Abstract
Toll-like receptors (TLRs) are key pattern recognition receptors that mediate innate immune responses to infection. However, uncontrolled TLR activation can lead to severe inflammatory disorders such as septic shock. The molecular mechanisms through which TLR responses are regulated are not fully understood. Here, we demonstrate an essential function of S100A10 in TLR signaling. S100A10 was constitutively expressed in macrophages, but was significantly downregulated upon TLR activation. S100A10-deficient macrophages were hyperresponsive to TLR stimulation, and S100A10-deficient mice were more sensitive to endotoxin-induced lethal shock and Escherichia coli-induced abdominal sepsis. Mechanistically, S100A10 regulated macrophage inflammatory responses by interfering with the appropriate recruitment and activation of the receptor-proximal signaling components and eventually inhibited TLR-triggered downstream signaling. These findings expand our understanding of TLR signaling and establish S100A10 as an essential negative regulator of TLR function and a potential therapeutic target for treating inflammatory diseases.
Keywords: Sepsis, Inflammation, TLR, S100A10, Innate immunity
Subject terms: Toll-like receptors, Infection
Introduction
Toll-like receptors (TLRs), the key pattern recognition receptors, play crucial roles in host defense against invading microbial pathogens by detecting conserved pathogen-associated molecular patterns (PAMPs). Recognition of PAMPs by TLRs initiates innate immune responses through the recruitment of the adaptors MyD88 or TRIF to induce downstream signaling cascades, which subsequently activates the transcription factors nuclear factor-κB (NF-κB) and activator protein-1 (AP-1) that are common to all TLRs, leading to the production of proinflammatory cytokines and type I interferons.1 Full activation of TLRs is essential for the initiation of the innate immune response and the enhancement of adaptive immunity to eliminate invading microorganisms. However, inappropriate activation or overactivation of TLR signaling may result in inflammatory diseases such as septic shock or autoimmune disorders.2 Thus, the negative regulation of TLR signaling is essential for avoiding excessive inflammatory immune responses and maintaining immune homeostasis. To date, numerous positive regulators have been identified as essential for the initiation and full activation of TLR responses, but inhibitors of TLR pathways need further investigation.3,4 Therefore, the identification of negative regulators and the detailed mechanisms underlying their role in TLR signaling remain to be fully elucidated.
S100A10, a unique member of the S100 EF-hand protein family, does not conform to other family members because it lacks a functional EF-hand Ca2+-binding domain, so its interaction with target proteins is Ca2+ independent. S100A10 was first identified within a heterotetrameric complex with annexin A2.5 Previous reports have shown that S100A10 tethers certain membrane proteins (i.e., serotonin 1B receptor, tissue-type plasminogen activator, sodium channel Nav1.8, actin-binding protein AHNAK) to annexin A2, thereby assisting their traffic to the plasma membrane and increasing their levels at the cell surface.6 One recent study demonstrated that S100A10 directly binds to the C-terminal cytoplasmic tail of CCR10, and this interaction regulates its cell surface presentation.7 CCR10, also known as orphan GPR-2, was identified as the specific receptor for the chemokine CCL27 and belongs to the chemokine receptor subfamily of G protein-coupled receptors. Hessner et al.7 found that S100A10 interacts physically with the carboxylic-terminal cytosolic tail within amino acid residues 326–340 of CCR10. In addition, annexin A2 was found to be associated with the CCR10-S100A10 complex, but annexin A2 did not directly interact with CCR10, indicating that S100A10 bears distinct binding sites for CCR10 and annexin A2 and that the S100A10 dimer might act as a linker between CCR10 and annexin A2 within the tripartite complex.7 Furthermore, S100A10 serves as an important regulator of inflammation, and abnormal expression of S100A10 has been found in patients with depression, irritable bowel syndrome, and Parkinson’s disease.8–11 In particular, it is intriguing that expression levels of S100A10 in peripheral blood leukocytes, including monocytes, natural killer cells, and CD8+ T cells, are altered in both depression and Parkinson’s disease.10 Although the functions of S100A10 have been extensively investigated in the central nervous system, its function in the immune system remains largely unknown.
A recent paper showed that S100A10 mediates macrophage recruitment in response to sterile inflammatory stimuli by activating promatrix metalloproteinase-9, which in turn promotes plasmin-dependent invasion in vitro and in vivo.12 However, the regulation of expression and other functions, such as the immune response of S100A10 in macrophages, remain unclear. Considering the critical roles of macrophages in the promotion of the innate immune response, we hypothesized that S100A10 could play an important role in regulating the innate inflammatory responses during sepsis. Using cells and mice with a target deletion of S100A10, we examined this hypothesis in vitro and in vivo. Our results uncover the crucial function of S100A10 as a negative regulator of the TLR pathways, which adds new insight into the regulation of innate immunity and TLR-triggered innate inflammatory responses.
Results
TLR stimulation significantly decreases S100A10 expression
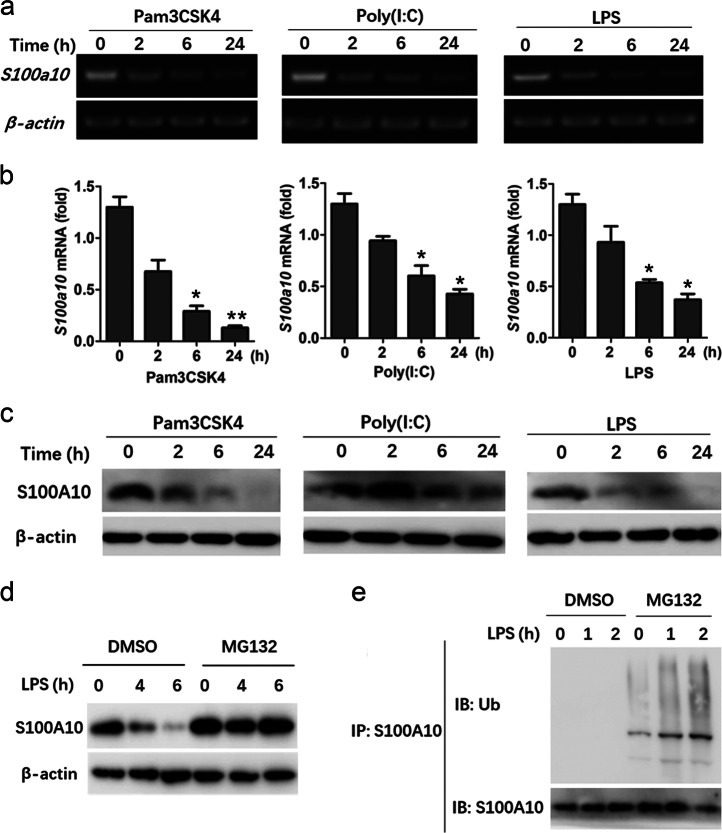
S100A10 is a member of the S100 protein family, which is composed of more than 20 members, but its biological function, especially in the immune response, remain largely unknown. Our initial analysis of the BioGPS database revealed that macrophages had an abundant expression of S100a10. To explore the potential functions of S100A10 in the immune system, we first examined its messenger RNA (mRNA) expression in murine bone marrow-derived macrophages (BMDMs) before and after stimulation with different TLR ligands, as many genes that are involved in the immune response undergo changes in the expression pattern after induction of inflammation. Upon stimulation with Pam3CSK4 (a TLR2 ligand), poly(I:C) (a TLR3 ligand), and lipopolysaccharide (LPS) (a TLR4 ligand), the expression of S100a10 mRNA was significantly decreased by all these ligands (Fig. 1a, b). S100A10 protein expression was also decreased after treatment with these ligands (Fig. 1c). Similar effects were observed in murine peritoneal macrophages after stimulation with LPS or Pam3CSK4 (Fig. S1a, b). These results indicate that TLR activation leads to the reduction of S100A10 expression in macrophages.
