Abstract
Purpose
Recent studies revealed divergent gene expression patterns in Ewing sarcoma (EwS) with canonical EWSR1–ETS gene fusions and undifferentiated round cell sarcomas (URCS) with EWSR1 rearrangements fused to the non-ETS gene NFATc2. Thus, the question arises whether the latter tumors really belong to EwS.
Methods
We collected five cases matching the group of URCS with EWSR1–NFATc2 fusion and performed DNA methylation and copy number profiling. Results were compared to methylation data of 30 EwS with various EWSR1–ETS fusions and one EwS with FUS–ERG fusion, 16 URCS with CIC rearrangement and 10 URCS with BCOR alteration and a total of 81 EWSR1-associated soft tissue sarcomas including 7 angiomatoid fibrous histiocytomas, 7 clear cell sarcomas of the soft tissue, 28 desmoplastic small round cell tumors, 10 extraskeletal myxoid chondrosarcomas and 29 myxoid liposarcomas.
Results
Unsupervised hierarchical clustering and t-distributed stochastic neighbor embedding analysis of DNA methylation data revealed a homogeneous methylation cluster for URCS with EWSR1–NFATc2 fusion, which clearly segregated from EwS and the other subtypes. Copy number profiles of EWSR1–NFATc2 cases showed recurrent losses on chromosome 9q and segmental gains on 20q13 and 22q12 involving the EWSR1 and NFATc2 loci, respectively.
Conclusion
In summary, URCS with EWSR1–NFATc2 fusion share a distinct DNA methylation signature and carry characteristic copy number alterations, which emphasizes that these sarcomas should be considered separately from EwS.
Electronic supplementary material
The online version of this article (10.1007/s00432-019-02895-2) contains supplementary material, which is available to authorized users.
Keywords: EWSR1, NFATc2, Ewing, Ewing like, DNA methylation
Introduction
Ewing sarcoma (EwS) is a highly malignant tumor predominantly affecting the bones of children, adolescents and young adults. EwS belongs to the group of undifferentiated round cell sarcoma (URCS) and represents one of the most frequently diagnosed malignant mesenchymal bone tumors, accounting for approximately 5–10% of all cases (Alava et al. 2013). However, 10–15% of EwS primarily arise within soft tissue (Sankar and Lessnick 2011).
Precise separation of EwS from URCS without knowledge of the molecular phenotype is challenging for pathologists, because most tumors lack unambiguous morphological features or a specific immunophenotype (Alava et al. 2013).
EwS is genetically characterized by EWSR1-ETS gene fusions (Grunewald et al. 2018). EWSR1–FLI1 represents the most common gene fusion in EwS (85%), followed by EWSR1-ERG (10%) (Delattre et al. 1992; Sorensen et al. 1994). Less frequently (< 1%), EWSR1 is fused to other ETS gene members such as ETV1, ETV4 and FEV (Jeon et al. 1995; Kaneko et al. 1996; Peter et al. 1997). In exceptionally rare EWSR1 wild-type cases, a FUS–ERG fusion has also been described (Chen et al. 2016; Shing et al. 2003).
Some URCS show a significant morphologic overlap with EwS. However, they lack the prototypic EWSR1–ETS or FUS–ERG fusions. These tumors were termed EwS-like URCS (Antonescu 2014). Through molecular studies investigating EwS-like URCS, it was possible to subcategorize this heterogeneous group of sarcomas further. More than 90% of EwS-like URCS carry either CIC–DUX4 fusions or BCOR alterations (Italiano et al. 2012; Pierron et al. 2012; Specht et al. 2016). The current concept proposes both BCOR- and CIC-positive URCS to be considered as stand-alone subtypes with distinct clinical features and differing biological behavior (Antonescu et al. 2017; Kao et al. 2018). Thus, precise subtyping of EwS-like URCS will soon become relevant in a clinical context.
