as two different redox states under control (DTT-free) conditions. PGDH1 was converted to a fully reduced state in the presence of DTT. It was thus suggested that PGDH1 can form at least one disulfide bond in the molecule. By contrast, the redox states of PGDH2 and PGDH3 were unaltered by DTT. We then examined the effects of DTT on PGDH enzyme activity (Fig. 3E). PGDH1 was largely activated by DTT, whereas PGDH2 and PGDH3 were not. Taken together, these results suggest that PGDH1 is a redox-sensitive protein whose activity is enhanced upon reduction, whereas PGDH2 and PGDH3 are not.
Cys86 and Cys102 are mainly involved in disulfide bond formation in PGDH1
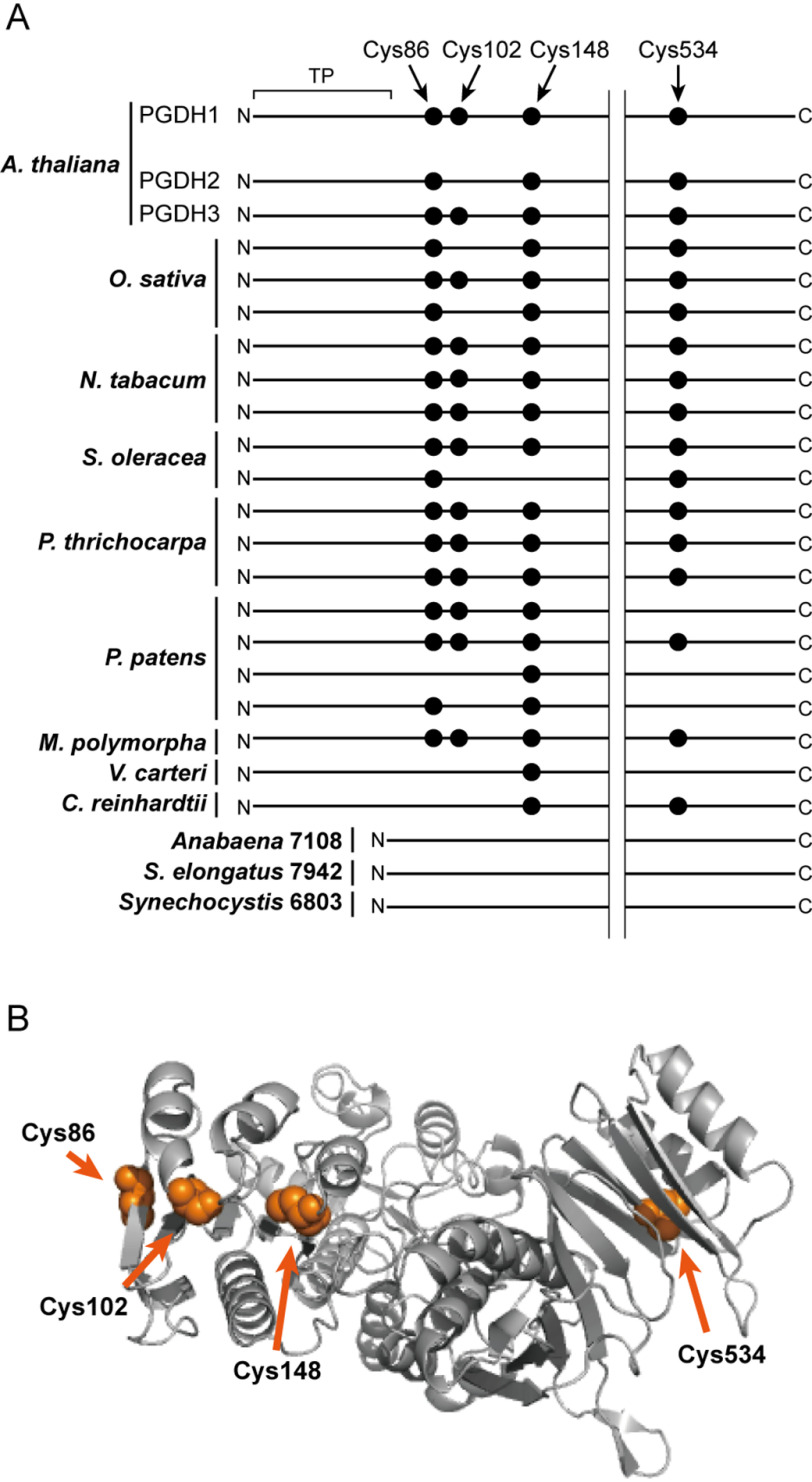
There are four Cys residues in Arabidopsis PGDH1 (Cys86, Cys102, Cys148, and Cys534; Fig. 4A, Fig. S2). Among these, only Cys102 is not found at the corresponding position of redox-insensitive PGDH2. A three-dimensional structure model of PGDH1 showed that Cys102 may be localized in spatial proximity to Cys86 and Cys148 (Fig. 4B, Fig. S3). We thus anticipated that Cys102 forms a disulfide bond with either Cys86 or Cys148.
Figure 4.