Fig. 1.
S100A10 expression is decreased upon TLR activation in bone marrow-derived macrophages. a, b Bone marrow-derived macrophages from WT mice were treated with or without Pam3CSK4 (50 ng/ml), poly(I:C) (10 μg/ml), or LPS (100 ng/ml) for the indicated times. S100a10 mRNA levels were determined by RT-PCR (a). Quantification of S100a10 mRNA expression by quantitative real-time PCR (b). c Bone marrow-derived macrophages from WT mice were treated as described in a, b, and S100A10 protein levels were determined by Western blot. d, e Bone marrow-derived macrophages from WT mice were pretreated with MG132 (10 μM) or DMSO control and then stimulated with or without LPS (100 ng/ml) for the indicated times. S100A10 protein levels were determined by Western blot (d). In addition, cell lysates were subjected to immunoprecipitation (IP) with an antibody against S100A10 and analyzed by Western blot for the indicated proteins (e). β-Actin was used as a loading control. Data are representative of three experiments (mean ± SEM of three samples in b; *P < 0.05; **P < 0.01). Similar results were obtained in three independent experiments in a, c–e
The S100A10 protein has been shown to be polyubiquitinated and degraded in endothelial cells via a proteasome-dependent mechanism.13 We therefore determined whether TLR ligands could also target the amount of S100A10 protein through the ubiquitin-proteasome system. Pretreatment of BMDMs with vehicle control and subsequent stimulation with LPS for 4 h resulted in a slight reduction of S100A10 and a marked decrease 6 h after stimulation (Fig. 1d). MG132, which is a reversible peptide aldehyde that blocks proteasome activity, reversed the ability of LPS to suppress S100A10 expression 4 and 6 h after stimulation (Fig. 1d). However, because prolonged MG132 treatment is toxic to cells, the effect of MG132 at 24 h noted before could not be analyzed. Nevertheless, it was obvious that the proteasome did affect the amount of S100A10 protein at earlier time points after LPS stimulation. Proteasomal degradation of target proteins depends on their polyubiquitination. Therefore, we examined the ability of the S100A10 protein to become ubiquitinated in response to LPS. Indeed, stimulation of MG132-pretreated BMDMs with LPS resulted in the appearance of the polyubiquitinated forms of S100A10 (Fig. 1e). Collectively, these data clearly demonstrate that, in addition to a reduction in the expression of S100a10 mRNA, LPS also induces the degradation of S100A10 by the proteasome, thus contributing to the loss of S100A10 protein in macrophages.
S100A10 deficiency increases TLR-triggered proinflammatory cytokine production in macrophages
S100A10 is constitutively expressed in macrophages, and TLR activation can decrease S100A10 expression, raising the possibility that S100A10 may be involved in the regulation of TLR-triggered inflammatory responses. To further elucidate the role of S100A10 in TLR-triggered innate inflammatory responses, we generated S100A10-deficient mice (called S100a10−/− here) by using the CRISPR-Cas9 strategy (Fig. S1c). The results from sequencing showed that a 7-bp deletion was observed in exon 2 of the S100a10 gene from positions 63 to 69 of the S100a10 open reading frame (ORF), which resulted in a frame shift and led to the degradation of mutant-transcribed S100a10 mRNA. Immunoblot analysis further confirmed a complete loss of S100A10 protein in the tissues, including the thymus, spleen, bone marrow, mesenteric lymph nodes, and peritoneal macrophages in S100a10−/− mice (Fig. S1d, e). S100a10−/− mice were born with the expected Mendelian ratio and showed no gross defects in growth and survival. The percentages and absolute numbers of myeloid cells, including macrophages, neutrophils, and dendritic cells in bone marrow, blood, and spleen, were comparable between wild-type (WT) and S100a10−/− mice (Fig. S2a, b). S100A10 deficiency did not affect the development of CD4+ and CD8+ T cells or natural regulatory T cells in the thymus, spleen, and mesenteric lymph nodes (Fig. S2c, d). In addition, the deficiency also did not affect the activation status of CD4+ and CD8+ T cells as observed by CD44 and CD62L staining (data not shown). BMDMs generated in vitro were also normal in S100a10−/− mice (Fig. S3a). Therefore, S100A10 deficiency did not affect the development or homeostasis of myeloid and lymphoid immune cell subsets. Furthermore, similar levels of other members of the S100 family were observed in BMDMs, peritoneal macrophages, and bone marrow-derived neutrophils in WT and S100a10−/− mice (Fig. S3), indicating that S100A10 deficiency did not affect the expression of these related genes.
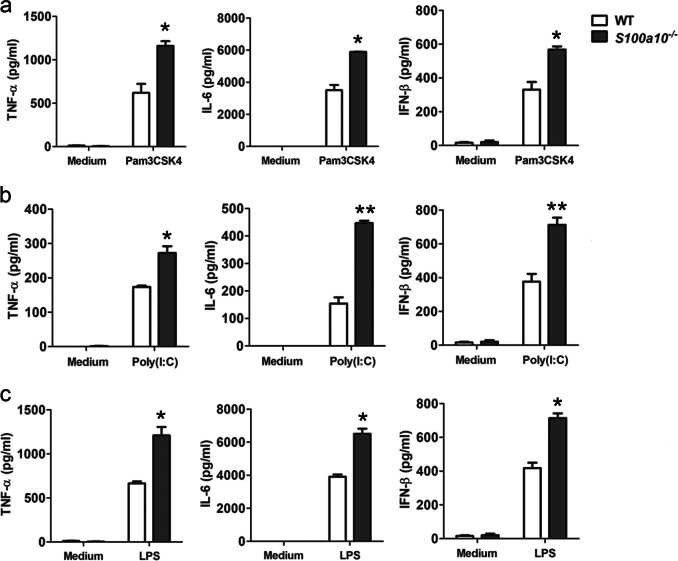
To determine the potential roles of S100A10 in innate immune responses to TLR ligands, we first studied S100A10 knockout cells in vitro. A variety of TLR agonists, including Pam3CSK4 (TLR2), poly(I:C) (TLR3), and LPS (TLR4), were used to stimulate naive BMDMs. Upon stimulation with these diverse agonists, S100a10−/− BMDMs produced significantly higher amounts of tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and interferon-β (IFN-β) compared with similarly treated WT controls (Fig. 2a–c). We also detected higher expressions of TNF-α, IL-6, IL-12, and IFN-β mRNA in S100a10−/− macrophages than in WT macrophages, in which TLR2, TLR3, and TLR4 were triggered (Fig. S4). Statistical analysis indicated significant differences between similarly treated WT and S100a10−/− cells. These findings show that S100A10 deficiency in macrophages enhances TLR-triggered production of proinflammatory cytokines and type I IFN, whereas it has no effect on macrophage development and differentiation.
Fig. 2.
