Abstract
Purpose
This study was conducted to analyze specific RNA expression profiles in gingival tissue and saliva samples in periodontitis patients and healthy individuals, and to determine their correlations in light of the potential use of microarray-based analyses of saliva samples as a periodontal monitoring tool.
Methods
Gingival tissue biopsies and saliva samples from 22 patients (12 with severe periodontitis and 10 with a healthy periodontium) were analyzed using transcriptomic microarray analysis. Differential gene expression was assessed, and pathway and clustering analyses were conducted for the samples. The correlations between the results for the gingival tissue and saliva samples were analyzed at both the gene and pathway levels.
Results
There were 621 differentially expressed genes (DEGs; 320 upregulated and 301 downregulated) in the gingival tissue samples of the periodontitis group, and 154 DEGs (44 upregulated and 110 downregulated) in the saliva samples. Nine of these genes overlapped between the sample types. The periodontitis patients formed a distinct cluster group based on gene expression profiles for both the tissue and saliva samples. Database for Annotation, Visualization and Integrated Discovery analysis revealed 159 enriched pathways from the tissue samples of the periodontitis patients, as well as 110 enriched pathways In the saliva samples. Thirty-four pathways overlapped between the sample types.
Conclusions
The present results indicate the possibility of using the salivary transcriptome to distinguish periodontitis patients from healthy individuals. Further work is required to enhance the extraction of available RNA from saliva samples.
Keywords: Microarray analysis, Periodontitis, Saliva, Transcriptome
Graphical Abstract
INTRODUCTION
The progression of periodontitis is characterized by a hyperinflammatory response to pathogens to which each patient has a different inherent susceptibility [1,2]. The progression rate of periodontitis varies, and can be clinically classified as rapid, moderate, or no progression according to the loss of attachment [3]. However, the clinical symptoms of periodontitis vary widely, and predicting the progression of the disease using clinical parameters is subject to several limitations [4]. This dilemma has prompted various attempts to diagnose or predict periodontitis by detecting tissue responses based on proteomics or genomics [5].
Transcriptome analysis is a powerful tool for detecting the overall pattern of tissue responses at the gene level, and it has been shown that gene expression profiles differ in the gingival tissue of periodontitis patients [6,7,8,9]. Offenbacher et al. [10] reported that the genes related to immune responses, neural processes, and epithelial defenses were selectively overexpressed during the induction of experimental gingivitis. Davanian et al. [11] also found that the genes involved in immune and inflammatory responses were overexpressed in periodontitis-affected tissue. Periodontitis can be diagnosed or detected based on these distinct expression patterns of periodontitis-affected tissue.
Even though a definitive diagnosis of periodontitis should be made by professional experts based on clinical parameters, monitoring periodontal conditions can have a critical impact on detecting disease onset and preventing disease progression from both the clinical and research perspectives. The differential gene expression pattern of in situ samples may provide clues for monitoring periodontitis, which is a chronic and unspecific disease. Most previous transcriptome studies have analyzed periodontal tissue samples, but repetitive sampling using periodontal tissue biopsies is problematic due to their inherent invasiveness and patient discomfort.
Saliva has recently entered the spotlight as a sampling source for genomic or proteomic research due to the convenience, safety, and noninvasiveness of collecting saliva samples from patients [12]. Mediators of tissue responses such as cytokines, enzymes, hormones, and antibodies are released into the oral fluid and can be used as a pool for biomarkers to detect tissue conditions [13]. Li et al. [14] reported that thousands of human messenger RNAs (mRNAs) are present in saliva, and that they can be isolated, amplified, and profiled from cell-free saliva. Several recent studies have also found that specific mRNAs in saliva may serve as biomarkers for detecting Sjögren syndrome [15], oral cancer [16], and other systemic diseases such as diabetes [17]. Therefore, saliva sampling could also be a candidate methodology for monitoring periodontal conditions, and could allow the onset of periodontitis to be detected using regular base-sequential sampling.
Previous studies of the salivary transcriptome have independently analyzed the transcriptome of saliva in patients, but did not compare the associations of salivary transcriptional profiles with those of tissue samples taken from the same individuals. The aim of this study was to determine the expression profiles of RNA in gingival tissue and saliva samples and to evaluate their correlations in order to assess the feasibility of using saliva samples in microarray analyses for monitoring periodontal conditions.
MATERIALS AND METHODS
Study design
Thirty-five patients (23 with severe periodontitis and 12 with a healthy periodontium) who visited the Department of Periodontology of the Yonsei University Dental Hospital for periodontal treatment (periodontitis patients) or a crown-lengthening procedure (healthy periodontium patients) between September 2014 and September 2015 were originally enrolled in this study. The experimental protocols and informed consent forms were designed according to the Declaration of Helsinki (Tokyo version, revisited in 2004) and Good Clinical Practice guidelines, and approved by the Institutional Review Board for Clinical Research of the Dental Hospital of Yonsei University (approval No. 2-2014-0026). The manuscript was prepared following the guidelines of the Strengthening the Reporting of Observational Studies in Epidemiology statement.
Saliva sampling and a gingival tissue biopsy were performed immediately before and during the periodontal procedures, respectively. Thirteen participants withdrew before sample acquisition; therefore, the gingival tissue biopsy and saliva collection were completed in a final total of 22 patients (12 with severe periodontitis and 10 with a healthy periodontium). RNA extraction and microarray analysis were conducted using the obtained gingival tissue and saliva samples.
