Abstract
Traditional techniques for cancer diagnosis, such as nuclear magnetic resonance, ultrasound and tissue analysis, require sophisticated devices and highly trained personnel, which are characterized by elevated operation costs. The use of biomarkers has emerged as an alternative for cancer diagnosis, prognosis and prediction because their measurement in tissues or fluids, such as blood, urine or saliva, is characterized by shorter processing times. However, the biomarkers used currently, and the techniques used for their measurement, including ELISA, western-blot, polymerase chain reaction (PCR) or immunohistochemistry, possess low sensitivity and specificity. Therefore, the search for new proteomic, genomic or immunological biomarkers and the development of new noninvasive, easier and cheaper techniques that meet the sensitivity and specificity criteria for the diagnosis, prognosis and prediction of this disease has become a relevant topic. The purpose of this review is to provide an overview about the search for new cancer biomarkers, including the strategies that must be followed to identify them, as well as presenting the latest advances in the development of biosensors that possess a high potential for cancer diagnosis, prognosis and prediction, mainly focusing on their relevance in lung, prostate and breast cancers.
Keywords: cancer diagnosis, prognosis, prediction, biomarker, proteomic, genomic, immunologic, biosensor
Introduction
Cancer is a disease that represents an enormous public health issue in both developed and developing countries. For 2018, it was estimated 18.1 million new cancer cases and 9.6 million cancer deaths worldwide. The 3 main cancers are lung, breast and prostate cancer.1 The development of new diagnostic tools for cancer detection is mainly justified because if cancer is detected early, it can be cured. A biomarker is an important tool in the detection and monitoring of cancer; these biomarkers include gene mutations, alterations in transcription or translation of genes, and/or protein products modifications.2 An advantage of biomarkers is that they can be used in the evaluation of the disease at different stages, serving as diagnosis, prognosis and prediction tools. Currently, the number of biomarkers in clinical use is limited. Examples of these biomarkers include prostate-specific antigen (PSA) for prostate cancer, carcinoembryonic antigen (CEA) for lung cancer, cancer antigen 125 (CA-125) for ovarian cancer and cancer antigen 15-3 (CA15-3) for breast cancer.3 Nevertheless, none of these biomarkers provides enough sensitivity and specificity information for an accurate diagnosis in the entire population. Thus, identify new biomarkers that enable the development of new diagnostic tools with high sensitivity and specificity is imperative. Current tools for cancer diagnosis using biomarkers are mainly based on immunohistochemistry, serological or PCR tests. A great disadvantage of immunohistochemistry is that it represents an invasive technique that depends on tissue samples, and, biomarker heterogeneity can exist inside the same tumor. Among serological tests such as western-blot, ELISA or PCR-based tests, the main disadvantage is that they suffer from technological limitations such as low detection and the use of expensive reagents in each assay.4 For these reasons, at present, there is a growing interest on biosensors development for early cancer diagnosis and prognosis, as well as for the prediction of cancer treatment responses, as they have shown a higher sensitivity. Furthermore, given that the measurements can be performed in real-time, the wait time for the result can be reduced.
Strategies for the Search of New Biomarkers
The following are the 5 different stages involved in understanding biomarker development: 1) an exploratory preclinical study aimed at identifying potential biomarkers; 2) the development of a test to identify and quantify the biomarker in the sample; 3) assays that evaluate the correlation with the disease; 4) prospective screening studies and 5) clinical impact studies.5-7 The first stages are critical because they allow the identification of potential biomarkers and discriminate those that do not meet the required characteristics. In the quest for new biomarkers, new -omics technologies have provided advances for the development of these markers. These technologies, including proteomics technologies such as 2D electrophoresis (2D-PAGE) and mass spectrometry, have facilitated the discovery of new biomarkers.3 Related to genomic technologies, whole-exome sequencing has allowed the comparison of gene mutations from different cancer types, enabling the identification of associations between gene mutations and the different kinds of tumors.8 The immune system plays a key role in cancer development and progression; therefore, the polarization to certain kinds of responses through different cytokine profiles or cell subpopulations can discriminate between healthy patients and cancer patients.9
Proteomics Approach and Potential Biomarkers
Differential expression of proteins can be used for the discovery and validation of new biomarkers.10 In conventional proteomics analysis, 2D-PAGE followed by mass spectrometry has been used as a primary technique for new biomarkers discovery during many years.2 Following this approach, many researches carried out in the last 2 decades has found proteins that could be used as potential biomarkers.11-14 Ummani et al,15 through 2D immunoblotting and mass spectrometry, found that PRDX6 and ANXA11 were strongly recognized in prostate cancer patient’s serum. Noteworthy, this group developed a test capable of discriminating between healthy and cancer patients with 90% and 100% sensitivity and specificity, respectively. A different method based on 2D-PAGE is 2D differential gel electrophoresis (2D-DIGE).16 In this method, different samples are labeled with fluorescent dyes, mixed and separated by 2D-PAGE. Then, laser scanning is used to observe the gel. Two advantages that 2D-DIGE offers over traditional 2D-PAGE are that 2D-DIGE reduces gel-gel variations and does not require a staining step. Xiao et al,17 using 2D-DIGE and mass spectrometry strategy, identified ANX1, HP, AZGP1 and calprotectin proteins as potential biomarkers in saliva samples from healthy controls and lung cancer patients. Even though the 2D-PAGE approach is effective, it has certain substantial limitations including being slow and laborious and showing limited sensitivity due to the high amounts of proteins needed for visualization on a gel.18 Latter problems associated with gel-based technologies originated the development of gel-free technologies such as labeling and targeted shotgun proteomics based on mass spectrometry analysis without the need for a previous gel electrophoresis step.19 An important step in such gel-free methods is the identification of accurate mass tags (AMTs) for protein determination,20 this step starts with a biological sample, a total lysate or a fractionated biochemical product. The general procedure to identify a protein begins with the tryptic digestion of complex protein mixtures. The tryptic peptides are separated by reverse high-pressure liquid chromatography (HPLC) and then analyzed by LC-MS/MS to obtain partial amino acid sequences. This information sequence is used to establish the identity of the parental protein from which all the peptides originate by searching for MS/MS spectral coincidences in an appropriate database.18 In a recent study, Kwon et at,21 using LC-MS/MS to identify AMT, investigated the differences in protein expression patterns between normal, prostate cancer and advanced prostate cancer tissues. Although finding 3 proteins differentially expressed; spermidine synthase (SMR), nucleolar and coiled-body phosphoprotein 1 (NOLC1) and protein prostacyclin synthase (PTGIS) as unknown candidate potential biomarkers, only spermidine synthase was increased in more advanced stages of prostate cancer, which make this protein a promising biomarker for prostate cancer prediction. A recent work, through LC-MS/MS, developed a multimarker test detecting apolipoprotein C-1 (APOCH1), carbonic anhydrase 1 (CAH1) and neural cell adhesion molecule L1-like protein (NCHL1) present in human plasma of breast cancer patients, such test showed diagnostic sensitivity, specificity and accuracy of 71.6%, 85.3%, and 77% respectively. Remarkably, this protein assay was able to discriminate other types of malignancies, suggesting that this assay was organ-specific.22
Sokolowska et al,23 through nanoLC-MS/MS, identified receptors for the tumor differentiation factor (TDF) expressed in human breast and prostate cancer cells, noteworthy, the receptors involved belonged to the heat shock 70-kDa family of proteins (HSP70), showing a relation between this family of proteins and cancer presence. Taking advantage that once a biomarker is found, it is possible to transfer to more simplified instrumentation, including western blot, ELISA, multiplex assays, immunohistochemistry and microarrays,24 heat shock proteins (HSPs) have gained attention as a promising tool in the diagnosis of cancer including prostate, breast, lung, ovarian and colorectal cancer.25 HSPs are classified in various families based on their molecular weight including HSP110, HSP90, HSP70, HSP60 and the small heat shock proteins groups.26 HSP70 has been considered a damage-associated molecular pattern that can stimulate a chronic inflammatory response after radiation-induced tumor cell death. That chronic inflammatory response has been shown to be related to tumor growth.27 In a recent research, serum levels of HSP70 were correlated with an unfavorable prognosis in breast cancer, since breast cancer patients with distant metastasis or recurrence showed higher serum levels than patients who remained free of the disease.28 HSP90α has recently been found to localize outside various cancer cells and, clinical trials have demonstrated that plasma HSP90α is a more accurate biomarker than CEA and CYFRA21 -1 in lung cancer.29 In that sense, Liu et al30 through a large-scale clinical study, validated HSP90α as a pan-cancer biomarker, showing a sensitivity of 81.33% and a sensitivity of 81.65% in a test cohort, and a sensitivity and sensibility of 81.72% and 81.03% respectively in a validation cohort.
Occludin and claudin proteins family (CLDN) have been associated with cell proliferation and differentiation.31 The expression of these proteins has been found altered in different tumor types.32,33 According to these, CLDN2, CLDN6, CLDN11, and CLDN14 have shown a prognostic potential in breast cancer, since the expression of these proteins in breast carcinoma evaluated by western blot and immunohistochemistry, has been found downregulated.34 Netrin-1 is a protein-related to tumorigenesis in bladder cancer (BC) which inhibits apoptotic pathways.35 El-Gamal et al36 through western blot, have shown that Netrin-1 levels in BC tissue could be a potential prediction marker since this biomarker predicts muscle invasion with 96% of sensibility. Something to highlight of this work, urinary levels of Netrin-1 were strongly correlated with tissue levels, showing the possibility for a non-invasive test for BC diagnosis. Based on the fact that detection of biomarkers in body fluids offers a huge advantage over the determination of these biomarkers in tumor tissue samples, Ma et al37 through a multiplexed assay for the determination of C-reactive protein (CRP), prolactin, hepatocyte growth factor (HGF) and autoantigen NY-ESO-1 in serum, was able to discriminate between healthy patients and those with different types of lung cancer. This panel of 4 biomarkers showed higher sensitivity and specificity compared with CEA.
Etheridge et al38 using seminal ejaculate, were able to detect the protein AMACR in clinically significant prostate cancer tumors by quantifying the protein in ELISA tests. Zhang et al,39 measuring urinary plasminogen and fibrinogen gamma chain levels in NSCLC patients and controls detected by ELISA, found that tissue levels and urinary levels were significantly elevated compared with controls, with an area under the ROC curve ranging from 0.827 to 0.947. Continuing with serological tests, by means of ELISA, Zheng et al40 used human epididymis 4 (HE4) and transthyretin (TTR) and found that HE4 exhibits a better performance in ovarian cancer diagnosis, even better than the performance of CA-125. Using immunohistochemistry, Guo et al41 analyzed samples from patients with non-small cell lung cancer (NSCLC) and associated the expression of the zinc finger protein ZNF71 with the response to chemotherapy, which showed enormous potential as a predictive biomarker; furthermore, given that the group that expressed this antigen at a higher concentration showed a higher survival rate, this protein has potential as a prognosis biomarker. Llie et al42 performed a multiplexed immunohistochemical assay on NSCLC samples and demonstrated that a panel of biomarkers, including TTF1, p40, PD-L1, and pan-keratin, as well as an additional panel focused on the molecular profile, including anti-ALK, anti-ROS1, and anti-BRAFV600E antibodies, were able to classify the tissues in different histotypes for diagnosis and immunophenotyping, helping to choose the therapeutic strategy.
