Abstract
The ventral hippocampus (vHPC) is a critical hub in networks that process emotional information. While recent studies have indicated that ventral CA1 (vCA1) projection neurons are functionally dissociable, the basic principles of how the inputs and outputs of vCA1 are organized remain unclear. Here we used viral and sequencing approaches to define the logic of the extended vCA1 circuit. Using high-throughput sequencing of genetically barcoded neurons (MAPseq) to map the axonal projections of thousands of vCA1 neurons, we identify a population of neurons that simultaneously broadcast information to multiple areas known to regulate the stress axis and approach/avoidance behavior. Through molecular profiling and viral input-output tracing of vCA1 projection neurons, we show how neurons with distinct projection targets may differ in their inputs and transcriptional signatures. These studies reveal novel organizational principles of the vHPC that may underlie its functional heterogeneity.
Introduction
Over the last decade, the ventral portion of the hippocampus (vHPC) has been increasingly recognized as a critical node in the extended limbic circuitry that controls motivated and emotional behavior.1 The nature of the region’s functions – e.g. controlling anxiety-related behaviors2–7, influencing reward seeking behavior2,8–10, and modulating the neuroendocrine response to stress11– have stoked considerable interest in vHPC’s potential contributions to mood and anxiety disorders.
The vHPC is highly functionally and anatomically heterogenous and sends outputs to numerous target regions. Studies using retrograde tracing approaches indicate that vHPC projection neurons are largely segregated, with only a small fraction of cells projecting to multiple downstream targets.4,12–15 This has led the hypothesis that divergent outputs from vHPC encode different kinds of stimuli and differentially contribute to distinct aspects of behavior. 2,4,13,15 For example, it has recently been shown that vCA1-lateral hypothalamus (LH) and vCA1-medial prefrontal cortex (mPFC) projecting neurons modulate anxiety-related behavior, 4,6,13,16 whereas vCA1- amygdala projections contribute to contextual fear encoding and renewal,15 and vCA1-nucleus accumbens (NAc) neurons encode reward-related information.2,8,9 In addition, cells that project to more than one region have also been suggested to have specialized roles. In rats, cells with collateral projections to NAc, mPFC and amygdala are, for example, most active during sharp wave ripples in a task dependent manner, and those projecting to both infralimbic and prelimbic regions of mPFC and those projecting to amygdala and mPFC are implicated in fear renewal.2,14,17
But while heterogeneity has been well-documented at the functional level, the basic principles of how inputs and outputs of the vCA1 are organized remain unclear. Here, we use a combination of genetic and anatomical tools to address the heterogeneity of vCA1 by asking three questions: 1) What proportion of individual vHPC neurons project to single vs multiple downstream targets? 2) Do neurons that project to different downstream targets have distinct presynaptic inputs? And 3) are vHPC neurons that project to different downstream areas transcriptionally distinct?
Results
MAPseq for high-throughput axonal tracing of vCA1 neurons
We first sought to understand the diversity of projection patterns of individual vHPC neurons, so as to extend earlier work that relied on either retrograde axonal tracing or single neuron tracing/reconstruction to map vHPC outputs.4,12–15,18,19 These studies were limited, respectively, by the number of fluorescent marker colors available to retrogradely label projection neurons , and the low-throughput nature of single neuron tracing/reconstruction. We instead performed high-throughput single neuron tracing using multiplexed analysis of projections by high throughput sequencing (MAPseq).20,21
We used viral vectors to label neurons in vCA1 with a library of random RNA sequences (barcodes) and allowed them to transport into the axonal processes, such that each neuron is expected to take up a unique but random barcode (see Methods and as previously published20,21). Then, we dissected seven known target regions19,22,23 and sequenced the barcodes present to determine which neurons projected to which region (Figure 1A). The relative abundance of each individual barcode was used as a proxy for projection strength (density of axons in the tissue) to that target. 20,21 Target regions were chosen for their putative roles in vHPC-mediated control of emotional behavior: mPFC2,6, NAc8,9, LH4, lateral septum (LS)13, bed nucleus of stria terminalis (BNST)24, basal amygdala (BA)4,12,15, and central amygdala (CeA)15 (Figure 1A,B).
Figure 1. High-throughput mapping of vCA1 projections using MAPseq.
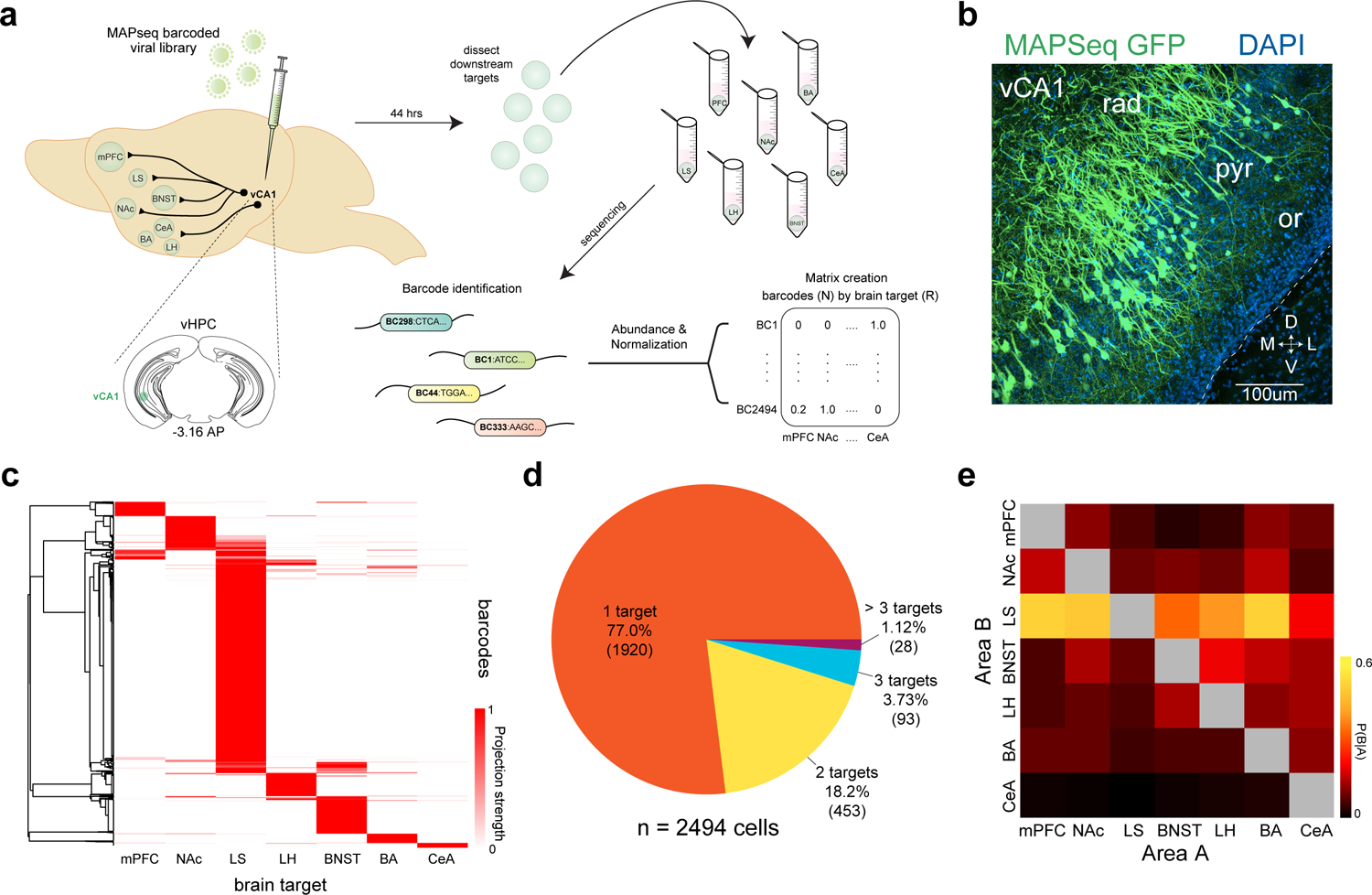
a. Experimental design. We mapped the projections from vCA1 to 7 downstream targets using MAPseq. Briefly, we injected a virus encoding random RNA sequences (barcodes) into vCA1 and allowed them to transport into the axonal processes. Then, we dissected seven known target regions and sequenced the barcodes present to determine which neurons projected to which region. The relative abundance of a barcode present in each area was used as a proxy for the relative projection strength, and an N X R matrix was generated for analysis (N-barcodes, R-brain regions). See Methods for details on each step of the experimental pipeline. b. expression of MAPseq GFP at the injection site in vCA1, representative from n=4 mice. c. Heatmap of projection strengths (from N X R matrix) of all 2494 vCA1 neurons (from 12 mice) to 7 targets mapped with MAPseq. Each row is an individual neuron’s projection strength to each of the 7 target regions, normalized to the maximal value in that row, resulting in a projection strength scale from 0 to 1 (see Methods). d. Distribution of number of projection targets of vCA1 neurons mapped. e. Heat map of conditional probability for two regions (see Methods), the proportion of cells projecting to area B within the subset of cells that project to area A.
We analyzed the axonal projection patterns of 2494 vCA1 neurons from 12 mice to the seven downstream target regions. We found that ~77% (1920/2494) of barcoded vHPC neurons projected to only one of the assayed target regions. And of the ~23% that projected to multiple targets, ~18.2% (453/2494) sent axons to two targets, ~3.7% (93/2494) to three targets, and ~1.1% (28/2494) to greater than three of the assayed targets (Figure 1C–D). Analysis of the conditional probability for two regions getting input from the same neurons suggested that BNST and LS were most likely to get input from cells targeting multiple regions (Figure 1E). To further explore the targeting patterns of neurons projecting to multiple downstream areas, we determined whether there were projection motifs that were over- or under- represented within our population of barcoded neurons. Here, we define a projection motif as a projection pattern shared by group of cells, regardless of the strength of each cell’s projection to any given target (all projection motifs in Supplementary Data Table 1). For example, a BNST-BA projection motif includes cells that only project to both the BNST and the BA, and not to any other assayed targets. We focused our analysis on those cells that projected to more than 1 target (Figure 2A). We compared our experimental data to a null model that assumes each neuron has no preferences among the possible motifs, and that its probability of projecting to one region is independent of its probability of projecting to another region (i.e. its projections are binomially distributed) (Figure 2B–C and see Supplementary Data Table 1 and Methods). We found 9 motifs were over-represented and 15 motifs that were under represented (Figure 2B–C). The most over-represented projection motifs were those cells that bifurcated to LS and NAc and those that bifurcated to LS and BNST. The most under-represented motifs were those that bifurcated to LH and BA and those that bifurcated to BA and CeA, which supported previous fluorescent retrograde tracing studies.4,15 For each projection motif, we plotted the projection strength (abundance of barcode sequences in a downstream target, which infers the axon density in the tissue21) for each cell, normalized by the cell’s maximum projection strength (Figure 2D–E and Extended Data Figure 1B). As in the heat map in Figure 1C, each line (different colors) is an individual neuron’s projection strength to each of the 7 target regions and is normalized to the maximal value in that row, resulting in a projection strength scale from 0 to 1. Within these motifs, neurons showed heterogeneity – as cells projected to each area with differing strengths (Figure 2D–E and Extended Data Figure 1B). These MAPseq data suggest significant heterogeneity among vCA1 projection neurons, as many vCA1 cells project to only one of the seven targets assayed and a significant proportion project to multiple targets in a non-random fashion.
Figure 2. vCA1 projection motifs.
