Abstract
Background:
Salmonella infection (salmonellosis) is a zoonotic bacterial disease. Widespread use of antibiotics in livestock and poultry production for different purposes such as treatment and growth promotion has led to the emergence of antibiotic-resistant Salmonella, causing treatment of Salmonella infections more difficult with each passing year.
Aims:
To determine the antibiotic resistance prevalence of Salmonella serotypes isolated from animals in different provinces of Iran.
Methods:
To find eligible articles, we searched the international and national electronic databases using appropriate keywords in English and Persian.
Results:
After applying predefined criteria, 54 articles reporting antibiotic resistance profiles of Salmonella serotypes were included. Salmonella isolates were mostly resistant against nalidixic acid (67%), tetracycline (66.9%), and streptomycin (49.6%), followed by trimethoprim/sulfamethoxazole (41.6%) and kanamycin (23.6%). The highest sensitivity was observed against imipenem, meropenem, and cefepime with 1.7%, 1.4%, and 1.9% of all isolates being resistant, respectively.
Conclusion:
Results revealed that the prevalence of resistant isolates to nalidixic acid, tetracycline and streptomycin is high and their use must be restricted. In addition, resistance to other antibiotics such as chloramphenicol, ampicillin, cephalothin, cefixime, and enrofloxacin is at an alarming level that calls for attention in the future infection control and antibiotic stewardship programs.
Key Words: Animals, Antibiotic resistance, Salmonella, Salmonellosis
Introduction
Salmonella are Gram-negative rod-shaped bacteria belonging to the Enterobacteriaceae. Salmonella genus comprises two species including S. enetrica and S. bongori. Salmonella enterica is the type species and is further divided into six subspecies including diarizonae, enterica, salamae, arizonae, indica, and houtenae (Su and Chiu, 2007 ▶). Based on Kauffmann-White scheme, which uses combination of H, O, and Vi surface antigens for serotyping, more than 2,600 unique serotypes have already been reported. The majority of identified serotypes belong to S. enterica subsp. enterica (Su et al., 2007 ▶; Gal-Mor et al., 2014 ▶). It has been shown that gastrointestinal tract of animals such as pigs, birds and cattle is the main reservoir of Salmonella serotypes. These bacteria can survive out of animal body and humans are mostly infected by contaminated foods such as meat, fruit and vegetables (Paniel and Noguer, 2019 ▶). Based on the severity of disease, infections caused by Salmonella serotypes are categorized into typhoidal and non-typhoidal infections. Infections caused by non-typhoidal Salmonella (Typhimurium) are usually self-restricted diseases in different mammals and birds, whereas typhoidal Salmonella (Typhi and Paratyphi) cause life-threatening diseases in human (Gal-Mor et al., 2014 ▶; GutVasiljevic et al., 2018 ▶; Paniel and Noguer, 2019 ▶). Salmonella infection (salmonellosis) is a zoonotic bacterial disease mainly transmitted to human by contaminated food and water. Poultry products and raw meat are known as the most important sources of Salmonella species transmitted to humans. In fact, the slaughtering process of food animals such as poultry and cattle is considered as one of the most important routes of Salmonella transmission to human. Salmonellosis affects intestine and the most important signs of infection are abdominal cramps, fever, vomiting and diarrhea (Gut et al., 2018 ▶; Paniel and Noguer, 2019 ▶). Widespread use of antibiotics in livestock and poultry production for different purposes such as treatment, growth promotion and prophylaxis has led to the emergence of antibiotic-resistant serotypes, causing treatment of Salmonella infections more difficult with each passing year (Rahmani et al., 2013 ▶). Owing to the importance of Salmonella serotypes in human public health and necessity of information about epidemiology of Salmonella antibiotic resistance patterns to launch infection control programs, the aim of present study was to determine antibiotic resistance profiles of Salmonella serotypes isolated from animals in Iran.
Materials and Methods
Search strategy
This study was carried out based on the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analysis) guidelines (Liberati et al., 2009 ▶). To find potentially eligible articles, we searched international and national electronic databases including Google Scholar (https://scholar.google.com), Scopus (https:// www.scopus.com), PubMed (https://www.ncbi.nlm.nih. gov/pubmed), ISI Web of Knowledge (https://www. webofknowledge.com(, Scientific Information Database (https://www.sid.ir), and Magiran (https://www.magiran. com). Based on MeSH terms, the following keywords and their combinations in Persian and English languages were used; Salmonella, antibiotic resistance, antimicrobial resistance, animal sources, drug resistance and Iran. To find missing articles, reference lists of eligible articles were manually searched. The last search was performed on September 2019.
Study selection, quality assessment and data extraction
Two independent researchers assessed the eligibility of articles. Joanna Briggs Institute (JBI) checklist was used to evaluate the quality of obtained papers (Munn et al., 2015 ▶). The titles and abstracts of articles were evaluated to find more relevant studies. We included studies reporting antibiotic resistance patterns of Salmonella serotypes isolated from animals. For each eligible article, first author name, provinces of study, antibiotic resistance method, sample origin and time of study were extracted. Additionally, articles were excluded if materials and methods were unclear, samples were collected from non-animal samples, non-original study, and full text of articles was not available.
Statistical analysis
We used Comprehensive Meta-Analysis (CMA) software (Biostat, Englewood, NJ, USA) for statistical analysis. Also, based on random- or fixed-effects models, data analysis was performed and the results were expressed as percentage and 95% confidence intervals (CIs). Heterogeneity among studies was also assessed using the I2 and Cochran Q test. In addition, funnel plots were generated to find potential publication bias (Smith et al., 1997).
