Abstract
Acute myeloid leukemia with normal karyotype (NK-AML) is a group of diseases with high heterogeneity and immunological processes are significantly associated with its initiation and development. The implication of the immunogenomic landscape in the prognosis of patients with NK-AML has remained largely elusive. In the present study, the expression profiles of immune-related genes (IRGs) were examined and their association with overall survival (OS) was determined in 60 patients with NK-AML from The Cancer Genome Atlas dataset and 104 patients from the Gene Expression Omnibus (GEO) dataset no. GSE71014. Univariate Cox regression analysis was used to identify 42 and 203 IRGs in the two respective cohorts, which were significantly associated with OS in NK-AML. A risk model was constructed based on the regression coefficient and expression values of nine survival-associated IRGs shared between the two datasets [zinc finger CCCH-type containing, antiviral 1 like; transferrin receptor; suppressor of cytokine signaling 1; ELAV like RNA binding protein 1; roundabout guidance receptor 3; unc-93 homolog B1, Toll-like receptor signaling regulator; protein tyrosine phosphatase non-receptor type 6; interleukin 2 receptor subunit alpha (IL2RA) and IL3RA]. Using this risk model, patients with NK-AML may be divided into high- and low-risk groups in prognostic predictions. The area under the receiver operating characteristic curve for predicting OS was 0.793. The prognostic role of this risk model was successfully verified in another independent cohort (GEO dataset no. GSE71014). The prognostic risk score was positively associated with age and fms related receptor tyrosine kinase 3 mutation and correlated with infiltration by T regulatory cells. In conclusion, the results of the present study provided an IRG score model for prognostic stratification of adult patients with NK-AML, as well as further insight into the implication of IRGs in NK-AML that may lead to the development of novel immunotherapy approaches for this disease.
Keywords: acute myeloid leukemia, normal karyotype, immune-related genes, prognostic model, survival
Introduction
Acute myeloid leukemia (AML) is a malignant clonal disease of hematopoietic stem cells; chromosomal aberrations have been recognized as the most important prognostic marker in patients with AML (1). Of all cases of AML, 40–50% have a normal karyotype (2). Recent technological advances in gene expression profiling have allowed for the identification of more aberrantly expressed genes and the assessment of their predictive value for survival of patients with AML with normal karyotype (NK-AML). The expression levels of genes such as neurocaldin delta, MAPK binding protein 1, copine 3 and RUNX family transcription factor 1 provide further references for predicting the prognosis of patients with NK-AML (3–6). Ahn et al (7) reported that in patients with NK-AML, the presence of mutated chromatin, RNA-splicing genes or both was associated with poor survival. In general, these studies (3–7) have indicated high genetic heterogeneity in NK-AML. Patients with NK-AML exhibit marked differences in terms of prognosis (8). While the significance of single gene expression in NK-AML progression remains restrictive, gene expression signature-derived scoring systems have a promising prognostic value in AML (9).
Previous studies have demonstrated that the immune system, consisting of immune-related genes (IRGs), tumor-infiltrating lymphocytes and cytokines, is vital during the initiation and progression of cancer. The immunological surveillance and immune escape in bone marrow microenvironment have an impact on the clinical outcome of patients (10). IRGs actively participate in the process of immune activity, involving antigen presentation, cytokine signaling or immune-checkpoint modulation (11). Several IRGs in the immune/inflammatory response, such as interleukin (IL)-10, IL-15, Toll-like receptor (TLR)8 and TRAIL, have been indicated as being involved in the proliferation and differentiation of AML cells (12–14). Increased soluble IL-2 receptor levels in adult AML are associated with reduced chemotherapy response and reduced overall survival time (15). In addition, clinical benefits have been observed with immune checkpoint therapies in patients with AML. Programmed cell death protein 1 and cytotoxic T-lymphocyte antigen 4 inhibitors may be effective in elderly patients with AML (16). CD163 has been identified as a potential therapeutic target (17). However, to the best of our knowledge, the clinical role, particularly the prognostic role of IRGs in NK-AML, has remained to be determined.
Based on the aforementioned studies on the influence of immunity factors on the prognosis of AML, the present study aimed to identify differentially expressed IRGs in patients with NK-AML and assess their utility as survival predictors. For this purpose, public data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases were subjected to bioinformatics analyses. Furthermore, given that integrated prognostic analysis of multiple immune genes is more meaningful compared with the predictive value of a single gene, the present study aimed to propose an integrative risk score model consisting of the IRGs associated with prognosis. Data of the immune status of patients with NK-AML enrolled in the present study from obtained from two databases and further investigated.
Materials and methods
Study design
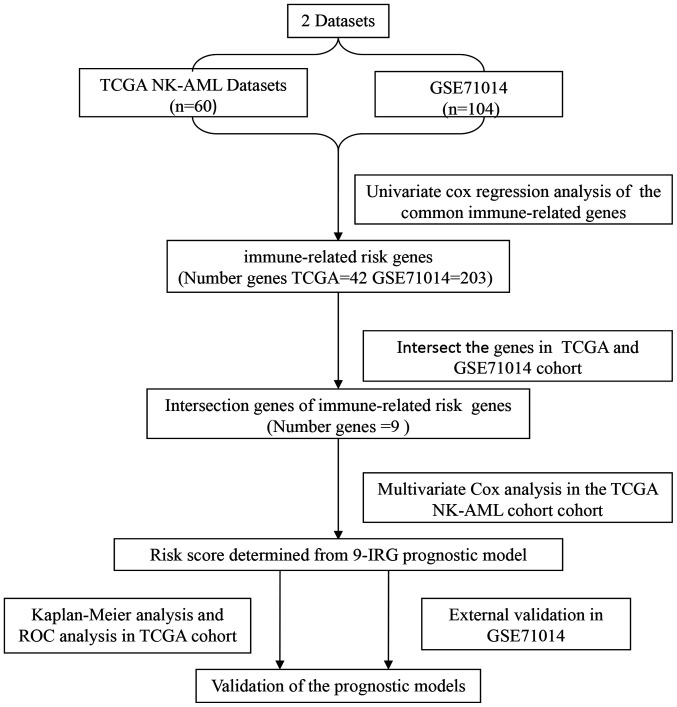
A flowchart illustrating the process of establishing the IRG risk score (IRGRS) model is presented in Fig. 1. The gene expression profiles and clinical data of patients with NK-AML from TCGA (https://tcga-data.nci.nih.gov/tcga/) and the GEO datasets (GSE71014) (https://ncbi.nlm.nih) were retrospectively analyzed. The expression levels of the common IRGs were screened from all protein-coding genes of the two datasets. Subsequently, the prognostic genes in TCGA and GEO datasets were identified by univariate Cox analysis. The survival-associated genes intersection of TCGA and GEO data were further analyzed to construct the prognostic model in the TCGA cohort. The GEO cohort (GSE71014) was employed to validate the immune-associated risk score model. Statistical analyses were performed using R software (version 3.5.3; http://www.r-project.org/) and Bioconductor (version 3.5.3; http://www.bioconductor.org/) was utilized for the bioinformatics analysis.
