Abstract
The majority of lizards classified in the superfamily Iguanoidea have an XX/XY sex-determination system in which sex-chromosomal linkage shows homology with chicken (Gallus gallus) chromosome 15 (GGA15). However, the genomics of sex chromosomes remain largely unexplored owing to the presence of homomorphic sex chromosomes in majority of the species. Recent advances in high-throughput genome complexity reduction sequencing provide an effective approach to the identification of sex-specific loci with both single-nucleotide polymorphisms (SNPs) and restriction fragment presence/absence (PA), and a better understanding of sex chromosome dynamics in Iguanoidea. In this study, we applied Diversity Arrays Technology (DArTseqTM) in 29 phenotypic sex assignments (14 males and 15 females) of green iguana (Iguana iguana). We confirmed a male heterogametic (XX/XY) sex determination mode in this species, identifying 29 perfectly sex-linked SNP/PA loci and 164 moderately sex-linked SNP/PA loci, providing evidence probably indicative of XY recombination. Three loci from among the perfectly sex-linked SNP/PA loci showed partial homology with several amniote sex chromosomal linkages. The results support the hypothesis of an ancestral super-sex chromosome with overlaps of partial sex-chromosomal linkages. However, only one locus among the moderately sex-linked loci showed homology with GGA15, which suggests that the specific region homologous to GGA15 was located outside the non-recombination region but in close proximity to this region of the sex chromosome in green iguana. Therefore, the location of GGA15 might be further from the putative sex-determination locus in green iguana. This is a paradigm shift in understanding linkages on homomorphic X and Y sex chromosomes. The DArTseq platform provides an easy-to-use strategy for future research on the evolution of sex chromosomes in Iguanoidea, particularly for non-model species with homomorphic or highly cryptic sex chromosomes.
Keywords: DArTseqTM, Iguanoidea, SNP, super-sex chromosome, non-recombination
Introduction
The genetic framework for sex organ differentiation is highly conserved, but a diversity of sex determination is observed in amniotes (O’Meally et al., 2012; Ezaz et al., 2017). The underlying mechanisms can broadly be divided into two main categories: genotypic sex determination and temperature-dependent sex determination. Diverse and rapid evolution of sex determination has, however, been reported even within closely related species (Inamdar et al., 2012; Altmanová et al., 2017). The turnover rate of sex-determination mechanisms differs among lineages (Pokorná and Kratochvíl, 2014; Rovatsos et al., 2014b, 2019; Altmanová et al., 2017; Natri et al., 2019). Squamate reptiles exhibit variability in sex-determination modes, with a rapid turnover of sex-determination mechanisms (Rovatsos et al., 2014b; Schartl et al., 2016; Ezaz et al., 2017; Singchat et al., 2020b). In addition to gecko lizards and skinks, multiple variants of sex chromosome systems have been identified in iguanian lizards, including male heterogamety (XX/XY) in the majority of iguanid lizards and female heterogamety (ZZ/ZW) in many agamid lizards (Ezaz et al., 2009a,b; Gamble, 2010; Alföldi et al., 2011; Rovatsos et al., 2014a; Altmanová et al., 2017; Matsubara et al., 2019; Srikulnath et al., 2019; Singchat et al., 2020b). The study of sex-determination systems and sex chromosomes in iguanian lizards has been difficult because many have homomorphic sex chromosomes that cannot be identified easily using conventional karyotyping (Brandley et al., 2006; Castiglia et al., 2013; Singchat et al., 2020b). The green anole (Anolis carolinensis, ACA), is a highly diversified lizard in the superfamily Iguanoidea. The green anole was the first non-avian reptile selected for whole-genome sequencing, which reveals that the species possesses a homomorphic XX/XY sex-chromosome system (Alföldi et al., 2011). The X chromosome of A. carolinensis (ACAX) was identified by observation of differences in fluorescent in situ hybridization (FISH) signals on microchromosomes between male and female individuals. The known X-linked region is probably homologous to chicken chromosome 15 (Gallus gallus, GGA15) (Alföldi et al., 2011). Studies of the partially comparative gene constituents of sex chromosomes between males and females using quantitative polymerase chain reaction (qPCR) analysis have revealed a pattern of different gene dosages in ACAX-linked regions in the majority of iguanoid lizards, which probably exhibit the XX/XY sex-determination system (Alföldi et al., 2011; Gamble et al., 2014; Rovatsos et al., 2014a,b; Altmanová et al., 2017). These results suggest that sex-chromosomal linkage homology is highly conserved in lizards of Iguanoidea.
Genome sequence analyses and cross-species chromosome mapping have, however, detected genomic convergence in which unrelated sex chromosomes share linkage homologies across distantly related lineages. This might involve genomic regions orthologous to squamate chromosome 2 (SR2) and snake W sex chromosomes (Matsubara et al., 2006, 2012, 2019; Srikulnath et al., 2009a,b, 2013, 2014, 2015; Alföldi et al., 2011; Pokorná et al., 2011; Uno et al., 2012; Matsuda et al., 2015; Ezaz et al., 2017; Singchat et al., 2018, 2020a,b). Comparison of genome-wide single-nucleotide polymorphisms (SNPs) between males and females of the Siamese cobra revealed female-specific loci that also show partial homology with a variety of amniote sex chromosomes, including the ACA X-linked region as GGA15 (Laopichienpong et al., 2020), supporting the hypothesis that the Siamese cobra W sex chromosome is derived from a larger ancestral amniote super-sex chromosome. Interestingly, the co-hybridization signals of the same sex-chromosomal linkage homology are also observed for different chromosomes, as seen in cDNA and bacterial artificial chromosome (BAC) chromosomal maps (Matsuda et al., 2005; Matsubara et al., 2006, 2012, 2019; Srikulnath et al., 2009a,b, 2013, 2014, 2015; Uno et al., 2012; Singchat et al., 2018, 2020a,b). Patterns of FISH signals for the same BACs on multiple different chromosome pairs might result from the presence of common genomic elements (O’Meally et al., 2010; Ezaz et al., 2017; Singchat et al., 2018, 2020a,b; Matsubara et al., 2019). These findings suggest the possibility that chromosomal rearrangement, such as fission or translocation, might have occurred in an ancestral amniote super-sex chromosome, which has been evolutionarily conserved after divergence ∼320 million years ago (MYA) (Hedges and Vidal, 2009). This has resulted in a variety of lineage-specific sex-chromosomal linkages among amniotes (O’Meally et al., 2012; Pokorná and Kratochvíl, 2014; Ezaz et al., 2017). It is therefore necessary to understand whether incidences of partial sex-chromosomal linkage observed on the sex chromosome of one lineage are also observed on both autosomes and/or sex chromosomes in a separate lineage, because this might influence molecular sexing based on sex-linked gene development (Singchat et al., 2018, 2020b; Matsubara et al., 2019). Taken together, this raises the questions of whether (a) a small number of partial gene constituents of sex-chromosomal linkage are reliable to prove sex-chromosomal linkage of each lineage based on the supposition of an ancestral super-sex chromosome, and (b) in addition to molecular dosage copy-number estimation and cytogenetic approaches, can comparison of genome sequences or genome-wide SNPs between males and females be used to investigate sex-chromosomal linkage?
For an improved understanding of the relationship of sex-chromosomal linkages and species that exhibit genotypic sex determination with homomorphic sex chromosomes, the green iguana (Iguana iguana) was selected as a suitable non-model species to determine sex identification and sex-linked markers (Rovatsos et al., 2014b). The green iguana is a large, arboreal, mostly herbivorous species of lizard in the Iguanoidea, and is found over a broad geographical area in South America (Falcón et al., 2013). Green iguana are sexually dimorphic in maturity, with external morphological traits such as femoral pores, heavy jowls, and a hemipenis bulge at the base of the tail (Kaplan, 1994; Rivas et al., 1996). The diploid chromosome number of the green iguana is 34, comprising six pairs of macrochromosomes and 11 pairs of microchromosomes. No difference in chromosome morphology between males and females is observed (Cohen et al., 1967; Singchat et al., 2020b); however, based on qPCR analyses, the green iguana is an XX/XY species and shares sex-chromosomal linkage across lizards in the Iguanoidea with GGA15, except for members of two genera of basilisks in the family Corytophanidae (Rovatsos et al., 2014a; Nielsen et al., 2019). These findings lead to our prediction that the X and Y sex chromosomes of the green iguana are microchromosomes and highly cryptic (i.e., morphologically poorly differentiated) (Singchat et al., 2020b). In light of this scenario, we tested the hypotheses that (i) the green iguana exhibits an XX/XY sex-chromosome system and harbors male-specific loci, and (ii) sex-chromosomal linkage homology should include a large proportion of the genomic segment GGA15. Recent genotyping-by-sequencing (GBS), using next-generation sequencing technologies such as Diversity Arrays Technology (DArTseqTM) developed by Diversity Arrays Technology, Pty Ltd. (Canberra, ACT, Australia), is an effective method with a key genome complexity reduction concept for identification of sex-linked loci in non-model species using SNP loci, to generate restriction site-specific presence/absence (PA) markers. This method is achieved through combining restriction enzymes that separate low copy sequences (most informative for marker discovery and typing) from the repetitive fraction of the genome. The markers generated by DArTseq reveal loci linked to a particular sex, thereby enabling accurate identification of loci tightly linked to the sex-determination region of the sex chromosomes. This is a useful molecular tool for disclosure of sex-determination modes in non-model species with cryptic sex chromosomes such as green iguana (Lambert et al., 2016). Other GBS technology such as RADseq used only one enzyme with different internal technical procedures and still retains more repetitive fractions (Gamble and Zarkower, 2014; Gamble et al., 2015). In this study, we addressed these two hypotheses using DArTseqTM. Findings provide novel insights into the evolutionary history of sex chromosomes, representing sex-chromosomal linkage in green iguana in relation to other amniotes. Identification of male-specific loci contributes to our understanding of sex-chromosome evolution.
Materials and Methods
Specimens and DNA Extraction
Twenty-nine individuals (14 males and 15 females) of adult green iguana (I. iguana) from several clutches were collected from Chonburi Iguana Farm, Thailand (13°25′02.4′′ N, 101°02′32.7′′ E). The sex of each green iguana was identified morphologically and confirmed by mating observation (Falcón et al., 2013). Animal care and all experimental procedures were approved by the Animal Experiment Committee, Kasetsart University, Thailand (approval no. ACKU61-SCI-023) and conducted in accordance with the Regulations on Animal Experiments at Kasetsart University. Blood samples were collected from the ventral tail vein using a 23-gauge needle attached to 3 ml disposable syringes. The syringes contained 10 mM ethylenediaminetetraacetic acid for DNA extraction, and total genomic DNA was extracted following the standard salting-out protocol as described previously (Supikamolseni et al., 2015). Each DNA sample was evaluated by gel electrophoresis for the presence of high-molecular-weight DNA and then stored at −20°C until required for DArTseqTM library construction and qPCR analysis.