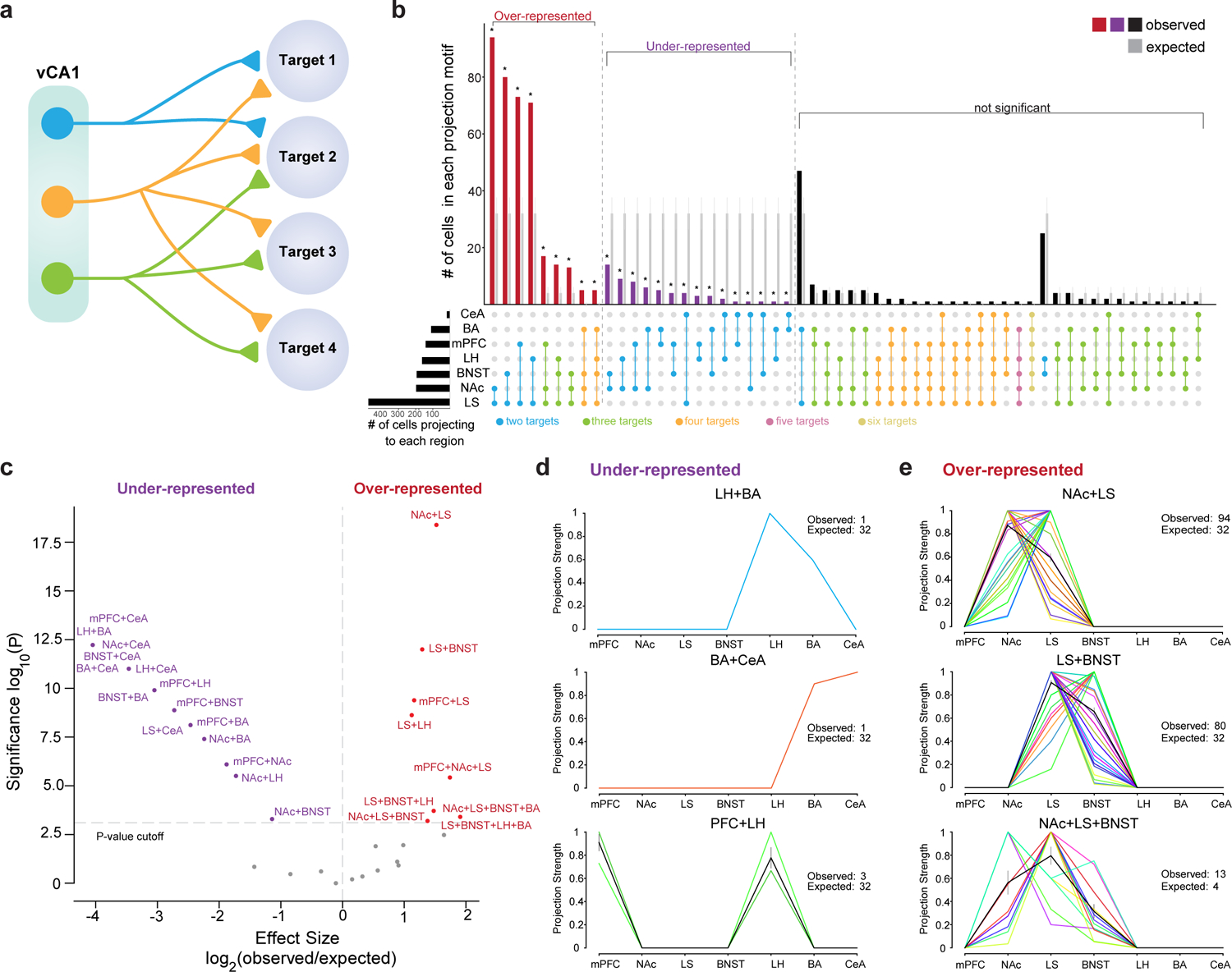
a. Schematic of different kinds of projection motifs, single target and multiple targets. A motif is defined as a projection pattern shared by group of cells. b. UpSet plot of the number of cells within each observed projection motif (red, purple and black bars represent observed motif numbers from n= 574 total cells with multiple targets, number of cells in each motif in Supplementary Data Table 1). We compared the observed motif number with expected from null model, grey bars represent expected counts, with the mean and standard deviation of the null binomial models, see Methods for details (binomial test, two-sided, p-value corrected for multiple comparisons using Bonferroni method, *Padj<0.05 after correction for multiple comparisons, P-values and effect sizes provided in Supplementary Data Table 1). c. Plot of statistical significance and effect size of over and under-represented projection motifs, see Methods for generation of the null model, (n= 574 cells, cell number in each motif, P-values and effect sizes provided in Supplementary Data Table 1.) d-e. Projection strengths of individual neurons within the under-represented group (d) and over-represented group (e). Each line (different color) is an individual neuron’s projection strength to each of the 7 target regions, normalized to the maximal value in that row, resulting in a projection strength scale from 0 to 1 (see Methods for normalization). Black line is mean projection strength for all neurons in that motif, and grey is SEM. Each motif is labeled above each graph. n = 1 LH+BA, 1 BA+CeA, 3 PFC+LH, 94 NAc+LS, 80 LS+BNST, 13 NAc+LS+BNST biologically independent cells, observed and expected numbers in inset of each panel, all other motifs in Extended data Figure 1.
Input-output viral tracing of vCA1 neurons that project to NAc, mPFC, LH, BA, adBNST, and LS
We next determined whether vHPC output neurons previously implicated in approach/avoidance behavior and fear learning receive long-range, extra-hippocampal inputs from similar or diverse upstream regions. Identifying vCA1 cells that extend axons to mPFC, NAc, LS, BNST, BA, LH – we catalogued these neurons’ patterns of long-range inputs using “tracing the relationship between input and output” (TRIO) method.25
Briefly, we injected a retrograde traveling Cre-recombinase (AAV2-retro-CAG-Cre26) into one of each of the six downstream target areas, and rabies helper virus into vCA1 (AAV1-synP-FLEX-TVA-P2A-EGFP-P2A-B19G27), followed three weeks later by RV-EnvA-∆G-mCherry into the same region of vCA1 (Figure 3A–C, Extended Data Figure 2F–G). We compared the sources of inputs to these six vCA1 output streams by brain-wide analysis of long-range, extra-hippocampal input neurons (Figure 4A, see Methods). Overall, these six groups of output neurons did not differ in their input patterns, as they received input from the same upstream areas (Figure 4A–B, Extended Data Figure 3A–E and Supplementary Data Table 2). However, there were a few differences in terms of the proportion of inputs from some areas to specific output neurons. First, while all projection neurons received thalamic input, most notably input from the paraventricular nucleus of the thalamus, TRIO suggested that vCA1-LH projection neurons receive proportionally more input from PVT than the projections to BA and mPFC (Figure 4C, Extended Data Figure 2A, Supplementary Data Table 2). Second, TRIO predicted that vCA1-BA projection neurons get proportionally more input from the basal forebrain, most notably from the nucleus of the diagonal band than vCA1-LS and vCA1-BNST projections (Figure 4C, Extended Data Figure 2E, Supplementary Data Table 2). And third, TRIO predicted that vCA1-BNST projection neurons get proportionally more input from the lateral amygdala that the other projections assayed (Figure 4C, Extended Data Figure 2B, Supplementary Data Table 2). These results suggest that these six vCA1 projection streams receive, in general, similar upstream inputs, but that there are a number of small biases in the strength of some of these inputs to some outputs.
Figure 3. Viral input-output tracing of vCA1.
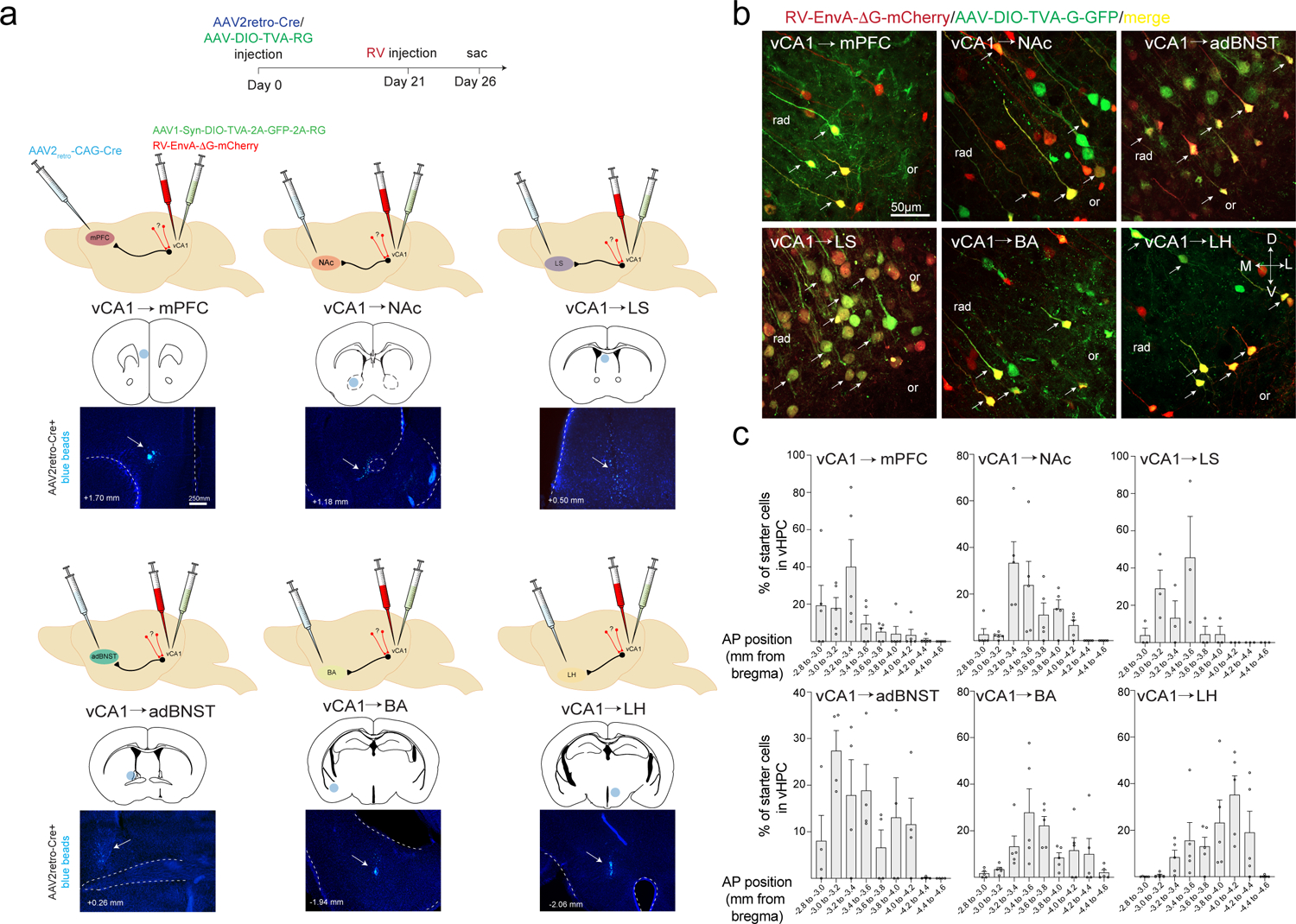
a. Schematics and timeline for input-output rabies tracing. We targeted six vCA1 outputs, the NAc, LH, BA, mPFC, adBNST and LS with injection of AAV2retro-Cre, (below each brain schematic is the injection sites labeled with blue beads, representative from n= n= 6 vCA1-mPFC, 8 vCA1-NAc, 9 vCA1-LH, 9 vCA1-BA, 4 vCA1-adBNST and 3 vCA1-LS mice,), and injected Cre-dependent rabies helper virus and EnvA-g deleted rabies virus expressing mCherry into vCA1 (see Methods for details). b. Starter cells defined by co-expression of GFP and mCherry in vCA1 in the 6 targeted output populations. Representative from n= 5 vCA1-BLA, 4 vCA1-adBNST, 3 vCA1-LS, 5 vCA1-mPFC, 5 vCA1-NAc and 5 vCA1-LH mice. c. Position of starter neurons counted along the AP axis of the vHPC for each projection population (n= 5 vCA1-BLA, 4 vCA1-adBNST, 3 vCA1-LS, 5 vCA1-mPFC, 5 vCA1-NAc and 5 vCA1-LH mice. Error bars represent SEM).
Figure 4. RV labeled inputs to vCA1 output neurons.
