Abstract
5-Fluorouracil (5-FU)-based chemotherapy is the first-line option for patients with advanced colorectal cancer (CRC). However, the development of chemoresistance is the primary cause of treatment failure. Halofuginone (HF), a small molecule alkaloid derived from febrifugine, has been demonstrated to exert strong anti-proliferative effects. However, to the best of our knowledge, whether HF inhibits the progression of 5-FU-resistant human CRC HCT-15/FU cells, and the underlying mechanisms, remain unknown. In the present study, the effects of HF on HCT-15/FU cells were assessed in vitro. The results revealed that HF inhibited HCT-15/FU cell viability as demonstrated by the MTT and colony formation assays. Following treatment of HCT-15/FU cells with HF, the migratory and invasive capacities of the cells were significantly decreased. MicroRNA (miRNA/miR)-sequencing data, subsequent miRNA trend analysis and reverse transcription-quantitative PCR all demonstrated that miR-132-3p expression was increased following treatment with HF in a dose-dependent manner. Western blot analysis indicated that following treatment with HF, the expression levels of proteins associated with proliferation, invasion and metastasis in cells were markedly downregulated. These results suggested that HF inhibited the proliferation, invasion and migration of HCT-15/FU cells by upregulating the expression levels of miR-132-3p. Therefore, miR-132-3p may serve as a molecular marker, which may be used to predict CRC resistance to 5-FU, and HF may serve as a novel clinical treatment for 5-FU-resistant CRC.
Keywords: halofuginone, colorectal cancer, microRNA-132-3p, progression, invasion, metastasis
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide (1). Due to the continuous development of endoscopic treatments, including endoscopic mucosal resection and endoscopic submucosal dissection, which allow radical resection of a CRC tumor without the need for open surgery (2–4), the 5-year survival rate of patients with early CRC is >90% (5,6). By contrast, for patients with advanced CRC with distant metastasis, 5-fluorouracil (5-FU)-based chemotherapy is the first-line option; however, the development of drug resistance is the primary cause of treatment failure, and the median survival time is <2 years (7). Therefore, the development of alternative approaches to overcome CRC resistance is urgently required.
Halofuginone (HF), a derivative of a traditional Chinese medicine and the effective constituent of febrifugine, has attracted increasing attention for its wide range of biological activities in malaria, autoimmune diseases, fibrosis and cancer (8–11). HF suppresses cancer cell proliferation and decreases tumor metastasis in hepatoma, melanoma and multiple myeloma in vitro (12–14). HF has already been trialed for use as an anticarcinogenic drug for advanced solid tumors in one phase I clinical trial (15). The trial found that the pharmacokinetics of HF were linear over the dose range studied, with a large interpatient variability. The dose-limiting toxicities of HF were nausea, vomiting and fatigue. The recommended dose for phase II studies of HF was 0.5 mg administered orally, once daily.
In the present study, the regulatory effects of HF on the proliferation, migration and invasion of 5-FU-resistant human CRC HCT-15/FU cells were assessed. Additionally, using bioinformatics analysis, alterations in the microRNA (miRNA/miR) expression profiles were determined following treatment with HF. The results of the present study highlight the potential of miR-132-3p as a potential therapeutic target, and the potential of HF as a novel therapeutic agent in 5-FU-resistant CRC.
Materials and methods
Chemicals and reagents
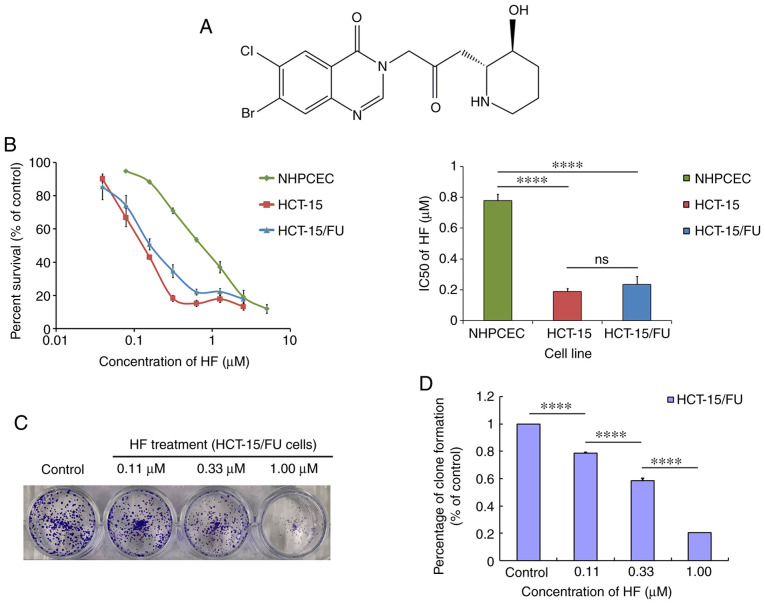
MTT was purchased from Sigma-Aldrich; Merck KGaA. Antibodies against β-catenin (cat. no. 8480), poly(ADP-ribose) polymerase (PARP; cat. no. 9532), ERK1/2 (cat. no. 4696), AKT (cat. no. 4691), Survivin (cat. no. 2808) and integrin β4 (ITGβ4; cat. no. 14803) were obtained from Cell Signaling Technology, Inc. An antibody against EGFR (cat. no. ab52894) was purchased from Abcam. GAPDH (1A6) monoclonal antibody (cat. no. MB001) and goat anti-mouse IgG (H+L)-HRP (cat. no. BS12478) were purchased from Bioworld Technology, Inc. Goat anti-rabbit IgG (H+L) HRP (cat. no. S0001) was purchased from Affinity Biosciences. All tissue culture reagents and other routine laboratory reagents were obtained from Guangzhou Whiga Technology Co., Ltd. HF (Fig. 1A), extracted from Dichroa febrifuga Lour of traditional Chinese herbs, with a purity of 99.05% was obtained from Selleck Chemicals (cat. no. S8144).
Figure 1.