Increased reactivity of S100A10-deficient macrophages to TLR stimulation. a–c Bone marrow-derived macrophages from WT and S100a10−/− mice (n = 3 per group) were treated with or without Pam3CSK4 (50 ng/ml), poly(I:C) (10 μg/ml), or LPS (100 ng/ml) for 4 h (TNF-α and IL-6) or 12 h (IFN-β). The concentrations of cytokines as indicated in supernatants were determined by ELISA. Data are representative of four experiments (mean ± SEM of three samples; *P < 0.05; **P < 0.01)
S100A10 deficiency enhances sensitivity to LPS and E. coli infection-induced septic inflammation
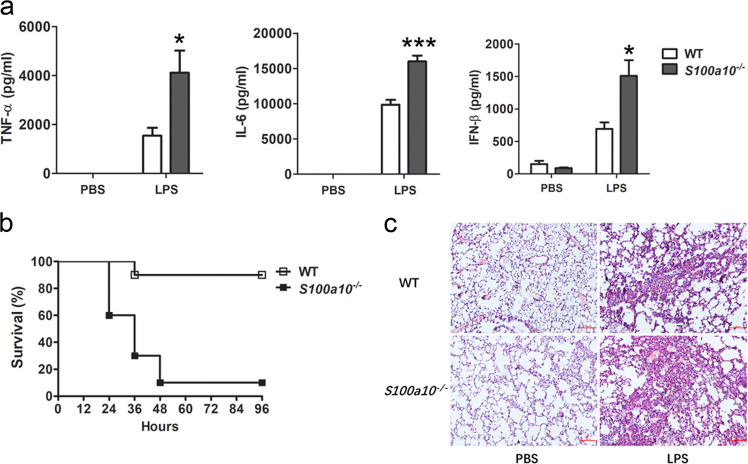
We next evaluated the physiological relevance of these findings. As described above, no apparent defect was detected for all myeloid and lymphoid immune cell types tested in S100a10−/− mice. Thus, the physiological difference(s) observed between WT and S100a10−/− mice should not be caused by changes in immune subpopulations. Because S100A10-deficient macrophages produced more TLR-induced cytokines than WT macrophages, we asked whether S100a10−/− mice were more susceptible to septic inflammation. To examine the function of S100A10 in the innate immune response in vivo, we challenged S100a10−/− mice with LPS or Gram-negative bacteria E. coli infection. We first studied LPS-induced sepsis in mice by injecting them intraperitoneally with a low dose of LPS. In response, S100a10−/− mice produced more TNF-α, IL-,6 and IFN-β in serum than WT control mice (Fig. 3a). Furthermore, after lethal challenge with LPS, we found that 70% of S100a10−/− mice died within 36 h and 90% of them died within 96 h, whereas 90% of WT mice survived the challenge (Fig. 3b). Consistent with that, we observed more severe infiltration of polymorphonuclear cells and interstitial pneumonitis in the lungs of S100a10−/− mice 6 h after LPS challenge (Fig. 3c).
Fig. 3.
S100a10−/− mice have more proinflammatory cytokine production when challenged with LPS and are more susceptible to endotoxic shock. a WT and S100a10−/− mice (n = 5 per group) were injected intraperitoneally with LPS (5 mg/kg). The concentrations of TNF-α, IL-6, and IFN-β in sera were measured by ELISA 6 h after LPS injection. b WT and S100a10−/− mice (n = 5 per group) were injected intraperitoneally with LPS (10 mg/kg). Survival was monitored until 96 h after the initiation of injection. c Hematoxylin and eosin staining of lungs from WT and S100a10-−/− mice 6 h after challenge with LPS. Original magnification, ×100. Data are representative of three experiments (mean ± SEM of five samples; *P < 0.05; ***P < 0.001)
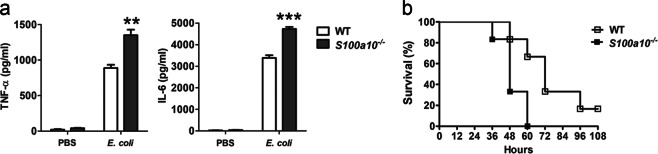
To further assess the role of S100A10 in host resistance to pathogen infection, we challenged WT and S100a10−/− mice intraperitoneally with intact Gram-negative E. coli, the most frequent cause of bacterial sepsis in humans. After infection with E. coli, the production of TNF-α and IL-6 in S100a10−/− mice was significantly greater than that of WT control mice (Fig. 4a). In addition, after lethal challenge with E. coli, 70% of mice succumbed to fatal shock within 48 h, and none survived at the end of the experiment. In contrast, 70% of WT mice were still alive 60 h after infection with E. coli, so S100a10−/− mice also displayed reduced survival (Fig. 4b). Taken together, these data suggest that S100a10−/− mice develop a more severe innate inflammatory response and are more susceptible to endotoxin-shock and E. coli-induced sepsis, which indicates that S100A10 may negatively regulate the TLR-triggered inflammatory response.
Fig. 4.
S100a10-−/− mice are more susceptible to E. coli infection. a WT and S100a10−/− mice (n = 5 per group) were injected intraperitoneally with E. coli (1 × 107 CFU). The concentrations of TNF-α and IL-6 in sera were measured by ELISA 4 h after injection. b WT and S100a10−/− mice (n = 5 per group) were injected intraperitoneally with E. coli (5 × 107 CFU). Survival was monitored until 108 h after the initiation of injection. Data are representative of three experiments (mean ± SEM of five samples; **P < 0.01; ***P < 0.001)
S100A10 is dispensable for macrophage phagocytosis
Activation of Rac1 and Cdc42, two important members of the Rho family of small GTPases, promotes actin polymerization, which is essential for macrophage phagocytosis. Previous studies demonstrated that S100A10 was involved in actin dynamics by promoting cell polarization, spreading, and migration via regulating Rac1 and Cdc42 activation.14,15 To determine whether S100A10 plays a role in macrophage phagocytosis, we performed experiments to examine the effect of S100A10 deficiency on scavenger receptor-mediated, FcγR receptor-mediated, and complement-mediated phagocytosis. In phagocytosis of fluorescein isothiocyanate (FITC)-labeled dextran, opsonized-latex beads, and opsonized zymosan, S100A10-deficient macrophages internalized particles comparably to WT control cells (Fig. S5a). We then examined the phagocytosis of FITC-labeled E. coli opsonized with normal mouse serum with S100A10-deficient macrophages and WT control macrophages. S100A10-deficient macrophages phagocytosed E. coli at a level that was similar to that of WT control cells (Fig. S5a). These results indicate that S100A10 does not play a role in phagocytosis in macrophages.
Enhanced TLR signaling in S100A10-deficient macrophages
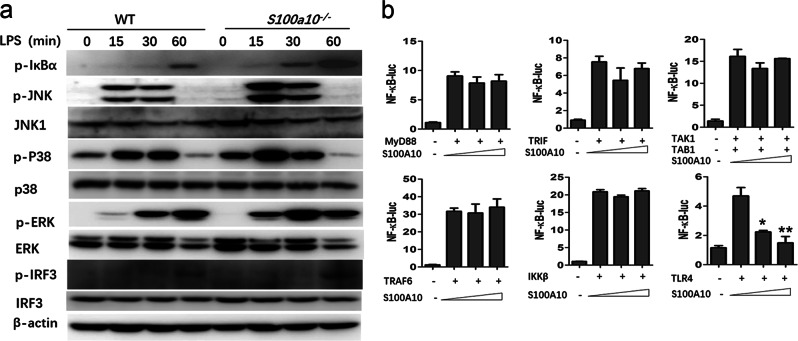
To investigate whether S100A10 deficiency intersected with the TLR signaling pathways in macrophages, we examined the activation kinetics of the mitogen-activated protein kinases (MAPKs) and transcription factor NF-κB pathways, which are downstream of TLR signaling. We observed enhanced phosphorylation of the kinases JNK, p38, and ERK and inhibitor IκBα in LPS-stimulated S100A10-deficient macrophages (Fig. 5a). We also observed more phosphorylation of the transcription factor IRF3 (Fig. 5a). These data suggest that S100A10 deficiency enhances TLR signaling in macrophages.
