Abstract
Background
Unveiling fungal genome structure and function reveals the potential biotechnological use of fungi. Trichoderma harzianum is a powerful CAZyme-producing fungus. We studied the genomic regions in T. harzianum IOC3844 containing CAZyme genes, transcription factors and transporters.
Results
We used bioinformatics tools to mine the T. harzianum genome for potential genomics, transcriptomics, and exoproteomics data and coexpression networks. The DNA was sequenced by PacBio SMRT technology for multiomics data analysis and integration. In total, 1676 genes were annotated in the genomic regions analyzed; 222 were identified as CAZymes in T. harzianum IOC3844. When comparing transcriptome data under cellulose or glucose conditions, 114 genes were differentially expressed in cellulose, with 51 being CAZymes. CLR2, a transcription factor physically and phylogenetically conserved in Trichoderma spp., was differentially expressed under cellulose conditions. The genes induced/repressed under cellulose conditions included those important for plant biomass degradation, including CIP2 of the CE15 family and a copper-dependent LPMO of the AA9 family.
Conclusions
Our results provide new insights into the relationship between genomic organization and hydrolytic enzyme expression and regulation in T. harzianum IOC3844. Our results can improve plant biomass degradation, which is fundamental for developing more efficient strains and/or enzymatic cocktails to produce hydrolytic enzymes.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-07158-w.
Keywords: Cellulose degradation, CAZymes, Genomic, Transcriptome, Fungi
Background
Trichoderma is a very diverse genus of fungi that produces enzymes applied in different areas; some strains are applied in biocontrol (T. atroviride, T. harzianum and T. virens) [1, 2], and others are specific for the biofuel technology (T. reesei and T. harzianum) [3, 4]. Different strains of T. harzianum have high cellulolytic activity, and the potential of these enzymes has been explored for applications in biomass degradation to the production of biofuels [4, 5].
Trichoderma harzianum is a common fungal species in soil and is used as a biological control against a variety of phytopathogenic fungi [6]. Despite that, T. harzianum capacity to biomass degradation is still poorly explored compared to that of other cellulolytic fungi. Due to the high cellulolytic activity some strains has shown considerable potential for application in plant biomass hydrolysis [4, 7, 8]. T. harzianum strains have potential for the production of an enzymatic/protein arsenal necessary for the complete hydrolysis of cellulosic compounds in fermentable sugars [5, 9–11].
Currently, the most-studied and widely used industrial-scale enzymes are produced by the fungus T. reesei and species from the Aspergillus genus. These organisms are the sources of most enzymes comprising enzymatic cocktails that are currently available on the market [12]. T. reesei is a widely studied fungus and is found in several works on genomics, transcriptomics, proteomics and metabolic engineering [3, 13–16]. Thus, increasing the number of studies related to the biotechnological potential of T. harzianum is necessary.
Different strains of a specific fungal species have different potentials for the degradation of plant biomass, and these differences may be associated with differences in the genome and regulation of CAZyme enzymes [17]. T. harzianum IOC3844 is a strain that showed high potential for the degradation of plant biomass in several studies [4, 10], demonstrated via the high expression of genes related to cellulose (CEL) and hemicellulose degradation [18], enzymatic activity [4, 17] and synthetic biology [19].
The three main groups involved in the hydrolysis of CEL are cellobiohydrolases, endo-β-1,4-glucanases and β-glucosidases. In addition, accessory enzymes such as copper-dependent lytic polysaccharide mono-oxygenases (LPMOs), cellulose-induced protein (CIP1 and CIP2) and swollenin also participate in this process [20–23].
One of the great challenges in understanding the molecular mechanism of biomass degradation is to capture how the transcription factors (TFs) act. Several fungal TFs have been related to the degradation of plant biomass, many of which belong to the binuclear zinc family [24]. Many TFs have been described as directly involved in the regulation of plant biomass [25]. This number has expanded rapidly in recent years, mainly due to the development of whole genome sequencing technologies associated with the exponential increase in the number of bioinformatics analysis tools that produced massive amounts of information and increased the numbers of identified genes [25, 26].
The present study aimed to analyze genomic regions with CAZyme genes using a bacterial artificial chromosome (BAC) library built in house [27] and to integrate these data with RNA-Seq, secretome data and coregulation networks. We sequenced a massive amount of DNA and used the information obtained to integrate genomic data (genomic regions containing CAZymes), expression patterns (transcriptome under degradation conditions), proteins (secretome by mass spectrometry) and systems biology (with gene regulatory networks) to obtain a broad and precise overview of the CEL degradation pathways. Our study characterized the main genes, accessory enzymes and regions involved in the degradation and regulation of hydrolytic enzymes. In addition, we analyzed the regulator cellulose degradation regulator 2 (CLR2) found in a cluster with other important enzymes. These results will be important for further studies on regulation and gene silencing.
Results
Genomic regions of T. harzianum IOC3844
In this study, a library of large genomic regions was used as a platform to search for genes of interest and to thoroughly study the genomic structure of T. harzianum IOC3844 (ThIOC3844) (Additional file 1: Fig. S1 and Additional file 2: Supplementary Table S1). Screening for genes of interest resulted in a total of 62 regions that contained CAZyme genes related to the degradation of plant biomass in the ThIOC3844 genome. Sequencing of these regions generated 5 Mb total of the estimated 40 Mb genome (Additional file 1: Supplementary Table S2 and S3). These regions ranged in size from 43 to 152 kb, enabling the prediction and annotation of 1676 gene models for this strain (Additional file 3: Supplementary Table S4). The average number of genes per region was 26 (Additional file 2: Supplementary Table S1).
The genome of T. reesei QM6a (PRJNA325840) was used to analyze the distribution of genes in ThIOC3844. This genome, composed of seven chromosomes with a total size of 34 Mb, was divided into 38 intervals (1 Mb) (Fig. 1). CAZyme genes annotated in ThIOC3844 were distributed throughout all T. reesei QM6a (TrQM6a) chromosomes. It was possible to map ThIOC3844 genes at all intervals in which the chromosomes were divided; mapping CAZyme genes was not possible in only four intervals.
Fig. 1.

Distribution of the T. harzianum IOC3844 genes on the 1 Mb intervals of the seven chromosomes of T. reesei QM6a. CAZyme genes of T. harzianum IOC3844 are in red, CAZyme genes of T. reesei are in blue, and all genes of T. harzianum IOC3844 are in green. Th: T. harzianum IOC3844; Tr: T. reesei QM6a
The genes were functionally annotated for the main gene ontologies: biological processes, cellular components and molecular functions (Fig. 2a and Additional file 1: Supplementary Fig. S2). We found 209 sequences related to hydrolytic activity, 139 related to transport proteins and 85 sequences related to the regulation of gene expression (possible TFs). In addition, a specific annotation was made for genes identified as enzymes, among which hydrolases (40%), oxidoreductases (25%), transferases (22%), lyases (6%), ligases (4%) and isomerases (3%) (Fig. 2b) were found. We also identified genes directly related to the degradation of CEL and hemicellulose, with activities of α-L-arabinofuranosidase (enzyme commission (EC) 3.2.1.55), endo-1,4-β-xylanases (EC 3.2.1.8), cellobiohydrolases (3.2.1.91), endo-β-1,4-glucanase (EC 3.2.1.4) and β-glucosidase (EC 3.2.1.21) (Fig. 2c and Additional file 4: Supplementary Table S5).
Fig. 2.
