Abstract
Type 1 cannabinoid receptor (CB1) antagonists have demonstrated promise for the treatment of obesity, liver disease, metabolic syndrome, and dyslipidemias. However, the inhibition of CB1 receptors in the central nervous system can produce adverse effects, including depression, anxiety, and suicidal ideation. Efforts are now underway to produce peripherally restricted CB1 antagonists to circumvent CNS-associated undesirable effects. In this study, a series of analogues were explored in which the 4-aminopiperidine group of compound 2 was replaced with aryl- and heteroaryl-substituted piperazine groups both with and without a spacer. This resulted in mildly basic, potent antagonists of human CB1 (hCB1). The 2-chlorobenzyl piperazine, 25, was found to be potent (Ki = 8 nM); to be >1000-fold selective for hCB1 over hCB2; to have no hERG liability; and to possess favorable ADME properties including high oral absorption and negligible CNS penetration. Compound 25 was tested in a mouse model of alcohol-induced liver steatosis and found to be efficacious. Taken together, 25 represents an exciting lead compound for further clinical development or refinement.
Graphical Abstract
INTRODUCTION
The endocannabinoid (EC) system, comprising endocannabinoids, receptors, transporters, and enzymes, is known to play a role in a large variety of physiological processes.1–3 Selective modulation of the EC system has potential therapeutic applications in a broad range of medical conditions. Human cannabinoid receptors hCB1 and hCB2 are G-protein-coupled receptors (GPCRs) whose primary function is to activate G proteins (Gi/o). The hCB1 receptor is present throughout the body and is highly expressed in the central nervous system (CNS). In contrast, hCB2 is primarily expressed in the immune system and nominally expressed in the CNS.1 The selective attenuation of peripheral-hCB1-receptor activity is a promising approach for treating diabetes, metabolic syndrome, dyslipidemias, and liver diseases such as nonalcoholic steatohepatitis (NASH) and alcoholic liver disease.4,5 Rimonabant (1, Figure 1), a CNS-penetrating hCB1-receptor inverse agonist developed by Sanofi-Aventis, was clinically approved for the treatment of obesity in Europe (marketed as Accomplia). It had to be withdrawn, however, because of adverse psychiatric episodes in some patients, including depression and suicidal ideation, resulting from the inhibition of hCB1 activity in the CNS.6 The adverse side effects seen with 1 precipitated the withdrawal of other CNS-penetrating hCB1 antagonists from clinical development, including otenabant (2), a compound developed by Pfizer. Currently, efforts are underway to produce peripherally selective CB1 antagonists that have limited brain penetration.7,8 Among these are candidate compounds, JD5037 and TM38837 are of particular note.9–11 The former is reportedly undergoing IND-enabling studies. The latter was tested in humans and demonstrated peripheral restriction, but its current development status is unknown. A thiophene carboxamide analogue of 1 has also been approved for phase 1 studies by the U.S. FDA.12
Figure 1.

Examples of clinical CB1 antagonists (1 and 2) and lead scaffolds 3 and 4.
Our previous approach to limiting CNS exposure using analogues of 2 focused on enhancing the topological polar surface area (TPSA) while eliminating the ability of the primary amide and ethyl amine to form a hydrogen-bonding pair, which has been proposed as the molecular basis for the high CNS permeability of 2.13,14 Using this strategy, we reported the discovery of a peripherally selective analogue, 3 (Figure 1). Compound 3 possessed excellent potency, reasonable hCB1 selectivity (~50-fold over hCB2), and most importantly, limited brain penetration (~5–7% of plasma concentration). Unfortunately, compound 3 suffered from limited aqueous solubility. Therefore, we continued to refine and produce analogues of 3 with better ADME properties, while attempting to further reduce brain penetration. We envisioned that the reintroduction of groups capable of being ionized at low pHs would have the benefit of providing the compounds with improved solubility in the stomach and upper gastrointestinal (GI) tract while still maintaining the potency and peripheral selectivity observed with 3.
Compound 2 contains a 4,4-disubstituted piperidine in the 6-position of the purine core. Conversion of the 4,4-disubstituted piperidine into a N-substituted piperazine afforded compound 4, which had good solubility in simulated gastric fluid but also possessed significant activity at hERG channels, which is associated with a risk of LQT syndrome.15 In addition, a close analogue of compound 4 (2,4-dichlorophenyl instead of 2-chlorophenyl at the 8-position of the purine ring) was reported to have peripheral selectivity, despite having no hydrogen-bond-donating groups. This led us to examine analogues of 2 in which the purine ring was functionalized with a piperazine that was then decorated with aryl or heteroaryl groups, both with and without intervening linkers. In this report, we describe our efforts to make analogues of 3 that resulted in 25, which has good potency, peripheral selectivity, oral bioavailability, and pharmacokinetic properties. Compound 25 also showed efficacy in a murine model of alcohol-induced liver steatosis.
CHEMICAL SYNTHESIS
The targeted compounds were prepared as shown in Scheme 1. Chloride 5 was prepared as previously reported15 and then reacted with commercially available piperazines to provide the target compounds in generally good yields (typically >50%).
Scheme 1. Synthesis of the Target Compoundsa.
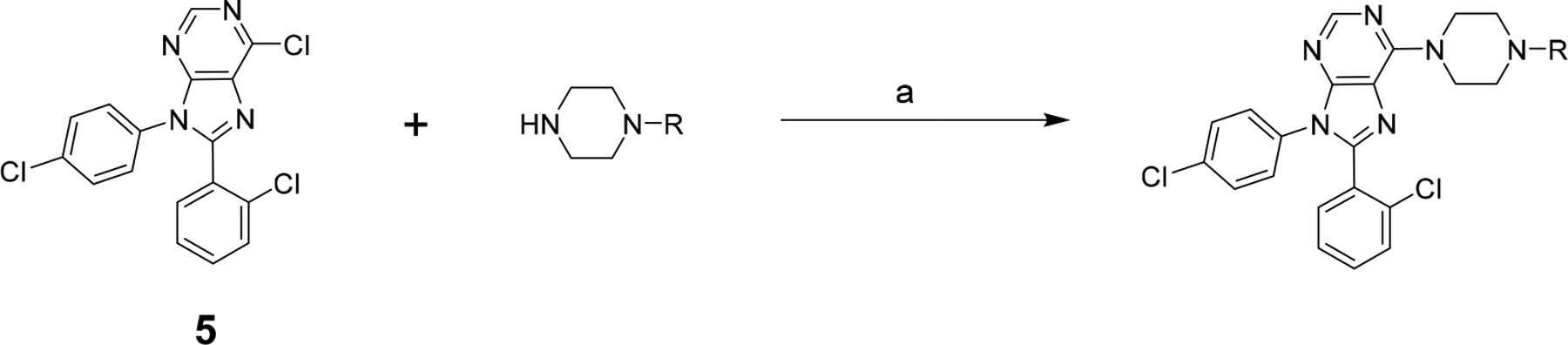
aReagents and conditions: (a) NEt3, EtOH, 80 °C, O/N.
RESULTS AND DISCUSSION
SAR Studies of CB1 Antagonists.
All the target compounds were evaluated in a FLIPR-based calcium-mobilization assay for hCB1 activity (Tables 1 and 2) as described in our previous publications.13,14 In general, those that demonstrated apparent-antagonist-dissociation-equilibrium constants (Ke) of <50 nM were further tested for their affinities to the hCB1 and hCB2 receptors using radioligand displacement of [3H]CP55940 in purified membrane fractions overexpressing either hCB1 or hCB2. The results were expressed as selectivity ratios (hCB2/hCB1). The most potent and hCB1-subtype-selective compounds were tested for peripheral selectivity by calculating percent apical (A) to basal (B) in an MDCK-mdr1 permeability assay, which was predictive of brain penetration.16 The compounds that showed limited transport from A to B were identified as desirable for peripheral selectivity.
Table 1.
In Vitro Data for the CB1 Antagonists with Aryl Rings Directly Connected to the Piperazines
| R | Ke hCB1 (nM) | Ki hCB1 (nM)a | Ki hCB2 (nM)a | selectivity Ki CB2/CB1 | MDCK-mdr1 A to B (%)b | |
|---|---|---|---|---|---|---|
| 2 | otenabantc | 0.2 | 0.7 | 7700 | 11000 | |
| 6 | Me | 120 | 3d | |||
| 7 | c-Hex | 17 | 2 | 110 | 64 | 2 |
| 8 | Ph | 5 | 2 | >10000 | >5000 | 0 |
| 9 | 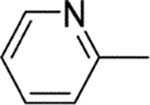 |
13 | 2 | 1400 | 700 | 0.06 |
| 10 | 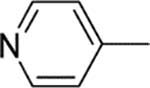 |
1200 | ||||
| 11 | 2-CN-Ph | 7 | 2 | >10000 | >5000 | 0.02 |
| 12 | 2-MeO-Ph | 16 | 2 | 5000 | 2500 | 0 |
| 13 | 3-MeO-Ph | 25 | ||||
| 14 | 3-HO-Ph | 48 | 25 | >10000 | >400 | 0.08 |
| 15 | 4-F-Ph | 3 | 2 | 5100 | 2600 | 2 |
| 16 | 4-CN-Ph | 4 | 2 | >10000 | >5000 | 0 |
| 17 | 4-MeO-Ph | 16 | 3 | >10000 | >3000 | 0.05 |
| 18 | 4-HO-Ph | 1400 |
Table 2.
In Vitro Data for the CB1 Antagonists with Alkyl Spacers
| R | Ke hCB1 (nM) | Ki hCB1 (nM)a | Ki hCB2 (nM)a | Selectivity Ki CB2/CB1 | MDCK-mdr1 A to B (%)b | |
|---|---|---|---|---|---|---|
| 19 | Bn | 17 | 4 | 430 | 110 | |
| 20 | 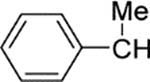 |
4 | 1.1 | 1300 | 1200 | 0 |
| 21 | 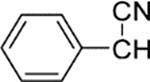 |
37 | 7 | 460 | 66 | |
| 22 | 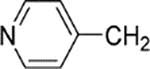 |
6 | 22 | 1700 | 77 | 4 |
| 23 |  |
0.5 | 12 | 1100 | 92 | 4 |
| 24 | 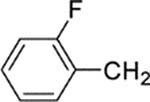 |
0.9 | 7 | 2300 | 330 | 2 |
| 25 | 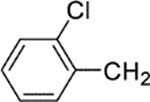 |
14 | 8 | >10000 | >1200 | 0 |
| 26 | 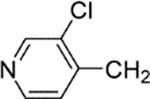 |
3 | 3 | >10000 | >3300 | 2 |
| 27 | 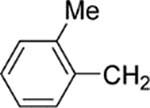 |
6 | 4 | 4600 | 1200 | |
| 28 |  |
5 | 5 | >10000 | >2000 | |
| 29 | 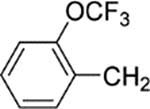 |
4 | 2 | >10000 | >5000 | |
| 30 | 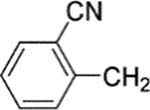 |
21 | 3 | >10000 | >3300 | 0.2 |
| 31 | 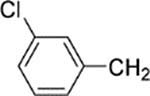 |
7 | 0.9 | >10000 | >10000 | 0 |
| 32 | 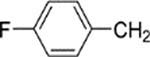 |
1 | 4 | >10000 | >2500 | 0 |
| 33 | 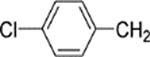 |
3 | 1 | >10000 | >10000 | 0 |
| 34 | 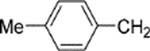 |
5 | 8 | >10000 | >1200 | 0 |
| 35 | 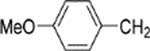 |
15 | 12 | >10000 | >800 | 0 |
| 36 |  |
42 | 8 | 990 | 120 | 7 |
| 37 | 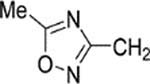 |
12 | 24 | 3600 | 150 | |
| 38 | PhCH2CH2 | 11 | 2 | 2100 | 1000 | 0 |
| 39 |  |
270 | 12 | 630 | 52 |
Displacement was measured using [3H]CP55940 in CHO-cell-membrane preparations overexpressing the hCB1 or hCB2 receptor.
Percent transported from the apical side (A) to the basal side (B).
