Abstract
Plant diseases caused by pathogens and pests are a constant threat to global food security. Direct crop losses and the measures used to control disease (e.g. application of pesticides) have significant agricultural, economic, and societal impacts. Therefore, it is essential that we understand the molecular mechanisms of the plant immune system, a system that allows plants to resist attack from a wide variety of organisms ranging from viruses to insects. Here, we provide a roadmap to plant immunity, with a focus on cell-surface and intracellular immune receptors. We describe how these receptors perceive signatures of pathogens and pests and initiate immune pathways. We merge existing concepts with new insights gained from recent breakthroughs on the structure and function of plant immune receptors, which have generated a shift in our understanding of cell-surface and intracellular immunity and the interplay between the two. Finally, we use our current understanding of plant immunity as context to discuss the potential of engineering the plant immune system with the aim of bolstering plant defenses against disease.
Keywords: plant immunity, cell-surface immunity, intracellular immunity, resistance engineering, effectors, nucleotide-binding leucine-rich repeat receptors (NLRs), receptor-like kinases (RLKs), receptor-like proteins (RLPs), plant defense, plant biochemistry, Nod-like receptor (NLR), cell surface receptor, cellular immune response
Plants suffer from disease. Their ability to respond to infection by microbial pathogens and pests is essential for survival. In agriculture, plant disease leads to loss in crop yield and can have devastating effects on both subsistence/small-holder and industrialized farming (1–3), with subsequent impact on food supply chains and prices. Plant diseases have also shaped our world, with perhaps the best-known example being the Irish potato famine in the mid-1800s, where potato late blight disease (caused by the filamentous plant pathogen Phytophthora infestans) contributed to mass emigration from Ireland (4).
As a rich source of nutrients, plants are the target of microbial pathogens and pests, including viruses, bacteria, filamentous pathogens (fungi and oomycetes), nematodes, and insects to complete their life cycle (5–8). Estimates of the impact of pre-harvest yield loss in crops due to disease vary, but at least 30% of global agricultural production is claimed annually (1). This can increase to 100% in localized outbreaks and represents a major contributor to food insecurity. In agriculture, plant diseases are largely controlled by chemicals, but this is unsustainable in the long-term due to environmental concerns and the necessity to rethink agricultural practices more generally in light of the climate emergency. Genetic forms of disease resistance offer the potential for environmentally friendly, low-input, sustainable agriculture (9). Over the last 25 years, remarkable progress has been made in our understanding of the molecular basis of plant disease resistance mechanisms. Plant immune receptors, encoded by resistance or “R” genes have been cloned and characterized and shown to be the genetic basis of disease resistance phenotypes used by plant breeders for >100 years. Recent studies have extended our knowledge to reveal our first insights into the structural basis of plant immune receptor function (10–19).
The immune system of plants shares similarities with the innate immune system of animals (20–22). But as plants lack an adaptive immune system, they rely solely on innate immunity to recognize microbial pathogens and pests. Conceptually, plant immunity can be divided into cell-surface and intracellular immunity (23). A full list of the structurally characterized immune receptors and associated ligands can be found in Table 1. Cell-surface immune receptors detect common signatures of pathogens or pests outside the host cell via extracellular domains (ECDs) and initiate cellular responses to resist infection via their intracellular kinase domains (KDs) (39). A subset of cell-surface immune receptors sense damaged “self” as a surrogate for the presence of pathogens or pests (15). Intracellular immune receptors detect signatures of adapted pathogens or pests (40). Typically, these signatures are translocated proteins known as “effectors,” which are delivered inside cells to modulate host physiology to promote colonization and proliferation (41, 42) (Fig. 1). Activation of intracellular immunity is generally considered a more robust response and can be associated with localized cell death that constrains the spread of infection. Although often presented as distinct signaling pathways, insights into how cell-surface and intracellular immune pathways in plants overlap and work synergistically to resist infection have recently begun to emerge (43, 44).
Table 1.
Structures of plant immune receptors or their domains covered in this review
| Receptor | Type: Cell-surface | Plant host | Ligand | Ligand type | Co-receptor | PDB code | References |
|---|---|---|---|---|---|---|---|
| FLS2 | LRR-RLK | Arabidopsis thaliana | flg22 | MAMP | BAK1 | 4MN8 | 12 |
| PEPR1 | LRR-RLK | A. thaliana | Atpep | DAMP | BAK1 | 5GR8 | 13 |
| CERK1 | LysM-RLK | A. thaliana | PGN | MAMP | LYM3/1 | 4EBY | 10 |
| SOBIR1 | LRR-RLK | A. thaliana | NAa | NA | LRR-RLP, BAK1 | 6R1H | 19 |
| BIR3 | Pseudokinase | A. thaliana | NA | NA | BRI1/S.E.RK1 | 6FG8 | 24 |
| BIK1 | RLCK | A. thaliana | NA | NA | BAK1, FLS2 | 5TOS | 25 |
| CEBiP | LysM-RLP | Oryza sativa | Chitin | MAMP | OsCERK1 | 5JCD, 5JCE | 26 |
| Receptor | Type: Intracellular | Plant host | Ligand(s) | Ligand type | Co-receptor | PDB code | References |
| MLA10 CC | CC-NLR | Hordeum vulgare | NA | NA | NA | 3QFL, 5T1Y | 27, 28 |
| Pikp-1 HMA | CC-NLR | O. sativa | AVR-PikD, AVR-PikE, AVR-PikA, AVR-Pia | MAX effector | Pikp-2 | 5A6W, 5A6P, 6G11, 6G10, 6Q76 | 11, 29, 30 |
| Pikm-1 HMA | CC-NLR | O. sativa | AVR-PikE, AVR-PikA, AVR-PikD | MAX effector | Pikm-2 | 6FUB, 6FUD, 6FU9 | 29 |
| Pia HMA | CC-NLR | O. sativa | Avr1-CO39 | MAX effector | RGA4 | 5ZNG, 5ZNE | 31 |
| RRS1 WRKY | TIR-NLR | A. thaliana | PopP2 | T3SE | RPS4 | 5W3X | 17 |
| ZAR1 | CC-NLR | A. thaliana | Avr-AC | T3SE | RKS1 | 6J5T, 6J6I, 6J5W, 6J5V | 14, 15 |
| RPS4 TIR | TIR-NLR | A. thaliana | NA | NA | RRS1 | 4C6T, 4C6R, | 16 |
| RRS1 TIR | TIR-NLR | A. thaliana | NA | NA | RPS4 | 4C6T,4C6S | 16 |
| SNC1 TIR | TIR-NLR | A. thaliana | NA | NA | NA | 5TEC | 32 |
| SNC1 TIR | TIR-NLR | A. thaliana | NA | NA | NA | 5H3C | 33 |
| Sr33 CC | CC-NLR | Aegilops tauschii | NA | NA | NA | 2NCG | 28 |
| RPP1 TIR | TIR-NLR | A. thaliana | NA | NA | NA | 5TEB | 32 |
| NRC1 NB-ARC | TIR-NLR | Solanum lycopersicum | NA | NA | NA | 6S2P | 34 |
| RUN1 TIR | TIR-NLR | Vitis rotundifolia | NA | NA | NA | 60OW | 35 |
| Rx CC | CC-NLR | Solanum tuberosum | NA | NA | RanGAP2 | 4M70 | 18 |
| RPV1 TIR | TIR-NLR | Vitis rotundifolia | NA | NA | NA | 5KU7 | 36 |
| L6 TIR | TIR-NLR | Linum usitatissimumm | NA | NA | NA | 3OZI | 37 |
| Pto | Kinase | Solanum pimpinellifolium | AvrPtoB | T3SE | Prf | 3HGK | 38 |
aNA, not applicable.
Figure 1.
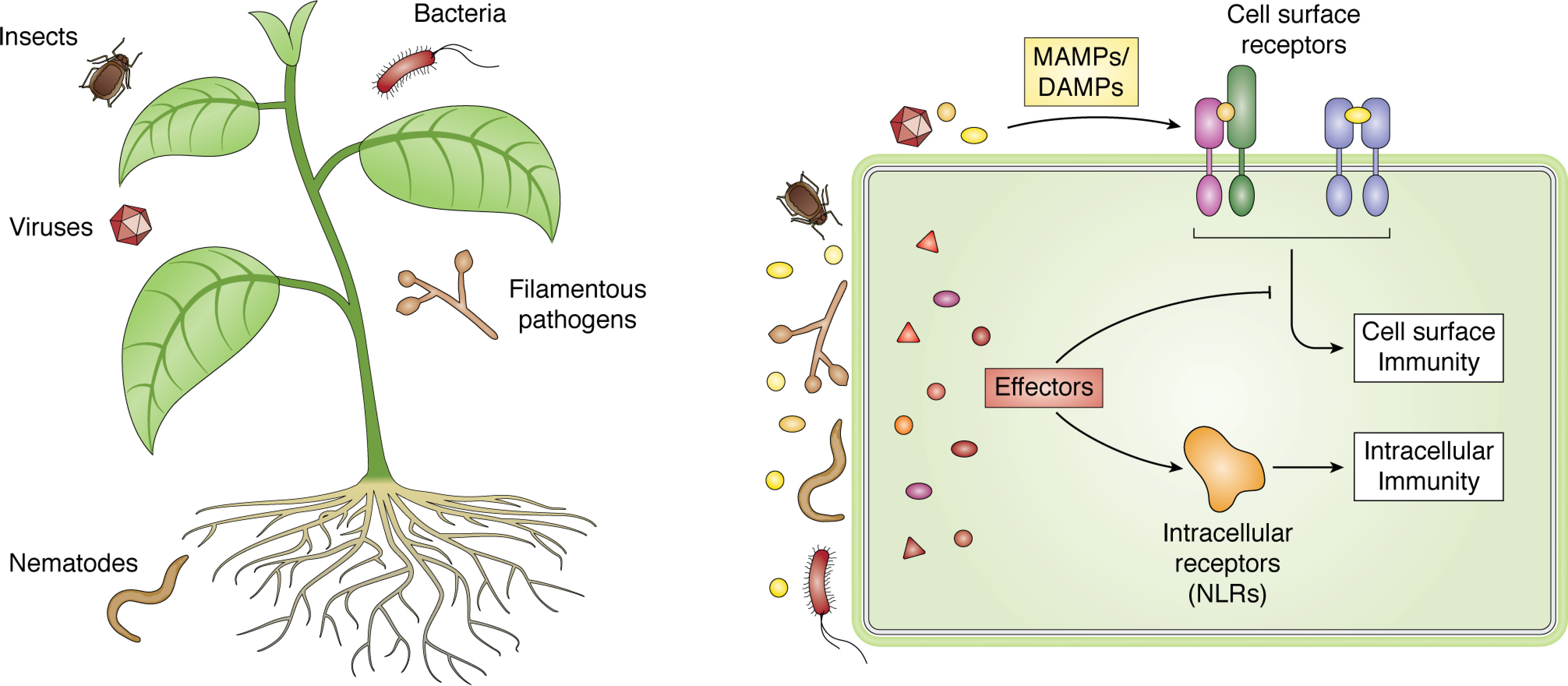
Plant immunity at a glance. Left, plants are the target of a variety of pathogens and pests that cause disease, via both their above-ground and underground structures. Right, pathogens/pests shed MAMPs or generate DAMPs that can be received by receptors to initiate cell-surface immunity. Pathogens/pests can deliver effectors to the outside (not shown here for simplicity) or inside of cells, where they can act on host systems to their benefit, including the suppression of signaling pathways downstream of cell-surface receptors. Effectors or their activities can be sensed by intracellular immune receptors (NLRs) to initiate intracellular immunity.
There are many excellent reviews covering plant immunity and its subversion by microbial pathogens and pests published over the last ∼15 years (21, 39, 45–53). Here, as part of this JBC “Plants in the Real World” thematic series, we provide an up-to-date overview of the general concepts of plant disease resistance mechanisms, with a focus on plant immune receptor function at the molecular level. We detail how these receptors perceive pathogen signatures at the cell surface and inside host cells and how this perception is translated into an immune response. This review summarizes the general concepts of plant immunity before providing in-depth analyses of the more recent breakthroughs that have greatly expanded our understanding of plant immune receptor function. Finally, in the context of current knowledge, we discuss how plant immune receptors could be engineered to deliver novel disease resistance properties to benefit global food security.
Effectors: Master manipulators of plant cells to promote infection
To best understand the interplay between the pathogens/pests and the plant immune system, we must first understand effectors and their role in promoting virulence. In the broadest definition, effectors are molecules used by a diverse array of organisms (including microbes, plants, and animals) to modulate the activity of another organism. In this review, we use the term “effectors” to define protein molecules secreted by microbial pathogens and pests to promote colonization of their plant hosts (53). These effectors can be delivered to the extracellular space or deployed to the inside of host cells.
Effector genes exist as large repertoires within pathogen genomes. They are among the most rapidly evolving genes in plant pathogens and can display high rates of nonsynonymous over synonymous mutations (54, 55). Selection for evasion of perception by the plant immune system is a major driver for adaptation, along with selection for favorable alleles that give a fitness benefit (56). Due to their sequence diversity, it is frequently challenging to identify effectors in pathogen/pest genomes or proteomes, although many bioinformatic tools exist to establish putative effector catalogues (57). Functional annotation of effectors is equally challenging. Whereas some effectors are enzymes, whose putative activity can be identified from sequence or structural analysis, many do not show sequence or structural homologies to help define function (49, 58). This often necessitates significant investment in research of a single protein to establish the molecular basis of activity (42). Such research will frequently address the host cell target of an effector, as this is often key to understanding its role in virulence. Some effectors converge on “hubs,” key components of host cell pathways, to modulate their function (59, 60). Such pathways include suppression of defense responses (Fig. 1) and redirection of host metabolism.
Effectors are also an Achilles' heel for the pathogen/pest. As signatures of non-self, they can be perceived by plant immune receptors at both the cell surface and inside cells. Intracellular perception of effectors or their activities is mediated and transduced by NLRs, as described elsewhere in this review.
Cell-surface immunity
Two major components of cell-surface immunity in plants are membrane-localized receptor-like kinases (RLKs) and receptor-like proteins (RLPs) that detect signatures of non-self as signs of infection (45). RLKs/RLPs also have other roles in plants, regulating self-incompatibility, growth and development, reproduction, response to abiotic stress, and symbiosis (45, 61–63). Also known as pattern recognition receptors (PRRs), cell-surface immune receptors monitor the extracellular environment for pathogen/pest invasion patterns (ligands known as MAMPs (microbial-associated molecular patterns) or DAMPs (damage-associated molecular patterns)) (64, 65). Frequently, ligand-sensing cell-surface receptors require co-receptors to transduce perception of non-self into a response (66, 67). Although proteinaceous receptors represent the major players in cell-surface immunity of plants, recent studies have highlighted an emerging role of membrane lipids in sensing infection (50).
Irrespective of their origin, invasion patterns recognized by cell-surface immune receptors tend to be evolutionarily constrained ligands derived from components essential to the fitness of the pathogen/pest. These essential components range from cell wall constituents or subunits of bacterial flagellin, to molecules secreted into the apoplast, to secreted proteins intended for the host cytosol (39). These specific ligands are perceived by cell-surface receptors at nanomolar concentrations and initiate signaling cascades, including production of reactive oxygen species (ROS), cytosolic Ca2+ bursts, activation of MAPKs, and changes in expression of various defense-related genes (64, 67, 68). Generally, cell-surface immune responses are considered less volatile when compared with intracellular immunity and do not result in host cell death to restrict infection. However, they constitute an effective host strategy against infection, leading to broad-spectrum resistance (69). This review focuses on the mechanisms of immune activation rather than the downstream effects of extracellular and intracellular immunity; for readers interested in the physiological effects of immune activation, we recommend relevant reviews (70–72).
Signaling cascades downstream of cell-surface immune receptors are major targets of pathogen/pest effector proteins, which interfere with these processes to benefit infection. It is also worth noting that many MAMPs are shared between pathogens and mutualistic microbes (62, 73), and as such it is important to understand how plants use extracellular immune receptors to distinguish between pathogens/pests and mutualists. In this review, we cover MAMP recognition from a pathogen/pest-detection perspective and would direct readers interested in plant-mutualist interaction to relevant reviews (62, 73).
