Summary
An essential event in gene regulation is the binding of a transcription factor (TF) to its target DNA. Models considering the interactions between the TF and the DNA geometry proved to be successful approaches to describe this binding event, while conserving data interpretability. However, a direct characterization of the DNA shape contribution to binding is still missing due to the lack of accurate and large-scale binding affinity data. Here, we use a binding assay we recently established to measure with high sensitivity the binding specificities of 13 Drosophila TFs, including dinucleotide dependencies to capture non-independent amino acid-base interactions. Correlating the binding affinities with all DNA shape features, we find that shape readout is widely used by these factors. A shape readout/TF-DNA complex structure analysis validates our approach while providing biological insights such as positively charged or highly polar amino acids often contact nucleotides that exhibit strong shape readout.
Subject Areas: Biomolecules, Molecular Biology, Molecular Mechanism of Gene Regulation
Graphical Abstract

Highlights
-
•
The DNA shape contribution to Drosophila TFs-DNA binding is directly characterized
-
•
Zeroth- and first-order TF-DNA binding specificities are measured with high accuracy
-
•
DNA shape readout is widely used by these TFs
-
•
A shape readout/structural correlation analysis provides biological insights
Biomolecules; Molecular Biology; Molecular Mechanism of Gene Regulation
Introduction
The binding of transcription factors (TFs) to specific DNA sequences is a key event for the regulation of gene expression. The features defining a binding site have been the focus of several decades of research starting from simple consensus motif binding sites, later replaced by probabilistic models of TF binding assuming that each base contributes independently to the overall affinity, the so-called position-specific weight matrices (PWMs) (Stormo et al., 1982). With the advent of high-throughput methods, binding specificities became available for thousands of TFs and it has become clear that more complex models for binding sites using non-independent nucleotide interactions lead to more accurate predictions than PWMs (Weirauch et al., 2013; Zhao and Stormo, 2011). Nucleotide correlations can originate from amino acids that contact multiple bases simultaneously or from stacking interactions that determine binding through DNA shape readout. Hence, although determining binding specificities is crucial to predict binding sites in the genome, such data alone are not sufficient to fully describe TF-DNA binding interactions as they do not provide insights about the mechanism the TF employs to bind to different DNA sequences. To elucidate how the TF “reads” the DNA is of paramount importance not only to improve algorithms predicting binding sites but also to refine our fundamental understanding of how TFs are recruited to specific DNA regulatory sequences.
To date, two distinct modes of protein-DNA recognition are known: base readout, which reflects the interplay at nucleobase-amino acid contacts mainly driven by the formation of hydrogen bonds, and shape readout, dominated by van der Waals interactions and electrostatic potentials (EPs), that recognizes the 3D structure of the DNA double helix. As a consequence, one can assume that, if the TF uses the shape readout, models incorporating DNA structural information should improve prediction of TF-DNA binding specificities. To test this hypothesis and thereby help model development, it would thus be highly desirable to (1) determine accurately TF-DNA binding specificities, including non-independent nucleotide interactions since deviations from linear binding can carry information about the influence of DNA shape, and (2) use these data to assess the contribution of DNA shape readout to the binding interaction.
Despite the availability of techniques able to measure protein-DNA interactions at high throughput such as protein binding microarray (PBM) (Berger et al., 2006), SELEX-seq (Rastogi et al., 2018) (Riley et al., 2014), and SMiLE-seq (Isakova et al., 2017), the accurate measurement of binding affinities remains problematic. Moreover, these methods require a resin- or filter-based selection step that introduces bias and/or use stringent washing protocols resulting in the loss of weak binders, which can lead to erroneously over-specific binding specificities (Jung et al., 2018). These limitations are critical, especially to determine higher-order binding interactions, which are intrinsically weak (Maerkl and Quake, 2007; Nutiu et al., 2011).
Evaluating the contribution to binding of DNA shape readout also poses challenges. First, although it had been known for a long time from crystal structures that TFs read out the DNA shape (see (Rohs et al., 2010) for review), it is still not possible to determine experimentally the DNA shape features at a large scale for any given DNA sequence. However, this would be necessary to quantitatively assess DNA shape influence on TF-DNA binding. This issue has been tackled by Zhou et al. who introduced “DNAShape” (Zhou et al., 2013), an algorithm that predicts structural DNA features from nucleotide sequences, considering at each DNA position a local 5-mers nucleotide environment. The original set of four geometric shape features was later completed by Li et al. (Li et al., 2017), who made tables available to calculate an expanded repertoire of 13 DNA shape features in total. Finally, Chiu et al. (Chiu et al., 2017) added in a comparable fashion the EP, which approximates the minor-groove EPs. The EP reflects the mean charge density of the DNA backbone sensed by positively charged amino acid residues of the binding protein.
Another difficulty to analyze the influence of DNA shape to binding is that, in spite of all the advances made possible by “DNAShape” and the succeeding studies, it is still not clear to what degree shape readout can be described as a function of the underling DNA sequence. It is indeed very difficult to tease apart whether a binding protein favors a given nucleotide sequence because it recognizes certain amino acids of this sequence or rather certain shapes features of the DNA helix. An important step was made with homeodomain TFs by Abe et al. (Abe et al., 2015), who were able to specifically remove the ability of the binding proteins to read a certain structural feature of DNA and to switch between different modes of DNA shape readouts. Another approach computationally dissects TF binding specificity in terms of base and shape readout (Rube et al., 2018). Remarkably, the authors determined that 92-99% of the variance in the shape features can be explained with a model considering only dinucleotides dependencies. They also found that interactions were much stronger between neighboring nucleotides than for non-adjacent positions, indicating that these dinucleotide features are the most important for binding. Hence, determining neighboring dinucleotide dependencies should be enough to capture most on the higher-order binding interactions.