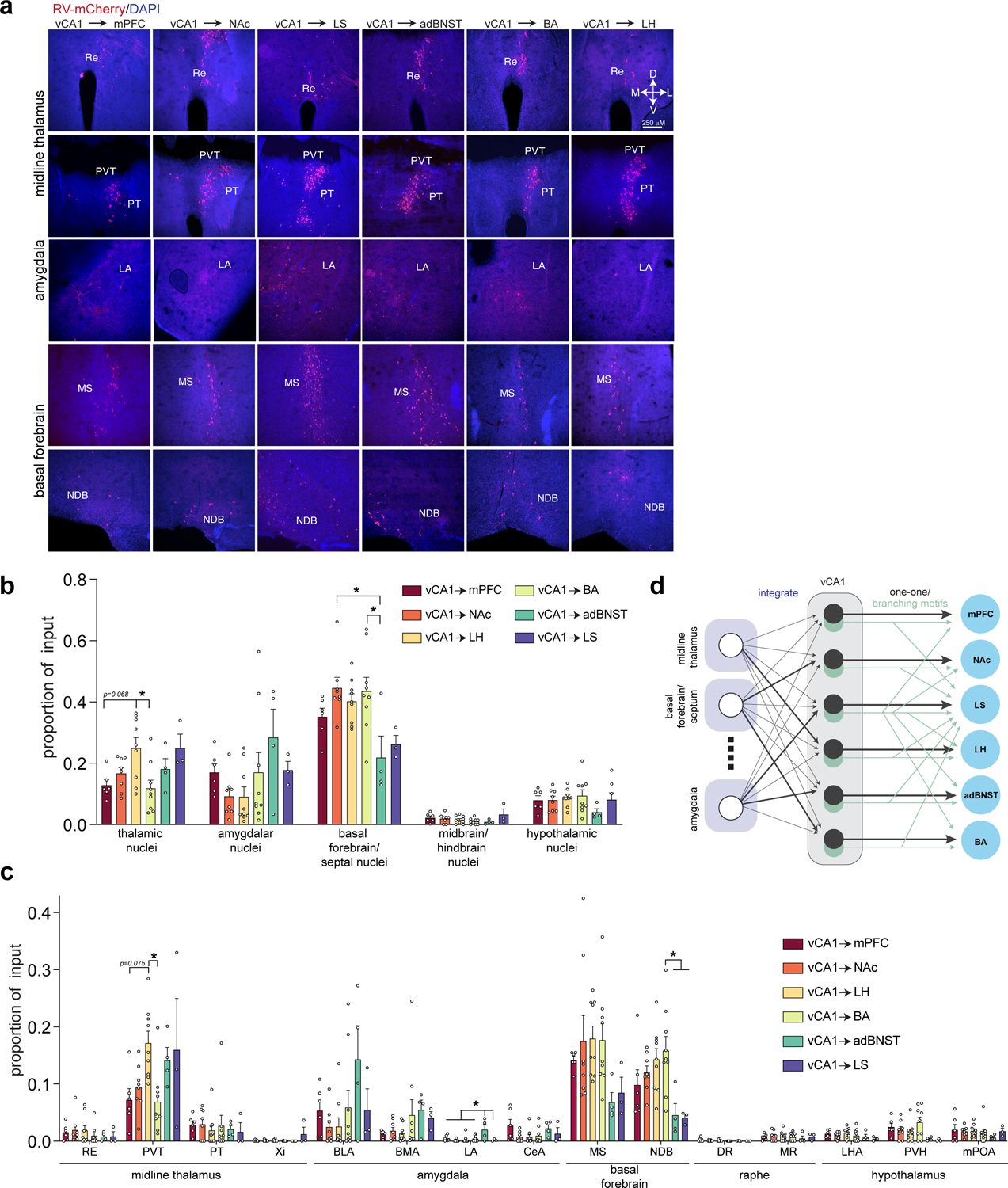
a. Representative images of extra-hippocampal long-range input neurons in the thalamus, amygdala and basal forebrain (from n= 6 vCA1-mPFC, 8 vCA1-NAc, 9 vCA1-LH, 9 vCA1-BA, 4 vCA1-adBNST and 3 vCA1-LS mice). b. Fraction of total extra-hippocampal input neurons found in thalamus, amygdala, basal forebrain, midbrain/hindbrain, and hypothalamus. One-way ANOVA, followed by post-hoc comparisons, with Holm-Sidak correction for multiple comparisons, all tests are two-sided. (*Padj<0.05 for post-hoc comparisons after correction for multiple comparisons. Thalamic nuclei: vCA1-LH vs. vCA1-BA, t33=3.652, Padj=0.0133, vCA1-mPFC vs. vCA1-LH t33=3.008, Padj=0.0679. Basal forebrain/septal nuclei: NAc vs. adBNST, t33= 3.649, Padj= 0.0134, BA vs. adBNST t33= 3.551, Padj= 0.0164). All statistical tests and P-values provided in Supplementary Data Table 2. c. Of the 5 areas in (b), we analyzed proportion of input to each projection population in 15 input areas using one-way ANOVAs, followed by post-hoc comparisons, with Holm-Sidak correction for multiple comparisons. All tests are two-sided. (*p<0.05 for post-hoc comparisons after correction for multiple comparisons. PVT: CA1-LH vs. vCA1-BA, t33= 3.421, Padj= 0.0249, vCA1-mPFC vs. vCA1-LH t33= 2.966, Padj= 0.0752. LA: mPFC vs. adBNST t33= 3.243, Padj= 0.0294, NAc vs. adBNST t33= 4.358, Padj= 0.0018 LH vs. adBNST t33= 4.013, Padj= 0.0045 BA vs. adBNST t33= 3.612, Padj= 0.0129, adBNST vs. LS t33= 3.320, Padj= 0.0261. NDB: BA vs. adBNST t33= 3.339, Padj= 0.0309 BA vs. LS t33= 3.133, Padj= 0.0495). See Supplementary Data Table 2 for proportion counts (mean +/− SEM) for assayed input regions, statistical tests and p-values. Error bars represent SEM. Abbreviations: RE-nucleus of reuniens, PVT-paraventricular nucleus of the thalamus, PT-parataenial nucleus, Xi-xiphoid thalamic nucleus, BLA-basolateral amygdala, BMA-basomedial amygdala, LA-lateral amygdala, CeA-central amygdala, MS-medial septum, NDB-diagonal band nucleus, DR-dorsal raphe nucleus, MR-median raphe, LHA-lateral hypothalamic area, PVH-paraventricular nucleus of hypothalamus, mPOA-medial preoptic area d. Schematic of input-output connection patterns in vCA1. vCA1 largely integrates input from upstream areas, with some biases in the proportion of input (darker lines) from thalamus, amygdala and basal forebrain. Then, this information is output to the 6 targets by cells that either form one-one connections (in black) with a given target, or those that branch to multiple downstream areas (in light green). See Figure 2 and Supplementary Data Table 1 for different branching motifs.
Molecular profiling of vCA1 neurons that project to NAc, mPFC, LH and BA
Finally, we determined whether vCA1 neurons projecting to four target areas, LH, NAc, BA and mPFC, differ in terms of the genes they express. To do so, we used transcriptional profiling methods that accessed translating mRNAs in vHPC neurons defined by their projection to one of these four targets. We labeled vHPC neurons that projected to mPFC, NAc, BA, and LH in different cohorts of Rosa26FsTRAP mice (using the same AAV2retro-Cre approach as with TRIO above) then used a translating ribosome affinity purification (TRAP) approach to profile translation in each of these populations (Figure 5A)28 . RNA sequencing (RNAseq) was then performed to identify differentially enriched mRNAs in the distinct vHPC projection neurons (Figure 5A, and Supplementary Data Tables 3–5).
Figure 5. Transcriptional profiling of vCA1 neurons defined by connectivity.
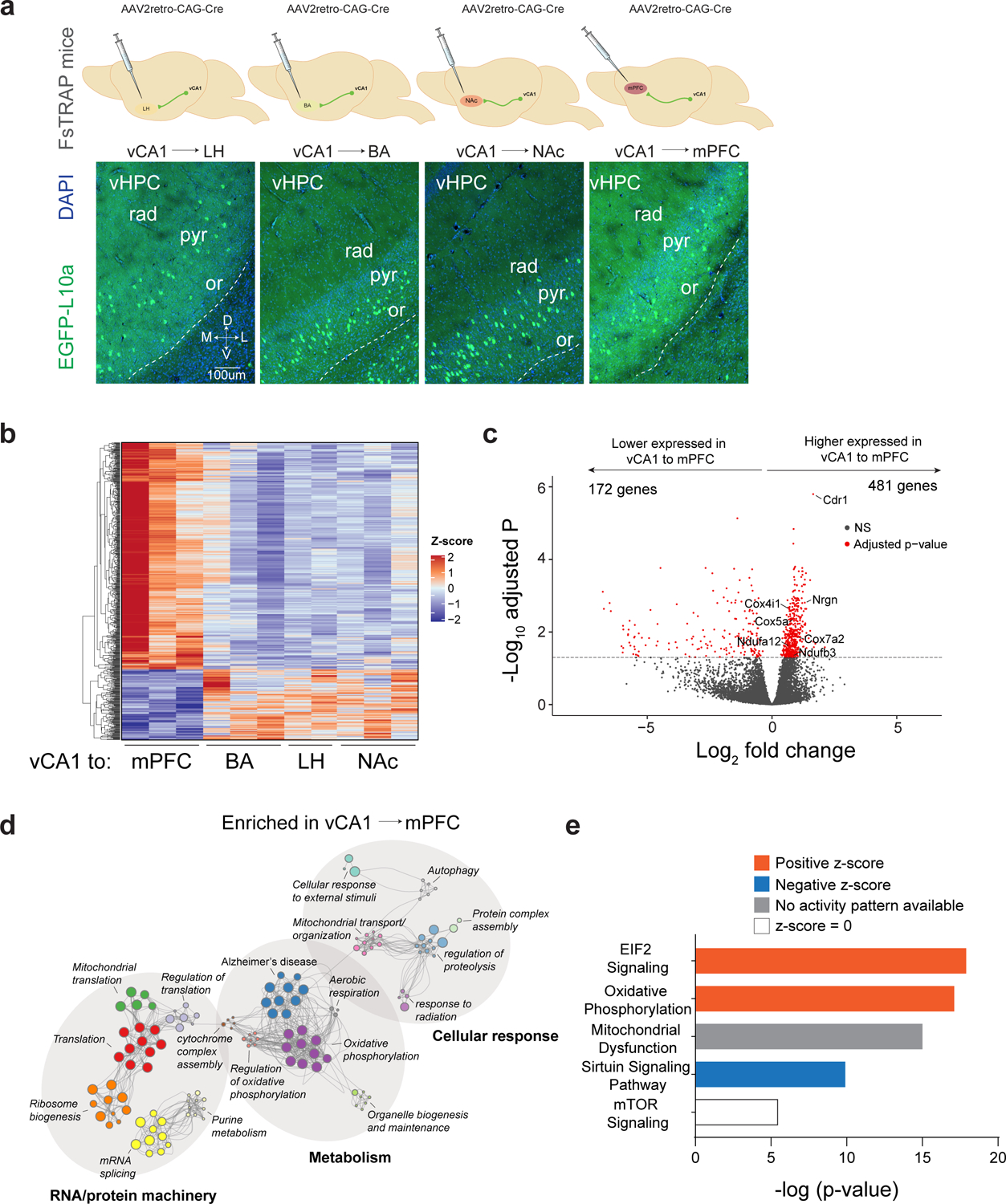
a. Experimental design. Rosa26fsTRAP mice were injected with AAV2retro-Cre into either the NAc, LH, BA or mPFC. Below. Expression of EGFP in vCA1 in mice injected in each pathway, representative from n=3 mice per pathway. vHPC was dissected and GFP IPs were performed for analysis of differential expression using RNAseq. (n=6 pooled ventral hippocampi for LH performed in duplicate, n=6 ventral hippocampi for BLA performed in triplicate, n=5 pooled ventral hippocampi for NAc performed in triplicate, n=6 ventral hippocampi for mPFC performed in triplicate. b. Heatmap of the differentially expressed genes from vCA1-mPFC vs vCA1-subcortical regions (significant genes pass threshold of adjusted p-value < 0.05 as determined by DESeq2; p-value acquired by Wald test, two-sided, corrected for multiple testing with Benjamini-Hochberg method, see Methods and Supplementary Data Table 4). c. Volcano plot depicting differentially expressed genes between vCA1-mPFC vs vCA1-subcortical regions (red dots; genes passing threshold of adjusted p-value < 0.05 as in b). All genes and p-values in Supplementary Data Table 4. d. Metascape enrichment network plot of genes enriched in mPFC relative to subcortical regions (genes passing threshold of adjusted p-value < 0.05 as in b; enriched ontology terms were determined with hypergeometric test, two-sided, and Benjamini-Hochberg multiple testing correction in Metascape, see Extended Table 5). e. Ingenuity Pathway Analysis (IPA) of canonical pathways for vCA1-mPFC vs. pooled vCA1-subcortical targets (genes passing threshold of adjusted p-value < 0.05 as in b; top 5 enriched pathways as determined by IPA with p-value < 0.05, see Supplementary Data Table 5). Colors indicate pathway activity (EIF2-eukaryotic Initiation Factor 2, mTOR- mammalian target of rapamycin).
Pairwise comparisons of vCA1 projections between each vCA1 projection neuron type revealed 24 genes that were significantly differentially expressed (Extended data Figure 3A and Supplementary Data Table 4). Interestingly however, these comparisons revealed a unique transcriptional profile in vCA1-mPFC projections relative to other subcortical projections (Figure 5B, Extended Data Figure 3A–B, Supplementary Data Table 4). Thus, we performed analysis of vCA1-mPFC vs. vCA1-subcortical regions, and found 653 genes that were differentially expressed (pAdj<0.05; Figure 5C, Supplementary Data Table 4). Gene ontology analysis of 481 mPFC-enriched genes revealed strong enrichment for metabolic genes, particularly genes involved in oxidative phosphorylation, that have also been implicated in neurodegenerative diseases such as Alzheimer’s Disease, Huntington’s Disease, and Parkinson’s Disease (Figure 5D–E and Supplementary Data Table 5).
