Abstract
Long noncoding RNAs (lncRNAs) are crucial in many cellular processes, yet relative few have been shown to regulate human cardiomyocyte differentiation. Here, we demonstrate an essential role of GATA6 antisense RNA 1 (GATA6-AS1) in cardiomyocyte differentiation from human pluripotent stem cells (hPSCs). GATA6-AS1 is adjacent to cardiac transcription factor GATA6. We found that GATA6-AS1 was nuclear-localized and transiently upregulated along with GATA6 during the early stage of cardiomyocyte differentiation. Knockdown of GATA6-AS1 did not affect undifferentiated cell pluripotency but inhibited cardiomyocyte differentiation, as indicated by no or few beating cardiomyocytes and reduced expression of cardiomyocyte-specific proteins. Upon cardiac induction, knockdown of GATA6-AS1 decreased GATA6 expression, altered Wnt-signaling gene expression and reduced mesoderm development. Further characterization of the intergenic region between genomic regions of GATA6-AS1 and GATA6 indicated that the expression of GATA6-AS1 and GATA6 were regulated by a bidirectional promoter within the intergenic region. Consistently, GATA6-AS1 and GATA6 were co-expressed in several human tissues including the heart, similar to the mirror expression pattern of GATA6-AS1 and GATA6 during cardiomyocyte differentiation. Overall, these findings reveal a previously unrecognized and functional role of lncRNA GATA6-AS1 in controlling human cardiomyocyte differentiation.
Keywords: cardiomyocyte, differentiation, hiPSC, hPSC, lncRNA, RNA-seq
INTRODUCTION
Human pluripotent stem cells (hPSCs) can give rise to spontaneously beating cardiomyocytes in vitro, a process that mirrors cardiogenesis. Similar to in vivo cardiogenesis, in vitro cardiomyocyte differentiation is a dynamic process, which is tightly regulated by signaling proteins and transcription factors (1). The improved understanding of molecular mechanisms controlling the differentiation of hPSCs to cardiomyocytes has made it possible to differentiate with encouraging efficiency, which is potentially a promising approach for regenerative therapy, disease modeling and other applications (2–5). Moreover, due to limited access to human embryonic tissues, studies on lineage commitment of hPSCs into cardiomyocytes present an alternative approach to study the molecular regulation of human-specific cardiogenesis events. In this regard, hPSCs provide a powerful and attractive tool to study the role of long non-coding RNAs (lncRNAs) in differentiation and development. Given that lncRNAs are less conserved among species compared with protein-coding genes, lncRNAs may have a species-specific role in many biological processes.
LncRNAs are transcripts longer than 200 nucleotides, which constitute a large part of the human genome with limited coding potential (6). LncRNAs are involved in various cellular processes such as genomic distribution, genomic imprinting, expression pattern, subcellular localization, chromatin modification, protein transport, and RNA alternative splicing (7–10). For example, co-expression of tissue-specific lncRNAs and protein-coding genes suggests a novel role of lncRNA-mediated gene regulation. In the brain, transient expression profiles of lncRNAs have been identified and functionally characterized for their involvement in the regulation of nearby protein-coding genes of neurological importance (11, 12). Similarly, a subset of lncRNAs in the lung play an important role in the regulation of adjacent transcription factors via Wnt/β-catenin signaling for lung endoderm morphogenesis (13). Moreover, lncRNA expression regulates many developmental genes during differentiation of human and mouse embryonic stem cells (hESCs/mESCs) (14, 15). However, despite these advancements, relatively few lncRNAs have been identified for their roles in cardiovascular development (16, 17).
Here, we report a functional role for a lncRNA, GATA6 antisense RNA 1 (GATA6-AS1), in hPSC cardiomyocyte differentiation. GATA family genes are known for their involvement in embryonic development and differentiation of germ layers. In particular, GATA6 is a direct target of cardiac transcription factor NKX2–5 during cardiogenesis (18). GATA6 expression is required for the cardiovascular development, and GATA6 mutations are associated with congenital heart diseases (19). Since GATA6-AS1 resides adjacent to GATA6 on the genome and since lncRNAs are known to have similar temporal expression to as well as regulatory roles on the neighboring genes, we first compared the expression patterns of GATA6 and GATA6-AS1 during hiPSC cardiomyocyte differentiation. As we found out that they displayed a similar temporal expression profile, we further asked whether GATA6-AS1 could affect the expression of GATA6 and thereby hPSC cardiomyocyte differentiation.
MATERIALS AND METHODS
Cell culture and cardiomyocyte differentiation
Undifferentiated IMR90 human induced pluripotent stem cells (hiPSCs) (WiCell, Madison, WI) (20), NKX2–5-eGFP (enhanced green fluorescent protein; eGFP) hESCs (21) and SCVI 273 hiPSCs (Stanford Cardiovascular Institute Biobank) were maintained in 6-well culture plates in a feeder-free condition (22) in mouse embryonic fibroblast-conditioned medium (MEF-CM) supplemented with basic fibroblast growth factor (bFGF) (8 ng/mL) or mTeSR1 (StemCell Technologies). When colonies occupied approximately 75–80% of the well surface area, cells were passaged to set up for cardiac differentiation. Cells were seeded at the density of 2×105 cells/cm2 into 12-well plates coated with Matrigel (BD Biosciences, San Jose, CA) and fed daily until compact colonies covered the wells. For growth factor-guided differentiation (3, 23), cells were treated with 100 ng/mL recombinant human activin A (R&D Systems, Minneapolis, MN) at the day of induction (day 0) in RPMI 1640 with 2% B27 minus insulin (Thermo Fisher Scientific, Waltham, MA). After 24 h (day 1), activin A was replaced with 10 ng/mL bone morphogenetic protein 4 (BMP-4) (R&D Systems, Minneapolis, MN) in RPMI 1640 with 2% B27 minus insulin. Then, induced cultures were incubated for an additional 4 days without any medium change. From day 5 onwards, the BMP-4-containing medium was replaced with 1 mL warm RPMI 1640 with 2% B27 with insulin (Thermo Fisher Scientific, Waltham, MA) and medium was replaced on alternate days. For differentiation using small molecules (4), dissociated single cells were seeded into Matrigel-coated plates in mTeSR1 medium and cultured until reaching ~85% confluence. On day 0, cultures were treated with 6 μM CHIR99021 (Selleckchem, Houston, TX) in RPMI 1640 supplemented with 2% B27 minus insulin for 48 h followed by removal of CHIR99021 with medium change. At day 3, cultures were treated with 5 μM IWR1 (Sigma-Aldrich, St. Louis, MO) in RPMI 1640 with 2% B27 minus insulin and incubated for another 2 days. At day 5, medium was replaced with RPMI 1640 medium supplemented with 2% B27 plus insulin and changed on alternate days afterward. Cells were monitored daily under the microscope for beating activity, which typically emerged after day 8.
Flow cytometry
Differentiation of hPSCs to cardiac cells was confirmed by intracellular staining of cardiomyocyte markers using flow cytometry. Differentiated culture at day 14 was harvested in 0.25% Trypsin/EDTA (Thermo Fisher Scientific, Waltham, MA) at 37o C for 10 min and subsequently neutralized by 10% FBS in DMEM. Cells were counted and 106 cells were first stained with ethidium monoazide (EMA, Thermo Fisher Scientific, Cat#E1374) to distinguish live/dead cells and further stained with either PE-conjugated mouse anti-cTnT (BD Bioscience, Cat#564767, Clone 13–11) or mouse monoclonal anti-α-actinin (Sigma-Aldrich, A7811) followed by secondary antibody against α-actinin. BD FACS Canto II was used for data acquisition by adjusting voltage and compensation using appropriate excitation and detection channels, i.e. FITC, PE and PerCP Cy5.5 for α-actinin, cTnT and EMA stained cells respectively. Forward versus side scatter quadrants were defined and at least 10,000 EMA negative (live) events were acquired. Finally, dot plots were generated upon data analysis using FlowJo software to show the purity of differentiated cardiomyocytes.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis
Total RNA was extracted from cells at several time-points during the course of differentiation according to the recommendations of the manufacturer using Aurum total RNA mini kit (Bio-Rad Laboratories, Inc., Hercules, CA). Individual RNA sample (1 μg) was reverse-transcribed by adding 100U of Superscript III enzyme and random primers in 20 μL reaction mixture containing Vilo reaction buffer as per manufacturer’s instruction in SuperScript® VILO™ cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA). Further, the reaction mixture was diluted 15 times to 300 μL and 2 μL cDNA was used for real-time PCR. Human specific PCR primers (Supplementary Table 1) were retrieved from the open access websites (http://primerdepot.nci.nih.gov/ or http://pga.mgh.harvard.edu/primerbank/). A unique primer sequence pair for the amplification spanning exon-intron junction of GATA6-AS1 was designed using Integrated DNA Technology (IDT) RT-PCR SciTools (https://www.idtdna.com/scitools/Applications/RealTimePCR/) and all primers were synthesized from IDT at 25 nmoles scales. RT-PCR reaction was performed, and the mRNA level in each sample was normalized to the GAPDH mRNA level.
qRT-PCR detection of cellular localization of GATA6-AS1
For cellular localization, cells were harvested and centrifuged at 200 g for 5 min, and cell pellets were washed once with PBS. Cellular fractionation was performed according to the manufacturer’s instruction using Paris kit (Ambion). RNA was extracted and then concentrated with LiCl and ethanol precipitation to remove carbohydrates and gross DNA contamination. LiCl also quantitatively precipitates RNA larger than 300 nucleotides. The precipitated pellet of each fraction was air-dried and dissolved in an equal volume of nuclease-free water. Further, an aliquot of RNA (10 μL) was converted to cDNA, and qRT-PCR was performed to compare the relative expression of GATA6-AS1 and GATA6 in the cytoplasmic and nuclear fractions.
