Abstract
African swine fever (ASF) is an infectious transboundary disease of domestic pigs and wild boar and spreading throughout Eurasia. There is no vaccine and treatment available. Complex immune escape strategies of African swine fever virus (ASFV) are crucial factors affecting immune prevention and vaccine development. MGF360 genes have been implicated in the modulation of the IFN-I response. The molecular mechanisms contributing to innate immunity are poorly understood. In this study, we demonstrated that ASFV MGF360-12L (MGF360 families 12L protein) significantly inhibited the mRNA transcription and promoter activity of IFN-β and NF-κB, accompanied by decreases of IRF3, STING, TBK1, ISG54, ISG56 and AP-1 mRNA transcription. Also, MGF360-12L might suppress the nuclear localization of p50 and p65 mediated by classical nuclear localization signal (NLS). Additionally, MGF360-12L could interact with KPNA2, KPNA3, and KPNA4, which interrupted the interaction between p65 and KPNA2, KPNA3, KPNA4. We further found that MGF360-12L could interfere with the NF-κB nuclear translocation by competitively inhibiting the interaction between NF-κB and nuclear transport proteins. These findings suggested that MGF360-12L could inhibit the IFN-I production by blocking the interaction of importin α and NF-κB signaling pathway, which might reveal a novel strategy for ASFV to escape the host innate immune response.
Electronic supplementary material
The online version of this article (10.1007/s12250-020-00304-4) contains supplementary material, which is available to authorized users.
Keywords: African swine fever virus (ASFV), MGF360-12L, IFN-I, NF-κB, Nuclear transport
Introduction
African swine fever (ASF) caused by African swine fever virus (ASFV) is a viral haemorrhagic disease of domestic and wild boars, and is currently spreading in Africa, Europe, and Asia, becoming a global threat with huge economic and ecological consequences (Dixon et al. 2019; Pikalo et al. 2019). ASFV can evade the host’s defense system to escape the native immunity which depends on complex interactions between virus and host. ASF is solely controlled through the application of strict sanitary measures in infected quarantine areas, and eradication is possible only by prevention or the slaughter of diseased animals (Cisek et al. 2016). The large genome and complex immune escape mechanism have challenged ASFV vaccine research.
ASFV, the only member of the Asfaviridae family (Alonso et al. 2018), is an envelope, cytoplasmic double-stranded DNA virus which encode more than 160 proteins. ASFV is divided into 24 genotypes based on the B646L gene, which encodes the capsid protein p72 (Rodriguez and Salas 2013). The genomes of different isolates vary in length from 170 to 190 kb (Dixon et al. 2013). ASFV has evolved multiple mechanisms for the manipulation of IFN responses, which has been utilized to evade the host’s innate and adaptive immune. For example, B119L, EP402R, L83L, DP148R proteins participate in immune escape (Monteagudo et al. 2017; Reis et al. 2017; Borca et al. 2018). The ASFV A238L, DP71L, EP153R, A224L and A179L are related to apoptosis of host cells (Revilla et al. 1997; Rodriguez et al. 2002; Hernaez et al. 2004; Hurtado et al. 2004; Barber et al. 2017), and pI215L is involved in interfering ubiquitination (Freitas et al. 2018). However, the function of many viral gene remains to be further clarified, particularly the members of multigene families (MGFs) (Tulman and Rock 2001; Correia et al. 2013).
ASFV MGFs are located in the left terminal 40 kb and right terminal 20 kb of the genome. Variation in the genomes of different ASFV isolates is most common due to gain or loss of MGFs 100, 110, 300, 360, 505/530 and p22 family (Dixon et al. 2013). ASFV has been reported to reduce the IFN-I response by targeting different intracellular signaling intermediates. A276R gene from MGF360 could inhibit IFN-β expression via both the TLR3 and the cytosolic pathways by targeting IRF3, but not IRF7 or NF-κB (Gallardo et al. 2018). However, the ASFV A528R from MGF505 inhibited the induction of both NF-κB and IRF3 branches of the IFN-I induction signaling pathway. The ASFV I329L gene was a functional viral Toll like receptor 3 (TLR3) homologue, which can inhibit the induction of IFN mediated by TIR domain-containing adaptor inducing interferon-β (TRIF) (Correia et al. 2013). DP96R was reported to negatively regulate IFN-I expression and NF-κB signaling by inhibiting both TANK binding kinase-1 (TBK1) and Inhibitor of kappa B kinase β (IKKβ) (O’Donnell et al. 2017; Wang et al. 2018). Studies demonstrated that deletion of 12L, a member of MGF360, resulted in induction of IFN-I (Reis et al. 2016). It has been hypothesized that MGF360-12L is a potential immune evasion protein. However, the mechanism of its action remains virtually unknown.
Escaping the host innate immune has a critical role in ASFV pathogenesis. IFN-Is are the earliest innate immune mediators against viral infection (Samuel 2001; Randall and Goodbourn 2008), which have been triggered by recognition of viral components on cell membrane and/or cytosolic pattern recognition receptors (PRRs) in infected cells (Henneke et al. 2002). IFN-β expression is regulated by transcription enhancers, including IRF3 and NF-κB (Kim and Maniatis 1997). The activation of NF-κB signaling pathway is relied on IκBα phosphorylation and degradation, which contributes to NF-κB nuclear translocation. The inhibition of NF-κB nuclear translocation results in escaping the host innate immune. It is showed that NF-κB nuclear translocation need specific carriers, which contain a nuclear localization signal (NLS) and are termed as karyopherins (including importin α and importin β). The N-terminus of the importin α protein contains the importin β binding (IBB) domain and auto-regulatory region, which can mimic the nuclear localization signal to block NLS binding site in normal circumstances. However, the NLS of exogenous cargo protein will compete to bind the NLS binding site of the importin α protein, which results in expose of auto-regulatory region and IBB domain. Then the IBB domain binds importin β protein, which contributes to nuclear localization of NLS-importin α/importin β trimer (Lange et al. 2007). Studies demonstrated that deletion of MGF360-12L can lead to induction of IFN-I (Reis et al. 2016), which indicated that MGF360-12L may be involved in the immune escape of the virus. However, the precise molecular mechanism for inhibition of MGF360-12L on the NF-κB signaling pathway is obscure.
In this study, we aimed to investigate the role of MGF360-12L in immune evasion. We found that the MGF360-12L impaired the capability of cells to produce IFN-β via inhibition of NF-κB nuclear translocation. MGF360-12L was demonstrated to bind to nuclear transport proteins, importin α2 (KPNA2), importin α4 (KPNA3) and importin α3 (KPNA4), which competitively blocked the interactions of KPNA2, KPNA3 and KPNA4 with p65. The findings might suggest a novel strategy by which MGF360-12L could subvert cellular innate immunity and evade host antiviral responses.