Results
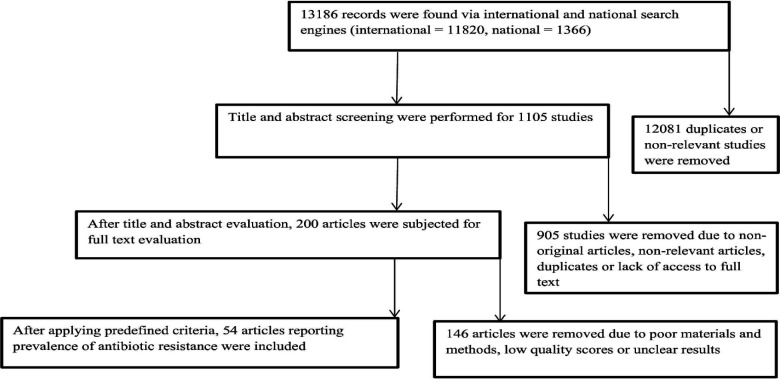
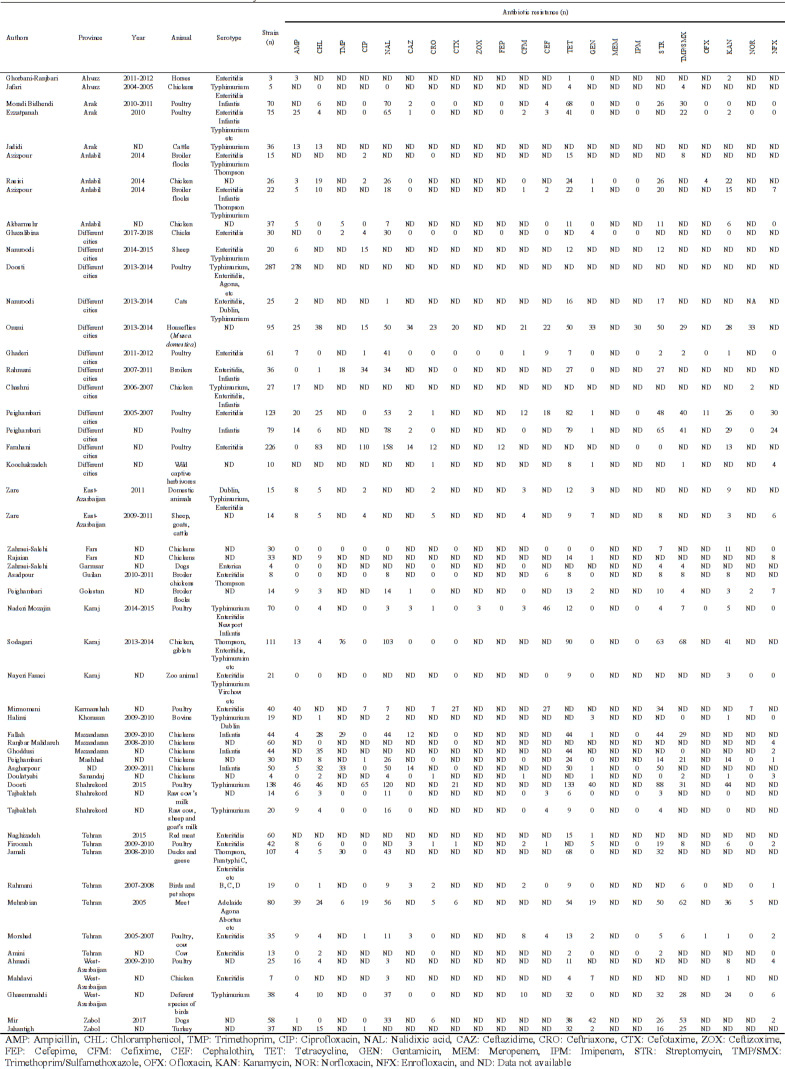
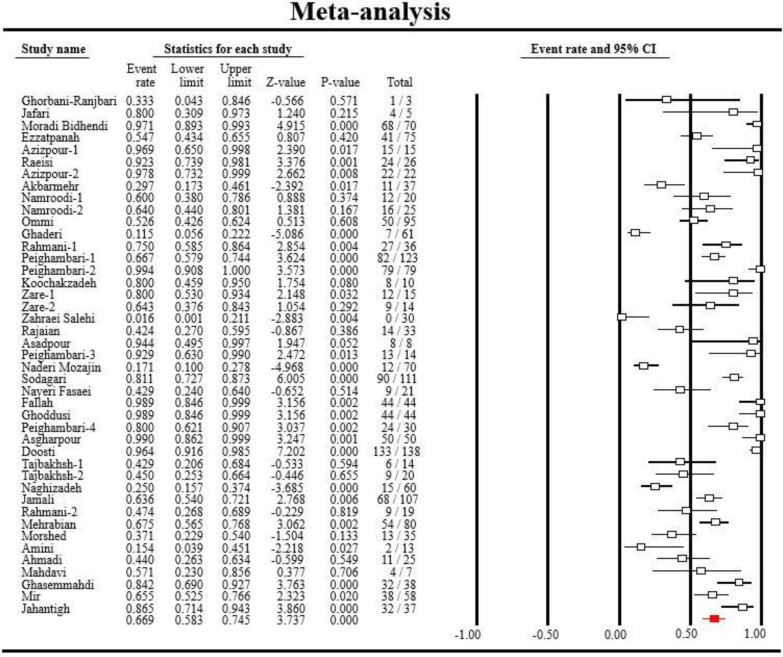
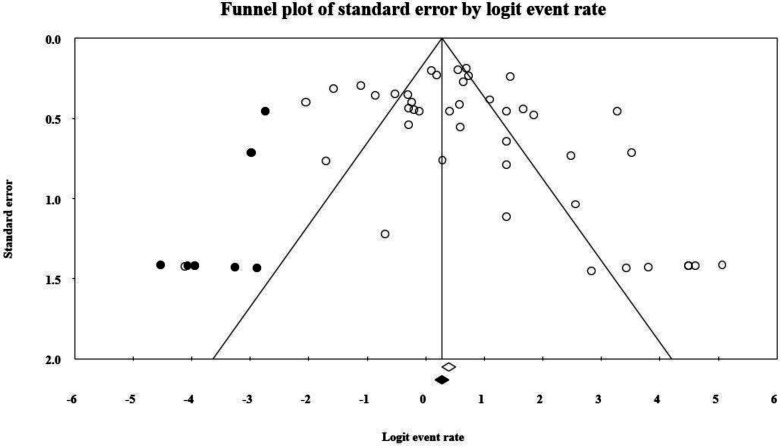
The detailed steps of process of study selection are shown in Fig. 1. Briefly, the electronic search engine resulted in 13186 potentially relevant records. Based on the screening of titles and abstracts, 12,986 articles were removed because they were duplicate or non-relevant and 200 were selected for full text checking to find eligible studies. After applying inclusion and exclusion criteria, 146 articles were removed due to their poor materials and methods or insufficient data or low-quality scores. Finally, 54 articles, reporting antibiotic resistance profiles of Salmonella serotypes were included in the study (Rajaeian et al., 2003 ▶; Salehi et al., 2005 ▶; Jafari et al., 2007 ▶; Mehrabian et al., 2007 ▶; Mirmomeni et al., 2007 ▶; Emadi et al., 2009 ▶; Morshed and Peighambari, 2010 ▶; Peighambari et al., 2011 ▶; Akbarmehr et al., 2012 ▶; Firoozeh et al., 2012 ▶; Jadidi et al., 2012 ▶; Tajbakhsh et al., 2012 ▶; Ahmadi et al., 2013 ▶; Ezatpanah et al., 2013 ▶; Fallah et al., 2013 ▶; Ghorbani-Ranjbary et al., 2013 ▶; Malidareh et al., 2013 ▶; Peighambari et al., 2013 ▶; Rahmani et al., 2013 ▶; Salehi et al., 2013 ▶; Tajbakhsh and Tajbakhsh, 2013 ▶; Asadpour et al., 2014 ▶; Asgharpour et al., 2014 ▶; Halimi et al., 2014 ▶; Jamali et al., 2014 ▶; Zare et al., 2014 ▶; Ghasemmahdi et al., 2015 ▶; Ghoddusi et al., 2015 ▶; Jahantigh et al., 2015 ▶; Oskouizadeh et al., 2015 ▶; Peighambari et al., 2015 ▶; Raeisi and Ghiamirad, 2015 ▶; Sodagari et al., 2015 ▶; Amini, 2016 ▶; Doosti et al., 2016a ▶, b; Ghaderi et al., 2016 ▶; Namroodi and Behine, 2016 ▶; Namroodi et al., 2016 ▶; Zare et al., 2016 ▶; Doulatyabi and Peyghambari, 2017 ▶; Fasaei et al., 2017 ▶; Moradi et al., 2017 ▶; Naghizadeh and Moradi, 2017 ▶; Ommi et al., 2017 ▶; Azizpour, 2018a ▶, b; Farahani et al., 2018 ▶; Mahdavi et al., 2018 ▶; Mir et al., 2018 ▶; Mozajin et al., 2018 ▶; Peighambari et al., 2018 ▶; Ghazalibina et al., 2019 ▶; Peighambari et al., 2019 ▶). Most of the reports were from Tehran and Ardabil provinces with seven and four studies, respectively (Table 1), followed by Arak (three studies), Mazandaran (three studies), and