HF exhibits potent cytotoxic effects on 5-FU-resistant human colorectal cancer HCT-15/FU cells. (A) Chemical structure of HF, rel-7-bromo-6-chloro-3-[3-[(2R,3S)-3-hydroxy-2-piperidinyl]-2-oxopropyl]-4(3H)-Quinazolinone; molecular weight, 414.68. (B) MTT assays were used to determine the effects of HF on HCT-15/FU cells, HCT-15 cells and NHPCECs. The cells were grown for 24 h and subsequently exposed to a range of concentrations of HF for 72 h. (C) HF inhibited the proliferation of HCT-15/FU cells as demonstrated by a colony formation assay. (D) Quantitative analysis of the colony formation assay. ****P<0.0001. HF, halofuginone; NHPCEC, normal human primary colonic epithelial cells; ns, no significance; FU, 5-fluorouracil.
Cell culture
The normal human primary colonic epithelial cells (NHPCECs; HUM-iCELL-d010), the 5-FU-resistant human CRC HCT-15/FU cell line (iCell-h073) and the corresponding 5-FU-sensitive HCT-15 cells (iCell-h072), the identities of which were confirmed using Short Tandem Repeat profiling, were purchased form iCell Bioscience, Inc. In detail, by gradually increasing the 5-FU concentration, HCT-15 cells were successfully induced to 5-FU-resistant cell lines and named HCT-15/FU. Specifically, HCT-15 cells in the logarithmic growth phase were seeded into 6-well plates. When the cell proliferation was stable and the confluence was ~80%, the medium was replaced with complete medium containing 7.5 µg/ml 5-FU. The drug concentration could be increased until the cells maintained normal growth at a concentration of 7.5 µg/ml for 4 days. The increasing drug concentrations were: 7.5, 10, 12.5, 15, 17.5 and 20 µg/ml. By day 208, the cells could be cultured in 20 µg/ml 5-FU-containing medium for 7 days, and the cells grew normally. At this point, HCT-15/FU cells were successfully constructed. 5-FU-resistant HCT-15/FU cells were authenticated by comparing their fold resistance with that of the parental 5-FU-sensitive HCT-15 cells (Fig. S1). The aforementioned two CRC cell lines were cultured in RPMI-1640 medium supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum (FBS) (Tianhang Biotech Co., Ltd.). NHPCECs were cultured in complete medium of the iCell primary epithelial cell culture system (PriMed-Icell-001; iCell Bioscience, Inc.). Both cultures were maintained at 37°C in a 5% CO2 incubator.
Cell cytotoxicity and colony formation assays
Cell cytotoxicity was evaluated using MTT assays as previously described (16). Specifically, 20 µl MTT (5 mg/ml) was added to each well for a 4-h reaction. Next, the supernatant was discarded, and the formed formazan product was dissolved in 100 µl DMSO/well. The optical density was detected at an absorbance wavelength of 540 nm using a Model 550 Microplate reader (Bio-Rad Laboratories, Inc.), with reference filter of 655 nm. Colony formation assays were performed to determine the cloning capability of HCT-15/FU cells. HCT-15/FU cells were treated with HF (0.11, 0.33, 1.00 µM) for 48 h. A total of 500 cells/well were seeded into 6-well plates and cultured for an additional 10 days in RPMI-1640 medium supplemented with 10% FBS at 37°C until most of the single colonies contained >50 cells. Subsequently, the colonies were fixed with 4% formaldehyde at room temperature, stained with 0.01% crystal violet for 30 min. Images were captured using ChemiDoc™ XRS+ (Bio-Rad Laboratories) (17).
Wound healing assay
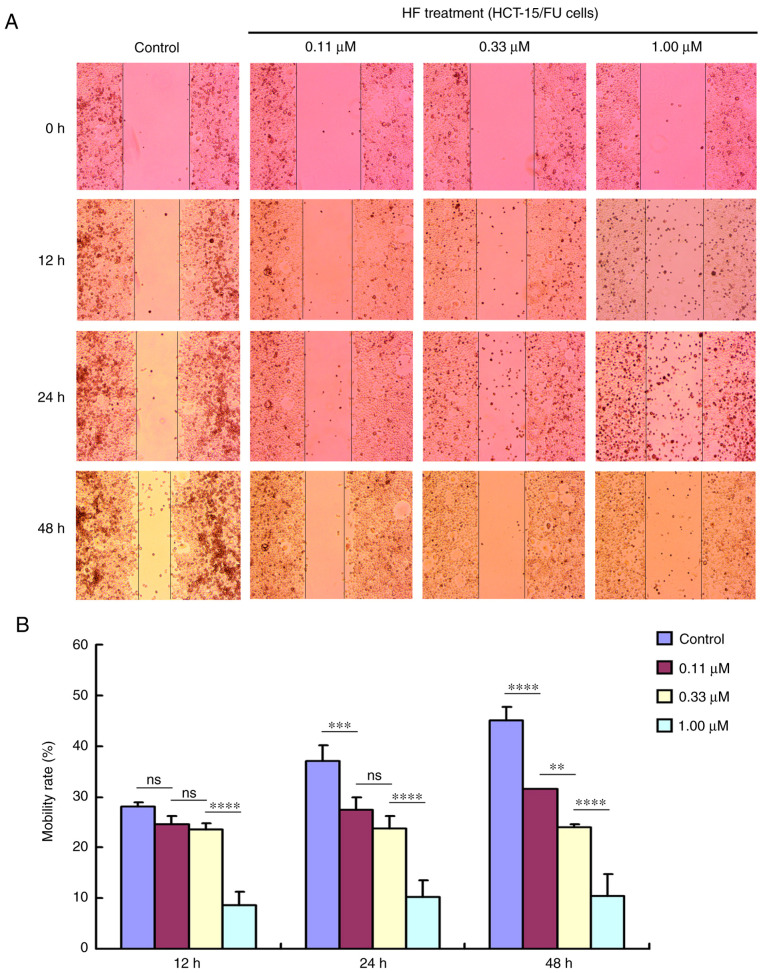
A wound healing assay was performed to determine cell migration in vitro. Briefly, cells were plated in 6-well plates and grown overnight to reach 100% confluence. Subsequently, a 1-ml pipette tip was used to scratch the cell monolayer, the wounded cell layer was washed with PBS to remove dead cells, and then FBS-free medium was added. The HCT-15/FU cells were incubated with HF (0.11, 0.33 and 1.00 µM) for 0, 12, 24 or 48 h. Images were captured using a light microscope (Olympus Corporation) at ×40 magnification. The surface area was calculated using ImageJ software (version 1.51j8; National Institutes of Health). The cell motility rate was calculated as follows: Cell motility rate (%) = migrated cell surface area/total surface area ×100.