Prediction of redox-active Cys residues in Arabidopsis PGDH1. A, simplified representation of Cys localization of PGDH1 and their conservation in photosynthetic organisms. There are four Cys residues in the A. thaliana PGDH1 mature protein region. The presence or absence of these Cys residues was compared with other PGDH. Other isoforms of A. thaliana PGDH (PGDH2 and PGDH3) and PGDH from other photosynthetic organisms, including Oryza sativa, Nicotiana tabacum, Spinacia oleracea, Populus thrichocarpa, Physcomitrella patens, Marchantia polymorpha, Volvox carteri, Chlamydomonas reinhardtii, Anabaena sp. PCC 7108, Synechococcus elongatus sp. PCC 7942, and Synechocystis sp. PCC 6803, were used for the comparison. The full alignment of PGDH amino acid sequences is shown in Fig. S2. TP, transit peptide. B, three-dimensional structure model of Arabidopsis PGDH1. The model was obtained as a homotetrameric form, but one chain is shown. Four Cys residues are shown by the orange spheres.
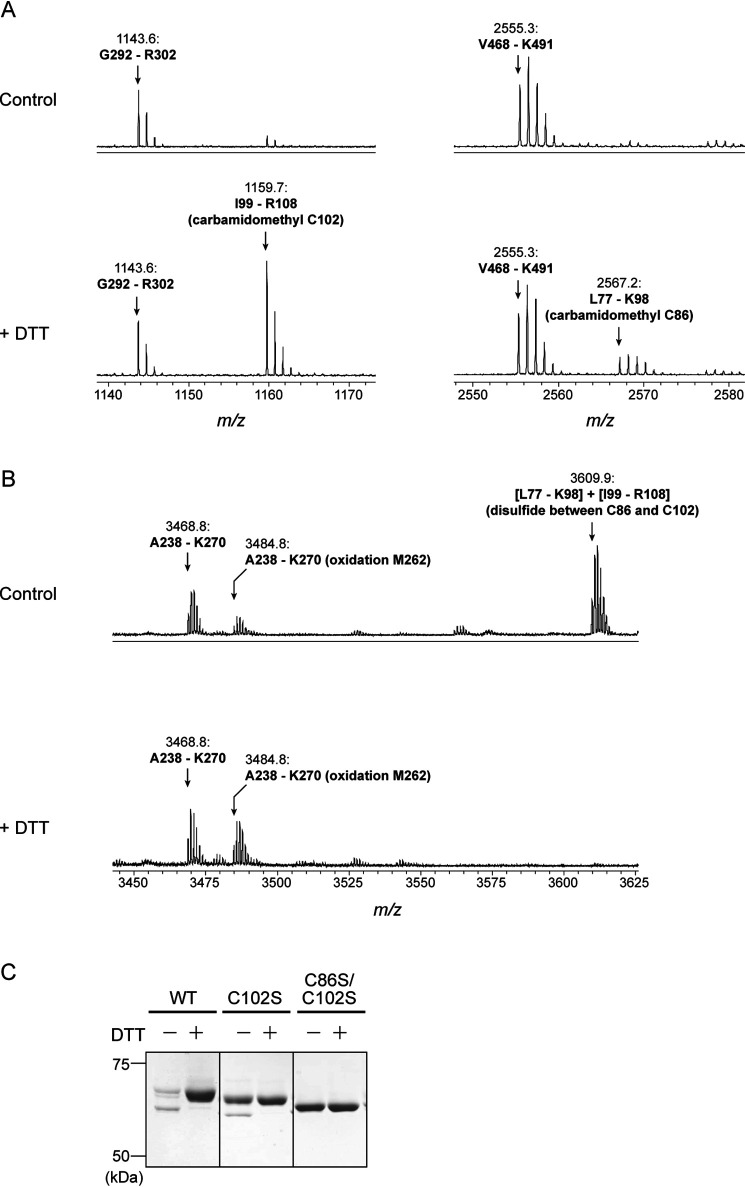
To directly evaluate this expectation, we performed peptide mapping analysis based on the MS. PGDH1 protein was in-gel digested using trypsin, and mass spectra of the resulting peptides were compared between control and DTT treatments. As expected, the overall mass spectra were apparently similar between the control and DTT-treated samples (Fig. S4), but a few differences were observed. Mass peaks of 1159.7 and 2567.2 were more dominant in the DTT-treated sample (Fig. 5A). Data query using the Mascot search engine indicated that these peaks corresponded to a tryptic peptide of Ile99–Arg108 (containing carbamidomethyl Cys102) and Leu77–Lys98 (containing carbamidomethyl Cys86) with a peptide tolerance of <50 ppm. By contrast, a mass peak of 3609.9 is specifically found in the control sample (Fig. 5B). It corresponded to a conjugated peptide of Leu77–Lys98 and Ile99–Arg108 linking with a disulfide bond. These results indicate that Cys86 and Cys102 are involved in disulfide bond formation in PGDH1.
Figure 5.
Identification of redox-active Cys residues in Arabidopsis PGDH1. A and B, peptide mapping analysis based on MS. Before the Cys alkylation and following in-gel digestion with trypsin, the protein sample was incubated in the absence (control) or presence of 10 mm DTT. Overall mass spectra are shown in Fig. S4. C, redox shift assay of PGDH1 WT and Cys-substituted mutants (C102S and C86S/C102S) using the thiol-modifying reagent. Each PGDH1 was incubated with or without 10 mm DTT for 15 min. PGDH was then labeled with the methyl-PEG24-maleimide and loaded on nonreducing SDS-PAGE.
