Abstract
Since gold nanoparticles (AuNPs) have great potential to bring improvements to the biomedical field, their impact on biological systems should be better understood, particularly in a long-term, using realistic doses of exposure. miRNAs are small noncoding RNAs that play key roles in the regulation of biological pathways, from development to cellular stress responses. In this study, we performed genome-wide microRNA (miRNA) expression profiling in primary human dermal fibroblasts 20 weeks after chronic and acute (non-chronic) treatments to four AuNPs with different shapes and surface chemistries at a low dose. The exposure condition and AuNP surface chemistry had a significant impact on the modulation of miRNA levels. In addition, a network-based analysis was employed to provide a more complex, systems-level perspective of the miRNA expression changes. In response to the stress caused by AuNPs, miRNA co-expression networks perturbed in cells under non-chronic exposure to AuNPs were enriched for target genes implicated in the suppression of proliferative pathways, possibly in attempt to restore cell homeostasis, while changes in miRNA co-expression networks enriched for target genes related to activation of proliferative and suppression of apoptotic pathways were observed in cells chronically exposed to one specific type of AuNPs. In this case, miRNA dysregulation might be contributing to enforce a new cell phenotype during stress. Our findings suggest that miRNAs exert critical roles in the cellular responses to the stress provoked by a low dose of NPs in a long term and provide a fertile ground for further targeted experimental studies.
Graphical Abstract
Herein, we identify the long-term genome-wide miRNA expression changes induced by a low dose of AuNPs, providing a systems-level perspective of these changes. The miRNA dysregulation, mostly implicated in the modulation of proliferative pathways, is a cellular response to the stress caused by AuNPs.
Introduction
Over the past decade, nanotechnology has evolved from fundamentals to applications. Gold nanoparticles (AuNPs) are being extensively explored in biomedicine. The combination of remarkable optical scattering/absorption properties, photothermal and photoacoustic capabilities, along with well-controlled synthesis procedures, make AuNPs very attractive for applications in sensing, molecular diagnostic, drug/gene delivery, therapeutics and theranostics.1 Furthermore, AuNPs are also present in consumer goods such as cosmetics, supplements, food packaging and beverages.2 By definition, nanoparticles are very small, have large surface areas, contain under-coordinated surface atoms and, therefore, might be more reactive than bulk materials. Thus, in parallel to the societal benefits promised by nanotechnology, some concerns have been expressed about the intersection of NPs with living systems.
In general, the biological responses to NPs have been assessed with the use of endpoint-based conventional toxicity tests for traditional chemicals, mostly at unrealistically high concentrations over short times. There is a clear gap in knowledge of the long-term effects of NPs on living systems considering data taken at realistic doses of exposure, under relevant environmental conditions. To shed light on this issue, our group investigated the in vitro long-term cellular responses (after 20 weeks) to well-characterized AuNPs (spheres and rods) with different surface chemistries at a very low dose (0.1nM) under chronic (20 weeks of continuous exposure) and non-chronic (24 h of exposure followed by NP-free cell media for the rest of the 20 weeks) conditions.3 We found that AuNPs were not cytotoxic at this dose in any condition, but they did perturb cell homeostasis by altering levels of stress response genes to different degrees: in general, while cells upon chronic exposure adaptatively responded to the continual stress, cells under non-chronic conditions showed a sustained endoplasmic reticulum (ER) stress, and activation of the unfolded protein response (UPR), an adaptative response aiming the restoration of cell homeostasis, suggesting that the non-chronic treated cells were still struggling to cope with perturbations caused by AuNPs stimulation 20 weeks before.3 The UPR activation trigger pro-survival signals to manage the stress but can also induce pro-death signals, depending on the cellular context. The unresolved ER stress is a concern since is implicated in the development of human diseases.4
Advanced tools to evaluate the NP-biological systems interactions have been proposed to provide insights into the molecular responses to NPs.5 These tools include omics-based and systems biology approaches (which involves the holistic analysis of complex interactions within biological systems) to detect molecular changes and perturbations in complex biological networks and pathways by toxicants, which may lead to potential adverse effects. miRNAs have been identified as critical regulators of biological pathways and play pivotal roles in cellular responses to environmental chemicals since miRNA target genes are preferentially regulated by those molecules in the human genome,6 highlighting the importance of miRNAs in the (nano) toxicology research. miRNAs are short non-coding RNAs that mediate the regulation of gene expression by inducing translation repression and/or degradation of mRNA targets.7 Each mRNA can be targeted by multiple miRNAs that act cooperatively in gene repression. One miRNA can potentially have hundreds of targets. Since miRNAs are virtually involved in the regulation of all cellular processes, from development to maintenance of homeostasis,8 miRNA dysregulation has been linked to many human diseases. In the last years, studies have associated the exposure to nanomaterials with changes in miRNA expression levels and corresponding biological pathways.9–18 However, few studies have addressed the impact of AuNPs on miRNA expression profile and they are limited to the evaluation of one AuNP type (citrate spheres) being mostly focused on the short-term effects of high doses of NPs.19–23
To assess the long-term changes in miRNA levels in response to one low dose of AuNPs, we performed genome-wide miRNA expression profiling in primary human dermal fibroblasts 20 weeks after chronic and non-chronic exposures to 0.1nM of AuNPs with different shapes and surface chemistries. Alongside the identification of differentially expressed miRNAs, Weighted Gene Correlation Network Analysis (WGCNA) was employed to gain a systems-level view of the miRNA changes after the AuNP exposure. The WGCNA is a systems biology approach used to explore the functionality of a transcriptome by considering correlations between gene transcripts, which are organized into functional clusters according to cellular processes/pathways.24 Further, bioinformatics analyses were carried out to identify miRNA targets and related biological pathways. Finally, we integrated the information obtained from the miRNome analysis with cellular parameters (e.g. cell proliferation, viability, and morphology) and gene expression data investigated previously in the same cells under the same conditions3 to provide a better understanding of the long-term cellular responses to AuNPs. To the best of our knowledge, no study has examined the long-term impact of AuNPs with different shapes and surface coatings on the miRNA modulation focusing on a realistic scenario of low dose, relevant exposure conditions, and using a systems biology approach.
Experimental
The AuNPs synthesis, surface modification, cell culture and cell treatment with AuNPs, and RNA isolation have already been described in more details elsewhere.3 Thus, only a brief description of these methodologies is presented here.
Gold nanoparticles synthesis, surface modification and characterization
The citrate spheres were prepared by a scale up Turkevich method. Briefly, 25mL of 0.01nM HAuCl4 solution (trihydrate, HAuCl4•3H2O, ≥99.9%, Sigma Aldrich) and 975 mL of ultrapure deionized water (18.2 MΩ, Barnstead NANOpure II) were combined and the solution was heated to boiling under constant stirring. Then, 20 mL of 5% (w/w) sodium citrate (tribasic dehydrate, C6H5Na3O7•2H2O, ≥99%, Sigma-Aldrich) was added. Additional 5 mL of 5% sodium citrate was added to the solution after 30 min. During this time, the solution turned wine-red. After cooling down to room temperature, the solution was cleaned up by centrifugation (8,000 rcf for 20 min). These gold nanospheres (average size: 18.4 nm ± 2.0 nm in diameter) bore a net negative charge due to adsorbed citrate ions.
The PAA spheres (poly(acrylic acid)-coated nanospheres) were prepared by wrapping citrate NPs using the typical layer-by-layer polyelectrolyte (PAH/PAA) procedures. Briefly, a solution of 25 mL of 0.01 M NaCl and 50 mL of 10 mg/mL PAH (poly(allylamine hydrochloride), M.W. 17,500 g/mol, Aldrich) (with 1 mM NaCl) was prepared and added to 1 L of citrate NPs under vigorous stirring. After centrifugation (8,000 rcf for 20 min), the pellet was redispersed in 1L of deionized water and the procedure was repeated a second time with 10 mg/mL PAA (sodium salt, M.W. 15,000 g/mol, Aldrich) (with 1 mM NaCl). The final solution was purified by centrifugation and resuspended in minimal water.
The synthesis of gold nanorods was performed by a scale-up seed-mediated silver-assisted method.25 In brief, 0.25 mL of 0.01 M HAuCl4 trihydrate and 9.75 mL of 0.1 M hexadecyltrimethylammonium bromide (CTAB for molecular biology, Sigma-Aldrich) were combined under vigorous stirring to make gold seeds. Then, 0.6 mL of freshly made 0.01 M sodium borohydride (NaBH4, Sigma-Aldrich) was added into the solution, resulting in a honey-brown solution. The 2 L batches of gold nanorods were prepared by mixing 16 mL of 0.01 M silver nitrate (AgNO3, 99.0%, Sigma-Aldrich), 1900 mL of 0.1 M CTAB solution, 100 mL of 0.01 M HAuCl4 and 11 mL of freshly made 0.1 M L-ascorbic acid (BioXtra, ≥99.0%, crystalline, Sigma) (the solution reverted to colorless). Finally, 2.4 mL of the gold seeds solution (prepared 1 h before) was added. Purification was done by centrifugation (13,500 rcf for 20 min, 2 x) and the pellet was resuspended in deionized water. Then, the as-made gold nanorods (46.0 nm ±4.1 nm; 15.9 nm ± 2.6 nm) were used to prepare PAA rods and PEG rods. A sequential layer-by-layer polyelectrolyte (PAA/PAH/PAA) procedure was used to make PAA rods. Briefly, a solution of 4mL of 0.01 M NaCl and 8 mL of 10 mg/mL PAA (with 1 mM NaCl) was prepared and added to 20 mL of gold nanorods. After 2 h on a shaker, the solution was centrifuged. This procedure was repeated 2 × more (once with PAH and another with PAA). After the final PAA coating, the solution was centrifuged and redispersed in minimal water. The PEG rods were made by mixing 20 mL of as-made nanorods (diluted to 40 mL in water) and 20 mL of 0.5 mM mPEG-SH (thiolated methoxy-PEG, M.W. 5,000 g/mol, Nanocs, stored at −20°C). Then, the solution was purified by centrifugation (8,000 rcf for 20 min, 2 x) before being redispersed in minimal water.
All AuNPs suspensions were gram-negative bacterial endotoxin-free determined by the Pierce™ LAL Chromogenic Endotoxin Quantitation Kit (Thermo Scientific, USA).
All AuNPs solutions (in water and cell media) were characterized by UV-Vis spectroscopy (Cary 500 Scan UV-vis-NIR spectrophotometer, Varian), dynamic light scattering (DLS), zeta potential measurements (ZetaPALS DLS/zeta potential analyzer, Brookhaven Instruments) and transmission electron microscopy (TEM, 2100 JEOL Cryo TEM) as previously reported3 (tables S1 and S2).The concentrations of AuNPs solutions were calculated using UV-vis spectroscopy.