In Table 1, the in vitro data for a set of aryl and heteroaryl compounds, in which the aryl ring was directly connected to the piperazine nitrogen, are reported. For comparison purposes, the methyl and cyclohexyl analogues are included. Methyl compound 6 demonstrated a Ke of 120 nM in the calcium hCB1 assay, but cyclohexyl compound 7 was about 10-fold more potent and had reasonable selectivity for hCB1 (~60-fold) in the binding assays. Compound 7, however, had ~2% permeability in the MDCK-mdr1 assay, which was deemed unacceptable for further development, as typically <1% permeability is considered acceptable for further development. Phenyl compound 8 was even more potent, with a Ke of 5 nM. It was also much more selective for hCB1 over hCB2 in the binding assay (>5000-fold selective for hCB1) than the cyclohexyl compound, 7. Phenyl analogue 8 performed well in the MDCK-mdr1 assay with no discernible transport, which predicted limited in vivo CNS penetration. This led us to investigate a broader set of compounds. Interestingly, the more polar 2-pyridyl compound, 9, retained similar hCB1 potency (Ke = 13 nM) and selectivity to those seen with phenyl analogue 8 and also showed limited permeability in the MDCK-mdr1 assay (% A to B = 0.06). The 4-pyridyl compound, 10, however, was much less potent at hCB1 (Ke = 1200 nM). Similarly, the 3-hydroxy compound, 14, had reasonably good potency (Ke = 48 nM), whereas the 4-hydroxy compound, 18, was only weakly active in the hCB1 functional calcium assay (Ke = 1400 nM). It appeared that increasing polarity in the 4-position was not well tolerated. In general, nonpolar substitutions, such as fluoro, cyano, and methoxy groups, were well tolerated in either the ortho, meta, or para positions (15–17), as demonstrated in Table 1. When tested in the MDCK-mdr1 assay, these compounds performed well with several demonstrating <1% A-to-B transport, predictive of limited in vivo brain penetration. The 2-fluoro- and 2-chlorophenylpiperazines were also prepared, but these had issues with solubility that resulted in their precipitation from the assay buffer, suggesting that solubility could be a limiting factor for certain aryl piperazines. We expected that adding an alkyl spacer would improve solubility by increasing the basicity of the piperazine and decreasing the rigidity of the piperazine substituent. This idea was explored in a series of analogues reported in Table 2.
Installing an alkyl spacer between the piperazine and the aryl group provided compounds that were more basic and were predicted to be generally more soluble (Table 2). The unsubstituted benzyl compound, 19, was both potent (Ke = 17 nM) and selective for hCB1 (~110-fold). α-Substitution to the nitrogen with a methyl, resulted in improved potency and selectivity (20, Ke = 4 nM, ~1200-fold selectivity). Adding an α-cyano group to increase the polarity resulted in compound 21, which still had reasonable potency (Ke = 37 nM), but its selectivity for hCB1 was reduced (~60-fold) compared with those of 19 and 20. Ortho, meta, and para substituents were well tolerated among the benzyl series (24–35). Increasing the spacer by one carbon provided phenethyl compound 38, which retained good potency (Ke = 11 nM) and was even more selective (2100-fold) than benzyl compound 19. A note of interest was that when the linker was one carbon, a 4-pyridyl group was well tolerated at hCB1 (22 and 23), but it was not well tolerated when a two-carbon linker was used (39). Five-membered heterocycles with one-carbon linkers (36 and 37) were also found to be potent (Ke = 42 and 12 nM, respectively). Several of these compounds (25, 30–35, and 38) showed no discernible penetration from the apical to the basal side and were deemed suitable for further evaluation. The heterocyclic analogues, however, displayed enhanced permeability in this assay (22, 23, and 36) and therefore were removed from further consideration.
Computational Modeling and Assessment of hCBI Antagonists.
Subsequent to the completion of this project, several crystal structures of hCB1 bound to small-molecule antagonists17,18 and agonists19 were solved. This afforded us the opportunity to evaluate our SAR using the recently published crystal structures. Our initial strategy was to reproduce the crystal-structure poses for AM6538 using Autodock VINA and AMBER16 molecular-mechanics-generalized-Born-surface-area (MMGBSA)-rescoring approaches, as described in the Experimental Section and the Supporting Information (Section S1).
Having validated our approach with the known crystal structures, we evaluated the binding modes for the compounds prepared during this investigation. Figure 2 shows the lowest-MMGBSA docked poses for the compounds in Table 1 in the hCB1-receptor environment with the protein visible (Figure 2A) and hidden (Figure 2B). The figure illustrates that no residues occlude one face of the 4-cyanophenyl in the 5TGZ crystal-structure coordinates (AMBER16 model). This highlights that alteration of the piperazine substituent preserves the positioning of the core scaffold, and it illustrates that variations in MMGBSA free-energy scores are due primarily to interactions at the tops of the TM helices. These substitutions are in an environment that is partially solvent exposed, as shown in Figure 3A. In this context, the alternately substituted piperazines can make favorable contacts with polar residues (e.g., His178, Ser123, Gln116, etc.) in addition to some extracellular solvent. As such, the ligand environment is quite hydrophobic in the diaryl regions of the ligands, but it is of mixed character in the region of SAR variation. Figure 3B shows an early molecular-dynamics snapshot of 16 in its docked (lowest-MMGBSA) pose with the hCB1-complex simulation in a membrane (1,2-dioleoyl-sn-glycero-3-phosphocho-line-DOPC)–KCl–H2O environment. During course of the initial 50–100 ns of the simulation, water diffuses into the extracellular N-terminal loop region, and by the extension of the SAR region into the extracellular milieu, the water interacts with the SAR regions of the ligands in Tables 1 and 2. Even at short time scales, water can percolate in from the DOPC/K+/Cl−/water hCB1 environment and contact the extracellular faces of the substituted piperazines.
Figure 2.
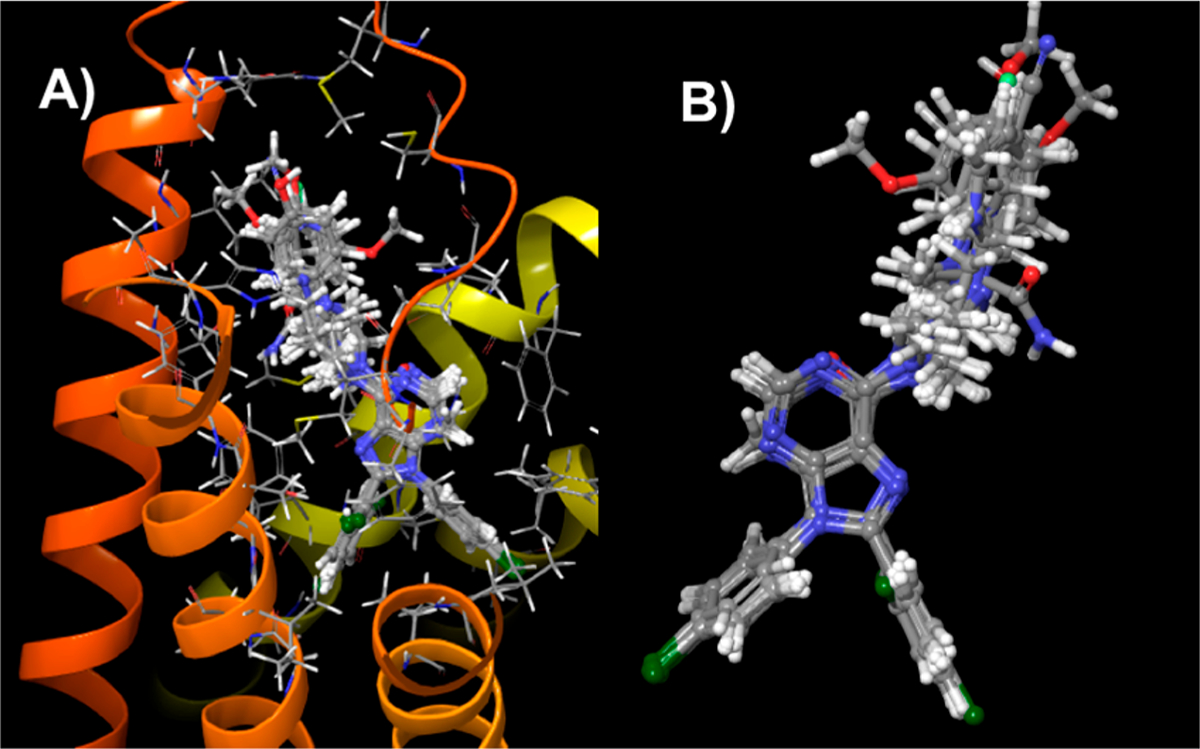
Docked poses for the compounds in Table 1 (2–18) in hCB1 (PDB ID: 5TGZ). (A) Compounds from Table 1 in their lowest-MMGBSA docked poses showing a common binding mode regarding the diphenyl purines. The alternately substituted piperazine substituents interact with the residues at the top of the helical bundle and can extend into the solvent-accessible loop domain. (B) Docked poses of the Table 1 compounds with the protein environment hidden, emphasizing the alternately substituted piperazine region.
Figure 3.
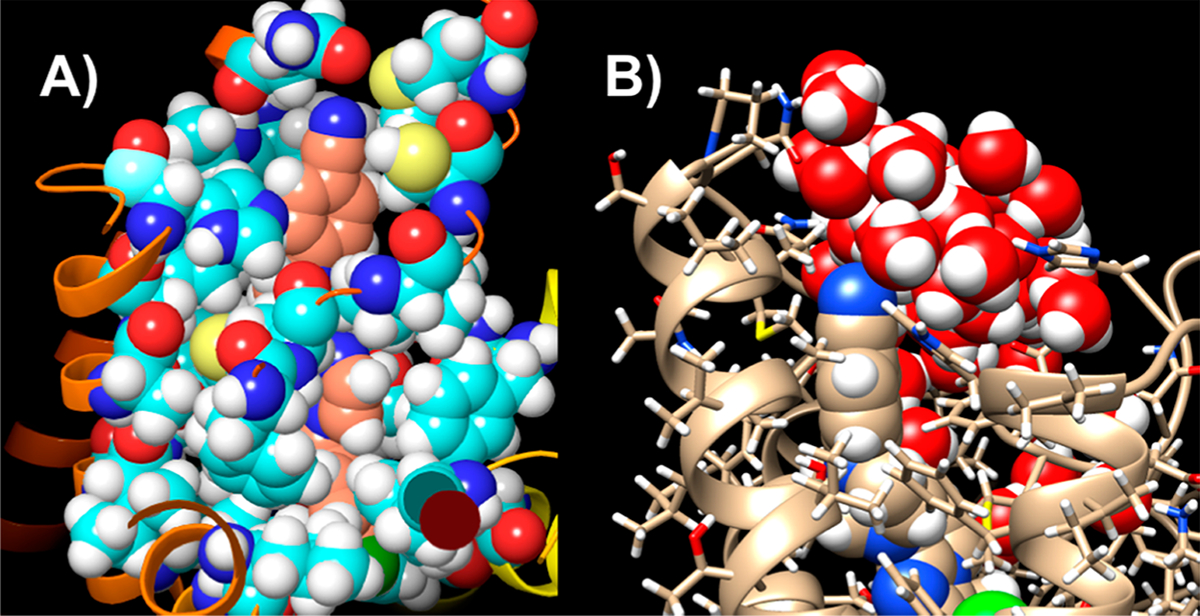
Overall positioning of the analogue modification from Table 1. (A) Compound 16 in its lowest-MMGBSA-scored pose with the hCB1 residues within 4 Å of the ligand displayed (PDB ID: 5TGZ). (B) Water contacting compound 16. Thus, 16 is partially solvent-accessible.
Correlating the computed MMGBSA free energetics of the Table 1 ligands in complex with the hCB1 receptor with the functional-antagonism values (ln Ke) demonstrated a moderate Pearson correlation (r = 0.61, Figure 4A). The correlation is modest, as indicated by the Pearson r value, but captures a reasonable antagonist-potency ranking. This moderate Pearson correlation suggested that other factors may be contributing to the variation in potency besides those included in the MMGBSA-free-energy-rescoring terms, such as configurational entropy (TΔS):
Figure 4.

Factors affecting antagonist potency. (A) MMGBSA scores for the compounds in Table 1 vs ln Ke. (B) DFT-calculated solvation energies vs ln Ke.
Figure 4B demonstrates that the ligands in Table 1 contact the extracellular aqueous environment and suggests that the exposure of the ligands to the more heavily aqueous environment at the extracellular face may influence antagonist potency. Figure 4B correlates ln Ke versus the quantum-chemically derived solvation values for the Table 1 ligands. Again, a reasonably good Pearson correlation (r = 0.7) demonstrates the importance of the desolvation energetics of the ligands upon the overall free energy estimated by the MMPBSA values.
The pattern exhibited throughout the SAR with the polar substituents added to the aryl piperazines, both with and without −CH2 spacers, is a competition between the cost of (partially) desolvating the ligand and finding complementary hCB1-orthosteric-site interactions given the position of the substitution. In cases where one introduces a polar substituent, giving rise to a large desolvation cost, but that substituent does not find a large compensatory interaction (e.g., hydrogen bonding) in the binding site, one will have diminished binding and a larger Ke (ln Ke). In cases where a polar substituent is both present and appears in an accessible configuration to a polar binding-site-side-chain partner, low Ke values are obtained. In the case of a modestly sized 4-F substitution in compound 15, with a computed B3LYP solvation energy of only −13.1 kcal/mol, one does not have to find a polar partner to compensate for the modest desolvation cost, and notably, the Ke value for 15, with 4-F-phenyl, is not very different from that of 8, which has phenyl (Ke = 3 and 5 nM, respectively). Compound 16 introduces a polar cyano group at the 4-position of the aryl ring, which has a sizable −17.3 kcal/mol solvation energy (optimized DFT estimate), but as Figure 3A shows, this substituent extends well into the region, entering the extracellular domain above the TM region with some exposure to Gln115 and Gln116, and makes ample contact with the diffusing solvent above the DOPC membrane; consequently, there is not a high desolvation cost, and it can find partial exposure to polar Gln groups at the top of helix 1. This compound is predicted to have modest affinity, as noted in our MMGBSA.