To gain further insight into the regulatory Cys residues in PGDH1, a site-directed mutagenesis analysis was conducted. We generated PGDH1 mutant proteins whose only Cys102 (C102S) or both Cys86 and Cys102 (C86S/C102S) were substituted to Ser (Fig. S1). The C102S mutant still showed DTT-dependent conversion of the redox state, although its conversion range became smaller (Fig. 5C). It is likely that, upon Cys102 mutation, another Cys (possibly Cys148 that lies close to Cys102; Fig. 4B, Fig. S3) forms a disulfide bond with Cys86. Such flexibility in disulfide bond formation is also reported in the fructose-1,6-bisphosphatase (44). By contrast, the C86S/C102S mutant completely lacked the ability of DTT-dependent redox shift. All these data suggest that Cys86 and Cys102 are mainly responsible for PGDH1 redox regulation. Alignment of PGDH from several photosynthetic organisms shows that this Cys pair is largely conserved in land plants, but not in algae or cyanobacteria (Fig. 4A, Fig. S2).
Some Trx subtypes in plastids support PGDH1 reductive activation
We finally studied the involvement of Trx and NTRC in PGDH1 redox regulation (Fig. 6, A–D). Four Trx isoforms (Trx-f1, Trx-m1, Trx-x, and Trx-y2) and NTRC from Arabidopsis were prepared. All Trx isoforms used here were previously shown to be capable of mediating redox regulation with different target selectivity (10, 11). Low concentrations of DTT (0.1 mm) had little impact on the PGDH1 redox state (Fig. 6A) and activity (Fig. 6B). When Trx-f1, Trx-m1, or Trx-y2 was added at 0.1 μm, PGDH1 reduction was promoted (Fig. 6A). Accordingly, the activity of PGDH1 was also enhanced (Fig. 6B). The effects of these Trx isoforms on PGDH1 were more pronounced when each Trx was added at 0.5 μm. By contrast, Trx-x hardly affected the PGDH1 redox state and enzyme activity. In the absence of DTT, all Trx isoforms failed to activate PGDH1 (Fig. S5). Experiments with PGDH1 C102S and C86S/C102S mutants confirmed the principal role of Cys86 and Cys102 for Trx-dependent redox regulation of PGDH1 (Fig. 6, C and D). NTRC was previously shown to efficiently transmit NADPH-derived reducing power to some proteins (13). However, it was not the case with PGDH1; NTRC was unable to promote PGDH1 reduction largely (Fig. 6A). These results suggest that selected types of plastidial Trx are effective in the reductive activation of PGDH1. By contrast, any Trx isoforms affected neither the redox state nor the activity of PGDH2 (Fig. 6E) and PGDH3 (Fig. 6F).
Figure 6.
Involvement of Trx in PGDH redox regulation. A, redox shift assay of PGDH1 using the thiol-modifying reagent. PGDH1 (1.2 μm) was incubated in the absence (control) or presence of 0.1 mm DTT or 0.5 mm NADPH for 30 min. Trx-f1, Trx-m1, Trx-x, Trx-y2, or NTRC were added at 0.1 or 0.5 μm. PGDH1 was then labeled with the maleimide-PEG11-biotin and loaded on nonreducing SDS-PAGE. Ox, oxidized form; Red, reduced form. B, enzyme activity measurement of PGDH1. PGDH1 was reacted as in A and its activity was then monitored. C, redox shift assay of PGDH1 WT and Cys-substituted mutants (C102S and C86S/C102S) using the thiol-modifying reagent. Each PGDH1 (1.2 μm) was incubated with or without 0.5 μm Trx in the presence of 0.1 mm DTT for 30 min. PGDH was then labeled with the methyl-PEG24-maleimide and loaded on nonreducing SDS-PAGE. D, enzyme activity measurement of PGDH1 WT and Cys-substituted mutants. PGDH1 WT and mutants were reacted in C and their activities were then monitored. Each activity is expressed as the relative value. E and F, redox shift assay and enzyme activity measurement of PGDH2 (E) and PGDH3 (F). Reaction conditions were same as A. Trx or NTRC was added at 0.5 μm. An asterisk in F indicates a band of NTRC. Data are shown as the mean ± S.D. (n = 3–4). B, E, and F, statistical analyses were performed using the Tukey-Kramer multiple comparison test. Different letters denote significant differences at p < 0.01. D, statistical analyses were performed using the Student's t test. N.S., not significant.
Discussion
Information on the redox-based regulatory network in plant cells has been increasingly expanding (16–22). Until now, several hundreds of proteins have been proposed to be redox-regulated in plants, based on large-scale proteomic approaches (34, 35). It should be noticed, however, that most of these are still potential, due to the lack of further studies for confirming the validity of redox regulation in detail; this fact largely hampers a consistent understanding of the redox-regulatory system. Using a biochemical procedure, we report here that PGDH is a previously uncharacterized redox-regulated protein.
PGDH has been found as one of the putative Trx-linked proteins by early proteomic studies (27, 30, 36). In accord with this, our sensitive analysis using an antibody indicated that PGDH can interact with Trx-f1 via the mixed disulfide bond (Fig. 2), firmly supporting a possibility for PGDH redox regulation. Indeed, one isoform of Arabidopsis PGDH, PGDH1, showed a clear shift in the redox state, coupled with changes in enzyme activity (Fig. 3, D and E). The Cys residues mainly responsible for the disulfide bond formation in PGDH1 were then determined (Cys86 and Cys102; Fig. 5). Land plants (from liverwort to vascular plants) widely share at least one PGDH isoform containing this Cys pair, whereas algae and cyanobacteria do not (Fig. 4A, Fig. S2). PGDH orthologs in nonphotosynthetic organisms also lack this Cys pair (45). PGDH redox regulation is thus considered as a unique strategy acquired during adaptation to terrestrial environments in photosynthetic organisms. Notably, another enzyme using 3-PGA as a substrate, 3-PGA kinase, is reported to be redox-regulated in algae (46, 47) and cyanobacteria (48). Cys residues essential for 3-PGA kinase redox regulation are not conserved in land plants (47). These findings highlight the changing regulatory modes of 3-PGA metabolism across photosynthetic organisms.