To further validate the enriched genes identified in our dataset we performed in situ hybridization to examine cerebellar degeneration-related antigen 1 (Cdr1) which was one of the top genes enriched in vCA1-mPFC neurons (Extended Data Figure 3C). Quantification of Cdr1 transcript in vCA1 of mice injected with a retrograde tracer (CTB-555) in the mPFC revealed the majority of vCA1-mPFC neurons expressed Cdr1. Notably, unsupervised clustering analysis suggested that this dataset was not able to resolve unique gene expression signatures for all of the vCA1 projection populations examined, thus, we cannot rule out potential transcriptomic differences amongst the subcortically projecting neurons (Extended Data Figure 3D–E). In summary, our transcriptomic data highlight a clear gene expression signature that distinguishes mPFC projecting vHPC neurons from subcortical projecting neurons.
Discussion
Together, these data enrich our understanding of the organization of the vCA1, showing how neurons there are differentially connected to upstream inputs, how activity there is disseminated to downstream areas and how neurons display subtly different molecular identities. We find that vCA1 projections show not only show a one-to-one connectivity with downstream areas, but that a significant fraction of neurons project to multiple downstream areas in a non-random fashion. Using rabies mediated input-output tracing of six vCA1 projection streams, we find that overall, these six groups of output neurons received a similar distribution of inputs from upstream subcortical areas, with a few differences in the proportion of those inputs. Finally, analysis of translating mRNAs found a gene expression signature that differentiates vCA1-mPFC projectors from those neurons that project to LH, BA and NAc.
Our MAPseq data describing the organization of vHPC outputs expands upon a number of previous studies using bulk axonal tracing, retrograde fluorescent tracers and single cell reconstruction to understand vCA14,14,15,17–19,22,23. While this work has provided considerable insight into the organization of vCA1, these techniques lack single cell specificity (in the case of bulk anterograde tracing), the number of colors available (in retrograde tracing), and single neuron reconstruction is labor intensive, low throughput and limited in the number of neurons reconstructed. With MAPseq we could scale up analysis of vCA1 in all these regards, by increasing the number of neurons and downstream targets that can be assayed simultaneously (seven targets and ~2500 neurons in this study) to perform statistical analysis on the fraction of neurons that project to more than one downstream target, allowing us to find some order in the organization of single neuron projection patterns. We have discovered two primary organizational principles, one that had been documented before, and one that is novel to this report. In line with previous studies of vHPC 4,12,14,15,17,29,30, we found a large fraction of cells project to one assayed area, while a smaller proportion project to multiple areas. In addition, our data presents the novel finding that those neurons that project to multiple areas do so in a non-random way (these projections are discussed in more detail below). We found that some projection patterns of single neurons were significantly more abundant than expected, while other targeting patterns were less abundant. This would indicate that there is some organization in vCA1 projection patterns, with some downstream areas receiving input from the same cells, while others receive input from largely distinct neurons. This non-random organization in broadcasting neurons has recently been described in the primary visual cortical outputs to higher order areas.20 However, it is important to note the limitations of MAPseq, namely the spatial resolution of the target dissection, and the lack of cell-type specific genetic information of the cells traced. Future studies using higher resolution dissection techniques and newer variants of sequencing-based tracing techniques (such as BARseq31) that provide cell type specificity will help in these regards.
Comparing our results to those using retrograde approaches in the vHPC finds a number of similarities, allowing us to both confirm and expand upon previous results. In the majority of these studies, two projection targets were assayed. For example, a recent study from our group analyzed vHPC neurons that projected to the LH and BA, and found, using retrograde tracers, that 3.4% of neurons sent projections to both targets.4 In line with this, our MAPseq data found 4.03% of neurons projecting to BA and LH target both areas (see Supplementary Data Table 1 for all motifs). Another recent study using retrograde tracing to analyze projections from vHPC (in ventral subiculum) to mPFC, LH and NAc found that 2% of neurons projected to both mPFC and LH, 5% to mPFC and NAc and 6% to NAc and LH.30 Again, this is strikingly similar to our MAPseq vCA1 data, where we found that 2.6% of neurons projected to mPFC/LH, 6.42% to mPFC/NAc and 4.22% to NAc/LH. We again found similarities comparing our amygdala results to a recent study using retrograde viral approaches that found 3% of vHPC neurons projected to both BA and CeA15 ; our MAPseq data found 2.3%. Finally comparing MAPseq data to a recent papers using HSV or CTB for mapping vHPC outputs to mPFC and BA that found ~9% of neurons projecting to both areas12,17, we found slightly less, 4.75% of cells projecting to both areas. In addition, a study using antidromic optogenetic stimulation for tagging vCA1 neurons found a small population that targeted mPFC, NAc and BA2, and again these projection cells were found in our dataset. One advantage of MAP-seq, is that it allowed us assay targeting of multiple downstream areas simultaneously. This allowed us to find that, in the neurons that projected to BA, mPFC, NAc and LH, the majority of cells that projected to two of these areas (as assayed by retrograde labeling in the studies above) also sent collaterals to at least one other area (see Supplementary Data Table 1 for counts of all projection motifs and relative enrichment of distinct motifs). Thus, understanding the heterogeneity vCA1 neuronal projection patterns and the logic of how they may be organized can aid in design of future functional experiments to further elucidate the function of vHPC.
Our dataset supported previous bulk anterograde tracing studies and single cell reconstruction studies indicating a dense projection from vCA1 to the LS.18,22,23 These studies suggested one of the main output routes of vCA1 neurons anterio-dorsally into the LS23, and one study traced neurons that bifurcated in the LS into the NAc and other association cortices18. Indeed, these projection motifs that included LS and at least one other target were found to be over-represented in our vCA1 dataset. Analysis of our MAPseq data found an overrepresentation of those vCA1 neurons that sent bifurcating projections to BNST and LS, LH and LS and trifurcating projections to all three of these downstream areas. From a functional perspective, this organization is intriguing, as modulation of distinct cell types in vCA1, LS, BNST and LH have all been shown to control distinct aspect of anxiety related behavior. For example, stimulation of the LS itself or cells in vHPC that project to the LS is anxiolytic, while inhibition of vHPC cells projecting to the LS is anxiogenic. 13,32 In addition, subpopulations of LS neurons are recruited by the open arms of the EPM and during periods of mobility in contexts previously associated with a footshock 32,33. In the adBNST, an area long known for its role in anxiety-related behavior 34, neurons have been found to be preferentially activated by safe areas, such as the closed arm of the EPM, while stimulation of distinct adBNST projections can reduce distinct anxiety-related features in mice (such as open arm time in the EPM, respiration rate and approach behaviors) 35. In addition, the BNST, LS and LH are all identified as potential relays between the vHPC and the PVH, the entry point to the hypothalamic-pituitary-adrenal axis through which the vHPC may modulate the neuroendocrine responses to stressful stimuli.36 Thus, studies aimed at understanding how information is encoded by these bifurcating and trifurcating vCA1 neurons, how they may modulate distinct features anxiety-related behavior, and how they may impact the HPA axis to control neuroendocrine responses to stressors will provide novel insight into the role of the vCA1 in the control of emotional behavior.
In addition, we found an overrepresentation of vCA1 neurons that project to the LS that bifurcate into either the mPFC or NAc. Recent studies shown that the vCA1 projection to mPFC modulates anxiety-related behavior and fear recall, while the vCA1 projection to NAc modulates reward seeking2,6,9,16,37. However, here we show a significant portion of these vCA1 neurons that project to NAc and mPFC also send a collateral to the LS. Targeting these bifurcating vCA1 neurons will provide insight into whether they have specialized functional roles that differ from vCA1 neurons with other targets, how they coordinate the activity in the LS, mPFC and NAc to regulate approach/avoidance decisions, and more generally, how stimuli of differing valences may be encoded within subpopulations of vCA1 neurons defined by their connectivity.
Our input-output tracing found that overall, vCA1 projection neurons receive similar upstream input, suggesting that vCA1 acts as a hub to integrate information and broadcast to multiple downstream areas (though in a non-random fashion as indicated by our MAPseq results, see Figure 4D). However, at the integration level, we found some subtle biases in the proportion of inputs to anatomically defined outputs. For example, vCA1-LH projections, when compared to vCA1-mPFC and vCA1-BA neurons, are differentially innervated by inputs from the PVT, while vCA1-BNST neurons receive more input from subregions of the amygdala. The PVT has been shown to be activated by numerous stimuli, including stressors, appetitive and aversive stimuli and changes in internal state38. However, what information the PVT transmits to vCA1, and how it may modulate the output of vCA1 generally, or vCA1-LH specifically, remains poorly understood. Recent studies have shown that the population of vCA1-LH neurons, when compared to vCA1-BA neurons, are enriched in neurons that encode features of anxiety-provoking environments such as the open arms of the elevated maze, or the center of an open field arena.4 Targeting the PVT-vCA1 connection will provide insight into how the PVT signal may be integrated at the level of the vCA1 to generate an output signal to drive avoidance-related behaviors. In addition, future studies will shed light onto how distinct vCA1 projection populations integrate short-range input (which was not assayed here), either from local inhibitory circuits, or intra-hippocampal excitatory afferents from CA3.
Finally, analysis of translating mRNAs revealed a number of genes that are differentially expressed in some vHPC neuronal subpopulations, with the vHPC-mPFC projection having a unique transcriptional profile. The dataset generated with the TRAP method used here did not resolve a unique gene expression signatures for each of the vCA1 projection populations, thus future studies employing alternative methods, such as single nucleus sequencing, may provide the resolution needed to distinguish between these subpopulations of pyramidal neurons. However, our analysis did find that when compared to subcortically projecting vCA1 neurons, vCA1-mPFC neurons were enriched for genes involved in metabolic and oxidative pathways. Future studies aimed at understanding the developmental trajectories, wiring properties or differential sensitivities to stress and damage will lend greater insight into the differences between vCA1-mPFC vs subcortically projecting neurons. In addition to Cdr1, we found that neurogranin (Nrgn) was one of the top genes enriched in vCA1-mPFC neurons. Nrgn has been associated with schizophrenia39, and its expression has been localized to the hippocampus, where it modulates synaptic plasticity, contextual and spatial learning and anxiety-related behavior.40–42 Future studies will shed light into how Nrgn levels vCA1-mPFC neurons are regulated by experience, and how Nrgn may differentially modulate information transfer between vCA1 to mPFC over other subcortical targets. These data not only provide future opportunities for targeting these classes of cells for visualization and manipulation, but by providing unique molecular signatures of cell-types with distinct functions, they may provide new avenues for testing new therapeutic targets.
METHODS
Mice
All procedures were conducted in accordance with the U.S. NIH Guide for the Care and Use of Laboratory Animals and the institutional Animal Care and Use Committees at UCSF. For rabies tracing and MAPseq experiments, male and female adult C57BL/6J mice were supplied by Jackson Laboratory and were used beginning at 8–12 weeks of age. Rosa26fsTRAP mice (JAX: 022367) were supplied by Jackson Laboratory and bred in house, with genotyping was performed as recommended by Jackson Laboratory via standard PCR of tail-derived genomic DNA. Mice were co-housed with littermates (2–5 per cage) in a temperature (22–24°C) and humidity (40–60%) controlled environment. Mice were maintained with unrestricted access to food and water on a 12-hour light/dark cycle, with tissue processed during the light phase. All subjects were randomly assigned to experimental conditions, with approximately equal numbers of male and female mice.
General Stereotaxic Surgical Procedures.
Animals were 8–12 weeks of age at time of viral injection surgery. Mice were anesthetized with 1.5% isoflurane with an oxygen flow rate of ~1 L / min, and head-fixed in a stereotactic frame (David Kopf, Tujunga, CA). Eyes were lubricated with an ophthalmic ointment, and body temperature was maintained at 34–37°C with a warm water re-circulator (Stryker, Kalamazoo, MI). Fur was shaved and incision site sterilized with isopropyl alcohol x3 and betadine solution x3 prior to beginning surgical procedures. Lidocaine HCl 2% solution was injected subcutaneously local to incision, and post-surgical analgesia was provided by meloxicam and slow-release buprenorphine. A craniotomy was made at injection site with a round 0.5 mm drill bit (David Kopf, Tujunga, CA). A Nanoject II syringe (Drummond Scientific, Broomall, PA) was used with a pulled glass pipette (tip width 20–30 microns) to inject viruses described below.
MAPseq
Sample Generation
Experiments were conducted as previously described 20,21. Adult male and female mice (n=12) were anesthetized and injected with MAPseq Sindbis viral barcode library (3×1010 GC/ml, diversity of 2×107 different barcode sequences, Cold Spring Harbor Laboratories) into the vCA1 region of vHPC at −3.20mm ML, −3.16mm AP, and −3.70 & −3.80mm DV from brain surface (with 96.6nL per DV site) for a total of 193.2nL virus.