Shahrekord (three studies) provinces. Disk diffusion was the most common method used for the determination of antibiotic resistance profiles of Salmonella serotypes isolated from animals in Iran. The majority of samples were collected from poultry, sheep and cow. Salmonella Typhimurium and Enteritidis were the most prevalent isolated serotypes (Table 1). The forest plot of resistance against tetracycline is shown in Fig. 2. As shown in Fig. 3, we observed some evidence of publication bias due to the asymmetric distribution of studies in funnel plots.
Fig. 1.
Study selection progress
Table 1.
characteristics of studies involved in this meta-analysis
Fig. 2.
The forest plot examining the overall prevalence of resistance to tetracycline on studies performed in Iran
Fig. 3.
Funnel plot with pseudo 95% confidence intervals demonstrating the effect sizes derived from each study
Prevalence of resistance against cephalosporins
Due to the presence of a high degree of heterogeneity among included studies, we used random-effects models for the meta-analysis of data on the prevalence of antibiotic resistance of Salmonella serotypes to cephalosporins except for ceftizoxime. The highest resistance rate was observed against cephalothin 13.5% (95% CI: 7.1-24.1; I2=88.8%; Q=153; P=0.00) and cefixime 9% (95% CI: 5.6-14; I2=63.6%; Q=52.2; P=0.00), followed by ceftazidime 5.5% (95% CI: 3-9.9; I2=82.2%; Q=118.6; P=0.00), ceftriaxone 5.2% (95% CI: 3.1-8.6; I2=68.2%; Q=81.9; P=0.00), cefotaxime 3.7% (95% CI: 1.6-8.6; I2=85.2%; Q=108.8; P=0.00), ceftizoxime 3.4% (95% CI: 1.2-9.2; I2=18.7%; Q=1.2; P=0.26), and cefepime 1.9% (95% CI: 0.7-5.3; I2=33.5%; Q=7.5; P=0.18).
Prevalence of resistance against quinolones
Similar to cephalosporins, a random-effects model was used for the meta-analysis of data on the prevalence of antibiotic resistance of Salmonella serotypes to quinolones. Antibiotic resistance patterns of Salmonella serotypes against quinolones were as follows; nalidixic acid 67% (95% CI: 56.1-76.3; I2=90.7%; Q=378.2; P=0.00), enrofloxacin 10.7% (95% CI: 6.8-16.6; I2=75.3%; Q=101.3; P=0.00), ciprofloxacin 7% (95% CI: 4-11.8; I2=87.6%; Q=267.8; P=0.00), norfloxacin 4.3% (95% CI: 1.8-9.9; I2=78.9%; Q=66.3; P=0.00), and ofloxacin 3.5% (95% CI: 1.4-8.6; I2=55.3%; Q=15.6; P=0.02).
