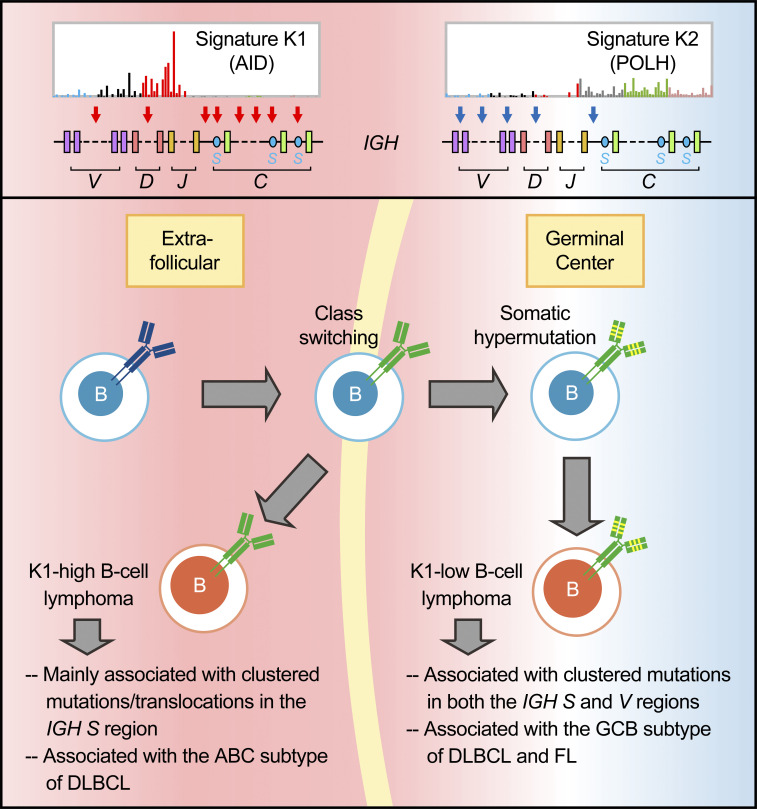
By using genomic and transcriptomic sequencing technologies, Ye et al. identify AID-dependent and/or polymerase η-associated mechanisms of mutagenesis in human germinal center–derived/related B cell lymphomas and link aberrant immunoglobulin gene diversification processes to different disease subtypes.
Abstract
Both somatic hypermutation (SHM) and class switch recombination (CSR) are initiated by activation-induced cytidine deaminase (AID). Dysregulation of these processes has been linked to B cell lymphomagenesis. Here we performed an in-depth analysis of diffuse large B cell lymphoma (DLBCL) and follicular lymphoma (FL) genomes. We characterized seven genomic mutational signatures, including two B cell tumor-specific signatures, one of which is novel and associated with aberrant SHM. We further identified two major mutational signatures (K1 and K2) of clustered mutations (kataegis) resulting from the activities of AID or error-prone DNA polymerase η, respectively. K1 was associated with the immunoglobulin (Ig) switch region mutations/translocations and the ABC subtype of DLBCL, whereas K2 was related to the Ig variable region mutations and the GCB subtype of DLBCL and FL. Similar patterns were also observed in chronic lymphocytic leukemia subtypes. Thus, alterations associated with aberrant CSR and SHM activities can be linked to distinct developmental paths for different subtypes of B cell lymphomas.
Graphical Abstract
Introduction
Two somatic DNA modification processes are required for Ig gene diversification and the production of functional antibodies: somatic hypermutation (SHM) and class switch recombination (CSR). SHM occurs in the dark zone of the germinal center (GC), where mutations are introduced in the Ig variable (V) regions at a high rate and may lead to increased affinity of the antibody produced (Victora and Nussenzweig, 2012). CSR was thought to take place in the light zone of the GC, which allows a previously rearranged Ig heavy-chain V domain to be expressed in association with a constant (C) region downstream of Cμ, leading to the production of different antibody classes, i.e., IgG, IgA, or IgE (Stavnezer et al., 2008). A recent study challenged this dogma and showed that CSR occurs during the initial T–B cell interaction before the formation of GC and SHM (Roco et al., 2019). Both SHM and CSR are initiated by activation-induced cytidine deaminase (AID; encoded by AICDA; Muramatsu et al., 2000), which deaminates cytosines into uracils upon recruitment to the V and switch (S) region sequences (Di Noia and Neuberger, 2007). The resulting uracils engage the activity of either the base-excision repair (BER) or the mismatch repair (MMR) pathway, creating nicks or double-strand breaks in the V or S regions to initiate SHM or CSR, respectively (Di Noia and Neuberger, 2007).
Diffuse large B cell lymphoma (DLBCL) and follicular lymphoma (FL) are the most common types of non-Hodgkin lymphomas, accounting for 30–40% and 22–25% of newly diagnosed cases, respectively (Rosenquist et al., 2017). DLBCL is a heterogeneous disease with two major subtypes as defined by gene expression profiling: the GC B cell–like (GCB) and activated B cell–like (ABC) subtypes (Alizadeh et al., 2000). FL is considered an indolent disease, but transformation to more aggressive malignancies, most commonly DLBCL, may occur in a subset of high-grade FLs (Lossos and Gascoyne, 2011). Both DLBCL and FL are believed to be derived from GC B cells (Küppers et al., 1999; Stevenson et al., 2001): GCB DLBCL and FL resemble B cells from the light zone of GC, whereas ABC DLBCL likely derives from B cells arrested during the early stage of plasma cell differentiation (Basso and Dalla-Favera, 2015).
AID expression has been detected in both DLBCL and FL (Hardianti et al., 2004; Lossos et al., 2004) and is highly correlated with the accumulation of genomic deoxyuridines in B cell lymphoma lines (Pettersen et al., 2015). Both DLBCL and FL have shown SHM-like mutations in a small set of non-Ig genes tested, including a number of proto-oncogenes and tumor suppressor genes (Halldórsdóttir et al., 2008; Pasqualucci et al., 2001). AID is also required for the generation of translocations to the Ig heavy chain (Igh) locus in mice, including Igh-Myc translocations in IL-6–induced mouse plasmacytomas (Ramiro et al., 2004) and Igh locus-specific translocations in mice with H2AX deficiency (Franco et al., 2006). Additionally, the expression of AID promotes chromosome translocations in normal mouse B cells, such as translocation between Myc and the Ig S regions (Ramiro et al., 2006). AID activity is furthermore shown to be required for the development of GC-related B cell lymphomas in mice (Pasqualucci et al., 2008).
In the last decade, high-throughput technologies have made it possible to study AID-targeted genes on a genome-wide scale. Based on the mutational pattern and the distance to the transcription start sites (TSS), potential AID-targeted genes were identified in DLBCL by whole-genome sequencing (WGS; Khodabakhshi et al., 2012). A further analysis of somatic (tumor-specific) mutations in 10 DLBCL genomes suggested that kataegis, which refers to localized clustered mutations (Nik-Zainal et al., 2012), are largely due to AID activity and are associated with B cell super enhancers (Qian et al., 2014). Additionally, a machine-learning algorithm, which was trained based on a deep-sequencing dataset, has been used to predict AID-targeted genes in nontransformed mouse B cells (Álvarez-Prado et al., 2018). Approximately 7% of those targets were found to be mutated in DLBCL. To further explore the mechanism underlying the mutagenesis of human B cell lymphomas, we performed a comprehensive analysis of mutational signatures and translocation patterns in DLBCL and FL based on whole-genome and transcriptome sequencing of a large number of tumor samples. Published datasets from DLBCL and chronic lymphocytic leukemia (CLL) genomes were also reanalyzed to validate our findings.
Results
Mutational landscapes and signatures in DLBCL and FL genomes
WGS data from 60 DLBCL and 22 FL tumor samples and their corresponding peripheral blood lymphocytes were analyzed, focusing on signatures of somatic mutations. In total, 778,301 somatic single base substitutions (SBSs) were identified in all samples, and on average, 3.82 (range, 0.10–10.12) and 1.37 (range, 0.10–3.10) somatic SBSs were detected per megabase pair (Mb) in the DLBCL and FL genomes, respectively. Additionally, 0.65 (range, 0.01–3.74) and 0.23 (range, 0.11–0.47) somatic insertions and deletions (indels) were detected per Mb in DLBCL and FL genomes, respectively. Microsatellite instability was estimated by using MSIseq (Huang et al., 2015), and 13 DLBCLs were identified as microsatellite instable (MSI), whereas the remaining DLBCLs and all FLs were characterized as microsatellite stable (MSS).
The total number of somatic SBSs (referred to henceforward as mutations) per genome and the corresponding clinical characteristics and mutation status in major DNA repair genes are summarized in Fig. 1 A based on the details presented in Table S1. Based on Mann-Whitney U tests, in DLBCL genomes, a significantly higher mutational load was associated with an older age at diagnosis (P = 0.0099), the GCB subtype (P = 0.0027), or MSI status (P = 0.0001). In addition, a higher number of mutations was observed in DLBCL samples with somatic mutations in key MMR or BER genes (P = 0.0036). In FL genomes, significantly higher mutation loads were associated with male sex in patients (P = 0.0364).
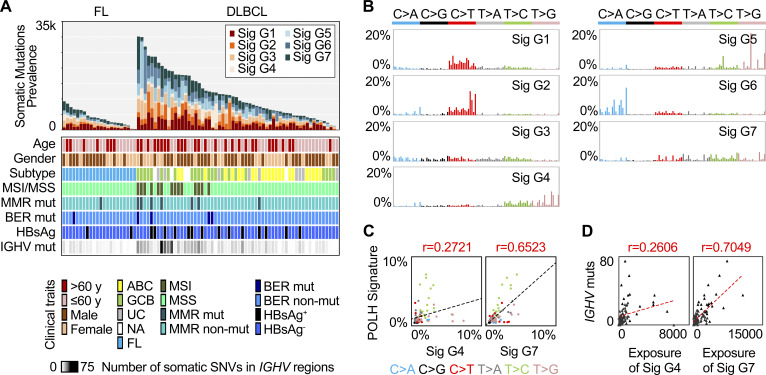
Figure 1.
Genomic mutational signatures in DLBCL and FL. (A) The somatic mutation prevalence (bar chart), the clinical traits (color heat map), and the number of somatic mutations in IGHV regions (gray heat map) for each sample. (B) Genomic mutational signatures from 60 DLBCL and 22 FL genomes. Each signature is displayed according to the 96 substitution classification defined by the substitution class (shown in different colors) and sequence context immediately 3′ and 5′ to the mutated base. The 96 possible mutated trinucleotides are on the x axis, and the frequency of the mutation type is shown on the y axis. (C) The correlation (PCC) with the spectrum of human POLH-induced mutations. (D) The correlation (PCC) of the mutation load in IGHV regions and the exposure of G4 or G7. HBsAg, hepatitis B virus surface antigen; mut, mutation; Sig, signature; UC, unclassified; NA, RNA not available.
Somatic mutations in the cancer genome may be a consequence of mutational processes of both endogenous and exogenous origin that operate in cancer cells and their precursors. Each mutational process, which involves different types of DNA damage and repair, may result in a characteristic mutational signature with unique combinations of substitution types (Helleday et al., 2014). The mutations in a given cancer genome may have been generated by several different mutational mechanisms, and mathematical methods have been developed to decipher mutational signatures based on mutational catalogs. To identify the mutational processes associated with DLBCL and FL, somatic mutations identified in the lymphoma genomes were first cataloged into 96 classes (see Materials and methods). Seven genomic signatures, referred to as G1–G7, were subsequently deciphered using a previously described method, SigProfiler (Fig. 1 B; Alexandrov et al., 2013b). G1 was characterized by dominant C>T transitions at NCG trinucleotides, which was similar to a previously described age-related signature, signature 1B (Fig. S1 A; Alexandrov et al., 2013a). As expected, a positive correlation between the exposure (the number of mutations attributed to the signature) of G1 and age at diagnosis was observed in our cohort (r = 0.3441, Pearson correlation coefficient; PCC). G2 was characterized by C>T transitions at TCN trinucleotides, resembling signature 2 in the database of Catalogue of Somatic Mutations in Cancer (COSMIC), which has been suggested to be associated with apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) family members (Nik-Zainal et al., 2012). Consistent with previous reports on other types of cancers (Burns et al., 2013; Roberts et al., 2013), the exposure of G2 was correlated with the expression of APOBEC3B in our lymphoma samples (r = 0.5437, PCC). G3, G5, and G6 were highly similar to COSMIC signatures 5, 17, and 18, which have unknown etiologies (Fig. S1 A).
