Abstract
Background:
Atopic dermatitis (AD) and food allergy (FA) are associated with skin barrier dysfunction.
Objective:
Skin biomarkers are needed for skin barrier interventions studies.
Methods:
In this study skin tape strip (STS) samples were collected from non-lesional skin of 62 AD FA+, AD FA− and nonatopic (NA) children for mass spectrometry proteomic analysis. TEWL and allergic sensitization were assessed. STS proteomic analysis results were validated in an independent cohort of 41 adults with AD with and without FA versus NA controls.
Results:
A group of 45 proteins was identified as a principal component 1 (PC1) with the highest expression in AD FA+ STS. This novel set of STS proteins was highly correlative to skin transepidermal water loss (TEWL) and allergic sensitization. PC1 proteins included keratin (KRT) intermediate filaments; proteins associated with inflammatory responses (S100 proteins, alarmins, protease inhibitors); glycolysis and anti-oxidant defense enzymes. Analysis of PC1 proteins expression in an independent adult AD cohort validated differential expression of STS PC1 proteins in the skin of adult AD patients with the history of clinical reactions to peanut.
Conclusion:
STS analysis of non-lesional skin of AD children identified a cluster of proteins with the highest expression in AD FA+ children. The differential expression of STS PC1 proteins was confirmed in a replicate cohort of adult AD patients with FA to peanut, suggesting a unique STS proteomic endotype for AD FA+ that persists into adulthood. Collectively, PC1 proteins are associated with abnormalities in skin barrier integrity and may increase the risk of epicutaneous sensitization to food allergens.
Keywords: atopic dermatitis, food allergy, skin barrier, proteomics, transepidermal water loss, IgE
Capsule summary:
A novel set of non-lesional skin proteins highly correlative to skin transepidermal water loss and allergic sensitization was identified in atopic dermatitis children. The results were validated in adults with atopic dermatitis and food allergy.
Introduction
The stratum corneum (SC) serves as a skin barrier, providing protection from potentially damaging external factors and controlling transcutaneous water loss. Transepidermal water loss (TEWL) measurement quantifies water diffusion across the SC and is commonly used for the physiologic assessment of skin barrier function1. In addition to basal TEWL to assess the undisturbed permeability of the skin barrier, TEWL measurements have also been conducted together with skin barrier perturbation using skin tape stripping (STS) to measure skin barrier integrity2. Healthy skin is not very sensitive to STS and can withstand mild perturbation, while disrupted skin, and skin with low structural integrity, exhibit greater changes in TEWL. The area under the curve for TEWL measurements done over a defined number of STS is used to assess the overall integrity of the SC. Skin barrier dysfunction and increased TEWL are major pathologic features of atopic dermatitis (AD)3–5. TEWL has been shown to correlate with AD severity6–8.
Epidermal proteins including filaggrin (FLG) are essential for the maintenance of the epidermal permeability barrier that prevents allergen penetration and loss of fluid5. Both FLG mutations and FLG inhibition by type 2 cytokines contribute to reduced FLG expression in AD skin9, 10. Several research groups have reported reduced FLG breakdown products and long-chain skin barrier lipid products in the skin of AD patients vs. non-atopic subjects using STS11, 12. Even in AD subjects who do not carry FLG gene mutations, significant changes in the skin expression of enzymes involved in lipid metabolism have been demonstrated13. These changes in the SC detectable by STS support the concept that skin barrier abnormalities in AD could allow penetration of environmental allergens and microbes and are accompanied by the release of epithelial-derived cytokines (e.g., thymic stromal lymphopoietin (TSLP), IL-25, IL-33), which drive type 2 immune responses5.
The utility of noninvasive approaches, such as TEWL measurement and STS protein and lipid profiling, to identify endotypes and skin biomarkers of clinical AD phenotypes is an area of active investigation14–17. However, the mechanisms underlying increased TEWL and changes in skin barrier function are not well understood. Identification of skin barrier components that control TEWL is critical for designing skin barrier interventions, to establish biomarkers and could facilitate the identification of AD subtypes.
In a recent prospective clinical study, funded by the NIH/NIAID Atopic Dermatitis Research Network (ADRN), we utilized a multi-omics approach with STS of non-lesional skin to show that children with AD and food allergy (FA) (AD FA+ children) represent a unique endotype that can be distinguished from AD children without FA (AD FA− children) and non-atopic (NA) subjects using a constellation of epithelial biomarkers including increased TEWL, low FLG breakdown products, changes in SC lamellar bilayer structure and lipid composition17. In this study, we report detailed information on STS proteomics analysis of this study cohort and examine associations of the proteomic data with TEWL and clinical allergy. The utility of STS proteomic analysis for the endotyping of AD FA+ and AD FA− patients is reported in this paper.
Methods
Study subjects.
In this study, we used data on 62 children from a cohort of patients from a prospective, clinical mechanistic study registered at ClinicalTrials.gov (identifier: NCT03168113). The details of the study cohort were reported earlier by our research group17. All AD participants had active skin disease diagnosed using published criteria18 and were stratified into 2 groups based on their FA status: 1) AD FA+ (n=21). These children were allergic to peanut with a peanut skin prick test wheal ≥ 8 mm, which has been reported to significantly correlate with immediate clinical reactions to oral peanut challenge19, as well as documentation of a previous positive oral food challenge to peanut or convincing history of an immediate allergic reaction to peanut. 2) AD FA− (n=19). These participants had no personal history of FA, as well as a negative skin prick test (wheal < 3 mm) to peanut, milk, egg, wheat, soy, shellfish mix (clam, crab, oyster, scallops, and shrimp), almond, English walnut, hazelnut, cashew, brazil nut, and sesame seed. 3) NA (n=22). NA controls were defined as those without a personal history of atopic diseases and negative skin prick tests to the same foods as well as common aeroallergens (cat, dog, dust mite, cockroach, mold mix, tree mix, grass mix, weeds mix). AD FA+ and AD FA− children were also skin tested for the aeroallergens and the information was recorded. The full inclusion and exclusion criteria for this study and study subject characteristics are detailed in our prior publication17. All participants avoided any treatments, baths, and skin creams or emollients which may potentially affect skin barrier function before skin sample collection.
Skin Disease Severity Assessments.
All AD FA+ and AD FA− patients in the study had active AD. None of the study subjects had an overt skin infection. AD severity was evaluated using the SCORing Atopic Dermatitis (SCORAD)20, and the Eczema Area and Severity Index (EASI)21. Persistence of AD was evaluated using the Nottingham Eczema Severity Score (NESS)22, a validated index incorporating clinical course over the past 12 months, disease intensity over the past 12 months and extent of body surface area involvement. Enrollment of AD FA+ and AD FA− participants was actively matched based on AD severity (mild or moderate) as determined by the NESS.
Skin Barrier Assessments.
Comparisons between groups for the TEWL Area Under the Curve (AUC), and TEWL at baseline before STS and after 5, 10, 15, and 20 tape strips were done on non-lesional skin of the forearm. TEWL was evaluated using the AquaFlux AF200 (Biox, London, UK) as previously described17, 18.
STS Collection.
Thirty D-Squame tape strips (22 mm diameter, CuDerm) were collected from non-lesional skin of the upper arms (forearm) using a D-Squame D500 pressure instrument to apply all STS with equivalent pressure (e.g. 225 g/cm2). Non-lesional sites selected for STS collection were at least 2 inches away from the lesion. Collected STS were stored individually in 12-well plates and kept frozen at −80°C prior to processing.
Sample Preparation for Global Proteomics Analysis.
For each sample collection, protein content was extracted from each of fifteen STS (every other STS to tape 30) with 50 mM ammonium bicarbonate, 1% SDS buffer supplemented with HALT™ protease inhibitors and 10 mM DTT. SDS was removed using a detergent removal spin column, protein concentrations were determined.
For proteomic analysis, 50 μg of protein from each sample were reduced in 5 mM DTT and alkylated in 10 mM iodoacetamide. The protein content was precipitated with cold (−20°C) 10% w/v trichloroacetic acid in acetone. The pellet was rinsed with cold acetone and dried. The dried protein pellet was dissolved in 5% trifluouroethanol (TFE) 50 mM TEAB and digested with 2 μg of Trypsin Lys-C mix overnight at 37°C. 10 μg of protein was labeled with Tandem mass tag (TMT) 10-plex reagents (ThermoFisher Scientific) for 2h at room temperature. The reaction was quenched with 1% ethanolamine17. The TMT labeled samples were combined into sets of nine samples containing samples from each study group. Each TMT set was step-fractionated on an Oasis plate (Waters) using 7 mM TEAB (pH ~8) in 5, 10, 25, and 70% acetonitrile. Each fraction was lyophilized, stored at −20°C and reconstituted in 0.1% formic acid, 2% acetonitrile prior to tandem mass spectrometry analysis.
LC-MS/MS.
Each fraction was separated over a 90 minute reversed phase gradient. Precursor ion scans spanning 400 – 1600 m/z were acquired at 120000 (m/z = 200) resolution every 3 seconds. Detected ions in the +2 to +6 charge states were individually isolated by the quadrupole in 0.4 m/z bins and fragmented in order of highest intensity by high energy collisional dissociation (38 normalized collision energy). Fragment ion scans were acquired at 50000 resolution (at m/z = 200) with automatic gain control set to 50000 and a maximum ion accumulation time of 86 ms. Fragmented ions were excluded from redundant fragmentation for 15 seconds.
Data analysis.