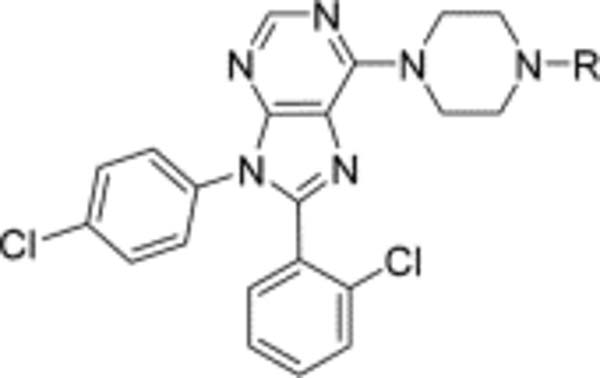
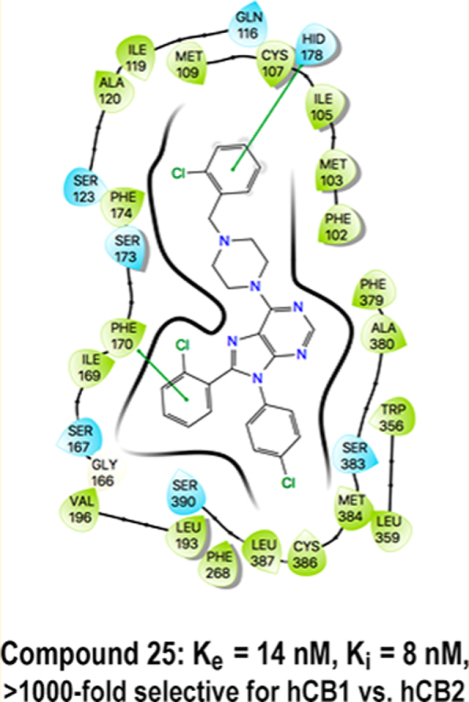
Compound 25 achieves its inhibitory potential (Ke = 14 nM) by the partial burial of its nonpolar 2-chlorophenyl into a favorable π-stacking hydrophobic interaction environment (HIS178/Phe174) at the top of helix 2. The desolvation cost of its terminal substituent is modest (−14.9 kcal/mol predicted solvation energy), and it has some solvent exposure. Although these facets lead to decreased inhibitory potency compared with that of its parent compound, 2 (Ke = 0.2 nM), with a more polar terminal substituent, it achieves greater selectivity against hCB2 with ample discrimination against the penetration of the BBB.
In summary, the SAR in Tables 1 and 2 are readily comprehensible utilizing the recent antagonist crystal structures and standard computational approaches. The alternately substituted piperazine groups evaluated in this manuscript do not occupy a buried hydrophobic pocket but extend into a region at the top of the transmembrane helices that interact with the aqueous environment. The trend in the ln Ke versus the binding free energies is a compensatory balance between polar hCB1-receptor interactions and solvent-exposed energetics.
Compound 25, an Inverse Agonist of hCB1.
Compound 25 was further characterized using the calcium-mobilization assay to establish whether this compound was a neutral antagonist or an inverse agonist of the hCB1 receptor. As indicated in Figure 5, 25 was an inverse agonist of hCB1 with an EC50 of ~462 nM. Compound 1 was used in this assay as a positive control. The maximal response of 25 was ~75% of 1.
Figure 5.
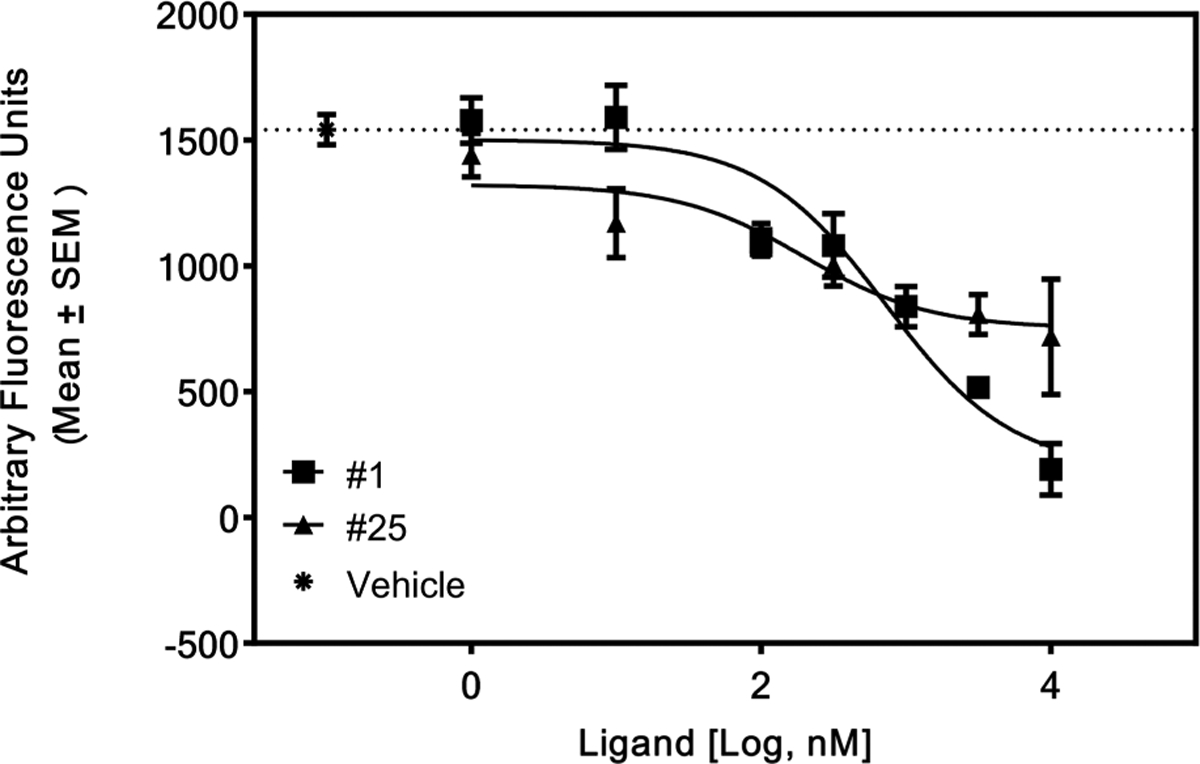
Compound 25 as an inverse agonist of hCB1. CHO-hCB1 cells were loaded with a calcium indicator dye for 60 min as described in the Experimental Section. The cells were then stimulated by various concentrations of each compound, and the fluorescence changes were recorded using a FLIPR Tetra (Molecular Devices) plate reader for 60 s. Cells stimulated with only the vehicle in the buffer served as a negative control. Compound 1 served as a positive control for this experiment.
hERG Activity and Stability in Human Microsomes and Aqueous-Solubility Determination.
The methyl piperazine, 6, was previously reported to have undesirable activity in a hERG binding assay and hence, we wanted to evaluate the most favorable compounds obtained from this study.15 We evaluated 25 in a [3H]astemizole-competition binding assay using HEK-hERG cell membranes.20 Compound 25 was found to have a Ki of >10 μM (Table 3). Thus, this compound is unlikely to cause LQT syndrome via a blockade of hERG. The other benzyl piperazines, 23 and 32, were also tested, and like 25, these did not demonstrate significant hERG interactions either (Ki >10 μM, data not shown).
Table 3.
hERG Activity, Microsomal Stability, and Solubility of 25
| hERG activity ([3H]astemizole displacement) | >10 μM |
| human-hepatic-microsomal stability | half-life: 90 min |
| clearance: 14 mL/min/kg | |
| aqueous solubility | pH 1.6: >200 μM |
| pH 7.4: <0.4 μM |
The stability of a compound in human liver microsomes is often predictive of human PK. As such, 25 was evaluated in human liver microsomes for stability and was found to have a half-life of 90 min and a clearance of 14 mL/min/kg (Table 3). Thus, 25 appeared to be suitable for further evaluation in vivo. Similarly, the aqueous solubility of a compound can be predictive of its absorption and distribution, and more soluble compounds are often better suited for clinical development. A kinetic solubility assay was utilized to assess the solubility of 25 (Table 3). The calculated aqueous solubility of 25 at an acidic pH of 1.6 was excellent (>200 μM), but it was poor at neutral pH (<0.4 μM at pH 7.4). This was expected for a basic, lipophilic compound. Thus, 25 was deemed to possess good ADME properties and was advanced to in vivo studies along with certain other candidates.
Pharmacokinetic Studies.
Select compounds were tested in rodents to assess their ability to be orally absorbed and kept out of the brain. Initial studies entailed the cassette dosing of rodents by oral gavage and consequently measuring the brain and plasma levels of the parent compound at 2 h postdosing (data not shown). This triage strategy enabled us to quickly narrow down the list of candidate compounds and focus on those that had brain/plasma ratios of <0.10 and reasonable oral absorption. The chloro-substituted piperazine, 25, performed well in initial studies and was evaluated in extended pharmacokinetic (PK) studies. For these studies, C57BL6 mice (~10 weeks old) were orally administered a single dose of the compound. Plasma, whole livers, and whole brains were taken from unperfused mice at 0.5, 1, 2, 4, 8, and 24 h postdosing.
The amount of parent compound was quantitated in each tissue using LC-MS. As demonstrated in Table 4 and Figure 6, piperazine 25 performed well in these PK studies. It was rapidly absorbed upon oral dosing, producing high plasma and liver concentrations. The plasma half-life of this compound was 5.4 h at the dosing concentration (Table 4), indicating that the drug was suitable for in vivo evaluations for efficacy studies. In addition, the hepatic concentrations at various time-points were also favorable, with a peak concentration of 2081 ng/mg at 0.5 h postdosing and a half-life of 5.3 h (Table 4 and Figure 6). The maximum unperfused brain/plasma ratio for this compound was 0.03, suggestive of very little brain penetration (Table 4). The other compounds tested either had higher levels of brain concentrations compared with that of 25 or were poorly absorbed upon oral dosing (data not shown). Furthermore, a 7 day repeated-dosing PK study of 25 (Supporting Materials, Table S2) indicated that this compound did not accumulate in the brain upon repeated dosing. In sharp contrast, PK analyses of 1 indicated that this reference compound had significant brain exposure upon oral dosing in mice (Supporting Materials, Figure S4) with higher brain concentrations as compared with plasma concentrations at certain time points.
Table 4.
Pharmacokinetic Properties of 25
| Tmax (h) | Cmax | T1/2 (h) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| po dose (mg/kg) | plasma | brain | liver | plasma (ng/mL) | brain (ng/g) | liver (ng/g) | plasma | brain | liver | brain/plasma ratio | brain/liver ratio |
| 10 | 0.5 | 0.5 | 0.5 | 507 | 17 | 2081 | 5.4 | 11.9 | 5.3 | 0.03 | 0.008 |
Figure 6.
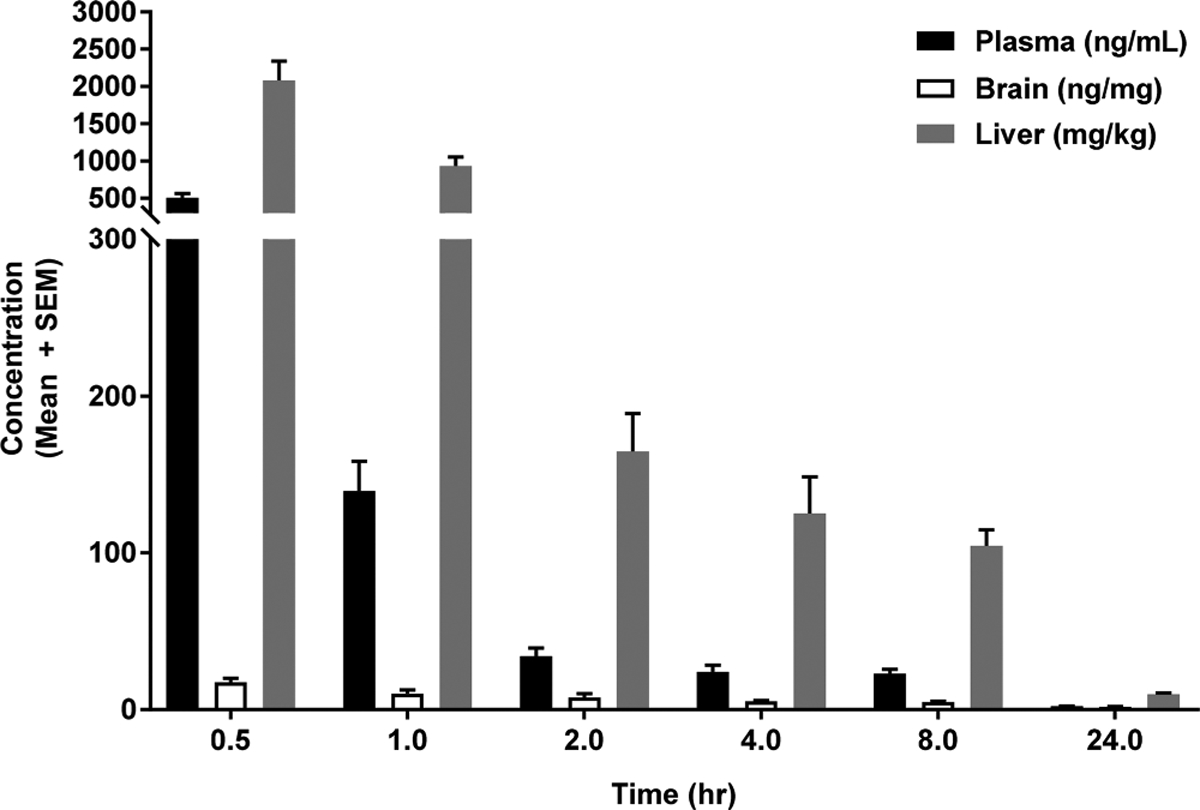
Pharmacokinetic evaluation of compound 25. C57BL6 mice (10 weeks) were orally dosed with 25 suspended in a suitable vehicle, as described in the Experimental Section. Animals were sacrificed at various time points, and tissues were collected. LC-MS was used to quantitate the tissue concentrations of 25 at each time point. Four mice were sacrificed at each time point for this study.