Carbapenems and aminoglycosides-resistant Salmonella
Antibiotic resistance profiles of Salmonella serotypes to carbapenems and aminoglycosides were assessed using either random- or fixed-effects models. The prevalence of resistance against imipenem and meropenem were 1.7% (95% CI: 0.2-11.3; I2=0.0%; Q=0.0; P=0.00), and 1.4% (95% CI: 0.4-4.6; I2=80.8%; Q=89; P=0.00), respectively. The highest resistance rate was seen against streptomycin and kanamycin with 49.6% (95% CI: 40.6-58.7; I2=87.7%; Q=278.1; P=0.00), and 23.6% (95% CI: 17.1-31.6; I2=85.7%; Q=217.7; P=0.00) of isolates being resistant, respectively. The lowest resistance rate was against gentamicin 6.3% (95% CI: 3.8-10.3; I2=82.8%; Q=232.9; P=0.00).
Prevalence of resistance against other antibiotics
The prevalence of resistant isolates to tetracycline, trimethoprim/sulfamethoxazole, ampicillin, trimethop-rim, and chloramphenicol were as follows: 66.9% (95% CI: 58.3-74.5; I2=87.2%; Q=330.4; P=0.00), 41.6% (95% CI: 31.4-52.5; I2=89.6%; Q=250.5; P=0.00), 21.9% (95% CI: 15.1-30.6; I2=89.7%; Q=390.6; P=0.00), 19.7% (95% CI: 9.8-35.8; I2=91.1%; Q=135.8; P=0.00), and 18.9% (95% CI: 14.1-24.9; I2=85.6%; Q=300.5; P=0.00), respectively. Meta-analysis of data on the prevalence of antibiotic-resistant Salmonella serotypes to these drugs was done using a random-effects model.
Discussion
Antimicrobial resistance is one of the most important public health concerns worldwide and contributes to the economic burden of both developed and developing countries through increasing the costs associated with treatment, hospitalization and infection control procedures (Antonelli et al., 2019 ▶). Salmonella isolates are among the most important food-borne pathogens that are predominantly transmitted to humans via consumption of eggs, dairy products and fresh fruits contaminated with animal faeces (Eng et al., 2015 ▶). In Iran, no comprehensive study is available on the prevalence of antibiotic-resistant Salmonella strains isolated from animals. Therefore, we conducted this study to investigate the antibiotic resistance profile of Salmonella serotypes isolated from animals in different provinces of Iran. In fact, resistance profiles of Salmonella serotypes against 22 antibiotics belonging to different classes such as beta-lactams, aminoglycosides and quinolones were investigated. Our results revealed that the majority of Salmonella strains were resistant to nalidixic acid, tetracycline and streptomycin, with 67%, 66.9% and 49.6% of all isolates being resistant, respectively. These resistance rates are in agreement with the results reported from United States, reporting 63% and 26% resistance to tetracycline and streptomycin, respectively (Velasquez et al., 2018 ▶). In Iran, tetracycline and quinolones are generally used in the livestock and poultry industries; therefore, this high frequency of resistance was predictable. In fact, antibiotic resistance profile of food-borne pathogens such as Salmonella serotypes can reflect the antibiotics used for animal treatment (Dallal et al., 2010 ▶). Tetracyclines, as protein synthesis inhibitors, bind to the small ribosomal subunit and prevent attachment of aminoacyl-tRNA to protein synthesis complex. Several mechanisms can confer resistance against tetracycline, including efflux pumps, mutations and enzymatic inactivation (Hao et al., 2016 ▶). Genes conferring resistance against tetracycline are often located in transferable genetic elements such as integrons and plasmids. Hence, it can be concluded that overuse of tetracycline might have resulted in the elimination of sensitive isolates and dissemination of resistant isolates harboring resistance genes (Hao et al., 2016 ▶). For several years, as the traditional first-line treatments, antimicrobial agents such as chloramphenicol, ampicillin and trimethoprim-sulfamethoxazole were used for Salmonella infection treatment. However, with the emergence of resistant isolates, the traditional antibiotics were replaced with cephalosporins (Eng et al., 2015 ▶). Based on the results of the present study, prevalence of isolates resistant to chloramphenicol, ampicillin and trimethoprim-sulfamethoxazole were 18.9%, 21.9% and 41.6%, respectively. These findings are in agreement with studies from Pakistan, India and Nepal, but in contrast to a previous study from Egypt (Ochiai et al., 2008 ▶; El-Sharkawy et al., 2017 ▶). Cephalosporins belong to the beta-lactam antibiotic family and inhibit bacterial growth by disruption of peptidoglycan. Cephalosporins have been extensively used in food animals, resulting in the development of antibiotic resistance in food-borne pathogens. Similar to the findings of a previous studies performed in different provinces of China, including Henan, Shaanxi, Fujian, Sichuan, Guangdong and Guangxi (Wang et al., 2015 ▶), our results revealed a relatively high susceptibility of Salmonella strains to cephalosporins, with prevalence of resistance against cephalothin 13.5%, cefixime 9%, ceftriaxone 5.2%, ceftizoxime 3.4% and cefotaxime 3.7%. Resistance to cephalosporins and carbapenems is often caused by beta-lactamase enzymes. Extended-spectrum beta-lactamases (ESBLs) are powerful group of enzymes that can hydrolyze the structure of beta-lactam antibiotics, especially new generation of cephalosporins and carbapenems (imipenem and meropenem). Based on their structure, these enzymes are divided into four groups: A, B, C and D (Bush and Jacoby, 2010 ▶). Because limited antibiotics are available for the treatment of infections caused by ceftazidime-, cefepime- and imipenem-resistant Salmonella, the prevalence of ESBLs-positive isolates is of critical importance. Although the prevalence of ESBLs-positive Salmonella is still rare, there has been an increasing trend in recent years that is alarming (Hur and Lee, 2012 ▶). In this study, we also investigated antibiotic resistance profile of Salmonella strains against aminoglycoside antibiotics. Unfortunately, based on our findings, resistance rates to streptomycin and kanamycin were high, with 49.6% and 23.6% of isolates being resistant, respectively. Aminoglycoside-modifying enzymes including aminoglycoside acetyltransferases, expressed by acc genes, and aminoglycoside phosphotransferases (strA, strB, aph(3)-Ib and aph(6)-Id) are the most important genes located in transferable genetic elements (integrons and plasmids), and are responsible for resistance against kanamycin and streptomycin (Vtnair et al., 2018 ▶). Unfortunately, the majority of resistance determinants, including those that confer resistance against β-lactam antibiotics, aminoglycosides, fluoroquinolones, chloramphenicol and tetracyclines, have been identified in various Salmonella serovars isolated from the food animals (Vtnair et al., 2018 ▶). The most important point is that these resistance genes are often located in the transferable genetic elements; therefore, misuse of antibiotics may lead to the elimination of susceptible isolates and increase the resistant population and then facilitate spread of multi-drug resistant Salmonella infections.