Inclusion and exclusion criteria
Patients underwent clinical and radiographic evaluations at their first visit. Those who showed generalized severe loss of the periodontium with familial aggregation were enrolled into the periodontitis group. The detailed inclusion criteria for the periodontitis group were as follows: (1) generalized signs of inflammation, such as gingival swelling, redness, and bleeding on probing; (2) generalized severe probing depth (>7 mm) and clinical attachment loss (>5 mm); (3) generalized severe alveolar bone loss on radiography (>50% of the root length); (4) angular bony defects at multiple sites; and (5) familial aggregation. Individuals with a healthy periodontium (henceforth referred to simply as “healthy patients”) were enrolled from patients requiring crown-lengthening procedures who had no sign of gingival/periodontal inflammation, no bleeding on probing, and no evidence of bone loss.
The following exclusion criteria were also applied: (1) current pregnancy or planning pregnancy, (2) any history of systemic diseases that may affect periodontal status (e.g., diabetes, malignancy, immune-related diseases, or respiratory diseases), (3) history of taking drugs that may affect periodontal status (e.g., nifedipine, phenytoin, or cyclosporine), (4) history of smoking (excluding smoking cessation more than 10 years previously), or (5) periodontitis patients with combined endodontic and periodontal lesions.
Collection of samples
Saliva samples
Saliva samples were acquired immediately before periodontal surgery in all patients. Patients were asked to refrain from eating and drinking for at least 30 minutes before saliva collection. Unstimulated saliva samples (5 mL) were collected by spitting [18] and immediately stored in RNA-preserving solution (ORAGENE-RNA RE-100, DNA Genotek, Ottawa, ON, Canada). The collection vial was stored for 24 hours at room temperature after sample acquisition, and then at −80°C until analysis.
Gingival tissue samples
The tissue biopsy procedure was performed during flap operation surgery in the periodontitis group. During the flap operation, samples were collected from gingival tissue with clinical attachment loss >5 mm, probing depth >7 mm, and the presence of bleeding on probing. The gingival tissue samples measured 3 mm2 and included connective tissue, epithelium, and granulation tissue. Tissue was harvested in the healthy group during crown-lengthening procedures. Healthy gingival tissue samples (3 mm2) were sampled from sites with a probing depth of <3 mm and without bleeding on probing. All of the gingival biopsy samples were immediately stored in RNAlater solution (RNAlater Stabilization Solution AM7020, Life Technologies, Carlsbad, CA, USA) at room temperature for 24 hours, and then stored at −80°C until analysis.
RNA isolation
Total RNA was isolated from the gingival tissue of each patient using TRIzol LS Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. RNA was isolated from the saliva using the Oragene RNA purification protocol with the Qiagen RNeasy Micro Kit (Qiagen, Valencia, CA, USA).
Quantity and quality of purified RNA
Quality and quantity control tests were applied to the isolated total RNA samples before performing the microarray analysis. The RNA concentration (in nanograms per microliter) and purity (absorbance ratios at 260 nm/280 nm and 260 nm/230 nm) were determined using a spectrophotometer (ND-1000, NanoDrop, Wilmington, DE, USA). The quality of the RNA was assessed using the Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) based on the RNA integrity number.
Microarray analysis
Total RNA was amplified and purified using a TargetAmp-Nano Labeling Kit for the Illumina Expression BeadChip device (EPICENTRE, Madison, WI, USA) to yield biotinylated complementary RNA (cRNA) according to the manufacturer's instructions. Briefly, 500 ng of total RNA was reverse-transcribed to cDNA using a T7 oligo (dT) primer. Second-strand cDNA was synthesized, transcribed in vitro, and labeled with biotin-NTP. After purification, the cRNA was quantified using a spectrophotometer (ND-1000, NanoDrop).
Biotinylated cRNA was prepared from 0.55 μg of total RNA using the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX, USA). Labeled cRNA samples (750 ng) were hybridized to individual Human HT-12 (version 4.0) Expression BeadChip devices for 17 hours at 58°C according to the manufacturer's instructions (Illumina, San Diego, CA, USA). The array signals were detected using Amersham fluorolink streptavidin-Cy3 (GE Healthcare Bio-Sciences, Little Chalfont, UK) following the bead-array manual. Arrays were scanned with an Illumina bead-array confocal scanner according to the manufacturer's instructions.
Raw data preparation and statistical analysis
The quality of hybridization and the overall chip performance were monitored by visual inspection of both internal quality control checks and the raw scanned data. Raw data were extracted using the software provided by the manufacturer (Illumina GenomeStudio, version 2011.1; Gene Expression Module, version 1.9.0). The measured intensities of the array probes were transformed by taking the logarithm and normalizing values into quartiles.
A comparative analysis was used to identify differences in the expression data in tissue and saliva samples between periodontitis and healthy patients. The statistical significance of relative differences in the expression data was determined using the independent t-test, with the null hypothesis that there would be no intergroup difference. The false discovery rate was controlled by adjusting the P value using the Benjamini-Hochberg algorithm. The cutoff criterion for probes was a change of at least twofold, with a P value of <0.05. Additionally, for each probe, the number of samples for which the P value of detection was ≥0.05 was considered to be the fail count. If the fail count exceeded 50% of the total number of samples, the probe was considered to be of low quality and was therefore excluded.
Unsupervised hierarchical clustering analysis was performed using complete linkage and a Euclidean distance metric with the expression profile of differentially expressed genes (DEGs). Pathway enrichment analysis for significant probes was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/home.jsp) [19]. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were used as functional annotation tools.
RESULTS
Patients and samples
The 12 patients in the periodontitis group comprised 6 male and 6 female aged 30 to 50 years, with a mean age of 42.16 years. The 10 patients in the healthy group comprised 7 male and 3 female aged 17 to 71 years, with a mean age of 43.7 years (Figure 1).
