Abstract
Stimulator of interferon genes (STING) has been shown to play a critical role in orchestrating immune responses to various pathogens through sensing cyclic dinucleotides. However, how STING regulates intestinal homeostasis is still not completely understood. In this study, we found that STING−/− mice were more susceptible to enteric infection with Citrobacter rodentium compared to WT mice evidenced by more severe intestinal inflammation and impaired bacterial clearance. STING−/− mice demonstrated lower expression of REG3γ, but not β-defensins and Cramp, in IECs. Consistently, STING−/− IECs showed reduced capacity to inhibit bacterial growth. STING agonists, both 10-carboxymethyl-9-acridanone (CMA) and 5,6-dimethylxanthenone-4-acetic acid (DMXAA), promoted REG3γ expression IECs. Furthermore, STING agonists promoted WT but not REG3γ-deficient IEC bacterial killing. Mechanistically, STING agonists activated STAT3 and promoted glycolysis in IECs. Inhibition of STAT3 pathway and glycolysis suppressed STING induced REG3γ production in IECs, and abrogated STING-mediated IEC killing of Citrobactor rodentium. Additionally, treatment with the STING ligand, 2,3-cGAMP, inhibited Citrobactor rodentium-induced colitis in vivo. Overall, STING promotes IEC REG3γ expression to inhibit enteric infection and intestinal inflammation, thus, maintaining the intestinal homeostasis.
Keywords: STING, REG3γ, IEC, Intestinal homeostasis
Introduction
The gastrointestinal (GI) tract is colonized by a heterogeneous, dynamic and densely populated microbial community. The gut microbiota are pertinent for normal gut function such as nutrient digestion and uptake; however, they also present a continuous threat to the intestinal barrier (1). In order to limit direct contact with the microbiota and prevent colonization by pathogens, the intestinal epithelial cells (IECs) produce an abundance of antimicrobial peptides (AMPs), which can rapidly kill or inactivate bacteria (2, 3). Several distinct families of AMPs have been identified, including defensins, cathelicidins and C-type lectins (e.g. the regenerating islet-derived protein (REG) family) (4), which contribute to host defense against microbiota (5). Among REG3 lectins, REG3γ is abundantly expressed in enterocytes of the intestine, which is promoted by the microbiota and upregulated during times of inflammation and infection (6–8). However, the mechanisms by which the microbiota regulates IEC production of AMPs, such as REG3γ, are still poorly understood.
Stimulator of interferon genes (STING), a cytoplasmic sensor for cyclic dinucleotides (CDNs), is an adaptor molecule for a number of intracellular DNA receptors. STING is crucial in host defense where its activation leads to enhanced expression of type I interferons (IFNs), which exert both antiviral and antimicrobial activities (9–11). Consequently, its dysregulation may lead to the development of autoimmune diseases (12, 13). Both Gram-positive and Gram-negative bacteria are known to induce aberrant activation of the STING pathway (9). Recent studies underscore the protective role of STING signaling in controlling intestinal inflammation and maintaining gut homeostasis (14). Given that abundance of microbiota-derived CDNs within the lumen of the GI tract (15), it is plausible that STING plays a crucial role in regulating intestinal inflammatory responses driven by microbiota. However, the potential roles of STING in regulating AMPs and controlling gut homeostasis have not been investigated.
In this study, we used in vitro and in vivo systems to investigate regulatory role of STING signaling in the expression of REG3γ in IECs, and the role of the STING-REG3γ axis in the maintenance of intestinal homeostasis. Our study demonstrates that STING activation promotes REG3γ expression in IECs and inhibits enteric bacterial growth and intestinal inflammation, thereby contributing to the maintenance of intestinal homeostasis.
Materials and Methods
Mice
C57BL/6 wild-type (WT) mice and C57BL/6-Tmem173gt (STING−/−) mice were purchased from the Jackson Laboratory. All mice were housed in the specific pathogen-free animal facility in the Animal Resource Center at UTMB. All described animal experiments were performed in accordance with protocols reviewed and approved by the Institutional Animal Care and Use Committees of UTMB.
Reagents
Recombinant mouse interferon-γ (Cat# 575306, Lot# B203843) was purchased from BioLegend. Culture medium RPMI 1640 (Cat# SH30027.01, Lot# AD16492263), penicillin/streptomycin (Cat# SV30010, Lot# J170049), and Percoll (Cat# 17089101, Lot# 10253323) were purchased from GE healthcare. ITS (Cat# 354350, Lot# 15918002), HEPES (Cat# 25–060-CI, Lot# 08617003), EDTA (Cat# 46–034-CI, Lot# 08816016), and Matrigel (Cat# 356231, Lot# 7142014) were purchased from Corning. DMEM/F12 (Cat# 12634–010, Lot# AC12854278), Lipofectamine RNAiMAX Transfection Reagent (Cat# 13778150, Lot# 1815569), Lipofectamine® 3000 Reagent and NuPAGEBis-Tris mini gels (Cat# NP0323PK2, Lot#17060670) were purchased from Thermo Fisher Scientific. STAT3 inhibitor HJC0152 was synthesized by Dr. Jia Zhou’s laboratory at University of Texas Medical Branch at Galveston following their reported procedures (16). U0126 (Cat# U120, Lot#078K4623V) was purchased from Sigma Aldrich. Predesigned siRNA directed against mouse STAT3 (Cat# SI01435287, Lot# 211309675), and negative control siRNA (Cat# 1027310, Lot# 208993226) were purchased from Qiagen. Western blot antibodies against phosphorylated STAT3 (Y705, Cat# 9145, Lot# 31), phosphorylated mTOR (S2448, Cat# 2971, Lot# 19), phosphorylated ERK1/2 (137F5, Cat# 4695, Lot# 8), β-actin (Cat# 8457, Lot# 7), and anti-rabbit secondary antibody conjugated with horseradish peroxidase (Cat# 7074, Lot# 28) were purchased from Cell Signaling Technology. Antibody against REG3γ was purchased from Abcam (Cat# ab198216, Lot# GR209823–25).
Citrobacter rodentium infections and STING agonist intervention
Citrobacter rodentium strain DBS100 (ATCC) was grown overnight in 4 mL of Luria-Bertani (LB) medium at 37 °C with shaking. Adult mice were challenged via oral gavage with 200 μl PBS containing 5 × 108 CFU of bacteria. Mice were monitored daily and weighed on specified days. At day 2, 4, 7 and 10 post infection, fecal samples were collected, weighed and homogenized in 5 mL sterile PBS. Serial dilutions of the fecal homogenates were plated on MacConkey agar and grown overnight at 37 °C. CFUs were counted and the results were expressed as CFU/g of feces.
For STING agonist intervention, 2’3’-cGAMP (0.5 mg/kg) or control PBS were administered via i.p. injection to Citrobacter rodentium-infected WT mice each day for 10 days. Mice were sacrificed by using CO2 asphyxiation on day 10.
Histology
Colon tissues were harvested and fixed in 10% buffered formalin. The tissues were then embedded in paraffin, cut into 5 μm sections and mounted on microslides. The slides were stained with hematoxylin and eosin (H&E) and scored by an expert pathologist in a blinded manner. According to previously published scoring system (17), each section was scored based on the following criteria: inflammation, dysplasia, hyperplasia, edema, crypt degeneration and epithelial damage. Both intensity and extent were scored for colitic lesions. Intensity was scored as: 0 (absent), 1 (mild), 2 (moderate), and 3 (severe): whereas extent was scored as: 1 (25%), 2 (50%), 3 (75%), and 4 (100% of the total tissue affected). The total lesion scores reflected the sum of the individual scores.