Figure 1.
Flowchart presenting the process of establishing the immune-related genes risk score model. TCGA, The Cancer Genome Atlas; ROC, receiver operating characteristic; NK-AML, acute myeloid leukemia with normal karyotype.
Patient data
The RNA-sequencing (RNA-seq) datasets of patients with AML, which had been experimentally determined using the Illumina HiSeq 2000 RNA Sequencing platform, were obtained from TCGA database (18). The corresponding clinical information was downloaded from the University of California Santa Cruz (UCSC) Xena site (https://xenabrowser.net/), including age, sex, leukemia French American British morphology code, the molecular analysis of nucleophosmin 1 (NPM1) and fms related receptor tyrosine kinase 3 (FLT3) mutations, survival time and outcome. Patients with normal karyotype were included and patients with acute promyelocytic leukemia were excluded. A total of 60 adult patients were selected from TCGA. There were 30 males (50.0%) and 30 females (50.0%) with a median age of 53.1 (range, 21–88) years. All gene expression levels were detected using the RNA-seq method in bone marrow cells of patients with NK-AML. The present study also included a total of 104 adult patients with NK-AML with a median age of 58 (range, 16–90) years from the GSE71014 dataset (19). To generate this dataset, cryopreserved bone marrow cells had been obtained from 104 patients with de novo AML and each sample was analyzed using the Illumina HumanHT-12 V4.0 expression beadchip (GPL10558). The gene expression profiles were acquired from the matched probes and were used for further analysis. In the present study, the IRGs were acquired from the ImmPort database, comprising 2,498 genes (https://immport.niaid.nih.gov) (20). The immune-associated genes that were upregulated or downregulated in TCGA and GSE71014 datasets was selected.
Construction and evaluation of the IRG risk score
The immune-associated genes in two datasets that were significantly associated with the overall survival of NK-AML were identified using univariate Cox analysis respectively (P≤0.005). The intersection of prognosis-associated IRGs in TCGA and GSE71014 datasets were selected and displayed in a Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/). The prognosis-associated IRGs were then submitted for multivariate analyses. Prognostic analysis was performed using the R package ‘survival’ (https://CRAN.R-project.org/package=survival; version 3.1–8). For the establishment of the prognostic score, the TCGA dataset was adopted. The IRGRS model was constructed by multiplying the gene expression levels of prognosis-associated IRGs with the corresponding Cox coefficient (coe), as follows: Risk score=∑ni IRGi × coei, where i represents a certain specific IRG and n is the number of all IRGs. This was accomplished using R package ‘states’ (https://github.com/andybega/states; version 0.2.2).
The risk score of each patient in the TCGA cohort was calculated according to this formula. The cut-off value was set as the median risk score and all patients were divided into high- and low-risk groups based on this value. Subsequently, it was investigated whether the survival times were different between the two groups. Kaplan-Meier curves were plotted for the OS of patients with high-and low- risk scores. Survival outcomes were compared using the log-rank test. The receiver operating characteristic (ROC) analysis was used to evaluate the predictive performance of the IRGRS model, making a distinction between the high-risk and low-risk group. Areas under the curve (AUCs), optimal threshold values, sensitivity and specificity, were determined using the R package ‘survivalROC’ (https://CRAN.R-project.org/package=survivalROC; version 1.0.3), to evaluate the significance of the survival difference between the two groups. In order to demonstrate whether the IRGRS may act as an independent risk factor associated with survival, the IRGRS for patients with NK-AML were assessed using univariate and multivariate Cox analyses which were adjusted for age, sex and the mutation of NPM1 and FLT3. Among them, 60 years was the cut-off for age grouping. In addition, the correlations between IRGRS and Tregs were investigated were investigated, which were assessed using Spearman's correlation.
Tumor immunity relevance of the IRGRS
Regarding the close association between the IRGs and immune cells of the bone marrow microenvironment, the immune landscape was assessed in the present study. The abundance of 22 types of infiltrating immune cells of each patient with NK-AML in the TCGA cohort was estimated by translating the expression levels of genes downloaded from the TCGA cohort into the relative proportion of immune cells. This was achieved with the CIBERSORT algorithm based on the deconvolution, using the ‘CIBERSORT’ R package (CIBERSORT R script v1.03; http://cibersort.stanford.edu/). This tool was developed by Newman et al (21) and has been validated to quantify the abundance of specific cell types successfully. Inaccurate samples were eliminated (P≥0.05). The 22 types of immune cells contain B, regulatory T cells (Tregs), CD4+T, CD8+ T, natural killer, mast, plasma and dendritic cells, neutrophils, eosinophils and macrophages. The association between IRGRS and immune cell infiltration level in NK-AML was analyzed in the TCGA dataset.
Validation of the immune-associated risk score model for NK-AML
In order to evaluate the robustness of the IRGRS in the prognostication of patients with NK-AML, this prognostic model was tested in the GEO dataset GSE71014 as an independent validation set, using the same analytical method used for the TCGA cohort. Patients were divided into high- and low-risk groups with the median risk score of the TCGA utilized as the cut-off value. The GSE71014 dataset was not able to be used to validate the association between the prognostic risk models and the clinical characteristics due to a lack of clinical parameters.