Molecular Sexing by Quantitative Real-Time PCR
Molecular sexing by qPCR was performed on five males and five females green iguanas selected randomly, following the method of Rovatsos et al. (2014b) based on comparison of differences in gene dosage between male and female genomes. Females (XX) have twice as many copies of genes linked to the X-specific portion of sex chromosomes as males (XY), whereas genes in autosomal or pseudoautosomal regions are present in equal copy numbers in both sexes. Four primer pairs for four X-linked loci of A. carolinensis (TMEM132D, CCDC92, ATP2A2, and PEBP1) were applied to identify sex assignment. The single-copy gene GAPDH, which was expected to be located on green iguana chromosome 3, corresponding to chicken chromosome 1, was used as the reference gene (Criscuolo et al., 2009; Singchat et al., 2020b) for normalization of gene dosages in the qPCR analyses (Supplementary Table S1). Each qPCR amplification was performed using 10 μl of 2 × KAPA SYBR® FAST qPCR Master Mix (Kapa Biosystems, Cape Town, South Africa), 0.25 μM primers, and 25 ng genomic DNA. Thermal cycling conditions were 95°C for 3 min, followed by 44 amplification cycles of 95°C for 15 s, 56°C for 30 s, and 72°C for 30 s, ending with a melting curve analysis to monitor for potential non-specific products. The dosage of each studied gene (R) was calculated from crossing point values and subsequently normalized to the dosage of the GAPDH gene from the same DNA sample, based on the equation: R = [2Cp gene/2Cp GAPDH]–1 (Cawthon, 2002). The relative gene dosage ratio (r) between the two sexes for each gene was calculated by dividing the average gene dosage in the male (Rmale) by the average dosage in the female (Rfemale), i.e., r = Rmale/Rfemale. A relative gene dosage ratio of 0.5 was expected for X-linked genes degenerated on the Y chromosome and 1.0 for autosomal or pseudoautosomal genes. Statistical differences in copy number between male and female green iguanas were examined using a Wilcoxon signed-rank test with R version 3.4.4 statistical software and the “stats” package (R Core Team, 2019).
Technical control qPCRs were run in triplicate for each primer set to ensure there was no contamination. Standard curves were produced for gene-specific sex primers and GAPDH primers using the pooled genomic DNA of 10 randomly selected green iguana individuals to ensure consistent rates of amplification over a wide range of DNA concentrations. Threefold serial dilutions were generated ranging from 25 to 0.00025 ng/μl, with five different concentrations providing a linear dynamic range (Supplementary Figure S1). According to best practice guidelines (Nolan et al., 2006), the reaction was considered in relation to a straight line with R2 exceeding 0.976 for TMEM132D, 0.98 for CCDC92, 0.9446 for ATP2A2, 0.967 for PEBP1, and 0.9879 for GAPDH, and fitted to the values obtained. The efficiency (E) of sex-specific primer amplification, calculated as E = -1 + 10(–1/slope), was 0.9169-2.0762, and the efficiency of GAPDH amplification was 1.0004 (Supplementary Table S2). To test intraplate repeatability, the intra-assay coefficient of variation (CV;%) was determined in triplicate for 10 samples. The percentage CV for each sample was calculated by determining the standard deviation (SD) of each technical triplicate Cp value for GAPDH (G) and sex-specific primers (S), then dividing by the triplicate mean and multiplying by 100. The intra-assay CV should be less than 10% (Schultheiss and Stanton, 2009; Hanneman et al., 2011; Singchat et al., 2019). In the present study, the intra-assay CVs were 1.6267–3.6803 and 1.9351% for G and S, respectively (Supplementary Table S2). All samples fell within the concentration range generated by the standard curve. The starting concentrations of S and the reference gene G were used to calculate the relative copy number and, in turn, the S/G ratio was calculated as the copy number of sex-specific primer repeats (Cawthon, 2002; Criscuolo et al., 2009). To test intra- and inter-plate repeatability, samples from 10 green iguana individuals were run in technical triplicate on three different plates in order to monitor plate-to-plate variation. The plate mean for each sample was calculated and subsequently the overall mean, SD, and CV were computed. Inter-assay CVs of less than 15% are generally acceptable (Schultheiss and Stanton, 2009; Hanneman et al., 2011). In the present study, the inter-assay CV (n = 3) was 3.43394%, whereas the intra-assay CV was 0.8696–4.2789 and 4.3715% for S and G, respectively (Supplementary Table S3).
Development of DArTseqTM
A detailed description of the DArTseqTM methodology is provided by Jaccoud et al. (2001) and often produces 69 bp sequences. Genotyping of multiple loci was performed using DArTseqTM for SNP loci and in silico DArT (presence/absence of restriction fragments in the representation; PA loci) to determine candidate sex-specific loci in male and female individuals. A set of green iguana specimens was used to develop the DArTseqTM arrays with approximately 100 ng genomic DNA from each individual. The DNA samples were subjected to digestion/ligation reactions as described by Kilian et al. (2012) and digested with PstI and a second restriction enzyme (SphI). Ligation reactions were then performed using two adaptors: a PstI-compatible adaptor consisting of an Illumina flow-cell attachment sequence, primer sequence, and a unique barcode sequence, and a SphI-compatible adaptor consisting of an Illumina flow-cell attachment region. Ligated fragments were then PCR amplified using an initial denaturation at 94°C for 1 min, followed by 30 cycles of 94°C for 20 s, 58°C for 30 s, and 72°C for 45 s, with a final extension step at 72°C for 7 min. Equimolar amounts of amplification products from each individual were pooled and subjected to Illumina’s proprietary cBot1 bridge PCR, followed by sequencing on an Illumina HiSeq 2000 platform. Single-read sequencing was run for 77 cycles.
Sequences were processed using proprietary DArTseqTM analytical pipelines (Ren et al., 2015). Initially, the HiSeq 2000 output (FASTQ file) was processed to filter poor-quality sequences. Two thresholds of quality were applied. For the barcode region (which allowed parsing of sequences into specific sample libraries), we applied stringent selection (minimum Phred pass score of 30, minimum pass percentage 75). For the remainder of the sequence, relaxed thresholds were applied (minimum Phred pass score 10, minimum pass percentage 50). This calling quality was assured by high average read depth per locus (average across all markers was over 30 reads/locus). These parameters provide the optimal filtering step for potential paralogs as they select markers with Mendelian patterns of behavior. Moreover, the sex-specific region of green iguana is expected to be very cryptic as homomorphic sex chromosomes. The whole read quality with low stringent thresholds must allow for all possible analyses. Approximately 2,000,000 sequences per individual were identified and used in marker calling. Finally, identical sequences were collapsed into “fastqcoll” files that were used in the secondary proprietary pipeline (DArTsoft14) for SNP and PA loci calling; DArTsoft14 implements a “reference-free” algorithm. All unique sequences from the set of fastqcoll files were identified and clustered by sequence similarity at a distance threshold of three base variations using an optimized (fast) clustering algorithm (in many cases more than 1 billion sequences are clustered within minutes). The sequence clusters were then parsed into SNP and in silico DArTseqTM markers utilizing a range of metadata parameters derived from the quantity and distribution of each sequence across all samples in the analysis. A high degree of technical replication was included in the DArTseqTM genotyping process, enabling reproducibility scores to be calculated for each candidate marker. Outputs by DArTsoft14 were then filtered on the basis of reproducibility values, average count for each sequence (sequencing depth), balance of average counts for each SNP allele, and call-rate (i.e., proportion of samples for which the marker was scored).
Marker Selection and DArTseqTM Analysis
Sex-specific loci were derived from analysis of SNP codominant markers and PA as dominant markers. The SNP data were coded as “0” for the reference allele homozygote (the most common allele), “1” for the SNP allele homozygote, “2” for the heterozygote, or “−” as the double null/null allele homozygote (absence of a SNP fragment in the genomic representation). The PA data were coded as “1” for presence, “0” for absence, or “−” for putative heterozygosity. For sex-linked markers in an XX/XY sex-determination system, reference alleles were located on the X-chromosome. In this study, “SNP alleles” were those that showed polymorphism relative to the reference allele. In an XX/XY system, SNP alleles should be those associated with the Y sex chromosome, and located in or near to the male-determination region if the allele is tightly Y-specific. If the two sex chromosomes are recombined, SNP alleles should appear occasionally on the X chromosome. Some males might then be homozygous for SNP alleles at particular loci, with females being heterozygous, exhibiting a copy of the SNP allele. The probability of a female being homozygous for a SNP allele was low. For evaluation of loci associated with an XX/XY system, SNP loci that maximized female homozygosity at the reference allele, maximized male heterozygosity, and minimized homozygosity for the SNP allele in both sexes were expected. Loci for which frequencies of female homozygosity for the reference allele were at least 70% were retained, while those for which homozygosity at the SNP allele was more than 30% with heterozygosity more than 30% were discarded. For males, loci for which frequency of homozygosity for the reference allele was at most 30% and heterozygosity was at least 70% were retained. However, this allowed sex-linked loci to show higher degrees of SNP allele homozygosity for males if recombination occurred. For PA markers, loci that had restriction fragments sequenced in at least 70% of males and not sequenced in at least 70% of females were selected. The SNP and PA loci sequenced for 80, 90, and 100% of males were also included in a separate data set. A filtering criterion of 100% probability was applied to define perfectly sex-linked loci, whereas 70–90% probabilities were deemed to define moderately sex-linked loci. A similar, but opposite, approach was performed to target loci with a ZZ/ZW system.
The Hamming distance was calculated to determine pairwise differences in the number of SNP and PA loci between male and female individuals using the “rdist” function of R version 3.5.1 statistical software (Nolan et al., 2006). Heatmaps were plotted using the “ggplot2” R package (Nolan et al., 2006). The Hamming distance represents the number of pairwise differences between all individuals across all loci. To examine the genetic association between each locus and phenotypic sex assignment from SNP and PA loci, the Cochran–Armitage trend test (CATT) was performed using the “catt” function of R version 3.5.1 implemented in the “HapEstXXR” package (Nolan et al., 2006; Sven and Klaus, 2019). The CATT results were similar to those of a chi-square test to assess whether the proportion of different genotypes followed the null expectation. Polymorphism information content (PIC), as an index for evaluation of the informativeness of SNP and PA loci, was calculated for each locus and ranged from 0 (fixation of one allele) to 0.5 (frequencies of both alleles are equal).
Estimation of Random Sex-Linkage
To estimate the probability that candidate loci might show random association with sex under a small sample size (Gamble et al., 2015) the formula Pi = 0.5n explains the probability of a locus being sex-linked by chance, where P is the probability for a given locus, i is sex-linked, 0.5 is the probability that either a male was either homozygous or a female was heterozygous at a given locus, and n is the number of individuals sequenced at the locus. After sequencing, we multiplied P by the number of high-quality SNPs produced to estimate the number of SNPs expected to exhibit a sex-linked pattern by chance.
Comparison of Potential Sex-Linked Loci
A Chi-square test was performed on the PA loci and on heterozygosity for SNP loci data to determine whether the four groups (100:0, 90:10, 80:20, and 70:30 of male:female ratios) had associated categorical variable. Differences in mean heterozygosity among the groups were determined using the Kruskal–Wallis test, with the Nemeyi test used for post hoc comparisons. All candidate loci were plotted against each individual using the “glPlot” function in the “dartR” R package (Gruber and Georges, 2019; R Core Team, 2019).
Homology Searching
For all sex-linked loci that attained the filtering criteria and showed a statistically significant association with sex phenotype, a Basic Local Alignment Search Tool (BLAST) search of the National Center for Biotechnology Information (NCBI) database was performed to investigate homologies of sex-linked SNP/PA loci against the green anole lizard (A. carolinensis) (AAWZ00000000.2) (Alföldi et al., 2011) and chicken (G. gallus) (AADN00000000.5) (International Chicken Genome Sequencing Consortium, 2004) reference genomes. Using the BLASTn program, all loci were then used to search the NCBI database2 and RepBase version 19.11 (Bao et al., 2015) (Genetic Information Research Institute3), a specialized nucleotide sequence collection for repeated or other significant sequences, which reports only E-values lower than 0.001 and queries coverage with similarity more than 70%.