Materials and Methods
Cell Culture and Transfection
HeLa cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 supplemented (Cat. no: SH30809.01, Hyclone, USA) with 10% fetal bovine serum (Cat. no: 10270-106, Gibco, USA), 1% penicillin–streptomycin solution (15140122, Gibco, USA) at 37 °C (Thermo Fisher Scientific, Loughborough, UK) in a humidified atmosphere of 5% CO2. Cells were seeded in 6- or 12-well plates (Corning, USA) at 1 × 105 or 1×104 cells/well, respectively, and then transfected with plasmid at a confluence of 70%–80%. HeLa cells were transfected using Lipofectamine 2000 reagent (Cat. no: 1756108, Invitrogen, USA), according to the manufacturer’s instructions.
Plasmid Construction
The MGF360-12L gene was amplified from pcDNA3.1-MGF360-12L and then cloned into pCAGGS-Flag vector. MGF360-12L (1–1053 nucleotides) and truncated mutants of the MGF360-12L (160–1053 nucleotides) were cloned into pCAGGS-Flag (NLS sequence position is predicted by cNLS mapper) using EcoRI (Cat. no: FD0274, Thermo Scientific™, USA) and XholI (Cat. no: FD0694, Thermo Scientific™, USA) sites, and named as pFlag-12L and pFlag-Δ12L respectively. pcDNA3.1-MGF360-12L was kindly provided by Jianzhong Zhu (Yangzhou University) and Xiaoyu Guo (Chinese Academy of Agricultural Sciences).
RNA Extraction and Quantitative PCR (qPCR)
Total RNA was extracted from the cells with Trizol reagent (Invitrogen, Carlsbad, CA, USA), and 1 μg RNA was reverse transcribed to cDNA using Hifair® II 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus) (Cat: no: 11123ES60, YEASEN, China). To determine the effects of MGF360-12L on the expression of IFN-β, IRF3, AP-1, NF-κB, ISG54, ISG56, STING and TBK1, HeLa cells in 6-well plates were transfected with 2.5 μg of the pCAGGS empty vector or a plasmid encoding the MGF360-12L. After 12 h, the cells were treated with poly (I: C) 12 h or TNFα 30 min. qPCR was performed using qPCR Super mix (Cat: no: 11201ES08, YEASEN, China) in Roche LightCycler480 qPCR system. The abundance of the individual mRNA transcripts was assayed in triplicate and normalized to GAPDH mRNA using 2−ΔΔCT method. All samples were performed in triplicate. The primers (synthesized in Qingke, China) used are listed in the Supplementary Table S1.
Dual-Luciferase Reporter (DLR) Assays
Cells were grown to 70%–80% confluency and co-transfected with plasmids IFN-β-Luc and pRL-TK by Lipofectamine 2000 reagent. Twelve hours later, cells were treated with poly (I: C) (5.0 μg/mL) (Cat. no: B5551, APEXBIO, USA). After 12 h stimulation, luciferase activities were measured with Dual Luciferase Assay Kit (Cat. no: E1910, Promega, USA) in a Microplate Luminometer.
Coimmunoprecipitation and Western Blotting
The cells were lysed in RIPA buffer (Cat. no: PP1202, Aidelai, China) containing protease inhibitor cocktail (Cat. no: G2006, Servicebio, China) for 60 min on ice. Protein A+G agarose beads (50 μL) (Cat. no: 36403ES03, YEASEN, China) were incubated with the indicated antibodies at 4 °C overnight. The cell lysate was added and incubated for another 6 h at 4 °C. The agarose beads were subsequently washed three times with RIPA buffer containing protease inhibitor cocktail. The cytoplasmic and nuclear proteins were extracted using Nuclear and Cytoplasmic Protein Extraction Kit (Cat. no: P0028, Beyotime, China). Protein concentrations were measured with a bicinchoninic acid (BCA) protein assay kit (Cat. no: PP0101, Aidelai, China). The proteins were separated by 8% or 12% polyacrylamide gel electrophoresis containing 0.1% SDS and transferred to PVDF membranes. The membranes were incubated for 2 h at room temperature in blocking buffer (20 mmol/L Tris-HCl, 137 mmol/L NaCl, pH 8.0, containing 0.1% Tween and 5% non-fat dry milk) and probed with antibodies against KPNA1 (18137-1-AP, Proteintech, China), KPNA2 (10819-1-AP, Proteintech, China), KPNA3 (abs118425, Absin, China), KPNA4 (12463-1-AP, Proteintech, China), KPNA5(13963-1-AP, Proteintech, China), KPNA6 (12366-2-AP, Proteintech, China), KPNB1 (10077-1-AP, Proteintech, China), Flag (20543-1-AP, Proteintech, China), Myc (60003-2-lg, Proteintech, China), p65 (10745-1-AP, Proteintech, China), p50 (14220-1-AP, Proteintech, China), IκBα (10268-1-AP, Proteintech, China), 1:2000 dilution; Histone H3 (1: 10000, GB13102-1, Servicebio, China), GAPDH (1: 5000, GB12002, Servicebio, China) antibodies at 4 °C overnight. After being washed three times with TBST (Tris Buffered Saline Tween), the membranes were incubated with HRP-conjugated anti-IgG (1: 5000, GB23303, Servicebio, China) at room temperature for 2 h. GAPDH or Histone H3 was used as internal controls. The reacted proteins were visualized using an electrochemiluminescence (ECL) system (Biotanon, China).
Immunofluorescence Assay
HeLa cells were cultured in 12-well plates and transfected with 1 μg pFlag-12L/pEGFP-C1-GST-2VSV40T (Zhao et al. 2019) /Empty vector, and after 24 h, cells were fixed with 4% paraformaldehyde at 37 °C for 20 min. Next, cells were incubated with PBS containing 1% Bovine Serum Albumin (BSA) and 1% TritonX-100 at 37 °C for 2 h, and then incubated with primary antibodies Anti-Flag (1: 100, 20543-1-AP, Proteintech, China), p65 (1: 100, 10745-1-AP, Proteintech, China), p50 (1: 100, 14220-1-AP, Proteintech, China) overnight at 4 °C. After three washes for 30 min, cells were further incubated with Alexa Fluor 488-conjugated affinipure goat anti-rabbit IgG (H+L) (1: 100, SA00013-2, Proteintech, China) or Alexa Fluor 594-conjugated affinipure goat anti-rabbit IgG (H+L) (1: 100, SA00013-4, Proteintech, China) for 1 h at room temperature, and the fluorescence was examined using a fluorescent microscopy (Zeiss) at a magnification of 40×.
Statistical Analysis
Data were expressed as the mean ± SEM of at least three independent experiments for each cellular experimental group. The results were from one representative experiment. Statistical analysis was performed by using Student’s t-test with Graphpad Prism version 5.0 (*P < 0.05; **P < 0.01; ***P < 0.001; ns indicates no significance).