Thus, gathering all these proteomic approaches, the biomarkers’ search has yielded potential cancer biomarkers that could be used in the development of new and more efficient diagnosis tools. Table 1 summarizes different protein biomarkers with potential in cancer diagnosis, prognosis, and prediction.
Table 1.
Cancer Associated Proteins With Diagnostic Potential.
| Proteomic marker | Cancer type | Clinical use | Detection method | References |
|---|---|---|---|---|
| PRDX6 and ANXA11 | Prostate | Diagnosis | 2D-PAGE | Ummani et al., 201515 |
| ANX1, HP, AZGP1 and calprotectin | Lung | Diagnosis | 2D-DIGE | Xiao et al., 201217 |
| SMR | Prostate | Prognosis | Mass spectrometry | Kwon et al., 202021 |
| APOCH1, CAH1 and NCHL1 | Breast | Diagnosis and prognosis | Mass spectrometry | Kim et al., 201922 |
| HSP70 | Breast | Prediction | ELISA | Rothammer et al., 201928 |
| HSP90α | Pan cancer | Diagnosis | ELISA and western blot | Liu et al., 201930 |
| CLDN2, CLDN6, CLDN11 and CLDN14 | Breast | Prognosis | Western blot | Jia et al., 201934 |
| Netrin-1 | Breast | Diagnosis | Western blot | El-Gamal et al., 202036 |
| CRP, prolactin, HGF and autoantigen NY-ESO-1 | Lung | Diagnosis | Multiplexed ELISA | Ma et al., 201637 |
| AMACR | Prostate | Diagnosis | ELISA | Etheridge et al., 201838 |
| Urinary plasminogen and fibrinogen gamma | Prostate | Diagnosis | ELISA | Zhang et al., 202039 |
| HE4 and TTR | Ovarian | Diagnosis | ELISA | Zheng et al., 201840 |
| ZNF71 | Lung | Prognosis | Immunohistochemistry | Guo et al., 201841 |
| TTF1, p40, PD-L1 | Lung | Diagnosis and prognosis | Immunohistochemistry | Llie et al., 201842 |
Genomics Approach
The advances in new DNA sequencing technologies have allowed the processing of thousands of different cancer types samples for the discovery of systemic mutations. This expanded aim, coupled with noteworthy progress in algorithms,43,44 has directly contributed to the characterization of significant functional mutations, genes, and pathways.45 Even though genetic mutations maintain a high frequency, most genetic mutations occur at an intermediate frequency (2-20%). Single nucleotide polymorphisms (SNPs) are important genetic biomarkers in humans that have functions in the beginning and progression of cancer, noteworthy, in addition to identifying mutations, classical procedures for detecting changes in the basal levels of expression or epigenetics features of different genes remain being used for the analysis of new candidates to evaluate their potential as a biomarker for cancer detection, or the announcement of prognosis of the diagnosed patient. Table 2 summarizes a set of different genes with biomarker properties for cancer diagnosis and prognosis reported recently.
Table 2.
Analysis of Cancer-Associated Genes Used as Biomarkers and With Potential Use in Biosensors.
| Genomic marker | Cancer type | Clinical use | Detection method | References |
|---|---|---|---|---|
| PD-L1 | Lung | Selection of patients for therapy | NGS-TMB determination | Cyriac and Gandhi, 201846 |
| STX2 | Lung, colorectal | Exome sequencing, qRT-PCR | Lawrence et al., 2014;47 Wang et al., 201848 | |
| ARHGAP35 | Lung, osteosarcoma | Risk associated to rs1052667 polymorphism detection | Exome sequencing, RNA sequencing, SNP Genotyping |
Kandoth et al., 201349; Lawrence et al., 201447; Zhao et al., 201450; Campbell et al., 201651 |
| PIP | Breast | qRT-PCR | Gangadharan et al., 201851 | |
| MSH2 | Hepatocellular carcinoma, lung | Risk associated to rs2303428 polymorphism detection |
SNP Genotyping | Zhu et al., 201852; Lo et al., 201153 |
| ACYP2 | Breast | Risk associated to rs1682111 and rs10439478 polymorphisms detection | SNP Genotyping | Wu et al., 201854 |
| NER genes ERCC1-5 RAD23B XPA XPC XPE |
Esophagus | Prognosis-associated to detection of rs3759497, rs3731054, rs2097215 and rs3916788 polymorphisms |
SNP Genotyping | Zhang et al.,201855 |
| POLK | Prostate, melanoma, lung, large intestine | Risk associated to detection of 9 different polymorphisms | SNP Genotyping and site directed mutagenesis | Antczak et al., 201856 |
| ERCC1 | Prostate | Cancer aggressiveness associated to detection of rs11615 polymorphism | SNP Genotyping | Henríquez-Hernández et al., 201457 |
| ATM | Prostate | Cancer aggressiveness associated to detection of rs17503908 polymorphism; risk associated to specific mutation | SNP Genotyping, Genomic data analysis | Henríquez-Hernández et al., 201457
Marshall et al., 201858 |
| BRCA1/2 | Prostate | Risk associated to specific mutation | SNP Genotyping | Marshall et al., 201858 |
| VTCN1 | Breast, lung | Microarray analysis | Akiyama et al., 201659 | |
| AKT1 PIK3CA PTEN TP53 |
Breast | Risk associated to specific mutation | NGS | Li et al., 201860 |
| RUNX1 | Breast | Poor prognosis | Tissue microarray/IHC | Ferrari et al., 201461 |
| miR-21 miR-210 |
Breast | Poor prognosis | Meta-analysis | Adhami et al., 201762 |
| miR-15b-5p miR-16-5p miR-20a-5p |
Lung | Diagnostic | qRT-PCR/ Fluorescence quantum dots liquid bead array | Fan et al., 201563 |
| miR-30b-5p miR-96-5p miR-182-5p miR-374b-5p miR-942-5p |
Breast | Diagnostic | Microarray/qRT-PCR | Zhang et al., 201764 |
| SCGB2A2 | Prostate | Risk associated to differentially methylated detection from biological samples | Bisulfite-converted sample DNA assay | Sherlock et al., 201465 |
| ZNF154 | Prostate | Prognosis | RNAseq/ NGS methylation detection | Zhang et al., 201866 |
| ADAMTS1 BNC1 |
Pancreas | Early detection | Bisulfite-converted sample DNA assay | Eissa et al., 201967 |
| CX43 | Lung | Poor prognosis | Tissue microarray/IHC | Aasen et al., 201968 |
In an ambitious effort, Cyriac et al46 carried on an in silico analysis on 3281 tumors of 12 different types of cancer and identified 127 genes involved in a wide range of processes, including regulatory mechanisms or transcriptional factors, histone modifiers, genome integrity, tyrosine/kinase receptor signaling, cell cycle, mitogen-activated protein kinases signaling (MAPK), phosphatidylinositol-3-OH kinase (PI(3)K) signaling, Wnt/β-catenin signaling, histones, ubiquitin-mediated proteolysis and splicing. Moreover, Lawrence et al47 used an algorithm with the ability to analyzing the cancer genomes of 21 different tumor types and found the association of 33 genes not previously reported as mutated genes in cancer that encode proteins with different functions such as anti-proliferation, proliferation, pro-apoptosis and chromatin regulation. Notably, Arg107 mutation in STX2 gene has been detected as a recurrent mutation in lung cancer patients,47 and upregulation of this gene has recently been ligated to colorectal cancer.48 Another mutation detected by exome sequencing with potential properties as clinical biomarker is the rs1052667 polymorphism in ARHGAP35 gene, whose product is a repressor of glucocorticoid receptor transcription,47,49 mutations determined in this gene by SNP genotyping and RNAseq have been associated with lung cancer and osteosarcoma.50,51
In a recent study using the bioinformatics platform tool BioXm, were identified 506 differentially expressed genes in breast cancer, including the genes BRCA1, BRCA2 and ERBB2; however, the PIP gene showed higher downregulation in cancer cell lines.69 DNA mismatching repairing (MMR) genes are key factors in genomic stability.70 As one of the most important MMR genes in humans, mut-S-homolog (MSH2) maintains genomic stability by repairing base pair mismatches. In that sense, Zhu et al52 found a relationship between SNP RR2303428 on MSH2 and hepatocellular cancer progression using peripheral blood. Interestingly, the same gene is also associated with lung cancer development.53 Moreover, in breast cancer, a report shows that the SNPs rs1682111 and rs10439478 in the ACYP2 gene are associated with an increased risk of breast cancer. Of note, the SNP-SNP interactions between both polymorphisms further increase the risk of breast cancer.54 The nucleotide excision repair pathway (NER) is one of the most important repairing mechanisms; polymorphisms harbored for genes in this pathway, such as ERCC1, ERCC2, ERCC3, ERCC4, ERCC5, RAD23B, XPA, XPC, and XPE, affected the survival of Chinese patients receiving chemotherapy as treatment for esophageal cancer,55 therefore, showing potential as prognostic biomarkers. By studying one-nucleotide changes in the translesion synthesis DNA polymerases from the Y family associated with breast cancer, Anctzak et al56 found that these polymerases showed different activities in the presence of damaged DNA, which can inhibit protein expression or lead to the expression of dysfunctional proteins; thus, these polymorphisms can be exploited as potential biomarkers. In prostate cancer, polymorphisms in the repair genes XRCC1, ERCC2, ERCC1, LIG4, and TP53 have been related with clinical variables such as tumor size and Gleason score, demonstrating that these polymorphisms could be potential prognostic biomarkers.57
Marshall et al58 correlated mutations in the repair genes ATM, BRCA1/2, CDK12, CHEK1/2, FANCA, FANCD2, FANCL, GEN1, NBN, PALB2, RAD51 and RAD51C with clinical factors for the purpose of obtaining a tool for a better stratification of patients to allow their oncologist to better choose prostate cancer treatments. Mutations in genes associated with the immune response, such as VTCN1, IL2RA, ULBP2, TREM1, MSR1, TNFSF9, and TNFRSF12A, are potential biomarkers for the response predictions to treatment with antibodies to anti-immune-check-point-proteins, such as programmed death-1 (PD-1), in different types of cancer.59 In breast cancer, AKT1, PIK3CA, PTEN, and TP53 gene mutations have shown potential for the development of a predictive tool based on the results of Li et al,60 whose deep mutational characterization of these genes showed valuable implications for the clinical handling and the design of tests. The RUNX1 gene is the most frequently mutated gene in human leukemia, and several studies have focused on its tumor-suppressive function.71 In triple-negative cancer, which represents between 15 and 20% of total breast cancers, tumors that express the RUNX1 gene constitutes a group of tumors with the poorest prognosis, suggesting that this gene could contribute in tumor progression61 and indicating its potential as a prognosis biomarker. Certain overexpressed genes could also be related to the risk of this disease. In that sense, in lung cancer, a 7 genes panel (ABCC4, CCL19, SLC39A8, CD27, FUT7, DAG1, and ZNF71) was able to predict the chemotherapy response and the course of the disease in NSCLC, exhibiting its potential as a predictive and prognostic tool. This panel also showed predictive potential for immunotherapy responses.41 MicroRNAs (miRNAs) are small, 18-25-nucleotide-long, non-coding RNA molecules that down-regulate the target mRNAs.72 Several types of research have been focusing on the expression profiles of microRNAs and their role in cancer diagnosis and prognosis.73,74 Some methods are nowadays available to profile the miRNAs in body fluids, i. e., qRT-PCR, miRNAs microarrays and deep sequencing.75 From numerous candidate miRNAs, one strategy to specify the most important is to identify intersections between miRNAs reported in not related researches. In that sense, Adhami et al,62 based on a PubMed search, performed a general systematic review regarding the published miRNA profiling studies, comparing the expression level between breast cancer and normal tissues. Noteworthy, they found 30 miRNAs differentially expressed, the most consistent differentially expressed miRNAs were miR-21, miR-210 and miR-145, showing potential as candidate biomarkers for breast cancer. Fan et al,63 using serum samples from a cohort of 94 NSCLC patients and 58 healthy volunteers for a miRNA profile training study through qRT-PCR, identified 7 miRNAs (miR-15b-5p, miR-16-5p, miR-17b-5p, miR-19-3p, miR-20a-5p, miR-28-3p y miR-92-3p) differentially expressed. Furthermore, to confirm the accuracy and specificity of these miRNAs, they carried out a validation study through nano-quantum dots microarray finding 5 (miR-16-5p, miR-17b-5p, miR-19-3p, miR-20a-5p y miR-92-3p) from the 7 miRNAs identified in the training study, showing that serum miRNA expression profile could serve as a non-invasive biomarker for NSCLC. In breast cancer, Zhang et al,64 based on a microarray screening for blood miRNA profile, followed by a validation qRT-PCR, found 5 upregulated miRNAs (miR-30b-5p, miR-96-5p, miR-182-5p, miR-374b-5p and miR-942-5p) in breast cancer patients compared with healthy controls. The detection of these 5 miRNA expression levels could significantly distinguish between breast cancer patients and healthy controls as shown by ROC curves, even in very early stages of the disease, showing its potential as early breast cancer diagnosis biomarker.
