Summary
Understanding plant adaptive responses to the space environment is a requisite for enabling space farming. Spaceflight produces deleterious effects on plant cells, particularly affecting ribosome biogenesis, a complex stress-sensitive process coordinated with cell division and differentiation, known to be activated by red light. Here, in a series of ground studies, we have used mutants from the two Arabidopsis nucleolin genes (NUC1 and NUC2, nucleolar regulators of ribosome biogenesis) to better understand their role in adaptive response mechanisms to stress on Earth. Thus, we show that nucleolin stress-related gene NUC2 can compensate for the environmental stress provided by darkness in nuc1 plants, whereas nuc2 plants are not able to provide a complete response to red light. These ground control findings, as part of the ESA/NASA Seedling Growth spaceflight experiments, will determine the basis for the identification of genetic backgrounds enabling an adaptive advantage for plants in future space experiments.
Subject Areas: Plant Biology, Omics, Space Sciences, Arabidopsis, Ground controls, Nucleolus, Stress response, RNA-Seq, Spaceflight
Graphical Abstract
Highlights
-
•
Ribosome synthesis is a target of spaceflight stressor effects on plant development
-
•
Nucleolin mutants promote a differential response to light/darkness stress
-
•
Red light and NUC2 may counteract the spaceflight alterations in gene expression
-
•
Ground controls are important for the interpretation of spaceflight -omics experiments
Plant Biology; Omics; Space Sciences
Introduction
Space exploration will soon include new human missions to the Moon as a first step in the human exploration of Mars. Recent studies in human (i.e., NASA Twin Study, Garrett-Bakelman et al., 2019) and mammalian systems (rodent missions, Beheshti et al., 2019; Ronca et al., 2019) are paving the way to understand the effects of the microgravity environment on human physiology, but human life in space will rely on the essential role of plants in bioregenerative life support systems (Zabel et al., 2016), not only as a source of water, food, and removal of CO2 but also by providing a terrestrial-like environment for the psychological well-being of astronauts.
Due to their sessile condition, plants have to promote adaptive responses to cope with changes in environmental conditions. Light plays multiple roles in the mechanisms of these adaptive responses. On the one hand, light is the source of energy by means of photosynthesis and regulates indirectly cell proliferation and cell growth via the central regulator TOR kinase through the expression of S-phase genes, and also ribosome biogenesis (Caldana et al., 2013; Xiong et al., 2013; Sablowski and Carnier Dornelas, 2014). In addition, light is a major driver in the establishment of the patterns of plant growth and development, by means of phototropism and photomorphogenesis. In playing this role, light is associated with gravity as one of the major tropistic cues, such that gravitropism and phototropism (and the interaction between them) are essential modulators of plant development (Vandenbrink et al., 2014). At the cellular and molecular levels, plant development relies on the activity of cell growth and proliferation taking place in the meristems, which supply differentiated cells and are influenced by the tropistic cues (Perrot-Rechenmann, 2010).
Darkness is indeed a stress condition for plants. Several articles describe that in the dark, apical meristem proliferation is arrested in the G1 and G2 phases of the cell cycle (López-Juez et al., 2008; Mohammed et al., 2017), but in the root meristem, light induces the production of flavonols and other metabolites leading to the reduction of cell proliferation (Silva-Navas et al., 2016). This reduction could be related to the sugar starvation that results from the inability to perform photosynthesis.
However, red light has a stimulating effect on ribosome biogenesis and cell proliferation. An increase in the mitotic index, in the expression of some regulators of these processes at both gene expression and protein levels, and in post-translational modifications of some protein factors has been described in plants irradiated with red light (Tong et al., 1997; Reichler et al., 2001).
The absence of gravity (weightlessness, or microgravity, as it exists in free-fall, or in spaceflight) is also, by itself, a stress condition for plants, and specifically for the functions of meristematic cells (Matía et al., 2010; Boucheron-Dubuisson et al., 2016). An experiment performed in the International Space Station (ISS) in which Arabidopsis thaliana seedlings grew for 4 days in darkness resulted in an increase in the rate of cell proliferation and a decrease in the cell growth rate, estimated by the activity of ribosome biogenesis in the nucleolus, compared with the 1g control (Matía et al., 2010). As the coordination of these two activities defines meristematic competence, the effect of the lack of tropistic stimuli, particularly of gravity signals, may result in serious alterations of the developmental pattern of the plant, as was also shown in simulated microgravity experiments (Boucheron-Dubuisson et al., 2016).
The Seedling Growth (SG) experiments, recently performed in the ISS, aimed at unraveling the link between phototropism and gravitropism, using the weightless environment of spaceflight (Vandenbrink et al., 2019; Herranz et al., 2019). In the first SG experiment, A. thaliana seedlings corresponding to wild-type (WT) ecotype Landsberg erecta (Ler) and two phytochrome mutants (phyA and phyB) known to be involved in phototropism (Kiss et al., 2003; Molas and Kiss, 2008) were grown in space for 6 days and photostimulated for the last 2 days, revealing differential blue and red light phototropism in space (Vandenbrink et al., 2016). The analysis of the expression of regulatory genes of cell cycle and ribosome biogenesis showed that red light irradiation produced a significant reversion of the uncoupling of cell proliferation and cell growth caused by microgravity in darkness (Matía et al., 2010) and, consequently, a compensation of the loss of meristematic competence (Valbuena et al., 2018).
In view of these results from our previous space experiments, we decided to focus on the cellular process of ribosome biogenesis for the successive spaceflight experiments of the SG project, with the purpose of testing the separate and synergistic effects of the light and gravity tropistic signals on specific molecular and cellular components of this process. Ribosome biogenesis, which takes place in the nucleolus, represents the most complex multi-step process that the cell must perform and is one of the most intricately regulated and controlled (Sáez-Vásquez and Medina, 2008; Pelletier et al., 2018). Therefore, its regulation must adapt to the environmental conditions in which the cell finds itself and coordinate with other cellular processes, such as cell division and differentiation. Several studies have described and used the nucleolus as a major stress sensor, using stress-induced changes in the organization and composition of this organelle (Mayer and Grummt, 2005; Boulon et al., 2010; Lewinska et al., 2010; Kalinina et al., 2018).
Briefly, ribosome biogenesis consists of the transcription of 45S rRNA genes (45S rDNA) containing the sequences of 18S, 5.8S, and 25S rRNAs, followed by the multi-step cleavage of 45S pre-rRNA to produce, in association with 5S rRNA and ribosomal proteins (RPs), the mature ribosomal subunits, which are then exported to the cytoplasm and assembled as mature ribosomes.