Results
Construction of the IRGRS model and internal validation
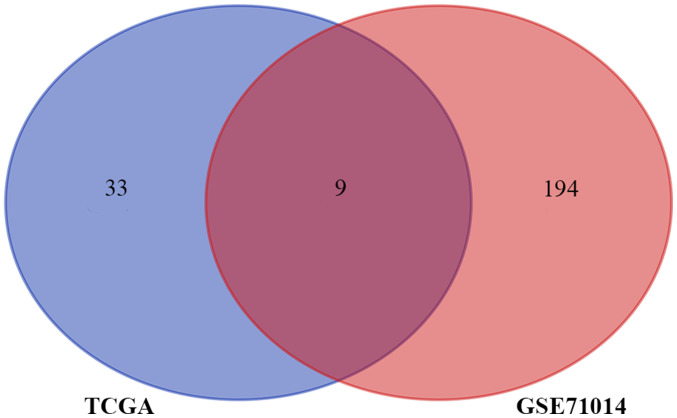
The aim of the present study was to construct an immune-associated prognostic model of NK-AML. A total of 164 patients with NK-AML were included in the two cohort studies. In the TCGA NK-AML cohort (n=60), 42 immune-associated prognostic genes were identified. In the 103 NK-AML samples of the GSE71014 cohort, the expression of 203 IRGs was significantly associated with OS (P<0.05). The Venn diagram represents the number of overlapping IRGs between the two cohorts (n=9; Fig. 2). These nine genes were zinc finger CCCH-type containing, antiviral 1 like (ZC3HAV1L), transferrin receptor (TFRC), suppressor of cytokine signaling 1 (SOCS1), ELAV like RNA binding protein 1 (ELAVL1), roundabout guidance receptor 3 (ROBO3), unc-93 homolog B1, TLR signaling regulator (UNC93B1), protein tyrosine phosphatase non-receptor type 6 (PTPN6), IL-2 receptor subunit alpha (IL2RA) and IL3RA. The hazard ratio (HR), 95% CI and P-value of univariate analyses for these nine IRGs in the TCGA cohort affecting OS are presented in Table I. In order to construct an IRG-based prognostic model to predict the risk of NK-AML, the nine prognostic IRGs were subjected to multivariate regression analysis using the Cox proportional hazards model for OS in the TCGA cohort. According to the corresponding coefficient value and expression value for each gene, the formula to calculate the risk score was as follows: IRGRS=[(−0.33298) × expression level of ZC3HAV1L] + [(−0.27376) × expression level of TFRC] + [(0.10993) × expression level of SOCS1] + [(−0.09726) × expression level of ELAVL1) + [(0.22322) × expression level of ROBO3] + [(−0.0229) × expression level of UNC93B1] + [(0.246114) × expression level of PTPN6] + [0.227133 × expression level of IL2RA] + [(0.217204) × expression level of IL3RA] (cut-off=1.006977).
Figure 2.
Venn diagram analysis of immune-related prognostic genes. TCGA, The Cancer Genome Atlas.
Table I.
Univariate Cox analysis for overall survival of the nine immune-related genes in The Cancer Genome Atlas cohort.
| Gene name | HR | 95% CI | P-value |
|---|---|---|---|
| ZC3HAV1L | 0.736 | 0.545–0.994 | 0.045 |
| TFRC | 0.633 | 0.450–0.889 | 0.008 |
| SOCS1 | 1.621 | 1.217–2.159 | 0.001 |
| ELAVL1 | 0.400 | 0.165–0.972 | 0.043 |
| ROBO3 | 1.329 | 1.042–1.695 | 0.022 |
| UNC93B1 | 1.494 | 1.098–2.034 | 0.011 |
| PTPN6 | 1.557 | 1.022–2.371 | 0.039 |
| IL2RA | 1.354 | 1.141–1.606 | 0.001 |
| IL3RA | 1.593 | 1.105–2.296 | 0.013 |
HR, hazard ratio; ZC3HAV1L, zinc finger CCCH-type containing, antiviral 1 like; TFRC, transferrin receptor; SOCS1, suppressor of cytokine signaling 1; ELAVL1, ELAV like RNA binding protein 1; ROBO3, roundabout guidance receptor 3; UNC93B1, unc-93 homolog B1, Toll-like receptor signaling regulator; PTPN6, protein tyrosine phosphatase non-receptor type 6; IL2RA, interleukin 2 receptor subunit alpha.
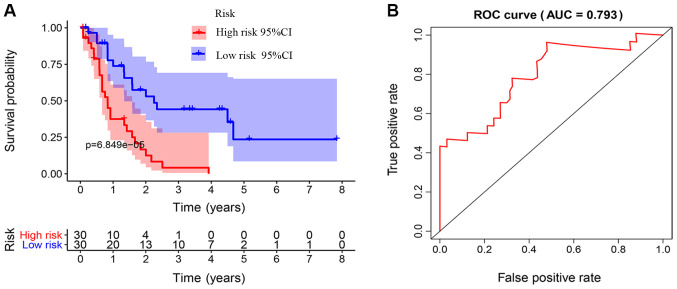
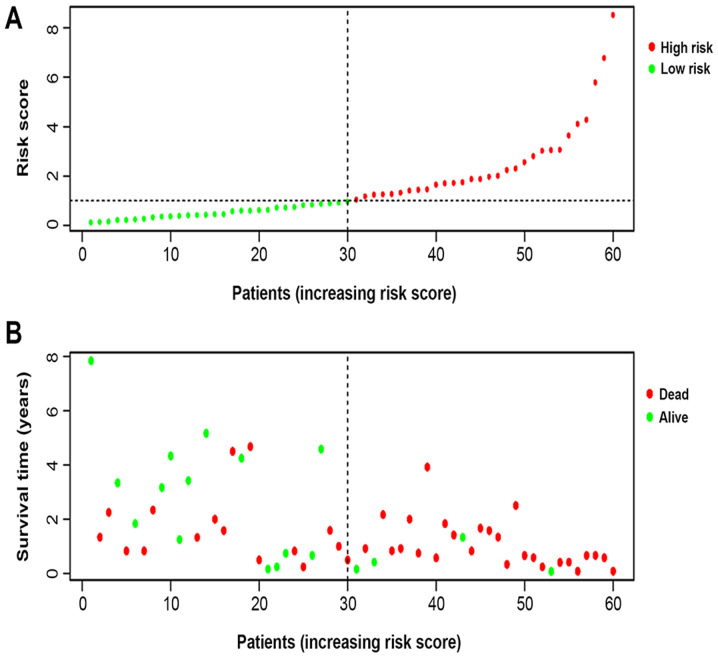
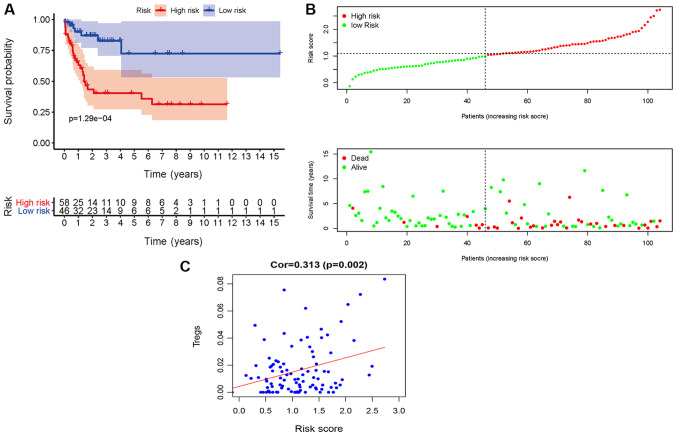