Cell culture and gold nanoparticles treatment
In brief, human dermal fibroblasts (HDF), primary cells obtained from normal human neonatal foreskin or adult skin (Sigma-Aldrich), were plated in 6-well plates (100,000 cells per well) and grown in Dulbecco’s Modified Eagle Medium (DMEM, without phenol red, Mediatech) supplemented with 10% Fetal Bovine Serum (FBS, Gemini Bio-Products) and 1% penicillin-streptomycin (Mediatech). These cells can be cultured at least 16 population doublings according to the manufacture. At each passage (once a week), cells were plated with NP-free media and allowed to adhere for 24 h. For chronic treatment with AuNPs, the cell media was replaced with media containing 0.1 nM AuNPs (~ 6 × 1010 NPs/mL) and cells were grown in the continuous presence of NPs for 20 weeks. For “non-chronic” treatment with AuNPs, cells were grown in the presence of 0.1 nM of NPs only during the first 24h. After this point, the cell media containing NPs was removed from non-chronic samples and replaced with NP-free media. Then, the non-chronic samples were cultured during the following 20 weeks in NP-free cell media such as the control cells (samples never exposed to NPs). The cell media on all samples was replaced four days after plating, with fresh 0.1 nM AuNP- containing media being added only to chronic samples. All experiments were performed in three biological replicates (for controls and per sample type: citrate spheres, PAA spheres, PAA rods and PEG rods; chronic and non-chronic exposures). No experiments were performed that contained equivalent amounts of free ligand (~1–10 nM) without the nanoparticles, as these molecular concentrations were expected to be below the threshold for cellular responses.
RNA isolation, miRNA library preparation and sequencing
The total RNA, including miRNAs, was isolated from HDF cells treated with 0.1 nM AuNPs under chronic and non-chronic conditions as well as from control cells after 20 weeks of culture using the AllPrep® DNA/RNA/miRNA universal kit (Qiagen, USA). Samples were then stored at −80 °C.
For the miRNA analysis, samples were quantified in Nanodrop 1000 (Thermo Scientific) and their quality checked by Agilent 2100 Bioanalyzer (Agilent Technologies, USA). The miRNA libraries were prepared using the NEXTflex Small RNA-Seq kit v3 (Bio Scientific) following the manufacturer’s instructions and 800 ng of total RNA was used as starting material. Briefly, after ligation of the 3’ adenylated and 5’adapters, the products were reverse transcribed and amplified by PCR (22 cycles) using different barcoded primers for each sample. The miRNA libraries were then size selected (~150 bp band) on 10% TBE-PAGE gel (BioRad) followed by overnight gel-elution and cleanup. Libraries were quantified with the Qubit dsDNA HS Assay kit (Thermo Fisher, USA) and validated by Agilent 2100 Bioanalyzer using the Bioanalyser High Sensitivity DNA assay (Agilent Technologies, USA) and Real-Time PCR (Applied Biosystems). Samples presenting low concentration (Qubit values <0.4 ng/μL) (poor-quality libraries) were not included in further analysis (control-replicate 2; citrate spheres chronic-replicate 1; PAA spheres chronic-replicate 3; PAA rods chronic-replicate 2). The high-quality barcoded miRNA libraries were then pooled and sequenced at the High-Throughput Sequencing and Genotyping Unit (Roy J Carver Biotechnology Center, UIUC) using the Illumina HiSeq4000 instrument with 50 nucleotide single-end reads.
Quality control, miRNA quantification and differential expression analysis
The sequencing data was extracted from FASTQ files. The average number of raw reads was about 14 million and the quality control of the data was assessed using the MultiQC tool.26 High-quality data was obtained by filtering the sequences based on defined cutoffs: low-quality sequence reads with a Phred score of less than 30 and a read-length shorter than 18 nucleotides were discarded and the adapters sequences were trimmed and removed. The Novoalign 3.06.03 (miRNA mode) was used to align the reads to the human miRNA mature sequences from miRbase V21 to match them to known miRNA sequences.27
The read counts for mature miRNAs were input into R (v 3.4.3) for quality control, pre-processing and statistical analysis using packages from Bioconductor,28 as indicated below. Two samples, one replicate of PEG rods chronic and one of PEG rods non-chronic, were deemed outliers from Principle Components Analysis clustering (data not shown) and were removed from further analysis. Normalization was done using the TMM (trimmed mean of M-values) method then log2-transformed with prior.count = 3 (hereafter referred to as logCPM).29 Any miRNA without logCPM > log2(1) in at least 2 samples were filtered out, leaving 991 miRNAs for analysis.
Differential miRNA expression was identified using the edgeR’s (v 3.4.1) likelihood ratio test method.30 The equivalent of a one-way ANOVA test was used to calculate global changes in miRNA expression across all nine groups (controls and AuNP-treated cells) and pairwise comparisons were made between each of the eight treated groups versus the control group. Additionally, pairwise comparisons were made between groups exposed to the same AuNP type in different conditions (chronic versus non-chronic). The Benjamini-Hochberg false discovery rate (FDR) method was used to adjust the raw P values.31 miRNAs presenting fold change (FC) ≥ 2 or ≤ −2 and an adjusted P value (FDR) < 0.05 were considered differentially expressed.
Quantitative real-time PCR
To validate the robustness of the data generated by sequencing, quantitative real-time PCR (qPCR) was performed on a set of miRNAs (hsa-miR-29b-1, hsa-miR-181a, hsa-miR-486) which presented significant sequencing-detected changes in their expression levels. Briefly, 10 ng of total RNA was reverse transcribed into cDNA using the TaqMan™ MicroRNA Reverse Transcription (RT) kit (Applied Biosystems, USA) following the manufacturer’s instructions. The qPCR reaction was carried out using TaqMan Universal Master Mix II (no UNG) (Applied Biosystems, USA), the cDNAs previously obtained, individual TaqMan™ MicroRNA assays (Applied Biosystems, USA) and nuclease-free water as recommend by the manufacturer on a Quanta Studio 7 instrument (Applied Biosystems, USA). The human U6B small nuclear RNA (RNU6B) was used as endogenous control for data normalization. Samples were run in triplicate for each miRNA along with no-template controls. Changes in miRNA relative expression levels were calculated using the 2−ΔΔCT method and differences were considered significant at P < 0.05.32
Weighted gene co-expression network analysis (WGCNA)
miRNA co-expression networks were built using the WGCNA package (v 1.61).33 We re-filtered the miRNAs using less rigorous criteria of logCPM (without any prior count added) > log2(1) in only one sample to include more miRNAs (1,130) in the pattern-based analysis. First, pairwise correlations between miRNAs were calculated using the biweight midcorrelation method. Next, pairwise topological overlap between miRNAs was calculated using a soft-thresholding power of 8 based on a fit to scale free topology. Then, we used the blockwiseModules() function with default parameters except corType = “bicor”, maxPOutliers = 0.1, power = 8, networkType = “signed hybrid”, deepSplit = 4, minModuleSize = 10, and mergeCutHeight = 0.2 to assign miRNAs to modules. The miRNAs expression data in each module was summarized by the parameter module eigengene (ME). The module membership, also known as eigengene-based connectivity (kME), was calculated to identify highly connected miRNAs within each module (hub miRNAs). A kME ≥ the median for each module presenting differences in the miRNA expression profile was used as a cut-off for hub miRNA selection (Turquoise: kME ≥ 0.68; Blue: kME ≥ 0.74; Yellow: kME ≥ 0.80; Green: kME ≥ 0.76; Red: kME ≥ 0.77; Pink: kME ≥ 0.76; Magenta: kME ≥ 0.81; Purple: kME ≥ 0.71; Green yellow: kME ≥ 0.69; Salmon: kME ≥ 0.77; Midnight blue: kME ≥ 0.65; Light cyan: kME ≥ 0.84). Pairwise comparisons among samples within the same module were performed using the one-way ANOVA test and adjusted P values (FDR) < 0.05 were considered significant.
Identification of predicted and validated miRNA targets and pathway enrichment analysis
The target genes of hub miRNAs were then bioinformatically predicted using miRWalk (2.0) (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) and only those targets commonly predicted by miRWalk (Predicted Target Module), miRDB, miRanda and TargetScan were filtered.34 In addition, miRNA target genes experimentally validated by reporter assay and western blot and/or qPCR methods (strong evidence of miRNA-target interaction) were selected from miRTarBase (release 7.0) (http://miRTarBase.mbc.nctu.edu.tw/).35
Both predicted and strongly validated targets that were hit by at least 2 or more miRNAs in each module were then considered in the enrichment analysis (≥ 2 in the green, pink, purple, green yellow, salmon, midnight blue and light cyan modules; ≥ 3 in the red module; ≥ 4 in the turquoise, blue, yellow and magenta modules, which presented the larger number of miRNA targets). The freely available enrichment analysis tool Enrichr (http://amp.pharm.mssm.edu/Enrichr) was used to identify overrepresented biological pathways (REACTOME, update 2016) related to the miRNA targets in each module (Fisher’s exact test, adjusted P value < 0.10).
The miRNA target genes filtered from those public databases were compared with differentially expressed genes (P value < 0.05) related to Stress and Toxicity pathways found in our previous study using the same samples to identify miRNA-mRNA target pairs.3
Results
miRNA changes in HDF cells exposed to a low dose of AuNPs
The overall workflow of our study is depicted in Fig. 1. The global changes in miRNA expression profiles in human dermal fibroblasts (HDF) exposed to AuNPs under both chronic and non-chronic conditions and controls (non-treated cells) were determined by next-generation sequencing (NGS). Well-characterized AuNPs presenting different surface chemistries, anionic citrate, anionic PAA and neutral PEG, considered some of the most innocuous coatings for NPs, and different shapes (spheres and rods) were used in this study.
Fig. 1. Flow chart of the overall approach.