Fig. 5.
S100A10 deficiency increases the MyD88- and TRIF-dependent activations of the NF-κB, MAPKs, and IRF3 pathways in macrophages. a Bone marrow-derived macrophages from WT and S100a10-−/− mice (n = 3 per group) were treated with or without LPS (100 ng/ml) for the indicated times. The phosphorylated (p-) and total NF-κB, MAPKs, and IRF3 signaling proteins in whole-cell lysates were determined by Western blot. b HEK293T cells were transiently transfected with MyD88, TRIF, TAK1 plus TAB1, TRAF6, IKKβ, or TLR4, together with NF-κB-luc reporter plasmid and increasing amounts of S100A10 plasmid, and luciferase activities were analyzed. Data are representative of three experiments (mean ± SEM of three samples in b; *P < 0.05, **P < 0.01). Similar results were obtained in three independent experiments in a
We next sought to determine at which level S100A10 inhibits TLR-induced innate immune signaling. TLR ligands ultimately induce MAPK and NF-κB activation via a complex cascade composed of multiple kinases and adaptors. To elucidate the molecular order and molecular target(s) of S100A10 in these two major signaling pathways, we examined the effects of S100A10 overexpression on NF-κB, AP-1, and IFN-β promoter activation mediated by various molecules by performing luciferase assays. We cotransfected HEK293T cells with MyD88, TRIF, TAK1/TAB1, TRAF6, IKKβ, or TLR4 to activate NF-κB-luc or AP-1-luc reporter genes. We found that the activation of NF-κB by MyD88, TRIF, TAK1/TAB1, TRAF6, and IKKβ and the activation of AP-1 by MyD88, TRIF, TAK1/TAB1, and TRAF6 were not inhibited by S100A10 (Fig. 5b and Fig. S5b). In contrast, S100A10 inhibited TLR4-mediated NF-κB and AP-1 activation in a dose-dependent manner (Fig. 5b and Fig. S5b). In addition, TRIF, TRAF3, and TBK1-induced IFN-β promoter activation was also not inhibited by S100A10 (Fig. S5c). Therefore, these data indicate that S100A10 may target proteins upstream of the common adaptors MyD88 and TRIF to inhibit signal transduction.
S100A10 competes with MyD88 or TRIF to interact with the TIR domain of TLR4
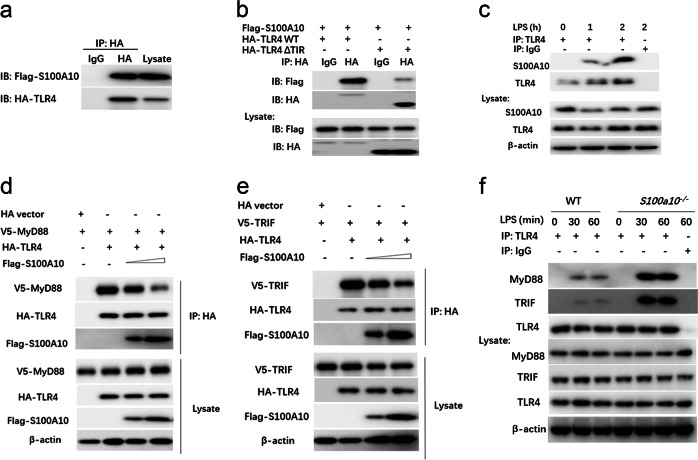
As our aforementioned results showed that S100A10 plays a general inhibitory role in LPS-mediated signaling, including two major signaling pathways, that is, the NF-κB and the MAPK pathways, we next determined whether S100A10 functioned at the receptor level. A previous study showed that S100A10 could increase the localization of the 5-hydroxytryptamine receptor at the cell surface.8 To understand the mechanisms by which S100A10 regulates TLR4 signaling, we first examined whether S100A10 could modulate the expression of TLR4, which is the functional receptor for LPS. We found no detectable difference in TLR4 surface expression levels between WT and S100A10-deficient macrophages at steady state (Fig. S5d). The downregulation of surface TLR4 in response to LPS challenge was also similar in WT and S100A10-deficient macrophages, which excluded the possibility that the enhancement of the TLR4 response achieved by an S100A10 deficiency was due simply to the higher expression of TLR4. Some S100 family proteins, including S100A8, S100A9, S100A4, and S100A12, are linked directly to the innate immune system and have been characterized as endogenous TLR4 ligands.16 Given that these family members exhibit a high degree of structural similarity, we next sought to determine whether S100A10 interacted with TLR4 and served as a cofactor in inhibiting TLR signaling by forming a complex. HA-TLR4 and Flag-S100A10 were cotransfected into HEK293T cells, and 24 h later after transfection, co-IP experiments were performed with HA antibody (Ab). We found that Flag-S100A10 was coprecipitated with HA-TLR4 (Fig. 6a). It is well known that S100A8 or S100A9, which is secreted by phagocytes, exerts both extracellular regulatory effects and a broad range of intracellular functions. However, in addition to having an intracellular distribution, S100A10 is present on the cellular surface of many cell types,17 suggesting that it is unlikely that S100A10 binds to the extracellular domain of TLR4. We assume that S100A10 may bind to the intracellular domain of TLR4. To test this possibility, we constructed a deletion mutant of the intracellular domain of TLR4 and transfected HEK293T cells with Flag-S100A10 together with HA-tagged constructs of full-length TLR4 or truncated TLR4 lacking the Toll/interleukin-1 receptor (TIR) homology domain. Flag-S100A10 was immunoprecipitated with both full-length and truncated TLR4, although much less Flag-S100A10 was associated with truncated TLR4 (Fig. 6b). We further extended our studies to investigate the interaction of endogenous S100A10 with TLR4 in macrophages. We detected strong LPS-dependent interactions between endogenous S100A10 and TLR4, demonstrating the recruitment of S100A10 to TLR4 in an LPS signal-dependent manner (Fig. 6c). Thus, these data suggest that S100A10 may function at the receptor level in regulating LPS-induced signaling.
Fig. 6.