Despite the recent progress in genomic characterization and biological understanding of URCS, some cases remain ambiguous. These URCS carry fusions between EWSR1 and members outside the ETS gene family, which often occur between EWSR1 and NFATc2 (Mastrangelo et al. 2000; Savola et al. 2009; Sumegi et al. 2011; Szuhai et al. 2009; Wang et al. 2007). The current 2013 World Health Organization (WHO) classification of soft tissue and bone tumors provisionally assigned these cases to EwS (Alava et al. 2013). However, recent gene expression-based studies observed major differences between URCS with EWSR1–NFATc2 fusion and EwS (Baldauf et al. 2018b; Specht et al. 2014; Watson et al. 2018). Also, EWSR1–NFATc2-positive sarcomas show a striking predominance of affecting mainly male adults (Grunewald et al. 2018). Thus, there is an ongoing debate whether URCS with EWSR1–NFATc2 fusion belong to EwS, or whether they represent a distinct entity (Baldauf et al. 2018a; Charville et al. 2018).
To address this issue further, we performed comparative, genome-wide DNA methylation profiling and cytogenetic analyses of five URCS with EWSR1–NFATc2 fusion.
Materials and methods
Sample selection
We collected five cases matching the group of URCS with EWSR1–NFATc2. The samples were retrieved from the Gerhard-Domagk Institute of Pathology of the University Hospital Münster (Germany), the Institute of Pathology of the University Hospital in Leiden (the Netherlands), the Institute of Pathology of the University Hospital in Basel (Switzerland) and the Department of Pathology of the University Hospital Virgen del Rocío in Seville (Spain). The EWSR1–NFATc2 fusion has been confirmed in four cases, among them two cases from the seminal paper by Szuhai and colleagues (Szuhai et al. 2009). In one previously published case, detection by next-generation sequencing and FISH analysis failed due to poor RNA quality, though the copy number profile revealed amplifications involving the EWSR1 locus (Koelsche et al. 2018a). The five URCS with EWSR1–NFATc2 were reviewed by H&E staining. Histological sections were scanned with the NanoZoomer 2.0-HT digital slide scanner (Hamamatsu Photonics, Hamamatsu City, Shizuoka Prefecture, Japan) and images were captured with the ImageScope software (Leica Biosystems, Buffalo Grove, IL, USA).
The control group included 31 EwS with EWSR1–FLI1 (n = 24), EWSR1–ERG (n = 4), EWSR1–ETV1 (n = 1), EWSR1–FEV (n = 1) and FUS–ERG (n = 1) gene fusions. Furthermore, 16 URCS with CIC rearrangement and 10 URCS with BCOR alteration were included.
In addition, the control group was expanded by including sarcomas that may comprise EWSR1-rearrangements, namely angiomatoid fibrous histiocytoma (n = 7), clear cell sarcoma of the soft tissue (n = 7), desmoplastic small round cell tumor (n = 28), extraskeletal myxoid chondrosarcoma (n = 10) and myxoid liposarcoma (n = 29). The methylation data of the control group and of two URCS with EWSR1–NFATc2 fusions have been previously reported (Koelsche et al. 2018a, b). Basic clinical information of the investigated cases is provided in Supplementary Table 1. This investigation was performed in accordance with the Declaration of Helsinki.
DNA isolation, genome-wide DNA methylation data generation and pre-processing
Representative tumor tissue with highest available tumor content was chosen for DNA extraction. The Maxwell® 16 FFPE Plus LEV DNA Kit or the Maxwell® 16 Tissue DNA Purification Kit (for frozen tissue) was applied on the automated Maxwell device (Promega, Madison, WI, USA) according to the manufacturer’s instructions. All tumors had a total amount of > 100 ng DNA and were suitable for the array-based DNA methylation analysis. All tumors were subjected to Illumina Infinium HumanMethylation450 (450 k) BeadChip or the successor EPIC/850 k BeadChip (Illumina, San Diego, USA) analysis at the Genomics and Proteomics Core Facility of the German Cancer Research Center (DKFZ) Heidelberg. DNA methylation data were normalized by performing background correction and dye bias correction (shifting of negative control probe mean intensity to zero and scaling of normalization control probe mean intensity to 20,000, respectively). Probes targeting sex chromosomes, probes containing multiple single nucleotide polymorphisms and those that could not be uniquely mapped were removed. Probes from the EPIC array were excluded if the predecessor Illumina Infinium 450 k BeadChip did not cover them, thereby making data generated by both 450 k and EPIC feasible for subsequent analyses. In total, 438,370 probes were kept for analysis.