Results
MGF360-12L Interferes with IFN-β Signaling Pathway
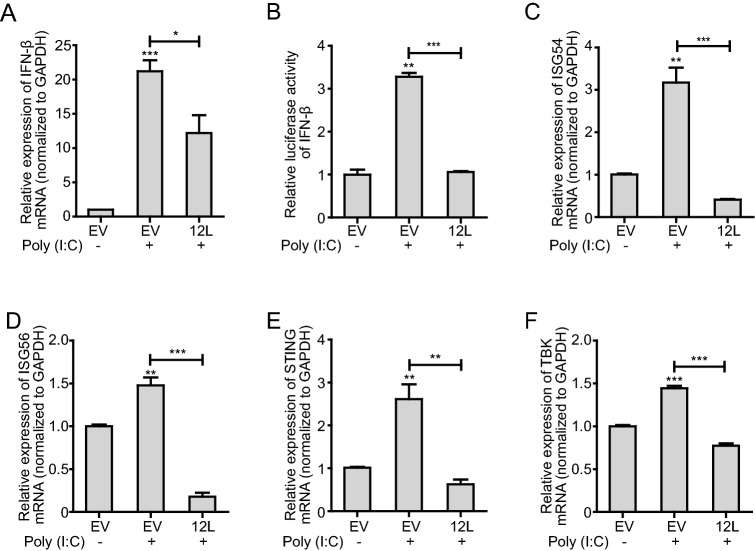
MGF360 genes have been implicated in the modulation of the IFN-I response (Reis et al. 2016). In order to confirm whether ASFV MGF360-12L involves in IFN-I signaling and modulates the induction of IFN-I, pFlag-12L was co-transfected with IFN-β-Luc reporter plasmid or pRL-TK plasmid respectively in HeLa cells. At 12 h post-transfection, cells were stimulated with poly (I: C) for 12 h, and subjected to qPCR assay to detect the mRNA transcription levels of key IFN-I signaling genes. Also, the promoter activity of IFN-β was investigated by using dual-luciferase reporter assays. Results showed that poly (I: C) treatment significantly increased IFN-β mRNA transcription (Fig. 1A) and IFN-β-Luc reporter activity (Fig. 1B). The mRNA transcription levels of key genes related to IFN-I signaling (ISG54, ISG56, STING, TBK1) were also elevated (Fig. 1C–1F). However, the IFN-β mRNA transcription and IFN-β-Luc reporter activity were blocked in cells transfected with pFlag-12L.
Fig. 1.
MGF360-12L interferes with IFN-β signaling pathway. 2.5 µg of plasmid encoding MGF360-12L or empty vector (EV) were transfected into HeLa cells. After 12 h, cells were left untreated or transfected with poly (I: C) (5 µg/mL) and incubated for another 12 h. qPCR was performed to measure IFN-β (A), ISG54 (C), ISG56 (D), STING (E), TBK1 (F) mRNA transcription. B 2.5 µg of plasmid encoding MGF360-12L or empty vector (EV) were co-transfected into HeLa cells with the IFN-β promoter-dependent reporter plasmid p125-Luc and with pRL-TK for normalization. After 12 h, cells were left untreated or transfected with poly (I: C) (5 µg/mL) and incubated for another 12 h. Luciferase activity was determined by dual-luciferase assay. Expression data were normalized to the expression of GAPDH. ***P < 0.001; **P < 0.01; *P < 0.05 (compared to cells transfected with empty vector) (n = 3).
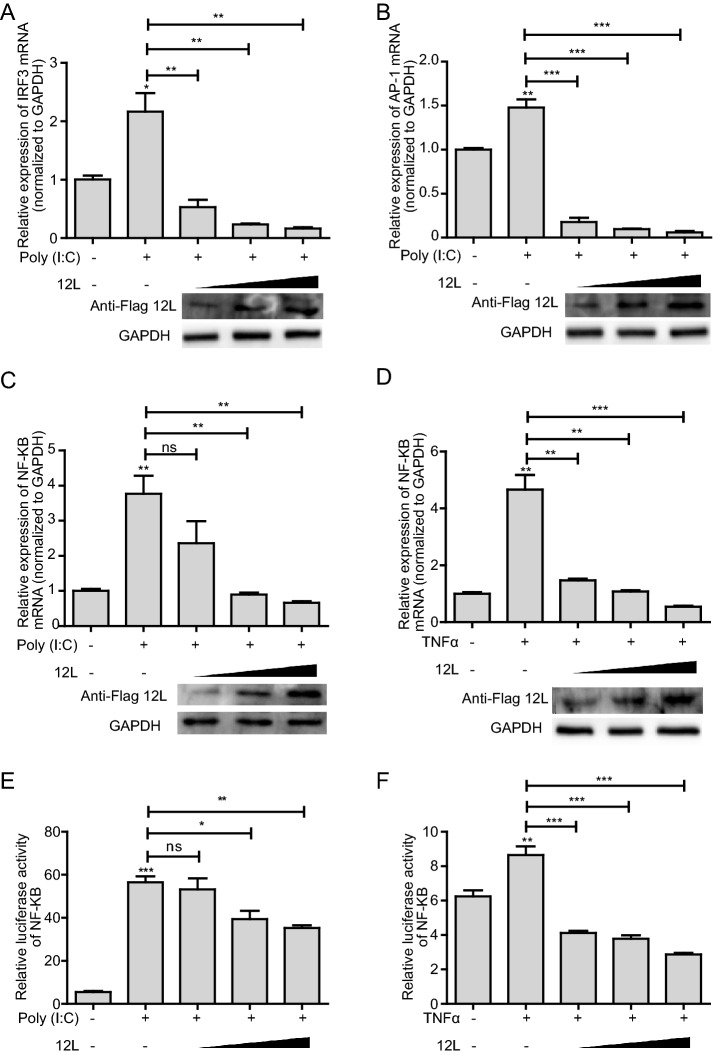
In order to further investigate the effect of MGF360-12L on IFN-I signaling pathway, HeLa cells were transfected with the increasing amounts of pFlag-12L. The expression of IFN-β transcription enhancing factors (IRF3, AP-1, and NF-κB) were analyzed by qPCR and dual-luciferase reporter assay. The results showed that MGF360-12L reduced the IRF3 (Fig. 2A), AP-1 (Fig. 2B), NF-κB (Fig. 2C–2F) expression in a dose-dependent manner after poly (I: C) or TNFα stimulation. These results confirmed the antagonistic property of MGF360-12L in IFN-β signaling pathway.
Fig. 2.
MGF360-12L inhibits poly (I: C)-induced production of IRF3, AP-1 and NF-κB. Increasing amounts (1.0 µg, 2.5 µg and 5.0 µg) of plasmid encoding MGF360-12L or empty vector (EV) were transfected into HeLa cells. 12 h later, cells were transfected with or without poly (I: C) (5 µg/mL) or TNFα (5 µg/mL). IRF3 (A), AP-1(B), NF-κB (C, E) mRNA level was measured by qPCR. D, F Increasing amounts (1.0 µg, 2.5 µg and 5.0 µg) of plasmid encoding MGF360-12L or empty vector (EV) were co-transfected into HeLa cells with the NF-κB-Luc and with pRL-TK for normalization. After 12 h, cells were left untreated or transfected with poly (I: C) (5 µg/mL) or TNFα (10 µg/mL) and incubated for another 30 min. Luciferase activity was determined by dual-luciferase assay. Expression data were normalized to the expression of GAPDH. ***P < 0.001; **P < 0.01; *P < 0.05 (compared to cells transfected with empty vector) (n = 3). The protein levels of MGF360-12L were determined by Western blotting.