Results
Comparison of Dosages of ACA X-Linked Genes by qPCR
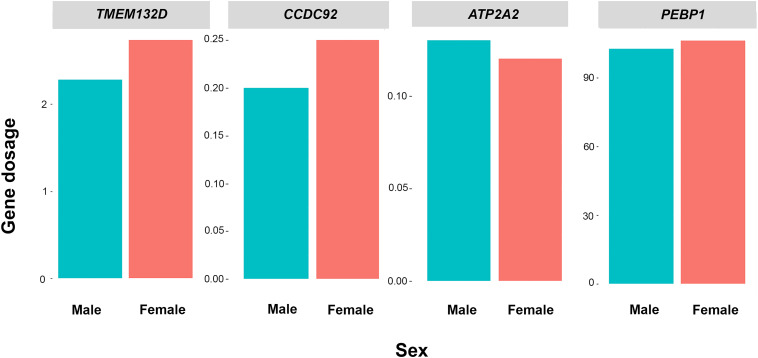
Comparison of the dosages of four genes (TMEM132D, CCDC92, ATP2A2, and PEBP1) among five males and five females by qPCR showed that the green iguana exhibited non-sex-specific dosages (Table 1, Figure 1 and Supplementary Tables S4–S7) for ACA X-linked genes, although sex-chromosomal homology was confirmed from the similarity to ACAX and correspondence to GGA15 (Rovatsos et al., 2014b).
TABLE 1.
Relative gene dosage ratios (r) among five males and five females individuals for four genes in green iguana (Iguana iguana).
| Primer pair | Gene symbol | Gene | Position in Anolis carolinensis | r | Wilcoxon signed-rank test |
| TMEM132D_1 | TMEM132D | Transmembrane protein 132D | X chromosome | 0.83 | W = 8, p = 0.7302 |
| CCDC92_1 | CCDC92 | Coiled-coil domain-containing protein 92 | X chromosome | 1.21 | W = 8, p = 1 |
| ATP2A2_1 | ATP2A2 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | X chromosome | 0.78 | W = 7, p = 1 |
| PEBP1_1 | PEBP1 | Phosphatidylethanolamine-binding protein 1 | X chromosome | 0.96 | W = 16, p = 0.5476 |
FIGURE 1.
Relative gene dosage ratios between male and female individuals of green iguana (Iguana iguana) for TMEM132D, CCDC92, ATP2A2, and PEBP1. Blue values indicate males and red values indicate females.
Sex-Determination System and Identification of Sex-Specific Loci in Green Iguana
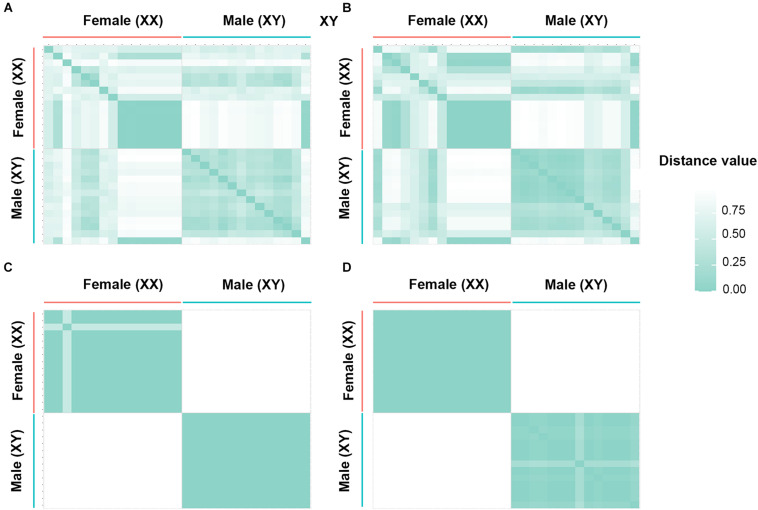
We sequenced 80,540 SNP loci and 83,292 PA loci. The PIC ranged from 0.49 to 0.50 for both SNP and PA markers. The overall distribution of PIC values was asymmetrical and skewed toward the higher values (Supplementary Figure S2). To determine whether XY or ZW determined the sex of green iguana individuals, we compared a number of SNPs and PAs after filtering with different male:female ratios as criteria. For the ZZ/ZW type of sex determination, filtering using the criterion of 30:70 male:female resulted in identification of 15 SNP loci and 457 PA loci as female-specific. The proportional pairwise Hamming distance between males and females using sex-specific SNP and PA loci (under the null exclusive model) showed within-sex distances of 0.499 ± 0.035 in males and 0.416 ± 0.036 in females for SNP loci, and 0.425 ± 0.033 in males and 0.410 ± 0.036 in females for PA loci. Between-sex distances showed 0.654 ± 0.021 for SNP loci and 0.649 ± 0.022 for PA loci. The CATT verified significant loci association with phenotypic sex for 15 SNP loci (χ2 = 5.00–9.00, p < 0.001) and 450 PA loci (χ2 = 5.00–16.00, p < 0.001). Three PA loci were specifically associated with females based on the criterion of 20:80 male:female, whereas no SNP loci were identified as female-specific. The proportional pairwise Hamming distance between males and females using sex-specific SNP and PA loci revealed within-sex distances of 0.341 ± 0.037 in males and 0.248 ± 0.042 in females for PA loci. Between-sex distances were 0.768 ± 0.023 for PA loci. The CATT verified a significant locus association with sex phenotype for three PA loci (χ2 = 13.00–16.00, p < 0.001). No SNP or PA loci associated with females were identified with the criteria of 10:90 or 0:100 male:female. Chi-square tests indicate that these filtering criteria showed no significant differences in males (χ2 = 1.1969, p = 0.5497) and females (χ2 = 0.83319, p = 0.6593) (Table 2 and Supplementary Figure S3).
TABLE 2.
DArT analysis for 14 males and 15 females individuals of green iguana (Iguana iguana) (ZZ/ZW sex-determination type).
|
20 males: 80 females |
30 males: 70 females |
|||
| PA1 | SNPs2 | PA1 | SNPs2 | |
| Total number of DArT analyses | 83,292 | 80,540 | 83,292 | 80,540 |
| Sex-specific loci | 3 | 0 | 457 | 15 |
| Polymorphic information content | 0.49–0.50 | – | 0.49–0.50 | 0.49–0.50 |
| Overall mean distance between males and females | 0.768 ± 0.023 | – | 0.649 ± 0.022 | 0.654 ± 0.021 |
| Overall mean distance within females | 0.248 ± 0.042 | – | 0.410 ± 0.036 | 0.416 ± 0.036 |
| Overall mean distance within males | 0.341 ± 0.037 | – | 0.425 ± 0.033 | 0.499 ± 0.035 |
1Presence–absence data. 2Single nucleotide polymorphism data.
By contrast, for the XX/XY type of sex determination, filtering using the criterion 70:30 male:female yielded 74 SNP loci and 908 PA loci as male-specific. The proportional pairwise Hamming distance between male and female green iguanas using sex-specific SNP and PA loci revealed within-sex distances of 0.429 ± 0.020 in males and 0.531 ± 0.030 in females for SNP loci, and 0.357 ± 0.025 in males and 0.423 ± 0.030 in females for PA loci. Between-sex distances were 0.654 ± 0.021 for SNP loci and 0.649 ± 0.022 for PA loci. The CATT verified a significant locus association with sex phenotype for 59 SNP loci (χ2 = 5.00–29.00, p < 0.001) and 902 PA loci (χ2 = 5.00–30.00, p < 0.001). Filtering with the criterion of 80:20 male:female yielded four SNP loci and 43 PA loci as male-specific. The Hamming distance between males and females using sex-specific SNP and PA loci revealed within-sex distances of 0.135 ± 0.019 in males and 0.357 ± 0.026 in females for SNP loci, and 0.135 ± 0.010 in males and 0.070 ± 0.007 in females for PA loci. Between-sex distances were 0.882 ± 0.013 for SNP loci and 0.940 ± 0.005 for PA loci. The CATT verified a significant locus association with sex phenotype for four SNP loci (χ2 = 7.00–30.00, p < 0.001) and 43 PA loci (χ2 = 13.00–30.00, p < 0.001). Filtering with the criterion 90:10 male:female identified two SNP loci and 33 PA loci as male-specific. The Hamming distance between males and females using sex-specific SNP and PA loci revealed within-sex distances of 0.000 ± 0.000 in males and 0.067 ± 0.017 in females for SNP loci, and 0.085 ± 0.008 in males and 0.000 ± 0.000 in females for PA loci. Between-sex distances were 1.000 ± 0.000 for SNP loci and 0.987 ± 0.003 for PA loci. The CATT verified a significant locus association with sex phenotype for two SNP loci (χ2 = 28.00–29.00, p < 0.001) and 33 PA loci (χ2 = 24.00–30.00, p < 0.001). Two SNP loci and 27 PA loci were associated with males based on the criterion of 100:0 male:female. The proportional pairwise Hamming distance between male and female green iguana using sex-specific SNP and PA loci revealed lower within-sex distances (0.000 ± 0.000 in males and 0.067 ± 0.017 in females for SNP loci, and 0.000 ± 0.000 in males and 0.058 ± 0.008 in females for PA loci). By contrast, higher between-sex distances of 1.000 ± 0.000 for both SNP and PA loci were observed. The CATT verified a significant locus association with sex phenotype for two SNP loci (χ2 = 28.00–29.00, p < 0.001) and 27 PA loci (χ2 = 27.00–30.00, p < 0.001) (Table 3, Figure 2 and Supplementary Figure S3).
TABLE 3.
DArT analysis for 14 males and 15 females individuals of green iguana (Iguana iguana) (XX/XY sex-determination type).
|
70 males: 30 females |
80 males: 20 females |
90 males: 10 females |
100 males: 0 females |
|||||
| PA1 | SNPs2 | PA1 | SNPs2 | PA1 | SNPs2 | PA1 | SNPs2 | |
| Total number of DArT analyses | 83,292 | 80,540 | 83,292 | 80,540 | 83,292 | 80,540 | 83,292 | 80,540 |
| Sex-specific loci | 902 | 59 | 43 | 4 | 33 | 2 | 27 | 2 |
| Polymorphic information content | 0.49–0.50 | 0.49–0.50 | 0.49–0.50 | 0.49–0.50 | 0.49–0.50 | 0.49–0.50 | 0.49–0.50 | 0.49–0.50 |
| Overall mean distance between males and females | 0.673 ± 0.018 | 0.712 ± 0.015 | 0.940 ± 0.005 | 0.882 ± 0.013 | 0.987 ± 0.003 | 1.000 ± 0.000 | 1.000 ± 0.000 | 1.000 ± 0.000 |
| Overall mean distance within females | 0.423 ± 0.030 | 0.531 ± 0.030 | 0.070 ± 0.007 | 0.357 ± 0.026 | 0 ± 0 | 0.067 ± 0.017 | 0.000 ± 0.000 | 0.067 ± 0.017 |
| Overall mean distance within males | 0.357 ± 0.025 | 0.429 ± 0.020 | 0.135 ± 0.010 | 0.135 ± 0.019 | 0.085 ± 0.008 | 0.000 ± 0.000 | 0.058 ± 0.008 | 0.000 ± 0.000 |
1Presence–absence data. 2Single nucleotide polymorphism data.
FIGURE 2.