CpG island methylation is a common epigenetic modification in tumors that leads to the inhibition of gene expression and functional loss.76 In that sense, a patent to detect methylations in SCGB2A2 and other genes was developed recently,65 leading to predict associated risks to the prostate cancer diagnosis. In a new study,66 differences between the methylation levels in genomic DNA from a prostate cancer tumor (PCa) and from a benign prostate tumor (BP) were analyzed. Using 1706 differentially expressed genes, they demonstrated that hypermethylation in the ZNF154 gene inhibits its expression and that this repression is associated with cancer progression. Also, it was recently suggested that methylated promoter detection of ADAMTS1 and BNC1 genes is useful for early detection of pancreatic cancer, greatly contributing to the early diagnosis of this aggressive cancer.67 Likewise, a novel biomarker detected in lung cancer patients is the expression of CX43, a gap junction protein without precedents in cancer implications, and whose presence is strongly related to a poor prognosis in those patients.68
Altogether, the genes and their specific characteristics and behaviors reviewed here may count to set a new group of cancer biomarkers with a high potential of use in biosensors development.
Immunological Approach and Biomarkers for cancer Disease
The release of proteins from tumors triggers an immune response in cancer patients. Responses to most tumor antigens are rarely observed in healthy individuals, making the humoral and cellular responses themselves a code that betrays the presence of underlying cancer. Immune responses show potential as clinical biomarkers because are easy to measure and are obtained in blood samples.77
Macrophages can be classified into the following 2 main groups: classic pathway-activated macrophages (M1) and alternative pathway-activated macrophages.78 Within M2 macrophages, there is a subpopulation known as M2d or tumor-associated macrophages (TAMs) that promotes tumor malignancy by facilitating angiogenesis and tumor growth. Therefore, the presence of these subpopulations could represent a discriminatory tool between healthy people and patients with cancer. Hegab et al79 used a murine model for lung adenocarcinoma to show that M2 macrophages support tumor growth by increasing tumor angiogenesis and proliferative capabilities. Of note, M2 macrophages showed immunosuppressive properties, as they were able to suppress the release of IFN-γ and to increase the production of L-arginase by T cells. However, infiltrating tumoral sites must be considered during the evaluation of M2 macrophages in lung cancer, since macrophages of different origins could show different biological and prognostic properties.80
Even though most research has been focused on the tumor-infiltrating cells, peripheral blood cell measurements could also serve as prognostic biomarkers. Zhang et al81 and Phan et al82 showed that the neutrophil-to-lymphocyte ratio (NLR) was an independent prognostic biomarker in NSCLC patients, where an elevated NLR was correlated with lower progression-free survival (PFS) and lower overall survival. Furthermore, not only cell populations but cytokines profiles, secreted chemokines and the receptors of both could also serve as diagnosis elements.83 Chemokines cover a group of approximately 50 small secreted proteins (8-14 kDa) that regulate cell traffic and are structurally like cytokines. In lung cancer, the chemokine CCL18 ligand is highly expressed in M2 macrophages. Schmid et al84 showed that a higher proportion of TAMs positives to CCL18 was related to a shorter survival rate by survival analysis; thus, this chemokine could be used as a prognosis biomarker. Schmid et al85 showed that elevated CCL18 levels are related to tumor size and poor prognosis in the NSCLC tumor microenvironment; moreover, this work also highlighted that serum levels of CCL18 were not related to either tumor size or prognosis. In contrast, Plönes et al86 measured and used CCL18 serum levels to discriminate between healthy controls and NSCLC patients, obtaining an area under the ROC curve of 0.968. It has recently been shown that there is an association between the chemokine CXCL10 and the recruitment of populations of T CD8+ CXCR3+ cells to the tumor site, which increases anti-tumor activities.87 Given that elevated levels of this chemokine are associated with a higher survival rate, this chemokine represents a potential prognosis biomarker. Other chemokines have been reported as potential biomarkers, Fujimoto et al88 analyzed the expression of CCL5 and CXCL9 in serum, stromal and cancer cells and found that serum levels of these chemokines might contribute to determine patient prognosis. High CXCL13 has also been associated with improved outcomes in the luminal-hEGFR2 subtype.89
IL-10 and IL-2, together with their receptor (IL-2 R), have been used in breast cancer to discriminate between malignant and benign tumors, showing great potential as diagnostic tools given the values obtained using ROC curves.90 Notably, these biomarkers were tested together with immune-check-point proteins such as PD-1 and cytotoxic T lymphocyte antigen 4 (CTLA-4). The PD-1 signaling pathway is an evasive strategy used by tumor cells that inhibits immunity by preventing chemotaxis, cell proliferation and release of cytokines by T cells. Dudnik et al91 evaluated the expression of the PD-1 ligand (PD-L1) in NSCLC containing a mutated BRAF gene. Together with the total mutational burden (TMB) and microsatellite instability status (MSI), this cancer ligand was associated with the response to treatment with immune-checkpoint inhibitors (ICPis). However, the high expression levels of PD-L1 were not a reliable predictive biomarker for ICPis.
Moreover, Krieger et al92 reported than TMB status and PD-L1 expression can be used for prediction response to checkpoint inhibitors and/or anti-cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) antibodies in NSCLC and melanoma, while a clear predictive trend was not identified in renal, breast, gastro or Merkel cell cancer. PD-1 can be expressed in activated T cells and is usually considered a marker for exhausted T cells. When this molecule is compromised by its ligand PD-L1, a ligand that can be expressed in tumor and immune cells, the function of T cells is inhibited.46 A different study performed after a vaccination cycle showed that the relatively high concentrations of PD-1+ and CD4+ cells obtained before and after the vaccination cycle were correlated with a higher survival rate in lung cancer93; thus, monitoring of these cell subpopulations also showed prognosis potential. El-Guindy et al94 measured PD-L1 expression and tumor-infiltrating lymphocyte (TIL) populations in NSCLC tumors and showed that elevated levels of PD-L1 correlated with low TILs, and in turn, both were associated with a lower survival rate, strengthening the potential of PD-L1 and cell populations as prognostic tools. Recently, Gonzalez-Ericcson et al95 reported the use of these biomarkers for optimal patient selection for immuno-therapeutic approaches. Morgan et al96 worked with metaplastic breast cancer samples and demonstrated a greater amount of CD163 in the stroma and PD-L1 in the tumor than TNBC, although more TNBC samples were positive for CD8 in the tumor than metaplastic breast cancer. In the same sense, PD-1 blockade in NSCLC led to enhanced IFN-γ production by CD8+ CD103+ TILs, suggesting that this immune cell subpopulation could be a predictive biomarker for immunotherapy based on PD-1 blockade97 and has been reported that cytokine signaling may be dysregulated in peripheral blood monocytes.97 Last year Li et al98 reported 4 immune-related genes (APOD, CXCL14, IL33, and LIFR) identified as biomarkers correlated with breast cancer prognosis. Their findings may provide different insights into prognostic monitoring of immune-related targets for breast cancer or can be served as a reference for further research and validation of biomarkers.
C-reactive protein (CRP) is an inflammatory biomarker that plays a key role in the innate immune response in humans; it is produced in response to inflammation, infection and tissue damage.99 Kaur et al100 measured the concentration of this biomarker in peripheral blood from breast cancer patients and found that elevated levels of CRP are associated with increased risk, recurrence, and mortality, thus showing its potential as a prognosis biomarker. Butyrophilin subfamily 3 member A2 (BTN3A2) was positively associated with better prognosis and could be served as a special diagnostic and independent prognostic marker for TNBC by regulating the T-cell receptor interaction and NF-κB signaling pathways.101 CCL20 and FOXP3 tumor-infiltrating lymphocytes may have synergistic effects, and their upregulated expressions may lead to immune evasion in breast cancer. Combinatorial immunotherapeutic approaches aiming at blocking CCL20 and depleting FOXP3 might improve therapeutic efficacy in breast cancer patients.102 Cerbelli et al103 showed that CD73 expression better predicts the response to neoadjuvant chemotherapy than stromal tumor-infiltrating lymphocytes in TNBC. The authors suggest than the characterization of both TILs and microenvironments could be a promising approach to personalize treatment.