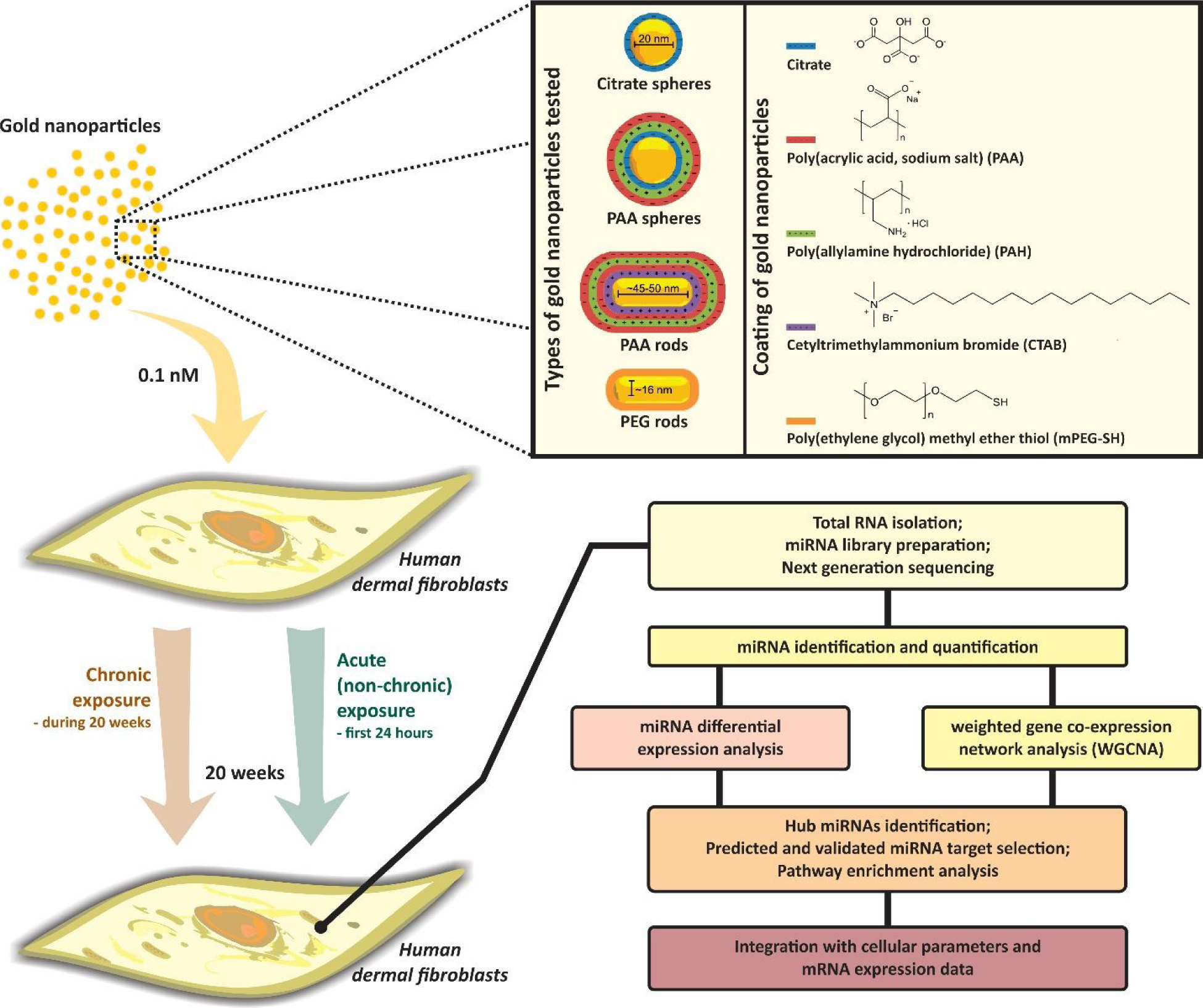
Human dermal fibroblasts were exposed to different types of well-characterized AuNPs at a low dose (0.1nM) under chronic and non-chronic conditions. After 20 weeks, genome-wide miRNA analysis was performed in cells exposed to AuNPs as well as in controls to assess the long-term changes in miRNA expression levels. Differentially expressed miRNAs were identified. Network-based analysis was also performed to gain a detailed, systems-level view of the miRNA changes. miRNA target genes were bioinfomatically selected and enrichment analysis were carried out to determine biological pathways perturbed by AuNPs exposure. Cellular parameters and mRNA expression data were integrated with miRNA data.
A total of 991 miRNAs were identified in our samples after quality control and reads alignment to human mature miRNA sequences from miRbase V21. A broad view of the miRNA expression patterns across samples was obtained using the Benjamini-Hochberg false discovery rate (FDR) set at 5% to adjust the raw P values and one hundred and thirty-four (134) miRNAs were found to be differentially expressed (DE) (FDR<0.05). Hierarchical clustering heatmaps of the DE miRNAs (Fig. 2A) revealed a complex miRNA response to AuNPs stimuli in HDF cells even for the low dose exposure. When grouped by chronic or non-chronic (Fig. 2A, left), we notice a clustering of DE miRNAs in samples exposed to AuNPs (PAA spheres, PAA rods and PEG rods) in a non-chronic way, suggesting some common miRNA dysregulation signatures. A clustering was less evident considering samples under chronic AuNPs exposure. Intriguingly, the miRNA expression profile of cells challenged with citrate spheres under the non-chronic condition grouped together with those found in samples chronically exposed to NPs (Fig. 2A, left). When samples were grouped by AuNP type (Fig. 2A, right), distinct miRNA expression patterns were observed between the exposure to the same particle type but under different conditions, particularly considering PAA rods and PEG rods NPs.
Fig. 2. miRNA expression changes after HDF cells exposure to AuNPs.
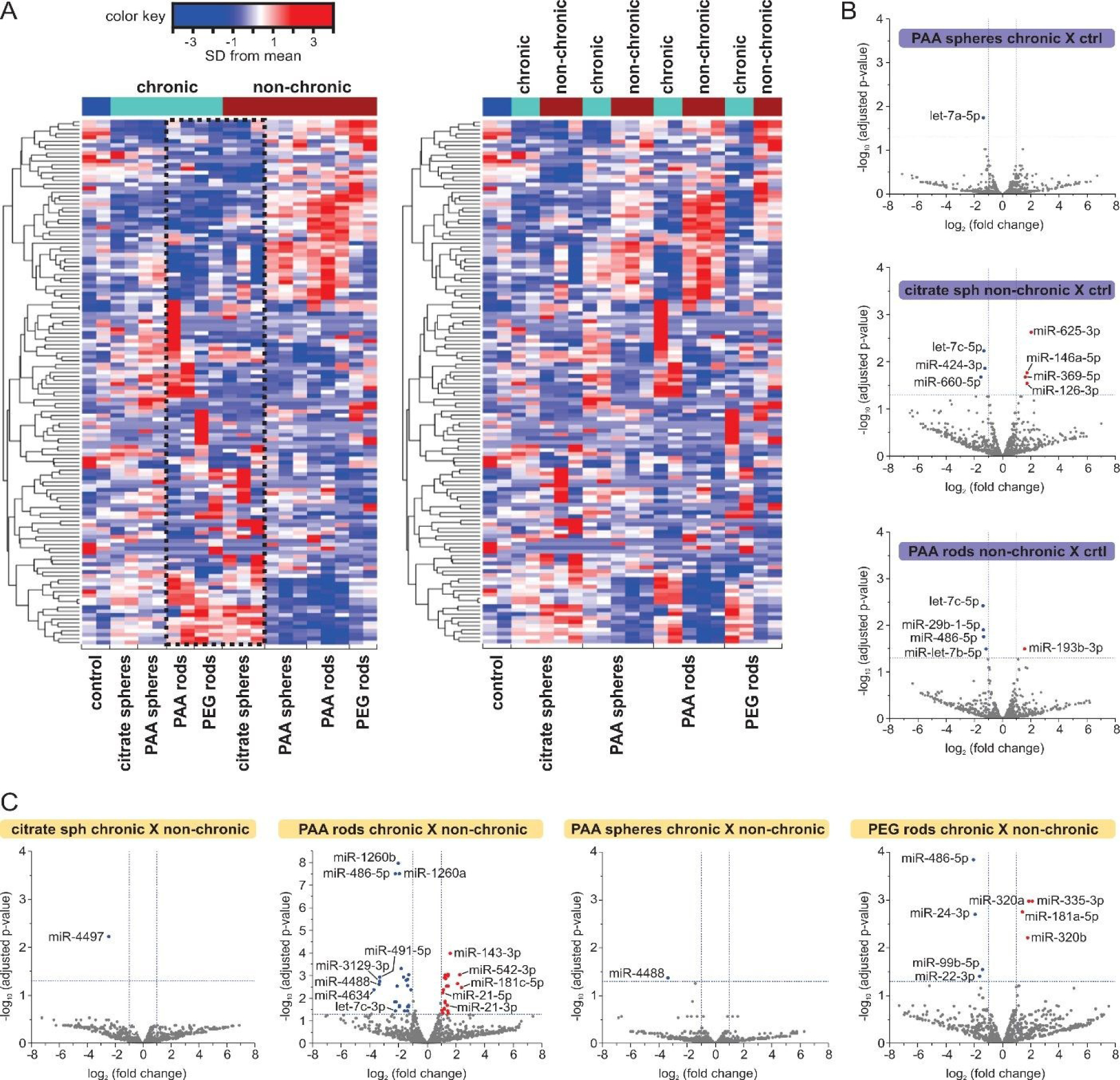
(A) Hierarchical clustering heatmaps showing changes in miRNA expression with samples grouped by exposure condition (left) and by AuNP type (right). Using a scale of standard deviations from the mean expression level, the change in expression level (one-way ANOVA, FDR<0.05) is shown as red (higher expression) or blue (lower expression) relative to the mean across all samples. Each row represents one miRNA and each column corresponds to one sample. The dotted line (2A, left) highlights the similarities between the miRNA expression profile in cells treated with citrate spheres under the non-chronic condition and samples chronically exposed to AuNPs. (B and C) Volcano plots of miRNA expression comparing cells treated with different types of AuNPs and controls (ctrl) (B), and cells treated with the same AuNP type under different conditions (C). Only comparisons showing DE miRNAs are presented. Blue dots represent miRNAs significantly downregulated while the red dots represent miRNAs significantly upregulated in treated samples (FC≥2 or≤−2, FDR<0.05).
To determine differences in the miRNA expression levels between groups, we performed pairwise analyses comparing HDF cells exposed to AuNPs with controls (ctrl) as well as comparing samples treated with the same AuNP type upon a different exposure condition (chronic or non-chronic). miRNAs were filtered with a more restrict cut-off criterion of fold change (FC) ≥ 2 or ≤2 and FDR < 0.05. The changes in miRNA expression are presented in Volcano plots (Fig. 2, B and C). The individual miRNAs with accompanying FC and raw and adjusted P values (FDR) are tabulated in tables S3 and S4.