Figure 1. Panoramic radiographic views of patients in (A) the periodontitis patient group and (B) the healthy patient group.
All of the tissue samples passed the RNA quality and quantity tests, whereas 8 of the saliva samples did not pass the tests (4 each from the periodontitis and healthy groups). The final microarray analysis was performed using 22 tissue samples (12 for periodontitis and 10 for healthy patients) and 14 saliva samples (8 for periodontitis and 6 for healthy patients).
DEG analysis of gingival tissue and saliva samples
In total, 621 genes were differentially expressed in the tissue samples: 320 were upregulated and 301 were downregulated in the periodontitis group compared to the healthy group (Supplementary Tables 1 and 2). The top 20 upregulated and downregulated genes are listed in Table 1. The top 20 upregulated genes included immune-response-related genes (CD79A, IGLL3, and TNFRSF17), a protease gene (MMP7), protein kinase genes (CAMK1G and CRKRS), and a cell-signaling-related gene (RGS1).
Table 1. Top 20 differentially expressed genes from each sample from periodontitis patients compared to healthy patients.
| Gene symbol | Definition | Fold change | P value | |
|---|---|---|---|---|
| Upregulated genes: gingival tissue | ||||
| CD79A | CD79a molecule, immunoglobulin-associated alpha | 10.70 | 4.38E-06 | |
| IGLL3 | Immunoglobulin lambda-like polypeptide 3 | 9.83 | 3.49E-05 | |
| JSRP1 | Junctional sarcoplasmic reticulum protein 1 | 9.05 | 4.86E-05 | |
| LOC649923 | Similar to Ig gamma-2 chain C region | 9.01 | 9.24E-04 | |
| CAMK1G | Calcium/calmodulin-dependent protein kinase IG | 8.99 | 4.01E-06 | |
| MMP7 | Matrix metallopeptidase 7 | 8.76 | 3.50E-05 | |
| LOC652694 | Similar to Ig kappa chain V-I region HK102 | 8.46 | 9.60E-05 | |
| FAM46C | Family with sequence similarity 46,member C | 7.70 | 7.30E-06 | |
| MGC29506 | Hypothetical protein MGC29506 (MGC29506) | 7.59 | 8.92E-05 | |
| LOC652102 | Ig heavy chain V-I region HG3 precursor | 7.53 | 3.16E-04 | |
| LOC647506 | Hypothetical LOC647506 | 7.44 | 4.67E-04 | |
| MMP7 | Matrix metallopeptidase 7 | 7.28 | 2.82E-05 | |
| LOC651751 | Similar to Ig kappa chain V-II region RPMI 6410 p | 7.07 | 1.10E-03 | |
| LOC647450 | Similar to Ig kappa chain V-I region HK101 | 6.98 | 1.21E-03 | |
| TNFRSF17 | Tumor necrosis factor receptor superfamily, member 17 | 6.81 | 1.75E-06 | |
| LOC652493 | Similar to Ig kappa chain V-I region HK102 | 6.45 | 1.69E-03 | |
| CRKRS | Cdc2-related kinase, arginine/serine-rich | 6.29 | 1.45E-06 | |
| RGS1 | Regulator of G-protein signaling 1 | 6.11 | 7.35E-06 | |
| RNF126P1 | Ring finger protein 126 pseudogene 1 | 6.11 | 1.29E-05 | |
| LOC642113 | Similar to Ig kappa chain V-III region HAH precursor | 5.93 | 2.92E-03 | |
| Downregulated genes: gingival tissue | ||||
| LOR | Loricrin | −22.22 | 6.52E-05 | |
| KRT1 | Keratin 1 | −20.30 | 3.89E-05 | |
| C6orf15 | Chromosome 6 open reading frame 15 | −13.06 | 4.69E-06 | |
| KRT2 | Keratin 2 | −12.62 | 5.18E-05 | |
| ASPRV1 | Aspartic peptidase, retroviral-like 1 | −11.42 | 7.54E-06 | |
| LCE2B | Late cornified envelope 2B | −10.52 | 2.20E-05 | |
| CDSN | Corneodesmosin | −9.91 | 7.31E-06 | |
| RPTN | Repetin (RPTN) | −9.89 | 1.26E-04 | |
| LCE2D | Late cornified envelope 2D | −9.45 | 1.16E-05 | |
| KRT76 | Keratin 76 | −9.25 | 1.07E-04 | |
| KPRP | Keratinocyte proline-rich protein | −8.83 | 3.07E-06 | |
| LCE2A | Late cornified envelope 2A | −8.65 | 6.05E-05 | |
| LCE2C | Late cornified envelope 2C | −8.54 | 1.15E-05 | |
| LCE3A | Late cornified envelope 3A | −8.19 | 6.30E-05 | |
| KRT3 | Keratin 3 (KRT3) | −7.29 | 1.31E-04 | |
| FLG | Filaggrin (FLG) | −6.58 | 3.58E-04 | |
| LCE3D | Late cornified envelope 3D | −6.56 | 3.34E-05 | |
| LY6G6C | Lymphocyte antigen 6 complex, locus G6C | −6.47 | 6.12E-07 | |
| LCE3E | Late cornified envelope 3E | −6.44 | 4.89E-06 | |
| SLURP1 | Secreted LY6/PLAUR domain containing 1 | −5.70 | 6.30E-07 | |
| Up-regulated genes: saliva | ||||
| VPS41 | Vacuolar protein sorting 41 homolog | 3.18 | 6.00E-03 | |