Recently were reported mutations of immune players as biomarkers, analyzed for specific human populations, Ahmad et al104 reported the TNF-a308 G/A change and its significant association with breast cancer patients from north India and over the major histocompatibility complex class I-related chain A (MICA). Ouni et al105 reported the MICA-129 Met/Val change as an inherited genetic biomarker contributing to an increased breast cancer risk in Tunisian women. Tumors are a heterogenous mix of different cells, with a wide array of metabolic, phenotypic and stemness-like properties, the immune responses or immune evasion that tumor cells induce at the host reflects these spectra. To select the most accurate immunological biomarker scheme for cancer diagnosis, recent research efforts focus on test combinations of immune markers, some of which are arranged in Table 3.
Table 3.
Recent Published Immune Biomarkers With Clinical Use.
| Immune marker | Cancer type | Clinical use | Detection method | References |
|---|---|---|---|---|
| M2 macrophages | Lung adenocarcinoma | Diagnosis and prognosis | Immunohistochemistry | Martínez F 200878
Hegab A 201879 Li Z 201880 |
| Neutrophil-to-lymphocyte ratio (NLR) | NSCLC | Prognosis and survival prediction | Blood cell count Kaplan-Meier analysis |
Zhang Y 201881
Phan T 201882 |
| CCL18 | NSCLC | Prognosis and survival prediction | ELISA | Plönes T 201286
Schmid S 201884 |
| IL-10 IL-2 IL-2R |
Breast | Diagnosis | qRT-PCR | Liu C 201890 |
| CRP | Breast | Diagnosis | ELISA | Kaur R 2019100 |
| TILs and PD-L1 | Breast NSCLC Lung |
Diagnosis | Immunohistochemistry H&E staining |
Gonzalez 202095
El-Guindy 201894 |
| TNF-a308 G/A | Breast | Significant association with breast cancer patients from north India. | PCR-RFLP | Ahmad MM 2020104 |
| MICA-129 Met/Val | Breast | An inherited genetic biomarker contributing to an increased breast cancer risk in Tunisian women. | Genotyped | Ouni N 2020105 |
| TMB status and PD-L1 expression | NSCLC Melanoma |
Prediction response to ICPis | Sequencing | Krieger T 202092 |
| IFNg | Breast | Cytokine signaling dysregulated | Phosphoflow cytometry | Wang L 202097 |
| CD163, PD-L1 and CD8 | Breast TNBC |
Breast cancer classification | Immunohistochemistry | Morgan E 202096 |
| CD73 | TNBC | Neoadjuvant chemotherapy response | Immunohistochemistry | Cerbelli B 2019103 |
| CCL20 and FOXP3 | Breast | Immune evasion in breast cancer. | Immunohistochemistry qRT-PCR |
Zhao X 2019102 |
| APOD, CXCL14, IL33 and LIFR | Breast | As biomarkers correlated with breast cancer prognosis | Weighted gene co-expression network analysis (WGCNA), single-sample gene set enrichment analysis (ssGSEA), multivariate COX analysis, least absolute shrinkage, and selection operator (LASSO), and support vector machine-recursive feature elimination (SVM-RFE) algorithm | Li J 201998 |
| CCL5 | Breast | Prognosis | Cytometric bead-based immunoassay Immunohistochemistry |
Fujimoto Y 202088 |
| CXCL9 and CXCL13 | Breast TNBC |
Prognosis | qRT-PCR | Razis E 202089 |
| BTN3A2 | TNBC | Prognosis | Expression in extensive cancers were analyzed with Oncomine and TIMER databases. | Cai P 2020101 |
Biosensors in the Diagnosis, Prognosis and Prediction of Cancer
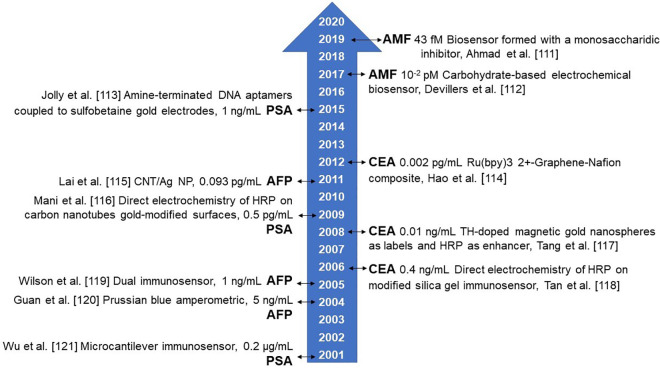
Conventional methods for cancer diagnosis include ultrasound, nuclear magnetic resonance and, biopsy, but these techniques require expensive equipment and highly qualified personnel, and, in the case of biopsy, is an invasive technique that represents a risk for the patient. Additionally, these techniques are inefficient in the early diagnosis of cancer since they depend on tumor phenotypic properties.106 Additionally, these techniques have a limit of detection close to 109 cells, growing as a single mass, thus being incapable of detecting cancer at early stages of the disease, since primary tumors are small.107 Biomarkers in body fluids represent a convenient, noninvasive and cheaper method for cancer diagnosis. Body fluids including serum or plasma, urine, saliva or sputum can be used in order to perform the biomarker detection. There have been considerable efforts in order to develop analytical assays for biomarkers detection. For protein biomarkers, the most common include western blotting, ELISA and mass spectrometry; for nucleic acids qRT-PCR, microarray, and next generation sequencing; and for cell populations flow cytometry and immunohistochemistry. Despite these techniques are in usage, they still have limitations like sophisticated analysis process, time-consuming operations and low sensitivity.108 New detection technologies need to be at least as sensitive as current technologies. The new methods based on biochemistry, immunology, and molecular biology are verified, developed and used in a continuous way in order to increase the sensibility of the biosensors, because of only trace levels of biomarkers exist in body fluids.109 In Figure 1 we represented a timeline for improved sensitivity of biosensors used to analyze representative biomarkers, as α-fetoprotein (AFP), Autocrine motility factor (AMF), carcinoembryonic antigen (CEA) and Prostate-specific antigen (PSA).
Figure 1.
Biosensors sensitivity timeline from 2001 to date for selected biomarkers, as alfa-fetoprotein (AFP), Autocrine motility factor (AMF), carcinoembryonic antigen (CEA) and Prostate-specific antigen (PSA). Abbreviations used: CNT/Ag NT, silver-nanoparticle enriched carbon nanotube and HRP Horseradish peroxidase.
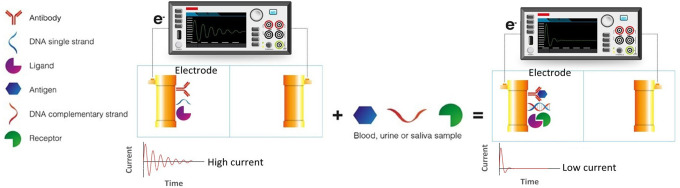
Currently, there is a growing interest in biosensors development for cancer diagnosis, prognosis, and prediction since these devices have shown real-time measurements and superior analytical performance. Since biosensors can detect minimal amounts of biomarkers in physiological samples, they can contribute to early cancer diagnosis.4 Biosensors offer flexibility related to biomarkers that can be used for their manufacturing, including antibodies, nucleic acids, or different specific recognizing molecules such as cytokines and chemokines. A biosensor is based on the interaction of the biomarker with a transducer, which converts the biological response produced into a signal that, depending on the transductor type, can be electrochemical, optical or a change in mass. Electrochemical biosensors (Figure 2) work analogous to an electrical circuit, converting the biosensor-biomarker interaction into a signal that can be translated as impedance, conductance, electrical current or potential. Electrochemical biosensors can be used for rapid biosensing and the measurement of key analytes in different cancer types using field-effect transistor (FET) techniques, square wave voltammograms (SWV), square wave stripping voltammetry (SWSV), electrochemical impedance spectroscopy (EIS), and cyclic voltammetry (CV).110
Figure 2.
Electrochemical biosensor. An increase in the electrode resistance owed to the interaction between a biomarker present in body fluids (antigen, complementary DNA strand or ligand) and its target molecule (antibody DNA single strand or receptor) attached to the electrode is measured as a change of current.
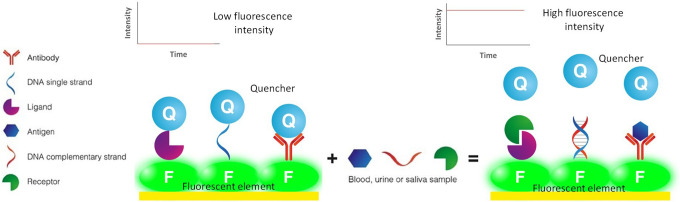
Most optical biosensors (Figure 3) for cancer diagnosis are based on phenomena such as fluorescence resonance energy transfer (FRET) or surface-enhanced Raman spectroscopy (SERS).4
Figure 3.
Optical biosensor. Owed to the interaction between a biomarker (antigen, complementary DNA strand or ligand) present in body fluids and its target molecule (antibody DNA single strand or receptor) attached to a florescence element, a quencher release occurs which translates in higher fluorescence.
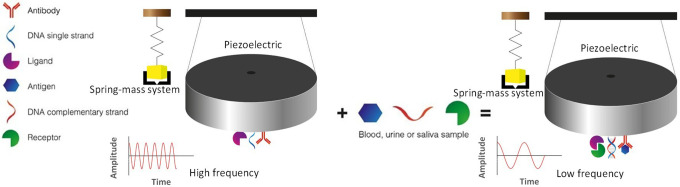
Biosensors for detecting changes in mass (Figure 4) make use of ultra-sensitive piezoelectric devices for detecting mass changes. These devices are analogous to a spring-mass mechanical system or electrical circuit with impedances, inductances, and capacitances. The changes in frequency within piezoelectric resonance gives a detectable signal due to an increase or decay in mass from biomarker interaction.
Figure 4.
Mass change biosensor. As a result of a change in mass in the piezoelectric device owed to the interaction between a biomarker (antigen, complementary DNA strand or ligand) present in body fluids and its target molecule (antibody DNA single strand or receptor) attached to the piezoelectric device a decay in frequency is recorded.