The Kaplan-Meier survival curves revealed that patients in the high-risk group exhibited a significantly lower OS rate than the low-risk group (P<0.01; Fig. 3A). The estimated 5-year OS rate was 8.0 vs. 54.9% for the high-risk and the low-risk group, respectively. The AUC of the ROC curve for OS prediction was 0.793, indicating a promising predictive value (Fig. 3B). In order to further investigate the impact of the IRGRS on the clinical outcomes, a risk curve was drawn. The prognostic model separates the NK-AML patients into two groups with discrete clinical outcomes (Fig. 4A). It also revealed that as the risk score increased, the survival time to decrease. (Fig. 4B). In the univariate regression analysis, the IRGRS was significantly associated with OS (HR, 1.749; 95% CI, 1.440–2.124; P<0.001). This result was consistent with that of the multivariate analysis (HR, 1.662; 95% CI, 1.321–2.091; P<0.001), which considered age, sex, the FAB classification, the type of NPM1 and the FLT3 genotype as co-effectors (Table II). These results demonstrated that the IRGRS is an independent predictor of OS in patients with NK-AML.
Figure 3.
Association of the IRGRS with survival. (A) Kaplan-Meier curves based on IRGRS in The Cancer Genome Atlas cohort. The blue and red shadows around the curves indicate the 95% confidence intervals. Patients with a high-risk score had unfavorable survival outcomes (P<0.001). (B) ROC curve of the IRGRS model for overall survival prediction. ROC, receiver operating characteristic; AUC, area under the ROC curve; IRGRS, immune-related genes risk score.
Figure 4.
Nine immune-related gene risk-score analysis of NK-AML patients in The Cancer Genome Atlas cohort. (A) Rank of risk score and distribution of groups, the X axis represented the number of patients, the Y axis represented their risk values and red and green points represent high and low risk, respectively. Each point represented one patient sorted by the rank of the risk score. (B) Survival status of patients in different groups. The below point showed risk score distribution, survival status and time until conclusion of the study in NK-AML patients. Red and green points represented dead and alive, respectively. NK-AML, acute myeloid leukemia with normal karyotype.
Table II.
Univariate and multivariate Cox analysis for overall survival of risk score and clinical parameters in The Cancer Genome Atlas cohort.
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age (≥60 years) | 1.023 | 1.004–1.041 | 0.016 | 1.018 | 0.998–1.038 | 0.074 |
| Sex (male) | 0.738 | 0.400–1.362 | 0.331 | 0.497 | 0.243–1.018 | 0.056 |
| Grade (0–7) | 1.160 | 0.954–1.411 | 0.136 | 0.987 | 0.778–1.252 | 0.913 |
| FLT3 (positive) | 3.004 | 1.578–5.719 | 0.001 | 3.434 | 1.469–8.028 | 0.004 |
| NPM1 (positive) | 0.918 | 0.497–1.694 | 0.784 | 0.391 | 0.185–0.828 | 0.014 |
| Risk score (>1.006977) | 1.749 | 1.440–2.125 | <0.001 | 1.662 | 1.321–2.091 | <0.001 |
HR, hazard ratio; FLT3, fms related receptor tyrosine kinase 3; NPM1, nucleophosmin 1.
Correlation of the IRGRS with clinical and immune characteristics
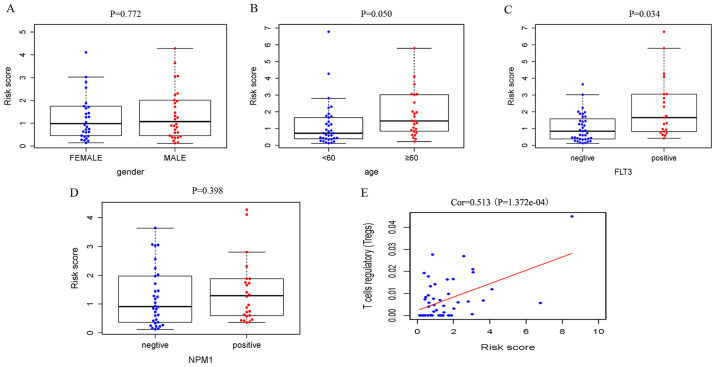
Relationships were analyzed between IRGRS model risk score and clinical characteristics, including age, sex and the mutations of NPM1 and FLT3. We also explored the relationships between nine prognostic IRGs expression and this clinical characteristics. (Table III). The risk scores in the elderly group and FLT3 mutant group were significantly higher compared with those in the non-elderly group and FLT3 wild-type group. It was revealed that a high risk score was associated with higher age (P=0.050) and a higher incidence of FLT3 mutation (P=0.034), and there was no significant association with sex (P=0.772) and NPM1 mutation (P=0.398; Fig. 5).
Table III.
