Abstract
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been a global public health emergency since December 2019, and so far, more than 980,000 people (until September 24, 2020) around the world have died. SARS-CoV-2 mimics the influenza virus regarding methods and modes of transmission, clinical features, related immune responses, and seasonal coincidence. Accordingly, co-infection by these viruses is imaginable because some studies have reported several cases with SARS-CoV-2 and influenza virus co-infection. Given the importance of the mentioned co-infection and the coming influenza season, it is essential to recognize the similarities and differences between the symptoms, immunopathogenesis and treatment of SARS-CoV-2 and influenza virus. Therefore, we reviewed the virology, clinical features, and immunopathogenesis of both influenza virus and SARS-CoV-2 and evaluated outcomes in cases with SARS-CoV-2 and influenza virus co-infection.
Keywords: Influenza, COVID-19, SARS-COV-2, Co-infection, Pathogenesis
1. Introduction
On December 31, 2019, the first report of a new respiratory disease with the severe acute respiratory syndrome (SARS)-like symptoms was reported from Wuhan, China, which led to the world's most massive pandemic of the century and one of the most significant health and economic threats by far. The rapid spread across the continents within a month prompted the World Health Organization (WHO) to declare a global health emergency [1]. Given the similarity to SARS and the first report in 2019, this new coronavirus-associated disease was named coronavirus disease 19 (COVID-19), and the virus was named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [2]. Coronaviruses are single-stranded positive-sense RNA viruses belonging to the family Coronaviridae that commonly infect vertebrates. Their human disease is usually manifested as a mild respiratory infection similar to the common cold or influenza. However, three coronavirus-associated fatal endemics have been reported in the last two decades, including SARS, Middle-East respiratory syndrome (MERS), and COVID-19 [[3], [4], [5]]. COVID-19 is now a major global health threat, which infected roughly >30.000.000 individuals with >950.000 deaths worldwide, until September 20, 2020. Approximately 20% of infected patients manifested acute respiratory distress syndrome (ARDS) and needed hospital intensive care [[6], [7], [8]]. The general mortality rate is about 3%, while it has been reported to be above 8% in people aged ≥70 years [9]. China, South Korea, and Iran, which were among the earliest countries declared the epidemics, have now substituted with the United States, India, Brazil, and Russia. Accordingly, the United States, Brazil, India, Mexico, and the United Kingdom have reported the highest rates of death [1,10]. Evidence showed that the most dramatic human deaths in the recent century were caused by the Spanish flu (H1N1 influenza A virus), which infected one-third of the world's population in 1918 and killed up to 50 million people [11,12]. Subsequent flu pandemics with lower mortality occurred in 1957, 1968, and 2009 [13]. According to the reports of WHO, influenza causes five million cases of severe illness worldwide annually and kills 650,000 people [14]. The Centers for Disease Control and Prevention (CDC) estimation for seasonal influenza in 2019–2020 in the United States was 38–54 million cases, with 400,000–730,000 severe cases and 24,000–62,000 deaths [15]. In the Middle-East; however, the prevalence of influenza varies between 5% in Qatar to 70% in Syria, with the mean of 10%. Iran has a prevalence of 18.5%, among which the healthcare workers, children younger than six years old, and Hajj pilgrims are more prone to have seasonal influenza [16,17]. Considering the constant antigenic changes of the influenza virus and the ability of swine and avian subtypes to infect humans, the risk of occurrence of a new epidemic or pandemic of influenza, similar to the 1918 pandemic, is highly predictable [18,19]. SARS-CoV-2 infection often occurs without any specific upper respiratory tract symptoms, and distinguishing it from other similar diseases is one of the main challenges in this disease [20]. In this regard, influenza infection may have similar symptoms to COVID-19 in different aspects [21]. The present lack of clinical knowledge about the SARS-CoV-2 infection and unstable and conflicting findings may be misleading in co-infection patient's diagnoses. On the other hand, co-infection can increase the severity of symptoms of both diseases in these cases, in which managing of this state is essential and critical in certain groups such as pregnant women, the elderly, and even children. Studies reported a high SARS-CoV-2 and influenza virus co-infection rate in children that highpoints the importance of COVID-19 screening, particularly during the prevalence of seasonal influenza and other respiratory disorders [22].
In this review article, we investigated influenza and COVID-19 co-infection outcomes regarding the clinical features, related immune responses, and seasonal coincidence. Additionally, therapeutic approaches that can reduce the severity of common symptoms in the co-infection have also been discussed.
2. Virology of influenza virus & SARS-CoV-2
2.1. Influenza virus
The influenza virus is an enveloped virus belonging to the Orthomyxoviridae family whose genetic content is negative single-stranded RNA(s). Influenza viruses comprise four distinct viruses, A, B, C, and D. Influenza A can infect a wide range of species, while the primary host of influenza B and C is human, and influenza D mainly infects cattle, goats and pigs [19,23]. The genome of influenza A and B viruses contain eight RNA segments capable of encoding 18 different proteins. However, influenza C and D lack the fifth segment, which is responsible for neuraminidase (NA) [24]. The fourth gene encodes hemagglutinin (HA), and the other genes encode basic and acidic polymerases, nucleoprotein, major matrix protein (M1), ion-channel matrix protein (M2), and non-structural proteins [24]. Influenza A viruses are categorized by surface HA, and NA glycoproteins found on the envelope of the virus in a way that the subtypes H1 to H18, known to date, are used for HA and N1 to N11 stands for subgroups of NA [25]. The initial step for infection is the attachment of the virus to the host cell; hence, the envelope glycoproteins (HA and NA) play crucial role in viral pathogenesis and stand as the primary target for neutralizing antibodies against the virus [18,26]. HA binds to the terminal glycosides of sialic acid on the surface of columnar epithelial cells in the upper respiratory tract (in human influenza types) and alveolar type II cells, as well as ocular epithelium (in avian influenza types) [26]. Those types of viruses which infect the upper respiratory tract have a high transmission capability but low virulence severity (such as H1N1), and those variants that infect the lower respiratory tract could result in more severe inflammation and complications, despite their lower contagious features (e.g., H5N1 and H7N9) [27,28]. Binding of the virus by HA glycoprotein to host cells is facilitated by M1 protein, which results in a fusion with the endoplasmic membrane and virion endocytosis. Virus entry into the cell nucleus occurs through the activation of the M2 protein. Endosomes, through the acidification of M2 protein, cause HA configuration changes, and HA2-mediated fusion of viral envelope with endosome membrane releases ribonucleoprotein (nucleocapsid), and then the ribonucleoprotein moves from the cytoplasm to the nucleus. Replication of the negative-sense RNAs of the virus requires RNA polymerases, which are carried with the virus into the host nucleus. Budding and release of newly assembled virions in order to infect surrounding cells are the functions of NA along with M1 and M2 proteins [27,29,30]. Nucleoprotein encapsidates the genetic content of virus, and non-structural proteins are mainly involved in host immune suppression and viral translation enhancement [30,31].
Cumulative mutations of the influenza virus cause minor changes in NA and HA glycoproteins, a phenomenon known as antigenic drift, that occurs in all influenza subtypes. Furthermore, the antigenic shift is due to gene rearrangement [19,32]. Evidence showed that a pandemic usually occurs following a lot of mutations in viruses to spread from one species to another [33]. However, antigenic drift does not generally cause pandemics because the population's immunity plays a selective role. The susceptibility of different hosts to a specific type of influenza virus depends on the virus binding ability to the host cell, virus genome replication, and escape from the host immune response [19,33].
2.2. SARS-CoV-2
Coronaviruses (CoVs) are known as one of the large family of enveloped viruses with a 26–32 kb single-stranded RNA genome that categorized into four types α, β, γ, and δ [34]. Human coronavirus NL63 and 229E belong to α type, and MERS-CoV, SARS-CoV, OC43, and HKU1 belong to β type [5,35]. In December 2019, the newly emerged SARS-CoV-2 was identified with 50% identity to the MERS-CoV genome sequence and 88% identity to two SARS-like coronaviruses genome sequences derived from bat's species, namely bat-SL-CoVZC45 and bat-SL-CoVZXC21 [36]. Recent investigations revealed that the SARS-CoV-2 genome consists of 14 open reading frames (ORFs) that encode about 27 proteins. ORF1a and ORF1ab are located at the 5′-end and encode the PP1a and PP1ab polyproteins [37]. Following proteolysis of these two polyproteins, 15 non-structural proteins (nsp1-10 and nsp12-16) are generated, responsible for viral replicase-transcriptase complex formation [38,39]. On the other hand, at the 3′-end of the genome, four structural proteins are encoded, including envelope (E), membrane (M), spike (S), and nucleocapsid (N), as well as eight accessory proteins, including ORF14, 9b, 8b, 7b, 7a, p6, and 3b [39,40]. However, the role of these eight accessory proteins is not yet fully understood. Comparative studies have shown that in some regions of the SARS-CoV-2 genome, significant differences can be detected when compared with the SARS-CoV genome. For instance, accessory protein 8a is present in the SARS-CoV virus sequence, whereas it is absent in the sequence of SARS-CoV-2 [41]. Overall, the amino acid sequences similarity of the SARS-CoV-2 and SARS-CoV is only 68% [36]. According to current studies, it appears that bats are the primary and natural hosts of SARS-CoV-2, and subsequently, the virus appears to has transmitted to the intermediate hosts, which can be pangolin or other wild animals and then transmitted from this intermediate host to human [42]. The latest evidence showed that the angiotensin-converting enzyme 2 (ACE2), CD147, dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN, CD209) and L-SIGN (CD209L) are recognized as the possible host cell entry receptors for SARS-CoV-2 [43,44]. Previous studies reported that ACE2, as the main receptor of SARS-CoVs, is highly expressed on alveolar type 2 epithelial cells [45,46]. In these infected cells, transmembrane serine proteases 2 and 11D (TMPRSS2, TMPRSS211D) are involved in the cleavage of S protein from SARS-CoV at R667 and R797 residues and induction of cell membrane fusion [47]. After membrane fusion, endocytosis is occurred with and without clathrin, which is led to virus entrance to the host cells [48]. The monitoring of infected VeroE6 cells revealed that the kinetic replication of SARS-CoV-2 and other SARS-CoVs occurs similarly. Finally, the budding of viruses happens in the endoplasmic reticulum-Golgi-cell membrane pathway [3].