Knockout of STAT3 and REG3γ Using CRISPR
LentiCRISPR vector (plasmid no. 52961; Addgene, Cambridge, MA) was used to knockdown STAT3 and REG3γ expression in MSIE cells. The design and cloning of the target guide RNA (gRNA) sequences were performed by following the Zhang laboratory’s protocol (http://www.genome-engineering.org) (18). Briefly, the suitable target sites for gRNA sequence against STAT3 and REG3γ were established using the CRISPR design tool software (http://crispr.mit.edu). Cas9 target sequences for the indicated genes were designed in http://www.genome-engineering.org. Then, gRNA (Integrated DNA Technologies) were synthesized and subcloned into the lentiCRISPR-v2 vector. The newly constructed lentiCRISPR plasmids were then transfected into normal MSIE cells. Following antibiotic positive selection, transfected cells were established as a stable cell line. gRNA oligo sequences for lentiCRISPR are listed in Supplementary Table 1.
Isolation of IEC
The entire intestines were opened longitudinally, cut into small fragments and washed with ice-cold phosphate-buffered saline (PBS). The intestinal fragments were placed in 50 ml tubes and incubated with 0.5 mM EDTA in PBS buffer containing 5% FBS at 37 °C for 40 min. The resulting buffer suspensions were passed through a 100 μm cell strainer to collect the cells. After washing with PBS, IECs that layered on the interface of 20%−40% gradient Percoll solution (Amersham Pharmacia Biotech) were collected.
Epithelial Cell Culture
Mouse small intestinal epithelial cells (MSIE) were cultured in RPMI 1640 medium supplemented with 5 U/ml murine gamma interferon (IFN-γ), 5% fetal bovine serum (FBS), ITS (5 μg/mL insulin, 5 μg/mL transferrin and 5 ng/mL selenous acid) and 100 U/mL penicillin/streptomycin at the permissive temperature of 33 °C. Before treatment with STING agonist, cells were starved in RPMI 1640 medium with 0.5% FBS for 16 hours at 37 °C. For IECs, cells were cultured in DMEM solution supplemented with 10% FBS at 37 °C and 5% CO2.
Western Blot
Total proteins of the cells were extracted using radioimmunoprecipitation assay buffer. Protein concentrations were determined using a BCA Protein Assay kit (Thermo Fisher Scientific) and normalized for all samples. Four micrograms of proteins were separated by NuPAGE Bis-Tris mini gel electrophoresis (Life Technologies, Carlsbad, CA, USA). Subsequently, the proteins were transferred onto polyvinylidene difluoride membranes (Thermo Fisher Scientific), blocked with 5% skimmed milk in TBST, and incubated overnight with anti-REG3γ, anti-phospho-mTOR, anti-phospho-STAT3, anti-phospho-ERK, and anti-β-actin primary Abs overnight. After three washes with TBST, the membranes were incubated with a horseradish peroxidase–conjugated anti-rabbit secondary antibody. Protein bands were visualized using enhanced chemiluminescence assay. ImageJ software was used for densitometry analysis of the western blot bands.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted with TRIzol reagent (Life Technologies; Carlsbad, CA) and quantified for cDNA synthesis. Quantitative real-time PCR was performed by using SYBR Green Gene Expression Assays (Bio-Rad, Hercules, CA, USA). GAPDH was used as the endogenous reference gene. The relative levels of target gene expression were calculated as a ratio relative to the GAPDH mRNA levels. Primers were listed in Supplementary Table 1.
Intestine Epithelial Cell Transfection
MSIE cells were starved in RPMI 1640 medium with 0.5% FBS for 16 hours at 37 °C and cultured in Gibco Opti-MEM medium. Cells were transfected with or without 1 μg/ml 2, 3-cGAMP using lipofectamine 3000 reagent. Cells were collected 48 hours post transfection.
In Vitro Killing of Bacteria by the Protein Extracts of Epithelial Cells
Primary intestinal epithelial cells isolated from the intestine of WT and STING−/− mice, as well as WT, STAT3 knockout and REG3γ knockout MSIE cells cultured with or without CMA for 24 hours, were used for the in vitro killing analysis. Briefly, the crude protein was extracted from cells in sterile PBS media supplemented with 1 mM PMSF. The concentration of the extracted proteins was normalized to 1.2 mg/mL. Appropriate aliquots of Citrobacter rodentium strain DBS100 (ATCC) or Escherichia coli O9:H4 (strain HS), which were kindly provided by Dr. Alfredo Torres of the University of Texas Medical Branch, with an initial OD600 value of 0.1–0.2, were added to the extracts of epithelial cells or control media. The bacterial suspensions were incubated at 37 °C for 18 hours under aerobic conditions, then diluted and transferred to solid MacConkey’s agar culture plates (Citrobacter rodentium) or Luria Broth’s agar culture plates (Escherichia coli HS) overnight for colony counting.
Cell metabolism measurement
MSIE cells (8K/well) were seeded into 96-well Seahorse plate. After 24 hours, cells were starved for 16 hours, and then treated with or without CMA for 6 hours. Cell ORC and EACR were then measured by Seahorse.
Statistical Analysis
All the data were presented as mean ± SD. Student’s t test was used for analysis the difference between two groups, and one-way ANOVA test were used for analysis the differences among groups more than two groups. Mann-Whitney U test was performed to determine differences in pathology scores. All the statistical analysis was performed using Prism 8. A p<0.05 was considered significant.
Results
STING−/− mice develop more severe colitis upon enteric infection by Citrobacter rodentium and demonstrate impaired control of bacteria
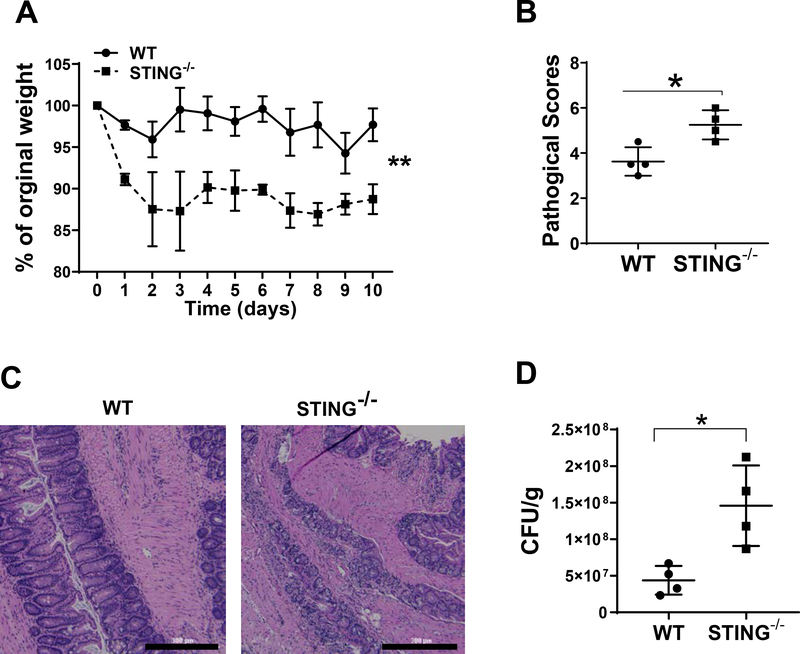
To study the effects of STING in regulating enteric infection, we infected WT and STING−/− mice orally with Citrobacter rodentium, an enteric bacterial strain similar to human enteropathogenic Escherichia coli (EPEC) associated with inflammatory bowel diseases (IBD) (19). Body weight changes were monitored daily. Feces were collected and cultured on MacConkey agar for bacteria counting. Mice were sacrificed on day 10 post infection. STING−/− mice displayed more severe colitis compared with WT mice, evidenced by increased weight loss and higher pathology scores (Fig. 1A–C). Furthermore, fecal Citrobacter rodentium counts were higher in the STING−/− mice (Fig. 1D), suggesting that STING promotes the clearance of Citrobacter rodentium in vivo.