Mouse Hypothermia Study.
In addition to showing low brain levels with PK experiments, it is important to establish that the compounds of interest do not affect a centrally mediated property of CB1 agonists, thus minimizing the risk of adverse effects. It is known that the CB1 agonist CP55940 induces a CNS-mediated reduction of body temperature in rodents.21 The CNS-penetrating CB1 antagonist, 2, is known to block this effect.22 A study of compound 25 in the rodent hypothermia assay showed little to no blocking of temperature drops when dosed orally at 10 mg/kg in mice (Figure 7). These data confirmed that the low brain penetration of 25 was not sufficient to block hypothermia mediated by centrally located CB1 receptors (Figure 7). The parent compound, 2 (30 mg/kg), however, blocked hypothermia, as expected, when used as a positive-control compound.
Figure 7.

Mouse hypothermia study of 25 and 2. Female C57BL6 mice (10 weeks) were dosed with a test agent (2, 25, or the vehicle) by oral gavage as indicated in the Experimental Section. Thirty minutes after being dosed with the inhibitor, the animals were administered CP55940 by ip injections. Rectal-temperature data were collected at regular intervals. Compound 2 blocked hypothermia induced by CP55940 (two-way ANOVA, p < 0.05) compared with the animals treated with the vehicle only, but this effect was absent in the animals dosed with 25.
Effects of 25 on Alcohol-Induced Liver Steatosis.
Past studies had indicated that the paracrine activation of CB1 on liver hepatocytes by 2-arachidonoylglycerol (2-AG), secreted by hepatic stellate cells (HSC) during alcoholic liver injury, promoted hepatic lipid accumulation and alcoholic steatosis (AS).23 The prototypical CB1 inverse agonist, 1, was effective at reducing AS by blocking the transcription of lipogenic genes activated by SREB-1C.23 Therefore, in vivo efficacy studies were undertaken in a mouse model of AS to assess whether 25 would be efficacious in blocking disease progression and development. Female C57BL6 mice were maintained on a liquid Lieber-DeCarli diet containing ethanol or a matched control diet without alcohol for 4 weeks. Compound 25 or the vehicle was administered to these rodents as described in the Experimental Section for the last 2 weeks. Figure 8 shows representative photomicrographs depicting lipid accumulation in liver slices of mice from various treatment groups. Lipid accumulation in the livers of the animals on the control diet (no ethanol) was minimal, as revealed through Oil Red O (ORO) staining (left panel in Figure 8). In contrast, the ethanol-containing diet caused significant accumulation of lipid droplets, as expected (center panel in Figure 8). Liver-histology and ORO-staining analyses showed obvious microsteatosis and macrosteatosis in the livers of all the ethanol-diet-fed mice compared with those of the control-diet-fed mice in the absence of drug treatment. Using the ethanol-diet-fed mice, the administration of compound 25 at a dose of 1.25 mg/kg twice daily by oral gavage significantly reduced hepatic lipid accumulation (right panel in Figure 8) compared with that in the vehicle control group (center panel in Figure 8). Additionally, an evaluation of the brains of the mice exposed to 25 at the end of this study indicated that there was no significant accumulation of the compound upon twice-daily repeated oral dosing for 14 days (Supporting Information Section S3). These findings support the efficacy of test compound 25 in limiting the progression of disease in a murine model of AS.
Figure 8.

Oil Red O staining of liver sections indicating a reduction of steatosis upon treatment with compound 25. (Top) Representative liver sections from mice receiving the control diet without ethanol and the vehicle (left panel), the ethanol-containing diet and the vehicle (center panel), or the ethanol-containing diet and 25 (right panel). (Bottom) Quantification (ImageJ software) of the lipid droplets in the liver sections indicating a statistically significant reduction of liver steatosis upon treatment with 25 (ANOVA, *p < 0.001 vs control and vehicle, #p < 0.01 vs ethanol and vehicle).
SUMMARY AND CONCLUSIONS
For peripherally restricted hCB1 antagonists, it is important to investigate their therapeutic value while minimizing their risk of adverse brain-mediated psychiatric disorders. Such compounds could become important tools in treating diabetes, metabolic syndrome, dyslipidemias, and liver diseases. Although several groups are actively working in this area, an advanced clinical candidate has yet to emerge.7 In this study, we investigated a series of compounds based on 2 in which the 4-aminopiper-idine group was replaced with a smaller piperazine group. The piperazine was used as a mildly basic linker to access a lipophilic binding site via functionalization with aryl or heteroaryl groups with and without linkers. Potent hCB1 antagonists and inverse agonists with excellent selectivity versus hCB2 were discovered. Positive MDCK-mdr1 results led us to test several compounds for peripheral restriction in rodents after oral dosing. Varying levels of peripheral restriction were observed. The 2-chlorobenzyl piperazine, 25, was found to have good peripheral restriction and to attain good plasma and hepatic levels. It is interesting to note that compound 25 bears neither a H-bond-donating group nor an additional polar group. The source of the peripheral restriction is probably the presence of a basic amine in combination with steric factors. Compound 25 performed as hoped in a pharmacological model of CB1-mediated CNS activity, failing to block hypothermia induced by a prototypical CB1 agonist in mice. This compound also had several other favorable properties, including good solubility at acidic pH, a lack of hERG interactions, and low clearance in human liver microsomes. Perhaps most interestingly, this compound was effective at reducing AS in a mouse model of liver injury induced by feeding the animals an ethanol-containing liquid diet. This compound and its analogues may be suitable for further clinical development. Over the years, several groups, including ours, have reported on the development of peripherally restricted CB1-receptor antagonists.7 The initial probelike molecules are now being refined in each successive generation to produce more mature lead structures that have favorable ADME and “drug-likeness”. The series of investigations reported in this paper provide support for the further development of this class of compounds.
EXPERIMENTAL SECTION
General Chemistry.
Compounds were characterized and their purities were established by a combination of MS, NMR, HPLC, and TLC analytical techniques described below. 1H spectra were recorded on a Bruker Avance DPX-300 (300 MHz) spectrometer and were determined in chloroform-d (7.26 ppm) or methanol-d4 (3.31 ppm) with tetramethylsilane (0.00 ppm) or the solvent peaks as the internal reference unless otherwise noted. Chemical shifts are reported in parts per million relative to the solvent signal, and coupling-constant (J) values are reported in hertz (Hz). Thin-layer chromatography (TLC) was performed on EMD precoated silica gel 60 F254 plates. TLC spots were visualized with UV light or I2 detection. Low-resolution mass spectra were obtained using a single quadrupole PE Sciex API 150EX (ESI). Unless stated otherwise, all test compounds were at least 95% pure as determined by HPLC. The HPLC method involved an Agilent-Varian system equipped with Prostar 210 pumps, a Prostar 335 Diode UV detector, and a Phenomenex Synergi 4 μm Hydro RP 80A C18 250 × 4.6 mm column with a 20 min gradient elution of 5–95% solvent B at 1 mL/min followed by 5 min at 95% solvent B. Solvent A was water with 0.1% TFA; solvent B was acetonitrile with 0.1% TFA and 5% water. Absorbance was monitored at 220 and 280 nm.
Testing for the Pan-Assay Interference (PAIN).
The compounds synthesized were considered low risk for PAIN as they were analogues of a previously well-characterized diphenyl purine scaffold specific for hCB1.15 Additionally, the compounds were manually inspected to identify structural similarities related to known PAIN compounds,24–26 and all the compounds were tested using the calcium-mobilization assay in parental cells without hCB1 to ensure specificity of action.
General Procedure for Reacting Piperazines with 5.
To a solution of chloropurine 5 (93.9 mg, 0.25 mmol) in EtOH (5 mL) was added TEA (3 equiv, 0.10 mL) and the appropriate piperazine (1.25 equiv, 0.31 mmol). The reaction vessel was capped, and the mixture stirred at 78 °C overnight. The resulting mixture was concentrated and chromatographed on a 4 g silica-gel column using a gradient of 0–100% EtOAc/hexanes. TLC and MS were used to identify the fractions with the desired products. The fractions were concentrated and dried under high vacuum overnight, thus providing the products as white and off-white solids.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-(4-methylpiperazin-1-yl)-9H-purine (6).
The title compound was prepared by the general method to provide 32 mg (29%) of a solid, mp 217–219 °C. 1H NMR (300 MHz, CDCl3) δ 8.39 (s, 1H), 7.49–7.56 (m, 1H), 7.29–7.44 (m, 5H), 7.15–7.23 (m, 2H), 4.40 (br s, 4H), 2.58 (t, J = 5.0 Hz, 4H), 2.36 (s, 3H). MS (m/z) 439.3 (M + 1). HPLC = 99% at 16.91 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-(4-cyclohexylpiperazin-1-yl)-9H-purine (7).
The title compound was prepared by the general method to provide 25 mg (61%) of a solid, mp 174–176 °C. 1H NMR (300 MHz, CDCl3) δ 8.38 (s, 1H), 7.49–7.56 (m, 1H), 7.29–7.43 (m, 5H), 7.15–7.24 (m, 2H), 4.37 (br s, 4H), 2.63–2.83 (m, 4H), 2.32 (br s, 1H), 1.71–1.97 (m, 4H), 1.64 (d, J = 11.7 Hz, 1H), 1.04–1.33 (m, 5H). MS (m/z) 507.1 (M + 1). HPLC ≥ 99% at 19.12 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-(4-phenylpiperazin-1-yl)-9H-purine (8).
The title compound was prepared by the general method to provide 54 mg (59%) of a solid, mp 199–201 °C. 1H NMR (300 MHz, CDCl3) δ 8.42 (s, 1H), 7.48–7.57 (m, 1H), 7.26–7.45 (m, 7H), 7.15–7.24 (m, 2H), 6.99 (d, J = 7.9 Hz, 2H), 6.90 (t, J = 7.3 Hz, 1H), 4.54 (br s, 4H), 3.25–3.47 (m, 4H). MS (m/z) 501.6 (M + 1). HPLC ≥ 99% at 22.47 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-(4-pyridin-2-ylpiperazin-1-yl)-9H-purine (9).
The title compound was prepared by the general method to provide 31 mg (45%) of a solid, mp 203–204 °C. 1H NMR (300 MHz, CDCl3) δ 8.42 (s, 1H), 8.18–8.29 (m, 1H), 7.47–7.58 (m, 2H), 7.30–7.42 (m, 5h), 7.15–7.24 (m, 2H), 6.61–6.76 (m, 2H), 4.51 (br s, 4H), 3.66–3.82 (m, 4H). MS (m/z) 502.6 (M + 1). HPLC = 99% at 18.05 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-(4-pyridin-4-ylpiperazin-1-yl)-9H-purine (10).
The title compound was prepared by the general method to provide 59 mg (47%) of a solid, mp 155–157 °C. 1H NMR (300 MHz, CDCl3) δ 8.42 (s, 1H), 8.31 (d, J = 6.4 Hz, 2H) 7.47–7.57 (m, 1H), 7.30–7.45 (m, 5H), 7.21 (d, J = 8.7 Hz, 2H), 6.71 (d, J = 6.5 Hz, 2H), 4.52 (br s, 4H), 3.38–3.70 (m, 4H). MS (m/z) 502.6 (M + 1). HPLC ≥ 99% at 17.87 min.
2-{4-[8-(2-Chlorophenyl)-9-(4-chlorophenyl)-9H-purin-6-yl]-piperazin-1-yl}benzonitrile (11).
The title compound was prepared by the general method to provide 59 mg (54%) of a solid, mp 168–169 °C. 1H NMR (300 MHz, CDCl3) δ 8.42 (s, 1H), 7.60 (dd, J = 1.5, 7.9 Hz, 1H), 7.46–7.57 (m, 2H), 7.29–7.44 (m, 5H), 7.16–7.25 (m, 2H), 6.98–7.11 (m, 2H), 4.60 (br s, 4H), 3.26–3.48 (m, 4H). MS (m/z) 526.5 (M + 1). HPLC ≥ 99% at 23.85 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(2-methoxyphenyl)-piperazin-1-yl]-9H-purine (12).
The title compound was prepared by the general method to provide 23 mg (17%) of a solid, mp 177–179 °C. 1H NMR (300 MHz, CDCl3) δ 8.41 (s, 1H), 7.49–7.59 (m, 1H), 7.30–7.41 (m, 5H), 7.17–7.24 (m, 2H), 7.00–7.09 (m, 1H), 6.85–6.99 (m, 3H), 4.57 (br s, 4H), 3.90 (s, 3h), 3.14–3.31 (m, 4h). MS (m/z) 531.5 (M + 1). HPLC = 99% at 20.81 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(3-methoxyphenyl)-piperazin-1-yl]-9H-purine (13).