Proteomic Biosensors
Proteomic biosensors are based on the recognition of molecular elements like antibodies, enzymes, and peptides. An et al111 developed a magneto-mediated electrochemical biosensor to sense the proteins MUC1, EpCAM, HER2, and CEA in the exosomes of breast cancer cells, showing potential for the determination of this proteins in breast cancer serum, which is promising for clinical diagnosis. Autocrine motility factor (AMF) or AMF receptor overexpression is closely related to tumor progression and malignity.112 By attaching the substrate for the enzyme glucose-phosphate-isomerase (GPI) in the surface of a gold electrode and given that AMF and GPI are identical and possess the same functions, Devillers et al113 developed a biosensor for the detection of AMF and its association with cancer progression that exhibits a minimal detection limit on the order of 10-2 pM. Using the same AMF protein, a more recent work reported an electrochemical biosensor capable to detect even lower concentrations, with a lower limit of 43 fM, however, this detection was performed in phosphate buffer and not in serum.114 The epithelial cell adhesion molecule (EpCAM) has been considered a tumor prognostic biomarker in different types of cancer.115 Using graphene quantum dots (GQDs) as a fluorescence emissary and molybdenum disulfide (MoS2) as a fluorescence quencher, Shi et al116 developed an optical biosensor based on FRET capable of detecting EpCAM in cancer cells with a pM level detection limit. Li et al117 used up-converting nanoparticles (UCPs) and palladium nanoparticles (PdNps) for fluorescence quenching and made a biosensor for CEA detection through an aptamer linkage on a FRET system. Of note, the detection limit in serum was on the order of 0.8 pg/mL. Su et al118 designed and constructed a dual biosensor for the detection of PSA and α-fetoprotein using a ceramic piezoelectric device by immobilizing antibodies against these antigens on the surface of gold electrodes. The sensitivity and specificity of this biosensor were on the order of pg/mL and were comparable to ELISA methods. Parathyroid hormone-related peptide has been associated with cancer metastasis in breast and prostate cancer.119 Crivianu-Gaita et al120 constructed a device based on acoustic waves by attaching anti-PTHrP antibodies to quartz disks treated with different linkers, achieving a sensitivity of 61 ng/mL. However, despite these results, we must consider that clinically relevant concentrations of this protein oscillate between 120 pg/mL and 14 ng/mL, which is lower than the limit detection of the device.
Sarcosine has been found to activate prostate cancer cells; therefore, its occurrence in blood and urine indicates malignity. Narwal et al121 constructed a biosensor capable of detecting sarcosine levels on the order of picograms with a chitosan electrode covered with copper nanoparticles (CuNps) and carbon nanotubes. This biosensor has enormous potential for prostate cancer detection and can maintain its stability during a long period, showing storage stability. A biosensor that could be used in order to detect different types of cancer is something desirable and achievable, since Rangel et al,122 continuing with previous work,123 developed a biosensor based on impedances capable of detecting the T antigen expression in serum samples of patients with different types of cancer, including breast, prostate and lung cancer, in which an increase in the expression of this antigen is well known.
Genomic Biosensors
Some genetic-approaching biosensors used for the detection of several types of cancer biomarkers have been developed recently. miRNAs, of which some examples of potential biomarkers are mentioned in Table 2, are easy to hybridize with specific DNA probes generating a signal that, depending on the method, can be detected and used for discriminating between a healthy or ill patient. Even though the concentration threshold of the biomarker in the biological sample that these biosensors are capable of detecting could become a limiting factor, devices with better resolution features are now appearing. To depict an important example, minimum concentrations from 10 fM of multiple target miRNAs involved in liver cancer were detected by a sensor equipped with a surface-enhanced Raman scattering (SERS) technology.124 By using a similar strategy, Ouyang et al125 reported the construction of an optical biosensor based on SERS able of detecting DNA methylation levels and reported a minimum detection limit of 0.2 pg/μL. Interestingly, this device was reusable, showing its practical relevance. Likewise, a dual-SERS biosensor capable of detect miRNAs from pancreatic cancer from exosomes and plasma is a promissory technology that matters for the early detection of this type of cancer.126 In this way, an electrochemical biosensor using a graphene electrode covered with gold nanoparticles (AuNps) was recently built,127 the device was able to detect mutations in the CYFRA-21 gene with a minimal DNA concentration of 10-2 pM, showing enormous potential in lung cancer diagnosis. Lung cancer patients with mutations in the gene for the epidermal-growth-factor-receptor (EGFR) are treated with molecules that inhibit tyrosine/kinases (TKIs) that promote apoptotic pathways; thus, these mutations could be a predictive biomarker for treatment.128 Also, Weng et al129 constructed an electrochemical biosensor capable of discriminating between different kinds of mutations on the EGFR gene. Notably, these mutations were corroborated through sequencing, confirming the applicability of this method. More recently, another device useful to detect the lung cancer related PIK3CA gene mutations was developed. By using a DNA probe coupled to an FMNs/MoS2 nanocomposite, this device detects the self-redox signal loss when the target DNA binds to the DNA probe and changes its conformation with a limit sensitivity of 1.2 × 10-17 mol/L.130 Purposely, with another strategy, PIK3CAH1047 R mutation was efficiently detected using a biosensor based in strand displacement amplification mediated by a restriction enzyme and a system of 4-way DNA junction with a low detection limit of mutation from biological samples of human serum.131
For breast cancer, an immobilized ssDNA probe attached to a poly (amidoamine) dendrimers matrix and coupled to the surface of an Au electrode was used for the construction of a new biosensor132; this device shows high sensitivity for DNA detection in the order of 1nM or less, and specifically is able to differentiate single-base mismatches of cancer biomarker BRAC1 gene. A biosensor with the accuracy to detect quantities from 1 pM of the p53 gene and its mutations was developed by Luo et al.133 In order to improve selectivity and sensitivity, this device uses a double level of biomarkers (a hairpin and an enzyme) and emits the test result in just 23 min, positioning it at a more acute level of detection compared to other biosensors with a single biomarker level. This apparatus constituting a new, fast and useful strategy for the clinical and non-invasive diagnosis of various types of cancer using body fluids. By the other hand, detection of liver cancer is the goal of other strategy utilized in a strip-style biosensor which combines the detection of SNP on CYP1A1 gene and the expression of the protein biomarker alpha-fetoprotein (AFP) with a sensitivity in the order of ng/mL.134 The same strategy for detection of SNPs on the CYP1A1 gene but without the involvement of AFP was published a couple of months later with the premise of detecting any SNP in no more than 10 minutes.135
Methylation of MGMT gene as a biomarker in head and neck cancer is the detection objective of a new genosensor built with DNA probes immobilized on a gold surface in combination with 11-mercaptoundecanoic acid (11-MUA), which uses electrochemical impedance spectroscopy; results obtained with this sensor indicated high selectivity for HN13 cells, a high degree MGMT methylation cell line, and good sensitivity in the range of pmol/L of methylated DNA.136
Advantageously in many cases, the use of these and other genetic biosensors increases the speed that diagnosis with traditional methods and devices can represent. However, many of these new biosensors include no reusable and expensive components that block the use in the clinic field, especially in countries with limitations to provide basic health services.
Novel Biosensors for Immune Biomarkers
Dysregulation of the chemokine system is implicated in several autoimmune and inflammatory diseases, as well as cancer. In 2015, Chen et al137 introduced a novel biosensor for simultaneous detection of multiple cytokines (IL-2, IL-4, IL-6, IL-10, TNFα, and interferon γ (IFNγ). Based on a microfluidic surface plasmon resonance (LSPR) chip, this sensor achieved a linear range detection between 5 and 20 pg/mL with only 1 µL of serum sample. Noteworthy, such biosensor showed an increase in sensitivity 10 times higher than conventional LSPR chips. Later, in 2018 Aydin et al138 constructed an electrochemical biosensor based on the immobilization of anti-IL-1β antibody on an indium tin oxide (ITO) electrode for interleukin 1β (IL-1β) detection in saliva and serum samples. Through EIS, CV and SFI, this electrochemical biosensor showed a detection limit of 7.5 fg/mL.
The use of a labeled secondary antibody (Ab2) specific to the target analyte is an alternative approach to improve the limit of detection for cancer biosensors. In that sense, Peng et al,139 using a Sandwich Nanoparticles Labeled Electrochemical Immunoassay (sECIA-NP) technique, developed an electrochemical ultrasensitive biosensor for IL-6 detection, reaching a detection limit of 0.1 pg/mL. Something to highlight, the biosensor showed good reproducibility and long term stability, 2 desirable features in this kind of devices.
Continuing with electrochemical biosensors, Chung et al140 immobilized the chemokine CXCR2 (C-X-C Motif Chemokine Receptor 2) over a nanocomposite film covered with gold nanoparticles (AuNPs). Among 3 ligands tested, CXCL5 (C-X-C Motif Chemokine Ligand 5) showed the strongest affinity to CXCR2, showing a detection limit of 0.078 ng/mL. Remarkably, the biosensor showed high sensitivity and specificity in human serum and colorectal cancer cells samples.
Conclusions
The public health problems presented by cancer requires societies to develop new tools for the detection of this disease during its initial stages when the probabilities of a cure are higher. However, patients who are already suffering from this disease and are following a treatment plan also require tools capable of predicting their responses to therapy and the course or advancement of the disease. In that sense, the search for new diagnostic, prognostic and predictive biomarkers from proteomics, genomic or immunological nature is relevant. On the other hand, the limitations related to the sensitivity and specificity of current methods using biomarkers, in addition to the high operation costs, make new technologies an attractive solution for resolving the many current issues that have been previously cited.
Traditional detection of cancer requires highly trained health personnel able to perform sophisticated and expensive tests, sometimes with very invasive procedures for patients. As an alternative to traditional methods, biosensors have shown enormous potential, especially given their higher sensitivity and specificity compared with traditional methods, achieving limits of detection on the order of pM or even fM. Furthermore, because biosensors are small and easy-to-use devices, they diminish operation costs and the sample processing time, therefore facilitating and streamlining cancer diagnosis, prognosis and prediction. Devices that sense cancer biomarkers from body fluids or from non-invasive-obtained biological samples represent in many cases an advantage, being able to obtain fast and reliable results; however, even though hundreds of biosensors have been built in recent years, their distribution is not cosmopolitan due to various factors such as: i) the cost of materials for their construction; ii) issues associated with copyright; iii) priorities of the health systems in each country; iv) limitations specific to the chosen biomarker, among others. Many of these challenges must be overcome before one of these devices could be used commercially, making cheaper the components for their construction and selecting highly sensitive and specific biomarkers; until now, none of the used biomarkers has gathered both. Nevertheless, the new advances in materials sciences, molecular biology, immunology, and artificial intelligence are continuously increasing the knowledge related with these devices, contributing to its successful development in a not so far future.
The search for cancer biomarkers has been an arduous task for decades by researchers in the area. More and more molecules of different nature are cataloged as markers associated with various types of this disease, and although a flood of data on this can be found in the literature, many new biomarkers are detected at every moment.
Cancer biomarkers can form a haystack where finding a useful “needle” can be difficult. Depending on the type of cancer, the known information on these markers can become more limited and selected to be sensed by new devices capable of diagnosing or generating prognosis. Because of this, the search for proteins, genes or components of the immune system that contribute to having a better point of care in this disease will continue to be in force.