3. Animal reservoirs of influenza virus and SARS-CoV-2
The emergence of SARS-CoV-2 from a seafood market in Wuhan, China, where various domestic and wild animals are sold, hypothesized animal source for SARS-CoV-2 [49]. Based on the bats’ role in the SARS-CoV and MERS-CoV pandemics and the close relations of SARS-CoV-2 with the other isolates of coronavirus isolated from bats, it is proposed that the SARS-CoV-2 is the result of sequential recombination among the former coronaviruses in bats [50,51]. Although the bats are the probable reservoir source of SARS-CoV-2, the intermediate hosts have not identified yet, and pangolins are reported as possible intermediate hosts, responsible for the zoonotic transmission to humans [52]. Tigers, lions, minks, pigs, and rabbits are the most common companion and zoo animals known as animal hosts [53]. Fortunately, many domestic and companion animals are less susceptible to SARS-CoV-2 compared to humans. The lower evolution of this virus than other viruses such as influenza, low level of ACE-2 receptor, and specific proteases in animals, especially domestic animals, have caused this virus to show less stability in animals than humans [54]. Cats and ferrets are susceptible to SARS-CoV-2, while it is shown that SARS-CoV-2 has low replication in chicken, duck, and pig [55].
A century after the influenza pandemic in 1918, it has been well established that human, avian, and swine are three important hosts to be susceptible to all four influenza genera. Various animal species reservoirs (domestic and wild) in influenza A, B, C, and D viruses not only making it difficult to overcome the disease, but also carries the risk of new pandemics. Focusing on the animal reservoir role on influenza A infection, poultry and pigs serve as important amplifying reservoirs for the virus spreading in man. Moreover, more than 100 types of influenza A infect most birds, pigs, horses, dogs, and seals [56]. Generally, influenza B virus (IBV) is mainly associated with human infection, but pigs, horses, and dogs are susceptible to IBV and may serve as a natural reservoir [[57], [58], [59], [60]]. Like the mentioned subtypes, the influenza C virus (ICV) has been known to infect domestic pigs and feral dogs [59] naturally.
Additionally, based on the recent studies, dromedary camels were suspected of serving as a newly documented host for ICV as they were found to harbor ICV antibodies [61]. The evidence on the carrier role of camels for human CoV infection accentuates the possible co-infection of SARS-CoV and influenza viruses through camels. According to genome sequencing analysis and serological study, it is indicated that the influenza D virus was detected in pigs, cattle, goats, and sheep [62,63]. These studies propose that influenza viruses may have a broader host range than previously thought, and pigs are of the most important reservoirs of influenza virus strains.
The possibility of SARS-CoV-2 transmission from pigs and the role of pigs as a predominant species in the influenza virus evolution raises the concern of co-infection possibility from pigs. Moreover, the evolution of a new virus comprising both corona and influenza in pigs is not impossible and needs to be investigated [50,55]. Hence, more caution should be considered towards pigs, which are in proximity to humans. Given the animal-human and human-human transmission of SARS-CoV-2, the human-animal transmission is also speculated, which could prolong the pandemics due to animal-human contacts [55].
Taken together, although the possibility of SARS-CoV-2 transmission from the domestic animals are much lower than wild ones, the proximity of such species and the high rate of influenza infections in them guarantee the further investigations and more cautions to prevent possible co-infection.
4. Clinical features of influenza virus & SARS-CoV-2
4.1. Influenza
In patients with influenza, symptoms frequently appear abruptly following a period of 2–5 days, and this can be a suitable discriminating feature between influenza and other viral infections of respiratory tract [64]. The most important symptom in these patients is fever, which occurs in more than 90% of them, along with other symptoms such as chills, body aches, sore throats, nasal congestion, fatigue, vomiting, abdominal pain, and diarrhea [65,66]. Studies have also shown that bacterial pneumonia, encephalitis, hemophagocytic syndrome, and myocarditis may also occur in these patients; however, these disorders are less common. Symptoms of the disease usually subside within 5–8 days in previously exposed healthy people while in young children, the elderly (aged ≥65 years), and immunocompromised patients, it can lead to more complicated conditions that might sometimes lead to pulmonary edema, ARDS, and death [[67], [68], [69]]. In these patients, diffuse alveolar hemorrhage (DAH) and extensive infiltration of lymphocytes and monocytes to the lung are typical hallmarks of ARDS and acute lung injury (ALI) [70,71]. Both the primary influenza pneumonia and secondary bacterial infection have almost the same radiographic infiltrates [72]. Primary influenza pneumonia is characterized by severe hypoxia, the existence of diffuse interstitial infiltrates in chest radiographs, and also high mortality rate [64,73]. However, radiographic diagnosis may not be valuable in the primary influenza pneumonia [72]. Evidence showed that primary influenza pneumonia most often occurs in patients with increased pulmonary vascular pressure, valvular heart disorders, and chronic inflammatory lung disease [65]. Previous investigations on pandemic influenza H1N1 in 2009 showed that the risk of developing primary influenza pneumonia was higher in pregnant women, particularly in the third trimester [74,75]. This severe and lethal disorder was associated with respiratory failure in the patients. In severe influenza, lymphopenia and failure of organs such as the heart and kidney have been reported. High-level replication of influenza virus in non-respiratory organ cells and dysregulation of the immune responses can play a pivotal role in the organ failures [70,76]. Clinical findings showed that 1–3 weeks after the incidence of influenza infection, secondary bacterial infections with Streptococcus pneumoniae or Staphylococcus aureus along with the accumulation of pus in the pleural cavity are common in infected patients [77]. Other disorders have also been reported in patients with influenza, including myelitis, post-infectious encephalitis, and Guillain-Barre’ syndrome [78,79]. Hence, due to the clinical features of influenza, special monitoring and care should be given to more susceptible individuals (Table 1 ).
Table 1.
Comparison of clinical symptoms of patients with COVID-19 and influenza.
| Clinical symptoms | |||
|---|---|---|---|
| Influenza | COVID-19 | Similarities | Differences |
| •Fever or feeling feverish/chills •Cough •Sore throat •Runny or stuffy nose •Muscle or body aches •Headaches •Fatigue (tiredness) •Vomiting and diarrhea |
•Fever or feeling feverish/chills •Dry cough •Sore throat •Runny or stuffy nose •Muscle or body aches •Headaches •Fatigue (tiredness) •Vomiting and diarrhea •Skin rash or discoloration of toes or fingers |
•Fever or feeling feverish/chills •Cough •Shortness of breath or difficulty breathing •Fatigue (tiredness) •Sore throat •Runny or stuffy nose •Muscle pain or body aches •Headache •Vomiting and diarrhea |
•Non-productive dry cough in SARS-CoV-2 infection •Change in or loss of taste or smell in SARS-CoV-2 infection •Skin rash or discoloration of toes or fingers in SARS-CoV-2 infection |
4.2. COVID-19
Current studies showed that in patients with COVID-19 incubation period is 1–14 days after exposure; however, in most infected patients, this period is about 3–7 days following exposure to SARS-COV-2 [80,81]. The severity of COVID-19 in infected patients can be mild, severe, or fulminant. According to the WHO and CDC, the COVID-19 symptoms vary depending on the severity of infection, which are mostly fever or chills, dry cough, fatigue, and myalgia. However, the body temperature of some patients with COVID-19 has been reported at 37.3 °C [[82], [83], [84]]. Occasionally, less frequent symptoms, such as sore throat, headache, gastrointestinal complications such as diarrhea and nausea, loss of smell or taste, conjunctivitis, rhinorrhea, skin rash, discoloration of toes or fingers, and venous thromboembolism have also been reported in these patients. The fatal symptoms of COVID-19 are shortness of breath, chest pressure and pain, loss of speech or movement, and organ failure [1,85]. One of the significant disorders of influenza and COVID-19 is ARDS, which in the SARS-COV-2 infected cases mostly occurs in the elderly (aged ≥60 years), or patients with hypertension, and diabetes mellitus. Recent studies have shown that ARDS usually occurs eight days after the onset of symptoms and almost immediately after the onset of dyspnea [86]. Acute cardiac injury and also arrhythmias are considered as other rare COVID-19 complications [87]. The average mortality rate has been reported by about 3%. However, according to studies around the world, this can vary from 0.7 to 8% based on different demographics, most commonly in elderly (aged ≥65 years) or patients with underlying severe medical conditions including diabetes mellitus, cardiovascular disease, hypertension, chronic kidney disease, cancer, and chronic lung disease [88,89]. Symptomatic COVID-19 infection in children seems to be infrequent, and when occurred, it is regularly mild, although cases with severe disease have been reported [90]. Besides, asymptomatic cases have also been diagnosed; however, their frequency is unidentified [91]. Laboratory findings also worsen with the severity of the COVID-19, and notably lymphopenia, increased liver enzymes, C-reactive protein (CRP), ferritin, creatine phosphokinase (CPK), lactate dehydrogenase (LDH), and D-dimer are prominent ones [92,93].
Additionally, there was a remarkable association between severe lymphopenia, high levels of D-dimer, and mortality [7]. Due to the increased inflammatory response induced by immune dysregulation in infected patients, cytokine release syndrome, or something similar has been reported [87]. Given the clinical features presented in these two sections, it can be concluded that there are several counterparts between the patients with influenza and COVID-19 that sometimes make the diagnosis difficult (Table 1). However, accurate laboratory techniques such as real-time PCR (RT-PCR) could be helpful in the differentiation and diagnosis of the infections [94].
