Arabidopsis TNI/UBP14 destabilizes AUX/IAA repressors and promotes auxin response by ubiquitin recycling.
Abstract
The ubiquitin-mediated proteasomal pathway regulates diverse cellular processes in plants by rapidly degrading target proteins, including the repressors of hormone signaling. Though ubiquitin proteases play a key role in this process by cleaving polyubiquitin chains to monomers, their function has not been studied in detail by mutational analysis. Here, we show that mutation in TARANI/UBIQUITIN-SPECIFIC PROTEASE14 (TNI/UBP14) leads to reduced auxin response and widespread auxin-related phenotypic defects in Arabidopsis (Arabidopsis thaliana). In a tni partial loss-of-function mutant that was originally isolated based on altered leaf shape, activity of the auxin-responsive reporters DR5::GUS, DR5::nYFP, and IAA2::GUS was reduced. Genetic interaction studies suggest that TNI is involved in auxin signaling and acts alongside TIR1, ARF7, and AUX1. Map-based cloning identified TNI as UBP14. Inefficient splicing of the mutant TNI transcript resulted in the formation of an inactive UBP14 protein, which led to accumulation of polyubiquitin chains and excess polyubiquitinated proteins in the mutant. In addition to the reduced auxin response, increased levels of DII:VENUS, IAA18:GUS, and HS::AXR3-NT:GUS were also observed in tni, perhaps due to inefficient polyubiquitin hydrolysis and proteasome-mediated degradation. Together, our study identifies a function for TNI/UBP14 in the auxin response through ubiquitin recycling.
In plants, response pathways of several major phytohormones rely on 26S proteasome-mediated protein degradation. For example, the negative regulators of auxin, GA, and jasmonic acid (JA) signaling pathways, such as AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA), DELLA, and JASMONATE-ZIM DOMAIN (JAZ), respectively, undergo polyubiquitination and are subsequently degraded by the 26S proteasome, resulting in a change in gene expression (Ruegger et al., 1998; Gray et al., 2001; Wang and Deng, 2011; Davière and Achard, 2013). The polyubiquitin chains generated upon target protein degradation are hydrolyzed into monoubiquitin by a group of processing enzymes known as deubiquitinases (DUBs; Yan et al., 2000; Callis, 2014; Majumdar and Nath, 2020). These proteases also hydrolyze ubiquitin polyproteins linked head to tail by an α-peptide bond and ubiquitin-ribosomal extension proteins into monoubiquitin (Callis, 2014). Thus, DUBs are implicated in ubiquitin recycling to accomplish diverse cellular functions. Domain organization, along with the catalytic residues, categorizes the DUBs into five families: UBIQUITIN SPECIFIC PROTEASES (UBPs), UBIQUITIN C-TERMINAL HYDROLASES, OVARIAN TUMOR PROTEASES, MACHADO-JOSEPH DOMAIN PROTEASES, and JAB1/MPN/MOV34 proteases (Yan et al., 2000; Isono and Nagel, 2014; Majumdar and Nath, 2020). Among these, UBPs comprise the largest family, with 27 members in Arabidopsis (Arabidopsis thaliana; Yan et al., 2000). The transfer DNA (T-DNA) knockout lines ubp14 and ubp19 show embryonic lethality, whereas ubp15 has narrow and serrated leaves (Liu et al., 2008). Single loss-of-function mutants of the remaining UBPs do not exhibit discernible phenotypic alteration, suggesting genetic redundancy (Liu et al., 2008). However, higher-order mutants exhibit defects in cell cycle progression, endoduplication, gametogenesis, meristem maintenance, and flowering time control (Liu et al., 2008; Xu et al., 2016, An et al., 2018). Most of these UBPs show in vitro deubiquitination activity against α-linked or iso-linked polyubiquitin chains and ribosomal extension proteins (Isono and Nagel, 2014; Majumdar and Nath, 2020).
Among the three phytohormones mentioned above, auxin is a key member that regulates a plethora of growth and developmental programs, including embryogenesis, organ morphogenesis, venation pattern, root development, and gravitropism (Hobbie et al., 2000; Swarup et al., 2005; ten Hove et al., 2015). Studies over the past few decades have characterized the auxin signal transduction pathway, which comprises the TRANSPORT INHIBITOR RECEPTOR1 (TIR1) auxin receptor, AUX/IAA inhibitor proteins, and AUXIN RESPONSE FACTOR (ARF) transcription factors (Ruegger et al., 1998; Reed, 2001). Several genetic and biochemical studies have emphasized the importance of auxin-dependent degradation of AUX/IAAs by SCFTIR1/AFB via the 26S proteasome-mediated degradation pathway for maintaining normal auxin response in Arabidopsis (Gray et al., 2001; Leyser, 2018). Hence, the auxin level of a given cell is translated into a response by activating a set of ARFs through their release from AUX/IAA repression. Gain-of-function mutations in the degron motif of AUX/IAAs enhance their stability, resulting in auxin-related defects in embryogenesis, vein patterns, lateral root formation, and apical dominance. These mutants include axr3-1, slr, shy2-2, crane-1/iaa18-1, and iaa28-1 (Leyser et al., 1996; Tian and Reed, 1999; Fukaki et al., 2002; Uehara et al., 2008; Ploense et al., 2009). Single loss-of-function mutants of AUX/IAAs do not exhibit visible phenotypic alterations, reflecting genetic redundancy (Okushima et al., 2005).
Perturbation in the components of the 26S proteasome pathway is expected to adversely affect the auxin, GA, and JA pathways, since their response is triggered by the degradation of their repressors. However, the functions of only a handful of these components have been analyzed by mutational studies. We previously reported the isolation and characterization of an Arabidopsis mutant named tarani (tni) with pleiotropic phenotypic defects including altered leaf shape (Karidas, 2014; Karidas et al., 2015). Here, we have identified TNI as UBP14, which is involved in ubiquitin recycling. Each of the null alleles of UBP14 reported earlier is embryonic lethal, rendering further studies of this gene in postembryonic development a difficult task (Doelling et al., 2001; Tzafrir et al., 2002). However, the hypomorphic tni allele allows us to study the function of UBP14 in postembryonic development. The only other reported allele of UBP14 with postembryonic viability is da3-1, which has defective nuclear ploidy and organ growth (Xu et al., 2016). By carrying out detailed phenotypic analysis of tni seedlings, we found that TNI/UBP14 is required for optimal auxin response in Arabidopsis. Homozygous tni plants showed diverse phenotypic aberrations including defective embryo pattern, tricotyledonous and rootless seedlings, altered root gravitropism, and fewer lateral roots. These defects are also found in mutants with perturbed auxin response. Ubiquitin recycling and turnover of several AUX/IAAs were perturbed in tni seedlings. Together, our results suggest that TNI/UBP14 maintains a balance between polyubiquitin and monoubiquitin, which is necessary for the turnover of auxin signaling repressors through the 26S proteasome and thus required for normal auxin response.