Similar to findings of this study, several independent studies in Iran showed that Salmonella strains isolated from human infections were mostly resistant against fluoroquinolones, tetracycline, streptomycin, chloram-phenicol and ceftizoxime (Eshraghi et al., 2010 ▶; Ranjbar et al., 2011 ▶; Khademi et al., 2020 ▶). Owing to the fact that Salmonella is able to establish zoonotic infections and also is able to acquire resistance genes from other enteric pathogens through mobile genetic elements such as integrons, plasmids and transposons, the misuse of antibiotics in food animal industries can lead to the spread of drug-resistant Salmonella infections (Ranjbar et al., 2010 ▶; Khademi et al., 2020 ▶).
To the best of the authors’ knowledge, this is the first comprehensive study on the prevalence of antibiotic resistance profile of Salmonella species isolated from animals. Our results revealed that resistance to nalidixic acid, tetracycline, streptomycin and kanamycin are high and their use must be restricted. Salmonella is one of the most important food-borne pathogens responsible for life-treating infections in humans. The routine practice of giving antibiotics to animal as a means of promoting growth and preventing disease creates selection pressure that results in the survival of antibiotic-resistant pathogens. Subsequently, these antibiotic-resistant pathogens can be transmitted to humans via direct contact with animals or even consumption of contaminated foods of animal origin. Therefore, appropriate use of antibiotics in veterinary medicine and animal feeding will reduce the emergence of multi-drug resistant infections.
References
- Ahmadi, M , Talebi, A , Mansouri, S , Rahman, B Antimicrobial resistance pattern and plasmid profile of Salmonella isolates from poultry. J. Vet. Microbiol. 2013;9:117–128. [Google Scholar]
- Akbarmehr, J Antimicrobial resistance in Salmonella isolated from broiler chicken carcasses. Afr. J. Microbiol. Res. 2012;6:1485–1488. [Google Scholar]
- Amini, K Prevalence of antibiotic resistance genes in Salmonella Enteritidis isolated from animal and human and determining their antibiotic resistance patterns. J. Comp. Pathobiol. 2016;12:1733–1740. [Google Scholar]
- Antonelli, P , Belluco, S , Mancin, M , Losasso, C , Ricci, A Genes conferring resistance to critically important antimicrobials in Salmonella enterica isolated from animals and food: a systematic review of the literature, 2013-2017. Res. Vet. Sci. 2019;126:59–67. doi: 10.1016/j.rvsc.2019.08.022. [DOI] [PubMed] [Google Scholar]
- Asadpour, Y , Mohammadi, M , Rasa, M Isolation, serotyping and antibiotic resistance of Salmonella isolated from chicken carcasses in Guilan province. Iran. Vet. J. 2014;9:5–13. [Google Scholar]
- Asgharpour, F , Rajabnia, R , Shahandashti, EF , Marashi, MA , Khalilian, M , Moulana, Z Investigation of class I integron in Salmonella Infantis and its association with drug resistance. Jundishapur J. Microbiol. 2014;7:1–5. doi: 10.5812/jjm.10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizpour, A Determining the antibiotic resistance patterns of isolated Salmonella from broiler flocks to 28 antimicrobial agents used in Iran. J. Comp. Pathobiol. 2018a;15:2411–2420. [Google Scholar]
- Azizpour, A A survey on prevalence of Salmonella Enteritidis and Salmonella Typhimurium serotypes in broiler flocks of Ardabil province and determination of their antibiotics resistance to five antibacterial agents widely used in the Iranian medical field. J. Health. 2018b;9:143–151. [Google Scholar]
- Bush, K , Jacoby, G Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallal, M , Doyle, M , Rezadehbashi, M , Dabiri, H , Sanaei, M , Modarresi, S Prevalence and antimicrobial resistance profiles of Salmonella serotypes, Campylobacter and Yersinia spp isolated from retail chicken and beef, Tehran, Iran. Food Cont. 2010;21:388–392. [Google Scholar]
- Doosti, A , Mahmoudi, E , Jami, M , Mokhtari-Farsani, A Prevalence of aadA1, aadA2, aadB, strA and strB genes and their associations with multidrug resistance phenotype in Salmonella Typhimurium isolated from poultry carcasses. Thai Vet. Med. 2016a;46:691–697. [Google Scholar]
- Doosti, A , Zohoor, A , Chehelgerdi, M , Mokhtari-Farsani, A Distribution of TEM-1 gene in Salmonella enterica isolated from poultry carcasses in Iran. Thai Vet. Med. 2016b;46:9–15. [Google Scholar]
- Doulatyabi, S , Peyghambari, S Survey of Salmonella infections in broiler farms around Sanandaj. Sci. J. Ilam Uni. Med. Sci. 2017;25:70–78. [Google Scholar]