Functional annotation of the genes predicted in the genomic regions of T. harzianum IOC3844. Annotation of genes for ontologies of biological processes, cellular components and molecular functions. a Distribution of enzymes annotated according to enzyme commission (b) and major enzyme commission (EC) related to cellulose and hemicellulose degradation (c)
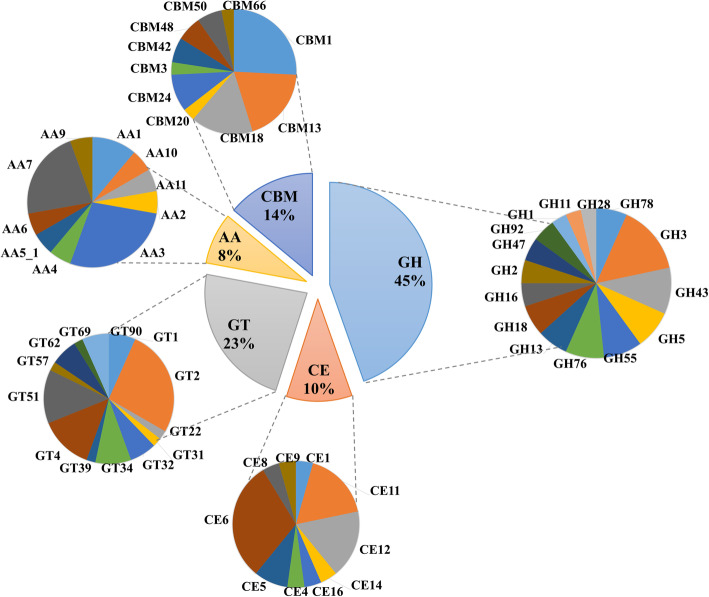
A total of 1676 genes/proteins were predicted (Additional file 3: Supplementary Table S4). Of these, 222 were annotated as CAZymes in ThIOC3844, including 45% annotated as glycoside hydrolases (GHs), 23% annotated as glycosyl transferases (GTs), 10% annotated as carbohydrate esterases (CEs), 8% annotated as auxiliary activities (AAs) and 14% annotated as carbohydrate-binding modules (CBMs) (Fig. 3 and Additional file 5: Supplementary Table S6). The GH class presented with the highest number of families, including GH2 (3 genes), GH7 (1 gene), GH3 (9 genes), GH5 (6 genes), GH12 (1 gene), GH18 (4 genes) and GH62 (1 gene).
Fig. 3.
CAZy classification of genes annotated in the genomic regions of T. harzianum IOC3844. GH: glycoside hydrolases; GT: glycosyl transferases; PLs: polysaccharide lyases; CEs: carbohydrate esterases; AA: auxiliary activities; CBM: carbohydrate-binding modules
Genomic comparison
For this analysis, we compared the genomic regions of ThIOC3844 against the entire genomes of different strains and species of the genus Trichoderma. Genomic comparison of the sequenced regions of ThIOC3844 with two other strains of the same species (T. harzianum B97 – ThB97 and T. harzianum – T6766) showed a higher similarity to ThB97 (99.25%) than to ThT6766 (91.61%). For the T. atroviride IMI206040 genome (TaIMI206040), the similarity to ThIOC3844 was 85.09%. For T. virens Gv29–8 (TvGv29–8), the similarity was 86.55%, and for T. reesei QM6a (TrQM6a), the similarity was 85.11%.
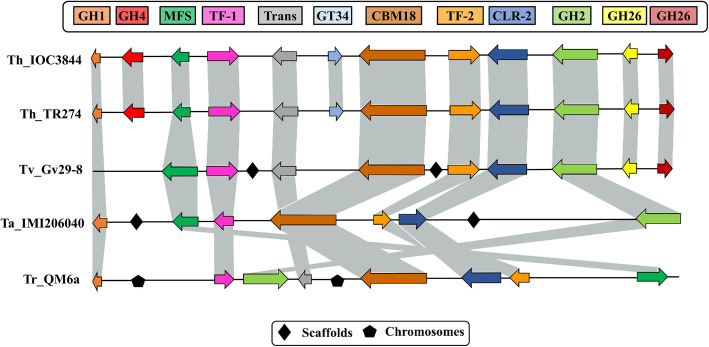
When we compared syntenic genes between groups of genes, a greater difference between T. harzianum, T. atroviride and T. reesei was observed. The T. harzianum TR274 (ThTR274) strain presented the same genomic organization gene profile as ThIOC3844. In TaIMI206040, four genes (GH4, transporter and two GH26) from the cluster were not found; for TvGv29–8, two genes were not found (GH1 and GH4). For T. reesei QM6a, three genes (GH4 and two GH26) were not found; in addition, the translocation of genes (MFS x GH2 and TF2 x CLR2) was found. The genes for the TF CLR2, putative TF TF2 and major facilitator superfamily (MFS) permease were maintained in all species analyzed. This result suggests a potential association between the regulation and expression of these genes (Fig. 4).
Fig. 4.
Comparison between the gene clusters of T. harzianum IOC3844 and those of other species of the genus Trichoderma spp. GH1: glycoside hydrolase 1; GH4: glycoside hydrolase 4; MFS: major facilitator superfamily permease; Trans: putative transporter; TF-1: putative transcription factor 1; GT38: glycosyl transferases 4; CBM18: carbohydrate-binding module 18; TF-2: putative transcription factor 2; CLR2: cellulose regulator 2; GH2: glycoside hydrolase 2; GH26: glycoside hydrolase 26; Th: T. harzianum; Tv: T. virens; Ta: T. atroviride; Tr: T. reesei
Expression determined by RNA-Seq and secreted proteins
All genes predicted in the genomic regions were analyzed according to the expression data by RNA-Seq (under CEL and glucose (GLU) degradation conditions) (Additional file 6: Supplementary Table S7), and secreted proteins were identified by mass spectrometry (LC-MS/MS) (Additional file 7, data obtained in a previous experiments of T. harzianum IOC3844 [18]). We found 114 genes with differential expression under CEL degradation conditions compared to GLU degradation conditions; among them, 51 were classified as CAZymes, such as beta-glucosidase of GH1 family (1.8-fold change - FC), LPMOs of the AA9 family (FC 5.0) and a hypothetical protein with CBM1 domain (FC 3.7). In addition, two differentially expressed TFs were identified, CLR2 (FC 1.6) and unidentified transcriptional regulator of zing finger – Zn2Cys6 (FC 2.3). Six transport proteins were also found (iron permease, MFS hexose transporter, siderophore transporter, ammonium permease, sugar transporter and siderophore iron transporter).
Among the genes annotated as CAZymes in ThIOC3844, 31 were found in the secretome of ThIOC3844 under CEL conditions, and the main families were GH3, GH12, CBM1, AA9, GH6/CBM1, GH45/CBM1, GH62 and GH5. In this analysis, we also used the expression levels of the secreted proteins. The gene with the highest transcripts per million (TPM) index (1567.4 TPM) was a cellobiohydrolase (EC 3.2.1.91) of the GH6 family. Our results indicate that genes with low expression levels are also important secreted enzymes (Table 1).
Table 1.