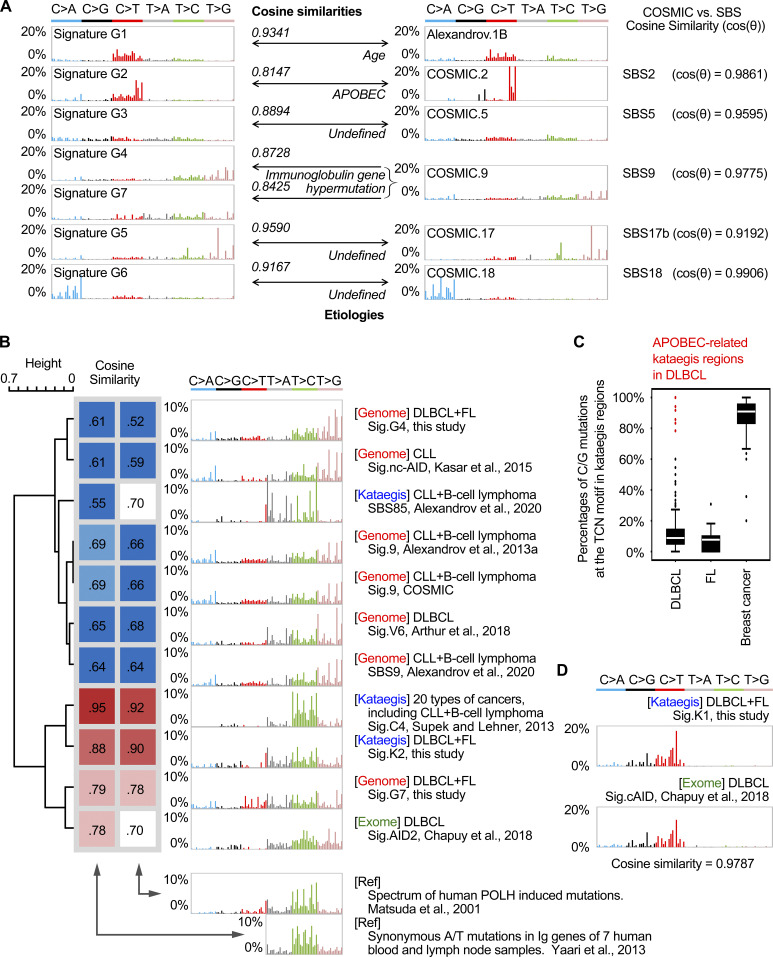
Figure S1.
Comparison of genomic and kataegis mutational signatures identified in DLBCL and FL with previously published mutational signatures. (A) Comparison of genomic signatures identified in this study with previously published mutational signatures. G1 to G7 can be assigned to the COSMIC signatures with high similarity. Both G4 and G7 were similar to COSMIC signature 9. Additionally, COSMIC signatures 2, 5, 9, 17, and 18 were highly similar to the recently published SBS signatures 2, 5, 9, 17b, and 18, respectively (Alexandrov et al., 2020). The similarities between the signatures were estimated by the cosine similarity. (B) Clustering of POLH-related signatures based on their correlations with known experimental POLH signatures. The cosine similarities to the two known spectra of POLH-related mutations (Matsuda et al., 2001; Yaari et al., 2013) were calculated, and the mutational signatures were clustered accordingly. For the study by Matsuda et al. (2001), all mutations were available for analysis. For the study by Yaari et al. (2013), only T mutations were available for analysis. The mutational signatures were identified on the genomic level (genome; Alexandrov et al., 2013a, Alexandrov et al., 2020; Arthur et al., 2018; Kasar et al., 2015), exonic level (exome; Chapuy et al., 2018), or in the kataegis regions (kataegis; Alexandrov et al., 2020; Supek and Lehner, 2017). Three signatures (G4, G7, and K2) from this study are included in the comparison. The color codes represent the cosine similarities between signatures. (C) The percentage of C/G mutations in the TCN motifs in kataegis identified from DLBCL, FL, and breast cancer genomes. Data from the DLBCL and FL genomes in this study and data from breast cancers from Alexandrov et al. (2013a) were analyzed and plotted. Outlier kataegis with abnormally high percentages of the TCN mutations in B cell malignancies were identified as APOBEC kataegis and are colored in red. Three of the APOBEC kataegis identified in DLBCLs overlapped with same percentage of TCN mutations. (D) Comparison of kataegis signature K1 identified in this study to a previously published mutational signature (cAID) based on exome sequencing (Chapuy et al., 2018). The cosine similarity between these two signatures was 0.9787. Ref, reference; Sig., signature.
G4 was characterized by T>S mutations (S = C/G, 72%) with enrichment at TW motifs (W = A/T, 54%). G7 was characterized by T>V mutations (V = A/C/G, 71%), likewise with enrichment at TW motifs (52%). Both signatures were highly similar to a B cell tumor–restricted signature, COSMIC signature 9 (COSMIC 9; Alexandrov et al., 2013a). However, G4 and G7 presented remarkable differences in both signature pattern (cosine similarity, cos(θ) = 0.6622) and sample exposure (r = 0.2677, PCC).
COSMIC 9 was inferred to be associated with error-prone DNA polymerase η (POLH) activity and SHM because of its mutation pattern (preference of T mutations at TW motifs) and B cell specificity (detected only in B cell lymphoma and CLL samples), and furthermore, POLH has been shown to be the main contributor to A/T mutations during SHM using mouse knockout models (Delbos et al., 2005). To investigate whether G4 and G7 are associated with POLH activity, we first directly compared them with the known human POLH-related mutational signatures. G7, but not G4, was highly correlated with the spectrum of POLH-induced mutations (Fig. 1 C; Matsuda et al., 2001) as well as the empirical estimates of the SHM spectrum from analysis of synonymous mutations in human Ig genes (Fig. S1 B; Yaari et al., 2013). Indeed, the correlation of G7 with the POLH-related signatures was stronger than that of COSMIC 9 (Fig. S1 B). We next asked whether G4 and G7 were associated with the mutation frequency in the V region genes of IGH (IGHV), one of the physiological SHM-targeted regions. The exposure of G7, but not G4, was highly correlated with the number of somatic mutations in IGHV genes (r = 0.7049 versus 0.2606, PCC; Fig. 1 D). The mutations in the IGHV genes only contributed to a very small proportion of G7 mutations (990/13,1327 = 0.7%). Furthermore, when considering the mutations outside the Ig loci, the correlation between the exposure of G7 and the number of mutations in IGHV genes remained strong (r = 0.6976, PCC). Taken together, in our DLBCL and FL cohort, we identified two genomic signatures that show similarity to the previously described B cell tumor–specific COSMIC 9, and we furthermore showed that G7, but not G4, is strongly associated with the POLH-induced mutation spectrum as well as the rate of mutations in the IGHV genes.
Mutational signatures of kataegis in DLBCL and FL
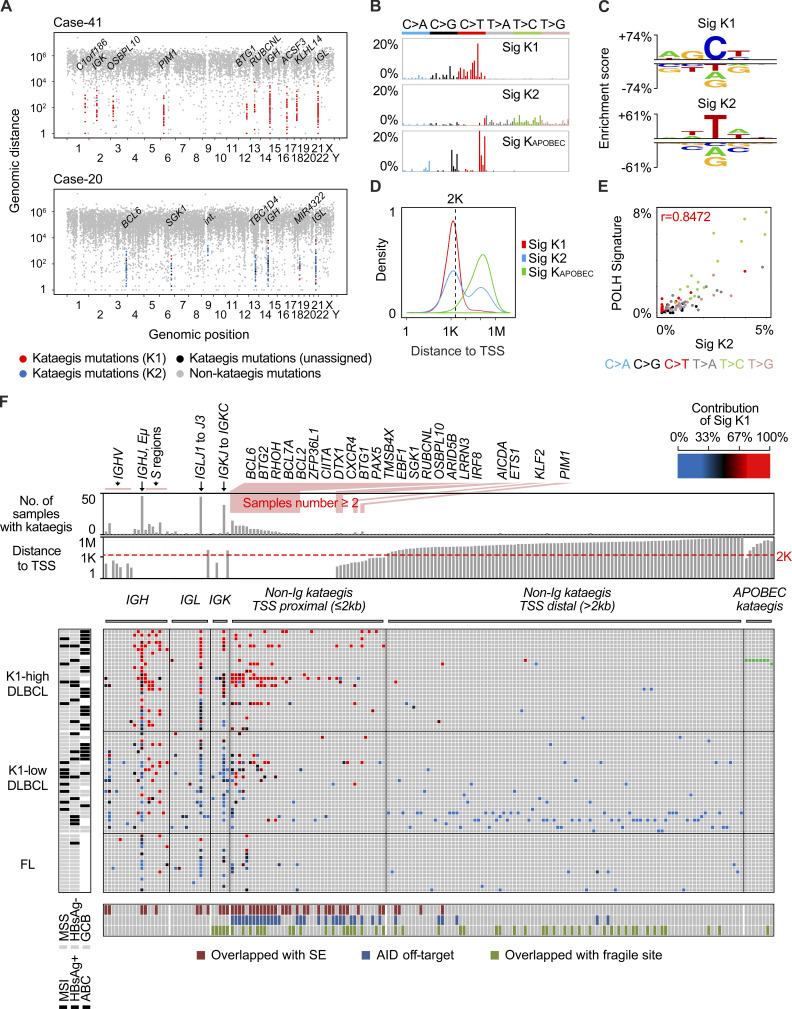
To further understand the mechanism underlying mutagenesis in B cell lymphoma, we next analyzed the mutational pattern of kataegis in the DLBCL and FL genomes. Kataegis was first described in breast cancer genome, which refers to those regional clustered C>T and C>G substitutions. These mutations often occur on the same DNA strand and are usually colocalized with somatic rearrangements. They have been linked to the activity of the APOBEC family of cytidine deaminases (Nik-Zainal et al., 2012). Further analysis of clustered mutations in tumor genomes from different types of cancers suggested that compared with nonclustered mutations, they may provide a more precise fingerprint of the mutagenic process (Supek and Lehner, 2017). In our DLBCL and FL dataset, 504 kataegis (Table S2) containing 10,223 mutations, accounting for ∼1.3% of the total somatic mutations, were identified. On average, 7.4 and 2.7 kataegis were identified for each DLBCL and FL genome, respectively. Kataegis identified from two represented DLBCL cases are illustrated in rainfall plots in Fig. 2 A.
Figure 2.