Unsupervised clustering, t-SNE analysis and cumulative copy number plotting
For unsupervised hierarchical clustering, we selected 10,000 probes that showed the highest median absolute deviation (MAD) across the beta values. Samples were hierarchically clustered using the Euclidean distance and Ward’s linkage method. Hierarchical clustering using Euclidean distance and complete linkage reordered methylation probes. The unscaled methylation levels were shown in a heat map from unmethylated state (blue color) to methylated state (red color). For unsupervised 2D representation of pairwise sample correlations, dimensionality reduction by t distributed stochastic neighbor embedding (t-SNE) was performed using the 10,000 most variable probes, a perplexity of 20 and 2500 iterations. Novel methylation groups were tested for stability by varying the number of the most variable probes. Copy number assessment was done based on methylation array data using the R-package conumee after additional baseline correction (https://github.com/dstichel/conumee).
Results
Study cohort
Five tumors from five patients matching the molecular criteria of URCS with EWSR1–NFATc2 fusion were included, four primary tumor samples and one tumor metastatic to the lung. Patients with EWSR1–NFATc2 positive URCS were older (median age 39 years; range 16–56 years) compared to 31 patients with EwS (median age 17 years, range 3–63 years), 10 patients with URCS with BCOR alteration (median age 14 years; range 0–22 years) and 16 patients with URCS with CIC rearrangement (median age 26 years; range 12–49 years). Clinical data of the five URCS with EWSR1–NFATc2 fusion are summarized in Table 1.
Table 1.
Clinical characteristics of undifferentiated round cell sarcomas with EWSR1–NFATc2 fusion
| Case ID | EWSR1–NFATc2 | Age at diagnosis | Gender | Location | Manifestation | References |
|---|---|---|---|---|---|---|
| 103662 | Non-determinable | 51 years | Male | Humerus | Primary | Koelsche et al. (2018a, b) |
| 128854 | Exon 8–Exon 3 | 39 years | Male | Lung (primary humerus) | Metastasis | Szuhai et al. (2009) |
| 110244 | Exon 8–Exon 3 | 56 years | Female | Femur | Primary | – |
| 128856 | Exon 8–Exon 3 | 16 years | Male | Femur | Primary | Szuhai et al. (2009) |
| 97480 | Exon 8–Exon 3 | 17 years | Female | Humerus | Primary | Koelsche et al. (2018a, b) |
Histologic features of undifferentiated round cell sarcomas with EWSR1–NFATc2 fusion
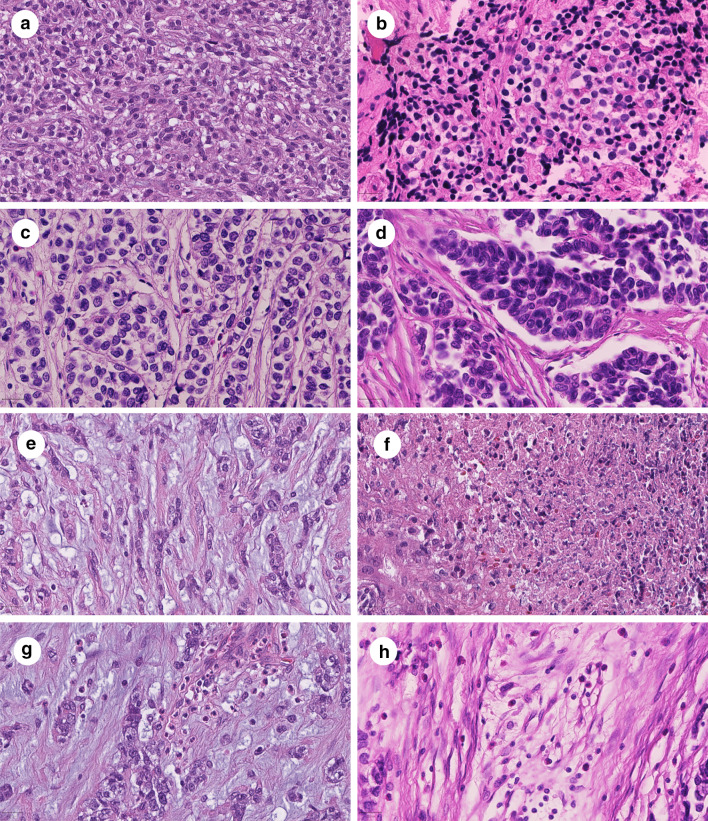
The tumor cells of URCS with EWSR1–NFATc2 fusion were round to polygonal with faint eosinophilic to clear cytoplasm, which sometimes appeared vacuolated. The tumor cell outlines were relatively ill defined, the nuclei were large with a dense chromatin pattern (Fig. 1a, b). In some areas, the tumor cells were separated by thin to coarse fibrous septae, which resulted in a nested pattern (Fig. 1c, d). Intriguingly, one case exhibited a predominant myxoid tumor matrix with tumor cells arranged in cords and groups therein (Fig. 1e). Foci of necrosis were present in two cases (Fig. 1f). Inflammatory bystander cells were composed of eosinophilic leukocytes (Fig. 1g, h).