Figure 1. STING−/− mice develop more severe colitis upon enteric infection of Citrobacter rodentium.
WT mice (n = 4) and STING−/− mice (n = 4) were given 200 μl PBS containing 5 × 108 CFU of Citrobacter rodentium by oral gavage. (A) weight changes were monitored daily. (B) Histological scores. (C) Representative histopathology of the colons from WT and STING−/− mice 10 days post-infection. Scale bar, 300μm. (D) Fecal colony-forming units (CFU) were quantified on day 7. *p < 0.05. Data were represented as means ± standard deviation. One representative of 3 experiments with similar results was shown.
STING−/− IECs show decreased REG3γ expression and impaired capacity in inhibiting bacterial growth
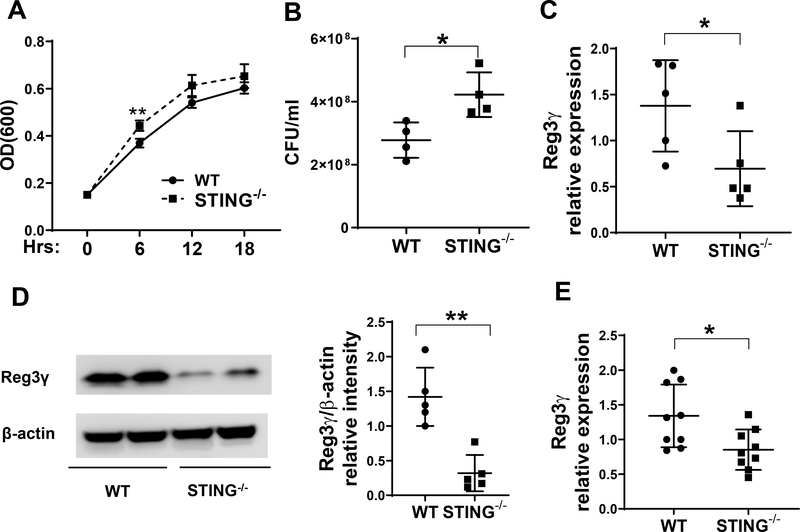
Since IECs play a central role in protecting intestinal mucosa from invasion by commensal microorganisms and pathogens (20), and STING, which is highly expressed in IECs, is an innate immune sensor for cyclic dinucleotides that can be produced by many bacteria species (21), we investigated whether STING in IECs regulates bacterial growth. Crude extracts of IECs from WT and STING−/− mice were cultured with Citrobacter rodentium. As shown in Fig. 2, the growth of Citrobacter rodentium was significantly inhibited by the extracts of WT IECs compared with those from STING−/− IECs (Fig. 2A–B), indicating that STING in IECs plays an essential role in inhibition of Citrobacter rodentium growth.
Figure 2. STING−/− IECs show decreased REG3γ expression and impaired capacity of inhibiting Citrobacter rodentium growth.
(A-B) Citrobacter rodentium was cultured with the crude extracts of IEC of WT and STING−/− mice. (A) The OD values (600 nm) were measured at different time points. (B) Bacteria culture suspensions were added to solid culture plates for colony counting overnight after treated with the extracts of IEC at 6 hours. (C-D) IECs were isolated from WT mice (n = 5) and STING−/− mice (n = 5) under steady conditions. The expressions of REG3γ was determined by qRT-PCR and normalized against GAPDH (C), and by Western blot with β-actin as the loading control (D). (E) WT (n = 9) and STING−/− mice (n = 9) were infected with Citrobacter rodentium by oral gavage. Primary IECs were isolated 10 days post infection of Citrobacter rodentium. The expressions of REG3γ was determined by qRT-PCR and normalized against GAPDH. *p < 0.05, **p < 0.01. Data are represented as means ± standard deviation. One representative of 3 experiments with similar results was shown.
As AMPs have been crucial in mediating IEC inhibition of bacteria (22), we next investigated whether STING regulates the production of AMPs. Under steady condition, REG3γ was significantly reduced at both the mRNA (Fig. 2C) and protein levels (Fig. 2D) in IECs of STING−/− mice compared with WT mice. However, there was no difference observed among other AMPs, such as β-defensin-2, 3 and cramp, between STING−/− IECs and WT IECs (Supplementary Fig. 1A–C). When infected with Citrobacter rodentium, REG3γ expression was significantly downregulated in IECs from STING−/− mice compared to those from the WT mice (Fig. 2E). As with baseline comparisons, expression of other AMPs, such as β-defensin-2, 3 and cramp, were similar between STING−/− IECs and WT IECs from infected mice (Supplementary Fig. 1D–F). These data indicate that IEC STING plays a protective function during enteric infection possibly through regulating the expression of REG3γ.
STING agonists enhance IEC production of Reg3γ and promote IEC to inhibit bacterial growth
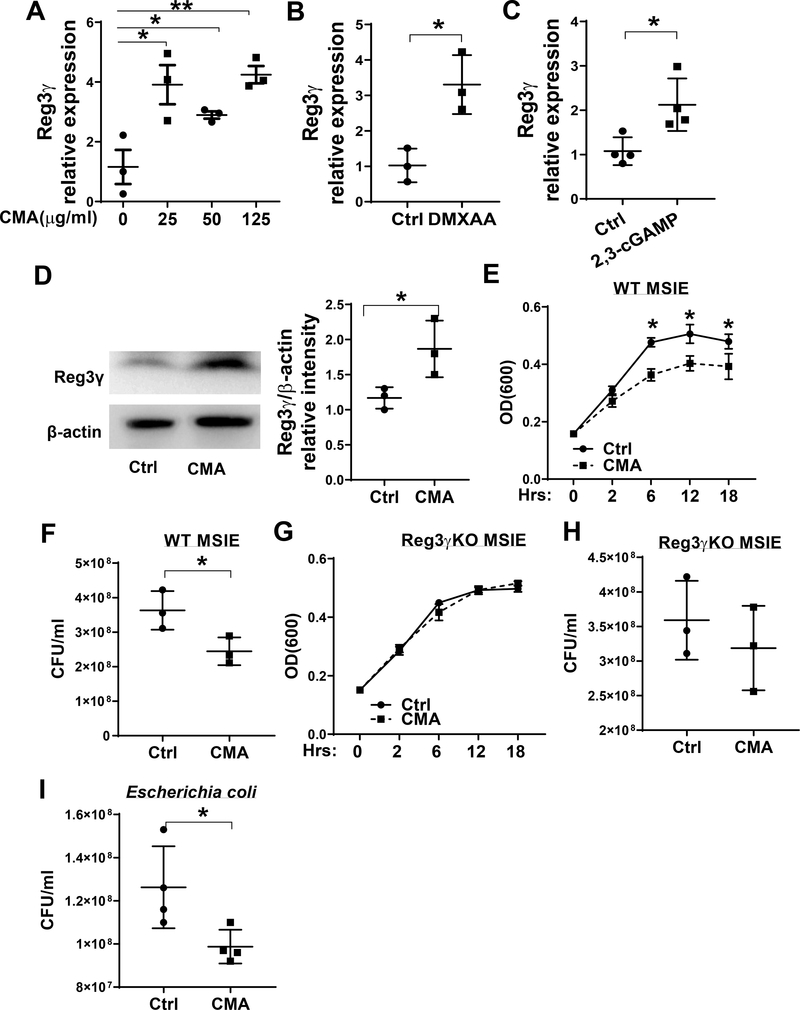
To investigate whether STING regulates IEC REG3γ production directly or indirectly, we stimulated MSIE, an immortalized epithelial cell line that retains properties of primary IECs (23) with different doses of CMA, a STING agonist, for 24 hours. The expression of REG3γ and other AMPs were assessed by qRT-PCR. We found that CMA promoted the expression of REG3γ (Fig. 3A), but not β-defensins-2, 3 and cramp (Supplementary Fig. 2A–C). To further confirm the results, we treated MSIE cells with another STING agonist, DMXAA. Similar to CMA, DMXAA increased REG3γ in MSIE cells (Fig. 3B). 2, 3-cGAMP, a STING agonist which works in both human and mice (24), also promoted REG3γ expression in MSIE cells (Fig. 3C). Additionally, CMA upregulated REG3γ in primary IECs (Fig. 3D). Next, we investigated whether STING affected the ability of IEC to inhibit bacterial growth. To do this, the cell extracts from WT MSIE cells treated with or without CMA for 24 hours were cultured with Citrobacter rodentium and the bacterial growth was monitored via OD600. We found that extracts from CMA treated cells promoted IEC inhibition of bacterial growth (Fig. 3E–F), and this effect was not due to CMA alone as CMA itself did not affect Citrobacter rodentium growth in the culture (Supplementary Fig. 3A–B). To investigate whether STING regulates the growth of intestinal commensal bacteria in addition to pathogens, Escherichia coli HS was also tested. We found that the extracts of CMA treated IECs suppressed Escherichia coli HS growth at higher levels compared with control IECs (Fig. 3I). Collectively, these data indicate the importance of STING in regulating growth of both commensal and pathogenic bacteria.