In addition to RPs, hundreds of non-ribosomal proteins (NRPs), or nucleolar proteins and small nucleolar RNAs are required for ribosome biogenesis, playing regulatory roles (Sáez-Vásquez and Delseny, 2019). Among NRPs, nucleolin is the most abundant protein of the nucleolus, where it plays a key role in the different steps involved in ribosome biogenesis, including RNA polymerase (Pol) I transcription, processing of pre-rRNA (Ginisty et al., 1999; Roger et al., 2003), and assembly and nucleocytoplasmic transport of ribosome particles (Bouvet et al., 1998). Moreover, nucleolin has been even implicated in other functions, with or without collateral relationship with ribosome biogenesis (Angelov et al., 2006; Ma et al., 2007; Mongelard and Bouvet, 2007; Stepiński, 2012).
Animal and yeast genomes encode a single nucleolin gene, whereas plants offer various examples of gene multiplicity. In A. thaliana, two genes encoding nucleolin proteins have been described: NUC1 and NUC2 (Pontvianne et al., 2007, 2010). The NUC1 gene is highly and ubiquitously expressed in normal growth conditions and NUC1 protein is required to inhibit NUC2 gene expression at the transcriptional level and may also influence the accumulation of NUC2 protein. In contrast, NUC2 is a functional protein-coding gene developmentally controlled in most plant tissues and organs (Durut et al., 2014). NUC1 and NUC2 proteins localize in the nucleolus.
Disruption of NUC1 gene (nuc1-2 mutant) leads to severe defects in plant growth and development. At the molecular level, this mutant produces NUC2 expression, nucleolus disorganization, rDNA (NOR) heterochromatin decondensation, pre-rRNA accumulation, de-repression of specific rDNA variants, and de-methylation of some sequences of rDNA (Pontvianne et al., 2010).
In contrast, disruption of NUC2 gene (nuc2-2 mutant) has much weaker effects. The nuc2-2 mutant seedlings grow quite similarly to WT plants, but flowering occurs later. Knockout of the NUC2 gene induces expression of some rDNA variants and hypermethylation of some sequences of pre-rRNA. In addition, NUC2 is required for the stability of rDNA variants' copy number (Durut et al., 2014).
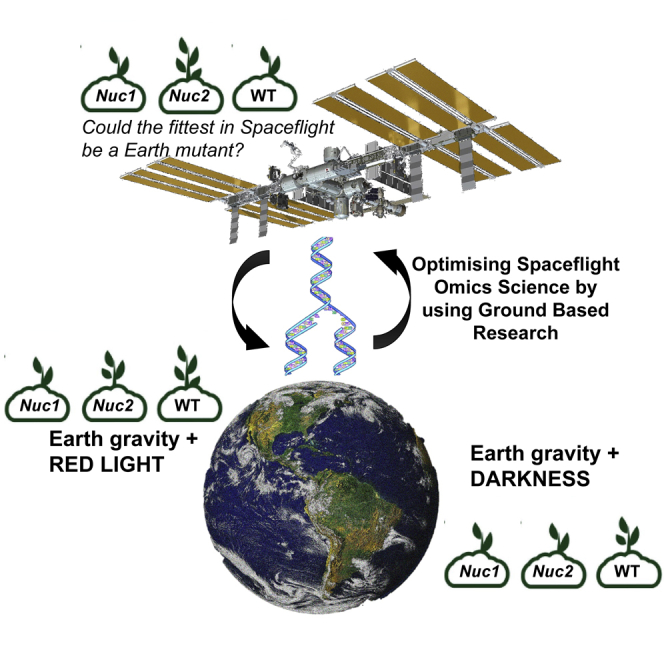
In this article, in a series of ground studies to complement space experiments, we have exposed A. thaliana (ecotype Columbia) WT, nuc1-2 and nuc2-2 seedlings to two different illumination regimes at the beginning of the plant development (6 days from germination). In addition to highlighting the importance of Earth reference controls in spaceflight -omics experiments, our goal was to better understand the responses of nucleolin mutants to red light stimulation for further uses in space experiments under altered gravity conditions. The differential adaptive responses to the light are shown, suggesting that stress-related mutants may show a reduced response to environmental stress. In the long term, these results may lead to more efficient agriculture if an exaggerated response may reduce plant development under suboptimal environmental conditions.
Results
Overall Transcriptional Profile Effects Confirm a Suitable Quality of Plant Material and Clustering of the Replicates
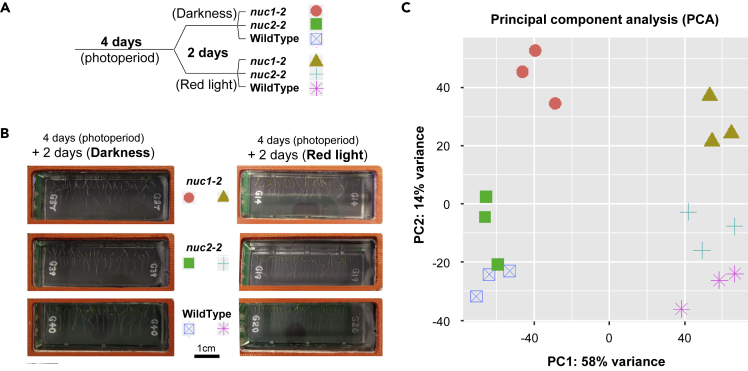
Two environmental conditions (light) are compared here, both of them as part of the SG Earth control reference experiment using the European Modular Cultivation System (EMCS). Seedlings were germinated in the same spaceflight hardware during 4 days with a day/night cycle of illumination, and then half of the samples were exposed to 2 days of darkness and the other group to 2 days of continuous red light (Figure 1A). After 6 days growth in the culture chamber (CC), the experiment was completed and the phenotypes of the three mutants were observed (Figure 1B, Supplemental Information, Figure S1). Seedling growth was homogeneous within each cassette, with clearly smaller seedlings in the case of the nuc1-2 mutant. No clear phenotypical differences were observed in the root system physiology between darkness- and red light-stimulated samples (including the absence of red phototropism at 1g conditions). The phenotype was compared with a previous experiment with the same duration and genotypes but in petri dishes and with a 6-day photoperiod illumination profile (Manzano et al., 2020 submitted). Despite the effects of the TROPI cassette volume, the overall phenotype of the seedlings is also very similar in both studies.
Figure 1.
The Seedling Growth 2 Ground Reference Experiment
(A) Experimental design including the illumination profile for each sample including color code used as key.
(B) Images of 6-day-old seedlings (WildType, nuc1-2, and nuc2-2, under the two illumination options) inside the CC just before collection for freezing (additional photos are provided as Supplemental InformationFigure S1). Scale bar, 1 cm (the gridded membrane has clearly defined grid lines spaced at 3.1 mm). The labels on the membrane represent the cassette # in the ground control.