Compared to controls, the non-chronic exposure to AuNPs led to a larger number of significant changes in miRNA expression levels (12 DE miRNAs versus 1 DE miRNA by the chronic counterpart) (Fig. 2B). Citrate spheres had the greatest impact on miRNA dysregulation under non-chronic conditions. Members of the hsa-let-7 (let-7) miRNA family were downregulated by the non-chronic exposure to both citrate spheres and PAA rods as well as by the chronic exposure to PAA spheres. The let-7 family is well-known to its essential role in cell cycle regulation and cell differentiation, presenting strong tumor suppressor activities.36 Moreover, the other significantly downregulated miRNAs by citrate spheres (hsa-miR-424–3p and hsa-miR-660–5p) and PAA rods (hsa-miR-29b-1–5p and hsa-miR-486–5p) exposures under non-chronic condition have been associated with modulation of proliferation in different cell types.37–39 More specifically in fibroblasts, while the downregulation of hsa-miR-424–3p seems to optimize the cell response to ER stress,40 decreased levels of hsa-miR-29b-1–5p and 486–5p were related to aberrant fibroblast proliferation during the pathogenesis of fibrosis.41,42 By contrast, the non-chronic exposure to PAA rods induced the expression of hsa-miR-193b-3p involved in inhibition of cell proliferation in human fibroblasts under oxidative stress, and apoptosis.43 On the contrary, the non-chronic exposure to citrate spheres led to upregulation of the hsa-miR-625–3p involved in a cell type-dependent induction or suppression of cell proliferation and survival,44 as well as upregulation of hsa-miR-146a-5p and hsa-126a-3p, negative regulators of the inflammatory response in fibroblasts,45 and hsa-miR-369–5p, involved in fibroblast proliferation.46
Differences in miRNA expression levels were more evident in the analyses comparing cells exposed to the same AuNP type under different conditions (chronic or non-chronic) (Fig. 2C). A total of 54 miRNAs was found to be dysregulated. While the comparison between samples exposed to citrate spheres chronic and non-chronic, and PAA spheres chronic and non-chronic showed changes in the expression of only 1 miRNA, remarkable differences in miRNA expression levels were noticed comparing samples treated with PAA rods in distinct ways: 44 DE miRNAs were identified. The most dysregulated miRNAs were hsa-miR-4634 (12.94-fold), hsa-miR-4488 (10.0-fold), hsa-miR-3129–3p (9.95-fold) and hsa-miR-491–5p (9.83-fold) (table S4). The functions of hsa-miR-4634, hsa-miR-3129–3p, and hsa-miR-4488 are still unclear; the hsa-miR-491–5p has been related to fibroblast senescence47 and serves mainly as tumor suppressor in cancer cells by inhibiting proliferation and inducing apoptosis.48 The list of altered miRNAs includes both well-known anti-proliferative miRNAs, such as hsa-miR-34b-3p and 34c-3p, hsa-let-7c-3p, hsa-miR-15b-5p, hsa-miR-486–5p, hsa-miR-143–3p, as well as miRNAs related to the induction of proliferation, such as hsa-miR-21–3p and 21–5p, and hsa-miR-17–5p (17~92 cluster), in fibroblasts and other cell types.49–52 The comparison between cells exposed to PEG rods under chronic and non-chronic conditions showed DE miRNAs also involved in cell proliferation suppression and/or apoptosis induction (hsa-miR-486–5p, hsa-miR-24–3p, hsa-miR-99b-5p, hsa-miR-22–3p, hsa-miR-320a and b, and hsa-miR-335–5p)53,54 and fibroblast senescence (hsa-miR-22–3p, hsa-miR-335–5p).55,56
Common sets of dysregulated miRNAs were identified: the hsa-miR-4488 was strongly downregulated in cells after the non-chronic exposure to PAA spheres and PAA rods compared to samples chronically treated with these AuNPs (Fig. 2C); the hsa-miR-486–5p was found downregulated after the treatment with PAA rods and PEG rods under non-chronic conditions whereas the upregulation of the hsa-miR-181 family (181a-5p, 181b-5p and 181c-5p), implicated in the regulation of inflammatory responses and senescence in fibroblasts, was observed (Fig. 2C).57
To validate some changes observed in miRNA expression levels after the cell exposure to AuNPs, quantitative real time PCR (qPCR) analysis was performed and confirmed the significant changes in the expression levels of hsa-miR-29b-1, hsa-miR-181a, hsa-miR-486 detected by sequencing (fig. S1), supporting the robustness and reproducibility of our miRNA-seq data.
Altogether, our results demonstrate that miRNAs mostly related to the modulation of cell proliferation are dysregulated after the cell exposure to AuNPs, and the effects of AuNPs on miRNA expression levels in HDF cells are dependent on both cell exposure condition and NP surface chemistry.
miRNA co-expression patterns perturbed by AuNPs at low dose
To further understand the impact of AuNPs exposure on miRNA expression changes at a system level, we performed the Weighted Gene Co-Expression Network Analysis (WGCNA). To ensure a more robust and meaningful network, we re-filtered our miRNAs to include more samples, considering now a total of 1,130 miRNAs (see Material and Methods). Widely used in genomic data analysis, the WGCNA is a systems biology approach that employs a cross-correlation method to identify modules of co-expressed genes (similar expression patterns) which may be functionally related, belong to the same biological pathway, or are likely co-regulated by a common set of transcription factors and/or chromatin regulators.24 We identified 16 modules labelled by different colors (fig. S2), summarized by module eigengene (ME) (Fig. 3, fig. S3). The grey color was reserved for unassigned miRNAs (do not belong to any module). The complete set of miRNAs in each module is listed in data file S1.
Fig. 3. Module eigengene (ME) heatmaps derived from WGCNA analysis.
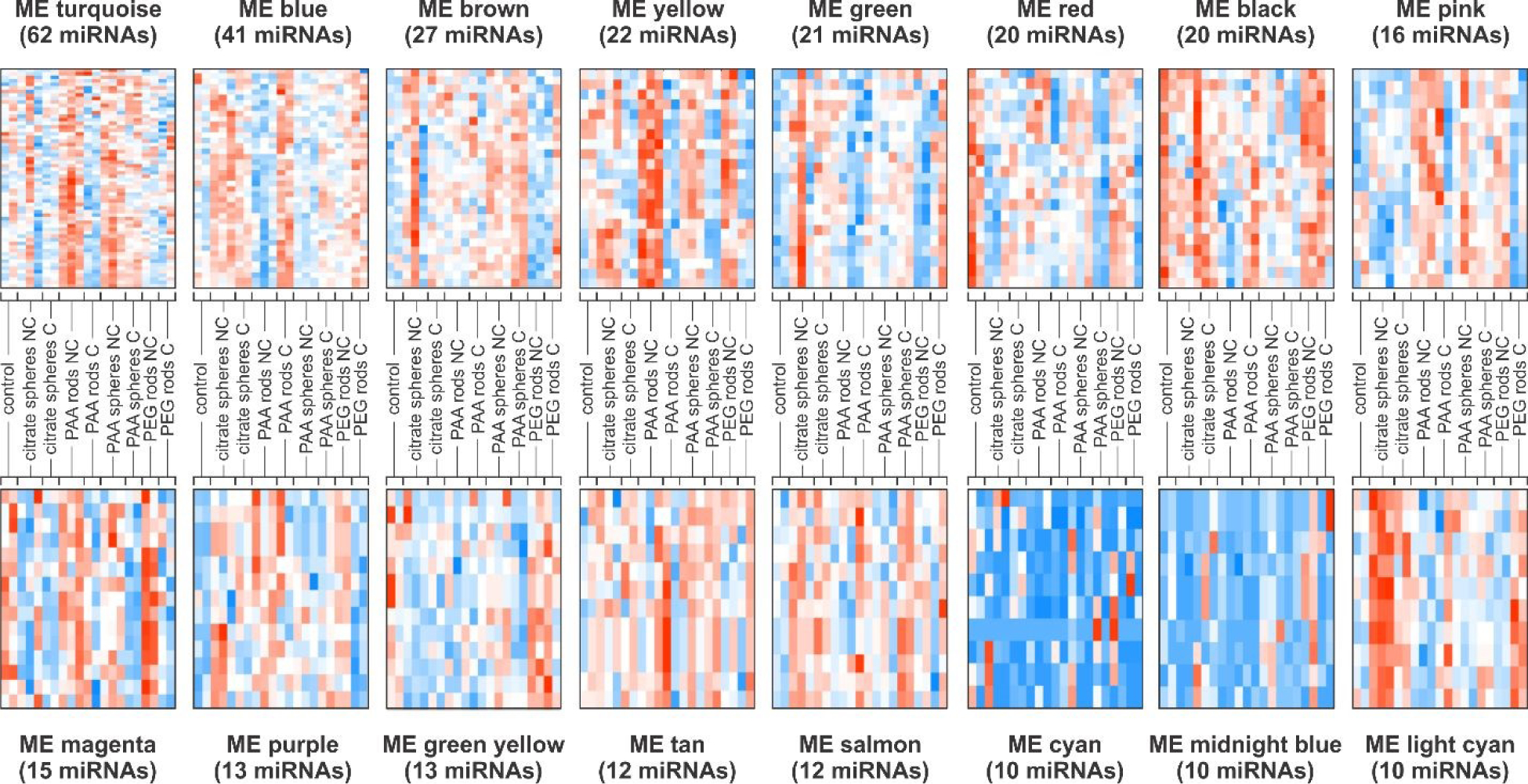
Each row represents one miRNA and each column corresponds to samples exposed to different AuNP types under chronic (C) and non-chronic (NC) conditions and controls. Red represents high expression and blue is for low expression.
We used a pairwise one-way ANOVA test to determine significant differences in the miRNA expression profiles between samples within the same module. The results are summarized in Table 1. Statistical differences were found in 12 modules. Overall, we observed that a single module was perturbed by different AuNP types, which may indicate dysregulation of common biological pathways by distinct particles. The miRNA profiles obtained from the chronic treatment with citrate spheres and PEG rods, and the non-chronic treatment with PAA spheres, did not change significantly from controls in any module. In contrast, the chronic exposure to PAA rods and spheres as well as the non-chronic exposure to citrate spheres, PAA rods and PEG rods led to perturbations in the miRNA expression profiles across different modules when compared with controls. Once again, the patterns observed after the non-chronic treatment with citrate spheres mostly followed the same trend (up or downregulation) as those observed in samples chronically exposed to AuNPs within the same module, independently of the NP type (except in the ME purple). Differences in the miRNA profiles of cells exposed to PAA spheres upon chronic and non-chronic conditions were evident in the yellow, red, magenta, green yellow and salmon modules whereas seven modules presented different patterns of miRNA expression between the chronic and non-chronic exposures to PEG rods. The miRNA expression profiles differed significantly between PAA rods chronic and non-chronic in the turquoise, blue and yellow modules. Indeed, 36 of the 44 DE miRNAs detected in our pairwise analysis comparing these two conditions are scattered across these 3 modules (15 DE miRNAs in the turquoise, 15 in the blue and 6 in the yellow modules). A clear effect of the exposure condition in the miRNA expression independently of the AuNP type can be observed in the yellow module which was upregulated in samples treated under non-chronic condition and downregulated by their chronic counterpart (Fig. 3).