Structural and functional diversity of ligand recognition by cell-surface receptors
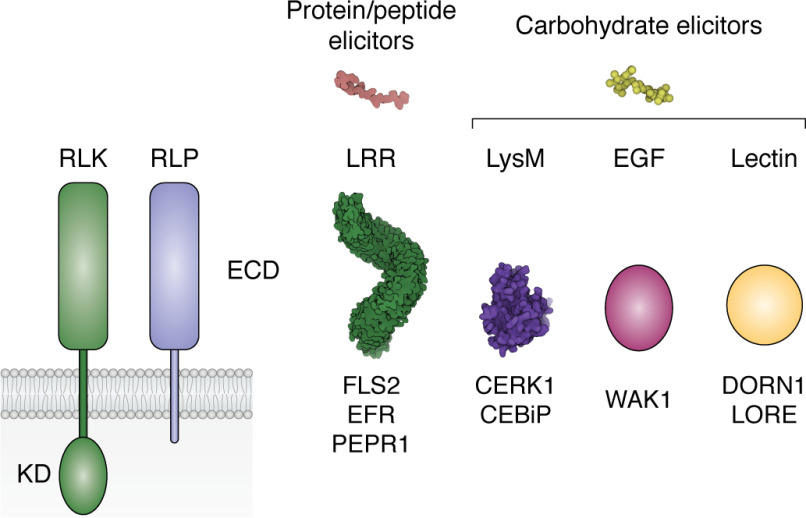
RLKs contain a variable extracellular domain that mediates ligand recognition, a single-pass transmembrane domain, and an intracellular KD that transduces the signal to downstream immune components (74) (Fig. 2). Most plant RLKs identified belong to the family of non-RD kinases (defined by the absence of conserved arginine in the catalytic loop) and often associate in dynamic complexes with membrane-bound RLKs that are functional RD kinases (such as BAK1 (BRASSINOSTEROID-INSENSITIVE 1–associated receptor kinase 1) and SERKs), which operate as co-receptors for perception to initiate immune signaling (75–77). Whereas RLPs exhibit a similar overall structure to RLKs, they only contain a short intracellular tail, lacking a kinase domain, and require a partner co-receptor to signal (63, 78).
Figure 2.
Diversity of cell-surface immune receptors. A schematic representation depicts the domain architecture of different classes of plant RLKs/RLPs. Surface representations are shown for those ECDs for which crystal structures are available. LRR, crystal structure of the ECD of Arabidopsis RLK FLS2, PDB entry 4MNA (green); LysM, crystal structure of the ECD of Arabidopsis RLK–CERK1, PDB entry 4EBY (purple).
Based on the type of ECD, RLKs and RLPs can be clustered into distinct subfamilies, including leucine-rich repeat (LRR), lysin motif (LysM), lectin, and epidermal growth factor (EGF) domain–containing receptors (66, 79, 80) (Fig. 2). The type of ECD mainly defines the nature of the ligand perceived by the RLK/RLPs; however, a few anomalies persist. Among the best-characterized cell-surface immune receptors are the Arabidopsis LRR-type RLKs, FLS2 (flagellin-sensitive 2) and EFR (elongation factor Tu (EF-Tu) receptor) (81, 82), and the LysM-type RLKs LYK5 (lysin motif receptor kinase 5) and CERK1 (chitin elicitor receptor kinase 1) (83, 84). FLS2 (Fig. 3) and EFR recognize peptide epitopes from the N termini of bacterial flagellin (flg22) and bacterial EF-Tu (elf18), respectively (90), whereas LYK5 and CERK1 bind fungal chitin oligomers (84).
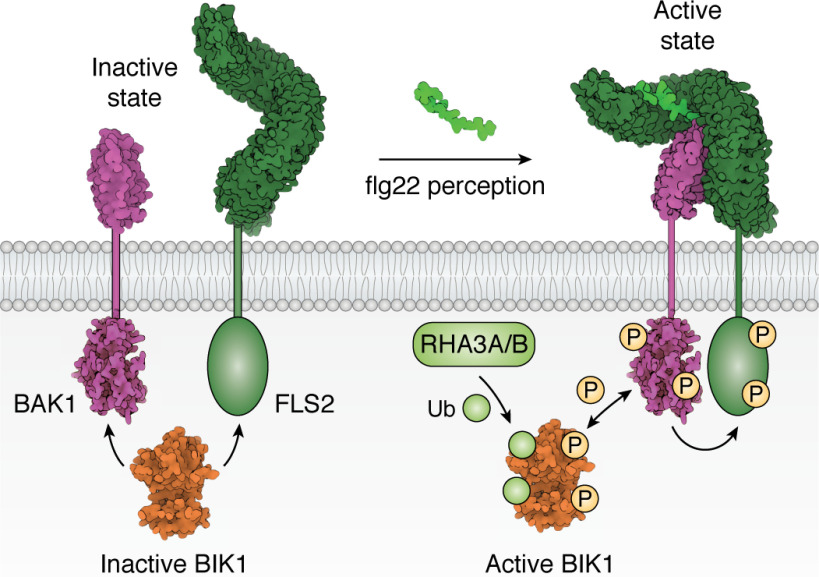
Figure 3.
A mechanistic view of flg22 sensing by FLS2. flg22 (light green) stabilizes the heterodimerization of FLS2 (dark green, PDB entries 4NMA and 4NM8) with BAK1 (purple, PDB entries 3ULZ and 4NM8) (82, 85, 86). Ligand perception leads to activation and phosphorylation of BIK1 (orange, PDB entry 5TOS) by BAK1 (87, 88). Following phosphorylation, BIK1 is monoubiquitinated (Ub) by the E3 ligases RHA3A/B. Monoubiquitinated BIK1 is then released from the FLS2–BAK1 complex and initiates ROS production and Ca2+ signaling through phosphorylation of plasma membrane–localized NADPH oxidases and cyclic nucleotide–gated channels (89). The bidirectional arrow indicates that both BIK1 and BAK1 can trans-phosphorylate each other.
Recognition of peptide/protein ligands
LRR-RLKs are a large subfamily of cell-surface receptors that preferentially bind peptides or proteins as ligands (91–93). In addition to the Arabidopsis FLS2 and EFR, LRR-RLKs from rice and solanaceous plants have been characterized. The rice cell-surface receptor Xa21 binds RaxX21-sY (a sulfated, 21-amino acid synthetic RaxX peptide), a tyrosine-sulfated protein from bacteria (94). Cell-surface receptors from tomato (CORE) and tobacco (NbCSPR) bind to conserved epitopes derived from bacterial cold shock protein (95–97). Likewise, Arabidopsis RLP23 binds the epitope nlp-20, a conserved peptide derived from ethylene-inducing peptide 1–like proteins of bacterial and filamentous pathogens (98).
Although not an LRR-RLK, the Arabidopsis cell-surface receptor FERONIA (FER) uses a tandem malectin-like ECD to perceive RALF1 (rapid alkalinization factor 1) peptides. RALF peptides are cysteine-rich peptides prevalent in the plant kingdom that regulate many aspects of plant life, such as reproduction, growth, responses to environment, and immunity (99, 100). Intriguingly, some functionally active RALF-like peptides have been characterized from fungal pathogens; however, the role of these RALF-like peptides in pathogenesis is unknown (101). In addition to MAMP ligands, some LRR-RLKs perceive proteinaceous DAMPs, such as Atpeps (plant elicitor peptides) and PIPs (PAMP-induced secreted peptides), respectively (102–105). Like LRR-RLKs, LRR-RLPs can also sense extracellular short peptide ligands; however, they can also sense larger extracellular proteinaceous ligands, such as apoplastic effectors. In tomato, the LRR-RLPs Cf-2/4/9 perceive apoplastic effectors Avr2, Avr4, and Avr9 from Cladosporium fulvum, respectively (106–110).
Recognition of carbohydrate/non-proteinaceous ligands
There are several different classes of receptor that are capable of sensing different carbohydrate ligands. LysM-RLKs/LysM-RLPs perceive carbohydrate MAMPs such as bacterial peptidoglycan (PGN), lipopolysaccharide (LPS), and fungal chitin (10, 83, 84, 111, 112). The ECD of the cell wall-associated kinase family (WAKs) comprise repeated EGF-like domains (113–116) that bind various types of pectins including pathogen/wound-induced short oligogalacturonic acid fragments (OG) as well as cell wall- associated longer pectins (116, 117). Intriguingly, lectin RLKs including structurally distinct lectin receptors - LORE (G-type lectins) and DORN1 (L-type lectins) senses non-carbohydrate ligands like low complexity bacterial metabolites such as bacterial medium-chain 3-hydroxy fatty acid (mc-3-OH-FA) (266) and extracellular ATP (e-ATP- as a DAMP signal) (118, 119) respectively, to trigger immune responses.
Ligand-induced homo/heterodimerization of cell-surface receptors
Plant cell-surface immune receptors function in complex with co-receptors and intracellular kinases to activate defense (65, 66, 78). The LRR-RLK BAK1 is the best-characterized co-receptor to date (13, 77, 120). BAK1 forms heterocomplexes with peptide-binding immunity-related LRR-RLKs, including FLS2 (Fig. 3), EFR, and PEPR1 (perception of the Arabidopsis danger signal peptide), and is required for immune signaling (12, 13, 85, 120, 121). Like BAK1, SOBIR1 (suppressor of Bir 1-1) is a regulatory LRR-RLK that serves as an adaptor for certain LRR-RLPs to trigger defense (122–124). Similar to LRR-RLKs, these RLP/adaptor complexes recruit BAK1 or other SERKs for signal transduction (125–127).
By contrast, the Arabidopsis carbohydrate-binding LysM-RLK CERK1 forms chitin-bridged homodimers (10). Homodimeric association has also been reported for the chitin-binding rice LysM-RLP CEBiP (128, 129), but the rice CEBiP can also form heterodimers with rice CERK1 (10, 130). Although oligomerization is important, the precise role of homo- or heterointeractions of LysM-RLK/RLPs in signaling recognition of chitin remains unclear (128).
RLCKs in downstream defense signaling
Ligand perception by plant cell-surface receptors typically results in homo- or heterodimerization that stimulates cis- and/or trans-phosphorylation of intracellular kinase domains (128). In turn, the kinase domains of cell-surface immune receptors activate receptor-like cytoplasmic kinases (RLCKs) to transduce immune signals (87, 131, 132).
The Arabidopsis RLCKs BIK1 (botrytis-induced kinase 1) and PBL (PBS1-like) proteins are substrates of distinct receptor/BAK1/CERK1 complexes at the cell surface (87, 88, 133). For example, in the absence of ligand, BIK1 interacts with BAK1 and associated cell-surface receptor kinase domains (Fig. 3). On ligand binding, a series of cis/trans-phosphorylation events promotes BIK1 dissociation from the complex (87, 88). BIK1 then activates various downstream immune signaling pathways, including ROS burst, Ca2+ accumulation, and mitogen-activated protein kinase pathways (134–136). Multiple RLCKs have been identified in plants that regulate a ROS burst by phosphorylating distinct sites in RBOHD (respiratory burst oxidase homolog protein D), a membrane-localized NADPH oxidase critical for ROS formation post-MAMP detection (25, 135, 137, 138).
Regulation of cell-surface immune responses
To prevent inappropriate signaling, the activity of plant cell-surface immune receptors is tightly controlled (139). Plants use various strategies to help maintain cell-surface receptors in an inactive state in the absence of ligand binding, including the regulation of phosphorylation status and ubiquitination by E3 ligases (139–142).
Phosphorylation is central to cell-surface immunity signaling cascades and is under tight regulation. Plants use phosphatases to negatively regulate cell-surface receptors to prevent the potentially harmful effects of autoinduction. For example, Arabidopsis PP2A (protein phosphatase 2A), a serine/threonine phosphatase, dephosphorylates BAK1/EFR to control defense signaling (143, 144). Similarly, PP2C38 regulates ligand-induced phosphorylation of BIK1, moderating signaling by this key transducer of cell-surface immunity (139). A second strategy to negatively regulate cell-surface immunity is the use of pseudokinases, such as BIR1 and BIR2, that are catalytically inactive but interact with BAK1 in its resting state, preventing the association of LRR-RLKs (145–147). Ligand binding relieves this inhibitory interaction, leading to the formation of activated immune complexes.
Regulation of immunity can also come from controlled degradation through ubiquitination. Two closely related E3-ubiquitin ligases, PUB25 and PUB26, together with both a calcium-dependent protein kinase CPK28 and a heterotrimeric G protein, form a regulatory module and maintain BIK1 homeostasis (148). Similarly, PUB12 and PUB13 polyubiquitinate and mediate degradation of ligand-bound FLS2 (149–151). Intriguingly, a recent study showed that monoubiquitination of BIK1 is necessary for its release from the FLS2/BAK1 complex and immune system activation (89). This demonstrates that a variety of post-translational modifications are important for both positive and negative regulation of cell-surface immune receptors.
In addition to regulating the pool of ligand-bound cell-surface receptors at the plasma membrane, plants also ensure the availability of ligand-free receptors for ongoing pathogen/pest perception. Cell-trafficking components, including SCD1 (DENN domain protein) (152, 153) and ESCRT-I (an endosomal sorting complex required for transport) (154, 155), are involved in delivering these receptors to the cell surface. Finally, it has been proposed that sets of cell-surface receptors may gather at discrete locations on membranes, forming discrete nano- or microdomains (156, 157). These nano-/microdomains are proposed to use similar downstream signaling components; however, different groupings of receptors would lead to different specificity in signal perception, resulting in different responses to stimuli. However, more work is needed to understand the specificity of these nano-/microdomains and how they are clustered into spatially distinct regions of the membrane (156, 157).
Next steps in understanding cell-surface immunity
Although hundreds of RLKs and RLPs have been identified in many plant species, only a subset have been characterized. The biological significance of the vast majority of these receptors remains elusive, and their underlying mechanism of ligand perception remains poorly understood. Understanding how cell-surface receptors with different ECDs perceive ligands will provide a foundation for engineering broad-spectrum resistance into crop plants (158, 159). Further, our understanding of how RLCKs coordinate their association with different receptors and facilitate distinct signaling outputs is a key challenge for the future. We have yet to understand whether activated cell-surface receptor complexes form higher-order supramolecular signaling units at the plasma membrane, what the molecular identity of these activated immune complex might be, and how they may differ across different ligand/cell-surface receptor pairs. Beyond this, we must endeavor to understand the determinants of specificity of plant cell-surface receptors for MAMPs, as this will provide insight into how plants distinguish the MAMPs of pathogenic microbes from those of the beneficial mutualistic microbes.
Case study 1: flg22 perception by the FLS2/BAK1 complex—an exemplar of ligand perception by cell-surface receptors
Genetic screens in Arabidopsis identified FLS2 as the gene that recognizes a conserved 22-amino acid N-terminal epitope (flg22) of bacterial flagellin to initiate cell-surface immunity (81, 82, 160). FLS2 belongs to the LRR-RLK class XII subfamily and shares homology with TLR5 (Toll-like receptor 5), an LRR-containing receptor that perceives flagellin in mammals (161, 162). Fig. 3 gives a detailed mechanistic view of how flg22 is perceived by FLS2.
Flagellin perception in Arabidopsis requires heterodimerization of FLS2 with BAK1 (82, 85, 86). The crystal structure of the ECDs of FLS2 and BAK1, in complex with flg22, revealed the structural basis of flg22 perception (12). The flg22 peptide is bound within the concave surface of the FLS2-ECD, via the leucine-rich repeat subunits LRR3 to LRR16. flg22 interactions with FLS2 are divided into two regions, separated by a kink in the peptide. The N-terminal seven amino acids of flg22 interact with LRRs 3–6, with the C-terminal 14 amino acids binding LRRs 7–16. Numerous hydrogen-bonding, electrostatic, and hydrophobic contacts are formed between flg22 and FLS2. Interactions between the FLS2 and BAK1-ECDs are both receptor- and flg22-mediated, but the peptide acts as a “molecular glue,” stabilizing the heterodimer.
In the absence of flg22, the Arabidopsis RLCK BIK1 can associate with the FLS2 and BAK1 kinase domains. Ligand perception leads to activation and phosphorylation of BIK1 by BAK1 (87, 88). Following phosphorylation, BIK1 is monoubiquitinated by the E3 ligases RHA3A/B (RING-H2 FINGER A3A/B). BIK1 has an N-terminal myristoylation motif, and plasma membrane localization of BIK1 is essential for ubiquitination. Monoubiquitinated BIK1 is then released from the FLS2–BAK1 complex and initiates ROS production and Ca2+ signaling through phosphorylation of plasma membrane-localized NADPH oxidases and cyclic nucleotide–gated channels (89).
Intracellular immunity
Intracellular immunity in plants is conferred by nucleotide-binding, leucine-rich repeat receptor proteins (NLRs). NLRs perceive the presence and/or activities of host-translocated effectors, leading to defense responses that may result in programmed cell death to limit the spread of infection (163). Prior to the molecular identification of NLR receptors and effectors, the genetic basis of what we now call intracellular immunity was established as the “gene-for-gene” model. The gene-for-gene model described a requirement for plants to utilize specialized immune receptors encoded by R (resistance) genes to counteract and respond to the effectors encoded by pathogen AVR (avirulence) genes (164).
NLRs comprise multiple domains with distinct functions
NLRs belong to the AAA+ class of “signal-transducing ATPases with numerous domains” (STAND) ATPases that share a conserved central nucleotide-binding domain across plant, animal, and fungal kingdoms (165). The STAND superfamily includes APAF1, the primary component of the mammalian apoptosome (166), and NLRC4 (NLR family CARD domain–containing protein 4) and NLRP3 (NLR family pyrin domain–containing 3), which are the best-characterized NLRs of the metazoan immune system (20, 167–171).