5. Immunopathological similarities between influenza virus and SARS-CoV-2
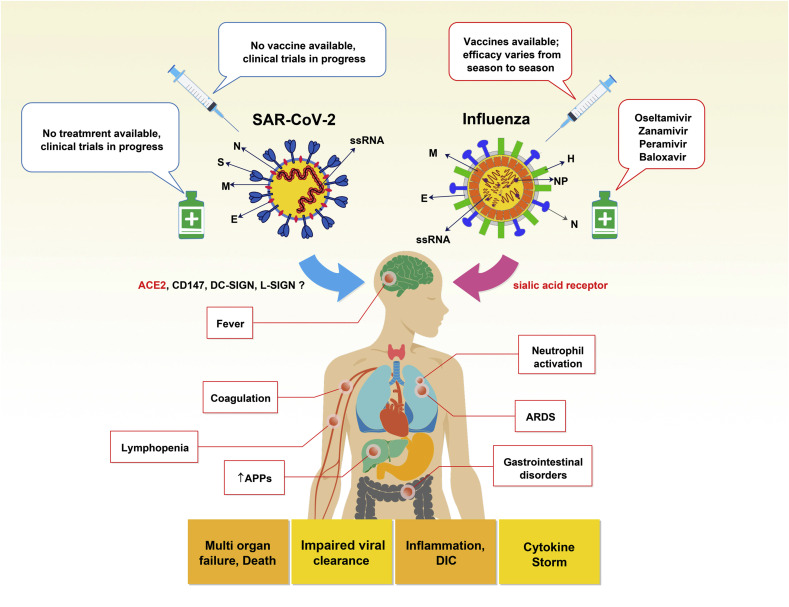
Numerous studies have elucidated the immunopathologic mechanisms in influenza. However, the possible mechanisms in COVID-19 are not well clarified yet. In this section, we will discuss the immunopathological similarities between influenza virus and SARS-CoV-2, which can help to understand and manage the pathological consequences of SARS-CoV-2 and influenza virus co-infection (Fig. 1 ).
Fig. 1.
Immunopathological similarities between influenza and COVID-19. The similarities between the two viruses are shown. Both infections eventually lead to pathological events and disorders such as fever, inflammation, lymphopenia, coagulation, DIC, gastrointestinal disorders, increased APPs, ARDS, multi-organ failure, and death. However, the severity of these symptoms can vary between the two infectious diseases. (SARS-CoV-2; severe acute respiratory syndrome coronavirus 2, S; spike, N; nucleoprotein, M; membrane, E; envelope), (Influenza, N; neuraminidase, H; hemagglutinin, M; membrane, E; envelope, NP; nucleoprotein), ss RNA; single strain RNA, ACE2; angiotensin-converting enzyme 2, DC-SIGN; dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin, APPs; acute phase proteins, ARDS; acute respiratory distress syndrome, DIC; disseminated intravascular coagulation.
5.1. Fever
Fever is one of the most common symptoms in patients with COVID-19 and influenza, which can occur without feeling the temperature rise on the skin [95,96]. Evidence suggests that fever is more common with viral infections, especially when it is unknown to the host. Following these types of infections, activated leukocytes can eliminate the infectious agent by producing the inflammatory and anti-inflammatory cytokines, which can display contradictory effects on the induction of fever [97]. Inflammatory cytokine levels such as IL-1β and IL-6, have been reported to increase significantly following anti-viral innate immune responses in patients with influenza or COVID-19, and these mediators play a crucial role in causing fever [98,99]. Cytokines can cross the blood-brain barrier and react with peripheral vagus nerve terminals, sending signals to the body's temperature control center in the hypothalamus. In response to these signals, the hypothalamus raises body temperature to control infection, which consequently causes shivering and contraction of the skin and blood vessels [100].
5.2. Cytokine storm
Cytokine storm syndrome (CSS) is an uncontrolled systemic inflammatory response following the production of high amounts of pro-inflammatory cytokines, usually after infection with pathogens possessing super-antigens. Patients with CSS are susceptible to multi-organ failure and death and require intensive care [101]. In respiratory viral infections such as COVID-19 and influenza, CSS is considered as one of the common causes of ARDS due to increased vascular permeability and perfusion of fluid and blood components into the alveoli. Evidence showed that the significant increase in cytokines including IL-1β, IL-4, IL-6, IL-10, IL-18, IL-33, TNF-α, and IFN-γ are implicated in CSS [96], most of which are found in laboratory findings obtained from COVID-19 patients, suggesting the CSS as the main consequence of SARS-CoV-2 infection [97]. The most prominent pro-inflammatory cytokine responsible for CSS complications is IL-6 that draws attention in COVID-19. IL-6 binds to either its soluble or membranous receptor, then interacts with gp130 to trigger several downstream signaling pathways, including JAK-STAT, MAPK, PI3K, and NOTCH [98]. The over-reaction of the immune response through cytokines, especially IL-6, causes a hyperinflammatory syndrome called secondary hemophagocytic lymphohistiocytosis (sHLH), leading to fatal multi-organ failure. Hence, a growing number of clinical trials on tocilizumab as an IL-6R blocker and JAK-STAT inhibitors and even glucocorticoids have been recently started to relieve the over-reaction of immune responses and CSS [68]. In addition to IL-6, stimulation of signaling pathways by TNF-α and members of the IL-1 family provoke an amplification loop, which results in nuclear factor-κB (NF-κB) activation and production of pro-inflammatory mediators. Besides, the activated NLRP3 inflammasome by M2 and viroporin 3a proteins of influenza virus and SARS-CoV-2 play a crucial role in prompting CSS through synthesizing IL-1β and IL-18 from their precursor proteins and stimulation of pyroptosis as an inflammatory programmed cell death process [99].
5.3. Tissue damage
Evidence showed that diffused alveolar disease could be detected in influenza-infected patients, while molecular tests confirming the presence of viral RNA are only positive in some of those patients [102]. Previous studies have shown that the cause of death in most people with influenza is dysregulated immune system responses and defects in the clearance of the virus [103,104]. An experimental study demonstrated that dysregulated cytokine and chemokine production and activation of neutrophils are involved in lethal forms of influenza infection [105]. To confirm the role of neutrophils in lung injury, studies have shown that by interfering or inhibiting chemotactic factors such as CXCL2 and CXCL10 (IP-10), locomotion of neutrophils is affected in the lungs resulting in a significant reduction of lung tissue damage [[105], [106], [107]]. Despite these findings, other studies have shown that complete neutrophil depletion worsens the condition of influenza patients, and the clearance of the virus is impaired. Given these contradictory findings, it may be concluded that neutrophils play a dual role in this disease. On the one hand, they help with clearance and reduce viral infection, and on the other hand, they cause tissue damage in these patients [108].
Recently, investigations on COVID-19 patients revealed a significant association between serum level of some inflammatory cytokines such as IL-6 and SARS-CoV-2 load. Also, there was a significant relationship between the mentioned viral load and ARDS and lung injury [109]. Cytokine storm caused by inflammatory cytokines and chemokines could be involved in multi-organ dysfunction and tissue damage in the heart, lungs, kidneys, and liver in COVID-19 patients [110]. Like inflammatory responses in influenza, these inflammatory mediators can recruit neutrophils and macrophages to the lung tissue resulting in hyaline membranes formation and diffuse alveolar damage in patients with COVID-19 [111]. Spleen atrophy and necrosis of lymph nodes have also been reported in patients with COVID-19, which can ultimately disrupt adaptive immune responses and reduce lymphocyte count [112].
5.4. Lymphopenia
Findings from previous studies indicated that in patients with influenza, there was a monocytosis along with lymphopenia (decrease in B and T lymphocytes number) and a normal or low total leukocyte count [[113], [114], [115]]. Previous experimental studies have seen a significant reduction in the number of T cells, along with a relatively mild decrease in B cells; however, seroconversion was confirmed in 90% of cases, indicating that their function has been maintained despite the reduction in B and T lymphocytes [116]. Cell migration from the circulation, cell death via necrosis or apoptosis, and suppression of hematopoiesis can consider as the main factors in reducing lymphocytes. In this regard, a study demonstrated that the influenza virus could infect CD34+ hematopoietic stem cells (HSCs) as well as mesenchymal stem cells and eventually induce their apoptosis. Apoptosis can also occur indirectly following HSCs CD34+ infection by secretion of paracrine factors [117].
Recently, it has also been shown that lymphopenia could be considered as a valuable indicator in patients with COVID-19. As discussed in influenza, several mechanisms and reasons could be involved in lymphopenia; nonetheless, in patients infected by SARS-CoV-2, hyper-production of inflammatory cytokines such as IL-6, the spleen and lymph nodes injury, decrease the expression of major histocompatibility complex (MHC) class II molecules in monocytes by IL-6, FAS molecule expression by infected host cells in different tissues, infected ACE2+CD68+CD169+ macrophages, and some other unknown mechanisms may play role in reducing the number of lymphocytes and weakening T and B-dependent immune responses during the disease [118,119]. It has been shown that IL-6 can decrease the MHC-II molecules in monocytes, and excessive activation of monocytes leads to an increase in IL-6 production as well as a decrease in the number of lymphocytes [120]. As mentioned before, in marginal sinuses lymph nodes and marginal zone of the spleen, infected ACE2+CD68+CD169+ macrophages that contained nucleoprotein antigen of SARS-COV-2 have been observed. These types of macrophages are able to produce high concentrations of IL-6, which ultimately causes lymphopenia and impaired viral clearance according to the mechanisms discussed [112]. However, due to the unknown nature of many of the COVID's aspects, further studies are needed to understand thoroughly the mechanisms by which the virus generates lymphopenia.
5.5. Coagulation
Impaired coagulation is considered as a common disorder in the pathogenesis of influenza, which is characterized by stimulation of lung endothelial, leakage of vessels, diffused intravascular coagulation and blockage in pulmonary arteries (micro-embolism). Evidences demonstrated that a dysregulated cellular and protein-dependent coagulation system, including endothelial cells, platelets, coagulation factors, fibrinolysis proteases and anticoagulants, could be involved in influenza pathogenesis resulting in an increased viral replication and subsequent destructive immune responses [121].
Previous studies indicated that influenza infections could increase the risk of venous thrombosis and atherothrombotic disorders through amplification in thrombin production, along with a reduction of C protein expression. In this regard, heparin therapy could be helpful in the prothrombotic state caused by influenza infection. An investigation also suggested that following influenza infection, a prothrombotic state occurred through the initiation of the coagulation cascade, regardless of direct cleavage of prothrombin into thrombin [122]. Also, it has been shown that following replication of the influenza virus in the lung, excessive inflammatory responses may participate in coagulation system dysfunction [[122], [123], [124]]. Pro-inflammatory cytokines could also play a crucial role in the activation of the coagulation cascade.