Proteins identified in genomic and in the T. harzianum IOC3844 secretome under cellulose growth conditions
| IDsa | Protein name | Secretome/UniProt ID | CAZy family | CEL (TPM) | GLU (TPM) |
|---|---|---|---|---|---|
| 1010 | Hypothetical protein | A0A0G0ALT6 | GH28 | 14.2 | 6.2 |
| 1043 | Cellulosome enzyme | A0A0G0A296 | GH30 | 35.6 | 11.5 |
| 1054 | Glycosyl hydrolase 10 | A0A0F9X8A4 | GH10 | 14.8 | 4.1 |
| 1075 | Glycosyl hydrolase 64 | A0A0F9ZIR5 | GH64 | 824.3 | 262.4 |
| 1095 | Glycosyl hydrolase 18 | A0A0F9ZHI0 | GH18 | 83.2 | 47.9 |
| 11 | Mutanase | A0A0F9XN06 | CBM24 | 2741.6 | 1452.9 |
| 1133 | Glycosyl hydrolase 12 | A0A0F9Y2E9 | GH12 | 1579.8 | 308.2 |
| 1150 | Glycosyl hydrolase 47 | A0A0F9WYR7 | GH47 | 83.9 | 74.6 |
| 1217 | Beta-mannosidase | A0A0F9ZDV4 | GH2 | 117.9 | 124.2 |
| 126 | Glycosyl hydrolase 76 | A0A0F9X1Q3 | GH76 | 616.7 | 375.4 |
| 1318 | Beta-xylosidase | A0A0G0A408 | GH3 | 172.3 | 125.2 |
| 1439 | Alpha-L-arabinofuranosidase B | A0A0G0A4Q2 | CBM42 | 450.4 | 343.5 |
| 1440 | Glycosyl hydrolase 3 | A0A0F9XRC5 | GH3 | 245.8 | 107.4 |
| 1498 | WSC domain-containing | A0A0F9ZXC9 | AA5_1 | 342.5 | 339.0 |
| 44 | Beta-1,3-glucanosyltransferase | A0A0F9ZKA8 | GH72 | 2431.7 | 3210.5 |
| 441 | Alpha-glucosidase | A0A0G0AG54 | GH31 | 2121.6 | 1655.2 |
| 559 | Alpha-1,2-mannosidase | A0A0G0ABI9 | GH92 | 226.9 | 153.4 |
| 666 | Glycosyl hydrolase 3 | A0A0F9XQT4 | GH3 | 77.2 | 43.0 |
| 667 | Hypothetical protein | A0A0G0AME2 | CBM1 | 874.9 | 142.8 |
| 668 | Glycosyl hydrolase 61 | A0A0F9XMI8 | AA9 | 3109.7 | 625.1 |
| 669 | Glycosyl hydrolase 16 | A0A0F9XP75 | CBM13 | 16.4 | 3.7 |
| 671 | Cytochrome P450 monooxygenase | A0A0G0A4Z5 | GT4 | 1569.5 | 1595.3 |
| 681 | Glycosyl hydrolase 11 | A0A0F9Y0Y9 | GH11/CBM1 | 4206.8 | 1316.1 |
| 741 | Endo-N-acetyl-beta-D-glucosaminidase | A0A0F9ZHA7 | GH18 | 3971.9 | 2328.0 |
| 759 | Hypothetical protein | A0A0F9ZJ74 | GH20 | 1184.8 | 1507.7 |
| 813 | Catalase peroxidase | A0A0F9X3Z8 | AA2 | 2677.7 | 2473.4 |
| 82 | Glycosyl hydrolase 6 | A0A0G0AEM7 | GH6/CBM1 | 5843.5 | 1567.4 |
| 842 | Hypothetical protein | A0A0F9XY55 | GH45/CBM1 | 41.3 | 15.3 |
| 9 | Glycosyl hydrolase 62 | A0A0F9X8Z0 | GH62 | 353.9 | 103.4 |
| 913 | Isoamyl alcohol oxidase | A0A0F9XC99 | AA7 | 39.3 | 13.2 |
| 918 | Hypothetical protein | A0A0F9XG06 | GH5_5 | 870.8 | 750.9 |
aThe annotated genes IDs can be found in Supplementary Table S4
CLR2 transcription factor
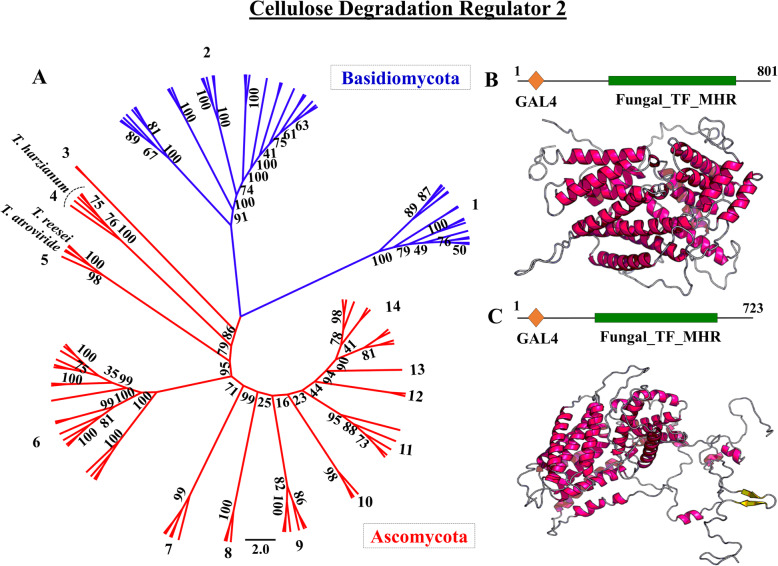
Phylogenetic analysis of the CLR2 factor showed a clear separation of this TF in relation to Basidiomycetes and Ascomycetes (Fig. 5a and Additional file 1: Supplementary Table S8). However, even within these groups, considerable phylogenetic diversity was observed among the species of analyzed fungi with a variety of clades within the same group. Different strains of T. harzianum grouped in a single clade with proximity to T. reesei and T. atroviride species. Our results show a wide range of functional varieties for CLR2, which may indicate different types of performance between species.
Fig. 5.
Molecular phylogeny of the CLR2 transcription factor in Ascomycota and Basidiomycota (a); in silico protein modeling for CLR2 in T. harzianum IOC3844 (b) and T. reesei QM6a (c)
Structural modeling analysis of the CLR2 protein of ThIOC3844 was performed using T. reesei as a reference. For both proteins, the best template was 6F07 (Saccharomyces cerevisiae), with e-values of 4.07e− 06 and 6.62e− 06 for ThIOC3844 (Fig. 5b) and T. reesei (Fig. 5c), respectively. Predictions of 1 and 3 protein domains were made for ThIOC3844 and T. reesei, respectively. For ThIOC3844, 59% of the residues were modeled, while T. reesei modeled 83% of residues. For ThIOC3844, the secondary structure prediction was 46% H (helix), 0% E (beta-sheet) and 53% C (loop), while solvent access had predictions of 56% E (exposed), 19% M (medium) and 23% B (buried).
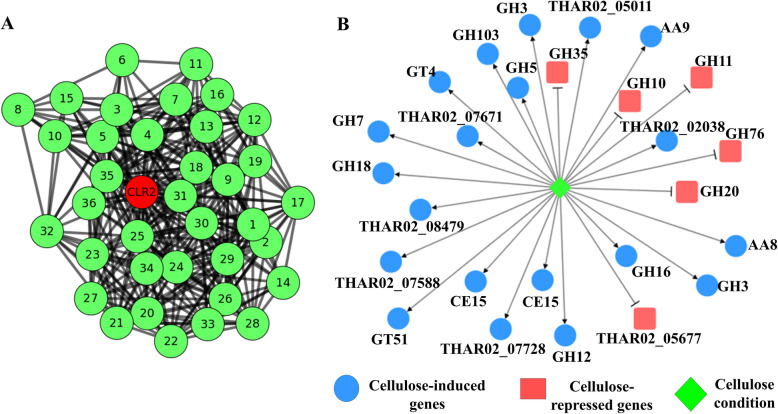
A coregulation network of genes directly related to the CLR2 regulator was constructed, to search for insights about other important proteins in the process of cellulases expression. We identified 36 genes directly linked to CLR2, of which 21 genes were annotated as hypothetical proteins. In addition, genes with known annotations were related to the process of gene expression, including initiation factors, kinases and helicases (Fig. 6a and Additional file 1: Supplementary Table S9).
Fig. 6.