Classically, plant NLRs comprise a C-terminal LRR domain; a central nucleotide-binding domain known as the NB-ARC (nucleotide-binding domain shared with APAF1, R gene products and CED4 (172); and a variable N-terminal module, which is typically either a TIR (Toll/interleukin-1 receptor/resistance), CC (coiled-coil) domain, or an RPW8 (resistance to powdery mildew 8)-like CC domain (CC-RPW8) (173). Interestingly, LRR domains appear in both cell-surface and intracellular immune receptors and are widely found to be ligand recognition motifs that mediate protein-protein interactions across kingdoms of life. The LRR domain has been implicated in effector recognition for some NLRs, although it is also likely to be important for autoinhibition of the receptor (20, 174, 175). The NB-ARC domain functions as a molecular switch, with effector perception relayed through this domain via nucleotide exchange (ADP for ATP) (48). The N-terminal domains are required for immunity and divide the NLRs into three major classes: TIR-NLRs, CC-NLRs, and RPW8-NLRs (21). Transient expression assays in plants have shown that the N-terminal domains can initiate cell death autonomously and in the absence of an effector (176). Recently, some NLRs have been shown to incorporate additional noncanonical domains into their architecture (177). Known as integrated domains (IDs), these domains can directly interact with effectors (11, 17, 178–180). Intriguingly, many NLR IDs share sequence/structural homology with established virulence-associated host targets of effectors, such as transcription factors or proteins important for cell homeostasis (181). Overall, the individual domains of plant NLRs function together to deliver an effective immune response against pathogen/pests.
Effector detection: Direct and indirect perception of effectors by plant NLRs
Conceptually, how plant NLRs perceive effectors has been grouped into three overarching models: the direct recognition model (non-ID), indirect recognition model (via guardees or decoys), and the integrated domain recognition model (via integration of effector targets as IDs into the NLR architecture) (Figs. 4A and 5).
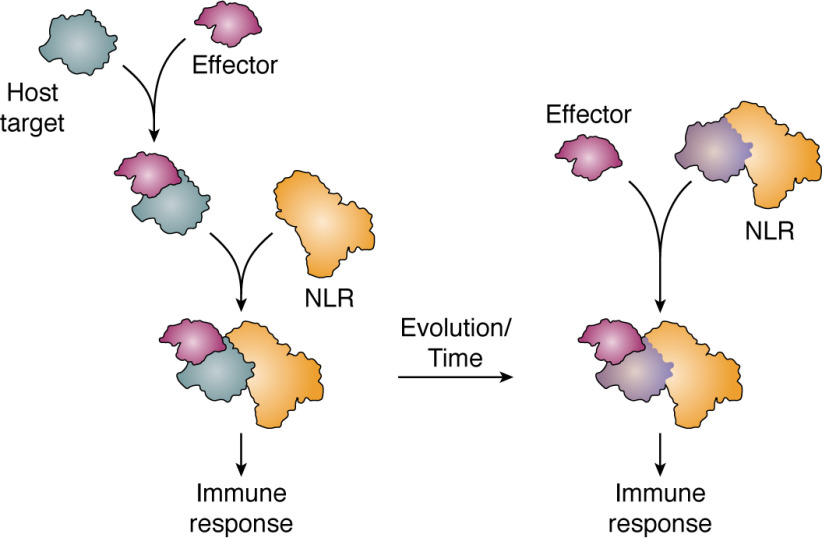
Figure 4.
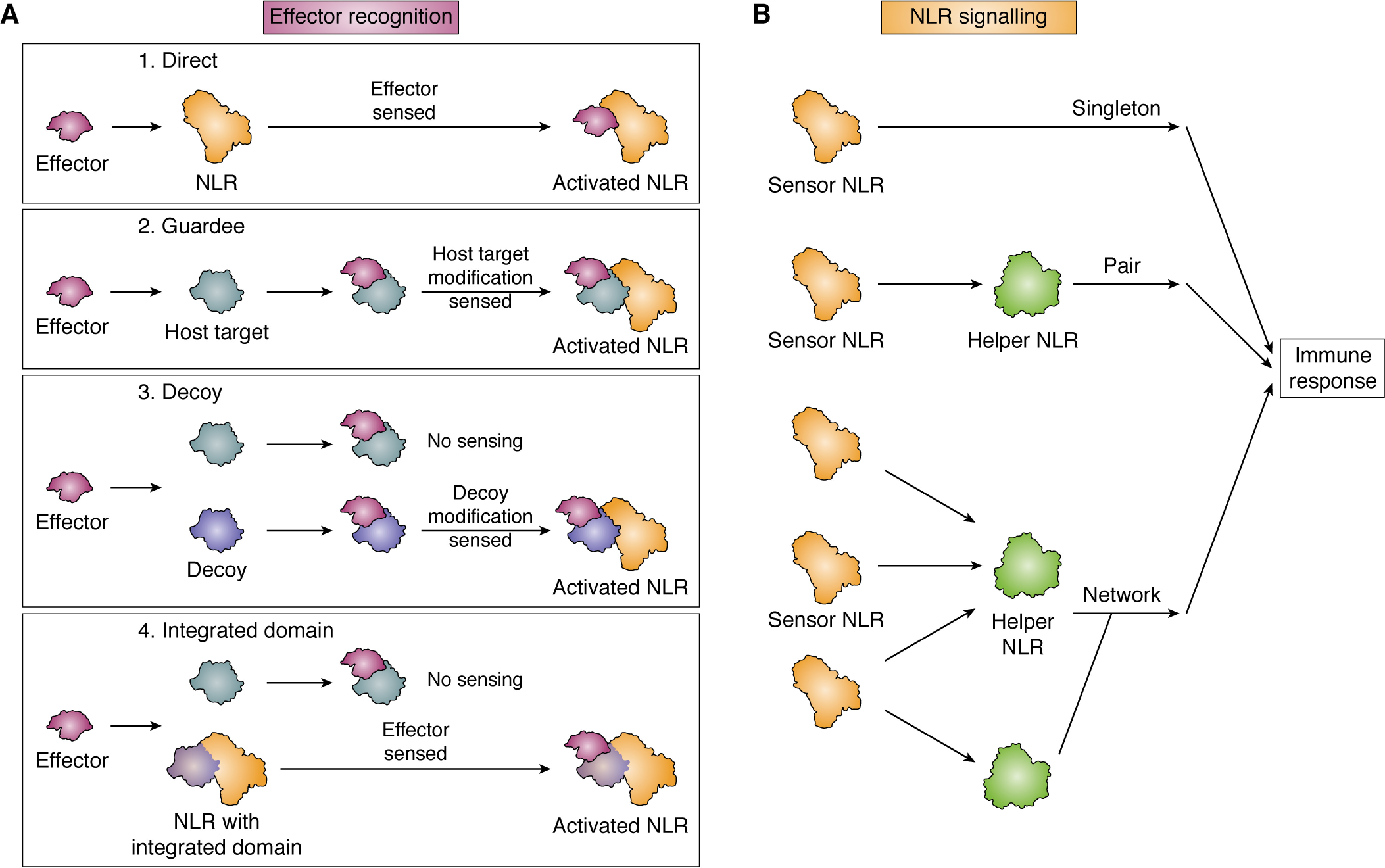
NLRs perceive effectors via distinct mechanisms and induce immune responses through different mechanisms. A, effector (purple) perception induces activation of the NLR (orange) via direct binding. NLRs can indirectly perceive and respond to effectors by monitoring modifications of a physiologically relevant host target (Guardee, gray) or a molecular mimic that likely resulted via gene duplication and is now only involved in immune signaling (Decoy, blue). NLRs can directly perceive and respond to effectors via NLR integrated domains (blue), which likely have their evolutionary origin in ancestral host targets of effectors. B, NLR singletons are able to initiate immune responses upon effector perception. Several sensor NLRs require downstream helper NLRs (green) to transduce effector perception into immune responses. NLRs can function in pairs or as part of interconnected networks.
Figure 5.
Incorporation of host targets in NLRs leads to the evolution of NLR with integrated domains. NLRs (orange) can sense changes in host proteins (gray) that are targeted by pathogen effector molecules (purple) and initiate defense signaling. Over time, some of these host proteins can be found integrated into the NLR core structure (blue), acting as the effector recognition domains for the NLR. Binding of an effector to the integrated domain of an NLR leads to initiation of defense responses.
Direct recognition
The LRR domain of NLR proteins has been implicated in direct interaction with effectors, as well as having a role in autoinhibition of receptor activity. Best-characterized in flax, this plant shows a variety of resistance phenotypes toward different strains of the flax stem rust pathogen (Melampsora lini) expressing different effector alleles (182). In particular, dissection of flax NLRs from the L resistance gene loci (encoding L5, L6, and L7 NLRs among others) and how they perceive alleles of the effector AVRL-567 revealed polymorphisms in the LRR region that underpin specificity (174, 183). Similarly, polymorphisms between the flax NLR variants P and P2 within the LRR domain determine different flax stem rust resistance specificities (184). Although genetic and biochemical evidence for effector perception by LRR domains is established, to date, the structural basis of such interactions has yet to be determined.
Indirect recognition
NLRs can act as “guards” for host proteins targeted by effectors (known as guardees (181)). Guard/guardee interactions can be divided into two models. In both models, the NLR monitors the biochemical status of the guardee (e.g. detecting post-translational modification or cleavage/degradation). In the guard model, the guardee is important for host cell function, whereas in the decoy model, the guardee is a mimic of an effector target but does not have a function outside of immunity.
RIN4 (RPM1 (resistance to Pseudomonas syringae pv. maculicola 1)-interacting protein 4) is a plasma membrane–localized negative regulator of plant immunity (185). This protein is a classic example of an effector “hub,” a host protein that is targeted by multiple effectors from different pathogens, and as a consequence, it is guarded by multiple NLRs (60). The Arabidopsis NLRs RPM1 and RPS2 (resistance to P. syringae 2) monitor the biochemical status of RIN4, detecting modifications, such as phosphorylation and degradation, that lead to activation of immunity (185, 186).
In tomato, Pto is a protein kinase that directly interacts with the NLR Prf (187, 188). Pto is a decoy that mimics the intracellular domains of cell-surface immune receptors (187, 188) and acts as a trap for effectors that pathogens have delivered to interfere with receptor signaling. Pto has no known function outside of this bait activity (189). Direct interactions between effectors and Pto lead to oligomerization of Prf and immune activation (188, 189).
Integrated domain model
The integrated domain model is an evolutionary innovation in plant NLRs where a domain that mimics an effector target is positioned in an NLR architecture, serving as a sensor domain by directly interacting with effectors (Figs. 4A and 5). A well-studied example of NLR IDs are the heavy metal–associated (HMA) domains of rice receptor proteins Pik-1 (Pyricularia oryzae resistance-k) and the Pia sensor NLR (RGA5; R-gene analog 5), which directly bind effectors of the fungal pathogen Magnaporthe oryzae (11, 178). Biochemical, structural, and in planta studies have shown how these HMA domains interact with pathogen effectors and demonstrate how different NLR variants perceive different alleles of the effectors (11, 29, 31, 190). Interestingly, a single integrated domain in an NLR can perceive multiple effectors. For example, the WRKY transcription factor–like domain of the Arabidopsis NLR RRS1 (resistance to Ralstonia solanacearum 1) interacts with two sequence-divergent and structurally divergent effectors (191). One of these effectors adopts a helix-loop-helix fold with an unknown virulence function (AvrRps4 (resistance to P. syringae 4); presumed to be a protein-protein interaction module) (17, 192), whereas a second is an acetyltransferase (PopP2) that acetylates both WRKY transcription factors and the RRS1-WRKY (17, 192). The structural basis of interaction between the RRS1-WRKY and PopP2 has been elucidated (17, 192), but the equivalent structure with AvrRps4 remains to be determined. The RRS1-WRKY case demonstrates the versatility of effector perception that integrated domains deliver to NLRs and suggests their utility for receptor engineering.
Case study 2: Integrated HMA domains—exemplars of integrated domains in NLRs
Many different types of proteins have been found as IDs in plant NLRs, and likely function in direct perception of effectors (193–197). The integrated HMA domains of the sensor NLRs of the rice Pik and Pia pairs are exemplars of IDs and serve as model systems for understanding the principles of effector perception by these domains (11, 29, 31, 178). Fig. 5 illustrates the integration of atypical domains into NLRs to facilitate effector perception.
The integrated HMA domains of Pik-1 and the Pia sensor (also known as RGA5) are likely derived from an expanded family of small plant proteins containing an HMA domain and, sometimes, a C-terminal isoprenylation motif (heavy metal–associated plant proteins (HPPs) or heavy metal-associated isoprenylated proteins (HIPPs) (198)). These proteins may have a role in abiotic stress and detoxification of heavy metals, such as copper or cadmium (198). Additionally, some of these proteins act as susceptibility factors (host targets that can be exploited to assist infection) for diverse pathogens (199–201). This suggests that HPPs/HIPPs may be effector hubs, repeatedly targeted by pathogens as part of infection strategies (59, 60). This provides an evolutionary context for their integration into NLRs as “baits” for triggering immunity (177).
In rice, integrated HMA domains can be found at the C terminus of the sensor NLR of Pia (178) and also between the CC and NB-ARC domain of the sensor NLR Pik-1 (11). Diversity in the location of domain integration implies that these were separate integration events.
The HMA domain of the Pia sensor binds two rice blast effectors, AVR-Pia and AVR1-CO39, whereas the Pik-HMA binds variants of the rice blast effector AVR-Pik (11, 29, 31, 190). Interestingly, these effectors bind to the Pia- and Pik-HMA domains via different interfaces, suggesting that they have independently evolved to target HMA domain–containing proteins, and rice has been able to use both of these interfaces to bait effectors and trigger immunity (31).
As a consequence of arms-race co-evolution with AVR-Pik effector variants (29, 202, 203), the HMA domain is the most variable domain region of the Pik NLRs (204), and the rice Pik receptors also exist as an allelic series with differential recognition specificity for effector variants (202). Biochemical and structural studies of the interaction between different AVR-Pik variants and two allelic HMA domains revealed how subtle changes in the effector/HMA-binding interface underpin variation in recognition specificity (29). Recently, the observation that subtle changes underpin specificity has been used in a proof-of-concept study to show that NLR IDs can be engineered to expand their recognition capacity to allelic effectors (205).
NLR activation
A general principle of NLR biology is that perception of effectors leads to conformational changes in the receptor. These changes can include domain rearrangements and oligomerization. Whereas the details depend on the mode of effector perception, nucleotide exchange (ADP for ATP) in the NB-ARC domain of NLRs is a factor for activation. Numerous studies have shown the importance of conserved sequences such as the “P-loop” and “MHD-like” motifs (a consensus sequence (methionine-histidine-aspartate) at the C terminus of ARC2) in nucleotide binding/exchange and NLR activation (206, 207).
Conformational change and/or oligomerization of NLRs perturb the N-terminal CC or TIR domains to initiate immune responses. Whereas recent studies have begun to shed light on the molecular basis of how these domains trigger immunity, whether these reflect general principles applicable for all NLRs remains to be determined. For example, for CC domains, the structure of the Arabidopsis NLR ZAR1 (HopZ-activated resistance 1) revealed a mechanism whereby oligomerization results in a “funnel” of the N-terminal helices, which then associate with membranes and may perturb cellular integrity (14, 15) (Fig. 6). A sequence motif within the N-terminal helix of some NLR CC domains has been associated with ZAR1-like cell death immunity, known as the MADA motif (a consensus MADAXVSFXVXKLXXLLXXEX sequence conserved at the N termini of NRC (NLR required for cell death) family proteins) (209). This suggests that a subset of NLRs may function in a manner similar to ZAR1. However, how CC-NLRs that do not contain this motif function to initiate immunity has yet to be determined.
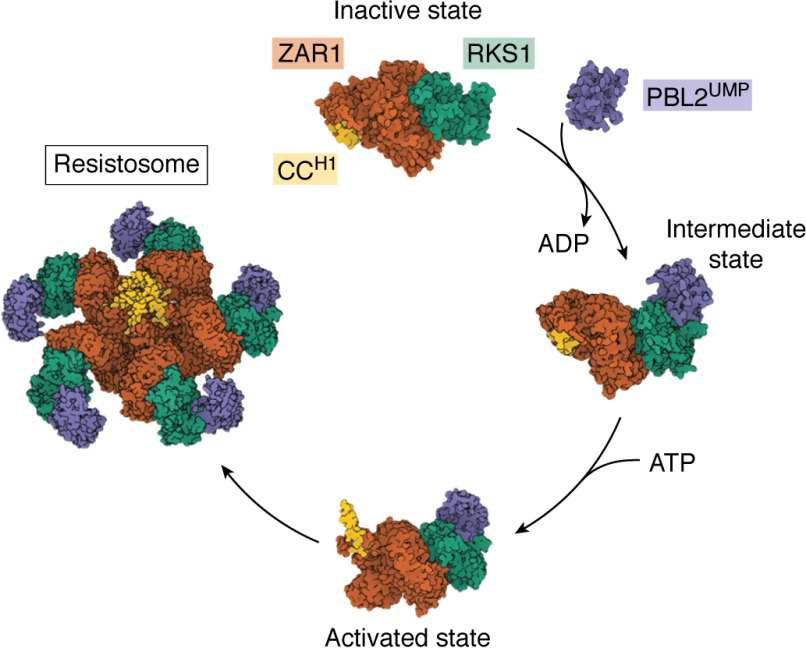
Figure 6.