Invasion assay
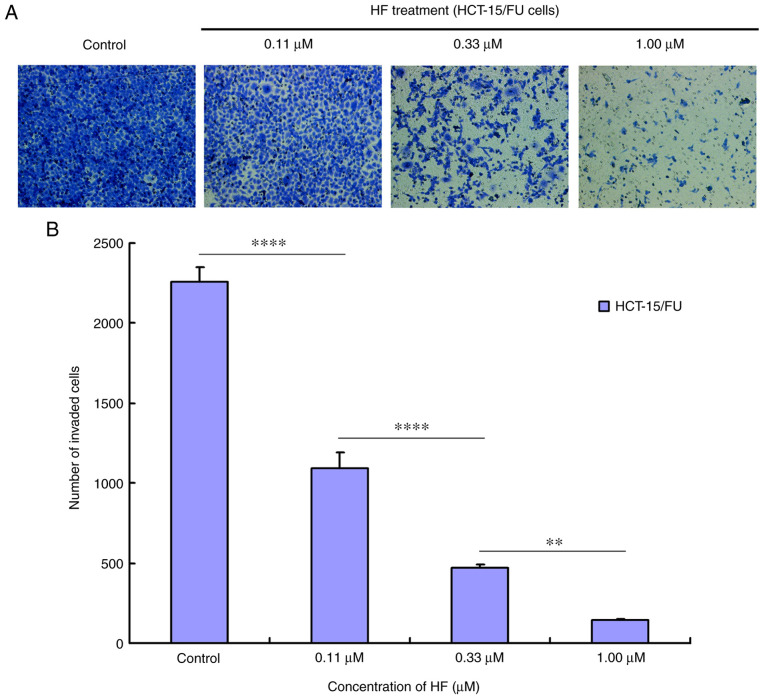
The bottom of the Transwell apparatus was pre-coated for 3 h with Matrigel (Corning Inc.), which was diluted 5 times with basal medium. HCT-15/FU cells were transferred into the upper chamber of the Transwell apparatus at a density of 2.5×104 cells/well in 200 µl serum-free RPMI-1640 medium, which contained the HF (0, 0.11, 0.33 and 1.00 µM). The lower chamber contained 600 µl RPMI-1640 medium with 10% FBS, which was a chemoattractant. After incubation for 48 h, cells that migrated into the lower chamber were fixed with methanol and stained with 0.05% crystal violet at room temperature for 30 min. Cells on the top layer were removed using a cotton swab, and images of the migrated cells were captured using an inverted microscope (Leica Microsystems) at ×200 magnification. Six random fields were imaged for quantification of migrated cells. The cell number that had invaded to the bottom chamber was calculated for each group by ImageJ software (version 1.51j8).
Western blotting
Cultured cells from different samples were collected and washed twice with ice-cold PBS. The pellet was vortexed and lysed in lysis buffer (Cell Signaling Technology, Inc.), supplementary with proteinase inhibitor cocktail (Cell Signaling Technology, Inc.) and phenylmethylsulfonyl fluoride (Cell Signaling Technology, Inc.). The protein concentration was determined using a BCA Protein assay kit (Thermo Fisher Scientific, Inc.). Proteins (25 µg/lane) were separated on 8–12% gels using SDS-PAGE and subsequently transferred onto PVDF membranes (EMD Millipore). The membranes were then blocked with TBST buffer [150 mmol/l NaCl, 20 mmol/l Tris-HCl (pH 7.4) and 0.4% (v/v) Tween-20] containing 5% dried non-fat milk, and subsequently incubated with primary antibodies overnight at 4°C. After being washed with TBST buffer three times, the membranes were incubated with horseradish peroxidase (HRP) conjugated secondary antibodies for 2 h at room temperature. Blots were visualized using a Western Lightning® Plus-ELC kit (PerkinElmer, Inc.) and analyzed by ChemiDoc™ XRS+ (Bio-Rad Laboratories, Inc.). Antibodies used in this study included: β-catenin (1:1,000 dilution), PARP (1:1,000 dilution), ERK1/2 (1:1,000 dilution), AKT (1:1,000 dilution), Survivin (1:1,000 dilution), ITGβ4 (1:1,000 dilution), EGFR (1:1,000 dilution), GAPDH (1:10,000 dilution), goat anti-mouse IgG (H+L) HRP (1:5,000 dilution) and goat anti-rabbit IgG (H+L) HRP (1:5,000 dilution).
Construction and sequencing of the miRNA library and subsequent miRNA trend analysis
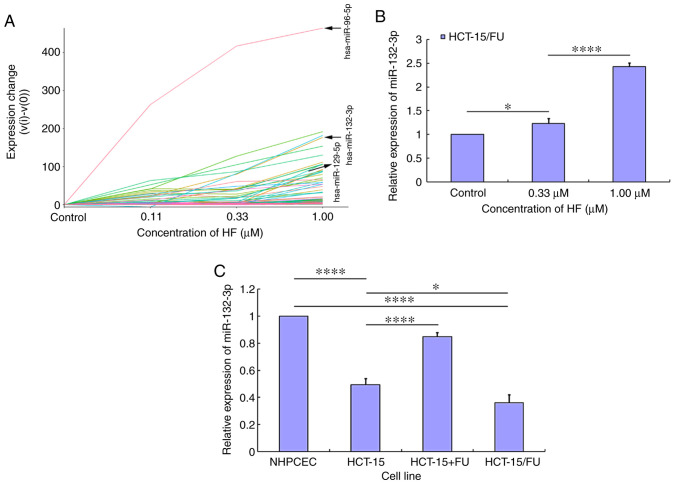
HCT-15/FU cells were treated with HF (0.11, 0.33 and 1.00 µM) for 48 h, and total RNA was extracted using TRIzol® (Thermo Fisher Scientific, Inc.). An miRNA sequencing library was constructed using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs Inc.; NEB #E7580L), which contains the adaptors, primers, enzymes and buffers required to convert small RNAs into indexed libraries for next generation sequencing on the Illumina platform. This construction was performed by KangChen Bio-tech Co., Ltd. Specifically, the experiment was performed according to the following steps: i) 3′-Adaptor ligation; ii) 5′-adaptor ligation; iii) cDNA synthesis; iv) PCR amplification; v) size selection of 135 to 155-bp PCR-amplified fragments (corresponding to 15 to 35-nt small RNAs). The libraries were denatured into single-stranded DNA molecules by 0.1 M NaOH, captured on Illumina flow cells (Illumina, Inc.), amplified in situ as clusters (TruSeq Rapid SR Cluster kit (#GD-402-4001; Illumina, Inc.) and finally sequenced for 51 cycles on an Illumina NextSeq 500 platform. Based on the miRNA sequencing results, the subsequent trend analysis was performed using Short Time-series Expression Miner software v1.3.8 (Kangchen BioTech Co., Ltd.). The miRNAs were assigned to the corresponding profile according to the number of genes assigned to that profile, the number of genes expected and the enrichment P-value.