- Egger, M , Smith, GD , Schneider, M , Minder, C Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharkawy, H , Tahoun, A , El-Gohary, A , El-Abasy, M , El-Khayat, F , Gillespie, T , El-Adawy, H Epidemiological, molecular characterization and antibiotic resistance of Salmonella enterica serovars isolated from chicken farms in Egypt. Gut Path. 2017;9:1–12. doi: 10.1186/s13099-017-0157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emadi, CS , Hasanzadeh, M , Bozorg, M , Mirzaei, S Characterization of the Salmonella isolates from backyard chickens in north of Iran, by serotyping, multiplex PCR and antibiotic resistance analysis. Arch. Razi Inst. 2009;7:35–41. [Google Scholar]
- Eng, SK , Pusparajah, P , Mutalib, N , Ser, H , Chan, KG , Lee, LH Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015;8:284–293. [Google Scholar]
- Eshraghi, S , Dalall, M , Fardsanei, F , Salehi, TZ , Ranjbar, R , Nikmanesh, B , Akbari, A Salmonella Enteritidis and antibiotic resistance patterns: a study on 1950 children with diarrhea. Tehran Uni. Med. J. 2010;67:876–882. [Google Scholar]
- Ezatpanah, E , Moradi, BS , Khaki, P , Ghaderi, R , Seyedan, JE , Moghtadaee, FS Isolation, serotyping and antibiotic-resistance pattern of isolated Salmonella from chicken of Arak. Iran. Vet. J. 2013;9:88–96. [Google Scholar]
- Fallah, SH , Asgharpour, F , Naderian, Z , Moulana, Z Isolation and determination of antibiotic resistance patterns in nontyphoid Salmonella spp isolated from chicken. Int. J. Enteric. Pathog. 2013;1:17–21. [Google Scholar]
- Farahani, RK , Ehsani, P , Ebrahimi-Rad, M , Khaledi, A Molecular detection, virulence genes, biofilm formation, and antibiotic resistance of Salmonella enterica serotype Enteritidis isolated from poultry and clinical samples. Jundishapur J. Microbiol. 2018;11:1–9. [Google Scholar]
- Fasaei, BN , Tamai, IA Detection of Salmonella spp from zoo animals in Iran, determination of serovars, antibiotic susceptibility and genotyping by RAPD-PCR. J. Hellenic Vet. Med. Soci. 2017;68:377–384. [Google Scholar]
- Firoozeh, F , Zahraei-Salehi, T , Shahcheraghi, F , Karimi, V , Aslani, MM Characterization of class I integrons among Salmonella enterica serovar Enteritidis isolated from humans and poultry. FEMS Immunol. Med. Microbiol. 2012;64:237–243. doi: 10.1111/j.1574-695X.2011.00883.x. [DOI] [PubMed] [Google Scholar]
- Gal-Mor, O , Boyle, EC , Grassl, GA Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 2014;5:1–10. doi: 10.3389/fmicb.2014.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaderi, R , Moradi, S Occurrence of multidrug-resistant Salmonella enterica serovar Enteritidis isolates from poultry in Iran. Arch. Razi Inst. 2016;71:43–49. [Google Scholar]
- Ghasemmahdi, H , Tajik, H , Moradi, M , Mardani, K , Modaresi, R , Badali, A , Dilmaghani, M Antibiotic resistance pattern and biofilm formation ability of clinically isolates of Salmonella enterica serotype Typhimurium. Int. J. Enteric. Pathog. 2015;3:1–6. [Google Scholar]
- Ghazalibina, M , Farahani, RK , Mansouri, S , Meskini, M , Farahani, AHK , Khaledi, A Molecular detection of antibiotic resistance genes, and class I, and II integrons in Salmonella Enteritidis isolated from Iranian one-day-old chicks. Gene Rep. 2019;16:1–8. [Google Scholar]
- Ghoddusi, A , Fasaei, BN , Karimi, V , Tamai, IA , Moulana, Z , Salehi, TZ Molecular identification of Salmonella Infantis isolated from backyard chickens and detection of their resistance genesby PCR. Iran. J. Vet. Res. 2015;16:293–297. [PMC free article] [PubMed] [Google Scholar]
- Ghorbani-Ranjbary, A , Ghorbani-Ranjbary, N , Ghorbani-Ranjbary, Z Survey on Salmonella infection horse rate in horses of some horse riding clubs Southern Iran. Am. J. Discovery Dev. 2013;3:181–187. [Google Scholar]
- Gut Abraham Majak , Vasiljevic, T , Yeager, T , Donkor, O Salmonella infection-prevention and treatment by antibiotics and probiotic yeasts: a review. Microbiology (Reading) 2018;164:1327–1344. doi: 10.1099/mic.0.000709. [DOI] [PubMed] [Google Scholar]
- Halimi, HA , Seifi, HA , Rad, M Bovine salmonellosis in Northeast of Iran: frequency, genetic fingerprinting and antimicrobial resistance patterns of Salmonella spp. Asian Pac. J. Trop. Biomed. 2014;4:1–7. doi: 10.1016/S2221-1691(14)60199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, H , Sander, P , Iqbal, Z , Wang, Y , Cheng, G , Yuan, Z The risk of some veterinary antimicrobial agents on public health associated with antimicrobial resistance and their molecular basis. Front. Microbiol. 2016;7:1–11. doi: 10.3389/fmicb.2016.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur, J , Jawale, C , Lee, JH Antimicrobial resistance of Salmonella isolated from food animals: a review. Food Res. Int. 2012;45:819–830. [Google Scholar]
- Jadidi, A , Hosseni, SD , Homayounimehr, A , Hamidi, A , Ghani, S , Rafiee, B Simple and rapid detection of Salmonella sp from cattle feces using polymerase chain reaction (PCR) in Iran. Afr. J. Microbiol. Res. 2012;6:5210–5214. [Google Scholar]
- Jafari, R , Ghorbanpour, M , Jaideri, A An investigation into Salmonella infection status in backyard chickens in Iran. Int. J. Poult. Sci. 2007;6:227–229. [Google Scholar]