Fig. 1.
Histologic features of undifferentiated round cell sarcomas with EWSR1–NFATc2 fusion. Undifferentiated round cell sarcomas with EWSR1–NFATc2 fusion were composed of relatively monotonous cells with a faint eosinophilic to clear cytoplasm and large, chromatin dense nuclei (a, b). The tumor matrix variably contained thin to coarse collagen bundles, which separated tumor cells and appeared as a nested growth pattern (c, d). One case presented with a myxoid tumor matrix (e). Focal necrosis was observed in two cases (f). Inflammatory cells were variably present and were composed of eosinophilic leukocytes (g, h). Magnification: 400-fold. Scale bars: 20 µm
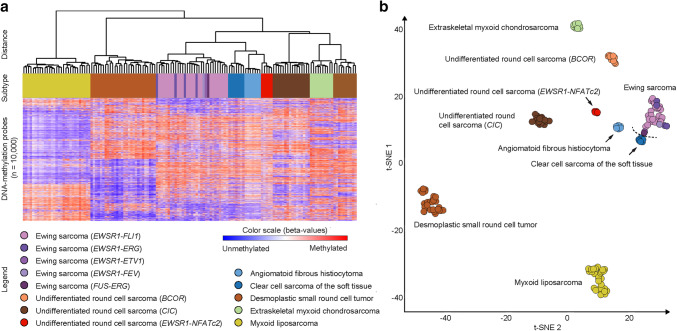
Distinct methylation signature in undifferentiated round cell sarcomas with EWSR1–NFATc2 fusion
URCS with EWSR1–NFATc2 fusion formed a homogeneous methylation class by both clustering (Fig. 2a) and t-SNE analyses (Fig. 2b), which also remained stable when varying the number of CpGs used for this analysis. The methylation profiles of URCS with EWSR1–NFATc2 fusion were distinct from the methylation class of EwS, which formed a homogeneous methylation cluster irrespective of their various TET–ETS gene fusion variants. URCS with CIC rearrangement, URCS with BCOR alteration and the tumor control subtypes angiomatoid fibrous histiocytoma, clear cell sarcoma of the soft tissue, desmoplastic small round cell tumor, extraskeletal myxoid chondrosarcoma and myxoid liposarcoma formed subtype-specific methylation classes, respectively.
Fig. 2.
Distinct DNA methylation patterns in undifferentiated round cell sarcomas with EWSR1–NFATc2 fusion. Unsupervised hierarchical clustering (a) and t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis (b) of DNA methylation data from undifferentiated round cell sarcoma variants and a reference set of prototypical soft tissue and bone sarcomas with frequent rearrangements involving gene members of the TET transcription factor family. The arrow points out the novel methylation group
Copy number analysis
We next analyzed the copy number profiles of URCS with EWSR1–NFATc2 (Fig. 3). A segmental gain on chromosome 22q12 involving the EWSR1 locus and losses on chromosome 9q were observed in all five cases. A segmental gain on chromosome 20q13 covering the NFATc2 locus was observed in four cases. None of these copy number alterations were present in the 31 EwS, 16 URCS with CIC rearrangement and 10 URCS with BCOR alteration.
Fig. 3.