Hamming distance between male and female individuals of green iguana (Iguana iguana). (A) Single-nucleotide polymorphism (SNP) loci filtered by the criterion of 70:30 (males:females), (B) restriction fragment presence/absence (PA) loci filtered by the criterion of 70:30 (males:females), (C) SNP loci filtered by the criterion of 0:100 (males:females), and (D) PA loci filtered by the criterion of 0:100 (males:females).
For the PA loci data, Chi-square tests showed that the filtering criteria yielded no significant differences in males (χ2 = 0.19, p = 0.98) and females (χ2 = 0.62, p = 0.89). However, for the SNP loci data, the filtering criteria yielded significantly different percentages of heterozygosity in both males and females (Kruskal–Wallis test, p ≤ 0.001). The 70:30 filtering criterion produced significantly lower heterozygosity than the other filtering criteria in males (Nemeyi test, p ≤ 0.001). In females, the 70:30 filtering criterion resulted in significantly lower heterozygosity than the other filtering criteria (p = 0.008), except for the 80:20 criterion (p = 0.45). A glPlot revealed that the sample group showed greater similarity between sexes when perfectly and moderately sex-linked loci were considered (Supplementary Figure S4).
Estimation of Random Sex-Linkage
Across a range of sample sizes and loci, 29 individuals were required to minimize the probability of selecting less than one spurious sex-linked marker. The probability Pi that a single locus exhibited a sex-linked pattern by chance was 1.86 × 10–9 from 173,144 loci (including SNP and PA loci), whereas the expected sex-linkage was estimated to be 3.23 × 10–4. Random sex-linked markers in green iguana were, therefore, more frequent than the expected values in this study.
Homology of Putative Sex-Specific Loci
Based on perfectly sex-linked loci, male sex-specific loci of green iguana showed sequence homology with the reference genomes of green anole and chicken, on the basis of global BLAST analyses of NCBI databases (accessed 22 March, 2020). We observed that six of 29 male-specific SNP/PA loci corresponded to GGA5, GGA18, GGA27, GGA28, and GGAZ (Supplementary Table S8), whereas three out of 29 SNP/PA loci were homologous with the putative candidate genes LIN52 (E-values 8 × 10–05 and similarity 72%), OGFOD3 (E-values 5.9 and similarity 23%), and RAPGEFL1 (E-values 5 × 10–07 and similarity 76%). In addition, four out of 29 SNP/PA loci were homologous to transposable elements consisting of one type of long terminal repeat (LTR) retrotransposons and two types of non-LTR retrotransposons (Supplementary Table S9).
Based on moderately sex-linked loci, we found 164 of 932 loci from SNP/PA male-specific loci corresponding to GGA1, GGA2, GGA3, GGA4, GGA5, GGA6, GGA7, GGA8, GGA9, GGA11, GGA12, GGA15, GGA17, GGA18, GGA20, GGA22, GGA26, GGA27, GGA33, and GGAZ (Supplementary Table S10), whereas 59 SNP/PA loci were homologous with putative functional genes. Moreover, 12 SNP loci and 126 PA loci were homologous to transposable elements consisting of four types of DNA transposons, one type of endogenous retroviruses, four types of LTR retrotransposons, and seven types of non-LTR retrotransposons (Supplementary Table S9).
Discussion
Evolution of sex chromosomes of lizards in Iguanoidea indicates linkage homology similar to ACAX, which is homologous to GGA15 (Alföldi et al., 2011; Rovatsos et al., 2014a,b; Ezaz et al., 2017). The majority of these sex chromosomes evolved before the divergence of iguanian lizards ∼73–93 MYA (Townsend et al., 2011; Zheng and Wiens, 2016), and after the divergence of Acrodonta and Pleurodonta ∼123–168 MYA given that Acrodonta possess different sex-determination systems (XX/XY and ZZ/ZW sex chromosomes) (Townsend et al., 2011; Rovatsos et al., 2014b, 2015; Zheng and Wiens, 2016; Srikulnath et al., 2019). It would be advantageous to investigate the function of sequences or genes located in the genomic block of GGA15 because it may contain the same sex-determination gene. Substantial efforts have been devoted to identification of the mechanisms of sex determination and sex chromosome evolution in lizards of the Iguanoidea (Alföldi et al., 2011; Gamble et al., 2014; Rovatsos et al., 2014a,b; Altmanová et al., 2017; Singchat et al., 2020b). Recent evidence based on comparison of partial gene content using a qPCR approach has shown that sex chromosomes of green iguana are highly conserved with many lizards of Iguanoidea (Rovatsos et al., 2014b). The results of qPCR molecular sexing conducted with five males and five females of green iguanas, following the method of Rovatsos et al. (2014b), reveal the copy number of non-sex-specific gene doses in specimens from the same species (I. iguana) used in the previous study (Rovatsos et al., 2014b). The efficiency of our qPCR system was also examined, and the results were under the standard baseline for qPCR prediction (Rutledge and Côté, 2003). This finding leads to the prediction that the green iguana in the two studies showed different linkages of sex-chromosome systems based on turnover from another autosomal pair. However, this scenario is difficult to envisage because the results of chromosome painting do not support sex-chromosome turnover in lizards of Iguanoidea (Lisachov et al., 2020). An additional possibility might be an intraspecific polymorphism in sex-determination systems, as in the case of the Japanese wrinkled frog Glandirana rugosa (Ogata et al., 2018). DArTseqTM technology is, therefore, an alternative approach as a paradigm shift for examining sex-determination systems and the genetic constitution of sex chromosomes. The high number of SNP/PA loci allowed us to predict sex-specific loci for the green iguana using a sample size of 29 individuals (14 males and 15 females). The probability that one locus would spuriously show sex linkage was 1.86 × 10–9. We sampled 3.23 × 10–4 sex-specific loci. Although this is likely to represent relatively small sample sizes, identification of any sex-linked loci by chance in the present samples was highly unlikely. Several cases found in the North American green frog (Rana clamitans) identified sex-linked loci using small sample sizes (Lambert et al., 2016). To our best knowledge, ours is the first report to demonstrate genome-wide SNP loci in relation to phenotypic sex assignment in green iguana. The X and Y sex chromosomes of green iguana probably differ in genetic composition from each other.
Confirmation of XY Sex Determination and Characterization of Male-Specific Sex Loci for Green Iguana
The present study identified 193 of 961 SNP/PA loci to be male-specific. Of these loci, 29 were perfectly linked with male determination (100:0, male:female) but not with female determination (0:100, male:female). Thus, across each identified locus all known sex individuals were present in all males and absent in all females. This finding confirms that green iguana exhibit an XX/XY sex-determination system (Rovatsos et al., 2014b). The perfectly sex-linked loci suggest the presence of a distinct male-specific region of the Y chromosome. The majority formed a haplotype that became fixed by the cooperative selection of favorable sex-linked chromosomes with hitchhiking effects on the Y sex chromosome and a focal variant (Ezaz et al., 2017). Chromosomes that carry a certain suite of sexually antagonistic genes are more likely to be recruited as sex chromosomes (Bachtrog et al., 2011). The acquisition of male-specific functions would set the stage for adaptive evolution involving sex-determination regions, and introduce strong selective constraints against the degeneration of Y-linked genes. Moreover, this region would represent the putative non-recombination region of sex chromosomes (Charlesworth, 2017). A BLAST search of the NCBI databases for homologous sequences detected three SNP/PA loci that encoded amino acids (LIN52, OGFOD3, and RAPGEFL1) in the tentative Y-specific fragments. However, there is no direct evidence that these genes are involved in squamate reptile sex determination or sex development. Given the relatively short sequence length of all loci, more detailed research is required to characterize fully these genes and their functionality in green iguana. Fragments of four SNP/PA loci were similar to transposable elements and showed 3.4 to 10.34% homology (Supplementary Table S8 and Supplementary Figure S3). The presence of transposable elements in non-recombination regions might regulate the gene expression pattern of the Y sex chromosome through interruption of the gene structure, resulting in gradual silencing and degeneration of the chromosome (Śliwińska et al., 2016).
The highly conserved karyotype of Iguanoid lizards, with homomorphic sex chromosomes frequently appearing on microchromosomes, suggests that the small proportional size of the putative non-recombination region of the X and Y chromosomes of green iguana might reflect the evolutionarily young status of the sex chromosomes, rather than turnover events with autosomes (Rovatsos et al., 2014b). At least a partially degenerate Y chromosome represents the ancestral situation for recently evolved iguanas. Kruskal–Wallis and Nemenyi tests reveal that a number of perfectly and moderately sex-linked loci were significantly different, especially under filtering criteria of 70:30 and 100:0 (male:female), suggesting that partial recombination of XY sex chromosomes clearly differed from the non-recombination region. Interestingly, sex-chromosome recombination is phenotypically, rather than genotypically, moderated in a male heterogametic species (Perrin, 2009; Sopniewski et al., 2019). In the present study, we observed that the 70:30 filtering criterion yielded significantly lower heterozygosity than the other filtering criteria in males, whereas in females heterozygosity with the 70:30 criterion was no different from that with the 80:20 filtering criterion, which indicates that the rate of recombination region is higher in females than males in green iguana. This suggests that sex-chromosome recombination is suppressed by ‘maleness’ rather than different sex chromosomes (Rodrigues et al., 2018; Sopniewski et al., 2019). A total of 932 out of 173,144 SNP/PA loci were, with regard to sex phenotype, completely discordant with the population of 14 males and 15 females individuals. This finding suggests that linkage might result in ambiguous genetic sexes for certain individuals, and 17.6% of SNP/PA loci showed homology with genes encoding amino acids. Preservation of such genes may be driven by selection in males to maintain two copies of dosage-sensitive genes, as observed in human X and Y chromosomes (Wilson Sayres and Makova, 2013; Wilson et al., 2020). A total of 14.8% of SNP/PA loci were homologous to transposable elements (Supplementary Table S4 and Supplementary Figure S3). The majority of loci involved Gypsy transposable elements, which are often distributed on sex chromosomes in medaka (Oryzias latipes), platyfish (Xiphophorus maculatus), guppy (Poecilia reticulata), Siamese cobra (Naja kaouthia), and tilapia (Herpin et al., 2010; Böhne et al., 2012; Chalopin et al., 2015; Laopichienpong et al., submitted data), whereas large numbers of short interspersed nuclear elements were detected on the Y sex chromosomes of frog (Hill et al., 2000; Luo et al., 2017).
