Abstract
Approximately 500,000 new head and neck squamous cell carcinoma (HNSCC) cases are detected every year around the world, and its incidence ranks sixth among all cancer types globally. Among these cases, oral squamous cell carcinoma (OSCC) and laryngeal squamous cell carcinoma (LSCC) are HNSCC subtypes with high incidence rates, especially in China. The present study examines the association between the apolipoprotein L1 (APOL1) mRNA and protein expression and clinical parameters in HNSCC. The two most common types (oral and larynx) of HNSCC were selected for subgroup analyses. Immunohistochemistry (IHC) was used to detect APOL1 protein expression levels in HNSCC clinical specimens. It was demonstrated that APOL1 protein expression in 221 cases of HNSCC was higher compared with that in normal tissues. Consistent upregulation of APOL1 protein was also found in subgroups of OSCC and LSCC. Through mining the ArrayExpress, The Cancer Genome Atlas and the Gene Expression Omnibus databases, microarrays and RNA sequencing data for HNSCC were retrieved, which were used to analyze APOL1 mRNA expression levels. The results showed that APOL1 expression was higher in both OSCC and LSCC subtypes, as well as in HNSCC, compared with that in non-cancerous squamous epithelium. The summary receiver operating characteristic analysis showed that APOL1 had potential as a diagnostic biomarker for HNSCC, OSCC and LSCC. Thus, upregulation of APOL1 may contribute to the tumorigenesis of HNSCC.
Keywords: apolipoprotein L1, head and neck squamous cell carcinoma, subgroups, clinical significance
Introduction
Head and neck squamous cell carcinoma (HNSCC), representing a group of tumors in the larynx, pharynx and oral cavity, is the sixth most common cancer in the world, with ~500,000 new cases worldwide each year (1,2). Among these cases, oral squamous cell carcinoma (OSCC) and laryngeal squamous cell carcinoma (LSCC) have a high incidence, especially in China (3,4), exhibiting high morbidity and mortality rates (5). According to GLOBOCAN 2012, the incidence and mortality rate of laryngeal cancer in China was 1.1/100,000 and 0.7/100,000, compared with the global incidence and mortality rate of which was 2.1/100,000 and 1.1/100,000, respectively (6). Oral cancer was also reported to be the 11th most common malignancy in the world in 2017 (7,8). HNSCC is associated with smoking and alcohol abuse (9), and it is also linked to infection with the human papillomavirus (10). In the occurrence and development of HNSCC, epigenetic events play an important role, including DNA methylation, post-translational covalent modification of histones, chromatin remodeling and the effects of non-coding RNA (11–14). Currently, an increasing number of splicing patterns have been found, such as the splicing variants of laminin subunit α3 (LAMA3), dystonin (DST), dedicator of cytokinesis (DOCK5), lysyl oxidase-like 2 (LOXL2) and gelsolin (GSN), and some studies have shown that these splicing variants can be used as cancer markers and potential therapeutic targets (15–18). Some of these variants could even become novel antigens for specific targeted therapy of HNSCC, such as the splicing variant of GSN, which was reported to be able to serve as a biomarker of HNSCC and a neoantigen for HNSCC treatment (18). In addition, HNSCC is characterized by clinical heterogeneity (19). For instance, patients with recurrent and/or metastatic HSNCC can be treated with cetuximab, an anti-EGFR antibody, but only ~13% of patients with metastasis respond to this therapy (20). In recent years, immune checkpoint inhibitors (ICIs), such as nivolumab and pembrolizumab, have shown certain potential in the treatment of HNSCC (21,22). However, primary resistance to ICIs emerges in the majority of patients, and the benefits of this therapy is therefore suboptimal (23) compared with the anti-programmed death-1 monoclonal antibody (24,25). Therefore, it is particularly important to identify novel predictive biomarkers to improve the screening of patients who can benefit from ICI treatment. At present, more and more evidence has shown that the progression and metastasis of HNSCC are associated with polygenic changes (26,27); thus, it is important to improve our understanding of the mechanisms underlying the pathogenesis of HNSCC to aid the identification of novel biomarkers.
Apolipoprotein L1 (APOL1) is located at chromosome 22q12.3 and APOL1 gene encodes a trypanolytic factor that dissolves pathogenic Trypanosoma brucei subspecies in humans and gorillas (28). Other studies have shown a link between non-diabetic nephropathy and variations in the APOL1 gene (29,30), which are also associated with atherosclerosis (31). APOL1 is one of six genes of the APOL gene family, which are a set of genomic hotspots for various diseases (32), including schizophrenia, cancer and chronic kidney disease (33,34). The APOL gene family is associated with programmed cell death and mainly encodes proteins to initiate apoptosis or autophagic death by increasing levels of free radicals, aggregating macromolecules, and inducing DNA damage and metabolic defects (35). Therefore, protein damage caused by apoptosis and autophagy is associated with the pathogenesis of several cancer types (36,37). However, there is no relevant literature to report the expression of APOL1 and its clinical significance in HNSCC, to the best of our knowledge.
By mining the high-throughput data from ArrayExpress, Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA), Human Protein Atlas (HPA), as well as using immunohistochemistry (IHC), the present study examined the associations between APOL1 mRNA and protein expression in HNSCC and related clinical parameters and prognoses. The two most common sites (oral and larynx) of HNSCC were selected for subgroup analyses (Fig. 1). Nasopharyngeal carcinoma was excluded from subgroup analysis due to its unique pathogenesis (38). The present results may improve our understanding of the association between HNSCC and APOL1 that has hitherto been lacking in oncology, and suggested that APOL1 is a promising biomarker that deserves further study.
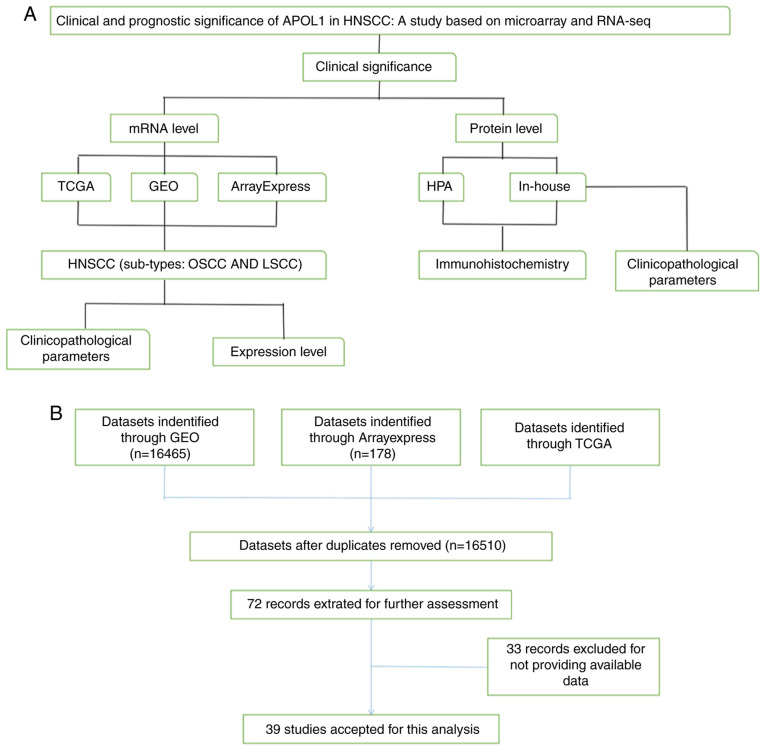
Figure 1.
Design and retrieval flow chart. (A) Flow chart of the design of the present study and (B) flow chart of the study search and selection proceed. APOL1, apolipoprotein L1; HNSCC, head and neck squamous cell carcinoma; OSCC, oral squamous cell carcinoma, LSCC, laryngeal squamous cell carcinoma; TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; HPA, Human Protein Atlas; RNA-seq; RNA sequencing.
Materials and methods
Study design
At the mRNA level, datasets, including 16,510 individual data points after removing duplicates, were obtained through TCGA (https://cancergenome.nih.gov/), GEO (http://www.ncbi.nlm.nih.gov/geo/), and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) databases. In total, 39 studies were used for the final analysis, which were divided into two subgroups: OSCC and LSCC, such as the datasets of GSE10121 (39), GSE13601 (40), GSE51985 (41). The control group included negative marginal tissue with squamous cells, healthy nasopharyngeal tissue and the epithelial tonsil samples. At the protein level, images of IHC staining results were obtained from the HPA (42–44). In-house validation with IHC was also used to verify the APOL1 mRNA expression data collected from the First Affiliated Hospital of Guangxi Medical University (Nanning, China). The inclusion criteria were: i) None of patients had received preoperative chemoradiotherapy and ii) all of the operative tissues were obtained during resection. The study exclusion criteria were as follows: i) Multiple primary tumors and ii) lack of clinicopathological data.
IHC
The HPA, through the use of IHC techniques, aims to provide information on the distribution of various kinds of human proteins in cells and tissues and the staining results of images that are fully representative (42–44). Thus, this database was used to collect images of IHC staining results for HNSCC. After obtaining approval from The Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (Nanning, China), as well as patient written consent, HNSCC, non-tumor tissues and corresponding clinical information were collected at the hospital between January 2017 and September 2018. IHC was then performed on these tissues, including 221 HNSCC and 24 non-tumor squamous epithelial tissues from 13 cases of the oral cavity, five of the larynx and six of the nasopharynx. All clinicopathological features were obtained to analyze the influence of the APOL1 protein expression on the occurrence and development of HNSCC.
Pathological sectioning was performed on both the HNSCC and non-cancerous tissues, which were then fixed in 4% formaldehyde solution for 24 h at room temperature, followed by being dehydrated with conventional gradient ethanol (80, 90, 95 and 100%) and embedded. Paraffin-embedded HNSCC and non-cancerous tissues were cut into 4-µm sections and heated at 75°C for 2 h. The sections were deparaffined with xylene and rehydrated with conventional gradient ethanol (100, 95, 85, 75 and 50%). Then these sections were washed twice under running water and boiled in EDTA antigen Retrieval Solution (pH=9.0) for 2.5 min in a pressure cooker for antigen retrieval. After that, the sections were immersed in 3% hydrogen peroxide for 5 min to block endogenous peroxidase activity and incubated with diluted anti-APOL1 antibody (1:100, cat. no. CL0171; Abcam) for 90 min at 37°C and with a secondary antibody for 25 min at room temperature. After the sections were incubated with DAB staining for 5 min at room temperature and counterstained with hematoxylin for 30 sec at room temperature. Finally, these sections were dehydrated with gradient ethanol, cleared by xylene, and sealed with resin. Images (magnification, ×100 and ×200) were captured using an optical microscope (Motic China Group Co., Ltd.). The score of the IHC staining results, including HPA and in-house IHC, was generated from the product of the proportion of stained cancerous cells among all cells (0, ≤5%; 1, 6–25%; 2, 26–50%; 3, 51–75%; 4, >75%) and the staining degree of the positive cancerous cells (0, no staining; 1, light staining; 2, moderate staining; 3, strong staining), which was assessed manually by two independent pathologists.