Acknowledgments
The authors thank Francisco Javier Ríos Fránquez for proof reading of this article.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following support for the research, authorship, and/or publication of this article: This study was supported by Secretaría de Energía (SENER) and Consejo Nacional de Ciencia y Tecnología (CONACYT); Program Ingeniería y Ciencia Aplicada al Sector Energético del Semidesierto del Norte del País (266492).
ORCID iD: Carlos A. Alba-Fierro  https://orcid.org/0000-0001-7915-5176
https://orcid.org/0000-0001-7915-5176
References
- 1. Bray F, Ferlay J, Soerjomatarum I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3(4):267–275. [DOI] [PubMed] [Google Scholar]
- 3. Chatterjee SK, Zetter BR. Cancer biomarkers: knowing the present and predicting the future. Future Oncol. 2005;3(1):37–50. [DOI] [PubMed] [Google Scholar]
- 4. Jayanthi VSPKSA, Das AB, Saxena U. Recent advances in biosensor development for the detection of cancer biomarkers. Biosens Bioelectron. 2017;91:15–23. [DOI] [PubMed] [Google Scholar]
- 5. Porto-Mascarenhas EC, Assad DX, Chardin H, et al. Salivary biomarkers in the diagnosis of breast cancer: a review. Crit Rev Oncol Hematol. 2017;110:62–73. [DOI] [PubMed] [Google Scholar]
- 6. Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93(14):1054–1061. [DOI] [PubMed] [Google Scholar]
- 7. Baker SG, Kramer BS, McIntosh M, Patterson BH, Shyr Y, Skates S. Evaluating markers for the early detection of cancer: overview of study designs and methods. Clin Trials. 2006;3(1):43–56. [DOI] [PubMed] [Google Scholar]
- 8. Diamandis EP. Present and future of cancer biomarkers. Clin Chem Lab Med. 2014;52(6):791–794. [DOI] [PubMed] [Google Scholar]
- 9. Rhee I. Diverse macrophages polarization in tumor microenvironment. Arch Pharm Res. 2016;39(11):1588–1596. [DOI] [PubMed] [Google Scholar]
- 10. Kulasingam V, Pavlou MP, Diamandis EP. Integrating high-throughput technologies in the quest for effective biomarkers for ovarian cancer. Nat Rev Cancer. 2010;10(5):371–378. [DOI] [PubMed] [Google Scholar]
- 11. Cui JF, Liu YK, Pan BS, et al. Differential proteomic analysis of human hepatocellular carcinoma cell line metastasis-associated proteins. J Cancer Res Clin Oncol. 2004;130(10):615–622. [DOI] [PubMed] [Google Scholar]
- 12. Charrier JP, Tournel C, Michel S, et al. Differential diagnosis of prostate cancer and benign prostate hyperplasia using two-dimensional electrophoresis. Electrophoresis. 2001;22(9):1861–1866. [DOI] [PubMed] [Google Scholar]
- 13. Chen G, Gharib TG, Huang CC, et al. Proteomic analysis of lung adenocarcinoma: identification of a highly expressed set of proteins in tumors. Clin Cancer Res. 2002;8(7):2298–2305. [PubMed] [Google Scholar]
- 14. Petricoin EF, Ardekani AM, Hitt BA, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359(9306):572–577. [DOI] [PubMed] [Google Scholar]
- 15. Ummanni R, Duscharla D, Barret C, et al. Prostate cancer-associated autoantibodies in serum against tumor-associated antigens as potential new biomarkers. J Proteomics. 2015;119:218–229. [DOI] [PubMed] [Google Scholar]
- 16. Kondo T. Cancer biomarker development and two dimensional difference gel electrophoresis (2D-DIGE). Biochim Biophys Acta Proteins Proteom. 2019;1867(1):2–8. [DOI] [PubMed] [Google Scholar]
- 17. Xiao H, Zhang L, Zhou H, Lee JM, Garon EB, Wong DT. Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol Cell proteomics. 2012;11(2):M111.012112 doi:10.1074/mcp.M111.012112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodland KD. Proteomics and cancer diagnosis: the potential of mass spectrometry. Clin Biochem. 2004;37(7):579–583. [DOI] [PubMed] [Google Scholar]
- 19. Cheung CHY, Juan HF. Quantitative proteomics in lung cancer. J Biomed Sci. 2017;24(1):37 doi:10.1186/s12929-017-0343-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conrads TP, Anderson GA, Veenstra TD, Pasa-Tolíc L, Smith RD. Utility of accurate mass tags for proteome-wide protein identification. Anal Chem. 2000;72(14):3349–3354. [DOI] [PubMed] [Google Scholar]
- 21. Kwon OK, Ha YS, NA AY, et al. Identification of novel prognosis and prediction markers in advanced prostate cancer tissues based on quantitative proteomics. Cancer Genomics Proteomics. 2020;17(2):195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim Y, Kang UB, Kim S, et al. A validation study of a multiple reaction monitoring-based proteomic assay to diagnose breast cancer. J Breast Cancer. 2019;22(4):579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sokolowska I, Woods AGN, Jayathirtha M, Darie CC. Role of mass spectrometry in investigating a novel protein: the example of tumor differentiation factor TDF. Adv Exp Med Bio.2019;1140:417–433. [DOI] [PubMed] [Google Scholar]
- 24. Tanase CP, Codrici E, Popescu ID, et al. Prostate cancer proteomics; current trends and future perspectives for biomarker discovery. Oncotarget. 2017;8(11):18497–18512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seigneuric R, Mjahed H, Gobbo J, et al. Heat shock proteins as danger signals for cancer detection. Front Oncol. 2011;1:37 doi:10.3389/fonc.2011.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saini J, Sharma PK. Clinical, prognostic and therapeutic significance of heat shock proteins in cancer. Curr Drug Targets. 2017;19(13):1478–1490. [DOI] [PubMed] [Google Scholar]
- 27. Hernandez C, Huebener P, Schwabe RF. Damage-associated molecular patterns in cancer: a double edge sword. Oncogene. 2016;35(46):5931–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rothammer A, Sage EK, Werner C, Combs SE Multhoff G. Increased heat shock protein 70 (Hsp70) serum levels and low NK cell counts after radiotherapy-potential markers for predicting breast cancer recurrence? Radiat Oncol. 2019;14(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi Y, Lou J. Plasma levels of heat shock protein 90 alpha associated with lung cancer development and treatment responses. Clin Cancer Res. 2014;20(23):6016–6022. [DOI] [PubMed] [Google Scholar]
- 30. Liu W, Li J, Zhang P, et al. A novel pan-cancer biomarker plasma heat shock protein 90alpha and its diagnosis determinants in clinic. Cancer Sci. 2019;110(9):2941–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. González-Mariscal L, Tapia R, Chamorro D. Crosstalk of tight junction components with signaling pathways. Biochim Biophys Acta. 2008;1778(3):729–756. [DOI] [PubMed] [Google Scholar]
- 32. Escudero-Esparza A, Jiang WG, Martin TA. The claudin family and its role in cancer and metastasis. Front Biosci (Landmark Ed). 2011;16:1069–1083. [DOI] [PubMed] [Google Scholar]
- 33. Zhou S, Piao X, Wang C, Wang R, Song Z. Identification of claudin-1, -3, -7 and -8 as prognostic markers in human laryngeal carcinoma. Mol Med Rep. 2019;20(1):393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jia H, Chai X, Li S, Wu D, Fan Z. Identification of claudin-2, -6, -11 and -14 as prognostic markers in human breast carcinoma. Int J Clin Exp Pathol. 2019;12(6):2195–2204. [PMC free article] [PubMed] [Google Scholar]
- 35. Arakawa H. Netrin-1 and its receptors in tumorigenesis. Nat Rev Cancer. 2004; 4(12):978–987. [DOI] [PubMed] [Google Scholar]
- 36. El-Gamal R, Mokhtar N, Ali-El-Dein B, Baiomy AA, Aboazma SM. Netrin-1: a new promising diagnostic marker for muscle invasion for bladder cancer. Urol Oncol. 2020;38(7):640.e1–640.e12. doi:10.1016/j.urolonc.2020.02.006 [DOI] [PubMed] [Google Scholar]
- 37. Ma S, Wang W, Xia B, et al. Multiplexed serum biomarkers for the detection of lung cancer. EbioMedicine. 2016;11:210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Etheridge T, Straus J, Ritter MA, Jarrard DF, Huang W. Semen AMACR protein as a novel method for detecting prostate cancer. Urol Oncol. 2018;36(12):532.e1–532.e7 doi:10.1016/j.urolonc.2018.09.010 [DOI] [PubMed] [Google Scholar]
- 39. Zhang W, Gao Z, Zeng G, et al. Clinical significance of urinary plasminogen and fibrinogen gamma chain as novel potential diagnostic markers for non-small-cell lung cancer. Clin Chim Acta. 2020;502:55–65. [DOI] [PubMed] [Google Scholar]
- 40. Zheng X, Chen S, Li L, et al. Evaluation of HE4 and TTR for diagnosis of ovarian cancer: comparison with CA-125. J Gynecol Obstet Hum Reprod. 2018;47(6):227–230. [DOI] [PubMed] [Google Scholar]
- 41. Guo NL, Dowlati A, Raese RA, et al. A predictive 7-gene assay and prognostic protein biomarkers for non-small cell lung cancer. EBioMedicine. 2018;32:102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Llie M, Beaulande M, Hamila M, Erb G, Hofman V, Hofman P. Automated chromogenic multiplexed immunohistochemistry assay for diagnosis and predictive biomarker testing in non-small cell lung cancer. Lung Cancer. 2018;124:90–94. [DOI] [PubMed] [Google Scholar]
- 43. Larson DE, Harris CC, Chen K, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28(3):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roth A, Ding J, Morin R, et al. JointSNVMix: a probabilistic model for accurate detection of somatic mutations in normal/tumour paired next-generation sequencing data. Bioinformatics. 2012;28(7):907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cyriac G, Gandhi L. Emerging biomarkers for immune checkpoint inhibition in lung cancer. Semin Cancer Biol. 2018;50(pt 2):269–277. [DOI] [PubMed] [Google Scholar]
- 47. Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Y, Xu H, Jiao H, et al. STX2 promotes colorectal cancer metastasis through a positive feedback loop that activates the NF-κB pathway. Cell Death Dis. 2018;9(6):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao J, Xu H, He M, Wang Z, Wu Y. Rho GTPase-activating protein 35 rs1052667 polymorphism and osteosarcoma risk and prognosis. BioMed Res Int. 2014;2014:396947: doi:10.1155/2014/396947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48(6): 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu X, Wang Z, Qiu X, et al. Rs2303428 of MSH2 is associated with hepatocellular carcinoma prognosis in Chinese population. DNA Cell Biol. 2018;37(7):634–641. [DOI] [PubMed] [Google Scholar]
- 53. Lo YL, Hsiao CF, Jou YS, et al. Polymorphisms of MLH1 and MSH2 genes and the risk of lung cancer among never smokers. Lung Cancer. 2011;72(3):280–286. [DOI] [PubMed] [Google Scholar]
- 54. Wu J, Wang S, Liu F, Li S. Single nucleotide polymorphism in the acilphosphatase 2 gene and the SNP-SNP interactions on the risk of breast cancer in Chinese Han women. Clin Breast Cancer. 2018;18(3):329–333. [DOI] [PubMed] [Google Scholar]
- 55. Zhang R, Zhou F, Cheng L, et al. Genetic variants in nucleotide excision repair pathways predict survival of esophageal squamous cell cancer patients receiving platinum-based chemotherapy. Mol Carcinog. 2018;57(11):1553–1565. [DOI] [PubMed] [Google Scholar]
- 56. Antczak NM, Walker AR, Stern HR, et al. Characterization of nine cancer-associated variants in human DNA polymerase. Chem Res Toxicol. 2018;31(8):697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Henríquez-Hernández LA, Valenciano A, Foro-Arnalot P, et al. Single nucleotide polymorphisms in DNA repair genes as risk factors associated to prostate cancer progression. BMC Med Genet. 2014;15:143 doi:10.1186/s12881-014-0143-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Marshall CH, Fu W, Wang H, Baras AS, Lotan TL, Antonarakis ES. Prevalence of DNA repair gene mutations in localized prostate cancer according to clinical and pathologic features: association of Gleason score and tumor stage. Prostate Cancer Prostatic Dis. 2019;22(1):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Akiyama Y, Kondou R, Iizuka A, et al. Immune response-associated gene analysis of 1,000 cancer patients using whole-exome sequencing and gene expression profiling—Project HOPE. Biomed Res. 2016;37(4):233–242. [DOI] [PubMed] [Google Scholar]
- 60. Li G, Guo X, Chen M, et al. Prevalence and spectrum of ANTK1, PIK3CA, PTEN and TP53 somatic mutations in Chinese breast cancer patients. PLoS One. 2018;13(9): e0203495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ferrari N, Mohammed ZM, Nixon C, et al. Expression of RUNX1 correlates with poor patient prognosis in triple negative breast cancer. PLoS One. 2014;9(6): e100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Adhami M, Haghdoost AA, Sadeghi B, Malekpour Afshar R. Candidate miRNAs in human breast cancer biomarkers: a systematic review. Breast Cancer. 2018;25(2):198–205. [DOI] [PubMed] [Google Scholar]
- 63. Fan L, Qi H, Teng J, et al. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumor Biol. 2016;37(6):7777–7784. [DOI] [PubMed] [Google Scholar]
- 64. Zhang K, Wang YW, Song Y, et al. Identification of microRNA biomarkers in the blood of breast cancer patients based on microRNA profiling. Gene. 2017;619:10–20. [DOI] [PubMed] [Google Scholar]
- 65. Sherlock GJ, Brooks JD, Myers RM, Kobayashi Y, Absher DM. U.S. Patent Application No. 14/008,480 2014.