(C) Principal-component analysis (PCA) of the 18 samples (three replicates per condition) using read counts data from FeatureCounts. This diagram gives an overview of the similarities and dissimilarities between samples and the experimental conditions' overall effects (see Data S1 for an RNA quality report on the red light samples). All replicates are consistently grouped according to their experimental conditions, and two clear PC1 (for illumination conditions during the last 2 days of growth: darkness and red light) and PC2 (for genetic background: WT, nuc1-2, and nuc2-2) are observed.
The quality of the replicates and overall similarities among the illumination regimes and different plants used is shown by principal-component analysis (PCA, Figure 1C and Supplemental Information, Data S1). First, a clear difference between the transcriptional profile of the WT, nuc1-2, and nuc2-2 plants exposed to darkness or red light photostimulation during the last 2 days of growth is shown by principal component 1 (PC1). Second, the differences between the genotypes can be observed in principal component 2 (PC2), where the nuc1-2 genotype has the most disrupted nucleolus phenotype, in that the nuc2-2 mutant is closer to the WT. The effect is clearer in the dark samples, because the red light stimulation produces more similarities among the three genotypes. Last, all the biological replicates included in the study cluster together according to the experimental condition, strengthening the statistical validity of the study. The use of three biological replicates (including up to 10 seedlings each) is enough considering the difficulties in increasing this number due to the availability of spaceflight research capabilities.
Global Effect of Red Light Photostimulation Differs in Each Arabidopsis thaliana Line
To understand the effect of the two lighting regimes (red light and darkness) on the transcriptional status, we compared seedlings with the differential illumination during the last 2 days of the plant growth period for each of the lines used (WT, nuc1-2, and nuc2-2).
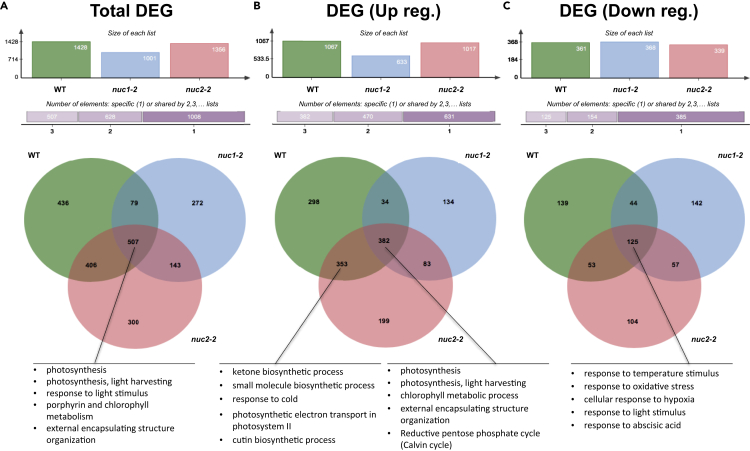
The number of differentially expressed genes (DEGs) in the WT is 1,428 genes, of which 1,067 have their expression activated and 361 have it repressed. In the nuc1-2 mutant, the total number of DEGs is lower (1,001), mainly at the expense of the up-regulated genes (633 are over-expressed and 368 are repressed). The number of DEGs from the comparison (red light versus darkness) in the nuc2-2 mutant is very similar as in the WT: a total of 1,017 up-regulated genes and 339 down-regulated genes, making a total of 1,365 DEGs (Figure 2).
Figure 2.
Differentially Expressed Genes (DEG p adj<0.05, Red Light Photostimulation versus Darkness) in WT and Nucleolin Mutants (nuc1-2 and nuc2-2)
(A) Venn diagram comparing all DEGs and the five most significant gene ontology (GO Enrichment) categories in the common DEG in the three genotypes.
(B) Venn diagram comparing up-regulated DEGs and the five most significant gene ontology (GO Enrichment) categories in the common DEGs in the WT and nuc2-2 genotypes and in the three genotypes, respectively.
(C) Venn diagram comparing down-regulated DEGs and the five most significant gene ontology (GO Enrichment) categories from common DEGs in the three genotypes.
Venn diagrams illustrate the group of DEGs that is affected in a single genotype or shared by others (Figure 2). Red light up-regulated genes affected only 298 genes in the WT, 134 genes in nuc1-2 mutant, and 199 genes for nuc2-2 mutant. On the other hand, the WT has 34 up-regulated genes in common with nuc1-2 but a large collection of 353 genes in common with nuc2-2 (note that the total number of up-regulated genes common for the three genotypes is 382), and only 83 genes for both nucleolin mutants (Figure 2B).
Among DEGs down-regulated by the red light compared with the dark, there is a similar number of unique DEGs for each of them; 139 genes for the WT, 142 genes for the nuc1-2 mutant, and 104 genes for the nuc2-2 mutant. In this case, the number of DEGs down-regulated in the three lines is 125 genes, without any of the pair comparisons reaching that level (the nuc1-2 and nuc2-2 mutants have 44 and 53 genes in common with WT, respectively, whereas the two nucleolin mutants share 57 down-regulated genes, Figure 2C).
In summary, the line with the least number of DEGs, when we compare plants illuminated with red light with those kept in darkness, is the nuc1-2 mutant. As the down-regulation response seems similar in all genotypes, the global effect is mainly due to a higher number of up-regulated DEGs in both nuc2-2 and WT genotypes. In fact, we can easily extrapolate from Figure 2B and compare the gene lists, including the common responses to red light (382 genes) and the list of up-regulated genes not detected in the nuc1-2 phenotype (353 genes). The main difference between these lists is shown by the functional analysis “GO Enrichment” included in Figure 2. All genotypes show the obvious photosynthesis/light-harvesting response together with increased Calvin cycle activity, but the nuc1-2 genotype is the only one lacking an increased secondary metabolism, together with a response to stress elements (cold) observed in both WT and nuc2-2. These results suggest that the presence of the NUC1 protein is necessary for the known effect of red light in the stimulation of cell proliferation, but the NUC2 protein is not an important player in that response.
A complementary functional analysis has been done using the full lists of up- or down-regulated genes in each genotype, obtained by means of a Heatmap GO Enrichment. Up-regulated gene lists reveal that red light photostimulation versus darkness mainly leads to an unequivocal activation of genes involved in different phases of photosynthesis, together with processes that involve external encapsulation, secondary metabolism, and drug catabolism in all three A. thaliana lines examined (Supplemental Information, Figure S2A). Other significantly affected categories are the GO involving cell wall modification, pathogenesis, responses to insects, and defense mechanisms to bacteria. In addition to the mentioned GO groups, red light activates different biosynthetic processes (ketone, cutin, and small molecules) and the response to stimuli, such as cold and UV light, in the WT and nuc2-2 mutant lines. Solely in the WT line, an enrichment in cell division-related genes (microtubule-based movement, cell cycle, meiotic cell cycle) is produced. In the case of down-regulated GO groups, red light photostimulation reduces expression of genes involved in the response to different stimuli (temperature, hypoxia, light, abscisic acid, oxidative stress) as well as the circadian rhythm of plants (Supplemental Information, Figure S2B). Additional down-regulated genes involved in the response to stress appear in WT and nuc2-2 comparison (heat and osmotic stress), in the WT and nuc1-2 comparison (cold acclimation, response to karrikin, hormone metabolism, and cell wall modification), or nuc1-2 only (response to gibberellin). The two nucleolin mutant lines share a repression of the amino acid metabolism (alpha-amino acid catabolic process and leucine degradation).