The activation of the ZAR1 immune receptor. ZAR1 (orange) is an Arabidopsis CC-NLR that forms complexes with pseudokinases, including ZED1 and RKS1 (green), to perceive effector activity (208). The ZAR1:RKS1 receptor complex guards the receptor-like cytoplasmic decoy kinase PBL2. Following uridylylation of PBL2 by the Xanthomonas campestris effector protein AvrAC, PBL2UMP (purple) binds to RKS1, activating ZAR1. Activated ZAR1 is then able to oligomerize into a pentameric wheel with the CC domains each contributing their H1 helix (yellow) to form a funnel-like structure.
Many TIR domain structures from plant NLRs have been determined (16, 32, 35–37) and revealed multiple mechanisms of self-association to form scaffolds for protein-protein interactions that may be important for immune activation (210). Recently, the TIR domains of several NLRs have been shown to catalyze the hydrolysis of NAD+, suggesting a new mechanism for TIR-NLR activity (35, 211). How NAD+ hydrolysis functions in plant immunity is currently unknown. Recently, the structure of the N. benthamiana TIR-NLR Roq1 was determined, marking the first structure of a TIR-NLR resistosome (212). The Roq1 structure provides insight into the novel recognition of its cognate effector, XopQ, through interaction with a unique integrated-like domain deemed the post-LRR domain. Furthermore, it verifies the importance of specific TIR domain self-association interfaces, alluding to self-association resulting in the opening of the TIR domain active site for NAD+ binding and hydrolysis.
NLRs function as singletons, pairs, and networks
To compensate for the lack of an adaptive immune system, plants have a diverse NLR repertoire, which has enabled functional specialization. This has resulted in the evolution of NLRs that function as singletons, in pairs, and as parts of interconnected networks (213–215) (Fig. 4B).
To date, several NLRs have been identified that appear to function autonomously, both sensing the presence of pathogens/pests and mounting a response. These are referred to as NLR singletons. Examples include NLRs of the mildew resistance locus A (MLA) in barley, Arabidopsis TIR-NLR RPP4 (recognition of Peronospora parasitica 4), and CC-NLR RPS2 (186, 216).
By contrast, other NLRs have specialized functions and can be broadly divided into two groups, sensors and helpers, and are generally referred to as NLR pairs (215). In these pairs, sensor NLRs perceive effectors, via the mechanisms discussed above, and helper NLRs are involved in converting effector perception into immune activation (181). NLR pairs can be genetically linked, often encoded at the same locus under the control of the same promoter. They are also always of the same class (CC-NLR or TIR-NLR). The best-characterized genetically linked sensor/helper paired NLRs are the Arabidopsis pair RRS1/RPS4, the rice pair Pik, and the rice pair Pia (also known as RGA5/RGA4). Intriguingly, each of the sensor NLRs of these pairs contains an integrated domain. General mechanisms for how paired NLRs function are based on models of suppression or receptor cooperation (217). The Pia and RRS1/RPS4 NLR pairs can be transiently expressed in tobacco leaves without clear cell death phenotypes. However, cell death phenotypes can be observed in tobacco leaves when RPS4 or the Pia helper NLRs are expressed without their cognate sensors or effectors. Co-expression of the RRS1 or Pia sensor NLRs suppresses the autoactive cell death phenotype of the helpers (218). Co-expression of the paired NLRs with their cognate effectors relieves this suppression, resulting in cell death. By contrast, expression of the Pik-2 helper NLR does not result in cell death, and co-expression of the Pik-1 sensor NLR and the cognate effector is required for cooperative cell death (11, 219).
However, a direct genetic link (head-to-head orientation or belonging to the same genetic loci) is not essential for NLR cooperation in immune activation, and some require complex NLR networks for function. The NLR “N-requirement gene 1” (NRG1), is required for the resistance to tobacco mosaic virus provided by the TIR-NLR, N (220). NRG1 is a member of the ADR1 (activated disease resistance 1) family of RPW8-NLRs, and since the discovery of NRG1, the RPW8-NLRs have been found to be important for the full function of many other CC-NLRs and TIR-NLRs (221, 222). Another NLR network has recently been uncovered in solanaceous plants. The NRCs are a phylogenetically distinct class of helper CC-NLRs consisting of functionally redundant paralogs (223). Sensor NLRs that require NRCs provide resistance to diverse pathogens and pests, including bacteria, oomycetes, nematodes, viruses, and insects (223). They display distinct specificities for different NRC helpers, with some sensors signaling through only one and others showing functional redundancy. Diversification of NLRs in the NRC network has allowed a varied arsenal of NLRs against a broad range of pathogens to have evolved.
Intriguingly, a new body of work has emerged, which has begun to uncover interplay between cell-surface and intracellular immunity (43, 44). These papers demonstrate that cell-surface immunity is required to potentiate intracellular immunity, enhancing NLR responses such as cell death. By contrast, NLR activity was shown to be important for cell-surface immunity receptor turnover, relieving attenuation of PRR signaling, and replenishing signaling components at the cell surface. These new findings open the door to further studies analyzing cross-network communications between cell-surface and intracellular immunity.
Case study 3: The structure of ZAR1—the first plant resistosome
Recently, cryo-EM structures of ZAR1 were solved in inactive and active states. These are the first structures of full-length plant NLRs to be determined, and they represent a major advance in our understanding of NLR biology (14, 15). In the inactive state, the LRR domain in the ZAR1:RKS1 (resistance-related kinase 1) receptor complex makes autoinhibitory contacts with both the NB-ARC and CC domains, and a molecule of ADP is bound within the NB-ARC domain. PBL2UMP binding to RKS1 induces a disorder-to-order transition of the RKS1 activation loop and a steric clash with the NB-ARC of ZAR1, which becomes displaced. This conformational change results in nucleotide exchange from bound ADP to ATP and stabilization of a structure primed for oligomerization with other activated RKS1:ZAR1 heterocomplexes. The pentamer that results from the oligomerization events is known as the “resistosome.” The conformational changes and oligomerization associated with PBL2UMP binding promote unfolding of the ZAR1 CC domain, releasing the N-terminal helix (H1) from a four-helical bundle. The released H1 helix then associates with the H1 helices of neighboring activated ZAR1 molecules, resulting in the formation of a funnel-like structure with a striking hydrophobic surface. There is evidence that the ZAR1 CC domain funnel is required for membrane association and that this membrane association is linked to induction of cell death, potentially through ion efflux or membrane perturbation (14, 15, 224).
As this review was being finalized, the structure of the Roq1 TIR-NLR from N. benthamiana was determined by cryo-EM (212). This structure provides a significant advance in our understanding of plant NLR immunity as it represents the first structure of a TIR-NLR resistosome and, as such, should be considered alongside this ZAR1 case study.
Next steps in understanding intracellular immunity
Despite recent advances, key questions on NLR function remain to be addressed. The ZAR1 and Roq1 structures have provided a wealth of information that has significantly expanded our understanding of plant NLR biology. However, it is as yet unclear how oligomerization of CC-NLRs into a resistosome mediates cell death. Furthermore, we lack structural information and evidence of resistosome formation for RPW8-NLRs. Even more perplexing is the role of TIR domain NADase activity and how it leads to the activation of RPW8-NLRs. Where we are beginning to generate a picture of the complex interactions between NLRs in plants, it is still unclear how one of the most primary interactions, effector detection, is mechanistically relayed from sensor NLRs to helper NLRs in pairs and networks. Each of these areas, among many others, requires further research to fully understand how NLRs provide resistance to pathogens/pests.
Engineering plant immunity
Since their discovery, cell-surface and intracellular immune receptors have been targets of biotechnological approaches to improve disease resistance in plants. These approaches have included different scales, from transferring genes encoding plant receptors between species to specific amino acid mutations to modulate effector binding or receptor activity (52, 225). Engineering requires a holistic view, incorporating a range of methods to deliver both broad and robust resistance. Broad low-level resistance is regularly found in nature; however, due to monoculture reducing natural diversity, bespoke resistance to specific pathogens is often more desirable. Whereas the GMO debate remains a focus of public discussion and governmental policy decisions, engineering and editing crop genomes offers potential solutions to food insecurity.
Engineering resistance by transferring genes
The transfer of traits conferring pathogen resistance is conceptually the most straightforward strategy to engineer disease immunity in plants. This method is used in classical plant breeding by selecting resistant phenotypes and crossing into desired cultivars. However, this approach is time-consuming and technically challenging (226). The recent development of new sequencing, phenotyping, and plant growth technologies has allowed researchers to overcome the limitations of this process (159, 227–229).
As plant cell-surface immune receptors tend to perceive common signatures of pathogens/pests and activate conserved signaling pathways in plants, they offer opportunities to transfer resistance between plant species. For example, the Arabidopsis EFR receptor is restricted to Brassicaceae species in nature but delivered novel resistance specificity against bacterial pathogens when it was transferred to tomato and rice (230–232). Similarly, transfer of the rice cell-surface receptor Xa21 to banana, sweet orange, or tomato increased resistance to Xanthomonas sp. (233–235). Further, an allelic FLS2 receptor from wild grape has been demonstrated to confer resistance to the crown-gall pathogen Agrobacterium tumefaciens when expressed in tobacco (236). Building on these advances, mining the diversity of cell-surface immune receptors with expanded recognition specificities from diverse plant species and their subsequent transfer to other plants, holds promise for engineering broad-spectrum disease resistance (230, 237, 238).
Recent advances in mining the genomes of wild plant species using new genomics technologies (228, 239–242) have allowed the rapid identification of candidate immune receptors for deployment in crops. Using such approaches, resistance to Asian soybean rust in soybean has been established by transferring an NLR from pigeon pea (243). Further, resistance has been shown against the potato late blight pathogen by introducing resistance genes from wild potato species (244, 245).
Plant intracellular NLR receptors are highly diverse (246–248) and often work together with other NLRs in pairs or networks. Therefore, NLRs frequently require a specific genetic background to provide effective disease resistance. As a consequence, the functional transfer of NLR receptors between species, or even cultivars of the same species, has proven challenging (249, 250).
Bespoke engineering of NLR responses
Based on recent advances in our understanding of the mechanisms of NLR function, a number of new approaches are being explored to enable more effective engineering of NLRs to help deliver disease resistance in target plants.
Domain exchange and mutagenesis
Domain exchange approaches between related NLRs have been explored for their potential to engineer disease resistance (251). Domain exchanges between the potato NLRs Rx and Gpa enabled the partial exchange of immune recognition from viruses to nematodes and vice versa (251). Autoactive and loss-of-function phenotypes were also observed in the chimeras and suggested that more subtle variations may have more potential.
High-throughput random mutagenesis of NLRs has been used to explore whether these receptors can be improved by enhancing their activation sensitivity or by expanding their recognition specificity. Following such approaches allowed expanded recognition of viruses by the NLR Rx (252, 253). This has been also applied to identify mutations that expanded the response of the potato NLR R3a and its tomato ortholog I-2 (254) to effectors from Phytophthora species (255). However, the translation of these expanded recognition phenotypes to disease resistance has remained limited (252–255). Recently, improved knowledge of how effectors, or effector activities, are directly perceived has inspired new methods of engineering.
Decoy engineering
Understanding how NLRs that guard host targets are activated can allow engineering of recognition specificity. The Arabidopsis NLR RPS5 perceives cleavage of the decoy kinase PBS1 by the P. syringae effector AvrPphB at a specific recognition sequence (256). Mutation of the recognition site in PBS1 to cleavage sequences recognized by other translocated pathogen proteases, including a second P. syringae effector, AvrRpt2, and the Nla protease from tobacco etch virus, switched the RPS5 recognition specificity (257). It is of special note that the latter switched RPS5 perception from bacteria to viruses. Although this approach is limited to pathogens that translocate proteases into the host, the widespread conservation of this protease recognition systems in crop plants (258, 259) has recently allowed engineering of disease resistance in soybean (256).
Integrated domains: New possibilities to engineer disease resistance
The discovery of integrated domains in plant NLRs opened new opportunities to understand and manipulate mechanisms of pathogen perception by intracellular immune receptors (177, 218, 260, 261) (Fig. 6). These domains have become a promising avenue for engineering disease resistance conferred by NLRs (205, 260, 261). As previously introduced, the allelic rice NLRs Pik perceive variants of the rice blast pathogen effector AVR-Pik by direct binding via an HMA domain (11, 29). Some natural effector variants are able to escape recognition by certain Pik NLR alleles, whereas other variants completely evade detection (29, 202, 262). Further, the binding of AVR-Pik effectors to the HMA is not in itself sufficient to activate immune signaling, and a threshold of binding needs to be reached to trigger immune responses (29) (Fig. 7A). An understanding of the biochemical and structural basis of different AVR-Pik/HMA interactions (29) has allowed the design of specific mutations that increase the binding affinity to effector alleles, expanding the recognition capability of the Pikp NLR (205) (Fig. 7B). This proof of concept demonstrated that NLR binding to effectors and the subsequent responses can be manipulated by rational design. Additional HMA domain engineering could now focus on extending perception of sequence-divergent rice blast effectors that also interact with HMAs, but at a different interface (30, 31, 190). Looking to the future, combining mutations in NLRs to both sensitize and lower the threshold to trigger immune responses, as discussed above (Fig. 7C), and directly increase binding affinity to effectors (Fig. 7D) is an exciting long-term goal for the field.
Figure 7.
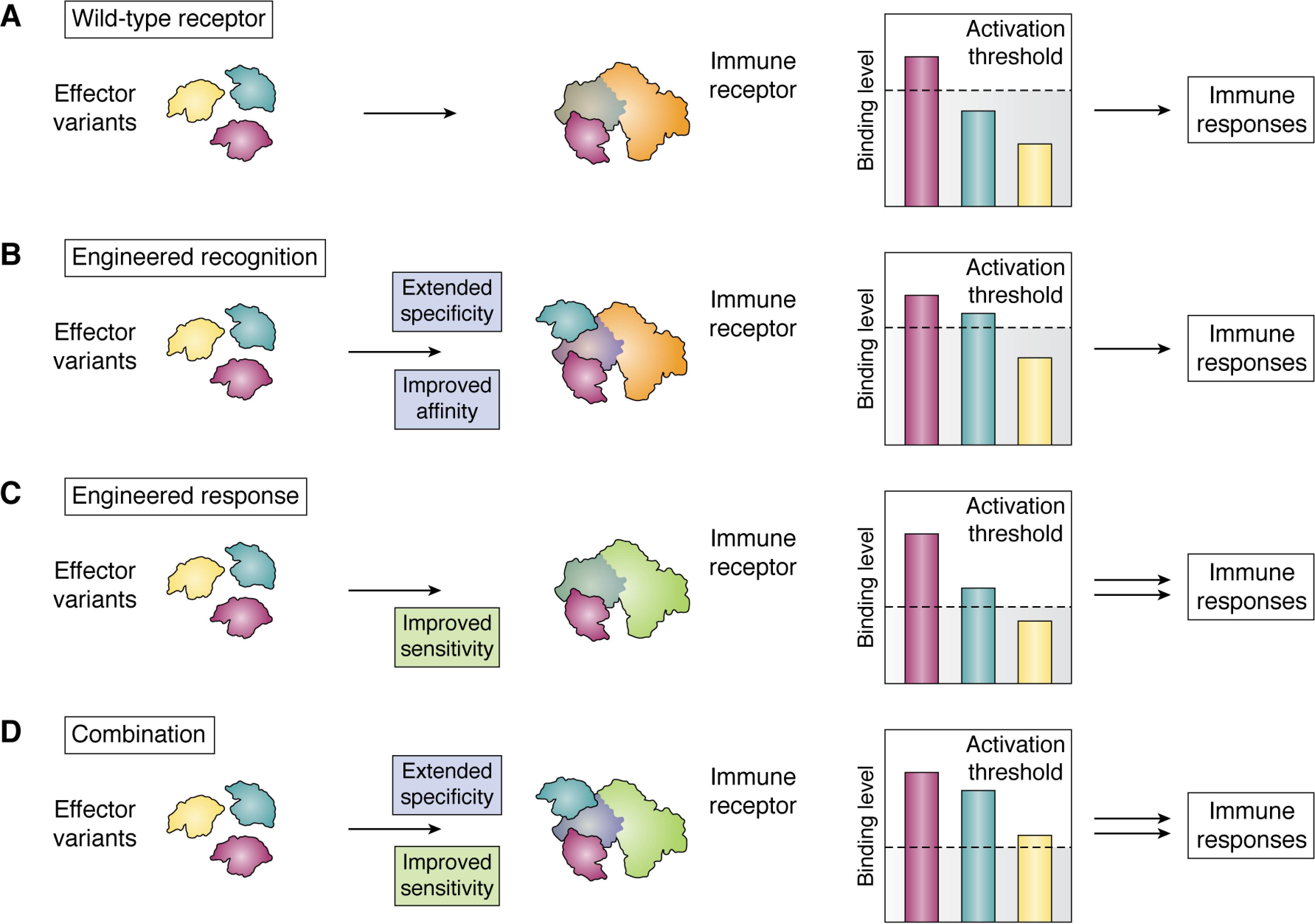
Alternative strategies for immune receptor engineering. A, plant immune receptors (orange/gray) bind natural variants of effector and ligands (purple/cyan/yellow) with different binding affinities (schematically depicted by the height of the colored bars). Only some binding events are of sufficient level to reach an activation threshold (represented by the dashed line), triggering immune responses. B, mutations in the receptor (gray to blue) can extend pathogen recognition by gaining or increasing binding to effectors and ligands, leading to immune responses to pathogens previously undetected. C, mutations in the immune receptors (orange to green) can lower the activation threshold, allowing for increased intensity of immune signaling. D, the combination of both mutations (green/blue receptor) that enhance recognition and immune responses can lead to wider and increased immune responses to effectors and ligands.