Microarray and RNA-sequencing data from TCGA, GEO and ArrayExpress databases
By mining TCGA, GEO and ArrayExpress databases, microarrays and RNA-sequencing (RNA-seq) data regarding HNSCC were retrieved. The search strategy was (head and neck OR HNSCC OR laryngeal OR nasopharyngeal OR pharyngeal OR oral OR LSCC OR NPC OR OSCC) AND (squamous cell carcinoma OR carcinoma OR tumor OR cancer OR neoplas* OR malignan*). The inclusion criteria for the microarray and RNA-seq data were as follows: i) Cancer samples came from HNSCC tissues; ii) the species was Homo sapien; iii) the included chips contained the expression APOL1; iv) the tissue sample sources were head and neck tissues and v) chip sequencing or other high-throughput detection methods were used. The downloaded data were normalized and log2 transformed. Basic information of the 39 microarrays of APOL1 expression profiling involved in the present study is summarized at Table I. There are 21 microarrays with APOL1 expression for OSCC, 4 for LSCC, 3 for NPC and 11 for unclassified HNSCC tissues. The mRNA expression of APOL1 in HNSCC and normal tissues were obtained from above databases.
Table I.
Basic information of the 39 microarrays of APOL1 expression profiling involved in the present study.
| Number of samples, n | APOL1 expression, mean ± SD | ||||||
|---|---|---|---|---|---|---|---|
| ID | Year | Tissue type | HNSCC | Normal | HNSCC | Normal | P-value |
| GSE10121 | 2008 | OSCC | 32 | 6 | 1.774±1.490 | −1.083±0.423 | <0.001 |
| GSE13601 | 2008 | OSCC | 31 | 26 | 7.917±1.284 | 5.466±1.271 | <0.001 |
| GSE19089 | 2009 | OSCC | 3 | 3 | 5.757±0.140 | 5.665±0.128 | 0.449 |
| GSE23558 | 2011 | OSCC | 27 | 5 | 2.107±1.454 | 0.022±1.329 | 0.006 |
| GSE25099 | 2011 | OSCC | 57 | 22 | 7.938±0.870 | 5.985±0.610 | <0.001 |
| GSE30784 | 2011 | OSCC | 167 | 45 | 8.522±1.349 | 5.970±0.951 | <0.001 |
| GSE31056 | 2011 | OSCC | 23 | 73 | 8.267±1.058 | 6.385±1.004 | <0.001 |
| GSE31853 | 2011 | OSCC | 8 | 3 | 6.027±0.948 | 3.933±1.275 | 0.015 |
| GSE34106 | 2012 | OSCC | 28 | 15 | 6.691±0.600 | 5.992±0.367 | <0.001 |
| GSE3524 | 2005 | OSCC | 16 | 4 | 0.054±1.165 | −1.842±1.222 | 0.010 |
| GSE36090 | 2012 | OSCC | 10 | 3 | 2.083±1.861 | 0.170±0.816 | 0.119 |
| GSE37991 | 2013 | OSCC | 40 | 40 | 7.546±0.334 | 8.205±0.566 | <0.001 |
| GSE55550 | 2014 | OSCC | 139 | 16 | 0.191±0.921 | −0.290±0.381 | <0.001 |
| GSE56532 | 2014 | OSCC | 10 | 6 | 10.080±1.340 | 7.661±0.735 | 0.001 |
| GSE6631 | 2007 | OSCC | 22 | 22 | 6.176±0.680 | 5.539±0.411 | 0.001 |
| GSE74530 | 2017 | OSCC | 6 | 6 | 7.033±1.548 | 4.744±0.268 | 0.015 |
| GSE9844 | 2008 | OSCC | 26 | 12 | 7.701±1.217 | 6.053±0.533 | <0.001 |
| GSE55549 | 2014 | OSCC | 8 | 4 | 0.158±0.666 | −0.268±0.424 | 0.276 |
| GSE78060 | 2017 | OSCC | 26 | 4 | 10.801±1.437 | 7.139±2.094 | <0.001 |
| GSE75539 | 2016 | OSCC | 7 | 8 | 6.43±0.258 | 6.143±0.181 | 0.026 |
| GSE75538 | 2016 | OSCC | 14 | 14 | 6.767±1.161 | 6.238±0.331 | 0.113 |
| GSE12452 | 2008 | NPC | 31 | 10 | 8.342±0.618 | 8.394±0.437 | 0.808 |
| GSE53819 | 2014 | NPC | 18 | 18 | 12.169±0.541 | 11.509±0.816 | 0.007 |
| GSE64634 | 2017 | NPC | 12 | 4 | 7.749±1.015 | 7.833±0.471 | 0.877 |
| GSE51985 | 2013 | LSCC | 10 | 10 | 31.337±43.389 | 18.568±26.459 | 0.437 |
| GSE59102 | 2014 | LSCC | 29 | 13 | 11.867±1.163 | 10.753±0.879 | 0.004 |
| GSE29330 | 2014 | LSCC | 13 | 5 | 8.322±1.548 | 6.486±0.619 | 0.002 |
| GSE84957 | 2016 | LSCC | 9 | 9 | 13.326±0.649 | 12.539±0.771 | 0.032 |
| GSE10774 | 2008 | Unclassified HNSCC | 10 | 4 | −0.812±0.853 | −1.780±0.366 | 0.012 |
| GSE13397 | 2009 | Unclassified HNSCC | 8 | 8 | 13.087±0.617 | 11.802±0.538 | <0.001 |
| GSE13398 | 2009 | Unclassified HNSCC | 8 | 8 | 1.484±0.604 | 0.363±0.376 | <0.001 |
| GSE33205 | 2013 | Unclassified HNSCC | 44 | 25 | 7.032±1.087 | 5.885±0.347 | <0.001 |
| GSE55546 | 2014 | Unclassified HNSCC | 12 | 4 | 0.395±0.947 | −0.358±0.414 | 0.153 |
| GSE58911 | 2014 | Unclassified HNSCC | 15 | 15 | 9.085±1.111 | 8.151±0.732 | 0.011 |
| GSE83519 | 2017 | Unclassified HNSCC | 22 | 22 | 2.672±0.236 | 2.781±0.180 | 0.090 |
| GSE107591 | 2017 | Unclassified HNSCC | 24 | 23 | 8.602±1.419 | 7.2±0.886 | <0.001 |
| GSE39400 | 2012 | Unclassified HNSCC | 28 | 11 | 6.585±0.287 | 6.365±0.235 | 0.030 |
| E-MTAB-1516 | NA | Unclassified HNSCC | 16 | 19 | −5.366±4.091 | −6.925±3.796 | 0.251 |
| TCGA | NA | Unclassified HNSCC | 502 | 44 | 4.383±0.106 | 4.257±0.104 | <0.001 |
HNSCC, head and neck squamous cell carcinoma; APOL1, apolipoprotein L1; SD, standard deviation; OSCC, oral squamous cell carcinoma; NPC, nasopharyngeal squamous cell carcinoma; LSCC, laryngeal squamous cell carcinoma; TCGA, The Cancer Genome Atlas; NA, not available.
Clinical parameter data of TCGA, GEO and ArrayExpress databases
Meanwhile, the information of corresponding clinicopathological parameters, such as sex, age, grade and Tumor-Node-Metastasis (TNM) stage (45), were extracted for further analysis and comparison. Kaplan-Meier estimator with log-rank tests were also used on the RNA-seq data to show the prognostic capability of APOL1 for HNSCC.
Statistical analysis
The appropriate statistical methods were applied to clarify the APOL1 expression in HNSCC and its two subgroups (OSCC and LSCC). SPSS 22.0 (IBM Corp) was used to carry out unpaired t-tests to investigate the differences in the APOL1 mRNA expression between tumor and normal tissues based on TCGA, GEO and ArrayExpress databases. Scatter diagrams and receiver operating characteristic (ROC) curves were drawn using GraphPad Prism version 7.0 (GraphPad Software). In addition, the summary receiver operating characteristic (sROC) curves were drawn using Stata 12 (StataCorp LLC). These were used to evaluate the capability of APOL1 to diagnose tumors, the accuracy of which was confirmed by the area under the curve (AUC). Comparing the APOL1 expression in tumor tissues with that in non-tumor tissues, standard mean differences (SMDs) and 95% confidence intervals (CIs) were used. The funnel plot and Begg's test were used to examine whether there was publication bias. χ2 and Fisher's exact tests were used to compare the IHC staining scores of APOL1 in HNSCC with those in the normal tissues and analyze the association between the clinicopathological parameters and expression of APOL1 proteins for 135 patients with HNSCC. χ2 tests were used when all theoretical frequency T≥5 and total sample size n≥40, while Fisher's test suitable for sample size n<40 or theoretical frequency T<1. Fagan's Tests were used to demonstrate the relationship between the former probability, the latter probability, the positive likelihood ratio and the negative likelihood ratio by taking APOL1 expression value as the diagnostic method and comparing with the gold standard for cancer diagnosis. If the heterogeneity test was P<0.05 or I2>50%, meaning the existence of heterogeneity, the random effects model was used. If the contrary was true, the fixed effects model was used. Some microarrays were excluded according to the following criteria: i) Microarrays with low expression, ii) microarrays with large confidence intervals (CIs) and iii) microarrays with no statistical significance. Meanwhile, Stata 12 (StataCorp) was also used to construct the forest plots, sensitivity analysis and to determine the positive or negative likelihood ratio and publication bias. P<0.05 was considered to indicate a statistically significant difference.
Results
APOL1 protein expression in OSCC detected by IHC and its association with clinicopathological parameters
In total, 135 cases of OSCC and 13 cases of normal oral tissues in-house were used to analyze the change of APOL1 expression in OSCC samples. Of the 135 cases with OSCC tissue samples, APOL1 was positively expressed in 61 cases (45.2%). Meanwhile, no positive results were observed in the 13 cases of non-cancerous oral tissues (Table II). It was demonstrated that the difference of APOL1 protein was statistically significant (P=0.001; Table II), which indicated a higher expression of APOL1 in OSCC tissues compared with in normal tissues. The clinicopathological features of the 135 OSCC are summarized in Table II. By analyzing the association between the expression of APOL1 and clinicopathological parameters in OSCC, it was revealed that there was a significant association between APOL1 and sex (P=0.015, Table II). No marked associations were observed between APOL1 expression and other clinical parameters. Thus, APOL1 protein expression was upregulated in OSCC tissues as compared to non-cancerous oral controls.