| ZNF786 | Zinc finger protein 786 | 2.81 | 2.61E-02 | |
| LOC389765 | Similar to KIF27C | 2.79 | 1.23E-02 | |
| ZNF738 | Misc_RNA | 2.65 | 1.40E-02 | |
| LOC100132585 | Hypothetical protein LOC100132585 | 2.59 | 1.58E-02 | |
| LOC100129502 | Hypothetical protein LOC100129502 | 2.57 | 1.09E-02 | |
| PNPT1 | Polyribonucleotide nucleotidyltransferase 1 | 2.54 | 1.02E-02 | |
| C15orf63 | Chromosome 15 open reading frame 63 | 2.49 | 2.83E-02 | |
| LOC100131718 | Misc_RNA | 2.48 | 3.52E-02 | |
| NLRP8 | NLR family, pyrin domain containing 8 | 2.46 | 2.04E-02 | |
| FLJ36131 | Hypothetical protein FLJ36131 | 2.41 | 2.22E-02 | |
| ZNF223 | Zinc finger protein 223 | 2.40 | 1.75E-02 | |
| SLC35E1 | Solute carrier family 35, member E1 | 2.39 | 3.21E-02 | |
| C5orf28 | Chromosome 5 open reading frame 28 | 2.36 | 1.69E-02 | |
| LOC644250 | Hypothetical protein LOC644250 | 2.33 | 1.75E-02 | |
| CCDC125 | Coiled-coil domain containing 125 | 2.33 | 9.45E-03 | |
| DUXAP3 | Double homeobox A pseudogene 3 | 2.33 | 4.95E-02 | |
| FAM73A | Family with sequence similarity 73, member A | 2.32 | 2.74E-02 | |
| FLJ40722 | Hypothetical protein FLJ40722 | 2.30 | 1.83E-02 | |
| N4BP2 | Nedd4 binding protein 2 | 2.25 | 1.08E-02 | |
| Down-regulated genes: saliva | ||||
| SPRR3 | Small proline-rich protein 3 | −15.26 | 3.10E-03 | |
| IL1B | Interleukin 1, beta | −14.12 | 1.33E-02 | |
| FTHL7 | Ferritin, heavy polypeptide-like 7 | −12.53 | 8.73E-03 | |
| FTHL16 | Misc_RNA | −10.08 | 7.71E-03 | |
| SPRR2D | Small proline-rich protein 2D | −9.91 | 4.98E-03 | |
| ACTB | Actin, beta | −9.89 | 3.44E-03 | |
| SPRR2A | Small proline-rich protein 2A | −9.67 | 5.49E-03 | |
| KRT13 | Keratin 13 | −9.55 | 4.85E-03 | |
| S100A9 | S100 calcium binding protein A9 | −8.39 | 1.01E-02 | |
| IL8 | Interleukin 8 | −8.19 | 2.95E-02 | |
| CRNN | Cornulin | −7.40 | 3.32E-03 | |
| ZFP36 | Zinc finger protein 36, C3H type | −6.46 | 1.67E-02 | |
| TPT1 | Tumor protein, translationally-controlled 1 | −6.45 | 1.67E-02 | |
| SPRR2F | Small proline-rich protein 2F | −6.21 | 1.70E-02 | |
| KRT6A | Keratin 6A | −6.03 | 1.72E-02 | |
| IL1RN | Interleukin 1 receptor antagonist | −5.89 | 4.54E-03 | |
| S100A8 | S100 calcium binding protein A8 | −5.72 | 2.97E-02 | |
| B2M | Beta-2-microglobulin | −5.64 | 1.74E-02 | |
| ACTG1 | Actin, gamma 1 | −5.59 | 2.57E-02 | |
| PLEK | Pleckstrin | −5.37 | 3.08E-02 | |
Upregulated genes in gingival tissue samples; downregulated genes in gingival tissue samples; upregulated genes in saliva samples; downregulated genes in saliva samples. Genes involved in overlapped enriched pathways between the results from the gingival tissue and saliva samples are gray-shaded.
Applying the same cutoff standard to the results for the saliva sample yielded 154 DEGs in the periodontitis group (44 upregulated and 110 downregulated DEGs listed in Supplementary Tables 3 and 4). The top-20 upregulated and downregulated genes are listed in Table 1; these genes included a cellular-transportation-related gene (VPS41) and an immune-system-related gene (NLRP8).
A comparison of DEGs in the gingival tissue and saliva samples revealed that there were 9 overlapping genes, five of which (SPRR2G, LOC643161, LOC647993, LOC647987, and CRCT1) were downregulated in both gingival tissue and saliva samples, while the other 4 genes (ALOX5, IL1B, SRGN, and RAC2) were upregulated in gingival tissue samples and downregulated in saliva samples.
Cluster analysis
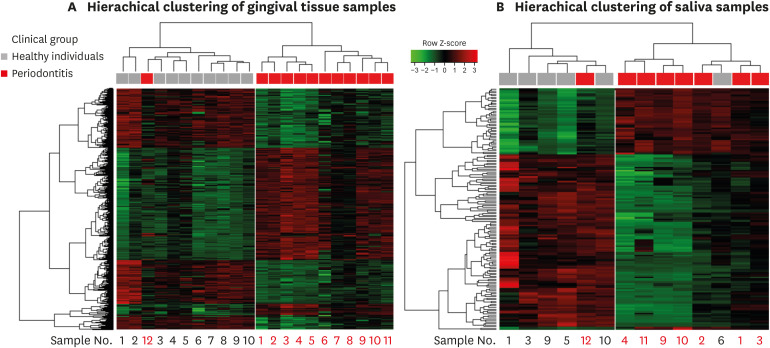
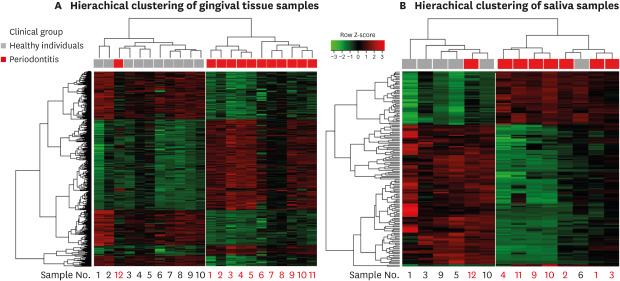
The results of the cluster analysis according to the similarity of the gene expression pattern of each sample using the total probe sets are shown in Figure 2. The branch heights in the horizontal dendrogram indicate similarity relationships between entities. Reading the dendrogram from top to bottom, it is divided into 2 cluster groups from the first branch at the top of the structure. Two different clusters were produced for each set of saliva and tissue samples.