Dissecting Transcriptional Status for Each Genotype for Illumination Conditions at Normal Earth Gravity
To better understand the differences in the transcriptional status of each genotype that could be attributed to the illumination conditions designed to be used in the SG spaceflight experiment, we performed a series of comparisons between the different lines, namely, nuc1-2 versus WT, nuc2-2 versus WT, and nuc1-2 versus nuc2-2, when they are either photostimulated with red light or kept in darkness for 48 h, after a growth period of 4 days under a 16 h/8 h white light photoperiod regime (regular growth condition).
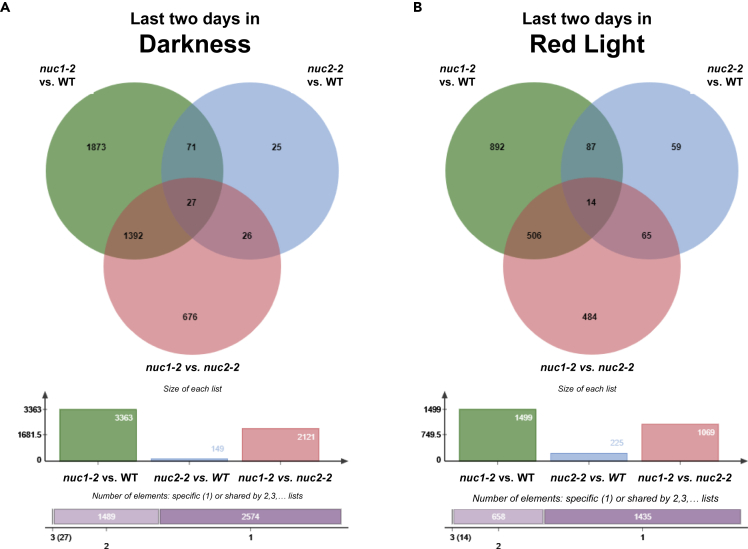
The total number of DEGs when comparing the severe mutant nuc1-2 with the WT is remarkable, with thousands of genes affected in the different illumination conditions. A similar result was obtained in the comparison of nuc1-2 with nuc2-2. This number was much higher in the comparisons of the nuc1-2 mutant with both the Col-0 and nuc2-2 mutant in darkness: 3,363 genes and 2,121 genes, respectively. Under red light stimulation, the numbers dropped to half: 1,499 genes compared with the WT and 1,069 compared with nuc2-2, respectively. In contrast, the small number of DEGs between the WT and nuc2-2 mild mutant (just 149 genes in darkness conditions) peaked to 225 when red light photostimulation was provided (Figure 3). The up- and down-regulated genes showed similar trends in this case (Supplemental Information, Figure S3).
Figure 3.
Differentially Expressed Genes (DEG p adj<0.05, in the Three Genotype Pair-Comparisons among WT, nuc1-2, and nuc2-2) within the Same Experimental Condition
(A) Venn diagram comparing all DEGs between plants exposed to darkness the last 2 days.
(B) Venn diagram comparing all DEGs between plants exposed to red light stimulation the last 2 days.
The differential numbers in DEGs can be assigned to particular GO groups. Ontology analysis shows that both comparisons involving the nuc1 mutant in the darkness produced an increase in the expression of genes mainly involved in cell division, such as cell cycle, meiotic cell cycle, microtubule cytoskeleton, nuclear chromosome segregation, cytokinesis, and cell cycle-G2/M transition (Supplemental Information, Figure S4A). Furthermore, in the nuc1-2 versus WT and nuc1-2 versus nuc2-2 comparisons, categories of responses to different stimuli, namely, responses to auxin, red light, gravity, and UV and ionizing radiations, also appeared overrepresented. In the comparison nuc1-2 versus nuc2-2 in darkness, most functional categories (GO terms) appearing up-regulated in the nuc1-2 mutant are related to the cell wall (cell wall macromolecule catabolic process, pectin biosynthetic process), the cuticle (cuticle development and cutin, suberin, and wax biosyntheses), and the plasma membrane (anchored component of plasma membrane, very-long-chain fatty acid biosynthetic process, transmembrane receptor protein kinase activity).
The up-regulated processes when red light is applied are quite different (Supplemental Information, Figure S4B). In the nuc1-2 versus WT and nuc1-2 versus nuc2-2 comparisons, DEGs appeared involved in cell wall (structural constituents of cell wall) and in development (positive regulation of growth, post-embryonic plant morphogenesis, cellular response to ethylene stimulus). In the nuc1-2 versus nuc2-2 comparison, many genes with a red-light-activated expression were involved in ribosome biogenesis (preribosome, maturation of SSU-rRNA, 90S preribosome, maturation of 5.8S RNA, ribosomal large subunit biogenesis).
The two nucleolin mutant lines relative to WT (nuc1-2 versus WT and nuc2-2 versus WT comparisons) in conditions of red light photoactivation have up-regulated genes involved in different response processes, such as the responses to hypoxia, drug, antibiotic, salicylic acid, and wounding. The number of response processes was increased in the nuc2-2 versus WT comparison with the GO categories: responses to cold, bacterium, and abscisic acid.
The identification of common gene categories down-regulated in both darkness and red light conditions was more challenging (Supplemental Information, Figures S4C and S4D). In darkness, nuc1-2 versus WT and nuc1-2 versus nuc2-2 comparisons were very similar and mainly related to developmental processes (regulation post-embryonic development, meristem development, regulation of seed development, plant organ senescence), mRNA quality (mRNA surveillance pathway), response to temperature (response to temperature stimulus, endopeptidase Clp complex, heat shock protein), and immune system (immune response, response to toxic substance, glutathione metabolism). In the comparisons indicated above, genes involved in long-day photoperiodism, flowering, rhythmic process, and response to light intensity were also less represented. In the same way, cellular responses to light stimulus category was over-represented in both nucleolin mutants with respect to WT and responses to light stimulus was a common GO category in all three comparisons. The gene categories repressed only in the nuc1-2 versus nuc2-2 comparison were involved in the spliceosome, protein demethylation, chromatin organization, mRNA binding, and histone acetyltransferase activity.