Fluorescence in situ hybridization
Sterile glass coverslips were placed into each well of a 24-well tissue-culture plate and dispensed with 1 mL/well of diluted Matrigel and incubated for overnight at 4oC. Then, cardiomyocyte differentiation was set up using the standard protocol and at day 5, cells were fixed with 4% paraformaldehyde (PFA; Sigma-Aldrich, St. Louis, MO) at room temperature for 10 min followed by washing two times with PBS and permeabilization with 70% ethanol 4°C for at least an hour. Cells were washed for 2–5 min with 1 mL of wash buffer and then incubated in 100 μL of nucleic acid probes diluted in the hybridization solution containing 1% (w/v) dextran sulfate, 2X saline-sodium citrate (SSC) and 1% formamide. The incubation was done in a dark humidified chamber at 37°C for 4 h. Cells were then washed and counterstained with DAPI (5 ng/mL) in wash buffer (2X SSC, 10% formamide). Cells were re-suspended in 2X SSC and GLOX buffer without enzymes was added for equilibration and incubation for 1–2 min. Cells were again re-suspended in the 100 μL GLOX buffer with enzymes (glucose oxidase and catalase) and imaging was performed. Note that the dried oligonucleotide probe was dissolved in 200 μL of probe buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) to create a probe stock at a total oligo concentration of 25 μM. All reagents were prepared in RNAse free water.
Gene knockdown of GATA6-AS1
HIV-based lentiviral vectors expressing the target-specific shRNA sequences against human GATA6-AS1 and a scrambled control were obtained from The RNAi Consortium (TRC, MISSION® TRC shRNA library, Sigma-Aldrich). The shRNA sequences against GATA6-AS1 were custom designed as following: shRNA1, CCTGGAGAGTTTCTGATATTT; and shRNA2, CCCAGGTAAATCCAAGTAAAT. These shRNAs did not target any other known genes in the bioinformatic validation by Sigma Aldrich.
Undifferentiated hPSCs were dissociated with Versene/EDTA (Thermo Fisher Scientific, Waltham, MA) and 5×105 cells were infected with concentrated lentivirus expressing either the control shRNA or GATA6-AS1 shRNA (pool of GATA6-AS1 shRNA1 and shRNA2) at 1 multiplicity of infection (MOI) and seeded into a Matrigel-coated 6-well plate in 1 mL MEF-CM or mTeSR1 medium containing 6 μg/mL polybrene (Sigma-Aldrich, St. Louis, MO) and 10 μM Rock inhibitor Y-27632 (Stemgent, Cambridge, MA). The next day, cells were replaced with fresh MEF-CM and allowed to expand for 3 days. Cells were then treated with 0.5 μg/mL puromycin in MEF-CM or mTeSR1 medium for an additional 7 days and antibiotics-selected hPSC pooled colonies were expanded to set up for cardiomyocyte differentiation.
Germ layer differentiation of hPSCs
Germ layer differentiation was performed as per the manufacturer’s instruction using Human Pluripotent Stem Cell Functional Identification Kit (R&D Systems, Minneapolis, MN). Control shRNA and GATA6-AS1 shRNA undifferentiated cultures were dissociated using Versene/EDTA (Thermo Fisher Scientific, Waltham, MA), and ~1×105 cells in 500 μL of MEF-CM supplemented with 10 ng/mL bFGF were seeded in 48-well Cultrex® Reduced Growth Factor Basement Membrane Extract, PathClear® (RGF/BME) overnight-coated culture plates. Cells were incubated overnight at 37°C and 5% CO2 to reach approximately 50% confluency. For ectoderm differentiation, cells were fed daily with 500 μL warm ectoderm differentiation base medium and on day 4 cells were fixed for immunocytochemistry (ICC) analysis. For mesoderm differentiation, cells were fed with 500 μL warm mesoderm differentiation base medium and incubated overnight. The differentiation medium was again replaced after 12 h and on day 3 cells were fixed for ICC analysis. For endoderm differentiation, cells were fed with 500 μL warm endoderm differentiation base medium I and incubated overnight. The differentiation medium was again replaced with differentiation base medium II on day 2 and day 3. Cells were fixed on day 4 for ICC analysis.
For ICC analysis, cells were washed twice with 500 μL PBS and fixed with 4% PFA in PBS for 20 min at room temperature. Next, the cells were washed 3 times with 1% BSA in PBS for 5 min. Further, the cells were permeabilized and blocked with 0.3% Triton X-100, 1% BSA, and 10% normal donkey serum in 500 μL PBS at room temperature for 45 min. Then, the cells were incubated overnight at 4oC in respective reconstituted primary antibody (OTX2, TBXT, and SOX17) in 300 μL PBS containing 0.3% Triton X-100, 1% BSA, and 10% normal donkey serum to a final concentration of 10 μg/mL. The cells were washed 3 times with 500 μL BSA (1% in PBS) for 5 min and incubated with 300 μL Alexa fluor 488 or Alexa fluor 594 conjugated donkey anti-goat secondary antibody at 1:1000 in PBS containing 1% BSA and incubated in the dark for 60 min at room temperature. The cells were then washed 3 times with 500 μL BSA (1% in PBS) for 5 min. Finally, nuclear staining was performed using Hoechst dye for 5 min in 300 μL PBS, washed 3 times with PBS, and visualized with a fluorescence microscope.
High-content imaging analysis by ArrayScan
High-content imaging analysis was performed to determine cardiomyocyte purity in the control shRNA and GATA6-AS1 shRNA cultures at differentiation day 14. Cells were dissociated using 0.25% Trypsin-EDTA and plated onto a Matrigel-coated 96-well culture plate at a density of 2×104 cells/well for the detection of NKX2–5 and α-actinin. The cells were maintained in the cardiomyocyte culture medium for 24 h to allow them to recover spontaneous beating. Cardiomyocyte culture medium was aspirated the next day and the cells were washed with PBS, fixed in 4% PFA and permeabilized with 0.1% Triton-X 100. The cells were then blocked with 5% normal goat serum in PBS at room temperature for 1 h and incubated overnight at 4°C with the primary antibodies against NKX2–5 (Cell Signaling; 1:1600) and α-actinin (Sigma-Aldrich; 1:500) diluted in 0.25% BSA/PBS for the purity assay. After incubation with the primary antibodies, the cells were washed twice with PBS and incubated with secondary antibodies Alexa Fluor 488 conjugated goat anti-mouse IgG1 (α-actinin; Life Technologies) and Alexa Fluor 594 conjugated goat anti-rabbit IgG (NKX2–5; Life Technologies), which were diluted at 1:500 in 0.25% BSA/PBS. The nuclei were counterstained with Hoechst in PBS and imaged using an ArrayScan™ XTI Live High Content Platform (Thermo Fisher Scientific).
High-content imaging analysis was also performed to determine the GATA6 protein level in the control shRNA and GATA6-AS1 shRNA cultures at differentiation days 0, 2, and 5. The immunostaining protocol used for GATA6 was the same as described for NKX2–5 and α-actinin. A GATA6 specific primary antibody (Cell Signaling, 5851T; 1:300) was used prior to the incubation with the secondary antibody Alexa Fluor 594 conjugated goat anti-rabbit IgG (Life Technologies; 1:500).
Images of immunocytochemistry were acquired and quantitatively analyzed using ArrayScan™ XTI Live High Content Platform. Twenty fields/well were imaged using a 10x objective. The Acquisition software Cellomics Scan (Thermo Fisher Scientific) was used to capture images and data analysis was performed using Cellomics View Software (Thermo Fisher Scientific). For the quantitation of NKX2–5 or GATA6, images were analyzed with a mask modifier for Hoechst and NKX2–5 or GATA6 positive cells restricted to the nucleus. The percentage of NKX2–5 positive cells per well in each treatment was used as a readout. For the quantitation of α-actinin, images were analyzed with a mask modifier for Hoechst restricted to the nucleus. α-actinin was quantified with a spot mask that extended 7 units from the nucleus. Spot threshold was set to 10 units and the detection limit was set at 25 units. The percentage of α-actinin-positive cells per well was used as a readout. For GATA6, the mean fluorescence intensity of GATA6-positive cells per well in each treatment was used as a readout.
RNA-seq analysis
RNA was isolated from the control shRNA and GATA6-AS1 shRNA cultures at differentiation day 2 and day 5 (n=3/group, total 12 samples) using Aurum total RNA mini kit (Bio-Rad) as per manufacturer’s instructions. At the High-throughput DNA Sequencing Core, Parker H. Petit Institute for Bioengineering and Bioscience of Georgia Tech, total RNA quality was tested using a 2100 Bioanalyzer, and RNA 6000 Nano Chip (Agilent Technologies). The Illumina TruSeq technology was then used to prepare RNA-Seq libraries, and next-generation sequencing was performed through an Illumina HiSeq 2500, 75bp paired-end sequencing. Total RNA reads were aligned to the reference human genome using gencode v28. Read counts per sample ranged from 20,008,004 to 2,804,638. Data analyses were carried out in R v3.4.0 (24) and DESeq2 R package was used for normalization and detection of differentially expressed genes (25, 26). The upregulated and downregulated genes were identified following Benjamini-Hochberg corrected p-value at a significance threshold of the -log10(p-value) >2.173 for samples at day 2 and >1.891 for samples at day 5 (27). Functional analysis of the up-/down-regulated differentially expressed genes was performed using the ToppFun software package (28).
qRT-PCR analysis of GATA6-AS1 and GATA6 expression in human tissues
Ambion’s FirstChoice® Human Total RNA Survey Panel consisting of pooled total RNA from 20 different normal human tissues was used to determine the expression of GATA6-AS1 and GATA6 in human tissues. Each RNA pool was comprised of at least 3 tissue donors and 1 μg RNA was used for cDNA synthesis followed by qRT-PCR analysis to determine the distribution of GATA6-AS1 and GATA6 transcripts in human tissues.