Other approaches: Controlled expression of autoactive NLRs
A further possibility for engineering disease resistance is to manipulate expression of NLRs. For example, the discovery of a mechanism controlling defense responses at transcriptional and translational level allowed the design of a pathogen-responsive expression cassette (263). Placing an autoactive NLR under control of this cassette generated an NLR-mediated plant defense system that does not rely on effector recognition. This conferred broad-spectrum resistance without a fitness cost (264), a defense-yield trade-off that can occur when engineering immunity (265).
Conclusion
Plant disease has shaped the natural and agricultural world. Crop losses due to disease and the emergence of resistant cultivars have been key events that have facilitated the way we breed and farm our food. Consequently, an understanding of the plant immune system is essential, as we attempt to develop new methods for disease control against a background of the climate emergency. Despite extensive studies, which we have reviewed here, further research is required to fully understand how the plant system works holistically to deliver disease resistance. Of the hundreds of cell-surface RLKs and RLPs identified, many of the biological functions and ligands of these receptors remain unknown. Furthermore, it is important to understand how plants distinguish the MAMPs of pathogens/pests from the MAMPs of the beneficial mutualist microbes. Determining the function of more of these cell-surface receptors will lead to new avenues for engineering resistance in crops. Similarly, advances in understanding NLR biology will help to better arm plants against pathogens and pests that evolve to circumvent cell-surface immunity. As we generate a better understanding of the complex interactions between plants and pathogens and pests, we can assemble the pieces to inform engineering of disease resistance, to produce more durable crops and help battle the food security problems of the future.
Acknowledgments
We thank Ruby O'Grady for the artwork and design inspiration for Fig. 1.
Funding and additional information—Research in the Banfield laboratory is supported by UKRI Biotechnology and Biological Sciences Research Council (BBSRC) Grants BB/P012574 and BB/M02198X; European Research Council Proposals 743165 and 669926; the John Innes Foundation; and UKRI BBSRC Norwich Research Park Biosciences Doctoral Training Partnership Grant BB/M011216/1.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ECD
- extracellular domain
- AVR
- avirulence gene
- CC
- coiled-coil
- CEBiP
- chitin elicitor–binding protein
- CORE
- cold shock protein receptor
- DAMP
- damage-associated molecular pattern
- EF-Tu
- elongation factor Tu
- EFR
- elongation factor Tu receptor
- EGF
- epidermal growth factor
- elf18
- N-acetylated peptide comprising the first 18 amino acids of bacterial elongation factor EF-Tu
- FER
- feronia
- flg22
- 22-amino acid epitope of bacterial flagellin
- HIPP
- heavy metal–associated isoprenylated plant protein
- HMA
- heavy metal–associated
- HPP
- heavy metal–associated plant protein
- ID
- integrated domain
- KD
- kinase domain
- LRR
- leucine-rich repeat
- LysM
- lysin motif
- MAMP
- microbial-associated molecular pattern
- MLA
- mildew resistance locus A
- NB-ARC
- nucleotide-binding domain shared with APAF1, R gene products, and CED4
- NLR
- nucleotide-binding leucine-rich repeat receptor
- NRC
- NLR required for cell death
- PBS1/2
- Arabidopsis AVRPPHB-susceptible 1/2
- PGN
- peptidoglycan
- PIP
- PAMP-induced secreted peptide
- PRR
- pattern recognition receptor
- RLCK
- receptor-like cytoplasmic kinase
- RLK
- receptor-like kinase
- RLP
- receptor-like protein
- ROS
- reactive oxygen species
- SERK
- somatic embryogenesis receptor kinase
- STAND
- signal transduction ATPases with numerous domains
- TIR
- Toll/interleukin-1 receptor
- WAK
- cell wall–associated kinase
- NLR
- nucleotide-binding leucine-rich repeat receptor
- PDB
- Protein Data Bank.
References
- 1. Savary S., Willocquet L., Pethybridge S. J., Esker P., McRoberts N., and Nelson A. (2019) The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 10.1038/s41559-018-0793-y [DOI] [PubMed] [Google Scholar]
- 2. Bebber D. P., and Gurr S. J. (2015) Crop-destroying fungal and oomycete pathogens challenge food security. Fungal Genet. Biol. 74, 62–64 10.1016/j.fgb.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 3. Fisher M. C., Henk D. A., Briggs C. J., Brownstein J. S., Madoff L. C., McCraw S. L., and Gurr S. J. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194 10.1038/nature10947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Turner R. S. (2005) After the famine: Plant pathology, Phytophthora infestans, and the late blight of potatoes, 1845–1960. Hist. Stud. Phys. Biol. Sci. 35, 341–370 10.1525/hsps.2005.35.2.341 [DOI] [Google Scholar]
- 5. Dean R., Van Kan J. A., Pretorius Z. A., Hammond-Kosack K. E., Di Pietro A., Spanu P. D., Rudd J. J., Dickman M., Kahmann R., Ellis J., and Foster G. D. (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamoun S., Furzer O., Jones J. D., Judelson H. S., Ali G. S., Dalio R. J., Roy S. G., Schena L., Zambounis A., Panabières F., Cahill D., Ruocco M., Figueiredo A., Chen X. R., Hulvey J., et al. (2015) The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434 10.1111/mpp.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mansfield J., Genin S., Magori S., Citovsky V., Sriariyanum M., Ronald P., Dow M., Verdier V., Beer S. V., Machado M. A., Toth I., Salmond G., and Foster G. D. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant. Pathol. 13, 614–629 10.1111/j.1364-3703.2012.00804.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scholthof K. B., Adkins S., Czosnek H., Palukaitis P., Jacquot E., Hohn T., Hohn B., Saunders K., Candresse T., Ahlquist P., Hemenway C., and Foster G. D. (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954 10.1111/j.1364-3703.2011.00752.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Esse H. P., Reuber T. L., and van der Does D. (2020) Genetic modification to improve disease resistance in crops. New Phytol. 225, 70–86 10.1111/nph.15967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu T., Liu Z., Song C., Hu Y., Han Z., She J., Fan F., Wang J., Jin C., Chang J., Zhou J.-M., and Chai J. (2012) Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164 10.1126/science.1218867 [DOI] [PubMed] [Google Scholar]
- 11. Maqbool A., Saitoh H., Franceschetti M., Stevenson C. E. M., Uemura A., Kanzaki H., Kamoun S., Terauchi R., and Banfield M. J. (2015) Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. Elife 4, e08709 10.7554/eLife.08709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun Y., Li L., Macho A. P., Han Z., Hu Z., Zipfel C., Zhou J.-M., and Chai J. (2013) Structural Basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628 10.1126/science.1243825 [DOI] [PubMed] [Google Scholar]
- 13. Tang J., Han Z., Sun Y., Zhang H., Gong X., and Chai J. (2015) Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 25, 110–120 10.1038/cr.2014.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang J., Hu M., Wang J., Qi J., Han Z., Wang G., Qi Y., Wang H.-W., Zhou J.-M., and Chai J. (2019) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870 10.1126/science.aav5870 [DOI] [PubMed] [Google Scholar]
- 15. Wang J., Wang J., Hu M., Wu S., Qi J., Wang G., Han Z., Qi Y., Gao N., Wang H.-W., Zhou J.-M., and Chai J. (2019) Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364, eaav5868 10.1126/science.aav5868 [DOI] [PubMed] [Google Scholar]
- 16. Williams S. J., Sohn K. H., Wan L., Bernoux M., Sarris P. F., Segonzac C., Ve T., Ma Y., Saucet S. B., Ericsson D. J., Casey L. W., Lonhienne T., Winzor D. J., Zhang X., Coerdt A., et al. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344, 299–303 10.1126/science.1247357 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Z.-M., Ma K.-W., Gao L., Hu Z., Schwizer S., Ma W., and Song J. (2017) Mechanism of host substrate acetylation by a YopJ family effector. Nat. Plants 3, 17115 10.1038/nplants.2017.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hao W., Collier S. M., Moffett P., and Chai J. (2013) Structural basis for the interaction between the potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating protein 2 (RanGAP2). J. Biol. Chem. 288, 35868–35876 10.1074/jbc.M113.517417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hohmann U., and Hothorn M. (2019) Crystal structure of the leucine-rich repeat ectodomain of the plant immune receptor kinase SOBIR1. Acta Crystallogr. D Struct. Biol. 75, 488–497 10.1107/S2059798319005291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bentham A., Burdett H., Anderson P. A., Williams S. J., and Kobe B. (2016) Animal NLRs provide structural insights into plant NLR function. Ann. Bot. 119, 698–702 10.1093/aob/mcw171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones J. D. G., Vance R. E., and Dangl J. L. (2016) Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395 10.1126/science.aaf6395 [DOI] [PubMed] [Google Scholar]
- 22. Meunier E., and Broz P. (2017) Evolutionary convergence and divergence in NLR function and structure. Trends Immunol. 38, 744–757 10.1016/j.it.2017.04.005 [DOI] [PubMed] [Google Scholar]
- 23. Wang W., Feng B., Zhou J.-M., and Tang D. (2020) Plant immune signaling: advancing on two frontiers. J. Integr. Plant Biol. 62, 2–24 10.1111/jipb.12898 [DOI] [PubMed] [Google Scholar]
- 24. Hohmann U., Nicolet J., Moretti A., Hothorn L. A., and Hothorn M. (2018) The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling. Nat. Plants 4, 345–351 10.1038/s41477-018-0150-9 [DOI] [PubMed] [Google Scholar]
- 25. Lal N. K., Nagalakshmi U., Hurlburt N. K., Flores R., Bak A., Sone P., Ma X., Song G., Walley J., Shan L., He P., Casteel C., Fisher A. J., and Dinesh-Kumar S. P. (2018) The receptor-like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe 23, 485–497.e5 10.1016/j.chom.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu S., Wang J., Han Z., Gong X., Zhang H., and Chai J. (2016) Molecular mechanism for fungal cell wall recognition by rice chitin receptor OsCEBiP. Structure 24, 1192–1200 10.1016/j.str.2016.04.014 [DOI] [PubMed] [Google Scholar]
- 27. Maekawa T., Cheng W., Spiridon L. N., Töller A., Lukasik E., Saijo Y., Liu P., Shen Q. H., Micluta M. A., Somssich I. E., Takken F. L. W., Petrescu A. J., Chai J., and Schulze-Lefert P. (2011) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9, 187–199 10.1016/j.chom.2011.02.008 [DOI] [PubMed] [Google Scholar]
- 28. Casey L. W., Lavrencic P., Bentham A. R., Cesari S., Ericsson D. J., Croll T., Turk D., Anderson P. A., Mark A. E., Dodds P. N., Mobli M., Kobe B., and Williams S. J. (2016) The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins. Proc. Natl. Acad. Sci. U. S. A. 113, 12856–12861 10.1073/pnas.1609922113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De la Concepcion J. C., Franceschetti M., Maqbool A., Saitoh H., Terauchi R., Kamoun S., and Banfield M. J. (2018) Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nat. Plants 4, 576–585 10.1038/s41477-018-0194-x [DOI] [PubMed] [Google Scholar]
- 30. Varden F. A., Saitoh H., Yoshino K., Franceschetti M., Kamoun S., Terauchi R., and Banfield M. J. (2019) Cross-reactivity of a rice NLR immune receptor to distinct effectors from the rice blast pathogen Magnaporthe oryzae provides partial disease resistance. J. Biol. Chem. 294, 13006–13016 10.1074/jbc.ra119.007730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo L., Cesari S., de Guillen K., Chalvon V., Mammri L., Ma M., Meusnier I., Bonnot F., Padilla A., Peng Y.-L., Liu J., and Kroj T. (2018) Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces. Proc. Natl. Acad. Sci. U. S. A. 115, 11637–11642 10.1073/pnas.1810705115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang X., Bernoux M., Bentham A. R., Newman T. E., Ve T., Casey L. W., Raaymakers T. M., Hu J., Croll T. I., Schreiber K. J., Staskawicz B. J., Anderson P. A., Sohn K. H., Williams S. J., Dodds P. N., et al. (2017) Multiple functional self-association interfaces in plant TIR domains. Proc. Natl. Acad. Sci. U. S. A. 114, E2046–E2052 10.1073/pnas.1621248114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hyun K-G., Lee Y., Yoon J., Yi H., and Song J.-J. (2016) Crystal structure of Arabidopsis thaliana SNC1 TIR domain. Biochem. Biophys. Res. Commun. 481, 146–152 10.1016/j.bbrc.2016.11.004 [DOI] [PubMed] [Google Scholar]
- 34. Steele J. F. C., Hughes R. K., and Banfield M. J. (2019) Structural and biochemical studies of an NB-ARC domain from a plant NLR immune receptor. PLoS ONE 14, e0221226 10.1371/journal.pone.0221226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Horsefield S., Burdett H., Zhang X., Manik M. K., Shi Y., Chen J., Qi T., Gilley J., Lai J.-S., Rank M. X., Casey L. W., Gu W., Ericsson D. J., Foley G., Hughes R. O., et al. (2019) NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365, 793–799 10.1126/science.aax1911 [DOI] [PubMed] [Google Scholar]
- 36. Williams S., YIn L., Foley G., Casey L., Outram M., Ericsson D., Lu J., Boden M., Dry I., and Kobe B. (2016) Structure and function of the TIR domain from the grape NLR protein RPV1. Front. Plant Sci. 7, 1850 10.3389/fpls.2016.01850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bernoux M., Ve T., Williams S., Warren C., Hatters D., Valkov E., Zhang X., Ellis J. G., Kobe B., and Dodds P. N. (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9, 200–211 10.1016/j.chom.2011.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dong J., Xiao F., Fan F., Gu L., Cang H., Martin G. B., and Chai J. (2009) Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto. Plant Cell 21, 1846–1859 10.1105/tpc.109.066878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kanyuka K., and Rudd J. J. (2019) Cell surface immune receptors: the guardians of the plant's extracellular spaces. Curr. Opin. Plant Biol. 50, 1–8 10.1016/j.pbi.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Wersch S., Tian L., Hoy R., and Li X. (2020) Plant NLRs: the whistleblowers of plant immunity. Plant Commun. 1, 100016 10.1016/j.xplc.2019.100016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Snelders N. C., Rovenich H., Petti G. C., Rocafort M., Vorholt J. A., Mesters J. R., Seidl M. F., Nijland R., and Thomma B. P. H. J. (2020) A plant pathogen utilizes effector proteins for microbiome manipulation. bioRxiv 10.1101/2020.01.30.926725 [DOI] [PubMed]
- 42. Varden F. A., De la Concepcion J. C., Maidment J. H. R., and Banfield M. J. (2017) Taking the stage: effectors in the spotlight. Curr. Opin. Plant Biol. 38, 25–33 10.1016/j.pbi.2017.04.013 [DOI] [PubMed] [Google Scholar]
- 43. Ngou B. P. M., Ahn H.-K., Ding P., and Jones J. D. (2020) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. bioRxiv 10.1101/2020.04.10.034173 [DOI] [PubMed]
- 44. Yuan M., Jiang Z., Bi G., Nomura K., Liu M., He S. Y., Zhou J.-M., and Xin X.-F. (2020) Pattern-recognition receptors are required for NLR-mediated plant immunity. bioRxiv 10.1101/2020.04.10.031294 [DOI] [PMC free article] [PubMed]
- 45. Jones J. D. G., and Dangl J. L. (2006) The plant immune system. Nature 444, 323–329 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 46. Chisholm S. T., Coaker G., Day B., and Staskawicz B. J. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814 10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 47. Takken F. L. W., Albrecht M., and Tameling W. I. L. (2006) Resistance proteins: molecular switches of plant defence. Curr. Opin. Plant Biol. 9, 383–390 10.1016/j.pbi.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 48. Takken F. L. W., and Goverse A. (2012) How to build a pathogen detector: structural basis of NB-LRR function. Curr. Opin. Plant Biol. 15, 375–384 10.1016/j.pbi.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 49. Wirthmueller L., Maqbool A., and Banfield M. J. (2013) On the front line: structural insights into plant-pathogen interactions. Nat. Rev. Microbiol. 11, 761–776 10.1038/nrmicro3118 [DOI] [PubMed] [Google Scholar]
- 50. Schellenberger R., Touchard M., Clément C., Baillieul F., Cordelier S., Crouzet J., and Dorey S. (2019) Apoplastic invasion patterns triggering plant immunity: plasma membrane sensing at the frontline. Mol. Plant Pathol. 20, 1602–1616 10.1111/mpp.12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kourelis J., and van der Hoorn R. A. L. (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299 10.1105/tpc.17.00579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tamborski J., and Krasileva K. V. (2020) Evolution of plant NLRs: from natural history to precise modifications. Annu. Rev. Plant Biol. 71, 355–378 10.1146/annurev-arplant-081519-035901 [DOI] [PubMed] [Google Scholar]
- 53. Hogenhout S. A., Van der Hoorn R. A. L., Terauchi R., and Kamoun S. (2009) Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 22, 115–122 10.1094/MPMI-22-2-0115 [DOI] [PubMed] [Google Scholar]