Figure 3. STING promotes IEC inhibition of bacterial growth through promoting REG3γ expression.
(A) MSIE cells (n = 3 /group) were treated with a series of doses of CMA for 24 hours. The expression of REG3γ was determined by qRT-PCR and normalized against GAPDH. (B) MSIE cells were treated with STING agonist DMXAA (1 μg/ml) for 24 hours, and the expression of REG3γ was determined by qRT-PCR and normalized against GAPDH. (C) MSIE cells were transfected with 2,3-cGAMP (1 μg/ml) for 48 hours, and the expression of REG3γ was determined by qRT-PCR and normalized against GAPDH. (D) Primary IECs of WT mice were treated with CMA (50 μg/ml) for 12 hours. The expression of REG3γ was determined by Western-blot with β-actin as the loading control. (E-H) WT MSIE cells and REG3γ KO MSIE cells were treated with or without CMA (50 μg/ml) for 24 hrs. The crude extracts were prepared and cultured with Citrobacter rodentium. The OD values (600 nm) were measured at different time points with the extracts of WT (E) and REG3γ KO (G) MISE cells. Bacterial culture suspensions were colonized to solid culture plates for colony counting after treated with the extracts of WT (F) and REG3γ KO (H) MISE cells. (H) *p < 0.05, **p < 0.01. (I) WT MSIE cells were treated with or without CMA (50 μg/ml) for 24 hrs. The crude extracts were prepared and cultured with Escherichia coli HS, and bacterial culture suspensions were colonized to solid culture plates for colony counting overnight. Data are represented as means ± standard deviation. One representative of 3–5 experiments with similar results was shown.
STING promotes IEC inhibition of bacterial growth through induction of REG3γ
To determine whether STING inhibits bacterial growth through upregulation of REG3γ, we generated REG3γ knockout (REG3γ KO) MSIE cells using CRISPR, and treated WT and Reg3γ KO MSIE cells with or without CMA. The decreased Reg3γ expression was confirmed in REG3γ KO MSIE cells (Supplementary Fig. 4). Cell extracts were then cultured with Citrobacter rodentium. While the extracts of the CMA-treated WT MSIE cells significantly reduced Citrobacter rodentium growth compared with control WT MSIE cells (Fig. 3E–F), CMA showed no effect on REG3γ KO MSIE cell to inhibit the Citrobacter rodentium growth (Fig. 3G and H), indicating STING promotes IEC inhibition of bacterial growth at least partially through upregulating IEC production of REG3γ.
STAT3 mediates STING promotion of IEC REG3γ production
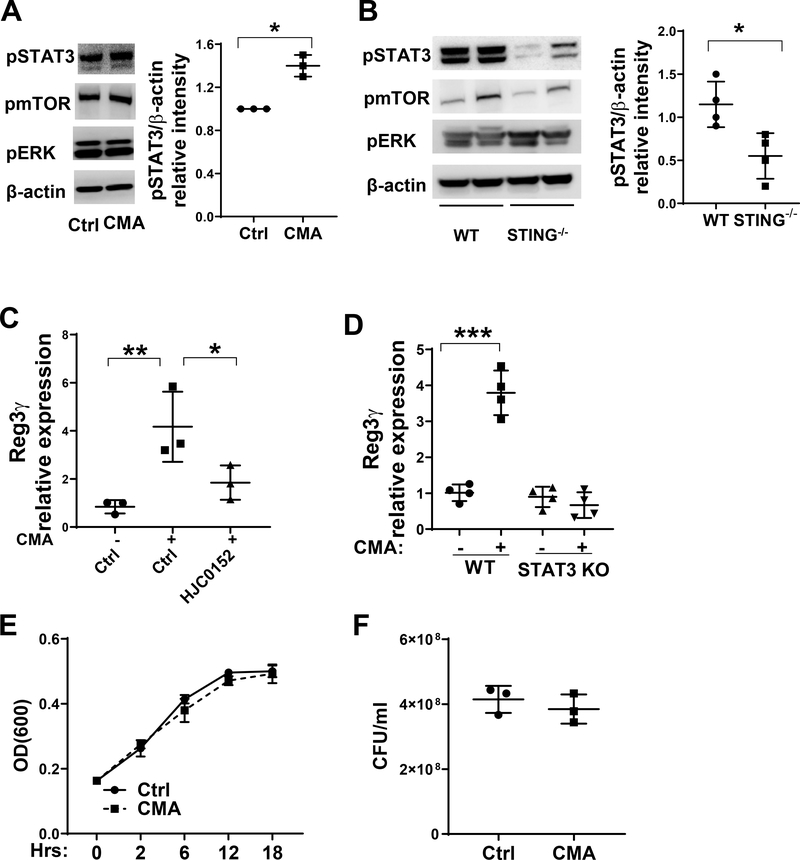
Several signal pathways, including STAT3, mTOR and ERK, have been shown to promote REG3γ production (25). To determine whether STING agonist activates STAT3, mTOR and ERK, and their roles in STING promotion of IEC REG3γ production, we cultured MSIE cells with or without STING agonist CMA. CMA enhanced p-STAT3, but not p-mTOR and p-ERK1/2, in IECs (Fig. 4A). To investigate whether STING activates STAT3 in IEC in vivo, we isolated the IECs from WT and STING−/− mice and measured their expression of p-STAT3, p-mTOR and p-ERK1/2. The expression of p-STAT3, but not p-mTOR and p-ERK1/2, was significantly downregulated in STING−/− mice (Fig. 4B). Collectively, these data indicate that STING promotes STAT3 activation in IECs.
Figure 4. STAT3 mediates STING-induced REG3γ in IEC.
(A) MSIE cells (n = 3/group) were treated with or without 50 μg/ml CMA for 5 min. The phosphorylated mTOR, ERK1/2 and STAT3 were determined by Western blot. (B) IECs were isolated from WT mice (n = 4) and STING−/− mice (n = 4), the phosphorylated mTOR, ERK1/2 and STAT3 were determined by Western blot. Data are reflective of 2 independent experiments. (C) MSIE cells (n = 3/group) were treated with 50 μg/ml CMA in the presence or absence of STAT3 inhibitor HJC0152. The expression of REG3γ was determined by qRT-PCR and normalized against GAPDH. (D) WT (n = 4) and STAT3 KO (n = 4) MSIE cells were treated with or without 50 μg/ml CMA for 24 hrs. The expression of REG3γ was determined by qRT-PCR and normalized against GAPDH. (E-F) Citrobacter rodentium were cultured with the crude extracts of STAT3 KO MSIE cells that were pretreated with or without CMA (n = 3/group) for 24 hrs. (E) The OD values (600 nm) were measured at different time points. (F) Bacteria culture suspensions were colonized to solid culture plates for colony counting overnight after treated with the extracts of IEC at 6 hours. *p < 0.05, **p < 0.01, ***p < 0.001. Data are represented as means ± standard deviation. One representative of 3 experiments with similar results was shown.