miRNA extraction and reverse transcription-quantitative (RT-q)PCR
Following treatment of HCT-15/FU cells with 0.33 and 1.00 µM HF for 48 h, total RNA was extracted using TRIzol® reagent (Thermo Fisher Scientific, Inc.). A Mir-X™ miRNA RT-qPCR SYBR kit (Takara Bio, Inc.) was used to reverse transcribe miRNAs into cDNA, and qPCR was performed according to the manufacturer's protocol. The thermocycling conditions were: 95°C for 10 sec; followed by 40 cycles of 95°C for 5 sec and 64°C for 35 sec; followed by 95°C for 30 sec and 55°C for 30 sec. U6 was used as the internal control. The primers were synthesized by Takara Bio, Inc. The sequences of the primers were: U6 forward, 5′-CGCAAGGATGACACG-3′ and reverse, 5′-GAGCAGGCTGGAGAA-3′; and miR-132-3p forward, 5′-TAACAGTCTACAGCCATGGTC-3′ and reverse, 5′-CAGTGCGTGTCGTGGAGT-3′. The relative expression levels of the genes were determined using the 2−ΔΔCq method (18).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 7 software (GraphPad Software, Inc.). Single factor variance analysis (one-way ANOVA) followed by Tukey's post-hoc test was used when multiple sets of data met the criteria for normality and the homogeneity of the variance. All results were presented as mean ± standard deviation (SD). P<0.05 was considered to indicate a statistically significant difference.
Results
HF is cytotoxic to HCT-15/FU cells
MTT assays were used to measure cell viability following treatment with HF. As shown in Fig. 1B, the IC50 values were 0.19±0.02 and 0.23±0.05 µmol/l for HCT-15 and HCT-15/FU cells, respectively (P>0.05), whereas for NHPCECs, the IC50 was 0.78±0.04 µmol/l. These data suggest that HF exhibited potent and similar cytotoxicity against both HCT-15/FU and HCT-15 cells. Compared with the two tumor cell lines, HF was less cytotoxic to NHPCECs (both P<0.05). Additionally, HF treatment inhibited the clone formation of HCT-15/FU cells as demonstrated by the colony formation assays (Fig. 1C and D).
HF inhibits migration and invasion of HCT-15/FU cells
Tumor cell migration and invasion promote tumor progression. To determine the effects of HF on migration and invasion, HCT-15/FU cells were treated with 0.11, 0.33 or 1.00 µmol/l HF for 48 h to assess changes in cell migration and invasion. The results demonstrated that HF inhibited HCT-15/FU cell migration (Fig. 2) and invasion (Fig. 3) in a dose-dependent manner. Notably, the results presented in Fig. 2 indicated that HF could also induce apoptosis of HCT-15/FU cells (consistent with the western blotting results for PARP protein). Cell mobility was quantified from living cells on the edge of the scratch. At 48 h after HF treatment, numerous apoptotic cells covered visible gaps and the edges of the scratch were not visible. Therefore, dead cells were removed to calculate cell mobility at 48 h.
Figure 2.
HF inhibits the migration of HCT-15/FU cells. (A) HCT-15/FU cell migration was inhibited by HF in a dose-dependent manner (×40 magnification). (B) Quantitative results of migration analysis. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01, ***P<0.001 and ****P<0.0001. HF, halofuginone; ns, no significance.
Figure 3.
HF inhibits the invasion of HCT-15/FU cells. (A) HCT-15/FU cell invasion was inhibited by HF in a dose-dependent manner (×200 magnification). (B) Quantitative results of the invasion analysis. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 and ****P<0.0001. HF, halofuginone.
miRNA trend analysis in HCT-15/FU cells treated with HF
Using miRNA sequencing analysis and subsequent miRNA trend analysis, the expression profiles of miRNAs in HCT-15/FU cells treated with HF were determined. The miRNA trend analysis results demonstrated that the expression levels of certain miRNAs were increased in a dose-dependent manner (Fig. 4A), and other miRNAs were assigned to the corresponding profile (Figs. S2 and S3). The labeled lines in Fig. 4A represent miR-96-5p, miR-132-3p and miR-129-5p. Subsequently, RT-qPCR was used to validate the expression levels of the aforementioned specific miRNAs. Following treatment with HF, only miR-132-3p expression was demonstrated to be increased in a dose-dependent manner (Fig. 4B), while the expression level of miR-96-5p and miR-129-5p changed irregularly (data not shown). To further elucidate the mechanism by which HF inhibited progression of HCT-15/FU cells, subsequent experiments focused on the role of miR-132-3p. miR-132-3p expression was lower in CRC cells (HCT-15 and HCT-15/FU cells) compared with in NHPCECs (P<0.01). When HCT-15 cells were treated with 5-FU, the expression levels of miR-132-3p were increased. Additionally, the expression levels of miR-132-3p were lower in HCT-15/FU cells compared with HCT-15 cells (P<0.05; Fig. 4C). These results suggest that HF may inhibit HCT-15/FU cell progression via upregulation of miR-132-3p expression.
Figure 4.