Figure 2. Hierarchical clustering results and heat map of the gene expression data from (A) gingival tissue samples and (B) saliva samples. The horizontal dendogram displays similarities between samples. Samples with similar patterns of gene expression are clustered together and there were 2 different cluster groups in each result. Under the dendogram, the clinical features of the samples are indicated by colored blocks. The samples obtained from the same patients were given the same number for gingival tissue and saliva sample listed under the heat map. The heat map is colored according to the relative expression of a probe, scaled by z-score normalized values.
The gene expression pattern in the tissue samples (Figure 2A) showed distinct clusters corresponding to specific clinical features. The saliva samples (Figure 2B) similarly generated 2 clustered groups of samples based on the gene expression patterns that corresponded to periodontitis and healthy patients. However, 2 exceptional samples were found in the clusters that did not match the clinical diagnoses: 1 tissue/saliva sample from a periodontitis patient and 1 saliva sample from a healthy patient.
Pathway enrichment analysis of DEGs
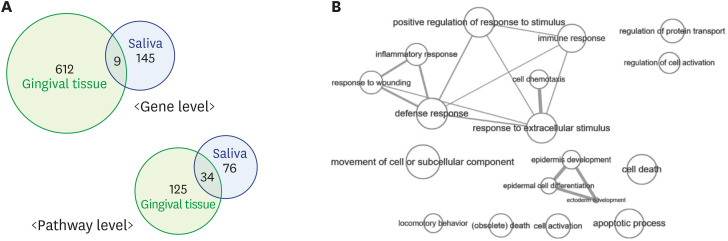
The DAVID analysis identified that 159 terms (146 GO terms and 13 KEGG terms) were enriched in periodontitis patients with P values of <0.05 in the results for the tissue samples, while 110 terms (104 GO terms and 6 KEGG terms) were enriched in the results for the saliva samples. There were 34 overlapping enriched pathways between the tissue and saliva samples (33 GO terms and 1 KEGG term) (Figure 3, Supplementary Table 5). These 34 overlapping pathways included immune responses, inflammatory responses, cytokine regulation, and response to wounding, and immune-response-related pathways were prominent (Figure 4).
Figure 3. (A) Venn diagrams showing the number of differentially expressed genes and enriched pathways of gingival tissue and saliva sample. The intersection of the 2 circles refers to genes and pathways commonly detected in the results from the gingival tissue and saliva samples. There were 9 overlapping genes and 34 overlapping pathways between the tissue and saliva samples. (B) Interactive maps of overlapping GO biological process terms between the tissue and saliva samples. The size of each node represents the frequency of GO terms in the underlying GO annotation database and the thickness of the lines indicates similarity between GO terms. The length of the lines is arbitrary. The data suggest that immune response-related terms were prominent in overlapping pathways. The maps were visualized using REVIGO (http://revigo.irb.hr).
GO: Gene Ontology.
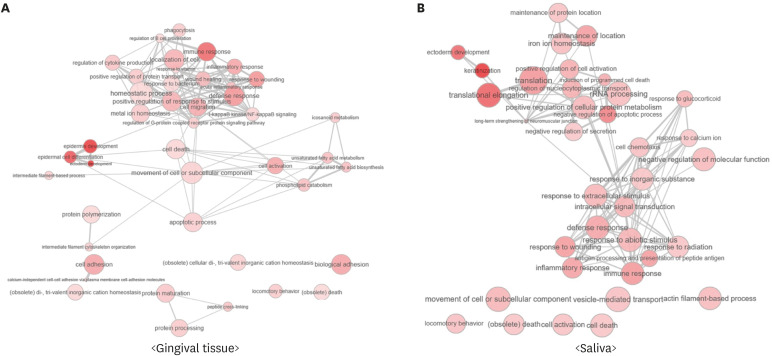
Figure 4. Interactive maps of significantly identified GO biological process terms from the results of (A) tissue samples and (B) saliva samples. The color of the nodes represents the P values in each data set (more intense colors indicate lower P values). The size of each node represents the frequency of GO terms in the underlying GO annotation database, and the thickness of the lines indicates similarity between GO terms. The length of the lines is arbitrary. The maps were visualized using REVIGO (http://revigo.irb.hr).
GO: Gene Ontology.
DISCUSSION
This study specifically compared gene expression in gingival tissue and saliva in both healthy and periodontitis patients with the aim of determining the feasibility of detecting pathological responses in tissue samples based on only saliva samples, and with the eventual goal of developing a noninvasive screening tool. Transcriptomic analysis showed that the RNA expression pattern of saliva could be discriminated between periodontitis and healthy patients, in a manner comparable to the results obtained for the tissue biopsy samples. The cluster analysis of gene expression revealed that periodontitis patients clustered together both in the tissue samples and in the saliva samples, which implies that salivary transcriptome data can be used to distinguish between periodontitis patients and healthy controls.