The mass spectrometry data were searched against the SwissProt Homo Sapien database (May 2019, 20,431 entries) with MASCOT 2.6.2 and Percolator re-scoring followed by quantitative analysis in Proteome Discoverer 2.2 (Thermo). This ensured that only human-derived proteins were analyzed. Spectra were matched to theoretical tryptic peptides using 5 parts-per-million (ppm) precursor and 0.01 Da fragment mass error tolerances and filtered to 1% false discovery rate (FDR). For quantitation, the identified spectra were filtered to exclude chimeric spectra with greater than 30% co-isolation interference, and peptides with deamidation or oxidation. Peptides were quantified based on the reporter ion signal to noise values. Each channel was adjusted for loading by leveling the summed peptide abundance to the channel with the highest summed peptide intensity. Proteins were quantified using only unique peptides, and the average protein abundance across all samples was scaled to 100. Fold changes and ANOVA (analysis of variance) significance values were calculated based on the scaled protein abundances. Protein grouping was performed according to molecular function and protein class using the Gene Ontology database (STRING version 11.0 [https://string-db.org/]).
FLG breakdown products and skin microbiome analysis.
In this manuscript the data on the FLG breakdown products and skin microbiome analysis collected for this study cohort was utilized17. For details of FLG breakdown products and microbiome analysis refer to previously published work17.
Statistical data analysis.
All statistical analyses were performed in the R programming environment23 and a p-value <0.05 was considered significant. Figures were created using the lattice package24.
The protein expression data set acquired in this study is high dimensional (n=149) and contains variables that correlate with each other which introduces multicollinearity in regression models. Multicollinearity, therefore, may interfere in determining the precise effect of each predictor and also tends to result in larger standard errors (and, hence, wider confidence intervals and bigger p-values). Given these considerations, we have used a principal component analysis in our study to reduce the set of 149 proteins into a few dimensions that reduce a large amount of the variability of the original variables. These dimensions, called components, have the properties of collecting highly correlated variables within each component and being uncorrelated with each other. The first principal component (PC1) explains the highest amount of the variability of the original data. Principal component analysis and correlation analysis was executed using the stats package23. The PC1 loadings were compared across the three patient groups with an ANOVA model including pair-wise comparisons using the emmeans package25.
To identify differentially expressed proteins between the patient groups, we conducted an analysis using the Bioconductor limma package26. This approach uses linear models to analyze the entire experiment as an integrated whole, rather than individual pair-wise comparisons. The model borrows information across proteins to smooth out variances and uses posterior variances in a classical t-test setting. The method is completely data-dependent and uses an empirical Bayes approach to estimate hyper-parameters. Contrasts were set up for pairwise comparisons between the patient groups. P-values were adjusted using the FDR correction method. A Venn diagram was constructed using the eulerr package27.
Correlation analysis of individual PC1 proteins with TEWL was performed. In addition, to examine whether STS PC1 proteomic data can predict TEWL, we developed a classification model using the Least Absolute Shrinkage and Selection Operator (LASSO) regression. LASSO regression is a type of linear regression that involves penalizing the absolute size of the regression coefficients. This technique reduces model complexity and prevents over-fitting, which may result from simple linear regression. We have incorporated 5-fold cross-validation as an internal validation strategy. The LASSO model identifies the smallest subset of proteins in the STS proteomics data set that are independent TEWL predictors. LASSO models were created using the glmnet package28.
To compare differences in demographics and severity characteristics between groups of correctly and incorrectly classified subjects resulting from the prediction model, chi-square tests were used for categorical variables, and the Wilcoxon test was used for the two-group comparisons.
Validation of proteomic findings in the independent patient cohort.
In this study, we have utilized an independent proteomic data set which examined non-lesional skin STS protein expression in adults with atopic dermatitis and NA controls. The patient cohort was a separate cohort gathered by the Atopic Dermatitis Research Network (ADRN). As part of the patients’ registry the study gathered data on patients’ history of clinical reactions to peanuts which included development of itchy skin rashes, swelling of the lips, tongue or eyes, breathing difficulties, gastrointestinal symptoms and anaphylactic reactions upon peanut consumption. Total serum IgE to peanut levels were analyzed. Twenty STS samples were collected from non-lesional skin of the volar surface of forearm at least 2 inches away from the lesion. Protein extracts were prepared from ten skin tape strips as described above and protein extracts were analyzed as the extracts from the pediatric cohort. STS PC1 protein expression was extracted from the data set. Protein IDs of PC1 proteins were used for data extraction and cumulative PC1 expression was compared between adult AD patients with and without history of allergic reactions to peanut. In addition, cumulative PC1 protein expression was compared between AD patients stratified by serum levels of total IgE to peanut.
Results
STS proteomics data principal component analysis
Protein extracts were prepared from STS samples of 62 study children. 371 individual proteins were detected in STS extracts by LC-MS/MS analysis. 149 of these proteins were quantified in STS extracts from all study subjects. Due to stochastic sampling of data-dependent mass spectrometry 222 proteins were only quantified in some TMT sets. No statistically significant differences were found in the expression of these 222 proteins between the NA, AD FA− versus AD FA+ subjects (p=0.64).
The 149 proteins that were detected in all of the STS extracts were further evaluated in an unsupervised principal component analysis. The leading principal component (PC1) explained 30% of the variance in the proteomics data and consisted of 45 proteins (see Table 1 for the list of these proteins). Importantly, as a group, these PC1 proteins were differentially expressed across the three patient groups in our cohort (Fig. 1), with the highest PC1 composite score for AD FA+ subjects, intermediate for AD FA− patients and the lowest PC1 score for NA controls (Fig. 1).
Table 1.
Proteins identified in skin tape strip samples from all study participants*
| A2ML1 | CAPNS2 | DSG1 | GSDMA | IL37 | KRT5 | NPC2 | PSMA1 | S100A8 | SPRR1A |
| ACPP | CASP14 | DSP | GSN | JUP | KRT77 | NPM1 | PSMA2 | S100A9 | SPRR2D |
| ACTB | CAT | ECM1 | GSS | KLK10 | KRT78 | NUDT5 | PSMA3 | SBSN | SSR4 |
| ALAD | CDSN | EEF1A1 | GSTP1 | KLK11 | KRT80 | NUTF2 | PSMA4 | SCPEP1 | TALDO1 |
| ALB | CPA4 | EEF2 | HAL | KLK5 | KRT9 | PARK7 | PSMA5 | SDR9C7 | TGM1 |
| ALDOA | CST6 | ENO1 | HEXB | KLK7 | LAMP1 | PEBP1 | PSMA6 | SERPINA12 | TGM3 |
| ALOX12B | CSTA | FLG | HIST1H2BK | KLK8 | LDHA | PIP | PSMA7 | SERPINB12 | TMED10 |
| ANXA2 | CTSA | FLG2 | HIST1H3F | KPRP | LGALS3 | PKM | PSMB1 | SERPINB3 | TPI1 |
| ARG1 | CTSD | GAPDH | HIST1H4A | KRT1 | LGALS7 | PLEC | PSMB3 | SERPINB6 | TUBA1B |
| ASAH1 | CTSH | GBA | HRNR | KRT10 | LYPD3 | POF1B | PSMB4 | SERPINB7 | TXN |
| ATP5G3 | CTSL2 | GDI2 | HSPA5 | KRT14 | LYZ | PRCP | PSMB5 | SERPINB8 | WFDC12 |
| AZGP1 | DCD | GGCT | HSPA8 | KRT16 | MDH2 | PRDX1 | PSMB7 | SFN | EPPK1 |
| BLMH | DMKN | GGH | HSPB1 | KRT17 | ME1 | PRDX2 | RNASE7 | SLURP1 | IgG heavy chain |
| CALML5 | DSC1 | GM2A | IDH1 | KRT2 | MUCL1 | PSAP | RPS27A | SOD1 | ALMS1 |
| CAPN1 | DSC3 | GRN | IGKC | KRT23 | NCCRP1 | PSAPL1 | S100A7 | SPINK5 |
371 individual proteins were detected in STS extracts by C-MS/MS analysis. 149 of these proteins were quantified in STS extracts from all study subjects and are summarized in this table. 45 proteins identified as PC1 in the principal component analysis of skin tape strip proteomics data are highlighted in grey.
Figure 1.