Screening of the miRNA expression profiles in HCT-15/FU cells treated with HF. (A) Using miRNA sequencing analysis and subsequent miRNA trend analysis screening, the expression profiles of human miRNAs in HCT-15/FU cells treated with HF were determined. The expression levels of certain miRNAs were increased in a dose-dependent manner. (B) miR-132-3p expression was increased in HCT-15/FU cells treated with HF in a dose-dependent manner. (C) miR-132-3p expression was downregulated in colorectal cancer HCT-15 and HCT-15/FU cells compared with in NHPCECs. Treatment of HCT15 cells with 5-FU increased the expression levels of miR-132-3p (HCT-15 versus HCT-15/FU) and miR-132-3p expression was lower in HCT-15/FU cells compared with in the corresponding sensitive HCT-15 cells. Experiments were performed in triplicate. *P<0.05 and ****P<0.0001. HF, halofuginone; miR/miRNA, microRNA; NHPCEC, normal human primary colonic epithelial cells; FU, 5-fluorouracil.
HF inhibits progression of HCT-15/FU cells through miR-132-3p-mediated targeting of proteins involved in proliferation, invasion and metastasis
Using TargetScan Human 7.2 prediction (http://www.targetscan.org/vert_72/), search results demonstrated that miR-132-3p might be predicted to target the 3′-untranslated regions (UTRs) of proteins involved in proliferation, invasion and metastasis, including MAPK1, MAPK3, MAPKBP1, AKT3 and integrin subunit α9. In the present study, western blotting results revealed that the protein expression levels of EGFR, AKT, ERK1/2, Survivin, β-Catenin, PARP and ITGβ4, which are associated with cell proliferation, apoptosis, invasion or metastasis, were decreased in HCT-15/FU cells treated with HF (Fig. 5). Overall, these results suggest that HF may inhibit HCT-15/FU cell progression via miR-132-3p-mediated targeting of proteins involved in proliferation, invasion and metastasis.
Figure 5.
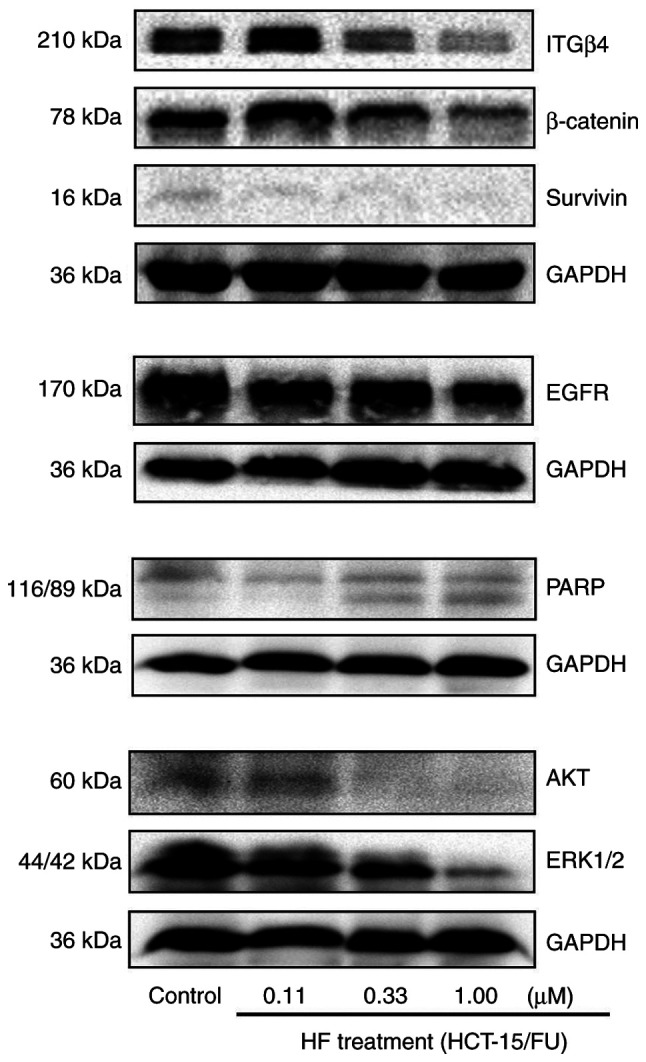
HF inhibits progression of HCT-15/FU cells via microRNA-132-3p-mediated targeting of proteins involved in proliferation, invasion and metastasis. Following treatment of HCT-15/FU cells with various concentrations of HF, the expression levels of proteins associated with cell proliferation, invasion and metastasis were detected. The protein expression levels of EGFR, AKT, ERK1/2 (MAPK1/2), Survivin, β-Catenin, PARP and ITGβ4 were downregulated. PARP, poly(ADP-ribose) polymerase; ITGβ4, Integrin β4; HF, halofuginone.
Discussion
HF has been demonstrated to exhibit anticancer activity via several mechanisms, including the induction of apoptosis, inhibition of proliferation and regulation of multiple downstream cancer-associated signaling molecules (19,20). Since not having more 5-FU-resistant CRC cell lines is a limitation, the present study detected the effects of HF on two pairs of cancer cell lines: KB and it corresponding resistant KBv200 cell line (data not shown), and HCT-15 and it corresponding resistant HCT-15/FU cell line. The results indicated that HF exhibited potent cytotoxicity against the two pairs of cells. To further elucidate the molecular mechanisms underlying the antitumor activity of HF, the effects of HF on the tumorigenic progression of human 5-FU-resistant HCT-15/FU cells were examined. MTT, colony formation, wound healing and invasion assays demonstrated that when HCT-15/FU cells were treated with HF, cell viability, migration and invasion were reduced, and western blotting indicated that the protein expression levels of EGFR, AKT, ERK1/2, Survivin, β-catenin, PARP and ITGβ4 were downregulated. Therefore, the effects of HF on HCT-15/FU cells may be attributed to two models: i) Inhibition of protein expression of EGFR, AKT, ERK1/2, Survivin, β-catenin, PARP, downstream of MAPK and/or PI3K/AKT signaling pathways, resulting in inhibition of HCT-15/FU cell proliferation and induction of apoptosis; and ii) inhibiting the protein expression of ITGβ4, thus inhibiting invasion and metastasis. Therefore, it was hypothesized that HF may exert beneficial effects in 5-FU-resistant CRC treatment.
miRNAs are small, endogenous single-stranded 21–23 nucleotide non-coding RNAs that serve as protooncogenes or tumor suppressor genes, and are abnormally expressed in a variety of tumors. Increasing bioinformatics evidence and subsequent functional assays have revealed that miRNAs serve key roles in numerous biological processes, including tumor occurrence, development and metastasis, in several types of cancer (21). miRNAs can bind to the 3′-UTR of target gene mRNAs and induce their post-transcriptional repression (22–24).