Copy number alterations in undifferentiated round cell sarcomas with EWSR1–NFATc2 fusion. Copy number profiles were generated using DNA methylation data. The arrows point out recurrent copy number alterations (a). Fluorescence in situ hybridization studies indicate a break and a copy number gain of EWSR1 (b)
Discussion
Genome-wide tumor DNA methylation patterns likely most often relate to the epigenetic state of their originating tissue and, therefore, can be used to distinguish molecular subgroups within morphologically homogeneous tumor groups. We here describe a distinct DNA methylation signature for URCS with EWSR1–NFATc2 fusion, which is clearly distinguishable from the DNA methylation signature of EwS. Accordingly, our results strongly support the concept of considering URCS with EWSR1–NFATc2 fusion separate from EwS. This concept has recently been fueled by gene expression studies, which revealed major differences between URCS with EWSR1–NFATc2 fusion and EwS with canonical TET–ETS gene fusions (Baldauf et al. 2018b; Watson et al. 2018).
Less than 20 URCS with EWSR1–NFATc2 fusion have been reported since their first description in 2009 (Antonescu 2014; Kinkor et al. 2014; Koelsche et al. 2018a; Machado et al. 2018; Romeo et al. 2012; Sadri et al. 2014; Szuhai et al. 2009; Watson et al. 2018). Overall, these cases share a significant pathological overlap with EwS, which retrospectively justifies their listing as variant EwS in the 2013 WHO classification of soft tissue and bone tumors (Alava et al. 2013).
The question of histiogenesis, however, has remained controversial in EWSR1–NFATC2 fused sarcomas ever since. Morphologically, URCS with EWSR1–NFATc2 fusion exhibit atypical features, e.g., presence of a hyaline stroma, a nested growth, enlarged cells with a clear cell appearance and nuclei with prominent nucleoli, characteristics that are not commonly seen in EwS (Folpe et al. 2005). Epithelial marker expression has been consistently described in URCS with EWSR1–NFATc2 fusion, which is rarely observed in EwS (Antonescu 2014; Folpe et al. 2005).
URCS with EWSR1–NFATc2 fusion carry a t(20;22)(q13;q12) chromosomal translocation (Szuhai et al. 2009). It is assumed that NFATc2, which acts as a transcription factor, recognizes similar core motifs compared to FLI1 and ERG (Sankar and Lessnick 2011). However, it has not been proven yet whether similar functional consequences may emerge from the EWSR1–NFATc2 fusion compared to TET–ETS gene fusions. Unique for the EWSR1–NFATc2 fusion is the amplification of the fusion partner NFATc2 on chromosome 20q13 and EWSR1 on chromosome 22q11 (Szuhai et al. 2009). This focal amplification leads to an increased number of EWSR1 probe signals, which may hamper fluorescence in situ hybridization evaluation or may be misjudged as an artifact (Machado et al. 2018). We observed the EWSR1 amplification in all five cases and the NFATc2 amplification in all but one case, which might have been missed due to relatively noisy signals in the copy number profile. It should be noted that besides the EWSR1 and NFATc2 loci, also the remaining CNV profiles of the EWSR1–NFATc2 sarcomas reported here are not typical for EwS (Grunewald et al. 2018).
From a clinical perspective, URCS with EWSR1–NFATc2 fusion predominantly occurred as intraosseous lesions, had a striking male predominance and occurred in patients with a slightly higher age at diagnosis compared to EwS (Antonescu 2014; Grunewald et al. 2018; Kinkor et al. 2014; Koelsche et al. 2018a; Machado et al. 2018; Romeo et al. 2012; Sadri et al. 2014; Szuhai et al. 2009; Watson et al. 2018). Surprisingly, two cases of our series were diagnosed in female patients, contradicting published data suggesting that these mainly intraosseous tumors affect male patients only. Our findings are in line with a recent summary reported in the literature, which indicated that EWSR1–NFATc2-positive sarcomas might also arise outside of bone and affect female patients (Grunewald et al. 2018).
Due to the exceptional rarity of URCS with EWSR1–NFATc2 fusion, clinical data regarding the biological behavior is very limited. In studies where follow-up data were available, these cases seemed to follow a more favorable clinical course when compared to classical EwS (Machado et al. 2018; Romeo et al. 2012). In our series, follow-up data were available for three patients, which responded to neoadjuvant chemotherapy and were subsequently treated with tumor resection. In line with previously published data, one of these patients had complete remission, the second patient, which had been primarily diagnosed with metastatic stage disease, is still alive 6 years after diagnosis and the third case had a local recurrence 2 years after diagnosis.