- Jahantigh, M , Jafari, SM , Rashki, A , Salari, S Prevalence and antibiotic resistance of Salmonella spp in Turkey. Open J. Med. Microbiol. 2015;5:113–117. [Google Scholar]
- Jamali, H , Radmehr, B , Ismail, S Prevalence and antimicrobial resistance of Listeria, Salmonella, and Yersinia species isolates in ducks and geese. Poultry Sci. 2014;93:1023–1030. doi: 10.3382/ps.2013-03699. [DOI] [PubMed] [Google Scholar]
- Khademi, F , Vaez, H , Ghanbari, F , Arzanlou, M , Mohammadshahi, J , Sahebkar, A Prevalence of fluoroquinolone-resistant Salmonella serotypes in Iran: a meta-analysis. Path. Glob. Health. 2020;114:16–29. doi: 10.1080/20477724.2020.1719701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati, A , Altman, DG , Tetzlaff, J , Mulrow, C , Gtzsche, PC , Ioannidis, JP , Moher, D The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6:1–28. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi, S , Azizi Dehbokri, M , Isazadeh, A Contamination of chicken meat with Salmonella spp distributed in mahabad city, Iran. Int. J. Enteric. Pathog. 2018;6:65–68. [Google Scholar]
- Malidareh, NR , Firouzi, S , Malidareh, NR , Habibi, H In vitro and in vivo susceptibility of Salmonella spp isolated from broiler chickens. Comp. Clin. Pathol. 2013;22:1065–1068. doi: 10.1007/s00580-012-1527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabian, S , Jaberi, E Isolasion, identification and antimicrobial resistance patterns of Salmonella from meat products in Tehran. Pak. J. Biol. Sci. 2007;10:122–126. doi: 10.3923/pjbs.2007.122.126. [DOI] [PubMed] [Google Scholar]
- Mir, R , Rashki Ghalehnoo, Z Frequency and antimicrobial resistance pattern of Salmonella spp in asymptomatic rural dog in Zabol. New Find. Vet. Microbiol. 2018;1:44–50. [Google Scholar]
- Mirmomeni, M , Colagar, AH , Ghazaey, S Molecular study of Salmonella Enteritidis in poultry samples by PCR, plasmid curing, antibiotic resistance and protein pattern analysis. Pak. J. Biol. Sci. 2007;10:1562–1570. doi: 10.3923/pjbs.2007.1562.1570. [DOI] [PubMed] [Google Scholar]
- Moradi, BS Investigation of antibiotic resistance in Salmonella Infantis isolated from poultry in Arak. Vet. Res. 2017;13:99–108. [Google Scholar]
- Morshed, R , Peighambari, SM Drug resistance, plasmid profile and random amplified polymorphic DNA analysis of Iranian isolates of Salmonella Enteritidis. New Microbiol. 2010;33:47–56. [PubMed] [Google Scholar]
- Mozajin, MN , Khaki, P , Noorbakhsh, F Antibiotic resistance of Salmonella enterica producing extended-spectrum β-lactamases (ESBLs) type CMY-2, in poultry. J. Gorgan Uni. Med. Sci. 2018;20:109–115. [Google Scholar]
- Munn, Z , Moola, S , Lisy, K , Riitano, D , Tufanaru, C Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and cumulative incidence data. Int. J. Evidence-Based Health. 2015;13:147–153. doi: 10.1097/XEB.0000000000000054. [DOI] [PubMed] [Google Scholar]
- Naghizadeh, S , Moradi, G Determination of antibiotic resistance and identification of tetracycline (tet) resistance genes in Salmonella Enteritidis strains isolated from food sources. Iran. J. Med. Microbiol. 2017;11:64–69. [Google Scholar]
- Namroodi, S , Behine, K Frequency, serovars and antibiotic resistant pattern of Salmonella spp isolated from sheep in golestan province, Iran. Iran. J. Rum. Res. 2016;4:21–36. [Google Scholar]
- Namroodi, S , Staji, H , Mazandarani, E Epidemiological survey of Salmonella in rural cats: a survey of serotype, presence of spv r and spv b genes, and antibiotic resistance pattern. Iran. J. Epidemiol. 2016;12:47–55. [Google Scholar]
- Ochiai, RL , Acosta, CJ , Danovaro-Holliday, M , Baiqing, D , Bhattacharya, SK , Agtini, MD , Shin, S A study of typhoid fever in five Asian countries: disease burden and implications for controls. Bull. World Health Organ. 2008;86:260–268. doi: 10.2471/BLT.06.039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ommi, D , Hemmatinezhad, B , Hafshejani, TT , Khamesipour, F Incidence and antimicrobial resistance of Campylobacter and Salmonella from houseflies (Musca domestica) in kitchens, farms, hospitals and slaughter houses. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2017;87:1285–1291. [Google Scholar]
- Oskouizadeh, K Detection of Salmonella spp from some wild captive herbivores in Iran and determination of serogroup, antibiotic susceptibility and presence of invA gene in the isolated strains. Arch. Razi Inst. 2015;70:81–87. [Google Scholar]
- Paniel, N , Noguer, T Detection of Salmonella in food matrices, from conventional methods to recent aptamer-sensing technologies. Foods. 2019;8:1–38. doi: 10.3390/foods8090371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peighambari, S , Akbarian, R , Morshed, R , Yazdani, A Characterization of Salmonella isolates from poultry sources in Iran. Iran. J. Vet. Med. 2013;7:35–41. [Google Scholar]