Possibility of an Amniote Super-Sex Chromosome
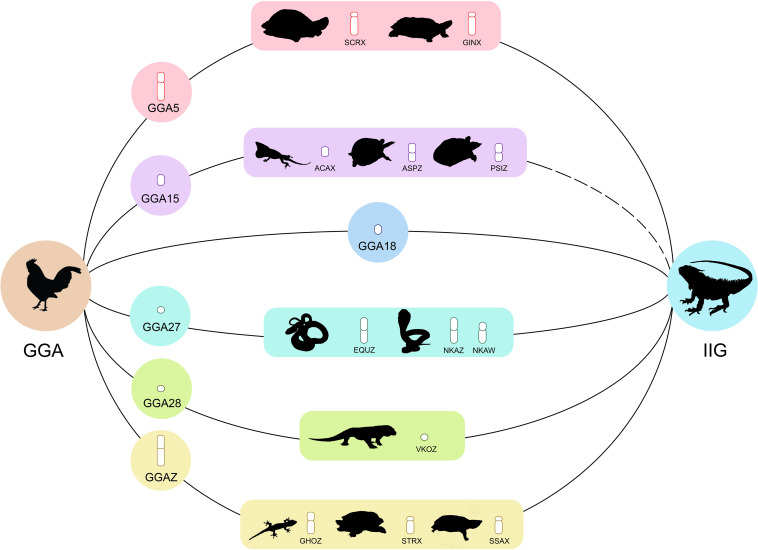
Using comparative gene mapping (i.e., chromosome mapping by means of a cytogenetic technique) and whole-genome sequencing, genomic convergence has been detected in which unrelated sex chromosomes share syntenies across distantly related taxa (Voss et al., 2001; Matsuda et al., 2005; Matsubara et al., 2006, 2012, 2019; Srikulnath et al., 2009a,b, 2013, 2014, 2015; Alföldi et al., 2011; Pokorná et al., 2011; Ezaz et al., 2013, 2017; Young et al., 2013; Gamble et al., 2017; Augstenová et al., 2018; Singchat et al., 2018, 2020a,b; Iannucci et al., 2019; Lind et al., 2019; Ahmad et al., 2020). The ACAX-specific region contains genomic content orthologous to genes on GGA15 (Alföldi et al., 2011), which is homologous to the Z chromosome of the Chinese softshell turtle (Pelodiscus sinensis) and the spiny softshell turtle (Apalone spinifera) (Kawagoshi et al., 2009, 2014). This result led to the prediction of a hypothetical larger ancestral amniote super-sex chromosome with overlaps of partial linkage homology among sex chromosomes of amniotes, whereby multiple translocations have then resulted in the appearance of various sex chromosomes in amniotes (Ezaz et al., 2017; Singchat et al., 2018, 2020a,b; Matsubara et al., 2019; Srikulnath et al., 2019; Ahmad et al., 2020). In the present study, six out of 29 SNP/PA loci that were perfectly sex-linked with phenotypic sex assignment showed partial homology with a number of amniote sex chromosomes (GGA5, GGA18, GGA27, GGA28, and GGAZ) (International Chicken Genome Sequencing Consortium, 2004) (Supplementary Figure S4). The micro-Z chromosomes of Varanus komodoensis correspond to GGA28 and the majority of snake Z chromosomes correspond to GGA2 and GGA27 (Pokorná and Kratochvíl, 2014; Singchat et al., 2018). The X chromosomes of the marsh turtle (Siebenrockiella crassicollis) and wood turtle (Glyptemys insculpta) correspond to GGA5 (Montiel et al., 2017; Singchat et al., 2018). Highly conserved linkage homology of the chicken Z chromosome has been observed for the X chromosomes of the Mexican giant musk turtle (Staurotypus triporcatus), the giant musk turtle (Staurotypus salvinii), and the Z chromosome of the Hokou gecko (Gekko hokouenesis) (Kawai et al., 2009; Kawagoshi et al., 2014; Srikulnath et al., 2015). These results suggest that male-specific loci of the Y sex chromosome in green iguana also share sex-chromosomal linkage homologies with unrelated sex-chromosome-linked regions of other amniotes. Comparative chromosome maps between squamate reptiles and birds using cross-species BAC FISH mapping revealed that GGA5 (CH261-161B22, BAC clone) and GGAZ (CH261-129A16, BAC clone) are mapped on different pairs of green iguana microchromosomes, whereas GGA5 (CH261-161B22, BAC clone) is homologous to the long arm of the W sex chromosome of N. kaouthia, Notechis scutatus and Daboia russelii, and chromosome 1 of Varanus salvator macromaculatus, and corresponds to squamate chromosome 2 (SR2) (Srikulnath et al., 2013; Singchat et al., 2018, 2020a,b). These results thus strongly support the hypothesis of an ancestral amniote super-sex chromosome that might have involved genomic regions orthologous to SR2 and the snake W sex chromosome (Ezaz et al., 2017; Singchat et al., 2018, 2020a,b; Matsubara et al., 2019; Ahmad et al., 2020). By contrast, we did not detect any evidence of loci in the non-recombination region homologous to GGA15; however, only one of 961 SNP/PA loci retained with the filtering criterion 90:10 male:female as moderately sex-linked loci was homologous to GGA15. This finding suggests that the specific region containing genetic constituents homologous to GGA15 tends to be located outside the non-recombination region but in close proximity to the non-recombination region of the sex chromosomes in green iguana. The location of GGA15 might therefore be more distant from the putative sex-determination locus in green iguana, differing from the previous hypothesis (Lambert et al., 2016; Rodrigues et al., 2018; Sopniewski et al., 2019; Ahmad et al., 2020). This becomes a paradigm shift in understanding sex-chromosomal linkages on homomorphic X and Y sex chromosomes. Remarkably, data from the Genomicus database (Nguyen et al., 20184) indicate that several genes from GGA15 are localized to autosomes in ACA (e.g., ACACB on ACA1 or SFI1 on ACA3) (Supplementary Figure S5). This might result from multiple translocations (insertions and/or fusions) that appeared during the 250 million years of divergence between ACA and GGA (Rovatsos et al., 2014a) (Figure 3), and might also have occurred in lizards of Iguanoidea. These hypotheses should be tested by observing the behavior of the sex chromosomes of green iguana during male meiosis with cDNA/BAC FISH mapping. Alternatively, partial linkage homology of green iguana Y sex chromosomes to several amniote sex chromosomes might represent random homologies, given that only small sets of genomic regions in a restricted set of species are involved. However, the main uncertainty in this scenario concerns the actual set of sex-chromosomal linkage homologies of green iguana. Additional evidence, such as whole-genome sequencing or molecular combing, is required to identify this linkage homology.
FIGURE 3.
Male-specific loci of green iguana (Iguana iguana) showing homologies with chicken (Gallus gallus, GGA) and other amniote sex chromosomes. Sex chromosomes of other amniotes shown are Komodo dragon (Varanus komodoensis, VKO), Japanese four-striped rat snake (Elaphe quadrivirgata, EQU), Monocled cobra (Naja kaouthia, NKA), Chinese softshell turtle (Pelodiscus sinensis, PSI), giant musk turtle (Staurotypus salvinii, SSA), Mexican musk turtle (Staurotypus triporcatus, STR), Wood turtle (Glyptemys insculpta, GIN), Spiny softshell turtle (Apalone spinifera, ASP), and Hokou gecko (Gekko hokouenesis, GHO).
How Many Functional Gene Markers Are Required for Sex Identification or Sex Chromosomal Linkage Investigation, If the Ancestral Super-Sex Chromosome Hypothesis Is Supported?
Molecular sexing is an attractive approach in evolutionary and ecological research as well as in conservation and breeding for accurate sex identification of adults in monomorphic or slightly sexually dimorphic species. It is also the only method capable of identifying the sex of embryos and juveniles before the development of primary and secondary sexual characteristics (Morinha et al., 2012). Sex chromosomes are conserved in birds, and the majority of mammals and snakes; therefore, established molecular sexing techniques are often applied throughout these groups (Fridolfsson and Ellegren, 1999; Fernando and Melnick, 2001; Morin et al., 2005; Brubaker et al., 2011; Morinha et al., 2012, 2015; Laopichienpong et al., 2017a,b; Tawichasri et al., 2017). However, no such widely applicable technique was thought possible for other amniotes because they show variability in sex-determination systems and/or homomorphic sex chromosomes (Ezaz et al., 2009b; Pokorná and Kratochvíl, 2009; Gamble, 2010; Sarre et al., 2011; Gamble et al., 2015). Comparative genomics have revealed that the ancestral X chromosome is conserved in lizards of Iguanoidea (Gamble et al., 2014; Rovatsos et al., 2014b; Altmanová et al., 2017). These studies suggest a common origin from the same ancestral pair of sex chromosomes, as evidenced by the sex-chromosomal linkage homology block composed of ACAX-specific genes, which has been detected in the sex chromosomes of various lineages (Alam et al., 2018). As mentioned above, the qPCR analysis in the present study was performed on the same species as those included in the study of Rovatsos et al. (2014b). No statistical differences in gene copy number were observed between males and females of green iguana. Ancestral X chromosomes of certain lizards in Iguanoidea underwent inversion (Gamble et al., 2014). It is possible that multiple independent diversification events in the analyzed region among green iguana individuals in the present study (i.e., intraspecific polymorphism) were associated with inversion of the ancestral sex-determination locus or translocation of the analyzed green iguana X-specific genes to the recombination region, as found in the one locus corresponding to GGA15. An additional possibility is the hypothesis of an ancestral super-sex chromosome as revealed by several segments involving sex-chromosomal linkages in several amniote species (Ezaz et al., 2017; Singchat et al., 2018, 2020a,b; Matsubara et al., 2019; Ahmad et al., 2020). The results of genome-wide SNP analysis, as is possible using DArTseqTM technology, provide a large number of sex-specific SNP/PA loci at perfectly and moderately sex-linked loci for identification of sex determination and sex-linked markers, although the method can only be applied to a small group of closely related species (Sopniewski et al., 2019). The use of a small number of genes to identify sex determination and sex-chromosomal linkages might result in errors because many homologous loci across amniote sex chromosomes were observed at Y-specific loci in the current study, while false positive qPCR results might often be obtained in different laboratories (Rovatsos et al., 2014b). Developmental molecular identification and analysis of sex-linked genetic markers based on a PCR approach are required to establish a genotyping tool for practical sexing of individuals in a commercial breeding program and allow accurate determination of sex in populations through insight into the evolutionary history of the Y chromosome (or the W chromosome in a female heterogametic species, such as occurs in snakes) (Laopichienpong et al., 2017a,b; Tawichasri et al., 2017).
Conclusion
We have successfully identified sex-specific loci on the Y sex chromosome of green iguana using DArTseqTM technology. We identified a notable number of perfectly sex-linked loci given the small portion of the genome. The results confirm the presence of XX/XY sex determination in green iguana. Several male-specific SNP/PA loci of green iguana had partial homology with a number of amniote sex chromosomes (GGA5, GGA18, GGA27, GGA28, and GGAZ), supporting the hypothesis of an ancestral amniote super-sex chromosome. However, this did not concur with the previous prediction hypothesized for a large proportion of the genomic segment GGA15. This represents a paradigm shift in the understanding of sex-chromosomal linkages on homomorphic X and Y sex chromosomes. It remains unclear whether the location of large genomic regions between X-specific and Y-specific fragments is associated with differentiation of sex chromosomes and sex-determination regions. It seems likely that these loci are sex-linked in other populations. A study population with a relatively even ratio of sexes or wild populations is thus required to minimize the probability of false-positives, although the number of samples needed may also depend on the amount of polymorphism present in the population, and will probably vary across species. Practically, a PCR-based approach should be developed from perfectly sex-linked SNP/PA loci as identified in this study, although failure of the PCR validation step has often been observed after DArTseqTM or RADseq bioinformatics analysis (Gamble et al., 2015). Chromosome mapping of sex-specific loci on mitotic and meiotic chromosomes using FISH is required to determine their positions on the sex chromosomes of green iguana. In addition, a high-quality complete genome assembly for green iguana is required to elucidate further the sex-determination mechanism. This approach will provide a solid basis for explaining the sex-determination mechanism and identifying potential sex-determination regions in iguana and other squamate reptiles.
Data Availability Statement
The full dataset and metadata from this publication are available from the Dryad Digital Repository. Dataset, https://doi.org/10.5061/dryad.j0zpc86b5.
Ethics Statement
The animal study was reviewed and approved by Animal care and all experimental procedures were approved by the Animal Experiment Committee, Kasetsart University, Thailand (approval no. ACKU61-SCI-023) and conducted in accordance with the Regulations on Animal Experiments at Kasetsart University.