Despite the growing evidence that URCS with EWSR1–NFATc2 fusion may be distinct from EwS, their origin has not yet been resolved. Some EWSR1–NFATc2 fused sarcomas exhibited morphological features matching those of myoepithelial tumors (Cohen et al. 2018; Romeo et al. 2012). Myoepithelial tumors typically arise in the soft parts, but skeletal counterparts have been described (Hornick and Fletcher 2003; Song et al. 2017). Histologically, most of these tumors show lobulated growth and are composed of cords or nests of epithelioid, ovoid, or spindle cells with a chondromyxoid or collagenous/hyalinized stroma (Hornick and Fletcher 2003). They strongly express epithelial markers and S-100 protein, often myogenic markers and in roughly half of the cases glial fibrillary acid protein (Jo and Fletcher 2015). This immunophenotype has not been described in URCS with EWSR1–NFATc2 fusion (Antonescu 2014; Kinkor et al. 2014; Koelsche et al. 2018a; Machado et al. 2018; Romeo et al. 2012; Sadri et al. 2014; Szuhai et al. 2009; Watson et al. 2018). However, myoepithelial tumors often carry EWSR1 gene fusions with a growing number of non-ETS fusion partner genes, e.g., POU5F1 (6p21), PBX1 (1q23), ZNF444 (19q23), ATF1 (12q13) and PBX3 (9q33) (Antonescu et al. 2011, 2010; Flucke et al. 2011). Most recently, a subcutaneous lesion with a phenotype matching with myoepithelial tumors has been described in a young female patient, which carried a EWSR1–NFATc2 fusion and additionally had amplifications in the EWSR1 and NFATc2 regions. This tumor was positive for epithelial markers, but interestingly lacked any myoepithelial marker expression like S-100, desmin or calponin, among others (Cohen et al. 2018).
Unfortunately, myoepithelial tumors were not part of this study and, therefore, could not be used for comparative purposes. However, it seems to become apparent that URCS with EWSR1–NFATc2 fusion is a multifaceted tumor, which is neither restricted to bone, nor exclusively occurs in male patients.
In conclusion, DNA methylation profiling segregates URCS with EWSR1–NFATc2 fusion from EwS with canonical TET–ETS fusions. It is, therefore, unlikely that URCS with EWSR1–NFATc2 and EwS share a common origin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (xlsx 19.142 KB)
Acknowledgements
We thank the Microarray Unit of the Genomics and Proteomics Core Facility, German Cancer Research Center (DKFZ), for providing excellent methylation services. We also thank the HUVR-IBiS Biobank (Andalusian Public Health System Biobank and ISCIII-Red de Biobancos PT17/0015/0041) for providing tissue.
Funding
The work was supported by the German Cancer Aid (Grant 70112499). The laboratory of T.G.P.G. was supported for this project by grants from the Dr. Leopold und Carmen Ellinger Stiftung and the German Cancer Aid (Grant 70112257).
Compliance with ethical standards
Conflict of interest
The authors state no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gunhild Mechtersheimer and Andreas von Deimling contributed equally.
Contributor Information
Christian Koelsche, Phone: +49(0)62215637882, Email: christian.koelsche@med.uni-heidelberg.de.
Andreas von Deimling, Phone: +49(0)6221564651, Email: andreas.vondeimling@med.uni-heidelberg.de.