MGF360-12L Blocks the Nuclear Translocation of NF-κB
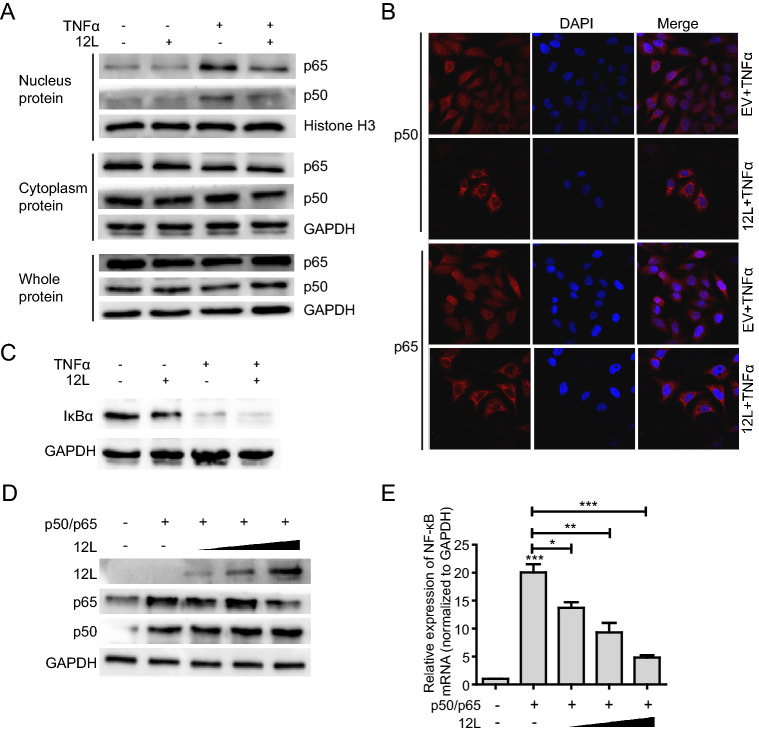
NF-κB nuclear translocation is the precondition of interferon production. Therefore, we checked the effect of MGF360-12L on NF-κB nuclear translocation. HeLa cells were transfected with pFlag-12L for 24 h and stimulated with TNFα for 30 min. Western blotting was performed to analyze the levels of NF-κB subunits (p50 and p65) in whole-cell lysates, cytoplasmic, and nuclear extracts. Results demonstrated that MGF360-12L did not affect the levels of p50 and p65 in whole-cell lysate; and there was no significant change in the cytoplasmic fraction. However, MGF360-12L transfection significantly reduced the expression of p50 and p65 in the nucleus after TNFα treatment (Fig. 3A). Meanwhile, we have explored nuclear translocation of IRF3 after MGF360-12L transfection. The data show that MGF360-12L can inhibit pIRF3 expression and nuclear translocation to suppress IRF3 signaling pathway (Supplementary Figure S1).
Fig. 3.
MGF360-12L inhibits NF-κB nuclear translocation independent of IκBα expression. A HeLa cells were transfected with a plasmid encoding MGF360-12L or with empty vector (EV). 24 h later, cells were transfected with or without TNFα (20 µg/mL) and incubated for another 30 min, followed by lysis. Cell lysates were separated into cytoplasmic and nuclear extracts, and the protein levels of p65 and p50 in the whole-cell lysate, cytoplasmic extract, and nuclear extract were analyzed by Western blotting. Protein levels of p65 and p50 were quantified by Western blotting and normalized to the the expression of of GAPDH or Histone H3. B HeLa cells were transfected with pFlag-12L (2.5 µg) or empty vector. 24 h later, cells were transfected with TNFα (20 µg/mL), At 30 min post, cells treated with TNFα were fixed and stained with DAPI (blue), anti-p65 (red), anti-p50 (red) antibody and examined by confocal microscope. Magnification, 40×. C HeLa cells were transfected with a plasmid encoding MGF360-12L or with empty vector. 24 h later, cells were transfected with or without TNFα. After incubation for another 30 min, the levels of IκBα were analyzed by Western blotting. D, E Increasing amounts (1.0 µg, 2.5 µg and 5.0 µg) of plasmid encoding MGF360-12L or empty vector (EV) were co-transfected into HeLa cells with plasmids expressing p50 and p65. At 24 h after transfection, cells were lysed and NF-κB mRNA expression was determined by qPCR. ***P < 0.001; **P < 0.01; *P < 0.05 (compared to cells transfected with TNFα and empty vector) (n = 3). MGF360-12L, p65 and p50 expression levels were analyzed by Western blotting and normalized to the expression of GAPDH.
To further explore the subcellular localization of p65 and p50 after MGF360-12L transfection, the transfected cells were inoculated with TNFα for 30 min, and the expression of p65 and p50 was detected by indirect immunofluorescence assay. Endogenous p65 and p50 protein was distributed diffusely in both cytoplasm and cell nuclei in mock-infected cells; in contrast, pFlag-12L transfection resulted in the accumulation of p65 and p50 around the nucleus and presented a characteristic packed tightly to the nuclear area (Fig. 3B). These observations revealed the capacity of MGF360-12L to inhibit the nuclear localization of p50 and p65.
MGF360-12L Inhibits NF-κB Nuclear Translocation Independent of IκBα Expression
IκBα is an inhibitory protein that binds to NF-κB transcription factors. Degradation of the IκB protein can result in NF-κB signaling activation by NF-κB NLSs. To investigate whether nuclear translocation of NF-κB regulated by MGF360-12L is related with IκBα, the expression of IκBα protein was examined after MGF360-12L transfection and TNFα stimulation. The results showed that the IκBα protein expression was significantly down-regulated after TNFα treatment, while the MGF360-12L transfection did not affect the degradation of IκBα (Fig. 3C), indicating that inhibition of NF-κB nuclear translocation by MGF360-12L might not depend on IκBα degradation. To further demonstrate the relationship between MGF360-12L and NF-κB inhibition, HeLa cells were co-transfected with different concentration plasmid of pFlag-12L, p65 or p50 plasmids (Ye et al. 2017). At 24 h after transfection qPCR was performed. MGF360-12L, p65 and p50 protein expression and NF-κB mRNA transcription expression were investigated. The data showed that p50 and p65 can be expressed with no significant difference after cells were co-transfected with pFlag-12L, pCDN3.1-p50 and pCDN3.1-p65 (Fig. 3D). However, NF-κB mRNA transcription was elevated by p65/p50 (Fig. 3E). Conversely, ectopic expression of different concentration pFlag-12L plasmid significantly reduced this induction with a dose-dependent manner (Fig. 3E). These data suggested that MGF360-12L might directly inhibit NF-κB nuclear localization independent of IκBα degradation.