Author Contributions
TK and KS drafted the manuscript and carried out the lab work. TK, NL, EK, and KS conceived the ideas and designed the methodology and participated in data analysis. All authors reviewed the data and the manuscript and gave final approval for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Werapat Kanakasai (Chonburi Iguana Farm) for specimen collection, and Kornsuang Jangtawan and Tulyawat Prasongnaneerut (ACCG, Kasetsart University, Thailand) for general discussion.
Funding. This study was financially supported by grants from the Center for Advanced Studies in Tropical Natural Resources, National Research University-Kasetsart University (CASTNAR, NRU-KU, Thailand) awarded to KS, the Capacity Building of KU Students on Internationalization Program (KUCSI) awarded to NL and KS, Thailand Research Fund (TRF) (nos. RSA6180075, PHD60I0014, and MSD60I0122) awarded to KS, WS, TK, and RP, Science Achievement Scholarship of Thailand (SAST) (no. 5917400296) from the Office of the Higher Education Commission, Thailand awarded to NL, and a University of Canberra internal research grant awarded to TE.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.556267/full#supplementary-material
Standard curves generated for (A) TMEM132D, (B) CCDC92, (C) ATP2A2, (D) PEBP1, and (E) GAPDH. Linear dynamic range from 25 to 0.00025 ng/μl genomic DNA concentration extracted from green iguana (Iguana iguana) (n = 10). Regression lines were calculated as R2 = 0.9446–0.9879, p < 0.001 for TMEM132D, CCDC92, ATP2A2, PEBP1, and GAPDH.
Density PIC plot for (A) single-nucleotide polymorphism (SNP) and (B) restriction fragment presence/absence (PA) loci.
Number of loci in different hypotheses of sex-determination systems after filtering using different male:female ratios as criteria. The x-axis indicates the number of loci and the y-axis indicates the filtering criteria. (A) Restriction fragment presence/absence (PA) loci and (B) single-nucleotide polymorphism loci.
An index of the two perfectly sex-aligned sequences and the additional 59 sex-related sequences created with the ‘glPlot’ function implemented in the “dartR” R package. Blue indicates homozygosity to the reference allele, purple is indicative of heterozygosity, and red indicates homozygosity to the alternate SNP-containing allele. Sequences two (right of the figure) show perfectly sex-linked sequences; females are indicated in blue and males are indicated in purple.
Segmental homology of green anole (Anolis carolinensis) and chicken (Gallus gallus). Segmental linkage homologies are derived from the Genomicus database (Louis et al., 2014). Arrow indicate chromosome 15 of chicken and autosome of anolis.
Primers used for the measurement of relative gene dosage by quantitative PCR analysis.
Inter- and intra-assay coefficient of variation (CV) and efficiency of five genes for dilution standard curve.
Inter- and intra-assay coefficient of variation (CV) and efficiency for five genes.
Relative gene dosage ratios between five males and five females individuals of green iguana (Iguana iguana) for TMEM132D.
Relative gene dosage ratios between five males and five females individuals of green iguana (Iguana iguana) for CCDC92.
Relative gene dosage ratios between five males and five females individuals of green iguana (Iguana iguana) for ATP2A2.
Relative gene dosage ratios between five males and five females individuals of green iguana (Iguana iguana) for PEBP1.
Gene function and pathway for single-nucleotide polymorphism (SNP) and restriction fragment presence/absence (PA) loci of green iguana (Iguana iguana) from a BLAST search of the green anole (Anolis carolinensis) genome (100:0, male:female).
Repeat searches for single-nucleotide polymorphism (SNP) and restriction fragment presence/absence (PA) loci of green iguana (Iguana iguana).
Gene function and pathway for single-nucleotide polymorphism (SNP) and restriction fragment presence/absence (PA) loci of green iguana (Iguana iguana) from a BLAST search of the green anole (Anolis carolinensis) genome (70:30, male:female).
References
- Ahmad S. F., Singchat W., Jehangir M., Panthum T., Srikulnath K. (2020). Consequence of paradigm shift with repeat landscapes in reptiles: powerful facilitators of chromosomal rearrangements for diversity and evolution (running title: genomic impact of repeats on chromosomal dynamics in reptiles). Genes 11:827 10.3390/genes11070827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S. M. I., Sarre S. D., Gleeson D., Georges A., Ezaz T. (2018). Did lizards follow unique pathways in sex chromosome evolution? Genes 9:239 10.3390/genes9050239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alföldi J., Palma F. D., Grabherr M., Williams C., Kong L., Mauceli E., et al. (2011). The genome of the Green anole lizard and a comparative analysis with birds and mammals. Nature 477 587–591. 10.1038/nature10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmanová M., Rovatsos M., Kratochvíl L., Pokorná M. J. (2017). All iguana families with the exception of basilisks share sex chromosomes. Zoology 126 98–102. 10.1016/j.zool.2017.11.007 [DOI] [PubMed] [Google Scholar]
- Augstenová B., Johnson P. M., Altmanová M., Frynta D., Rovatsos M., Kratochvíl L. (2018). ZW, XY, and yet ZW: sex chromosome evolution in snakes even more complicated. Evolution 72 1701–1707. 10.1111/evo.13543 [DOI] [PubMed] [Google Scholar]
- Bachtrog D., Kirkpatrick M., Mank J. E., McDaniel S. F., Pires C. J., Rice W., et al. (2011). Are all sex chromosomes created equal? Trends Genet. 27 350–357. 10.1016/j.tig.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Bao W., Kojima K. K., Kohany O. (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6:11 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhne A., Zhou Q., Darras A., Schmidt C., Schartl M., Galiana-Arnoux D., et al. (2012). Zisupton–a novel superfamily of DNA transposable elements recently active in fish. Biol. Evol. 29 631–645. 10.1093/molbev/msr208 [DOI] [PubMed] [Google Scholar]
- Brandley M. C., Wynn A., Queiroz K. (2006). Karyotype and relationships of Anolis desechensis. J. Herpetol. 40 136–139. 10.1670/98-05N.1 [DOI] [Google Scholar]
- Brubaker J. L., Karouna-Renier N. K., Chen Y., Jenko K., Sprague D. T., Henry P. F. P. (2011). A non-invasive, direct real-time PCR method for sex determination in multiple avian species. Mol. Ecol. Resour. 11 415–417. 10.1111/j.1755-0998.2010.02951.x [DOI] [PubMed] [Google Scholar]
- Castiglia R., Flores-Villela O., Bezerra A. M. R., Muñoz A., Gornung E. (2013). Pattern of chromosomal changes in ‘beta’ Anolis (Norops group) (Squamata: Polychrotidae) depicted by an ancestral state analysis. Zool. Stud. 52:60 10.1186/1810-522X-52-60 [DOI] [Google Scholar]
- Cawthon R. M. (2002). Telomere measurement by quantitative PCR. Nucleic Acids Res. 30:e47 10.1093/nar/30.10.e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalopin D., Volff J. N., Galiana D., Anderson J. L., Schartl M. (2015). Transposable elements and early evolution of sex chromosomes in fish. Chromosome Res. 23 545–560. 10.1007/s10577-015-9490-8 [DOI] [PubMed] [Google Scholar]
- Charlesworth D. (2017). Evolution of recombination rates between sex chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160456 10.1098/rstb.2016.0456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. M., Huang C. C., Clark H. F. (1967). The somatic chromosomes of 3 lizard species: Gekko gecko, Iguana iguana, and Crotaphytus collaris. Experientia 23 769–771. 10.1007/bf02154168 [DOI] [PubMed] [Google Scholar]
- Criscuolo F., Bize P., Nasir L., Metcalfe N. B. (2009). Real-time quantitative PCR assay for measurement of avian telomeres. J. Avian Biol. 40 342–347. 10.1111/j.1600-048X.2008.04623.x [DOI] [Google Scholar]
- Ezaz T., Azad B., O’Meally D., Young M., Matsubara K., Edwards M. J., et al. (2013). Sequence and gene content of a large fragment of a lizard sex chromosome and evaluation of candidate sex differentiating gene R–spondin 1. BMC Genomics 14:899 10.1186/1471-2164-14-899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaz T., Quinn A. E., Sarre S. D., O’Meally D., Georges A., Graves J. A. (2009a). Molecular marker suggests rapid changes of sex-determining mechanisms in Australian dragon lizards. Chromosome Res. 17 91–98. 10.1007/s10577-008-9019-5 [DOI] [PubMed] [Google Scholar]
- Ezaz T., Sarre S. D., O’Meally D., Graves J. A., Georges A. (2009b). Sex chromosome evolution in lizards: independent origins and rapid transitions. Cytogenet. Genome Res. 127 249–260. 10.1159/000300507 [DOI] [PubMed] [Google Scholar]
- Ezaz T. M., Srikulnath K., Graves J. A. (2017). Origin of amniote sex chromosomes: an ancestral super–sex chromosome, or common requirements. J. Hered. 108 94–105. 10.1093/jhered/esw053 [DOI] [PubMed] [Google Scholar]
- Falcón W., Ackerman J. D., Recart W., Daehler C. C. (2013). Biology and impacts of Pacific island invasive species. 10. Iguana iguana, the Green iguana (Squamata: Iguanidae). Pac. Sci. 2 157–186. 10.2984/67.2.2 [DOI] [Google Scholar]
- Fernando P., Melnick D. J. (2001). Molecular sexing eutherian mammals. Mol. Ecol. Resour. 1 350–353. 10.1046/j.1471-8278.2001.00112.x [DOI] [Google Scholar]
- Fridolfsson A. K., Ellegren H. (1999). A simple and universal method for molecular sexing of non-ratite birds. J. Avian Biol. 30 116–121. 10.2307/3677252 [DOI] [Google Scholar]
- Gamble T. (2010). A review of sex determining mechanisms in Geckos (Gekkota: Squamata). Sex. Dev. 4 88–103. 10.1159/000289578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T., Castoe T. A., Nielsen S. V., Banks J. L., Card D. C., Schield D. R., et al. (2017). The discovery of XY sex chromosomes in a Boa and Python. Curr. Biol. 27 2148–2153. 10.1016/j.cub.2017.06.010 [DOI] [PubMed] [Google Scholar]
- Gamble T., Coryell J., Lynch J., Ezaz T., Scantlebury D. P., Zarkower D. (2015). Restriction site-associated DNA sequences (RAD-seq) reveals an extraordinary number of transitions among Gecko sex-determining systems. Mol. Biol. Evol. 32 1296–1309. 10.1093/molbev/msv023 [DOI] [PubMed] [Google Scholar]
- Gamble T., Geneva A. J., Glor R. E., Zarkower D. (2014). Anolis sex chromosomes are derived from a single ancestral pair. Evolution 68 1027–1041. 10.1111/evo.12328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T., Zarkower D. (2014). Identification of sex-specific molecular markers using restriction site-associated DNA sequencing. Mol. Ecol. Res. 14 902–913. 10.1111/1755-0998.12237 [DOI] [PubMed] [Google Scholar]
- Gruber B., Georges A. (2019). DartR: Importing and Analysing SNP and Silicodart Data Generated by Genome-Wide Restriction Fragment Analysis. Available online at: https://cran.r-project.org/web/packages/dartR/index.html (accessed January 22, 2020). [Google Scholar]
- Hanneman S. K., Cox C. D., Green K. E., Kang D. H. (2011). Estimating intra- and inter–assay variability in salivary cortisol. Biol. Res. Nurs. 13 243–250. 10.1177/1099800411404061 [DOI] [PubMed] [Google Scholar]