- 54. Dong S., Raffaele S., and Kamoun S. (2015) The two-speed genomes of filamentous pathogens: waltz with plants. Curr. Opin. Genet. Dev. 35, 57–65 10.1016/j.gde.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 55. Raffaele S., Farrer R. A., Cano L. M., Studholme D. J., MacLean D., Thines M., Jiang R. H., Zody M. C., Kunjeti S. G., Donofrio N. M., Meyers B. C., Nusbaum C., and Kamoun S. (2010) Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330, 1540–1543 10.1126/science.1193070 [DOI] [PubMed] [Google Scholar]
- 56. Allen R. L., Bittner-Eddy P. D., Grenville-Briggs L. J., Meitz J. C., Rehmany A. P., Rose L. E., and Beynon J. L. (2004) Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306, 1957–1960 10.1126/science.1104022 [DOI] [PubMed] [Google Scholar]
- 57. Sperschneider J. (2019) Machine learning in plant–pathogen interactions: empowering biological predictions from field scale to genome scale. New Phytol. 10.1111/nph.15771 10.1111/nph.15771 [DOI] [PubMed] [Google Scholar]
- 58. Franceschetti M., Maqbool A., Jiménez-Dalmaroni M. J., Pennington H. G., Kamoun S., and Banfield M. J. (2017) Effectors of filamentous plant pathogens: commonalities amid diversity. Microbiol. Mol. Biol. Rev. 81, e00066–16 10.1128/MMBR.00066-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weßling R., Epple P., Altmann S., He Y., Yang L., Henz S. R., McDonald N., Wiley K., Bader K. C., Gläßer C., Mukhtar M. S., Haigis S., Ghamsari L., Stephens A. E., Ecker J. R., et al. (2014) Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16, 364–375 10.1016/j.chom.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mukhtar M. S., Carvunis A. R., Dreze M., Epple P., Steinbrenner J., Moore J., Tasan M., Galli M., Hao T., Nishimura M. T., Pevzner S. J., Donovan S. E., Ghamsari L., Santhanam B., Romero V., et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333, 596–601 10.1126/science.1203659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sanabria N., Goring D., Nürnberger T., and Dubery I. (2008) Self/nonself perception and recognition mechanisms in plants: a comparison of self-incompatibility and innate immunity. New Phytol. 178, 503–514 10.1111/j.1469-8137.2008.02403.x [DOI] [PubMed] [Google Scholar]
- 62. Antolín-Llovera M., Petutsching E. K., Ried M. K., Lipka V., Nürnberger T., Robatzek S., and Parniske M. (2014) Knowing your friends and foes—plant receptor-like kinases as initiators of symbiosis or defence. New Phytol. 204, 791–802 10.1111/nph.13117 [DOI] [PubMed] [Google Scholar]
- 63. Jamieson P. A., Shan L., and He P. (2018) Plant cell surface molecular cypher: receptor-like proteins and their roles in immunity and development. Plant Sci. 274, 242–251 10.1016/j.plantsci.2018.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boller T., and Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- 65. Saijo Y., Loo E. P-I., and Yasuda S. (2018) Pattern recognition receptors and signaling in plant–microbe interactions. Plant J. 93, 592–613 10.1111/tpj.13808 [DOI] [PubMed] [Google Scholar]
- 66. Böhm H., Albert I., Fan L., Reinhard A., and Nürnberger T. (2014) Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 20, 47–54 10.1016/j.pbi.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 67. Macho A. P., and Zipfel C. (2014) Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272 10.1016/j.molcel.2014.03.028 [DOI] [PubMed] [Google Scholar]
- 68. Dodds P. N., and Rathjen J. P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548 10.1038/nrg2812 [DOI] [PubMed] [Google Scholar]
- 69. Boutrot F., and Zipfel C. (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286 10.1146/annurev-phyto-080614-120106 [DOI] [PubMed] [Google Scholar]
- 70. Lamb C., and Dixon R. A. (1997) The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275 10.1146/annurev.arplant.48.1.251 [DOI] [PubMed] [Google Scholar]
- 71. Balint-Kurti P. (2019) The plant hypersensitive response: concepts, control and consequences. Mol. Plant Pathol. 20, 1163–1178 10.1111/mpp.12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Greenberg J. T. (1997) Programmed cell death in plant-pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 525–545 10.1146/annurev.arplant.48.1.525 [DOI] [PubMed] [Google Scholar]
- 73. Zipfel C., and Oldroyd G. E. (2017) Plant signalling in symbiosis and immunity. Nature 543, 328–336 10.1038/nature22009 [DOI] [PubMed] [Google Scholar]
- 74. Wang J., and Chai J. (2020) Structural insights into the plant immune receptors PRRs and NLRs. Plant Physiol. 182, 1566–1581 10.1104/pp.19.01252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dardick C., Schwessinger B., and Ronald P. (2012) Non-arginine-aspartate (non-RD) kinases are associated with innate immune receptors that recognize conserved microbial signatures. Curr. Opin. Plant Biol. 15, 358–366 10.1016/j.pbi.2012.05.002 [DOI] [PubMed] [Google Scholar]
- 76. Ma X., Xu G., He P., and Shan L. (2016) SERKing coreceptors for receptors. Trends Plant Sci. 21, 1017–1033 10.1016/j.tplants.2016.08.014 [DOI] [PubMed] [Google Scholar]
- 77. Gao X., Ruan X., Sun Y., Wang X., and Feng B. (2018) BAKing up to survive a battle: functional dynamics of BAK1 in plant programmed cell death. Front. Plant Sci. 9, 1913 10.3389/fpls.2018.01913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Burkart R. C., and Stahl Y. (2017) Dynamic complexity: plant receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 40, 15–21 10.1016/j.pbi.2017.06.016 [DOI] [PubMed] [Google Scholar]
- 79. Wu Y., and Zhou J.-M. (2013) Receptor-like kinases in plant innate immunity. J. Integr. Plant Biol. 55, 1271–1286 10.1111/jipb.12123 [DOI] [PubMed] [Google Scholar]
- 80. Zipfel C. (2014) Plant pattern-recognition receptors. Trends Immunol. 35, 345–351 10.1016/j.it.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 81. Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., and Felix G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16, 3496–3507 10.1105/tpc.104.026765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J. D. G., Felix G., and Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- 83. Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., and Shibuya N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 104, 19613–19618 10.1073/pnas.0705147104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cao Y., Liang Y., Tanaka K., Nguyen C. T., Jedrzejczak R. P., Joachimiak A., and Stacey G. (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 3, e03766 10.7554/eLife.03766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schulze B., Mentzel T., Jehle A. K., Mueller K., Beeler S., Boller T., Felix G., and Chinchilla D. (2010) Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444–9451 10.1074/jbc.M109.096842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Heese A., Hann D. R., Gimenez-Ibanez S., Jones A. M. E., He K., Li J., Schroeder J. I., Peck S. C., and Rathjen J. P. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. U. S. A. 104, 12217–12222 10.1073/pnas.0705306104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lu D., Wu S., Gao X., Zhang Y., Shan L., and He P. (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. U. S. A. 107, 496–501 10.1073/pnas.0909705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang J., Li W., Xiang T., Liu Z., Laluk K., Ding X., Zou Y., Gao M., Zhang X., Chen S., Mengiste T., Zhang Y., and Zhou J.-M. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290–301 10.1016/j.chom.2010.03.007 [DOI] [PubMed] [Google Scholar]
- 89. Ma X., Claus L. A. N., Leslie M. E., Tao K., Wu Z., Liu J., Yu X., Li B., Zhou J., Savatin D. V., Peng J., Tyler B. M., Heese A., Russinova E., He P., et al. (2020) Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 581, 199–203 10.1038/s41586-020-2210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D. G., Boller T., and Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- 91. Albert M. (2013) Peptides as triggers of plant defence. J. Exp. Bot. 64, 5269–5279 10.1093/jxb/ert275 [DOI] [PubMed] [Google Scholar]
- 92. Mott G. A., Middleton M. A., Desveaux D., and Guttman D. S. (2014) Peptides and small molecules of the plant-pathogen apoplastic arena. Front. Plant Sci. 5, 677 10.3389/fpls.2014.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Smakowska-Luzan E., Mott G. A., Parys K., Stegmann M., Howton T. C., Layeghifard M., Neuhold J., Lehner A., Kong J., Grünwald K., Weinberger N., Satbhai S. B., Mayer D., Busch W., Madalinski M., et al. (2018) An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553, 342–346 10.1038/nature25184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pruitt R. N., Schwessinger B., Joe A., Thomas N., Liu F., Albert M., Robinson M. R., Chan L. J. G., Luu D. D., Chen H., Bahar O., Daudi A., De Vleesschauwer D., Caddell D., Zhang W., et al. (2015) The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci. Adv. 1, e1500245 10.1126/sciadv.1500245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Felix G., and Boller T. (2003) Molecular sensing of bacteria in plants: the highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 278, 6201–6208 10.1074/jbc.M209880200 [DOI] [PubMed] [Google Scholar]
- 96. Wang L., Albert M., Einig E., Fürst U., Krust D., and Felix G. (2016) The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat. Plants 2, 16185 10.1038/nplants.2016.185 [DOI] [PubMed] [Google Scholar]
- 97. Wei Y., Caceres-Moreno C., Jimenez-Gongora T., Wang K., Sang Y., Lozano-Duran R., and Macho A. P. (2018) The Ralstonia solanacearum csp22 peptide, but not flagellin-derived peptides, is perceived by plants from the Solanaceae family. Plant Biotechnol. J. 16, 1349–1362 10.1111/pbi.12874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Albert I., Böhm H., Albert M., Feiler C. E., Imkampe J., Wallmeroth N., Brancato C., Raaymakers T. M., Oome S., Zhang H., Krol E., Grefen C., Gust A. A., Chai J., Hedrich R., et al. (2015) An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1, 15140 10.1038/nplants.2015.140 [DOI] [PubMed] [Google Scholar]
- 99. Haruta M., Sabat G., Stecker K., Minkoff B. B., and Sussman M. R. (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411 10.1126/science.1244454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Xiao Y., Stegmann M., Han Z., DeFalco T. A., Parys K., Xu L., Belkhadir Y., Zipfel C., and Chai J. (2019) Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572, 270–274 10.1038/s41586-019-1409-7 [DOI] [PubMed] [Google Scholar]
- 101. Thynne E., Saur I. M. L., Simbaqueba J., Ogilvie H. A., Gonzalez-Cendales Y., Mead O., Taranto A., Catanzariti A.-M., McDonald M. C., Schwessinger B., Jones D. A., Rathjen J. P., and Solomon P. S. (2017) Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Mol. Plant Pathol. 18, 811–824 10.1111/mpp.12444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Krol E., Mentzel T., Chinchilla D., Boller T., Felix G., Kemmerling B., Postel S., Arents M., Jeworutzki E., Al-Rasheid K. A. S., Becker D., and Hedrich R. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285, 13471–13479 10.1074/jbc.M109.097394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Yamaguchi Y., Huffaker A., Bryan A. C., Tax F. E., and Ryan C. A. (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508 10.1105/tpc.109.068874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hou S., Wang X., Chen D., Yang X., Wang M., Turrà D., Di Pietro A., and Zhang W. (2014) The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog. 10, e1004331 10.1371/journal.ppat.1004331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hou S., Liu Z., Shen H., and Wu D. (2019) Damage-associated molecular pattern-triggered immunity in plants. Front. Plant Sci. 10, 646 10.3389/fpls.2019.00646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Dixon M. S., Jones D. A., Keddie J. S., Thomas C. M., Harrison K., and Jones J. D. G. (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84, 451–459 10.1016/s0092-8674(00)81290-8 [DOI] [PubMed] [Google Scholar]
- 107. Dixon M. S., Hatzixanthis K., Jones D. A., Harrison K., and Jones J. D. G. (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10, 1915–1925 10.1105/tpc.10.11.1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Krüger J., Thomas C. M., Golstein C., Dixon M. S., Smoker M., Tang S., Mulder L., and Jones J. D. G. (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296, 744–747 10.1126/science.1069288 [DOI] [PubMed] [Google Scholar]
- 109. Luderer R., Takken F. L. W., Wit P. J. G. M. D., and Joosten M. H. A. J. (2002) Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45, 875–884 10.1046/j.1365-2958.2002.03060.x [DOI] [PubMed] [Google Scholar]
- 110. Rooney H. C. E., van T., Klooster J. W., van der Hoorn R. A. L., Joosten M. H. A. J., Jones J. D. G., and de Wit P. J. G. M. (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308, 1783 10.1126/science.1111404 [DOI] [PubMed] [Google Scholar]
- 111. Wan J., Zhang X.-C., Neece D., Ramonell K. M., Clough S., Kim S.-Y., Stacey M. G., and Stacey G. (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20, 471–481 10.1105/tpc.107.056754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gust A. A. (2015) Peptidoglycan perception in plants. PLoS Pathog. 11, e1005275 10.1371/journal.ppat.1005275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Verica J. A., and He Z.-H. (2002) The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 129, 455–459 10.1104/pp.011028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Brutus A., Sicilia F., Macone A., Cervone F., and De Lorenzo G. (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U. S. A. 107, 9452–9457 10.1073/pnas.1000675107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kohorn B. D. (2016) Cell wall-associated kinases and pectin perception. J. Exp. Bot. 67, 489–494 10.1093/jxb/erv467 [DOI] [PubMed] [Google Scholar]
- 116. Souza C. D A., Li S., Lin A. Z., Boutrot F., Grossmann G., Zipfel C., and Somerville S. C. (2017) Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiol. 173, 2383–2398 10.1104/pp.16.01680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kohorn B. D., and Kohorn S. L. (2012) The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3, 88 10.3389/fpls.2012.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Choi J., Tanaka K., Cao Y., Qi Y., Qiu J., Liang Y., Lee S. Y., and Stacey G. (2014) Identification of a plant receptor for extracellular ATP. Science 343, 290–294 10.1126/science.343.6168.290 [DOI] [PubMed] [Google Scholar]
- 119. Tanaka K., Choi J., Cao Y., and Stacey G. (2014) Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Front. Plant Sci. 5, 446 10.3389/fpls.2014.00446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Yasuda S., Okada K., and Saijo Y. (2017) A look at plant immunity through the window of the multitasking coreceptor BAK1. Curr. Opin. Plant Biol. 38, 10–18 10.1016/j.pbi.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 121. Chinchilla D., Shan L., He P., de Vries S., and Kemmerling B. (2009) One for all: the receptor-associated kinase BAK1. Trends Plant Sci. 14, 535–541 10.1016/j.tplants.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gust A. A., and Felix G. (2014) Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 21, 104–111 10.1016/j.pbi.2014.07.007 [DOI] [PubMed] [Google Scholar]
- 123. Liebrand T. W. H., van den Burg H. A., and Joosten M. H. A. J. (2014) Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 19, 123–132 10.1016/j.tplants.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 124. van der Burgh A. M., Postma J., Robatzek S., and Joosten M. H. A. J. (2019) Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol. 20, 410–422 10.1111/mpp.12767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tör M., Lotze M. T., and Holton N. (2009) Receptor-mediated signalling in plants: molecular patterns and programmes. J. Exp. Bot. 60, 3645–3654 10.1093/jxb/erp233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Postma J., Liebrand T. W. H., Bi G., Evrard A., Bye R. R., Mbengue M., Kuhn H., Joosten M. H. A. J., and Robatzek S. (2016) Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol. 210, 627–642 10.1111/nph.13802 [DOI] [PubMed] [Google Scholar]
- 127. Domazakis E., Wouters D., Visser R. G. F., Kamoun S., Joosten M. H. A. J., and Vleeshouwers V. G. A. A. (2018) The ELR-SOBIR1 complex functions as a two-component receptor-like kinase to mount defense against Phytophthora infestans. Mol. Plant Microbe Interact. 31, 795–802 10.1094/MPMI-09-17-0217-R [DOI] [PubMed] [Google Scholar]
- 128. Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., and Shibuya N. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214 10.1111/j.1365-313X.2010.04324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Squeglia F., Berisio R., Shibuya N., and Kaku H. (2017) Defense against pathogens: structural insights into the mechanism of chitin induced activation of innate immunity. Curr. Med. Chem. 24, 3980–3986 10.2174/0929867323666161221124345 [DOI] [PubMed] [Google Scholar]