The gene categories with repressed expression under red light for any of the comparisons (nuc1-2 versus WT and nuc1-2 versus nuc2-2) were related to mRNA splicing (alternative mRNA splicing via spliceosome), light responses (response to light stimulus, photoperiodism flowering, circadian rhythm), photosynthesis (chlorophyll biosynthesis process, chloroplast envelope), and the immune system (immune system process, defense response to bacterium, regulation of salicylic acid metabolic process, Supplemental Information, Figure S4D).
Protein-Protein Interaction Networks Helps to Visualize the Differential Transcriptional State of nuc1-2 Genotype in 1g Control References
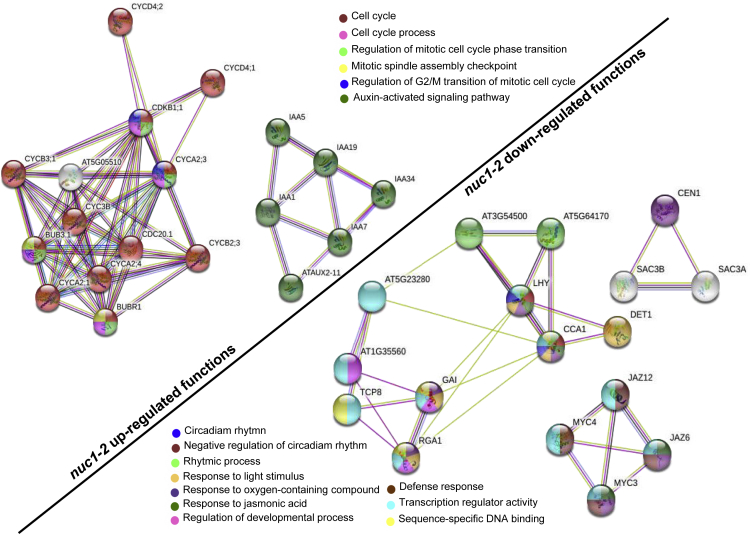
Enrichment-PPI analysis (protein-protein interaction network) also showed that, in darkness, the mutant nuc1-2 has up-regulated the expression of genes involved in the regulation of G2/M transition of the mitotic cell cycle, mitotic spindle assembly checkpoint, and auxin-activated signaling pathway, with respect to both the WT and the mutant nuc2-2 (Figure 4). In the down-regulation side, nuc1-2 mutant, when compared with WT, showed down-regulated genes involved in transcription regulator activity, sequence-specific DNA binding, defense response, and regulation of developmental process. In addition, the nuc1-2 mutant has genes with repressed expression in darkness condition related to negative regulation of circadian rhythm, responses to light stimulus, and responses to jasmonic acid, when compared with WT and nuc2-2 mutant (Figure 4). These results were in agreement with functional GO analysis (Supplemental Information, Figures S2 and S4).
Figure 4.
Protein-Protein Interaction Network of Nuc1-2 Differential Response under Darkness Conditions
Enrichment network visualization for results from the over-represented gene lists in Nuc1-2 versus the other genotypes. Gene in each node is represented by a chart indicating the functional category to which each one belongs (following the color legend). The networks show that processes such as cell cycle (left part or up-regulated genes), circadian rhythms, and stress responses (right part or down-regulated genes) are already affected in the 1×g control conditions as a reference for the Seedling Growth spaceflight experiments.
Discussion
Results from Spaceflight Experiments Can Have Several Limitations that Are Not Often Present in Other Biological -Omics Works
Major constraints of spaceflight experiments are the reduced amount of material and reproducibility, but other issues include the storage of samples before and after the experiment execution in space, causing staggered preparation and processing of the samples (Millar et al., 2010; Correll et al., 2013). In this work, we have shown that, using the spaceflight hardware and procedures, we can perform a transcriptomic analysis with enough reliability to describe the differential transcriptional state of the nucleolin mutants under different light conditions. Three replicates could be considered low for current standards for animal -omics studies in Earth, but in our experiment each replica already represents the average of 10 plants. We show here that the replicates expected to be collected from the spaceflight experiment can be estimated to be sufficient for performing a sequencing study that allows clustering of the different genotypes and environmental conditions (Figure 1).
Due to logistical considerations, a specific constraint for plant space biology is the limited number of different mutants/genotypes that can be used in a spaceflight experiment in true microgravity. We have to be sure that the mutants of choice will provide valuable information. In that sense, the use of nucleolin mutants is a promising choice to provide insight into both the cell cycle and stress response mechanisms, apart from the ribosome biogenesis in which this protein is directly involved. These processes are known to be recurrently affected in space -omics experiments with plants (Choi et al., 2019; Ferl et al., 2015; Herranz et al., 2019; Johnson et al., 2017; Kruse et al., 2017; Paul et al., 2013, 2017). The analysis performed here allows us to know the specific functions assumed by the two nucleolin proteins of A. thaliana (NUC1 and NUC2) in different environmental conditions, such as red light photostimulation and darkness, affecting tropistic stimuli. This information can be considered as the 1g reference control required to obtain the best possible understanding of the changes produced by the spaceflight conditions on plant development, and how the gravitational stress could be counteracted by changing phototropic stimuli. A longer-term goal is to use this knowledge for plant cultivation into bioregenerative life support systems.
The Transcriptomic Baseline for Nucleolin Mutants Is Different under Red Light Photostimulation and Darkness
Red light illumination mainly produces the activation of genes involved in photosynthesis, cell wall modification, drug catabolism, pathogenesis, and biotic stress response. Conversely, different abiotic stimuli responses (temperature, hypoxia, light, abscisic acid, oxidative stress) are down-regulated by the photostimulation treatment, as well as the circadian rhythm of plants. This transcriptional effect is similar in the three lines studied (Col-0 WT, nuc1-2, and nuc2-2), suggesting that it may be independent of the nucleolin protein functions. Therefore, the interpretation of transcriptional effects on plants' grown during spaceflight will be straightforward. In contrast, red light illumination produces a down-regulation of genes involved in the response to karritin, cold acclimation, and hormone metabolism in WT and nuc1-2, indicating that the NUC2 protein, and/or other proteins up-regulated in nuc1-2 (Pontvianne et al., 2007), could participate in these functions. In addition, in the nuc1-2 mutant, genes coding for response to gibberellin with the red light appear repressed. Gibberellin is a hormone involved in seed germination, bud and fruit formation, shoot longitudinal growth, and axial organ elongation (Hedden and Sponsel, 2015), which could support the relationship of defective plant growth and development with NUC1 gene disruption.