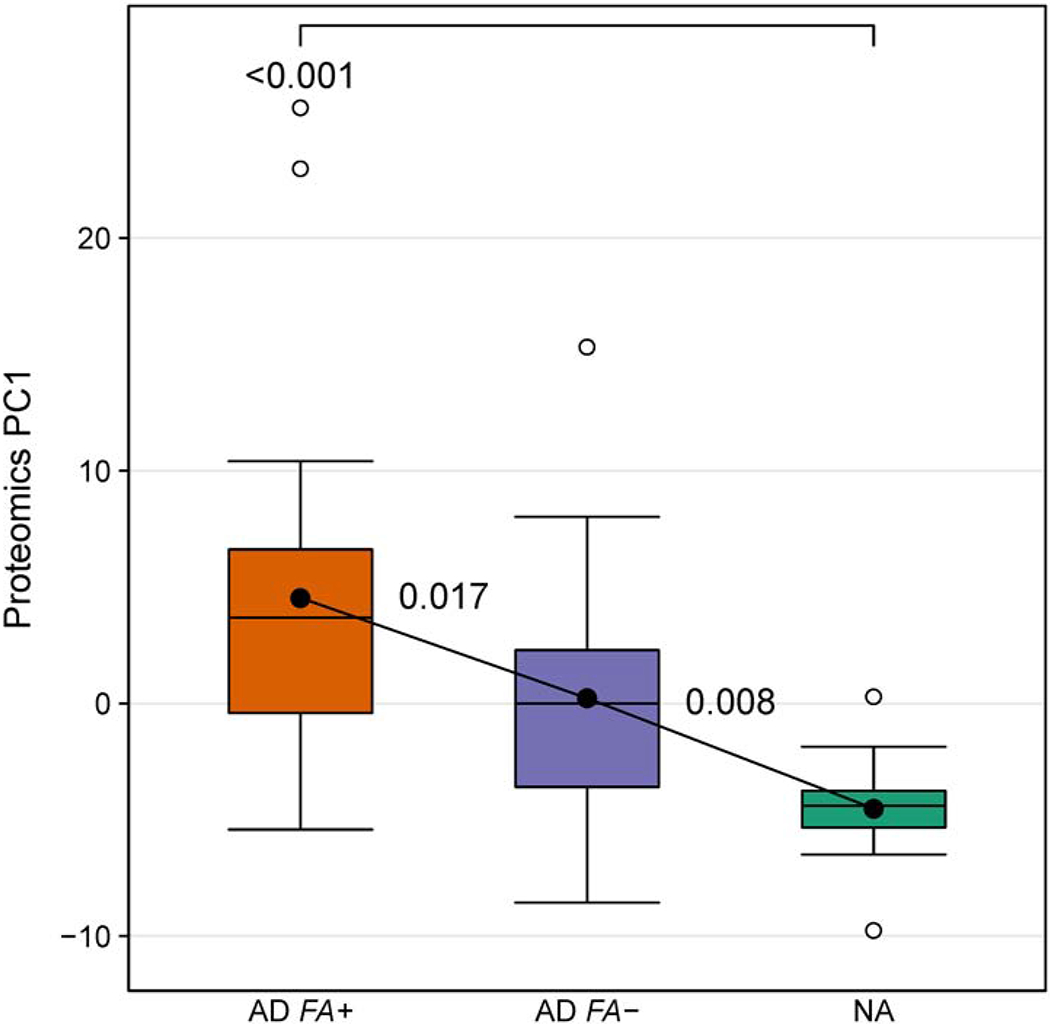
The unsupervised principal component analysis of proteins detected in the skin tape strip samples from AD FA+, AD FA− and NA children identifies a group of principal component 1 proteins (PC1) that are differentially expressed among the three study groups. In this and all other figures with boxplots solid horizontal line and filled circle within each box represent the median and mean, respectively, the box margins are the interquartile range (50% of the observations), whisker lines extend for 1.5 times the interquartile range, and observations outside the whisker are marked by an open circle. The annotations are the p values from pairwise comparisons between groups obtained from a one-way analysis of variance (ANOVA).
Relationship between STS PC1 proteins and skin TEWL
We have previously reported increased TEWL in the skin of AD FA+ patients17. Given that PC1 proteins had the highest expression in AD FA+ subjects, we evaluated the relationship between skin PC1 protein expression and skin TEWL. Importantly, we observed a significant positive correlation between PC1 protein expression and the cumulative TEWL estimate over 20 STS (TEWL AUC) (Fig. 2A). In our recent study, we reported dissociation in TEWL measurements between AD FA+ versus AD FA− and NA controls with tape stripping, with significant enhancement in skin TEWL at STS 15 and 20 in AD FA+ subjects as compared to AD FA− subjects and NA controls17. Using TEWL data measurement collected at STS 15, we observed a significant positive correlation between skin PC1 protein expression and skin TEWL at tape 15 (Fig. 2B).
Figure 2.
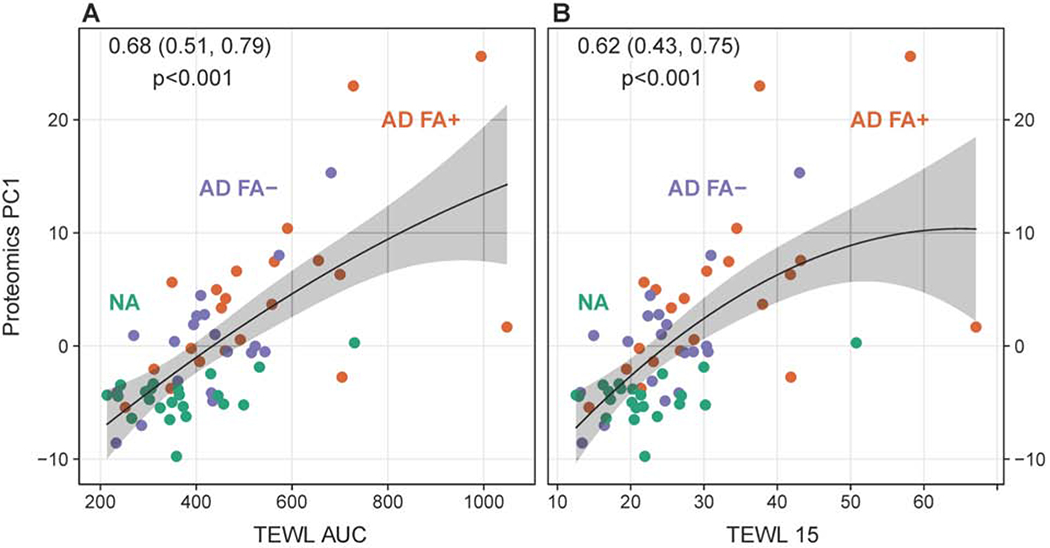
The associations of skin transepidermal water loss (TEWL) with skin tape strip proteomic PC1. The associations for the TEWL area under the curve (TEWL AUC) (A) and TEWL 15 (TEWL measurement in the skin after 15 skin tape strips) (B) are shown. In this and all other figures with correlation analysis correlation coefficients with 95% confidence interval and associated p values are shown in the upper part of the graphs.
Relationship between STS PC1 proteins and parameters of allergic sensitization in AD FA+ and AD FA− subjects
We have evaluated a relationship between cumulative STS PC1 protein expression and total serum IgE in AD patients as a marker of allergic sensitization. For this analysis, two of AD groups were combined. A significant positive correlation between the cumulative expression level of STS PC1 proteins and total serum IgE was found (Table 2). We further examined relationships between STS PC1 protein expression and IgE to foods (peanut, milk, egg) and aeroallergens (Phadiatop IgE) in AD groups. A significant correlation between serum levels of IgE to peanuts and PC1 protein skin expression was observed, while a trend for a positive correlation between serum IgE to milk and egg and PC1 was found. A significant correlation between the aeroallergen panel, Phadiatop IgE, and PC1 skin expression was also identified (Table 2). No association between STS PC1 expression and aforementioned serum IgE parameters was found if the data were analyzed separately for AD FA+ and AD FA− groups (data not shown).
Table 2.
Total serum IgE, food-specific IgE and aeroallergen IgE correlations with PC1 proteins among AD FA+ and AD FA− children (N=40)
| Variable | N | Correlation (95% CI) w/PC1 | p-value |
|---|---|---|---|
| Total IgE log10 (kU/L) | 40 | 0.35 (0.04, 0.59) | 0.028 |
| IgE a-Peanut log10 (kUA/L) | 40 | 0.33 (0.02, 0.58) | 0.037 |
| IgE a-Milk log10 (kUA/L) | 40 | 0.29 (−0.02, 0.55) | 0.068 |
| IgE a-Egg log10 (kUA/L) | 40 | 0.23 (−0.09, 0.50) | 0.20 |
| Phadiatop (PAU/L) * | 40 | 0.32 (0.01, 0.57) | 0.047 |
Phadiatop IgE analysis was restricted to aeroallergens.
Skin prick test assessment confirmed a significant correlation between the number of positive allergen skin prick tests and STS PC1 skin protein expression (Table 3). A positive correlation between the number of skin tests to foods and PC1 was found. Similarly, a positive correlation between the number of positive skin prick tests to aeroallergens and PC1 was observed (Table 3). No association between number of positive skin tests and PC1 expression was observed if the data was analyzed separately for AD FA+ and AD FA− groups (data not shown).
Table 3.
Skin test associations with PC1 proteins among AD FA+, AD FA−, and NA children (N=62)
| Variable | N† | Correlation (95% CI) w/PC1 | p-value |
|---|---|---|---|
| # Positive Skin Tests to Allergens (20) | 62 | 0.54 (0.33, 0.69) | 0.001 |
| # Positive Skin Tests to Foods (12) | 62 | 0.48 (0.26, 0.65) | 0.001 |
| # Positive Skin Tests to Aeroallergens (8) | 62 | 0.52 (0.31, 0.68) | 0.001 |
Differential expression of PC1 proteins among AD FA+, AD FA− and NA subjects.
The expression of each of the 45 PC1 proteins was compared across the three study groups and a summary of results is included in Figure 3.
Figure 3.
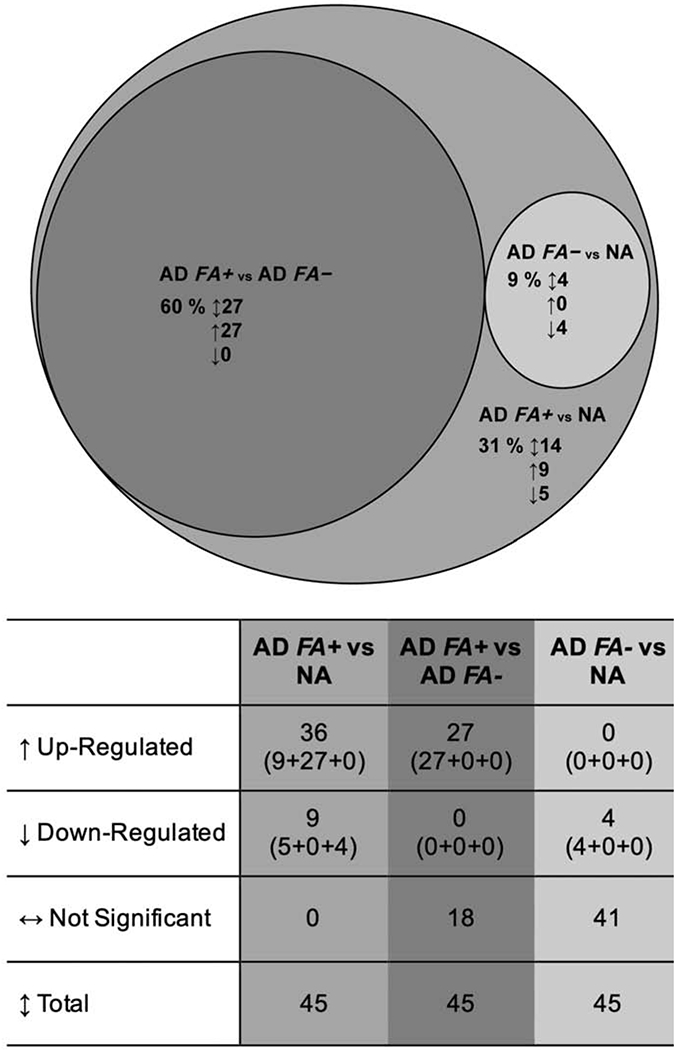
Differential expression of the 45 PC1 proteins in skin tape strip samples from AD FA+, AD FA− and NA children. The large circle illustrates the number of proteins that were differentially expressed between AD FA+ and NA subjects. Circles of smaller diameter reflect the number of proteins that were differentially expressed between AD FA+ and AD FA− subjects (medium dark grey circle) and between AD FA− and NA subjects (small light grey circle). 4 proteins were decreased in all AD in comparison to NA.