Interestingly, IL-1β and TNF-α, despite their inflammatory role, play an anti-coagulation role during viral infection, whereas IL-6 can initiate coagulation through interfering in the creation of tissue factor and thrombin [[125], [126], [127]]. Thrombin, through the proteinase-activated receptor-1 (PAR-1), can participate in various mechanisms such as coagulation and inflammation. In a homeostatic mechanism and to reduce inflammation, anticoagulant molecules such as protein C and antithrombin III regulate the production of thrombin. However, during a viral infection such as SARS-CoV-2 or influenza, these molecules become defective due to uncontrolled inflammation, and eventually, coagulation and inflammatory pathways are stimulated, resulting in diffused intravascular coagulation, micro thrombosis and multi-organ failure [89,128,129].
6. Influenza virus and SARS-CoV-2 immune evasion mechanisms
6.1. Influenza immune escape mechanisms
Influenza viruses use different mechanisms to escape the antiviral immune system responses. For instance, NS1 protein is involved in the provoking of host anti-viral responses via inhibition of RIG-I receptor signaling, tripartite motif-containing protein 25 (TRIM25), interferon regulatory factor 3 (IRF-3), NF-κB, ATF-2/c-Jun, IFN-β promoter, as well as negative interfering effects on DCs maturation, and specific CD8+ T cells induction. NS1 also modifies host cell gene expression by binding to CPSF30 and interfering with the mRNA export machinery [130]. Other influenza proteins, including PB2 and PB1-F2, also can reduce the production of IFN-β that is considered as one of the most effective anti-viral cytokines in the innate immune responses. PA-X protein of influenza also can suppress the expression of cellular regulator genes which are responsible for initiation of the immune response [131]. In addition, following the binding of influenza M2 protein to p58IPK-hsp40 complex, the release of p58IPK and protein synthesis could be repressed and finally, host cell apoptosis processes might be initiated and viral particles released from mentioned apoptotic cells. After encapsidating influenza, NP protein reduces the formation of dsRNA, which could prevent the recognition of viral RNA by cytosolic PRR. The influenza virus also stimulates the expression of suppressor of cytokine signaling (SOCS) proteins, which inhibit activation of IFN α/β receptor signaling in the JAK/STAT pathway. Besides, the virus can reduce cytotoxicity and impair the recognition of infected cells via consecutive mutations in the HA protein as well as direct infection of NK cells [132].
Moreover, it is well documented that the antigenic shift and drift of the influenza virus cause the escape of virus from specifically produced antibodies. Also, virus surface proteins are not detectable by neutralizing antibodies, which can also be considered part of the virus escape mechanisms [132].
6.2. SARS-CoV-2 immune escape mechanisms
Some evidences showed that, SARS-CoV similar to influenza and other viruses uses almost identical strategies to escape host antiviral responses. One of the main mechanisms of escape by viruses, especially SARS-CoV, is the inhibition of IFN production as an anti-viral cytokine [133,134]. Membrane proteins of SARS-CoV can suppress adaptor molecules in the downstream of the IRFs signaling pathway, resulting in decreased IRF3/IRF7 transcription factor translocation and expression [135]. It is well known that cathepsins and proteasomes play the crucial roles in the MHC-I and -II pathways and processing of viral protein antigens. In contrast to this host defense process, the virus reduces the expression of cathepsins and proteasomes molecules, which ultimately reduces the expression of MHC-I molecules on the surface of the infected cells. Another evasion strategy is the direct infection of immune cells such as DCs, which can be led to the disruption of maturation and development process of these cells and the lack of effective presentation of viral antigens to T cells [136,137]. As mentioned before, SARS-CoV can stimulate inflammatory cytokines, which are involved in forming the cytokine storm as a destructive inflammatory immune response following viral infection [101,138,139]. In the context of cellular immune responses, viral mechanisms can also exhaust virus specific CD8+ T cells, which can be characterized by the overexpression of T cell immunoglobulin and mucin domain-containing protein 3 (TIM3) and programmed cell death protein 1 (PD-1) on the surface of theses immune cells [140]. As the overall and since the aforementioned mechanisms were lessons learned from SARS-CoV, the escape mechanisms of SARS-CoV-2 are probably very similar, although further studies are needed to clarify SARS-CoV-2 escape strategies.
6.3. Reported SARS-CoV-2 and influenza virus co-infections
Viral infections that affect the respiratory system prompt patients to co-infections resulting in higher disease severity and mortality. Recently, there have been reports from several countries of co-infection with SARS-CoV-2 and influenza virus, suggesting that the COVID-19 may be associated with other infectious diseases, including influenza, and challenge the diagnosis and treatment of the disease. In this section, the studies conducted in this field have been briefly reviewed.
A study in China reported a 69-year-old man case with SARS-CoV-2 and influenza virus co-infection. The patients had not any underlying disease. At the time of admission, cough and fever were reported; also, CT findings revealed a ground-glass consolidation lesion in the right inferior lobe of his lungs. Leukopenia was not reported, and his real-time reverse transcription-PCR (RT-PCR) test was negative for SARS-CoV-2, but Xpert Flu/RSV Xpress assay results showed that the nasopharyngeal swab sample was positive for influenza A but negative for SARS-CoV-2. After one week, the patient returned with persistent fever and worsening dyspnea, and the results of the RT-PCR test for positive SARS as well as leukopenia were reported [141]. One of the most common problems in SARS-CoV-2 molecular tests is the lack of criteria for the adequacy of the sample taken from the patient, which may have affected the results of the study. The findings of the study revealed that further challenges to diagnosis remain, mainly when the results of the patient's diagnostic test are negative for SARS-CoV-2 but positive for other viruses.
Another study from Iran which conducted on four patients with pneumonia symptoms and suspicious for SARS-CoV-2 infection reported that following nasopharyngeal and oropharyngeal throat swab samples examination by RT-PCR for SARS-CoV-2, influenza viruses, as well as all the respiratory panel findings, showed that all four patients were co-infected by SARS-CoV-2 and influenza virus. The results of this study show the high importance of performing molecular tests such as RT-PCR in patients with suspected COVID-19 during the pandemic, even if these patients do not have symptoms of other viral infections related to the respiratory tract [142]. An investigation in Spain, four patients with persistent non-productive cough, fever, as well as dyspnea, were studied, in which all of them had an underlying medical condition such as diabetes and severe kidney disorder. Rapid nucleic acid amplification assay for influenza showed that all four patients were positive for influenza type A and B. Additionally, RT-PCR data showed that all patient samples were positive for SARS-CoV-2 and co-infection was confirmed [143]. A study in Eiju General Hospital, Tokyo, Japan reported a 78-year-old woman with dyslipidemia, hypothyroidism, cough, 37.7 °C body temperature, malaise, anorexia, along with weight loss. COVID-19 was proposed regarding the travel history, prolonged clinical symptoms, and findings of her chest X-ray and CT. RT-PCR results showed that sputum specimen was positive for SARS-CoV-2, and rapid influenza test also was confirmed influenza A virus infection. The authors stated that discrimination between COVID-19 and influenza is complicated regarding the only on clinical indicators [21]. A 66-year-old African-American woman with fever (38.9 °C), non-productive cough, anorexia, shortness of breath, along with underlying diseases including chronic kidney disease, diabetes, coronary artery disease, and hypertension were also studied, and clinical symptoms, as well as laboratory tests results, confirmed that she had COVID-19 and influenza co-infection [144]. In a study from Wuhan, among 115 pneumonia patients with SARS-CoV-2 infection, five patients were positive for the influenza virus. Co-infected patients with SARS-CoV-2 and influenza virus have symptoms such as fever, cough, fatigue, and headache. In this report, patients had unusual symptoms such as a nasal tampon, pharyngalgia, diarrhea, and mild hemoptysis, which are not common in patients with COVID-19 and should be considered in co-infected patients. During the admission, all patients had lymphopenia; however, the total lymphocyte count was gradually raised during the remission phase. This study revealed that the laboratory findings were similar to those before stated in patients with only SARS-CoV-2 infection [7,145]. Also, the analysis of radiographs of patients showed that there was no significant difference between patients with COVID-19 and co-infected patients. Dyspnea was realized in all co-infected patients, and only 15.7% of the patients with SARS-CoV-2 infection were with severe disease afterward admission to the hospital. Accordingly, probably, patients with SARS-CoV-2 and influenza virus co-infection did not experience more severe conditions compared with patients who have one of these viral infections [96].
In a study from Germany, A 4-month-old infant was reported with cough, fever (38.2 °C). The history of the mentioned patients showed that the child's grandmother was positive for COVID-19, and her last contact with the patient was eight days before the onset of symptoms. Data obtained from RT-PCR approved SARS-CoV-2 infection, while CO2 retention was not realized in blood gas analysis. Oseltamivir was prescribed for five days for the mentioned patient. To date, there is no apparent reason why children show little or no symptoms following infection with SARS-CoV-2. Nonetheless, studies show that this could be due to insufficient expression of the ACE-2 receptor, as well as its low affinity for binding to virus proteins. As a result, weaker inflammatory responses are formed in the body and cause less tissue damage [146].
Interestingly, an investigation suggested a protective effect of co-infection with influenza for COVID-19 patients and reported that co-infection with the influenza virus fatality rate was decreased remarkably than non-influenza patients. The authors stated that co-infection with the influenza virus acts as a protective occurrence for patients with COVID-19 [147]. However, this theory needs further study, because according to the data presented, sometimes the opposite has happened, and some co-infected patients have worsened their disease.