Next, we investigated whether the STAT3 pathway mediates STING induction of Reg3γ in IECs. We treated MSIE cells with CMA in the presence or absence of the STAT3 inhibitor, HJC0152 (16, 26). The CMA-induced REG3γ expression in MSIE cells was significantly reduced after treatment with STAT3 inhibitor (Fig. 4C). To further confirm the role of STAT3 in mediating STING-induced REG3γ expression in IEC, we knocked out STAT3 in MSIE cells by CRISPR, and then treated the cells with CMA. CMA did not promote REG3γ expression in STAT3 KO MSIE cells (Fig. 4D). These data indicated that STAT3 pathway mediates STING-induced REG3γ expression in IECs.
To investigate the role of STAT3 in STING-promoted IEC inhibition of bacterial growth, we treated WT and STAT3 KO MSIE cells with or without CMA for 24 hours. Citrobactor rodentium was cultured with the extracts of the cells in vitro. Consistently, extracts from CMA-treated STAT3 KO MSIE cells were unable to inhibit bacterial growth (Fig. 4E–F). Together, these data indicate that STAT3 mediates STING-induced IEC bacterial clearance through promoting REG3γ expression.
Glycolysis is involved in STING induction of REG3γ in IECs
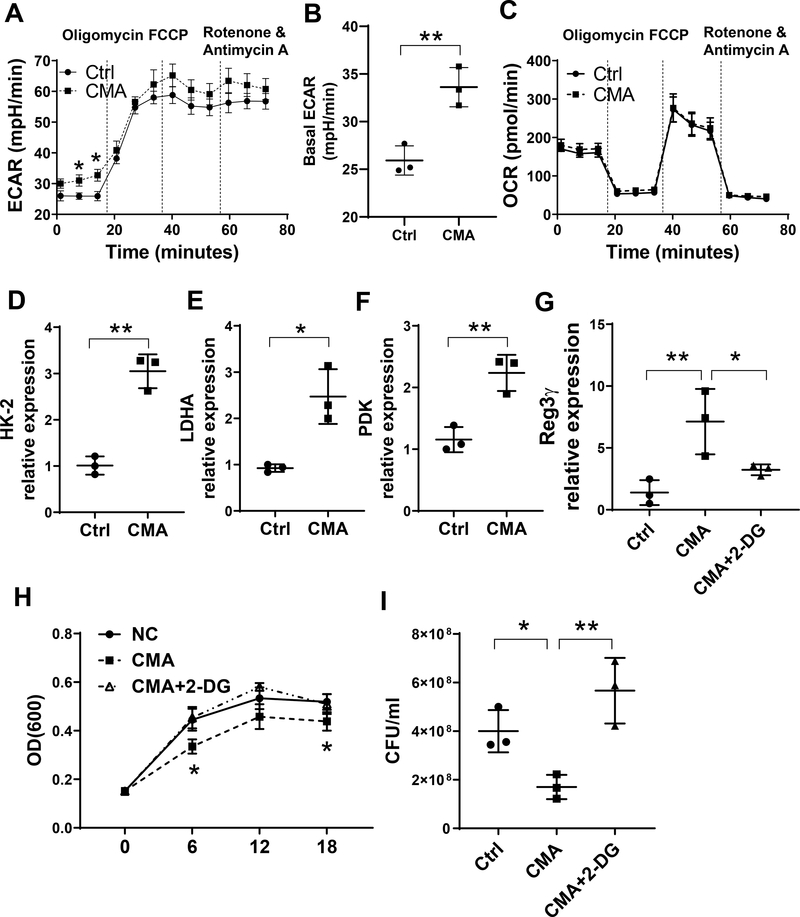
It has been shown recently that metabolism plays unique and intricate roles in many cells. However, whether STING regulates IEC metabolism remains unknown. To determine the effect of STING signaling in IEC metabolism, we treated MSIE cells with or without CMA for 24 hours and measured their respiratory and glycolytic capacity by Seahorse. CMA treatment increased Extracellular Acidification Rate (ECAR), an indicator of glycolysis (Fig. 5A–B) but did not affect Oxygen Consumption Rate (OCR) (Fig. 5C), suggesting that STING promotes glycolysis but not oxidation in IECs. Consistently, CMA increased the expression of glycolysis-related genes, including HK-2, LDHA, and HDK (Fig. 5D–F). To investigate whether glycolysis regulates STING induction of REG3γ, we treated MSIE cells with or without 2-Deoxy-D-glucose (2DG), a glycolysis inhibitor (27), in the presence of CMA. Treatment with 2DG suppressed CMA-induced REG3γ (Fig. 5G). Furthermore, treatment with 2DG reduced STING-induced inhibition of Citrobacter rodentium growth by IECs (Fig. 5H–I). These data indicate that glycolysis is critical in regulating STING induction of REG3γ and control of bacterial growth in IECs.
Figure 5. Glycolysis is involved in STING induction of REG3γ in IEC.
(A-C) MSIE cells (n = 3/group) were treated with or without 50 μg/ml CMA for 24 hrs. The levels of ECAR (A-B) and OCR (C) were measured by Seahorse. (D-F) MSIE cells (n = 3 /group) were treated with or without 50 μg/ml CMA for 24 hrs. The expression of HK-2 (D), LDHA (E), and PDK (F) was determined by qRT-PCR and normalized against GAPDH. (G) MSIE cells (n = 3/group) were treated with or without 50 μg/ml CMA in the presence or absence of 2-DG (1 μM) for 24 hrs, REG3γ was determined by qRT-PCR and normalized against GAPDH. (H-I) MSIE cells were treated with or without 50 μg/ml CMA in the presence or absence of 2-DG (1 μM) for 24 hrs. Citrobacter rodentium were cultured with the crude extracts of the cells. (H) The OD values (600 nm) were measured at different time points. (I) Bacterial culture suspensions were colonized to solid culture plates for colony counting overnight after treated with the extracts of IEC at 6 hrs. *p < 0.05, **p < 0.01. Data are represented as means ± standard deviation. One representative of 2–3 experiments with similar results was shown.