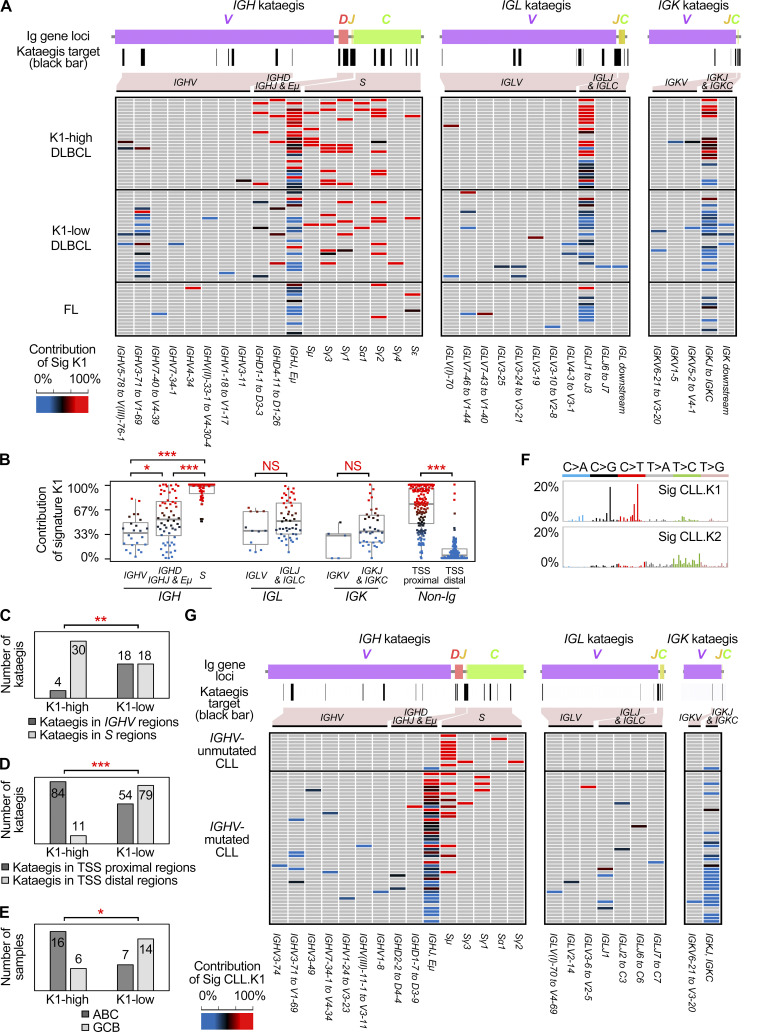
Mutational signatures of kataegis in DLBCL and FL. (A) Rainfall plots of somatic mutations from two representative samples. Each dot represents a somatic mutation, and they are ordered on the x axis based on their genomic locations, from chromosome 1 to Y. The intermutation distance is plotted on the y axis (log10 transformed). Kataegis are colored based on their assignment of specific signatures: red (K1), blue (K2), or black (could not be assigned). Mutations in nonkataegis regions are colored in gray. Selected genes targeted by kataegis are highlighted. Int, intergenic region. (B) Three mutational signatures of kataegis were identified in the DLBCL and FL genomes. The x or y axis represents 96 mutation types and the percentage of mutations attributed to a specific mutation type, respectively. (C) Preference of sequence motifs for mutations assigned to K1 and K2. Each motif contains the mutation and two 5′ and two 3′ residues. For each residue, if a base type is significantly enriched or depleted (t test, P < 0.05) as compared with randomly selected control sequences, a symbol of this base type is shown. The heights of symbols represent the degree of either enrichment (positive y axis) or depletion (negative y axis) for each base type. (D) The distribution of distances between mutations and their nearest TSS. (E) The correlation (PCC) between K2 and the spectrum of human POLH-induced mutations. (F) The main matrix shows the distribution of kataegis in DLBCL and FL genomes. Only the samples with at least one kataegis are shown. Each row represents a sample, and each column represents a genomic region with kataegis. Each brick in the matrix represents a kataegis and is colored based on the contribution of K1 (see the color bar), or KAPOBEC (green); no kataegis (gray). The rows of the matrix were first grouped by DLBCL and FL and then sorted by K1 contribution, which is calculated as the average contribution of K1 for all kataegis in each sample. The DLBCLs were further divided into two groups based on K1 contribution. The columns of the matrix were grouped by kataegis in Ig loci, non-Ig (TSS proximal) and non-Ig locus (TSS distal) loci, and the APOBEC kataegis. Regions in the Ig loci were sorted from the V to the C regions. The targets in non-Ig loci are sorted by their distance to TSS. The status of subtypes, HBV infection, and MSI for each sample are shown on the left. For each targeted region, the overlaps with B cell super enhancers, reported AID off-targets, and human chromosome fragile sites are indicated at the bottom. Bars on the top show the number of samples in each kataegis targeted region and the distance to TSS for each target. E, enhancer; HBsAg, hepatitis B virus surface antigen; SE, super enhancer; Sig, signature.
As APOBEC deamination is one of the contributors to genome-wide mutations in our cohort (G2), we first tested whether some of the C mutations in the kataegis identified could potentially be due to APOBEC activities. A small number of APOBEC-related kataegis (n = 8, 2% of total) from two samples were indeed identified in our cohort based on their preference for C mutations at TCN motifs (Fig. S1 C and Table S2; Nik-Zainal et al., 2016). Accordingly, these two samples also had the highest exposure of G2 among all samples. The signature accumulated from these eight kataegis (as KAPOBEC) is highly similar to the APOBEC signature described previously (Alexandrov et al., 2013a; cos(θ) = 0.9403). After removal of the mutations belonging to these APOBEC-related kataegis, the SigProfiler pipeline was applied, and two main mutational signatures (referred to as K1 and K2) were extracted from the remaining 496 kataegis (Fig. 2 B).
To identify the etiologies of K1 and K2, we first assigned the mutations that are highly likely to belong to either of the signatures. In total, 9,149 (90.8%) of the mutations in the kataegis regions were assigned, including 4,598 (45.6%) mutations to K1 and 4,551 (45.2%) mutations to K2. The K1 mutations were dominated by C>T and C>G substitutions and were highly enriched in the motif WRCY (W = A/T, R = A/G, Y = C/T), a well-known hotspot for SHM (Rogozin and Kolchanov, 1992), or WRC, the preferred AID motif (P < 10−4, t test; Fig. 2 C; Pham et al., 2003). Compared with the KAPOBEC mutations, mutations assigned to K1 were highly enriched within 2 kb from the TSS (P < 10−15, Mann–Whitney U test; Fig. 2 D), which indicates the involvement of AID off-target activities (Pasqualucci et al., 2001). On the other hand, the mutations assigned to K2 were highly enriched in the POLH motif (TW; P < 10−4, t test; Fig. 2 C; Rogozin et al., 2001). Furthermore, K2 was more similar to G7 than to G4 (cos(θ) = 0.8780 versus 0.6219), and its exposure was also highly correlated with G7 but not to G4 (r = 0.6677 versus 0.2100, PCC). Finally, K2 showed even higher similarity than G7 to the POLH-related signatures (Fig. 2 E and Fig. S1 B). Thus, although the mutations assigned to K2 account for only a small fraction of somatic mutations in the lymphoma genomes (0.6%), they represent POLH activity more closely than those belonging to G7. Moreover, there were two peaks of K2 mutations, one major peak (n = 2,799) proximal to TSS and one minor peak (n = 1,752) distal to the TSS (Fig. 2 D). The number of mutations in the major peak, but not the minor peak, is highly correlated with the number of somatic mutations in the IGHV loci (r = 0.5790 versus 0.2101, PCC).
Taken together, mutations assigned to the K1 are clearly associated with AID deamination and resemble phase I SHM: direct replication over AID-induced U:G lesions, resulting in “AID fingerprints,” i.e., C>T transitions in the WRCY motifs, or removal of the uracil by BER enzyme UNG (uracil DNA glycosylase) followed by replication, generating C to T/G transitions/transversions. Most of K2 mutations (the major peak) are likely to result from additional repair by POLH after the initial AID-induced lesions and thus resemble phase II SHM: the MMR pathway recruits error-prone POLH to generate T mutations at TW motifs. The remaining K2 mutations (the minor peak) are also contributed by POLH but seem to be independent of AID deamination and the SHM process.
K1- or K2-dominant kataegis in the Ig S or V regions, respectively
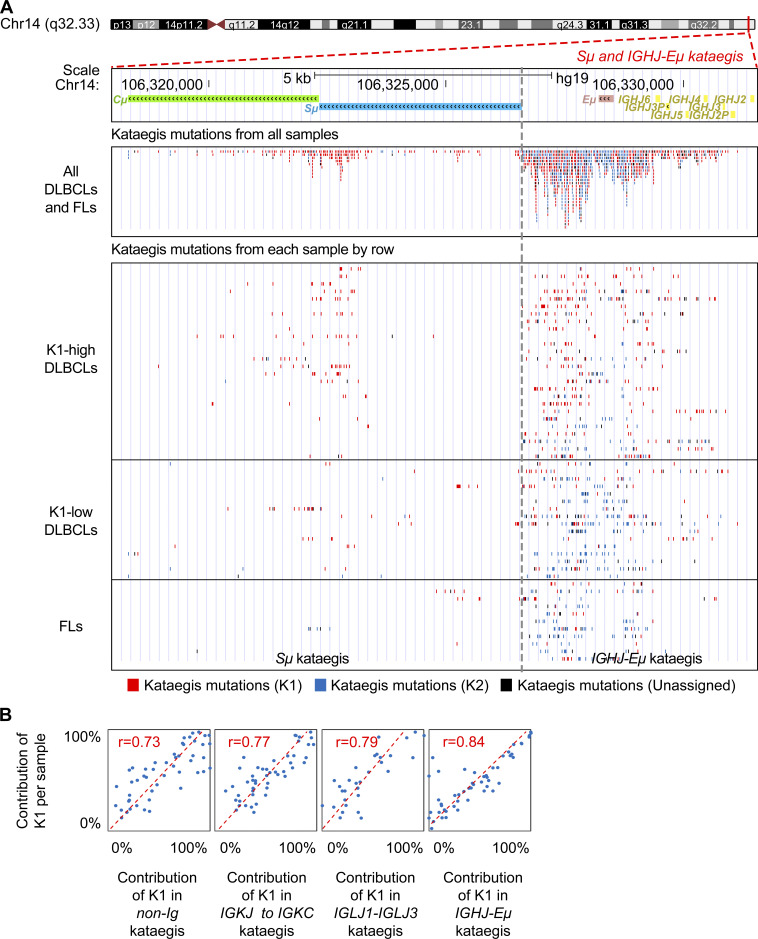
We next estimated the relative contribution of K1 and K2 mutations to each kataegis identified in DLBCLs and FLs: for a given kataegis, if the contribution of K1 is greater than 67%, i.e., at least twofold higher compared with K2, it was referred to as K1-dominant kataegis; K2-dominant kataegis were assigned similarly. In total, 242 (48%) kataegis were identified at the Ig loci. Within the IGH locus, in addition to several IGHV and IGHD genes, the sequences from IGHJ to upstream of the Sμ region, including the intronic enhancer (Eμ; IGHJ-Eμ) and several S regions (Sμ, Sγ1, Sγ2, and Sγ3), were frequent sites of kataegis (Fig. 2 F and Fig. 3 A). The vast majority of kataegis in the S regions were K1-dominant, whereas kataegis in IGHV regions were more related to K2 mutations (Fig. 3 B). Kataegis identified in the sequences across the IGHJ and Sμ were initially counted together due to their closely adjacent positions in the genome. However, when these regions were dissected more closely, kataegis in the Sμ region were mainly contributed by K1 mutations, whereas kataegis in the IGHJ-Eμ regions as well as IGHD regions were contributed by K1 and/or K2 mutations, with an overall intermediate contribution of K1 (Fig. 3 B and Fig. S2 A). Within the Ig light chain λ (IGL) and κ (IGK) loci, the sequences across IGLJ1 and IGLJ3, overlapping with the locus of IGLL5, and the region from IGKJ to IGKC were frequent sites for kataegis (Fig. 3 A). These highly targeted regions in the light chain loci, similar to the IGHJ-Eμ region, were contributed by K1 and/or K2 mutations (Fig. 3 B) and overlapped with B cell superenhancers (Fig. 2 F). The IGLV and IGKV genes were comparatively less targeted, and kataegis in these regions were mainly contributed by K2 mutations (Fig. 3 B).
Figure 3.
Distribution of kataegis in Ig loci in DLBCL, FL, and CLL. (A) A higher resolution of distribution of kataegis in the Ig loci in the DLBCL and FL genomes. The plot on the top of the main matrix proportionally shows the order of the Ig genes. The positions of kataegis in the Ig loci are indicated by black bars. The name of each region with kataegis is shown at the bottom. (B) The contribution of K1 for the kataegis in different parts of genome. Each dot represents a kataegis and is colored according to the K1 contribution (see the color bar). Mann–Whitney test; *, P < 0.05; ***, P < 0.0005. (C) Number of kataegis at the IGHV and S loci in different groups. (D) Number of non-Ig kataegis in the TSS proximal and distal regions in different groups. (E) Number of ABC and GCB samples in different groups. Statistics used in C to E, Fisher’s exact test; *, P < 0.05; **, P < 0.005; ***, P < 0.0005. (F) Two mutational signatures of kataegis were identified in 153 previously published CLL genomes. Legends are as described for Fig. 2 B. (G) The distribution of kataegis in the Ig loci of the CLL genomes. Only the CLLs with at least one Ig kataegis and the subtype information are shown. Legends are as described for A. Sig, signature.
Figure S2.