Some studies have also examined the prevalence of other infectious pathogens in COVID-19 patients, which has yielded exciting results. Wang et al. reported that among 104 COVID-19 patients, 2.88% were co-infected with other coronaviruses, and 1.94% were co-infected with influenza A [148]. Another study reported that 94.2% of patients with COVID-19 were co-infected with other bacteria, viruses, and fungi pathogens. They found a few co-infected patients with influenza A and B, whereas there were no patients with other coronavirus co-infection. These outcomes specified that the co-infections with mentioned viruses are not common in patients with SARS-CoV-2 infection; however, regarding the prevalence of the seasonal influenza risk of co-infections could upsurge in patients with COVID-19 [149]. No deaths were reported in patients with the co-infection, and only three patients were transferred to the ICU, but no significant differences were found in the clinical and laboratory findings of these patients and other patients. The study claims that there is no considerable difference between patients with COVID-19 and those who were infected with other pathogens in addition to SARS-CoV-2 [149].
Taken together, it appears that during the high prevalence of seasonal influenza, discriminating influenza from SARS-CoV-2 is difficult because there were several similarities regarding the fever, non-productive cough, and dyspnea between COVID-19 and influenza patients. In this regard, RT-PCR and the nucleic acid test should be performed on all suspected patients. In this way, the co-infections will be identified. Besides, mentioned patients should be isolated due to inhibiting infection transmission to other people. Consequently, there would be a positive effect on SARS-CoV-2 and influenza virus co-infection prevention [150].
7. Diagnosis and treatment key points
Regarding the similar symptoms of influenza and COVID-19, the diagnosis and differentiation of co-infections are very critical in the management of disease and treatment because some associated infections can be treated with anti-viral drugs or antibiotics.
Approved diagnostic tests currently available for the diagnosis of influenza viruses are divided into two general categories: 1) molecular assays such as rapid molecular assay, RT-PCR, and other nucleic acid amplification assays; and 2) antibody-based assays such as immunofluorescence and rapid influenza diagnostic tests [151,152] (Table 2 ).
Table 2.
Comparison of diagnosis methods of influenza and COVID-19.
| Diagnosis methods | Advantages | Disadvantages | Ref | |
|---|---|---|---|---|
| Influenza | Rapid influenza diagnostic tests (RIDTs) | •Quick outcomes in <15 min •Simple to perform •Available for office/bedside use •RIDTs with CLIA waived can be utilized in settings that include point-of-care •High specificity |
•Sub-optimal test sensitivity, false-negative results are common •Influenza B antigens detection sensitivity is lower compared with influenza A antigens •false-positive results •All RIDTs could not discriminate between influenza A or B viruses •Do not give information about influenza A virus or specific virus strain •RIDTs are not able to differentiate between novel influenza A virus and seasonal influenza A infections |
[[171], [172], [173]] |
| Rapid molecular assay and RT-PCR | •Quick outcomes in approximately 15–30 min to less than 1.5 h •High sensitivity and specificity •Low false positive or false negative result •Some molecular assays can differentiate between specific influenza A virus subtypes •RT-PCR results could be obtained between 4 and 8 h |
•Results of some RT-PCR and other molecular assays may not be available in a clinically relevant time frame to inform clinical management decisions •Low availability of RT-PCR facilities •Respiratory specimens may need to be sent to a state public health laboratory or commercial laboratory for RT-PCR •Although the test time is low, the actual time to receive results may be substantially longer •Most FDA-cleared molecular assays are not approved to test lower respiratory tract specimens •High costs of the tests •All molecular assays may not specifically identify all currently circulating influenza A virus subtypes •Problem with the assessment of the accuracy of all available RT-PCR and molecular assays •Rapid molecular assays have lower sensitivity than RT-PCR |
||
| Immunofluorescence (DFA and IFA) | •Antigen detection assays •Outcomes in approximately 2–4 h •High specificity •Both direct (DFA) and indirect fluorescent antibody (IFA) staining assays are available to detect influenza A and B viral antigens in respiratory tract specimens •Can be used as an RIDT with an analyzer device to produce results in approximately 15 min |
•A fluorescent microscope is required •Moderate sensitivity •Subtyping or further identification of influenza A virus is not possible by immunofluorescence assays. |
||
| Serologic (antibody detection) | •Antibody assessments may be used in conjunction with viral detection tests to support the clinical assessment of persons who present late in their illnesses •High availability compared with molecular tests •Low costs |
•The FDA has not authorized antibody testing to diagnose influenza •The CDC does not currently recommend using antibody testing for the diagnosis of any infection |
||
| COVID-19 | Real-time PCR | •Authorized assays for viral testing include those that detect COVID-19 nucleic acid or antigen •Viral (nucleic acid or antigen) tests check samples from the respiratory system (such as nasal or oral swabs) or saliva to determine whether COVID-19 is present •High sensitivity and specificity •Low false positive or false negative result •RT-PCR results could be obtained between 4 and 8 h |
•Results of some RT-PCR and other molecular assays may not be available in a clinically relevant time frame to inform clinical management decisions •Low availability of RT-PCR facilities •Respiratory specimens may need to be sent to a state public health laboratory or commercial laboratory for RT-PCR •Although the test time is low, the actual time to receive results may be substantially longer •Most FDA-cleared molecular assays are not approved to test lower respiratory tract specimens •High costs of the tests •Problem with the assessment of the accuracy of all available RT-PCR and molecular assays |
[83,174] |
| Serologic (antibody detection) | •Antibody assessments may be used in conjunction with viral detection tests to support the clinical assessment of persons who present late in their illnesses •High availability compared with molecular tests •Low costs |
•The FDA has not authorized antibody testing to diagnose influenza •The CDC does not currently recommend using antibody testing for the diagnosis of any infection |
||
In the case of SARS-CoV-2 detection, since it has recently been identified, diagnostic tests with high sensitivity and specificity have not yet been introduced. It has been suggested that the presence of viral nuclear acid in specimens such as urine and serum were negative, regardless of disease severity in patients with COVID-19. Serological tests in COVID-19 patients, unlike those with influenza, are not widely used in the early phase of the disease because the titers of IgM and IgG antibodies begin to rise a few days to weeks after the virus entrance to the body. On the other hand, cross-reaction with the antigens of other coronaviruses can call into question the results of such tests. Based on the CDC recommendation, a respiratory tract specimen (preferably a nasopharyngeal flock swab) should be examined for the presence of SARS-CoV-2 nucleic acid using the RT-PCR method.
Additionally, CT scan (as a high sensitivity but low specificity test), along with inflammatory biomarkers such as LDH and CRP, can help diagnose and monitor the course of treatment in COVID-19 patients, despite their non-specificity. To confirm that the virus has cleared from the body of previously infected patients with SARS-CoV-2, at least two samples obtained from the upper respiratory tract should be negative for the presence of SARS-CoV-2 at 24-h intervals [153]. Studies have shown that molecular tests such as RT-PCR and also rapid diagnostic tests are more commonly used to detect SARS-CoV-2 infection, but the high rate false-negative results sometimes making it difficult to diagnose [141]. It seems that due to the high probability of co-infection, especially in the influenza season, diagnostic tests related to both viruses should be performed for all suspected and symptomatic cases [96]. Therefore, in order to diagnose co-infections, and due to the similarity of the clinical and analytical courses in COVID-19 and influenza infections, it is recommended to perform specific tests to differentiate between these two viral infections as well as other pathogens.
There are two groups of approved drugs for controlling influenza, including Adamantanes (Amantadines and Rimantadines) and neuraminidase inhibitors (oseltamivir, zanamivir, laninamivir, and peramivir). However, sometimes influenza viruses are resistant to these anti-viral drugs [154,155]. In several studies on patients with a SARS-CoV-2 and influenza virus co-infection, the oseltamivir has been used to treat the influenza infection because it can reduce the duration of the disease with a lower risk of otitis media in children [21,96,144,146,156,157]. However, this drug has caused gastrointestinal disorders such as diarrhea in some patients and other adverse effects [158] (Table 3 ).
Table 3.
More common treatments for influenza and COVID-19.
| Treatments | Adverse effects and Challenges | Ref | |
|---|---|---|---|
| Influenza | Amantadines/Rimantadines | •CNS and gastrointestinal side effects •The incidence of CNS side effects, including nervousness, anxiety, difficulty concentrating, and lightheadedness is higher among persons taking amantadine compared to those taking rimantadine •Serious side effects (e.g., marked behavioral changes, delirium, hallucinations, agitation, and seizures) |
[176] |
| Oseltamivir | •Nausea, vomiting, sporadic, transient neuropsychiatric events (self-injury or delirium) | [176] | |
| Zanamivir | •Allergic reactions: oropharyngeal or facial edema •Diarrhea, nausea, sinusitis, nasal signs and symptoms, bronchitis, cough, headache, dizziness, and ear, nose and throat infections |
[176] | |
| Laninamivir | •Psychiatric disorders (abnormal behavior, etc.), gastrointestinal disorders (diarrhea, nausea, etc.) and nervous system disorders (dizziness, etc.) •No serious ADRs occurred |
[[175], [176]] | |
| COVID-19 | Tocilizumab | •Injection site reaction •New bacterial, viral or fungal infections (Severe liver failure due to herpes simplex virus 1 reactivation, leading to death) •Elevation of liver enzyme levels (dose-dependent) •Neutropenia or thrombocytopenia (uncommon) •Not approved for the treatment of patients with COVID-19 •High cost |
[[83], [177],,178] |
| Siltuximab | •Serious risk of infections •Rash, severe bacterial infections in long-term dosing (once every three weeks) •High cost •Not approved for the treatment of patients with COVID-19 |
[83,178] | |
| HCQ | •Prolongation of QTc interval, Torsades de Pointes, AV block, ventricular arrhythmia •Gastrointestinal effects (e.g., nausea, vomiting, diarrhea) •Hepatitis •Hypoglycemia •Myopathy •Anxiety, agitation, hallucinations, psychosis •Allergic reaction/rash •Given the risk of heart rhythm problems, the FDA cautions against using HCQ to treat COVID-19 outside of a hospital or a clinical trial |
[83] | |
| Remdesivir | •Only for use in hospitalized patients with COVID-19 who require supplemental oxygen but who do not require oxygen delivery through a high-flow device, noninvasive ventilation, invasive mechanical ventilation, or extracorporeal membrane oxygenation (ECMO) •Gastrointestinal symptoms (e.g., nausea, vomiting) •Elevated transaminase levels •Increase in prothrombin time (without a change in the international normalized ratio) •Renal toxicity (SBECD accumulation) •Supplies are limited |
[83] | |
| Azithromycin | •Gastrointestinal effects (e.g., diarrhea, nausea, vomiting) • Hepatotoxicity •Prolongation of QTc interval when prescribed with HCQ |
[83] | |
| Lopinavir/Ritonavir | •Gastrointestinal effects (e.g., nausea, vomiting, diarrhea) •Transaminase elevation •QTc interval prolongation and Torsades de Pointes •PR interval prolongation •Unknown optimal drug concentrations that can inhibit the SARS-CoV-2 proteases •Did not show efficacy in a moderately sized randomized controlled trial in COVID-19 patients |
[83] |
A study revealed that intensive care unit (ICU), extracorporeal membrane oxygenation, and the invasive ventilator were not used in enrolled patients with COVID-19 and influenza co-infection. On the other hand, almost all mentioned patients recovered oseltamivir as well as other anti-viral/antibacterial drugs, oxygen inhalation and were leaving the hospital after treatment without death [96]. A specific drug has not yet been introduced to treat patients with COVID-19, and studies are ongoing. Although drugs such as tocilizumab [[159], [160], [161], [162], [163]], siltuximab [164,165], hydroxychloroquine [166,167], remdesivir [168], lopinavir/ritonavir [169], and azithromycin [170] have been used to treat the disease; however, they have had conflicting results and require further study [83] (Table 3).