S100A10 interacts with TLR4 and affects its association with adaptor proteins. a HEK293T cells were transiently transfected with expression plasmids for Flag-tagged S100A10 and HA-tagged TLR4 as indicated. Twenty-four hours later, cell lysates were prepared and immunoprecipitated (IP) with anti-HA or control IgG. The precipitates and whole-cell lysates were subjected to immunoblot (IB) with antibodies for the indicated antigens. b HEK293T cells were transiently transfected with plasmids expressing Flag-tagged S100A10 and either HA-tagged full-length TLR4 (HA-TLR4 WT) or HA-tagged truncated TLR4 lacking TIR (HA-TLR4-ΔTIR); IP with anti-HA or control IgG was followed by IB with anti-Flag and anti-HA antibodies. c Bone marrow-derived macrophages from WT (n = 3) mice were stimulated with LPS for the indicated time periods. Cell lysates were then subjected to IP with anti-TLR4 or control IgG, followed by IB analysis with anti-S100A10, anti-TLR4, and anti-β-actin antibodies. d, e HEK293T cells were transiently transfected with plasmids expressing HA-tagged TLR4 or HA-tagged empty vector, V5-tagged MyD88 (d) or V5-tagged TRIF (e) together with increasing amounts of Flag-tagged S100A10; IP with anti-HA was followed by IB with anti-Flag, anti-V5, anti-HA, and anti-β-actin antibodies. f Bone marrow-derived macrophages from WT and S100a10−/− mice (n = 3 per group) were treated with or without LPS (100 ng/ml) for the indicated times. Cell lysates were then subjected to IP with anti-TLR4 or control IgG, followed by IB analysis with anti-TLR4, anti-MyD88, anti-TRIF, and anti-β-actin antibodies. Similar results were obtained in three independent experiments
All TLR family members signal via a conserved TIR domain in the cytosolic region, which recruits TIR domain-containing adaptor molecules, including MyD88 and TRIF, and activates NF-κB and MAPK common signaling pathways. Considering that the interaction of S100A10 with TLR4 was promoted by LPS treatment, we next explored whether S100A10 inhibited LPS-mediated signaling by interfering with the association between TLR4 and adaptors. HEK293T cells were cotransfected with HA-TLR4, V5-MyD88, or V5-TRIF and increasing doses of Flag-S100A10 plasmids. Twenty-four hours after transfection, coimmunoprecipitation experiments were performed with HA antibody. As shown in Fig. 6d, e, coimmunoprecipitation analysis confirmed the interaction between ectopically expressed S100A10 and TLR4, and S100A10 dose-dependently inhibited the association between TLR4 and MyD88 or TRIF. Previous studies demonstrated that LPS treatment increased the interaction not only between TLR4 and MyD88 but also between TLR4 and TRIF in macrophages.18 Next, we performed coimmunoprecipitation assays in macrophages with an endogenous TLR4 antibody. As shown in Fig. 6f, LPS stimulation induced an interaction between TLR4 and MyD88 or TRIF, and these interactions were markedly enhanced in S100A10-deficient macrophages. Taken together, these data suggest that S100A10 binds to the TIR domain of TLR4 and interferes with the association of TLR4 with MyD88 or TRIF, thereby inhibiting TLR4 activation and downstream signaling.
Discussion
Macrophages are centrally involved in the promotion of TLR-triggered innate immune responses and in the pathogenesis of sepsis. The results reported here indicate that S100A10 functions as an inhibitory modulator of TLR-mediated macrophage activation. S100A10 deficiency in macrophages enhances the TLR-mediated secretion of proinflammatory cytokines and type I interferon, as well as signaling transduction. Accordingly, S100A10-deficient mice are more susceptible to LPS-induced septic shock and E. coli challenge compared to WT mice. S100A10 is constitutively expressed at a high level in macrophages and is distributed in the cytoplasm at steady state. The recognition of PAMPs by TLRs leads to the latter recruiting the key adaptors MyD88 or TRIF to initiate downstream signaling transduction while at the same time inducing S100A10 to competitively interact with the cytoplasmic TIR domain of TLRs to inhibit the interaction of TLRs with MyD88 or TRIF, which may provide an effective way to avoid overactivation. After full activation, proteasomal degradation activated by TLR signaling can degrade ubiquitinated S100A10 protein in the cells. Thus, S100A10 serves as a negative regulator of TLR-triggered innate inflammatory responses.
S100A10 negatively regulates TLR signaling pathways, as revealed by the inhibition of cytokine production and the promotion of the inactivation of transcription factors (Figs. 3 and 5). Moreover, the effect of S100A10 is exerted at the initial signaling events, that is, TLR4-MyD88 or TLR4-TRIF complex formation (Fig. 6). Emerging evidence indicates that several molecules act as negative regulators in multiple TLR signaling pathways and exert their effects at the level of receptors or adaptors.19,20 For example, IRF4 is induced by TLR activation and competes with IRF5 for binding to MyD88, resulting in the shutdown of the expression of IRF5-dependent genes.21,22 TRAM adaptor with GOLD domain (TAG), a variant of TRAM, competes with TRAM for TRIF binding and inhibits the TRIF-dependent pathway.23 ST2 negatively regulates TLR2 and TLR4 signaling but not TLR3 signaling, which is not completely similar to the effects of S100A10.24 Although ST2 inhibits TLR4 signaling by sequestration of the adaptor MyD88, but not of TRIF,24 S100A10 suppresses the LPS-induced interaction between TLR4 and MyD88 and between TLR4 and TRIF. Thus, the mechanisms of negative regulation by different regulators of the TLR signaling pathways are dependent not only on the type of adaptor protein as well as transcription factors but also on the specificity of TLRs.
S100A10 exerts its negative regulatory effect on TLR signaling pathways through its direct effect on immediate signaling events, including signal-triggered NF-κB, MAPKs, and IRF3 activation. Nevertheless, the details of the mechanism by which S100A10 exerts its effect on signal transduction are not yet clear. Given that S100A10 is a dimeric protein composed of only two 11-kDa subunits, this protein does not have the classical TIR domain, which often mediates the interaction with TIR domain-containing target receptors. However, LPS treatment promotes the association of S100A10 with TLR4 and may also lead to the formation of a complex between S100A10 and MyD88 or TRIF. We propose that S100A10 may negatively regulate TLR pathways via its interaction with the TLR complex. Upon LPS treatment, receptor-proximal signaling components, including MyD88 and TRIF, are recruited to TLR4 to form a receptor complex. After their appropriate activation at the receptor complex, these key signaling adaptors are released from the receptor to interact and activate downstream signaling molecules. S100A10 may negatively regulate the TLR pathways by interfering with the appropriate recruitment and activation of the receptor-proximal signaling components (e.g., MyD88 and TRIF) or by promoting the dissociation of the activated signaling components from the receptor complex.
The S100 proteins, belonging to a calcium-binding cytosolic protein family, have a broad range of intracellular and extracellular functions. At least four S100 proteins are specifically linked to innate immune functions by their expression in cells of myeloid origin via interactions with different receptor systems. In particular, S100A8 and S100A9 are endogenous ligands of TLR4 and are important in the pathogenesis of sepsis upstream of TNF-α action.25 S100A12 has been shown to exhibit its proinflammatory activities via interaction with the multiligand receptors RAGE and TLR4, acting as an amplifier of innate immunity during early inflammation and the development of sepsis.26,27 Furthermore, S100A4 has been identified as a marker of a specific subset of inflammatory macrophages in liver injury and fibrosis,28 with its effect likely mediated by upregulation of c-Myb.29 Recently, S100A11 was identified as the alarmin protein, which induces a potent chemokine response to Toxoplasma gondii by engaging RAGE and regulates monocyte recruitment.30 However, unlike these members, S100A10 is Ca2+ insensitive due to substitutions in both EF-hand regions, so its binding to cellular target proteins is Ca2+-independent. Many reports have shown that S100A10-annexin A2 heterotetramers positively regulate the expression of a number of cell surface receptors and ion channels.6 However, inconsistent with previous studies, we found no correlation between S100A10 and cell surface expression levels of TLR4 before and after stimulation, raising the possibility that the regulatory role of S100A10 on TLR-triggered responses is S100A10-annexin A2 heterotetramer independent. Indeed, Swisher et al.31 found that extracellular annexin A2 could modulate macrophage activation through TLR4. However, annexin A2 has different or additional requirements for TLR4 signaling compared with the classical ligand LPS, indicating that annexin A2 may provide other ways for the discrete regulation of TLR responses.31 A more recent study demonstrated that S100A10 interacts physically with the cytosolic tail of chemokine receptor CCR10.7 Notably, a similar interaction was observed between S100A10 and the cytosolic TIR domain of TLR4, but the binding site located within the TIR domain needs to be further investigated. Given that the effect of S100A10 on the TLR-mediated response may be annexin A2 independent, S100A10 may bear distinct binding sites for TLR4 and annexin A2, and the site that is most likely covered by annexin A2 may not be required for TLR4 binding. In addition, future studies also need to confirm whether cAMP/protein kinase A is involved in these processes.