Several previous transcriptome analyses using tissue samples found that patients with periodontitis were clustered together [8,11,20], with gingival tissue from periodontitis patients showing a significantly different gene expression pattern that formed distinct clusters compared to that of healthy patients. Kebschull et al. [8] also demonstrated similarly clustered results based on DEGs between diseased and healthy periodontal tissues regardless of the phenotype of periodontitis. The present study included periodontitis patients who showed familial aggregation and clear clinical features of severe periodontal destruction, and produced a specific expression pattern of genes that was in agreement with previous studies. However, this is the first study to obtain clustering results from the salivary transcriptome in periodontitis patients in addition to analyzing tissue samples. Our clustering results showed that the transcriptome of saliva samples could be used to distinguish periodontitis patients from healthy patients. Based on hierarchical clustering and the DEGs between saliva samples from diseased and healthy patients, salivary RNA expression could provide clues for identifying systemic disease [21] and salivary disease [22].
Among the genes that were significantly overexpressed in the tissue samples, the TNFRSF17 gene was noteworthy, because its product specifically binds to the tumor necrosis factor family and plays roles in the immune responses, especially signal transduction, B-cell development, and cell differentiation. These results also support reports of a relation between upregulation of the TNFRSF17 gene and the periodontitis phenotype [11,23,24]. Another meaningful gene found in the present study was MMP7, which is strongly correlated with the tissue inflammation level. MMP7 plays important roles in bone resorption and remodeling, and the possibility of detecting periodontitis based on the upregulation of MMP7 has been reported previously. These significant genes in the present data from periodontal tissue support the findings of previous functional studies of periodontitis, as well as whole-genome and microarray studies.
At the functional pathway level, 34 enriched pathways overlapped in the gingival tissue and saliva samples obtained from periodontitis patients compared to healthy patients, and 12 of those pathways were related to the immune responses. Although different sets of genes were detected in the tissue and saliva samples, it is meaningful that the immune-response pathway was commonly detected at the functional level. Similar results have also been found in other studies of the salivary transcriptomes of periodontitis patients, with cellular immunity pathways being significantly upregulated [25]. Other studies of periodontal tissue samples have found that important roles are played by immune-response pathways, such as the leukocyte extravasation signaling pathway [23], the pathway involving the chemotaxis and transendothelial migration of leukocytes [10], and immune-system processes including defense responses [11]. Chronic inflammation due to persistent infection of the periodontium causes hyperactivation of immune responses (both innate and adaptive). Changes in the homeostasis of immune responses around the periodontium may affect various sources of salivary mRNA [26]. In detail, gingival crevicular fluid is directly affected by the state of the tissue [27], and major salivary glands may be affected by circulating inflammatory mediators [26,28].
The present results revealed only small numbers of overlapping DEGs between periodontitis and healthy patients (1.5% from the entire DEG lists of the tissue samples and 5.8% from the saliva samples). This limitation might have been caused by the total amount of mRNA detected from the whole saliva being less than that detected from the tissue samples, due to the nature of saliva and limitations of the present analytical technology (Figure 5). The present results are in agreement with those of a previous study showing only small amounts of total RNA, with wide variations and contamination from bacterial sources, in microarray analyses of samples taken from patients with chronic periodontitis [29]. Salivary mRNA is stabilized by interactions with salivary proteins and apoptotic bodies [30], and RNases and proteases from microbial/human sources can rapidly degrade mRNA. These features have prompted the developed of various collection methods and RNase inhibitors [14,15,31], but there are no standardized methods for obtaining acceptable amounts of mRNAs from saliva and for their amplification [32].
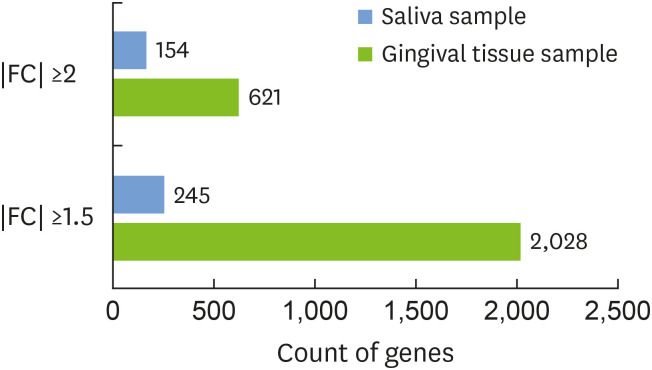
Figure 5. The number of differentially expressed genes detected with different FCs (FC ≥2 or ≥1.5 in absolute value) is indicated by a horizontal bar. The results for each type of sample are shown in different colors: gingival tissue (blue) and saliva samples (green).
FC: fold change.
Of particular note, 4 genes (ALOX5, IL1B, SRGN, RAC2) overlapped between the saliva and tissue samples, but with the opposite expression pattern. These findings clearly demonstrate limitations or hurdles to be overcome in the degree to which saliva samples can be currently used as a diagnostic tool based on specific biomarkers. The scant quantity of the transcriptome from the periodontal pocket in saliva samples and the unclear pathophysiological classification of periodontitis may cause noise within the data, inducing discordant results. In addition, the expression levels of several genes, such as IL1B and ALOX5, have been found to show a temporal pattern during the process of inflammation [33,34]. Periodontitis has multiple pathological steps, with repeated cycles of rest and active phases, and further studies are needed on the pathophysiological mechanism and the specific gene expression patterns to clarify a specific biomarker. However, the present study identified specific clustered patterns of gene expression in severe periodontitis based on the analysis of either saliva samples or tissue samples. In addition, there were more overlapping results in the pathway enrichment analysis between gingival tissue and saliva samples than in the individual gene-level analysis, as overlap was found for 12.8% of all enriched pathways from the tissue samples and for 22.5% of enriched pathways from the saliva samples of periodontitis patients. Therefore, further studies focusing on the pathways and changes in their categorical genes would be helpful for identifying specific markers for use in diagnosis and disease monitoring.