The titer and volume of Sindbis virus used is similar to that previously published, in which single cell analysis indicated that on average each neuron expressed one barcode, with a small fraction expressing more than one. 21 However, as discussed at length previously20,21,43, if neurons express more than one barcode, this would not change the distribution of projection patterns of assayed neurons. While the total number of traced neurons would be overestimated, the relative abundance of each projection motif type and bulk connection strength would not be changed. In addition, using our known barcode diversity (2×107) and a range of estimated values infected cells (from lower bound of 4000, see Over and under-represented motifs below, to an upper bound of 200,000 which assumes that 50% of all pyramidal cells in the entire hemispheres were labeled44 (which is an overestimation, as our injections were localized to a small portion of vCA1), we predict that >99% of cells would be labeled uniquely, if fraction of unique labeled cells F=(1-(1/N))k−1 where N is the barcode diversity and k is the number of infected neurons21.
44–46 hours after surgery, mice were rapidly decapitated, and brains were flash frozen in a slurry of 2-methylbutane (Fisher Scientific) on dry ice and stored in −80ºC until processing. Brains were then embedded in O.C.T. (Fisher Scientific) and sectioned coronally on an HM525 cryostat (Fisher Scientific) with 100–200μm thickness of: target areas (PFC, NAc, LS, BNST, LH, BLA, and CeA), negative control area (DLS) and the source region of interest (vCA1) were mounted on SuperFrostPlus (Fisher Scientific) slides and kept on dry ice. Brain punches of target areas were collected (while avoiding fiber tracts) on dry ice with a chilled 500μm puncher (Electron Microscopy Instruments), which was cleaned between each brain areas with 100% ethanol, and samples were stored in 1.5ml microcentrifuge tubes from each mouse and kept on dry ice until processing. Each sample was then homogenized in 400μL of TRIzol (Thermofisher) and vortexed followed by a quick spin then kept on dry ice before shipping to MAPseq Core Facility for barcode extraction and sequencing (Cold Spring Harbor Laboratories).20,21
Barcode extraction and sequencing was done previously described.20 In brief, total RNA was extracted from each sample using TRIzol reagent (Thermo Fisher) and sample RNA with spike-in RNA was mixed (obtained by in vitro transcription of a double-stranded ultramer with sequence 5′-GTCATGATCATAATACGACTCACTATAGGGGACGAGCTGTACAAGTAAACGCGTAATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNNNNNNNNNNNNNNNNNNATCAGTCATCGGAGCGGCCGCTACCTAATTGCCGTCGTGAGGTACGACCACCGCTAGCTGTACA-3′ (IDT)30) and reverse transcribed using a gene specific primer 5′-CTTGGCACCCGAGAATTCCANNNNNNNNNNNNxrefXXTGTACAGCTAGCGGTGGTCG-3′, where X is one of >300 true-seq-like sample-specific identifiers and N12 is the unique molecular identifier, and SuperscriptIV Reverse Transcriptase (Thermo Fisher, with manufacturer instructions). All first-strand cDNAs were pooled and purified using SPRI beads (Beckman Coulter) to produce double-stranded cDNA. Samples were treated with ExonucleaseI (NEB) and performed two rounds of nested PCR using primers 5′-CTGTACAAGTAAACGCGTAATG-3′ and 5′-CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ for the first PCR and primers 5′-AATGATACGGCGACCACCGA-3′ and 5′-CAAGCAGAAGACGGCATACGA-3′ for the second PCR using Accuprime Pfx polymerase (Thermo Fisher). Finally, PCR amplicons were gel extracted using Qiagen MinElute Gel extraction kit and library sequenced on a Illumina NextSeq500, set at high-output run and paired-end 36, using the SBS3T sequencing primer for paired-end 1 and the Illumina small RNA sequencing primer 2 for paired-end 2.
All preprocessing of sequencing data was performed at the MAPseq Core Facility at Cold Spring Harbor Laboratories exactly as described in Kebshull 2016, Supplementary note 421. All sequencing was done blinded to sample identity. Briefly, Illumina sequencing results (.fastq files) were merged into one file that contained paired end 1 (barcode sequence) and paired end 2 (the 12-nt unique molecular identifier (UMI) and 8-nt slice-specific identifier (SSI)), so that each line contained corresponded to a single read containing the 30-nt barcode, the 2-nt pyrimidine anchor (YY), the 12-nt UMI and the 8-nt SSI. Reads were de-multiplexed based on SSI using , fastx_barcode_splitter tool (http://hannonlab.cshl.edu/fastx_toolkit/commandline.html#fastx_barcode_splitter_usage) and filtered to remove ambiguous bases, and collapsed to unique sequences and sorted. Next, a threshold was selected for how many reads a sequence has to be used in analysis. As in Kebshull et. al 201621, a minimum read threshold was manually selected to remove the long tail of the sequence rank profile of the Illumina results so as to avoid contamination with PCR and sequencing errors. The remaining reads were collapsed after removal of the 12-nt of the UMI to convert reads into counts. Spike-in molecules, 24-nt barcodes followed by the sequence ATCAGTCA, were split from the virally expressed barcodes and processed separately. Error correction was performed using the short read aligner bowtie, which generated all possible alignments of barcodes sequences (>1 counts allowing up to 3 mismatches). All barcode sequences that mapped to each other were found, and low complexity sequences were removed by filtering barcodes with stretches of more than 6 identical nucleotides.
Raw barcodes reads were then normalized by relative number of spike-in RNAs and organized into an N x R matrix with N barcodes detected in R possible regions. The N barcodes is a proxy for the number of cells sending projections in the source region, in the text, we refer to each row as a cell. Each value in a row indicates the number (non-negative integer values) of detected barcodes in that region, which corresponds to the strength of the projection (density of axons) to that region. Matrices were concatenated for all mice to obtain a single larger matrix. In order to limit analysis only to cells that project to at least one region, we removed rows that contained all zeros. We threshold-filtered data based on presence of 10-fold enrichment of source (vCA1) barcode reads compared to at least one target area and removal of any barcode with a read count in the negative target area (DLS) prior to further analyses. Lastly, in order to compare projection patterns between neurons on the same scale, we normalize each row in the N x R matrix to that row’s maximum value, such that raw barcode counts are rescaled between zero and one.
MAPseq data analysis
Conditional probability
To calculate the conditional probability , for a pair of regions A and B, we first found the number of cells that project to region A, inclusive (i.e. cells that may project to A alone or A with projections to other regions as well), denoted . We then found the subset of cells in this group that projects to region B, inclusive (i.e. cells that may project to A and B, and may or may not have projections to additional regions), denoted . The conditional probability is the proportion of cells projecting to B within the subset of cells that project to A, i.e.
Over and under-represented motifs
In order to quantify the significantly over and under-represented projection motifs in our data, we developed a null model with which to compare. First, we note that our data is a matrix of neurons by target regions, and when analyzing projection motifs we first binarized this matrix to only consider whether or not a neuron projects to a particular target, regardless of its projection strength. This binarized matrix is then interpreted as a bipartite graph where the neurons form one node set and the target regions form the other node set and values of 1 in the matrix indicate an edge between nodes.45
We then constructed our null model as an Erdős–Rényi random (bipartite) graph where edge formation is determined by a constant probability following a binomial distribution43,46. This results in a null model where neurons are assumed to not have any intrinsic preference for projecting to particular target regions and where the probability of projecting to one region is conditionally independent from that of another region.
The probability of edge formation, denoted , determines the edge density of the null model, which we assumed is equal to our empirical data. In generating a random graph using , some simulated neurons will have zero projections to any of the target regions, however, due to the nature of MAPseq, the empirical data only includes neurons that have at least one projection to the target regions sampled. In order to properly model our data, we need to know the total number of neurons, including those that had no projections to one of our target regions. We can estimate this by noting that , where denotes the number of neurons in our empirical data and refers to the total number of neurons, such that would be the number of neurons with zero projections.20
First, we infer from the empirical data by assuming a binomial model and recalling that the probability of at least one projection is one minus the probability of no projections:
Where refers the probability of a neuron projecting to region among the total regions. We define , where is the number of neurons in the empirical data that project to region . By substitution we get:
We now have a polynomial with one unknown, , and we can solve this using any root solving algorithm. Knowing and , we can then solve for :
This is because the number of neurons with one at least projection using the null model with must reproduce the number of neurons in the empirical data which are also filtered to only include those with at least one projection. This equation is also a polynomial with one unknown, , and again we can solve it numerically using a root solving algorithm. In our data, , and we computed . Using and we can compute the expected counts for each motif, e.g. the expected counts for each single-projector motif under the null model is computed as , since there are 7 target regions. We then compute the two-sided p-values for our observed counts under the null binomial model using the binomial test.
k-means clustering
We performed k-means clustering on the normalized data matrix. We chose the k parameter (number of clusters) using the “elbow method,” (https://arxiv.org/abs/1205.1117) in which we plot the cluster inertia versus increasing trials of k from 1 to 15, and choose the k at which the inertia first begins to plateau. The inertia is the within-cluster sum of squared distances between each point in the cluster and its centroid. For the full set of 7 regions, we found to be near-optimal. Each centroid’s coordinates represent the representative (of the cluster) projection strengths for the corresponding region.
We plotted the centroids as a dot plot (i.e. grid-structured scatter plot) where each row is a centroid (cluster) and the color of the dots along each column correspond to the centroid’s coordinate value. Each coordinate value corresponds to a projection strength to a region, so a bright yellow dot in row 1 and column 2 represents a strong projection to that region for cluster 1. The k-means results demonstrate that the cells are well separated in the original high-dimensional space by the region to which they maximally project. All MAPseq data was analyzed using custom scripts written in Python (available at github.com/mkheirbek).
Input-Output Rabies Tracing.
Surgeries.
Male and female mice were unilaterally injected with AAV2retro-CAG-Cre (2.8 × 1012 vg/mL, UNC Vector Core) with 1:50 Fluoromax beads (ThermoFisher) targeted one of the six downstream projections from vHPC: mPFC, NAc, LH, adBNST, LS and BA. If the blue beads were not found in the target area, the animal was excluded from analysis. For the mPFC, 96.6nL were injected at 1.7mm AP, −0.3mm ML, −2.75mm DV, then another 64.4nL were injected at −2.5mm DV, NAc condition, 161nL were injected at 1.2mm AP, −1.1mm ML, −4.6mm DV, LH condition, 64.4nL were injected at each DV coordinate: −2.0mm AP, −0.75mm ML, −5.25mm DV, −5.0mm DV, and −4.75mm DV, and BA condition, 96.6nL were injected at −1.7mm & −2.0mm AP, −3.0mm ML, −5.5mm DV then another 64.4nL were injected at −5.25mm DV; adBNST condition, 128.8nL were injected at: 0.2mm AP, −1mm ML, −4.3mm DV, −4.15mm DV, −4.0mm DV; and LS condition, 128.8nL were injected at: 0.5mm AP, −0.35mm ML, −3.24mm DV, −3mm DV, −2.75mm DV. Followed by an injection of AAV1-synP-FLEX-TVA-P2A-EGFP-P2A-B19G (3.9 × 1012 vg/mL, UNC Vector Core) targeting the vCA1 region of vHPC with 64.4nL at −3.25mm ML, −3.14mm AP, −3.85mm DV, 64.4nL at −3.55mm DV, and finally 32.2nL at −3.35mm DV. Two to three weeks later, mice were then injected with 64.4nL of RV-EnvA-∆G-mCherry (3.79 × 108 vg/mL, Salk) into vHPC at 3.25mm ML, −3.14mm AP, −3.55mm DV.
Tissue Processing.
Five days after RV-EnvA-∆G-mCherry injection mice were deeply anesthetized with ketamine (80mg/kg) intraperitoneally and transcardially perfused with 0.01M PBS followed by 4% paraformaldehyde (PFA). Brains were extracted and post-fixed in 4% PFA overnight, followed by cryoprotectant with 30% sucrose for at least 24 hours. Brains were then serially sectioned at 40μm coronal sections with a Leica SM2000R freezing microtome then consecutive sections were mounted on Superfrost Plus slides (Fisher Scientific) and coverslipped with DAPI-Fluoromount-G Clear Mounting Media (Fisher Scientific).
Imaging
Whole-brain images were acquired of every section at 10X magnification with an Aperio Versa Slide Scanner (Leica Biosystems). Individual sections on each slide were isolated then registered to the Allen Brain Atlas using NeuroInfo software with Brainmaker module (MBF Bioscience) and all RV+ neurons outside of the hippocampus (with the exception of contralateral CA3)were mapped and counted. In ventral hippocampal sections, cells co-expressing mCherry and EGFP (starter cells) were imaged with a CSU-W1 spinning disk widefield confocal microscope (Nikon Imaging Center, UCSF) at 40X magnification, then registered to the Allen Brain Atlas using NeuroInfo (MBF Bioscience) and counted.