Table 1:
Intramodular pairwise comparisons (statistically significant differences).
| Module | Comparison | Fold Change | FDR* |
|---|---|---|---|
| Turquoise | PAA rods chronic × non-chronic | 1.48 | 0.005 |
| Blue | CTRL × citrate spheres non-chronic | 1.24 | 0.017 |
| CTRL × PAA rods chronic | 1.34 | 0.010 | |
| PAA rods chronic × non-chronic | −1.59 | 0.005 | |
| Yellow | CTRL × PAA rods chronic | −1.19 | 0.040 |
| CTRL × PAA rods non-chronic | 1.28 | 0.004 | |
| PAA rods chronic × non-chronic | 1.53 | 7.91E-06 | |
| PAA spheres chronic × non-chronic | 1.29 | 0.003 | |
| PEG rods chronic × non-chronic | 1.36 | 0.001 | |
| Green | PEG rods chronic × non-chronic | −1.36 | 0.042 |
| Red | CTRL × citrate spheres non-chronic | −1.38 | 0.014 |
| CTRL × PAA rods chronic | −1.43 | 0.016 | |
| CTRL × PAA spheres chronic | −1.60 | 0.003 | |
| PAA spheres chronic × non-chronic | 1.36 | 0.028 | |
| Pink | PEG rods chronic × non-chronic | 1.38 | 0.030 |
| Magenta | CTRL × PAA spheres chronic | −1.35 | 0.025 |
| PAA spheres chronic × non-chronic | 1.28 | 0.037 | |
| PEG rods chronic × non-chronic | 1.42 | 0.010 | |
| Purple | CTRL × citrate spheres non-chronic | 1.38 | 0.012 |
| CTRL × PAA rods chronic | 1.31 | 0.040 | |
| CTRL × PAA rods non-chronic | 1.45 | 0.004 | |
| CTRL × PEG rods non-chronic | 1.45 | 0.016 | |
| PEG rods chronic × non-chronic | 1.46 | 0.007 | |
| Green yellow | CTRL × citrate spheres non-chronic | −1.33 | 0.020 |
| CTRL × PAA spheres chronic | −1.44 | 0.014 | |
| PAA spheres chronic × non-chronic | 1.31 | 0.035 | |
| Salmon | CTRL × PAA spheres chronic | 1.43 | 0.025 |
| PAA spheres chronic × non-chronic | −1.39 | 0.029 | |
| Midnight blue | PEG rods chronic × non-chronic | 1.37 | 0.028 |
| Light cyan | CTRL × citrate spheres non-chronic | 1.43 | 0.003 |
| PEG rods chronic × non-chronic | −1.33 | 0.012 |
One-way ANOVA
Hub miRNAs identification and target genes prediction by module
The WGCNA method applied in our study is also a useful tool to find the key drivers (i.e. hub miRNAs) within modules. miRNAs that share the largest number intramodular connections with others are called hub miRNAs since they play pivotal roles in the biological function of co-expression networks. Thus, a set of highly connected miRNAs (hub miRNAs) was identified within each of the 12 modules perturbed by AuNPs based on their module membership scores (kME) (Fig. 4, data file S2). Strikingly, 43 of the 62 DE miRNAs found in our samples were listed as hub miRNAs within different modules. Furthermore, the miRNAs with the highest degree of connection in the turquoise, blue, yellow, green, magenta and light cyan modules were also found to be DE in our cells, suggesting their critical function in the cellular responses to AuNPs (data file S2).
Fig. 4. Interaction networks of hub miRNAs by module.
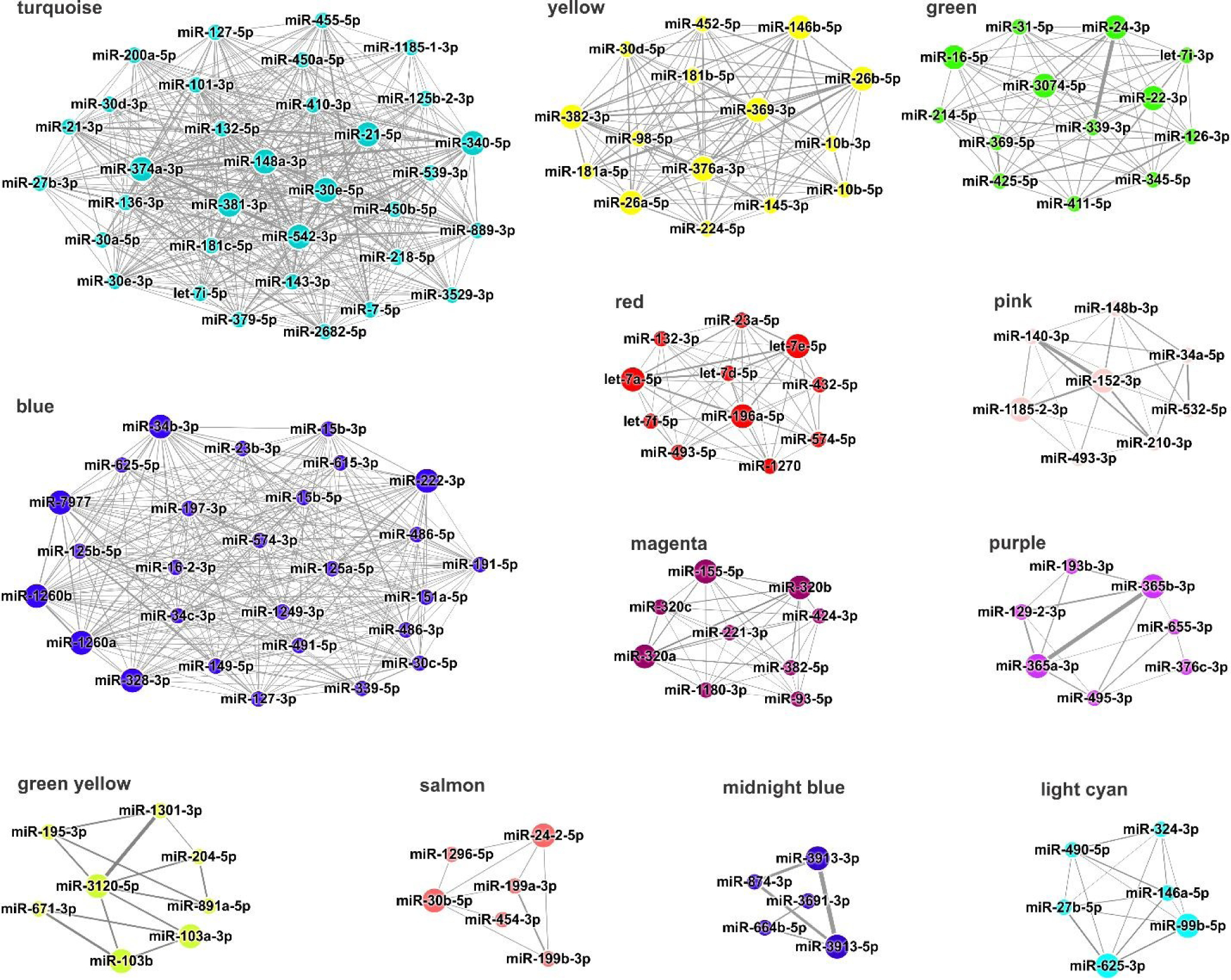
Edge thickness is proportional to the correlation between interacting miRNAs. miRNAs with the highest intramodular degree of connection are highlighted (bigger node size). The networks were generated using Cytoscape v 3.5.1.
The biological function of miRNAs is dictated by the genes (mRNAs) they regulate. Hence, for the functional characterization of co-expressed miRNAs in each module, experimentally validated (strong experimental evidence from miRTarBase 7.0) as well as targets bioinformatically predicted by four well-known miRNA target prediction databases accessed via miRWalk 2.0 (see Experimental section) were identified (data file S3). To reduce false positive results generated by the target prediction algorithms, only targets commonly predicted by all four prediction databases were selected. Moreover, we considered only genes targeted by at least 2 or more miRNAs since the combination of multiple miRNAs at the same target may determines the most dramatic changes in its expression and, consequently, the biological effect.
To verify the validity of the bioinformatically determined data, the set of miRNA target genes identified in each module was then compared with the set of differentially expressed genes (P value <0.05) related to cell stress and toxicity pathways (cell death, unfolded protein response, oxidative stress, etc) previously found in the same samples.3 Considering the commonly accepted role of miRNAs to negatively regulate gene expression, a total of 72 inversely correlated miRNA-mRNA target pairs was identified across modules (data file S4). However, since our previous gene expression data was limited to the evaluation of only 72 genes and miRNA-mRNA target pairs were not observed in some modules perturbed by AuNPs, the further pathway enrichment analysis was not restricted to the matched miRNA targets identified, but instead, the whole set of target genes rigorously selected using bioinformatics tools was considered.
Biological pathway enrichment analysis
To interpret each module in a biological context, we further conducted pathway enrichment analysis considering genes targeted by the hub miRNAs. Employing the REACTOME pathway database through Enrichr’s interface, biological pathways related to signal transduction, metabolism of proteins, gene expression regulation, programmed cell death, cell senescence, cell differentiation, etc., were found overrepresented in cells after AuNPs treatment. The most significantly biological pathways enriched in each of the 12 modules are presented in Fig. 5. The complete lists of the significantly enriched pathways along with their respective adjusted P values and miRNA target genes by module are shown in data file S5.
Fig. 5. The most significantly enriched biological pathways associated to hub miRNA target genes by module.