MGF360-12L Interrupts the Nuclear Translocation of NF-κB by Targeting Importin α
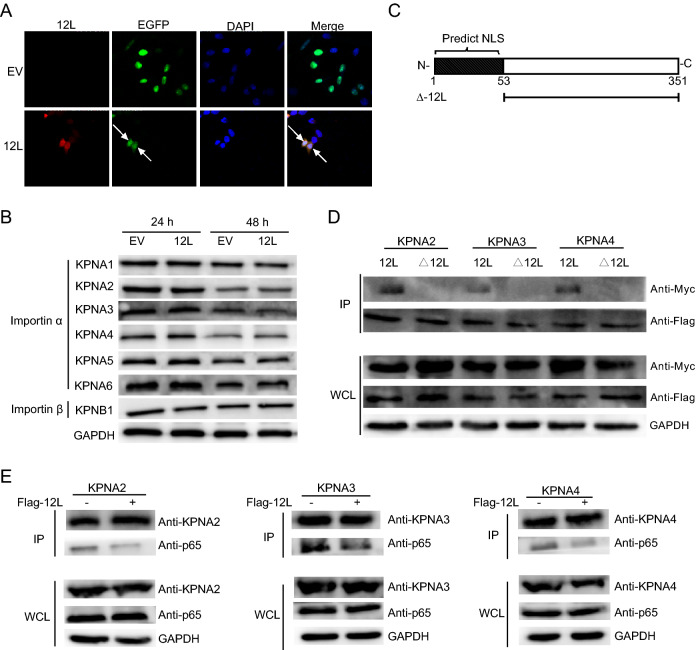
NLS is essential for the transport of active NF-κB complexes into the nucleus, which contributes to regulate the expression of cellular and viral genes. In order to investigate whether MGF360-12L had effect on the classical nuclear entry pathway, the plasmids expressing the classical NLS EGFP tandem protein (pEGFP-C1-GST-2VSV40T) (Zhao et al. 2019) were co-transfected with MGF360-12L plasmids in HeLa cells. After transfection 36 h, indirect immunofluorescence assay was performed. In control group, the EGFP proteins with SV40 Tag NLS located in the nucleus completely, and no EGFP can be observed in the cytoplasm. However, the green fluorescence diffusely distributed in the cytoplasm in MGF360-12L transfected cells (Fig. 4A). It indicated that MGF360-12L could inhibit the nuclear transport of proteins mediated by classical NLS.
Fig. 4.
MGF360-12L may interact with KPNA2, KPNA3 and KPNA4 and disrupt the interaction between p65 and KPNA2, KPNA3, KPNA4. A The plasmid encoding MGF360-12L or empty vector (EV) were co-transfected into HeLa cells with plasmids EGFP-C1-GST-2VSV40T. After transfection 36 h, cells were fixed and stained with DAPI (blue), EGFP (green), anti-Flag (red) antibody and examined by confocal microscope. Magnification, 40×. B HeLa cells were transfected with ASFV pFlag-12L plasmid or empty vector. At 24 h and 48 h after transfection, collection cells, and the expression levels of endogenous KPNA1, KPNA2, KPNA3, KPNA4, KPNA5, KPNA6, and KPNB1 were analyzed by Western blotting. C Schematic diagram of NSL-deleted 12L. D HeLa cells were co-transfected with pFlag-12L or pFlag-Δ12L plasmid and the pMyc-KPNA2, pMyc-KPNA3 or pMyc-KPNA4. At 36 h after transfection, immunoprecipitation (IP) was performed with whole-cell lysates by using anti-Flag antibody. E HeLa cells were co-transfected with the pMyc-KPNA2, pMyc-KPNA3 or pMyc-KPNA4 and pFlag-12L plasmid or empty vector. At 24 h after transfection, cells were treated with TNFα (5 µg/mL) for 30 min and collected. Immunoprecipitation was performed with KPNA2, KPNA3 or KPNA4 antibodies, and Western blotting was performed with antibodies against KPNA2, KPNA3 or KPNA4, Flag and p65.
The shuttle of NF-κB between the nucleus and the cytoplasm through a nuclear-pore complexs (NPCs) depends on importin-cargo nuclear complex and nuclear export of importin (Poon and Jans 2005; Stewart 2007). Nuclear transport proteins include importin α (KPNA1–6) and importin β (KPNB1). To investigate whether MGF360-12L might target the nuclear transport proteins, we first detected the expression levels of importin α (KPNA1–4) and importin β (KPNB1) in HeLa cells after transfected with pFlag-12L plasmid or blank vector (Fig. 4B). The data showed that there was no significant difference of importin α and importin β expression after MGF360-12L transfection, which means MGF360-12L doesn’t affect the expression of importin proteins. Thus, we speculated that MGF360-12L might inhibit the nuclear transportation of NF-κB by competitively interacting with importin α. We used cNLS Mapper software to predict the NLS region in MGF360-12L (Fig. 4C), and constructed NSL-deleted mutant MGF360-Δ12L. HeLa cells were co-transfected with Myc-tagged KPNA2, KPNA3, KPNA4 plasmids (Ye et al. 2017) and pFlag-12L or pFlag-Δ12L. Immunoprecipitation assay revealed that MGF360-12L interacted with KPNA2, KPNA3, and KPNA4 (Fig. 4D), and no interaction was observed between MGF360-Δ12L and KPNAs.
To explore whether MGF360-12L may interrupt the nuclear translocation of p65 by affecting the nuclear transport proteins, co-immunoprecipitations were performed with subsequent Western blotting using p65 antibody to detect the interaction of p65 with KPNA2, KPNA3 and KPNA4. HeLa cells were co-transfected with pFlag-12L plasmid and the Myc-KPNA2, Myc-KPNA3 or Myc-KPNA4 plasmid following by TNFα treatment. Cell lysates were collected for immunoprecipitation at 36 h after transfection. Co-immunoprecipitations and western blotting were performed. The results revealed that the interaction between p65 and KPNA2, KPNA3, KPNA4 was weakened followed by MGF360-12L transfection (Fig. 4E), which indicates that MGF360-12L competitively interrupted the interaction of p65 with KPNA2, KPNA3 and KPNA4. Together, these findings demonstrated that MGF360-12L interrupts the nuclear translocation of NF-κB by targeting KPNA2, KPNA3, and KPNA4.
Overexpression of KPNA2, KPNA3, KPNA4 Restore the MGF360-12L Induced IFN-β Suppression
To further confirm that MGF360-12L inhibited the activation of IFN-I by targeting KPNA2, KPNA3 and KPNA4. Overexpressing plasmids of KPNA2, KPNA3 and KPNA4 were constructed to test whether they can reverse the inhibition of IFN-β expression induced by MGF360-12L. The plasmids of KPNA2, KPNA3 or KPNA4 were co-transfected into HeLa cells with pFlag-12L or empty vector plasmid to measure the expression of IFN-β mRNA using qPCR. Result showed that overexpression of KPNA2, KPNA3 or KPNA4 can restore the expression of IFN-β inhibited by MGF360-12L (Fig. 5). The results indicated that MGF360-12L might attenuate the IFN-β expression by targeting KPNA2, KPNA3 and KPNA4 to escape the immunity.
Fig. 5.