For each brain, the number of input neurons in a specific brain region was normalized to the total number of input neurons (RV+) counted across the brain outside of the hippocampus. All data collection and counts were made blinded to the experimental group. The mean and standard error of the mean are listed in Supplementary Data Table 3 for each brain region. For analysis one- way ANOVAs were run and those regions with P<0.05 were run with multiple t-tests with Holm-Sidak correction for multiple comparisons. All statistical results for significant effects are in Supplementary Data Table 4. Data was analyzed using Prism 8 (GraphPad).
Projection TRAP-RNAseq Profiling.
Brain Tissue Generation.
Adult FsTRAP heterozygous (B6.129S4-Gt(ROSA)26Sortm1(CAG-EGFP/Rpl10a,-birA)Wtp/J, Rosa26fsTRAP, stock #022367, Jackson Laboratory) mice were injected with AAV2retro-CAG-Cre (2.8 × 1012 vg/mL, UNC Vector core) with the same coordinates and parameters in mPFC, NAc, LH, and BA as described above in Input-output Rabies Tracing. Three to four weeks later, mice were rapidly decapitated and bilateral ventral hippocampi were microdissected on ice (by peeling away the hippocampus from cortex via the hippocampal fissure, then removing the dentate gyrus and CA3, and then dissecting the ventral portion of CA1/subiculum for analysis), and placed in ice-cold Buffer B (1xHBSS, 4mM NaHCO3, 2.5mM HEPES, 35mM Glucose) with 100mg/mL cycloheximide (Sigma Aldrich). Ventral hippocampi from male and female mice were pooled for each projection target (mPFC, NAc, LH, or BA) in duplicate or triplicate. Processing of the samples was as done as described in Ekstrand et al. 2014.28 Briefly, samples were homogenized via glass homogenizer (Kimble Kots 20) in 1.5mL of ice-cold Buffer C (10mM HEPES, 150mM KCl, 5mM MgCl2) with 0.5mM DTT (Sigma), 80 U/mL RNasin Plus (Promega), 40U/mL Superase-In (Life Technologies), 100 mg/mL cycloheximide, protease inhibitor cocktail (Roche) and 100ng/mL GFP-Trap Protein (ChromoTek), following with variable-speed homogenizer (Glas-Col) at 4ºC, three times at 250rpm and ten times at 750rpm. Samples were then transferred into new centrifuge tubes and centrifuged at 2,000 x g for 10 minutes at 4ºC, followed by addition of 140μL of 10% IGEPAL CA-630 (NP-40; Sigma Aldrich) and DHPC (100mg/0.69 mL; Avanti Polar Lipids). Solutions were then mixed before centrifugation at 17,000 x g for 15 minutes at 4ºC, and supernatant solution was transferred to new microcentrifuge tubes. Beads and GFP antibody were prepared by washing 300μL of Streptavidin MyOne T1 Dynabeads (Invitrogen) twice with PBS on a magnetic rack and loaded with biotinylated Protein L (Thermo Scientific) for 35 minutes at room temperature. Bead mixture was then blocked with five washes of 3% IgG-free and protease-free BSA (Jackson ImmunoResearch) in PBS and resuspended in Buffer A (10mM HEPES, 150mM KCl, 5mM MgCl2, 1% NP-40) with 100mg monocolonal anti-GFP antibody (Doyle et al., 2008) at room temperature. Finally, before use anti-GFP + bead mixture was washed twice in Buffer A with 0.5mM DTT, 80 U/mL RNasin Plus, and 100mg/ml cycloheximide then samples were added and incubated for 40 minutes at 4ºC. Sample mixture was washed three times with Buffer D (10mM HEPES, 350mM KCl, 5mM MgCl2, 1% NP-40), 0.5mM DTT, 80U/mL RNasin Plus, and 100mg/mL cycloheximide, then moved to a new tube before a fourth wash was performed. 100μL of Lysis Buffer was then added to each sample and purified with RNeasy Micro Kit (Qiagen) and stored at −80ºC before being sent for sequencing at the Functional Genomics Core at UCSF.
RNAseq and analysis.
RNA sequencing library was generated from SMART-seq.v4 Ultra Low Input RNA Kit (Takara Bio) and Nextera XT DNA Library preparation kit with multiplexing primers (Illumina) via manufactures’ protocols. All sequencing was done blinded to sample identity. Fragment size distribution and concentration were measured with 5200 Fragment Analyzer System and DNA high-sensitivity chip (Agilent). Libraries were multiplexed at 6 per flow-cell lane and sequenced on HiSeq-4000 to generate single end 50 base pair reads following manufacturer’s protocol (Illumina). Unique transcript reads were aligned to mouse genome (vGRCm38.78) with STAR v.2.5.2b49 using --quantMode GeneCounts, and soft clipping the 3’ adapter: GATCGGAAGAGCACACGTCTGAACTCCAGTCAC, with the following added settings: --outFilterMismatchNoverLmax = 0.04, outFilterMismatchNmax=999, alignSJDBoverhangMin=1, outFilterMultimapNmax=1, alignIntronMin=20, alignIntronMax=1000000, alignMatesGapMax= 1000000. Aligned reads were counted using HTseq with default parameters47,48.
Count normalization and differential gene expression analysis was performed using DESeq250 using default parameters as described in the vignette (http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html). All raw counts were used as input, no additional filtering was applied. See Supplementary Data Table 4 for all genes, normalized counts, fold changes, raw and corrected P-values. Genes were deemed significant when adjusted p-value < 0.05. Volcano plots for the different comparisons were made using EnhancedVolcano package (R package version 1.4.0, https://github.com/kevinblighe/EnhancedVolcano.) The ComplexHeatmap package49 was used to depict differentially expressed genes from mPFC vs subcortical regions (normalized counts from DESeq2 were scaled to −2 to +2). Gene ontology term enrichment analysis was run on genes higher expressed in mPFC vs subcortical regions using Metascape.50 In addition, all differentially expressed genes between mPFC vs subcortical regions were run through Ingenuity Pathway Analysis (Qiagen) to define activated/inhibited Canonical Pathways.
Clustering plots in Extended data figure 3 were plotted using the following R packages: PCA: prcomp v3.6.3 for calculating PCA, ggplot2 v3.3.0 for plotting, hierarchical clustering QC heatmap: pheatmap v1.0.12 for plotting and clustering.
RNAscope
Reagents were purchased from ACDBio. CDR1 probe was warmed for 10 minutes at 40C, and then cooled to room temperature. RNAScope Fluorescent Multiplex Reagents (Amp 1–4 FL) were equilibrated to room temperature. Fixed tissues were transferred briefly to RNAase-free dH20 and mounted onto slides. Hydrophobic barriers were drawn around each section. Slides were left to dry in the dark at room temperature for at least 30 minutes. Slides were then dipped rapidly in 100% EtOH and left to air dry for several minutes. Slides were transferred to a humidity tray. 2–4 drops of protease IV were added and sections were left to incubate for 30 minutes at room temperature. Afterwards, slides were dipped rapidly in a Coplin jar with RNAase-free dH2O. 4 drops of CDR1 probe were added to cover each section, and then slides were incubated for 2 hours at 40C in humidified tray. Slides were washed in 1X wash buffer twice for 2 minutes at room temperature. 4 drops of Amp 1-FL were added to cover each section, and the slides were then incubated for 30 min at 40C in a humidified tray. Slides were washed in 1X wash buffer twice for 2 minutes at room temperature. This process was repeated for Amp 2–4 FL, except incubation periods were 15 min, 30 min, and 15 min respectively. The color module chosen for Amp 4FL was Alt A-F(C2 in far red). After final wash of 1X wash buffer, slides were coverslipped with ProLong Gold Antifade Mountant. Slides were imaged using a Nikon Ti confocal, and counts were taken at 20X in a 1.3mm2 field of view (9 FOVs from 2 mice).
Statistical analysis
No statistical methods were used to predetermine sample size a priori, but the sample sizes used were similar to those reported in previous studies21,21,25,28. Data distribution was assumed to be normal, but this was not formally tested. For identification of over- and under-represented motifs from MAPseq data, we compared observed counts to expected from the null model using a two-sided binomial test with P-value corrected for multiple comparisons using Bonferroni method. P-values for all motifs provided in Supplementary Data Table 1. For input-output tracing, data was analyzed one-way ANOVAs, followed by post-hoc comparisons, with Holm-Sidak correction for multiple comparisons, all tests two-sided. For all dot plots, data are presented as the mean ± SEM. All statistical tests and P-values for rabies input-output tracing provided in Supplementary Data Table 2. For RNAseq data, differential gene expression analysis was performed using DESeq250 using parameters as described in the vignette (http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html). Briefly, P-value was acquired by Wald test, two-sided, corrected for multiple comparisins with Benjamini-Hochberg method. Prism 9 (GraphPad), Matlab (MathWorks) and Python (SciPy) were used for all data analyses.
Extended Data
Extended Data Fig. 1. Clustering MAPseq data and projection strengths of individual vCA1 neurons.
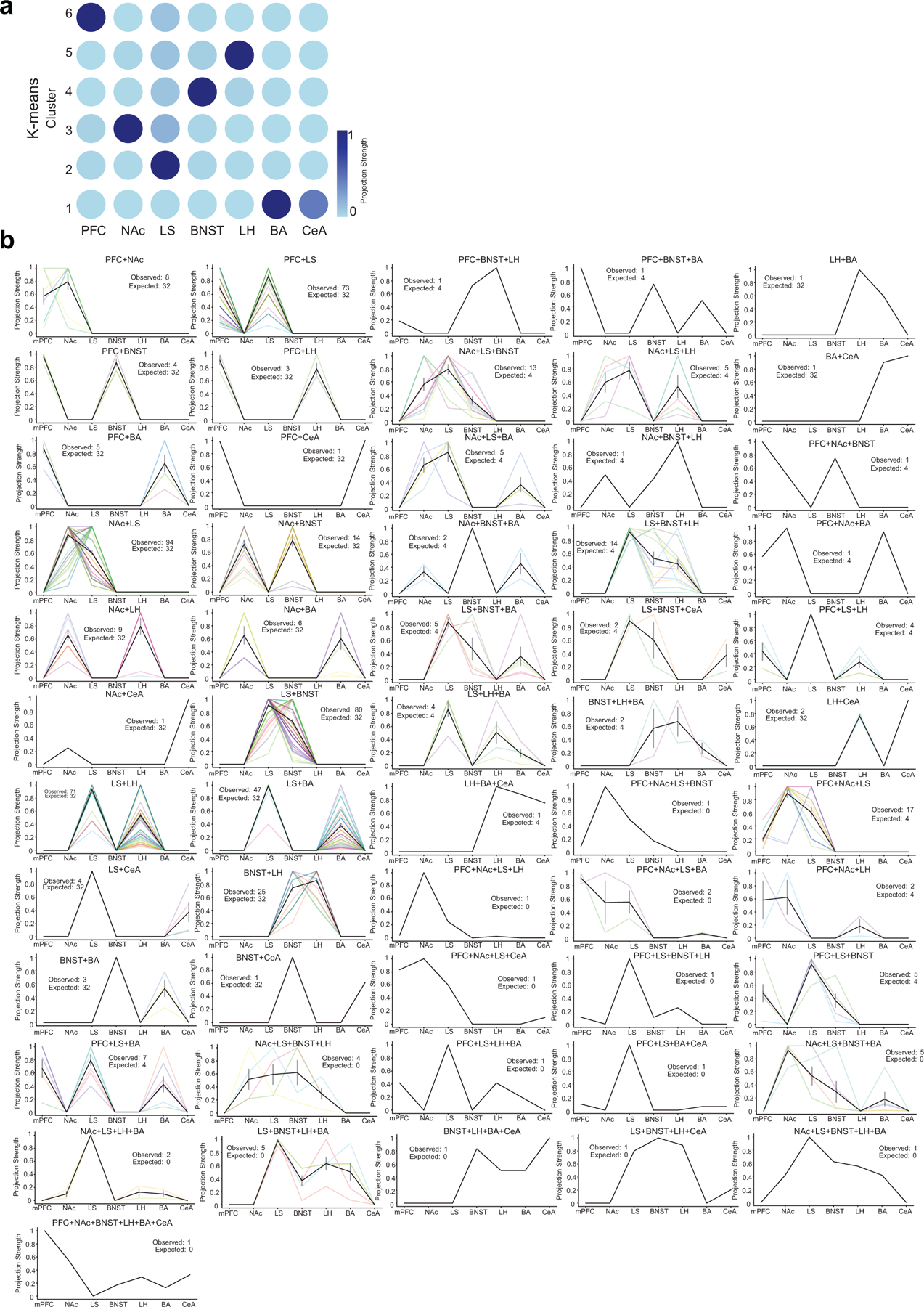
a. k-means clustering of MAPseq data. Clusters are plotted as a grid-structured scatter plot where each row is a cluster and the color of the dots along each column correspond to the normalized projection strength to a region b. Plots of normalized projection strength of all multiple projection barcode motifs from MAPseq data. As in Figure 2 each line (different color) is an individual neuron’s projection strength to each of the 7 target regions, normalized to the maximal value in that row, resulting in a projection strength scale from 0 to 1 (see Methods). Black line is mean projection strength for all neurons in that motif, and grey is SEM. Exact cell number for each motif is provided in the inset (observed and expected from null model, see Methods), and in Extended Data Table 2.