In vitro characterization of GATA6-AS1 and GATA6 promoter
Five pairs of primers with their restriction endonuclease (BglII or MluI) sites at 5’-end were designed for the PCR amplification of the intergenic region between GATA6-AS1 and GATA6 and the intergenic region containing part of the exon1 of GATA6-AS1 and/or GATA6 (Supplementary Table 2). These amplified fragments were subsequently cloned into the pLightSwitch promoter vector in both forward and reverse orientations. The presence of the PCR product in the cloned plasmids was confirmed by colony PCR followed by mini-preparations of plasmid DNA. The plasmid DNA thus obtained was restriction enzyme digested with BglII and MluI and checked on agarose gel to confirm the right size of the insert.
For promoter reporter assay, HeLa cells were plated into 96-well tissue culture plates at 25,000 cells per well in 100 μL to obtain ~70–90% confluency on the following day. Subsequently, they were transfected in triplicate with a mixture of 100 ng plasmid and 0.5 μL Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA) in the Opti-MEM™ medium (Thermo Fisher Scientific, Waltham, MA). After 24 h, the cells were incubated with 100 μL of luciferase substrate (SwitchGear Genomics) for 30 min and luciferase activities were assayed in a black 96-well plate luminometer according to the manufacturer’s protocol. The promoter strength of each DNA fragment was determined by calculating the relative luciferase signal as compared with the empty vector.
Statistical Analysis
Unpaired t-test was used to compare the control shRNA group with the GATA6-AS1 shRNA group from at least 3 biological samples for each group. Data were presented as mean ± standard deviation.
RESULTS
GATA6-AS1 expression is transiently upregulated during the early stage of cardiomyocyte differentiation
GATA6-AS1 is a previously characterized 1788 nucleotide long sequence (accession no. NR_102763.1) on chromosome 18 located next to GATA6, an important regulator of cardiovascular development (Fig. 1A). It is not conservative with the mouse counterpart (29), doesn’t contain any tandem repeats and has little protein-coding capacity (coding potential score: 1.43409) although it has the potential of being adenylated (Supplementary Results).
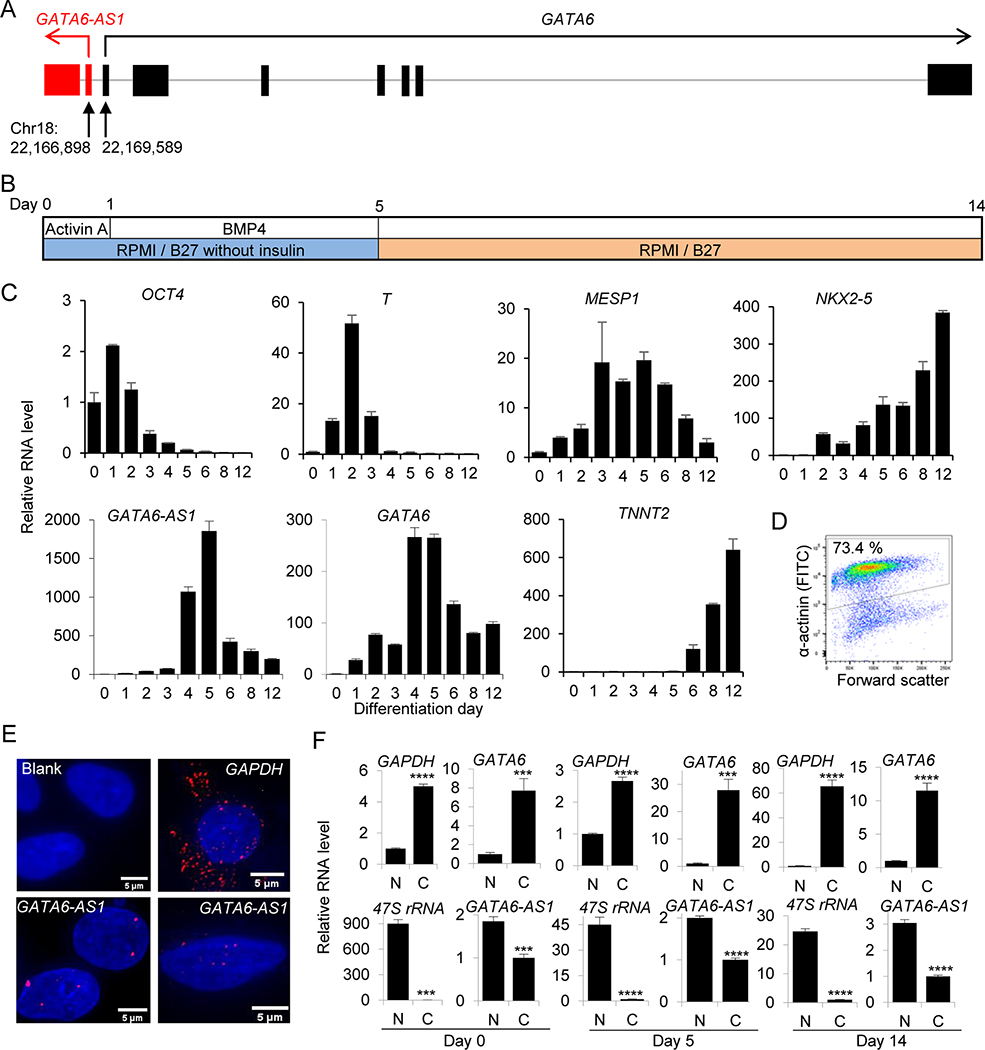
Figure 1. Expression of lncRNA GATA6-AS1 during differentiation from hPSCs.
(A) The schematic diagram of the genomic organization of GATA6-AS1 and GATA6 represented by lines and exons are shown as vertical bars. Red and black arrows indicate the direction of transcription of GATA6-AS1 and GATA6 respectively. (B) Cardiomyocyte differentiation procedure. Undifferentiated cells were induced to differentiate by the treatment with activin A at day 0 followed by BMP4 at day 1 in RPMI/B27 insulin-free medium and maintained in RPMI/B27 after day 5. Cells were collected at several time-points for the analysis of gene expression and harvested at day 14 for the assessment of cardiomyocyte purity. (C) Quantitative reverse transcriptase PCR (qRT-PCR) analysis of genes in differentiation cultures. The expression of genes associated with pluripotent stem cells (OCT4), mesoderm (T and MESP1) and cardiac (NKX2–5 and cTnT) along with genes of interest (GATA6 and lncRNA GATA6-AS1) for time-points covering day 0 to day 12. Gene expression was normalized to the level of undifferentiated cells at day 0. The mean fold difference and standard deviation in expression for each time-point were calculated using three replicates. (D) Flow cytometry analysis of differentiated cells at day 14 showing 73.4 % α-actinin positivity. (E) RNA FISH was performed at day 5 and GATA6-AS1 signal (red dot) was localized in the nucleus; DAPI (blue) was used as nuclear stain. Blank as negative control and GAPDH was used as cytoplasmic localization control. Scale bar = 5 μm. (F) hPSCs at days 0, 5 and 14 were fractionated into nuclear and cytoplasmic fractions followed by RNA extraction and qRT-PCR of GATA6-AS1 and GATA6 along with GAPDH and 47S rRNA as cytoplasmic and nuclear fraction controls, respectively. Gene expression of GAPDH and GATA6 was normalized to the level of nuclear fraction whereas RNA level of GATA6-AS1 and 47S rRNA was normalized to cytoplasmic fraction. The mean fold difference and standard deviation in gene expression for each fraction were calculated by using three replicates. N, nuclear; C, cytoplasmic. ***, P<0.001; ****, P<0.0001.
We first analyzed the gene expression profile during cardiomyocyte differentiation using IMR90-hiPSCs starting induction at day 0 with Activin A and BMP 4 at day 1 in RPMI-B27 without insulin medium. The induction medium was replaced with RPMI-B27 at day 5 and the cells were harvested at day 14 (Fig. 1B). Time-point qRT-PCR analysis of differentiated cells showed the decreased expression of the stem cell marker OCT4 from day 0 to 12, whereas mesendodermal markers T (brachyury) and MESP1 showed maximum expression at day 2 and day 5 respectively. The expression of other cardiac genes, NKX2–5 and cTnT, kept on increasing and reached their highest level at day 12. Both GATA6-AS1 and GATA6 reached their maximum at day 5 during differentiation (Fig. 1C).
In this differentiation culture, the cardiomyocyte positivity based on α-actinin staining was 73.4 % at day 14 (Fig. 1D). When we repeated time-point gene expression analysis in another cell line (NKX2–5-eGFP hESC), we observed similar expression profiles for these genes. Both GATA6-AS1 and GATA6 peaked at day 5 and day 6, and expressions of GATA6-AS1 and GATA6 were >9,000- and >20,000- fold higher respectively as compared to day 0 (Supplementary Fig. 1A). Expression of cardiac genes NKX2–5 and cTnT reached to the maximum at day 14; this differentiation culture contained ~80% (α-actinin+ cells) cardiomyocytes (Supplementary Fig. 1A, B).
Next, we examined the localization of GATA6-AS1 in cells at differentiation day 5 by RNA FISH. Following labeling with a mixture of 45 probes against GATA6-AS1 (Supplementary Table 3), fluorescence signals of the probes were detected in the DAPI-positive nuclear area (Fig. 1E). In contrast, the fluorescence signal of GAPDH was detected mostly in the cytoplasm area (Fig. 1E). The nuclear localization of GATA6-AS1 was further confirmed by a cell fractionation assay. Transcript levels of GATA6-AS1 and GATA6 at days 0, 5 and 14 were analyzed in nuclear and cytoplasmic fractions. Similar to the levels of 47S rRNA (a control for nuclear RNA), the expression of GATA6-AS1 was much higher in the nuclear fraction than in the cytoplasmic fraction, whereas the RNA levels of GAPDH and GATA6 were several folds higher in the cytoplasmic fraction than in the nuclear fraction (Fig. 1F). These results indicate that GATA6-AS1 could be a crucial lncRNA for controlling cardiomyocyte differentiation from hPSCs.