8. Concluding remarks
According to the findings of studies conducted on co-infected patients with COVID-19 and influenza, several of the clinical and laboratory findings are similar in both infections, making it challenging to diagnose these co-infected patients. Dedicated specific assays for influenza viruses and SARS-CoV-2 is recommended in all suspected patients. Furthermore, vaccination and immunization against influenza at a suitable time is also necessary to prevent co-infection because, in some cases, it can worsen the patient's symptoms, clinical condition, and even death. It is also proposed that SARS-CoV-2 may be underdiagnosed because of false-negative results for co-infection with other viral respiratory infections. It is suggested that broader viral assessments be needed to improve clinical management decisions and outcomes.
Declaration of competing interest
All of the authors declare no conflict of interest.
Acknowledgments
Iran University of Medical Sciences supported this project.
References
- 1.Coronavirus Disease (COVID-19) Pandemic. 9.19. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 Available from: [Google Scholar]
- 2.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E., et al. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zolfaghari Emameh R., Nosrati H., Taheri R.A. Combination of biodata mining and computational modelling in identification and characterization of ORF1ab polyprotein of SARS-CoV-2 isolated from Oronasopharynx of an Iranian patient. Biol. Proced. Online. 2020;22:1–8. doi: 10.1186/s12575-020-00121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z., et al. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. CMAJ. 2020;41(2):145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 9.Baud D., et al. Real estimates of mortality following COVID-19 infection. Lancet Infect. Dis. 2020;20(7):773. doi: 10.1016/S1473-3099(20)30195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zolfaghari Emameh R., Falak R., Bahreini E. Application of system biology to explore the association of neprilysin, angiotensin-converting enzyme 2 (ACE2), and carbonic anhydrase (CA) in pathogenesis of SARS-CoV-2. Biol. Proced. Online. 2020;22(1):1–9. doi: 10.1186/s12575-020-00124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taubenberger J.K., Kash J.C., Morens D.M. The 1918 influenza pandemic: 100 years of questions answered and unanswered. Sci. Transl. Med. 2019;11(502):eaau5485. doi: 10.1126/scitranslmed.aau5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taubenberger J.K., Morens D.M. 1918 Influenza: the mother of all pandemics. Emerg. Infect. Dis. 2006;12(1):15. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jester B., Uyeki T., Jernigan D. Readiness for responding to a severe pandemic 100 years after 1918. Am. J. Epidemiol. 2018;187(12):2596–2602. doi: 10.1093/aje/kwy165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berry J.D., et al. Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology. MAbs. 2010;2:53–66. doi: 10.4161/mabs.2.1.10788. 1st ed, Taylor & Francis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention . 2019-2020 U.S. Flu Season: Preliminary In-Season Burden Estimates. CDC; 2020. National Center for Immunization and Respiratory Diseases (NCIRD) [Google Scholar]
- 16.Moghoofei M., et al. Prevalence of influenza A infection in the Middle‐East: a systematic review and meta‐analysis. Clin. Respir. J. 2018;12(5):1787–1801. doi: 10.1111/crj.12758. [DOI] [PubMed] [Google Scholar]
- 17.Esghaei M., et al. Trends in surveillance data of influenza virus in Tehran before decreasing dispatch of Iranian Hajj pilgrims to Mecca. Med. J. Islam. Repub. Iran. 2018;32:41. doi: 10.14196/mjiri.32.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Gruta N.L., et al. A question of self‐preservation: immunopathology in influenza virus infection. Immunol. Cell Biol. 2007;85(2):85–92. doi: 10.1038/sj.icb.7100026. [DOI] [PubMed] [Google Scholar]
- 19.Long J.S., et al. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019;17(2):67–81. doi: 10.1038/s41579-018-0115-z. [DOI] [PubMed] [Google Scholar]
- 20.Parikh S.R., et al. Pediatric otolaryngology divisional and institutional preparatory response at Seattle Children’s Hospital after COVID-19 regional exposure. Otolaryngol. Head Neck Surg. 2020;162(8):800–803. doi: 10.1177/0194599820919748. [DOI] [PubMed] [Google Scholar]
- 21.Azekawa S., et al. IDCases; 2020. Co-infection with SARS-CoV-2 and Influenza A Virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Q., et al. Co-infection and other clinical characteristics of COVID-19 in children. Pediatrics. 2020;146(1) doi: 10.1542/peds.2020-0961. [DOI] [PubMed] [Google Scholar]
- 23.Zhai S.-L., et al. Influenza D virus in animal species in Guangdong Province, southern China. Emerg. Infect. Dis. 2017;23(8):1392. doi: 10.3201/eid2308.170059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasin A., et al. Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res. 2014;185:53–63. doi: 10.1016/j.virusres.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 25.Niu P., et al. Ultrapotent human neutralizing antibody repertoires against Middle East respiratory syndrome coronavirus from a recovered patient. J. Infect. Dis. 2018;218(8):1249–1260. doi: 10.1093/infdis/jiy311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumlin U., et al. Sialic acid tissue distribution and influenza virus tropism. Influenza and other respiratory viruses. 2008;2(5):147–154. doi: 10.1111/j.1750-2659.2008.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camp J.V., et al. Lower respiratory tract infection of the ferret by 2009 H1N1 pandemic influenza A virus triggers biphasic, systemic, and local recruitment of neutrophils. J. Virol. 2015;89(17):8733–8748. doi: 10.1128/JVI.00817-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansfield K.G. Viral tropism and the pathogenesis of influenza in the Mammalian host. Am. J. Pathol. 2007;171(4):1089. doi: 10.2353/ajpath.2007.070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herold S., et al. Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur. Respir. J. 2015;45(5):1463–1478. doi: 10.1183/09031936.00186214. [DOI] [PubMed] [Google Scholar]
- 30.Rossman J.S., Lamb R.A. Influenza virus assembly and budding. Virology. 2011;411(2):229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaisri U., Chaicumpa W. Evolution of therapeutic antibodies, influenza virus biology, influenza, and influenza immunotherapy. BioMed Res. Int. 2018 doi: 10.1155/2018/9747549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wikramaratna P.S., et al. The antigenic evolution of influenza: drift or thrift? Phil. Trans. Biol. Sci. 2013;368(1614) doi: 10.1098/rstb.2012.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taubenberger J.K., Kash J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7(6):440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su S., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu R., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu A., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microb. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. 3rd ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fehr A.R., Perlman S. Coronaviruses. Springer; 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bzowka M., et al. Structural and evolutionary analysis indicate that the SARS-CoV-2 Mpro is a challenging target for small-molecule inhibitor design. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21093099. 9th ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., et al. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharmaceut. Anal. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan S., et al. Emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus 2: biology and therapeutic options. J. Clin. Microbiol. 2020;58(5) doi: 10.1128/JCM.00187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L., et al. Origin and evolution of the 2019 novel Coronavirus. Clin. Infect. Dis. 2020;71:882–883. doi: 10.1093/cid/ciaa112. 15th ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K., et al. 2020. SARS-CoV-2 Invades Host Cells via a Novel Route: CD147-Spike Protein. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan R., et al. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bi Q., et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet. 2020;20:911–919. doi: 10.1016/S1473-3099(20)30287-5. 8th ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H., et al. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020:1–5. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng T., et al. 2020. The Insert Sequence in SARS-CoV-2 Enhances Spike Protein Cleavage by TMPRSS. BioRxiv. [Google Scholar]
- 48.Wang H., et al. SARS coronavirus entry into host cells through a novel clathrin-and caveolae-independent endocytic pathway. Cell Res. 2008;18(2):290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitworth J. COVID-19: a fast evolving pandemic. Trans. R. Soc. Trop. Med. Hyg. 2020;114(4):241. doi: 10.1093/trstmh/traa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malik Y.S., et al. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 2020;40(1):68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abdullahi I.N., et al. Exploring the genetics, ecology of SARS-COV-2 and climatic factors as possible control strategies against COVID-19. Inf. Med. 2020;28(2):166–173. [PubMed] [Google Scholar]
- 53.Abdel-Moneim A.S., Abdelwhab E.M. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens. 2020;9(7):529. doi: 10.3390/pathogens9070529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He Y., et al. Pathogenesis of osteoarthritis: risk factors, regulatory pathways in chondrocytes, and experimental models. Biology. 2020;9(8):194. doi: 10.3390/biology9080194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tiwari R., et al. COVID-19: animals, veterinary and zoonotic links. Vet. Q. 2020:1–22. doi: 10.1080/01652176.2020.1766725. (just-accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bailey E.S., et al. The continual threat of influenza virus infections at the human–animal interface. Evol. Med. Public Health. 2018:192–198. doi: 10.1093/emph/eoy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osterhaus A., et al. Influenza B virus in seals. Science. 2000;288(5468):1051–1053. doi: 10.1126/science.288.5468.1051. [DOI] [PubMed] [Google Scholar]
- 58.Ran Z., et al. Domestic pigs are susceptible to infection with influenza B viruses. J. Virol. 2015;89(9):4818–4826. doi: 10.1128/JVI.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawano J., et al. Distribution of antibodies in animals against influenza B and C viruses. Jpn. J. Vet. Res. 1978;26(3–4):74–80. [PubMed] [Google Scholar]