Studies have demonstrated that miRNAs regulate the expression of various genes associated with human cancer (25,26). Several other studies have revealed that miRNAs may serve as biomarkers of CRC, and the majority of these miRNAs have a tumor suppressor role (27,28). miR-132 has been demonstrated to regulate cancer via regulation of several cellular behaviors, including tumorigenesis, proliferation, resistance and progression (29). Previous studies have indicated that miR-132 exerts tumor-suppressing functions in various types of cancer, including prostate cancer, thyroid cancer, pancreatic cancer, cervical cancer and renal carcinoma (30–35). Furthermore, miR-132 affects drug resistance. Studies have demonstrated that overexpression of miR-132 enhances the chemotherapy or radiotherapy sensitivity of CRC, ovarian cancer, nasopharyngeal carcinoma and cervical cancer (36–39). However, the biological functions of miR-132-3p in 5-FU-resistant CRC remain poorly understood, which largely limits its potential as a biomarker of resistance. In the present study, miR-132-3p expression was determined to be downregulated in CRC cells compared with NHPCECs. Additionally, the levels of miR-132-3p were lower in HCT-15/FU cells compared with the sensitive HCT-15 cells. Therefore, it was hypothesized that miR-132-3p may be involved not only in the tumorigenesis, but also in the process of CRC resistance to 5-FU. Overall, the present study identified that miR-132-3p may regulate CRC resistance to 5-FU.
Studies have demonstrated that natural compounds, including curcumin, tectorigenin and avenanthramide A can regulate the expression of numerous miRNAs, which increases the sensitivity of cancer cells to conventional chemotherapeutics and thereby effectively suppresses tumor cell proliferation (40–42). Previous studies have demonstrated that anticarcinogenic effects of HF on various types of cancer (43–45). In order to determine the mechanisms by which HF affected HCT-15/FU cells, bioinformatics analysis, miRNA-sequencing data, subsequent miRNA trend analysis and RT-qPCR verification were used, and it was demonstrated that miR-132-3p expression was increased in a dose-dependent manner.
ERK1/2 are critical molecules in the MAPK signaling pathway, which is an important pathway involved in tumor proliferation (46). Using TargetScan prediction software, it was suggested that miR-132-3p bound to the 3′-UTR of MAPK1 (ERK1). Consistent with this prediction, the present study demonstrated that HF treatment increased miR-132-3p expression and reduced the protein expression levels of ERK1/2. Therefore, it was hypothesized that miR-132-3p served as a tumor suppressor in HCT-15/FU cells via reduction of ERK1/2 expression. Additionally, western blotting suggested that the protein expression levels of EGFR, AKT, Survivin, β-Catenin, PARP and ITGβ4 were reduced following treatment of HCT-15/FU cells with HF. In addition, cell cycle-related proteins are closely associated with tumor growth pathways. In our other work, after HCT15/FU cells were added to HF, it was demonstrated using label-free quantitative proteomics that the expression levels of some cell cycle-related proteins, such as CDK2 and CDK11B, were altered (data not shown). Overall, these results suggested that HF inhibited proliferation of HCT-15/FU cells via upregulation of miR-132-3p-mediated targeting of proteins involved in proliferation, apoptosis, invasion and metastasis, providing a novel perspective on anti-5-FU-resistant CRC. Both miR-132-3p and cancer progression-related proteins may potentially serve as 5-FU-resistant biomarkers and/or targets for developing CRC therapeutics. These results suggest that HF and its derivatives may serve as novel therapeutics for treatment of 5-FU-resistant CRC.
In conclusion, the present study demonstrated that downregulated expression of miR-132-3p in HCT-15/FU cells promoted CRC resistance to 5-FU, and thus subsequent progression. miR-132-3p expression was increased in HCT-15/FU cells treated with HF in a dose-dependent manner. HF inhibited proliferation of HCT-15/FU cells via upregulation of miR-132-3p, which in turn targeted genes associated with cancer progression. To the best of our knowledge, the present study was the first to report the relationship among HF treatment, miR-132-3p and cancer progression-related proteins in HCT-15/FU cells, and HF and its derivatives may be a novel therapeutic agent for treatment of 5-FU-resistant CRC.
Supplementary Material
Acknowledgements
The authors would like to thank Professor Jianye Zhang, Dr Jiajun Li and Dr Qiaoru Guo (affiliated to both to the Guangdong Provincial Key Laboratory of Molecular Target and Clinical Pharmacology, School of Pharmaceutical Sciences and to the Fifth Affiliated Hospital, Guangzhou Medical University, Guangzhou, China) for their assistance with the statistical analysis.