Subnetwork of CLR2 transcription factors and related genes (a) and network of induced (blue) and repressed (red) genes under cellulose conditions (b). CLR2: cellulose regulator 2; GH: glycoside hydrolases; GT: glycosyl transferases; AA: auxiliary activities
Network of induced/repressed genes in cellulose
Using the gene expression data of the secreted proteins, a Bayesian network of induced/repressed genes was constructed based on the CEL growth conditions for T. harzianum IOC3844 (Fig. 6b). The major genes that were induced under this condition belong to the GH7 (exoglucanase), GH5 (endo-β-1,4-glucanase), GH3 (β-glucosidase), GH12 (murein transglycosylase), CE15 (CIP2), AA9 (LPMO) and AA8 (hypothetical protein) families. In addition, seven genes that were not classified as CAZymes were also induced under CEL conditions. The families of repressed genes were GH10 (glycoside hydrolase 10 family endo-1,4-β-xylanase), GH11 (glycoside hydrolase 11 family endo-1,4-β-xylanase), GH76 (alcohol dehydrogenase 1), GH20 (β-N-acetylhexosaminidase) and GH35 (glycoside hydrolase 35).
Discussion
In the present study, an integrative multiomics approach was used to mine CAZyme-rich regions of ThIOC3884. BAC clones were selected, sequenced and used in comparative analyses focusing on the expression profiles via RNA-Seq and the secretome, under different fungal growth conditions. This allowed the discovery of important genes/proteins mechanisms related to plant biomass degradation (Additional file 1: Supplementary Fig. S3).
The vast majority of enzymes that are important for the degradation of plant biomass are already known [28–30]. The current challenge is determining how enzymes are regulated and the genetic mechanism of their activation. Thus, many studies with cellulolytic fungi have focused on TFs, accessory enzymes, transporters and the mechanism by which different types of biomass affects cellulase and hemicellulose regulation [25, 31–33]. Other studies have already shown the potential of T. harzianum for the degradation of plant biomass. This is the first study that integrates results from different biotechnological approaches and focuses on the prediction of the most important enzymes and TFs used by T. harzianum IOC3844 to degrade CEL.
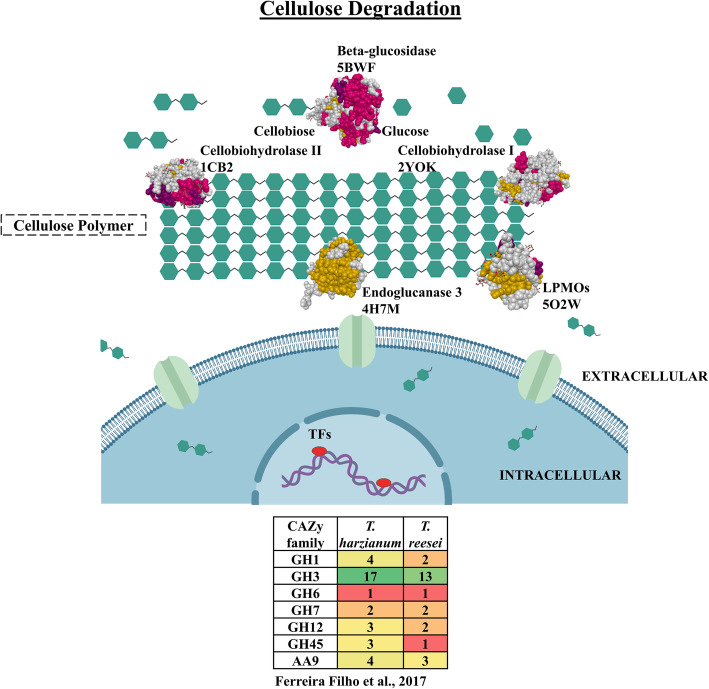
The molecular process of CEL degradation is extremely complex and involves hydrolytic enzymes acting on the extracellular medium, carrier proteins and TFs (Fig. 7). For T. harzianum and T. reesei, the major CAZy families related to CEL degradation were identified in the genome (GH1, GH3, GH6, GH7, GH12, GH45 and AA9) [34], and the three-dimensional structures of cellulases have already been solved. Despite that, many key proteins in the degradation process still not well known, as well as transporters and TFs related to the regulation of related enzymes.
Fig. 7.
Molecular scheme of the enzymatic model in the degradation of cellulose in Trichoderma spp. Enzymes and PDB code: beta-glucosidase (5BWF), cellobiohydrolase I (2YOK), cellobiohydrolase II (1CB2), endoglucanase 3 (4H7M), copper-dependent lytic polysaccharide mono-oxygenases (LPMOs) (5O2W)
The study of genomic regions is an important tool that provides a global view of the important genes and regulatory regions of a genome [27, 35]. The genomes of few strains of T. harzianum are available [36, 37]. A complete genome draft sequenced in 1572 scaffolds is available for T. harzianum T6776 [36]; however, little is known about the ThIOC3844 genome, and as this strain has potential for hydrolytic enzymes, more genomic information regarding CAZyme sequences is needed. Our strategy herein aimed to use large genomic regions and integrate these data with other genetic information.
Although the T. harzianum T6776 genome is available, it is divided into several small-sized scaffolds [36], which makes impossible in some cases to study gene clusters, regulatory and promoter regions. However, sequencing several regions of the T. harzianum IOC3844 genome with single-molecule real-time (SMRT) technology made possible to obtain large scaffolds with reliable assembly. Our sequencing approach allowed to determine the main regions containing important CAZymes involved in the degradation of CEL and hemicellulose, beyond the study of these genes’ clusters.
In the used approach, we chose to select regions of interest instead of sequencing the complete T. harzianum IOC3844 genome; this complex approach was only scientifically and economically viable due to previous studies in which we obtained information on the key enzymes in this strain related to the biomass degradation process and optimized an approach for rapid selection and sequencing of large genomic regions for fungi and plants [27, 38]. In this way, we were able to carry out genomic enrichment work quickly and at low cost.
A large number of fungal genomes have already been used as a platform to search for new genes related to the degradation of biomass. It is the case of T. reesei QM6a, which has a finalized genome divided into seven chromosomes [39]. Our study results with the genomic regions of ThIOC3844 showed a large number of enzymes classified as CAZymes as well as TFs and transporters in clusters in the genome, important information for future studies on genetic modification of this lineage.
Analyzing the expression levels of determined genes under certain conditions is an important step to understand how transcription is affected in a specific biological condition [12]; however, a direct relationship does not always exist between the gene being highly expressed and the proteins that are important/active in the extracellular medium. Thus, in this work, in addition to studying the genes most expressed in the genomic regions, we also searched for those with a confirmed presence in the fungal secretome under CEL degradation conditions. Our results described the key CAZy families to the degradation of CEL, with high levels of expression and a positive presence as secreted proteins.
Genomic comparison is a powerful tool for understanding differences and evolutionary dynamics among related species [40–42]. Our data show a high similarity among different strains of T. harzianum (IOC3844, B97 and T6776), which indicates that differences in enzyme production and efficiency may be related more to gene regulation mechanisms than to differences in the sequence itself. In addition, synteny analysis showed a greater difference in relation to the T. reesei genome, which can be explained by the loss of genes and genomic modifications carried out in T.reesei lineages that potentially increases the production of enzymes related to plant biomass degradation [13, 43].
Despite the fact that genomic sequences of different T. harzianum strains are highly similar, studies from our group showed that stains differences can be observed in terms of regulation of CEL degradation-related genes and in the enzyme activity profiles [17, 18]. Therefore, punctual genomic differences between T. harzianum strains (such as SNPs in introns and gene regions) can play an important role in the efficiency of strains.
The CLR2 TF was described as an important regulator of cellulase expression on Neurospora crassa [25]; however, CLR2 functional role in fungi of the genus Trichoderma, including T. reesei, remains unclear [15, 44]. In the ThIOC3844 genome, we found a cluster with the CLR2 TF in association with other putative TFs, CAZymes, transporters and MFS permease. The same behavior was found for the T. reesei CLR2 TF, which has physical proximity and is coexpressed with a sugar transporter [32, 45]. These results indicate a mechanism for joint regulation and expression of this TF with transporters related to biomass degradation. RNA-Seq data showed differential expression of CLR2 under CEL conditions. In this way, we analyzed the coregulation network of the CLR2 regulator. The present study illuminates unclear areas of the genomic organization, expression, and putative regulation of CLR2 in T. harzianum.