All 45 PC1 proteins identified in STS analysis were differentially expressed in AD FA+ subjects when compared to the NA controls; specifically, the expression of 36 proteins was up-regulated and 9 proteins were down-regulated in AD FA+ STS protein extracts. Of these 36 up-regulated proteins in AD FA+ STS, 27 were also significantly increased, when comparing the STS protein extracts from AD FA+ subjects to the AD FA− STS (Suppl. Fig. 1). Of the 9 down-regulated proteins in the comparison of the AD FA+ versus the NA STS (Suppl. Fig. 2), the expression of 4 proteins (N-acylsphingosine aminohidrolase 1, acid ceramidase (ASAH1), cystatin A (CSTA), keratin (KRT) 77 (KRT77), serine protease inhibitor 12 (SERPINB12)) was also significantly decreased, when comparing the AD FA− STS with the NA only STS protein extracts (Suppl. Fig. 3).
Of the 45 PC1 proteins, a total of 14 (9 up-regulated, 5 down-regulated) proteins were significantly expressed in the comparison of AD FA+ STS with NA STS protein extracts, exclusively (i.e. that do not overlap with any other group comparisons (Suppl. Fig. 2)).
Functional analysis of proteins in PC1 cluster
Functional analysis of the PC1 proteins identified in STS proteomics analysis revealed three major functional groups of proteins: 1) A group of keratin intermediate filaments; 2) Proteins associated with inflammatory response (S100 proteins, alarmins, protease inhibitors); 3) Glycolysis and oxidative stress response proteins (glycolytic enzymes, oxidative stress response enzymes) (Fig. 4). The expression of these proteins was altered in non-lesional skin STS samples from AD FA+ patients.
Figure 4.
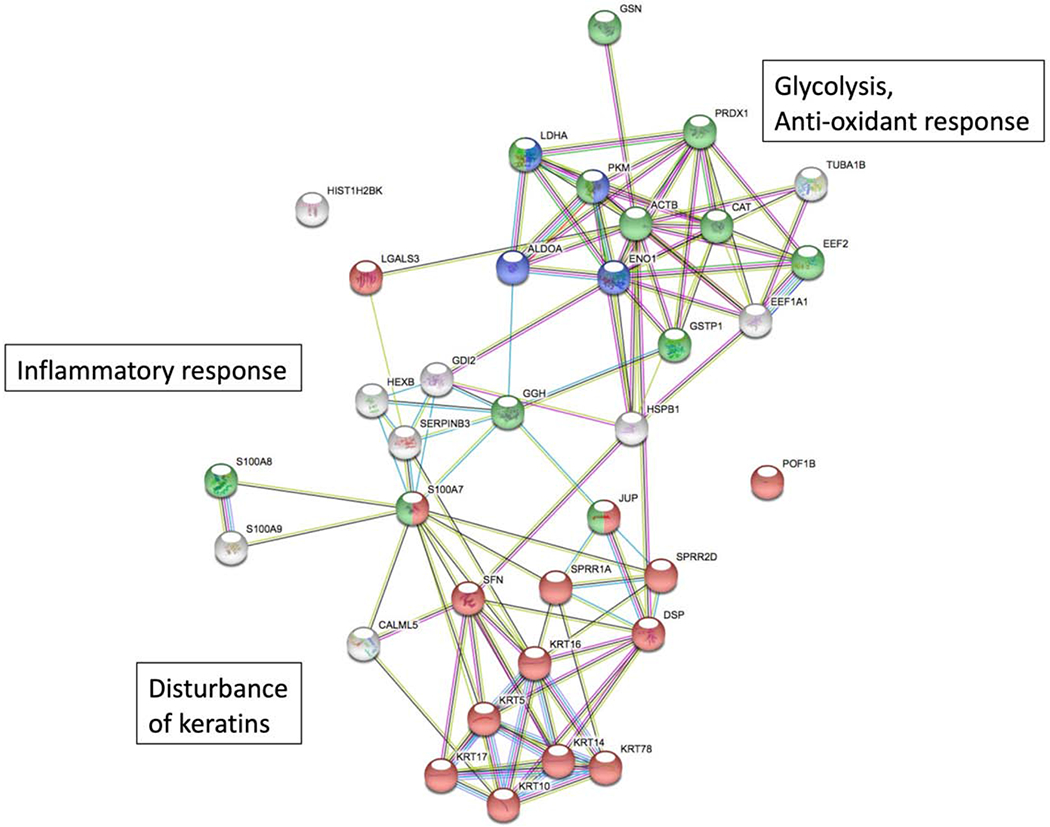
Functional analysis of STS PC1 proteins. The schematic represents proteins (individual nodes) and functional groups of proteins. Differentially expressed proteins were classified with the GO terms “cytokeratins, keratinocyte development” (n=11), “inflammatory response” (n=6), “glycolysis” (n=4), and “anti-oxidant response”’ (n=4). Linear connectors indicate evidence for association in published data sets (STRING11.0; accessed September 20, 2019)
In the stratified epithelia, keratins are expressed in a specific pattern tightly regulated by the differentiation program of the tissue29, 30. The expression of several keratins was found to be selectively altered only in STS samples from AD FA+ subjects. An increase in the levels of KRT16, a keratin associated with epidermal proliferation31, was observed in AD FA+ STS as compared to AD FA− and NA STS (Suppl. Fig. 1), KRT17, another keratin reported to be induced by inflammatory cytokines or in response to wound healing and oxidative or UV stress, was found increased in AD FA+ STS only (Suppl. Fig. 2)29. At the same time, the cornified envelop of AD FA+ skin was enriched in keratins that are normally found in the basal layer of epidermis, i.e. KRT5 and KRT14 (Suppl. Fig. 1), suggesting that the keratinocyte layers are not fully differentiated in AD FA+ skin. The expression of keratins that are part of epidermal maturation and differentiation was decreased in AD FA+ skin (KRT10, KRT78) (Suppl. Fig. 2).
In contrast, the expression of KRT77 was equally decreased in AD FA+ and AD FASTS samples as compared to NA STS (Suppl. Fig. 3). KRT77 has been reported to be weakly expressed by suprabasal cells in human epidermis32 and is highly expressed by the epithelial cells of sweat gland ducts33. Rearrangement of keratins in AD FA+ skin SC and to some extent in AD FA− skin SC may result in tissue fragility and reduced mechanical stress resilience of the skin29.
Concomitantly, a significant increase in expression of S100 calcium binding protein A family proteins (S100A7, S100A8, S100A9) in AD FA+ STS was observed (Suppl. Fig. 1). An increase in alarmins, like heat shock protein family B (small) member 1 (HSPB1), galectins (LGALS3, etc.) was found in AD FA+ STS (Suppl. Fig. 1). The increase in these proteins, in general, reflects ongoing inflammatory response in the skin. Importantly, the expression of protease inhibitors was altered in AD FA+ and AD FA− STS, with increased serine proteinase inhibitor glade B (SERPINB) member 3 (SERPINB3) in AD FA+ STS only and decreased SERPINB member 12 (SERPINB12) and CSTA in both AD FA+ and AD FA− STS (Suppl. Fig. 1, 3). These protease inhibitors are critical in maintaining the activity of skin proteases, like papain-like proteases (SERPINB3), trypsin, plasmin (SERPINB12), and thiol proteinases (CSTA). SERPINB12 is abundant at epithelial surfaces and, unlike other protease inhibitors, functions as an intracellular protease inhibitor34, 35.
Increased levels of glycolytic enzymes were found in STS of AD FA+ patients, including enolase 1 (ENO1), lactic dehydrogenase (LDHA) (as compared to both AD FA− and NA STS [Suppl. Fig. 1]), aldolase, fructose-bisphosphate A (ALDOA), and pyruvate kinase M1/2 (PKM) (as compared to NA STS only [Suppl. Fig. 2]), suggesting an increased energy requirement in AD FA+ STS. Lastly, a significant increase in glutathione S-transferase Pi 1 (GSTP1) (as compared to AD FA− and NA STS), peroxiredoxin 1 (PRDX1) and a decrease in catalase (CAT) (as compared to NA STS only) were found in AD FA+ skin tape strips, supporting a demand on antioxidant defense in the AD FA+ skin with perturbed barrier (Suppl. Fig. 1, 2).
In summary, among the STS PC1 proteins the expression of keratins, proteases, inflammatory mediators, alarmins, glycolytic enzymes and anti-oxidant defense proteins was found to be selectively altered in AD FA+ STS samples in comparison to AD FA− and NA STS samples. The expression of four proteins, KRT77, protease inhibitors SERPINB12, CSTA, and acid ceramidase ASAH1 was significantly decreased in non-lesional skin STS samples from all AD patients, irrespective of FA.