Funding
This work was supported by grants from the Fund project of Inner Mongolia Autonomous Region Applied Technology Research and Development (2018–3rd batch-1; Wuhai Municipal People's Hospital), the National Natural Science Foundation of China (grant no. 81902152), and the Fund of Shanxi Province Higher Education Technology Innovation Project (grant no. 2019L0753).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YYY designed the study. CW and JBZ wrote the manuscript and performed the MTT, reverse transcription-quantitative PCR and western blotting assays. XJG and XW performed the migration and invasion assays. WZ and XLW performed miRNA trend analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chiba H, Takahashi A, Inamori M, Goto T, Ohata K, Matsuhashi N, Nakajima A. Early colon cancer presenting as intussusception and successfully treated using endoscopic submucosal dissection. Endoscopy. 2014;46:E326–E327. doi: 10.1055/s-0034-1377224. [DOI] [PubMed] [Google Scholar]
- 3.Kiriyama S, Saito Y, Yamamoto S, Soetikno R, Matsuda T, Nakajima T, Kuwano H. Comparison of endoscopic submucosal dissection with laparoscopic-assisted colorectal surgery for early-stage colorectal cancer: A retrospective analysis. Endoscopy. 2012;44:102410–102430. doi: 10.1055/s-0032-1310259. [DOI] [PubMed] [Google Scholar]
- 4.Kudo S. Endoscopic mucosal resection of flat and depressed types of early colorectal cancer. Endoscopy. 1993;25:455–461. doi: 10.1055/s-2007-1010367. [DOI] [PubMed] [Google Scholar]
- 5.Yang D, Othman M, Draganov PV. Endoscopic mucosal resection vs endoscopic submucosal dissection for barrett's esophagus and colorectal neoplasia. Clin Gastroenterol Hepatol. 2019;17:1019–1028. doi: 10.1016/j.cgh.2018.09.030. [DOI] [PubMed] [Google Scholar]
- 6.Dumoulin FL, Hildenbrand R. Endoscopic resection techniques for colorectal neoplasia: Current developments. World J Gastroenterol. 2019;25:300–307. doi: 10.3748/wjg.v25.i3.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fakih MG. Metastatic colorectal cancer: Current state and future directions. J Clin Oncol. 2015;33:1809–1824. doi: 10.1200/JCO.2014.59.7633. [DOI] [PubMed] [Google Scholar]
- 8.Jiang S, Zeng Q, Gettayacamin M, Tungtaeng A, Wannaying S, Lim A, Hansukjariya P, Okunji CO, Zhu S, Fang D. Antimalarial activities and therapeutic properties of febrifugine analogs. Antimicrob Agents Chemother. 2005;49:1169–1176. doi: 10.1128/AAC.49.3.1169-1176.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pines M, Snyder D, Yarkoni S, Nagler A. Halofuginone to treat fibrosis in chronic graft-versus-host disease and scleroderma. Biol Blood Marrow Transplant. 2003;9:417–425. doi: 10.1016/S1083-8791(03)00151-4. [DOI] [PubMed] [Google Scholar]
- 10.Pines M, Nagler A. Halofuginone: A novel antifibrotic therapy. Gen Pharmacol. 1998;30:445–450. doi: 10.1016/S0306-3623(97)00307-8. [DOI] [PubMed] [Google Scholar]
- 11.Xia X, Wang L, Zhang X, Wang S, Lei L, Cheng L, Xu Y, Sun Y, Hang B, Zhang G, et al. Halofuginone-induced autophagy suppresses the migration and invasion of MCF-7 cells via regulation of STMN1 and p53. J Cell Biochem. 2018;119:4009–4020. doi: 10.1002/jcb.26559. [DOI] [PubMed] [Google Scholar]
- 12.Nagler A, Ohana M, Shibolet O, Shapira MY, Alper R, Vlodavsky I, Pines M, Ilan Y. Suppression of hepatocellular carcinoma growth in mice by the alkaloid coccidiostat halofuginone. Eur J Cancer. 2004;40:1397–1403. doi: 10.1016/j.ejca.2003.11.036. [DOI] [PubMed] [Google Scholar]
- 13.Juárez P, Mohammad KS, Yin JJ, Fournier PG, McKenna RC, Davis HW, Peng XH, Niewolna M, Javelaud D, Chirgwin JM, et al. Halofuginone inhibits the establishment and progression of melanoma bone metastases. Cancer Res. 2012;72:6247–6256. doi: 10.1158/0008-5472.CAN-12-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leiba M, Jakubikova J, Klippel S, Mitsiades CS, Hideshima T, Tai YT, Leiba A, Pines M, Richardson PG, Nagler A, Anderson KC. Halofuginone inhibits multiple myeloma growth in vitro and in vivo and enhances cytotoxicity of conventional and novel agents. Br J Haematol. 2012;157:718–731. doi: 10.1111/j.1365-2141.2012.09120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Jonge MJ, Dumez H, Verweij J, Yarkoni S, Snyder D, Lacombe D, Marréaud S, Yamaguchi T, Punt CJ, van Oosterom A, EORTC New Drug Development Group (NDDG) Phase I and pharmacokinetic study of halofuginone, an oral quinazolinone derivative in patients with advanced solid tumours. Eur J Cancer. 2006;42:1768–1774. doi: 10.1016/j.ejca.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 16.Yan YY, Bi H, Zhang W, Wen Q, Liu H, Li JX, Zhang HZ, Zhang YX, Li JS. Downregulation and subcellular distribution of HER2 involved in MDA-MB-453 breast cancer cell apoptosis induced by lapatinib/celastrol combination. J BUON. 2017;22:644–651. [PubMed] [Google Scholar]
- 17.Wei M, Li J, Qiu J, Yan Y, Wang H, Wu Z, Liu Y, Shen X, Su C, Guo Q, et al. Costunolide induces apoptosis and inhibits migration and invasion in H1299 lung cancer cells. Oncol Rep. 2020;43:1986–1994. doi: 10.3892/or.2020.7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Assis PA, De Figueiredo-Pontes LL, Lima AS, Leão V, Cândido L, Pintão CT, Garcia AB, Saggioro FP, Panepucci RA, Chahud F, et al. halofuginone inhibits phosphorylation of SMAD-2 reducing angiogenesis and leukemia burden in an acute promyelocytic leukemia mouse model. J Exp Clin Cancer Res. 2015;34:65. doi: 10.1186/s13046-015-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Figueiredo-Pontes LL, Assis PA, Santana-Lemos BA, Jácomo RH, Lima AS, Garcia AB, Thomé CH, Araújo AG, Panepucci RA, Zago MA, et al. Halofuginone has anti-proliferative effects in acute promyelocytic leukemia by modulating the transforming growth factor beta signaling pathway. PLoS One. 2011;6:e26713. doi: 10.1371/journal.pone.0026713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babaei K, Shams S, Keymoradzadeh A, Vahidi S, Hamami P, Khaksar R, Norollahi SE, Samadani AA. An insight of microRNAs performance in carcinogenesis and tumorigenesis; an overview of cancer therapy. Life Sci. 2020;240:117077. doi: 10.1016/j.lfs.2019.117077. [DOI] [PubMed] [Google Scholar]