Unfortunately, although these studies shed new light on the role of DNA shape in TF-DNA recognition, they were limited to the analysis of only a few factors and used only four different shape features. This was due to the lack of quantitative data on higher-order binding specificities and to the lack of tables to calculate other shape features. Thus, a more comprehensive analysis of TF-DNA binding – especially including higher-order dependencies – is urgently needed to better understand TF-DNA binding in general and to what extent DNA shape features are recognized by TFs in particular.
Recently, we presented high-performance fluorescence anisotropy (HiP-FA) (Jung et al., 2018, Jung et al., 2019), a method that determines TF-DNA binding energies directly in solution with high sensitivity and at a large scale and allows for measuring the affinity of a TF to any given DNA sequence. These features make HiP-FA an ideal tool to measure TF-DNA binding specificities, in particular the higher-order dependencies since these interactions are generally weak and their accurate measurement is both difficult and indispensable.
Here, we used HiP-FA to measure binding energies for 13 TFs of the Drosophila segmentation gene network belonging to 8 different binding domain families. We determined their 0th order of binding specificities taking only into account independent base contributions (PWM) and their first order of binding specificities accounting for dinucleotide dependencies represented by the dinucleotide position weight matrices (DPWMs). In this work, we define DPWMs as being the scoring matrices characterizing the deviations in the dinucleotide binding energies compared to pure PWMs (Transparent Methods). Correlating our affinity data with the 13 known DNA shape features and the EP, we found that nearly all our factors extensively use shape readout for DNA recognition, independently of the binding domain family. For 11 TFs for which structural information is available, we examined the correlations between their nuclear magnetic resonance (NMR)/co-crystal structures or structures of analog proteins obtained by homology-based modeling and the shape attributes obtained from our analysis. Finally, we ran a cluster analysis to test if certain shape features tend to co-occur in the DNA shape readout used by our TFs.
Results
Determination of the TF-DNA Binding Specificities and Overall Strategy for the Analysis
In a previous work (Jung et al., 2018), we have already presented the PWMs determined by HiP-FA for the 13 selected factors. We have also validated our method for determining binding affinities using two orthogonal assays, electrophoretic mobility shift assay and microscale thermophoresis. Finally, we have demonstrated that our PWMs were superior to those obtained with other methods (bacterial one hybrid or DNase footprinting) in predicting ChiP-seq data and when used in a thermodynamic model for predicting gene expression in Drosophila embryos (Jung et al., 2018). Herein, we extended our method to capture potential higher-order TF-DNA interactions by measuring the binding affinities of all mononucleotide and neighboring dinucleotide mutations (Figure 1A) in the core of each TF-DNA consensus binding sequence (6 positions for GATAe, 7 for all the other TFs). For 6 factors, we measured duplicates or triplicates to check reproducibility, leading to in total ∼1600 individual titration curves. We analyzed the data in two steps: first, we used the binding affinities to determine the PWMs and the DPWMs. Importantly, in the analysis procedure, we developed an algorithm (PySite; https://github.com/Reutern/PySite) to correct for the energy contribution of off-target binding sites that might be created by chance in dinucleotide mutations (Figure 1B and Transparent Methods). Second, we assessed the influence of the shape of DNA around the core binding site on the TF-DNA binding strength. For all dinucleotide mutations, we calculated the 13 shape features and the EP at each position in the binding site using available look-up tables (Chiu et al., 2017; Li et al., 2017; Zhou et al., 2013). We then applied a robust linear regression algorithm (Transparent Methods) to correlate, at a given position, the values of each shape feature with the binding energies measured for all tested mutations of the binding site (Figure 1C; see below for details).
Figure 1.
Experimental and Data Analysis Strategies
(A) Sequence design and measurement of binding energies by HiP-FA. A consensus sequence is mutated with all possible mononucleotide and dinucleotide mutations. The individual TF-DNA binding energies are measured using a robotic system and an automated custom-modified fluorescence microscope; the titration binding curves are reconstructed and analyzed following the HiP-FA procedure (Jung et al., 2018, Jung et al., 2019).
(B) Data analysis. After an off-target removal procedure (Transparent Methods), the binding energies are used to determine the 0th order of binding (PWMs) and the first order of binding (DPWMs), as shown for the TF Bcd. The DPWMs exhibits the mutual information (MI, a metric similar to the information content IC but for dinucleotide representation), which is not included in the simple linear PWM.
(C) Analysis of the DNA shape readout contribution. The sensitivity to DNA shape is analyzed following the subsequent steps: the DNA shape features are calculated using look-up tables (Chiu et al., 2017; Li et al., 2017; Zhou et al., 2013). The sensitivity to shape readout (termed shape readout value) is plotted per position against the binding energies (lower panel of c), and a robust linear regression is performed (Transparent Methods). Besides the fit (blue line), the steepest (gray dashed line) and the least steep fit (purple dashed line) are estimated using the confidence intervals provided by the robust linear regression. To make a conservative choice, the least steep slope is taken as the shape readout value. The shape readout values of all features and positions are depicted in the lower right panel for Bcd.