Coregulation networks provide insights into how genes correlate and interact with each other [46, 47]. We identified 36 genes directly associated with the CLR2 regulatory factor; these genes may be important in the regulatory process of this factor, which is linked to the expression of cellulases in other filamentous fungi. Techniques such as gene knockout can further validate the functional or synergistic importance of these genes with key TFs for the expression of genes related to the degradation of plant biomass.
Conclusions
Our results present an innovative approach of using different types of omics data to search for new important genes and genetic regulation mechanisms during the process of CEL degradation. We found several TFs, accessory enzymes, and transporters in the genomic regions of ThIOC3844 that may be important for the expression/secretion of CAZyme genes. Among these, CLR2, CIP2 and LPMOs are promising candidates for further study. Our results indicate that the CRL2 regulator matches all the requirements for involvement in CEL degradation by T. harzianum. In addition, the use of coregulation networks makes it possible to understand the relationship between genes and to find new targets for biochemical characterization. The results allowed the identification of important genetic regions, key genes and functional proteins, and this information can be used for the development and improvement of enzymatic hydrolysis technology for the bioethanol industry.
Methods
T. Harzianum strain and genomic resources
T. harzianum IOC3844 (ThIOC3844) was obtained from Institute Oswaldo Cruz (IOC, Rio de Janeiro, Brazil) and deposited on the Brazilian Collection of Environment and Industrial Microorganisms (CBMAI, Campinas, Brazil). A library of BACs consisting of 5760 clones previously constructed for this fungal strain [27] was used to search for genomic regions. The genomic sequences of T. harzianum T6776 (PRJNA252551), T. reesei QM6a (PRJNA325840), T. atroviride IMI206040 (PRJNA19867) and T. virens Gv29–8 (PRJNA19983) were used for comparison with ThIOC3844.
BAC library screening for gene selection in T. harzianum IOC3844
We designed primers for 62 target CAZyme genes (Additional file 2: Supplementary Table S1) using transcriptome data [4] to search for positive BAC clones that contain genes previously selected. A pool of clones from each plate that made up the BAC library was used for selection (the complete BAC library consisted of fifteen 384-well plates), and pools with clones of all columns of the plates (24 columns on each plate) were also used. In this way, three selections were made to find the positive clone, searches in the plate pool, in the column pool and in the positive column, to obtain the coordinates of the positive clones. The plate and column pools were amplified using the Illustra GenomiPhi HY DNA Amplification Kit (GE Healthcare Life Sciences, UK) according to the manufacturer’s instructions. The screening reactions for the search for positive clones were performed via PCR using the CFX384 Touch Real-Time PCR Detection System (Bio-Rad).
Single-molecule real-time (SMRT) sequencing and assembly
Libraries for sequencing were prepared according to the Pacific Biosciences (PacBio) protocol, and sequencing was performed at the Arizona Genomics Institute (AGI; Tucson, USA) using a SMRT DNA sequencing system available from PacBio (PacBio RSII platform). The sequences were deposited into the NCBI SRA databank under bioproject number PRJNA647392. De novo assembly was performed with the PacBio Corrected Reads (PBcR) pipeline implemented as part of Wgs-assembler v8.3rc2 [48] and Celera Assembler [49]. The contigs obtained with the assemblers were subjected to error correction with pbalign (v0.2). The PacBio reads were aligned using the BLASR algorithm [50], and assembly polishing was performed with the Quiver tool (accession numbers MK861589-MK861650 and Additional file 1: Supplementary Table S2 and S3) [51].
Gene prediction and functional annotation
The FGENESH tool was used for initial gene prediction analysis [52], followed by manual correction with the T. harzianum T6776 and T. reesei QM6a gene models. Annotations of the ontologies were performed with Blast2GO [53]. InterPro protein domains were predicted using InterProScan (http://www.ebi.ac.uk/interpro/) [54]. Information derived from the CAZy database was downloaded for each CAZyme family (www.cazy.org). The protein sequences of T. harzianum IOC3844 were used as queries in basic local alignment search tool (BLASTp) searches against the locally built CAZyme BLAST database. Only BLAST matches showing an e-value less than e-10, identity greater than 30% (the identity value allows capture of the similarity between proteins that have conserved functional domains, as is the case for proteins with CBM domains), and queries covering more than 70% of the sequence length were retained and classified as GHs, GTs, polysaccharide lyases (PLs), CEs, CBMs or AAs according to the CAZyme catalytic activities.
Genomic comparison in Trichoderma spp.
The genomes of Trichoderma (T. harzianum, T. virens, T. atroviride and T. reesei) were compared with that of T. harzianum IOC3844 using global alignment through Nucmer (−maxmatch), which is part of the software package MUMmmer 3.23 [55]. Delta-filter (−q), show-coords (−rcl), and DNADIFF (standard parameters) were used for filtering, obtaining the mapping coordinates and generating the statistical report in the alignment, respectively. SimpleSynteny software (https://www.dveltri.com/simplesynteny/) [56] was used to compare a cluster of 12 genes (in physical proximity to the CLR2 TF) among different species of Trichoderma spp.
Phylogenetic analysis and structural modeling of CLR2
The CLR2 sequences of ThIOC3844, T. reesei QM6a, T. atroviride, T. virens and other species of fungi were used as the basis for constructing the phylogenetic trees. These fungi were divided into Ascomycetes and Basidiomycetes. The sequences were aligned using ClustalW [57] and analyzed with Molecular Evolutionary Genetics Analysis (MEGA) software v7.0 (https://www.megasoftware.net/) [58]. The phylogenetic analyses were performed in MEGA7 using the maximum likelihood (ML) [59] method of inference based on the Jones-Taylor-Thornton (JTT) matrix-based model and 1000 bootstrap replicates [60]. Pairwise deletion was employed to address alignment gaps and missing data. The trees were visualized and edited using the FigTree program (http://tree.bio.ed.ac.uk/software/figtree/). In silico modeling of the CLR2 domain was performed using RaptorX protein structure prediction software (http://raptorx.uchicago.edu/) [61].
RNA-Seq and secreted protein analysis
The ThIOC3844 expression levels were analyzed using RNA-Seq data (PRJNA336221) obtained from a previous study in which the transcripts were obtained following growth of the fungus on two different carbon sources, CEL and GLU [18]. The reads from the RNA-Seq library were mapped against those of the ThIOC3844 genes using the CLC Genomics Workbench (QIAGEN, Aarhus, Denmark) [62]. The expression values are expressed in reads per kilobase of exon model per million mapped reads (RPKM), and the normalized value for each sample was calculated in TPM. For the analysis of differential expression, the following parameters were used: fold change (FC) greater than or equal to 1.5 and a p-value lower than 0.05. Secreted proteins were analyzed using a Blastp search of the annotated proteins in ThIOC3844 against a local protein database (Additional file 7) generated by a previous study that determined the proteins secreted by the fungus under CEL and GLU conditions using a liquid chromatography tandem mass spectrometry (LC-MS/MS) technique [18].
Gene regulatory network
The gene regulatory networks were assembled from the reference mapped RNA-Seq data using each set of biological triplicates for the CEL and GLU conditions [18]. The interaction between the genes was obtained by calculating Pearson’s correlation for each pair of genes. The induction and repression networks were constructed based on the expression data of a set of genes that were identified in the secretome of the CEL growth condition by the Bayesian inference method [63]. If the secreted protein was present in the condition, it was assigned a value of one. If the secreted protein was absent, it was assigned a value of zero. The treatment conditions were considered regulators of the network to detect the direct relationships between the conditions and the genes. Thus, the Bayesian network represents the relationships among the conditions, gene expression, and secreted proteins. Cytoscape software v 3.4.042 [64] (https://cytoscape.org/) was used for data analysis and construction of the CLR2 subnetwork.