The overall contribution of K1 for a given sample is highly correlated to its contribution to the different regions within the same sample, except S regions. (A) The mutation pattern and distribution of kataegis in the Sμ and IGHJ-Eμ regions. Top: For the region of interest, the gene structure and location on the chromosome are shown. The kataegis mutations are shown together for all samples (middle) or separately by each sample (bottom). Each dot represents a kataegis mutation. The red and blue dots represent mutations belonging to K1 and K2, respectively. The black dots represent unclassified mutations to either K1 or K2. For the bottom panel, each row represents a sample. The samples are in the same order as in Fig. 2 F and are grouped as K1-high DLBCL, K1-low DLBCL, and FL. Mutations are grouped in either the Sμ region (left) or IGHJ-Eμ region (right). The kataegis across both regions were initially identified together due to their closely adjacent positions in the genome. To reveal the characteristics of kataegis for both parts, kataegis located in the Sμ and IGHJ-Eμ regions were divided for further analysis. For the divided kataegis, only those with at least 10 mutations were counted for further analysis. (B) The contribution of K1 for a given sample highly correlated to its contribution to different regions, including the most targeted IGHJ-Eμ, IGLJ1-IGLJ3, and IGKJ to IGKC regions, as well as the non-Ig regions. Correlation was calculated by the PCC.
SHM-like mutations have previously been identified in Sμ regions in both mouse (Nagaoka et al., 2002) and human B cells (Pan-Hammarström et al., 2003). Here, for the first time, we characterized the pattern of a large number of somatic mutations in the S donor as well as the S acceptor regions in human B cell lymphoma samples. We showed that in contrast to those in the V regions, clustered mutations in the S regions, presumably associated with aberrant CSR activity, are K1-dominant, i.e., contributed by AID and BER, without the influence of the additional DNA repair mechanism required in SHM, i.e., MMR/POLH.
K1- or K2-dominant kataegis in non-Ig loci
Outside the Ig loci, we identified altogether 262 kataegis, a small number of which (n = 8) displayed the KAPOBEC signature (Fig. 2 F). The majority of the non-Ig kataegis (n = 254) were contributed by K1 and/or K2, affecting 143 genomic regions. Kataegis located in regions proximal (<2 kb) to the TSS were mainly K1-dominant, but they could also be contributed by both signatures or K2-dominant, whereas kataegis in the regions distal to the TSS were mainly K2-dominant (Fig. 3 B). Furthermore, kataegis regions proximal to TSS were more frequently associated with super enhancers (77.9% versus 3.8%, P < 1e-16, Fisher’s exact test, two-tailed test unless specified), and more often reported as AID off-targets in human or mouse studies (83.2% versus 4.8%, P < 1e-16, Fisher’s exact test; Fig. 2 F; Álvarez-Prado et al., 2018; Hakim et al., 2012; Jiang et al., 2012; Khodabakhshi et al., 2012; Liu et al., 2008; Pasqualucci et al., 2001). Thus, most of the non-Ig kataegis localized in TSS proximal regions likely resulted from AID-initiated off-targeting activity, and can be either K1- or K2-dominant, whereas those distal of TSSs were K2-dominant and seemed to be contributed by AID-independent POLH activity.
Altogether, 178 nonsilent mutations were detected in kataegis in non-Ig loci, of which 97.2% (173/178) were TSS proximal regions. These nonsilent mutations were detected in 25 DLBCLs and 6 FLs, and recurrent mutations were observed in BCL2, BTG2, CXCR4, ZFP36L1, BTG1, DTX1, PIM1, and SGK1. Thus, most mutations in kataegis that may contribute to lymphomagenesis likely result from AID off-targeting events.
K1-high DLBCL is mainly associated with kataegis in the S regions and AID off-targets
The overall contribution of K1 for a given sample is highly correlated to its contribution to the different regions within the same sample, including the most targeted IGHJ-Eμ, IGLJ1–IGLJ3, and IGKJ to IGKC regions, and the non-Ig regions (Fig. 2 F and Fig. S2 B). This can also be visualized in the rainfall plots from two representative DLBCL cases where kataegis mutations from case 41 and case 20 were mostly assigned to either K1 or K2, respectively (Fig. 2 A). Thus, kataegis from the same lymphoma sample often showed a dominant contribution of either K1 or K2 signature. The notable exceptions were kataegis at S regions, which were almost always K1-dominant, and to some extent the V regions, which were often K2-dominant. To further investigate this feature, we divided DLBCL samples into K1-high (n = 28, K1 contribution = 75.5 ± 13.1%) and K1-low (n = 28, K1 contribution = 35.5 ± 15.2%) groups. Fewer kataegis were observed in FL, and the overall K1 contribution (n = 16, K1 contribution = 40.1 ± 19.9%) was similar to that of the K1-low DLBCLs.
For the K1-high group, a larger number of kataegis were observed in the S regions as compared with the IGHV regions, whereas within the K1-low group, a similar number of kataegis were identified in these regions (Fig. 3 C). Indeed, IGHV kataegis were mainly found in the K1-low samples (P = 0.0007, Mann–Whitney U test; Fig. 3 C). For the non-Ig loci within the K1-high group, a larger number of kataegis were identified in the TSS proximal regions, presumably AID off-targets, while for the K1-low group, kataegis were found both in the TSS proximal and distal regions (Fig. 3 D). When comparing the K1-high and K1-low groups, kataegis located in TSS proximal regions were significantly enriched in the K1-high DLBCLs (P < 0.0001, Mann–Whitney U test; Fig. 3 D).
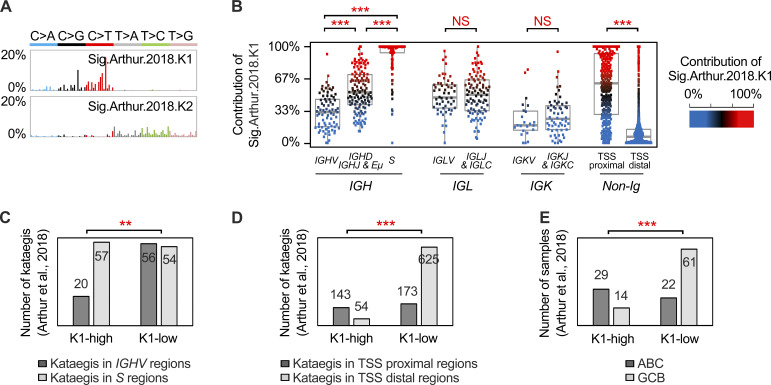
We next reanalyzed the WGS data from a previously published DLBCL cohort (n = 153; Arthur et al., 2018) and characterized the pattern of kataegis using the method described in this study. Altogether, 1,538 kataegis were identified, and two kataegis signatures that were highly similar to K1 and K2 were depicted (Fig. S3 A), with a largely similar distribution of K1- and K2-dominant kataegis (Fig. S3, B–D).
Figure S3.
The features of kataegis signatures in DLBCL samples from Arthur et al. (2018). (A) Two mutational signatures of kataegis were identified in 153 DLBCL from Arthur et al. (2018). Sig.Authur.2018.K1 and Sig.Authur.2018.K2 were highly similar to K1 (cosine similarity = 0.9809) and K2 cCosine similarity = 0.9861) from our study, respectively. (B) The relative contribution of Sig.Authur.2018.K1 for the kataegis located in different parts of the Ig locus. Each dot represents a kataegis. The color of each dot is based on the contribution of Sig.Authur.2018.K1 (from red to black to blue). Mann–Whitney U test; ***, P < 0.0005. (C) For the Sig.Authur.2018.K1-high DLBCLs group, a larger number of kataegis was observed in the S regions compared with the IGHV regions, whereas within the K1-low group, a similar number of kataegis was identified in these regions. A significantly lower number of kataegis in IGHV regions was observed in the Sig.Authur.2018.K1-high versus Sig.Authur.2018.K1-low group. (D) Non-Ig kataegis were enriched in the TSS proximal regions in Sig.Authur.2018.K1-high DLBCLs. (E) ABC samples were enriched in the Sig.Authur.2018.K1-high DLBCLs. Fisher’s exact test for C–E; **, P < 0.005; ***, P < 0.0005. Sig., signature.
In summary, DLBCL can be grouped based on K1-K2 dominance. K1-high DLBCLs seem to mainly associate with clustered mutations generated in the S regions and AID off-targeted regions, whereas K1-low DLBCLs can have cluster mutations in both S and V regions, as well as in AID-targeted and nontargeted non-Ig regions.
K1-high DLBCL is associated with the ABC subtype
We next compared the clinical and pathological features of K1-high and K1-low groups (Table 1). The MSI status and hepatitis B virus (HBV) infection status did not affect the distribution of K1-high or K1-low samples (Fig. 2 F). Samples with the ABC subtype were, however, significantly enriched in the K1-high group, whereas samples with the GCB subtype were more likely in the K1-low group (Fig. 3 E; P = 0.0148, Fisher’s exact test). The association of the K1-high group and ABC subtype was validated in the published DLBCL cohort (Fig. S3 E; P < 0.0001; Fisher’s exact test).
Table 1. Clinical characterization of Sig K1-high/low DLBCL samples.
| Sig K1-high | Sig K1-low | P valuea | |
|---|---|---|---|
| No. of patients | 28 | 28 | |
| Age, yr | 0.2847 | ||
| >60 (%) | 16 (57.1) | 11 (39.3) | |
| ≤60 (%) | 12 (42.9) | 17 (60.7) | |
| Gender | 0.1707 | ||
| Male (%) | 14 (50.0) | 20 (71.4) | |
| Female (%) | 14 (50.0) | 8 (28.6) | |
| Subtypeb | 0.0148 | ||
| ABC (%) | 16 (72.7) | 7 (33.3) | |
| GCB (%) | 6 (27.3) | 14 (66.7) | |
| Stage | 1.0000 | ||
| I–II (%) | 12 (42.9) | 13 (46.4) | |
| III–IV (%) | 16 (57.1) | 15 (53.6) | |
| Primary or relapse | 0.3516 | ||
| Primary (%) | 24 (85.7) | 27 (96.4) | |
| Relapse (%) | 4 (14.3) | 1 (3.6) | |
| HBsAg status | 0.7585 | ||
| Positive (%) | 6 (21.4) | 8 (28.6) | |
| Negative (%) | 22 (78.6) | 20 (71.4) | |
| MSIseq | 0.5279 | ||
| MSI (%) | 5 (17.9) | 8 (28.6) | |
| MSS (%) | 23 (82.1) | 20 (71.4) | |
| Treatmentb | 0.2238 | ||
| CHOP (%) | 5 (18.5) | 9 (32.1) | |
| RCHOP (%) | 22 (81.5) | 17 (60.7) |
Values are reported as n (%) of patients unless indicated otherwise. CHOP, a chemotherapy combination of cyclophosphamide, doxorubicin hydrochloride, vincristine (Oncovin, Vincasar PFS), and prednisolone; HBsAg, hepatitis B virus surface antigen; RCHOP, a chemotherapy combination of rituximab (Rituxan) and CHOP; Sig, signature.
Fisher's exact test was used for comparison. Significant value (P < 0.05) is highlighted in bold.
The calculation was based on 43 or 53 samples with available data.
K1-dominant kataegis identified in the S regions of the IGHV-unmutated group of CLL
CLL is one of the most common leukemia in adults in Western countries, characterized by the clonal expansion of mature CD5+ B cells (Hallek et al., 2018). Two major subsets can be divided based on the mutation status of IGHV genes, IGHV-mutated or IGHV-unmutated, and the latter is associated with a poor disease outcome (Damle et al., 1999). “Canonical” and “non-canonical” AID activities have been suggested to be responsible for the respective clustered and unclustered mutations in the CLL genomes (Kasar et al., 2015). To further dissect the mutagenesis mechanism in B cell tumors, WGS data from 153 CLL genomes from two previous studies (Alexandrov et al., 2013a; Puente et al., 2015) were reanalyzed. In total, 148 kataegis were identified, including 138 in Ig loci and 10 in non-Ig regions. Two main mutational signatures (termed CLL.K1 and CLL.K2) were extracted from these kataegis, which were highly similar with K1 (cos(θ) = 0.8515) and K2 (cos(θ) = 0.9252), respectively (Fig. 3 F).
Kataegis were identified in the Ig loci in 20% IGHV-unmutated CLLs and in 94% IGHV-mutated CLLs. Kataegis in the IGHV-unmutated group were almost exclusively located in the S regions and were CLL.K1-dominant, reminiscent of the K1-high DLBCL group. One notable difference was that the most targeted regions in the Ig loci in DLBCL, i.e., the IGHJ-Eμ, IGLJ1–IGLJ3, and IGKJ to IGKC regions, were almost not targeted by kataegis in the IGHV-unmutated CLLs. Kataegis in the IGHV-mutated group showed a more similar pattern to that observed in the K1-low DLBCL group (Fig. 3 G).