Although our results suggest that Cys86 and Cys102 are key to redox regulation of PGDH1, other factors are likely to affect its regulatory property. Despite having this Cys pair, PGDH3 was found to be redox-insensitive (Fig. 3, D and E, Fig. 6F). It seems possible that some amino acids specific to PGDH3 have an inhibitory effect on disulfide bond formation, but future studies are warranted to elucidate the underlying mechanisms in detail. Determining the PGDH1 structures in the oxidized and reduced forms, followed by the comparison of spatial arrangement of amino acids, will be largely helpful for its elucidation.
We demonstrated that Trx-f, Trx-m, and Trx-y assist the reductive activation of PGDH1 (Fig. 6, A and B). There is little consistent molecular property among these three Trx subtypes. For example, the midpoint redox potential is substantially different (Trx-f1, −321 mV; Trx-m1, −335 mV; and Trx-y2, −295 mV (at pH 7.5)) (10, 11). Recently, we identified the determinant amino acid residues for Trx-f–specific functions (49), but they appear to have little impact on PDGH1 redox regulation. Further studies are needed to gain deeper mechanistic insight into divergent or redundant Trx functions. Nevertheless, it is reasonable to consider that, in contrast to other proteins (e.g. fructose-1,6-bisphosphatase specifically reduced by Trx-f (7, 10)), PGDH1 is redox-regulated with less Trx selectivity.
Our in vitro data raise a new question: how is PGDH redox-regulated in vivo? In Arabidopsis, both PGDH1 and several Trx isoforms were shown to be widely expressed in multiple tissues (39, 41, 50), supporting the idea of their cross-talk in vivo. In photosynthetic tissues, redox-sensitive PGDH is thought to be reductively activated in response to light via the Fd/FTR/Trx pathway. The in vitro PGDH activity is enhanced under alkaline conditions (45, 51), which may be advantageous for PGDH up-regulation in the light (via an increase in stromal pH). On the other hand, physiological studies have proposed that the PGDH-mediated phosphorylated pathway has only a minor role in Ser biosynthesis when Ser is produced via the photorespiratory glycolate pathway (37, 38). Therefore, the extent to which reductive activation of PGDH is physiologically important in illuminated chloroplasts is unclear. Instead, the phosphorylated pathway becomes more important under conditions where the photorespiratory cycle does not function (e.g. under high CO2 conditions or in nonphotosynthetic tissues). The Trx-based redox-regulatory system is assumed to work also in nonphotosynthetic plastids (30), but its mode of action remains elusive. Further studies should be directed to clarify this point and the linkage to PGDH regulation. In this regard, it is worth noting that PGDH was identified as being Trx-linked in germinating Medicago truncatula seeds (36).
In this study, we provide a biochemical insight into PGDH redox regulation. Besides this regulatory way, PGDH activity is allosterically affected by several amino acids (e.g. inhibited by Ser in a feedback manner) (39, 45, 51). Furthermore, PGDH is controlled at a transcriptional level. Namely, gene expression of each PGDH isoform is differentially induced or suppressed by light/dark transitions, high CO2, and salinity stress (39, 41, 52, 53). Taken together, it is likely that PGDH functions are dynamically and sophisticatedly regulated at several levels, determining the rates of the phosphorylated pathway and the resulting Ser biosynthesis. Elucidating its overall regulatory mechanisms and their interplay is of importance as a next step.
Experimental procedures
Preparation of expression plasmids
Total RNA was isolated from A. thaliana as described previously (54) and used as a template for RT-PCR. The gene fragment encoding the predicted mature protein region of PGDH1 (Lys61–Leu603), PGDH2 (Lys82–Leu624), or PGDH3 (Lys46–Leu588) was amplified. The oligonucleotide primers were designed to express objective protein as the His-tagged form at the N-terminal end. The amplified DNA was incorporated into the pET-23d expression vector (Novagen) using the Hot Fusion method (55). The sequences of PGDH expression plasmids were confirmed to be correct by DNA sequencing (3730xl DNA Analyzer; Applied Biosystems). The other expression plasmids (for Trx isoforms and NTRC) were prepared previously (10, 11, 13).
Site-directed mutagenesis
Point mutations in PGDH1 (Cys86 to Ser; Cys102 to Ser) were introduced using the PrimeSTAR Mutagenesis Basal Kit (Takara) according to the manufacturer's instructions. The following oligonucleotide primers were used for site-directed mutagenesis: for Cys86 to Ser, 5′-GTCGACTCTTCGTATAACATGACTCCT-3′ (forward) and 5′-ATACGAAGAGTCGACATTCGCGACATC-3′ (reverse); for Cys102 to Ser, 5′-TCGCTCTCTGACGCCTTGATCGTGAGG-3′ (forward) and 5′-GGCGTCAGAGAGCGAGATCTTAATGTT-3′ (reverse). The mutated codons are underlined.