Zeroth- and First-Order Binding Specificities for the Drosophila TFs
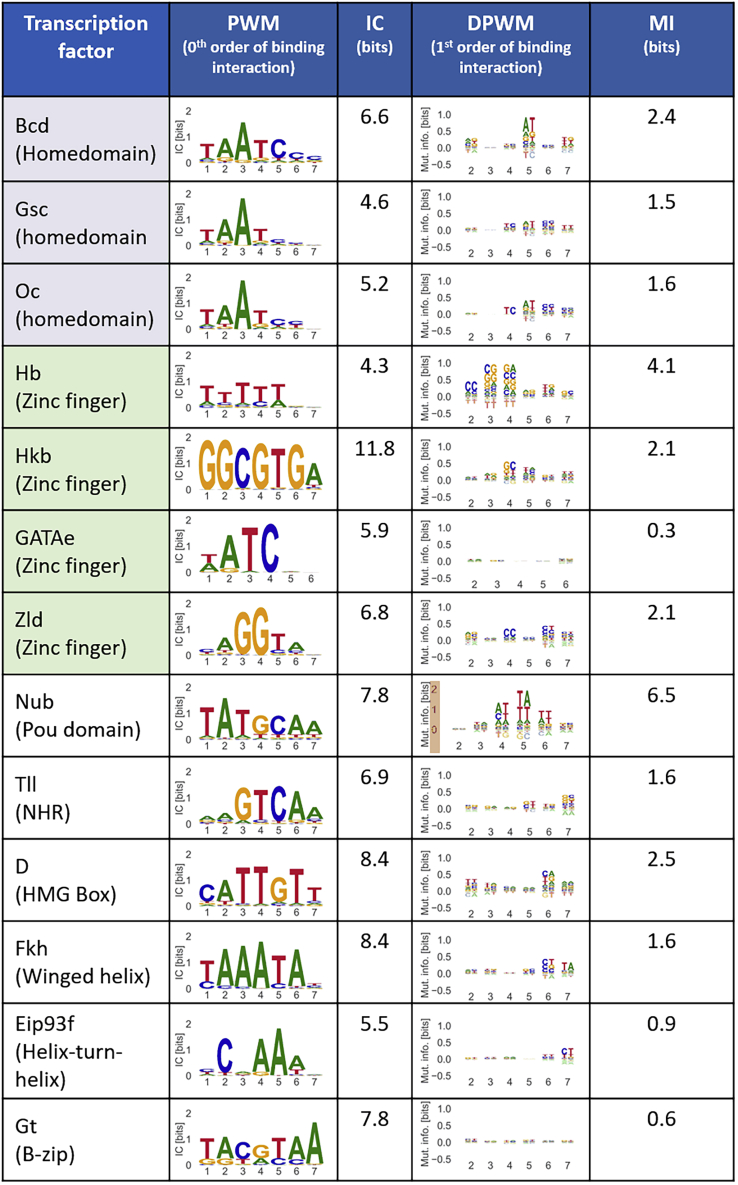
We used our measured binding affinities to determine the PWMs and DPWMs (Data S1) of the factors (Figure 2 and Transparent Methods). Overall, the PWMs are similar and largely share the same consensus sequences with those obtained by other methods, but they generally present a lower specificity (measured by their information content [IC]) as already discussed in our previous work (Jung et al., 2018). In contrast, our DPWMs show fewer but more preferred dinucleotides (as indicated by higher mutual information [MI] (Transparent Methods), a metric similar to IC but for dinucleotide representation) compared to computationally derived scoring matrices including nucleotides (Siebert and Soding, 2016) or obtained using SMiLE-seq data (Rube et al., 2018). The low noise present in our binding specificities can be visually appreciated by comparing with the logos obtained from SMiLE-seq data for Bicoid (Bcd) (Rube et al., 2018) (Figure S2), to our knowledge the only factor among our TFs with an already known higher-order specificities based on binding affinities. For example, at position 5 of the HiP-FA Bcd DPWM (corresponding to the dinucleotide mutations between positions 4 and 5 in the PWM; Figure 2), the four pairs AT, AG, GT, and CA have a cumulated MI of nearly 1 bit, thereby dominating over the other 11 possible dinucleotide mutations. Another more direct way to assess the effect of non-independent binding is to compare our experimentally determined affinities with predicted values assuming purely linear base contribution (Figure S1). Many dinucleotide mutation sequences disagree with measured values (defined as lying within 3σ of the measured values), confirming the presence of non-independent amino acid-nucleotide interactions.
Figure 2.
Overview of the PWMs and DPWMs for all the Investigated TFs
In the DPWMs, the heights of the dinucleotide letters represent the mutual information (MI) between two positions for the first order of binding. The total information content (IC) and MI are indicated in the right hand side columns for the PWM and DPWMs, respectively. Homeodomain factors and zinc fingers are grouped by color. Average PWMs and DPWMs are shown when replicate measurements were performed.
For all factors, we observe that the 0th order contribution to binding dominates over the first order, as indicated by the higher ICs of the specificity logos (6.9 bits on average for the 0th order compared to 2.1 bits MI for the first order; Figure 2). This was expected since the simple PWM model has proven to capture most of the sequence preferences for numerous TFs (Stormo et al., 1982; Zhao and Stormo, 2011). Surprisingly, the DPWMs of nearly all our TFs (with the exceptions of GATAe and Gt) show a high contribution to the overall binding specificities, revealed by their relatively high total MI (>∼1 bit), above our threshold for significant MI (0.03 bits per nucleotide positions, corresponding to ∼0.2 bits for the total MI of our DPWM logos; see Transparent Methods). Several studies already emphasized the importance of neighboring nucleotides in the prediction of TF binding (Nitta et al., 2015; Siebert and Soding, 2016; Zhao et al., 2012) but only for a few factors. The sensitivity of the HiP-FA assay enables us to accurately resolve weak – but measurable – binding events and their deviations from a purely linear binding of independent bases.
Noteworthy, the three members of the homeobox family (Bcd, Gsc, and Oc; Figure 2) have resembling PWMs and DPWMs, reflecting the similarity of their binding domains. This observation is in line with previous works describing the high similarity between homeodomain TFs' PWMs (Affolter et al., 2008). Our DPWMs clearly show that this similarity holds also at the first order of binding. A closer inspection, however, reveals the presence of subtle differences in specificities. At position 5 of the three DPWMs, although the preferred dinucleotides are very resembling (AT being the strongest positive deviation from linear binding, TC being the strongest repulsive one), the corresponding MIs differ substantially between the three TFs (for the positive MI: 0.76 bits for Bcd, 0.42 for Oc, and 0.27 for Gsc). In addition, Bcd differs at position 2 in its DPWM from the two other factors with its relatively high MI (0.35 compared with 0.08 for Gsc and 0.04 for Oc). Although these differences are small, their concerted effect might be important to allow these homeodomains to execute their distinct biological functions.