Table II.
The clinicopathological parameters and expression of APOL1 protein for 135 patients with oral squamous cell carcinoma.
| APOL1 expression, n | |||
|---|---|---|---|
| Clinicopathological parameter | High | Low | P-value |
| Cancerous cases vs. non-cancerous cases | 0.001 | ||
| Cancerous cases | 61 | 74 | |
| Non-cancerous cases | 0 | 13 | |
| Sex | |||
| Male | 53 | 51 | 0.015 |
| Female | 8 | 23 | |
| Age, years | |||
| ≥60 | 24 | 31 | 0.728 |
| <60 | 37 | 42 | |
| Grade | |||
| 1–2 | 25 | 35 | 0.491 |
| 3–4 | 36 | 39 | |
| Stage | |||
| I–II | 25 | 35 | 0.491 |
| III–IV | 36 | 39 | |
| Pathological T | |||
| T1-T2 | 43 | 15 | 0.840 |
| T3-T4 | 59 | 18 | |
| Pathological N | |||
| N0 | 34 | 45 | 0.601 |
| N1 | 27 | 29 | |
| Pathological M | |||
| M0 | 61 | 73 | 0.456 |
| M1 | 0 | 1 | |
APOL1, apolipoprotein L1; T, tumor; N, node; M, metastasis; Grade, clinical pathological grade; Stage, TNM stage.
Analysis of APOL1 mRNA expression in OSCC
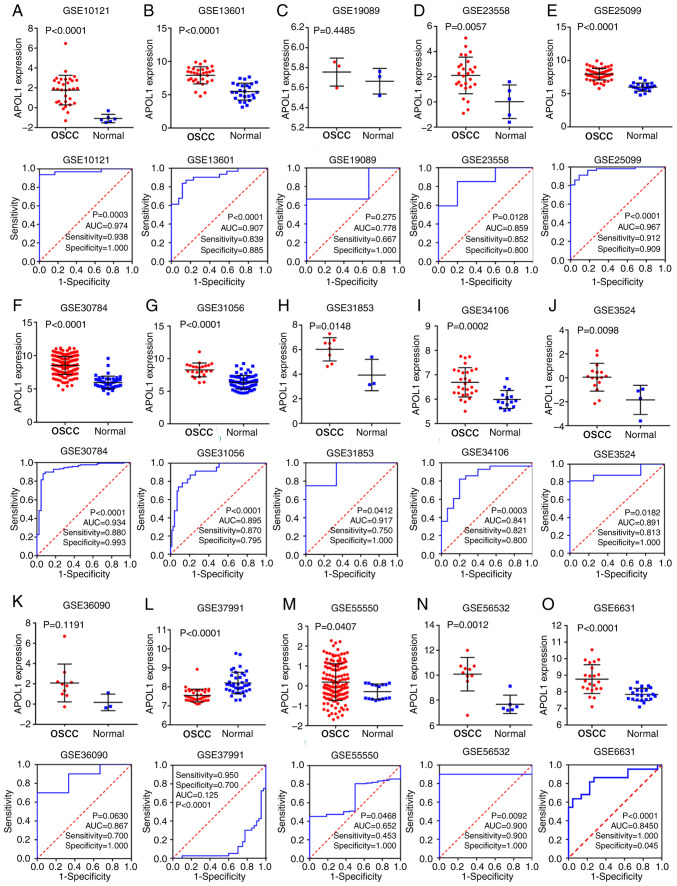
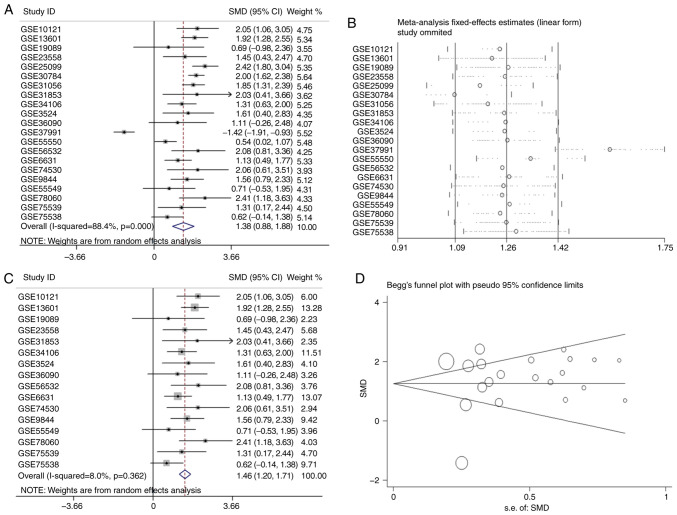
TCGA, GEO and ArrayExpress databases were searched and 21 microarrays containing APOL1 expression profile were performed with OSCC tissues (Table I). Of the 21 microarrays (Fig. 2A-O and 3A-F), 20 showed that APOL1 was highly expressed in OSCC tissues compared with in normal tissues. Statistically significant differences were found in 17 out of 21 microarrays, while four microarrays (GSE19089, GSE36090, GSE55549 and GSE75538 had no statistical significance in the change of expression level of APOL1 between the OSCC tissues and control group. Analysis revealed that the overall SMD was 1.38 (95% CI, 0.88–1.88) (Fig. 4A). In other words, the expression of APOL1 was stronger in OSCC tissues compared with in normal tissues because the SMD>0 and 0 were not included in the 95% CI (Fig. 4A). The heterogeneity test was applied, and the outcome demonstrated that the analysis was heterogeneous via the random effects model (Pheterogeneity<0.0001; I2=88.4%; Fig. 4A). The sensitivity analysis was then conducted (Fig. 4B). After removing five microarrays (GSE25099, GSE30784, GSE31056, GSE37991 and GSE55550), the SMD was 1.46 (95% CI, 1.21–1.69); no significant heterogeneity was found (the fixed effects model: Pheterogeneity=0.362; I2=8.0%; Fig. 4C). According to the funnel plot and the results of Begg's test (P=0.608; Fig. 4D), no publication bias was observed. Thus, the mRNA expression of APOL1 in OSCC tissues was higher than in normal tissues, which was consistent with the protein level.
Figure 2.
mRNA APOL1 expression in OSCC tissue and the corresponding ROC curves. (A-O) Expression data of APOL1 and corresponding ROC curves in OSCC tissues in 15 microarrays from Gene Expression Omnibus datasets. APOL1, apolipoprotein L1; ROC, receiver operating characteristic; OSCC, oral squamous cell carcinoma; AUC, area under the curve.
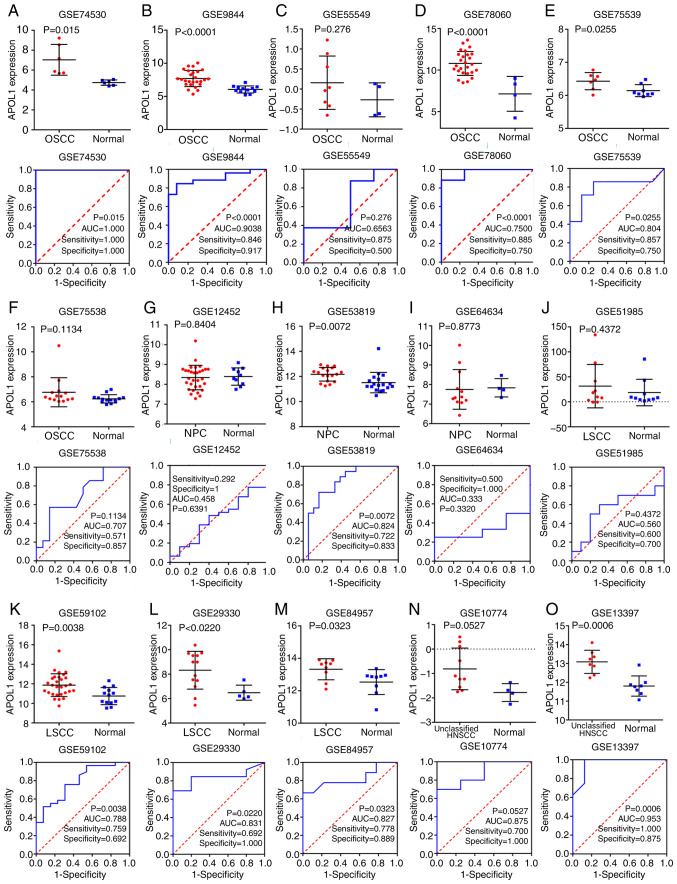
Figure 3.
mRNA APOL1 expression in HNSCC and normal tissues. (A-F) Expression data of APOL1 and corresponding ROC curves in OSCC tissues in six microarrays from GEO datasets. (G-I) Expression data of APOL1 and corresponding ROC curves in NPC tissues in three microarrays from GEO datasets. (J-M) Expression data of APOL1 and corresponding ROC curves in LSCC tissues in four microarrays from GEO datasets. (N and O) Expression data of APOL1 and corresponding ROC curves in unclassified subgroups of HNSCC in two microarrays from GEO datasets. APOL1, apolipoprotein L1; ROC, receiver operating characteristic; HNSCC, head and neck squamous cell carcinoma; OSCC, oral squamous cell carcinoma; LSCC, laryngeal squamous cell carcinoma; NPC, nasopharyngeal carcinoma; AUC, area under the curve.
Figure 4.
Forest plots and funnel plot of APOL1 expression between OSCC and non-tumor tissues. (A) Forest plots of studies evaluating standard mean difference of APOL1 between OSCC and non-tumor tissues based on GEO datasets. (B) Sensitivity analysis and the forest plot after sensitivity analysis; sensitivity analysis of analysis of the expression levels of APOL1 in OSCC and non-tumor tissues based on GEO datasets. (C) Forest plots of APOL1 expression between OSCC and non-tumor tissues after removing the following studies: GSE25099, GSE30784, GSE31056, GSE37991and GSE55550. (D) Funnel plot for publication bias test after analysis of the expression level of APOL1 in OSCC, The size represented by the Begg's plot circle is the weight of each data concentration. APOL1, apolipoprotein L1; OSCC, oral squamous cell carcinoma; GEO, Gene Expression Omnibus.