The present study identified specific clustering patterns of gene expression in both saliva samples and tissue samples. Based on these results, saliva samples can be considered as candidate biospecimens for monitoring periodontitis at the level of immunity-related pathways. These findings represent an initial demonstration of the possibility of using saliva samples for transcriptome-based periodontal monitoring; however, further studies are needed to determine how to enhance the yield of RNA from saliva samples and how to perform pathway-specific analyses.
Footnotes
Funding: This study was supported by a faculty research grant of Yonsei University College of Dentistry (6-2014-0176).
- Conceptualization: Jung-Seok Lee.
- Formal analysis: Yoon-Sun Jeon.
- Investigation: Ji-Hyun Lee, Jae-Kook Cha.
- Methodology: Seong-Ho Choi.
- Project administration: Jung-Seok Lee.
- Writing - original draft: Yoon-Sun Jeon, Jung-Seok Lee.
- Writing - review & editing: Yoon-Sun Jeon, Jae-Kook Cha, Seong-Ho Choi, Ji-Hyun Lee, Jung-Seok Lee.
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
SUPPLEMENTARY MATERIALS
Full list of upregulated genes in the results for gingival tissue samples from periodontitis patients compared to healthy patients
Full list of downregulated genes in the results for gingival tissue samples from periodontitis patients compared to healthy patients
Full list of upregulated genes in the results for saliva samples from periodontitis patients compared to healthy patients
Full list of downregulated genes in the results for saliva samples from periodontitis patients compared to healthy patients
List of overlapping pathways between gingival tissue and saliva samples according to the DAVID results
References
- 1.Al-Yahfoufi Z, Mombelli A, Wicki A, Lang NP. The effect of plaque control in subjects with shallow pockets and high prevalence of periodontal pathogens. J Clin Periodontol. 1995;22:78–84. doi: 10.1111/j.1600-051x.1995.tb01774.x. [DOI] [PubMed] [Google Scholar]
- 2.Trombelli L, Tatakis DN, Scapoli C, Bottega S, Orlandini E, Tosi M. Modulation of clinical expression of plaque-induced gingivitis. II. Identification of “high-responder” and “low-responder” subjects. J Clin Periodontol. 2004;31:239–252. doi: 10.1111/j.1600-051x.2004.00478.x. [DOI] [PubMed] [Google Scholar]
- 3.Löe H, Anerud A, Boysen H, Morrison E. Natural history of periodontal disease in man. Rapid, moderate and no loss of attachment in Sri Lankan laborers 14 to 46 years of age. J Clin Periodontol. 1986;13:431–445. doi: 10.1111/j.1600-051x.1986.tb01487.x. [DOI] [PubMed] [Google Scholar]
- 4.Halazonetis TD, Haffajee AD, Socransky SS. Relationship of clinical parameters to attachment loss in subsets of subjects with destructive periodontal diseases. J Clin Periodontol. 1989;16:563–568. doi: 10.1111/j.1600-051x.1989.tb02138.x. [DOI] [PubMed] [Google Scholar]
- 5.Gonçalves LDR, Soares MR, Nogueira FCS, Garcia C, Camisasca DR, Domont G, et al. Comparative proteomic analysis of whole saliva from chronic periodontitis patients. J Proteomics. 2010;73:1334–1341. doi: 10.1016/j.jprot.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, et al. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79:2112–2124. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jönsson D, Ramberg P, Demmer RT, Kebschull M, Dahlén G, Papapanou PN. Gingival tissue transcriptomes in experimental gingivitis. J Clin Periodontol. 2011;38:599–611. doi: 10.1111/j.1600-051X.2011.01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kebschull M, Guarnieri P, Demmer RT, Boulesteix AL, Pavlidis P, Papapanou PN. Molecular differences between chronic and aggressive periodontitis. J Dent Res. 2013;92:1081–1088. doi: 10.1177/0022034513506011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taiete T, Casarin RCV, Ruiz KGS, Nociti FH, Jr, Sallum EA, Casati MZ. Transcriptome of healthy gingival tissue from edentulous sites in patients with a history of generalized aggressive periodontitis. J Periodontol. 2018;89:93–104. doi: 10.1902/jop.2017.170221. [DOI] [PubMed] [Google Scholar]
- 10.Offenbacher S, Barros SP, Paquette DW, Winston JL, Biesbrock AR, Thomason RG, et al. Gingival transcriptome patterns during induction and resolution of experimental gingivitis in humans. J Periodontol. 2009;80:1963–1982. doi: 10.1902/jop.2009.080645. [DOI] [PubMed] [Google Scholar]
- 11.Davanian H, Stranneheim H, Båge T, Lagervall M, Jansson L, Lundeberg J, et al. Gene expression profiles in paired gingival biopsies from periodontitis-affected and healthy tissues revealed by massively parallel sequencing. PLoS One. 2012;7:e46440. doi: 10.1371/journal.pone.0046440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaczor-Urbanowicz KE, Martin Carreras-Presas C, Aro K, Tu M, Garcia-Godoy F, Wong DT. Saliva diagnostics - current views and directions. Exp Biol Med (Maywood) 2017;242:459–472. doi: 10.1177/1535370216681550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Henson BS, Camargo PM, Wong DT. The clinical value of salivary biomarkers for periodontal disease. Periodontol 2000. 2009;51:25–37. doi: 10.1111/j.1600-0757.2009.00315.x. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Zhou X, St John MAR, Wong DTW. RNA profiling of cell-free saliva using microarray technology. J Dent Res. 2004;83:199–203. doi: 10.1177/154405910408300303. [DOI] [PubMed] [Google Scholar]