To conclude, our sensitive measurements of binding affinities provide us with refined binding specificities for our TFs, including first-order binding interactions.
DNA Shape Readout Is Used by Most of the Investigated TFs
The fact that most of the variance in DNA shape is encoded in dinucleotides (Rube et al., 2018) encouraged us to tackle the question to which extent TF-DNA binding is driven by DNA shape. To this end, we calculated the 13 geometric shape features and the EP at each position for all tested DNA sequences and determined their influence on our binding energies. For a given factor, we evaluate whether the change in binding energies correlates with a feature of interest when a base at a certain position and/or at a neighboring position deviates from the consensus sequence. For example, in the case of Bcd, at position 4, the binding energy decreases over an amplitude of ∼4 (normalized) when the relative minor groove width (MGW) increases from ∼0.2 to ∼0.8 Å (Figure 1C). The sensitivity to DNA shape readout is determined by a robust linear fitting procedure (Transparent Methods) to minimize the effect of extreme values (identified as outliers by the algorithm) and to provide a confidence interval to the resulting fitting parameters. The slope of the robust linear fitting provides an estimate of how much the binding of the TF at the particular position is influenced by the local DNA shape. On the following, we define the “shape readout value” as being this slope after normalization using for the binding energies their z/standard score and for the DNA shape features their amplitude (details about the normalization procedures in Transparent Methods). The shape readout value profiles allow one to compare the shape sensitivity of the free binding energy for the different shape features along the TF-DNA interface, while providing an intuitive metric of their deviation from their “average” behavior. For each TF, we applied this analysis procedure for every shape feature and at all base positions along the core DNA binding sequence (Figure 3 and Data S1). The reproducibility of the shape readout values among replicates was high with a mean squared Pearson coefficient R2 = 0.78 for the 6 factors having duplicates or triplicates (Figure S3).
Figure 3.
Overview of DNA Shape Sensitivities for the investigated TFs
The stacked shape readout values are plotted for each feature at each position (intra-base pair features) or between two positions (inter-base pair features). To facilitate the comparison with Figure 2, the positions are also labeled with their respective nucleobase at this position of the consensus sequence. The legend for the respective features is in the lower right corner. Homeodomain TFs (blue background color) and zinc finger TFs (green) are grouped together. The significance levels are indicated for each shape readout value bar with a hashing code indicated in the right bottom corner (see Transparent Methods for details). Average shape sensitivity plots are shown when replicate measurements were performed. Overall, there is a widespread use of the DNA shape readout by our TFs.
The shape sensitivity plots reveal a widespread use of DNA shape readout for all our TFs (Figure 3), with strong differences in the shape readout values between factors and at different base positions for a given factor. Remarkably, the members of the homeodomain family (blue box in Figure 3) behave the same with similar shape sensitivity plots (discussed in details below), as already observed for the PWMs and DPWMs. This does not hold true for the zinc finger family (green box) or for the other factors with different binding domains for which the shape sensitivity plots exhibit various patterns along the DNA binding sequences. Other studies have reported that zinc fingers are diverse in their binding behavior, in contrast to other TF families (Kribelbauer et al., 2019; Rohs et al., 2010). Interestingly, we found that in the center of the binding sites of GATAe and Zelda (Zld) (positions 3 and 4 for both factors in the shape sensitivity plots), the shape readout values are very low, as discussed below in more details for GATAe. At these positions, the sequence logos have a high IC, as indicated by the prominent TC and GG bases in the PWMs of GATAe and Zld (Figure 2). Conversely, shape features become important where sequence information is not well defined, like for GATAe at positions 5 and 6 and for Zld at positions 1 and 7. This phenomenon has already been reported for other factors (Abe et al., 2015). Interestingly, we observe a similar phenomenon for the side chains of most of our factors, for the three homeodomains and for Hb, Tll, Fkh, and Eip93f. In these cases, shape features contain more information for binding than sequence alone in the side chains.
Correlation between the DNA Shape Sensitivity of the TFs and Structural Information
We next wondered whether the observed shape readout values can be related to protein structures as interactions between the TF and its target DNA (Figure 4). Unfortunately, structural information is only available for the homeodomain TF Bcd, which has an NMR structure (Baird-Titus et al., 2006), and for the other homeodomains Gcs and Oc sharing very similar protein structure and binding specificities. For the other factors, we thus sought for experimental structure of homologous TFs using protein homology-based modeling (Kelley et al., 2015) (Figure S4). In this section, we will focus on the homeodomains, on a B-ZIP Gt homolog (Pap1) (Fujii et al., 2000), and on a zinc finger GATAe mouse homolog (GATA3) (Bates et al., 2008).
Figure 4.
Correlation between DNA Shape Readout and Structural Information
Homeodomains TFs.
(A) Shape readout value profile for Bcd. Positions with strong shape readout highlighted with blue rectangles.
(B) Residue contacts map for Bcd (obtained using the DNAproDB database (Sagendorf et al., 2019), details in Figure S4). Interaction between bases and positively charged residues highlighted in yellow.
(C) Crystal structure of Bcd (pdb-ID: 1ZQ3) (Baird-Titus et al., 2006). Base contacts with the recognition helix in red, with the N-terminal tail in blue. The bluearrow points at the position where the binding domain contacts the narrowing minor groove.
(D) Shape readout values for the three homeodomain TFs at position TAAT of the consensus sequence (position 4 of the corresponding PWMs in Figure 2). In addition to being very similar, all three homeodomains show a strong readout of the minor groove at this position.