Supplementary information
Additional file 1 Fig. S1. Screening genes of interest in the genomic library of T. harzianum IOC3844 by qPCR (a); read size sequenced using PacBio technology (b); genes clustered in a genomic region of T. harzianum (c). Fig. S2. Distribution of the main GO terms of the annotated genes in T. harzianum IOC3844. Fig. S3. Pipeline approach for the analyses used in this study of genes and genomes in T. harzianum. Supplementary Table S2. Assembly parameters of a set of sequenced genomic regions using PacBio technology. Supplementary Table S3. Comparison of genomic data among different species of Trichoderma spp. Supplementary Table S8. Description of the species used for the phylogenetic analysis of the transcription factor CLR2. Supplementary Table S9. Description of the genes found in the coregulation networks.
Additional file 2 Supplementary Table S1. Description of the genomic regions sequenced in T. harzianum IOC3844.
Additional file 3 Supplementary Table S4. Annotation of all genes predicted in T. harzianum IOC3844.
Additional file 4 Supplementary Table S5. Description of the EC codes for T. harzianum IOC3844 genes.
Additional file 5 Supplementary Table S6. Description of the CAZyme genes in T. harzianum IOC3844.
Additional file 6 Supplementary Table S7. Expression levels of the genes annotated in T. harzianum IOC3844 by means of RNA-Seq.
Additional file 7 FASTA sequences of proteins secreted in T. harzianum IOC3844 under cellulose conditions developed in the work of Horta et al., 2018.
Acknowledgments
We thank Hélène Bergès, Elisa Prat, Nadège Arnal, Arnaud Bellec and Sonia Vautrin (French Plant Genomic Resources Center – CNRGV, French National Research Institute for Agriculture, Food and Environment - INRA, France) for their helpful insights, for performing the BAC vector extraction and for their contributions to the BAC library construction and manipulation. We also thank David Kudrna (Arizona Genomics Institute – The University of Arizona, USA), who assisted with the PacBio BAC sequencing. This manuscript was previously posted to bioRxiv: https://www.biorxiv.org/content/10.1101/731323v1.
Abbreviations
- AA
Auxiliary activities
- B
Buried
- BAC
Bacterial artificial chromosome
- BLAST
Basic local alignment search tool
- bp
Base pair
- BRENDA
Braunschweig enzyme database
- C
Loop
- CAZymes
Carbohydrate-active enzymes
- CBMAI
Brazilian collection of environment and industrial microorganisms
- CBM
Carbohydrate-binding module
- CE
Carbohydrate esterases
- CEL
Cellulose
- CIP1
Cellulose-induced protein 1
- CIP2
Cellulose-induced protein 2
- CLR2
Cellulose degradation regulator 2
- DNA
Deoxyribonucleic acid
- E
Beta-sheet
- EC
Enzyme commission number
- Ex
Exposed
- FC
Fold change
- GH
Glycoside hydrolases
- GLU
Glucose
- GO
Gene ontology
- GT
Glycosyltransferases
- H
Helix
- JTT
Jones-Taylor-Thornton
- kb
Kilobases
- LPMO
Lytic polysaccharide monooxygenase
- M
Medium
- Mb
Megabase
- MEGA
Molecular evolutionary genetics analysis
- MFS
Major facilitator superfamily permease
- ML
Maximum likelihood
- PacBio
Pacific biosciences
- PBcR
PacBio corrected reads
- PCR
Polymerase chain reaction
- PL
Polysaccharide lyases
- RNA
Ribonucleic acid
- RNA-Seq
RNA sequencing
- RPKM
Reads per kilobase of exon model per million mapped reads
- SMRT
Single-molecule real-time
- TaIMI206040
T. atroviride IMI206040
- TF
Transcription factor
- ThB97
T. harzianum B97
- ThIOC3844
Trichoderma harzianum IOC-3844
- ThTR274
T. harzianum TR274
- Th6766
T. harzianum
- TPM
Transcripts per million
- TrQM6a
T. reesei QM6a
- TvGv29–8
T. virens Gv29–8
Authors’ contributions
APS and JAFF designed the study. JAFF, MACH, CAS, DAA, JSM, DAS and AC performed the research. JAFF, MACH, CAS, DAA, NFM and CBCS analyzed the data. JAFF, MACH, CAS and APS wrote the paper. All authors critically read the text and approved the manuscript.
Funding
This work was supported by grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2015/09202–0 and 2018/19660–4), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) Computational Biology Program (CBP: Process number 88882.160095/2013–01) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq: Process number 312777/2018–3). JAFF received a PhD fellowship from CNPq (170565/2017–3) and a PD fellowship from CAPES (CBP - 88887.334235/2019–00); MACH received a PD fellowship from FAPESP (2014/18856–1) and an SWE PD fellowship from CAPES, Computational Biology Program; CAS received a PD fellowship from FAPESP (2016/19775–0) and an SWE PD fellowship from CAPES (CBP); DAA received a partial MS fellowship from FAPESP (17/17782–2) and a partial MS fellowship from CAPES (CBP); NFM received a PD fellowship from CNPq and CAPES (CBP); JSM received a PD fellowship from CAPES (CBP); CBCS received a PD fellowship from FAPESP (17/26781–0) and CAPES (CBP); and APS is the recipient of a research fellowship from CNPq (312777/2018–3). The funding bodies played no role in the design of the study, analysis and interpretation of the data or writing of the manuscript.
Availability of data and materials
The raw data of the genomic regions (PacBio reads) can be found by the accession number PRJNA647392. The raw data of the RNA-Seq (Illumina reads) can be found by the accession number PRJNA336221. Assemblies and annotations (Additional file 3: Supplementary Table S4) from the genomic regions were submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MK861589-MK861650 (Additional file 2: Supplementary Table S1 and Additional file 3: Supplementary Table S4).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lombardi N, Vitale S, Turrà D, Reverberi M, Fanelli C, Vinale F, Marra R, Ruocco M, Pascale A, d’Errico G. Root exudates of stressed plants stimulate and attract Trichoderma soil fungi. Mol Plant-Microbe Interact. 2018;31(10):982–994. doi: 10.1094/MPMI-12-17-0310-R. [DOI] [PubMed] [Google Scholar]
- 2.Marra R, Lombardi N, d’Errico G, Troisi J, Scala G, Vinale F, Woo SL, Bonanomi G, Lorito M. Application of Trichoderma strains and metabolites enhances soybean productivity and nutrient content. J Agric Food Chem. 2019;67(7):1814–1822. doi: 10.1021/acs.jafc.8b06503. [DOI] [PubMed] [Google Scholar]
- 3.Li Y-H, Zhang X-Y, Zhang F, Peng L-C, Zhang D-B, Kondo A, Bai F-W, Zhao X-Q. Optimization of cellulolytic enzyme components through engineering Trichoderma reesei and on-site fermentation using the soluble inducer for cellulosic ethanol production from corn Stover. Biotechnol Biofuels. 2018;11(1):49. doi: 10.1186/s13068-018-1048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horta MAC, Vicentini R, da Silva DP, Laborda P, Crucello A, Freitas S, Kuroshu RM, Polikarpov I, da Cruz Pradella JG, Souza AP. Transcriptome profile of Trichoderma harzianum IOC-3844 induced by sugarcane bagasse. PloS One. 2014;9(2):e88689. [DOI] [PMC free article] [PubMed]
- 5.de Castro AM, Pedro KCNR, da Cruz JC, Ferreira MC, Leite SGF, Pereira N. Trichoderma harzianum IOC-4038: a promising strain for the production of a cellulolytic complex with significant β-glucosidase activity from sugarcane bagasse cellulignin. Appl Biochem Biotechnol. 2010;162(7):2111–2122. doi: 10.1007/s12010-010-8986-0. [DOI] [PubMed] [Google Scholar]
- 6.Elad Y, Chet I, Katan J. Trichoderma harzianum: a biocontrol agent effective against Sclerotium rolfsii and Rhizoctonia solani. Phytopathology. 1980;70(2):119–121. doi: 10.1094/Phyto-70-119. [DOI] [Google Scholar]