- 22.An X, Sarmiento C, Tan T, Zhu H. Regulation of multidrug resistance by microRNAs in anti-cancer therapy. Acta Pharm Sin B. 2017;7:38–51. doi: 10.1016/j.apsb.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of Primary microRNAs by the microprocessor Complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Li D, Zhang Y, Ding Z, Zheng Y, Chen S, Wan Y. Integrative analysis of mRNA and miRNA expression profiles reveals seven potential diagnostic biomarkers for nonsmall cell lung cancer. Oncol Rep. 2020;43:99–112. doi: 10.3892/or.2019.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung G, Hernández-Illán E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17:111–130. doi: 10.1038/s41575-019-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, Zheng Y, Lei W, Xiang L, Chen M. miR-1307-3p overexpression inhibits cell proliferation and promotes cell apoptosis by targeting ISM1 in colon cancer. Mol Cell Probes. 2019;48:101445. doi: 10.1016/j.mcp.2019.101445. [DOI] [PubMed] [Google Scholar]
- 28.Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Zhu Q, Lu L, Liu Y. MiR-132 inhibits migration and invasion and increases chemosensitivity of cisplatin-resistant oral squamous cell carcinoma cells via targeting TGF-β1. Bioengineered. 2020;11:91–102. doi: 10.1080/21655979.2019.1710925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li SL, Sui Y, Sun J, Jiang TQ, Dong G. Identification of tumor suppressive role of microRNA-132 and its target gene in tumorigenesis of prostate cancer. Int J Mol Med. 2018;41:2429–2433. doi: 10.3892/ijmm.2018.3421. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Li M, Zhou H, Zhang L. miR-132 targets FOXA1 and exerts tumor-suppressing functions in thyroid cancer. Oncol Res. 2019;27:431–437. doi: 10.3727/096504018X15201058168730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan S, Ebeling MC, Chauhan N, Thompson PA, Gara RK, Ganju A, Yallapu MM, Behrman SW, Zhao H, Zafar N, et al. Ormeloxifene suppresses desmoplasia and enhances sensitivity of gemcitabine in pancreatic cancer. Cancer Res. 2015;75:2292–2304. doi: 10.1158/0008-5472.CAN-14-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Zhu H, Wang Y, Song Y, Zhang P, Wang Z, Gao J, Li Z, Du Y. MicroRNA-132 plays an independent prognostic role in pancreatic ductal adenocarcinoma and acts as a tumor suppressor. Technol Cancer Res Treat. 2019;18:1533033818824314. doi: 10.1177/1533033818824314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao JL, Zhang L, Guo X, Wang JH, Zhou W, Liu M, Li X, Tang H. miR-212/132 downregulates SMAD2 expression to suppress the G1/S phase transition of the cell cycle and the epithelial to mesenchymal transition in cervical cancer cells. IUBMB life. 2015;67:380–394. doi: 10.1002/iub.1381. [DOI] [PubMed] [Google Scholar]
- 35.Yu Y, Lu W, Zhou X, Huang H, Shen S, Guo L. MicroRNA-132 suppresses migration and invasion of renal carcinoma cells. J Clin Lab Anal. 2020;34:e22969. doi: 10.1002/jcla.22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Zhang M. miR-132 Regulates adriamycin resistance in colorectal cancer cells through targeting extracellular signal-regulated kinase 1. Cancer Biother Radiopharm. 2019;34:398–404. doi: 10.1089/cbr.2018.2749. [DOI] [PubMed] [Google Scholar]
- 37.Zhang XL, Sun BL, Tian SX, Li L, Zhao YC, Shi PP. MicroRNA-132 reverses cisplatin resistance and metastasis in ovarian cancer by the targeted regulation on Bmi-1. Eur Rev Med Pharmacol Sci. 2019;23:3635–3644. doi: 10.26355/eurrev_201905_17787. [DOI] [PubMed] [Google Scholar]
- 38.Li YL, Zhao YG, Chen B, Li XF. MicroRNA-132 sensitizes nasopharyngeal carcinoma cells to cisplatin through regulation of forkhead box A1 protein. Pharmazie. 2016;71:715–718. doi: 10.1691/ph.2016.6764. [DOI] [PubMed] [Google Scholar]
- 39.Liu GF, Zhang SH, Li XF, Cao LY, Fu ZZ, Yu SN. Overexpression of microRNA-132 enhances the radiosensitivity of cervical cancer cells by down-regulating Bmi-1. Oncotarget. 2017;8:80757–80769. doi: 10.18632/oncotarget.20358. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Saini S, Arora S, Majid S, Shahryari V, Chen Y, Deng G, Yamamura S, Ueno K, Dahiya R. Curcumin modulates microRNA-203-mediated regulation of the Src-Akt axis in bladder cancer. Cancer Prev Res (Phila) 2011;4:1698–1709. doi: 10.1158/1940-6207.CAPR-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Liu X, Chen S, Wu J, Ye X, Xu L, Chen H, Zhang D, Tan R, Wang Y. Tectorigenin inhibits the in vitro proliferation and enhances miR-338* expression of pulmonary fibroblasts in rats with idiopathic pulmonary fibrosis. J Ethnopharmacol. 2010;131:165–173. doi: 10.1016/j.jep.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 42.Fu R, Yang P, Sajid A, Li Z. Avenanthramide A induces cellular senescence via miR-129-3p/Pirh2/p53 signaling pathway to suppress colon cancer growth. J Agric Food Chem. 2019;67:4808–4816. doi: 10.1021/acs.jafc.9b00833. [DOI] [PubMed] [Google Scholar]
- 43.Xia X, Wang X, Zhang S, Zheng Y, Wang L, Xu Y, Hang B, Sun Y, Lei L, Bai Y, Hu J. miR-31 shuttled by halofuginone-induced exosomes suppresses MFC-7 cell proliferation by modulating the HDAC2/cell cycle signaling axis. J Cell Physiol. 2019;234:18970–18984. doi: 10.1002/jcp.28537. [DOI] [PubMed] [Google Scholar]
- 44.Demiroglu-Zergeroglu A, Turhal G, Topal H, Ceylan H, Donbaloglu F, Karadeniz Cerit K, Odongo RR. Anticarcinogenic effects of halofuginone on lung-derived cancer cells. Cell Biol Int. 2020;44:1934–1944. doi: 10.1002/cbin.11399. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Xie Z, Lu H. Significance of halofuginone in esophageal squamous carcinoma cell apoptosis through HIF-1α-FOXO3a pathway. Life Sci. 2020;257:118104. doi: 10.1016/j.lfs.2020.118104. [DOI] [PubMed] [Google Scholar]
- 46.Robert Roskoski Jr. Targeting ERK1/2 protein-serine/threonine kinases in human cancers. Pharmacol Res. 2019;142:151–168. doi: 10.1016/j.phrs.2019.01.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.