(E) MGW profile along the binding sequence for the consensus binding sequence used for the homeodomains. It exhibits a minimum value at position TAAT (red arrow).
B-ZIP TF Gt.
(F) Shape readout values for Gt. The black rectangle indicates positions with highly symmetrical shape readout values around the middle vertical axis (added to all three panels at the same position).
(G) Crystal structure of a similar B-ZIP TF (pdb-ID: 1GD2) (Fujii et al., 2000) with the same core consensus sequence as Gt. The black box indicates the region of high mirror symmetry around the black axis. Base contacts highlighted in red.
(H) Gt's PWM, the first position augmented with data from Jung et al. (Jung et al., 2018). The entire PWM is highly symmetrical.
Zinc finger TF GATAe.
(I) Shape readout values for GATAe. Positions with strong (1, blue) and weak (4, orange) shape readout values are indicated at the x axis.
(J) Their corresponding positions in the protein structure of a similar GATA TF (pdb-ID: 3DFV) (Bates et al., 2008) at both sides. The perspective shows a position with pronounced contacts to the DNA's phosphate backbone and minor groove. Base contacts in red. All crystal structures were produced using the DNAproDB portal (Sagendorf et al., 2019).
As for other homeodomain proteins in complex with their DNA targets, the recognition helix of Bcd (in red in Figure 4C) is thought to be engaged in base readout of the major groove, whereas the N-terminal tail (in blue) is involved in shape readout of the minor groove (Baird-Titus et al., 2006; Dror et al., 2014; Yang et al., 2017). Although little is known about the relationship between structural features and binding affinities, it has been shown that a narrow MGW can enhance the negative EP in the minor groove (Chang et al., 2013; Rohs et al., 2009), which can attract positively charged amino acids such as arginine (R), lysine (K), and histidine (H), considering the latter can be protonated. We thus focused on nucleotide positions with strong shape readout values (highlighted with blue rectangles in Figure 4A), in particular with significant shape readout values for the features MGW and EP, and sought the presence of contacts with R, K, and H residues at these positions. The residue contact map of Bcd (Figure 4B) shows the individual nucleotide-residue interactions, DNA secondary structure, protein secondary structure, and DNA interaction moieties (Figure S4 for details). One can observe multiple interactions (highlighted in yellow) between arginine (R4, R55, and R56) or lysine (K51 and K55) amino acids, and nucleotides positions with strong shape readout values (positions 1, 2, and 4, highlighted in blue in Figure 4A) exhibiting significant MWG and EP shape readout values. Another study (Dror et al., 2014) that analyzed 168 mouse homeodomains using PBM data found a significantly high correlation between the positively charged R or K residues of the N-terminal tail with the minor groove. Our data confirm the interaction between the R4 residue of the N-terminal tail (indicated by the blue arrow in Figure 4C) and the T and A nucleotides at position 1 and 2, respectively, exhibiting high and significant MWG shape readout value and EP (interactions shown in yellow). In addition, we observe another nucleotide T (position 4, highlighted with the right blue rectangle) with strong shape readout and that contacts an arginine (R55) and a lysine residue (K51), both belonging to the recognition α-helix. Interestingly, the A at position 3 (black arrows in Figure 4A) with a very low shape readout interacts only with the non-charged asparagine and the hydrophobic isoleucine residues (N52 and I48, respectively; black arrows in Figure 4B). This prompted us to evaluate the frequency of contacts with positive residues for all our factors for which we found a structure of a homologous protein (Figure S4). Remarkably, we found for the strong shape readout positions much more frequent nucleotide interactions with R, K, or H (64% of the contacts) than for the other positions (28% of the contacts). Hence, these results generalize the contribution of the positively charged amino acids to the shape readout to other DNA secondary structures (such as α-helixes) and to other binding domains. Interestingly, we also found for the POU domain Nub (Figure S4E) that positively charged residues can strongly contribute not only to DNA shape readout but also to non-charged but highly polar amino acids (glutamine, threonine, and serine in this case).
As an additional validation for our analysis, although the DNA shape features ProT, Roll, HelT, and MGW have been quantitatively investigated for Bcd by Rube et al. (Rube et al., 2018), the MGW was the only shape feature with significant shape sensitivity coefficients in their study. For a detailed comparison, we plotted our shape readout values for all positions of the MGW against the corresponding shape sensitivity coefficients determined by Rube et al. (Rube et al., 2018) (Figure S5). We obtained an excellent correlation (R2 = 0.99) for the subset of coefficients that Rube et al. found to be significant. Note that our shape readout values were also significant (p < 0.05) at these positions. Remarkably, we also found significant correlations for additional features and for the other homeodomains, like at position 4 where the MGW exhibits a local minimum (Figures 4D and 4E) for Stretch and the EP for the three homedomain proteins, as well as for ProT (Gsc) and Buckle (Bcd and Gsc). This shows that the TF reads multiple DNA shape features at this position. As mentioned above, the reproducibility of the shape feature values is remarkable among the different homeodomains (Figure 4D). Given the sequence similarity between the three proteins, it is not surprising to find a similar shape readout for most features, which also speaks for the high reproducibility of our measurements.
Another pertinent example is the TF Giant (Gt) belonging to the family of B-ZIP proteins (Figures 4F–4H and S4F). Members of this family approach the DNA like a scissor, with two alpha helices contacting the major groove from two opposing sites (Figure 4G). Interestingly, the same mirror symmetry with a mirror plane between position 3 and 4 (C and G) is found in the PWM (Figure 4H) and partially in the shape sensitivity plot (Figure 4F). The shape readout values of both inter- and intra-base pair features between positions 2 and 5 show a highly symmetrical pattern, in line with the binding mode of B-ZIP proteins (Figure 4H and the residue contact map in Figure S4F). This pattern, although conserved in the PWM, is not maintained at the side positions in the shape readout values, probably due to the fact that the DNA has more flexibility outside of the B-ZIP's scissor and the TF has less contact to its minor groove and backbone (Figure S4F).