- 66. Zhang W, Shu P, Wang S, et al. ZNF154 is a promising diagnosis biomarker and predicts biochemical recurrence in prostate cancer. Gene. 2018;675:136–143. [DOI] [PubMed] [Google Scholar]
- 67. Eissa MA, Lerner L, Abdelfatah E, et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin Epigenetics. 2019;11(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aasen T, Sansano I, Montero MÁ, et al. Insight into the role and regulation of gap junction genes in lung cancer and identification of nuclear Cx43 as a putative biomarker of poor prognosis. Cancers. 2019;11(3): 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gangadharan A, Nyirenda T, Patel K, et al. Prolactin induced protein (PIP) is a potential biomarker for early stage and malignant breast cancer. Breast. 2018;39:101–109. [DOI] [PubMed] [Google Scholar]
- 70. Campbell PT, Curtin K, Ulrich CM, et al. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut. 2009;58(5):661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lam K, Zhang DE. RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis. Front Biosci. 2012;17:1120–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. [DOI] [PubMed] [Google Scholar]
- 73. Zhang K, Zhang Y, Liu C, Xiong Y, Zhang J. MicroRNAs in the diagnosis and prognosis of breast cancer and their therapeutic potential. Int J Oncol. 2014;45(3):950–958. [DOI] [PubMed] [Google Scholar]
- 74. Wang YW, Shi DB, Chen X, Gao C, Gao P. Clinicopathological significance of microRNA-214 in gastric cancer and its effect on cell biological behavior. PLoS One. 2014;9(3): e91307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chakraborty C, Das S. Profiling cell-free and circulating miRNA: a clinical diagnostic tool for different cancers. Tumor Biol. 2016;37(5):5705–5714. [DOI] [PubMed] [Google Scholar]
- 76. Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res. 2005;4(4):1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. [DOI] [PubMed] [Google Scholar]
- 79. Hegab AE, Ozaki M, Kagawa S, et al. Tumor associated macrophages support the growth of FGF9-induced lung adenocarcinoma by multiple mechanisms. Lung Cancer. 2018;119:25–35. [DOI] [PubMed] [Google Scholar]
- 80. Li Z, Maeda D, Yoshida M, et al. The intratumoral distribution influences the prognostic impact of CD68- and CD204- positive macrophages in non-small cell lung cancer. Lung Cancer. 2018;123:127–135. [DOI] [PubMed] [Google Scholar]
- 81. Zhang Y, Feng YC, Zhu HG, et al. The peripheral blood-to-lymphocyte ratio is a prognostic predictor for survival of EGFR-mutant nonsmall cell lung cancer patients treated with EGFR-TKIs. Medicine. 2018;97(30): e11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Phan TT, Ho TT, Nguyen HT, Nguyen HT, Tran TB, Nguyen ST. The prognostic impact of neutrophil to lymphocyte ratio in advanced non-small cell lung cancer patients treated with EGFR TKI. Int J Gen Med. 2018;11:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tamura R, Tanaka T, Yamamoto Y, Akasaki Y, Sasaki H. Dual role of macrophage in tumor immunity. Immunotherapy. 2018:10(10):899–909. [DOI] [PubMed] [Google Scholar]
- 84. Schmid S, Csanadi A, Kozhuharov N, et al. CC-chemokine ligand 18 is an independent prognostic marker in lymph node-positive non-small cell lung cancer. Anticancer Res. 2018;38(7):3913–3918. [DOI] [PubMed] [Google Scholar]
- 85. Schmid S, Le UT, Haager B, et al. Local concentrations of CC-chemokine-ligand 18 correlate with tumor size in non-small cell lung cancer and are elevated in lymph node-positive disease. Anticancer Res. 2016:36(9):4667–4672. [DOI] [PubMed] [Google Scholar]
- 86. Plönes T, Krohn A, Burger M, et al. Serum level of CC-chemokine ligand 18 is increased in patients with non-small-cell lung cancer and correlates with survival time in adenocarcinomas. PLoS One. 2012;7(7): e41746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Karin N. Chemokines and cancer: new immune checkpoints for cancer therapy. Curr Opin Immunol. 2018;51:140–145. [DOI] [PubMed] [Google Scholar]
- 88. Fujimoto Y, Inoue N, Morimoto K, et al. Significant association between high serum CCL5 levels and better disease-free survival of patients with early breast cancer. Cancer Sci. 2020;111(1):209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Razis E, Kalogeras KT, Kotsantis I, et al. The role of CXCL13 and CXCL9 in early breast cancer. Clin Breast Cancer. 2020;20(1): e36–e53. [DOI] [PubMed] [Google Scholar]
- 90. Liu C, Sun B, Xu B, et al. A panel containing PD-1, IL-2Rα, IL-10, and CA15-3 as a biomarker to discriminate breast cancer from benign breast disease. Cancer Manag Res. 2018;10:1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dudnik E, Peled N, Nechushtan H, et al. BRAF mutant lung cancer: PD-L1 expression, tumor mutational burden, microsatellite instability status and response to immune check-point inhibitors. J Thorac Oncol. 2018;13(8):1128–1137. [DOI] [PubMed] [Google Scholar]
- 92. Krieger T, Pearson I, Bell J, Doherty J, Robbins P. Targeted literature review on use of tumor mutational burden status and programmed cell death ligand 1 expression to predict outcomes of checkpoint inhibitor treatment. Diagn Pathol. 2020;15(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Waki K, Yamada T, Yoshiyama K, et al. PD-1 expression on peripheral blood T-cell subsets correlates with prognosis in non-small cell lung cancer. Cancer Sci. 2014;105(10):1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. El-Guindy DM, Helal DS, Sabry NM, Abo El-Nasr M. Programmed cell death ligand-1 (PD-L1) expression combined with CD8 tumor infiltrating lymphocytes density in non-small cell lung cancer patients. J Egypt Natl Canc Inst. 2018;30(4):125–131. [DOI] [PubMed] [Google Scholar]
- 95. Gonzalez-Ericsson PI, Stovgaard ES, Sua LF, et al. The path to a better biomarker: application of a risk management framework for the implementation of PD-L1 and TILs as immuno-oncology biomarkers into breast cancer clinical trials and daily practice. J Pathol. 2020;250(5):667–684. doi:10.1002/path.5406 [DOI] [PubMed] [Google Scholar]
- 96. Morgan E, Suresh A, Ganju A, et al. Assessment of outcomes and novel immune biomarkers in metaplastic breast cancer: a single institution retrospective study. World J Surg Oncol. 2020;18(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang L, Simons DL, Lu X, et al. Breast cancer induces systemic immune changes on cytokine signaling in peripheral blood monocytes and lymphocytes. EBioMedicine. 2020;52:102631 doi:10.1016/j.ebiom.2020.102631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li J, Liu C, Chen Y, et al. Tumor characterization in breast cancer identifies immune-relevant gene signatures associated with prognosis. Front Genet. 2019;10:1119 doi:10.3389/fgene.2019.01119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Favaro E, Amadori A, Indraccolo S. Cellular interactions in the vascular niche: implications in the regulation of tumor dormancy. APMIS. 2008;116(7-8):648–659. [DOI] [PubMed] [Google Scholar]
- 100. Kaur RP, Rubal, Banipal RPS, Vashistha R, Dhiman M, Munshi A. Association of elevated levels of C-reactive protein with breast cancer, breast cancer subtypes and poor outcome. Curr Probl Cancer. 2019;43(2):123–129. [DOI] [PubMed] [Google Scholar]
- 101. Cai P, Lu Z, Wu J, et al. BTN3A2 serves as a prognostic marker and favors immune infiltration in triple-negative breast cancer. J Cell Biochem. 2020;121(3):2643–2654. [DOI] [PubMed] [Google Scholar]
- 102. Zhao X, Li Y, Wang X, et al. Synergistic association of FOXP3+ tumor infiltrating lymphocytes with CCL20 expressions with poor prognosis of primary breast cancer: a retrospective cohort study. Medicine (Baltimore), 2019;98(50):e18403 doi:10.1097/MD.0000000000018403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cerbelli B, Botticelli A, Pisano A, et al. CD73 expression and pathologic response to neoadjuvant chemotherapy in triple negative breast cancer. Virchows Arch. 2019;476(4):569–576. doi:10.1007/s00428-019-02722-6 [DOI] [PubMed] [Google Scholar]
- 104. Ahmad MM, Parveen F, Akhter N, Siddiqui JA, Shukla NK, Husain SA. Genetic polymorphism in TNF-α-308 G/A and TNF-β +252 A/G, as prognostic biomarker in breast cancer patients among Indian population. Asian Pac J Cancer Prev. 2020;21(2):301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ouni N, Ben Chaaben A, Ayari F, et al. MICA-129 Met/Val polymorphism could be a genetic biomarker for familial breast cancer in the Tunisian population. Int J Immunogenet. 2020. doi:10.1111/iji.12480 [DOI] [PubMed] [Google Scholar]