Protein expression and purification
Each PGDH expression plasmid was transformed into the E. coli Rosetta (DE3) pLysS competent cells. The transformed cells were cultured at 37 °C until A600 reached 0.2–0.3. For PGDH1 and PGDH2, the expression was induced by the addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside, followed by overnight culture at 20 °C. For PGDH3, the expression was slowly induced without adding isopropyl 1-thio-β-d-galactopyranoside, because it rendered most of PGDH3 proteins insoluble. The cells were disrupted by sonication. After centrifugation (125,000 × g for 40 min), the resulting supernatant was used to purify the protein. The His-tagged PGDH protein was purified using a nickel-nitrilotriacetic acid affinity column as described previously (56). Other recombinant proteins (Trx isoforms and NTRC) were prepared as described previously (10, 11, 13). The protein concentration of PGDH was determined by Bradford assay with BSA as standard.
Screening for Trx-f1- or NTRC-interacting proteins
The method of Trx affinity chromatography (25) was applied for screening Trx-f1- or NTRC-interacting proteins from spinach chloroplasts and PGDH-transformed E. coli cells. Arabidopsis Trx-f1 and NTRC were prepared in the form of monocysteinic variants. NTRC has two redox-active Cys pairs (in NTR domain and Trx domain). Here, the pair in Trx domain was mutated into a single Cys. The detailed procedures are available in Refs. 13 and 32. Binding of the PGDH protein was tested using an antibody raised against Arabidopsis PGDH2. This antibody reacted to all of recombinant PGDH1, PGDH2, and PGDH3 proteins, but with different affinities (Fig. S6).
PGDH redox shift assay
For the reducing reaction, PGDH was reacted at 25 °C in a solution containing 50 mm Tris-HCl (pH 7.5) and 50 mm NaCl. Information on protein concentrations, DTT concentrations, or reaction periods are described in each figure.
PGDH redox states were determined by discriminating the thiol status with the use of thiol-modifying reagents described in each figure. These reagents change protein mobility on the SDS-PAGE, allowing the determination of protein redox states with an observable band shift. Following the reducing reaction, proteins were precipitated with 10% (w/v) TCA and then washed with ice-cold acetone. The precipitated proteins were labeled with the thiol-modifying reagents described in each figure for 1 h at room temperature. Proteins were subjected to nonreducing SDS-PAGE and stained with Coomassie Brilliant Blue R-250.
Measurement of PGDH activity
PGDH activity was measured at 25 °C in solution containing 50 mm Tris-HCl (pH 8.8), 600 mm NaCl, 5 mm 3-PGA, 1 mm NAD+, and 60 nm PGDH. Activity was monitored as an increase in absorbance at 340 nm due to NAD+ reduction. A molar extinction coefficient for NADH of 6.2 mm−1 cm−1 was used for calculating the amounts of catalyzed NAD+.
Peptide mapping analysis using MS
After nonreducing SDS-PAGE, a Coomassie Brilliant Blue-stained protein band of PGDH1 was excised from the gel and fully destained with 50 mm NH4HCO3 and 50% (v/v) acetonitrile. For DTT treatment, the gel slice was incubated with 100 mm NH4HCO3 and 10 mm DTT at 56 °C for 1 h. The free thiols were then alkylated using iodoacetamide. After being dried completely, the gel slice was incubated with 50 mm NH4HCO3 containing 20 ng/µl of trypsin (Promega) at 37 °C overnight. The resulting peptides were then extracted from the gel with 0.1% (v/v) TFA and 50–75% (v/v) acetonitrile. The peptide sample was mixed with the matrix solution (α-cyano-4-hydroxycinnamic acid) on a MALDI plate (MTP 384 target plate ground steel BC, Bruker Daltonics). Mass spectra were obtained using MALDI-TOF MS (UltrafleXtreme; Bruker Daltonics).
Modeling of PGDH1 three-dimensional structure
The three-dimensional structure model of PGDH was constructed using the Swiss-Model software (57). The crystal structure of Mycobacterium tuberculosis PGDH (PDB code 3DDN) was used as a template (58).
Statistical analysis
Statistical analyses were performed with Microsoft Excel software for the Student's t test, and SPSS 12.0J software (SPSS Inc.) for the Tukey-Kramer multiple comparison test.
Data availability
All data are contained within the article.
Supplementary Material
Acknowledgments
We thank the Biomaterial Analysis Division, Tokyo Institute of Technology for supporting DNA sequencing analysis and the Suzukakedai Materials Analysis Division, Tokyo Institute of Technology, for supporting MS analysis. This study was supported by Dynamic Alliance for Open Innovation Bridging Human, Environment and Materials.
This article contains supporting information.
Author contributions—K. Y. and K. O. conceptualization; K. Y. investigation; K. Y. writing-original draft; K. Y., K. O., M. Y. H., and T. H. writing-review and editing; M. Y. H. and T. H. supervision.
Funding and additional information—This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 19H03241 (to K. Y.) and 16H06556 (to K. Y. and T. H.), Sumitomo Foundation Grant 180881 (to K. Y.), the Yoshinori Ohsumi Fund for Fundamental Research (to K. Y.), and the Tokyo Tech Challenging Research Award (to K. Y.).
Conflict of interest—The authors declare no conflict of interest with the contents of this article.
- Trx
- thioredoxin
- Fd
- ferredoxin
- FTR
- Fd-Trx reductase
- NTRC
- NADPH-Trx reductase C
- NTR
- NADPH-Trx reductase
- PGDH
- 3-phosphoglycerate dehydrogenase
- 3-PGA
- 3-phosphoglycerate.