Association between immune indices and demographic features and FLT3 and NPM1 mutation.
| Age, ≥60 vs. <60 years | Sex, male vs. female | FLT3, mutant vs. wildtype | NPM1, mutant vs. wildtype | |||||
|---|---|---|---|---|---|---|---|---|
| Gene | t | P-value | t | P-value | t | P-value | t | P-value |
| ZC3HAV1L | 1.879 | 0.067 | −0.355 | 0.724 | 0.721 | 0.475 | 1.359 | 0.180 |
| TFRC | 0.394 | 0.695 | 0.030 | 0.976 | 3.277 | 0.002 | 3.796 | <0.001 |
| SOCS1 | −2.86 | 0.006 | 0.752 | 0.455 | −1.579 | 0.123 | −0.678 | 0.501 |
| ELAVL1 | 1.332 | 0.191 | −1.349 | 0.183 | 1.790 | 0.080 | 1.595 | 0.116 |
| ROBO3 | −2.543 | 0.014 | −1.776 | 0.081 | −0.875 | 0.386 | 1.916 | 0.060 |
| UNC93B1 | −1.733 | 0.089 | −0.498 | 0.621 | −1.266 | 0.213 | −0.484 | 0.630 |
| PTPN6 | −1.341 | 0.186 | −0.046 | 0.964 | −1.611 | 0.116 | −1.736 | 0.089 |
| IL2RA | −1.142 | 0.258 | 0.373 | 0.710 | −2.494 | 0.018 | −0.68 | 0.500 |
| IL3RA | −0.851 | 0.399 | 0.604 | 0.549 | −0.526 | 0.601 | 1.019 | 0.312 |
| Risk score | −2.02 | 0.050 | 0.291 | 0.772 | −2.208 | 0.034 | −0.852 | 0.398 |
Mutations in NPM1 and FLT3 and age ≥60 were associated with overall survival. Patients were divided into two groups according to age (≥60 vs. <60), Sex (male vs. female), FLT3 (mutant vs. wildtype), NPM1 (mutant vs. wildtype). t: t value of Student's t-test; P: P-value of Student's t-test; FLT3, fms related receptor tyrosine kinase 3; NPM1, nucleophosmin 1; ZC3HAV1L, zinc finger CCCH-type containing, antiviral 1 like; TFRC, transferrin receptor; SOCS1, suppressor of cytokine signaling 1; ELAVL1, ELAV like RNA binding protein 1; ROBO3, roundabout guidance receptor 3; UNC93B1, unc-93 homolog B1, Toll-like receptor signaling regulator; PTPN6, protein tyrosine phosphatase non-receptor type 6; IL2RA, interleukin 2 receptor subunit alpha.
Figure 5.
Associations between the immune-related genes risk score and clinical factors. (A) Distribution of the risk score in female and male group. (B) Distribution of the risk score for the ≥60 years old and <60 years old group. (C and D) Distribution of the risk score in different mutation status of FLT3 and NPM1. Data presented as box plots, showing the lower line representing 25th percentile, highest line representing the 75th percentile. The horizontal line in the middle is the median (50th percentile). Dots past the ± bars indicate outliers outside of 1.5× the interquartile range. (E) The correlation between Tregs and the risk score was performed by using Pearson correlation analysis. FLT3, fms related receptor tyrosine kinase 3; Tregs, T-regulatory cells; NPM1, nucleophosmin 1; Corr, correlation coefficient.
For the immunity relevance analysis of the IRGRS, 50 samples were available from the TCGA database, whose relative content of immune cells had been precisely estimated. A correlation analysis of the ratio of infiltrating immune cells and the IRGRS was performed (Table IV). In TCGA, significant correlations were identified between the IRGRS and monocytes (P=0.014), NK resting cells (P=0.028) and Tregs (P<0.001). As presented in Figs. 5E and 6C, a significantly positive correlation was observed between the amount of Tregs and the IRGRS in both TCGA and GEO databases (P<0.05).
Table IV.
Correlation between immune scores and immune cell fractions.
| TCGA cohort | GEO cohort | |||
|---|---|---|---|---|
| Immune cell type | Coef | P-value | Coef | P-value |
| B memory cells | −0.200 | 0.163 | 0.083 | 0.421 |
| Macrophages M0 | −0.166 | 0.250 | 0.006 | 0.957 |
| Macrophages M1 | −0.084 | 0.562 | −0.096 | 0.347 |
| Macrophages M2 | −0.040 | 0.780 | −0.105 | 0.306 |
| Monocytes | 0.345 | 0.014 | 0.086 | 0.403 |
| NK activated cells | 0.050 | 0.729 | 0.138 | 0.179 |
| NK resting cells | −0.311 | 0.028 | −0.001 | 0.993 |
| Plasma cells | −0.247 | 0.084 | −0.209 | 0.039 |
| Activated CD4+ | −0.082 | 0.572 | 0.081 | 0.431 |
| T memory cells | ||||
| Resting CD4+ | −0.133 | 0.356 | −0.115 | 0.263 |
| T memory cells | ||||
| CD4+ naïve T cells | −0.124 | 0.391 | −0.074 | 0.474 |
| CD8+ T cells | −0.011 | 0.941 | 0.047 | 0.647 |
| T follicular helper cells | −0.127 | 0.380 | −0.040 | 0.697 |
| T γ Δ cells | −0.084 | 0.561 | −0.308 | 0.002 |
| T regulatory cells | 0.513 | <0.001 | 0.313 | 0.002 |
| B naïve cells | 0.009 | 0.952 | −0.003 | 0.978 |
| Activated dendritic cells | −0.121 | 0.238 | ||
| Resting dendritic cells | 0.268 | 0.008 | ||
| Eosinophils | 0.038 | 0.708 | ||
| Activated mast cells | −0.165 | 0.105 | ||
| Resting mast cells | −0.046 | 0.653 | ||
| Neutrophils | −0.009 | 0.932 | ||
Coef, correlation coefficient; NK, natural killer; TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus.
Figure 6.
Validation of the IRGRS model in the dataset GSE71014. (A) Survival analysis based on IRGRS. Survival curve showed that patients with high-risk score were correlated with a trend toward worse survival outcomes. (B) Distribution of the risk score and the associated survival data and mRNA expression heatmap. The x-axis represented the number of patients. Each column represented the same patient corresponded to the below point showing risk score distribution, survival status and time in NK-AML patients. (C) Associations between the IRGRS and the infiltrating Tregs.