Contribution of individual PC1 proteins to skin TEWL
Importantly, all of the 45 proteins that make up PC1 had a significant correlation with TEWL AUC (Suppl. Table 1). SERPINB3 expression in the skin had the highest positive correlation with TEWL AUC, while KRT10 expression in skin samples had the highest negative correlation with TEWL AUC in our data set (Suppl. Table 1).
Using a statistical method for the determination of the relative importance of individual PC1 proteins to skin TEWL, we were able to quantify the independent effect of each of the 45 PC1 proteins on skin TEWL AUC. The LASSO regularized regression model selected nine out of the 45 proteins that were highly predictive for TEWL AUC: SERPINB3, gelsolin (GSN), KRT77, tubulin alpha 1b (TUBA1B), ALDOA, histone H2B type 1-K (HIST1H2BK), KRT16, S100A8, premature ovarian failure protein 1B (POF1B) (in decreasing order of the absolute penalized regression coefficient) (Fig. 5A). Selected proteins had the largest contribution to R2 in independently explaining TEWL AUC as an outcome variable, i.e. selected proteins were weighted equally (unrelated to the order or position of the predictor in the model statement) and were found to be independent from other proteins selected by the model, suggesting that these proteins out of 45 proteins analyzed are the top representatives of several independent pathways/processes that contribute to skin TEWL. SERPINB3 (Fig. 5B), GSN (Fig. 5C) and KRT77 (Fig. 5D) were also identified as top two positive and one negative correlates of skin TEWL AUC.
Figure 5.
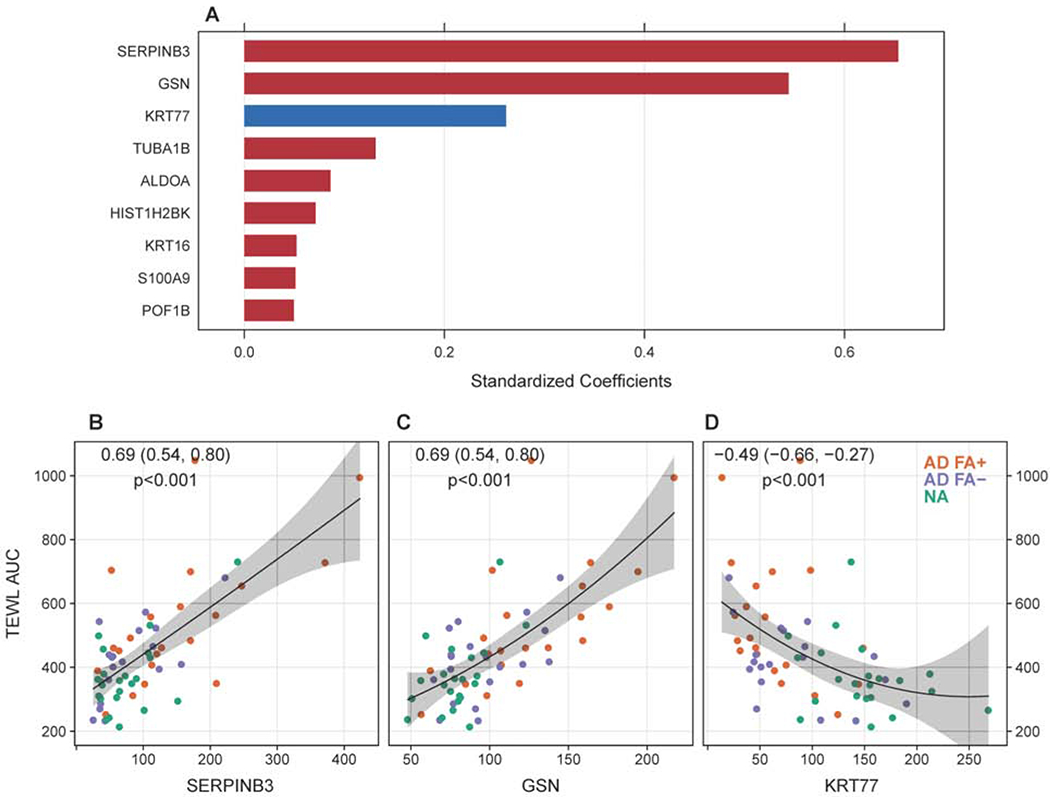
Summary of the top PC1 proteins with the greatest contribution to skin TEWL. (A) Top nine PC1 proteins identified by the classification model (LASSO generalized linear model, with 5-fold cross-validation) based on their contribution to skin TEWL. A univariate correlation analysis between SERPINB3 (B), GSN (C) and KRT77 (D) expression in skin tape strip samples and skin TEWL in the study cohort.
We have previously determined a significant inverse correlation between the levels of FLG breakdown products pyroglutamic acid (PCA) and urocanic acid (UCA) in non-lesional skin STS of this study cohort and skin TEWL17. SERPINB3 and GSN had a significant inverse correlation with the levels of PCA (Suppl. Fig. 4A, B) and UCA (Suppl. Fig. 4D, E) in non-lesional skin of the study subjects, while KRT77 had a significant positive correlation with PCA and UCA levels in the non-lesional skin (Suppl. Fig. 4C, F).
Microbial dysbiosis and Staphylococcus aureus colonization have been implicated in regulation of skin barrier function regulation in AD skin36. Skin microbiome analysis of this study cohort has been recently reported17. We, therefore, examined the relationship between S. hominis and S. aureus expression in the skin of the study subjects and PC1 protein expression in STS (Suppl. Fig. 5). A significant positive correlation between S. aureus skin levels and PC1 protein expression was found (Suppl. Fig. 5B).
Validation of non-lesional STS PC1 in an independent patient cohort.
Our data suggest that despite the normal appearance of AD non-lesional skin in the pediatric cohort, structural and pro-inflammatory changes were already present in the skin of these patients. In particular, the most significant changes in STS protein composition were observed in AD FA+ STS samples.
In the next set of experiments, we examined whether the identified differences in PC1 protein expression in non-lesional skin STS proteomic analysis could be confirmed in an independent cohort of AD patients. We studied the non-lesional STS proteome from a separate cohort of adult AD patients enrolled in the ADRN. STS samples were collected from non-lesional skin of 28 adults with AD and 13 NA controls (see Table 4 for study subject’s characteristics). STS proteomic analysis was done and the expression of 45 PC1 proteins previously identified in the pediatric cohort STS analysis was analyzed from these samples. In this cohort, the clinical history of allergic reactions to peanuts was collected and we were able to subgroup AD patients into AD FA+ or AD FA− based on the clinical history of allergic reactions to peanuts. Using these definitions, we have found that the PC1 proteins previously identified in the pediatric cohort were also differentially expressed in non-lesional skin STS samples from AD adults with a history of clinical reactions to peanuts vs. adults with AD and no history of peanut allergy vs. NA controls. Importantly, the cumulative expression of the previously identified PC1 proteins was the highest in adults with AD and peanut allergy and significantly different from adults with AD without peanut allergy and NA controls (Fig. 6A). As in the pediatric cohort, the expression of STS PC1 proteins was intermediate in adults with AD without peanut allergy, and was the lowest in NA adults (Fig. 6A).
Table 4.
Clinical characteristics of the adult validation cohort used in STS proteomic analysis.
| Characteristic | AD FA+* | AD FA− | NA | p value |
|---|---|---|---|---|
|
Age (years) Mean (SD) |
33.0 (9.5) |
36.9 (11.9) |
41.0 (8.4) |
0.16 |
|
Gender, n(%) Female Male |
9 (75.0) 3 (25.0) |
11 (61.1) 7 (39.9) |
10 (76.9) 3 (24.1) |
0.57 |
|
Race, n(%) Caucasian Black or African American |
10 (83.3) 2 (16.6) |
16 (88.9) 2 (11.1) |
10 (76.9) 3 (24.1) |
0.67 |
|
EASI score Mean (SD) |
17.3 (10.8) |
16.4 (13.9) |
0.85 |
|
|
Total serum IgE, kU/L Mean (SD) |
1368 (1332) |
2475 (7373) |
35.6 (40.5) |
<0.0001 |
|
Total IgE to peanut, kUA/L Mean (SD) |
9.4 (14.9) |
4.4 (16.4) |
0.11 (0.03) |
<0.0001 |
Patients were defined as AD FA+ based on the history of clinical reactions to peanut.
Figure 6.
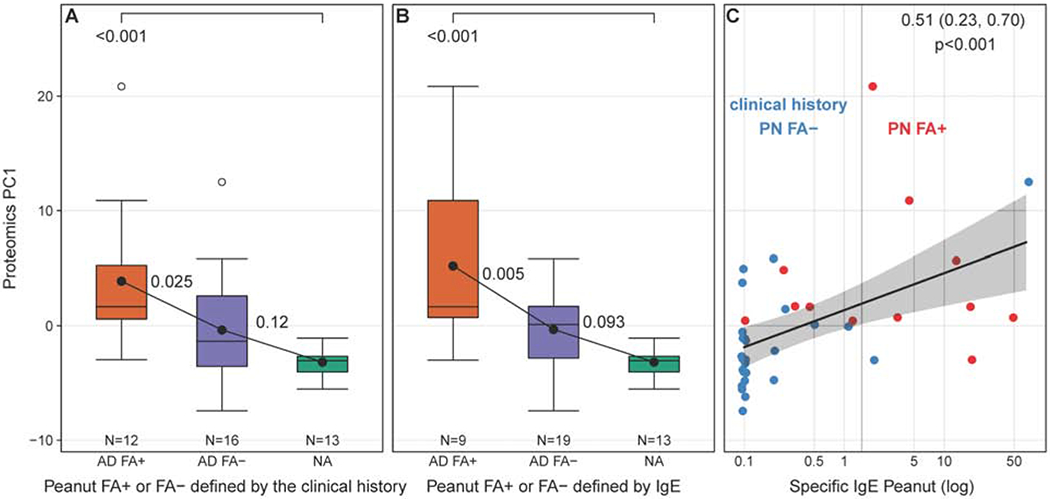
Validation of the PC1 results in STS analysis of non-lesional skin samples from the independent cohort of adults with AD with and without FA to peanut and NA controls. (A) The cumulative expression of PC1 proteins defined in the pediatric STS analysis was found to be differentially expressed in the proteomic analysis of STS from adults with AD and a history of clinical reactions to peanut, as compared to AD adults with no clinical history of allergic reaction to peanut and NA controls. (B) Expression of PC1 proteins was the highest in AD adults with the positive total IgE to peanut defined by the cutoff above 2kUA/L. (C) A significant positive correlation between total serum IgE levels to peanut and STS PC1 expresion was observed.