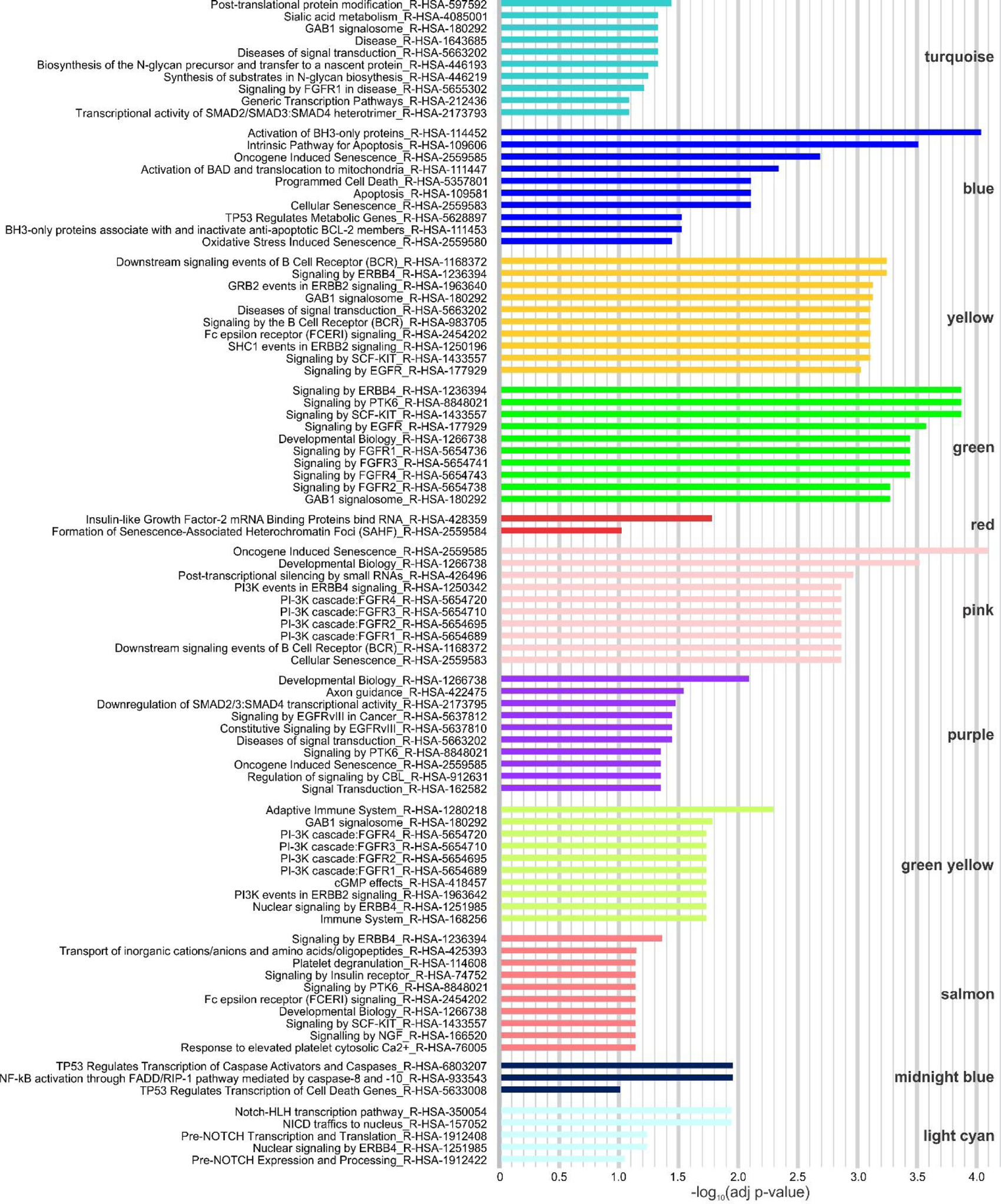
REACTOME pathway database (update 2016). Adjusted P value < 0.1 was considered as cutoff for significance.
Pathways related to post-translational protein modification were among the most enriched in the turquoise module, particularly pathways related to the asparagine-linked (N-linked) glycosylation process, the most prominent protein modification of eukaryotic cells which controls the folding and functional properties of proteins.58 The miRNA target genes of the blue module were mainly associated to cell death process by apoptosis (mitochondrial perturbation). Inversely correlated miRNA-mRNA pairs involving two important pro-apoptotic genes (both downregulated in our previous gene expression analysis) were found: TP53 was the target of hsa-miR-125a, 125b, 151a, 30c, 491, and BNIP3L was the target of hsa-miR-30c and 34b, all hub miRNAs in the blue module (data file S4). Moreover, the upregulation of hsa-miR-222–3p, the top hub miRNA in this module, was recently associated with inhibition of apoptosis in fibroblasts.59 Signaling pathways mediated by receptor tyrosine kinases (RTKs) such as KIT, IGF1R (Insulin-like Growth Factor 1 Receptor) and the RTK families ERBB (ERBB2, ERBB4 and EGFR - Epidermal Growth Factor Receptor) and FGFR (Fibroblast Growth Factor Receptors 1,2,3,4) were highly significantly overrepresented in the yellow and green modules. Notably, the enrichment for RTKs-related pathways was also observed in other 6 modules (turquoise, blue, pink, purple, green yellow and salmon) (Fig. 5, data file S5). The downstream cascades activated by RTKs, like MAPK/ERK and PI3K/AKT are related to the regulation of cell proliferation and differentiation, cell survival, cell metabolism, and other critical cellular processes.60 Other signal transduction pathways such as SMAD2/3:SMAD4 transcription activity (TGF-β signaling), implicated in the control of fibroblast proliferation and transformation61 (turquoise and purple modules), Insulin-like Growth Factor-2 (IGF-2) mRNA binding proteins (IGF2BPs/IMPs/VICKZs)62 (red module), and Notch pathway63 (light cyan module) were also found overrepresented. The pink module was highly related to cellular senescence, the most significantly enriched pathway found in our analysis. Interestingly, the top hub miRNA in this module, hsa-miR-152–3p (data file S2), has been related to the induction of senescence in human dermal fibroblasts.57 Enrichment for senescence pathways was also observed in the blue, green, red, and purple modules. No biological pathways were significantly enriched for miRNA target genes of the magenta module. The purple module was mainly related to developmental biology, particularly axon guidance (related to cell differentiation). In addition to the RTK cascades, as highlighted above, the green yellow module was highly related to immune response. Pathways involved in nonapoptotic functions of caspases 8 and 10 mediating the activation of NF-κB,64 a nuclear factor implicated in cell proliferation and survival, cell stress responses and immune response, and transcriptional regulation of cell death genes, such as caspase 10, by TP53 were enriched in the midnight blue module.
Discussion
In our genome-wide analysis, we identified the long-term perturbations in the miRNome induced by the exposure of non-transformed human fibroblasts to a very low dose of different types (shape and surface chemistry) of AuNPs under relevant exposure conditions (chronic and non-chronic). In addition, we used a network-based approach (WGCNA) to provide a more complex, systems-level perspective of the miRNA expression changes leading to important insights into the biological processes underlying the cellular responses to AuNPs exposure.
Here, we report the significant role of cell exposure condition in miRNA dysregulation in the long term, with the non-chronic exposure to 0.1nM of AuNPs having a wider impact in the modulation of miRNA levels in HDF cells. We found that miRNAs mostly implied in controlling cell proliferation were substantially dysregulated in our samples. Altered levels of miRNAs related to cell proliferation pathways have been reported after short-term cell exposure to different NPs, including citrate AuNPs.10,22 In our previous work, we showed that the non-chronic cell exposure to our AuNPs induced ER stress in HDF.3 ER stress is implicated in ROS production, involved in the activation of proliferative signaling pathways.65 Tsai and co-authors reported a significant increase of ROS and activation of proteins in RTK pathways associated with the ER stress response in cells treated with a low dose of “naked” AuNPs (no surface coating).66 Our previous analysis has shown no increase in cell proliferation rates in HDF cells after the treatment with any AuNP under non-chronic exposure.3 Indeed, at the systems-level perspective, the non-chronic exposure led to significant perturbations in pathways related to modulation of cell proliferation across virtually all WGCNA modules (Table 1, Fig. 5) with cells under this exposure condition potentially suppressing proliferative signaling pathways, as a general trend, through miRNAs. This was well exemplified in the yellow module (Fig. 3), enriched for RTK signaling pathways, where co-expressed miRNAs were upregulated in cells under non-chronic exposure, independently of the AuNP type (exposure condition effect). The upregulation of most hub miRNAs in this module, including the top 4 hub miRNAs, hsa-miR-26b-5p, 376a-3p, 146b-5p, 369–3p, as well as hub miRNAs found DE (data file S2) have been associated with inhibition of fibroblast proliferation.57,67–72 miRNAs exert a profound influence in the response to stress in developed tissues, suggesting their critical function in the maintenance of homeostasis.8 Thus, we speculate that, in this case, the miRNA dysregulation leading to inhibition of proliferative signals is a cellular response to the stress provoked by the non-chronic exposure to AuNPs in attempt to restore cell hemostasis.