RESULTS
Altered Auxin-Related Phenotype in tni
The tni mutant with altered leaf shape was originally isolated in a forward genetic screen (Karidas et al., 2015). Detailed characterization of tni revealed multiple defects in embryonic and postembryonic development (Fig. 1). The tni embryo exhibited an aberrant early cell division pattern (Fig. 1, A–C). In the wild type, the apical cell of the single-cell proembryo undergoes vertical division to form the two-cell proembryo and the basal cell undergoes a series of anticlinal divisions to form the suspensor (Boscá et al., 2011). Whereas the apical cell of each of the wild-type embryos studied (n = 30) underwent vertical division, a noticeable fraction of tni embryos showed horizontal (∼21%, n = 87) or oblique (∼8%, n = 87) division (Fig. 1A; Supplemental Fig. S1, A and B). In addition, the topmost suspensor cell in ∼36% of tni embryos (n = 38) underwent periclinal division, which was never observed in Columbia (Col-0; n = 30; Fig. 1B).Such defects are also observed in the auxin-signaling mutant bodenlos (bdl) and the higher-order polar auxin-transport mutant pin2pin3pin4pin7 (pin-formed; Hamann et al., 1999; Blilou et al., 2005; Křeček et al., 2009). The hypophysis, the precursor of root stem cell initials, undergoes asymmetric division during the dermatogen stage of embryogenesis and forms a lens-shaped cell, which is incorporated into the embryo at the globular stage (Scheres et al., 1994). A normal lens-shaped cell was observed in all the Col-0 globular embryos (n = 30), whereas ∼7% of tni embryos (n = 95) did not form a proper lens-shaped cell (Fig. 1C). The lack of such formative division, which is necessary for specifying root stem cell initials, is expected to result in rootless seedlings. We observed ∼13% rootless seedlings in tni (n = 407), which was never seen in Col-0 (n = 127; Fig. 1D). Perturbed auxin signaling in the gain-of-function mutant bdl and the loss-of-function mutant monopteros (mp) also results in rootless seedlings (Hamann et al., 1999, 2002).
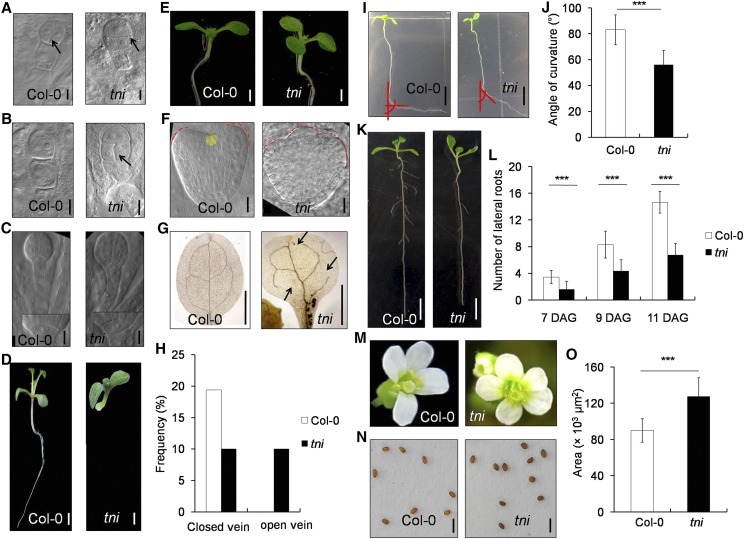
Figure 1.
Auxin-related phenotypes in tni. A, A two-cell proembryo. Arrows highlight vertical (Col-0) and horizontal (tni) cell division. B, A single-cell proembryo. The arrow indicates a periclinal cell division defect in the tni basal cell. C, Globular stage embryo. Normal (Col-0) and aberrant (tni) lens-shaped cells are magnified in the insets. Scale bars = 10 μm. D, Normal (Col-0) and rootless (tni) seedlings. Scale bars = 1 mm. The image was digitally extracted to remove the background. E, Seven-day-old seedlings with two (Col-0) or three (tni) cotyledons. Scale bars = 1 mm. The photograph was taken in a black background. F, Heart stage embryo with two (Col-0) or three (tni) cotyledon primordials (red dotted line). The Col-0 meristem is falsely colored yellow. Scale bars = 10 μm. G, Cotyledons from 7-d-old seedlings are cleared to highlight veins. Open venation in tni is marked by arrows. Scale bars = 1 mm. H, Percentage of cotyledons (n = 103 in Col-0 and 90 in tni) showing closed or open veins. I, Effect of gravistimulation on primary roots of 7-d-old seedlings. Angles of root bending are indicated by red lines. Scale bars = 5 mm. J, Average angle of curvature of primary roots (n = 52 for Col-0 and 43 for tni) following gravistimulation as in I. Error bars represent the sd. Asterisks indicate significance using unpaired Student’s t test (***P < 0.0001). K, Eleven-day-old seedlings. Scale bars = 5 mm. L, Average number of lateral roots (n = 10–13) in seedlings at the indicated DAG. Error bars represent the sd. Asterisks indicate significance using unpaired Student’s t test (***P < 0.0001). M, Open flowers showing variation in petal number in the tni mutant. The image was digitally extracted to remove the background. N, Mature seeds. Scale bars = 1 mm. O, Average seed area (n = 15). Error bars represent the sd. Asterisks indicate significance using unpaired Student’s t test (***P < 0.0001).
The postembryonic developmental defects in tni included tricotyledonous seedlings, reduced complexity in cotyledon venation, defects in root gravitropic response, fewer lateral roots, larger seeds, and an increase in petal number (Fig. 1, E–O). We estimated that ∼4% of tni seedlings (n = 306) form three cotyledons (Fig. 1E), which was never observed in Col-0. To address the origin of this phenotype, we examined the transition-state embryos when cotyledon primordia are initiated (ten Hove et al., 2015). Col-0 embryos always formed two cotyledon primordia whereas tni embryos occasionally produced three (Fig. 1F). The tricotyledonous phenotype is also observed in pin-formed 1 (pin1) loss-of-function mutant with perturbed auxin transport (Krecek et al., 2009). Mature tni cotyledons showed fewer complete areoles (closed veins) as compared to four complete areoles produced by the Col-0 cotyledons (Fig. 1, G and H). Nearly 20% of Col-0 cotyledons (n = 103) showed maximal complexity with four complete areoles and the remaining cotyledons had fewer than four areoles with some open vasculature (Fig. 1H; Supplemental Fig. S1, C and D; Sieburth, 1999). By contrast, only ∼10% (n = 90) of tni cotyledons formed four areoles and another ∼10% exhibited an open-top vein defect that was never seen in Col-0 cotyledons (Fig. 1H). Such a venation defect is also observed in the auxin-resistant mutant6 (axr6) mutant (Hobbie et al., 2000).