Validation in the GEO database
Finally, the predictive model based on the IRGRS for NK-AML was further validated in the GSE71014 cohort. Application of the model to the validation cohort in a survival analysis was able to distinctly discriminate patients with different survival time according to their risk score value (Fig. 6A and B). Furthermore, in this cohort, Tregs were positively correlated with the IRGRS (P=0.002; Fig. 6C). Plasma cells, T γ Δ cells and resting dendritic cells were also correlated with the IRGRS. Taken together, the data of the present study indicated that the predictive IRGRS model was of great prognostic value in NK-AML.
Discussion
The significance of IRGs in AML progression and immunotherapeutics have been reported previously (22,23). However, to the best of our knowledge, no comprehensive, genome-wide profiling analysis of the prognostic significance of all IRGs in NK-AML has so far been performed. Thus, the expression levels of IRGs in 164 patients with NK-AML were analyzed in the present study and the prognostic value of IRGs was subsequently assessed. A total of 42 and 203 IRGs with prognostic value were identified from the TCGA and GEO cohort, respectively. Between the two cohorts, nine overlapping IRGs with prognostic value in NK-AML were identified. Due to the limited power of individual indicators to predict prognosis, multivariate Cox regression analysis was used to analyze the prognostic significance of nine IRGs and their combination in NK-AML. The prognostic assessment model used in the present study was combined based on the regression coefficient and expression value of each of those nine IRGs. The AUC of the ROC curve was 0.793, suggesting moderate potential for evaluating the prognosis of patients with NK-AML based on the IRGRS model. The present study then validated the predictive capability of the risk model in another independent cohort, namely the 104 patients from GSE71014. The subgroup of patients with NK-AML with high risk scores exhibited poor survival. The IRGs-based risk score was demonstrated to be of high prognostic value to predict the survival of patients with NK-AML in the clinic and it provided valuable biomarkers for this disease.
In addition, the present study also determined that the nine IRGs constituting the model exhibited distinct risk-associated patterns, including 6 relatively high-risk genes (SOCS1, ROBO3, UNC93B1, PTPN6, IL2RA and IL3RA) and 3 relatively low-risk genes (ZC3HAV1L, TFRC and UNC93B1). To the best of our knowledge, the ZC3HAV1L gene has not been extensively investigated; whereas its paralog ZC3HAV1 has been reported to participate in the innate antiviral immune process (24). TFRC, a gene that takes part in iron uptake, has the role of regulating the proliferation of T and B cells. Although it is not considered relevant to the prognosis of numerous different types of cancer, the prognostic value of TRFC in AML remains controversial (25–27). SOCS1 is involved in the negative regulation of cytokines in the JAK/STAT3 signaling pathway. A study by Hou et al (28) revealed that its high expression was indicative of unfavorable survival prognosis. ELAVL1, encoding RNA-binding proteins, has been studied in a variety of tumor types. It has been demonstrated to regulate the expression of tumor protein 53 to mediate anti-proliferative activity and has an important role in the differentiation of myeloid cells in AML (29). ROBO3, an immunoglobulin transmembrane receptor gene, is involved in the SLIT/ROBO pathway associated with the development and progression of certain types of cancer, where AML is also included (30,31). UNC9381 was reported to be implicated in the innate and adaptive immune response by regulating the TLR pathway, whose activation has been observed in tumors (32,33). To the best of our knowledge, the involvement of the UNC9381 gene in AML has not been previously reported. PTPN6 is known to regulate processes of cell proliferation and differentiation, the mitotic cycle and oncogenic transformation. The implication of PTPN6 in AML has gradually attracted increasing attention. It is primarily expressed in hematopoietic cells and may have a marked effect on the pathogenesis of leukemia (34). IL2RA is a receptor for IL-2, which is involved in the regulation of immune tolerance by controlling the activity of Tregs, whose expression was recognized as a poor prognostic marker of leukemia (15,35,36). IL3RA, also named CD123, regulates the development of hematopoietic and immune cells. Its high expression has been indicated to be associated with poor prognosis in AML (37). However, the underlying molecular mechanisms of the nine IRGs corresponding to their potential clinical value in NK-AML require further investigation.
Poor prognosis in elderly patients and patients with FLT3 mutations has been widely recognized (38). Of note, the results of the present study revealed that the IRG model was associated with age and the FLT3 mutation. An obviously higher proportion of older age over 60 and FLT3 mutation in patients with NK-AML was observed in the higher risk score group. Furthermore, the IRG model may not only serve as a prognostic indicator but also as an immune status indicator. The present study investigated the associations between the IRG model and immune cell infiltration in order to reflect the status of the immune microenvironment of NK-AML. In TCGA cohort, the levels of infiltrating monocytes and Tregs were positively correlated with IRGRS, and NK resting cells were negatively correlated with IRGRS. In the GEO cohort, resting dendritic cells, T γ Δ cells and Tregs were positively correlated with IRGRS, whilst plasma cells and T γ Δ cells were negatively correlated with IRGRS. The level of infiltrating Tregs was significantly positively correlated with IRGRS in two cohorts. The percentage of circulating Tregs was higher in patients with AML compared with that in normal controls and Tregs accumulating in the peripheral circulation vigorously mediate immune suppression (38). Patients with lower Treg frequency at diagnosis have a better response to induction chemotherapy (16,39). It has been reported that an increased amount of Tregs is associated with an elevated risk of relapse in AML (40). The results of the present study also suggested that the IRGRS model has the potential to serve as a predictor for increased immune cell infiltration. The role of immune cells in AML has remained to be fully investigated. Radpour et al (41) indicated that CD8+ T cells expand stem and progenitor cells in low- but not high-risk AML. Immune cells with statistical differences only in the TCGA or GEO cohort could not determine their correlation with risk models. The role of immune cells in acute myeloid leukemia still requires further studies with larger samples. The preliminary observation of the present study may provide a perspective for further investigation, which is required in the future.
However, there were certain limitations to the present study. First, the potential mechanism of these nine IRGs and Treg cell-mediated immune infiltration in NK-AML progression remains elusive and further in vitro or in vivo experiments are required to clarify this. Furthermore, transcriptome analysis is not able to reflect the global immune status. In addition, the validity of the prognostic model based on the nine IRGs identified in the present study requires to be verified using more independent samples.
In conclusion, in the present study, the clinical and prognostic value of IRGs in NK-AML was systematically analyzed. An IRG-based prognostic evaluation model was constructed with a moderate level of performance in the prognostication of patients with NK-AML. The results of the present study provide novel approaches for immunotherapy for patients with NK-AML.