The title compound was prepared by the general method to provide 77 mg (85%) of a solid, mp 171–172 °C. 1H NMR (300 MHz, CDCl3) δ 8.41 (s, 1H), 7.49–7.58 (m, 1H), 7.30–7.43 (m, 5H), 7.15–7.24 (m, 3H), 6.60 (dd, J = 2.0, 8.0 Hz, 1H), 6.52 (t, J = 2.3 Hz, 1H), 6.46 (dd, J = 2.0, 8.0 Hz, 1H), 4,53 (br s, 4H), 3.80 (s, 3H), 3.29–3.42 (m, 4H), 1.58 (s, 2H). MS (m/z) 531.4 (M + 1). HPLC = 95% at 22.73 min.
3-{4-[8-(2-Chlorophenyl)-9-(4-chlorophenyl)-9H-purin-6-yl]-piperazin-1-yl}phenol (14).
The title compound was prepared by the general method to provide 22 mg (33%) of a solid, mp 281–283 °C. 1H NMR (300 MHz, CDCl3) δ 8.42 (s, 1H), 7.50–7.57 (m, 1H), 7.30–7.45 (m, 5H), 7.17–7.24 (m, 2H), 7.13 (t, J = 8.1 Hz, 1H), 6.56 (dd, J = 1.9, 8.2 Hz, 1H), 6.43 (t, J = 2.2 Hz, 1H), 6.35 (dd, J = 2.0, 7.9 Hz, 1H), 5.11 (br s, 1H), 4.52 (br s, 4H), 3.24–3.43 (m, 4H). MS (m/z) 517.4 (M + 1). HPLC ≥ 99% at 19.57 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(4-fluorophenyl)-piperazin-1-yl]-9H-purine (15).
The title compound was prepared by the general method to provide 22 mg (17%) of a solid, mp 180–182 °C. 1H NMR (300 MHz, CDCl3) δ 8.42 (s, 1H), 7.49–7.58 (m, 1H), 7.30–7.45 (m, 5H), 7.16–7.24 (m, 2H), 6.88–7.07 (m, 4H), 4.53 (br s, 4H), 3.17–3.35 (m, 4H). MS (m/z) 519.6 (M + 1). HPLC ≥ 99% at 23.31 min.
4-{4-[8-(2-Chlorophenyl)-9-(4-chlorophenyl)-9H-purin-6-yl]-piperazin-1-yl}benzonitrile (16).
The title compound was prepared by the general method to provide 24 mg (30%) of a solid, mp 253–254 °C. 1H NMR (300 MHz, CDCl3) δ 8.43 (s, 1H), 7.47–7.59 (m, 3H), 7.28–7.46 (m, 5H), 7.14–7.24 (m, 2H), 6.91 (d, J = 9.0 Hz, 2H), 4.53 (br s, 4H), 3.41–3.62 (m, 4H). MS (m/z) 526.3 (M + 1). HPLC = 97% at 22.99 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(4-methoxyphenyl)-piperazin-1-yl]-9H-purine (17).
The title compound was prepared by the general method to provide 23 mg (42%) of a solid, mp 185–186 °C. 1H NMR (300 MHz, CDCl3) δ 8.41 (s, 1H), 7.48–7.60 (m, 1H), 7.30–7.44 (m, 5H), 7.16–7.24 (m, 2H), 6.91–7.02 (m, 2H), 6.80–6.91 (m, 2H), 4.54 (br s, 4H), 3.78 (s, 3h), 3.13–3.34 (m, 4h). MS (m/z) 531.4 (M + 1). HPLC ≥ 99% at 20.17 min.
4-{4-[8-(2-Chlorophenyl)-9-(4-chlorophenyl)-9H-purin-6-yl]-piperazin-1-yl}phenol (18).
The title compound was prepared by the general procedure to provide 8.3 mg (6%) of a solid, mp 298–299 °C. 1H NMR (300 MHz, CDCl3) δ 8.41 (s, 1H), 7.49–7.56 (m, 1H), 7.29–7.42 (m, 5H), 7.16–7.24 (m, 2H), 6.86–6.96 (m, 2H), 6.73–6.83 (m, 2H), 4.53 (br s, 4H), 3.14–3.30 (m, 4H). MS (m/z) 517.6 (M + 1). HPLC = 98% at 17.99 min.
6-[4-Benzylpiperazin-1-yl]-8-(2-chlorophenyl)-9-(4-chlorophenyl)-9H-purine (19).
The title compound was prepared by the general method to provide 60 mg (46%) of a solid, mp 94–95 °C. 1H NMR (300 MHz, CDCl3) δ 8.37 (s, 1H), 7.50 (d, J = 6.6 Hz, 1H), 7.27–7.42 (m, 11H), 7.19 (d, J = 8.7 Hz, 2H), 4.38 (br s, 4H), 3.57 (s, 2H) 2.61 (t, J = 4.9 Hz, 4H). MS (m/z) 515.3 (M + 1). HPLC = 94% at 18.80 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(1-phenylethyl)-piperazin-1-yl]-9H-purine (20).
The title compound was prepared by the general method to provide 55 mg (49%) of a solid, mp 171–172 °C. 1H NMR (300 MHz, CDCl3) δ 8.36 (s, 1H), 7.44–7.55 (m, 1H), 7.27–7.40 (m, 9H), 7.13–7.22 (m, 2H), 4.35 (br s, 3H), 3.44 (q, J = 6.7 Hz, 1H), 2.59 (ddt, J = 5.0, 11.2, 16.5 Hz, 4H), 1.41 (d, J = 6.7 Hz, 3H). MS (m/z) 529.7 (M + 1). HPLC ≥ 99% at 19.44 min.
{4-[8-(2-Chlorophenyl)-9-(4-chlorophenyl)-9H-purin-6-yl]-piperazin-1-yl}(phenyl)acetonitrile (21).
The title compound was prepared by the general method to provide 66.7 mg (49%) of a solid, mp 108–110 °C. 1H NMR (300 MHz, CDCl3) δ 8.39 (s, 1H), 7.47–7.62 (m, 3H), 7.28–7.47 (m, 9H), 7.16–7.25 (m, 2H), 4.94 (s, 1H), 4.43 (br s, 4H), 2.74 (t, J = 5.0 Hz, 4H). 13C NMR (75 MHz, CDCl3) δ 153.8, 153.1, 152.4, 145.9, 134.3, 134.2, 133.0, 132.6, 132.5, 131.4, 130.0, 129.6, 129.4, 129.1, 129.1, 128.9, 128.0, 128.0, 127.8, 126.9, 119.8, 115.2, 62.3, 49.8, 45.0. MS (m/z) 540.1 (M + 1). HPLC = 98% at 22.80 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(pyridin-4-ylmethyl)-piperazin-1-yl]-9H-purine (22).
The title compound was prepared by the general method to provide 106 mg (82%) of a solid, mp 92–93 °C. 1H NMR (300 MHz, CDCl3) δ 8.56 (d, J = 5.8 Hz, 2H), 8.38 (s, 1H), 7.47–7.56 (m, 1H), 7.26–7.42 (m, 7H), 7.14–7.23 (m, 2H), 4.40 (br s, 4H), 3.57 (s, 2h), 2.61 (t, J = 4.9 Hz, 4H). MS (m/z) 516.4 (M + 1). HPLC ≥ 99% at 15.76 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(1-pyridin-4-ylethyl)-piperazin-1-yl]-9H-purine (23).
The title compound was prepared by the general method to provide 72 mg (54%) of a solid, mp 105–107 °C. 1H NMR (300 MHz, CDCl3) δ 8.55 (d, J = 6.0 Hz, 2H), 8.37 (s, 1H), 7.44–7.53 (m, 1H), 7.25–7.42 (m, 7H), 7.13–7.23 (m, 2H), 4.37 (br s, 4H), 3.45 (q, J = 6.7 Hz, 1H), 2.45–2.73 (m, 4H), 1.38 (d, J = 6.7 Hz, 3H). MS (m/z) 530.6 (M + 1). HPLC = 98% at 15.87 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(2-fluorobenzyl)-piperazin-1-yl]-9H-purine (24).
The title compound was prepared by the general method to provide 94 mg (70%) of a solid, mp 78–80 °C. 1H NMR (300 MHz, CDCl3) δ 8.37 (s, 1H), 7.47–7.57 (m, 1H), 7.23–7.44 (m, 7H), 7.15–7.22 (m, 2H), 6.95–7.15 (m, 2H), 4.39 (br s, 4H), 3.66 (s, 2H), 2.65 (t, J = 4.9 Hz, 4H). MS (m/z) 533.2 (m + 1). HPLC ≥ 99% at 18.80 min.
6-[4-(2-Chlorobenzyl)piperazin-1-yl]-8-(2-chlorophenyl)-9-(4-chlorophenyl)-9H-purine (25).
The title compound was prepared by the general method to provide 113 mg (82%) of a solid, mp 78–80 °C. 1H NMR (300 MHz, CDCl3) δ 8.38 (s, 1H), 7.51 (d, J = 6.8 Hz, 2H), 7.29–7.41 (m, 6H), 7.115–7.28 (m, 4H), 4.40 (br s, 4H), 3.69 (s, 2H), 2.68 (t, J = 4.9 Hz, 4H). MS (m/z) 549.4 (M + 1). HPLC ≥ 99% at 19.35 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-{4-[(3-chloropyridin-4-yl)methyl]piperazin-1-yl}−9H-purine (26).
The title compound was prepared by the general method to provide 46 mg (33%) of a solid, mp 84–86 °C. 1H NMR (300 MHz, CDCl3) δ 8.56 (s, 1H), 8.48 (d, J = 5.0 Hz, 1H), 8.39 (s, 1H), 7.46–7.61 (m, 2H), 7.29–7.43 (m, 5H), 7.13–7.24 (m, 2H), 4.42 (br s, 4H), 3.69 (s, 2h), 2.69 (t, J = 4.9 Hz, 4H). 13C NMR (75 MHz, CDCl3) δ 158.8, 153.9, 153.2, 152.2, 145.6, 139.1, 134.2, 133.1, 132.5, 131.4, 131.2, 130.0, 129.7, 129.4, 128.3, 128.0, 126.9, 119.7, 116.1, 110.4, 60.4, 55.2, 53.2, 19.5. MS (m/z) 552.3 (M + 1). HPLC = 98% at 18.45 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(2-methylbenzyl)-piperazin-1-yl]-9H-purine (27).
The title compound was prepared by the general method to provide 116 mg (87%) of a solid, mp 97–98 °C. 1H NMR (300 MHz, CDCl3) δ 8.38 (s, 1H), 7.45–7.56 (m, 1H), 7.25–7.42 (m, 6H), 7.09–7.23 (m, 5H), 4.36 (br s, 4H), 3.51 (s, 2H), 2.60 (t, J = 4.9 Hz, 4H), 2.39 (s, 3H). MS (m/z) 529.6 (M + 1). HPLC ≥ 99% at 19.28 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-{4-[2-(trifluoromethyl)-benzyl]piperazin-1-yl}−9H-purine (28).
The title compound was prepared by the general method to provide 102 mg (70%) of a solid, mp 119–121 °C. 1H NMR (300 MHz, CDCl3) δ 8.39 (s, 1H), 7.86 (d, J = 7.8 Hz, 1H), 7.64 (d, J = 7.8 Hz, 1H), 7.44–7.58 (m, 2h), 7.28–7.42 (m, 6h), 7.15–7.25 (m, 2H), 4.40 (br s, 4H), 3.73 (s, 2h), 2.65 (t, J = 4.9 Hz, 4H). 13C NMR (75 MHz, CDCl3) S 153.9, 153.2, 152.2, 145.6, 137.0, 134.2, 134.2, 134.1, 133.1, 1321.5, 131.3, 130.0, 129.7, 129.4, 128.9, 128.4, 128.0, 126.9, 119.7, 56.5, 53.1, 45.4. 19F NMR (282 MHz, CDCl3) δ −59.13. MS (m/z) 583.9 (M + 1). HPLC ≥ 99% at 19.83 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-{4-[2-(trifluoromethoxy)benzyl]piperazin-1-yl}−9H-purine (29).
The title compound was prepared by the general method to provide 81 mg (54%) of a solid, mp 110–111 °C. 1H NMR (300 MHz, CDCl3) δ 8.38 (s, 1H), 7.54–7.66 (m, 1H), 7.45–7.54 (m, 1H), 7.23–7.4–3 (m, 8H), 7.15–7.22 (m, 2H), 4.39 (br s, 4H), 3.65 (s, 2H), 2.64 (t, J = 4.9 Hz, 4H). 13C NMR (75 MHz, CDCl3) δ 153.9, 153.2, 152.3, 148.0, 147.9, 145.6, 134.2, 134.2, 133.1, 132.5, 131.4, 131.1, 130.7, 130.0, 129.7, 129.4, 128.4, 128.0, 126.9, 126.7, 122.3, 120.6, 120.6, 119.7, 118.9, 56.0, 53.1, 45.3. 19F NMR (282 MHz, CDCl3) δ −56.91. MS (m/z) 599.6 (M + 1). HPLC = 95% at 20.15 min.