The tni mutant exhibited defects in subaerial organs as well (Fig. 1, I–L). Primary roots in tni seedlings showed defects in gravitropic response (Fig. 1I). Col-0 primary roots responded to gravistimulation by bending toward gravity at an average angle of 83.1° ± 11.50° (n = 52; Fig. 1J). By contrast, the tni roots bent only by 56.1° ± 11.0° (n = 43) under similar experimental conditions, indicating a reduced gravitropic response. The tni seedlings also produced fewer lateral roots (Fig. 1, K and L). In Col-0 seedlings, the number of emerged lateral roots steadily increased from 7 to 11 d after germination (DAG; Fig. 1L). Though a similar trend was observed in the tni seedlings, the number remained ∼50% lower than that in Col-0 at all the growth stages measured. Other mutants reported to have fewer lateral roots with perturbed auxin transport and signaling include tir1-1, auxin1-7 (aux1-7), and arf7-1 (Ruegger et al., 1998; Marchant et al., 2002; Okushima et al., 2007).
The tni mutant also showed an altered floral phenotype. Whereas all Col-0 flowers (n = 142) formed four petals, the petal number in ∼58% of tni flowers (n = 140) increased to five or six (Fig. 1M). This phenotype is similar to the weaker allele of polar auxin-transport mutant pin1-5 (Yamaguchi et al., 2014). In addition, tni plants produced bigger seeds with an ∼30% increase in seed area compared to the wild type (Fig. 1, N and O). Bigger seeds are seen in the mutant of ARF2, a negative regulator of cell division and expansion in Arabidopsis (Schruff et al., 2006).
Reduced Auxin Response in tni
The altered phenotype in tni described above, also seen in several auxin-pathway mutants (Bennett et al., 1995; Ruegger et al., 1998; Marchant et al., 2002; Swarup et al., 2005), suggests a possible involvement of TNI in the auxin pathway. Therefore, we compared auxin responses in Col-0 and tni seedlings using the auxin-responsive DR5::GUS, DR5::nYFP, and IAA2::GUS reporter lines (Ulmasov et al., 1997; Marchant et al., 2002; Mähönen et al., 2014). GUS assay of 3-d-old DR5::GUS seedlings showed strong GUS activity throughout the cotyledon margin, with the highest signal at the tip, indicative of auxin maxima (Fig. 2A; Sabatini et al., 1999; Mattsson et al., 2003). However, no distinct auxin maxima were detected in the DR5::GUS tni cotyledons, which exhibited an overall reduction in GUS signal except at the margin. A similar reduction in auxin maxima was observed at the tip of emerging leaves, primary roots, and lateral roots of DR5::GUS tni seedlings (Fig. 2, B–D). Comparison of the fluorescence signal in DR5::nYFP Col-0 and DR5::nYFP tni also revealed reduced auxin maxima in the mutant at the cotyledon tips (Fig. 2E) and root tips (Fig. 2F), whereas the N-terminal YFP fragment (nYFP) signal was increased in the tni columella cells, which appeared to have bigger nuclei (Fig. 2F). Quantitative analysis of DR5 activity by western blot analysis using anti-GFP antibody showed reduced DR5::nYFP signal in the DR5::nYFP tni seedlings compared to DR5::nYFP (Fig. 2G). Similarly, the GUS activity was reduced by nearly half in DR5::GUS tni seedlings compared to DR5::GUS seedlings (Fig. 2H). These results suggest that auxin response is reduced in the tni mutant.
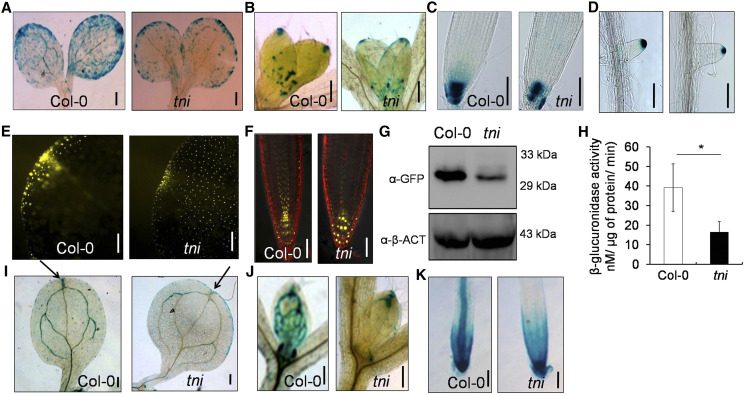
Figure 2.
Auxin response in tni. A to D, DR5::GUS activity in the cotyledons (A), first leaf pair (B), primary root tips (C), and lateral root tips (D) of 3- (A) and 7-d-old (B–D) DR5::GUS (Col-0) and DR5::GUS tni seedlings. Scale bars = 200 μm (A and B) and 100 μm (C and D). E and F, DR5::nYFP signal at the tips of cotyledons of 5-d-old seedlings (E) and primary roots (F) of 7- to 8-d-old DR5::nYFP (Col-0) and DR5::nYFP tni seedlings. Scale bars = 50 μm. Root samples in F were stained with propidium iodide. G, Western blots of the total proteins from 7-d-old DR5::nYFP (Col-0) and DR5::nYFP tni seedlings. α-β-ACTIN (α-β-ACT) was used as a control. H, GUS activity estimated in total extracts of DR5::GUS (Col-0) and DR5::nYFP tni seedlings. Averages of three biological replicates are shown. Error bars represent the sd. Asterisk indicates significance using unpaired Student’s t test (*P = 0.0406). I to K, IAA2::GUS activity in the cotyledons (I), first leaf pairs (J) and primary roots (K) of 7-d-old IAA2::GUS (Col-0) and IAA2::GUS tni seedlings. Arrows in I indicate IAA2::GUS activity at cotyledon tips. Scale bars = 200 μm (I and J) and 100 μm (K).
IAA2 is an immediate auxin-responsive gene whose induction depends on the endogenous auxin level. The IAA2::GUS signal was detected primarily in the vasculature of Col-0 cotyledons, leaves, and primary roots, and in the root meristematic region (Fig. 2, I–K), which is consistent with previous reports (Marchant et al., 2002). By contrast, vascular IAA2::GUS activity in tni was reduced in each of these organs, with expression limited only to their tips. In addition, ectopic expression was observed in the tni cotyledon margin (Fig. 2I).