Acknowledgements
Not applicable.
Funding
The present study was funded by Youth Support Project of Luhe Hospital (grant no. LHYH2019-JC06).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request. The raw datasets used during the current study are available in the corresponding repositories, including the TCGA (https://tcga-data.nci.nih.gov/tcga/), GEO (ncbi.nlm.nih.gov/geo/), UCSC Xena (https://xenabrowser.net/) and the ImmPort database (https://immport.niaid.nih.gov).
Authors' contributions
XD and DZ conceived and designed the study. XD, XC, YoZ, XZ and JZ analyzed the data. DZ, TC, and YuZ analyzed the data and generated the figures. XD and HZ made contributions to analysis and interpretation of data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. The medical research council adult and children's leukaemia working parties. Blood. 1998;92:2322–2333. doi: 10.1182/blood.V92.7.2322. [DOI] [PubMed] [Google Scholar]
- 2.Kihara R, Nagata Y, Kiyoi H, Kato T, Yamamoto E, Suzuki K, Chen F, Asou N, Ohtake S, Miyawaki S, et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia. 2014;28:1586–1595. doi: 10.1038/leu.2014.55. [DOI] [PubMed] [Google Scholar]
- 3.Song Y, Zhang W, He X, Liu X, Yang P, Wang J, Hu K, Liu W, Zhang X, Jing H, Yuan X. High NCALD expression predicts poor prognosis of cytogenetic normal acute myeloid leukemia. J Transl Med. 2019;17:166. doi: 10.1186/s12967-019-1904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu L, Shi J, Hu K, Wang J, Wang W, Ke X. Mitogen-activated protein kinase binding protein 1 (MAPKBP1) is an unfavorable prognostic biomarker in cytogenetically normal acute myeloid leukemia. Oncotarget. 2015;6:8144–8154. doi: 10.18632/oncotarget.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu L, Fu H, Qiao J, Pang Y, Xu K, Zhou L, Wu Q, Li Z, Ke X, Xu K, Shi J. High expression of CPNE3 predicts adverse prognosis in acute myeloid leukemia. Cancer Sci. 2017;108:1850–1857. doi: 10.1111/cas.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu L, Fu H, Tian L, Xu K, Hu K, Wang J, Wang J, Jing H, Shi J, Ke X. High expression of RUNX1 is associated with poorer outcomes in cytogenetically normal acute myeloid leukemia. Oncotarget. 2016;7:15828–15839. doi: 10.18632/oncotarget.7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn JS, Kim HJ, Kim YK, Lee SS, Ahn SY, Jung SH, Yang DH, Lee JJ, Park HJ, Lee JY, et al. Assessment of a new genomic classification system in acute myeloid leukemia with a normal karyotype. Oncotarget. 2017;9:4961–4968. doi: 10.18632/oncotarget.23575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, Paietta E, Willman CL, Head DR, Rowe JM, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group study. Blood. 2000;96:4075–4083. doi: 10.1182/blood.V96.13.4075.h8004075_4075_4083. [DOI] [PubMed] [Google Scholar]
- 9.Bullinger L, Döhner K, Bair E, Fröhling S, Schlenk RF, Tibshirani R, Döhner H, Pollack JR. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 10.Teague RM, Kline J. Immune evasion in acute myeloid leukemia: Current concepts and future directions. J Immunother Cancer. 2013;1:1. doi: 10.1186/2051-1426-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Austin R, Smyth MJ, Lane SW. Harnessing the immune system in acute myeloid leukaemia. Crit Rev Oncol Hematol. 2016;103:62–77. doi: 10.1016/j.critrevonc.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 12.Nishioka C, Ikezoe T, Pan B, Xu K, Yokoyama A. MicroRNA-9 plays a role in interleukin-10-mediated expression of E-cadherin in acute myelogenous leukemia cells. Cancer Sci. 2017;108:685–695. doi: 10.1111/cas.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Correa B, Bergua JM, Pera A, Campos C, Arcos MJ, Bañas H, Duran E, Solana R, Tarazona R. In vitro culture with interleukin-15 leads to expression of activating receptors and recovery of natural killer cell function in acute myeloid leukemia patients. Front Immunol. 2017;8:931. doi: 10.3389/fimmu.2017.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia S, Verga JA, Stephansky J, Pastika TJ, Story JL, et al. Single-cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019;176:1265–1281. doi: 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakase K, Kita K, Kyo T, Tsuji K, Katayama N. High serum levels of soluble interleukin-2 receptor in acute myeloid leukemia: Correlation with poor prognosis and CD4 expression on blasT cells. Cancer Epidemiol. 2012;36:e306–e309. doi: 10.1016/j.canep.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Q, Munger ME, Highfill SL, Tolar J, Weigel BJ, Riddle M, Sharpe AH, Vallera DA, Azuma M, Levine BL, et al. Program death-1 signaling and regulatory T cells collaborate to resist the function of adoptively transferred cytotoxic T lymphocytes in advanced acute myeloid leukemia. Blood. 2010;116:2484–2493. doi: 10.1182/blood-2010-03-275446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachli EB, Schaer DJ, Walter RB, Fehr J, Schoedon G. Functional expression of the CD163 scavenger receptor on acute myeloid leukemia cells of monocytic lineage. J Leukoc Biol. 2006;79:312–318. doi: 10.1189/jlb.0605309. [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Network. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ, Jr, Laird PW, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang MK, Chiu YC, Chou WC, Hou HA, Tseng MH, Kuo YY, Chen Y, Chuang EY, Tien HF. An mRNA expression signature for prognostication in de novo acute myeloid leukemia patients with normal karyotype. Oncotarget. 2015;6:39098–39110. doi: 10.18632/oncotarget.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J, et al. ImmPort: Disseminating data to the public for the future of immunology. Immunol Res. 2014;58:234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 21.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramzi M, Khalafi-Nezhad A, IravaniSaadi M, Jowkar Z. Association between TLR2 and TLR4 expression and response to induction therapy in acute myeloid leukemia patients. Int J Hematol Oncol Stem Cell Res. 2018;12:303–312. [PMC free article] [PubMed] [Google Scholar]