2-({4-[8-(2-chlorophenyl)-9-(4-chlorophenyl)-9H-purin-6-yl]-piperazin-1-yl}methyl)benzonitrile (30).
The title compound was prepared by the general method to provide 125 mg (92%) of a solid, mp 127–128 °C. 1H NMR (300 MHz, CDCl3) δ 8.38 (s, 1H), 7.66 (d, J = 7.8 Hz, 1H), 7.56–7.63 (m, 2H), 7.46–7.53 (m, 1H), 7.29–7.43 (m, 6H), 7.13–7.25 (m, 2H), 4.40 (br s, 4H), 3.77 (s, 2H), 2.68 (t, J = 4.9 Hz, 4H). 13C NMR (75 MHz, CDCl3) δ 153.9, 153.2, 152.3, 145.7, 142.0, 134.2, 134.2, 133.1, 133.0, 132.6, 132.5, 131.4, 130.1, 130.0, 129.6, 129.4, 128.0, 127.7, 126.9, 119.8, 117.7, 113.2, 60.5, 53.1, MS (m/z) 540.2 (M + 1). HPLC ≥ 99% at 19.67 min.
6-[4-(3-Chlorobenzyl)piperazin-1-yl]-8-(2-chlorophenyl)-9-(4-chlorophenyl)-9H-purine (31).
The title compound was prepared by the general method to provide 105 mg (77%) of a solid, mp 120–122 °C. 1H NMR (300 MHz, CDCl3) δ 8.38 (s, 1H), 7.45–7.56 (m, 1H), 7.28–7.41 (m, 6H), 7.13–7.25 (m, 5H), 4.39 (br s, 4H), 3.53 (s, 2H), 2.60 (t, J = 4.90 Hz, 4H). MS (m/z) 549.2 (M + 1). MS (m/z) 549.2 (M + 1). HPLC ≥ 99% at 19.55 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(4-fluorobenzyl)-piperazin-1-yl]-9H-purine (32).
The title compound was prepared by the general method to provide 109 mg (82%) of a solid, mp 78–80 °C. 1H NMR (300 MHz, CDCl3) δ 8.38 (s, 1H), 7.47–7.55 (m, 1H), 7.27–7.42 (m, 7H), 7.16–7.24 (m, 2H), 7.01 (t, J = 8.7 Hz, 2H), 4.38 (br s, 4H), 3.52 (s, 2H), 2.58 (t, J = 4.9 Hz, 4h). MS (m/z) 533.4 (M + 1). HPLC ≥ 99% at 18.90 min.
6-[4-(4-Chlorobenzyl)piperazin-1-yl]-8-(2-chlorophenyl)-9-(4-chlorophenyl)-9H-purine (33).
The title compound was prepared by the general method to provide 37 mg (28%) of a solid, mp 172–174 °C. 1H NMR (300 MHz, CDCl3) δ 8.42 (s, 1H), 7.48–7.57 (m, 1H), 7.30–7.42 (m, 5H), 7.15–7.26 (m, 4H), 6.83–6.95 (m, 2H), 4.53 (br s, 4H), 3.24–3.41 (m, 4H). MS (m/z) 537.2 (M + 1). HPLC = 92% at 21.45 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(4-methylbenzyl)-piperazin-1-yl]-9H-purine (34).
The title compound was prepared by the general method to provide 110 mg (83%) of a solid, mp 104–105 °C. 1H NMR (300 MHz, CDCl3) δ 8.37 (s, 1H), 7.45–7.56 (m, 1H), 7.27–7.41 (m, 5H), 7.07–7.25 (m, 6H), 4.38 (br s, 4H), 3.53 (s, 2H), 2.59 (t, J = 4.9 Hz, 4H), 2.33 (s, 3H). MS (m/z) 529.7 (M + 1). HPLC ≥ 99% at 19.53 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(4-methoxybenzyl)-piperazin-1-yl]-9H-purine (35).
The title compound was prepared by the general method to provide 102 mg (75%) of a solid, mp 138–139 °C. 1H NMR (300 MHz, CDCl3) δ 8.37 (s, 1H), 7.45–7.56 (m, 1H), 7.28–7.42 (m, 5H), 7.25 (d, J = 8.1 Hz, 2H), 7.19 (d, J = 8.8 Hz, 2H), 6.86 (d, J = 8.6 Hz, 2H), 4.37 (br s, 4H), 3.80 (s, 3H),3.51 (s, 2h), 2.59 (t, J = 4.9 Hz, 4H). MS (m/z) 545.5 (M + 1). HPLC = 96% at 19.08 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-{4-[(1,5-dimethyl-1H-pyrazol-4-yl)methyl]piperazin-1-yl}−9H-purine (36).
The title compound was prepared by the general method to provide 144 mg (105%) of a solid, mp 138–139 °C. 1H NMR (300 MHz, CDCl3) δ 8.37 (s, 1H), 7.45–7.57 (m, 1H), 7.27–7.42 (m, 6H), 7.19 (d, J = 8.7 Hz, 2H), 4.37 (br s, 4H), 3.77 (s, 3H), 3.41 (s, 2H), 2.57 (t, J = 4.8 Hz, 4h), 2.24 (s, 3H). 13C NMR (75 MHz, CDCl3) δ 153.8, 153.1, 152.2, 145.6, 139.2, 137.0, 134.2, 134.2, 133.1, 132.5, 131.4, 130.0, 129.6, 129.3, 128.0, 126.9, 119.7, 114.0, 52.7, 52.4, 45.3, 36.2, 9.6. MS (m/z) 533.3 (M + 1). HPLC = 99% at 17.99 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-{4-[(5-methyl-1,2,4-oxadiazol-3-yl)methyl]piperazin-1-yl}−9H-purine (37).
The title compound was prepared by the general method to provide 96 mg (74%) of a pale-yellow film. 1H NMR (300 MHz, CDCl3) δ 8.38 (s, 1H), 7.46–7.56 (m, 1H), 7.29–7.43 (m, 5H), 7.19 (d, J = 8.7 Hz, 2H), 4.44 (br s, 4H), 3.74 (s, 2H), 2.74 (t, J = 4.9 Hz, 4H), 2.60 (s, 3h). 13C NMR (75 MHz, CDCl3) δ 176.8, 167.2, 153.8, 53.1, 152.3, 145.7, 134.2, 133.0, 132.5, 131.4,130.0, 129.6, 129.4, 128.0, 126.9, 119.7, 53.2, 52.9, 45.0, 12.3. MS (m/z) 521.4 (M + 1). HPLC = 99% at 17.88 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(2-phenylethyl)-piperazin-1-yl]-9H-purine (38).
The title compound was prepared by the general method to provide 56 mg (42%) of a solid, mp 156–157 °C. 1H NMR (300 MHz, CDCl3) δ 8.39 (s, 1H), 7.49–7.56 (m, 1H), 7.26–7.43 (m, 7H), 7.14–7.25 (m, 5H), 4.42 (br s, 4H), 2.80–2.94 (m, 2H), 2.60–2.77 (m, 6H). MS (m/z) 529.7 (M + 1). HPLC ≥ 99% at 19.53 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(2-pyridin-4-ylethyl)-piperazin-1-yl]-9H-purine (39).
The title compound was prepared by the general method to provide 39 mg (30%) of a solid, mp 142–143 °C. 1H NMR (300 MHz, CDCl3) δ 8.51 (d, J = 5.9 Hz, 2H), 8.39 (s, 1H), 7.46–7.57 (m, 1H), 7.29–7.41 (m, 5H), 7.10–7.23 (m, 4H), 4.41 (br s, 4H), 2.79–2.93 (m, 2H), 2.61–2.75 (m, 6H). MS (m/z) 530.6 (M + 1). HPLC = 97% at 15.21 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(2-fluorophenyl)-piperazin-1-yl]-9H-purine (40).
The title compound was prepared by the general method to provide 50 mg (39%) of a solid, mp 211–212 °C. 1H NMR (300 MHz, CDCl3) δ 8.42 (s, 1H), 7.49–7.58 (m, 1H), 7.30–7.41 (m, 5H), 7.16–7.24 (m, 2H), 6.89–7.14 (m, 4H), 4.56 (br s, 4H), 3.17–3.34 (m, 4H). MS (m/z) 519.4 (M + 1). HPLC ≥ 99% at 24.56 min.
8-(2-Chlorophenyl)-9-(4-chlorophenyl)-6-[4-(2-chlorophenyl)-piperazin-1-yl]-9H-purine (41).
The title compound was prepared by the general method to provide 67 mg (50%) of a solid, mp 205–209 °C. 1H NMR (300 MHz, CDCl3) δ 8.42 (s, 1H), 7.50–7.58 (m, 1H), 7.30–7.45 (m, 6H), 7.16–7.25 (m, 3H), 6.96–7.11 (m, 2H), 4.57 (br s, 4H), 3.13–3.33 (m, 4H). MS (m/z) 537.1 (M + 1). HPLC = 99% at 24.48 min.
hCBI Calcium-Mobilization and Radioligand-Displacement Assays.
Each compound was biologically characterized using a functional fluorescent hCB1-activated Gαq16-coupled intracellular-calcium-mobilization assay in CHO-K1 cells, as described in our previous publications, and apparent-affinity (Ke) values were determined.13,14 Briefly, CHO-K1 cells were engineered to coexpress human CB1 and Gαq16. Activation of CB1 by an agonist then led to the generation of inositol phosphatase 3 (IP3) and the activation of IP3 receptors, which led to the mobilization of intracellular calcium. Calcium flux was monitored in a 96-well format using the fluorescent dye calcein-4 AM in an automated plate reader (FLIPR, Molecular Devices). The antagonism of a test compound was measured by its ability to shift the concentration-response curve of the synthetic CB1 agonist CP55940 rightwards using the equation:
where DR is the EC50 ratio of CP55940 in the presence or absence of a test agent.
For some inverse agonist assays, cells were loaded with calcein-4 AM as described below and directly stimulated with various concentrations of a test agent for 90 s. Decreases in basal fluorescence were used in these assays to calculate EC50 values.
Further characterization of select compounds was performed using the radioligand displacement of [3H]CP55940, and the equilibrium-dissociation-constant values (Ki) were determined as described previously.13,14 The selectivities of these compounds for hCB1 versus hCB2 were also determined by obtaining the Ki values with membranes of CHO cells overexpressing either receptor. The data reported are average values from three to six measurements typically with <30% standard errors.
Radioligand-Displacement Assay for hERG Interaction.
Competition binding with [3H]astemizole was used to measure hERG interactions as previously described.20 Preparations of membranes overexpressing hERG were purchased from PerkinElmer. The binding assays were performed for 60 min using 4 μg of hERG-expressing membranes, ~3 nM [3H]astemizole, and various concentrations of the test agents in a pH 7.4 buffer (10 mM HEPES, 130 mM NaCl, 5 mM KCl, 0.8 mM MgCl2, 1 mM NaEDTA, 10 mM glucose, and 0.1% BSA). Binding was terminated by rapid filtration onto GF/B fiber filtermats presoaked in 0.3% polyethylenimine, followed by rapid washing (6 × 2 mL) with an ice-cold pH 7.4 buffer (25 mM Tris-HCl, 130 mM NaCl, 5 mM KCl, 0.8 mM MgCl2, 0.05 mM CaCl2, and 0.1% BSA) using a Brandel harvester. The filters were dried and counted after the addition of a scintillant. The data were analyzed using nonlinear regression (GraphPad Prism) and Ki values. All experiments were performed at least twice in duplicate, and the data reported are the mean values.
MDCK-mdr1 Permeability Assays.
MDCK-mdr1 cells obtained from The Netherlands Cancer Institute were grown on Transwell-type filters (Corning) for 4 days to confluence in DMEM/F12 media containing 10% fetal bovine serum and antibiotics. Compounds were added to the apical sides at concentrations of 10 μM in a transport buffer comprising 1× Hank’s balanced salt solution with 25 mM d-glucose and buffered with HEPES to pH 7.4. The samples were incubated for 1 h at 37 °C and carefully collected from both the apical and basal sides of the filters. The compounds selected for the MDCK-mdr1 cell assays were infused on an Applied Biosystems API-4000 mass spectrometer in order to optimize them for analysis using multiple-reaction monitoring (MRM), as previously described.27 Chromatography was conducted with an Agilent 1100 binary pump with a flow rate of 0.5 mL/min. The mobile-phase solvents were 0.1% formic acid in water (A) and 0.1% formic acid in methanol (B). The solvent conditions were 10% B for 1 min, followed by a gradient to 95% B over 5 min. The data reported are average values from two to three measurements.
Hepatic-Microsomal-Stability Studies.
Human-microsomal-stability assays were performed as described previously.14 Briefly, the test compounds were incubated at 1 μM final concentrations with 0.5 mg/mL pooled human liver microsomes from 200 unidentified donors (Xenotech, LLC) in 100 mM phosphate buffer (pH 7.4) containing 3 mM MgCl2, 1 mM nicotinamide adenine dinucleotide phosphate (NADPH), 5 mM uridine diphosphate glucuronic acid (UDPGA), and 50 μg/mL alamethicin. Triplicate samples were incubated for up to 120 min. Samples were removed at regular intervals. The reactions were terminated by the addition of three volumes of methanol and processed as described for the MDCK-mdr1 assays, but standard curves were prepared in blank matrixes for each compound for the quantitative assessments. Intrinsic clearance rates were calculated according to the half-life method of Obach et al.28 The data reported are average values from three measurements.