To reveal the capacity of the APOL1 expression to diagnose patients with OSCC, the sROC was used to analyze the AUC and 95% CI. As Fig. 5A shows, the AUC of APOL1 was 0.93 (95% CI, 0.79–0.90). The pre-test probability, post-test probability positive and post-test probability negative were 20, 63 and 4%, respectively (Fig. 5B), with a sensitivity of 0.85 (95% CI, 0.79–0.90) and a specificity of 0.88 (95% CI, 0.81–0.92) (Fig. 5C and D, respectively). Hence, the results indicated that the upregulation of APOL1 mRNA has potential as a marker for differentiating OSCC from non-cancerous oral epithelium.
Figure 5.
Analysis of the mRNA expression of APOL1 in OSCC. (A) sROC analysis of APOL1 for discriminating OSCC from normal tissues based on GEO datasets. (B) Prior probability and post-probability positive and negative of the included studies. The prior probability, post probability positive and negative reached 20, 63 and 4%, respectively. (C and D) Sensitivity and specificity values of the included studies. The sensitivity and specificity values of the included studies were 0.85 (95% CI, 0.79–0.90) and 0.88 (95% CI, 0.81-.92), respectively. Oral squamous cell carcinoma (OSCC). APOL1, apolipoprotein L1; sROC, Summary receiver operating characteristic; LR, likelihood ratio; GEO, Gene Expression Omnibus; OSCC, oral squamous cell carcinoma; CI, confidence interval.
Association between APOL1 protein expression and the clinicopathologic parameters of LSCC
A total of 49 cases of LSCC tissues and 5 cases of normal laryngeal tissues were collected in-house. Of the 49 LSCC tissues collected, 45 were positive for APOL1, accounting for 91.8% (Table III). Meanwhile, all normal laryngeal tissues showed a low expression. Statistical calculation revealed that APOL1 levels were higher in LSCC tissues compared with normal tissues (P<0.0001; Table III). The association between APOL1 expression levels and clinicopathological parameters were analyzed but no significant relationships were observed (Table III). So, the expression of APOL1 protein was increased in LSCC tissues as compared to non-cancerous controls.
Table III.
Clinicopathological parameters and APOL1 expression of 49 patients with LSCC.
| APOL1 expression, n | |||
|---|---|---|---|
| Clinicopathological parameter | High | Low | P-value |
| Cancerous cases vs. non-cancerous cases | |||
| Cancerous cases | 45 | 4 | <0.0001 |
| Non-cancerous cases | 0 | 5 | |
| Sex | |||
| Male | 44 | 4 | 0.9180 |
| Female | 1 | 0 | |
| Age, years | |||
| ≥60 | 20 | 0 | 0.1350 |
| <60 | 25 | 4 | |
| Grade | |||
| 1–2 | 29 | 4 | 0.1930 |
| 3–4 | 16 | 0 | |
| Stage | |||
| I–II | 15 | 0 | 0.2190 |
| III–IV | 30 | 4 | |
| Pathological T | |||
| T1-T2 | 28 | 0 | 0.0600 |
| T3-T4 | 16 | 3 | |
| Pathological N | |||
| N0 | 22 | 0 | 0.0830 |
| N1 | 23 | 4 | |
| Pathological M | – | ||
| M0 | 45 | 4 | |
| M1 | 0 | 0 | |
LSCC, laryngeal squamous cell carcinoma; IHC, immunohistochemistry; T, tumor; N, node; M, metastasis; APOL1, apolipoprotein L1. Grade, clinical pathological grade; Stage, TNM stage
Analysis of mRNA expression of APOL1 in LSCC
Through deep mining the GEO, TCGA and ArrayExpress, finally the number of microarrays of APOL1 expression in LSCC and normal tissues was four (Table I). All of them showed that APOL1 expression was higher in LSCC tissues compared with in normal tissues (Fig. 3J-M). Besides, three out of four microarrays of LSCC showed that the differences were significant according to Table I. The SMD was 0.92 (95% CI, 0.48–1.36) (Fig. 6A). As there was no obvious heterogeneity (Pheterogeneity=0.512; I2=0.0%, Fig. 6A), the fixed effects model was used. Similarly, according to the funnel plot and the results of Begg's test (P=0.308, Fig. 6B), no publication bias was observed for the analysis of APOL1 expression.
Figure 6.
Analysis of the mRNA expression of APOL1 in LSCC. (A) Forest plots of studies evaluating standard mean difference of APOL1 between LSCC and non-tumor group. (B) Funnel plot for publication bias test after analysis of the expression level of APOL1 in LSCC, The size represented by the Begg's plot circle is the weight of each data concentration. APOL1, apolipoprotein L1; LSCC, laryngeal squamous cell carcinoma; GEO, Gene Expression Omnibus.
In addition, as shown in Fig. 7A, APOL1 had a notable discriminatory capability for LSCC due to the AUC of the APOL1 upregulation being 0.82 (95% CI, 0.78–0.85). The pre-test probability, post-test probability positive, and post-test probability negative were 20, 45 and 8%, respectively (Fig. 7B), with the sensitivity being 0.72 (95% CI, 0.60–0.82) and the specificity 0.78 (95% CI, 0.62–0.89) (Fig. 7C and D, respectively). So it was shown that the mRNA expression of APOL1 in LSCC tissues was higher than in normal tissues, which was in line with the APOL1 protein level.
Figure 7.
sROC curves analysis of APOL1 for discriminating LSCC from normal tissues, prior probability post-probability, positive and negative of the included studies and sensitivity and specificity values. (A) The sROC analysis of APOL1 for discriminating LSCC from normal tissues based on GEO datasets. (B) Prior probability and post-probability positive and negative of the included studies. The prior probability, post probability positive and negative reached 20, 45 and 8%, respectively. (C and D) Sensitivity and specificity values of the included studies. The sensitivity and specificity values of the included studies were 0.72 (95% CI, 0.60–0.82) and 0.78 (95% CI, 0.62–0.89), respectively. APOL1, apolipoprotein L1; GEO, Gene Expression Omnibus; LSCC, laryngeal squamous cell carcinoma; sROC, summary receiver operating characteristic; LR, likelihood ratio; CI, confidence interval.
APOL1 protein level in HNSCC
Overall, the present study analyzed 221 cases of HNSCC (including OSCC, LSCC, NPC and some HNSCC without specified sites) and 24 cases of normal head and neck tissues in-house. So, the entire HNSCC group included the OSCC, LSCC, and non-classified HNSCC groups, which was analyzed as a whole in order to observe the association between the APOL1 and the entire HNSCC tissues in present study. The IHC results and the information of the patients in-house were also evaluated. Of the 221 cases, APOL1 was highly expressed in 126 (57%), whereas levels of APOL1 in the normal tissues showed lower expression. The Fisher's exact test showed that the proportion of APOL1 in tumor tissues was significantly higher compared with in non-cancer tissues (P<0.0001; Table IV). The HPA database was used to verify the expression of APOL1 (Fig. 8A-C). After analyzing the association between APOL1 expression and the clinical parameters, it was reported that there was a significant difference in terms of sex (P=0.004), tumor pathological grade (P<0.0001) and pathological T stage (P=0.019) (Table IV). So the APOL1 protein level was generally upregulated in HNSCC tissues than normal tissues.
Table IV.
The clinicopathological parameters and APOL1 expression of 221 patients with HNSCC.
| APOL1 expression, n | |||
|---|---|---|---|
| Clinicopathological parameter | High | Low | P-value |
| Cancerous cases vs. non-cancerous cases | |||
| Cancerous cases | 126 | 95 | <0.0001 |
| Non-cancerous cases | 0 | 24 | |
| Sex | |||
| Male | 114 | 70 | 0.0040 |
| Female | 12 | 25 | |
| Age, years | |||
| ≥60 | 54 | 38 | 0.6850 |
| <60 | 72 | 57 | |
| Grade | |||
| 1–2 | 89 | 79 | <0.0001 |
| 3–4 | 39 | 14 | |
| Stage | |||
| I–II | 50 | 39 | 0.0870 |
| III–IV | 76 | 54 | |
| Pathological T | |||
| T1-T2 | 85 | 66 | 0.0190 |
| T3-T4 | 41 | 27 | |
| Pathological N | |||
| N0 | 69 | 54 | 0.7630 |
| N1 | 57 | 39 | |
| Pathological M | 0.4270 | ||
| M0 | 126 | 93 | |
| M1 | 0 | 0 | |
APOL1, apolipoprotein L1; HNSCC, head and neck squamous cell carcinoma; T, tumor; N, node; M, metastasis. Grade, clinical pathological grade; Stage, TNM stage.
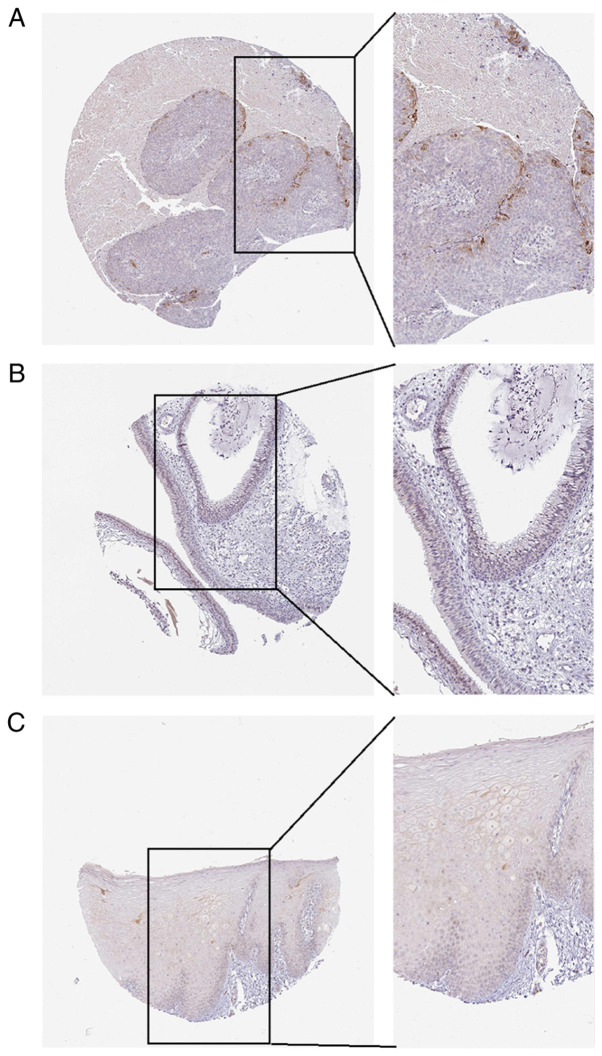
Figure 8.