- 106. Altintas Z, Uludag Y, Gurbuz Y, Tothill IE. Surface plasmon resonance based immunosensor for the detection of the cancer biomarker carcinoembryonic antigen. Talanta. 2011;86:377–383. [DOI] [PubMed] [Google Scholar]
- 107. Silva ACQ, Vilela C, Santos HA, Silvestre AJD, Ferire CSR. Recent trends on the development of systems for cancer diagnosis and treatment by microfluidic technology. Applied Materials Today. 2020;18:100450 doi:10.1016/j.apmt.2019.100450 [Google Scholar]
- 108. Shao B, Xiao Z. Recent achievements in exosomal biomarkers detections by nano-materials optical biosensors. Anal Chim Acta. 2020;1114:74–84. doi:10.1016/j.aca.2020.02.041 [DOI] [PubMed] [Google Scholar]
- 109. Khanmohammadi A, Aghaie A, Vahedi E, et al. Electrochemical biosensors for the detection of lung cancer biomarkers: a review. Talanta. 2020;206:120251 doi:10.1016/j.talanta.2019.120251 [DOI] [PubMed] [Google Scholar]
- 110. Ranjan R, Esimbekova EN, Kratasyuk VA. Rapid biosensing tools for cancer biomarkers. Biosens Bioelectron. 2017;87:918–930. [DOI] [PubMed] [Google Scholar]
- 111. An Y, Li R, Zhang F, He P. Magneto-mediated electrochemical sensor for simultaneous analysis of breast cancer exosomal proteins. Anal Chem. 2020;92(7):5404–5410. doi:10.1021/acs.analchem.0c00106 [DOI] [PubMed] [Google Scholar]
- 112. Watanabe H, Carmi P, Hogan V, et al. Purification of human tumor cell autocrine motility factor and molecular cloning of its receptor. J Biol Chem. 1991;266(20):13442–13448. [PubMed] [Google Scholar]
- 113. Devillers M, Ahmad L, Korri-Youssoufi H, Salmon L. Carbohydrate-based electrochemical biosensor for detection of a cancer biomarker in human plasma. Biosens Bioelectron. 2017;96:178–185. [DOI] [PubMed] [Google Scholar]
- 114. Ahmad L, Salmon L, Korri-Youssoufi H. Electrochemical detection of the human cancer biomarker “autocrine motility factor-phosphoglucose isomerase” based on a biosensor formed with a monosaccharidic inhibitor. Sensor Actuat B-Chem. 2019;299:126933 doi:10.1016/j.snb.2019.126933 [Google Scholar]
- 115. Baeuerle PA, Gires O. EpCAM (CD36) finding its role in cancer. Br J Cancer. 2007;96(3):417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shi J, Lyu J, Tian F, Yang M. A fluorescence turn-on biosensor based on graphene quantum dots (GQDS) and molybdenum disulfide (MoS2) nanosheets for epithelial cell adhesion molecule (EpCAM) detection. Biosens Bioelectron. 2017;93:182–188. [DOI] [PubMed] [Google Scholar]
- 117. Li H, Shi L, Sun D, Li P, Liu Z. Fluorescence resonance energy transfer biosensor between upconverting nanoparticles and palladium nanoparticles for ultrasensitive CEA detection. Biosens Bioelectron. 2016;86:791–798. [DOI] [PubMed] [Google Scholar]
- 118. Su L, Zou L, Fong CC, et al. Detection of cancer biomarkers by piezoelectric biosensor using PZT ceramic resonator as the transducer. Biosens Bioelectron. 2013;46:155–161. [DOI] [PubMed] [Google Scholar]
- 119. Nishihara M, Kanematsu T, Taguchi T, Razzaque MS. PTHrP and tumorigenesis: is there a role in prognosis? Ann N Y Acad Sci. 2007;1117:385–392. [DOI] [PubMed] [Google Scholar]
- 120. Crivianu-Gaita V, Aamer M, Posaratnanathan RT, Romaschin A, Thompson M. Acoustic wave biosensor for the detection of the breast and prostate cancer metastasis biomarker protein PTHrP. Biosens Bioelectron. 2016;78:92–99. [DOI] [PubMed] [Google Scholar]
- 121. Narwal V, Kumar P, Joon P, Pundir CS. Fabrication of an amperometric sarcosine biosensor based on sarcosine oxidase/chitosan/CuNPs/c-MWCNT/Au electrode for detection of prostate cancer. Enzyme Microb Technol. 2018;113:44–51. [DOI] [PubMed] [Google Scholar]
- 122. Rangel MGH, Silva MLS. Detection of the cancer-associated T antigen using an Arachys hypogaea agglutinin biosensor. Biosens Bioelectron. 2019;141:111401 doi:10.1016/j.bios.2019.111401 [DOI] [PubMed] [Google Scholar]
- 123. Silva MLS, Rangel MGH. A Vicial Villosa agglutinin biosensor for cancer-associated tn antigen. Sensor Actuat B-Chem. 2017;252:777–784. [Google Scholar]
- 124. Zhou W, Tian YF, Yin BC Ye BC. Simultaneous surface-enhanced Raman spectroscopy detection of multiplexed microRNA biomarkers. Anal Chem. 2017;89(11):6120–6128. [DOI] [PubMed] [Google Scholar]
- 125. Ouyang L, Hu Y, Zhu L, Cheng GJ, Irudayaraj J. A reusable laser wrapped graphene-Ag array based SERS sensor for trace detection of genomic DNA methylation. Biosens Bioelectron. 2017;92:755–762. [DOI] [PubMed] [Google Scholar]
- 126. Pang Y, Wang C, Lu L, Wang C, Sun Z, Xiao R. Dual-SERS biosensor for one-step detection of microRNAs in exosome and residual plasma of blood samples for diagnosing pancreatic cancer. Biosens Bioelectron. 2019;130:204–213. [DOI] [PubMed] [Google Scholar]
- 127. Chen M, Wang Y, Su H, et al. Three-dimensional electrochemical DNA biosensor based on 3D graphene-Ag nanoparticles for sensitive detection of CYFRA21 -1 in non-small cell lung cancer. Sensor Actuat B-Chem. 2018;255(pt 3):2910–2918. [Google Scholar]
- 128. Douillard JY, Shepherd FA, Hirsh V, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non–small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol. 2010;28(5):744–752 [DOI] [PubMed] [Google Scholar]
- 129. Weng XH, Xu XW, Wang CL, et al. Genotyping of common EGFR mutations in lung cancer patients by electrochemical biosensor. J Pharm Biomed Anal. 2018;150:176–182. [DOI] [PubMed] [Google Scholar]
- 130. Zhang W, Yang J Wu D. Surface-functionalized MoS2 nanosheets sensor for direct electrochemical detection of PIK3CA gene related to lung cancer. J Electrochem Soc. 2020;167(2):027501. [Google Scholar]
- 131. Wang T, Peng Q, Guo B, et al. An integrated electrochemical biosensor based on target-triggered strand displacement amplification and “four-way” DNA junction towards ultrasensitive detection of PIK3CA gene mutation. Biosens Bioelectron. 2020;150:111954 doi:10.1016/j.bios.2019.111954 [DOI] [PubMed] [Google Scholar]
- 132. Senel M, Dervisevic M, Kokkokoğlu F. Electrochemical DNA biosensors for label-free breast cancer gene marker detection. Anal Bioanal Chem. 2019:411(13):2925–2935. [DOI] [PubMed] [Google Scholar]
- 133. Luo Z, Xu Y, Huang Z, et al. A rapid, adaptative DNA biosensor based on molecular beacon-concatenated dual signal amplification strategies for ultrasensitive detection of p53 gene and cancer cells. Talanta. 2020;210:120638 doi:10.1016/j.talanta.2019.120638 [DOI] [PubMed] [Google Scholar]
- 134. Ma LH, Wang HB, Zhang T, et al. Visual simultaneous detection of single nucleotide polymorphism of tumor susceptibility gene and marker alpha-fetoprotein based on double-labeled colloidal gold probe with lateral flow strip biosensor. Sensor Actuat B-Chem. 2019;298:126819 doi:10.1016/j.snb.2019.126819 [Google Scholar]
- 135. Wang HB, Ma LH, Zhang T, Huang KC, Zhao YD, Liu TC. Simple and accurate visual detection of single nucleotide polymorphism based on colloidal gold nucleic acid strip biosensor and primer-specific PCR. Anal Chim Acta. 2020;1093:106–114. [DOI] [PubMed] [Google Scholar]
- 136. Carr O, Raymundo-Pereira PA, Shimizu FM, et al. Genosensor made with a self-assembled monolayer matrix to detect MGMT gene methylation in head and neck cancer cell lines. Talanta. 2020;210:120609 doi:10.1016/j.talanta.2019.120609 [DOI] [PubMed] [Google Scholar]
- 137. Chen P., Chung M. T., McHugh W., et al. Multiplex serum cytokine immunoassay using nanoplasmonic biosensor microarrays. ACS Nano. 2015;9(4),4173–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Aydın EB, Sezgintürk MK. A disposable and ultrasensitive ITO based biosensor modified by 6-phosphonohexanoic acid for electrochemical sensing of IL-1β in human serum and saliva. Anal Chim Acta. 2018;18;1039:41–50. [DOI] [PubMed] [Google Scholar]
- 139. Peng J., Feng L.N., Ren Z.J., Jiang L.P., Zhu J.J. Synthesis of silver nanoparticle–hollow titanium phosphate sphere hybrid as a label for ultrasensitive electrochemical detection of human interleukin-6. Small. 2011;7(20):2921–2928. [DOI] [PubMed] [Google Scholar]
- 140. Chung S, Chandra P, Koo JP, Shim YB. Development of a bifunctional nanobiosensor for screening and detection of chemokine ligand in colorectal cancer cell line. Biosens Bioelectron. 2018;100:396–403. [DOI] [PubMed] [Google Scholar]