- 15.Hu S, Wang J, Meijer J, Ieong S, Xie Y, Yu T, et al. Salivary proteomic and genomic biomarkers for primary Sjögren's syndrome. Arthritis Rheum. 2007;56:3588–3600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10:8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 17.Javaid MA, Ahmed AS, Durand R, Tran SD. Saliva as a diagnostic tool for oral and systemic diseases. J Oral Biol Craniofac Res. 2016;6:66–75. doi: 10.1016/j.jobcr.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navazesh M. Methods for collecting saliva. Ann N Y Acad Sci. 1993;694:72–77. doi: 10.1111/j.1749-6632.1993.tb18343.x. [DOI] [PubMed] [Google Scholar]
- 19.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker ST, Beck-Broichsitter BE, Graetz C, Dörfer CE, Wiltfang J, Häsler R. Peri-implantitis versus periodontitis: functional differences indicated by transcriptome profiling. Clin Implant Dent Relat Res. 2014;16:401–411. doi: 10.1111/cid.12001. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, Park NH, et al. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology. 2010;138:949–957.e1-7. doi: 10.1053/j.gastro.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hjelmervik TOR, Petersen K, Jonassen I, Jonsson R, Bolstad AI. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjögren's syndrome patients from healthy control subjects. Arthritis Rheum. 2005;52:1534–1544. doi: 10.1002/art.21006. [DOI] [PubMed] [Google Scholar]
- 23.Guzeldemir-Akcakanat E, Sunnetci-Akkoyunlu D, Orucguney B, Cine N, Kan B, Yılmaz EB, et al. Gene-expression profiles in generalized aggressive periodontitis: a gene network-based microarray analysis. J Periodontol. 2016;87:58–65. doi: 10.1902/jop.2015.150175. [DOI] [PubMed] [Google Scholar]
- 24.Guo X, Wang Y, Wang C, Chen J. Identification of several hub-genes associated with periodontitis using integrated microarray analysis. Mol Med Rep. 2015;11:2541–2547. doi: 10.3892/mmr.2014.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B, Lin T, He H. Comparative analysis of blood and saliva expression profiles in chronic and refractory periodontitis patients. BMC Oral Health. 2015;15:166. doi: 10.1186/s12903-015-0150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giannobile WV, Beikler T, Kinney JS, Ramseier CA, Morelli T, Wong DT. Saliva as a diagnostic tool for periodontal disease: current state and future directions. Periodontol 2000. 2009;50:52–64. doi: 10.1111/j.1600-0757.2008.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loos BG, Tjoa S. Host-derived diagnostic markers for periodontitis: do they exist in gingival crevice fluid? Periodontol 2000. 2005;39:53–72. doi: 10.1111/j.1600-0757.2005.00129.x. [DOI] [PubMed] [Google Scholar]
- 28.Teng YT, Sodek J, McCulloch CA. Gingival crevicular fluid gelatinase and its relationship to periodontal disease in human subjects. J Periodontal Res. 1992;27:544–552. doi: 10.1111/j.1600-0765.1992.tb01830.x. [DOI] [PubMed] [Google Scholar]
- 29.Hidayat MF, Milne T, Cullinan MP, Seymour GJ. Feasibility of the salivary transcriptome as a novel biomarker in determining disease susceptibility. J Periodontal Res. 2018;53:369–377. doi: 10.1111/jre.12522. [DOI] [PubMed] [Google Scholar]
- 30.Hasselmann DO, Rappl G, Tilgen W, Reinhold U. Extracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serum. Clin Chem. 2001;47:1488–1489. [PubMed] [Google Scholar]
- 31.Park NJ, Yu T, Nabili V, Brinkman BM, Henry S, Wang J, et al. RNAprotect saliva: an optimal room- temperature stabilization reagent for the salivary transcriptome. Clin Chem. 2006;52:2303–2304. doi: 10.1373/clinchem.2006.075598. [DOI] [PubMed] [Google Scholar]
- 32.Park NJ, Zhou X, Yu T, Brinkman BM, Zimmermann BG, Palanisamy V, et al. Characterization of salivary RNA by cDNA library analysis. Arch Oral Biol. 2007;52:30–35. doi: 10.1016/j.archoralbio.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belstrøm D, Damgaard C, Könönen E, Gürsoy M, Holmstrup P, Gürsoy UK. Salivary cytokine levels in early gingival inflammation. J Oral Microbiol. 2017;9:1364101. doi: 10.1080/20002297.2017.1364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paula-Silva FWG, Petean IBF, da Silva LAB, Faccioli LH. Dual role of 5-lipoxygenase in osteoclastogenesis in bacterial-induced apical periodontitis. J Endod. 2016;42:447–454. doi: 10.1016/j.joen.2015.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full list of upregulated genes in the results for gingival tissue samples from periodontitis patients compared to healthy patients
Full list of downregulated genes in the results for gingival tissue samples from periodontitis patients compared to healthy patients
Full list of upregulated genes in the results for saliva samples from periodontitis patients compared to healthy patients
Full list of downregulated genes in the results for saliva samples from periodontitis patients compared to healthy patients
List of overlapping pathways between gingival tissue and saliva samples according to the DAVID results