The first characterization of S100A10 function in macrophages was reported by O’Connell et al.12 They used a thioglycolate (TG)-induced peritonitis model to elucidate the potential role of S100A10 in the regulation of peritonitis-dependent macrophage migration and found that S100a10−/− mice displayed severely compromised macrophage recruitment in response to inflammatory stress.12 The authors concluded that S100A10 is required for macrophage invasion into the peritoneal cavity in this noninfectious inflammation model (i.e., TG-induced peritonitis).12 We confirmed this phenotype by using the recently generated S100A10-deficient mice in our laboratory with the CRISPR-Cas9 targeting strategy (data not shown). As previously reported, TG broth, a kind of inflammogen that induces inflammation, causes a milder and longer-lasting accumulation of macrophages.32 However, the inflammation induced by thioglycolate is confined primarily to the peritoneal cavity. In the present study, we sought to determine the contribution of S100A10 to the inflammatory responses induced by the bacterial product LPS or intact bacteria E. coli. Intraperitoneal administration of LPS or E. coli induces not only local inflammation in the peritoneal cavity but also significant systemic inflammatory effects, which well mimics inflammatory responses to clinical infection processes. Our data argue against the established inflammatory role of S100A10 during the inflammatory response.33 Indeed, our study supports the notion that S100A10 serves as an anti-inflammatory protein. On the basis of our findings, combined with the previous report that S100A10 is required for macrophage recruitment in response to inflammatory stimuli, we propose a regulatory model that is carried out by S100A10 during the inflammatory responses: S100A10 positively regulates macrophage migration via an interaction with annexin A2 and negatively regulates macrophage immune responses via an interaction with the TLR signaling complex. The S100A10-based regulatory outcome guarantees effective recruitment of inflammatory cells to the site of an infection as well as an adequate host defense against invading microorganisms. However, we do not have evidence that these proteins are in a single complex. Therefore, by integrating different signaling pathways that play related roles, S100A10 may more effectively control complex cellular processes such as migration and survival than by targeting only a single pathway.
Taken together, we have demonstrated for the first time that S100A10 negatively regulates TLR signaling pathways likely by interfering with the appropriate recruitment and activation of the receptor-proximal signaling components, resulting in suppression of downstream signaling and cytokine production. Although further studies are needed to elucidate the precise mechanisms of S100A10 action, our results reveal that S100A10 is a novel candidate for a negative regulator of the TLR signaling pathways. This finding advances our understanding of the mechanisms of inflammatory diseases, such as sepsis, a leading cause of death worldwide that has little effective treatment, and it may lead to the development of S100A10-based strategies for treating inflammatory diseases.
Methods
Mice
S100a10−/− mice on a C57BL/6J background were generated with CRISPR/Cas9-mediated genome editing by Cyagen Biosciences Inc. (Guangzhou, China). S100a10−/− mice bearing a 7-bp deletion in exon 2, resulting in the degradation of truncated S100a10 mRNA, were identified with primers flanking the break sites. The sequences of the primers were 5′- CCCCCAAACTGAGCCTTAGA-3′ (forward) and 5′-GAACTCCCGTTCCATGAGCA-3′ (reverse). The sequence-validated F0 generation mice mated with WT C57BL/6J mice to produce positive F1 generation heterozygous mice. Heterozygous mice were then bred to obtain viable homozygous mice. All mice used were age- and sex matched and 8–12 weeks old and were maintained under pathogen-free conditions in the Xinxiang Medical University Animal Care Facilities. All animal procedures were preapproved by the Institutional Animal Care and Use Committee of Xinxiang Medical University.
Reagents and antibodies
LPS (E. coli O111:B4) and poly(I:C) were purchased from Sigma-Aldrich. Pam3CSK4 was purchased from InvivoGen. MG132 was obtained from Merck-Calbiochem. Escherichia coli O111:B4 was obtained from the China Center for Type Culture Collection. Anti-p-IκBα (9246), anti-p-p38 (9215), anti-p-JNK (9251), anti-p-ERK (4377), and anti-p-IRF3 (Ser 396, 29047) were purchased from Cell Signaling Technology. Anti-p38 (A11340), anti-JNK1 (A0288), anti-TLR4 (A5258), anti-MyD88 (A0980), anti-TRIF (A1155), and anti-ERK (A10613) antibodies were purchased from Abclonal Technology. Anti-S100A10 (11250-1-AP), anti-IRF3 (11312-1-AP), and anti-β-actin (66009-1-Ig) were purchased from Proteintech. Anti-Ub (sc-8017) and protein A/G agarose (sc-2003) used for immunoprecipitation were purchased from Santa Cruz Biotechnology. Anti-Flag (F3165) was purchased from Sigma. Anti-HA (901513, previously Covance catalog# MMS-101R) and anti-V5 (680601) were obtained from BioLegend. Fluorochrome-conjugated anti-CD4 (GK1.5), anti-CD8 (53-6.7), anti-B220 (RA3-6B2), anti-Ly-6G (1A8), anti-CD11b (M1/70), anti-CD11c (N418), anti-TLR4 (SA15-21), anti-F4/80 (BM8), anti-CD44 (IM7), and anti-CD62L (MEL-14) antibodies were purchased from BioLegend. Fluorochrome-conjugated anti-CD25 (PC61.5) and Foxp3 (FJK-16s) antibodies were purchased from eBioscience.
Endotoxin-shock model and E. coli infection
For the endotoxin-shock mouse model, mice were injected intraperitoneally with LPS from E. coli 055:B5 LPS (L2880, Sigma) at 10 mg/kg, as described previously.34 For E. coli infection, serotype O111:B4 in mid-logarithmic growth was collected, counted on agar plates, and then resuspended in sterile phosphate-buffered saline (PBS). Mice were injected intraperitoneally with 200 μl bacterial suspension (5 × 107 colony-forming units) and were monitored for survival rates. The concentrations of TNF-α, IL-6, and IFN-β in sera were measured by enzyme-linked immunosorbent assay (ELISA). Lungs were collected for hematoxylin and eosin (HE) staining in mice given an intraperitoneal injection of LPS (5 mg/kg) or E. coli (1 × 107 colony-forming units).
Plasmid constructs
Human S100A10 ORF (NM_002966) was obtained from Vigene Biosciences and subcloned into a pcDNA3.1 eukaryotic expression vector (Invitrogen) with a Flag tag at the C terminus. Recombinant vectors encoding TRAF6 and TRAF3 were constructed by PCR-based amplification of complementary DNA (cDNA) from THP-1 cells and then cloned into a pcDNA3.0 eukaryotic expression vector (Invitrogen) with an HA tag at the C terminus. The deletion mutant of HA-tagged TLR4 was generated by PCR. The primers used for amplification of the sequence encoding TLR4-ΔTIR were 5′-CTGGATGGTAAATCATGGAATCCAGAAGGA-3′ and 5′-GATGTTTTCACCTCTACCATACTTTATGCA-3′. All constructs were confirmed by DNA sequencing. TLR4, MyD88, TRIF, TBK1, and IKKβ expression plasmids and IFN-β reporter plasmids were gifts from Dr. Peihui Wang (Shandong University, Shandong, China). TAK1 and TAB1 expression plasmids and NF-κB reporter plasmids were provided by Dr. Wei Zhao (Shandong University, Shandong, China). The AP-1 reporter plasmid was purchased from Beyotime Biotechnology.