Solubility Determination.
For these experiments, 10 mM DMSO stocks of the compounds were directly diluted into 10 mM phosphate buffer at pH 7.4 or 1.4 and shaken for 90 min at room temperature using a plate mixer. The final concentrations of DMSO were 1%. After the incubations were complete, the samples were filtered through 0.4 μm filter plates (Multiscreen, Millipore). The filtrates were carefully collected in a 96-well plate. The analysis of the compounds was performed by LC/MS using previously available methods, and the concentrations were determined by fitting unknown concentrations to a six-point standard curve for each compound using GraphPad Prism. The data are reported as mean values from three determinations.
Pharmacokinetic Testing.
Female C57BL6 mice were procured from Jackson Laboratories at 8 weeks of age. The doses were formulated in 0.5% sodium carboxymethylcellulose with 1% NMP and 0.3% Tween 80 at 10 mg/kg. Animals were sacrificed at various time points, and samples were removed. Pharmacokinetic analyses were performed as described in our previous publications using Phoenix WinNonlin (Certara).13,14
Hypothermia Mouse Study.
Female C57BL6 mice were procured from Jackson Laboratories at 8 weeks of age. After an acclimation period of 2 weeks, the mice were dosed with either the vehicle or a test agent by oral gavage and allowed to rest for 30 min. The full cannabinoid-receptor agonist, CP55940, was then administered to each animal at concentrations of 0.56 mg/kg via ip injections dissolved in corn oil. The rectal temperature of each animal was recorded at regular intervals for up to 4 h after CP55940 administration. Six animals were used per group, and the data are reported as means and standard deviations calculated using GraphPad Prism. Two-way ANOVA analyses were also performed between groups using the same software.
Alcohol-Induced-Hepatic-Steatosis Study.
Alcoholic steatosis of the liver was induced using the approach of Lieber and DeCarli.29 Pathogen-free female C57BL/6J mice, aged 9 weeks (Jackson Laboratories) and weighing 19–21 g, were used in this study. Animals were housed in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited, specific-pathogen-free, environmentally controlled facility at Mispro Biotech Services. All experimental procedures were conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC). The C57BL/6 mice were offered a solid PicoLab Certified Rodent LabDiet 5053 (pellet) ad libitum during the acclimation period, after which all the animals were switched to a Lieber-DeCarli 82’ Control (Catalogue No. F1259SP, BioServ) diet, which was nutritionally complete. Control and ethanol (Catalogue No. F1258SP, BioServ) liquid diets were prepared daily as per the instructions provided by the manufacturer. Once on the liquid diet for 3 days, ethanol was introduced progressively from 1 to 5% (v/v) over 15 days to the scheduled animals. The animals were maintained on the liquid diet for 4 additional weeks after reaching the final concentration of ethanol and administered the test article for the last 2 weeks to evaluate its efficacy in limiting or reversing alcoholic steatosis (AS). The test compound, 25, was formulated as a suspension in 0.5% sodium carboxymethylcellulose with 1% N-methyl-2-pyrrolidone (NMP) and 0.3% Tween 80 as the vehicle. The piperazine, 25, was administered to the mice twice daily by oral gavage at a concentration of 1.25 mg/kg on the basis of the data from the PK experiments and preliminary studies. Control animals were administered the vehicle alone. Pairwise-feeding within groups was conducted to adjust for caloric intake between groups.
Following the final day of the liquid diet and oral dosing administration, the animals were euthanized and necropsied to collect tissue samples. Blood was collected from all the animals via cardiac punctures at the time of the scheduled necropsy into SAI Infusion Technologies collection microtubes with lithium heparin as the anticoagulant and centrifuged at approximately 2800g for approximately 10 min at 4 °C in order to obtain the plasma within 1 h of collection. For each animal, the liver was removed and weighed, after which a section of the median lobe of the liver was embedded in optimal-cutting-temperature solution (OCT, Tissue-Plus) and preserved in a base mold (24 × 24 × 5 mm, Fisherbrand) for sectioning, so that the degree of steatosis, the degree of liver damage, and general cytotoxicity could be determined.
For the histopathology studies, Oil Red O (ORO) staining was performed on the harvested liver tissue to assess the degree of liver steatosis in the ethanol- and control-diet-fed mouse groups. The OCT-embedded liver sections were stained with ORO and examined under a light microscope. Digitized images of the mouse liver sections (10 μm in thickness) were assessed semiquantitatively for the degree of steatosis using ImageJ software (NIH). Briefly, a 5 in2 digital grid (ImageJ Command Menu: Plugin-Analyze-Grid) was placed on each image wherein the lipid droplets in six grids per image were counted, and the average number of oil droplets was recorded for each animal (n = 6 per group). The results have been reported as means ± SD. Statistical significance was determined through ANOVA between the treatment and control groups using Graph Pad Prism 7.0 (GraphPad Software Inc.) at a confidence level of p < 0.001.
Computational Methods.
Model Preparation from Antagonist-CB1 Crystal-Structure Complexes.
Subsequent to the completion of the SAR for this project, several crystal structures of CB1 bound to small-molecule antagonists17,18 and agonists19 have been solved as chimeric entities with additional thermostabilizing mutations and N- and C-terminal suitable truncations. Shao and co-workers obtained diffraction-quality crystals by employing lipidic-cubic-phase crystallization and replacing the third intracellular loop (ICL3) with a thermostable Purococcus abyssi glycogen synthase (PGS) domain, introducing a T210A thermostabilizing mutation, eliminating the first 89 N-terminal residues, and truncating the C-terminal residues following residue 421. These researchers reported the co-complex of taranabant bound to CB1 (5U09). In the same time frame, Hua and co-workers18 solved the structure of AM6538 (Ki = 5.1 nM) bound to CB1 by introducing flavodoxin as a stabilizing fusion partner within the ICL3 at V306 and P332; truncating the N- and C-termini by 98 and 58 residues, respectively; and introducing T210A, E272K, T283V, and R340G mutations to improve the expression and thermostability (5TGZ). The two antagonist crystal structures are in agreement within crystallographic solution uncertainties, with some differences in diffraction and resolution in the N-terminal regions. We reproduced the docking results in both models and initially achieved comparable results for both AM6538 and taranabant using both structures when we employed Autodock VINA and AMBER16 molecular-mechanics-generalized-Born-surface-area (MMGBSA) rescoring, described below. Although we used the 5TGZ and 5TGV chimeric forms for the initial docking and MMGBSA-rescoring studies below, we have, in selected cases, transformed the docked poses into a full N- and C-terminal model with all the thermostabilizing mutations reversed, and we have equilibrated the docked-ligand-CB1 poses in a DOPC/K+/Cl−/water model. All mentions of poses or docked poses refer to configurations obtained by AMBER16 energy-minimized or simulated annealing (MD 300 K).
Docking.
Both Autodock Vina30 and Schrodinger GLIDE-XP31 docking approaches were employed in a large region near the tops of the transmembrane helical domains, inclusive of the crystal structures’ ligand (orthosteric) binding sites. The docking box employed for VINA was ca. 20 × 20 × 20 Å3 in spatial extent; the default box size employed in computing the GLIDE-XP docking grid was based on one of the largest ligand sizes, but it was chosen in accord with the inclusion of the orthosteric region out to the N-terminal and extracellular domains. Our own use of Autodock VINA relied on the VINA scoring function to filter and obtain reasonable docking poses, which optimized facets present in the scoring function, such as hydrophobic contacts, hydrogen bonds, and lack of steric clash. Although the informatics-based scoring function is quite good for most targets, we view all docking primarily as a means of “filtering” or obtaining the most plausible collection of poses from a large number of sampled docked configurations. For this reason, we collected up to 20 poses per ligand with a high “exhaustiveness” setting of 40–80. The GLIDE-XP workflow and scoring function likewise was used to obtain up to 10 poses per ligand. The GLIDE-XP scoring function contains terms such as desolvation lipophilic and hydrophobic contacts and cavity costs in addition to physics-based Coulombic and van der Waals interactions, desolvation, and ligand strain; as such, it is a hybrid physics- and informatics-based scoring function.
For the initial docking and MMGBSA-rescoring phase, we chose to use the 5TGZ structure, including the ICL3 insertion and stabilizing mutations. We developed an AMBER16 all-H model employing the AMBER14SB force field. The initial structure prepared with TLEAP was energy-minimized for 8000 steps of a conjugate gradient that followed an initial minimization of the 400 steepest descents, which removed the initial bad contacts. The point of using the analogue of the 5TGZ structure was to initially explore the SAR poses in the context of the crystallographic coordinates before reverse modeling to get the “natural” sequence.
MMGBSA-Min and MMGBSA-SA.
Although both of these docking approaches do find physiologically relevant poses within the top 5–20 poses, our experience is that quite often the top-ranking docking-function-scored pose is not the crystallographically observed pose in both in-house studies of relevant targets with structural solutions (GPCR and globular proteins and enzymes) as well our own and others published PDB-Bind assessments.32 We find that (a) most frequently, the lowest- or second-lowest-MMGBSA-scored pose has the lowest RMSD to the known crystallographic solution, and (b) the best MMGBSA pose gives the best affinity correlations.33–35 In order to provide a common platform for MMGBSA and MMPBSA rescoring of all the docking approaches, we employed a combination of AMBER16 software in conjunction with our own C++ and bash-script base to provide an automated workflow for picking up the docked poses for all the ligands (in mol2 format), performing ligand formal-charge perception (C++ and OpenBabel open-source), computing charges on the basis of either AM1-BCC or other ab initio QM approaches, minimizing the energy of all the poses in complex with the receptor protein, optionally employing a GPU-CPU hybrid MD-simulated annealing procedure (300 K MD followed by energy minimization), and finally computing the MMGBSA or MMPBSA score. Given that we chose flexible-ligand-rigid-receptor docking (perhaps with the allowance for hydroxyl rotation) as an initial step, our philosophy was that we wanted to allow at most quick MDs (i.e., 20–80 ps) before reminimizing the complex and performing MMGBSA or MMPBSA scoring. We wanted to explore the minor relaxations and responses of the protein to the flexible-ligand docked pose rather than an extensive MD exploration. In this way, one explores “local” energy wells not involving large-amplitude ligands or protein motion away from the original docked pose. In cases where we wanted to explore the impacts of larger receptor deformations, we employed MD-snapshot sampled receptor-ensemble docking. Our experience using normal mode and quasi-harmonic-entropy-estimate corrections suggests that although it can be helpful in more accurately ranking ligands and providing more sensible total-free-energy estimates36 (for comparison with the experiment), this quite often introduces additional error sources (in sampling) while attempting to get more physical estimates of the free energy of the bound pose. We employed the approximation by using the energy-minimized receptor coordinates obtained from the receptor-complex pose as the receptor structure, rather than from a separate APO simulation of CB1.
Several approaches were used to perform MMGBSA rescoring, including igb = 1 with mbondi generalized Born radii as well as igb = 8 with mbondi3 radii; no significant differences in conclusions were found for this work. The MMGBSA-simulated-annealing protocol we employed was based on a 2000 step minimization of the complex and 20 ps of MD, followed by 2000 steps of reminimization. We collected five samples (one every 4 ps) during the MD phase, and these snapshots were re-energy-minimized and used to compute the MMGBSA average score for the pose.
Limited MD Exploration of the Relaxation of Loops and Water-Accessible Domains.
In selected cases in this work, we chose to explore the equilibration of the VINA- and MMGBSA-rescored poses of the antagonists in the CB1 orthosteric binding site, and we employed a DOPC/K+/Cl−/TIPS3P/water environment. For this purpose, the VINA+MMGBSA pose was employed in a full N-terminal and C-terminal representation of CB1, with the thermostabilizing mutations reverted back to the normal sequence,37 the ICL3 insertion removed, and an equilibrated loop model used as the start of the simulations that included the Cys257/264 disulfide bond. An initial 50 × 50 × 50 Å3 DOPC membrane was constructed, including 696 DOPC lipids prior to deletions based on atomic overlaps, along with 192 K+/Cl− ion pairs and 64 505 water molecules. The Sali MODELER338 and SCRWL39 rotamer modified initial model of the docked complex was equilibrated in the DOPC-membrane/K+/Cl−/water environment, and all the DOPC/K+/Cl−/water overlaps with the CB1-receptor complex were deleted. The Walker group LIPID1440 model was used for the DOPC representation. The same GAFF/AM1-BCC representation of the ligand was employed in the membrane-complex simulations as that employed in the MMGBSA-rescoring treatment. The complex model immersed in the DOPC/K+/Cl−/water model was then equilibrated for 150–500 ns for the mere examination of the equilibrated-loop-water environment proximate to the selected ligands. Although it was obvious from the initial docking that the SAR extension region should have some solvent accessibility, with solvent percolating into the upper regions of the N-terminal end and loops, these GPU-accelerated simulations verified this facet, as shown in selected figures in this paper. The initial AM6538 simulation has been analyzed at both the ASP338-R214 salt bridge and at the Y200-W356-W279 aromatic toggle region and found to be, in fact, stabilized by the antagonist compared with either APO or the ongoing simulations on CB1 agonists. Such results, although they are beyond the scope of this paper, did establish the rational nature of the starting MMGBSA-docking-rescored poses and the short simulations based on them to illustrate the nature of the SAR environment in a membrane-ion-water environment. Such results amplify our conclusions based on the MMGBSA and QM-desolvation analyses.