The histochemical analysis of DR5::GUS and IAA2::GUS described above, together with the DR5::nYFP expression data, suggests that auxin response is reduced in tni. The data set of an earlier microarray experiment carried out on young tni leaves (Karidas et al., 2015) identified 29 auxin-related genes that were differentially expressed >2-fold (16 downregulated and 13 upregulated) in tni compared to the wild type (Supplemental Fig. S2A). These genes are predicted to regulate auxin biosynthesis, transport, or signaling. Many of these transcripts are also altered in seedlings externally treated with IAA (Supplemental Fig. S2B). Taken together, these data suggest that TNI is required to maintain normal auxin response in Arabidopsis.
Altered Sensitivity of tni to External Auxin Manipulation
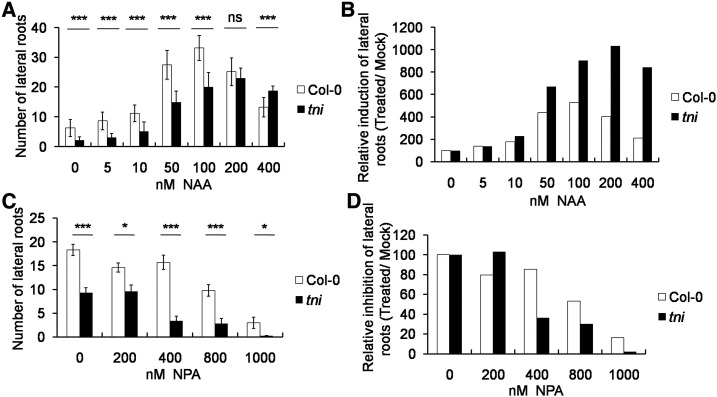
Altered auxin response in tni could be due to perturbed auxin level or signaling. To test this, we compared the sensitivity of Col-0 and tni seedlings to exogenous administration of the synthetic auxin 1-naphthaleneacetic acid (NAA). Since auxin is known to stimulate lateral root formation in a dose-dependent manner (Ivanchenko et al., 2010), we used the number of lateral roots as a readout of auxin sensitivity. In Col-0, the number of lateral roots progressively increased with increased concentrations of NAA up to 100 nm, followed by a decrease with a further increase in NAA concentration (Fig. 3, A and B), thus forming a characteristic bell-shaped auxin-response curve (Ivanchenko et al., 2010). Though a similar trend was observed for tni roots, the peak response in tni was achieved at a NAA concentration (200 nm) that was twice that required for Col-0 (Fig. 3B), consistent with its reduced auxin-response phenotypes (Fig. 2). Conversely, tni showed increased sensitivity toward the polar auxin transport inhibitor N-1-naphthylphthalamic acid (NPA), which blocks lateral root initiation by reducing the IAA level at the basal root meristem (Casimiro et al., 2001). The total number of lateral roots in Col-0 remained unaltered up to400 nm NPA, beyond which the value declined to ∼20% at 1 μm concentration (Fig. 3, C and D). In tni, the lateral root number reduced to <40% at 400 nm NPA and to nearly zero at 1 μm concentration. Taken together, these results suggest a reduced endogenous auxin response in tni roots.
Figure 3.
Auxin sensitivity of the tni mutant. A and B, Average number of lateral roots of 7-d-old seedlings grown in the presence of NAA (A) and their relative increase (B). C and D, Average number of lateral roots in 9-d-old seedlings treated with NPA (C) and their relative decrease (D).; Error bars represent the sd (n = 12–15 [A and C]). Asterisks indicate significance using unpaired Student’s t test (***P ≤ 0.0001 and *P < 0.006). ns, Not significant.
Genetic Interaction between tni and Mutants with Auxin-Related Growth Defects
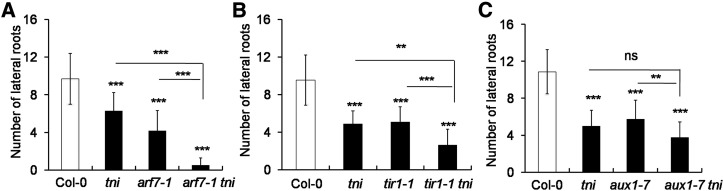
To assess the genetic link between TNI and the auxin pathway, we crossed tni with mutants defective in auxin signaling and transport, namely, arf7-1, tir1-1, and aux1-7, and studied the phenotypes of the double homozygous lines. ARF7 and ARF19 redundantly promote lateral root formation and the arf7-1single mutant produces fewer lateral roots (Okushima et al., 2005, 2007). Since arf19-1 is a weak allele and the arf7-1 arf19-1 double mutant totally lacks lateral roots, we studied the genetic interaction of tni with arf7-1. Lateral root formation was severely reduced in the arf7-1 tni double mutant compared to the parental lines (Fig. 4A). Fewer lateral roots formed in the auxin-receptor mutant tir1-1 (Ruegger et al., 1998), and even fewer in the tir1-1 tni double mutant, compared to Col-0 (Fig. 4B). AUX1 encodes an auxin-influx carrier that promotes lateral root formation by facilitating the distribution of auxin from leaf to root, and aux1-7 seedlings make fewer lateral roots and lateral root primordia (Marchant et al., 2002). Phenotypic analysis showed that the aux1-7 tni mutant had fewer lateral roots than either of the parents (Fig. 4C). Since the mutant alleles used in these genetic interaction studies were weak in nature, one interpretation of these results is that TNI works in the auxin-response pathway.
Figure 4.
Genetic interaction between tni and mutants with auxin-related defects. A to C, Graphs show the average number of lateral roots (n = 10–15) of 9-d-old seedlings. Error bars represent the sd. Asterisks indicate significance using unpaired Student’s t test (***P < 0.0001 and **P = 0.0084). ns, Not significant.