Cell culture and transfection
BMDMs were generated as described previously.35 The purity of the BMDMs was >95%, as determined by CD11b+ and F4/80+ flow cytometry staining. Bone marrow-derived mature neutrophils were purified by Percoll gradient centrifugation as described previously.35 The purity of the neutrophils was ~85%, as assessed by CD11b+ and Ly-6G+ flow cytometry staining. Thioglycolate-elicited peritoneal macrophages were prepared as described previously.36 The HEK293T cell line was obtained from American Type Culture Collection. Primary macrophages and HEK293T cells were maintained in complete Dulbecco’s modified Eagle’s medium supplemented with 10% (vol/vol), fetal bovine serum (FBS) (Gibco), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Gibco). HEK293T cells were transiently transfected with plasmids using PEI reagent according to the manufacturer’s instructions (Polysciences).
ELISA and real-time PCR
The concentrations of TNF-α and IL-6 in sera or in cell culture supernatants were determined with ELISA kits (eBioscience). The concentration of IFN-β in sera or in cell culture supernatants was measured with an ELISA kit (BioLegend) according to the manufacturer’s instructions. Total RNA was extracted with TRIzol reagent (Invitrogen), as described previously.37 Briefly, 500 ng of total RNA were reverse transcribed using RT Master Mix (Takara). Real-time quantitative PCR was carried out in an Applied Biosystems 7500 System with TB Green Premix Ex Taq II (Takara). The relative changes in gene expression were analyzed by the 2−ΔΔct method, and a melting-curve analysis was performed to ensure the specificity of the products. Each sample was run in triplicate. The relative changes in gene expression were calculated using β-actin as the loading control. The specific primers used are shown in Supplemental Table 1 and were synthesized by GENEWIZ.
Immunoprecipitation and Western blot analysis
Cells were lysed with cell lysis buffer containing 1% (vol/vol) Nonidet P-40, 50 mM Tris-HCl (pH 7.6), 50 mM EDTA, 150 mM NaCl, and protease inhibitor cocktail tablets (Roche). The lysates were cleared by centrifugation for 15 min at 14,000 × g and incubated with 1 μg monoclonal anti-Flag or 1 μg monoclonal anti-HA together with protein A/G Plus-agarose Immunoprecipitation reagent (Santa Cruz) at 4 °C overnight. After incubation, the beads were washed three times with lysis buffer and boiled for 10 min in 30 μl 2× sodium dodecyl sulfate (SDS) sample buffer. For endogenous immunoprecipitation experiments, the cell lysates were incubated with the indicated antibodies, as described above. For Western blot analysis, immunoprecipitates or whole-cell lysates were loaded and subjected to SDS-polyacrylamide gel electrophoresis, transferred onto polyvinylidene difluoride membranes (Millipore), and then blotted by an Amersham Imager 600RGB detection system (GE Healthcare), as described previously.36,38
Flow cytometry
Immune organs including bone marrow, thymus, spleen, and lymph nodes or BMDMs were collected to obtain single-cell suspensions following standard procedures, as described previously.39 Cells were stained with fluorochrome-conjugated antibodies for CD4, CD8, B220, Ly-6G, CD11b, CD11c, TLR4, F4/80, CD25, CD44, Foxp3, and CD62L in 2% FBS-PBS at 4 °C for 20 min and washed with PBS. Intracellular staining for Foxp3 was performed using a Foxp3 Staining Buffer Set (eBioscience). For the gating strategy, forward scatter/side scatter was initially applied to the gate for live cells and then used for the antibodies with specific fluorochromes to make the subsequent gates. Data were acquired by flow cytometry using a guava easyCyte™ HT System (Millipore) and analyzed with guavaSoft™ software (Millipore).
Luciferase assay
HEK293T cells were seeded into 24-well plates and transfected with 200 ng NF-κB, AP-1, or IFN-β luciferase reporter plasmids, 20 ng pRL-TK Renilla luciferase internal control plasmids together with increasing amounts of S100A10 (0, 200, and 400 ng) expression plasmids, and indicated adaptor plasmids using PEI transfection reagent. Total amounts of DNA were maintained constantly by supplementing with a pcDNA3.1 empty vector. Twenty-four hours after transfection, cells were collected, and luciferase activities were measured with a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Data were acquired by GloMax EXPLORER (Promega) and were normalized for transfection efficiency by dividing firefly luciferase activity with that of Renilla luciferase.
Phagocytosis assays
For flow cytometry-based measurement of phagocytosis, a total of one million WT or S100a10−/− BMDMs were seeded in 12-well plates. FcγR-mediated phagocytosis was performed using fresh mouse serum-opsonized FITC-labeled latex beads (2 μm, Sigma) at a macrophage/target ratio of 1:10. Complement-mediated phagocytosis was performed using fresh mouse serum-opsonized FITC-labeled zymosan particles (Molecular Probes) at a macrophage/target ratio of 1:20. Samples were incubated for 15 min at 4 °C for binding, followed by 30 min at 37 °C or 4 °C for phagocytosis. To analyze the phagocytosis of bacteria, FITC-labeled E. coli (Molecular Probes) was opsonized with normal mouse serum (Invitrogen), mixed with BMDMs at a macrophage/target ratio of 1:30, and incubated at 37°C or 4°C for 30 min. After incubation, plates were rapidly washed twice with ice-cold PBS. The fluorescence of extracellular particles was quenched by replacement of the medium with 0.2% Trypan blue (Sigma) in PBS, and cells were collected with 5 mM EDTA-PBS. Cells were then fixed with 2% paraformaldehyde in PBS, stained with Percpcy5.5-conjugated anti-CD11b, and analyzed by flow cytometry. Phagocytosis of dextran was performed as described previously.16,35 Briefly, FITC-dextran (Sigma) was added to the cell suspension at a final concentration of 1 mg/ml. The cells were incubated for 30 min at 37 °C or 4°C, washed with ice-cold PBS, and analyzed by flow cytometry.
Statistical analysis
All quantitative data are presented as the means ± SEM of two or three experiments. The survival curves were plotted according to the Kaplan–Meier method and compared by the log-rank test. A two-tailed Student’s t test was used for all other cases, with a p value <0.05 considered statistically significant. All statistical analyses were performed with Prism 5.0 for Windows (GraphPad Software).
Supplementary information
Acknowledgements
This work was funded by the National Natural Science Foundation of China (Grant No. 81871309) and the Program for Ph.D. Starting Research Funding from Xinxiang Medical University (Grant No. 505248). We thank Drs. Bo Yang, Yun Zhang, Qianqian Zheng, Liangwei Duan, Chunlei Guo, Jie Wang, Zhiguo Niu, and Hui Liu for reagents and/or valuable advice. We are also grateful to Drs. Wei Zhao (Shandong University) and Peihui Wang (Shandong University) for providing plasmids and to Dr. Youhai Chen (University of Pennsylvania) for critical reading and editing of the manuscript.
Author contributions
H.W. and Y.L. conceived the study and wrote the manuscript; Y.L. and M.H. performed most of the experiments and analyzed the data. H.L. performed some of the in vivo experiments. Y.N. performed the histopathology study on the lung samples. J.G. and Y.L. generated and supplied S100a10−/− mice. W.Z. provided expertise and advice. H.W. oversaw the project.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yunwei Lou, Meijuan Han
Supplementary information
The online version of this article (10.1038/s41423-019-0278-1) contains supplementary material.