The interaction of STAT3 and glycolysis affects STING induction of REG3γ in IECs
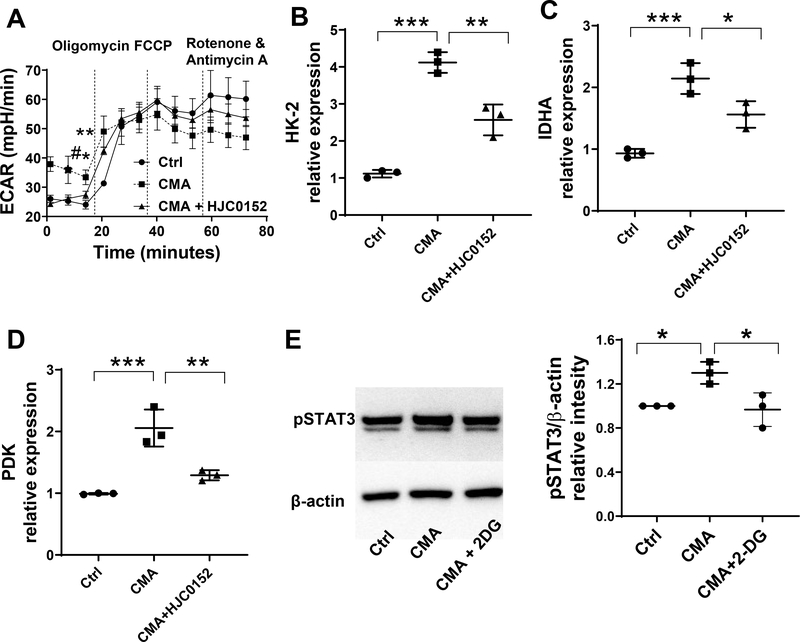
Next, we investigated whether STING induced glycolysis and STAT3 activation affect each other. First, we treated MSIE cells with CMA in the presence or absence of STAT3 inhibitor, HJC0152, for 24 hours, and then measured glycolysis by Seahorse. As shown in Fig. 6A, treatment with HJC0152 abrogated the effects of CMA on ECAR. Furthermore, HJC0152 suppressed expression of the CMA-induced glycolysis-related genes (Fig. 6B–D). To determine whether glycolysis regulates STING-induced STAT3 activation, we treated MSIE cells with or without 2DG in the presence of CMA. Treatment with 2DG inhibited the activation of STAT3 (Fig. 6E). Taken together, these data indicate that the interaction of STAT3 and glycolysis affects STING induction of REG3γ in IECs.
Figure 6. STAT3 and glycolysis interact each other in IECs upon STING stimulation.
(A) MSIE cells (n = 3/group) were treated with or without 50 μg/ml CMA in the presence or absence of STAT3 inhibitor HJC0152 for 24 hrs. ECAR was measured by Seahorse. (B-D) The expression of HK-2 (B), LDHA (C), and PDK (D) was determined by qRT-PCR and normalized against GAPDH. (E) MSIE cells (n = 3/group) were treated with or without 50 μg/ml CMA in the presence or absence of 2-DG (1 μM), and the protein levels of phosphorylated STAT3 were determined by Western blot. One representative of 2–3 experiments with similar results was shown.
STING protects the intestines from Citrobacter rodentium-induced inflammation
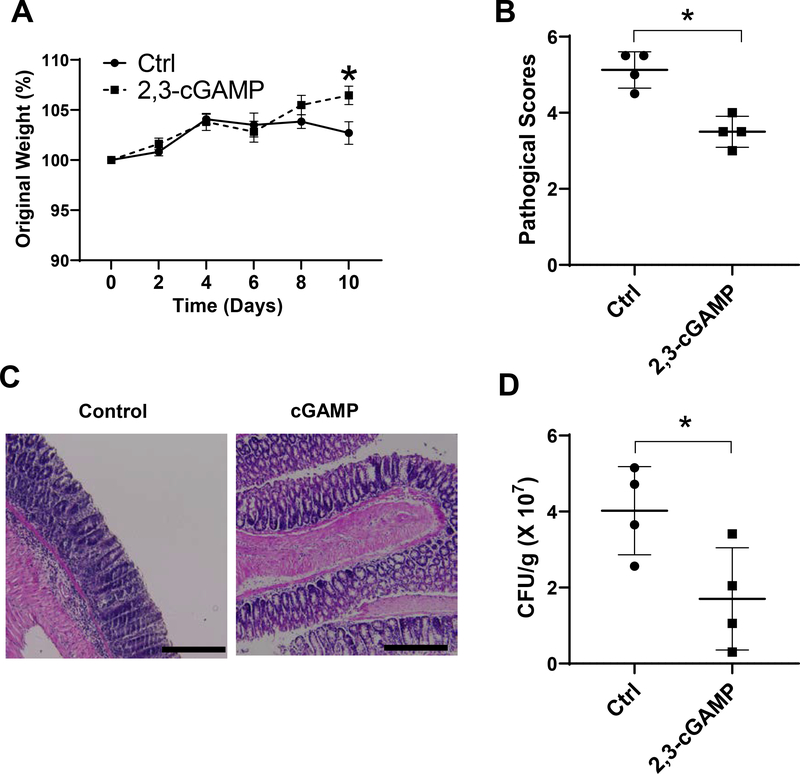
To investigate whether STING inhibits Citrobacter rodentium infection and intestinal inflammation, we infected WT mice with Citrobacter rodentium, and administered mice daily with 0.5 mg/kg 2’3’-cGAMP, a STING agonist used in both human and mice (24). Mice weights changes were monitored daily, and feces were collected on day 7 for analysis of bacteria clearance. The mice were sacrificed 10 days post infection, and histopathology of the colon and cecum was examined. Treatment with STING agonist 2’3’-cGAMP inhibited Citrobacter rodentium infection and colitis development, as evidenced by less weight loss (Fig. 7A), less severe intestine inflammation (Fig. 7B and C), and increased Citrobacter rodentium clearance (Fig. 7D).
Figure 7. STING agonist 2,3-cGAMP protects the intestines from Citrobacter rodentium infection in vivo.
WT B6 mice were given 5 × 108 CFU of bacteria via oral garage. The mice were administered daily with 0.5 mg/kg 2’3’-cGAMP or PBS as control. (A) Changes in body weight were monitored daily. (B) Representative histopathology of the colon from WT and STING−/− mice on day 10 post-infection. Scale bar, 300μm. (C) Pathology scores. (D) Fecal colony-forming units (CFU) were quantified on day 7. *p < 0.05. Data were represented as means ± standard deviation. One representative of 2 experiments with similar results was shown.
Discussion
Gut microbiota are continuous sources of CDNs (15). STING, a cytoplasmic sensor for microbial nucleic acid, is expressed in IEC (28), maintains the surveillance of cytoplasmic CDNs and is involved in gut mucosal immune responses (29). Genome-wide association studies (GWAS) have identified that several DNA sensor-encoding genes (including AIM2, IFI16, ZBP1 and DDX41), which are correlated with STING, are associated with IBD (30). Although STING has been shown to be involved in host defense against viral, bacterial, and eukaryotic pathogens (31, 32), it is unclear how STING regulates IEC, thus contributes to the maintenance of intestinal homeostasis. In this report, by using genetic manipulated STING knockout mice and the cell lines with specific gene knockout by CRISPR in combination with enteric infection and bacterial killing in vitro, we demonstrated that STING deficiency impaired IEC production of REG3γ, a crucial AMP in controlling microbiota and enteric pathogens. STING promoted IEC antimicrobial activity by promoting IEC production of REG3γ. Furthermore, administration of STING agonist inhibited enteric infection and intestinal inflammation. This finding indicates that STING is critical in inhibiting intestinal inflammation, thus, provides the great insights into developing STING agonists as potential therapeutics in treating patients with IBD.
Intestinal epithelial cells form a physiochemical barrier that separates the gut microbial community from the host’s internal milieu. Defective interactions between the gut microbiota and intestinal epithelium can lead to sustained inflammation and development of chronic diseases including IBD (33, 34). We found that STING−/− IECs showed impaired control of bacterial growth. Additionally, STING agonists promoted IEC to inhibit bacterial growth, indicating STING is critical in the maintenance of intestinal homeostasis. To cope with the continuous and complex intestinal microbial challenge, the epithelial cells produce a diverse array of AMPs (33), including REG3γ, which are important components of intestinal mucosal innate immunity, and play essential roles in maintaining intestinal homeostasis. REG3γ is primarily expressed in enterocytes and secreted apically into the intestinal lumen (7). It has been shown that REG3γ expression is low in germ-free mice and significantly upregulated during the initial establishment of the gut microbiota (35), indicating a crucial role of microbiota in IEC production of REG3γ. REG3γ has also been essential for enforcing the physical separation of microbiota and the intestinal mucosa (34, 36). In current study, we found that REG3γ expression, but not expression of other AMPs, was lower in STING−/− IECs, and STING agonists increased IEC production of REG3γ. A previous report demonstrated an altered gut microbiota in STING−/− mice (14). Interestingly, STING agonist promoted WT IECs but failed to promote REG3γKO IECs to control bacterial growth, suggesting that IEC expression of STING protects the host from enteric pathogen attack at least partially through promoting REG3γ production, which provides a novel regulatory mechanism in mucosal protection and intestinal homeostasis.