TNI Encodes UBP14
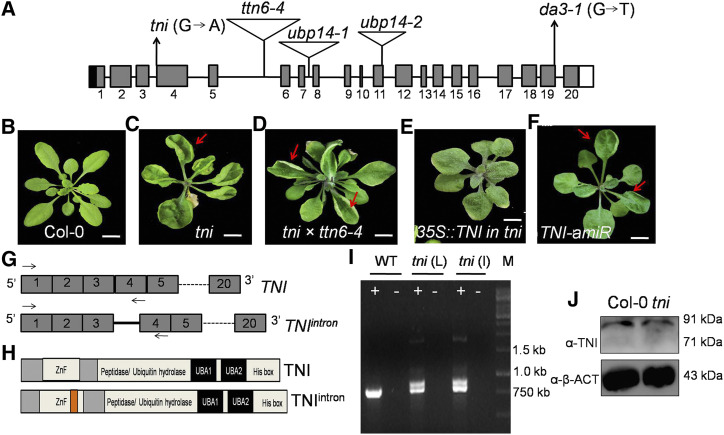
Using a map-based cloning approach, we delimited the tni locus to a 65-kb-long genomic region with the help of 927 recombinant mutant plants in a mapping population (see “Materials and Methods”). Sequencing of the protein-coding genes within this interval identified an exonic G to A transition in the At3G20630 locus (Fig. 5A; Supplemental Fig.S3, A and B). This mutation mapped at the canonical 3′ splice acceptor site at the junction of intron 3 and exon 4. At3G20630 is predicted to encode UBP14, a ZnF DUB protein involved in ubiquitin recycling (Xu et al., 2016; Doelling et al., 2001). Several alleles of At3G20630 have been previously described (Fig. 5A), most of which show embryonic lethality (Majumdar and Nath, 2020). To further examine tni identity, we performed an allelism test with titan6-4 (ttn6-4), a known allele of UBP14 (Tzafrir et al., 2002). When tni/tni plants were crossed to ttn6-4/+ heterozygous plants (which resembled Col-0), 12 out of 42 F1 individuals produced cup-shaped rosette leaves (Fig. 5, B–D), whereas tni/+ plants always resembled Col-0, suggesting that tni is allelic to ttn6. Further, the cup-shaped phenotype of tni leaves was partly rescued by overexpressing the wild-type TNI transcript; three of 14 hygromycin-resistant transformants recovered and produced flat rosette leaves, though these leaves were rounder than the Col-0 leaves (Fig. 5E). In addition, expressing an artificial microRNA targeting the wild-type TNI transcript under the constitutive RPS5a promoter in the Col-0 background partially recapitulated the tni phenotype (Fig. 5F); 3 of 25 hygromycin-resistant transformants recovered produced rosette leaves with weak cup-shaped lamina.
Figure 5.
Cloning of TNI. A, Schematic representation of the TNI/UBP14 locus showing the 5′ untranslated region (black box), exons (gray box), introns (black line), and the 3′ untranslated region (white box). Positions of T-DNA insertions in various mutant alleles are shown by inverted triangles. The G→A transition at the third intron-exon junction in the tni allele and the G to T substitution at the nineteenth exon-intron junction in the da3-1 allele are indicated. B to F, Thirty-day-old rosettes of wild-type (B), tni (C), tni × ttn6-4 (+/−) F1 plants (D), tni plants overexpressing TNI transcript (E), and Col-0 plants expressing artificial microRNA (amiR) against the TNI transcript (F). Red arrows indicate leaf curvature. Scale bars = 0.5 cm. Rosette images were digitally extracted to remove the background. G, Schematic representations of the predicted wild-type TNI and the mutant TNIintron transcripts. Note the retention of intron 3 (solid line) in TNIintron. Arrows indicate the positions of the primers used for RT-PCR analysis. Dotted lines indicate continuity of exons. H, Domain architecture of TNI and TNIintron proteins. The orange box indicates a 34-amino acid insertion encoded by intron 3. I, Ethidium bromide-stained agarose gel showing the products of RT-PCR on total RNA from Col-0 (WT), mature tni leaves (tni L), and tni inflorescence (tni I). Plus and minus signs indicate cDNA and RNA, respectively, as PCR templates, and M indicates a DNA marker. J, Western blot of total protein extracted from 7-d-old seedlings using antibody raised against a peptide corresponding to residues 156 to 174 of TNI. α-β-ACTIN (α-β-ACT) was used as loading control.
The tni Locus Encodes TNIintron, an Aberrant UBP14
The wild-type TNI locus, consisting of 20 exons and 19 introns, is predicted to encode an 88-kD protein product (Fig. 5A; Supplemental Fig. S3C). If the G to A transition interferes in splicing, the tni locus is predicted to produce an additional aberrant transcript (TNIintron), whereby 102 nucleotides corresponding to intron 3 are retained in frame in the wild-type transcript (Fig. 5G).Whereas reverse transcription PCR (RT-PCR) analysis with primers flanking intron 3 (Fig. 5G) detected a single product of 750 bp in both Col-0 and tni, an additional band corresponding to retention of intron 3 was detected in the tni samples (Fig. 5I). The intensity of these two bands in the tni mutant, which corresponded to TNI and TNIinron transcripts, seemed comparable, indicating nearly equal abundance of the two transcripts. Transgenic Col-0 plants expressing the RPS5a::TNIintron cassette produced cup-shaped leaves in the T1 generation (Supplemental Fig. S3E); 2 of 107 independent insertion lines produced all cup-shaped leaves, whereas another 32 lines showed at least one cup-shaped leaf. This genetic evidence suggests an association between retention of TNI intron 3 and the tni phenotype.
Western blot analysis using an antibody generated against a 19-residue polypeptide within the ZnF domain detected a single band in both Col-0 and tni seedlings that corresponded to the endogenous TNI (see “Materials and Methods”; Fig. 5J; Supplemental Fig. S3F). This antibody also detected the GST-TNI fusion protein but failed to recognize a truncated TNI that lacked the ZnF domain (Supplemental Fig. S3, G and H), suggesting that the antibody is specific to TNI. The TNIintron transcript is likely to encode a full-length TNI protein wherein the ZnF domain is disrupted by an insertion of an additional 34 residues (Fig. 5H; Supplemental Fig. S3D). Since both wild-type and mutant transcripts were detected in equal proportion in tni (Fig. 5I), it is likely that both TNI (88 kD) and its mutant variant TNIintron (92 kD) are translated in tni (Fig. 5J), though they cannot be resolved in western blot analysis due to their similar Mr values. Indeed, the anti-GST antibody could not distinguish between recombinant GST-TNI and GST-TNIintron in western blot analysis (Supplemental Fig. S3H).