Knockdown of GATA6-AS1 inhibits cardiomyocyte differentiation from hPSCs
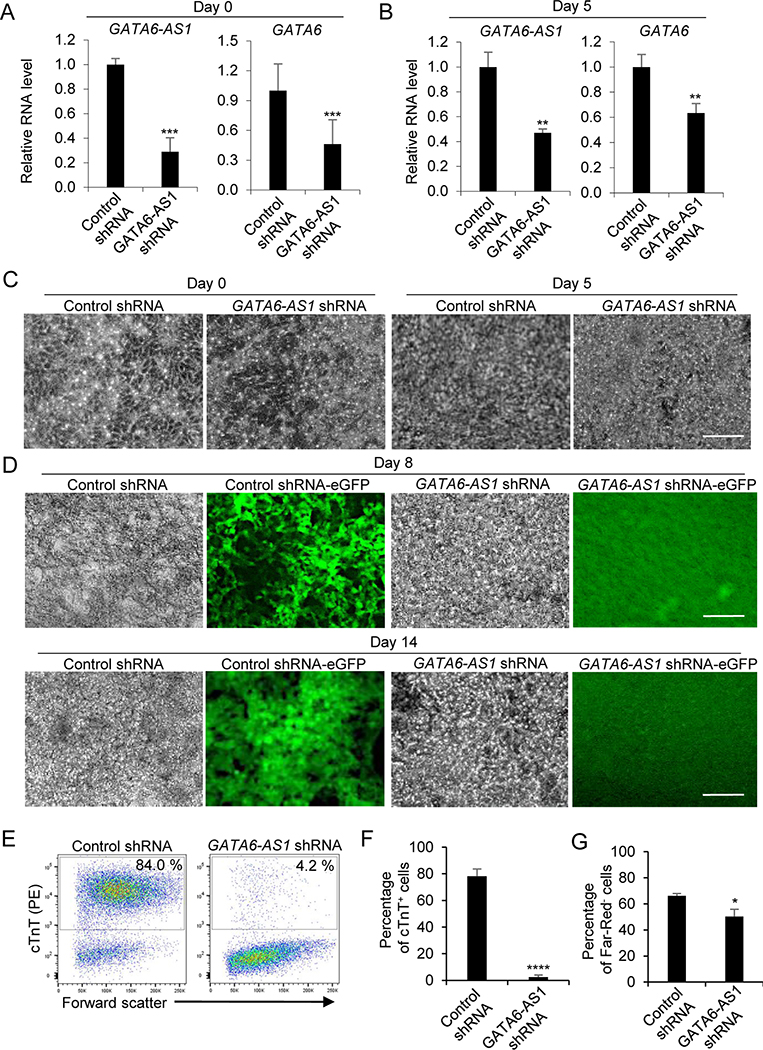
We next investigated the functional role of GATA6-AS1 during the differentiation of cardiomyocytes from hESCs with NKX2–5 promoter-driven eGFP by knockdown of GATA6-AS1 using short hairpin RNA (shRNA). Cells were transfected with a lentivirus expressing the GATA6-AS1 shRNA or a scrambled control shRNA. Stable clones were expanded after puromycin antibiotic selection and subjected to growth factor-guided cardiomyocyte differentiation. The relative RNA levels of GATA6-AS1 were significantly lower in the GATA6-AS1 shRNA cultures than in the control shRNA cultures at day 0 and day 5 (Fig. 2A). Interestingly, the relative RNA level of GATA6 was also lower in the GATA6-AS1 shRNA cultures than in the control shRNA cultures at day 0 and day 5 (Fig. 2B). Similar to the control shRNA cultures, the GATA6-AS1 shRNA cultures at day 0 and day 5 showed typical morphologies with tightly packed stem cells at day 0 and differentiated cells at day 5 (Fig. 2C). However, the control shRNA cultures differentiated into eGFP+ contractile areas by day 8 but no such eGFP+ contractile area was observed in the GATA6-AS1 shRNA cultures. Similarly, tightly packed beating cardiomyocytes with eGFP+ fluorescence were observed from the control shRNA cultures but not from the GATA6-AS1 shRNA cultures at day 14 (Fig. 2D). Based on the flow cytometry analysis of cardiomyocyte marker cTnT, only ~4% cTnT+ cells were detected in GATA6-AS1 shRNA cultures compared with ~84% in the control shRNA cultures (Fig. 2E, F). Viability detected by Far-Red-negative cells at day 14 in GATA6-AS1 cultures were significantly lower compared with the control shRNA cultures (Fig. 2G). These results suggest that knockdown of GATA6-AS1 in hESCs inhibits cardiomyocyte differentiation.
Figure 2. Knockdown of GATA6-AS1 inhibits the differentiation of cardiomyocytes induced by growth factors.
Control shRNA or GATA6-AS1 shRNA NKX2–5-eGFP hESCs were induced for cardiomyocyte differentiation using growth factors activin A and BMP4. (A, B) qRT-PCR analysis of GATA6-AS1 and GATA6 expression at day 0 and day 5. Data are normalized with amount of GAPDH mRNA relative to the corresponding value for cells transduced with the control shRNA. The mean fold difference and standard deviation in expression were calculated using three replicates. (C) Cell morphology of the control shRNA culture and the GATA6-AS1 shRNA culture during cardiomyocyte differentiation at day 0 and day 5. Scale bars = 200 μm. (D) Cell morphology and expression of NKX2–5-eGFP in the control shRNA culture and the GATA6-AS1 shRNA culture during cardiomyocyte differentiation at day 8 and day 14. Scale bars = 200 μm. (E) Flow cytometry analysis of differentiation cultures detected by α-actinin at day 14 in the control shRNA culture and the GATA6-AS1 shRNA culture. (F) Summary of the purity of cardiomyocytes from triplicate wells of the control shRNA culture and the GATA6-AS1 shRNA culture. (G) Summary of the viability of cardiomyocytes from triplicate wells of the control shRNA culture and the GATA6-AS1 shRNA culture. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001.
To further confirm the role of GATA6-AS1 in cardiomyocyte differentiation, we repeated GATA6-AS1 knockdown in IMR90 hiPSCs and examined cardiomyocyte differentiation by growth factor induction. Similar to the findings in hESCs, expression of both GATA6-AS1 and GATA6 was significantly downregulated in the GATA6-AS1 shRNA hiPSC culture compared with the control shRNA culture at both day 2 and day 5 (Supplementary Fig. 2A, B). The levels of GATA6-AS1 and GATA6 in the GATA6-AS1 knockdown cultures at day 5 was lower than the basal levels at day 2 (Supplementary Fig. 2A, B). The morphology of cells at the early stage of differentiation was similar between the GATA6-AS1 shRNA culture and the control shRNA culture. However, robust beating cardiomyocytes appeared in the control shRNA culture at day 10 but no or only few cardiomyocyte patches were seen in the GATA6-AS1 shRNA culture (Supplementary Fig. 2C, D). The defect of cardiomyocyte differentiation in the GATA6-AS1 shRNA culture was further confirmed by the quantitative analysis of cardiomyocyte purity by flow cytometry of cTnT. High purity of cardiomyocytes (~70% cTnT+ cells) was detected in the control shRNA culture whereas the GATA6-AS1 shRNA culture generated only ~10% cTnT+ cells (Supplementary Fig. 2E, F). These results show knockdown of GATA6-AS1 inhibits differentiation of cardiomyocyte from hiPSCs.
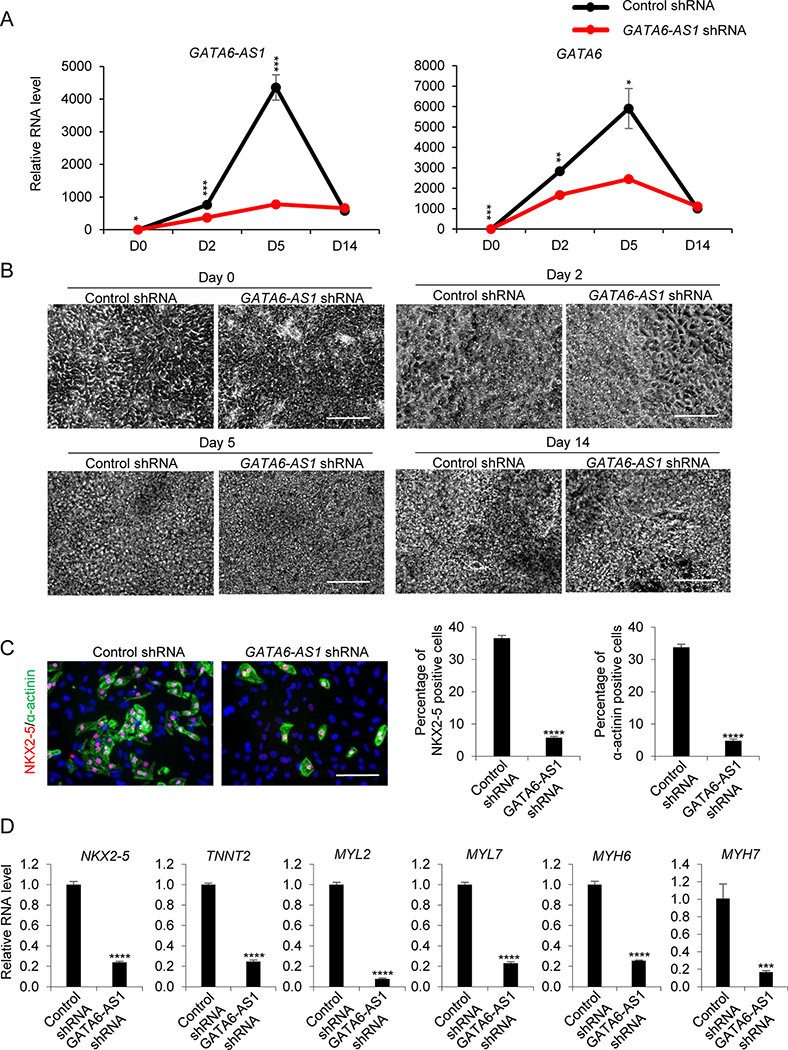
To examine if the effect of GATA6-AS1 knockdown in cardiomyocyte differentiation is differentiation method-independent. We generated GATA6-AS1 knockdown stable cells in SCVI 273 hiPSCs and examined cardiomyocyte differentiation using small molecules targeting Wnt signaling (4). The GATA6-AS1 knockdown culture had significantly reduced expression of GATA6-AS1 and GATA6 at differentiation days 0, 2 and 5 (Fig. 3A). High-content ArrayScan imaging analysis showed that GATA6 protein was detectable at days 2 and 5 (Supplementary Fig. 3A). There was no significant difference in GATA6 between the control shRNA culture and the GATA6-AS1 shRNA culture at day 2; however, the level of GTAT6 significantly decreased in the GATA6-AS1 shRNA culture compared with the control shRNA culture at day 5 (Supplementary Fig. 3B).