K1-high DLBCL is associated with translocations resulting from aberrant CSR activity
Structural variations (SVs), including large deletions, amplifications, inversions, and translocations, were next characterized in the DLBCL and FL genomes. In total, 2,475 somatic SVs, including 348 translocations, were identified. 91 of the 348 translocations, including all IGH translocations (n = 55), were verified by Sanger sequencing. For DLBCL, an average of 45 (range, 6–177) somatic SVs per sample were detected, including an average of 6 (range, 0–26) translocations. For FL, an average of 17 (range, 3–68) somatic SVs per sample were identified, including an average of 4 (range, 0–18) translocations.
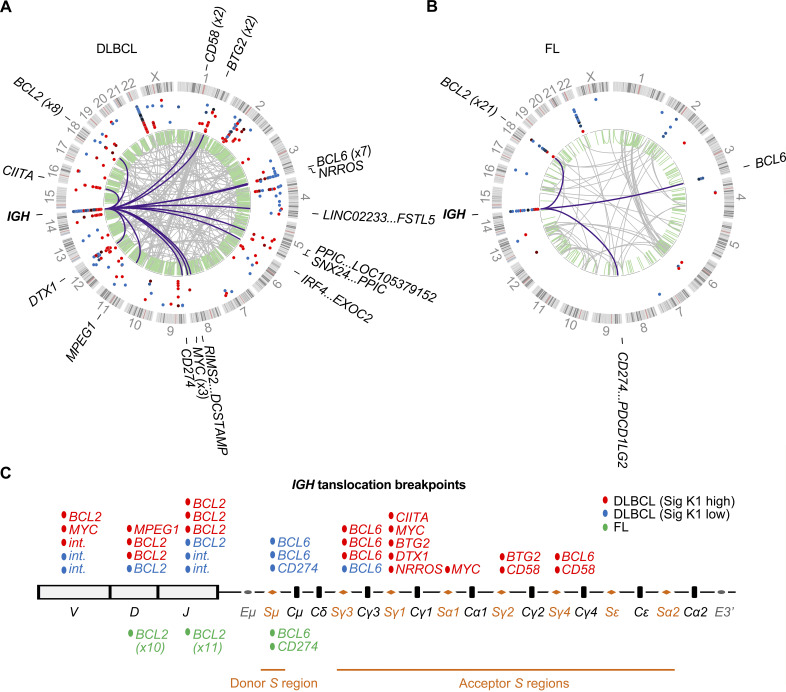
In the DLBCL samples, 437 kataegis and 263 translocations were identified, including 32 IGH translocations, 3 IGL translocations, and 228 non-Ig translocations (Fig. 4 A). Only 20 (5%) kataegis were colocalized with the translocation breakpoints in the same sample, and these breakpoints were located either in the Ig loci or in the corresponding translocation partner genes, which were all known AID off-targets. In the FL cases, 59 kataegis and 85 translocations were identified, including 23 translocations in the IGH region and 1 translocation in the IGK region (Fig. 4 B). Of these, nine (15%) kataegis were colocalized with translocation breakpoints, of which eight were in the IGH region. Thus, excluding those located in the IGH or AID off-targeted regions, the majority of the kataegis identified in DLBCL and FL did not colocalize with the genomic rearrangement events.
Figure 4.
SVs in DLBCL and FL genomes. (A and B) The identified kataegis (dots on the outer circle) and SVs (lines in the inner circle) in FLs (A) and DLBCLs (B). The kataegis are colored based on the contribution of K1 as in Fig. 2 F. The SVs are colored purple (IGH translocations), gray (nonIGH translocations), or green (intrachromosomal SVs). The partner genes for IGH translocations are labeled outside the circles. (C) Translocation breakpoints in the IGH loci were further mapped into V, D, J, or S regions, and the corresponding translocation partners are shown above (DLBCL) or below (FL) the IGH region schema. Sig, signature; int., intergenic regions.
In DLBCL, 47% of IGH translocations involved IGHV, IGHD, or IGHJ regions, and half of these (8/15) had BCL2 as the partner (Fig. 4, A and C). The remaining IGH translocations involved S regions in either the donor (Sμ, n = 3) or the acceptor S regions (n = 14). Notably, 13 of 14 translocations involved in the acceptor S regions were from K1-high samples, further suggesting an association between K1-high DLBCLs and aberrant CSR. In FL, most (21/23) translocations joined IGHD or IGHJ to BCL2 (Fig. 4, B and C), which have been proposed to occur during D-J or V(D)J rearrangements in preB cells (Bakhshi et al., 1987). The other two translocations joined Sμ to BCL6 or CD274. Both of them were from relapsed FL patients, suggesting that Sμ region translocations might be a key driver in FL relapse or transformation. There were no translocations in FL related to the acceptor S regions.
Discussion
A B cell tumor–specific signature, COSMIC 9, or SBS9 (Alexandrov et al., 2020), was first identified from a mixed set of B cell lymphoma and CLL samples (Alexandrov et al., 2013a). Later, focusing on DLBCL (Arthur et al., 2018) or CLL (Kasar et al., 2015), respectively, genomic mutational signatures highly similar to COSMIC 9 were identified, referred to as V6 or nc-AID. COSMIC 9 and related signatures were attributed to SHM and POLH activities as they were B cell tumor–restricted and showed a characteristic preference of T mutations in the TW motif. However, approximately half of the T mutations in COSMIC 9 were T>G mutations, whereas T mutations introduced by human POLH (Matsuda et al., 2001), or during SHM (Yaari et al., 2013), were dominated by T>C substitutions. A recent study based on whole-exome sequencing (WES) of DLBCL samples identified yet another signature, AID2, which is similar to COSMIC 9 but has more T>C mutations (Chapuy et al., 2018). However, the T mutations from AID2 were not enriched at TW motifs to the same extent as other signatures, potentially due to the limitation of using WES data. In this study, seven genomic signatures were identified from DLBCLs and FLs. Among these, G4 and G7 showed a high similarity to COSMIC 9. Upon further dissecting the mechanism underlying these B cell tumor–specific signatures, we showed that G7, compared with G4 and all previously described genomic signatures, had higher similarity with the mutation spectrum of human POLH and the bona fide SHM pattern. Furthermore, we showed that G7, but not G4, correlated with the rate of mutations at the IGHV locus, thus clearly linking a genome-wide mutational signature to the aberrant SHM process.
The difference in mutational signatures identified from different studies is probably due to the sequencing platform used, number and type of lymphoma samples analyzed, and analytic methods applied. COSMIC 9 or other previously described related signatures are likely to be a mixture of G4 and G7. The etiology of G4 remains unclear. Two additional error-prone polymerases have been implicated in SHM: Pol ζ (Saribasak et al., 2012) and Rev1 (Jansen et al., 2006). However, neither Pol ζ nor REV1 seems to be associated with the pattern of G4. Additionally, only 23% of lymphoma samples had substantial exposure to G4 (i.e., with >10% mutations assigned to G4). Thus, G4 may reflect some activities of POLH or other unknown enzymes but seems to be independent of SHM and only contributes to a subset of B cell lymphomas.
Focusing on kataegis, three signatures were subsequently identified in this study, including two major signatures associated with AID (K1) and POLH (K2) activities and one minor signature associated with APOBEC (KAPOBEC). Seven of the eight KAPOBEC-related kataegis were derived from one HBV-positive sample, which may reflect fingerprints of the antiviral activity of the APOBEC enzymes (Janahi and McGarvey, 2013). K1 was similar to the canonical AID signature identified by WES (Fig. S1 D; Chapuy et al., 2018) and extended our previous observation that kataegis in DLBCL are largely due to AID activity (Qian et al., 2014), whereas K2 is novel for DLBCL and FL. The POLH-related signature has previously been extracted from clustered mutations in several types of cancers (Supek and Lehner, 2017). Here, we dissected the mutations assigned to the K2 and discovered that most of these K2 mutations (the major peak) were likely due to additional repair by POLH after initial AID-induced lesions.
Theoretically, kataegis in the IGHV regions can be difficult to distinguish from real SHM events. However, most, if not all, kataegic mutations identified in the Ig loci represent a process that is associated with aberrant AID/SHM/CSR activities, and they had distinct features compared with those generated during the physiological SHM process. First, the kataegis in Ig loci in tumor samples were often located on the same DNA strand, which suggests that the mutations occurred in a processive manner within a short time. During SHM, however, the mutations accumulate with time and in a stepwise manner, with only a few unlinked mutations being fixed per cell division (Casellas et al., 2016). Second, kataegis in the S regions were clearly K1-dominant, representing the AID fingerprints, and are distinct from the SHM spectrum. Third, the targeted regions of kataegis mutations in the Ig loci are different from those targeted during the normal SHM process, as most targeted sequences for kataegis overlap with super enhancers and do not fall within the IGHV coding regions. Finally, when the analysis only included kataegis mutations in the non-Ig regions, we could identify two mutational signatures that were highly similar to K1/K2, suggesting that similar mechanisms underlie the clustered mutations within and outside the Ig loci. It is important to point out that we have illustrated that the kataegis identified in B cell lymphomas have unique features compared with those observed from non–B cell tumors, such as breast cancer. First, AID instead of APOBEC is the major driver for kataegis in B cell lymphoma. Second, for POLH-related K2 mutations, there are two groups, one of which is associated with AID activity, which has not been identified in non–B cell tumors. Third, most kataegis in B cell lymphoma do not colocalize with somatic translocations, with the exception of those in the IGH region and AID off-targets.
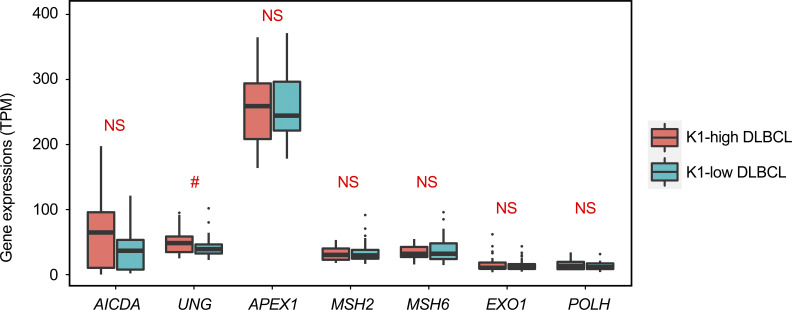
Our analyses of the mutational signatures and IGH translocation profiles suggest a link between the aberrant CSR events and the K1-high group of DLBCL (ABC subtype–enriched). This is consistent with the previous finding that the ABC subtype of DLBCL is associated with a higher frequency of internal deletions within the Sμ or Sγ regions as well as illegitimate CSR (detected by Southern blot analysis; Lenz et al., 2007). As kataegis in the S and V regions were mainly K1- or K2-dominant, respectively, it is likely that the S regions preferentially recruit AID/BER, while V regions preferentially recruit MMR/POLH-related factors. One interesting possibility is thus that the expression levels of corresponding DNA repair genes are different between the K1-high and K1-low DLBCL samples. However, we did not find major differences in the expression of AID–encoding gene (AICDA), key BER (UNG and APEX1) and MMR (MSH2, MSH6, and EXO1) genes, as well as POLH gene between the groups (Fig. S4). Furthermore, the number of kataegis in the S regions was similar between the two groups, and they were K1-dominant in all samples. Thus, another possibility is that the aberrant CSR events can occur independently from SHM and probably at a different time point or location, which is supported by a recent study demonstrating that CSR occurs before GC formation and SHM (Roco et al., 2019). It is possible that at least some of the K1-high or ABC subtype of DLBCLs originated from activated B cells that have mainly been exposed to aberrant CSR activity, with a relatively low level of mutations in the IGHV regions, and are programmed to differentiate into extrafollicular plasma cells without entering the GC reaction (Higgins et al., 2019). In contrast, most of the GCB subtype of DLBCLs derived from B cells that have entered a GC reaction and have been exposed to both aberrant CSR (possibly first) and SHM activities. FL had fewer mutations in both S and IGHV regions but generally shared similar features with the K1-low or GCB subtype of DLBCL, suggesting a similar cellular origin, i.e., B cells that have entered GC reactions.
Figure S4.