Cell Type-Specific TNI Activity Regulates Lateral Root Formation
UBP14 has been reported to be detected ubiquitously in all tissue types (Doelling et al., 2001; Xu et al., 2016). Consistently, GUS activity was detected throughout the seedlings in all five independent pTNI::GUS reporter lines that we generated (Supplemental Fig. S4). In primary roots, more intense GUS signal was detected in discrete pericycle cells that initiate lateral roots (De Smet, 2012). Thus, promotion of lateral roots by TNI (Fig. 1K and L) could be due to its local expression in roots or to the systemic effect of auxin response. To test this, we manipulated TNI expression in the lateral root initials of transgenic plants and studied its effect on lateral roots. When we expressed an artificial microRNA against TNI under the truncated PLETHORA7 promoter (pPLT7) that is active specifically in lateral root primordia (Prasad et al., 2011), lateral root number was reduced by 45% to 80% in six independent pPLT7::TNI-amiR transgenic lines in the T2 generation. A reduced number of lateral roots was also observed in two homozygous pPLT7::TNI-amiR transgenic lines established in the T3 generation (Supplemental Fig. S5, A–C). When TNIintron was expressed in the lateral root initials, the number of lateral roots was reduced to ∼50% in a homozygous pPLT7::TNIintron transgenic line (Supplemental Fig. S5, B and D), a reduction that is similar to what was observed in the tni allele (Fig. 1L). These results suggest that the cause of fewer lateral roots in tni is the local loss of UBP14 activity in the lateral root initials.
TNIintron Lacks Deubiquitinase Activity
To test whether the mutant protein retains enzymatic activity, we compared TNI with TNIintron through an in vitro deubiquitination assay (Rao-Naik et al., 2000; Doelling et al., 2001). Recombinant TNI efficiently cleaved 2- to 7-mer Lys-48-linked polyubiquitin substrates into monoubiquitin (Supplemental Fig. S6A). However, TNIintron, as well as the catalytically inactive UBP14C317S control protein (Doelling et al., 2001), failed to cleave polyubiquitin substrates. TNIintron and TNIC317S also failed to cleave UBQ10-encoded α-linked hexa-ubiquitin chains in Escherichia coli cells (Rao-Naik et al., 2000), whereas hexa-ubiquitin substrate was completely cleaved into di- and monoubiquitin forms by TNI and UBP14 from yeast, a functional homolog of TNI (Supplemental Fig. S6B; Amerik et al., 1997). A similar in vivo deubiquitination assay in E. coli cells expressing α-linked, His-tagged tetra-ubiquitin (Rao-Naik et al., 2000) also showed that TNI, but not TNIintron or TNIC317S, cleaves the substrate into His-tagged di- and monoubiquitin products as detected by anti-His antibody (Supplemental Fig. S6C). Together, these results show that TNIintron is catalytically inactive toward iso-linked and α-linked polyubiquitin substrates.
Increased Accumulation of Polyubiquitin and Polyubiquitinated Proteins in tni
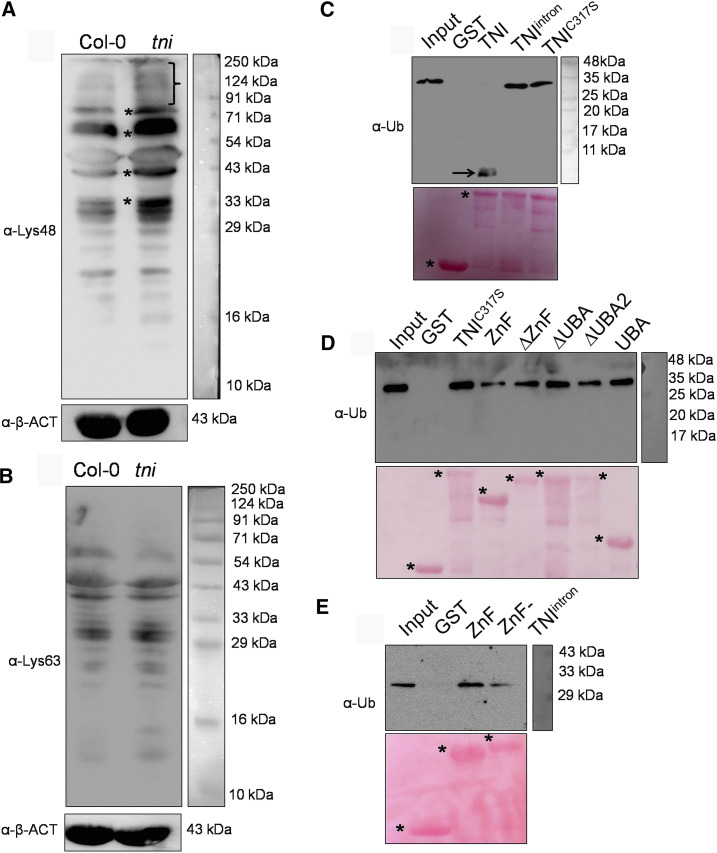
Western blot analysis of the total protein samples from Col-0 and tni plants using antiubiquitin antibody showed reduced monoubiquitin and an excess accumulation of polyubiquitinated proteins in tni (Supplemental Fig. S7, A and B). In line with the lack of catalytic activity in TNIintron, free polyubiquitin chains were detected only in tni plants (Supplemental Fig. S7A), as reported earlier for other ubp14 alleles (Doelling et al., 2001; Xu et al., 2016). In Arabidopsis, the most abundant Lys-48-linked polyubiquitin is implicated in protein turnover through the 26S proteasome, whereas Lys-63-linked polyubiquitin imparts a nondegradative fate to the cellular proteins (Kim et al., 2013; Mevissen and Komander, 2017). Western blot analysis of the total protein extracts using linkage-specific antibodies showed a higher abundance of Lys-48-linked polyubiquitinated proteins, and not Lys-63-linked proteins, in tni seedlings relative to Col-0 (Fig. 6, A and B), suggesting that TNI is involved in the turnover of cellular proteins by ubiquitin-26S proteasomal degradation. These ubiquitin antibodies did not recognize monoubiquitin because it lacks such linkages.
Figure 6.
Linkage specificity and ubiquitin binding by TNI. A and B, Western blots of total proteins extracted from 7-d-old seedlings probed with anti-ubiquitin antibody specific to Lys-48 (α-Lys-48; A) or Lys-63 (α-Lys-63; B) linkage. The bracket and asterisks (A) indicate a smear of Lys-48-linked polyubiquitinated proteins, and α-β-ACTIN (α-β-ACT) served as the loading control. C to E, Antiubiquitin (α-Ub) western blots of Lys-48-linked tetra-ubiquitin substrate incubated with recombinant, GST-tagged full-length (TNI, TNIintron, and TNIC317S [C and D]), and truncated (ZnF, ΔZnF, ΔUBA, ΔUBA2, UBA, and ZnF-TNIintron [D and E]) forms of TNI protein immobilized on glutathione beads. Lys-48-linked tetra-ubiquitin substrate alone (Input) and recombinant GST protein served as positive and negative controls, respectively. The arrow in C indicates a monoubiquitin product. Ponceau-stained membranes shown below the western blots served as the loading control. Asterisks denote the recombinant proteins used for the assays.