Figure 3. Knockdown of GATA6-AS1 inhibits the differentiation of cardiomyocytes induced by small molecules.
The control shRNA or GATA6-AS1 shRNA SCVI273 hiPSCs were induced for cardiomyocyte differentiation using small molecules CHIR990021 and IWR1. (A) qRT-PCR analysis of of GATA6-AS1 and GATA6 expression at differentiation days 0, 2 and 5. The mean fold difference and standard deviation in gene expression were calculated using three replicates. (B) Cell morphology of the control shRNA culture and the GATA6-AS1 shRNA culture during cardiomyocyte differentiation at days 0, 2, 5 and 14. Scale bars = 200 μm. (C) High-content imaging analysis of the differentiated cells detected by NNX2–5 and α-actinin at day 14 in the control shRNA culture and the GATA6-AS1 shRNA culture. Summary of the purity of cardiomyocytes from 12 wells of the control shRNA culture and the GATA6-AS1 shRNA culture. Scale bars = 50 μm. (D) The expression of genes associated with cardiomyocytes was analyzed by qRT-PCR in the control shRNA culture and the GATA6-AS1 shRNA culture at day 14. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001.
Similar to growth factor-guided differentiation, the morphology of the GATA6-AS1 knockdown culture was indistinguishable from the control shRNA culture at the early stage of differentiation (Fig. 3B). Cellular proliferation of the control shRNA culture and the GATA6-AS1 shRNA culture based on cell counting showed no significant difference at differentiation day 0, 2 and 5 (Supplementary Fig. 3C). At the late stage of differentiation, beating cardiomyocytes in the control shRNA culture were more abundant than in the GATA6-AS1 shRNA culture (Supplementary Movies S1 and S2). These beating cardiomyocytes were positive for cardiomyocyte-associated proteins NKX2–5 and α-actinin (Fig. 3C). The control shRNA culture generated ~37% NKX2–5+ cells and ~34% α-actinin+ cells while the GATA6-AS1 knockdown culture contained only ~6% NKX2–5+ cells and ~5% α-actinin+ cells as analyzed by high-content imaging using ArrayScan (Fig. 3C). Furthermore, the expression of cardiomyocyte-associated genes, including NKX2–5, TNNT2, MYL2, MYL7, MYH6 and MYH7, was reduced in the GATA6-AS1 shRNA culture compared with the control shRNA culture (Fig. 3D). These findings suggest that GATA6-AS1 is essential for cardiomyocyte differentiation and this defect as a result of GATA6-AS1 knockdown is differentiation method-independent.
Taken together, these results indicate that lncRNA GATA6-AS1 plays a functional role in controlling cardiomyocyte differentiation.
Knockdown of GATA6-AS1 does not affect the pluripotency of hPSCs
To confirm that GATA6-AS1 knockdown did not influence the basic properties of hPSCs, we compared the control shRNA culture and the GATA6-AS1 shRNA culture and verified if they were truly pluripotent stem cells that can differentiate into cells of the three germ layers. Upon differentiation induction using the Human Pluripotent Stem Cell Functional Identification Kit (R&D Systems) which contains three medium formations designed for specific differentiation of hPSCs into cells of three germ layers respectively, the control shRNA culture and the GATA6-AS1 shRNA culture differentiated efficiently into cells of all three germ layers as indicated by the expression of markers associated with ectoderm (OTX2), mesoderm (TBXT), and endoderm (SOX17) in differentiation cultures (Supplementary Fig. 4A). Further, mRNA levels of pluripotent stem cell markers OCT4, SOX2, NANOG and LIN28 were detected at similar levels in both the undifferentiated control shRNA culture and the undifferentiated GATA6-AS1 shRNA cultures (Supplementary Fig. 4B). These results indicate that both the control shRNA culture and the GATA6-AS1 shRNA culture have comparable ability to differentiate into cells of all three germ layers and knockdown of GATA6-AS1 does not compromise the pluripotency of hPSCs.
Knockdown of GATA6-AS1 downregulates the expression of mesoderm markers and alters the expression of canonical and non-canonical Wnt signaling-related genes
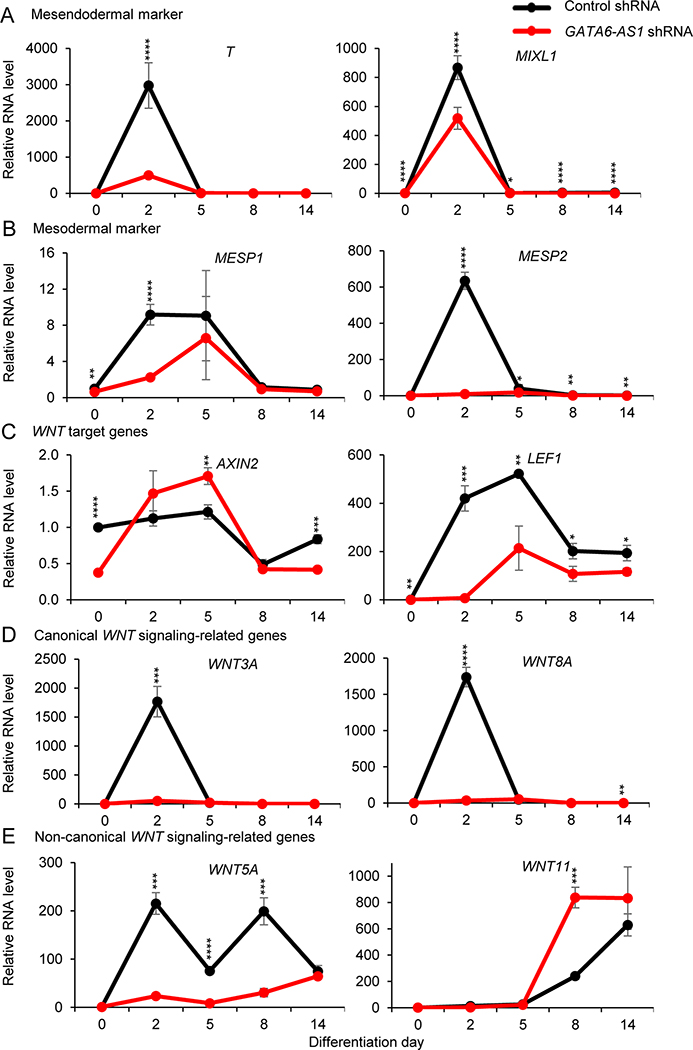
Cardiomyocyte differentiation from hPSCs is regulated by the temporal expression of many genes associated with mesodermal patterning and Wnt signaling (30). The typical peak expression of mesendodermal markers (T and MIXL1) at day 2 was reduced in the GATA6-AS1 shRNA culture compared with the control shRNA culture (Fig. 4A). Similarly, the GATA6-AS1 shRNA culture had lower levels of mesodermal markers (MESP1 and MESP2) at day 2 than the control shRNA culture (Fig. 4B). Further, GATA6-AS1 knockdown also altered the expression of the Wnt signaling genes that are known to drive the differentiation and development of hPSCs into cardiomyocytes. Wnt target genes AXIN2 at day 2 and LEF1 at days 2 and 5 were significantly downregulated in the GATA6-AS1 shRNA culture compared with the control shRNA culture (Fig. 4C). Expression of the canonical Wnt signaling-related genes (WNT3A and WNT8A) peaked at day 2 in the control shRNA culture but significantly downregulated in the GATA6-AS1 shRNA culture (Fig. 4D). The expression of the non-canonical Wnt signaling genes, WNT5A (days 2, 5 and 8) and WNT11 (day 8), was significantly changed in the GATA6-AS1 shRNA culture compared with the control shRNA culture (Fig. 4E). Overall, these results suggest that the downregulation of GATA6-AS1 abrogates mesoderm patterning and alters the expression of the Wnt signaling genes at the early stage of cardiomyocyte differentiation, providing a possible mechanism on how GATA6-AS1 knockdown affects cardiomyocyte differentiation from hPSCs.
Figure 4. Knockdown of GATA6-AS1 inhibits mesendoderm and mesoderm induction and alters the expression of Wnt signaling genes.
The expression of cardiac mesodermal and Wnt signaling markers was analyzed by qRT-PCR in the control shRNA and GATA6-AS1 shRNA NKX2–5-eGFP hESC cultures at differentiation days 0, 2, 5, 8 and 14: (A) mesendodermal markers T and MIXL1; (B) mesodermal markers MESP1 and MESP2; (C) Wnt target genes AXIN2 and LEF1; (D) canonical Wnt-signaling genes WNT3A and WNT8A; and (E) non-canonical Wnt signaling genes WNT5A and WNT11. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001.