Although the pathways in activation of dendritic cells (DC) upon microbial DNA recognition has been reported (37), the signaling pathways mediating STING-induced REG3γ expression in IECs remain largely unknown. It has been reported that STING signaling promoted STAT3 phosphorylation in DC upon cytosolic DNA stimulation, and released both pro- and anti-inflammatory cytokines (38). STAT3 mediated REG3γ expression in lung epithelial cells in response to Staphylococcus aureus infection (25), and enhanced the expression of several AMPs, including REG3β and REG3γ, induced by IL-6 in urothelial cells during urinary tract infection (39). In the current study, we demonstrated that STING agonist activated STAT3, but not mTOR and Erk, which have been shown to be involved in regulating REG3γ expression in IECs by other stimuli (40, 41). STING deficiency downregulated STAT3 activation in IECs. Importantly, inhibition of STAT3 compromised the STING induction of REG3γ production and suppressed STING inhibition of bacterial growth. Taken together, these data indicated that STAT3 mediates STING-induced REG3γ expression in IECs.
Accumulating evidence indicates a crucial role of metabolism in regulation of different cell functions, including IECs (42, 43). Intestinal microbiota, which can trigger STING activation, is tightly related with host metabolism (14, 44). Additionally, metabolism is important in regulating colitis (45, 46). However, how microbiota affects IEC cell metabolism is still largely unknown. In the current study, we found that STING agonist promoted IEC glycolysis but not oxidation. Inhibition of glycolysis by 2-DG suppressed REG3γ production induced by STING agonist and inhibited IECs to control bacterial growth, indicating that glycolysis is involved in STING induction of REG3γ and STING inhibition of bacterial growth. Furthermore, STAT3 and glycolysis acted in concert with each other to regulate STING induction of REG3γ, which is consistent with previous studies (47–49). Considering that STING mediates IEC glycolysis, which may influence glucose concentration in conditioned medium to affect bacteria growth, we did not use the conditioned medium for in vitro bacterial killing experiments. In addition, there are higher levels of REG3γ in IEC extracts, which reduces the demand of cell numbers used. Therefore, we chose IEC extracts instead of conditioned medium for in vitro bacterial killing experiments in this study.
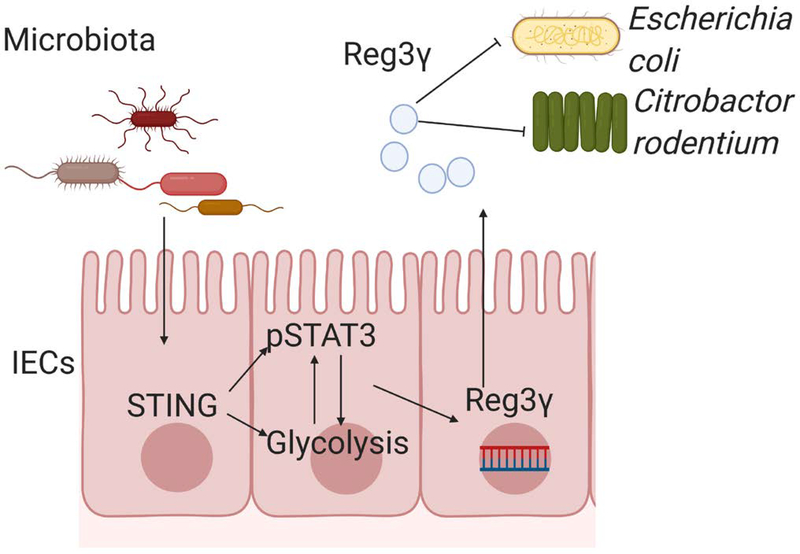
In summary, our study demonstrates that STING, which is a cytoplasmic DNA sensor, promotes IEC expression of REG3γ through STAT3 activation and glycolysis, and has an essential role in host defense against the overwhelming invasion of gut microbiota and enteric pathogens (Fig. 8). Furthermore, strategies that trigger STING signaling may promote intestinal AMPs production to regulate intestinal homeostasis in the host.
Figure 8. STING regulates intestinal bacteria growth through upregulating REG3γ in IECs.
Microbiota-derived CDNs activate STING pathway in IECs, which upregulates REG3γ through interaction of glycolysis and STAT3 pathway. STING induction of REG3γ inhibits intestinal pathogenic and commensal bacteria (i.e., Citrobacter rodentium and Escherichia coli HS).
Supplementary Material
Acknowledgement
This work was supported by National Institutes of Health grants DK105585, DK112436, DK125011 and AI150210, and University of Texas System STARs award (YC), and McLaughlin postdoctoral Fellowship, UTMB (WY). Dr. Alfredo Torres kindly provided us the Escherichia coli O9:H4 (strain HS). Fig. 8 was created with BioRender.com.
Nonstandard Abbreviation
- STING
Stimulator of interferon genes STING
- WT
Wild-type
- AMP
antimicrobial peptides
- IL
Interleukin
- IEC
intestinal epithelial cells
- REG
regenerating islet-derived protein
- CMA
10-carboxymethyl-9-acridanone
- DMXAA
5,6-dimethylxanthenone-4-acetic acid
- CDNs
cyclic dinucleotides
- IFNs
type I interferons
- EPEC
Escherichia coli
- IBD
inflammatory bowel diseases
- ECAR
Extracellular Acidification Rate
- OCR
Oxygen Consumption Rate
- 2-DG
2-Deoxy-D-glucose
Footnotes
Conflict of Interest Statement
No authors have conflicting financial interests.