- 7.da Silva DP, Farinas CS, da Silva MR, Azzoni SF, da Cruz Pradella JG. Use of a new Trichoderma harzianum strain isolated from the Amazon rainforest with pretreated sugar cane bagasse for on-site cellulase production. Bioresour Technol. 2012;107:517–521. doi: 10.1016/j.biortech.2011.12.048. [DOI] [PubMed] [Google Scholar]
- 8.da Silva DP, Rodrigues GN, Zubieta MP, Ramoni J, Codima CA, Lima DJ, Farinas CS, da Cruz Pradella JG, Seiboth B. The relation between xyr1 overexpression in Trichoderma harzianum and sugarcane bagasse saccharification performance. J Biotechnol. 2017;246:24–32. doi: 10.1016/j.jbiotec.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira Filho JA, Horta MAC, Beloti LL, dos Santos CA, de Souza AP. Carbohydrate-active enzymes in Trichoderma harzianum: a bioinformatic analysis bioprospecting for key enzymes for the biofuels industry. BMC Genomics. 2017;18(1):779. doi: 10.1186/s12864-017-4181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Castro AM, Ferreira MC, da Cruz JC, Pedro KCNR, Carvalho DF, Leite SGF, Pereira N. High-yield endoglucanase production by Trichoderma harzianum IOC-3844 cultivated in pretreated sugarcane mill byproduct. Enzyme Res. 2010;v.2010:1–8. [DOI] [PMC free article] [PubMed]
- 11.Santos CA, Zanphorlin LM, Crucello A, Tonoli CC, Ruller R, Horta MA, Murakami MT, de Souza AP. Crystal structure and biochemical characterization of the recombinant ThBgl, a GH1 β-glucosidase overexpressed in Trichoderma harzianum under biomass degradation conditions. Biotechnol Biofuels. 2016;9(1):71. doi: 10.1186/s13068-016-0487-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bischof RH, Ramoni J, Seiboth B. Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb Cell Factories. 2016;15(1):106. doi: 10.1186/s12934-016-0507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 14.Jun H, Kieselbach T, Jönsson LJ. Enzyme production by filamentous fungi: analysis of the secretome of Trichoderma reesei grown on unconventional carbon source. Microb Cell Factories. 2011;10(1):68. doi: 10.1186/1475-2859-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Häkkinen M, Arvas M, Oja M, Aro N, Penttilä M, Saloheimo M, Pakula TM. Re-annotation of the CAZy genes of Trichoderma reesei and transcription in the presence of lignocellulosic substrates. Microb Cell Factories. 2012;11:134. doi: 10.1186/1475-2859-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvira P, Gyalai-Korpos M, Barta Z, Oliva JM, Réczey K, Ballesteros M. Production and hydrolytic efficiency of enzymes from Trichoderma reese iRUTC30 using steam pretreated wheat straw as carbon source. J Chem Technol Biotechnol. 2013;88(6):1150–1156. doi: 10.1002/jctb.3955. [DOI] [Google Scholar]
- 17.Almeida DA, MAC H, Ferreira Filho JA, Murad NF, de Souza AP. The synergistic actions of hydrolytic genes in coexpression networks reveal the potential of Trichoderma harzianum for cellulose degradation. bioRxiv. 2020;1:1–41. [DOI] [PubMed]
- 18.Horta MAC, Ferreira Filho JA, Murad NF, de Oliveira SE, dos Santos CA, Mendes JS, Brandão MM, Azzoni SF, de Souza AP. Network of proteins, enzymes and genes linked to biomass degradation shared by Trichoderma species. Sci Rep. 2018;8(1):1–11. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos CA, Morais MA, Terrett OM, Lyczakowski JJ, Zanphorlin LM, Ferreira-Filho JA, Tonoli CC, Murakami MT, Dupree P, Souza AP. An engineered GH1 β-glucosidase displays enhanced glucose tolerance and increased sugar release from lignocellulosic materials. Sci Rep. 2019;9(1):1–10. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bischof R, Fourtis L, Limbeck A, Gamauf C, Seiboth B, Kubicek CP. Comparative analysis of the Trichoderma reesei transcriptome during growth on the cellulase inducing substrates wheat straw and lactose. Biotechnol Biofuels. 2013;6(1):127. doi: 10.1186/1754-6834-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta VK, Steindorff AS, de Paula RG, Silva-Rocha R, Mach-Aigner AR, Mach RL, Silva RN. The post-genomic era of Trichoderma reesei: what's next? Trends Biotechnol. 2016;34(12):970–982. doi: 10.1016/j.tibtech.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Santos CA, Ferreira-Filho JA, O’Donovan A, Gupta VK, Tuohy MG, Souza AP. Production of a recombinant swollenin from Trichoderma harzianum in Escherichia coli and its potential synergistic role in biomass degradation. Microb Cell Factories. 2017;16(1):83. doi: 10.1186/s12934-017-0697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villares A, Moreau C, Bennati-Granier C, Garajova S, Foucat L, Falourd X, Saake B, Berrin J-G, Cathala B. Lytic polysaccharide monooxygenases disrupt the cellulose fibers structure. Sci Rep. 2017;7:40262. doi: 10.1038/srep40262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benocci T, Aguilar-Pontes MV, Zhou M, Seiboth B, de Vries RP. Regulators of plant biomass degradation in Ascomycetous fungi. Biotechnol Biofuels. 2017;10(1):152. doi: 10.1186/s13068-017-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, Glass NL. Conserved and essential transcription factors for cellulase gene expression in ascomycete Fungi. Proc Natl Acad Sci U S A. 2012;109(19):7397–7402. doi: 10.1073/pnas.1200785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Xue Y, Liu J, Gan L, Long M. ACE3 as a master transcriptional factor regulates cellulase and xylanase production in Trichoderma orientalis EU7-22. BioResources. 2018;13(3):6790–6801. [Google Scholar]
- 27.Crucello A, Sforca DA, Horta MAC, dos Santos CA, Viana AJC, Beloti LL, de Toledo MAS, Vincentz M, Kuroshu RM, de Souza AP. Analysis of genomic regions of Trichoderma harzianum IOC-3844 related to biomass degradation. PloS One. 2015;10(4):e0122122. [DOI] [PMC free article] [PubMed]
- 28.Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315(5813):804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 29.van den Brink J, de Vries RP. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. 2011;91(6):1477–1492. doi: 10.1007/s00253-011-3473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Dyk JS, Pletschke BI. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes—factors affecting enzymes, conversion and synergy. Biotechnol Adv. 2012;30(6):1458–1480. doi: 10.1016/j.biotechadv.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Nitta M, Furukawa T, Shida Y, Mori K, Kuhara S, Morikawa Y, Ogasawara W. A new Zn (II) 2Cys6-type transcription factor BglR regulates β-glucosidase expression in Trichoderma reesei. Fungal Genet Biol. 2012;49(5):388–397. doi: 10.1016/j.fgb.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Häkkinen M, Valkonen MJ, Westerholm-Parvinen A, Aro N, Arvas M, Vitikainen M, Penttilä M, Saloheimo M, Pakula TM. Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol Biofuels. 2014;7(1):14. doi: 10.1186/1754-6834-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westereng B, Loose JS, Vaaje-Kolstad G, Aachmann FL, Sørlie M, Eijsink VG. Analytical tools for characterizing cellulose-active lytic polysaccharide monooxygenases (LPMOs) Methods Mol Biol. 1796;2018:219–246. doi: 10.1007/978-1-4939-7877-9_16. [DOI] [PubMed] [Google Scholar]