- Hedges S. B., Vidal N. (2009). Lizards, Snakes, and Amphisbaenians (Squamata) in the Timetree of Life. Oxford: Oxford University Press, 383–389. [Google Scholar]
- Herpin A., Braasch I., Kraeussling M., Schmidt C., Thoma E. C., Nakamura S., et al. (2010). Transcriptional rewiring of the sex determining dmrt1 gene duplicate by transposable elements. PLoS Genet. 6:e1000844 10.1371/journal.pgen.1000844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E. W., Jobling M. A., Bradley D. G. (2000). Y–chromosome variation and Irish origins. Nature 404 351–352. 10.1038/35006158 [DOI] [PubMed] [Google Scholar]
- Iannucci A., Altmanová M., Ciofi C., Ferguson-Smith M., Milan M., Pereira J. C., et al. (2019). Conserved sex chromosomes and karyotype evolution in Monitor lizards (Varanidae). Heredity 123 215–227. 10.1038/s41437-018-0179-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamdar D. L. S., Vani V., Seshagiri P. B. (2012). A tropical oviparous lizard, Calotes versicolor, exhibiting a potentially novel FMFM pattern of temperature-dependent sex determination. J. Exp. Zool. A Ecol. Genet. Physiol. 317 32–46. 10.1002/jez.718 [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium (2004). Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 433 695–716. 10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Jaccoud D., Peng K., Feinstein D., Kilian A. (2001). Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic. acids research 29:E25 10.1093/nar/29.4.e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M. (1994). Sexing Green Iguanas. Available online at: http://www.anapsid.org/iguana/sexingigs.html (accessed February 02, 2020). [Google Scholar]
- Kawagoshi T., Uno Y., Matsubara K., Matsuda Y., Nishida C. (2009). The ZW micro-sex chromosomes of the Chinese soft-shelled turtle (Pelodiscus sinensis Trionychidae, Testudines) have the same origin as chicken chromosome 15. Cytogenet. Genome Res. 125 125–131. 10.1159/000227837 [DOI] [PubMed] [Google Scholar]
- Kawagoshi T., Uno Y., Nishida C., Matsuda Y. (2014). The Staurotypus turtles and aves share the same origin of sex chromosomes but evolved different types of heterogametic sex determination. PLoS One 9:e105315 10.1371/journal.pone.0105315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai A., Ishijima J., Nishida C., Kosaka A., Ota H., Kohno S., et al. (2009). The ZW sex chromosomes of Gekko hokouenesis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma 118 43–51. 10.1007/s00412-008-0176-2 [DOI] [PubMed] [Google Scholar]
- Kilian A., Wenzl P., Huttner E., Carling J., Xia L., Blois H., et al. (2012). Diversity arrays technology: a generic genome profiling technology on open platforms. Methods Mol. Biol. 888 67–89. 10.1007/978-1-61779-870-2_5 [DOI] [PubMed] [Google Scholar]
- Lambert M. R., Skelly D. K., Ezaz T. (2016). Sex-linked markers in the North American green frog (Rana clamitans) developed using DArTseq provide early insight into sex chromosome evolution. BMC Genomics 17:844 10.1186/s12864-016-3209-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laopichienpong N., Muangmai N., Chanhome L., Suntrarachun S., Twilprawat P., Peyachoknagul S., et al. (2017a). Evolutionary dynamics of the gametologous CTNNB1 gene on the Z and W chromosomes of snakes. J. Hered. 108 142–151. 10.1093/jhered/esw074 [DOI] [PubMed] [Google Scholar]
- Laopichienpong N., Tawichasri P., Chanhome L., Phatcharakullawarawat R., Singchat W., Kantachumpoo A., et al. (2017b). A novel method of caenophidian snake sex identification using molecular markers based on two gametologous genes. Ecol. Evol. 7 4661–4669. 10.1002/ece3.3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laopichienpong N., Kraichak E., Singchat W., Sillapaprayoon S., Muangmai N., Suntrarachun S., et al. (2020). Genome-wide SNP analysis of Siamese cobra (Naja kaouthia) reveals the molecular basis of transitions between Z and W sex chromosomes and supports the presence of an ancestral super-sex chromosome in amniotes. Genomics. 10.1016/j.ygeno.2020.09.058 [DOI] [PubMed] [Google Scholar]
- Lind A. L., Lai Y. Y. Y., Mostovoy Y., Holloway A. K., Iannucci A., Mak A. C. Y., et al. (2019). Genome of the Komodo dragon reveals adaptations in the cardiovascular and chemosensory systems of monitor lizards. Nat. Ecol. Evol. 3 1241–1252. 10.1038/s41559-019-0945-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisachov A. P., Giovannotti M., Pereira J. C., Andreyushkova D. A., Romanenko S. A., Ferguson-Smith M. A., et al. (2020). Chromosome painting does not support a sex chromosome turnover in Lacerta agilis linnaeus. Cytogenet. Genome Res. 160 134–140. 10.1159/000506321 [DOI] [PubMed] [Google Scholar]
- Louis A., Nguyen N. T. T., Muffato M., Roest Crollius H. (2014). Genomicus update 2015: karyoView and MatrixView provide a genome-wide perspective to multispecies comparative genomics. Nucleic Acids Res. 43 D682–D689. 10.1093/nar/gku1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Li Y., Xia L., Hu W., Gao W., Guo L., et al. (2017). Analysis of the expression patterns of the novel large multigene TRIM gene family (finTRIM) in zebrafish. Fish Shellfish Immunol. 66 224–230. 10.1016/j.fsi.2017.04.024 [DOI] [PubMed] [Google Scholar]
- Matsubara K., Kuraku S., Tarui H., Nishimura O., Nishida C., Agata K., et al. (2012). Intra-genomic GC heterogeneity in sauropsids: evolutionary insights from cDNA mapping and GC3 profiling in snake. BMC Genomics 13:604 10.1186/1471-2164-13-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K., O’Meally D., Sarre S. D., Georges A., Srikulnath K., Ezaz T. (2019). ZW sex chromosomes in Australian dragon lizards (Agamidae) originated from a Combination of duplication and translocation in the nucleolar organising region. Genes 10:861 10.3390/genes10110861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K., Tarui H., Toriba M., Yamada K., Nishida C., Agata K., et al. (2006). Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl. Acad. Sci. U.S.A. 103 18190–18195. 10.1073/pnas.0605274103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y., Nishida-Umehara C., Tarui H., Kuroiwa A., Yamada K., Isobe T., et al. (2005). Highly conserved linkage homology between birds and turtles: bird and turtle chromosomes are precise counterparts of each other. Chromosome Res. 13 601–615. 10.1007/s10577-005-0986-5 [DOI] [PubMed] [Google Scholar]
- Matsuda Y., Uno Y., Kondo M., Gilchrist M. J., Zorn A. M., Rokhsar D. S., et al. (2015). A new nomenclature of Xenopus laevis chromosomes based on the phylogenetic relationship to Silurana/Xenopus tropicalis. Cytogenet. Genome Res. 145 187–191. 10.1159/000381292 [DOI] [PubMed] [Google Scholar]
- Montiel E. E., Badenhorst D., Tamplin J., Burke R. L., Valenzuela N. (2017). Discovery of the youngest sex chromosomes reveals first case of convergent co-option of ancestral autosomes in turtles. Chromosoma 126 105–113. 10.1007/s00412-016-0576-7 [DOI] [PubMed] [Google Scholar]
- Morin P. A., Nestler A., Rubio-Cisneros N. T., Robertson K. M., Mesnick S. L. (2005). Interfamilial characterization of a region of the ZFX and ZFY genes facilitates sex determination in cetaceans and other mammals. Mol. Ecol. 14 3275–3286. 10.1111/j.1365-294X.2005.02651.x [DOI] [PubMed] [Google Scholar]
- Morinha F., Cabral J. A., Bastos E. (2012). Molecular sexing of birds: a comparative review of polymerase chain reaction (PCR)-based methods. Theriogenology 78 703–714. 10.1016/j.theriogenology.2012.04.015 [DOI] [PubMed] [Google Scholar]
- Morinha F., Cabral J. A., Martins S., Cruz E., Alvura N., Nunese P., et al. (2015). (R)evolution in the molecular sexing of ratite birds: identification and analysis of new candidate sex-linked markers. Avian Biol. Res. 8 145–159. 10.3184/175815515X14380769320720 [DOI] [Google Scholar]
- Natri H. M., Merilä J., Shikano T. (2019). The evolution of sex determination associated with a chromosomal inversion. Nat. Commun. 10:145 10.1038/s41467-018-08014-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N. T. T., Vincens P., Crollius H., Genomicus A. L. (2018). Genomicus 2018: karyotype evolutionary trees and on-the-fly synteny computing. Nucleic Acids Res. 46 D816–D822. 10.1093/nar/gkx1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S. V., Guzmán-Méndez I. A., Gamble T., Blumer M., Pinto B. J., Kratochvíl L., et al. (2019). Escaping the evolutionary trap? Sex chromosome turnover in basilisks and related lizards (Corytophanidae: Squamata). Biol. Lett. 15:20190498 10.1098/rsbl.2019.0498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T., Hands R. E., Bustin S. A. (2006). Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1 1559–1582. 10.1038/nprot.2006.236 [DOI] [PubMed] [Google Scholar]
- Ogata M., Lambert M., Ezaz T., Miura I. (2018). Reconstruction of female heterogamety from admixture of XX-XY and ZZ-ZW sex-chromosome systems within a frog species. Mol. Ecol. 27 4078–4089. 10.1111/mec.14831 [DOI] [PubMed] [Google Scholar]
- O’Meally D., Ezaz T., Georges A., Sarre S. D., Graves J. A. M. (2012). Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res. 20 7–19. 10.1007/s10577-011-9266-8 [DOI] [PubMed] [Google Scholar]
- O’Meally D., Patel H. R., Stiglec R., Sarre S. D., Georges A., Graves J. A. M., et al. (2010). Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. 18 787–800. 10.1007/s10577-010-9152-9 [DOI] [PubMed] [Google Scholar]
- Perrin N. (2009). Sex reversal: a fountain of youth for sex chromosomes? Evolution 63 3043–3049. 10.1111/j.1558-5646.2009.00837.x [DOI] [PubMed] [Google Scholar]
- Pokorná J. M., Kratochvíl L. (2014). What was the ancestral sex-determining mechanism in amniote vertebrates? Biol. Rev. 91 1–12. 10.1111/brv.12156 [DOI] [PubMed] [Google Scholar]
- Pokorná M. J., Kratochvíl L. (2009). Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool. J. Linn. Soc. 156 168–183. 10.1111/j.1096-3642.2008.00481.x [DOI] [Google Scholar]
- Pokorná M. J., Kratochvíl L., Kejnovskı E. (2011). Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and Lacertidae: Eremias velox). BMC Genet. 12:90 10.1186/1471-2156-12-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ren R., Ray R., Li P., Xu J., Zhang M., Liu G., Yao, et al. (2015). Construction of a high-density DArTseq SNP-based genetic map and identification of genomic regions with segregation distortion in a genetic population derived from a cross between feral and cultivated-type watermelon. Mol. Gen. Genomics. 290 1457–1470. 10.1007/s00438-015-0997-7 [DOI] [PubMed] [Google Scholar]
- Rivas J., Rivas J. A., Avila T. A., Avila T. Á. (1996). Sex identification in Juvenile green iguanas (Iguana iguana) by cloacal analysis. Copeia 1996 219–221. 10.2307/1446964 [DOI] [Google Scholar]
- Rodrigues N., Studer T., Dufresnes C., Perrin N. (2018). Sex-chromosome recombination in common frogs brings water to the fountainof-youth. Mol. Biol. Evol. 35 942–948. 10.1093/molbev/msy008 [DOI] [PubMed] [Google Scholar]