Extended Data Fig. 2. Brainwide extra-hippocampal input to vCA1 neurons that project to LH, BA, NAc, adBNST, LS and mPFC.
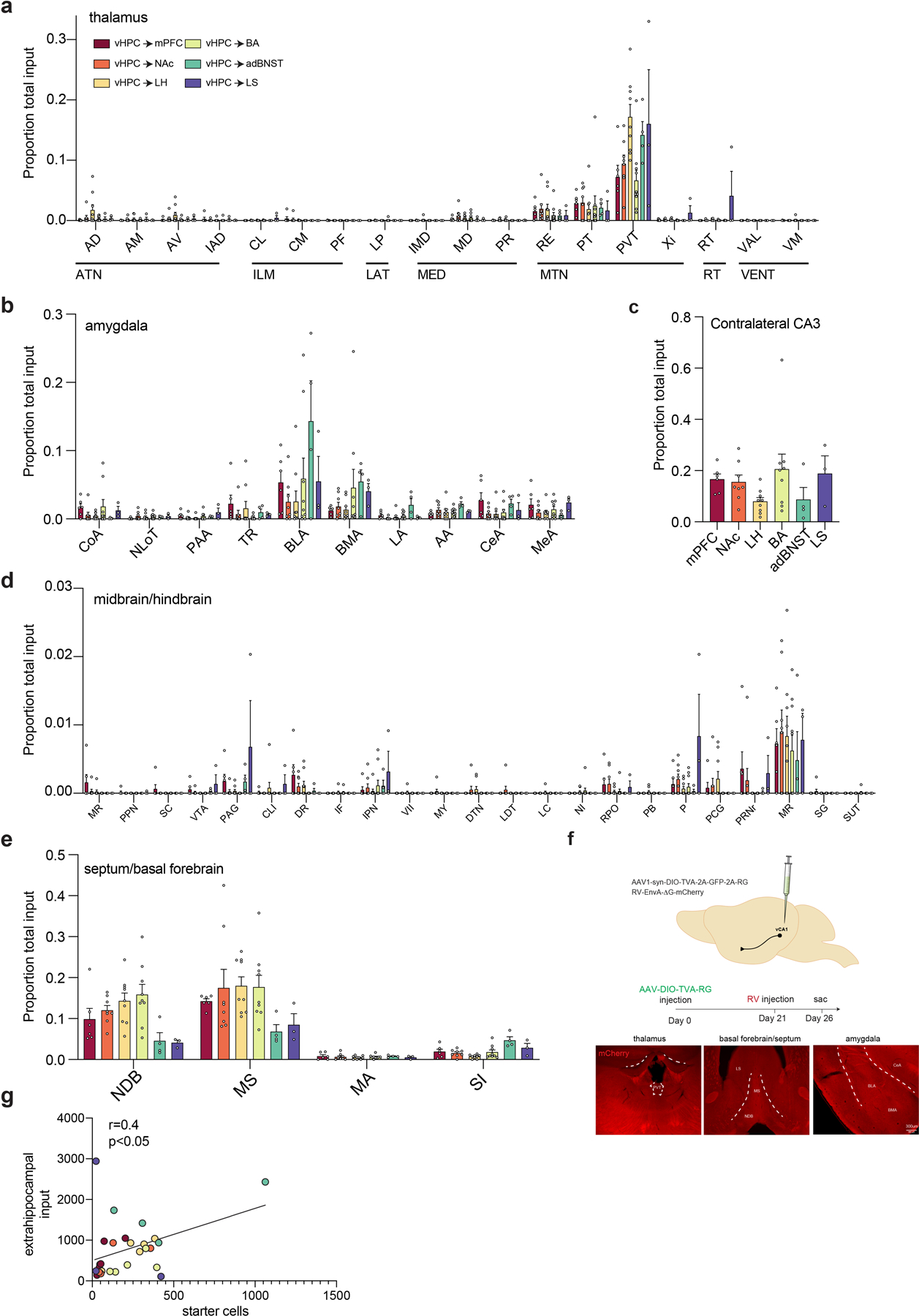
a–d. Fraction of extrahippocampal input from nuclei in the (a) thalamus, (b) amygdala, (c) contralateral CA3, (d) midbrain/hindbrain and (e) basal forebrain/septum. See Methods for all abbreviations. n= 6 vCA1-mPFC, 8 vCA1-NAc, 9 vCA1-LH, 9 vCA1-BA, 4 vCA1-adBNST and 3 vCA1-LS mice . Error bars represent standard error of the mean. See Extended data table 2 for proportion counts (mean +/− SEM) for all assayed input regions and all statistical values. f. Controls for non-specific rabies infection. No long-range inputs were labeled in mice injected with Cre-dependent rabies helper virus and EnvA-G deleted rabies mCherry virus in the vCA1 subregion of vHPC in the absence of AAV2retro Cre injection (n=2 mice, no long-range input cells detected). right, image of red channel from thalamus, basal forebrain and amygdala of control mice. g. Number of extrahippocampal inputs correlates with the number of starter cells in RV samples (Pearson’s r=0.4035, two-tailed, p=0.037). AA-anterior amygdala, AD-anterodorsal nucleus, AM-anteromedial nucleus, BLA-basolateral amygdala, BMA-basomedial amygdala, CeA-central amygdala, CL-central lateral nucleus of the thalamus, CLI-central linear nucleus raphe, CM-central medial nucleus of the thalamus, CoA-cortical amygdala, DR-dorsal raphe nucleus, DTN-dorsal tegmental nucleus, IAD-interanterodorsal nucleus, IF-interfascicular nucleus raphe, IMD-Intermediodorsal nucleus of the thalamus, IPN-interpeduncular nucleus, LA-lateral amygdala, LC-locus coeruleus, LDT-laterodorsal tegmental nucleus, LHA-lateral hypothalamic area, LP-lateral posterior nucleus of the thalamus, LS-lateral septum, MR-median raphe, MA-magnocellular nucleus, MD-intermediodorsal nucleus of the thalamus, mPOA-medial preoptic area, MS-medial septum, MY-medulla, NDB-diagonal band nucleus, NI-nucleus incertus, NLoT-nucleus of lateral olfactory tract, P-pons, PAA-piriform-amygdalar area, PAG-periaqueductal gray, PB-parabrachial nucleus, PCG-pontine central gray, PF-parafascicular nucleus, PPN-pedunculopontine nucleus, PR-perireunensis nucleus, PRNr-pontine reticular nucleus, PT-parataenial nucleus, PVH-paraventricular nucleus of hypothalamus, PVT, paraventricular nucleus of the thalamus, RE-nucleus of reuniens, RPO-nucleus raphe pontis, RT- reticular nucleus of the thalamus, SC- superior colliculus, SG-supragenual nucleus, SI- substantia innominate, SUT-supratrigeminal nucleus, TR-piriform transition area, VAL-ventral anterior-lateral complex of the thalamus, VII-facial motor nucleus, VM-ventral medial nucleus of the thalamus, VTA-ventral tegmental, Xi-xiphoid thalamic nucleus
Extended Data Fig. 3. Transcriptional profiling of vCA1 projection neurons.
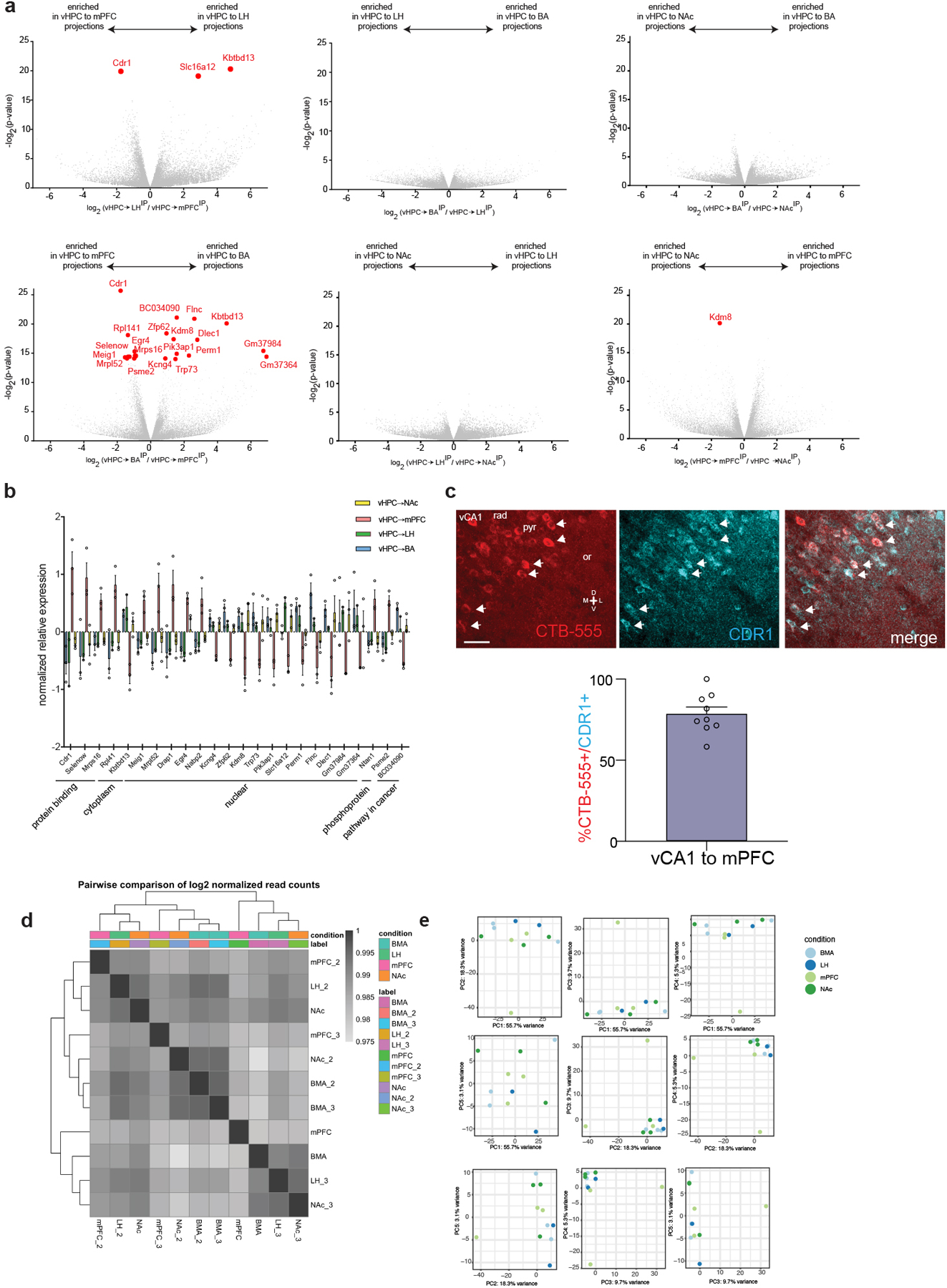
a. Volcano plots of differentially expressed genes for each set of pairwise comparisons. Red dots indicate differentially expressed genes that passed p<0.05 cutoff using Wald test, two-sided, and after correction for multiple comparisons with Benjamini Hochberg. All exact p-values provided in Extended Data Table 4. b. Normalized relative expression of the all 24 genes that passed the significance threshold for differential expression in pairwise comparisons . n=3 vHPC to mPFC replicates, 2 vHPC to LH replicates, 3 vHPC to NAc replicates and 3 vHPC to BA replicates. Error bars indicate SEM. c. RNAscope of CDR1 transcript in vCA1 neurons defined by their projection to the mPFC. Arrowheads indicate co-labeled neurons. Scale bar 40μm. Right. Quantification of overlap in CTB labeled and CDR1 labeled cells, n=9 FOVs from 2 mice, error bars indicate SEM. d. Heatmap correlations of total log2 normalized read counts from individual sample sets used for profiling experiments. e. Principal components plots of 500 most variable genes in the dataset, showing first 5 PCs.
Supplementary Material
Supplementary Table 1 MAPseq motif counts. Sheet 1: Co-projection motif counts and percentages (within a motif) from MAPseq dataset.
Sheet 2: Observed and expected motif counts from MAPseq dataset with effect size and p-values.
Supplementary Table 2 Input-output rabies tracing data Sheet 1: Proportion of inputs to mPFC-, NAc-, LH-, BA-, adBNST and LS projecting vHPC neurons.