The expression of a set of genes related to AID, BER, and MMR. The expression of each gene for each sample was calculated by TPM as described in the Materials and methods. The expression levels of seven genes in K1-high and K1-low groups are shown as box plots, including AICDA, key BER genes (UNG and APEX1), key MMR genes (MSH2, MSH6, EXO1), and POLH. Each box is from the first quartile to the third quartile of the dataset, with a horizontal line extending through the box at the median. Values out of 1.5 times of interquartile range from each quartile were counted as outliers, which were labeled as separated dots in the plot. The P value obtained using the Mann–Whitney U test for each gene between two groups is shown in the figure. #, 0.05 < P < 0.1.
AID positivity predicted unmutated IGHV status (Heintel et al., 2004), and high AID expression has been shown to be restricted to a subpopulation of CLL cells with unmutated IGHV genes and ongoing CSR (Palacios et al., 2010). A recent WGS study also suggested that ongoing canonical AID (c-AID) activity is enriched in IGHV-unmutated cases (Kasar et al., 2015). The discovery of K1-dominant kataegis in S regions in a subset (20%) of IGHV-unmutated group of CLL further support these earlier observations that CSR activity is associated with this group of CLL patients with a more aggressive disease. It may also support an extrafollicular origin of this subgroup of CLLs, which further suggests that aberrant CSR and SHM activities can be linked to distinct developmental paths for different subtypes of B cell lymphomas. Further studies with larger cohorts of samples will be required to validate our findings.
In summary, we characterized the mutational signatures at the genomic level for two major types of GC-related B cell lymphomas, DLBCL and FL. We furthermore mapped the IGH-associated translocations identified from DLBCL and FL to a base pair resolution. Our study supports the critical role of AID dysregulation in B cell lymphomagenesis and also provides new insights into the molecular and cellular origins of different subtypes of GC-derived/related B cell lymphomas.
Materials and methods
Patients and samples
Samples from patients were described previously (de Miranda et al., 2013, 2014; Georgiou et al., 2016; Ren et al., 2018) and are summarized in Table S1. Detection of HBV surface antigen was performed as a routine blood test in all patients as described previously (Ren et al., 2018). Informed consent was obtained from all patients, and the institutional review boards at Tianjin Medical University Cancer Hospital and the Karolinska Institutet approved the study.
Genomic DNA was extracted from frozen tumor biopsies and the respective peripheral blood lymphocyte samples derived from patients using the DNeasy Blood & Tissue Mini Kit (Qiagen) following the manufacturer’s instructions. Total RNA was extracted from tumor samples using TRIzol reagent (Invitrogen).
WGS
WGS was performed using Illumina’s HiSeq 2000 or HiSeq X-Ten platform (BGI-Shenzhen) for 47 pairs of DLBCL and 22 pairs of FL samples and the Complete Genomics (CG) platform (BGI-Shenzhen) for the remaining 13 pairs of DLBCL samples as described previously (Ren et al., 2018). For the Illumina platforms, 2–3 µg of genomic DNA from each sample was fragmented using a Covarias sonication system to a mean size of 500 bp. After fragmentation, libraries were constructed according to the Illumina Paired-End protocol and sequenced on the HiSeq 2000 or X-Ten platform using 2 × 90-bp or 2 × 150-bp paired-end reads. Library construction and WGS of paired-end clones performed by CG were described previously (Drmanac et al., 2010; Lee et al., 2010; Roach et al., 2010).
Calling of single-nucleotide variants (SNVs), SBSs, and indels
For the Illumina platform, the sequencing reads containing adaptor sequences, low-quality reads (no-call positions >10%), and low-quality bases (>50% bases with quality <5) were removed. The high-quality paired-end reads were then gap-aligned to the human reference genome (hg19) using Burrows-Wheeler Aligner (BWA; Li and Durbin, 2009). After fixing mate information and adding read group information, Picard (v1.54; http://picard.sourceforge.net/) was used to mark duplicate reads caused by PCR. Local realignment of the BWA-aligned reads was subsequently performed by using the Genome Analysis Toolkit (McKenna et al., 2010). Somatic SNVs were detected using Varscan2 (Koboldt et al., 2012). SNVs were kept for further analysis if (1) there were at least three variant supporting reads in the tumor; (2) there was at least one supporting read for the reference allele on the plus and minus strands, as well as for the variant allele on the plus and minus strands; (3) the variant base quality, mapping quality, and position on read were significantly >15 (P < 0.06), 30 (P < 0.05), and 5 (P < 0.20), respectively (Mann–Whitney U test); (4) the distances between the variant and indels or repeat region were >20 and >5, respectively; (5) the variant frequency in tumor ≥10% or the variant frequency in tumor <10% if the variant frequency in normal tissue = 0; and (6) the variant frequency in normal tissue ≤2% or variant frequency in normal tissue >2% if the variant frequency in the tumor was 10 times higher than in the normal tissue. Somatic indels were detected with Platypus (Rimmer et al., 2014). CG Analysis Tools v2.0 was used to identify high-confidence SNVs and indels (Carnevali et al., 2012). SBSs were identified as the SNVs without any other adjacent SNVs in the same sample. The SBSs called by both platforms showed similar numbers and patterns. The indels called by both platforms also showed similar patterns. The number of indels called by the Illumina platform was significantly higher than the number called by the CG platform, which is consistent with a previous study (Lam et al., 2011). Thus, for the indel-related analysis (e.g., MSI analysis), the data from two platforms were used separately.
Variation filtering
Before further analysis, filtering was performed to remove potential residual germline mutations and technology-specific sequencing artifacts as previously described (Alexandrov et al., 2013a). Residual germline variations were removed by filtering against the complete list of germline mutations from the dbSNP (Sherry et al., 2001), the 1000 Genomes Project (Abecasis et al., 2012), the National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project (Fu et al., 2013), and the 69 Complete Genomics Panel (http://www.completegenomics.com/public-data/69-Genomes/). Any variation detected in at least two control samples from those corresponding platforms was defined as a technology-specific sequencing artifact and was removed. The remaining somatic mutations were used for further analysis.
All nonsilent, somatic mutations in the coding genome were manually checked using the Integrative Genomics Viewer. Furthermore, we have previously selected 135 mutations, of which 132 could be validated by targeted resequencing (Ren et al., 2018). Moreover, 77 of a selected 78 mutations were validated by Sanger sequencing.
Analysis of MSI status
MSIseq, which implements a decision tree classifier with a machine-learning framework, was applied to evaluate the MSI status of each DLBCL sample (Huang et al., 2015). The SNVs and micro-indels were prepared as recommended. The classification was performed using the default classifier of NGSclassifier. As Illumina and CG data showed different number ranges for indels, the MSI statuses of these two datasets were identified separately as modified Z scores of S.ind >1.25. 14 samples from our previous study were used to validate this identification (de Miranda et al., 2013). MMR somatic mutations were detected in 5 of 16 (31%) MSI samples. In comparison, MMR somatic mutations were detected in only 2 of 44 (5%) MSS samples, which is significantly lower than the value in MSI samples. No significant difference was observed between the Illumina and CG data (P = 0.2244, Fisher's exact test).
Mutational signature analysis
Mutational signature analysis was performed using the nonnegative matrix factorization–based method, SigProfiler (Alexandrov et al., 2013a). Mutational signatures were extracted from the WGS and kataegis data of 60 DLBCL and 22 FL tumors as follows: (1) somatic SBSs of each sample were classified into 96 possible mutated trinucleotides, as 6 types of substitution (C:G>A:T, C:G>G:C, C:G>T:A, T:A>A:T, T:A>C:G and T:A>G:C) × 4 types of 5′ base × 4 types of 3′ base, to generate a mutational catalog. Then, the prevalence of somatic mutations in each sample was calculated for each type of substitution; (2) signatures generated from the mutational catalog were deciphered by the mutational signature framework; and (3) the number of signatures extracted (N) was determined as described previously (Alexandrov et al., 2013b). Nonnegative matrix factorizations were performed iteratively 20 times for different values of N (1–15). The reproducibility and average reconstruction error were evaluated for each N. Finally, N was determined as 7 for our cohort of samples as it resulted in relatively fewer errors and high reproducibility (>90%). Reference signatures were cited from a previous study (Alexandrov et al., 2013a) and the COSMIC database (http://cancer.sanger.ac.uk/cosmic/signatures). Cosine similarity, cos(θ), was used to estimate the similarity between signatures. PCC (r) was used to estimate the association between signature exposure and the proposed etiology.
Detection of clustered somatic mutations (kataegis)
Kataegis were detected as previously described (Qian et al., 2014): (1) the abnormal distance line (ADL, one tenth the average distance of adjacent somatic mutations) was calculated for each tumor sample; (2) for every 10 adjacent mutations located within 10 kb of each other, the numbers of intermutation distances above and below the ADL were counted; (3) the adjacent set of 10 mutations was considered as a kataegis if the fraction of intermutation distances below the ADL was significantly different from that observed across all mutations in that sample (P < 0.0001, Fisher’s exact test, one-tailed test); and (4) overlapping kataegis were further merged if the resulting P value for the merged region was still <0.0001.
Construction of the experimental POLH-induced mutation signature
Experimental data on synthetic errors generated by POLH were extracted from previous studies (Matsuda et al., 2001; Yaari et al., 2013). All the mutations from Matsuda et al. (2001) were available for analysis. A mutational signature of POLH-induced errors was constructed by mapping all mutations into corresponding trinucleotide templates. The trinucleotide template frequency was normalized using the observed trinucleotide frequency in the selected region on M13mp2 DNA to the human genome. Only T mutations from Yaari et al. (2013) were available for analysis. As the original template was not available, the signature was directly adopted from the published summary (Supek and Lehner, 2017).
Assign single-base substitutions to specific signatures
The probability of each mutation from a given signature was calculated as previously described (Kasar et al., 2015). Briefly, each mutation was annotated by both signature process and exposure matrixes. The likelihood of a mutation generated by a specific signature was calculated by the proportion of this mutation in the signature times the exposure of this signature in the sample. If the probability that a mutation was generated by a given signature was >0.75, which means that the possibility of one signature was at least threefold higher than any other signature, then the mutation was annotated as generated by that signature.
Motif analysis of kataegis mutations
A web-based application, “two sample logo,” was used to identify the preference of motifs of kataegis mutations (Vacic et al., 2006). Mutations assigned to a given signature were first selected. Five residues, including two upstream and two downstream residues of each mutation, were then exacted to generate the case set. The control set consists of 5,000 randomly selected sequences with 5-bp length. Statistical significance was calculated for each residue at each position in the motif sequences from case and control sets. The null hypothesis was that the residue was generated according to the same distribution in both sets, and the P value was calculated using the t test. The statistically significant symbols were plotted using the size of the symbol that was proportional to the difference between the two sets. Residues were separated as enriched (positive y axis) and depleted (negative y axis) in the case set.
Transcriptome analysis by RNA sequencing (RNA-Seq)
Transcriptome sequencing of lymphoma samples has been described previously (Ren et al., 2018). Briefly, 2 µg of total RNA from each sample was used for transcriptome analysis as previously described (Wu et al., 2015). Raw sequencing reads with adaptors, with >10% unknown bases, or with >50% low-quality bases in one read, were filtered out. Clean data were aligned to the genome reference by BWA and to the gene reference by Bowtie2 (Langmead and Salzberg, 2012). Fewer than five mismatches were allowed for each read in the alignment. The gene expression level was calculated by using the transcripts per million (TPM) method. The RemoveBatchEffect command from the R package Limma was used to remove any potential batch effect (Ritchie et al., 2015).
DLBCL subtype classification and validation
DLBCL subtypes were identified using an RNA-Seq–based method (Reddy et al., 2017). Log2-transformed TPMs were z-normalized across the genes. The ABC and GCB scores were calculated for each sample by taking the average of the z-scores for ABC and GCB genes. The RNA-Seq subtype score was then calculated by taking the difference between the ABC score and the GCB score. Any sample with RNA-Seq subtype score >0.25 and GCB score <0.75 was defined as ABC. Any sample with RNA-Seq subtype score less than −0.25 and ABC score <0.75 was defined as GCB. The remaining samples were defined as the unclassified group. Among 55 samples with RNA-Seq data available, 23 ABC, 23 GCB, and 9 unclassified subtypes were identified. As a validation, the RNA-Seq subtype score was significantly correlated with the relative expression of ABC genes over GCB genes using the Lymph2Cx gene set (rspearman = 0.9439, P < 10−26, Spearman’s correlation; Scott et al., 2014) and was consistent with the Hans GCB versus non-GCB classification by immunohistochemistry (P = 0.0767, Mann–Whitney U test; Hans et al., 2004).