- Rovatsos M., Altmanová M., Pokorná M. J., Kratochvíl L. (2014a). Novel X-linked genes revealed by quantitative polymerase chain reaction in the green anole, Anolis carolinensis. G3 4 2107–2113. 10.1534/g3.114.014084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M., Farkaèová K., Altmanová M., Pokorná J. M., Kratochvíl L. (2019). The rise and fall of differentiated sex chromosomes in geckos. Mol. Ecol. 12 3042–3052. 10.1111/mec.15126 [DOI] [PubMed] [Google Scholar]
- Rovatsos M., Johnson Pokorná M., Altmanová M., Kratochvíl L. (2015). Female heterogamety in Madagascar chameleons (Squamata: Chamaeleonidae: Furcifer): differentiation of sex and neo-sex chromosomes. Sci. Rep. 5:13196 10.1038/srep13196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M., Pokorná M., Altmanová M., Kratochvíl L. (2014b). Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biol. Lett. 10:20131093 10.1098/rsbl.2013.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge R. G., Côté C. (2003). Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res. 31:e93 10.1093/nar/gng093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarre S. D., Ezaz T., Georges A. (2011). Transitions between sex-determining systems in reptiles and amphibians. Annu. Rev. Genomics Hum. Genet. 12 391–406. 10.1146/annurev-genom-082410-101518 [DOI] [PubMed] [Google Scholar]
- Schartl M., Larue L., Goda M., Bosenberg M. W., Hashimoto H., Kelsh R. N. (2016). What is a vertebrate pigment cell? Pigment Cell Melanoma Res. 29 8–14. 10.1111/pcmr.12409 [DOI] [PubMed] [Google Scholar]
- Schultheiss O. C., Stanton S. J. (2009). “Assessment of salivary hormones,” in Methods in Social Neuroscience, eds Harmon-Jones E., Beer J. S. (New York, NY: Guilford; ), 17–44. [Google Scholar]
- Singchat W., Ahmad S. F., Sillapaprayoon S., Muangmai N., Duengkae P., Peyachoknagul S., et al. (2020a). Partial amniote sex chromosomal linkage homologies shared on snake W sex chromosomes support the ancestral super-sex chromosome evolution in amniotes. Front. Genet. 11:948 10.3389/fgene.2020.00948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singchat W., Kraichak E., Tawichasri P., Tawan T., Suntronpong A., Sillapaprayoon S., et al. (2019). Dynamics of telomere length in captive Siamese cobra (Naja kaouthia) related to age and sex. Ecol. Evol. 9 6366–6377. 10.1002/ece3.5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singchat W., O’Connor R. E., Tawichasri P., Suntronpong A., Sillapaprayoon S., Suntrarachun S., et al. (2018). Chromosome map of the Siamese cobra: Did partial synteny of sex chromosomes in the amniote represent “a hypothetical ancestral super-sex chromosome” or random distribution? BMC Genomics 19:939 10.1186/s12864-018-5293-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singchat W., Sillapaprayoon S., Muangmai N., Baicharoen S., Indananda C., Duengkae P., et al. (2020b). Do sex chromosomes of snakes, monitor lizards, and iguanian lizards result from multiple fission of an “ancestral amniote super-sex chromosome”? Chromosome Res. 28 209–228. 10.1007/s10577-020-09631-4 [DOI] [PubMed] [Google Scholar]
- Śliwińska E. B., Martyka R., Tryjanowski P. (2016). Evolutionary interaction between W/Y chromosome and transposable elements. Genetica 144 267–278. 10.1007/s10709-016-9895-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopniewski J., Shams F., Scheele B. C., Kefford B. J., Ezaz T. (2019). Identifying sex-linked markers in Litoria aurea: a novel approach to understanding sex chromosome evolution in an amphibian. Sci. Rep. 9:16591 10.1038/s41598-019-52970-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikulnath K., Azad B., Singchat W., Ezaz T. (2019). Distribution and amplification of interstitial telomeric sequences (ITSs) in Australian dragon lizards support frequent chromosome fusions in Iguania. PLoS One 14:e0212683 10.1371/journal.pone.0212683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikulnath K., Matsubara K., Uno Y., Nishida C., Olsson M., Matsuda Y. (2014). Identification of the linkage group of the Z sex chromosomes of the Sand lizard (Lacerta agilis, Lacertidae) and elucidation of karyotype evolution in lacertid lizards. Chromosoma 123 563–575. 10.1007/s00412-014-0467-8 [DOI] [PubMed] [Google Scholar]
- Srikulnath K., Matsubara K., Uno Y., Thongpan A., Suputtitada S., Apisitwanich S., et al. (2009a). Karyological characterization of the. Cytogenet. Genome Res. 125 213–223. 10.1159/000230005 [DOI] [PubMed] [Google Scholar]
- Srikulnath K., Nishida C., Matsubara K., Uno Y., Thongpan A., Suputtitada S., et al. (2009b). Karyotypic evolution in squamate reptiles: comparative gene mapping revealed highly conserved linkage homology between the Butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Lacertilia) and the Japanese four-striped rat snake (Elaphe quadrivirgata, Colubridae, Serpentes). Chromosome Res. 17 975–986. 10.1007/s10577-009-9101-7 [DOI] [PubMed] [Google Scholar]
- Srikulnath K., Uno Y., Nishida C., Matsuda Y. (2013). Karyotype evolution in monitor lizards: cross-species chromosome mapping of cDNA reveals highly conserved synteny and gene order in the Toxicofera clade. Chromosome Res. 21 805–819. 10.1007/s00412-014-0467-8 [DOI] [PubMed] [Google Scholar]
- Srikulnath K., Uno Y., Nishida C., Ota H., Matsuda Y. (2015). Karyotype reorganization in the Hokou gecko (Gekko hokouensis, Gekkonidae): the process of microchromosome disappearance in Gekkota. PLoS One 10:e0134829 10.1371/journal.pone.0134829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supikamolseni A., Ngaoburanawit N., Sumontha M., Chanhome L., Suntrarachun S., Peyachoknagul S., et al. (2015). Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify seven snake groups for which antivenom is available in Thailand. Genet. Mol. Res. 14 13981–13997. 10.4238/2015.October.29.18 [DOI] [PubMed] [Google Scholar]
- Sven K., Klaus R. (2019). HapEstXXR: Multi-Locus Stepwise Regression. Available online at: https://rdrr.io/cran/HapEstXXR/man/coding.baseline.allele.html (accessed February 12, 2020). [Google Scholar]
- Tawichasri P., Laopichienpong N., Chanhome L., Phatcharakullawarawat R., Singchat W., Koomgun T., et al. (2017). Using blood and noninvasive shed skin samples to identify sex of caenophidian snakes based on multiplex PCR assay. Zool. Anz. 271 6–14. 10.1016/j.jcz.2017.11.003 [DOI] [Google Scholar]
- Townsend T. M., Mulcahy D. G., Noonan B. P., Sites J. W., Jr., Kuczynski C. A., Wiens J. J., et al. (2011). Phylogeny of iguanian lizards inferred from 29 nuclear loci, and a comparison of concatenated and species-tree approaches for an ancient, rapid radiation. Mol. Phylogenet. Evol. 61 363–380. 10.1016/j.ympev.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Uno Y., Nishida C., Tarui H., Ishishita S., Takagi C., Nishimura O., et al. (2012). Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS One 7:53027 10.1371/journal.pone.0053027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss S. R., Smith J. J., Gardiner D. M., Parichy D. M. (2001). Conserved vertebrate chromosome segments in the large salamander genome. Genetics 158 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J., Staley J. M., Wyckoff G. J. (2020). Extinction of chromosomes due to specialization is a universal occurrence. Sci. Rep. 10:2170 10.1038/s41598-020-58997-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Sayres M. A., Makova K. D. (2013). Gene survival and death on the human Y chromosome. Mol. Biol. Evol. 30 781–787. 10.1093/molbev/mss267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. J., O’Meally D., Sarre S. D., Georges A., Ezaz T. (2013). Molecular cytogenetic map of the Central bearded dragon, Pogona vitticeps (Squamata: Agamidae). Chromosome Res. 21 361–374. 10.1007/s10577-013-9362-z [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wiens J. J. (2016). Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 94 537–547. 10.1016/j.ympev.2015.10.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Standard curves generated for (A) TMEM132D, (B) CCDC92, (C) ATP2A2, (D) PEBP1, and (E) GAPDH. Linear dynamic range from 25 to 0.00025 ng/μl genomic DNA concentration extracted from green iguana (Iguana iguana) (n = 10). Regression lines were calculated as R2 = 0.9446–0.9879, p < 0.001 for TMEM132D, CCDC92, ATP2A2, PEBP1, and GAPDH.
Density PIC plot for (A) single-nucleotide polymorphism (SNP) and (B) restriction fragment presence/absence (PA) loci.
Number of loci in different hypotheses of sex-determination systems after filtering using different male:female ratios as criteria. The x-axis indicates the number of loci and the y-axis indicates the filtering criteria. (A) Restriction fragment presence/absence (PA) loci and (B) single-nucleotide polymorphism loci.
An index of the two perfectly sex-aligned sequences and the additional 59 sex-related sequences created with the ‘glPlot’ function implemented in the “dartR” R package. Blue indicates homozygosity to the reference allele, purple is indicative of heterozygosity, and red indicates homozygosity to the alternate SNP-containing allele. Sequences two (right of the figure) show perfectly sex-linked sequences; females are indicated in blue and males are indicated in purple.
Segmental homology of green anole (Anolis carolinensis) and chicken (Gallus gallus). Segmental linkage homologies are derived from the Genomicus database (Louis et al., 2014). Arrow indicate chromosome 15 of chicken and autosome of anolis.
Primers used for the measurement of relative gene dosage by quantitative PCR analysis.
Inter- and intra-assay coefficient of variation (CV) and efficiency of five genes for dilution standard curve.
Inter- and intra-assay coefficient of variation (CV) and efficiency for five genes.
Relative gene dosage ratios between five males and five females individuals of green iguana (Iguana iguana) for TMEM132D.
Relative gene dosage ratios between five males and five females individuals of green iguana (Iguana iguana) for CCDC92.
Relative gene dosage ratios between five males and five females individuals of green iguana (Iguana iguana) for ATP2A2.
Relative gene dosage ratios between five males and five females individuals of green iguana (Iguana iguana) for PEBP1.
Gene function and pathway for single-nucleotide polymorphism (SNP) and restriction fragment presence/absence (PA) loci of green iguana (Iguana iguana) from a BLAST search of the green anole (Anolis carolinensis) genome (100:0, male:female).
Repeat searches for single-nucleotide polymorphism (SNP) and restriction fragment presence/absence (PA) loci of green iguana (Iguana iguana).
Gene function and pathway for single-nucleotide polymorphism (SNP) and restriction fragment presence/absence (PA) loci of green iguana (Iguana iguana) from a BLAST search of the green anole (Anolis carolinensis) genome (70:30, male:female).
Data Availability Statement
The full dataset and metadata from this publication are available from the Dryad Digital Repository. Dataset, https://doi.org/10.5061/dryad.j0zpc86b5.