- 130. Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., and Shibuya N. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U. S. A. 103, 11086–11091 10.1073/pnas.0508882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lin W., Ma X., Shan L., and He P. (2013) Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J. Integr. Plant Biol. 55, 1188–1197 10.1111/jipb.12071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Liang X., and Zhou J.-M. (2018) Receptor-like cytoplasmic kinases: central players in plant receptor kinase–mediated signaling. Annu. Rev. Plant Biol. 69, 267–299 10.1146/annurev-arplant-042817-040540 [DOI] [PubMed] [Google Scholar]
- 133. Veronese P., Nakagami H., Bluhm B., AbuQamar S., Chen X., Salmeron J., Dietrich R. A., Hirt H., and Mengiste T. (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273 10.1105/tpc.105.035576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ranf S., Eschen-Lippold L., Fröhlich K., Westphal L., Scheel D., and Lee J. (2014) Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol. 14, 374 10.1186/s12870-014-0374-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Kadota Y., Shirasu K., and Zipfel C. (2015) Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56, 1472–1480 10.1093/pcp/pcv063 [DOI] [PubMed] [Google Scholar]
- 136. Monaghan J., Matschi S., Romeis T., and Zipfel C. (2015) The calcium-dependent protein kinase CPK28 negatively regulates the BIK1-mediated PAMP-induced calcium burst. Plant Signal. Behav. 10, e1018497 10.1080/15592324.2015.1018497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., Chen S., and Zhou J.-M. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338 10.1016/j.chom.2014.02.009 [DOI] [PubMed] [Google Scholar]
- 138. Qi J., Wang J., Gong Z., and Zhou J.-M. (2017) Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100 10.1016/j.pbi.2017.04.022 [DOI] [PubMed] [Google Scholar]
- 139. Couto D., and Zipfel C. (2016) Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 10.1038/nri.2016.77 [DOI] [PubMed] [Google Scholar]
- 140. Gómez-Gómez L., Bauer Z., and Boller T. (2001) Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13, 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- 141. Ding Z., Wang H., Liang X., Morris E. R., Gallazzi F., Pandit S., Skolnick J., Walker J. C., and Van Doren S. R. (2007) Phosphoprotein and phosphopeptide interactions with the FHA domain from Arabidopsis kinase-associated protein phosphatase. Biochemistry 46, 2684–2696 10.1021/bi061763n [DOI] [PubMed] [Google Scholar]
- 142. Park C.-J., Caddell D. F., and Ronald P. C. (2012) Protein phosphorylation in plant immunity: insights into the regulation of pattern recognition receptor-mediated signaling. Front. Plant Sci. 3, 177 10.3389/fpls.2012.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Segonzac C., Macho A. P., Sanmartín M., Ntoukakis V., Sánchez-Serrano J. J., and Zipfel C. (2014) Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 33, 2069–2079 10.15252/embj.201488698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Durian G., Rahikainen M., Alegre S., Brosché M., and Kangasjärvi S. (2016) Protein phosphatase 2A in the regulatory network underlying biotic stress resistance in plants. Front. Plant Sci. 7, 812 10.3389/fpls.2016.00812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Halter T., Imkampe J., Blaum B. S., Stehle T., and Kemmerling B. (2014) BIR2 affects complex formation of BAK1 with ligand binding receptors in plant defense. Plant Signal. Behav. 9, e28944 10.4161/psb.28944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Halter T., Imkampe J., Mazzotta S., Wierzba M., Postel S., Bücherl C., Kiefer C., Stahl M., Chinchilla D., Wang X., Nürnberger T., Zipfel C., Clouse S., Borst J. W., Boeren S., et al. (2014) The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24, 134–143 10.1016/j.cub.2013.11.047 [DOI] [PubMed] [Google Scholar]
- 147. Liu Y., Huang X., Li M., He P., and Zhang Y. (2016) Loss-of-function of Arabidopsis receptor-like kinase BIR1 activates cell death and defense responses mediated by BAK1 and SOBIR1. New Phytol. 212, 637–645 10.1111/nph.14072 [DOI] [PubMed] [Google Scholar]
- 148. Wang J., Grubb L. E., Wang J., Liang X., Li L., Gao C., Ma M., Feng F., Li M., Li L., Zhang X., Yu F., Xie Q., Chen S., Zipfel C., et al. (2018) A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 69, 493–504.e6 10.1016/j.molcel.2017.12.026 [DOI] [PubMed] [Google Scholar]
- 149. Robatzek S., Chinchilla D., and Boller T. (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542 10.1101/gad.366506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T. P., He P., and Shan L. (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332, 1439–1442 10.1126/science.1204903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Smith J. M., Salamango D. J., Leslie M. E., Collins C. A., and Heese A. (2014) Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol. 164, 440–454 10.1104/pp.113.229179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Korasick D. A., McMichael C., Walker K. A., Anderson J. C., Bednarek S. Y., and Heese A. (2010) Novel functions of stomatal cytokinesis-defective 1 (SCD1) in innate immune responses against bacteria. J. Biol. Chem. 285, 23342–23350 10.1074/jbc.M109.090787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. McMichael C. M., Reynolds G. D., Koch L. M., Wang C., Jiang N., Nadeau J., Sack F. D., Gelderman M. B., Pan J., and Bednarek S. Y. (2013) Mediation of clathrin-dependent trafficking during cytokinesis and cell expansion by Arabidopsis stomatal cytokinesis defective proteins. Plant Cell 25, 3910 10.1105/tpc.113.115162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Spallek T., Beck M., Ben Khaled S., Salomon S., Bourdais G., Schellmann S., and Robatzek S. (2013) ESCRT-I mediates FLS2 endosomal sorting and plant immunity. PLoS Genet. 9, e1004035 10.1371/journal.pgen.1004035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Schuh A. L., and Audhya A. (2014) The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit. Rev. Biochem. Mol. Biol. 49, 242–261 10.3109/10409238.2014.881777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Ben Khaled S., Postma J., and Robatzek S. (2015) A moving view: subcellular trafficking processes in pattern recognition receptor–triggered plant immunity. Annu. Rev. Phytopathol. 53, 379–402 10.1146/annurev-phyto-080614-120347 [DOI] [PubMed] [Google Scholar]
- 157. Bücherl C. A., Jarsch I. K., Schudoma C., Segonzac C., Mbengue M., Robatzek S., MacLean D., Ott T., and Zipfel C. (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 6, e25114 10.7554/eLife.25114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Dong O. X., and Ronald P. C. (2019) Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiol. 180, 26–38 10.1104/pp.18.01224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Rodriguez-Moreno L., Song Y., and Thomma B. P. H. J. (2017) Transfer and engineering of immune receptors to improve recognition capacities in crops. Curr. Opin. Plant Biol. 38, 42–49 10.1016/j.pbi.2017.04.010 [DOI] [PubMed] [Google Scholar]
- 160. Gómez-Gómez L., and Boller T. (2000) FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011 10.1016/s1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- 161. Shiu S.-H., and Bleecker A. B. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U. S. A. 98, 10763–10768 10.1073/pnas.181141598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., and Aderem A. (2001) The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 10.1038/35074106 [DOI] [PubMed] [Google Scholar]
- 163. Dangl J. L., and Jones J. D. G. (2001) Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 10.1038/35081161 [DOI] [PubMed] [Google Scholar]
- 164. Flor H. H. (1971) Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296 10.1146/annurev.py.09.090171.001423 [DOI] [Google Scholar]
- 165. Takken F. L. W., and Tameling W. I. L. (2009) To nibble at plant resistance proteins. Science 324, 744–746 10.1126/science.1171666 [DOI] [PubMed] [Google Scholar]
- 166. Zou H., Li Y., Liu X., and Wang X. (1999) An APAF-1·cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 274, 11549–11556 10.1074/jbc.274.17.11549 [DOI] [PubMed] [Google Scholar]
- 167. Duncan J. A., Bergstralh D. T., Wang Y., Willingham S. B., Ye Z., Zimmermann A. G., and Ting J. P.-Y. (2007) Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl. Acad. Sci. U. S. A. 104, 8041–8046 10.1073/pnas.0611496104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Poyet J. L., Srinivasula S. M., Tnani M., Razmara M., Fernandes-Alnemri T., and Alnemri E. S. (2001) Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276, 28309–28313 10.1074/jbc.C100250200 [DOI] [PubMed] [Google Scholar]
- 169. Sharif H., Wang L., Wang W. L., Magupalli V. G., Andreeva L., Qiao Q., Hauenstein A. V., Wu Z., Núñez G., Mao Y., and Wu H. (2019) Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570, 338–343 10.1038/s41586-019-1295-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Tenthorey J. L., Haloupek N., López-Blanco J. R., Grob P., Adamson E., Hartenian E., Lind N. A., Bourgeois N. M., Chacón P., Nogales E., and Vance R. E. (2017) The structural basis of flagellin detection by NAIP5: a strategy to limit pathogen immune evasion. Science 358, 888–893 10.1126/science.aao1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Zhang L., Chen S., Ruan J., Wu J., Tong A. B., Yin Q., Li Y., David L., Lu A., Wang W. L., Marks C., Ouyang Q., Zhang X., Mao Y., and Wu H. (2015) Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409 10.1126/science.aac5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Van der Biezen E. A., and Jones J. D. (1998) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456 10.1016/S0968-0004(98)01311-5 [DOI] [PubMed] [Google Scholar]
- 173. Duxbury Z., Ma Y., Furzer O. J., Huh S. U., Cevik V., Jones J. D. G., and Sarris P. F. (2016) Pathogen perception by NLRs in plants and animals: parallel worlds. BioEssays 38, 769–781 10.1002/bies.201600046 [DOI] [PubMed] [Google Scholar]
- 174. Dodds P. N., Lawrence G. J., Catanzariti A.-M., Ayliffe M. A., and Ellis J. G. (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16, 755–768 10.1105/tpc.020040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Faustin B., Lartigue L., Bruey J. M., Luciano F., Sergienko E., Bailly-Maitre B., Volkmann N., Hanein D., Rouiller I., and Reed J. C. (2007) Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell 25, 713–724 10.1016/j.molcel.2007.01.032 [DOI] [PubMed] [Google Scholar]
- 176. Zhang X., Dodds P. N., and Bernoux M. (2017) What do we know about NOD-like receptors in plant immunity? Annu. Rev. Phytopathol. 55, 205–229 10.1146/annurev-phyto-080516-035250 [DOI] [PubMed] [Google Scholar]
- 177. Cesari S., Bernoux M., Moncuquet P., Kroj T., and Dodds P. N. (2014) A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy hypothesis. Front. Plant Sci. 5, 606 10.3389/fpls.2014.00606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Cesari S., Thilliez G., Ribot C., Chalvon V., Michel C., Jauneau A., Rivas S., Alaux L., Kanzaki H., Okuyama Y., Morel J. B., Fournier E., Tharreau D., Terauchi R., and Kroj T. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481 10.1105/tpc.112.107201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Sarris P. F., Duxbury Z., Huh S. U., Ma Y., Segonzac C., Sklenar J., Derbyshire P., Cevik V., Rallapalli G., Saucet S. B., Wirthmueller L., Menke F. L. H., Sohn K. H., and Jones J. D. G. (2015) A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100 10.1016/j.cell.2015.04.024 [DOI] [PubMed] [Google Scholar]
- 180. Le Roux C., Huet G., Jauneau A., Camborde L., Tremousaygue D., Kraut A., Zhou B., Levaillant M., Adachi H., Yoshioka H., Raffaele S., Berthome R., Coute Y., Parker J. E., and Deslandes L. (2015) A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161, 1074–1088 10.1016/j.cell.2015.04.025 [DOI] [PubMed] [Google Scholar]
- 181. Cesari S. (2018) Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 219, 17–24 10.1111/nph.14877 [DOI] [PubMed] [Google Scholar]
- 182. Islam M. R., and Mayo G. M. E. (1990) A compendium on host genes in flax conferring resistance to flax rust. Plant Breed. 104, 89–100 10.1111/j.1439-0523.1990.tb00409.x [DOI] [Google Scholar]
- 183. Ellis J. G., Lawrence G. J., Luck J. E., and Dodds P. N. (1999) Identification of regions in alleles of the flax rust resistance gene that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506 10.2307/3870876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Dodds P. N., Lawrence G. J., and Ellis J. G. (2001) Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13, 163–178 10.2307/3871161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Mackey D., Holt B. F. 3rd, Wiig A., and Dangl J. L. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754 10.1016/S0092-8674(02)00661-X [DOI] [PubMed] [Google Scholar]
- 186. Mackey D., Belkhadir Y., Alonso J. M., Ecker J. R., and Dangl J. L. (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389 10.1016/S0092-8674(03)00040-0 [DOI] [PubMed] [Google Scholar]
- 187. Mucyn T. S., Clemente A., Andriotis V. M. E., Balmuth A. L., Oldroyd G. E. D., Staskawicz B. J., and Rathjen J. P. (2006) The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18, 2792–2806 10.1105/tpc.106.044016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Mucyn T. S., Wu A.-J., Balmuth A. L., Arasteh J. M., and Rathjen J. P. (2009) Regulation of tomato Prf by Pto-like protein kinases. Mol. Plant Microbe Interact. 22, 391–401 10.1094/MPMI-22-4-0391 [DOI] [PubMed] [Google Scholar]
- 189. Ntoukakis V., Saur I. M., Conlan B., and Rathjen J. P. (2014) The changing of the guard: the Pto/Prf receptor complex of tomato and pathogen recognition. Curr. Opin. Plant Biol. 20, 69–74 10.1016/j.pbi.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 190. Ortiz D., de Guillen K., Cesari S., Chalvon V., Gracy J., Padilla A., and Kroj T. (2017) Recognition of the Magnaporthe oryzae effector AVR-Pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell 29, 156–168 10.1105/tpc.16.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Narusaka M., Shirasu K., Noutoshi Y., Kubo Y., Shiraishi T., Iwabuchi M., and Narusaka Y. (2009) RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226 10.1111/j.1365-313X.2009.03949.x [DOI] [PubMed] [Google Scholar]
- 192. Sohn K. H., Hughes R. K., Piquerez S. J., Jones J. D. G., and Banfield M. J. (2012) Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proc. Natl. Acad. Sci. U. S. A. 109, 16371–16376 10.1073/pnas.1212332109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Steuernagel B., Witek K., Krattinger S. G., Ramirez-Gonzalez R. H., Schoonbeek H-J., Yu G., Baggs E., Witek A., Yadav I., Krasileva K. V., Jones J. D., Uauy C., Keller B., Ridout C. J., and Wulff B. B. (2020) The NLR-Annotator tool enables annotation of the intracellular immune receptor repertoire. Plant Physiol. 183, 468–482 10.1104/pp.19.01273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Bailey P. C., Schudoma C., Jackson W., Baggs E., Dagdas G., Haerty W., Moscou M., and Krasileva K. V. (2018) Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 19, 23 10.1186/s13059-018-1392-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Sarris P. F., Cevik V., Dagdas G., Jones J. D. G., and Krasileva K. V. (2016) Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 14, 8 10.1186/s12915-016-0228-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Kroj T., Chanclud E., Michel-Romiti C., Grand X., and Morel J. B. (2016) Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210, 618–626 10.1111/nph.13869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Wang L., Zhao L., Zhang X., Zhang Q., Jia Y., Wang G., Li S., Tian D., Li W.-H., and Yang S. (2019) Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc. Natl. Acad. Sci. U. S. A. 116, 18479–18487 10.1073/pnas.1910229116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. de Abreu-Neto J. B., Turchetto-Zolet A. C., de Oliveira L. F. V., Bodanese Zanettini M. H., and Margis-Pinheiro M. (2013) Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS J. 280, 1604–1616 10.1111/febs.12159 [DOI] [PubMed] [Google Scholar]
- 199. Cowan G. H., Roberts A. G., Jones S., Kumar P., Kalyandurg P. B., Gil J. F., Savenkov E. I., Hemsley P. A., and Torrance L. (2018) Potato mop-top virus co-opts the stress sensor HIPP26 for long-distance movement. Plant Physiol. 176, 2052–2070 10.1104/pp.17.01698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Radakovic Z. S., Anjam M. S., Escobar E., Chopra D., Cabrera J., Silva A. C., Escobar C., Sobczak M., Grundler F. M. W., and Siddique S. (2018) Arabidopsis HIPP27 is a host susceptibility gene for the beet cyst nematode Heterodera schachtii. Mol. Plant Pathol. 19, 1917–1928 10.1111/mpp.12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Fukuoka S., Saka N., Koga H., Ono K., Shimizu T., Ebana K., Hayashi N., Takahashi A., Hirochika H., Okuno K., and Yano M. (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325, 998–1001 10.1126/science.1175550 [DOI] [PubMed] [Google Scholar]