The APOL1 expression in normal tissues and HNSCC tissues from the Human Protein Atlas database (antibody CAB056156). (A) Poorly differentiated metastasis of HNSCC to lymph nodes for APOL1. (B) Normal nasopharynx tissues low expression for APOL1. (C) Normal oral mucosa negative tissues for APOL1. Magnification, ×100 and ×200. HNSCC, head and neck squamous cell carcinoma; APOL1, apolipoprotein L1.
Clinical significance of APOL1 mRNA expression in HNSCC
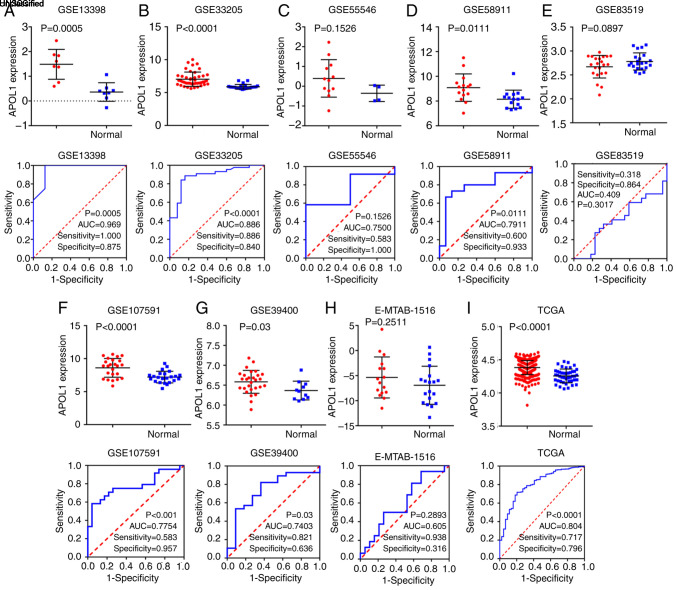
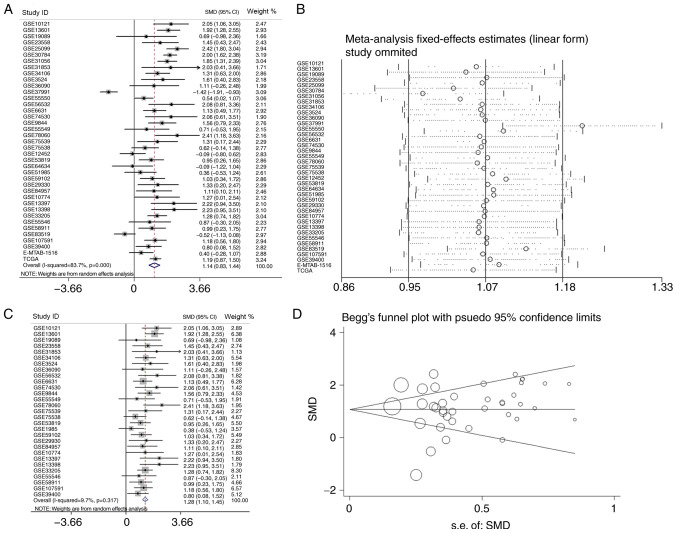
In the present study, a total of 39 microarrays and RNA-seq data regarding HNSCC and normal tissues were analyzed. Among them, 21 OSCC and four LSCC microarrays were used for subgroup analysis as aforementioned. Three of the remaining microarrays were nasopharyngeal carcinoma (NPC), the pathogenesis of which is different from that of other HNSCCs (38), and 11 were not classified any of the HNSCC subgroups according to the TCGA, GEO and ArrayExpress databases; thus, these were grouped into the overall HNSCC group. The 39 studies are listed in Table I. Of the three microarrays of APOL1 expression profiling in NPC tissues, one NPC microarray showed that the difference was significant (P=0.007) according to the Table I. And combining all of the aforementioned studies, it was reported that, in the 39 microarrays, the expression of APOL1 in HNSCC tissues was higher compared with that in non-cancerous tissues (Figs. 2, 3 and 9). This difference was statistically significant in 29 out of 39 microarrays (P<0.05; Table I). However, in the remaining 10 microarrays (GSE19089, GSE55549, GSE75538, GSE12452, GSE64634, GSE51985, GSE55546, GSE83519, GSE36090 and E-MTAB-1516), there was no statistical significance in the APOL1 expression between the HNSCC and normal tissues. The SMD was 1.14 (95% CI, 0.83–1.44) (Fig. 10A), which confirmed that APOL1 expression was higher in HNSCC tissues compared with the corresponding control group. The heterogeneity test was applied, and the outcome demonstrated that the analysis was heterogeneous using the random effects model (Pheterogeneity<0.0001; I2=83.7%; Fig. 10A). The sensitivity analysis was then conducted (Fig. 10B). To determine the datasets with heterogeneity, the 10 microarrays (GSE25099, GSE30784, GSE31056, GSE37991, GSE55550, GSE12452, GSE64634, GSE83519, E-MTAB-1516 and all TCGA datasets) were removed. After the removal of 10 microarrays, which had the potential to produce heterogeneity, the SMD was 1.28 (95% CI, 1.10–1.45) and we found no obvious heterogeneity among them (the fixed effects model: Pheterogeneity=0.317; I2=9.7%; Fig. 10C). According to the funnel plot, the results of Begg's test (P=0.468, Fig. 10D), no publication bias was identified. So it was consistently shown that the expression of APOL1 mRNA in the HNSCC was higher than in the normal tissues.
Figure 9.
APOL1 expression and ROC curves in unclassified subgroup HNSCC tissues. (A-I) Expression data of APOL1 and corresponding ROC curves in unclassified subgroups HNSCC tissues in nine microarrays from TCGA, ArrayExpress and GEO datasets. APOL1, apolipoprotein L1; TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; ROC, receiver operating characteristic; HNSCC, head and neck squamous cell carcinoma; AUC, area under the curve.
Figure 10.
The analysis of the mRNA expression of APOL1 in HNSCC. (A) Forest plots of studies evaluating standard mean difference of APOL1 between HNSCC and non-tumor group. (B) Sensitivity analysis of the expression level of APOL1 in HNSCC and non-tumor group. (C) Forest plots of APOL1 expression between HNSCC and non-tumor group after removing the following studies: GSE25099, GSE30784, GSE31056, GSE37991, GSE55550, GSE12452, GSE64634, GSE83519, E-MTAB-1516 and TCGA. (D) Funnel plot for publication bias test after analysis of the expression level of APOL1 in HNSCC, The size represented by the Begg's plot circle is the weight of each data concentration. HNSCC, head and neck squamous cell carcinoma; APOL1, apolipoprotein L1; SMD, standardized mean difference; CI, confidence interval.
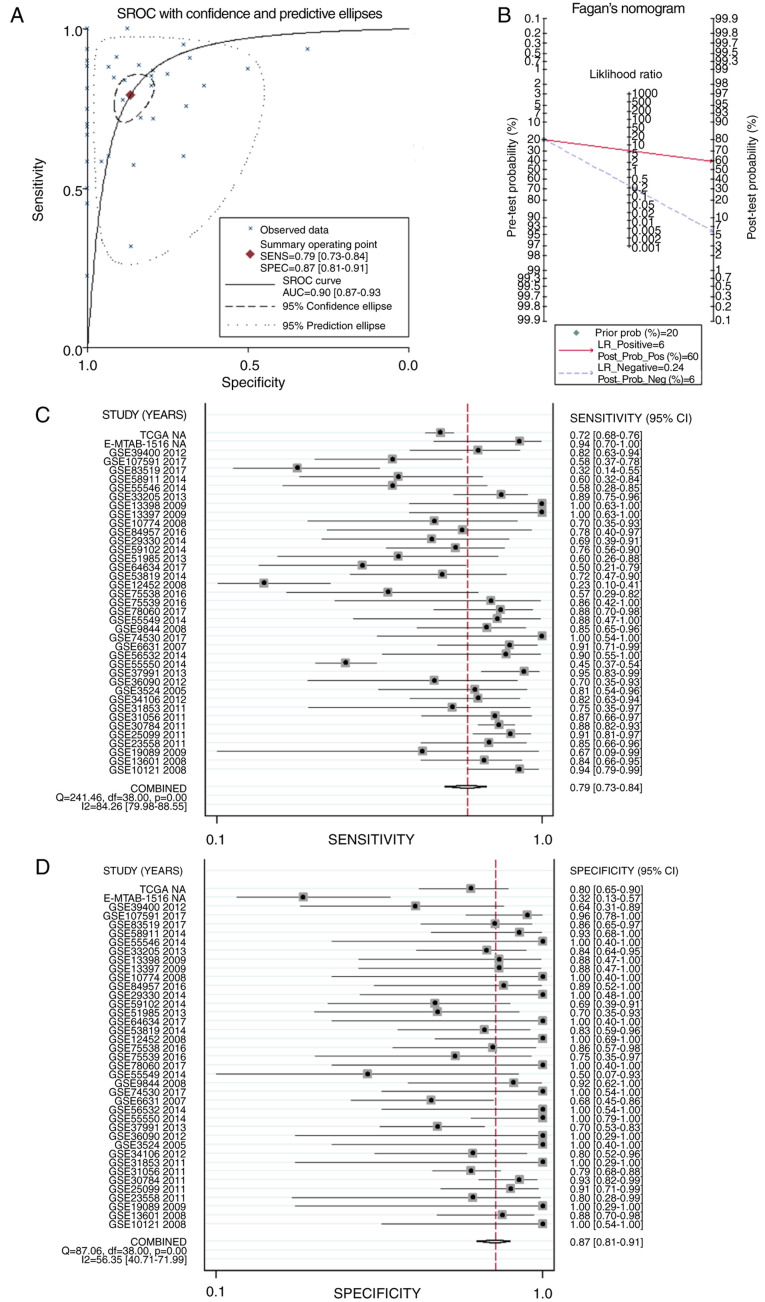
sROC was used to analyze the value of APOL1 overexpression regarding the diagnostic potential of HNSCC (AUC, 0.90; 95% CI, 0.87–0.93; Fig. 11A). The pre-test probability, post-test probability positive and post-test probability negative were 20, 60 and 6%, respectively (Fig. 11B), with a sensitivity of 0.79 (95% CI, 0.73–0.84) and a specificity of 0.87 (95% CI, 0.81–0.91) (Fig. 11C and D, respectively), thereby indicating the notable discriminatory capability of high APOL1 level for differentiating HNSCC from non-cancerous lesions.