Exposure to the dark during the last 2 days of cultivation of seedlings promotes gene categories mainly involved in processes of cell division and in the response to different stimuli (red light, UV, ionizing radiation, gravity, auxins). They are common in nuc1-2 versus WT and nuc1-2 versus nuc2-2 comparisons and unique in nuc1-2 versus WT comparison. These observations could indicate that, in a stress condition such as darkness, the NUC2 protein is capable of rescuing functions that the NUC1 protein performs under normal growth conditions. Indeed, a role of nucleolin in mitosis has been reported (de Carcer and Medina, 1999; Ma et al., 2007). Therefore, in the nuc1 mutant, the NUC2 protein would be capable of performing functions necessary for plant survival, such as ribosome biogenesis regulation (Pontvianne et al., 2007), cell proliferation, and DNA repair. It is important to note that cell division GO was only significantly up represented in the WT when red light was provided (Figure 2B). In contrast, the gene categories down-regulated in darkness are mainly related to developmental processes, mRNA quality, response to temperature, and immune system. This may indicate that in the dark stress condition, the NUC2 protein is not capable of supplying the function of the NUC1 protein in these processes. These results, together with those previously described, indicate that the functional rescue of NUC1 by NUC2, when NUC1 is not expressed, would be only partial, which may be due to the structural difference between both proteins, including longer N-terminal acidic domain and less-conserved GAR domain I in the C terminal (Durut and Sáez-Vásquez, 2015).
Differential Role of Nucleolin Mutants' Transcriptomic Baseline May Offer New Insight in Spaceflight Experiments
Differential transcriptional response is observed when comparing the two nucleolin mutants. The categories of genes activated differentially in both mutants (nuc1-2 versus nuc2-2 comparison) are mainly involved in the cell wall and membrane systems. This finding is potentially interesting in space -omics to discriminate gravitropism from graviresistance mechanisms (Herranz and Medina, 2014) and could be related to the phenotype described in the nuc1-2 mutant, in which a reduction in the cell number and a disorganization in every cell layer is observed in transversal sections of primary leaves (Pontvianne et al., 2007). Moreover, the gene categories repressed only in the above-mentioned comparison are involved in the spliceosome, protein demethylation, chromatin organization, mRNA binding, and histone acetyltransferase activity, which could be related to the described antagonist activity of these two proteins in pre-rRNA methylation and with their role in chromatin remodeling, RNA Pol I transcription, mRNA stability, and RNA/DNA metabolism (Pontvianne et al., 2010; Durut et al., 2014).
In the nuc1-2 versus nuc2-2 comparison under red light photostimulation, the ribosome biogenesis GO shows the most significant enrichment within the up-regulated genes (Supplemental Information, Figure S4B). This result indicates that the described activating effect of red light in this process depends more on the expression of NUC2 protein than on the NUC1 expression. Ultimately, the two nucleolin mutant lines, compared with WT, have genes up-regulated by red light involved in different response processes, such as the response to hypoxia, drug, antibiotic, salicylic acid, and wounding. These response processes are extended in the nuc2-2 versus WT comparison, including response to cold, bacteria, and abscisic acid. These results indicate that even though both nucleolar proteins, NUC1 and NUC2, are involved in response processes, the implication of NUC2 is higher as to the number of processes. Therefore, NUC2 has a greater response capacity to environmental signals, such as illumination with red light, thus being more sensitive.
A major challenge to be overcome in spaceflight -omics research is to differentiate and characterize which of the spaceflight environmental conditions (including not only microgravity and cosmic radiation but also the constraints imposed to the upload/download and storage processes in orbit, the gaseous atmosphere, among other factors) are responsible for each of the observed alterations in the transcriptome. According to our results, space -omics researchers need to pay special attention to the analysis of ground reference controls on Earth before conclusions can be made on the results obtained in spaceflight. In our particular example, the two nucleolin mutants are the perfect complement to the WT genotype to study how the NUC2 protein can replace the essential functions of NUC1 in orbit, and how light can modulate it, even rescuing WT transcriptional profiles in the nuc1-2 mutant. The fact that nuc2-2 mutant shows an increase in stress response GO when exposed to red light connects the red light stimulus with the modulation of the stress response. In parallel, the cell cycle and ribosome biogenesis functions required to be restored in microgravity are also light dependent in both the nuc1-2 mutant (in Earth, as shown here) and the WT plants in orbit (Valbuena et al., 2018).
Limitations of the Study
Due to logistical constraints, this study lacks a continuously illuminated control set of samples performed simultaneously due to the absence of those samples in the spaceflight experiment as well as the reference experiment. The next step in this research is to analyze the samples from the space experiment in microgravity and the partial gravity conditions produced by the centrifuge in the EMCS on the ISS. We anticipate confirming that the duplicated nucleolin gene system works during spaceflight will lead us to discover novel mechanisms for plant adaptation to spaceflight conditions. Thus, this research will be eventually translated into better crops for life support systems in the human spaceflight ventures of the 21st century.
Resource Availability
Lead Contact
Further information and requests for materials should be directed to and will be fulfilled by the lead contact, Raúl Herranz (r.herranz@csic.es)
Materials Availability
Materials generated in this study are available from the lead contact with a completed materials transfer agreement.
Data and Code Availability
The GLDS-313 datasets generated during this study have been deposited and it is available at GENELAB repository (Ray et al., 2019). Original data have been deposited to (GENELAB: https://doi.org/10.26030/0g0m-dj21, https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-313)
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We want to acknowledge the collaboration and support of many people who have contributed to the success of the experiments of the “Seedling Growth” project whose results are reported in this paper. This includes payload developers (Airbus), ESA and NASA managers, scientific collaborators (Drs. Eugenie Carnero-Diaz, Richard E. Edelmann, Joshua P. Vandenbrink, Malgorzata Ciska, Katherine D.L. Millar, and Miguel Angel Valbuena), and the astronauts who performed the experiments on board the International Space Station as well as the EMCS managers (N-USOC) who performed the ground control operations. This work was supported by the Agencia Estatal de Investigación of the Spanish Ministry of Science and Innovation, Grants #ESP2015-64323-R and #RTI2018-099309-B-I00 (co-funded by EU-ERDF) to F.J.M., by pre-doctoral fellowships to A.M. and A.V. from the Spanish National Program for Young Researchers Training (MINECO, Ref. BES-2013-063933, BES-2016-077976), and the Seedling Growth Project to the ISS LSRA2009-0932/1177, a shared project of ESA-ELIPS Program and NASA. J.Z.K. is funded by Grants NNX12A065G and 80NSSC17K0546. These results are related to the Space Omics TT funded by the European Space Agency contract ESA 4000131202/20/NL/PG to R.H.
Author Contributions
Conceptualization, R.H. and F.J.M.; Methodology and Investigation, A.V., A.M., and R.H.; Writing – Original Draft, A.M. and R.H.; Writing – Review & Editing, A.V., F.J.M., J.Z.K., and J.S.-V.; Supervision and Funding Acquisition, F.J.M., J.Z.K., and J.S.-V.
Declaration of Interests
The authors declare no competing interests.
Published: November 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101686.
Supplemental Information
GO terms that show a significant enrichment (log q-value < 0.05) for any of the DEG lists that appear in the main article (Figure 2) and the Supplemental Information (Figures S1 and S3) are provided.