Separately, serum analysis for total IgE to peanuts was done and adult study participants were sub grouped into AD patients with peanut allergy or AD patients with no peanut allergy using 2 kUA/L of total serum IgE to peanut as a cutoff. Again, the expression of STS PC1 proteins in adult AD patients with serum IgE levels to peanut above 2 kUA/L was significantly higher than in adult AD patients with serum IgE to peanut below this cutoff (Fig. 6B). Lastly, a positive correlation between total serum IgE levels to peanut and STS PC1 expression in this adult independent cohort was found (Fig. 6C).
Discussion
In this study, STS proteomics analysis identified abnormalities in skin barrier function of AD children, and adults with and without FA that can be noninvasively assessed using skin tape strips. We report several novel findings. First, in this study we took a novel bioinformatics approach to STS proteomic data analysis, and instead of using a predetermined list of skin biomarkers, we applied an unbiased principal component analysis to the list of 150 proteins that was found to be expressed by all of the non-lesional skin samples. Through this approach we determined a group of 45 proteins, Principal Component 1 (PC1), that depicts major differences between the three study groups, i.e. NA controls, AD FA− and AD FA+ subjects (30% of the variance explained). Importantly, the expression of this group of proteins in the skin samples was found to be a significant positive correlate of skin TEWL in these patients. Second, the proteins that make up PC1 provide novel insights into biology of AD and FA. All of the proteins that make up PC1 were found to significantly correlate with skin TEWL. Through a multivariant analysis we determined the top nine contributors to TEWL, with SERPINB3, gelsolin (GSN) and KRT77 identified to have the greatest impact on skin TEWL. Third, according to the data presented by our group recently, TEWL alone was not a sufficient parameter to differentiate non-lesional skin of AD FA− patients from NA controls17. In this manuscript, we observed that skin expression of PC1 proteins can distinguish non-lesional skin samples between all three study groups. Lastly and noteworthy, the findings of the study were confirmed and validated in an independent cohort of adults with AD with the history of clinical reactions to peanut. The expression of PC1 proteins was the highest in the skin of AD patients with the history of FA to peanuts, intermediate in AD patients and the lowest in NA controls. This is an important and novel finding, as it suggests that AD patients with FA are a unique endotype which persists into adulthood. Importantly, the cumulative expression of PC1 proteins in all AD study patients had a positive correlation with serum IgE and sensitization to food and aeroallergens, supporting the notion that compromised skin barrier could contribute to the allergen penetration and allergic sensitization37.
Collected data suggest that TEWL abnormalities in the skin stem from disorganization in expression of skin KRT intermediate filaments with increased expression of KRTs associated with epidermal proliferation, wound healing and oxidative stress responses (KRT16, KRT17)29,31, increased detection of immature KRTs that normally are associated with basal skin layer (KRT4, KRT1529, 30), with concomitant decrease in KRTs that are part of epidermal maturation and differentiation (KRT10, KRT7829, 30). KRT intermediate filaments protect the epidermis against mechanical force, support strong adhesion, help barrier formation, and regulate growth. Mice lacking all type I or type II KRTs display severe barrier defects and fragile skin, leading to perinatal mortality, if fully penetrant38. Comparative proteomics of cornified envelopes from these mice collected prenatally demonstrates that absence of KRT intermediate filaments causes dysregulation of many cornified envelop constituents38, with some of these features also observed in our proteomic analysis, including the increase in desmosomal proteins desmoplakin (DSP) and junction plakoglobin (JUP), deregulated expression of protease inhibitors (SERPINs), increased expression of epidermal alarmins (annexin A2 (ANXA2), HSPB1, S100 proteins, galectins), activation of targets that are involved in antioxidant defense in the skin, like small proline rich protein 2D (SPRR2D)39. Of note, the absence of KRT intermediate filaments puts excessive stress on mitochondria to supply energy for increased mechanical stress resistance and cell proliferation38. Notably, in our study among PC1 proteins, an increased expression of glycolysis enzymes (ALDOA, ENO1, PKM, LDHA) and deregulated expression of antioxidant defense proteins (GSTP1, CAT, PRDX1) was found. We propose that observed changes in expression of glycolytic enzymes, antioxidant defense proteins, proteases and inflammatory mediators in AD STS samples is likely a compensatory mechanism to counterbalance initial changes and disorganization of KRT intermediate filaments under the influence of type 2 cytokine environment in AD skin.
We report here that all of the 45 proteins that make up PC1 had a significant correlation with TEWL. SERPINB3 expression in the skin had the highest positive correlation with TEWL, while KRT10 expression in skin samples had a highest negative correlation with TEWL in our data set. The expression of both proteins has been previously shown to be regulated by IL-4/IL-1340, 41, as SERPINB3 has been found to be directly induced by IL-4/IL-13 in bronchial epithelial cells40, while KRT10 was reported to be inhibited by IL-4 in keratinocytes41, suggesting the involvement of type 2 inflammation in AD skin in regulation of skin barrier function5. In addition, a multivariant analysis identified nine proteins expressed in PC1 as TEWL predictors, with SERPINB3, GSN and KRT77 identified as top independent predictors of TEWL. The data acquired suggests that type 2 cytokines are only one of several potential pathways involved in regulation of skin TEWL. We propose that STS protein analysis can provide broad knowledge about biochemical pathways involved in TEWL and skin barrier, including (but not limited to) an alternative and independent, minimally invasive skin targeted approach for the readout of type 2 inflammation in AD skin.
Serine proteases are critical for epidermal barrier homeostasis, and their aberrant expression and/or activity is associated with chronic skin diseases. Elevated levels of the serine protease inhibitors SERPINB3 and SERPINB4 have been found in AD skin42. In experimental mouse model allergen exposure induced SERPINB3 expression in the skin, along with increased TEWL, epidermal thickness, and skin inflammation, all of which were attenuated in the absence of SERPINB3 43. Attenuated TEWL correlated with decreased expression of the pro-inflammatory marker S100A8. RNA-seq analysis following allergen exposure identified a network of pro-inflammatory genes induced in wild-type mice that was absent in SERPINB3 -null mice, suggesting that that SERPINB3 contributes to early inflammatory responses in the skin following allergen exposure, disrupting skin barrier. The involvement of other factors, including microbes, environmental pollutants, detergents, etc., in regulation of SERPINB3 expression in the skin is unexplored.
KRT10 deficiency was previously shown to result in profound changes in permeability barrier function44. Baseline TEWL in skin with normal appearance of newborn homozygous KRT10 deficient mice was increased 8-fold compared with wild type controls44. Adult heterozygotes exhibited delayed barrier repair after experimental barrier disruption. SC hydration was reduced in homozygous and heterozygous mice44. Skin fragility in KRT10 knockout mice is suggested to be a consequence of two complementing mechanisms, namely, a decrease of normal KRT1/KRT10 filaments and an increase in KRT6/KRT16 with a poor filament-forming capacity45. KRT10 knockout mice displayed hyperproliferation of basal keratinocytes, with the induction of c-Myc, cyclin D1, and 14-3-3σ (stratifin, SFN)46. The study suggested a direct involvement of K10 in cell cycle control46. Of note, in our study KRT16 and SFN were also identified as PC1 proteins, were significantly increased in AD skin and had a significant positive correlation to TEWL.
Of interest, KRT77 is highly expressed in luminal duct cells of eccrine sweat glands in the skin, including intraepidermal duct region33, but only weakly expressed by suprabasal epithelia32. An impaired sweating ability is observed in patients with AD47–49. The mechanisms of decreased sweat production in AD include obstruction of sweat pores by keratin plugs, sweat production and secretion abnormalities from sweat glands; and sweat leakage into surrounding tissues47. Decreased sweating exacerbates the symptoms of dermatitis, as it prolongs heat retention, skin dryness, and increased susceptibility to infection47. The decreased levels of KRT77 in STS of AD patients identified in our study and significant negative correlation with TEWL may suggest previously unrecognized decreased eccrine sweat glands density in AD skin. Of interest, reduced sweat production, sweat glands obstruction and abnormalities in sweat gland morphology have been found in the footpads of flg mutant mice as compared to wild type mice50, also suggesting a potential regulation of sweat glands by flg. Our data demonstrates a significant positive correlation between KRT77 levels in STS and levels of FLG in the same skin site as determined by FLG breakdown products PCA and UCA.
The role of GSN in TEWL regulation has not been previously reported. GSN acts as a key regulator of actin filament assembly and disassembly, binds to the ends of actin filaments and prevents monomer exchange (end-blocking or capping)51. Moreover, GSN has also been suggested to exert multifunctional roles inside the cell, functioning as transcription or apoptosis regulator51. GSN and other actin associated proteins are reported to be less abundant in suprabasal cells than in basal cells in the epidermis52. On the contrary, increased GSN expression in AD STS was found in our study, suggesting abnormal epidermal differentiation in AD skin. Loss of integrins occurs during keratinocyte differentiation and changes in actin cytoskeleton through the action of cytoplasmic proteins that control actin assembly have been proposed to be involved in signaling between extracellular matrix bound integrins and cell nuclei53, 54. Further studies are required to examine the role of altered GSN-regulated actin assembly/integrin signal transduction in AD skin in epidermal differentiation and skin barrier homeostasis.