- Peighambari, S , Morshed, R , Baziar, M , Sharifi, A , Sadrzadeh, A Salmonellosis in broiler flocks of Golestan province: frequency, serogroups and drug resistance patterns of Salmonella isolates. New Find. Vet. Microbiol. 2018;1:70–78. [Google Scholar]
- Peighambari, S , Qorbaniun, E , Morshed, R , Haghbin Nazarpak, H A survey on Salmonella infection in broiler farms around Mashhad city: determination of serogroups and antimicrobial resistance pattern of the Salmonella isolates. Iran. Vet. J. 2019;15:34–43. [Google Scholar]
- Peighambari, S , Sorahi, NM , Morshed, R Detection of Salmonellaenterica serovar Infantis among serogroup C Salmonella isolates from poultry using PCR and determination of drug resistance patterns. Iran. Vet. J. 2015;11:53–60. [Google Scholar]
- Peighambari, S , Yazdani, A , Hojjati, P Salmonella infection in birds kept in parks and pet shops in Tehran, Iran. Iran. J. Vet. Med. 2011;5:145–148. [Google Scholar]
- Raeisi, E , Ghiamirad, M Survey on prevalence of Salmonella serogroups and antibiotics susceptibility pattern in chicken meat in Ardabil, Iran. J. Ardabil Uni. Med. Sci. 2015;15:320–329. [Google Scholar]
- Rahmani, M , Peighambari, SM , Svendsen, CA , Cavaco, LM , Agers, Y , Hendriksen, RS Molecular clonality and antimicrobial resistance in Salmonella enterica serovars Enteritidis and Infantis from broilers in three Northern regions of Iran. BMC Vet. Res. 2013;9:66–76. doi: 10.1186/1746-6148-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaeian, H , Firouzi, R , Jalaei, J , Heydari, DF Antibiotic resistance of several common bacterial species isolated from chickens in Shiraz area. J. Vet. Med. 2003;58:223–226. [Google Scholar]
- Ranjbar, R , Giammanco, GM , Aleo, A , Plano, MRA , Naghoni, A , Owlia, P , Mammina, C Characterization of the first extended-spectrum β-lactamase–producing nontyphoidal Salmonella strains isolated in Tehran, Iran. Foodborne Pathog. Dis. 2010;7:91–95. doi: 10.1089/fpd.2009.0382. [DOI] [PubMed] [Google Scholar]
- Ranjbar, R , Giammanco, GM , Farshad, S , Owlia, P , Aleo, A , Mammina, C Serotypes, antibiotic resistance, and class 1 integrons in Salmonella isolates from pediatric cases of enteritis in Tehran, Iran. Foodborne Pathog. Dis. 2011;8:547–553. doi: 10.1089/fpd.2010.0736. [DOI] [PubMed] [Google Scholar]
- Salehi, TZ , Badouei, MA , Madadgar, O , Ghiasi, SR , Tamai, IA Shepherd dogs as a common source for Salmonella enterica serovar Reading in Garmsar, Iran. Turk. J. Vet. Anim. Sci. 2013;37:102–105. [Google Scholar]
- Salehi, TZ , Mahzounieh, M , Saeedzadeh, A The isolation of antibiotic-resistant Salmonella from intestine and liver of poultry in Shiraz province of Iran. Int. J. Poult. Sci. 2005;4:320–322. [Google Scholar]
- Sodagari, HR , Mashak, Z Prevalence and antimicrobial resistance of Salmonella serotypes isolated from retail chicken meat and giblets in Iran. J. Infect. Dev. Count. 2015;9:463–469. doi: 10.3855/jidc.5945. [DOI] [PubMed] [Google Scholar]
- Su, L , Chiu, C Salmonella: clinical importance and evolution of nomenclature. Chang Gung Med. J. 2007;30:210–219. [PubMed] [Google Scholar]
- Tajbakhsh, F , Tajbakhsh, E Determination of antibiotic resistance in Salmonella spp isolated from raw cow, sheep and goat’s milk in Chaharmahal Va Bakhtiyari Provience, Iran. Glob. Vet. 2013;10:681–685. [Google Scholar]
- Tajbakhsh, F , Tajbakhsh, E , Momeni, M , Rahimi, E , Sohrabi, R Occurrence and antibiotic resistance of Salmonella spp isolated from raw cow’s milk from Shahahrekord, Iran. Int. J. Microbiol. Res. 2012;3:242–245. [Google Scholar]
- Velasquez, C , Macklin, K , Kumar, S , Bailey, M , Ebner, P , Oliver, H , Singh, M Prevalence andantimicrobial resistance patterns of Salmonella isolated from poultry farms in southeastern United States. Poult. Sci. 2018;97:2144–2152. doi: 10.3382/ps/pex449. [DOI] [PubMed] [Google Scholar]
- Vtnair, D , Venkitanarayanan, K , Kollanoor Johny, A Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods. 2018;7:1–24. doi: 10.3390/foods7100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y , Yang, B , Wu, Y , Zhang, Z , Meng, X , Xi, M , Wang, D Molecular characterization of Salmonella enterica serovar Enteritidis on retail raw poultry in six provinces and two national cities in China. Food Microbiol. 2015;46:74–80. doi: 10.1016/j.fm.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Zare, P , Ghorbani-Choboghlo, H , Jaberi, S , Razzaghi, S , Mirzaei, M , Mafuni, K Occurrence and antimicrobial resistance of Salmonella spp and Escherichia coli isolates in apparently healthy slaughtered cattle, sheep and goats in East Azarbaijan province. Int. J. Ent. Pathog. 2014;2:1–4. [Google Scholar]
- Zare, P , Ghorbani-Choboghlo, H , Tolouei, M , Hadavi, J Evaluation of the occurrence of Salmonella serovars and its antibiotic susceptibility in apparently healthy domestic animals in rural areas of East Azerbaijan province. Iran. J. Med. Microbiol. 2016;10:88–92. [Google Scholar]