References
- Angelov D., Bondarenko V.A., Almagro S., Menoni H., Mongélard F., Hans F., Mietton F., Studitsky V.M., Hamiche A., Dimitrov S., Bouvet P. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beheshti A., Shirazi-Fard Y., Choi S., Berrios D., Gebre S.G., Galazka J.M., Costes S.V. Exploring the effects of spaceflight on mouse physiology using the open access NASA genelab platform. J. Vis. Exp. 2019;143:1–11. doi: 10.3791/58447. [DOI] [PubMed] [Google Scholar]
- Boucheron-Dubuisson E., Manzano A.I., Le Disquet I., Matia I., Saez-Vasquez J., van Loon J.J., Herranz R., Carnero-Diaz E., Medina F.J. Functional alterations of root meristematic cells of Arabidopsis thaliana induced by a simulated microgravity environment. J. Plant Physiol. 2016;207:30–41. doi: 10.1016/j.jplph.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P., Diaz J.J., Kindbeiter K., Madjar J.J., Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- de Carcer G., Medina F.J. Simultaneous localization of transcription and early processing markers allows dissection of functional domains in the plant cell nucleolus. J. Struct. Biol. 1999;128:139–151. doi: 10.1006/jsbi.1999.4187. [DOI] [PubMed] [Google Scholar]
- Caldana C., Li Y., Leisse A., Zhang Y., Bartholomaeus L., Fernie A.R., Willmitzer L., Giavalisco P. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 2013;73:897–909. doi: 10.1111/tpj.12080. [DOI] [PubMed] [Google Scholar]
- Choi W.G., Barker R.J., Kim S.H., Swanson S.J., Gilroy S. Variation in the transcriptome of different ecotypes of Arabidopsis thaliana reveals signatures of oxidative stress in plant responses to spaceflight. Am. J. Bot. 2019;106:123–136. doi: 10.1002/ajb2.1223. [DOI] [PubMed] [Google Scholar]
- Correll M.J., Pyle T.P., Millar K.D., Sun Y., Yao J., Edelmann R.E., Kiss J.Z. Transcriptome analyses of Arabidopsis thaliana seedlings grown in space: implications for gravity-responsive genes. Planta. 2013;238:519–533. doi: 10.1007/s00425-013-1909-x. [DOI] [PubMed] [Google Scholar]
- Durut N., Abou-Ellail M., Pontvianne F., Das S., Kojima H., Ukai S., de Bures A., Comella P., Nidelet S., Rialle S. A duplicated NUCLEOLIN gene with antagonistic activity is required for chromatin organization of silent 45S rDNA in Arabidopsis. Plant Cell. 2014;26:1330–1344. doi: 10.1105/tpc.114.123893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durut N., Sáez-Vásquez J. Nucleolin: dual roles in rDNA chromatin transcription. Gene. 2015;556:7–12. doi: 10.1016/j.gene.2014.09.023. [DOI] [PubMed] [Google Scholar]
- Ferl R.J., Koh J., Denison F., Paul A.L. Spaceflight induces specific alterations in the proteomes of Arabidopsis. Astrobiology. 2015;15:32–56. doi: 10.1089/ast.2014.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Bakelman F.E., Darshi M., Green S.J., Gur R.C., Lin L., Macias B.R., McKenna M.J., Meydan C., Mishra T., Nasrini J. The NASA twins study: a multidimensional analysis of a year-long human spaceflight. Science. 2019;364 doi: 10.1126/science.aau8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H., Sicard H., Roger B., Bouvet P. Structure and functions of nucleolin. J. Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Hedden P., Sponsel V. A century of gibberellin research. J. Plant Growth Regul. 2015;34:740–760. doi: 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz R., Vandenbrink J.P., Villacampa A., Manzano A., Poehlman W.L., Feltus F.A., Kiss J.Z., Medina F.J. RNAseq analysis of the response of Arabidopsis thaliana to fractional gravity under blue-light stimulation during spaceflight. Front. Plant Sci. 2019;10:1–11. doi: 10.3389/fpls.2019.01529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz R., Medina F.J. Cell proliferation and plant development under novel altered gravity environments. Plant Biol. 2014;16:23–30. doi: 10.1111/plb.12103. [DOI] [PubMed] [Google Scholar]
- Johnson C.M., Subramanian A., Pattahil S., Correll M.J., Kiss J.Z. Comparative transcriptomics indicate changes in cell wall organization and stress response in seedlings during spaceflight. Am. J. Bot. 2017;104:1219–1231. doi: 10.3732/ajb.1700079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina N.O., Makarova S., Makhotenko A., Love A.J., Taliansky M. The multiple functions of the nucleolus in plant development, disease and stress responses. Front. Plant Sci. 2018;9:1–19. doi: 10.3389/fpls.2018.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss J.Z., Mullen J.L., Correll M.J., Hangarter R.P. Phytochromes A and B mediate red-light-induced positive phototropism in roots. Plant Physiol. 2003;131:1411–1417. doi: 10.1104/pp.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse C.P.S., Basu P., Luesse D.R., Wyatt S.E. Transcriptome and proteome responses in RNAlater preserved tissue of Arabidopsis thaliana. PLoS One. 2017;12:1–10. doi: 10.1371/journal.pone.0175943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinska A., Wnuk M., Grzelak A., Bartosz G. Nucleolus as an oxidative stress sensor in the yeast Saccharomyces cerevisiae. Redox Rep. 2010;15:87–96. doi: 10.1179/174329210X12650506623366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E., Dillon E., Magyar Z., Khan S., Hazeldine S., de Jager S.M., Murray J.A.H., Beemster G.T.S., Bögre L., Shanahan H. Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell. 2008;20:947–968. doi: 10.1105/tpc.107.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Matsunaga S., Takata H., Ono-Maniwa R., Uchiyama S., Fukui K. Nucleolin functions in nucleolus formation and chromosome congression. J. Cell Sci. 2007;120:2091–2105. doi: 10.1242/jcs.008771. [DOI] [PubMed] [Google Scholar]
- Manzano A., Pereda-Loth V., de Bures A., Sáez-Vásquez J., Herranz R., Medina F.J. Interaction of light and gravity signals as a mechanism of counteracting alterations caused by simulated microgravity in proliferating plant cells. Am. J. Bot. 2020 doi: 10.21203/rs.3.rs-29236/v1. Under review. [DOI] [PubMed] [Google Scholar]
- Matía I., González-Camacho F., Herranz R., Kiss J.K., Gasset G., van Loon J.J.W.A., Marco R., Medina F.J. Plant cell proliferation and growth are altered by microgravity conditions in spaceflight. J. Plant Physiol. 2010;167:184–193. doi: 10.1016/j.jplph.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Mayer C., Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2005;4:1036–1038. doi: 10.4161/cc.4.8.1925. [DOI] [PubMed] [Google Scholar]
- Millar K.D., Kumar P., Correll M.J., Mullen J.L., Hangarter R.P., Edelmann R.E., Kiss J.Z. A novel phototropic response to red light is revealed in microgravity. New Phytol. 2010;186:648–656. doi: 10.1111/j.1469-8137.2010.03211.x. [DOI] [PubMed] [Google Scholar]