- 202. Kanzaki H., Yoshida K., Saitoh H., Fujisaki K., Hirabuchi A., Alaux L., Fournier E., Tharreau D., and Terauchi R. (2012) Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 72, 894–907 10.1111/j.1365-313X.2012.05110.x [DOI] [PubMed] [Google Scholar]
- 203. Yoshida K., Saitoh H., Fujisawa S., Kanzaki H., Matsumura H., Yoshida K., Tosa Y., Chuma I., Takano Y., Win J., Kamoun S., and Terauchi R. (2009) Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21, 1573–1591 10.1105/tpc.109.066324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Costanzo S., and Jia Y. (2010) Sequence variation at the rice blast resistance gene Pi-km locus: Implications for the development of allele specific markers. Plant Sci. 178, 523–530 10.1016/j.plantsci.2010.02.014 [DOI] [Google Scholar]
- 205. De la Concepcion J. C., Franceschetti M., MacLean D., Terauchi R., Kamoun S., and Banfield M. J. (2019) Protein engineering expands the effector recognition profile of a rice NLR immune receptor. Elife 8, e47713 10.7554/eLife.47713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Williams S. J., Sornaraj P., deCourcy-Ireland E., Menz R. I., Kobe B., Ellis J. G., Dodds P. N., and Anderson P. A. (2011) An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol. Plant Microbe Interact. 24, 897–906 10.1094/MPMI-03-11-0052 [DOI] [PubMed] [Google Scholar]
- 207. Tameling W. I. L., Vossen J. H., Albrecht M., Lengauer T., Berden J. A., Haring M. A., Cornelissen B. J. C., and Takken F. L. W. (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140, 1233–1245 10.1104/pp.105.073510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Baudin M., Hassan J. A., Schreiber K. J., and Lewis J. D. (2017) Analysis of the ZAR1 immune complex reveals determinants for immunity and molecular interactions. Plant Physiol. 174, 2038–2053 10.1104/pp.17.00441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Adachi H., Contreras M. P., Harant A., Wu C. H., Derevnina L., Sakai T., Duggan C., Moratto E., Bozkurt T. O., Maqbool A., Win J., and Kamoun S. (2019) An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. Elife 8, e49956 10.7554/eLife.49956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Ve T., Williams S. J., and Kobe B. (2015) Structure and function of Toll/interleukin-1 receptor/resistance protein (TIR) domains. Apoptosis 20, 250–261 10.1007/s10495-014-1064-2 [DOI] [PubMed] [Google Scholar]
- 211. Wan L., Essuman K., Anderson R. G., Sasaki Y., Monteiro F., Chung E.-H., Osborne Nishimura E., DiAntonio A., Milbrandt J., Dangl J. L., and Nishimura M. T. (2019) TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365, 799–803 10.1126/science.aax1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Martin R., Qi T., Zhang H., Liu F., King M., Toth C., Nogales E., and Staskawicz B. J. (2020) Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ. bioRxiv 10.1101/2020.08.13.246413 [DOI] [PMC free article] [PubMed]
- 213. Wu C. H., Derevnina L., and Kamoun S. (2018) Receptor networks underpin plant immunity. Science 360, 1300–1301 10.1126/science.aat2623 [DOI] [PubMed] [Google Scholar]
- 214. Adachi H., Derevnina L., and Kamoun S. (2019) NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 50, 121–131 10.1016/j.pbi.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 215. Jubic L. M., Saile S., Furzer O. J., El Kasmi F., and Dangl J. L. (2019) Help wanted: helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 50, 82–94 10.1016/j.pbi.2019.03.013 [DOI] [PubMed] [Google Scholar]
- 216. van der Biezen E. A., Freddie C. T., Kahn K., Parker J. E., and Jones J. D. (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29, 439–451 10.1046/j.0960-7412.2001.01229.x [DOI] [PubMed] [Google Scholar]
- 217. Białas A., Zess E. K., De la Concepcion J. C., Franceschetti M., Pennington H. G., Yoshida K., Upson J. L., Chanclud E., Wu C.-H., Langner T., Maqbool A., Varden F. A., Derevnina L., Belhaj K., Fujisaki K., et al. (2018) Lessons in effector and NLR biology of plant-microbe systems. Mol. Plant Microbe Interact. 31, 34–45 10.1094/MPMI-08-17-0196-FI [DOI] [PubMed] [Google Scholar]
- 218. Césari S., Kanzaki H., Fujiwara T., Bernoux M., Chalvon V., Kawano Y., Shimamoto K., Dodds P., Terauchi R., and Kroj T. (2014) The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33, 1941–1959 10.15252/embj.201487923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Zdrzałek R., Kamoun S., Terauchi R., Saitoh H., and Banfield M. J. (2020) The rice NLR pair Pikp-1/Pikp-2 initiates cell death through receptor cooperation rather than negative regulation. bioRxiv 10.1101/2020.06.20.162834 [DOI] [PMC free article] [PubMed]
- 220. Peart J. R., Mestre P., Lu R., Malcuit I., and Baulcombe D. C. (2005) NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 15, 968–973 10.1016/j.cub.2005.04.053 [DOI] [PubMed] [Google Scholar]
- 221. Castel B., Ngou P. M., Cevik V., Redkar A., Kim D. S., Yang Y., Ding P., and Jones J. D. G. (2019) Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 222, 966–980 10.1111/nph.15659 [DOI] [PubMed] [Google Scholar]
- 222. Feehan J. M., Castel B., Bentham A. R., and Jones J. D. G. (2020) Plant NLRs get by with a little help from their friends. Curr. Opin. Plant Biol. 56, 99–108 10.1016/j.pbi.2020.04.006 [DOI] [PubMed] [Google Scholar]
- 223. Wu C.-H., Abd-El-Haliem A., Bozkurt T. O., Belhaj K., Terauchi R., Vossen J. H., and Kamoun S. (2017) NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. U. S. A. 114, 8113–8118 10.1073/pnas.1702041114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Burdett H., Bentham A. R., Williams S. J., Dodds P. N., Anderson P. A., Banfield M. J., and Kobe B. (2019) The plant resistosome: structural insights into immune signaling. Cell Host Microbe 26, 193–201 10.1016/j.chom.2019.07.020 [DOI] [PubMed] [Google Scholar]
- 225. Monteiro F., and Nishimura M. T. (2018) Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Annu. Rev. Phytopathol. 56, 243–267 10.1146/annurev-phyto-080417-045817 [DOI] [PubMed] [Google Scholar]
- 226. Ahmar S., Gill R. A., Jung K. H., Faheem A., Qasim M. U., Mubeen M., and Zhou W. (2020) Conventional and molecular techniques from simple breeding to speed breeding in crop plants: recent advances and future outlook. Int. J. Mol. Sci. 21, 2590 10.3390/ijms21072590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Kumar K., Gambhir G., Dass A., Tripathi A. K., Singh A., Jha A. K., Yadava P., Choudhary M., and Rakshit S. (2020) Genetically modified crops: current status and future prospects. Planta 251, 91 10.1007/s00425-020-03372-8 [DOI] [PubMed] [Google Scholar]
- 228. Hickey L. T., A N. H., Robinson H., Jackson S. A., Leal-Bertioli S. C. M., Tester M., Gao C., Godwin I. D., Hayes B. J., and Wulff B. B. H. (2019) Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754 10.1038/s41587-019-0152-9 [DOI] [PubMed] [Google Scholar]
- 229. Chakraborty J., Ghosh P., and Das S. (2018) Autoimmunity in plants. Planta 248, 751–767 10.1007/s00425-018-2956-0 [DOI] [PubMed] [Google Scholar]
- 230. Lacombe S., Rougon-Cardoso A., Sherwood E., Peeters N., Dahlbeck D., van Esse H. P., Smoker M., Rallapalli G., Thomma B. P. H. J., Staskawicz B., Jones J. D. G., and Zipfel C. (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369 10.1038/nbt.1613 [DOI] [PubMed] [Google Scholar]
- 231. Kunwar S., Iriarte F., Fan Q., Evaristo da Silva E., Ritchie L., Nguyen N. S., Freeman J. H., Stall R. E., Jones J. B., Minsavage G. V., Colee J., Scott J. W., Vallad G. E., Zipfel C., Horvath D., et al. (2018) Transgenic expression of EFR and Bs2 genes for field management of bacterial wilt and bacterial spot of tomato. Phytopathology 108, 1402–1411 10.1094/PHYTO-12-17-0424-R [DOI] [PubMed] [Google Scholar]
- 232. Schoonbeek H. J., Wang H. H., Stefanato F. L., Craze M., Bowden S., Wallington E., Zipfel C., and Ridout C. J. (2015) Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 206, 606–613 10.1111/nph.13356 [DOI] [PubMed] [Google Scholar]
- 233. Tripathi J. N., Lorenzen J., Bahar O., Ronald P., and Tripathi L. (2014) Transgenic expression of the rice Xa21 pattern-recognition receptor in banana (Musa sp.) confers resistance to Xanthomonas campestris pv. musacearum. Plant Biotechnol. J. 12, 663–673 10.1111/pbi.12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Mendes B. M. J., Cardoso S. C., Boscariol-Camargo R. L., Cruz R. B., Filho F. A. A. M., and Filho A. B. (2010) Reduction in susceptibility to Xanthomonas axonopodis pv. citri in transgenic Citrus sinensis expressing the rice Xa21 gene. Plant Pathol. 59, 68–75 10.1111/j.1365-3059.2009.02148.x [DOI] [Google Scholar]
- 235. Afroz A., Chaudhry Z., Rashid U., Muhammad Ali G., Nazir F., Iqbal J., and Rashid Khan M. (2011) Enhanced resistance against bacterial wilt in transgenic tomato (Lycopersicon esculentum) lines expressing the Xa21 gene. Plant Cell Tissue Organ Culture 104, 227–237 10.1007/s11240-010-9825-2 [DOI] [Google Scholar]
- 236. Fürst U., Zeng Y., Albert M., Witte A. K., Fliegmann J., and Felix G. (2020) Perception of Agrobacterium tumefaciens flagellin by FLS2(XL) confers resistance to crown gall disease. Nat. Plants 6, 22–27 10.1038/s41477-019-0578-6 [DOI] [PubMed] [Google Scholar]
- 237. Piquerez S. J. M., Harvey S. E., Beynon J. L., and Ntoukakis V. (2014) Improving crop disease resistance: lessons from research on Arabidopsis and tomato. Front. Plant Sci. 5, 671 10.3389/fpls.2014.00671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Tian J., Xu G., and Yuan M. (2020) Towards engineering broad-spectrum disease-resistant crops. Trends Plant Sci. 25, 424–427 10.1016/j.tplants.2020.02.012 [DOI] [PubMed] [Google Scholar]
- 239. Arora S., Steuernagel B., Gaurav K., Chandramohan S., Long Y., Matny O., Johnson R., Enk J., Periyannan S., Singh N., Asyraf Md Hatta M., Athiyannan N., Cheema J., Yu G., Kangara N., et al. (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 37, 139–143 10.1038/s41587-018-0007-9 [DOI] [PubMed] [Google Scholar]
- 240. Steuernagel B., Periyannan S. K., Hernández-Pinzón I., Witek K., Rouse M. N., Yu G., Hatta A., Ayliffe M., Bariana H., Jones J. D., Lagudah E. S., and Wulff B. B. (2016) Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655 10.1038/nbt.3543 [DOI] [PubMed] [Google Scholar]
- 241. Bevan M. W., Uauy C., Wulff B. B. H., Zhou J., Krasileva K., and Clark M. D. (2017) Genomic innovation for crop improvement. Nature 543, 346–354 10.1038/nature22011 [DOI] [PubMed] [Google Scholar]
- 242. Jupe F., Witek K., Verweij W., Sliwka J., Pritchard L., Etherington G. J., Maclean D., Cock P. J., Leggett R. M., Bryan G. J., Cardle L., Hein I., and Jones J. D. (2013) Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 76, 530–544 10.1111/tpj.12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Kawashima C. G., Guimarães G. A., Nogueira S. R., MacLean D., Cook D. R., Steuernagel B., Baek J., Bouyioukos C., Melo B. D V. A., Tristão G., de Oliveira J. C., Rauscher G., Mittal S., Panichelli L., Bacot K., et al. (2016) A pigeonpea gene confers resistance to Asian soybean rust in soybean. Nat. Biotechnol. 34, 661–665 10.1038/nbt.3554 [DOI] [PubMed] [Google Scholar]
- 244. Ghislain M., Byarugaba A. A., Magembe E., Njoroge A., Rivera C., Román M. L., Tovar J. C., Gamboa S., Forbes G. A., Kreuze J. F., Barekye A., and Kiggundu A. (2019) Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol. J. 17, 1119–1129 10.1111/pbi.13042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Witek K., Jupe F., Witek A. I., Baker D., Clark M. D., and Jones J. D. (2016) Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat. Biotechnol. 34, 656–660 10.1038/nbt.3540 [DOI] [PubMed] [Google Scholar]
- 246. Meyers B. C., Kozik A., Griego A., Kuang H. H., and Michelmore R. W. (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834 10.1105/tpc.009308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Cao F. Y., Yoshioka K., and Desveaux D. (2011) The roles of ABA in plant-pathogen interactions. J. Plant Res. 124, 489–499 10.1007/s10265-011-0409-y [DOI] [PubMed] [Google Scholar]
- 248. Van de Weyer A.-L., Monteiro F., Furzer O. J., Nishimura M. T., Cevik V., Witek K., Jones J. D. G., Dangl J. L., Weigel D., and Bemm F. (2019) A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell 178, 1260–1272.e14 10.1016/j.cell.2019.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Hurni S., Brunner S., Stirnweis D., Herren G., Peditto D., McIntosh R. A., and Keller B. (2014) The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J. 79, 904–913 10.1111/tpj.12593 [DOI] [PubMed] [Google Scholar]
- 250. Stirnweis D., Milani S. D., Brunner S., Herren G., Buchmann G., Peditto D., Jordan T., and Keller B. (2014) Suppression among alleles encoding nucleotide-binding–leucine-rich repeat resistance proteins interferes with resistance in F1 hybrid and allele-pyramided wheat plants. Plant J. 79, 893–903 10.1111/tpj.12592 [DOI] [PubMed] [Google Scholar]
- 251. Slootweg E., Koropacka K., Roosien J., Dees R., Overmars H., Lankhorst R. K., van Schaik C., Pomp R., Bouwman L., Helder J., Schots A., Bakker J., Smant G., and Goverse A. (2017) Sequence exchange between homologous NB-LRR genes converts virus resistance into nematode resistance, and vice versa. Plant Physiol. 175, 498–510 10.1104/pp.17.00485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Farnham G., and Baulcombe D. C. (2006) Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc. Natl. Acad. Sci. U. S. A. 103, 18828–18833 10.1073/pnas.0605777103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Harris C. J., Slootweg E. J., Goverse A., and Baulcombe D. C. (2013) Stepwise artificial evolution of a plant disease resistance gene. Proc. Natl. Acad. Sci. U. S. A. 110, 21189–21194 10.1073/pnas.1311134110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Giannakopoulou A., Steele J. F., Segretin M. E., Bozkurt T. O., Zhou J., Robatzek S., Banfield M. J., Pais M., and Kamoun S. (2015) Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 28, 1316–1329 10.1094/MPMI-07-15-0147-R [DOI] [PubMed] [Google Scholar]
- 255. Segretin M. E., Pais M., Franceschetti M., Chaparro-Garcia A., Bos J. I., Banfield M. J., and Kamoun S. (2014) Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol. Plant Microbe Interact. 27, 624–637 10.1094/MPMI-02-14-0040-R [DOI] [PubMed] [Google Scholar]
- 256. Pottinger S. E., Bak A., Margets A., Helm M., Tang L., Casteel C., and Innes R. W. (2020) Optimizing the PBS1 decoy system to confer resistance to potyvirus infection in Arabidopsis and soybean. Mol. Plant Microbe Interact. 33, 932–944 10.1094/mpmi-07-19-0190-r [DOI] [PubMed] [Google Scholar]
- 257. Kim S. H., Qi D., Ashfield T., Helm M., and Innes R. W. (2016) Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351, 684–687 10.1126/science.aad3436 [DOI] [PubMed] [Google Scholar]
- 258. Carter M. E., Helm M., Chapman A. V. E., Wan E., Restrepo Sierra A. M., Innes R. W., Bogdanove A. J., and Wise R. P. (2019) Convergent evolution of effector protease recognition by Arabidopsis and barley. Mol. Plant Microbe Interact. 32, 550–565 10.1094/MPMI-07-18-0202-FI [DOI] [PubMed] [Google Scholar]
- 259. Helm M., Qi M., Sarkar S., Yu H., Whitham S. A., and Innes R. W. (2019) Engineering a decoy substrate in soybean to enable recognition of the soybean mosaic virus NIa protease. Mol. Plant Microbe Interact. 32, 760–769 10.1094/MPMI-12-18-0324-R [DOI] [PubMed] [Google Scholar]
- 260. Ellis J. G. (2016) Integrated decoys and effector traps: how to catch a plant pathogen. BMC Biol. 14, 13 10.1186/s12915-016-0235-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Malik S., and Van der Hoorn R. A. (2016) Inspirational decoys: a new hunt for effector targets. New Phytol. 210, 371–373 10.1111/nph.13917 [DOI] [PubMed] [Google Scholar]
- 262. Longya A., Chaipanya C., Franceschetti M., Maidment J. H. R., Banfield M. J., and Jantasuriyarat C. (2019) Gene duplication and mutation in the emergence of a novel aggressive allele of the AVR-Pik effector in the rice blast fungus. Mol. Plant Microbe Interact. 32, 740–749 10.1094/MPMI-09-18-0245-R [DOI] [PubMed] [Google Scholar]
- 263. Xu G., Yuan M., Ai C., Liu L., Zhuang E., Karapetyan S., Wang S., and Dong X. (2017) uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494 10.1038/nature22372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Xu G., Greene G. H., Yoo H., Liu L., Marqués J., Motley J., and Dong X. (2017) Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545, 487–490 10.1038/nature22371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Huot B., Yao J., Montgomery B. L., and He S. Y. (2014) Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287 10.1093/mp/ssu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Kutschera A., Dawid C., Gisch N., Schmid C., Raasch L., Gerster T., Schäffer M., Smakowska-Luzan E., Belkhadir Y., Vlot A. C., Chandler C. E., Schellenberger R., Schwudke D., Ernst R. K., Dorey S., et al. (2019) Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science. 364, 178–181 10.1126/science.aau1279 [DOI] [PubMed] [Google Scholar]