We also determined a positive correlation between S. aureus expression in the skin of the study subjects and PC1 protein expression in STS. The data suggests that S. aureus can be a contributing factor involved in the regulation of the PC1 proteins expression in the skin. It is possible that observed association is a consequence of type 2 inflammation in AD skin, which supports microbial dysbiosis and S. aureus colonization5. The direct role of S. aureus in AD development remains controversial55.
TEWL as a research tool enables noninvasive measurement of skin barrier function. TEWL elevation may precede clinical manifestation of eczema, suggesting that TEWL measurement may be useful in guiding AD prevention strategies. Recent studies have demonstrated that TEWL measured during the first days of life can predict the development of AD in infancy, independent of FLG status56,57. These findings suggest that TEWL could potentially be used to identify neonates at increased risk of AD and help guide prevention strategies. However, the variability and insensitivity of TEWL measurements contributes to the lack of precision for individual subjects. Our data propose that STS proteomic assessment can also provide a minimally invasive assessment of the skin in critical disease groups and can provide insights into the structural and inflammatory changes that are ongoing in atopic skin.
Importantly, we were able to validate the findings of a unique proteomic profile of pediatric AD patients with FA in an independent cohort of adults with AD with the history of clinical reactions to peanut and positive total serum IgE levels to peanut. As part of patients’ registry, the data on clinical history of allergic reactions to foods, including peanuts was gathered and serum IgE levels to peanut, egg and milk were collected. STS PC1 protein expression in this cohort was examined in two ways, first, by subgrouping patients based on prior history of clinical reactions to peanuts, second, based on total serum IgE levels to peanut. Both strategies for AD patient stratification determined that skin expression of PC1 proteins previously identified in children with AD and FA to peanut was also significantly increased in AD adults with allergic sensitization to peanuts. This is an important finding, as it suggests that AD patients with FA are a unique endotype as a unique proteomic profile of this group can be seen in STS proteomic analysis and it is similar in children and adults with AD and FA. We have previously shown that alterations in lipids, gene expression and skin protein levels in non-lesional skin, detected by STS, individually distinguish AD FA+ from AD FA− patients17. The comprehensive STS proteomic analysis conducted in this study provides further evidence that non-lesional skin distinguishes AD FA+ as a unique phenotype.
Identification of patients who are prone to development of AD, FA, and environmental allergy prior to clinical manifestation of these diseases in early childhood is of tremendous value as this is the group that is at high risk to undergo the atopic march58. Early prognosis for patients at risk may direct these patients for specific treatment strategies and early intervention59. In the current study, all non-lesional proteomic skin analyses were done in patients with existing AD, and, therefore, the validation of this established set of proteins for TEWL assessment and AD and FA prediction in subjects prior to the establishment of clinical features of AD is warranted in future birth cohort studies.
In conclusion, in this study we identified STS PC1 proteins that demonstrated a significant positive correlation with TEWL and allergic sensitization, suggesting that skin expression of these proteins is involved in the determination of skin structures that may be associated with skin barrier function. We propose that altered expression of these skin proteins may support penetration of allergens through a damaged barrier and also may support consequent type 2 inflammatory response that ensures after penetration, suggesting potential new targets for future early interventions to prevent AD and FA.
Supplementary Material
Clinical Implications:
Skin tape proteomics analysis identified abnormalities in barrier function of atopic dermatitis children and adults with and without food allergy that can be noninvasively assessed using skin tape strips.
Acknowledgments:
The authors would like to thank the nursing staff (Susan Leung, Caroline Bronchick, Shirley Palombi) and coordinators (Marco Ramirez-Gama and Shannon Garcia) of the Clinical Translational Research Center at National Jewish Health for their work on patient recruitment, patient clinical characterization, and STS sample collection. The authors also thank Ms. Brittany Richers for her assistance with the preparation of STS protein extracts for LC-MS analysis. This work was supported by the NIH/NIAID Atopic Dermatitis Research Network U19AI117673 and UM2AI117870, the Edelstein Family Chair of Pediatric Allergy and UL1 TR-002535 from the National Center for Research Resources (NCRR/NIH).
Funding sources: This work was supported by NIH/NIAID Atopic Dermatitis Research Network U19AI117673 and UM2AI117870, the Edelstein Family Chair of Pediatric Allergy and UL1 TR-002535 from the National Center for Research Resources (NCRR/NIH).
Abbreviations
- AD
Atopic dermatitis
- ADRN
Atopic Dermatitis Research Network
- ALDOA
aldolase, fructose-bisphosphate A
- ANOVA
analysis of variance
- ANXA2
annexin A2
- ASAH1
N-acylsphingosine aminohidrolase 1, acid ceramidase
- AUC
area under the curve
- CAT
catalase
- CSTA
cystatin A
- DSP
desmoplakin
- EASI
Eczema Area and Severity Index
- ENO1
enolase 1
- FA
Food allergy
- FDR
false discovery rate
- FLG
filaggrin
- GSN
gelsolin
- GSTP1
glutathione S-transferase Pi 1
- HIST1H2BK
histone H2B type 1-K
- HSPB1
heat shock protein family B (small) member 1
- IL
interleukin
- JUP
junction plakoglobin
- KRT
keratin
- LASSO
Least Absolute Shrinkage and Selection Operator
- LC-MS
liquid chromatography mass spectrometry
- LDHA
lactic dehydrogenase
- LGALS
lectin galactoside-binding, soluble; galectin
- NA
nonatopic
- NESS
Nottingham Eczema Severity Score
- PC1
principal component 1
- PCA
pyroglutamic acid
- POF1B
premature ovarian failure protein 1B
- PRDX
peroxiredoxin
- PKM
pyruvate kinase M1/2
- S100A
S100 calcium binding protein A
- SC
Stratum corneum
- SCORAD
SCORing Atopic Dermatitis
- SERPINB
serine proteinase inhibitor glade B
- SFN
stratifin
- SPRR2D
small proline rich protein 2D
- STS
skin tape strip
- TEWL
transepidermal water loss
- TSLP
thymic stromal lymphopoietin
- TUBA1B
tubulin alpha 1b
- UCA
urocanic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fluhr JW, Feingold KR, Elias PM. Transepidermal water loss reflects permeability barrier status: validation in human and rodent in vivo and ex vivo models. Exp Dermatol 2006; 15:483–92. [DOI] [PubMed] [Google Scholar]
- 2.Alexander H, Brown S, Danby S, Flohr C. Research Techniques Made Simple: Transepidermal Water Loss Measurement as a Research Tool. J Invest Dermatol 2018; 138:2295–300 e1. [DOI] [PubMed] [Google Scholar]
- 3.Elias PM. Skin barrier function. Curr Allergy Asthma Rep 2008; 8:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flohr C, England K, Radulovic S, McLean WH, Campbel LE, Barker J, et al. Filaggrin loss-of-function mutations are associated with early-onset eczema, eczema severity and transepidermal water loss at 3 months of age. Br J Dermatol 2010; 163:1333–6. [DOI] [PubMed] [Google Scholar]
- 5.Goleva E, Berdyshev E, Leung DY. Epithelial barrier repair and prevention of allergy. J Clin Invest 2019; 129:1463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson EL, Villarreal M, Jepson B, Rafaels N, David G, Hanifin J, et al. Patients with Atopic Dermatitis Colonized with Staphylococcus aureus Have a Distinct Phenotype and Endotype. J Invest Dermatol 2018; 138:2224–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamlin SL, Kao J, Frieden IJ, Sheu MY, Fowler AJ, Fluhr JW, et al. Ceramide-dominant barrier repair lipids alleviate childhood atopic dermatitis: changes in barrier function provide a sensitive indicator of disease activity. J Am Acad Dermatol 2002; 47:198–208. [DOI] [PubMed] [Google Scholar]
- 8.Flohr C, Perkin M, Logan K, Marrs T, Radulovic S, Campbell LE, et al. Atopic dermatitis and disease severity are the main risk factors for food sensitization in exclusively breastfed infants. J Invest Dermatol 2014; 134:345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drislane C, Irvine AD. The role of filaggrin in atopic dermatitis and allergic disease. Ann Allergy Asthma Immunol 2020; 124:36–43. [DOI] [PubMed] [Google Scholar]
- 10.Howell MD, Kim BE, Gao P, Grant AV, Boguniewicz M, DeBenedetto A, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol 2009; 124:R7–R12. [DOI] [PubMed] [Google Scholar]