- Mohammed B., Bilooei S.F., Dóczi R., Grove E., Railo S., Palme K., Ditengou F.A., Bögre L., López-Juez E. Converging energy and hormonal signalling control meristem activity, leaf initiation and growth. Plant Physiol. 2017;176:1365–1381. doi: 10.1104/pp.17.01730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molas M.L., Kiss J.Z. PKS1 plays a role in red-light-based positive phototropism in roots. Plant Cell Environ. 2008;31:842–849. doi: 10.1111/j.1365-3040.2008.01797.x. [DOI] [PubMed] [Google Scholar]
- Mongelard F., Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Paul A.L., Zupanska A.K., Schultz E.R., Ferl R.J. Organ-specific remodeling of the Arabidopsis transcriptome in response to spaceflight. BMC Plant Biol. 2013;13:112. doi: 10.1186/1471-2229-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A.L., Sng N.J., Zupanska A.K., Krishnamurthy A., Schultz E.R., Ferl R.J. Genetic dissection of the Arabidopsis spaceflight transcriptome: are some responses dispensable for the physiological adaptation of plants to spaceflight? PLoS One. 2017;12:1–24. doi: 10.1371/journal.pone.0180186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., Thomas G., Volarević S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat. Rev. Cancer. 2018;18:51–63. doi: 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. Cellular responses to auxin: division versus expansion. Cold Spring Harb. Perspect. Biol. 2010;2:1–15. doi: 10.1101/cshperspect.a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Matía I., Douet J., Tourmente S., Medina F.J., Echeverria M., Sáez-Vásquez J. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol. Biol. Cell. 2007;18:369–379. doi: 10.1091/mbc.E06-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Abou-Ellail M., Douet J., Comella P., Matía I., Chandrasekhara C., DeBures A., Blevins T., Cooke R., Medina F.J. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 2010;6:1–13. doi: 10.1371/journal.pgen.1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S., Gebre S., Fogle H., Berrios D.C., Tran P.B., Galazka J.M., Coste S.V. GeneLab: omics database for spaceflight experiments. Bioinformatics. 2019;35:1753–1759. doi: 10.1093/bioinformatics/bty884. [DOI] [PubMed] [Google Scholar]
- Reichler S.A., Balk J., Brown M.E., Woodruff K., Clark G.B., Roux S.J. Light differentially regulates cell division and the mRNA abundance of pea nucleolin during de-etiolation. Plant Physiol. 2001;125:339–350. doi: 10.1104/pp.125.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger B., Moisand A., Amalric F., Bouvet P. Nucleolin provides a link between RNA polymerase I transcription and pre-ribosome assembly. Chromosoma. 2003;111:399–407. doi: 10.1007/s00412-002-0221-5. [DOI] [PubMed] [Google Scholar]
- Ronca A.E., Moyer E.L., Talyansky Y., Lowe M., Padmanabhan S., Choi S., Gong C., Cadena S.M., Stodieck L., Globus R.K. Behavior of mice aboard the international space station. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-40789-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R., Carnier Dornelas M. Interplay between cell growth and cell cycle in plants. J. Exp. Bot. 2014;65:2703–2714. doi: 10.1093/jxb/ert354. [DOI] [PubMed] [Google Scholar]
- Sáez-Vásquez J., Delseny M. Ribosome biogenesis in plants: from functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell. 2019;31:1945–1967. doi: 10.1105/tpc.18.00874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Vásquez J., Medina F.J. The plant nucleolus. Adv. Bot. Res. 2008;47:1–46. [Google Scholar]
- Silva-Navas J., Moreno-Risueño M.A., Manzano C., Téllez-Robledo B., Navarro-Neila S., Carrasco V., Pollmann S., Gallego F.J., del Pozo J.C. Flavonols mediate root phototropism and growth through regulation of proliferation-to-differentiation transition. Plant Cell. 2016;28:1372–1387. doi: 10.1105/tpc.15.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepiński D. Nucleolin level in plant root meristematic cells under chilling stress and recovery. Micron. 2012;43:870–875. doi: 10.1016/j.micron.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Tong C.G., Reichler S., Blumentahal S., Balk J., Hsieh H.L., Roux S.J. Light regulation of the abundance of mRNA encoding a nucleolin-like protein localized in the nucleoli of pea nuclei. Plant Physiol. 1997;114:643–652. doi: 10.1104/pp.114.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valbuena M.A., Manzano A., Vandenbrink J.P., Pereda-Loth V., Carnero-Diaz E., Edelmann R.E., Kiss J.Z., Herranz R., Medina F.J. The combined effects of real or simulated microgravity and red-light photoactivation on plant root meristematic cells. Planta. 2018;248:691–704. doi: 10.1007/s00425-018-2930-x. [DOI] [PubMed] [Google Scholar]
- Vandenbrink J.P., Kiss J.Z., Herranz R., Medina F.J. Light and gravity signals synergize in modulating plant development. Front. Plant Sci. 2014;5:1–18. doi: 10.3389/fpls.2014.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbrink J.P., Herranz R., Medina F.J., Edelmann R.E., Kiss J.Z. A novel blue-light phototropic response is revealed in roots of Arabidopsis thaliana in microgravity. Planta. 2016;244:1201–1215. doi: 10.1007/s00425-016-2581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbrink J.P., Herranz R., Poehlman W., Feltus F.A., Villacampa A., Ciska M., Medina F.J., Kiss J.Z. RNA-seq analyses of Arabidopsis thaliana seedlings after exposure to blue-light phototropic stimuli in microgravity. Am. J. Bot. 2019;106:1466–1476. doi: 10.1002/ajb2.1384. [DOI] [PubMed] [Google Scholar]
- Xiong Y., McCormack M., Li L., Hall Q., Xiang C., Sheen J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496:181–186. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel P., Bamsey M., Schubert D., Tajmar M. Review and analysis of over 40 years of space plant growth systems. Life Sci. Space Res. 2016;10:1–16. doi: 10.1016/j.lssr.2016.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GO terms that show a significant enrichment (log q-value < 0.05) for any of the DEG lists that appear in the main article (Figure 2) and the Supplemental Information (Figures S1 and S3) are provided.
Data Availability Statement
The GLDS-313 datasets generated during this study have been deposited and it is available at GENELAB repository (Ray et al., 2019). Original data have been deposited to (GENELAB: https://doi.org/10.26030/0g0m-dj21, https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-313)