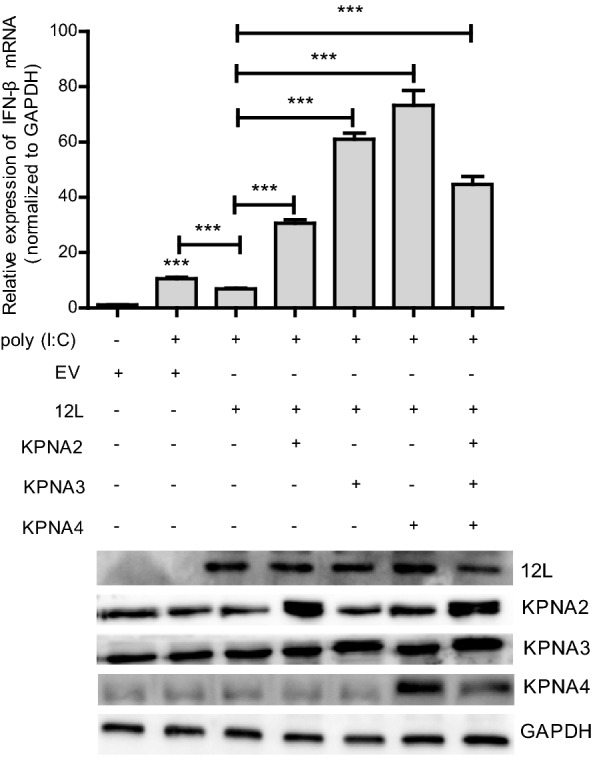
Overexpression of KPNA2, KPNA3, KPNA4 restores the MGF360-12L induced IFN-β suppression. (Top) HeLa cells were transfected or co-transfected with pFlag-12L or pMyc-KPNA2, pMyc-KPNA3, pMyc-KPNA4. After transfection 12 h, cells were left untreated or transfected with poly (I:C) (5 µg/mL). After 12 h, the mRNA levels of IFN-β were detected by qPCR. *** P < 0.001 compared to cells transfected with poly (I: C), empty vector or pFlag-12L (n = 3). (Bottom) MGF360-12L expression levels were analyzed by Western blotting.
Discussion
ASFV causes a lethal hemorrhagic disease in swine and wild boars with a fatality rate close to 100% (Galindo and Alonso 2017). There is no effective vaccine developed to prevent ASF. ASFV can evade the host’s defense system to escape the native immunity depending on complex interactions of virus and host factors (Golding et al. 2016). IFN-I is a crucial component of the innate response to viral infection (Golding et al. 2016), which are produced following recognition of microbial products by cell surface and intracellular pattern recognition receptors, and contributed to innate immunity by inducing production of anti-viral proteins to eliminate invasive pathogens. IFN-Is expression can be triggered by the collaboration of NF-κB, IRF3 and Ap-1 (Kim and Maniatis 1997). Activation of NF-κB, IRF3 and AP-1 engagement with IFN-β enhancer are needed for maximal levels of IFN-I expression contributing to an important defense against viral infections (Panne 2008). ASFV has been reported sensitivity to IFN-Is (Portugal et al. 2018). MGF360 and MGF 530 genes related with length variations in the genome have been implicated in the modulation of IFN-Is (Burrage et al. 2004). MGFs protein including A276R, A528R, I329L, DP96L can inhibit IFN signaling by targeting TLR3, IRF3, cGAS-STING (Correia et al. 2013; Golding et al. 2016; Wang et al. 2018). In this study, results demonstrated that MGF360-12L significantly inhibited IFN-β and NF-κB mRNA transcription and promoter activity following poly (I: C) or TNFα stimulation, accompanied by a decrease of IRF3, AP-1 mRNA expression, suggesting that MGF360-12L is able to inhibit IFN-Is expression.
NF-κB functions as nuclear transcription factor that regulates the expression of genes influencing a broad range of biological processes including immunity, inflammation, stress responses, development, and lymphoid organogenesis. NF-κB can be activated after exposure to pathogens, pathogen-associated molecular patterns and various cytokines, which results in post-translational modifications (phosphorylation, acetylation, glycosylation) and translocation to the nucleus (Rahman and McFadden 2011). In this study, we confirmed that MGF360-12L significantly inhibited NF-κB mRNA transcription and promoter activity. We speculated that MGF360-12L could inhibit the nuclear translocation of NF-κB. Results indicated that there was no significant change of p50 and p65 in cytoplasm. However, MGF360-12L transfection significantly reduced the expression of p50 and p65 in the nucleus after TNFα treatment, and the inhibition of p50 and p65 mRNA transcription was showed the dos-dependent manner after MGF360-12L overexpression. It is showed that NF-κB can be inhibited in non-stimulated cells through cytoplasmic retention by IκB which can block the interaction of NF-κB and importin α (Beg et al. 1992). IκB degradation results in the complex of NF-κB and importin gene to enter the nucleus and NF-κB functions as a transcription factor initiating the target genes expression. Our findings suggested that MGF360-12L did not alter the degradation of IκBα mediated by TNFα, suggesting that MGF360-12L inhibits NF-κB nuclear translocation independent of IκBα degradation.
NF-κB nuclear translocation is not only related to IκBα degradation, but also relevant to interaction with Karyopherins. Karyopherins including karyopherin-α (KPNA) and karyopherin-β (KPNB) is a nuclear transport receptor that is responsible for transporting its specific cargo proteins, which harbor nuclear localization signals (NLSs) through the nuclear pore complex (NPCs) (Beaudet et al. 2020; Hazawa et al. 2020). Some viruses, such as Japanese encephalitis virus (JEV) (Ye et al. 2017), Ebola virus (EBOV) (Reid et al. 2006), pelargonium line pattern virus (Perez-Canamas and Hernandez 2018), hantavirus (HTNV) (Taylor et al. 2009), prevent nuclear import of cellular cargo molecules by binding importin family proteins or down-regulating importin alpha expression. Porcine reproductive and respiratory syndrome virus (PRRSV) induces KPNA1 degradation and inhibits ISGF3 Nucleation (Wang et al. 2013). Importin-α3, one of the main NF-κB transporters, is abundantly expressed in the mammalian respiratory tract. Importin-α3-deficient mice showed reduced NF-κB-activated antiviral gene expression and increased influenza lethality (Thiele et al. 2020). The capsid protein of venezuelan equine encephalitis virus (VEEV) forms a tetramer complex with the nuclear export receptor CRM1 and the nuclear import receptor importin α/β, which accumulates in the central channel of the nuclear pore, preventing different nuclear proteins-mediated nuclear input (Atasheva et al. 2010). It has been reported that KPNA3 and KPNA4 can directly combine with NF-κB p50 and p65 protein nuclear localization signals (NLSs) to mediate nuclear transport (Fagerlund et al. 2005). In our experiment, a correlation between the inhibitory capacity of MGF360-12L and its interaction with KPNA2, KPNA3 and KPNA4 was confirmed by a binding competition assay. We showed that MGF360-12L inhibited nuclear import of NF-κB by inhibiting KPNA2, KPNA3 and KPNA4 interacting with p65, thereby inhibiting NF-κB transcriptional activity and preventing host antiviral response. Overexpression of KPNA2, KPNA3, KPNA4 might restore the inhibitory effect of MGF360-12L on IFN-β transcription.