References
- 1. Holmgren A. (1985) Thioredoxin. Annu. Rev. Biochem. 54, 237–271 10.1146/annurev.bi.54.070185.001321 [DOI] [PubMed] [Google Scholar]
- 2. Jacquot J. P., Lancelin J. M., and Meyer Y. (1997) Thioredoxins: structure and function in plant cells. New Phytol. 136, 543–570 10.1046/j.1469-8137.1997.00784.x [DOI] [PubMed] [Google Scholar]
- 3. Buchanan B. B. (1980) Role of light in the regulation of chloroplast enzymes. Annu. Rev. Plant Physiol. 31, 341–374 10.1146/annurev.pp.31.060180.002013 [DOI] [Google Scholar]
- 4. Buchanan B. B., Schürmann P., Wolosiuk R. A., and Jacquot J. P. (2002) The ferredoxin/thioredoxin system: from discovery to molecular structures and beyond. Photosynth. Res. 73, 215–222 [DOI] [PubMed] [Google Scholar]
- 5. Lemaire S. D., Michelet L., Zaffagnini M., Massot V., and Issakidis-Bourguet E. (2007) Thioredoxins in chloroplasts. Curr. Genet. 51, 343–365 10.1007/s00294-007-0128-z [DOI] [PubMed] [Google Scholar]
- 6. Serrato A. J., Fernandez-Trijueque J., Barajas-Lopez J. D., Chueca A., and Sahrawy M. (2013) Plastid thioredoxins: a “one-for-all” redox-signaling system in plants. Front. Plant Sci. 4, 463 10.3389/fpls.2013.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collin V., Issakidis-Bourguet E., Marchand C., Hirasawa M., Lancelin J. M., Knaff D. B., and Miginiac-Maslow M. (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J. Biol. Chem. 278, 23747–23752 10.1074/jbc.M302077200 [DOI] [PubMed] [Google Scholar]
- 8. Michelet L., Zaffagnini M., Marchand C., Collin V., Decottignies P., Tsan P., Lancelin J. M., Trost P., Miginiac-Maslow M., Noctor G., and Lemaire S. D. (2005) Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proc. Natl. Acad. Sci. U.S.A. 102, 16478–16483 10.1073/pnas.0507498102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Toivola J., Nikkanen L., Dahlstrom K. M., Salminen T. A., Lepisto A., Vignols H. F., and Rintamaki E. (2013) Overexpression of chloroplast NADPH-dependent thioredoxin reductase in Arabidopsis enhances leaf growth and elucidates in vivo function of reductase and thioredoxin domains. Front. Plant Sci. 4, 389 10.3389/fpls.2013.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoshida K., Hara S., and Hisabori T. (2015) Thioredoxin selectivity for thiol-based redox regulation of target proteins in chloroplasts. J. Biol. Chem. 290, 14278–14288 10.1074/jbc.M115.647545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshida K., and Hisabori T. (2017) Distinct electron transfer from ferredoxin-thioredoxin reductase to multiple thioredoxin isoforms in chloroplasts. Biochem. J. 474, 1347–1360 10.1042/BCJ20161089 [DOI] [PubMed] [Google Scholar]
- 12. Serrato A. J., Pérez-Ruiz J. M., Spinola M. C., and Cejudo F. J. (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J. Biol. Chem. 279, 43821–43827 10.1074/jbc.M404696200 [DOI] [PubMed] [Google Scholar]
- 13. Yoshida K., and Hisabori T. (2016) Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc. Natl. Acad. Sci. U.S.A. 113, E3967–E3976 10.1073/pnas.1604101113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pérez-Ruiz J. M., Naranjo B., Ojeda V., Guinea M., and Cejudo F. J. (2017) NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proc. Natl. Acad. Sci. U.S.A. 114, 12069–12074 10.1073/pnas.1706003114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nikkanen L., Toivola J., Trotta A., Diaz M. G., Tikkanen M., Aro E. M., and Rintamäki E. (2018) Regulation of cyclic electron flow by chloroplast NADPH-dependent thioredoxin system. Plant Direct 2, e00093 10.1002/pld3.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geigenberger P., Thormählen I., Daloso D. M., and Fernie A. R. (2017) The unprecedented versatility of the plant thioredoxin system. Trends Plant Sci 22, 249–262 10.1016/j.tplants.2016.12.008 [DOI] [PubMed] [Google Scholar]
- 17. Gütle D. D., Roret T., Hecker A., Reski R., and Jacquot J. P. (2017) Dithiol disulfide exchange in redox regulation of chloroplast enzymes in response to evolutionary and structural constraints. Plant Sci. 255, 1–11 10.1016/j.plantsci.2016.11.003 [DOI] [PubMed] [Google Scholar]
- 18. Knuesting J., and Scheibe R. (2018) Small molecules govern thiol redox switches. Trends Plant Sci. 23, 769–782 10.1016/j.tplants.2018.06.007 [DOI] [PubMed] [Google Scholar]
- 19. Cejudo F. J., Ojeda V., Delgado-Requerey V., Gonzalez M., and Pérez-Ruiz J. M. (2019) Chloroplast redox regulatory mechanisms in plant adaptation to light and darkness. Front. Plant Sci. 10, 380 10.3389/fpls.2019.00380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nikkanen L., and Rintamäki E. (2019) Chloroplast thioredoxin systems dynamically regulate photosynthesis in plants. Biochem. J. 476, 1159–1172 10.1042/BCJ20180707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoshida K., Yokochi Y., and Hisabori T. (2019) New light on chloroplast redox regulation: molecular mechanism of protein thiol oxidation. Front. Plant Sci. 10, 1534 10.3389/fpls.2019.01534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zaffagnini M., Fermani S., Marchand C. H., Costa A., Sparla F., Rouhier N., Geigenberger P., Lemaire S. D., and Trost P. (2019) Redox homeostasis in photosynthetic organisms: novel and established thiol-based molecular mechanisms. Antioxid. Redox Signal. 31, 155–210 10.1089/ars.2018.7617 [DOI] [PubMed] [Google Scholar]