- 60.Chang C., et al. Influenza virus isolations from dogs during a human epidemic in Taiwan. Int. J. Zoonoses. 1976;3:61–64. 1st ed. [PubMed] [Google Scholar]
- 61.Salem E., et al. Serologic evidence for influenza C and D virus among ruminants and camelids, Africa, 1991–2015. Emerg. Infect. Dis. 2017;23(9):1556. doi: 10.3201/eid2309.170342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuanji G., et al. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J. Gen. Virol. 1983;64(1):177–182. doi: 10.1099/0022-1317-64-1-177. [DOI] [PubMed] [Google Scholar]
- 63.Ferguson L., et al. Pathogenesis of influenza D virus in cattle. J. Virol. 2016;90(12):5636–5642. doi: 10.1128/JVI.03122-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Treanor J.J. Churchill Livingstone; Philadelphia: 2010. Influenza Viruses, Including Avian Influenza and Swine Influenza. Principles and Practice of Infectious Diseases; pp. 2265–2288. [Google Scholar]
- 65.Clark N.M., Lynch J.P. Influenza: epidemiology, clinical features, therapy, and prevention. Semin. Respir. Crit. Care Med. 2011;32:373–392. doi: 10.1055/s-0031-1283278. 4th ed. [DOI] [PubMed] [Google Scholar]
- 66.Collins J.P., et al. Clinical features and outcomes of immunocompromised children hospitalized with laboratory-confirmed influenza in the United States, 2011–2015. J. Pediatr. Infect. Dis. Soc. 2019;8(6):539–549. doi: 10.1093/jpids/piy101. [DOI] [PubMed] [Google Scholar]
- 67.Kim Y.-J., Boeckh M., Englund J.A. Community respiratory virus infections in immunocompromised patients: hematopoietic stem cell and solid organ transplant recipients, and individuals with human immunodeficiency virus infection. Semin. Respir. Crit. Care Med. 2007;28:222–242. doi: 10.1055/s-2007-976494. 2nd ed. [DOI] [PubMed] [Google Scholar]
- 68.Narasaraju T., et al. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am. J. Pathol. 2011;179(1):199–210. doi: 10.1016/j.ajpath.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoshi S.-L., et al. Cost–effectiveness analysis of influenza vaccination for people aged 65 and over in Japan. Vaccine. 2007;25(35):6511–6521. doi: 10.1016/j.vaccine.2007.05.067. [DOI] [PubMed] [Google Scholar]
- 70.Hsieh Y.-C., et al. Influenza pandemics: past, present and future. J. Formos. Med. Assoc. 2006;105(1):1–6. doi: 10.1016/S0929-6646(09)60102-9. [DOI] [PubMed] [Google Scholar]
- 71.Julkunen I., et al. Inflammatory responses in influenza A virus infection. Vaccine. 2000;19:S32–S37. doi: 10.1016/s0264-410x(00)00275-9. [DOI] [PubMed] [Google Scholar]
- 72.Song J.Y., et al. Clinical, laboratory and radiologic characteristics of 2009 pandemic influenza A/H1N1 pneumonia: primary influenza pneumonia versus concomitant/secondary bacterial pneumonia. Influenza and other respiratory viruses. 2011;5(6):e535–e543. doi: 10.1111/j.1750-2659.2011.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Presanis A.M., et al. The severity of pandemic H1N1 influenza in the United States, from April to July 2009: a Bayesian analysis. PLoS Med. 2009;6(12) doi: 10.1371/journal.pmed.1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Louie J.K., et al. Severe 2009 H1N1 influenza in pregnant and postpartum women in California. N. Engl. J. Med. 2010;362(1):27–35. doi: 10.1056/NEJMoa0910444. [DOI] [PubMed] [Google Scholar]
- 75.Neuzil K.M., et al. Impact of influenza on acute cardiopulmonary hospitalizations in pregnant women. Am. J. Epidemiol. 1998;148(11):1094–1102. doi: 10.1093/oxfordjournals.aje.a009587. [DOI] [PubMed] [Google Scholar]
- 76.De Jong M.D., et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006;12(10):1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shrestha S., et al. Time and dose-dependent risk of pneumococcal pneumonia following influenza: a model for within-host interaction between influenza and Streptococcus pneumoniae. J. R. Soc. Interface. 2013;10(86):20130233. doi: 10.1098/rsif.2013.0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noor A., Gradidge E. A case of reye syndrome caused by influenza A virus. Ochsner J. 2018;18(4):425–427. doi: 10.31486/toj.18.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bengualid V., Berger J. Neurologic complications of acute influenza in adults: case report and review of the literature. J Neurosci Clin Res. 2017;2(1):2. [Google Scholar]
- 80.Li Q., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guan W.-j., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Symptoms of Coronavirus. 9.19.2020. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html Available from:
- 83.COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. Natl. Inst. Health. . Available from: https://www.covid19treatmentguidelines.nih.gov/.
- 84.Bertoletti L., et al. Venous thromboembolism and covid-19. Respir. Med. Res. 2020;78:100759. doi: 10.1016/j.resmer.2020.100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Coronavirus (COVID-19) SYMPTOMS and TESTING. 9.19.2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 Available from:
- 86.Wang D., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mehta P., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McIntosh, K., M.S. Hirsch, and A. Bloom, Coronavirus Disease 2019 (COVID-19). UpToDate. Hirsch MS, Bloom A (Eds.). Accessed 03, 2020, 5.
- 89.Zhou F., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu W., et al. Detection of covid-19 in children in early january 2020 in Wuhan, China. N. Engl. J. Med. 2020;382:1370–1371. doi: 10.1056/NEJMc2003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Y.-C., et al. A locally transmitted case of SARS-CoV-2 infection in Taiwan. N. Engl. J. Med. 2020;382(11):1070–1072. doi: 10.1056/NEJMc2001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu C., et al. JAMA internal medicine; 2020. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shi S., et al. JAMA Cardiology; 2020. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patel A., Jernigan D.B. Initial public health response and interim clinical guidance for the 2019 novel coronavirus outbreak-United States, December 31, 2019-February 4, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:140–146. doi: 10.15585/mmwr.mm6905e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guieu J.D., Hellon R.F. The chill sensation in fever. Pflügers Archiv. 1980;384(1):103–104. doi: 10.1007/BF00589522. [DOI] [PubMed] [Google Scholar]
- 96.Ding Q., et al. The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J. Med. Virol. 2020 doi: 10.1002/jmv.25781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Conti B., et al. Cytokines and fever. Front. Biosci. 2004;9(12):1433–1449. doi: 10.2741/1341. [DOI] [PubMed] [Google Scholar]
- 98.Leon L.R. Invited review: cytokine regulation of fever: studies using gene knockout mice. J. Appl. Physiol. 2002;92(6):2648–2655. doi: 10.1152/japplphysiol.01005.2001. [DOI] [PubMed] [Google Scholar]
- 99.Mehta P., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eccles R. Mechanisms of symptoms of the common cold and influenza. Br. J. Hosp. Med. 2005;68(2):71–75. doi: 10.12968/hmed.2007.68.2.22824. 2007. [DOI] [PubMed] [Google Scholar]
- 101.Zolfaghari Emameh R., et al. Expansion of single cell transcriptomics data of SARS-CoV infection in human bronchial epithelial cells to COVID-19. Biol. Proced. Online. 2020;22(16) doi: 10.1186/s12575-020-00127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bautista E., et al. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N. Engl. J. Med. 2010;362(18):1708–1719. doi: 10.1056/NEJMra1000449. [DOI] [PubMed] [Google Scholar]
- 103.Iwasaki A., Pillai P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014;14(5):315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Unkel B., et al. Alveolar epithelial cells orchestrate DC function in murine viral pneumonia. J. Clin. Invest. 2012;122(10):3652–3664. doi: 10.1172/JCI62139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brandes M., et al. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154(1):197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sakai S., et al. Therapeutic effect of anti-macrophage inflammatory protein 2 antibody on influenza virus-induced pneumonia in mice. J. Virol. 2000;74(5):2472–2476. doi: 10.1128/jvi.74.5.2472-2476.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ichikawa A., et al. CXCL10-CXCR3 enhances the development of neutrophil-mediated fulminant lung injury of viral and nonviral origin. Am. J. Respir. Crit. Care Med. 2013;187(1):65–77. doi: 10.1164/rccm.201203-0508OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tate M.D., et al. The role of neutrophils during mild and severe influenza virus infections of mice. PloS One. 2011;6(3) doi: 10.1371/journal.pone.0017618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen X., et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Farnoosh G., et al. Are Iranian sulfur mustard-exposed survivors more vulnerable to SARS-CoV-2: some similarity in their pathogenesis. Disaster Med. Public Health Prep. 2020:1–12. doi: 10.1017/dmp.2020.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Magro G. Cytokine: X; 2020. SARS-CoV-2 and COVID-19: is interleukin-6 (IL-6) the’culprit lesion’of ARDS onset? What is there besides Tocilizumab? SGP130Fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Park M.D. Macrophages: a Trojan Horse in COVID-19? 2020. Nat. Rev. Immunol. 2020;20(351) doi: 10.1038/s41577-020-0317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Merekoulias G., et al. Lymphocyte to monocyte ratio as a screening tool for influenza. PLoS currents. 2010;2 doi: 10.1371/currents.rrn1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Criswell B.S., et al. The lymphocyte response to influenza in humans. Am. Rev. Respir. Dis. 1979;120(3):700–704. doi: 10.1164/arrd.1979.120.3.700. [DOI] [PubMed] [Google Scholar]
- 115.Dolin R., et al. Cell-mediated immune responses in humans after induced infection with influenza A virus. J. Infect. Dis. 1977;135(5):714–719. doi: 10.1093/infdis/135.5.714. [DOI] [PubMed] [Google Scholar]
- 116.Faguet G.B. The effect of killed influenza virus vaccine on the kinetics of normal human lymphocytes. J. Infect. Dis. 1981;143(2):252–258. doi: 10.1093/infdis/143.2.252. [DOI] [PubMed] [Google Scholar]
- 117.Thanunchai M., et al. Tropism of avian influenza A (H5N1) virus to mesenchymal stem cells and CD34+ hematopoietic stem cells. PloS One. 2013;8(12) doi: 10.1371/journal.pone.0081805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tan L., et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal transduction and targeted therapy. 2020;5(1):1–3. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liao Y.-C., et al. IL-19 induces production of IL-6 and TNF-α and results in cell apoptosis through TNF-α. J. Immunol. 2002;169(8):4288–4297. doi: 10.4049/jimmunol.169.8.4288. [DOI] [PubMed] [Google Scholar]