To determine whether proteasomal activity is altered in tni, we compared the sensitivity of Col-0 and tni seedlings to MG132, a proteasome inhibitor. The polyubiquitinated protein signal was more intense in tni than in Col-0 at 0, 0.1, and 0.2 mm MG132 (Supplemental Fig. S7, C and D), suggesting that tni cells are hypersensitive to perturbation of proteasomal activity. Interestingly, MG132 treatment did not alter the steady-state level of free polyubiquitin chains in tni (Supplemental Fig. S7C), suggesting that they accumulate due to their inefficient hydrolysis by TNIintron. Indeed, incubation of total protein extract from tni seedlings with recombinant TNI, but not with TNIintron or TNIC317S, resulted in the disappearance of the free polyubiquitin chains with concomitant accumulation of monoubiquitin (Supplemental Fig. S8), resulting in a western blot profile somewhat similar to the Col-0 profile (Supplemental Fig. S7A).
TNIintron Binds to Polyubiquitin Substrate
To examine whether the 34-residue insertion in TNIintron interferes with its substrate-binding ability, we compared the interaction of various forms of TNI with Lys-48-linked tetra-ubiquitin (Fig. 6, C and D). Bead-bound GST-tagged recombinant TNI, TNIintron, and TNIC317S were incubated with Lys-48-linked tetra-ubiquitin substrate and the precipitate was analyzed through western blot analysis using anti-ubiquitin antibody. Each of the three forms of the protein bound to the substrate, whereas only TNI cleaved the tetra-ubiquitin into monoubiquitin (Fig. 6C). Thus, retention of intron 3 in TNIintron does not interfere with substrate binding.
To map the polyubiquitin binding domains of TNI, we generated and expressed five truncated forms of TNI in E. coli (Supplemental Fig. S9): (1) N-terminal ZnF domain alone (ZnF); (2) TNI without ZnF (ΔZnF); (3) TNI without UBA domains (ΔUBA); (4) TNI without UBA2 (ΔUBA2); and (5) only the two UBA domains (UBA). Since full-length TNI efficiently hydrolyzed polyubiquitin into monomer (Fig. 6C) and therefore cannot be used as a positive control for the binding study, we instead used TNIC317S (Fig. 6C). In western blot analysis of the in vitro substrate-binding assay, all the truncated forms of TNI and TNIC317S bound to the Lys-48-linked tetra-ubiquitin substrate with varying efficiency (Fig. 6D). The ZnF domain of the TNIintron protein in isolation (here called ZnF-TNIintron) also bound to the tetra-ubiquitin substrate (Fig. 6E), albeit with reduced efficiency, suggesting that its disruption does not affect its substrate-binding capability. Thus, the inability of TNIintron to hydrolyze polyubiquitin substrate is perhaps due to an overall conformational change rendering the catalytic domain inactive.
Increased Abundance of AUX/IAA Transcriptional Repressors in tni
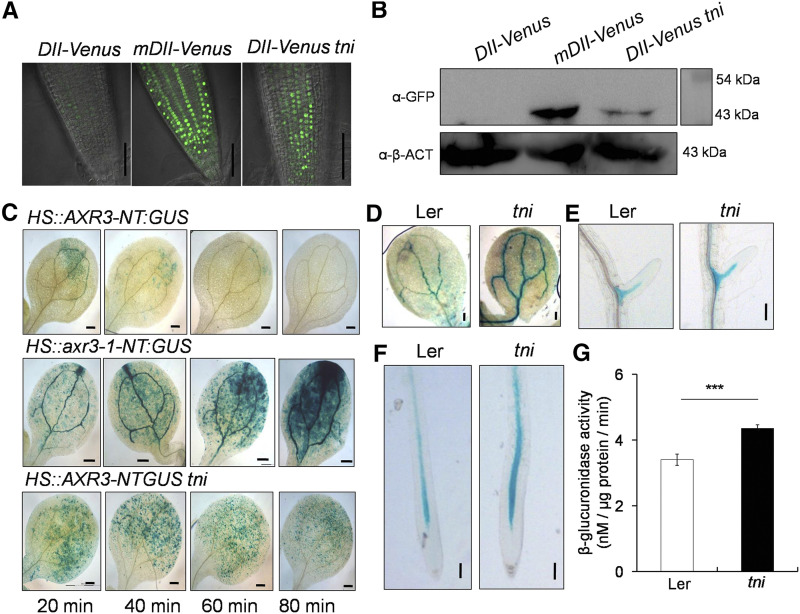
Reduced 26S-proteasome activity in tni may lead to an array of auxin-related growth defects by stabilizing AUX/IAA repressors, similar to what is found in their gain-of-function mutants (Hamann et al., 1999; Tian and Reed, 1999; Uehara et al., 2008). To test this, we compared Domain II (DII):VENUS signals, a readout of AUX/IAA level, of tni and the wild type (Fig. 7A). A weak VENUS signal was detected in Col-0 primary roots due to rapid turnover of DII:VENUS, which is suggestive of high auxin activity. Col-0 roots expressing a mutant, nondegradable form of the protein, mDII:VENUS, showed strong and widespread VENUS signal, which is consistent with previous reports (Brunoud et al., 2012). The primary roots of DII:VENUS tni also showed VENUS signal stronger than Col-0. Western blot analysis of protein extracts from 7-d-old seedlings using anti-GFP antibody further confirmed that the level of DII:VENUS was indeed more abundant in tni seedlings than in Col-0 (Fig. 7B).
Figure 7.
Stabilization of AUX/IAAs in tni. A, DII:VENUS signal in the primary roots of 7-d-old Col-0 (DII:VENUS and mDII:VENUS) and tni (DII:VENUS tni) seedlings. The strong signal of mDII:VENUS, a nondegradable version of the DII domain, served as a positive control. Scale bars = 50 μm. B, Anti-GFP western blot of total protein extracts from 7-d-old Col-0 (DII:VENUS and mDII:VENUS) and tni (DII:VENUS tni) seedlings. α-β-ACTIN (α-β-ACT) served as the internal control. Numbers indicate molecular-weight markers. C, GUS activity in cotyledons of 7-d-old seedlings in response to increasing durations of heat shock. The nondegradable form, axr3-1-NT (middle), served as a positive control. NT, N-terminal domain. Scale bars = 200 μm. D to F, IAA18:GUS signal in a cotyledon vein (D), a lateral root (E), and the primary root (F) of IAA18:GUS (Ler)and IAA18:GUS tni seedlings. Scale bars = 200 μm (D) and 100 μm (E and F). G, GUS activity in IAA18:GUS (Ler) and IAA18:GUS tni (tni) seedlings. Averages of triplicate biological replicates are shown. Error bars represent the sd. Asterisks indicate significance using unpaired Student’s t test (***P < 0.0001).