In summary, our data demonstrated that MGF360-12L significantly inhibited IFN-I production and the nuclear localization of p50 and p65. Also, MGF360-12L might bind importins to interrupt the interaction of p65 with KPNA2, KPNA3, KPNA4 and importins is NLS dependent. Additionally, MGF360-12L can competitively inhibit the interaction of NF-κB and nuclear transporters, and thus interfere with the nuclear transport of NF-κB (Fig. 6). These findings suggested that MGF360-12L could inhibit the IFN-I production by blocking the interrelation of importin α and NF-κB signaling pathway, which might reveal a novel strategy for ASFV to escape the host innate immune response.
Fig. 6.
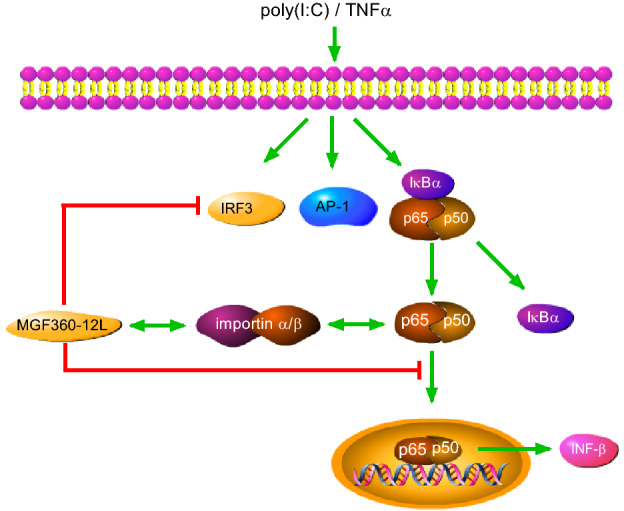
The mechanisms of MGF360-12L-mediated antagonism of IFN-I-dependent signaling in poly (I: C) or TNFα stimulated cells. MGF360-12L interferes with the nuclear translocation of NF-κB induced by poly (I: C) or TNFα by competitively inhibiting the interaction of p65 with nuclear transport proteins. MGF360-12L might suppress the induction of IFN-I in host cells via this mechanism. These findings revealed a novel strategy and insight for ASFV to evade the host innate immune response.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was sponsored by National key Research and Development Program (Grant No. 2017YFD0502301), and National College Students’ innovation and entrepreneurship training program (201910504017).
Author Contributions
YZ and HD conceived and designed the study. YZ, ZG, TB, CZ, LH performed the experiments. YZ, HD analyzed the experimental data. YZ, HD wrote the paper. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Alonso C, Borca M, Dixon L, Revilla Y, Rodriguez F, Escribano JM, Ictv Report C. ICTV virus taxonomy profile: Asfarviridae. J Gen Virol. 2018;99:613–614. doi: 10.1099/jgv.0.001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasheva S, Fish A, Fornerod M, Frolova EI. Venezuelan equine Encephalitis virus capsid protein forms a tetrameric complex with CRM1 and importin alpha/beta that obstructs nuclear pore complex function. J Virol. 2010;84:4158–4171. doi: 10.1128/JVI.02554-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber C, Netherton C, Goatley L, Moon A, Goodbourn S, Dixon L. Identification of residues within the African swine fever virus DP71L protein required for dephosphorylation of translation initiation factor eIF2alpha and inhibiting activation of pro-apoptotic CHOP. Virology. 2017;504:107–113. doi: 10.1016/j.virol.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudet D, Pham N, Skaik N, Piekny A. Importin-binding mediates the intramolecular regulation of anillin during cytokinesis. Mol Biol Cell. 2020 doi: 10.1091/mbc.E20-01-0006:mbcE20010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA, Baldwin AS., Jr I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- Borca MV, O’Donnell V, Holinka LG, Ramirez-Medina E, Clark BA, Vuono EA, Berggren K, Alfano M, Carey LB, Richt JA, Risatti GR, Gladue DP. The L83L ORF of African swine fever virus strain Georgia encodes for a non-essential gene that interacts with the host protein IL-1beta. Virus Res. 2018;249:116–123. doi: 10.1016/j.virusres.2018.03.017. [DOI] [PubMed] [Google Scholar]
- Burrage TG, Lu Z, Neilan JG, Rock DL, Zsak L. African swine fever virus multigene family 360 genes affect virus replication and generalization of infection in Ornithodoros porcinus ticks. J Virol. 2004;78:2445–2453. doi: 10.1128/JVI.78.5.2445-2453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek AA, Dabrowska I, Gregorczyk KP, Wyzewski Z. African swine fever virus: a new old enemy of Europe. Ann Parasitol. 2016;62:161–167. doi: 10.17420/ap6203.49. [DOI] [PubMed] [Google Scholar]
- Correia S, Ventura S, Parkhouse RM. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 2013;173:87–100. doi: 10.1016/j.virusres.2012.10.013. [DOI] [PubMed] [Google Scholar]
- Dixon LK, Chapman DA, Netherton CL, Upton C. African swine fever virus replication and genomics. Virus Res. 2013;173:3–14. doi: 10.1016/j.virusres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Dixon LK, Sun H, Roberts H. African swine fever. Antiviral Res. 2019;165:34–41. doi: 10.1016/j.antiviral.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Fagerlund R, Kinnunen L, Kohler M, Julkunen I, Melen K. NF-{kappa}B is transported into the nucleus by importin {alpha}3 and importin {alpha}4. J Biol Chem. 2005;280:15942–15951. doi: 10.1074/jbc.M500814200. [DOI] [PubMed] [Google Scholar]
- Freitas FB, Frouco G, Martins C, Ferreira F. African swine fever virus encodes for an E2-ubiquitin conjugating enzyme that is mono- and di-ubiquitinated and required for viral replication cycle. Sci Rep. 2018;8:3471. doi: 10.1038/s41598-018-21872-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo I, Alonso C. African swine fever virus: a review. Viruses. 2017;9:103. doi: 10.3390/v9050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo C, Sanchez EG, Perez-Nunez D, Nogal M, de Leon P, Carrascosa AL, Nieto R, Soler A, Arias ML, Revilla Y. African swine fever virus (ASFV) protection mediated by NH/P68 and NH/P68 recombinant live-attenuated viruses. Vaccine. 2018;36:2694–2704. doi: 10.1016/j.vaccine.2018.03.040. [DOI] [PubMed] [Google Scholar]
- Golding JP, Goatley L, Goodbourn S, Dixon LK, Taylor G, Netherton CL. Sensitivity of African swine fever virus to type I interferon is linked to genes within multigene families 360 and 505. Virology. 2016;493:154–161. doi: 10.1016/j.virol.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazawa M, Sakai K, Kobayashi A, Yoshino H. Disease-specific alteration of karyopherin-α subtype establishes feed-forward oncogenic signaling in head and neck squamous cell carcinoma. Oncogene. 2020;39:2212–2223. doi: 10.1038/s41388-019-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke P, Takeuchi O, Malley R, Lien E, Ingalls RR, Freeman MW, Mayadas T, Nizet V, Akira S, Kasper DL, Golenbock DT. Cellular activation, phagocytosis, and bactericidal activity against group B streptococcus involve parallel myeloid differentiation factor 88-dependent and independent signaling pathways. J Immunol. 2002;169:3970–3977. doi: 10.4049/jimmunol.169.7.3970. [DOI] [PubMed] [Google Scholar]