At last, we examined the zinc finger protein GATAe (Figures 4I–4J and S4D). Zinc fingers contact the DNA at two opposing strands with three contacts being at one strand (positions 4 to 2, ATC in the case of GATAe) and another at the opposing strand (position 1, T) (Fedotova et al., 2017). There are multiple contacts at position 1 (blue circles in Figure 4J and the residue contacts map in Figure S4D) between the TF and the DNA backbone, which match the high shape readout values at this position (in total 14.1 AU, absolute sum). The contacts between the TF and minor groove or DNA backbone decrease moving toward the central binding site, as seen in the crystal structure (orange circle in Figures 4J and S4D). The sum of the shape readout values shows a similar behavior, a decreasing overall shape sensitivity going from position 1 to 4 (black arrow in Figure 4I). At position 4 (yellow circle), one can observe contacts in the structure exclusively to bases in the major groove (yellow circle in Figure 4J), and the sum of the shape readout values is reduced to a minimum (1.8 AU). It was recently reported that metazoan zinc fingers tend to establish several contacts to the DNA backbone (Najafabadi et al., 2017), possibly permitting DNA shape readout at these positions.
In overall, this shape/structure analysis validates our approach, while bringing biological insights about the relationship between shape readout values and residue contacts with the TF-DNA interface.
Shape Readout Values/TF Clustering
Finally, we asked if TFs use predominantly certain shape features to bind to DNA. To test whether shape features tend to co-occur in the shape readout, we performed two distinct cluster analysis of the shape readout values matrix (Figure S6 and Transparent Methods): (1) we clustered the different features with respect to their feature readout by the TF (vertical lines) and (2) its reverse – a clustering of the TFs versus their shape readout for each feature as a matrix including all nucleotide positions (horizontal lines).
The TF clustering indicates that the different binding proteins show little similarity in their use of the shape features except for the homeodomains, which was expected. In contrast, the clustering of the shape features reflects structural dependencies between shape features. There are three distinct clusters of shape features, which may be related to biophysical properties of the DNA and its interplay with the binding protein, such as bends or kinks (details in caption of Figure S6). For instance, the first cluster consists of slide, helix twist, roll, and MGW. These features were reported to correlate the most with each other both in bound and unbound DNA (El Hassan and Calladine, 1995) (Stella et al., 2010) and are read out concertedly in DNA-protein complexes (Suzuki et al., 1997). Thus, the cluster analysis confirms properties of the features' interdependencies in shape readout.
Discussion
HiP-FA constitutes a powerful tool to quantify TF-DNA binding specificity, especially the non-independent interactions requiring to be determined with high accuracy. The throughput of the method is not sufficient to discover de novo shape motifs or to explore the large sequence space possible with sequencing-based methods like HT-SELEX or SMiLE-seq. However, this is not a major limitation since the prior knowledge that HiP-FA requires (some information about the TF's binding preferences) is known for many TFs, and dinucleotide mutations are sufficient to cover most of the non-independent amino acid-nucleotide interactions. It would also be straightforward to extend the measurements in the flanking regions of the core binding motif. A comparison of the different approaches and a summary of the results obtained as far as DNA shape readout analysis is concerned can be found in Table S1.
Our approach consisting in measuring binding energies of a complete set of dinucleotide mutations is more direct than the one used by Rube et al. (Rube et al., 2018) that requires a prior analysis with the “No Read Left Behind” (Rastogi et al., 2018) algorithm to derive affinities from high-throughput data. In addition, our downstream analysis of shape sensitivity, which employs a robust linear regression algorithm, uses fewer parameters and provides directly an interpretable characterization of shape sensitivity. However, it is not possible to distinguish between base and shape readout directly. In the analysis procedure, we cannot exclude the possibility of energy changes due to base readout, leading to an incidental correlation between binding energies and shape features. We reason that this apparent contribution of shape features may average out in the linear fitting procedure and, as a consequence, will not lead to a strong correlation bias in the robust linear regression. This assumption is supported by the following: (1) our estimation of the shape sensitivity is in excellent agreement with the one obtained by the more complex algorithm elaborated by Rube et al. (Figures S2 and S5), (2) our shape readout values can be related to structural features of the factors (Figures 4 and S4), (3) the clustering of the shape feature readout rediscovers already known interdependencies between shape features (Figure S6). Not all shape features are recognized independently by the TFs. The different groups in our clustering might represent a specific DNA conformation which is read out by a TF, rather than the readout of several independent DNA features. These conformations might play an important role for the binding behavior of several TFs.
By combining directly TF-DNA binding affinities, DNA shape features, and structural information, we gained insights into their correlation, a debated topic due to their intrinsic covariation. Importantly, our results suggest that DNA shape readout is widespread among our TFs. The extended use of DNA shape readout by TFs has become increasingly apparent over the past years (Chiu et al., 2017; Mathelier et al., 2016; Pal et al., 2019; Rube et al., 2018; Samee et al., 2019; Yang et al., 2017; Zhou et al., 2015), which comes as no surprise considering that the number of van der Waals interactions enabling shape readout account for two-third of the protein-DNA interactions (Rube et al., 2018). The correlation analysis of the shape readout values with protein-DNA complex structures allows us to generalize the influence of the charged amino acids on the shape readout that has been described so far only for homeodomains in the minor groove region of the DNA. We observe this effect to other DNA secondary structures (such as α-helixes) and to other binding domains. In addition, for the POU domain Nub we identify non-charged but polar residues that can also lead to a strong DNA shape readout. To the best of our knowledge, these effects on DNA shape readout have not been reported. The difficulty to detect the effects of charged and non-charged residues, especially in the major groove, is that they are obscured by the interactions involved in the base readout. Our analysis was able to resolve even subtle effects due to the high sensitivity of the binding affinity measurements, and our shape analysis was able to deconvolve, to some extent, shape from base readout.