Identification and validation of SVs
For the 69 samples sequenced by the Illumina platform, somatic SVs were called by both manta algorithms (Chen et al., 2016) and SeekSV (Liang et al., 2017). To validate SVs, they were first double-checked by the Integrative Genomics Viewer, and then were further tested by PCR and Sanger sequencing. Primers mapping to both sides of the SVs were designed and used for breakpoint-specific PCR amplification on both tumor and normal DNAs. The gel bands with the expected size were recovered for Sanger sequencing. Somatic SVs were defined as validated if target bands were amplified only in tumor samples and confirmed by Sanger sequencing. In total, 2,507 SVs (345 translocations) and 28,543 SVs (1,271 translocations) were called by the manta and SeekSV pipelines, respectively, from which 687 SVs were selected for validation, including 635 translocations and 52 intrachromosomal SVs. For the 635 translocations, 23 were called by manta only, of which 21 (91%) were successfully validated. 53 were called by both manta and SeekSV, of which 37 (70%) were successfully validated. A total of 559 were called only by SeekSV, of which 60 (11%) were successfully validated. Thus, the manta algorithms performed much better than SeekSV in our cohort. However, for the 55 validated IGH translocations, close to half (n = 23, 42%) were called only by SeekSV. Thus, by focusing on IGH translocations, a reliable somatic SV dataset was constructed by combining all manta results and validated IGH translocations from the SeekSV results. All the SVs validated as germlines were removed. If both sides of two SVs were localized within 1,000 bp, only one was counted. Finally, 2,475 SVs remained for further analysis. All the breakpoints were annotated by ANNOVAR (Wang et al., 2010).
Data deposition
WGS and RNA-Seq data were deposited in the China National GeneBank Sequence Archive of China National GeneBank Database with accession nos. CNP0001228 and CNP0001220.
Reference databases
Gene coordinates, functions, and TSSs were annotated from National Center for Biotechnology Information RefSeq (O’Leary et al., 2016). The super enhancers of tonsil B cells (ID: CB5_H3K27Ac) were downloaded from SEdb (Jiang et al., 2019). Human chromosomal fragile sites were downloaded from HumCFS (Kumar et al., 2019). The data of 153 DLBCLs and 153 CLLs were downloaded from the supplementary tables of Alexandrov et al. (2013a), Arthur et al. (2018), and Puente et al. (2015). Among CLL samples, 69 were IGHV-unmutated, and 52 were IGHV-mutated. The remaining 32 samples that lacked subtype information were also included to empower the mutational signature analysis.
Online supplemental material
A comparison of genomic signatures identified in DLBCL and FL with the previously published mutational signatures is shown in Fig. S1 A. A comparison of POLH-related signatures is shown in Fig. S1 B. The identification of APOBEC-related kataegis is illustrated in Fig. S1 C. A comparison of AID-related signatures is shown in Fig. S1 D. The mutation pattern and distribution in kataegis regions in the Sμ and IGHJ-Eμ regions are shown in Fig. S2 A. The correlation of the overall contribution of K1 for a given sample and its contribution to different regions of the Ig and non-Ig loci is shown in Fig. S2 B. Mutational signatures identified from kataegis in a published DLBCL cohort (Arthur et al., 2018) are shown in Fig. S3. The expression level of a set of genes related to AID, BER, and MMR in K1-high and K1-low groups is shown in Fig. S4. Clinical and sample information of the DLBCL and FL cohort are described in Table S1. Details of all kataegis identified in the DLBCL and FL genomes are presented in Table S2.
Supplementary Material
describes the clinical and sample information of the DLBCL and FL cohort.
presents details of all kataegis identified in the DLBCL and FL genomes.
Acknowledgments
We thank L. Chen, A. Zaravinos, K. Georgiou, R. Caridha, and Y. Su for help with the analysis or validation experiments.
This work was supported by Cancerfonden (Swedish Cancer Society), the Swedish Research Council, the Swedish Childhood Cancer fund, the National Natural Science Foundation of China (81670184), the Joint Research Initiative of the School of Medicine, Shanghai Jiao Tong University, the Radiumhemmet research fund, the Center for Innovative Medicine, the Guangdong Enterprise Key Laboratory of Human Disease Genomics (2020B1212070028), and the China National GeneBank.
Author contributions: X. Ye designed the study, performed the data analysis regarding mutation signatures and translocations, and wrote the manuscript; H. Zhang and W. Li provided patient samples and clinical data; W. Ren and X. Wang prepared the tumor samples; D. Liu, X. Li, Y. Hou, S. Zhu, and K. Wu performed the sequencing data analysis; W. Li and W. Ren performed validation experiments; F-L. Meng, L-S. Yeap, and R. Casellas supervised the data analysis; and Q. Pan-Hammarström designed and supervised the study and wrote the manuscript.
References
- Abecasis, G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., and McVean G.A.. 1000 Genomes Project Consortium . 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491:56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov, L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., et al. ICGC PedBrain . 2013a Signatures of mutational processes in human cancer. Nature. 500:415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov, L.B., Nik-Zainal S., Wedge D.C., Campbell P.J., and Stratton M.R.. 2013b Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 3:246–259. 10.1016/j.celrep.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov, L.B., Kim J., Haradhvala N.J., Huang M.N., Tian Ng A.W., Wu Y., Boot A., Covington K.R., Gordenin D.A., Bergstrom E.N., et al. PCAWG Consortium . 2020. The repertoire of mutational signatures in human cancer. Nature. 578:94–101. 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh, A.A., Eisen M.B., Davis R.E., Ma C., Lossos I.S., Rosenwald A., Boldrick J.C., Sabet H., Tran T., Yu X., et al. . 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403:503–511. 10.1038/35000501 [DOI] [PubMed] [Google Scholar]
- Álvarez-Prado, A.F., Pérez-Durán P., Pérez-García A., Benguria A., Torroja C., de Yébenes V.G., and Ramiro A.R.. 2018. A broad atlas of somatic hypermutation allows prediction of activation-induced deaminase targets. J. Exp. Med. 215:761–771. 10.1084/jem.20171738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur, S.E., Jiang A., Grande B.M., Alcaide M., Cojocaru R., Rushton C.K., Mottok A., Hilton L.K., Lat P.K., Zhao E.Y., et al. . 2018. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat. Commun. 9:4001 10.1038/s41467-018-06354-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshi, A., Wright J.J., Graninger W., Seto M., Owens J., Cossman J., Jensen J.P., Goldman P., and Korsmeyer S.J.. 1987. Mechanism of the t(14;18) chromosomal translocation: structural analysis of both derivative 14 and 18 reciprocal partners. Proc. Natl. Acad. Sci. USA. 84:2396–2400. 10.1073/pnas.84.8.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso, K., and Dalla-Favera R.. 2015. Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 15:172–184. 10.1038/nri3814 [DOI] [PubMed] [Google Scholar]
- Burns, M.B., Temiz N.A., and Harris R.S.. 2013. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 45:977–983. 10.1038/ng.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevali, P., Baccash J., Halpern A.L., Nazarenko I., Nilsen G.B., Pant K.P., Ebert J.C., Brownley A., Morenzoni M., Karpinchyk V., et al. . 2012. Computational techniques for human genome resequencing using mated gapped reads. J. Comput. Biol. 19:279–292. 10.1089/cmb.2011.0201 [DOI] [PubMed] [Google Scholar]
- Casellas, R., Basu U., Yewdell W.T., Chaudhuri J., Robbiani D.F., and Di Noia J.M.. 2016. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat. Rev. Immunol. 16:164–176. 10.1038/nri.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy, B., Stewart C., Dunford A.J., Kim J., Kamburov A., Redd R.A., Lawrence M.S., Roemer M.G.M., Li A.J., Ziepert M., et al. . 2018. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 24:679–690. 10.1038/s41591-018-0016-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., and Saunders C.T.. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 32:1220–1222. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- Damle, R.N., Wasil T., Fais F., Ghiotto F., Valetto A., Allen S.L., Buchbinder A., Budman D., Dittmar K., Kolitz J., et al. . 1999. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 94:1840–1847. 10.1182/blood.V94.6.1840 [DOI] [PubMed] [Google Scholar]
- de Miranda, N.F., Peng R., Georgiou K., Wu C., Falk Sörqvist E., Berglund M., Chen L., Gao Z., Lagerstedt K., Lisboa S., et al. . 2013. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J. Exp. Med. 210:1729–1742. 10.1084/jem.20122842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Miranda, N.F., Georgiou K., Chen L., Wu C., Gao Z., Zaravinos A., Lisboa S., Enblad G., Teixeira M.R., Zeng Y., et al. . 2014. Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood. 124:2544–2553. 10.1182/blood-2013-12-546309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbos, F., De Smet A., Faili A., Aoufouchi S., Weill J.C., and Reynaud C.A.. 2005. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 201:1191–1196. 10.1084/jem.20050292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia, J.M., and Neuberger M.S.. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22. 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Drmanac, R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G., et al. . 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 327:78–81. 10.1126/science.1181498 [DOI] [PubMed] [Google Scholar]
- Franco, S., Gostissa M., Zha S., Lombard D.B., Murphy M.M., Zarrin A.A., Yan C., Tepsuporn S., Morales J.C., Adams M.M., et al. . 2006. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 21:201–214. 10.1016/j.molcel.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Fu, W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., et al. NHLBI Exome Sequencing Project . 2013. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 493:216–220. 10.1038/nature11690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou, K., Chen L., Berglund M., Ren W., de Miranda N.F., Lisboa S., Fangazio M., Zhu S., Hou Y., Wu K., et al. . 2016. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood. 127:3026–3034. 10.1182/blood-2015-12-686550 [DOI] [PubMed] [Google Scholar]
- Hakim, O., Resch W., Yamane A., Klein I., Kieffer-Kwon K.R., Jankovic M., Oliveira T., Bothmer A., Voss T.C., Ansarah-Sobrinho C., et al. . 2012. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 484:69–74. 10.1038/nature10909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halldórsdóttir, A.M., Frühwirth M., Deutsch A., Aigelsreiter A., Beham-Schmid C., Agnarsson B.A., Neumeister P., and Richard Burack W.. 2008. Quantifying the role of aberrant somatic hypermutation in transformation of follicular lymphoma. Leuk. Res. 32:1015–1021. 10.1016/j.leukres.2007.11.028 [DOI] [PubMed] [Google Scholar]
- Hallek, M., Shanafelt T.D., and Eichhorst B.. 2018. Chronic lymphocytic leukaemia. Lancet. 391:1524–1537. 10.1016/S0140-6736(18)30422-7 [DOI] [PubMed] [Google Scholar]
- Hans, C.P., Weisenburger D.D., Greiner T.C., Gascoyne R.D., Delabie J., Ott G., Müller-Hermelink H.K., Campo E., Braziel R.M., Jaffe E.S., et al. . 2004. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 103:275–282. 10.1182/blood-2003-05-1545 [DOI] [PubMed] [Google Scholar]
- Hardianti, M.S., Tatsumi E., Syampurnawati M., Furuta K., Saigo K., Nakamachi Y., Kumagai S., Ohno H., Tanabe S., Uchida M., and Yasuda N.. 2004. Activation-induced cytidine deaminase expression in follicular lymphoma: association between AID expression and ongoing mutation in FL. Leukemia. 18:826–831. 10.1038/sj.leu.2403323 [DOI] [PubMed] [Google Scholar]
- Heintel, D., Kroemer E., Kienle D., Schwarzinger I., Gleiss A., Schwarzmeier J., Marculescu R., Le T., Mannhalter C., Gaiger A., et al. German CLL Study Group . 2004. High expression of activation-induced cytidine deaminase (AID) mRNA is associated with unmutated IGVH gene status and unfavourable cytogenetic aberrations in patients with chronic lymphocytic leukaemia. Leukemia. 18:756–762. 10.1038/sj.leu.2403294 [DOI] [PubMed] [Google Scholar]
- Helleday, T., Eshtad S., and Nik-Zainal S.. 2014. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 15:585–598. 10.1038/nrg3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, B.W., McHeyzer-Williams L.J., and McHeyzer-Williams M.G.. 2019. Programming Isotype-Specific Plasma Cell Function. Trends Immunol. 40:345–357. 10.1016/j.it.2019.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M.N., McPherson J.R., Cutcutache I., Teh B.T., Tan P., and Rozen S.G.. 2015. MSIseq: Software for Assessing Microsatellite Instability from Catalogs of Somatic Mutations. Sci. Rep. 5:13321 10.1038/srep13321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janahi, E.M., and McGarvey M.J.. 2013. The inhibition of hepatitis B virus by APOBEC cytidine deaminases. J. Viral Hepat. 20:821–828. 10.1111/jvh.12192 [DOI] [PubMed] [Google Scholar]