Figure 11.
sROC curves analysis of APOL1 for discriminating HNSCC from normal tissues, prior probability post-probability, positive and negative of the included studies and sensitivity and specificity values. (A) sROC curves analysis of APOL1 for discriminating HNSCC from normal tissues based on GEO datasets. (B) Prior probability and post-probability positive and negative of the included studies. The prior probability, post probability positive and negative reached 20, 60 and 6%, respectively. (C and D) Sensitivity and specificity values of the included studies. The sensitivity and specificity values of the included studies were 0.79 (95% CI, 0.73–0.84) and 0.87 (95% CI, 0.81–0.91), respectively. HNSCC, head and neck squamous cell carcinoma; APOL1, apolipoprotein L1; LR, likelihood ratio; sROC, summary receiver operating characteristic; CI, confidence interval.
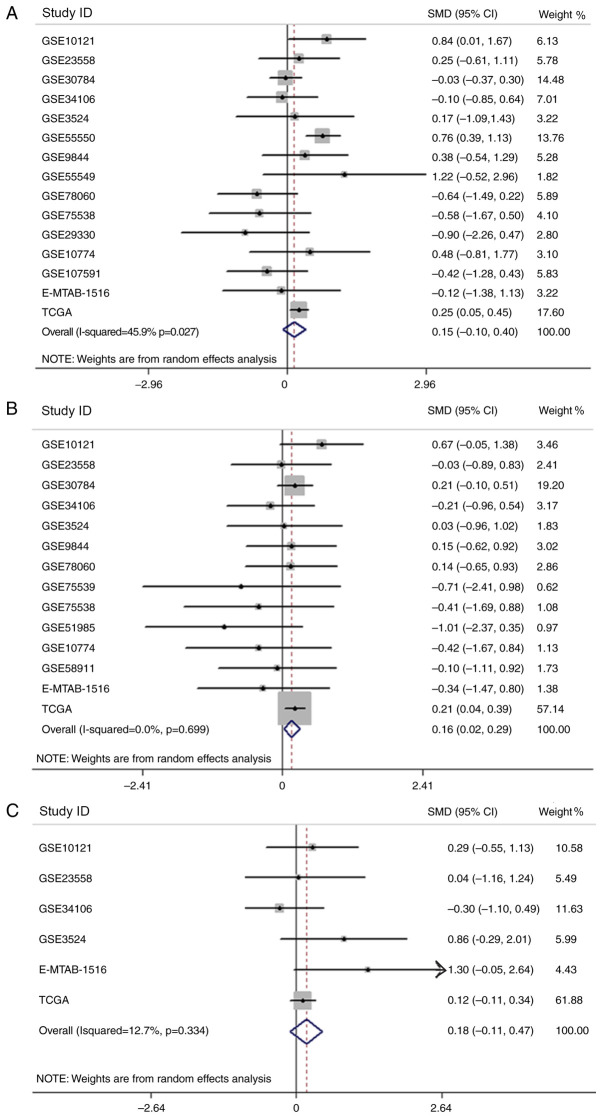
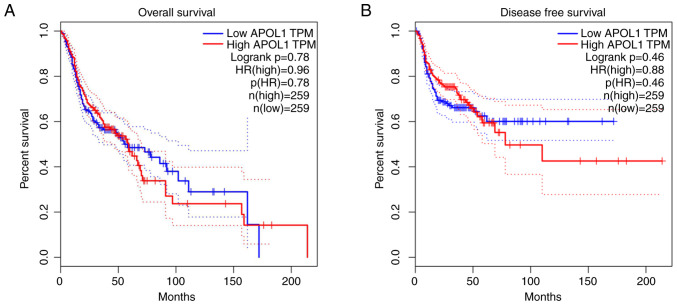
After analyzing the association between the expression of APOL1 and clinicopathological parameters in HNSCC, relevant clinical information from the chips included in the current study were extracted and SMD analysis was performed in terms of sex, age and pathological staging (Fig. 12A-C). However, only the results concerning age were statistically significant (P<0.0001), and the expression of APOLI in patients >60 years old was higher compared with that in patients <60 years old (Fig. 12B). Using the Kaplan-Meier estimator, it was revealed that there was no significant difference in terms of survival regarding APOL1 expression (Fig. 13).
Figure 12.
The clinical significance of the mRNA expression of APOL1 in HNSCC. (A) Forest plots of studies evaluating SMD of APOL1 expression between the male and female group. (B) Forest plots of studies evaluating SMD of APOL1 expression between patients <60 and ≥60 years old. (C) Forest plots of studies evaluating SMD of APOL1 expression between stage I–II and III–IV. APOL1, apolipoprotein L1; HNSCC, head and neck squamous cell carcinoma; SMD, standardized mean difference; CI, confidence interval.
Figure 13.
The Kaplan-Meier survival curve of APOL1 in head and neck squamous cell carcinoma by RNA sequencing. (A) Overall survival and (B) disease-free survival. APOL1, apolipoprotein L1; TPM, transcripts per million; HR, hazard ratio.
Discussion
The human APOL1 gene is located at chromosome 22q13 (28). APOL1 induces inflammatory cytokines (for example IFN and TNF) and the tumor suppressor gene p53 (46). Alterations in APOL1 function can lead to the occurrence of cancer, chronic nephropathy, atherosclerosis and other diseases (32–34). It has been reported that autophagy is a cellular pathway that transmits cytoplasmic components to lysosomes for degradation and recycling (47). APOL1 is a Bcl-2 homologous domain 3 monolipid-binding protein, which can maintain sufficient autophagy flux of podocytes under normal physiological conditions. When APOL1 is overexpressed, it can enhance the autophagy ability of podocytes, liver cancer cells and melanoma cells (48–50). A previous study has shown that among the seven exons of APOL1, exon 4 is the main determinant of APOL1G0 cytotoxicity (51). This lysosome-related autophagy is triggered by several pathways. First, APOL1 disrupts intracellular vesicle transport by interfering with autophagosome maturation, and APOL1 can be inserted into the plasma membrane to form pH-sensitive cation channels, which can change the pH value of endocytosis and enhance the activity of trypsin decomposition (46,48). Second, APOL1 may interact with GTPase or SNARE proteins, thus interfering with autophagy maturation and lysosome fusion (52). APOL1 renal risk alleles (G1 and G2), also called renal risk variants (RRVs), inhibit the APOL1-miR193a axis of differentiated podocytes (DPDs) (53), resulting in a decrease in the assembly of PI3KC3-ATG14L and PI3KC3-anti-UV radiation resistance associated gene complexes needed for autophagy nucleus formation and maturation. This is a separate pathway by which APOL1 can trigger lysosome-mediated autophagy (54). Third, APOL1RRVs can affect the permeability of intracellular and plasma membrane (55), and activate the opening of mitochondrial pores and form oligomers, resulting in cytotoxicity (56). The process of the activation of the cGAS/IFI16-STING pathway begins by binding of cytosolic nsDNA by cyclic GMP-AMP synthase (cGAS) and interferon-inducible protein 16 (IFI16), which activates stimulator of interferon genes (STING), and the cGAS/IFI16-STING pathway finally promotes the transcription of APOL1 through signaling cascade (57). The expression of APOL1 induced by nucleosome-associated double-stranded DNA fragments is mediated by the IFN β-dependent signaling pathway, which is triggered by the activation of the cGAS/IFI16-STING pathway (57). It has been demonstrated that a high level of APOL1 expression can cause autophagy and autophagy-related cell death in various tissues that are associated with the occurrence of cancer, cardiomyopathy, obesity and neurodegenerative diseases (58,59). Autophagy plays a dual role oncogenesis and the development of tumors (60,61). In normal physiological conditions, autophagy helps to degrade denatured proteins and damaged organelles, which aids in preventing cells from becoming cancerous (62). However, when tumor cells are formed, adjacent normal cells can also provide nutrients for the growth of tumors through autophagy (63). These factors led to the hypothesis that APOL1 may be associated with the development of HNSCC. Based on the present results, it was speculated that the expression of APOL1 would make it more easily to promote the development of HNSCC through inducing autophagy, which leads to tumor growth and invasion.
The present study collected a mass of chips by mining TCGA, ArrayExpress and GEO databases. The HPA was also used to verify differences in the APOL1 protein expression between HNSCC and normal tissues. HNSCC tissues and normal tissues were also collected in-house for IHC analysis. The current study had several highlights. First of all, a total of 39 sets of microarrays and RNA-seq data were collected to show the mRNA level of APOL1, and 221 cases of HNSCC tissues and 24 cases of normal head and neck tissues were used in IHC detection for protein level. Secondly, several methods were used to analyze APOL1 expression in HNSCC tissues, such as IHC on tissue microarrays, gene microarrays, RNA-sequencing, and also the integrated analysis. Therefore, the association between the HNSCC tissues and the expression of APOL1 could be observed at both the mRNA and protein levels in multiple cohorts using multiple detection methods. This provided more evidence demonstrating APOL1 upregulation in HNSCC than results from a single cohort and single detection method approach would.
In order to make the study more applicable, OSCC and LSCC cases were selected for HNSCC subgroup analysis. By applying IHC to analyze the protein expression levels of APOL1 in 135 cases of OSCC tissues, it was demonstrated that the protein content of APOL1 in OSCC was higher compared with that in healthy tissues. However, after analyzing the association between APOL1 expression and the clinicopathological parameters in OSCC, significant differences were only identified in terms of sex. In the analysis of APOL1 mRNA expression in OSCC, APOL1 was expressed at significantly higher levels in OSCC tissues compared with in normal tissues. In addition, the results of sROC, pre-test probability, post-test probability positive and post-test probability negative indicated that APOL1 had a notable discriminatory capability for OSCC.
By applying IHC to analyze the protein expression levels of APOL1 in 49 cases of LSCC tissues, it was revealed that the protein expression levels of APOL1 in LSCC tissues were significantly higher compared with those in normal tissues. The association between APOL1 expression levels and clinicopathological parameters (including sex, age, tumor grade and clinical stage) were investigated but no significant differences were observed. In the analysis of APOL1 mRNA expression in LSCC microarrays, APOL1 levels were significantly higher in LSCC tissues compared with those in normal tissues. In addition, the results of sROC, the pre-test probability, the post-test probability positive and the post-test probability negative indicated that APOL1 had a notable discriminatory capability for LSCC.
It has been reported that APOL1 expression is higher in papillary thyroid carcinoma (PTC) and renal cell carcinoma (RCC) tissues compared with respective normal tissues (64,65). However, Liu et al (34) demonstrated that the expression of APOL1 in paracancerous tissues is greater compared with that in pancreatic cancer (PC) tissues (34). This may be because PTC is derived from squamous epithelial cells. Similarly, OSCC and LSCC are also derived from squamous epithelial cells, therefore the expression of APOL1 in cancer may be dependent on the cell type. However, the dominant histological type of PC was pancreatic ductal adenocarcinoma, derived from ductal or acinar cells, and RCC originates in proximal renal tubular epithelial cells (65,66).