- 34.Filho JAF, Horta MAC, Beloti LL, dos Santos CA, de Souza AP. Carbohydrate-active enzymes in Trichoderma harzianum: a bioinformatic analysis bioprospecting for key enzymes for the biofuels industry. BMC Genomics. 2017;18(1):779. doi: 10.1186/s12864-017-4181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toyotome T, Hamada S, Yamaguchi S, Takahashi H, Kondoh D, Takino M, Kanesaki Y, Kamei K. Comparative genome analysis of Aspergillus flavus clinically isolated in Japan. DNA Res. 2019;26(1):95–103. doi: 10.1093/dnares/dsy041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baroncelli R, Piaggeschi G, Fiorini L, Bertolini E, Zapparata A, Pè ME, Sarrocco S, Vannacci G. Draft whole-genome sequence of the biocontrol agent Trichoderma harzianum T6776. Genome Announc. 2015;3(3):e00647–e00615. doi: 10.1128/genomeA.00647-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubicek CP, Steindorff AS, Chenthamara K, Manganiello G, Henrissat B, Zhang J, Cai F, Kopchinskiy AG, Kubicek EM, Kuo A, et al. Evolution and comparative genomics of the most common Trichoderma species. BMC Genomics. 2019;20(1):485. doi: 10.1186/s12864-019-5680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sforça DA, Vautrin S, Cardoso-Silva CB, Mancini MC, MVRd C, Pereira GS, Conte M, Bellec A, Dahmer N, Fourment J. Gene duplication in the sugarcane genome: a case study of allele interactions and evolutionary patterns in two genic regions. Front Plant Sci. 2019;10:553. doi: 10.3389/fpls.2019.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W-C, Huang C-H, Chen C-L, Chuang Y-C, Tung S-Y, Wang T-F. Trichoderma reesei complete genome sequence, repeat-induced point mutation, and partitioning of CAZyme gene clusters. Biotechnol Biofuels. 2017;10:170. doi: 10.1186/s13068-017-0825-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuan CS, Yew SM, Toh YF, Chan CL, Ngeow YF, Lee KW, Na SL, Yee W-Y, Hoh C-C, Ng KP. Dissecting the fungal biology of Bipolaris papendorfii: from phylogenetic to comparative genomic analysis. DNA Res. 2015;22(3):219–232. doi: 10.1093/dnares/dsv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haitjema CH, Gilmore SP, Henske JK, Solomon KV, de Groot R, Kuo A, Mondo SJ, Salamov AA, LaButti K, Zhao Z, et al. A parts list for fungal cellulosomes revealed by comparative genomics. Nat Microbiol. 2017;2:17087. doi: 10.1038/nmicrobiol.2017.87. [DOI] [PubMed] [Google Scholar]
- 42.Wang R, Dong L, He R, Wang Q, Chen Y, Qu L, Zhang Y-A. Comparative genomic analyses reveal the features for adaptation to nematodes in Fungi. DNA Res. 2018;25(3):245–256. doi: 10.1093/dnares/dsx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie B-B, Qin Q-L, Shi M, Chen L-L, Shu Y-L, Luo Y, Wang X-W, Rong J-C, Gong Z-T, Li D, et al. Comparative genomics provide insights into evolution of Trichoderma nutrition style. Genome Biol Evol. 2014;6(2):379–390. doi: 10.1093/gbe/evu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hassan L, Lin L, Sorek H, Sperl LE, Goudoulas T, Hagn F, Germann N, Tian C, Benz JP. Crosstalk of cellulose and mannan perception pathways leads to inhibition of cellulase production in several Filamentous fungi. mBio. 2019;10(4):e00277–e00219. doi: 10.1128/mBio.00277-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanova C, Ramoni J, Aouam T, Frischmann A, Seiboth B, Baker SE, Le Crom S, Lemoine S, Margeot A, Bidard F. Genome sequencing and transcriptome analysis of Trichoderma reesei QM9978 strain reveals a distal chromosome translocation to be responsible for loss of vib1 expression and loss of cellulase induction. Biotechnol Biofuels. 2017;10:209. doi: 10.1186/s13068-017-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawler K, Hammond-Kosack K, Brazma A, Coulson RM. Genomic clustering and co-regulation of transcriptional networks in the pathogenic fungus Fusarium graminearum. BMC Syst Biol. 2013;7(1):52. doi: 10.1186/1752-0509-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.dos Santos CL, Pedersoli WR, Antoniêto ACC, Steindorff AS, Silva-Rocha R, Martinez-Rossi NM, Rossi A, Brown NA, Goldman GH, Faça VM. Comparative metabolism of cellulose, sophorose and glucose in Trichoderma reesei using high-throughput genomic and proteomic analyses. Biotechnology Biofuels. 2014;7(1):41. doi: 10.1186/1754-6834-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berlin K, Koren S, Chin C-S, Drake JP, Landolin JM, Phillippy AM. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol. 2015;33:623–630. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- 49.Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KHJ, Remington KA, et al. A whole-genome assembly of Drosophila. Science. 2000;287(5461):2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 50.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13(1):238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 52.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10(4):516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 54.Mitchell A, Chang H-Y, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43(Database issue):D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veltri D, Wight MM, Crouch JA. Simple synteny: a web-based tool for visualization of microsynteny across multiple species. Nucleic Acids Res. 2016;44(W1):W41–W45. doi: 10.1093/nar/gkw330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 60.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 61.Källberg M, Margaryan G, Wang S, Ma J, Xu J. RaptorX server: a resource for template-based protein structure modeling. In: Protein Structure Prediction. New York, NY: Humana Press, 2014. p. 17–27. [DOI] [PubMed]
- 62.CLC Genomics Workbench 9.0. Qiagen (Aarhus A/S): Manual for CLC genomics workbench 9.0 windows, Mac OS X and Linux Denmark. 2016.
- 63.Wilczynski B, Dojer N. BNFinder: exact and efficient method for learning bayesian networks. Bioinformatics. 2009;25(2):286–287. doi: 10.1093/bioinformatics/btn505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Fig. S1. Screening genes of interest in the genomic library of T. harzianum IOC3844 by qPCR (a); read size sequenced using PacBio technology (b); genes clustered in a genomic region of T. harzianum (c). Fig. S2. Distribution of the main GO terms of the annotated genes in T. harzianum IOC3844. Fig. S3. Pipeline approach for the analyses used in this study of genes and genomes in T. harzianum. Supplementary Table S2. Assembly parameters of a set of sequenced genomic regions using PacBio technology. Supplementary Table S3. Comparison of genomic data among different species of Trichoderma spp. Supplementary Table S8. Description of the species used for the phylogenetic analysis of the transcription factor CLR2. Supplementary Table S9. Description of the genes found in the coregulation networks.
Additional file 2 Supplementary Table S1. Description of the genomic regions sequenced in T. harzianum IOC3844.
Additional file 3 Supplementary Table S4. Annotation of all genes predicted in T. harzianum IOC3844.
Additional file 4 Supplementary Table S5. Description of the EC codes for T. harzianum IOC3844 genes.
Additional file 5 Supplementary Table S6. Description of the CAZyme genes in T. harzianum IOC3844.
Additional file 6 Supplementary Table S7. Expression levels of the genes annotated in T. harzianum IOC3844 by means of RNA-Seq.
Additional file 7 FASTA sequences of proteins secreted in T. harzianum IOC3844 under cellulose conditions developed in the work of Horta et al., 2018.
Data Availability Statement
The raw data of the genomic regions (PacBio reads) can be found by the accession number PRJNA647392. The raw data of the RNA-Seq (Illumina reads) can be found by the accession number PRJNA336221. Assemblies and annotations (Additional file 3: Supplementary Table S4) from the genomic regions were submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MK861589-MK861650 (Additional file 2: Supplementary Table S1 and Additional file 3: Supplementary Table S4).