References
- 1.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 2.Marshak-Rothstein A, Rifkin IR. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu. Rev. Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- 3.Liew FY, Xu D, Brint EK, O’Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 4.Qian C, Cao X. Regulation of Toll-like receptor signaling pathways in innate immune responses. Ann. NY Acad. Sci. 2013;1283:67–74. doi: 10.1111/j.1749-6632.2012.06786.x. [DOI] [PubMed] [Google Scholar]
- 5.Gerke V, Weber K. The regulatory chain in the p36-kd substrate complex of viral tyrosine-specific protein kinases is related in sequence to the S-100 protein of glial cells. EMBO J. 1985;4:2917–2920. doi: 10.1002/j.1460-2075.1985.tb04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donato R, et al. Functions of S100 proteins. Curr. Mol. Med. 2013;13:24–57. doi: 10.2174/156652413804486214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hessner F, et al. CC chemokine receptor 10 cell surface presentation in melanocytes is regulated by the novel interaction partner S100A10. Sci. Rep. 2016;6:22649. doi: 10.1038/srep22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svenningsson P, et al. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311:77–80. doi: 10.1126/science.1117571. [DOI] [PubMed] [Google Scholar]
- 9.Camilleri M, et al. Alterations in expression of p11 and SERT in mucosal biopsy specimens of patients with irritable bowel syndrome. Gastroenterology. 2007;132:17–25. doi: 10.1053/j.gastro.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green H, et al. Alterations of p11 in brain tissue and peripheral blood leukocytes in Parkinson’s disease. Proc. Natl. Acad. Sci. USA. 2017;114:2735–2740. doi: 10.1073/pnas.1621218114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo J, et al. Probable involvement of p11 with interferon alpha induced depression. Sci. Rep. 2016;6:17029. doi: 10.1038/srep17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Connell PA, Surette AP, Liwski RS, Svenningsson P, Waisman DM. S100A10 regulates plasminogen-dependent macrophage invasion. Blood. 2010;116:1136–1146. doi: 10.1182/blood-2010-01-264754. [DOI] [PubMed] [Google Scholar]
- 13.He KL, et al. Endothelial cell annexin A2 regulates polyubiquitination and degradation of its binding partner S100A10/p11. J. Biol. Chem. 2008;283:19192–19200. doi: 10.1074/jbc.M800100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rescher U, Gerke V. S100A10/p11: family, friends and functions. Pflug. Arch. 2008;455:575–582. doi: 10.1007/s00424-007-0313-4. [DOI] [PubMed] [Google Scholar]
- 15.Sayeed S, et al. S100A10 is required for the organization of actin stress fibers and promotion of cell spreading. Mol. Cell. Biochem. 2013;374:105–111. doi: 10.1007/s11010-012-1509-2. [DOI] [PubMed] [Google Scholar]
- 16.Holzinger D, Tenbrock K, Roth J. Alarmins of the S100-family in juvenile autoimmune and auto-inflammatory diseases. Front. Immunol. 2019;10:182. doi: 10.3389/fimmu.2019.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Fogg DK, Waisman DM. RNA interference-mediated silencing of the S100A10 gene attenuates plasmin generation and invasiveness of Colo 222 colorectal cancer cells. J. Biol. Chem. 2004;279:2053–2062. doi: 10.1074/jbc.M310357200. [DOI] [PubMed] [Google Scholar]
- 18.Nakahira K, et al. Carbon monoxide differentially inhibits TLR signaling pathways by regulating ROS-induced trafficking of TLRs to lipid rafts. J. Exp. Med. 2006;203:2377–2389. doi: 10.1084/jem.20060845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 2012;33:449–458. doi: 10.1016/j.it.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Brown J, Wang H, Hajishengallis GN, Martin M. TLR-signaling networks: an integration of adaptor molecules, kinases, and cross-talk. J. Dent. Res. 2011;90:417–427. doi: 10.1177/0022034510381264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takaoka A, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 22.Negishi H, et al. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc. Natl. Acad. Sci. USA. 2005;102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palsson-McDermott EM, et al. TAG, a splice variant of the adaptor TRAM, negatively regulates the adaptor MyD88-independent TLR4 pathway. Nat. Immunol. 2009;10:579–586. doi: 10.1038/ni.1727. [DOI] [PubMed] [Google Scholar]
- 24.Brint EK, et al. ST2 is an inhibitor of interleukin 1 receptor and Toll-like receptor 4 signaling and maintains endotoxin tolerance. Nat. Immunol. 2004;5:373–379. doi: 10.1038/ni1050. [DOI] [PubMed] [Google Scholar]
- 25.Vogl T, et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat. Med. 2007;13:1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann MA, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/S0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 27.Foell D, et al. Proinflammatory S100A12 can activate human monocytes via Toll-like receptor 4. Am. J. Respir. Crit. Care Med. 2013;187:1324–1334. doi: 10.1164/rccm.201209-1602OC. [DOI] [PubMed] [Google Scholar]
- 28.Österreicher CH, et al. Fibroblast-specific protein 1 identifies an inflammatory subpopulation of macrophages in the liver. Proc. Natl. Acad. Sci. USA. 2011;108:308–313. doi: 10.1073/pnas.1017547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, et al. S100A4 promotes liver fibrosis via activation of hepatic stellate cells. J. Hepatol. 2015;62:156–164. doi: 10.1016/j.jhep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 30.Safronova A, et al. Alarmin S100A11 initiates a chemokine response to the human pathogen Toxoplasma gondii. Nat. Immunol. 2019;20:64–72. doi: 10.1038/s41590-018-0250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swisher JF, Burton N, Bacot SM, Vogel SN, Feldman GM. Annexin A2 tetramer activates human and murine macrophages through TLR4. Blood. 2010;115:549–558. doi: 10.1182/blood-2009-06-226944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ajuebor MN, et al. Role of resident peritoneal macrophages and mast cells in chemokine production and neutrophil migration in acute inflammation: evidence for an inhibitory loop involving endogenous IL-10. J. Immunol. 1999;162:1685–1691. [PubMed] [Google Scholar]
- 33.Miles LA, Parmer RJ. S100A10: a complex inflammatory role. Blood. 2010;116:1022–1024. doi: 10.1182/blood-2010-05-284083. [DOI] [PubMed] [Google Scholar]
- 34.Xu S, et al. Constitutive MHC class I molecules negatively regulate TLR-triggered inflammatory responses via the Fps-SHP-2 pathway. Nat. Immunol. 2012;13:551–559. doi: 10.1038/ni.2283. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, et al. TIPE2 protein serves as a negative regulator of phagocytosis and oxidative burst during infection. Proc. Natl. Acad. Sci. USA. 2012;109:15413–15418. doi: 10.1073/pnas.1204525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou Y, et al. Enhanced atherosclerosis in TIPE2-deficient mice is associated with increased macrophage responses to oxidized low-density lipoprotein. J. Immunol. 2013;191:4849–4857. doi: 10.4049/jimmunol.1300053. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, et al. Ku70 senses HTLV-1 DNA and modulates HTLV-1 replication. J. Immunol. 2017;199:2475–2482. doi: 10.4049/jimmunol.1700111. [DOI] [PubMed] [Google Scholar]
- 38.Huai W, et al. Aryl hydrocarbon receptor negatively regulates NLRP3 inflammasome activity by inhibiting NLRP3 transcription. Nat. Commun. 2014;5:4738. doi: 10.1038/ncomms5738. [DOI] [PubMed] [Google Scholar]
- 39.Sun H, et al. Exacerbated experimental colitis in TNFAIP8-deficient mice. J. Immunol. 2015;194:5736–5742. doi: 10.4049/jimmunol.1401986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.