QM-Solvation Calculations.
Schrodinger Jaguar (V9.7) DFT computations were performed on each of the ligands; a B3LYP hybrid nonlocal functional with the ligands optimized was employed, and the solvation was evaluated using Schrodinger’s Poisson-Boltzmann solver. A modest 6–31G** split-valence set was employed for all the calculations and given optimal performance in Friesner’s pseudospectral DFT/Jaguar method.37 The reference-optimized ligand energy was employed to compute the solvation energies, default optimization, and single-point SCF; optimization and gradient criteria were employed. Upon examination, the solvation energy evaluated in the guise of AMBER16 was found to be in accord (correlated) with the QM-solvation estimates.
Supplementary Material
ACKNOWLEDGMENTS
We express our gratitude to the NIDA drug-supply program for providing the radiolabeled probes and to Dr. Brian Thomas for supplying the CB1 cells. We thank Keith Warner, Taylor Rosa, Elaine Gay, Aliah Hackney, and Melody Markley for excellent technical help. This research was funded by research grants AA022235 and DK100414 to R.M. from the NIH.
ABBREVIATIONS USED
- BBB
blood-brain barrier
- CB1
cannabinoid receptor 1
- CB2
cannabinoid receptor 2
- CHO-K1
Chinese hamster ovary cells
- CNS
central nervous system
- EtOH
ethanol
- EtOAc
ethyl acetate
- Ke
apparent-affinity constant
- MDCK-mdr1
Madin-Darby canine kidney cells transfected with the human MDR1 gene
- IP3
inositol phosphatase 3
- MRM
multiple-reaction monitoring
- rt
room temperature
- TEA
triethylamine
- TFA
trifluoroacetic acid
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.7b01820.
Crystal structure model docking of AM6538 and AMBER16 free energy (MMGBSA) scoring of residue-ligand interactions, repeat-dosing pharmacokinetic study of 25, brain concentration of 25 after 2 weeks of dosing in the alcoholic-steatosis study, and pharmacokinetic assessment of rimonabant (PDF)
Molecular-formula strings (CSV)
The authors declare no competing financial interest.
Atomic coordinates for docking studies using the chimeric protein representation will be provided upon request by the authors but were based on AMBER16 and/or OPSL3 prepared all-hydrogen representations. Coordinates for molecular dynamic snapshot representations will be provided upon request.
REFERENCES
- (1).Howlett AC Pharmacology of cannabinoid receptors. Annu. Rev. Pharmacol. Toxicol 1995, 35, 607–634. [DOI] [PubMed] [Google Scholar]
- (2).Pacher P; Kunos G Modulating the endocannabinoid system in human health and disease-successes and failures. FEBS J. 2013, 280, 1918–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Lu Y; Anderson HD Cannabinoid signaling in health and disease. Can. J. Physiol. Pharmacol 2017, 95, 311–327. [DOI] [PubMed] [Google Scholar]
- (4).Kunos G; Osei-Hyiaman D; Batkai S; Sharkey KA; Makriyannis A Should peripheral CB(1) cannabinoid receptors be selectively targeted for therapeutic gain? Trends Pharmacol. Sci 2009, 30, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Tam J; Liu J; Mukhopadhyay B; Cinar R; Godlewski G; Kunos G Endocannabinoids in liver disease. Hepatology 2011, 53, 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Christensen R; Kristensen PK; Bartels EM; Bliddal H; Astrup A Efficacy and safety of the weight-loss drug rimonabant: a meta-analysis of randomised trials. Lancet 2007, 370, 1706–1713. [DOI] [PubMed] [Google Scholar]
- (7).Chorvat RJ Peripherally restricted CB1 receptor blockers. Bioorg. Med. Chem. Lett 2013, 23, 4751–4760. [DOI] [PubMed] [Google Scholar]
- (8).Kunos G; Tam J The case for peripheral CB(1) receptor blockade in the treatment of visceral obesity and its cardiometabolic complications. Br. J. Pharmacol 2011, 163, 1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Klumpers LE; Fridberg M; de Kam ML; Little PB; Jensen NO; Kleinloog HD; Elling CE; van Gerven JM Peripheral selectivity of the novel cannabinoid receptor antagonist TM38837 in healthy subjects. Br. J. Clin. Pharmacol 2013, 76, 846–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Takano A; Gulyas B; Varnas K; Little PB; Noerregaard PK; Jensen NO; Elling CE; Halldin C Low brain CB1 receptor occupancy by a second generation CB1 receptor antagonist TM38837 in comparison with rimonabant in nonhuman primates: a PET study. Synapse 2014, 68, 89–97. [DOI] [PubMed] [Google Scholar]
- (11).Tam J; Cinar R; Liu J; Godlewski G; Wesley D; Jourdan T; Szanda G; Mukhopadhyay B; Chedester L; Liow JS; Innis RB; Cheng K; Rice KC; Deschamps JR; Chorvat RJ; McElroy JF; Kunos G Peripheral cannabinoid-1 receptor inverse agonism reduces obesity by reversing leptin resistance. Cell Metab. 2012, 16, 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chang CP; Wu CH; Song JS; Chou MC; Wong YC; Lin Y; Yeh TK; Sadani AA; Ou MH; Chen KH; Chen PH; Kuo PC; Tseng CT; Chang KH; Tseng SL; Chao YS; Hung MS; Shia KS Discovery of 1-(2,4-dichlorophenyl)-N-(piperidin-1-yl)-4-((pyrrolidine-1-sulfonamido)methyl)-5-(5-((4-(trifluoromethyl)phenyl)ethynyl)thiophene-2-yl)-1H-pyrazole-3-carboxamide as a novel peripherally restricted cannabinoid-1 receptor antagonist with significant weight-loss efficacy in diet-induced obese mice. J. Med. Chem 2013, 56, 9920–9933. [DOI] [PubMed] [Google Scholar]
- (13).Fulp A; Bortoff K; Zhang Y; Seltzman H; Mathews J; Snyder R; Fennell T; Maitra R Diphenyl purine derivatives as peripherally selective cannabinoid receptor 1 antagonists. J. Med. Chem 2012, 55, 10022–10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Fulp A; Bortoff K; Seltzman H; Zhang Y; Mathews J; Snyder R; Fennell T; Maitra R Design and synthesis of cannabinoid receptor 1 antagonists for peripheral selectivity. J. Med. Chem 2012, 55, 2820–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Griffith DA; Hadcock JR; Black SC; Iredale PA; Carpino PA; DaSilva-Jardine P; Day R; DiBrino J; Dow RL; Landis MS; O’Connor RE; Scott DO Discovery of 1-[9-(4-chlorophenyl)-8-(2-chlorophenyl)-9H-purin-6-yl]-4-ethylaminopiperi-dine-4-carboxylic acid amide hydrochloride (CP-945,598), a novel, potent, and selective cannabinoid type 1 receptor antagonist. J. Med. Chem 2009, 52, 234–237. [DOI] [PubMed] [Google Scholar]
- (16).Wang Q; Rager JD; Weinstein K; Kardos PS; Dobson GL; Li J; Hidalgo IJ Evaluation of the MDR-MDCK cell line as a permeability screen for the blood-brain barrier. Int. J. Pharm 2005, 288, 349–359. [DOI] [PubMed] [Google Scholar]
- (17).Shao Z; Yin J; Chapman K; Grzemska M; Clark L; Wang J; Rosenbaum DM High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature 2016, 540, 602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hua T; Vemuri K; Pu M; Qu L; Han GW; Wu Y; Zhao S; Shui W; Li S; Korde A; Laprairie RB; Stahl EL; Ho JH; Zvonok N; Zhou H; Kufareva I; Wu B; Zhao Q; Hanson MA; Bohn LM; Makriyannis A; Stevens RC; Liu ZJ Crystal structure of the human cannabinoid receptor CB1. Cell 2016, 167, 750–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hua T; Vemuri K; Nikas SP; Laprairie RB; Wu Y; Qu L; Pu M; Korde A; Jiang S; Ho JH; Han GW; Ding K; Li X; Liu H; Hanson MA; Zhao S; Bohn LM; Makriyannis A; Stevens RC; Liu ZJ Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature 2017, 547, 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Chiu PJ; Marcoe KF; Bounds SE; Lin CH; Feng JJ; Lin A; Cheng FC; Crumb WJ; Mitchell R Validation of a [3H]astemizole binding assay in HEK293 cells expressing HERG K+ channels. J. Pharmacol. Sci 2004, 95, 311–319. [DOI] [PubMed] [Google Scholar]
- (21).Chaperon F; Thiebot MH Behavioral effects of cannabinoid agents in animals. Crit. Rev. Neurobiol 1999, 13, 243–281. [DOI] [PubMed] [Google Scholar]
- (22).Hadcock JR; Griffith DA; Iredale PA; Carpino PA; Dow RL; Black SC; O’Connor R; Gautreau D; Lizano JS; Ward K; Hargrove DM; Kelly-Sullivan D; Scott DO In vitro and in vivo pharmacology of CP-945,598, a potent and selective cannabinoid CB(1) receptor antagonist for the management of obesity. Biochem. Biophys. Res. Commun 2010, 394, 366–371. [DOI] [PubMed] [Google Scholar]
- (23).Jeong WI; Osei-Hyiaman D; Park O; Liu J; Batkai S; Mukhopadhyay P; Horiguchi N; Harvey-White J; Marsicano G; Lutz B; Gao B; Kunos G Paracrine activation of hepatic CB1 receptors by stellate cell-derived endocannabinoids mediates alcoholic fatty liver. Cell Metab. 2008, 7, 227–235. [DOI] [PubMed] [Google Scholar]
- (24).Baell JB; Holloway GA New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem 2010, 53, 2719–2740. [DOI] [PubMed] [Google Scholar]
- (25).Baell JB; Nissink JWM Seven Year Itch: Pan-assay interference compounds (PAINS) in 2017-utility and limitations. ACS Chem. Biol 2018, 13, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dahlin JL; Walters MA How to triage PAINS-full research. Assay Drug Dev. Technol 2016, 14, 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Fulp A; Bortoff K; Zhang Y; Seltzman H; Snyder R; Maitra R Towards rational design of cannabinoid receptor 1 (CB1) antagonists for peripheral selectivity. Bioorg. Med. Chem. Lett 2011, 21, 5711–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Obach RS; Baxter JG; Liston TE; Silber BM; Jones BC; MacIntyre F; Rance DJ; Wastall P The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J. Pharmacol. Exp. Ther 1997, 283, 46–58. [PubMed] [Google Scholar]
- (29).Lieber CS; DeCarli LM Liquid diet technique of ethanol administration: 1989 update. Alcohol Alcohol. 1989, 24, 197–211. [PubMed] [Google Scholar]
- (30).Trott O; Olson AJ AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem 2010, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Friesner RA; Murphy RB; Repasky MP; Frye LL; Greenwood JR; Halgren TA; Sanschagrin PC; Mainz DT Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem 2006, 49, 6177–6196. [DOI] [PubMed] [Google Scholar]
- (32).Greenidge PA; Kramer C; Mozziconacci JC; Sherman W Improving docking results via reranking of ensembles of ligand poses in multiple X-ray protein conformations with MM-GBSA. J. Chem. Inf. Model 2014, 54, 2697–2717. [DOI] [PubMed] [Google Scholar]
- (33).Zhang X; Wong SE; Lightstone FC Toward fully automated high performance computing drug discovery: a massively parallel virtual screening pipeline for docking and molecular mechanics/generalized born surface area rescoring to improve enrichment. J. Chem. Inf. Model 2014, 54, 324–337. [DOI] [PubMed] [Google Scholar]
- (34).Greenidge PA; Kramer C; Mozziconacci JC; Wolf RM MM/GBSA binding energy prediction on the PDBbind data set: successes, failures, and directions for further improvement. J. Chem. Inf. Model 2013, 53, 201–209. [DOI] [PubMed] [Google Scholar]
- (35).Hou T; Wang J; Li Y; Wang W Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J. Comput. Chem 2011, 32, 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Harris DL; Park JY; Gruenke L; Waskell L Theoretical study of the ligand-CYP2B4 complexes: effect of structure on binding free energies and heme spin state. Proteins: Struct., Fund. Genet 2004, 55, 895–914. [DOI] [PubMed] [Google Scholar]
- (37).Bochevarov AD; Harder E; Hughes TF; Greenwood JR; Braden DA; Philipp DM; Rinaldo D; Halls MD; Zhang J; Friesner RA Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem 2013, 113, 2110–2142. [Google Scholar]
- (38).Sali A; Blundell TL Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol 1993, 234, 779–815. [DOI] [PubMed] [Google Scholar]
- (39).Wang Q; Canutescu AA; Dunbrack RL, Jr SCWRL and MolIDE: computer programs for side-chain conformation prediction and homology modeling. Nat. Protoc 2008, 3, 1832–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Dickson CJ; Madej BD; Skjevik AA; Betz RM; Teigen K; Gould IR; Walker RC Lipid14: The amber lipid force field. J. Chem. Theory Comput 2014, 10, 865–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.