Under non-chronic conditions, the cell treatment with citrate spheres surprisingly presented the greatest impact on miRNA expression levels compared to controls, suggesting that the AuNP surface chemistry also had a large effect in miRNA expression changes. Generally, citrate is considered a relatively benign surface chemistry to cells. Perturbations in three miRNA co-expression modules, red, green yellow and light cyan, enriched for pathways related to the modulation of cell proliferation, were noticed. The downregulation of hsa-miR-132–3p, let-7a-5p, and 196a-5p, all hub miRNAs in the red module, and the upregulation of their target CDKN1A (p21) (FC: 1.40, P value: 0.016; unpublished observation), associated with inhibition of cell proliferation, were validated in our cells under the non-chronic exposure to citrate spheres (data file S4). Moreover, miRNA-mRNA pairs were also found in the light cyan module: the upregulation of hsa-miR-146a-5p by citrate spheres, non-chronic treatment, and concomitant downregulation of its targets TLR-4 and NFAT5,3 related to cell cycle progression and inflammation. These findings strongly suggest that cells are activating anti-proliferative programs through miRNAs. The suppression of HDF proliferation associated with significant changes in miRNA levels after a short-term treatment with 20 nm-citrate spheres in a high dose (200 μM) was shown previously.22
Interestingly, the global miRNA expression pattern observed for the non-chronic treatment with citrate spheres presented more similarities to the ones found in cells chronically treated with AuNPs (Fig. 2A) and the WGCNA showed miRNA co-expression patterns following the same trend as observed particularly in samples chronically exposed to PAA Rods. This is well-represented in some modules such as in the blue one (Table 1), enriched for intrinsic mitochondrial apoptotic pathway, which plays a major role in the cell death triggered by metal-based NP-induced ROS.66,73 As discussed above, the upregulation of miRNAs in this module by the non-chronic citrate spheres and the chronic PAA rods exposures was concomitant with the inhibition of two pro-apoptotic target genes, TP53 and BNIP3L, in the same samples. These data, along with our previous results showing no significant cell death after the non-chronic exposure to citrate spheres or the chronic exposure to PAA rods compared to controls, suggest that the inhibition of apoptosis play a pivotal role in the cellular response to these AuNPs in such exposure conditions. As mentioned previously, ER stress was observed at the mRNA level in all non-chronic treated samples but also in samples chronically treated with PAA rods.3 It was demonstrated that the ER stress can lead to a decrease in the expression of tumor suppressor genes such as TP53, target of five different hub miRNAs in the blue module, preventing apoptosis and promoting cell survival in attempt to restore cell homeostasis.74 In line with our data, the suppression of apoptosis in HDF cells was observed after the acute treatment with citrate spheres (200 μM), via pathways regulation by miRNAs.22
Profound differences in miRNA expression between samples treated with the same AuNP type but under distinct exposure conditions were observed. This was particularly true considering the exposure to PAA rods, the most endocytosed AuNP type by our cells,3 indicating distinct mechanisms of cell response to these AuNPs in an exposure condition-dependent way. At the systems-level perspective, differences in miRNA co-expression profile between these two treatments were highlighted in the turquoise, blue and yellow modules. The turquoise module was mainly enriched for miRNA target genes involved in the protein modification pathways, in particular, N-linked glycosylation, implicated in the protein folding process in the ER lumen and critical to cell homeostasis.58 Alterations in this pathway are common features of cancer cells.75 The downregulation of co-expressed miRNAs in the yellow module and upregulation of the blue module by the chronic cell exposure to PAA rods compared to both non-chronic exposure and controls suggest that the treatment with these NPs under chronic condition might stimulates cell proliferation and survival pathways with concomitant repression of apoptosis, as discussed above. Notably, the chronic exposure to PAA rods also downregulated other genes associated with cell cycle control and apoptosis (GADD45A and TNFRSF10A) in addition to TP53 and BNIP3L.3 These observations are compatible to those described during the carcinogenesis process. In response to stress, cells either reestablish their homeostasis or assume an altered state (“new homeostasis”) in the new environment, and miRNAs play a key role in mediating cell decision.8 Thus, our data suggest that the miRNA modulation occurred in response to the chronic exposure to PAA rods (continual stress) might be enforcing a new state of our cells. However, no changes in cell proliferation was yet observed in our samples. Of note, we previously shown that both chronic and non-chronic treatments with PAA rods evoked oxidative stress-related responses in HDF cells without a concomitant upregulation of the protective antioxidant genes present in our array (such as NQO1 and GSTP1), increasing the probability of cell injury.3
The two different exposure conditions for PEG rods, the least endocytosed AuNP type but found free in the cytoplasm of HDF cells,3 also elicited great differences in the miRNA expression profiles. PEGylated NPs are popularly thought to resist protein adsorption and be biocompatible.76 However, PEGylated gold NPs indeed cause measurable cellular responses in vitro and in vivo.3,77 A total of seven miRNA co-expression modules was perturbed by PEG rods, including the pink module, strongly enriched for target genes related to senescence (Table 1, Fig. 5). Our miRNA DE analysis showed a 4.4x increase in the hsa-miR-335–5p levels in cells under non-chronic exposure to PEG rods compared their chronic counterpart (Fig. 2, table S4). This miRNA is associated with the senescence-associated secretory phenotype (SASP) development in human fibroblasts by inducing the inflammatory cytokine IL-6, and other SASP markers.56 Interestingly, we found the IL-6 gene highly expressed in our cells after the non-chronic exposure to PEG rods, as well as increased levels of the chemokine CCL2 gene, another component of the SASP, and the DDIT3 gene (also known as CHOP),3 related to UPR-mediated cell senescence in response to prolonged ER stress.78 In addition, cells under this treatment presented a significant increase in size,3 a morphological change feature of senescence, and a slight increase in the expression levels of CDKN1A (p21) (FC: 1.3, P value: 0.04, unpublished observation), a cell-cycle inhibitor often upregulated in senescent cells. Senescence is a mechanism of cell response to stress (genotoxic, oxidative, oncogenic, and others) activated when cells are unable to cope (adapt) with severe perturbations of homeostasis, adopting a state of proliferative inactivation, and other characteristics,79 as discussed above. However, no suppression of cell proliferation was yet noticed in our cells non-chronically exposed to PEG rods after 20 weeks.3 Our network-based analysis revealed that the non-chronic exposure to PEG rods elicited the overexpression of miRNAs in the pink module (Table 1). Notably, the top 3 hub miRNAs in this module, hsa-miR-152-3p, 1185-2-3p and 34a-5p, have been reported to promote cellular senescence in fibroblasts.57,80,81 Furthermore, the same exposure led to downregulation of miRNAs in the green module which has the hsa-miR-24–3p as the top hub miRNA. Interestingly, decreased levels of this miRNA, also found DE in the comparison PEG rods chronic × non-chronic, has been associated with senescence phenotype in fibroblasts.82 Despite our data provide evidences implying a link between the non-chronic exposure to PEG rods and the senescence program in HDF cells in a long-term, further investigation is needed to clarify this effect.
Conclusions
Taken together, our data revealed that HDF cells exposed to AuNPs upon non-chronic condition presented greater dysregulation in the miRNA levels than the chronic one, despite the time of cell exposure (only 24h) and the low dose applied. These cells were under ER stress after 20 weeks of the exposure to AuNPs.3 At the systems-level perspective, our findings suggest that the alterations in miRNA levels represent a cell strategy to cope with the changes in homeostasis after the non-chronic treatment with AuNPs, mostly by suppressing proliferative signaling pathways. Indeed, no significant alterations in cell proliferation, a possible ER stress outcome,65 were observed in our cells.
Furthermore, we showed that the surface chemistry plays a critical role in the miRNA dysregulation and, therefore, in the pathways perturbed by AuNPs. Considering the non-chronic cell exposure, citrate spheres led to the highest number of miRNA changes compared to any other AuNP type, indicating the big impact of these NPs on cell homeostasis. Some mechanisms of cellular response to the NPs exposure were mostly correlated to a specific AuNP surface chemistry. For instance, the non-chronic treatment with citrate was related to antiapoptotic pathways whereas PEG-coated rods were possibly activating senescence pathway. Different mechanisms of cell response are most likely to be related to the intensity/duration of ER stress in the NPs treated samples.
The observation that cells chronically exposed to PAA rods are potentially suppressing apoptosis pathways while inducing proliferative pathways is a concern and needs to be further explored. Here, the miRNA dysregulation might be contributing to cells reprogram their gene expression patterns during continual stress and reach a new state. High amounts of PAA rods were found accumulated in HDF cells under chronic exposure after 20 weeks, inducing oxidative stress without eliciting antioxidant defense,3 which increases the chances of cell damage.
Collectively, our study provides new insights into the molecular changes underlying the long-term cellular responses to a low dose of AuNPs through genome-wide miRNA profiling combined with a systems biology approach. We suggest that miRNAs play critical roles in the cellular responses to the stress provoked by AuNP stimuli. The exposure condition and surface chemistry had a significant impact on the miRNA changes and in the mechanisms of cellular response to AuNPs. Our findings can be used as a guide to further experimental studies to validate specific mechanisms/pathways perturbed by NPs, providing a fertile ground for testing more molecular-level hypotheses on how exactly NPs exert their effects in biological systems.
Supplementary Material
Acknowledgments
We would like to acknowledge the High-Performance Biological Computing (HPCBio) at Roy J. Carver Biotechnology Center for the miRNA sequencing data analyses. We thank Prof. Isaac Cann (Institute of Genomic Biology) for sharing the thermocycler and UV light instrument in the miRNA library preparation; Prof. Hee-Sun Han (Department of Chemistry) for sharing the thermocycler in the sequencing-data validation experiments; Dr. Chris Fields and Dr. Jenny Drnevich Zadeh (HPCBio) for helpful discussions; Dr. Tassia Mangetti for her help with the Cytoscape software. This work was supported by the NIH (R01 GM125845) and the University of Illinois at Urbana-Champaign.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest
Electronic supplementary information (ESI) available: supporting figures and tables
References
- 1.Falagan-Lotsch P, Grzincic EM and Murphy CJ, Bioconjug. Chem, 2017, 28, 135–152. [DOI] [PubMed] [Google Scholar]