- 11.Kezic S, O’Regan GM, Yau N, Sandilands A, Chen H, Campbell LE, et al. Levels of filaggrin degradation products are influenced by both filaggrin genotype and atopic dermatitis severity. Allergy 2011; 66:934–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, Villarreal M, Stewart S, Choi J, Ganguli-Indra G, Babineau DC, et al. Altered composition of epidermal lipids correlates with Staphylococcus aureus colonization status in atopic dermatitis. Br J Dermatol 2017; 177:e125–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole C, Kroboth K, Schurch NJ, Sandilands A, Sherstnev A, O’Regan GM, et al. Filaggrin-stratified transcriptomic analysis of pediatric skin identifies mechanistic pathways in patients with atopic dermatitis. J Allergy Clin Immunol 2014; 134:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berdyshev E, Goleva E, Bronova I, Dyjack N, Rios C, Jung J, et al. Lipid abnormalities in atopic skin are driven by type 2 cytokines. JCI Insight 2018; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broccardo CJ, Mahaffey S, Schwarz J, Wruck L, David G, Schlievert PM, et al. Comparative proteomic profiling of patients with atopic dermatitis based on history of eczema herpeticum infection and Staphylococcus aureus colonization. J Allergy Clin Immunol 2011; 127:186–93, 93, e1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyjack N, Goleva E, Rios C, Kim BE, Bin L, Taylor P, et al. Minimally invasive skin tape strip RNA sequencing identifies novel characteristics of the type 2-high atopic dermatitis disease endotype. J Allergy Clin Immunol 2018; 141:1298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung DYM, Calatroni A, Zaramela LS, LeBeau PK, Dyjack N, Brar K, et al. The nonlesional skin surface distinguishes atopic dermatitis with food allergy as a unique endotype. Sci Transl Med 2019; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck LA, Boguniewicz M, Hata T, Schneider LC, Hanifin J, Gallo R, et al. Phenotype of atopic dermatitis subjects with a history of eczema herpeticum. J Allergy Clin Immunol 2009; 124:260–9, 9, e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts G, Lack G. Diagnosing peanut allergy with skin prick and specific IgE testing. J Allergy Clin Immunol 2005; 115:1291–6. [DOI] [PubMed] [Google Scholar]
- 20.Severity scoring of atopic dermatitis: the SCORAD index. Consensus Report of the European Task Force on Atopic Dermatitis. Dermatology 1993; 186:23–31. [DOI] [PubMed] [Google Scholar]
- 21.Leshem YA, Hajar T, Hanifin JM, Simpson EL. What the Eczema Area and Severity Index score tells us about the severity of atopic dermatitis: an interpretability study. Br J Dermatol 2015; 172:1353–7. [DOI] [PubMed] [Google Scholar]
- 22.Emerson RM, Charman CR, Williams HC. The Nottingham Eczema Severity Score: preliminary refinement of the Rajka and Langeland grading. Br J Dermatol 2000; 142:288–97. [DOI] [PubMed] [Google Scholar]
- 23.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; . Vienna, Austria: 2018. [Google Scholar]
- 24.Sarkar D Lattice: Multivariate Data Visualization with R. New York: Springer 2008. [Google Scholar]
- 25.Lenth R emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.4. 2019. [Google Scholar]
- 26.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsson J eulerr: Area-Proportional Euler and Venn Diagrams with Ellipses. R package version 6.0.0. 2019. [Google Scholar]
- 28.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010; 33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 29.Haines RL, Lane EB. Keratins and disease at a glance. J Cell Sci 2012; 125:3923–8. [DOI] [PubMed] [Google Scholar]
- 30.Bragulla HH, Homberger DG. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J Anat 2009; 214:516–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paladini RD, Coulombe PA. Directed expression of keratin 16 to the progenitor basal cells of transgenic mouse skin delays skin maturation. J Cell Biol 1998; 142:1035–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edqvist PH, Fagerberg L, Hallstrom BM, Danielsson A, Edlund K, Uhlen M, et al. Expression of human skin-specific genes defined by transcriptomics and antibody-based profiling. J Histochem Cytochem 2015; 63:129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langbein L, Rogers MA, Praetzel S, Cribier B, Peltre B, Gassler N, et al. Characterization of a novel human type II epithelial keratin K1b, specifically expressed in eccrine sweat glands. J Invest Dermatol 2005; 125:428–44. [DOI] [PubMed] [Google Scholar]
- 34.Niehaus JZ, Good M, Jackson LE, Ozolek JA, Silverman GA, Luke CJ. Human SERPINB12 Is an Abundant Intracellular Serpin Expressed in Most Surface and Glandular Epithelia. J Histochem Cytochem 2015; 63:854–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niehaus JZ, Miedel MT, Good M, Wyatt AN, Pak SC, Silverman GA, et al. SERPINB12 Is a Slow-Binding Inhibitor of Granzyme A and Hepsin. Biochemistry 2015; 54:6756–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakatsuji T, Gallo RL. The role of the skin microbiome in atopic dermatitis. Ann Allergy Asthma Immunol 2019; 122:263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W, Freeland DMH, Nadeau KC. Food allergy: immune mechanisms, diagnosis and immunotherapy. Nat Rev Immunol 2016; 16:751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar V, Bouameur JE, Bar J, Rice RH, Hornig-Do HT, Roop DR, et al. A keratin scaffold regulates epidermal barrier formation, mitochondrial lipid composition, and activity. J Cell Biol 2015; 211:1057–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schafer M, Farwanah H, Willrodt AH, Huebner AJ, Sandhoff K, Roop D, et al. Nrf2 links epidermal barrier function with antioxidant defense. EMBO Mol Med 2012; 4:364–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray R, Choi M, Zhang Z, Silverman GA, Askew D, Mukherjee AB. Uteroglobin suppresses SCCA gene expression associated with allergic asthma. J Biol Chem 2005; 280:9761–4. [DOI] [PubMed] [Google Scholar]
- 41.Omori-Miyake M, Yamashita M, Tsunemi Y, Kawashima M, Yagi J. In vitro assessment of IL-4- or IL-13-mediated changes in the structural components of keratinocytes in mice and humans. J Invest Dermatol 2014; 134:1342–50. [DOI] [PubMed] [Google Scholar]
- 42.Kawashima H, Nishimata S, Kashiwagi Y, Numabe H, Sasamoto M, Iwatsubo H, et al. Squamous cell carcinoma-related antigen in children with atopic dermatitis. Pediatr Int 2000; 42:448–50. [DOI] [PubMed] [Google Scholar]
- 43.Sivaprasad U, Kinker KG, Ericksen MB, Lindsey M, Gibson AM, Bass SA, et al. SERPINB3/B4 contributes to early inflammation and barrier dysfunction in an experimental murine model of atopic dermatitis. J Invest Dermatol 2015; 135:160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen JM, Schutze S, Neumann C, Proksch E. Impaired cutaneous permeability barrier function, skin hydration, and sphingomyelinase activity in keratin 10 deficient mice. J Invest Dermatol 2000; 115:708–13. [DOI] [PubMed] [Google Scholar]
- 45.Reichelt J, Bauer C, Porter R, Lane E, Magin V. Out of balance: consequences of a partial keratin 10 knockout. J Cell Sci 1997; 110 ( Pt 18):2175–86. [DOI] [PubMed] [Google Scholar]
- 46.Reichelt J, Magin TM. Hyperproliferation, induction of c-Myc and 14-3-3sigma, but no cell fragility in keratin-10-null mice. J Cell Sci 2002; 115:2639–50. [DOI] [PubMed] [Google Scholar]
- 47.Murota H, Yamaga K, Ono E, Katayama I. Sweat in the pathogenesis of atopic dermatitis. Allergol Int 2018; 67:455–9. [DOI] [PubMed] [Google Scholar]
- 48.Murota H, Matsui S, Ono E, Kijima A, Kikuta J, Ishii M, et al. Sweat, the driving force behind normal skin: an emerging perspective on functional biology and regulatory mechanisms. J Dermatol Sci 2015; 77:3–10. [DOI] [PubMed] [Google Scholar]
- 49.Hendricks AJ, Vaughn AR, Clark AK, Yosipovitch G, Shi VY. Sweat mechanisms and dysfunctions in atopic dermatitis. J Dermatol Sci 2018; 89:105–11. [DOI] [PubMed] [Google Scholar]
- 50.Rerknimitr P, Tanizaki H, Yamamoto Y, Amano W, Nakajima S, Nakashima C, et al. Decreased Filaggrin Level May Lead to Sweat Duct Obstruction in Filaggrin Mutant Mice. J Invest Dermatol 2017; 137:248–51. [DOI] [PubMed] [Google Scholar]
- 51.Feldt J, Schicht M, Garreis F, Welss J, Schneider UW, Paulsen F. Structure, regulation and related diseases of the actin-binding protein gelsolin. Expert Rev Mol Med 2019; 20:e7. [DOI] [PubMed] [Google Scholar]
- 52.Kubler MD, Jordan PW, O’Neill CH, Watt FM. Changes in the abundance and distribution of actin and associated proteins during terminal differentiation of human epidermal keratinocytes. J Cell Sci 1991; 100 (Pt 1): 153–65. [DOI] [PubMed] [Google Scholar]
- 53.Kenny FN, Connelly JT. Integrin-mediated adhesion and mechano-sensing in cutaneous wound healing. Cell Tissue Res 2015; 360:571–82. [DOI] [PubMed] [Google Scholar]
- 54.Noethel B, Ramms L, Dreissen G, Hoffmann M, Springer R, Rubsam M, et al. Transition of responsive mechanosensitive elements from focal adhesions to adherens junctions on epithelial differentiation. Mol Biol Cell 2018; 29:2317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams MR, Gallo RL. Evidence that Human Skin Microbiome Dysbiosis Promotes Atopic Dermatitis. J Invest Dermatol 2017; 137:2460–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelleher MM, Dunn-Galvin A, Gray C, Murray DM, Kiely M, Kenny L, et al. Skin barrier impairment at birth predicts food allergy at 2 years of age. J Allergy Clin Immunol 2016; 137:1111–6 e8. [DOI] [PubMed] [Google Scholar]
- 57.Horimukai K, Morita K, Narita M, Kondo M, Kabashima S, Inoue E, et al. Transepidermal water loss measurement during infancy can predict the subsequent development of atopic dermatitis regardless of filaggrin mutations. Allergol Int 2016; 65:103–8. [DOI] [PubMed] [Google Scholar]
- 58.Davidson WF, Leung DYM, Beck LA, Berin CM, Boguniewicz M, Busse WW, et al. Report from the National Institute of Allergy and Infectious Diseases workshop on “Atopic dermatitis and the atopic march: Mechanisms and interventions”. J Allergy Clin Immunol 2019; 143:894–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lowe AJ, Leung DYM, Tang MLK, Su JC, Allen KJ. The skin as a target for prevention of the atopic march. Ann Allergy Asthma Immunol 2018; 120:145–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