There were several similarities in the expression of APOL1 in OSCC and LSCC subgroups. First, by applying IHC to measure the levels of APOL1, it was reported that APOL1 was significantly expressed in OSCC and LSCC tumor tissues compared with respective normal tissues. Second, analysis of APOL1 mRNA expression revealed that APOL1 levels were higher in tumor tissues compared with normal tissues. Third, no significant difference was found between APOL1 expression levels and certain clinicopathological parameters, including age, tumor grade and TNM stage (45) for both subtypes. Fourth, APOL1 had a valuable discriminatory capability for OSCC and LSCC to be separated from non-cancerous SCC tissues.
According to Table III, of the three microarrays of APOL1 expression profiling in NPC tissues, one NPC microarray showed that the upregulation of APOL1 was significant (P=0.007). The result was quite different from that of OSCC and LSCC. It has been shown that the initiation and progress of NPC is closely associated with the status of Epstein-Barr virus (EBV) infection (67). Some EBV gene products are involved in tumor progression and play a vital part in epithelial-mesenchymal transition, angiogenesis and metastasis in NPC (67,68). The APOL1 gene may have a limited contribution to the initiation and progress of NPC.
By analyzing the data from the HPA database and our IHC results, it was observed that the content of APOL1 protein in tumor tissue higher compared with that in non-tumor tissue. In the analysis of the mRNA expression of APOL1 in HNSCC by microarrays and RNA-seq, the same pattern was observation. The results of sROC, the pre-test probability, the post-test probability positive and the post-test probability negative indicated that APOL1 upregulation had potential diagnostic value for HNSCC. When overexpressed, APOL1 induces autophagy and autophagy-related cell death (65). Through autophagy, adjacent normal cells are degraded and provide nutrition for the growing tumor (63).
Despite the aforementioned findings, the present study has some limitations. For example, we did not perform in vitro studies to investigate the mechanisms underpinning APOL1 function in HNSCC cells, and the sample size used in present study could be enlarged. The pathological mechanism of the disease is still not clear, and further research is underway. However, the present results may aid the development of potential interventions in the future. High expression of APOL1 may directly or indirectly result in the occurrence of HNSCC, therefore APOL1 is expected to be a biomarker for HNSCC. The present results may help improve our understanding of the occurrence and prognosis of HNSCC and provide new avenues for future research.
In conclusion, compared with non-cancerous head and neck tissues, APOL1 expression was significantly elevated in most HNSCC tissues. This indicated that APOL1 might play a vital role in the oncogenesis and development of HNSCC. The present study is the first to identify an association between HNSCC and APOL1, to the best of our knowledge. The current findings also suggest that APOL1 has the potential to be a promising biomarker, which warrants further study. However, further research is needed to resolve the molecular mechanisms underpinning the function of APOL1 in HNSCC.
Acknowledgements
Not applicable.
Funding
The study was supported by the Guangxi Medical and Health Appropriate Technology Development, and Popularization and Application Project (grant no. S2017020) and the Promoting Project of Basic Capacity for Young and Middle-aged University Teachers in Guangxi, China (grant no. 2018KY0123).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the TCGA (http://cancergenome.nih.gov/), the GEO (https://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) data portals. The in-house IHC data from the present study can be acquired from the correspondence author on reasonable request.
Authors' contributions
XGZ, YL, MXL, GSL, XYC, YXY and YYQ collected data from public datasets, analyzed the data and performed the statistical analysis. HPL, YWD and ZGH performed in-house IHC experiments. FZ and GC participated in the conception and design of the study and in manuscript correction. MM, KLZ, HD and ZXW conceived and designed the study and assisted in the drafting of the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
This research program was approved by The Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (Nanning, China). All participants signed informed consent forms.
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Huang C, Cintra M, Brennan K, Zhou M, Colevas AD, Fischbein N, Zhu S, Gevaert O. Development and validation of radiomic signatures of head and neck squamous cell carcinoma molecular features and subtypes. EBioMedicine. 2019;45:70–80. doi: 10.1016/j.ebiom.2019.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Q, Wang C, Li B, Kim K, Li J, Mao M, Qin L, Li H, Huang X, Xing R, et al. The impact of age on oral squamous cell carcinoma: A longitudinal cohort study of 2,782 patients. Oral Dis. 2019;25:730–741. doi: 10.1111/odi.13015. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Zhao Q, Ding G, Zhu Y, Li W, Chen W. Incidence and mortality of laryngeal cancer in China, 2008–2012. Chin J Cancer Res. 2018;30:299–306. doi: 10.21147/j.issn.1000-9604.2018.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin Y, Yang Y. Bioinformatics-based discovery of PYGM and TNNC2 as potential biomarkers of head and neck squamous cell carcinoma. Biosci Rep Jul. 2019;39:BSR20191612. doi: 10.1042/BSR20191612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 7.Ghantous Y, Abu Elnaaj I. Global incidence and risk factors of oral cancer. Harefuah. 2017;156:645–649. (In Hebrew) [PubMed] [Google Scholar]
- 8.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 9.Zhou C, Shen Z, Ye D, Li Q, Deng H, Liu H, Li J. The association and clinical significance of CDKN2A promoter methylation in head and neck squamous cell carcinoma: A meta-analysis. Cell Physiol Biochem. 2018;50:868–882. doi: 10.1159/000494473. [DOI] [PubMed] [Google Scholar]
- 10.Stjernstrøm KD, Jensen JS, Jakobsen KK, Grønhøj C, von Buchwald C. Current status of human papillomavirus positivity in oropharyngeal squamous cell carcinoma in Europe: A systematic review. Acta Otolaryngol. 2019;139:1112–1116. doi: 10.1080/00016489.2019.1669820. [DOI] [PubMed] [Google Scholar]
- 11.Gaździcka J, Gołąbek K, Strzelczyk JK, Ostrowska Z. Epigenetic modifications in head and neck cancer. Biochem Genet. 2020;58:213–244. doi: 10.1007/s10528-019-09941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhat S, Kabekkodu SP, Jayaprakash C, Radhakrishnan R, Ray S, Satyamoorthy K. Gene promoter-associated CpG island hypermethylation in squamous cell carcinoma of the tongue. Virchows Arch. 2017;470:445–454. doi: 10.1007/s00428-017-2094-2. [DOI] [PubMed] [Google Scholar]
- 13.Ferlazzo N, Currò M, Zinellu A, Caccamo D, Isola G, Ventura V, Carru C, Matarese G, Ientile R. Influence of MTHFR genetic background on p16 and MGMT methylation in oral squamous cell cancer. Int J Mol Sci. 2017;18:724. doi: 10.3390/ijms18040724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, Zheng M, Ren Y. Overexpression of TRMT12 may independently predict poor overall survival in patients with head and neck squamous cell carcinoma. OncoTargets Ther. 2019;12:7269–7279. doi: 10.2147/OTT.S212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Guo T, Xu G, Sakai A, Ren S, Fukusumi T, Ando M, Sadat S, Saito Y, et al. Characterization of alternative splicing events in HPV-negative head and neck squamous cell carcinoma identifies an oncogenic DOCK5 variant. Clin Cancer Res. 2018;24:5123–5132. doi: 10.1158/1078-0432.CCR-18-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, Ochs MF, Ahn SM, Hennessey P, Tan M, Soudry E, Gaykalova DA, Uemura M, Brait M, Shao C, et al. Expression microarray analysis reveals alternative splicing of LAMA3 and DST genes in head and neck squamous cell carcinoma. PLoS One. 2014;9:e91263. doi: 10.1371/journal.pone.0091263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C, Guo T, Sakai A, Ren S, Fukusumi T, Ando M, Sadat S, Saito Y, Califano JA. A novel splice variant of LOXL2 promotes progression of human papillomavirus-negative head and neck squamous cell carcinoma. Cancer. 2020;126:737–748. doi: 10.1002/cncr.32610. [DOI] [PubMed] [Google Scholar]
- 18.Kelley DZ, Flam EL, Guo T, Danilova LV, Zamuner FT, Bohrson C, Considine M, Windsor EJ, Bishop JA, Zhang C, et al. Functional characterization of alternatively spliced GSN in head and neck squamous cell carcinoma. Transl Res. 2018;202:109–119. doi: 10.1016/j.trsl.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li ZX, Zheng ZQ, Wei ZH, Zhang LL, Li F, Lin L, Liu RQ, Huang XD, Lv JW, Chen FP, et al. Comprehensive characterization of the alternative splicing landscape in head and neck squamous cell carcinoma reveals novel events associated with tumorigenesis and the immune microenvironment. Theranostics. 2019;9:7648–7665. doi: 10.7150/thno.36585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Licitra L, Mesia R, Rivera F, Remenár É, Hitt R, Erfán J, Rottey S, Kawecki A, Zabolotnyy D, Benasso M, et al. Evaluation of EGFR gene copy number as a predictive biomarker for the efficacy of cetuximab in combination with chemotherapy in the first-line treatment of recurrent and/or metastatic squamous cell carcinoma of the head and neck: EXTREME study. Ann Oncol. 2011;22:1078–1087. doi: 10.1093/annonc/mdq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferris RL, Blumenschein G, Jr, Fayette J, Guigay J, Colevas AD, Licitra L, Harrington KJ, Kasper S, Vokes EE, Even C, et al. Nivolumab vs investigator's choice in recurrent or metastatic squamous cell carcinoma of the head and neck: 2-year long-term survival update of CheckMate 141 with analyses by tumor PD-L1 expression. Oral Oncol. 2018;81:45–51. doi: 10.1016/j.oraloncology.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauml J, Seiwert TY, Pfister DG, Worden F, Liu SV, Gilbert J, Saba NF, Weiss J, Wirth L, Sukari A, et al. Pembrolizumab for platinum- and cetuximab-refractory head and neck cancer: Results from a single-arm, phase II study. J Clin Oncol. 2017;35:1542–1549. doi: 10.1200/JCO.2016.70.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napolitano M, Schipilliti FM, Trudu L, Bertolini F. Immunotherapy in head and neck cancer: The great challenge of patient selection. Crit Rev Oncol Hematol. 2019;144:102829. doi: 10.1016/j.critrevonc.2019.102829. [DOI] [PubMed] [Google Scholar]