References
- Antonescu C (2014) Round cell sarcomas beyond Ewing: emerging entities. Histopathology 64:26–37. 10.1111/his.12281 [DOI] [PubMed] [Google Scholar]
- Antonescu CR et al (2010) EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer 49:1114–1124. 10.1002/gcc.20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR et al (2011) EWSR1-ATF1 fusion is a novel and consistent finding in hyalinizing clear-cell carcinoma of salivary gland. Genes Chromosomes Cancer 50:559–570. 10.1002/gcc.20881 [DOI] [PubMed] [Google Scholar]
- Antonescu CR et al (2017) Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: a clinicopathologic and molecular study of 115 cases. Am J Surg Pathol 41:941–949. 10.1097/PAS.0000000000000846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf MC et al (2018a) Are EWSR1-NFATc2-positive sarcomas really Ewing. sarcomas? Mod Pathol 31:997–999. 10.1038/s41379-018-0009-7 [DOI] [PubMed] [Google Scholar]
- Baldauf MC et al (2018b) Robust diagnosis of Ewing sarcoma by immunohistochemical detection of super-enhancer-driven EWSR1-ETS targets. Oncotarget 9:1587–1601. 10.18632/oncotarget.20098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charville GW et al (2018) PAX7 expression in sarcomas bearing the EWSR1-NFATC2 translocation. Mod Pathol. 10.1038/s41379-018-0095-6 [DOI] [PubMed] [Google Scholar]
- Chen S, Deniz K, Sung YS, Zhang L, Dry S, Antonescu CR (2016) Ewing sarcoma with ERG gene rearrangements: a molecular study focusing on the prevalence of FUS-ERG and common pitfalls in detecting EWSR1-ERG fusions by FISH. Genes Chromosomes Cancer 55:340–349. 10.1002/gcc.22336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JN, Sabnis AJ, Krings G, Cho SJ, Horvai AE, Davis JL (2018) EWSR1-NFATC2 gene fusion in a soft tissue tumor with epithelioid round cell morphology and abundant stroma: a case report and review of the literature. Hum Pathol. 10.1016/j.humpath.2018.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alava E, Lessnick SL, Sorensen PH (2013) Ewing Sarcoma. In: Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F (eds) WHO classification of tumours of soft tissue and bone. International Agency for Research on Cancer (IARC), Lyon, pp 305–310 [Google Scholar]
- Delattre O et al (1992) Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359:162–165. 10.1038/359162a0 [DOI] [PubMed] [Google Scholar]
- Flucke U, Palmedo G, Blankenhorn N, Slootweg PJ, Kutzner H, Mentzel T (2011) EWSR1 gene rearrangement occurs in a subset of cutaneous myoepithelial tumors: a study of 18. cases Mod Pathol 24:1444–1450. 10.1038/modpathol.2011.108 [DOI] [PubMed] [Google Scholar]
- Folpe AL, Goldblum JR, Rubin BP, Shehata BM, Liu W, Dei Tos AP, Weiss SW (2005) Morphologic and immunophenotypic diversity in Ewing family tumors: a study of 66 genetically confirmed cases. Am J Surg Pathol 29:1025–1033 [PubMed] [Google Scholar]
- Grunewald TGP et al (2018) Ewing sarcoma. Nat Rev Dis Primers 4:5. 10.1038/s41572-018-0003-x [DOI] [PubMed] [Google Scholar]
- Hornick JL, Fletcher CD (2003) Myoepithelial tumors of soft tissue: a clinicopathologic and immunohistochemical study of 101 cases with evaluation of prognostic parameters. Am. J Surg Pathol 27:1183–1196 [DOI] [PubMed] [Google Scholar]
- Italiano A, Sung YS, Zhang L, Singer S, Maki RG, Coindre JM, Antonescu CR (2012) High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer 51:207–218. 10.1002/gcc.20945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon IS, Davis JN, Braun BS, Sublett JE, Roussel MF, Denny CT, Shapiro DN (1995) A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene 10:1229–1234 [PubMed] [Google Scholar]
- Jo VY, Fletcher CD (2015) Myoepithelial neoplasms of soft tissue: an updated review of the clinicopathologic, immunophenotypic, and genetic features. Head Neck Pathol 9:32–38. 10.1007/s12105-015-0618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y et al (1996) Fusion of an ETS-family gene, EIAF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy Genes Chromosomes Cancer 15:115–121 10.1002/(SICI)1098-2264(199602)15:2%3C115::AID-GCC6%3E3.0.CO;2-6 [DOI] [PubMed] [Google Scholar]
- Kao YC et al (2018) BCOR-CCNB3 fusion positive sarcomas: a clinicopathologic and molecular analysis of 36 cases with comparison to morphologic spectrum and clinical behavior of other round cell sarcomas. Am J Surg Pathol 42:604–615. 10.1097/PAS.0000000000000965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkor Z, Vanecek T, Svajdler M Jr, Mukensnabl P, Vesely K, Baxa J, Kokavec M (2014) Where does Ewing sarcoma end and begin-two cases of unusual bone tumors with t(20;22)(EWSR1-NFATc2) alteration. Cesk Patol 50:87–91 [PubMed] [Google Scholar]