- 23. Hisabori T., Sunamura E., Kim Y., and Konno H. (2013) The chloroplast ATP synthase features the characteristic redox regulation machinery. Antioxid. Redox Signal. 19, 1846–1854 10.1089/ars.2012.5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Michelet L., Zaffagnini M., Morisse S., Sparla F., Perez-Perez M. E., Francia F., Danon A., Marchand C. H., Fermani S., Trost P., and Lemaire S. D. (2013) Redox regulation of the Calvin-Benson cycle: something old, something new. Front. Plant Sci. 4, 470 10.3389/fpls.2013.00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Motohashi K., Kondoh A., Stumpp M. T., and Hisabori T. (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 98, 11224–11229 10.1073/pnas.191282098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yano H., Wong J. H., Lee Y. M., Cho M. J., and Buchanan B. B. (2001) A strategy for the identification of proteins targeted by thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 98, 4794–4799 10.1073/pnas.071041998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Balmer Y., Koller A., del Val G., Manieri W., Schürmann P., and Buchanan B. B. (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. U.S.A. 100, 370–375 10.1073/pnas.232703799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marchand C., Le Maréchal P., Meyer Y., Miginiac-Maslow M., Issakidis-Bourguet E., and Decottignies P. (2004) New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics 4, 2696–2706 10.1002/pmic.200400805 [DOI] [PubMed] [Google Scholar]
- 29. Marchand C., Le Maréchal P., Meyer Y., and Decottignies P. (2006) Comparative proteomic approaches for the isolation of proteins interacting with thioredoxin. Proteomics 6, 6528–6537 10.1002/pmic.200600443 [DOI] [PubMed] [Google Scholar]
- 30. Balmer Y., Vensel W. H., Cai N., Manieri W., Schürmann P., Hurkman W. J., and Buchanan B. B. (2006) A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc. Natl. Acad. Sci. U.S.A. 103, 2988–2993 10.1073/pnas.0511040103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Balmer Y., Vensel W. H., Tanaka C. K., Hurkman W. J., Gelhaye E., Rouhier N., Jacquot J. P., Manieri W., Schürmann P., Droux M., and Buchanan B. B. (2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 101, 2642–2647 10.1073/pnas.0308583101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoshida K., Noguchi K., Motohashi K., and Hisabori T. (2013) Systematic exploration of thioredoxin target proteins in plant mitochondria. Plant Cell Physiol. 54, 875–892 10.1093/pcp/pct037 [DOI] [PubMed] [Google Scholar]
- 33. Yamazaki D., Motohashi K., Kasama T., Hara Y., and Hisabori T. (2004) Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol. 45, 18–27 10.1093/pcp/pch019 [DOI] [PubMed] [Google Scholar]
- 34. Lindahl M., and Kieselbach T. (2009) Disulfide proteomes and interactions with thioredoxin on the track toward understanding redox regulation in chloroplasts and cyanobacteria. J. Proteomics 72, 416–438 10.1016/j.jprot.2009.01.003 [DOI] [PubMed] [Google Scholar]
- 35. Montrichard F., Alkhalfioui F., Yano H., Vensel W. H., Hurkman W. J., and Buchanan B. B. (2009) Thioredoxin targets in plants: the first 30 years. J. Proteomics 72, 452–474 10.1016/j.jprot.2008.12.002 [DOI] [PubMed] [Google Scholar]
- 36. Alkhalfioui F., Renard M., Vensel W. H., Wong J., Tanaka C. K., Hurkman W. J., Buchanan B. B., and Montrichard F. (2007) Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiol. 144, 1559–1579 10.1104/pp.107.098103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ros R., Cascales-Miñana B., Segura J., Anoman A. D., Toujani W., Flores-Tornero M., Rosa-Tellez S., and Muñoz-Bertomeu J. (2013) Serine biosynthesis by photorespiratory and non-photorespiratory pathways: an interesting interplay with unknown regulatory networks. Plant Biol. 15, 707–712 10.1111/j.1438-8677.2012.00682.x [DOI] [PubMed] [Google Scholar]
- 38. Ros R., Muñoz-Bertomeu J., and Krueger S. (2014) Serine in plants: biosynthesis, metabolism, and functions. Trends Plant Sci. 19, 564–569 10.1016/j.tplants.2014.06.003 [DOI] [PubMed] [Google Scholar]