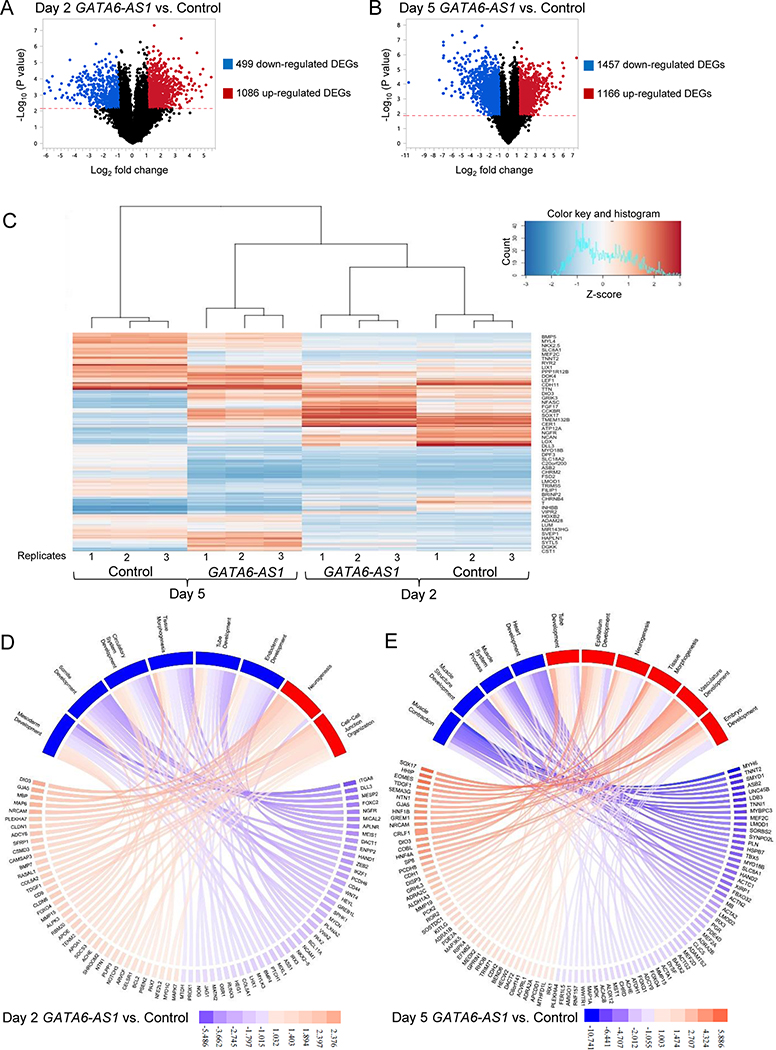
To further examine the effect of GATA6-AS1 knockdown during cardiomyocyte differentiation, we compared the genome-wide gene expression profile of the GATA6-AS1 knockdown culture with that of the control shRNA culture at cardiomyocyte differentiation days 2 and 5 by RNA-seq (n=3 for each sample type). As expected, the principal components analysis showed that the gene expression profiles of three replicates of each sample type were similar to one another, and that the gene expression profiles of GATA6-AS1 knockdown were different from those of the control shRNA culture at the same time points (Supplementary Fig. 5). In addition, the difference between day 2 and day 5 showed the highest level of separation in the dataset (Supplementary Fig. 5). At day 2, GATA6-AS1 knockdown resulted in 499 down-regulated and 1086 up-regulated differentially expressed genes (set at -log10 p-value = 1.996; log2 changes <−1 or >1) (Fig. 5A). At day 5, GATA6-AS1 knockdown resulted in 1457 down-regulated and 1166 up-regulated genes (Fig. 5B). The differential expression of the top 100 variable genes across all samples indicated that the gene expression profiles of the GATA6-AS1 shRNA culture at day 5 were similar to from those of the control shRNA culture and the GATA6-AS1 shRNA culture at day 2 (Fig. 5C), suggesting a developmental defect and delay in the GATA6-AS1 shRNA culture.
Figure 5. Differential gene expression in day 2 and day 5 samples from the GATA6-AS1 knockdown culture compared with those from the control shRNA culture.
(A, B) Volcano plot portrays log2 fold change of the GATA6-AS1 knockdown samples vs. the control shRNA samples on day 2 and day 5. The grey dots signify genes that were above the -log10(p-value) calculated using a False Discovery Rate [-log10(p-value)>2.173 on day 2 and -log10(p-value)>1.891 on day 5] with log2 fold changes of >1 or <−1. (C) Heatmap showing the expression of the top 100 differentially expressed genes across all 12 samples. (D, E) Pathway plots of significantly downregulated and upregulated pathways for the GATA6-AS1 knockdown culture on day 2 and day 5. The color keys show the log2 fold changes in gene expression. Pathways upregulated and downregulated in the GATA6-AS1 knockdown culture are shown in red and blue, respectively. Pathways were determined by entering all of the differentially expressed genes found in Panels A and B and inputted into ToppFun. Genes shown in Panel D were selected on their significant p-value to represent the overall log differential of all the genes in the pathway. The average of the log differential of the ten genes represented in Panel D is equal to the average of the log differential of all the genes contributing to the pathway. Genes in Panel E were similarly selected
Among the top downregulated genes at day 2, many of them were associated with mesoderm induction and development, such as T, MESP1 and MEIS1 (Supplementary Table 4). In addition, GATA6-AS1 knockdown also dramatically reduced the expression of the Wnt singling molecules such as LEF1, RSPO3 and WNT5A at day 2 (Supplementary Table 4). GATA6-AS1 knockdown however increased the expression of genes associated with neuron differentiation, including MAP6, GDNF, PDGFB, FOXA1 and PAX7 at day 2. At day 5, GATA6-AS1 knockdown resulted in a dramatic decrease in the expression of genes associated with cardiac development including cardiac transcription factors (NKX2–5 and MEF2C), a transcriptional repressor (SMYD1), cardiac structural proteins (MYL4, TNNT2, and TTN), calcium handling proteins (SLC8A1 and RYR2), as well as proteins that are highly expressed in muscle (LMOD1, MYBPC3 and UNC45B) (Supplementary Table 5). However, GATA6-AS1 knockdown increased the expression of genes associated with endoderm differentiation including SOX17, HNF1B and HNF4A at day 5 (Supplementary Table 5). These results are consistent with the observed defects in mesoderm induction and cardiomyocyte differentiation from the GATA6-AS1 knockdown hPSCs.
Pathway analysis based on two subsets of up and down-regulated genes for each timepoint were then analyzed through toppFun gene enrichment analysis program that identifies pathways over-represented in the list of differentially expressed genes. At day 2, the top significantly down-regulated gene ontology families by GATA6-AS1 knockdown included genes involved in tissue morphogenesis, tube development, somite development, heart development, circulatory system development, mesoderm development and endoderm development (Fig. 5D and Supplementary Table 6). GATA6-AS1 knockdown significantly increased gene ontology families of generation of neurons, neurogenesis, neuron differentiation, neuron development, neuron projection development and cell-cell junction organization (Fig. 5D and Supplementary Table 6). At day 5, GATA6-AS1 knockdown significantly inhibited gene ontology families including the biological processes of muscle structure development, muscle system process, heart development, muscle contraction, muscle tissue development, and circulatory system development (Fig. 5E and Supplementary Table 7). The ontology families of the biological processes that were up-regulated in the GATA6-AS1 knockdown culture at day 5 included tube development, epithelium development, tissue morphogenesis, embryo development, neurogenesis and vasculature development (Fig. 5E and Supplementary Table 7).
We next examined whether the expression of GATA6 target genes (http://amp.pharm.mssm.edu/Harmonizome/gene_set/GATA6/TRANSFAC+Curated+Transcription+Factor+Targets) was affected by GATA6-AS1 knockdown. Among the 258 known GATA6 target genes, 73 and 101 genes were affected at differentiation day 2 and day 5, respectively. Many of these differentially expressed GATA6 target genes are known to be associated with cardiogenesis. For example, cardiac mesoderm marker PDGFRA was downregulated in the GATA6-AS1 shRNA culture at both day 2 and day 5 (Supplementary Table 8). Cardiac markers TBX5, MEF2C and MYBPC3 were also downregulated in the GATA6-AS1 shRNA culture at day 5 (Supplementary Table 9).
Many differentially expressed genes affected by GATA6-AS1 knockdown overlapped between day 2 and day 5 (Supplementary Tables 10 and 11). The overlapped upregulated genes included genes associated with neuronal and endoderm differentiation, such as SOX17, HNF1B and NRCAM (Supplementary Table 10), and the overlapped downregulated genes included genes associated with cardiac development, such as WNT5A, PDGFRA, MEIS1, NKX2.5, IRX5, CACNA1C and KCNA5 (Supplementary Table 11).
These results are consistent with the observation that GATA6-AS1 knockdown inhibits mesoderm induction and cardiomyocyte differentiation in hPSCs.