- Jansen, J.G., Langerak P., Tsaalbi-Shtylik A., van den Berk P., Jacobs H., and de Wind N.. 2006. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 203:319–323. 10.1084/jem.20052227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., Soong T.D., Wang L., Melnick A.M., and Elemento O.. 2012. Genome-wide detection of genes targeted by non-Ig somatic hypermutation in lymphoma. PLoS One. 7:e40332 10.1371/journal.pone.0040332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., Qian F., Bai X., Liu Y., Wang Q., Ai B., Han X., Shi S., Zhang J., Li X., et al. . 2019. SEdb: a comprehensive human super-enhancer database. Nucleic Acids Res. 47(D1):D235–D243. 10.1093/nar/gky1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasar, S., Kim J., Improgo R., Tiao G., Polak P., Haradhvala N., Lawrence M.S., Kiezun A., Fernandes S.M., Bahl S., et al. . 2015. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nat. Commun. 6:8866 10.1038/ncomms9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodabakhshi, A.H., Morin R.D., Fejes A.P., Mungall A.J., Mungall K.L., Bolger-Munro M., Johnson N.A., Connors J.M., Gascoyne R.D., Marra M.A., et al. . 2012. Recurrent targets of aberrant somatic hypermutation in lymphoma. Oncotarget. 3:1308–1319. 10.18632/oncotarget.653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt, D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., and Wilson R.K.. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22:568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R., Nagpal G., Kumar V., Usmani S.S., Agrawal P., and Raghava G.P.S.. 2019. HumCFS: a database of fragile sites in human chromosomes. BMC Genomics. 19(S9, Suppl 9):985 10.1186/s12864-018-5330-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küppers, R., Klein U., Hansmann M.L., and Rajewsky K.. 1999. Cellular origin of human B-cell lymphomas. N. Engl. J. Med. 341:1520–1529. 10.1056/NEJM199911113412007 [DOI] [PubMed] [Google Scholar]
- Lam, H.Y., Clark M.J., Chen R., Chen R., Natsoulis G., O’Huallachain M., Dewey F.E., Habegger L., Ashley E.A., Gerstein M.B., et al. . 2011. Performance comparison of whole-genome sequencing platforms. Nat. Biotechnol. 30:78–82. 10.1038/nbt.2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W., Jiang Z., Liu J., Haverty P.M., Guan Y., Stinson J., Yue P., Zhang Y., Pant K.P., Bhatt D., et al. . 2010. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 465:473–477. 10.1038/nature09004 [DOI] [PubMed] [Google Scholar]
- Lenz, G., Nagel I., Siebert R., Roschke A.V., Sanger W., Wright G.W., Dave S.S., Tan B., Zhao H., Rosenwald A., et al. . 2007. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J. Exp. Med. 204:633–643. 10.1084/jem.20062041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y., Qiu K., Liao B., Zhu W., Huang X., Li L., Chen X., and Li K.. 2017. Seeksv: an accurate tool for somatic structural variation and virus integration detection. Bioinformatics. 33:184–191. 10.1093/bioinformatics/btw591 [DOI] [PubMed] [Google Scholar]
- Liu, M., Duke J.L., Richter D.J., Vinuesa C.G., Goodnow C.C., Kleinstein S.H., and Schatz D.G.. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845. 10.1038/nature06547 [DOI] [PubMed] [Google Scholar]
- Lossos, I.S., and Gascoyne R.D.. 2011. Transformation of follicular lymphoma. Best Pract. Res. Clin. Haematol. 24:147–163. 10.1016/j.beha.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossos, I.S., Levy R., and Alizadeh A.A.. 2004. AID is expressed in germinal center B-cell-like and activated B-cell-like diffuse large-cell lymphomas and is not correlated with intraclonal heterogeneity. Leukemia. 18:1775–1779. 10.1038/sj.leu.2403488 [DOI] [PubMed] [Google Scholar]
- Matsuda, T., Bebenek K., Masutani C., Rogozin I.B., Hanaoka F., and Kunkel T.A.. 2001. Error rate and specificity of human and murine DNA polymerase eta. J. Mol. Biol. 312:335–346. 10.1006/jmbi.2001.4937 [DOI] [PubMed] [Google Scholar]
- McKenna, A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M.A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu, M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., and Honjo T.. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Nagaoka, H., Muramatsu M., Yamamura N., Kinoshita K., and Honjo T.. 2002. Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Smu region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J. Exp. Med. 195:529–534. 10.1084/jem.20012144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal, S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., et al. Breast Cancer Working Group of the International Cancer Genome Consortium . 2012. Mutational processes molding the genomes of 21 breast cancers. Cell. 149:979–993. 10.1016/j.cell.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal, S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C., et al. . 2016. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 534:47–54. 10.1038/nature17676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary, N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. . 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44(D1):D733–D745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, F., Moreno P., Morande P., Abreu C., Correa A., Porro V., Landoni A.I., Gabus R., Giordano M., Dighiero G., et al. . 2010. High expression of AID and active class switch recombination might account for a more aggressive disease in unmutated CLL patients: link with an activated microenvironment in CLL disease. Blood. 115:4488–4496. 10.1182/blood-2009-12-257758 [DOI] [PubMed] [Google Scholar]
- Pan-Hammarström, Q., Dai S., Zhao Y., van Dijk-Härd I.F., Gatti R.A., Børresen-Dale A.L., and Hammarström L.. 2003. ATM is not required in somatic hypermutation of VH, but is involved in the introduction of mutations in the switch mu region. J. Immunol. 170:3707–3716. 10.4049/jimmunol.170.7.3707 [DOI] [PubMed] [Google Scholar]
- Pasqualucci, L., Neumeister P., Goossens T., Nanjangud G., Chaganti R.S., Küppers R., and Dalla-Favera R.. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 412:341–346. 10.1038/35085588 [DOI] [PubMed] [Google Scholar]
- Pasqualucci, L., Bhagat G., Jankovic M., Compagno M., Smith P., Muramatsu M., Honjo T., Morse H.C. III, Nussenzweig M.C., and Dalla-Favera R.. 2008. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 40:108–112. 10.1038/ng.2007.35 [DOI] [PubMed] [Google Scholar]
- Pettersen, H.S., Galashevskaya A., Doseth B., Sousa M.M., Sarno A., Visnes T., Aas P.A., Liabakk N.B., Slupphaug G., Sætrom P., et al. . 2015. AID expression in B-cell lymphomas causes accumulation of genomic uracil and a distinct AID mutational signature. DNA Repair (Amst.). 25:60–71. 10.1016/j.dnarep.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Pham, P., Bransteitter R., Petruska J., and Goodman M.F.. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 424:103–107. 10.1038/nature01760 [DOI] [PubMed] [Google Scholar]
- Puente, X.S., Beà S., Valdés-Mas R., Villamor N., Gutiérrez-Abril J., Martín-Subero J.I., Munar M., Rubio-Pérez C., Jares P., Aymerich M., et al. . 2015. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 526:519–524. 10.1038/nature14666 [DOI] [PubMed] [Google Scholar]
- Qian, J., Wang Q., Dose M., Pruett N., Kieffer-Kwon K.R., Resch W., Liang G., Tang Z., Mathé E., Benner C., et al. . 2014. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 159:1524–1537. 10.1016/j.cell.2014.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro, A.R., Jankovic M., Eisenreich T., Difilippantonio S., Chen-Kiang S., Muramatsu M., Honjo T., Nussenzweig A., and Nussenzweig M.C.. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438. 10.1016/j.cell.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Ramiro, A.R., Jankovic M., Callen E., Difilippantonio S., Chen H.T., McBride K.M., Eisenreich T.R., Chen J., Dickins R.A., Lowe S.W., et al. . 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109. 10.1038/nature04495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, A., Zhang J., Davis N.S., Moffitt A.B., Love C.L., Waldrop A., Leppa S., Pasanen A., Meriranta L., Karjalainen-Lindsberg M.L., et al. . 2017. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell. 171:481–494.e15. 10.1016/j.cell.2017.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, W., Ye X., Su H., Li W., Liu D., Pirmoradian M., Wang X., Zhang B., Zhang Q., Chen L., et al. . 2018. Genetic landscape of hepatitis B virus-associated diffuse large B-cell lymphoma. Blood. 131:2670–2681. 10.1182/blood-2017-11-817601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer, A., Phan H., Mathieson I., Iqbal Z., Twigg S.R.F., Wilkie A.O.M., McVean G., Lunter G., and Lunter G.. WGS500 Consortium . 2014. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 46:912–918. 10.1038/ng.3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach, J.C., Glusman G., Smit A.F., Huff C.D., Hubley R., Shannon P.T., Rowen L., Pant K.P., Goodman N., Bamshad M., et al. . 2010. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 328:636–639. 10.1126/science.1186802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G., et al. . 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45:970–976. 10.1038/ng.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roco, J.A., Mesin L., Binder S.C., Nefzger C., Gonzalez-Figueroa P., Canete P.F., Ellyard J., Shen Q., Robert P.A., Cappello J., et al. . 2019. Class-Switch Recombination Occurs Infrequently in Germinal Centers. Immunity. 51:337–350.e7. 10.1016/j.immuni.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin, I.B., and Kolchanov N.A.. 1992. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta. 1171:11–18. 10.1016/0167-4781(92)90134-L [DOI] [PubMed] [Google Scholar]
- Rogozin, I.B., Pavlov Y.I., Bebenek K., Matsuda T., and Kunkel T.A.. 2001. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat. Immunol. 2:530–536. 10.1038/88732 [DOI] [PubMed] [Google Scholar]
- Rosenquist, R., Beà S., Du M.Q., Nadel B., and Pan-Hammarström Q.. 2017. Genetic landscape and deregulated pathways in B-cell lymphoid malignancies. J. Intern. Med. 282:371–394. 10.1111/joim.12633 [DOI] [PubMed] [Google Scholar]
- Saribasak, H., Maul R.W., Cao Z., Yang W.W., Schenten D., Kracker S., and Gearhart P.J.. 2012. DNA polymerase ζ generates tandem mutations in immunoglobulin variable regions. J. Exp. Med. 209:1075–1081. 10.1084/jem.20112234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, D.W., Wright G.W., Williams P.M., Lih C.J., Walsh W., Jaffe E.S., Rosenwald A., Campo E., Chan W.C., Connors J.M., et al. . 2014. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 123:1214–1217. 10.1182/blood-2013-11-536433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry, S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., and Sirotkin K.. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29:308–311. 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer, J., Guikema J.E., and Schrader C.E.. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292. 10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, F.K., Sahota S.S., Ottensmeier C.H., Zhu D., Forconi F., and Hamblin T.J.. 2001. The occurrence and significance of V gene mutations in B cell-derived human malignancy. Adv. Cancer Res. 83:81–116. 10.1016/S0065-230X(01)83004-9 [DOI] [PubMed] [Google Scholar]
- Supek, F., and Lehner B.. 2017. Clustered Mutation Signatures Reveal that Error-Prone DNA Repair Targets Mutations to Active Genes. Cell. 170:534–547.e23. 10.1016/j.cell.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Vacic, V., Iakoucheva L.M., and Radivojac P.. 2006. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 22:1536–1537. 10.1093/bioinformatics/btl151 [DOI] [PubMed] [Google Scholar]
- Victora, G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Wang, K., Li M., and Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Zhang X., Li F., Xiao D., Hou Y., Zhu S., Liu D., Ye X., Ye M., Yang J., et al. . 2015. Frequent alterations in cytoskeleton remodelling genes in primary and metastatic lung adenocarcinomas. Nat. Commun. 6:10131 10.1038/ncomms10131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaari, G., Vander Heiden J.A., Uduman M., Gadala-Maria D., Gupta N., Stern J.N., O’Connor K.C., Hafler D.A., Laserson U., Vigneault F., and Kleinstein S.H.. 2013. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front. Immunol. 4:358 10.3389/fimmu.2013.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
describes the clinical and sample information of the DLBCL and FL cohort.
presents details of all kataegis identified in the DLBCL and FL genomes.