- Koelsche C et al (2018a) Array-based DNA-methylation profiling in sarcomas with small blue round cell histology provides valuable diagnostic information. Mod Pathol 31:1246–1256. 10.1038/s41379-018-0045-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelsche C et al (2018b) Primary intracranial spindle cell sarcoma with rhabdomyosarcoma-like features share a highly distinct methylation profile and DICER1 mutations. Acta Neuropathol 136:327–337. 10.1007/s00401-018-1871-6 [DOI] [PubMed] [Google Scholar]
- Machado I et al (2018) Review with novel markers facilitates precise categorization of 41 cases of diagnostically challenging, “undifferentiated small round cell tumors”. A clinicopathologic, immunophenotypic and molecular analysis. Ann Diagn Pathol 34:1–12. 10.1016/j.anndiagpath.2017.11.011 [DOI] [PubMed] [Google Scholar]
- Mastrangelo T et al (2000) A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene 19:3799–3804. 10.1038/sj.onc.1203762 [DOI] [PubMed] [Google Scholar]
- Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O (1997) A new member of the ETS family fused to EWS in Ewing tumors. Oncogene 14:1159–1164. 10.1038/sj.onc.1200933 [DOI] [PubMed] [Google Scholar]
- Pierron G et al (2012) A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet 44:461–466. 10.1038/ng.1107 [DOI] [PubMed] [Google Scholar]
- Romeo S et al (2012) Malignant fibrous histiocytoma and fibrosarcoma of bone: a re-assessment in the light of currently employed morphological immunohistochemical molecular approaches. Virchows Arch 461:561–570. 10.1007/s00428-012-1306-z [DOI] [PubMed] [Google Scholar]
- Sadri N et al (2014) Malignant round cell tumor of bone with EWSR1-NFATC2 gene fusion. Virchows Arch 465:233–239. 10.1007/s00428-014-1613-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar S, Lessnick SL (2011) Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet 204:351–365. 10.1016/j.cancergen.2011.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savola S et al (2009) Combined use of expression and CGH arrays pinpoints novel candidate genes in Ewing sarcoma family of tumors. BMC Cancer 9:17. 10.1186/1471-2407-9-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shing DC et al (2003) FUS/ERG gene fusions in Ewing’s tumors. Cancer Res 63:4568–4576 [PubMed] [Google Scholar]
- Song W, Flucke U, Suurmeijer AJH (2017) Myoepithelial tumors of bone. Surg Pathol Clin 10:657–674. 10.1016/j.path.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT (1994) A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor. ERG Nat Genet 6:146–151. 10.1038/ng0294-146 [DOI] [PubMed] [Google Scholar]
- Specht K, Sung YS, Zhang L, Richter GH, Fletcher CD, Antonescu CR (2014) Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas: further evidence toward distinct pathologic entities. Genes Chromosomes Cancer 53:622–633. 10.1002/gcc.22172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht K et al (2016) Novel BCOR-MAML3 and ZC3H7B-BCOR Gene Fusions in Undifferentiated Small Blue Round Cell Sarcomas Am. J Surg Pathol 40:433–442. 10.1097/PAS.0000000000000591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumegi J, Nishio J, Nelson M, Frayer RW, Perry D, Bridge JA (2011) A novel t(4;22)(q31;q12) produces an EWSR1-SMARCA5 fusion in extraskeletal Ewing sarcoma/primitive neuroectodermal tumor. Mod Pathol 24:333–342. 10.1038/modpathol.2010.201 [DOI] [PubMed] [Google Scholar]
- Szuhai K, Ijszenga M, de Jong D, Karseladze A, Tanke HJ, Hogendoorn PC (2009) The NFATc2 gene is involved in a novel cloned translocation in a Ewing sarcoma variant that couples its function in immunology to oncology. Clin Cancer Res 15:2259–2268. 10.1158/1078-0432.CCR-08-2184 [DOI] [PubMed] [Google Scholar]
- Wang L, Bhargava R, Zheng T, Wexler L, Collins MH, Roulston D, Ladanyi M (2007) Undifferentiated small round cell sarcomas with rare EWS gene fusions: identification of a novel EWS-SP3 fusion and of additional cases with the EWS-ETV1 and EWS-FEV fusions. J Mol Diagn 9:498–509. 10.2353/jmoldx.2007.070053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson S et al (2018) Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol 245:29–40. 10.1002/path.5053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (xlsx 19.142 KB)