References
- 1.Kau AL, Ahern PP, Griffin NW, Goodman AL, and Gordon JI (2011) Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goto Y, and Ivanov II. (2013) Intestinal epithelial cells as mediators of the commensal-host immune crosstalk. Immunol Cell Biol 91, 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun M, He C, Cong Y, and Liu Z (2015) Regulatory immune cells in regulation of intestinal inflammatory response to microbiota. Mucosal Immunol 8, 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukherjee S, and Hooper LV (2015) Antimicrobial defense of the intestine. Immunity 42, 28–39 [DOI] [PubMed] [Google Scholar]
- 5.Lai Y, and Gallo RL (2009) AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol 30, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallo RL, and Hooper LV (2012) Epithelial antimicrobial defence of the skin and intestine. Nature reviews. Immunology 12, 503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin JH, and Seeley RJ (2019) Reg3 Proteins as Gut Hormones? Endocrinology 160, 1506–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa H, Fukushima K, Naito H, Funayama Y, Unno M, Takahashi K, Kitayama T, Matsuno S, Ohtani H, Takasawa S, Okamoto H, and Sasaki I (2003) Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflammatory bowel diseases 9, 162–170 [DOI] [PubMed] [Google Scholar]
- 9.Barber GN (2015) STING: infection, inflammation and cancer. Nature reviews. Immunology 15, 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Q, Knight PH, Ren Y, Ren H, Zheng J, Wu X, Ren J, and Sawyer RG (2019) The emerging role of stimulator of interferons genes signaling in sepsis: Inflammation, autophagy, and cell death. Acta Physiol (Oxf) 225, e13194. [DOI] [PubMed] [Google Scholar]
- 11.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, and Rice CM (2014) Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, and Vance RE (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa H, and Barber GN (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canesso MCC, Lemos L, Neves TC, Marim FM, Castro TBR, Veloso É, Queiroz CP, Ahn J, Santiago HC, Martins FS, Alves-Silva J, Ferreira E, Cara DC, Vieira AT, Barber GN, Oliveira SC, and Faria AMC (2018) The cytosolic sensor STING is required for intestinal homeostasis and control of inflammation. Mucosal immunology 11, 820–834 [DOI] [PubMed] [Google Scholar]
- 15.Danilchanka O, and Mekalanos JJ (2013) Cyclic dinucleotides and the innate immune response. Cell 154, 962–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Yang Z, Ding C, Chu L, Zhang Y, Terry K, Liu H, Shen Q, and Zhou J (2013) Discovery of O-Alkylamino Tethered Niclosamide Derivatives as Potent and Orally Bioavailable Anticancer Agents. ACS medicinal chemistry letters 4, 180–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iqbal N, Oliver JR, Wagner FH, Lazenby AS, Elson CO, and Weaver CT (2002) T helper 1 and T helper 2 cells are pathogenic in an antigen-specific model of colitis. J Exp Med 195, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanjana NE, Shalem O, and Zhang F (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, and Weaver CT (2006) Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 441, 231–234 [DOI] [PubMed] [Google Scholar]
- 20.Fukata M, and Arditi M (2013) The role of pattern recognition receptors in intestinal inflammation. Mucosal Immunol 6, 451–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marinho FV, Benmerzoug S, Oliveira SC, Ryffel B, and Quesniaux VFJ (2017) The Emerging Roles of STING in Bacterial Infections. Trends Microbiol 25, 906–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bevins CL, and Salzman NH (2011) Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nature reviews. Microbiology 9, 356–368 [DOI] [PubMed] [Google Scholar]
- 23.Whitehead RH, and Robinson PS (2009) Establishment of conditionally immortalized epithelial cell lines from the intestinal tissue of adult normal and transgenic mice. Am J Physiol Gastrointest Liver Physiol 296, G455–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, and Chen ZJ (2013) Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Molecular cell 51, 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi SM, McAleer JP, Zheng M, Pociask DA, Kaplan MH, Qin S, Reinhart TA, and Kolls JK (2013) Innate Stat3-mediated induction of the antimicrobial protein Reg3gamma is required for host defense against MRSA pneumonia. J Exp Med 210, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Zhu T, Xu Y, Wu C, Chen J, Ren Y, Kong L, Sun S, Guo W, Wang Y, Jing C, Dong J, Zhou J, Zhang L, Shen Q, and Zhou X (2019) A novel STAT3 inhibitor, HJC0152, exerts potent antitumor activity in glioblastoma. American journal of cancer research 9, 699–713 [PMC free article] [PubMed] [Google Scholar]
- 27.Huang CC, Wang SY, Lin LL, Wang PW, Chen TY, Hsu WM, Lin TK, Liou CW, and Chuang JH (2015) Glycolytic inhibitor 2-deoxyglucose simultaneously targets cancer and endothelial cells to suppress neuroblastoma growth in mice. Disease models & mechanisms 8, 1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa H, Ma Z, and Barber GN (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, and Barber GN (2013) STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell 50, 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taboada B, Verde C, and Merino E (2010) High accuracy operon prediction method based on STRING database scores. Nucleic Acids Res 38, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahn J, and Barber GN (2019) STING signaling and host defense against microbial infection. Exp Mol Med 51, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, and Wang C (2016) The emerging roles of the STING adaptor protein in immunity and diseases. Immunology 147, 285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zasloff M (2019) Antimicrobial Peptides of Multicellular Organisms: My Perspective. Adv Exp Med Biol 1117, 3–6 [DOI] [PubMed] [Google Scholar]
- 34.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, and Hooper LV (2011) The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El Aidy S, van Baarlen P, Derrien M, Lindenbergh-Kortleve DJ, Hooiveld G, Levenez F, Dore J, Dekker J, Samsom JN, Nieuwenhuis EE, and Kleerebezem M (2012) Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol 5, 567–579 [DOI] [PubMed] [Google Scholar]
- 36.Loonen LM, Stolte EH, Jaklofsky MT, Meijerink M, Dekker J, van Baarlen P, and Wells JM (2014) REG3gamma-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol 7, 939–947 [DOI] [PubMed] [Google Scholar]
- 37.Liu S, Zhang Y, Ren J, and Li J (2015) Microbial DNA recognition by cGAS-STING and other sensors in dendritic cells in inflammatory bowel diseases. Inflammatory bowel diseases 21, 901–911 [DOI] [PubMed] [Google Scholar]
- 38.Ahn J, Son S, Oliveira SC, and Barber GN (2017) STING-Dependent Signaling Underlies IL-10 Controlled Inflammatory Colitis. Cell reports 21, 3873–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ching CB, Gupta S, Li B, Cortado H, Mayne N, Jackson AR, McHugh KM, and Becknell B (2018) Interleukin-6/Stat3 signaling has an essential role in the host antimicrobial response to urinary tract infection. Kidney Int 93, 1320–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y, Chen F, Wu W, Sun M, Bilotta AJ, Yao S, Xiao Y, Huang X, Eaves-Pyles TD, Golovko G, Fofanov Y, D’Souza W, Zhao Q, Liu Z, and Cong Y (2018) GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal immunology 11, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao Y, Huang X, Zhao Y, Chen F, Sun M, Yang W, Chen L, Yao S, Peniche A, Dann SM, Sun J, Golovko G, Fofanov Y, Miao Y, Liu Z, Chen D, and Cong Y (2019) Interleukin-33 Promotes REG3gamma Expression in Intestinal Epithelial Cells and Regulates Gut Microbiota. Cellular and molecular gastroenterology and hepatology 8, 21–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rambold AS, and Pearce EL (2018) Mitochondrial Dynamics at the Interface of Immune Cell Metabolism and Function. Trends in immunology 39, 6–18 [DOI] [PubMed] [Google Scholar]
- 43.Cairns RA, Harris IS, and Mak TW (2011) Regulation of cancer cell metabolism. Nature reviews. Cancer 11, 85–95 [DOI] [PubMed] [Google Scholar]
- 44.Blaut M, and Clavel T (2007) Metabolic diversity of the intestinal microbiota: implications for health and disease. The Journal of nutrition 137, 751S–755S [DOI] [PubMed] [Google Scholar]
- 45.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay JM, Langella P, Xavier RJ, and Sokol H (2016) CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nature medicine 22, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duffy MM, Regan MC, Ravichandran P, O’Keane C, Harrington MG, Fitzpatrick JM, and O’Connell PR (1998) Mucosal metabolism in ulcerative colitis and Crohn’s disease. Diseases of the colon and rectum 41, 1399–1405 [DOI] [PubMed] [Google Scholar]
- 47.Douros JD, Baltzegar DA, Reading BJ, Seale AP, Lerner DT, Grau EG, and Borski RJ (2018) Leptin Stimulates Cellular Glycolysis Through a STAT3 Dependent Mechanism in Tilapia. Frontiers in endocrinology 9, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li M, Jin R, Wang W, Zhang T, Sang J, Li N, Han Q, Zhao W, Li C, and Liu Z (2017) STAT3 regulates glycolysis via targeting hexokinase 2 in hepatocellular carcinoma cells. Oncotarget 8, 24777–24784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sola-Penna M, Paixao LP, Branco JR, Ochioni AC, Albanese JM, Mundim DM, Baptista-de-Souza D, Figueiredo CP, Coelho WS, Marcondes MC, and Zancan P (2020) Serotonin activates glycolysis and mitochondria biogenesis in human breast cancer cells through activation of the Jak1/STAT3/ERK1/2 and adenylate cyclase/PKA, respectively. British journal of cancer 122, 194–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.