Expression of GATA6-AS1 and GATA6 is regulated by bidirectional promoter
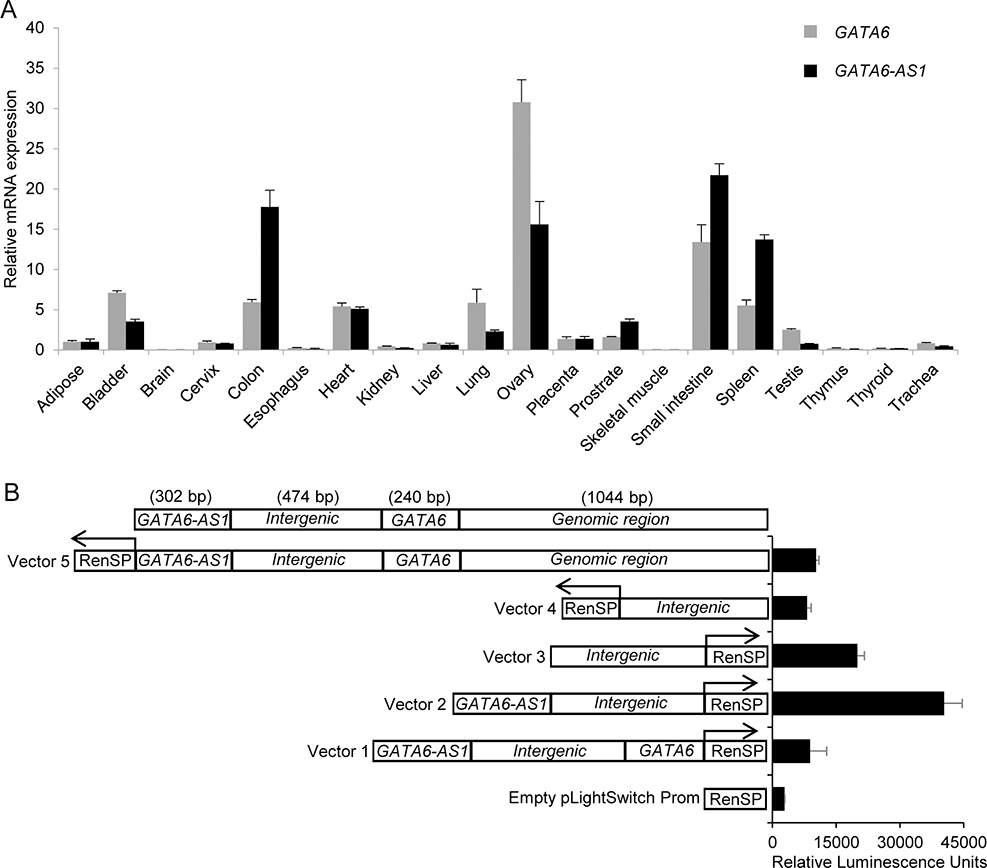
Since knockdown of GATA6-AS1 impairs the expression of GATA6, mesoderm induction and cardiomyocyte differentiation, we further investigated the relationship of GATA6-AS1 and GATA6. LncRNAs typically exhibit tissue-specific expression patterns (31). Given the genomic loci of GATA6-AS1 and GATA6, we speculated that GATA6-AS1 and GATA6 may be co-expressed in a tissue-specific manner. We investigated tissue expression patterns of GATA6-AS1 and GATA6 using Ambion’s FirstChoice® Human Total RNA Survey Panel consisting of pooled total RNA from 20 different normal human tissues. Each pool was comprised of at least 3 tissue donors and 1 μg RNA was used for cDNA synthesis followed by RT-PCR analyses to determine the distribution of GATA6-AS1 and GATA6 transcripts in human tissues. The transcripts of both GATA6-AS1 and GATA6 appeared to be co-expressing in all tissues including at a level similar in the heart; and the tissue-specific expression pattern of GATA6-AS1 almost mirrored the expression pattern of GATA6 (Fig. 6A). Like GATA6, GATA6-AS1 was expressed at relatively high levels in colon, ovary, small intestine, and spleen, moderate levels in heart, lung, placenta and prostrate, and little or no expression in adipose, brain, cervix, esophagus, kidney, liver, skeletal muscle, testis, thymus, thyroid and trachea. These expression patterns suggest co-expression of GATA6-AS1 and GATA6 in various tissues including the heart.
Figure 6. Characterization of GATA6 and GATA6-AS1 expression.
(A) Expression of GATA6 and GATA6-AS1 in multiple human tissues detected by qRT-PCR and normalized by GAPDH expression. The relative mRNA expression levels were normalized to that in adipose tissue. The mean fold difference and standard deviation in gene expression in each tissue were calculated by using three replicates. (B) Detection of promoter activity of the intergenic region. HeLa cells were transiently transfected with vectors with luciferase reporter driven by an inserted sequence containing different combinations of intergenic region with part of the GATA6 and GATA6-AS1 genomic sequence upstream of GATA6 cloned in either the forward or reverse upstream orientation of. The y-axis of the graph represents the relative luciferase activity (RLU), with the transfected constructs shown on the x-axis. The empty pLight switch prom vector represents the luciferase vector lacking a promoter. Arrows indicate the direction of RenSP luciferase reporter gene.
The co-expression of lncRNA along with the transcription of its neighboring protein-coding genes can be initiated in both directions by a promotor located between the lncRNA and the protein-coding gene (32). Such expression of lncRNA influences the expression of its neighboring protein-coding genes, revealing a new layer of transcriptional regulation (33). We tested the possible bidirectional promoter activity regulating the transcription of GATA6-AS1 and GATA6 in the intergenic region between genomic regions of GATA6-AS1 and GATA6. We cloned the intergenic region into pLightSwitch promoter vector in forward and reverse directions. Since possible enhancers or other regulatory sequences may reside near the intergenic region, we also constructed vectors containing the intergenic region with extra adjacent sequences of GATA6-AS1 and GATA6. We then evaluated the activity of the promoter based on luciferase activity after the transfection of the vectors into HeLa cells. The relative luciferase activity in cells transfected with vectors containing intergenic region in both directions was higher when compared with cells transfected with empty LightSwith promoter vector without the insertion (Fig. 6B). These results suggest that the intergenic regions have bidirectional promoter activity that may control the co-expression of GATA6-AS1 and GATA6.
DISCUSSION
Taken together, our results indicate a role of lncRNA GATA6-AS1 in the differentiation of cardiomyocytes from hPSCs. Similar to its adjacent gene that encodes cardiac transcription factor GATA6, GATA6-AS1 is actively transcribed and transiently upregulated with peak expression at the early stage (days 4–6) during cardiomyocyte differentiation from hPSCs. The expression of GATA6-AS1 and GATA6 appears to be controlled by a bidirectional promoter within the intergenic region between the genomic loci of GATA6-AS1 and GATA6. Consistently, the expression of GATA6-AS1 is correlated with that of GATA6 in several human tissues including the heart. Furthermore, knockdown of GATA6-AS1 in hPSCs down-regulated the expression of GATA6, GATA6-target genes and genes associated with mesendoderm, mesoderm and Wnt signaling, resulting in significantly lower levels of cardiomyocyte differentiation.
Our observation that knockdown of GATA6-AS1 impairs the expression of GATA6 and GATA6-target genes are consistent with the roles of lncRNAs in gene regulation. GATA6-AS1 epigenetically regulates endothelial gene expression and cellular function; however silencing of GATA6-AS1 did not affect GATA6 expression suggesting a non-functional role of cis-regulatory element in HUVECs in hypoxic condition (29). Down-regulation of a number of other lncRNAs have been reported to result in the downregulation of protein-coding genes adjacent to these lncRNAs (34). Specifically, these lncRNAs are coordinately transcribed from a unique locus together with their genomically adjacent protein-coding genes in the opposite direction (34, 35). These lncRNAs often exhibit expression profiles similar to their protein-coding partners, which could be controlled by bidirectional promoters which initiate the expression of two transcripts in close proximity but in opposite directions (32, 36). Such a coordinated expression pattern is observed in our study for GATA6-AS1 and GATA6 in human tissues and cardiomyocyte differentiation of hPSCs. However, the expression pattern of GATA6 and adjacent lncRNAs is different between mice and human. In mice, lncRNA LL33 resides next to the GATA6 locus exhibits a distinct expression pattern compared with GATA6 in the developing heart (13). LncRNA LL33 is expressed late in the developing heart after E18.5 whereas GATA6 is expressed in the heart at all stages of cardiac development (13). The difference between human GATA6-AS1 and mouse LL33 may reflect the species-specific role and regulation of lncRNAs, highlighting the need to investigate lncRNAs in a human system. Given that species differences are prominent for lncRNAs, studies on hPSC differentiation are useful to discover the expression and function of human-specific lncRNAs. Such studies are particularly significant since lncRNAs are involved in not only cardiac development but also pathophysiology of human heart diseases (37, 38).
The regulation of GATA6 by GATA6-AS1 as observed in our study also provides a rationale for the defect of cardiomyocyte differentiation in the GATA6-AS1 knockdown cells. In mice, GATA6 is both necessary and sufficient for regulating the cardiac differentiated gene expression (39–41). In humans, GATA6 mutations have also been identified in patients with congenital heart defects (19). In addition, our findings on the alteration in the expression of genes associated with the Wnt signaling caused by GATA6-AS1 knockdown are in line with the link of GATA transcription factors with the Wnt signaling. During heart development, GATA transcription factors integrate canonical and non-canonical Wnt signaling in regulating cardiogenesis (42). In mice, Wnt acts in a feedforward transcriptional loop with GATA6 to drive the expansion and differentiation of posterior cardiac progenitors (43). In hPSCs, cardiac induction can be accomplished through temporal activation and inhibition of the Wnt signaling (4), and the expression of the Wnt target and signaling genes is temporally regulated during the cardiovascular lineage commitment of hPSCs (44). Our study indicates that knockdown of GATA6-AS1 affects the expression of not only GATA6 but also the Wnt target and signaling genes, suggesting an important link between GATA6-AS1 with GATA6 and the Wnt signaling.
Our findings support that lncRNAs can be indispensable players for cardiac specification and differentiation (reviewed in (45)). In mice, a lncRNA, linc1405 mediates cardiac mesoderm specification and promotes cardiogenesis (46). Knockdown of lnc1405 significantly inhibited cardiac differentiation of mouse PSCs (46). A mouse lateral mesoderm-specific lncRNA Fendrr is essential for proper heart development and plays an essential role in fine-tuning the regulatory networks which control the fate of lateral mesoderm derivatives (47). Similarly, a mouse heart-specific lncRNA Braveheart is essential for the activation of cardiac transcription factors and further commitment of mouse PSCs to cardiomyocytes (17). In contrast, a human lncRNA Heartbrake is a negative regulator of cardiomyocyte differentiation and the overexpression of Heartbrake represses cardiomyocyte differentiation from hiPSCs by counteracting MIR1 (48).