- 23.Yan H, Qu J, Cao W, Liu Y, Zheng G, Zhang E, Cai Z. Identification of prognostic genes in the acute myeloid leukemia immune microenvironment based on TCGA data analysis. Cancer Immunol Immunother. 2019;68:1971–1978. doi: 10.1007/s00262-019-02408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 25.Wu B, Shi N, Sun L, Liu L. Clinical value of high expression level of CD71 in acute myeloid leukemia. Neoplasma. 2016;63:809–815. doi: 10.4149/neo_2016_519. [DOI] [PubMed] [Google Scholar]
- 26.Kollia P, Samara M, Stamatopoulos K, Belessi C, Stavroyianni N, Tsompanakou A, Athanasiadou A, Vamvakopoulos N, Laoutaris N, Anagnostopoulos A, Fassas A. Molecular evidence for transferrin receptor 2 expression in all FAB subtypes of acute myeloid leukemia. Leuk Res. 2003;27:1101–1103. doi: 10.1016/S0145-2126(03)00100-0. [DOI] [PubMed] [Google Scholar]
- 27.Kollia P, Stavroyianni N, Stamatopoulos K, Zoi K, Viniou N, Mantzourani M, Noguchi CT, Paterakis G, Abazis D, Pangalos C, et al. Molecular analysis of transferrin receptor mRNA expression in acute myeloid leukaemia. Br J Haematol. 2001;115:19–24. doi: 10.1046/j.1365-2141.2001.03065.x. [DOI] [PubMed] [Google Scholar]
- 28.Hou HA, Lu JW, Lin TY, Tsai CH, Chou WC, Lin CC, Kuo YY, Liu CY, Tseng MH, Chiang YC, et al. Clinico-biological significance of suppressor of cytokine signaling 1 expression in acute myeloid leukemia. Blood Cancer J. 2017;7:e588. doi: 10.1038/bcj.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Annabi B, Currie JC, Moghrabi A, Béliveau R. Inhibition of HuR and MMP-9 expression in macrophage-differentiated HL-60 myeloid leukemia cells by green tea polyphenol EGCg. Leuk Res. 2007;31:1277–1284. doi: 10.1016/j.leukres.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Han S, Cao C, Tang T, Lu C, Xu J, Wang S, Xue L, Zhang X, Li M. ROBO3 promotes growth and metastasis of pancreatic carcinoma. Cancer Lett. 2015;366:61–70. doi: 10.1016/j.canlet.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Narayan G, Goparaju C, Arias-Pulido H, Kaufmann AM, Schneider A, Dürst M, Mansukhani M, Pothuri B, Murty VV. Promoter hypermethylation-mediated inactivation of multiple Slit-Robo pathway genes in cervical cancer progression. Mol Cancer. 2006;5:16. doi: 10.1186/1476-4598-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koehn J, Huesken D, Jaritz M, Rot A, Zurini M, Dwertmann A, Beutler B, Korthäuer U. Assessing the function of human UNC-93B in toll-like receptor signaling and major histocompatibility complex II response. Hum Immunol. 2007;68:871–878. doi: 10.1016/j.humimm.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Goto Y, Arigami T, Kitago M, Nguyen SL, Narita N, Ferrone S, Morton DL, Irie RF, Hoon DS. Activation of toll-like receptors 2, 3, and 4 on human melanoma cells induces inflammatory factors. Mol Cancer Ther. 2008;7:3642–3653. doi: 10.1158/1535-7163.MCT-08-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beghini A, Ripamonti CB, Peterlongo P, Roversi G, Cairoli R, Morra E, Larizza L. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet. 2000;9:2297–2304. doi: 10.1093/oxfordjournals.hmg.a018921. [DOI] [PubMed] [Google Scholar]
- 35.Terwijn M, Feller N, van Rhenen A, Kelder A, Westra G, Zweegman S, Ossenkoppele G, Schuurhuis GJ. Interleukin-2 receptor alpha-chain (CD25) expression on leukaemic blasts is predictive for outcome and level of residual disease in AML. Eur J Cancer. 2009;45:1692–1699. doi: 10.1016/j.ejca.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 36.Fujiwara SI, Muroi K, Yamamoto C, Hatano K, Okazuka K, Sato K, Oh I, Ohmine K, Suzuki T, Ozawa K. CD25 as an adverse prognostic factor in elderly patients with acute myeloid leukemia. Hematology. 2017;22:347–353. doi: 10.1080/10245332.2016.1276240. [DOI] [PubMed] [Google Scholar]
- 37.Testa U, Riccioni R, Diverio D, Rossini A, Coco FL, Peschle C. Interleukin-3 receptor in acute leukemia. Leukemia. 2004;18:219–226. doi: 10.1038/sj.leu.2403224. [DOI] [PubMed] [Google Scholar]
- 38.Fey MF, Buske C, ESMO Guidelines Working Group Acute myeloblastic leukaemias in adult patients: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):vi138–vi143. doi: 10.1093/annonc/mdt320. [DOI] [PubMed] [Google Scholar]
- 39.Szczepanski MJ, Szajnik M, Czystowska M, Mandapathil M, Strauss L, Welsh A, Foon KA, Whiteside TL, Boyiadzis M. Increased frequency and suppression by regulatory T cells in patients with acute myelogenous leukemia. Clin Cancer Res. 2009;15:3325–3332. doi: 10.1158/1078-0432.CCR-08-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams P, Basu S, Garcia-Manero G, Hourigan CS, Oetjen KA, Cortes JE, Ravandi F, Jabbour EJ, Al-Hamal Z, Konopleva M, et al. The distribution of T-cell subsets and the expression of immune checkpoint receptors and ligands in patients with newly diagnosed and relapsed acute myeloid leukemia. Cancer. 2019;125:1470–1481. doi: 10.1002/cncr.31896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radpour R, Riether C, Simillion C, Höpner S, Bruggmann R, Ochsenbein AF. CD8+ T cells expand stem and progenitor cells in favorable but not adverse risk acute myeloid leukemia. Leukemia. 2019;33:2379–2392. doi: 10.1038/s41375-019-0441-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request. The raw datasets used during the current study are available in the corresponding repositories, including the TCGA (https://tcga-data.nci.nih.gov/tcga/), GEO (ncbi.nlm.nih.gov/geo/), UCSC Xena (https://xenabrowser.net/) and the ImmPort database (https://immport.niaid.nih.gov).