Sheet 2: Statistical results for TRIO dataset
Supplementary Table 5 Metascape GO Metabolism table
Supplementary Table 3 RNAseq mapping, alignment and raw counts data Sheet 1:Table containing summary information regarding the mapping of each RNAseq sample.
Sheet 2: Table containing summary information regarding counts of alignment types from each RNAseq sample.
Sheet 3: Table of all RNAseq samples and their raw (unnormalized) read counts per gene.
Supplementary Table 4 RNAseq differential expression data. Sheet 1: All genes from TRAP dataset with counts, fold changes and p-values.
Sheet 2: Differentially expressed genes for vHPC-mPFC vs vHPC-subcortical targets
Acknowledgements
We thank J. Jimenez and L. Drew for feedback on the manuscript, A. Zador for initial guidance on MAPseq, H. Zhan and the CSHL MAPseq core facility, and Z. Knight, C. Zimmerman and D. Lieb for advice on TRAP experiments. A.V.M is supported by the Pew Charitable Trusts, NIMH (R01MH119349 and DP2MH116507), the Brain and Behavior Research Foundation, and the Burroughs Welcome Fund. M.A.K. is supported by NIMH (R01 MH108623, R01 MH111754, R01 MH117961), a One Mind Rising Star Award, the Human Frontier Science Program, the Pew Charitable Trusts, the Esther A. and Joseph Klingenstein Fund, the McKnight Memory and Cognitive Disorders Award and The Ray and Dagmar Dolby Family Fund.
Footnotes
Competing interests
Authors declare no competing interests.
Data availability.
RNA sequencing data is available in the NCBI’s Gene Expression Omnibus, accession # GSE150869, and MAPseq data is available at NLM Sequence Read Archive BioProject: PRJNA633836. RNAseq differential expression data is provided in Supplementary Data Table 4. All MAPseq source data can be downloaded at github.com/mkheirbek. MAPseq motif counts provided in Supplementary Data Table 1. All counts for input-output rabies tracing (n, mean, SEM) provided in Supplementary Data Table 2.
Code availability
All MAPseq data and code for analysis is posted on the Kheirbek lab GitHub site (github.com/mkheirbek).
Accession codes
MAPseq data Accession number: PRJNA633836, RNA-seq data Accession number: GSE150869
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
References
- 1.Fanselow MS & Dong HW Are the dorsal and ventral hippocampus functionally distinct structures? Neuron 65, 7–19, doi: 10.1016/j.neuron.2009.11.031 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciocchi S, Passecker J, Malagon-Vina H, Mikus N & Klausberger T Brain computation. Selective information routing by ventral hippocampal CA1 projection neurons. Science 348, 560–563, doi: 10.1126/science.aaa3245348/6234/560 [pii] (2015). [DOI] [PubMed] [Google Scholar]
- 3.Felix-Ortiz AC et al. BLA to vHPC inputs modulate anxiety-related behaviors. Neuron 79, 658–664, doi: 10.1016/j.neuron.2013.06.016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jimenez JC et al. Anxiety Cells in a Hippocampal-Hypothalamic Circuit. Neuron 97, 670–683 e676, doi: 10.1016/j.neuron.2018.01.016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kjelstrup KG et al. Reduced fear expression after lesions of the ventral hippocampus. Proceedings of the National Academy of Sciences of the United States of America 99, 10825–10830, doi: 10.1073/pnas.152112399152112399 [pii] (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padilla-Coreano N et al. Direct Ventral Hippocampal-Prefrontal Input Is Required for Anxiety-Related Neural Activity and Behavior. Neuron 89, 857–866, doi: 10.1016/j.neuron.2016.01.011S0896-6273(16)00012-X [pii] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kheirbek MA et al. Differential control of learning and anxiety along the dorsoventral axis of the dentate gyrus. Neuron 77, 955–968, doi: 10.1016/j.neuron.2012.12.038S0896-6273(13)00046-9 [pii] (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britt JP et al. Synaptic and behavioral profile of multiple glutamatergic inputs to the nucleus accumbens. Neuron 76, 790–803, doi: 10.1016/j.neuron.2012.09.040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeGates TA et al. Reward behaviour is regulated by the strength of hippocampus-nucleus accumbens synapses. Nature 564, 258–262, doi: 10.1038/s41586-018-0740-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reed SJ et al. Coordinated Reductions in Excitatory Input to the Nucleus Accumbens Underlie Food Consumption. Neuron 99, 1260–1273 e1264, doi: 10.1016/j.neuron.2018.07.051 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Jacobson L & Sapolsky R The role of the hippocampus in feedback regulation of the hypothalamic-pituitary-adrenocortical axis. Endocr Rev 12, 118–134, doi: 10.1210/edrv-12-2-118 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Kim WB & Cho JH Synaptic Targeting of Double-Projecting Ventral CA1 Hippocampal Neurons to the Medial Prefrontal Cortex and Basal Amygdala. J Neurosci 37, 4868–4882, doi: 10.1523/JNEUROSCI.3579-16.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parfitt GM et al. Bidirectional Control of Anxiety-Related Behaviors in Mice: Role of Inputs Arising from the Ventral Hippocampus to the Lateral Septum and Medial Prefrontal Cortex. Neuropsychopharmacology 42, 1715–1728, doi: 10.1038/npp.2017.56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Jin J & Maren S Renewal of extinguished fear activates ventral hippocampal neurons projecting to the prelimbic and infralimbic cortices in rats. Neurobiol Learn Mem 134 Pt A, 38–43, doi: 10.1016/j.nlm.2016.04.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu C et al. Distinct Hippocampal Pathways Mediate Dissociable Roles of Context in Memory Retrieval. Cell 167, 961–972 e916, doi: 10.1016/j.cell.2016.09.051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padilla-Coreano N et al. Hippocampal-Prefrontal Theta Transmission Regulates Avoidance Behavior. Neuron 104, 601–610 e604, doi: 10.1016/j.neuron.2019.08.006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin J & Maren S Fear renewal preferentially activates ventral hippocampal neurons projecting to both amygdala and prefrontal cortex in rats. Sci Rep 5, 8388, doi: 10.1038/srep08388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arszovszki A, Borhegyi Z & Klausberger T Three axonal projection routes of individual pyramidal cells in the ventral CA1 hippocampus. Front Neuroanat 8, 53, doi: 10.3389/fnana.2014.00053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bienkowski MS et al. Integration of gene expression and brain-wide connectivity reveals the multiscale organization of mouse hippocampal networks. Nat Neurosci 21, 1628–1643, doi: 10.1038/s41593-018-0241-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y et al. The logic of single-cell projections from visual cortex. Nature 556, 51–56, doi: 10.1038/nature26159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kebschull JM et al. High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron 91, 975–987, doi: 10.1016/j.neuron.2016.07.036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cenquizca LA & Swanson LW Analysis of direct hippocampal cortical field CA1 axonal projections to diencephalon in the rat. The Journal of comparative neurology 497, 101–114, doi: 10.1002/cne.20985 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cenquizca LA & Swanson LW Spatial organization of direct hippocampal field CA1 axonal projections to the rest of the cerebral cortex. Brain research reviews 56, 1–26, doi: 10.1016/j.brainresrev.2007.05.002 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glangetas C et al. NMDA-receptor-dependent plasticity in the bed nucleus of the stria terminalis triggers long-term anxiolysis. Nat Commun 8, 14456, doi: 10.1038/ncomms14456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz LA et al. Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature 524, 88–92, doi: 10.1038/nature14600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tervo DG et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382, doi: 10.1016/j.neuron.2016.09.021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohara K et al. Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat Neurosci 17, 269–279, doi: 10.1038/nn.3614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ekstrand MI et al. Molecular profiling of neurons based on connectivity. Cell 157, 1230–1242, doi: 10.1016/j.cell.2014.03.059 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cembrowski MS & Spruston N Heterogeneity within classical cell types is the rule: lessons from hippocampal pyramidal neurons. Nature reviews. Neuroscience 20, 193–204, doi: 10.1038/s41583-019-0125-5 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Wee RWS & MacAskill AF Biased Connectivity of Brain-wide Inputs to Ventral Subiculum Output Neurons. Cell Rep 30, 3644–3654 e3646, doi: 10.1016/j.celrep.2020.02.093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X et al. High-Throughput Mapping of Long-Range Neuronal Projection Using In Situ Sequencing. Cell 179, 772–786 e719, doi: 10.1016/j.cell.2019.09.023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas E, Burock D, Knudsen K, Deterding E & Yadin E Single unit activity in the lateral septum and central nucleus of the amygdala in the elevated plus-maze: a model of exposure therapy? Neurosci Lett 548, 269–274, doi: 10.1016/j.neulet.2013.05.078 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Besnard A et al. Dorsolateral septum somatostatin interneurons gate mobility to calibrate context-specific behavioral fear responses. Nature neuroscience 22, 436–446, doi: 10.1038/s41593-018-0330-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lebow MA & Chen A Overshadowed by the amygdala: the bed nucleus of the stria terminalis emerges as key to psychiatric disorders. Molecular psychiatry 21, 450–463, doi: 10.1038/mp.2016.1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SY et al. Diverging neural pathways assemble a behavioural state from separable features in anxiety. Nature 496, 219–223, doi: 10.1038/nature12018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cullinan WE, Herman JP & Watson SJ Ventral subicular interaction with the hypothalamic paraventricular nucleus: evidence for a relay in the bed nucleus of the stria terminalis. The Journal of comparative neurology 332, 1–20, doi: 10.1002/cne.903320102 (1993). [DOI] [PubMed] [Google Scholar]
- 37.Hallock HL et al. Molecularly-Defined Hippocampal Inputs Regulate Population Dynamics in the Prelimbic Cortex to Suppress Context Fear Memory Retrieval. Biol Psychiatry, doi: 10.1016/j.biopsych.2020.04.014 (2020). [DOI] [PMC free article] [PubMed]
- 38.Hsu DT, Kirouac GJ, Zubieta JK & Bhatnagar S Contributions of the paraventricular thalamic nucleus in the regulation of stress, motivation, and mood. Front Behav Neurosci 8, 73, doi: 10.3389/fnbeh.2014.00073 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruano D et al. Association of the gene encoding neurogranin with schizophrenia in males. J Psychiatr Res 42, 125–133, doi: 10.1016/j.jpsychires.2006.10.008 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Huang KP et al. Neurogranin/RC3 enhances long-term potentiation and learning by promoting calcium-mediated signaling. J Neurosci 24, 10660–10669, doi: 10.1523/JNEUROSCI.2213-04.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones KJ et al. Rapid, experience-dependent translation of neurogranin enables memory encoding. Proc Natl Acad Sci U S A 115, E5805–E5814, doi: 10.1073/pnas.1716750115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyakawa T et al. Neurogranin null mutant mice display performance deficits on spatial learning tasks with anxiety related components. Hippocampus 11, 763–775, doi: 10.1002/hipo.1092 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Huang L et al. BRICseq bridges brain-wide interregional connectivity to neural activity and gene expression in single animals. bioRxiv, doi: 10.1101/422477 (2019). [DOI]
- 44.Abusaad I et al. Stereological estimation of the total number of neurons in the murine hippocampus using the optical disector. The Journal of comparative neurology 408, 560–566, doi: (1999). [DOI] [PubMed] [Google Scholar]
- 45.Pavlopoulos GA et al. Bipartite graphs in systems biology and medicine: a survey of methods and applications. Gigascience 7, 1–31, doi: 10.1093/gigascience/giy014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miele V, Matias C, Robin S & Dray S Nine quick tips for analyzing network data. PLoS Comput Biol 15, e1007434, doi: 10.1371/journal.pcbi.1007434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, doi: 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930, doi: 10.1093/bioinformatics/btt656 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Gu Z, Eils R & Schlesner M Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849, doi: 10.1093/bioinformatics/btw313 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10, 1523, doi: 10.1038/s41467-019-09234-6 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 MAPseq motif counts. Sheet 1: Co-projection motif counts and percentages (within a motif) from MAPseq dataset.
Sheet 2: Observed and expected motif counts from MAPseq dataset with effect size and p-values.
Supplementary Table 2 Input-output rabies tracing data Sheet 1: Proportion of inputs to mPFC-, NAc-, LH-, BA-, adBNST and LS projecting vHPC neurons.
Sheet 2: Statistical results for TRIO dataset
Supplementary Table 5 Metascape GO Metabolism table
Supplementary Table 3 RNAseq mapping, alignment and raw counts data Sheet 1:Table containing summary information regarding the mapping of each RNAseq sample.
Sheet 2: Table containing summary information regarding counts of alignment types from each RNAseq sample.
Sheet 3: Table of all RNAseq samples and their raw (unnormalized) read counts per gene.
Supplementary Table 4 RNAseq differential expression data. Sheet 1: All genes from TRAP dataset with counts, fold changes and p-values.
Sheet 2: Differentially expressed genes for vHPC-mPFC vs vHPC-subcortical targets