In summary, our findings provide evidence for the involvement of lncRNA GATA6-AS1 in cardiomyocyte differentiation as well as potential insights in the molecular regulation of cardiogenesis by linking the expression of GATA6-AS1 with GATA6 and the Wnt signaling.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs. A. Elefanty, E. Stanley, and D. Elliot from the Murdoch Children’s Research Institute, Royal Children’s Hospital, Australia for kindly sharing NKX2–5-eGFP hESCs. We thank the flow cytometry core facility of the Emory+Children’s Pediatric Research Center, and S. Biliya from the core facilities at the Parker H. Petit Institute for Bioengineering and Bioscience at the Georgia Institute of Technology for the use of their shared equipment, services and expertise. This study was supported in part by CASIS (GA-2017–266), NIH/NIAAA (R21AA025723) and NIH/NHLBI (R01HL136345) and the Center for Pediatric Technology at Emory University/Georgia Institute of Technology.
ABBREVIATIONS
- GATA6-AS1
GATA6 antisense RNA 1
- eGFP
enhanced green fluorescent protein
- lncRNA
long noncoding RNA
- hiPSC
human induced pluripotent stem cell
- hPSC
human pluripotent stem cell
- MOI
multiplicity of infection
- PFA
paraformaldehyde
- qRT-PCR
quantitative RT-PCR
- RNA-seq
RNA-sequencing
- shRNA
short hairpin RNA
- SSC
saline-sodium citrate
Footnotes
SUPPLEMENTARY INFORMATION
Supplementary information includes 5 Supplementary Figures, 11 Supplementary Tables and 2 Supplementary Movies.
REFERENCES
- 1.Murry CE, and Keller G (2008) Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680 [DOI] [PubMed] [Google Scholar]
- 2.Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, and Vunjak-Novakovic G (2018) Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, and Murry CE (2007) Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 25, 1015–1024 [DOI] [PubMed] [Google Scholar]
- 4.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, and Palecek SP (2012) Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 109, E1848–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, and Wu JC (2014) Chemically defined generation of human cardiomyocytes. Nat Methods 11, 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marques AC, and Ponting CP (2009) Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol 10, R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilusz JE, Sunwoo H, and Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23, 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, and Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, and Guigo R (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mondal T, Rasmussen M, Pandey GK, Isaksson A, and Kanduri C (2010) Characterization of the RNA content of chromatin. Genome Res 20, 899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, and Mattick JS (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A 105, 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponjavic J, Oliver PL, Lunter G, and Ponting CP (2009) Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet 5, e1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM, and Morrisey EE (2014) Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev 28, 1363–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinger ME, Pang KC, Mercer TR, and Mattick JS (2008) Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 4, e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, and Young RA (2013) Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A 110, 2876–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Li Y, Lin B, Sheng Y, and Yang L (2017) HBL1 Is a Human Long Noncoding RNA that Modulates Cardiomyocyte Development from Pluripotent Stem Cells by Counteracting MIR1. Dev Cell 42, 333–348 e335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, and Boyer LA (2013) Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molkentin JD, Antos C, Mercer B, Taigen T, Miano JM, and Olson EN (2000) Direct activation of a GATA6 cardiac enhancer by Nkx2.5: evidence for a reinforcing regulatory network of Nkx2.5 and GATA transcription factors in the developing heart. Dev Biol 217, 301–309 [DOI] [PubMed] [Google Scholar]
- 19.Kodo K, Nishizawa T, Furutani M, Arai S, Yamamura E, Joo K, Takahashi T, Matsuoka R, and Yamagishi H (2009) GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc Natl Acad Sci U S A 106, 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, and Thomson JA (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 21.Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, Skelton RJ, Ward-van Oostwaard D, Lim SM, Khammy O, Li X, Hawes SM, Davis RP, Goulburn AL, Passier R, Prall OW, Haynes JM, Pouton CW, Kaye DM, Mummery CL, Elefanty AG, and Stanley EG (2011) NKX2–5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods 8, 1037–1040 [DOI] [PubMed] [Google Scholar]
- 22.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, and Carpenter MK (2001) Feeder-free growth of undifferentiated human embryonic stem cells. Nature Biotech. 19, 971–974 [DOI] [PubMed] [Google Scholar]
- 23.Jha R, Wu Q, Singh M, Preininger MK, Han P, Ding G, Cho HC, Jo H, Maher KO, Wagner MB, and Xu C (2016) Simulated Microgravity and 3D Culture Enhance Induction, Viability, Proliferation and Differentiation of Cardiac Progenitors from Human Pluripotent Stem Cells. Sci Rep 6, 30956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Development Core Team. (2014) R: A Language and Environment for Statistical Computing., R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 25.Anders S, and Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, and Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, and Hochberg Y (1995) Controlling False Discovery Rate: A practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society B 57, 289–300 [Google Scholar]
- 28.Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann P, Jae N, Knau A, Glaser SF, Fouani Y, Rossbach O, Kruger M, John D, Bindereif A, Grote P, Boon RA, and Dimmeler S (2018) The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat Commun 9, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q, Jiang C, Xu J, Zhao MT, Van Bortle K, Cheng X, Wang G, Chang HY, Wu JC, and Snyder MP (2017) Genome-Wide Temporal Profiling of Transcriptome and Open Chromatin of Early Cardiomyocyte Differentiation Derived From hiPSCs and hESCs. Circ Res 121, 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, Sarkar MK, Li B, Ding J, Voorhees JJ, Kang HM, Nair RP, Chinnaiyan AM, Abecasis GR, and Elder JT (2015) Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol 16, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, and Myers RM (2004) An abundance of bidirectional promoters in the human genome. Genome Res 14, 62–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Britanova O, Akopov S, Lukyanov S, Gruss P, and Tarabykin V (2005) Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci 21, 658–668 [DOI] [PubMed] [Google Scholar]
- 34.Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, Shao W, Lu Z, Li H, Na J, Tang F, Wang J, Zhang YE, and Shen X (2016) Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell 18, 637–652 [DOI] [PubMed] [Google Scholar]
- 35.Frank S, Ahuja G, Bartsch D, Russ N, Yao W, Kuo JC, Derks JP, Akhade VS, Kargapolova Y, Georgomanolis T, Messling JE, Gramm M, Brant L, Rehimi R, Vargas NE, Kuroczik A, Yang TP, Sahito RGA, Franzen J, Hescheler J, Sachinidis A, Peifer M, Rada-Iglesias A, Kanduri M, Costa IG, Kanduri C, Papantonis A, and Kurian L (2019) yylncT Defines a Class of Divergently Transcribed lncRNAs and Safeguards the T-mediated Mesodermal Commitment of Human PSCs. Cell Stem Cell 24, 318–327 e318 [DOI] [PubMed] [Google Scholar]
- 36.Engstrom PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, Brozzi A, Luzi L, Tan SL, Yang L, Kunarso G, Ng EL, Batalov S, Wahlestedt C, Kai C, Kawai J, Carninci P, Hayashizaki Y, Wells C, Bajic VB, Orlando V, Reid JF, Lenhard B, and Lipovich L (2006) Complex Loci in human and mouse genomes. PLoS Genet 2, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono K, Kuwabara Y, Horie T, and Kimura T (2018) Long Non-Coding RNAs as Key Regulators of Cardiovascular Diseases. Circ J 82, 1231–1236 [DOI] [PubMed] [Google Scholar]
- 38.Schonrock N, Harvey RP, and Mattick JS (2012) Long noncoding RNAs in cardiac development and pathophysiology. Circ Res 111, 1349–1362 [DOI] [PubMed] [Google Scholar]
- 39.Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, and Duncan SA (2005) GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol 25, 2622–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, and Duncan SA (2008) Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol 317, 614–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Berlo JH, Elrod JW, van den Hoogenhof MM, York AJ, Aronow BJ, Duncan SA, and Molkentin JD (2010) The transcription factor GATA-6 regulates pathological cardiac hypertrophy. Circ Res 107, 1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afouda BA, Martin J, Liu F, Ciau-Uitz A, Patient R, and Hoppler S (2008) GATA transcription factors integrate Wnt signalling during heart development. Development 135, 3185–3190 [DOI] [PubMed] [Google Scholar]
- 43.Tian Y, Yuan L, Goss AM, Wang T, Yang J, Lepore JJ, Zhou D, Schwartz RJ, Patel V, Cohen ED, and Morrisey EE (2010) Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Dev Cell 18, 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jha R, Singh M, Wu Q, Gentillon C, Preininger MK, and Xu C (2017) Downregulation of LGR5 Expression Inhibits Cardiomyocyte Differentiation and Potentiates Endothelial Differentiation from Human Pluripotent Stem Cells. Stem Cell Reports 9, 513–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ounzain S, Burdet F, Ibberson M, and Pedrazzini T (2015) Discovery and functional characterization of cardiovascular long noncoding RNAs. J Mol Cell Cardiol 89, 17–26 [DOI] [PubMed] [Google Scholar]
- 46.Guo X, Xu Y, Wang Z, Wu Y, Chen J, Wang G, Lu C, Jia W, Xi J, Zhu S, Jiapaer Z, Wan X, Liu Z, Gao S, and Kang J (2018) A Linc1405/Eomes Complex Promotes Cardiac Mesoderm Specification and Cardiogenesis. Cell Stem Cell 22, 893–908 e896 [DOI] [PubMed] [Google Scholar]
- 47.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, and Herrmann BG (2013) The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24, 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Li Y, Lin B, Sheng Y, and Yang L (2017) HBL1 Is a Human Long Noncoding RNA that Modulates Cardiomyocyte Development from Pluripotent Stem Cells by Counteracting MIR1. Dev Cell 43, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.