- 2.Vance ME, Kuiken T, Vejerano EP, McGinnis SP, Hochella MF and Hull DR, Beilstein J. Nanotechnol, 2015, 6, 1769–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falagan-Lotsch P, Grzincic EM and Murphy CJ, Proc. Natl. Acad. Sci, 2016, 113, 13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown MK and Naidoo N, Front. Physiol, 2012, 3, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fadeel B, Farcal L, Hardy B, Vázquez-Campos S, Hristozov D, Marcomini A, Lynch I, Valsami-Jones E, Alenius H and Savolainen K, Nat. Nanotechnol, 2018, 13, 537–543. [DOI] [PubMed] [Google Scholar]
- 6.Wu X and Song Y, BMC Genomics, 2011, 12, 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP, Cell, 2004, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 8.Leung AKL and Sharp PA, Mol. Cell, 2010, 40, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halappanavar S, Jackson P, Williams A, Jensen KA, Hougaard K, Vogel U, Yauk C and Wallin H, Environ. Mol. Mutagen, 2011, 52, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Wang H, Qi Y, Tu J, Bai Y, Tian T, Huang N, Wang Y, Xiong F, Lu Z and Xiao Z, Biomaterials, 2011, 32, 9021–9030. [DOI] [PubMed] [Google Scholar]
- 11.Sun B, Liu R, Ye N and Xiao ZD, PLoS One, 2015, 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dymacek J, Snyder-Talkington BN, Porter DW, Mercer RR, Wolfarth MG, Castranova V, Qian Y and Guo NL, Toxicol. Sci, 2015, 144, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thai SF, Wallace KA, Jones CP, Ren H, Prasad RY, Ward WO, Kohan MJ and Blackman CF, J. Nanosci. Nanotechnol, 2015, 15, 492–503. [DOI] [PubMed] [Google Scholar]
- 14.Grogg MW, Braydich-Stolle LK, Maurer-Gardner EI, Hill NT, Sakaram S, Kadakia MP and Hussain SM, Toxicol. Res. (Camb), 2016, 5, 1733–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh JH, Son MY, Choi MS, Kim S, young Choi A, Lee HA, Kim KS, Kim J, Song CW and Yoon S, Toxicol. Appl. Pharmacol, 2016, 299, 8–23. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y, Lü X and Lü X, J. Biomed. Nanotechnol, 2018, 14, 2042–2055. [DOI] [PubMed] [Google Scholar]
- 17.Scala G, Kinaret P, Marwah V, Sund J, Fortino V and Greco D, NanoImpact, 2018, 11, 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventura C, Vieira L, Silva C, Sousa-Uva A and Silva MJ, Toxicol. Lett, 2020, 328, 7–18. [DOI] [PubMed] [Google Scholar]
- 19.Ng CT, Dheen ST, Yip WCG, Ong CN, Bay BH and Lanry Yung LY, Biomaterials, 2011, 32, 7609–7615. [DOI] [PubMed] [Google Scholar]
- 20.Chew WS, Poh KW, Siddiqi NJ, Alhomida AS, Yu LE and Ong WY, Biomarkers, 2012, 17, 750–757. [DOI] [PubMed] [Google Scholar]
- 21.Balansky R, Longobardi M, Ganchev G, Iltcheva M, Nedyalkov N, Atanasov P, Toshkova R, De Flora S and Izzotti A, Mutat. Res. - Fundam. Mol. Mech. Mutagen, 2013, 751–752, 42–48. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Lü X, Qu Y, Yang Y and Wu S, Biomaterials, 2015, 37, 13–24. [DOI] [PubMed] [Google Scholar]
- 23.Brzóska K, Gradzka I and Kruszewski M, Materials (Basel)., 2019, 12, 1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langfelder P and Horvath S, BMC Syst. Biol, 2007, 1, 54–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sau TK and Murphy CJ, Langmuir, 2004, 20, 6414–6420. [DOI] [PubMed] [Google Scholar]
- 26.Ewels P, Magnusson M, Lundin S and Käller M, Bioinformatics, 2016, 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozomara A and Griffiths-Jones S, Nucleic Acids Res, , DOI: 10.1093/nar/gkt1181. [DOI] [Google Scholar]
- 28.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MaCdonald J, Obenchain V, Oles̈ AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron Land Morgan M, Nat. Methods, 2015, 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MD and Oshlack A, Genome Biol, 2010, 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MD, McCarthy DJ and Smyth GK, Bioinformatics, 2009, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y and Hochberg Y, J. R. Stat. Soc. Ser. B, 1995, 57, 289–300. [Google Scholar]
- 32.Livak KJ and Schmittgen TD, Methods, 2001, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 33.Langfelder P and Horvath S, BMC Bioinformatics, 2008, 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dweep H and Gretz N, Nat. Methods, 2015, 12, 697–697. [DOI] [PubMed] [Google Scholar]
- 35.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, Chiew MY, Tai CS, Wei TY, Tsai TR, Huang HT, Wang CY, Wu HY, Ho SY, Chen PR, Chuang CH, Hsieh PJ, Wu YS, Chen WL, Li MJ, Wu YC, Huang XY, Ng FL, Buddhakosai W, Huang PC, Lan KC, Huang CY, Weng SL, Cheng YN, Liang C, Hsu WL and Da Huang H, Nucleic Acids Res, 2018, 46, D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Han S, Kwon CS and Lee D, Protein Cell, 2016, 7, 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, Xie X, Zhou C and Lu W, Oncogene, 2013, 32, 976–987. [DOI] [PubMed] [Google Scholar]
- 38.Peng Y, Dai Y, Hitchcock C, Yang X, Kassis ES, Liu L, Luo Z, Sun H-L, Cui R, Wei H, Kim T, Lee TJ, Jeon Y-J, Nuovo GJ, Volinia S, He Q, Yu J, Nana-Sinkam P and Croce CM, Proc. Natl. Acad. Sci, 2013, 110, 15043–15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J-R, Yan B, Guo Q, Fu F, Wang Z, Yin Z and Wei Y, Onco. Targets. Ther, 2015, 8, 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta A, Hossain MM, Read DE, Hetz C, Samali A and Gupta S, Sci. Rep, 2015, 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV and Lü J, Am. J. Respir. Cell Mol. Biol, 2011, 45, 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji X, Wu B, Fan J, Han R, Luo C, Wang T, Yang J, Han L, Zhu B, Wei D, Chen J and Ni C, Sci. Rep, , DOI: 10.1038/srep14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewinska A, Adamczyk-Grochala J, Kwasniewicz E, Deregowska A, Semik E, Zabek T and Wnuk M, Redox Biol, 2018, 14, 20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng H, Ma R, Wang Q, Zhang P, Li D, Wang Q, Wang J, Li H, Liu H and Wang Z, Oncotarget, 2015, 6, 27805–27815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giuliani A, Prattichizzo F, Micolucci L, Ceriello A, Procopio AD and Rippo MR, Mediators Inflamm, 2017, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, Benes V, Blake J, Pfister S, Eckstein V and Ho AD, PLoS One, 2008, 3, e2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhahbi JM, Atamna H, Boffelli D, Magis W, Spindler SR and Martin DIK, PLoS One, , DOI: 10.1371/journal.pone.0020509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun R, Liu Z, Tong D, Yang Y, Guo B, Wang X, Zhao L and Huang C, Cell Death Dis, , DOI: 10.1038/cddis.2017.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, Xiao C, Hu WY, Han J and Sun P, Cancer Res, 2010, 70, 8547–8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Felice B, Manfellotto F, Garbi C, Santoriello M and Nacca M, Mol. Med. Rep, 2018, 17, 7081–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lang A, Grether-Beck S, Singh M, Kuck F, Jakob S, Kefalas A, Altinoluk-Hambüchen S, Graffmann N, Schneider M, Lindecke A, Brenden H, Felsner I, Ezzahoini H, Marini A, Weinhold S, Vierkötter A, Tigges J, Schmidt S, Stühler K, Köhrer K, Uhrberg M, Haendeler J, Krutmann J and Piekorz RP, Aging (Albany. NY)., 2016, 8, 484–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savardashtaki A, Shabaninejad Z, Movahedpour A, Sahebnasagh R, Mirzaei H and Hamblin MR, Epigenomics, 2019, 11, 1627–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xishan Z, Ziying L, Jing D and Gang L, Sci. Rep, 2015, 5, 12460–12470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Kang H, Rho JG, Kim C, Tak H, Lee H, Ji E, Ahn S, Shin AR, Il Cho H, Huh YH, Song WK, Kim W and Lee EK, Sci. Rep, 2017, 7, 44847–44856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, Matsunaga J, Takahashi R. u., Takata T, Shimamoto A, Ochiya T and Tahara H, J. Cell Biol, 2011, 193, 409–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabir TD, Leigh RJ, Tasena H, Mellone M, Coletta RD, Parkinson EK, Prime SS, Thomas GJ, Paterson IC, Zhou D, McCall J, Speight PM and Lambert DW, Aging (Albany. NY)., 2016, 8, 1608–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mancini M, Saintigny G, Mahé C, Annicchiarico-Petruzzelli M, Melino G and Candi E, Aging (Albany. NY)., 2012, 4, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Banerjee DK, Biochim. Biophys. Acta - Gen. Subj, 2012, 1820, 1338–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Hong WL, Li ZM, Zhang QY and Zeng K, Aesthetic Plast. Surg, 2020, 1–9. [DOI] [PubMed] [Google Scholar]
- 60.Lemmon MA and Schlessinger J, Cell, 2010, 141, 1117–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhowmick N, Chytil A, Plieth D, Gorska A, Dumont N, Shappell S, Washington M, Neilson E and Moses H, Science (80-. ), 2004, 303, 848–851. [DOI] [PubMed] [Google Scholar]
- 62.Cao J, Mu Q and Huang H, Stem Cells Int, 2018, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bray SJ, Nat. Rev. Mol. Cell Biol, 2016, 17, 722–735. [DOI] [PubMed] [Google Scholar]
- 64.Shikama Y, Yamada M and Miyashita T, Eur. J. Immunol, 2003, 33, 1998–2006. [DOI] [PubMed] [Google Scholar]
- 65.Schieber M and Chandel NS, Curr. Biol, 2014, 24, R453–R462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsai YY, Huang YH, Chao YL, Hu KY, Te Chin L, Chou SH, Hour AL, Der Yao Y, Tu CS, Liang YJ, Tsai CY, Wu HY, Tan SW and Chen HM, ACS Nano, 2011, 5, 9354–9369. [DOI] [PubMed] [Google Scholar]
- 67.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Orjalo AV, Rodier F, Lithgow GJ and Campisi J, Aging (Albany NY), 2009, 1, 402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harada M, Luo X, Qi XY, Tadevosyan A, Maguy A, Ordog B, Ledoux J, Kato T, Naud P, Voigt N, Shi Y, Kamiya K, Murohara T, Kodama I, Tardif JC, Schotten U, Van Wagoner DR, Dobrev D and Nattel S, Circulation, 2012, 126, 2051–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, Zhang Z, Lin QX, Zheng XL, Min-Yang, Wu SL, Cheng JDand Shan ZX, Sci. Rep, 2017, 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng R, Dang R, Zhou Y, Ding M and Hua H, Hum. Cell, 2017, 30, 192–200. [DOI] [PubMed] [Google Scholar]
- 71.Tao H, Dai C, Ding JF, Yang JJ, Ding XS, Xu SS and Shi KH, Toxicology, 2018, 410, 182–192. [DOI] [PubMed] [Google Scholar]
- 72.Dai G, Yao X, Zhang Y, Gu J, Geng Y, Xue F and Zhang J, Bull. Cancer, 2018, 105, 336–349. [DOI] [PubMed] [Google Scholar]
- 73.Wang X, Chang CH, Jiang J, Liu X, Li J, Liu Q, Liao YP, Li L, Nel AE and Xia T, Small, 2020, 2000528, 1–15. [Google Scholar]
- 74.Pluquet O, Qu L-K, Baltzis D and Koromilas AE, Mol. Cell. Biol, 2005, 25, 9392–9405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scott E and Munkley J, Int. J. Mol. Sci, 2019, 20, 1389–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walkey CD, Olsen JB, Guo H, Emili A and Chan WCW, J. Am. Chem. Soc, 2012, 134, 2139–2147. [DOI] [PubMed] [Google Scholar]
- 77.Al-Harbi NS, Alrashood ST, Siddiqi NJ, Arafah MM, Ekhzaimy A and Khan HA, Nanomedicine (Lond), 2020, 15, 289–302. [DOI] [PubMed] [Google Scholar]
- 78.Denoyelle C, Abou-Rjaily G, Bezrookove V, Verhaegen M, Johnson TM, Fullen DR, Pointer JN, Gruber SB, Su LD, Nikiforov MA, Kaufman RJ, Bastian BC and Soengas MS, Nat. Cell Biol, 2006, 8, 1053–1063. [DOI] [PubMed] [Google Scholar]
- 79.Hernandez-Segura A, Nehme J and Demaria M, Trends Cell Biol, 2018, 28, 436–453. [DOI] [PubMed] [Google Scholar]
- 80.Marasa BS, Srikantan S, Martindale JL, Kim MM, Lee EK, Gorospe M and Abdelmohsen K, Aging (Albany. NY), 2010, 2, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cui H, Ge J, Xie N, Banerjee S, Zhou Y, Antony VB, Thannickal VJ and Liu G, Am. J. Respir. Cell Mol. Biol, 2017, 56, 168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, Navarro F, Dykxhoorn D, Lieberman J and Gorospe M, PLoS One, 2008, 3, e1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