- 120.Giamarellos-Bourboulis E.J., et al. Cell host & microbe; 2020. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang Y., Tang H. Aberrant coagulation causes a hyper-inflammatory response in severe influenza pneumonia. Cell. Mol. Immunol. 2016;13(4):432–442. doi: 10.1038/cmi.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Keller T.T., et al. Effects on coagulation and fibrinolysis induced by influenza in mice with a reduced capacity to generate activated protein C and a deficiency in plasminogen activator inhibitor type 1. Circ. Res. 2006;99(11):1261–1269. doi: 10.1161/01.RES.0000250834.29108.1a. [DOI] [PubMed] [Google Scholar]
- 123.van der Sluijs K.F., et al. Toll-like receptor 4 is not involved in host defense against respiratory tract infection with Sendai virus. Immunol. Lett. 2003;89(2–3):201–206. doi: 10.1016/S0165-2478(03)00138-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van der Sluijs K.F., et al. IL-12 deficiency transiently improves viral clearance during the late phase of respiratory tract infection with influenza A virus in mice. Antivir. Res. 2006;70(2):75–84. doi: 10.1016/j.antiviral.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Levi M., van der Poll T. Two-way interactions between inflammation and coagulation. Trends Cardiovasc. Med. 2005;15(7):254–259. doi: 10.1016/j.tcm.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 126.Van der Poll T., et al. Elimination of interleukin 6 attenuates coagulation activation in experimental endotoxemia in chimpanzees. J. Exp. Med. 1994;179(4):1253–1259. doi: 10.1084/jem.179.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Levi M., et al. The cytokine‐mediated imbalance between coagulant and anticoagulant mechanisms in sepsis and endotoxaemia. Eur. J. Clin. Invest. 1997;27(1):3–9. doi: 10.1046/j.1365-2362.1997.570614.x. [DOI] [PubMed] [Google Scholar]
- 128.Tang N., et al. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemostasis. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.García-Sastre A., et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252(2):324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 131.Guilligay D., et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 2008;15(5):500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 132.Van de Sandt C.E., Kreijtz J.H., Rimmelzwaan G.F. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses. 2012;4(9):1438–1476. doi: 10.3390/v4091438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hu Y., et al. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 2017;91(8) doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Siu K.-L., et al. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3· TANK· TBK1/IKKε complex. J. Biol. Chem. 2009;284(24):16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Keam S., et al. Immunopathology and immunotherapeutic strategies in severe acute respiratory syndrome coronavirus 2 infection. Rev. Med. Virol. 2020 doi: 10.1002/rmv.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hu W., et al. SARS-CoV regulates immune function-related gene expression in human monocytic cells. Viral Immunol. 2012;25(4):277–288. doi: 10.1089/vim.2011.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Spiegel M., et al. Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J. Gen. Virol. 2006;87(7):1953–1960. doi: 10.1099/vir.0.81624-0. [DOI] [PubMed] [Google Scholar]
- 138.Abbasifard M., Khorramdelazad H. The Bio-mission of Interleukin-6 in the pathogenesis of COVID-19: a brief look at potential therapeutic tactics. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jafarzadeh A., et al. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Diao B., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu X., et al. Co-infection with SARS-CoV-2 and influenza A virus in patient with pneumonia, China. Emerg. Infect. Dis. 2020;26(6):1324. doi: 10.3201/eid2606.200299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Khodamoradi Z., Moghadami M., Lotfi M. 2020. Co-infection of Coronavirus Disease 2019 and Influenza: a Report from Iran. [DOI] [PubMed] [Google Scholar]
- 143.Cuadrado-Payán E., et al. SARS-CoV-2 and influenza virus co-infection. Lancet. 2020;395(10236):e84. doi: 10.1016/S0140-6736(20)31052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Konala V.M., et al. Co-infection with influenza A and COVID-19. European Journal of Case Reports in Internal Medicine. 2020;7(5) doi: 10.12890/2020_001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang D., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wehl G., Laible M., Rauchenzauner M. Klinische Pädiatrie; 2020. Co-infection of SARS CoV-2 and influenza A in a pediatric patient in Germany. [DOI] [PubMed] [Google Scholar]
- 147.Wang, G., et al., Is Co-Infection With Influenza Virus a Protective Factor of COVID-19? 2020.
- 148.Wang M., et al. medRxiv; 2020. Clinical Diagnosis of 8274 Samples with 2019-novel Coronavirus in Wuhan. [Google Scholar]
- 149.Zhu X., et al. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005. doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wu D., et al. Positive effects of COVID-19 control measures on influenza prevention. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Merckx J., et al. Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: a systematic review and meta-analysis. Ann. Intern. Med. 2017;167(6):394–409. doi: 10.7326/M17-0848. [DOI] [PubMed] [Google Scholar]
- 152.Bruning A.H., et al. Rapid tests for influenza, respiratory syncytial virus, and other respiratory viruses: a systematic review and meta-analysis. Clin. Infect. Dis. 2017;65(6):1026–1032. doi: 10.1093/cid/cix461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cheng M.P., et al. Diagnostic testing for severe acute respiratory syndrome–related coronavirus-2: a narrative review. Ann. Intern. Med. 2020 doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hussain M., et al. Drug resistance in influenza A virus: the epidemiology and management. Infect. Drug Resist. 2017;10:121. doi: 10.2147/IDR.S105473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Samson M., et al. Influenza virus resistance to neuraminidase inhibitors. Antivir. Res. 2013;98(2):174–185. doi: 10.1016/j.antiviral.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 156.Malosh R.E., et al. Efficacy and safety of oseltamivir in children: systematic review and individual patient data meta-analysis of randomized controlled trials. Clin. Infect. Dis. 2018;66(10):1492–1500. doi: 10.1093/cid/cix1040. [DOI] [PubMed] [Google Scholar]
- 157.Jing R., et al. 2020. Co-infection of COVID-19 and Influenza A in A Hemodialysis Patient: A Case Report. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang H., et al. BioRxiv; 2020. The Digestive System Is a Potential Route of 2019-nCov Infection: a Bioinformatics Analysis Based on Single-Cell Transcriptomes. [Google Scholar]
- 159.Xu X., et al. ChinaXiv preprint; 2020. Effective Treatment of Severe COVID-19 Patients with Tocilizumab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fu B., Xu X., Wei H. Why tocilizumab could be an effective treatment for severe COVID-19? J. Transl. Med. 2020;18(1):1–5. doi: 10.1186/s12967-020-02339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Toniati P., et al. Tocilizumab for the treatment of severe COVID-19 pneumonia with hyperinflammatory syndrome and acute respiratory failure: a single center study of 100 patients in Brescia, Italy. Autoimmun. Rev. 2020:102568. doi: 10.1016/j.autrev.2020.102568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Klopfenstein T., et al. Tocilizumab therapy reduced intensive care unit admissions and/or mortality in COVID-19 patients. Médecine et Maladies Infectieuses. 2020 doi: 10.1016/j.medmal.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Issa N., et al. Feasibility of tocilizumab in ICU patients with COVID‐19. J. Med. Virol. 2020 doi: 10.1002/jmv.26110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gritti G., et al. MedRxiv; 2020. Use of Siltuximab in Patients with COVID-19 Pneumonia Requiring Ventilatory Support. [Google Scholar]
- 165.Palanques-Pastor T., López-Briz E., Andrés J.L.P. Involvement of interleukin 6 in SARS-CoV-2 infection: siltuximab as a therapeutic option against COVID-19. Eur. J. Hosp. Pharm. 2020 doi: 10.1136/ejhpharm-2020-002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Bioscience trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 167.Colson P., et al. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020:105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Grein J., et al. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382(24):2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cao B., et al. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gautret P., et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 171.Information on Rapid Molecular Assays, RT-PCR, and Other Molecular Assays for Diagnosis of Influenza Virus Infection. 9.19. 2020. https://www.cdc.gov/flu/professionals/diagnosis/molecular-assays.htm#advantage-disadvantage Available from: [Google Scholar]
- 172.Rapid Influenza Diagnostic Tests. 9.19. 2020. https://www.cdc.gov/flu/professionals/diagnosis/clinician_guidance_ridt.htm Available from: [Google Scholar]
- 173.Overview of Influenza Testing Methods. 9.19.2020. https://www.cdc.gov/flu/professionals/diagnosis/overview-testing-methods.htm Available from:
- 174.CDC Diagnostic Tests for COVID-19. 9.19. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/testing.html Available from: [Google Scholar]
- 175.Kashiwagi S., et al. Safety of the long-acting neuraminidase inhibitor laninamivir octanoate hydrate in post-marketing surveillance. Int. J. Antimicrob. Agents. 2012;40(5):381–388. doi: 10.1016/j.ijantimicag.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 176.Influenza Antiviral Medications . 2020. Summary for Clinicians. 9.18.https://www.cdc.gov/flu/professionals/antivirals/summary-clinicians.htm Available from: [Google Scholar]
- 177.Guaraldi G., et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. The Lancet Rheumatology. 2020;2(8):e474–e484. doi: 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Atal S., Fatima Z. IL-6 inhibitors in the treatment of serious COVID-19: a promising therapy? Pharmaceut. Med. 2020:1–9. doi: 10.1007/s40290-020-00342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]