- Hernaez B, Diaz-Gil G, Garcia-Gallo M, Ignacio Quetglas J, Rodriguez-Crespo I, Dixon L, Escribano JM, Alonso C. The African swine fever virus dynein-binding protein p54 induces infected cell apoptosis. FEBS Lett. 2004;569:224–228. doi: 10.1016/j.febslet.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Hurtado C, Granja AG, Bustos MJ, Nogal ML, Gonzalez de Buitrago G, de Yebenes VG, Salas ML, Revilla Y, Carrascosa AL. The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology. 2004;326:160–170. doi: 10.1016/j.virol.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Kim TK, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteagudo PL, Lacasta A, Lopez E, Bosch L, Collado J, Pina-Pedrero S, Correa-Fiz F, Accensi F, Navas MJ, Vidal E, Bustos MJ, Rodriguez JM, Gallei A, Nikolin V, Salas ML, Rodriguez F. BA71DeltaCD2: a new recombinant live attenuated African swine fever virus with cross-protective capabilities. J Virol. 2017;91:e01058-17. doi: 10.1128/JVI.01058-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell V, Risatti GR, Holinka LG, Krug PW, Carlson J, Velazquez-Salinas L, Azzinaro PA, Gladue DP, Borca MV. Simultaneous deletion of the 9GL and UK Genes From The African swine fever virus Georgia 2007 isolate offers increased safety and protection against homologous challenge. J Virol. 2017;91:e01760-16. doi: 10.1128/JVI.01760-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D. The enhanceosome. Curr Opin Struct Biol. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Perez-Canamas M, Hernandez C. New insights into the nucleolar localization of a plant RNA virus-encoded protein that acts in both RNA packaging and RNA silencing suppression: involvement of importins alpha and relevance for viral infection. Mol Plant–Microbe Interact. 2018;31:1134–1144. doi: 10.1094/MPMI-02-18-0050-R. [DOI] [PubMed] [Google Scholar]
- Pikalo J, Zani L, Huhr J, Beer M, Blome S. Pathogenesis of African swine fever in domestic pigs and European wild boar—lessons learned from recent animal trials. Virus Res. 2019 doi: 10.1016/j.virusres.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Poon IK, Jans DA. Regulation of nuclear transport: Central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Portugal R, Leitao A, Martins C. Modulation of type I interferon signaling by African swine fever virus (ASFV) of different virulence L60 and NHV in macrophage host cells. Vet Microbiol. 2018;216:132–141. doi: 10.1016/j.vetmic.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Rahman MM, McFadden G. Modulation of NF-kappaB signalling by microbial pathogens. Nat Rev Microbiol. 2011;9:291–306. doi: 10.1038/nrmicro2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Reid SP, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, Volchkov VE, Nichol ST, Basler CF. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis AL, Abrams CC, Goatley LC, Netherton C, Chapman DG, Sanchez-Cordon P, Dixon LK. Deletion of African swine fever virus interferon inhibitors from the genome of a virulent isolate reduces virulence in domestic pigs and induces a protective response. Vaccine. 2016;34:4698–4705. doi: 10.1016/j.vaccine.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis AL, Goatley LC, Jabbar T, Sanchez-Cordon PJ, Netherton CL, Chapman DAG, Dixon LK. Deletion of the African swine fever virus gene DP148R does not reduce virus replication in culture but reduces virus virulence in pigs and induces high levels of protection against challenge. J Virol. 2017;91:01428. doi: 10.1128/JVI.01428-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla Y, Cebrian A, Baixeras E, Martinez C, Vinuela E, Salas ML. Inhibition of apoptosis by the African swine fever virus Bcl-2 homologue: role of the BH1 domain. Virology. 1997;228:400–404. doi: 10.1006/viro.1996.8395. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Nogal ML, Carrascosa AL, Salas ML, Fresno M, Revilla Y. African swine fever virus IAP-like protein induces the activation of nuclear factor kappa B. J Virol. 2002;76:3936–3942. doi: 10.1128/JVI.76.8.3936-3942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JM, Salas ML. African swine fever virus transcription. Virus Res. 2013;173:15–28. doi: 10.1016/j.virusres.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Taylor SL, Frias-Staheli N, Garcia-Sastre A, Schmaljohn CS. Hantaan virus nucleocapsid protein binds to importin alpha proteins and inhibits tumor necrosis factor alpha-induced activation of nuclear factor kappa B. J Virol. 2009;83:1271–1279. doi: 10.1128/JVI.00986-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele S, Stanelle-Bertram S, Beck S, Kouassi NM, Zickler M, Müller M, Tuku B, Resa-Infante P, van Riel D, Alawi M, Günther T, Rother F, Hügel S, Reimering S, McHardy A, Grundhoff A, Brune W, Osterhaus A, Bader M, Hartmann E, Gabriel G. Cellular importin-α3 expression dynamics in the lung regulate antiviral response pathways against influenza A virus infection. Cell Rep. 2020;31:107549. doi: 10.1016/j.celrep.2020.107549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulman ER, Rock DL. Novel virulence and host range genes of African swine fever virus. Curr Opin Microbiol. 2001;4:456–461. doi: 10.1016/s1369-5274(00)00235-6. [DOI] [PubMed] [Google Scholar]
- Wang R, Nan Y, Yu Y, Zhang YJ. Porcine reproductive and respiratory syndrome virus Nsp1beta inhibits interferon-activated JAK/STAT signal transduction by inducing karyopherin-alpha1 degradation. J Virol. 2013;87:5219–5228. doi: 10.1128/JVI.02643-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wu J, Wu Y, Chen H, Zhang S, Li J, Xin T, Jia H, Hou S, Jiang Y, Zhu H, Guo X. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochem Biophys Res Commun. 2018;506:437–443. doi: 10.1016/j.bbrc.2018.10.103. [DOI] [PubMed] [Google Scholar]
- Ye J, Chen Z, Li Y, Zhao Z, He W, Zohaib A, Song Y, Deng C, Zhang B, Chen H, Cao S. Japanese encephalitis virus NS5 inhibits type i interferon (IFN) production by blocking the nuclear translocation of IFN regulatory factor 3 and NF-kappaB. J Virol. 2017;91:e00039-17. doi: 10.1128/JVI.00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tao M, Han W, Fan Z, Imran M, Cao S, Ye J. Nuclear localization of Zika virus NS5 contributes to suppression of type I interferon production and response. J Gen Virol. 2019 doi: 10.1099/jgv.0.001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.