In summary, we determined the binding specificities for 13 Drosophila TFs including first-order dependencies, provided insights into the correlation between their binding affinities to DNA and the shape features of the DNA helix, and gave structural insights in the shape readout. Our method could easily be extended to more factors and to different organisms to provide a refined catalog of TF-DNA shape readout landscapes.
Limitations of the Study
Although our HiP-FA assay allows us to determine accurately binding affinities at a relatively large scale, we cannot cover the whole sequence space as high-throughput methods do. To restrict the number of measurements, we thus focussed on the core binding motif of the TFs, and to all mononucleotide and dinucleotides mutations of the consensus sequence rather that all possible mutations. This should however cover most of the TF-DNA interactions since it has been shown that dinucleotide models explain >92% of the variance for the MGW, ProT, Roll, and HelT shape features (Rube et al., 2018). In addition, our analysis based on the direct correlation between binding affinities and shape features can only indirectly and partially tease apart the respective contributions of base and DNA shape readouts. Note that how to achieve the deconvolution between base and shape readouts is a longstanding issue in the field.
Resource Availability
Lead Contact
Dr. Christophe Jung
phone. +49 (0)89 2180 71101
fax. +49 (0)89 2180 71105
email: jung@genzentrum.lmu.de.
Materials Availability
All plasmids generated in this study for the expression of the TF's binding domains are available from the Lead Contact without restriction. This study did not generate new unique reagents.
Data and Code Availibility
The PWMs, DPWMs, and shape readout values for all factors and shape features are available as Data S1. The Python3 code of PySite is available on Github (https://github.com/Reutern/PySite). All the other Python3 codes used for data analysis are available upon request.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We dedicate this publication to the memory of Prof. Ulrike Gaul who deceased after a long illness during the review of the manuscript. This work was supported by the DFG (large equipment grant for automated system), the Sonderforschungsbereich SFB646, the Center for Integrated Protein Science Munich (CIPSM), and the Graduate School of Quantitative Biosciences Munich (QBM). U.G. acknowledges support by the Humboldt Foundation (Alexander von Humboldt-Professorship). The authors want to thank Peter Bandilla for his help with the robotics and Stefano Ceolin for reading the manuscript and for fruitful discussions about the experiments and the analysis. We also particularly thank Ulrich Unnerstall for the numerous and useful discussions about the project.
Author Contributions
M.S., C.J., and U.G. developed the project; M.S. and C.J. designed the experiments; M.S. and C.L. performed the experiments; M.S., M.R., and C.J. performed data analysis; and M.S. and C.J. wrote the manuscript.
Declaration of Interests
The authors declare no conflict of interests.
Published: November 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101694.
Supplemental Information
References
- Abe N., Dror I., Yang L., Slattery M., Zhou T., Bussemaker H.J., Rohs R., Mann R.S. Deconvolving the recognition of DNA shape from sequence. Cell. 2015;161:307–318. doi: 10.1016/j.cell.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affolter M., Slattery M., Mann R.S. A lexicon for homeodomain-DNA recognition. Cell. 2008;133:1133–1135. doi: 10.1016/j.cell.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Baird-Titus J.M., Clark-Baldwin K., Dave V., Caperelli C.A., Ma J., Rance M. The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site. J. Mol. Biol. 2006;356:1137–1151. doi: 10.1016/j.jmb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Bates D.L., Chen Y., Kim G., Guo L., Chen L. Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J. Mol. Biol. 2008;381:1292–1306. doi: 10.1016/j.jmb.2008.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Philippakis A.A., Qureshi A.M., He F.S., Estep P.W., Iii, Bulyk M.L. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotech. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.P., Xu M., Machado A.C., Yu X.J., Rohs R., Chen X.S. Mechanism of origin DNA recognition and assembly of an initiator-helicase complex by SV40 large tumor antigen. Cell Rep. 2013;3:1117–1127. doi: 10.1016/j.celrep.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu T.P., Rao S., Mann R.S., Honig B., Rohs R. Genome-wide prediction of minor-groove electrostatic potential enables biophysical modeling of protein-DNA binding. Nucleic Acids Res. 2017;45:12565–12576. doi: 10.1093/nar/gkx915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror I., Zhou T., Mandel-Gutfreund Y., Rohs R. Covariation between homeodomain transcription factors and the shape of their DNA binding sites. Nucleic Acids Res. 2014;42:430–441. doi: 10.1093/nar/gkt862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hassan M.A., Calladine C.R. The assessment of the geometry of dinucleotide steps in double-helical DNA; a new local calculation scheme. J. Mol. Biol. 1995;251:648–664. doi: 10.1006/jmbi.1995.0462. [DOI] [PubMed] [Google Scholar]
- Fedotova A.A., Bonchuk A.N., Mogila V.A., Georgiev P.G. C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Nat. 2017;9:47–58. [PMC free article] [PubMed] [Google Scholar]
- Fujii Y., Shimizu T., Toda T., Yanagida M., Hakoshima T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 2000;7:889–893. doi: 10.1038/82822. [DOI] [PubMed] [Google Scholar]
- Isakova A., Groux R., Imbeault M., Rainer P., Alpern D., Dainese R., Ambrosini G., Trono D., Bucher P., Deplancke B. SMiLE-seq identifies binding motifs of single and dimeric transcription factors. Nat. Methods. 2017;14:316–322. doi: 10.1038/nmeth.4143. [DOI] [PubMed] [Google Scholar]