- 39. Benstein R. M., Ludewig K., Wulfert S., Wittek S., Gigolashvili T., Frerigmann H., Gierth M., Flügge U. I., and Krueger S. (2013) Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25, 5011–5029 10.1105/tpc.113.118992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cascales-Miñana B., Muñoz-Bertomeu J., Flores-Tornero M., Anoman A. D., Pertusa J., Alaiz M., Osorio S., Fernie A. R., Segura J., and Ros R. (2013) The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell 25, 2084–2101 10.1105/tpc.113.112359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Toujani W., Muñoz-Bertomeu J., Flores-Tornero M., Rosa-Tellez S., Anoman A. D., Alseekh S., Fernie A. R., and Ros R. (2013) Functional characterization of the plastidial 3-phosphoglycerate dehydrogenase family in Arabidopsis. Plant Physiol. 163, 1164–1178 10.1104/pp.113.226720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grant G. A. (2018) D-3-Phosphoglycerate dehydrogenase. Front. Mol. Biosci. 5, 110 10.3389/fmolb.2018.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hisabori T., Hara S., Fujii T., Yamazaki D., Hosoya-Matsuda N., and Motohashi K. (2005) Thioredoxin affinity chromatography: a useful method for further understanding the thioredoxin network. J. Exp. Bot. 56, 1463–1468 10.1093/jxb/eri170 [DOI] [PubMed] [Google Scholar]
- 44. Jacquot J. P., Lopez-Jaramillo J., Miginiac-Maslow M., Lemaire S., Cherfils J., Chueca A., and Lopez-Gorge J. (1997) Cysteine-153 is required for redox regulation of pea chloroplast fructose-1,6-bisphosphatase. FEBS Lett. 401, 143–147 10.1016/S0014-5793(96)01459-7 [DOI] [PubMed] [Google Scholar]
- 45. Okamura E., and Hirai M. Y. (2017) Novel regulatory mechanism of serine biosynthesis associated with 3-phosphoglycerate dehydrogenase in Arabidopsis thaliana. Sci. Rep. 7, 3533 10.1038/s41598-017-03807-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bosco M. B., Aleanzi M. C., and Iglesias A. A. (2012) Plastidic phosphoglycerate kinase from Phaeodactylum tricornutum: on the critical role of cysteine residues for the enzyme function. Protist 163, 188–203 10.1016/j.protis.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 47. Morisse S., Michelet L., Bedhomme M., Marchand C. H., Calvaresi M., Trost P., Fermani S., Zaffagnini M., and Lemaire S. D. (2014) Thioredoxin-dependent redox regulation of chloroplastic phosphoglycerate kinase from Chlamydomonas reinhardtii. J. Biol. Chem. 289, 30012–30024 10.1074/jbc.M114.597997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tsukamoto Y., Fukushima Y., Hara S., and Hisabori T. (2013) Redox control of the activity of phosphoglycerate kinase in Synechocystis sp. PCC6803. Plant Cell Physiol. 54, 484–491 10.1093/pcp/pct002 [DOI] [PubMed] [Google Scholar]
- 49. Yokochi Y., Sugiura K., Takemura K., Yoshida K., Hara S., Wakabayashi K. I., Kitao A., and Hisabori T. (2019) Impact of key residues within chloroplast thioredoxin-f on recognition for reduction and oxidation of target proteins. J. Biol. Chem. 294, 17437–17450 10.1074/jbc.RA119.010401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Belin C., Bashandy T., Cela J., Delorme-Hinoux V., Riondet C., and Reichheld J. P. (2015) A comprehensive study of thiol reduction gene expression under stress conditions in Arabidopsis thaliana. Plant Cell Environ. 38, 299–314 10.1111/pce.12276 [DOI] [PubMed] [Google Scholar]
- 51. Akashi H., Okamura E., Nishihama R., Kohchi T., and Hirai M. Y. (2018) Identification and biochemical characterization of the serine biosynthetic enzyme 3-phosphoglycerate dehydrogenase in Marchantia polymorpha. Front. Plant Sci. 9, 956 10.3389/fpls.2018.00956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ho C. L., Noji M., Saito M., and Saito K. (1999) Regulation of serine biosynthesis in Arabidopsis: crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J. Biol. Chem. 274, 397–402 10.1074/jbc.274.1.397 [DOI] [PubMed] [Google Scholar]
- 53. Rosa-Telléz S., Anoman A. D., Alcántara-Enguidanos A., Garza-Aguirre R. A., Alseekh S., and Ros R. (2020) PGDH family genes differentially affect Arabidopsis tolerance to salt stress. Plant Sci. 290, 110284 10.1016/j.plantsci.2019.110284 [DOI] [PubMed] [Google Scholar]
- 54. Yoshida K., and Noguchi K. (2009) Differential gene expression profiles of the mitochondrial respiratory components in illuminated Arabidopsis leaves. Plant Cell Physiol. 50, 1449–1462 10.1093/pcp/pcp090 [DOI] [PubMed] [Google Scholar]
- 55. Fu C., Donovan W. P., Shikapwashya-Hasser O., Ye X., and Cole R. H. (2014) Hot Fusion: an efficient method to clone multiple DNA fragments as well as inverted repeats without ligase. PLoS ONE 9, e115318 10.1371/journal.pone.0115318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yoshida K., Matsuoka Y., Hara S., Konno H., and Hisabori T. (2014) Distinct redox behaviors of chloroplast thiol enzymes and their relationships with photosynthetic electron transport in Arabidopsis thaliana. Plant Cell Physiol. 55, 1415–1425 10.1093/pcp/pcu066 [DOI] [PubMed] [Google Scholar]
- 57. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22, 195–201 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 58. Dey S., Burton R. L., Grant G. A., and Sacchettini J. C. (2008) Structural analysis of substrate and effector binding in Mycobacterium tuberculosis d-3-phosphoglycerate dehydrogenase. Biochemistry 47, 8271–8282 10.1021/bi800212b [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.