- 24.Kao HF, Lou PJ. Immune checkpoint inhibitors for head and neck squamous cell carcinoma: Current landscape and future directions. Head Neck. 2019;41(Suppl 1):4–18. doi: 10.1002/hed.25930. [DOI] [PubMed] [Google Scholar]
- 25.Ferris RL, Blumenschein G, Jr, Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Lippman SM, Lee JJ, Yang H, Khuri FR, Kim E, Lin J, Chang DW, Lotan R, Hong WK, et al. Genetic variations in regulator of G-protein signaling genes as susceptibility loci for second primary tumor/recurrence in head and neck squamous cell carcinoma. Carcinogenesis. 2010;31:1755–1761. doi: 10.1093/carcin/bgq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Spitz MR, Lee JJ, Lippman SM, Ye Y, Yang H, Khuri FR, Kim E, Gu J, Lotan R, et al. Novel susceptibility loci for second primary tumors/recurrence in head and neck cancer patients: Large-scale evaluation of genetic variants. Cancer Prev Res (Phila) 2009;2:617–624. doi: 10.1158/1940-6207.CAPR-09-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M, Winkler CA, Kopp J, Rotimi C, Adeyemo A, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci USA. 2014;111:E2130–E2139. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riella C, Siemens TA, Wang M, Campos RP, Moraes TP, Riella LV, Friedman DJ, Riella MC, Pollak MR. APOL1-associated kidney disease in Brazil. Kidney Int Rep. 2019;4:923–929. doi: 10.1016/j.ekir.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freedman BI, Spainhour M, Hicks PJ, Turner J, Robertson J, Langefeld CD, Murea M, Divers J. Nephropathy progression in African Americans with a family history of ESKD: Implications for Clinical Trials in APOL1-associated nephropathy. Am J Kidney Dis. 2019;74:284–286. doi: 10.1053/j.ajkd.2019.03.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franceschini N, Kopp JB, Barac A, Martin LW, Li Y, Qian H, Reiner AP, Pollak M, Wallace RB, Rosamond WD, et al. Association of APOL1 with heart failure with preserved ejection fraction in postmenopausal African American women. JAMA Cardiol. 2018;3:712–720. doi: 10.1001/jamacardio.2018.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontaine F, Lecordier L, Vanwalleghem G, Uzureau P, Van Reet N, Fontaine M, Tebabi P, Vanhollebeke B, Büscher P, Pérez-Morga D, et al. APOLs with low pH dependence can kill all African trypanosomes. Nat Microbiol. 2017;2:1500–1506. doi: 10.1038/s41564-017-0034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto K, Rausch JW, Wakashin H, Fu Y, Chung JY, Dummer PD, Shin MK, Chandra P, Suzuki K, Shrivastav S, et al. APOL1 risk allele RNA contributes to renal toxicity by activating protein kinase R. Commun Biol. 2018;1:188. doi: 10.1038/s42003-018-0188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Zheng W, Wang W, Shen H, Liu L, Lou W, Wang X, Yang P. A new panel of pancreatic cancer biomarkers discovered using a mass spectrometry-based pipeline. Br J Cancer. 2017;117:1846–1854. doi: 10.1038/bjc.2017.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Toole JF, Bruggeman LA, Madhavan S, Sedor JR. The cell biology of APOL1. Semin Nephrol. 2017;37:538–545. doi: 10.1016/j.semnephrol.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kardideh B, Samimi Z, Norooznezhad F, Kiani S, Mansouri K. Autophagy, cancer and angiogenesis: Where is the link? Cell Biosci. 2019;9:65. doi: 10.1186/s13578-019-0327-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Codogno P, Morel E. FOXO3a provides a quickstep from autophagy inhibition to apoptosis in cancer therapy. Dev Cell. 2018;44:537–539. doi: 10.1016/j.devcel.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 38.Tao Q, Chan AT. Nasopharyngeal carcinoma: Molecular pathogenesis and therapeutic developments. Expert Rev Mol Med. 2007;9:1–24. doi: 10.1017/S1462399407000312. [DOI] [PubMed] [Google Scholar]
- 39.Sticht C, Freier K, Knöpfle K, Flechtenmacher C, Pungs S, Hofele C, Hahn M, Joos S, Lichter P. Activation of MAP kinase signaling through ERK5 but not ERK1 expression is associated with lymph node metastases in oral squamous cell carcinoma (OSCC) Neoplasia. 2008;10:462–470. doi: 10.1593/neo.08164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estilo CL, O-charoenrat P, Talbot S, Socci ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y, Boyle JO, et al. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lian M, Fang J, Han D, Ma H, Feng L, Wang R, Yang F. Microarray gene expression analysis of tumorigenesis and regional lymph node metastasis in laryngeal squamous cell carcinoma. PLoS One. 2013;8:e84854. doi: 10.1371/journal.pone.0084854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 43.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 44.Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Blal HA, Alm T, Asplund A, Björk L, Breckels LM, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 45.Huang SH, O'Sullivan B. Overview of the 8th edition TNM classification for head and neck cancer. Curr Treat Options Oncol. 2017;18:40. doi: 10.1007/s11864-017-0484-y. [DOI] [PubMed] [Google Scholar]
- 46.Zhaorigetu S, Wan G, Kaini R, Jiang Z, Hu CA. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4:1079–1082. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho CJ, Gorski SM. Molecular nechanisms underlying autophagy-mediated treatment resistance in Cancer. Cancers (Basel) 2019;11:1775. doi: 10.3390/cancers11111775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Toole JF, Schilling W, Kunze D, Madhavan SM, Konieczkowski M, Gu Y, Luo L, Wu Z, Bruggeman LA, Sedor JR. ApoL1 overexpression drives variant-independent cytotoxicity. J Am Soc Nephrol. 2018;29:869–879. doi: 10.1681/ASN.2016121322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kruzel-Davila E, Shemer R, Ofir A, Bavli-Kertselli I, Darlyuk-Saadon I, Oren-Giladi P, Wasser WG, Magen D, Zaknoun E, Schuldiner M, et al. APOL1-mediated cell injury involves disruption of conserved trafficking processes. J Am Soc Nephrol. 2017;28:1117–1130. doi: 10.1681/ASN.2016050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng D, Weckerle A, Yu Y, Ma L, Zhu X, Murea M, Freedman BI, Parks JS, Shelness GS. Biogenesis and cytotoxicity of APOL1 renal risk variant proteins in hepatocytes and hepatoma cells. J Lipid Res. 2015;56:1583–1593. doi: 10.1194/jlr.M059733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khatua AK, Cheatham AM, Kruzel ED, Singhal PC, Skorecki K, Popik W. Exon 4-encoded sequence is a major determinant of cytotoxicity of apolipoprotein L1. Am J Physiol Cell Physiol. 2015;309:C22–C37. doi: 10.1152/ajpcell.00384.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beckerman P, Bi-Karchin J, Park AS, Qiu C, Dummer PD, Soomro I, Boustany-Kari CM, Pullen SS, Miner JH, Hu CA, et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat Med. 2017;23:429–438. doi: 10.1038/nm.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar V, Paliwal N, Ayasolla K, Vashistha H, Jha A, Chandel N, Chowdhary S, Saleem MA, Malhotra A, Chander PN, et al. Disruption of APOL1-miR193a axis induces disorganization of podocyte actin cytoskeleton. Sci Rep. 2019;9:3582. doi: 10.1038/s41598-019-39376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar V, Ayasolla K, Jha A, Mishra A, Vashistha H, Lan X, Qayyum M, Chinnapaka S, Purohit R, Mikulak J, et al. Disrupted apolipoprotein L1-miR193a axis dedifferentiates podocytes through autophagy blockade in an APOL1 risk milieu. Am J Physiol Cell Physiol. 2019;317:C209–C225. doi: 10.1152/ajpcell.00538.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruno J, Pozzi N, Oliva J, Edwards JC. Apolipoprotein L1 confers pH-switchable ion permeability to phospholipid vesicles. J Biol Chem. 2017;292:18344–18353. doi: 10.1074/jbc.M117.813444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah SS, Lannon H, Dias L, Zhang JY, Alper SL, Pollak MR, Friedman DJ. APOL1 kidney risk variants induce cell death via mitochondrial translocation and opening of the mitochondrial permeability transition pore. J Am Soc Nephrol. 2019;30:2355–2368. doi: 10.1681/ASN.2019020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis SE, Khatua AK, Popik W. Nucleosomal dsDNA Stimulates APOL1 expression in human cultured podocytes by activating the cGAS/IFI16-STING signaling pathway. Sci Rep. 2019;9:15485. doi: 10.1038/s41598-019-51998-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 59.Kundu M, Thompson CB. Autophagy: Basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 60.Antunes F, Erustes AG, Costa AJ, Nascimento AC, Bincoletto C, Ureshino RP, Pereira GJ, Smaili SS. Autophagy and intermittent fasting: The connection for cancer therapy? Clinics (Sao Paulo) 2018;73(Suppl 1):e814s. doi: 10.6061/clinics/2018/e814s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salminen A, Kaarniranta K, Kauppinen A. Beclin 1 interactome controls the crosstalk between apoptosis, autophagy and inflammasome activation: Impact on the aging process. Ageing Res Rev. 2013;12:520–534. doi: 10.1016/j.arr.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 62.Yu L, Chen Y, Tooze SA. Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 2018;14:207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levy JM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chidiac M, Fayyad-Kazan M, Daher J, Poelvoorde P, Bar I, Maenhaut C, Delrée P, Badran B, Vanhamme L. ApolipoproteinL1 is expressed in papillary thyroid carcinomas. Pathol Res Pract. 2016;212:631–635. doi: 10.1016/j.prp.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 65.Hu CA, Klopfer EI, Ray PE. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett. 2012;586:947–955. doi: 10.1016/j.febslet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Y, Liu J, Nipper M, Wang P. Ductal vs. acinar? Recent insights into identifying cell lineage of pancreatic ductal adenocarcinoma. Ann Pancreat Cancer. 2019 Jun 17; doi: 10.21037/apc.2019.06.03. (Epub ahead of print). doi: 10.21037/apc.2019.06.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang J, Jia L, Tsang CM, Tsao SW. EBV infection and glucose metabolism in nasopharyngeal carcinoma. Adv Exp Med Biol. 2017;1018:75–90. doi: 10.1007/978-981-10-5765-6_6. [DOI] [PubMed] [Google Scholar]
- 68.Elgui de Oliveira D, Müller-Coan BG, Pagano JS. Viral carcinogenesis beyond malignant Transformation: EBV in the progression of human cancers. Trends Microbiol. 2016;24:649–664. doi: 10.1016/j.tim.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the TCGA (http://cancergenome.nih.gov/), the GEO (https://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) data portals. The in-house IHC data from the present study can be acquired from the correspondence author on reasonable request.