- Jung C., Bandilla P., von Reutern M., Schnepf M., Rieder S., Unnerstall U., Gaul U. True equilibrium measurement of transcription factor-DNA binding affinities using automated polarization microscopy. Nat. Commun. 2018;9:1605. doi: 10.1038/s41467-018-03977-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Schnepf M., Bandilla P., Unnerstall U., Gaul U. High sensitivity measurement of transcription factor-DNA binding affinities by competitive titration using fluorescence microscopy. JoVE. 2019:e58763. doi: 10.3791/58763. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kribelbauer J.F., Rastogi C., Bussemaker H.J., Mann R.S. Low-affinity binding sites and the transcription factor specificity paradox in eukaryotes. Annu. Rev. Cell Dev. Biol. 2019;35:357–379. doi: 10.1146/annurev-cellbio-100617-062719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sagendorf J.M., Chiu T.P., Pasi M., Perez A., Rohs R. Expanding the repertoire of DNA shape features for genome-scale studies of transcription factor binding. Nucleic Acids Res. 2017;45:12877–12887. doi: 10.1093/nar/gkx1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerkl S.J., Quake S.R. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- Mathelier A., Xin B., Chiu T.-P., Yang L., Rohs R., Wasserman W.W. DNA shape features improve transcription factor binding site predictions in vivo. Cell Syst. 2016;3:278–286.e4. doi: 10.1016/j.cels.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi H.S., Garton M., Weirauch M.T., Mnaimneh S., Yang A., Kim P.M., Hughes T.R. Non-base-contacting residues enable kaleidoscopic evolution of metazoan C2H2 zinc finger DNA binding. Genome Biol. 2017;18:167. doi: 10.1186/s13059-017-1287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta K.R., Jolma A., Yin Y., Morgunova E., Kivioja T., Akhtar J., Hens K., Toivonen J., Deplancke B., Furlong E.E. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. Elife. 2015;4:e04837. doi: 10.7554/eLife.04837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutiu R., Friedman R.C., Luo S., Khrebtukova I., Silva D., Li R., Zhang L., Schroth G.P., Burge C.B. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat. Biotechnol. 2011;29:659–664. doi: 10.1038/nbt.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Hoinka J., Przytycka T.M. Co-SELECT reveals sequence non-specific contribution of DNA shape to transcription factor binding in vitro. Nucleic Acids Res. 2019;47:6632–6641. doi: 10.1093/nar/gkz540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi C., Rube H.T., Kribelbauer J.F., Crocker J., Loker R.E., Martini G.D., Laptenko O., Freed-Pastor W.A., Prives C., Stern D.L. Accurate and sensitive quantification of protein-DNA binding affinity. Proc. Natl. Acad. Sci. U S A. 2018;115:E3692–E3701. doi: 10.1073/pnas.1714376115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T.R., Slattery M., Abe N., Rastogi C., Liu D., Mann R.S., Bussemaker H.J. SELEX-seq: a method for characterizing the complete repertoire of binding site preferences for transcription factor complexes. Methods Mol. Biol. (Clifton, NJ) 2014;1196:255–278. doi: 10.1007/978-1-4939-1242-1_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rube H.T., Rastogi C., Kribelbauer J.F., Bussemaker H.J. A unified approach for quantifying and interpreting DNA shape readout by transcription factors. Mol. Syst. Biol. 2018;14:e7902. doi: 10.15252/msb.20177902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagendorf J., Markarian N., Berman H., Rohs R. DNAproDB: an expanded database and web-based tool for structural analysis of DNA-protein complexes. Nucleic Acids Res. 2019;48:D277–D287. doi: 10.1093/nar/gkz889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samee M.A.H., Bruneau B.G., Pollard K.S. A de novo shape motif discovery algorithm reveals preferences of transcription factors for DNA shape beyond sequence motifs. Cell Syst. 2019;8:27–42.e26. doi: 10.1016/j.cels.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert M., Soding J. Bayesian Markov models consistently outperform PWMs at predicting motifs in nucleotide sequences. Nucleic Acids Res. 2016;44:6055–6069. doi: 10.1093/nar/gkw521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S., Cascio D., Johnson R.C. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010;24:814–826. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G.D., Schneider T.D., Gold L., Ehrenfeucht A. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Amano N., Kakinuma J., Tateno M. Use of a 3D structure data base for understanding sequence-dependent conformational aspects of DNA11Edited by B. Honig. J. Mol. Biol. 1997;274:421–435. doi: 10.1006/jmbi.1997.1406. [DOI] [PubMed] [Google Scholar]
- Weirauch M.T., Cote A., Norel R., Annala M., Zhao Y., Riley T.R., Saez-Rodriguez J., Cokelaer T., Vedenko A., Talukder S. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol. 2013;31:126–134. doi: 10.1038/nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Orenstein Y., Jolma A., Yin Y., Taipale J., Shamir R., Rohs R. Transcription factor family-specific DNA shape readout revealed by quantitative specificity models. Mol. Syst. Biol. 2017;13:910. doi: 10.15252/msb.20167238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ruan S., Pandey M., Stormo G.D. Improved models for transcription factor binding site identification using nonindependent interactions. Genetics. 2012;191:781–790. doi: 10.1534/genetics.112.138685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Stormo G.D. Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nat. Biotechnol. 2011;29:480–483. doi: 10.1038/nbt.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Shen N., Yang L., Abe N., Horton J., Mann R.S., Bussemaker H.J., Gordan R., Rohs R. Quantitative modeling of transcription factor binding specificities using DNA shape. Proc. Natl. Acad. Sci. U S A. 2015;112:4654–4659. doi: 10.1073/pnas.1422023112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Yang L., Lu Y., Dror I., Dantas Machado A.C., Ghane T., Di Felice R., Rohs R. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013;41:W56–W62. doi: 10.1093/nar/gkt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.