The above result suggests that the DII domain, and hence some of the AUX/IAA repressors, are stabilized in tni roots. To test this, we compared the AXR3/IAA17 levels in Col-0 and tni. The heat-inducible HS::AXR3-NT:GUS reporter line has been extensively used to monitor IAA17 turnover in various mutants in which protein degradation by 26S proteasome is affected (Gray et al., 2001). We detected a weak and patchy GUS signal 20 min after heat shock treatment of the HS::AXR3-NT:GUS seedlings, but it disappeared within 80 min (Fig. 7C, top), suggesting efficient turnover of the protein in Col-0. The nondegradable form of the protein, axr3-1-NT:GUS, was detected in large amounts in the HS::axr3-1-NT:GUS line at 20 min and continued to accumulate, producing a stronger GUS signal after 80 min (Fig. 7C, middle). Similarly, the GUS signal in HS::AXR3-NT:GUS tni cotyledons also accumulated with more intensity than in Col-0, and it decreased at a slower pace, retaining a considerable signal even after 80 min of heat shock induction (Fig. 7C, bottom).
As in tni, reduced auxin sensitivity effects, including fewer lateral roots, are also seen in the iaa18-1 allele, where IAA18 is stabilized (Uehara et al., 2008; Ploense et al., 2009). To determine whether IAA18 is stabilized in the tni mutant, we compared IAA18:GUS activity in tni and Landsberg erecta (Ler) seedlings. GUS signal was detected in the vasculature of cotyledons, lateral roots, and primary roots in the IAA18:GUS seedlings (Fig. 7, D–F). Though the pattern of the signal remained more or less similar in the vasculature of the IAA18:GUS tni seedlings compared to Ler seedlings, its intensity increased, suggesting that IAA18 is more abundant in the mutant. Quantification of GUS activity also showed an increase in IAA18:GUS signal in tni plants (Fig. 7G). Taken together, the above observations suggest that TNI is required for turnover of certain AUX/IAA proteins.
DISCUSSION
The tni allele of UBP14 is recessive and likely hypomorphic in nature as its knockout alleles show embryonic lethality. The previously reported ttn6-4 allele, which has a deletion of 400 bp resulting in the elimination of exons 6 and 7, causes defective embryos arrested at the globular stage (Tzafrir et al., 2002). T-DNA insertions in intron 7 of ubp14-1 and intron 11 of ubp14-2 alleles also yield null mutants with similar embryo phenotypes (Doelling et al., 2001). The tni allele also exhibited partial embryo lethality resulting in reduced seed setting (Karidas, 2014). The aborted embryos of ubp14-1 and ubp14-2 have higher levels of polyubiquitin and polyubiquitinated proteins (Doelling et al., 2001), implying that UBP14 is required for ubiquitin recycling, which is crucial for the progression of embryo development. Our results show that the tni plants also have elevated accumulation of unanchored polyubiquitin chains, as well as polyubiquitinated proteins, with a concomitant reduction in monoubiquitin (Supplemental Fig. S7, A and B). Thus, TNI/UBP14 is also involved in ubiquitin recycling during postembryonic development. Improper ubiquitin recycling results in multiple growth and developmental defects. The massive increase in total ubiquitin in tni can perhaps be explained by the reduction in the steady-state level of monoubiquitin, which is known to trigger increased ubiquitin biosynthesis (Park and Ryu, 2014). Indeed, the polyubiquitin biosynthetic genes UBQ13 and UBQ14 are 2-fold upregulated in tni, as revealed by an earlier microarray data set (Karidas et al., 2015). Detection of monoubiquitin in tni in low abundance implies that partial deubiquitination activity is retained in the mutant, whereas monoubiquitin was not detected in the null alleles of ubp14 (Doelling et al., 2001).
Since two transcripts corresponding to wild-type TNI and aberrant TNIintron were detected in almost equal abundance in tni plants (Fig. 5I), it is likely that both normal and aberrant protein forms are expressed at comparable levels, assuming that their translation efficiency is similar. Perhaps the lack of catalytic activity of half of the protein pool (TNIintron) results in an inefficient turnover of polyubiquitin into monoubiquitin. Recently, it was shown that the da3-1 allele of UBP14 has a G to T transversion at the 5′ exon-intron boundary of the last intron, generating a premature stop codon (Xu et al., 2016). The protein encoded by the da3-1 locus is catalytically inactive since it lacks the C-terminal His box essential for catalysis. Our results show that UBP14 with a disrupted ZnF domain (TNIintron) is also catalytically inactive. However, inactive TNIintron efficiently binds to the polyubiquitin substrates in vitro, suggesting that the mutant protein sequesters some of the cellular targets and acts as a dominant negative form of wild-type TNI. Sequestration of the substrates in the pPLT7::TNIintron transgenic line perhaps results in fewer lateral roots, an effect similar to target stabilization in the tni mutant due to the compromised activity of TNI.
Considering that UBPs maintain the steady-state level of monoubiquitin, which is an essential substrate for marking the target proteins for degradation, perturbation in their function is likely to affect multiple aspects of plant biology. Consequently, mutational analysis of 27 Arabidopsis UBPs revealed their diverse functions in embryogenesis, leaf development, and organ size control (Doelling et al., 2001; Liu et al., 2008; Majumdar and Nath, 2020). Complete loss of UBP14 and UBP19 results in embryo lethality, whereas disruption of the deubiquitination activity of UBP14 in the hypomorphic da3-1 allele results in increased ploidy and enlarged organs (Xu et al., 2016). UBP26 deubiquitinates H2B, required for gene silencing, in the nucleus, and its loss severely affects seed development (Sridhar et al., 2007; Luo et al., 2008). Mutant alleles of UBP15 produce narrow, serrated leaves (Liu et al., 2008). Apart from these few examples, single mutants of other UBPs do not show discernible phenotypic changes, suggesting functional redundancy. For example, UBP12 and UBP13 together promote root meristem development by stabilizing ROOT MERISTEM GROWTH FACTOR1 receptor (RGFR1) and RGFR2 (An et al., 2018). Besides, they remove ubiquitin marks from polyubiquitinated MYC2 in vitro and promote the JA response (Jeong et al., 2017). These recent studies demonstrate the substrate specificity of UBPs and their preeminent role in plant development (Majumdar and Nath, 2020). Therefore, it is not surprising to see multiple phenotypic defects in the tni/ubp14 mutant.
Most phenotypic defects in tni resembled those with compromised auxin response. For example, the rootless phenotype of tni is similar to that of bdl/IAA12 gain-of-function and mp/arf5 loss-of-function mutants (Hamann et al., 1999, 2002).The stabilized form of IAA12 in the bdl mutant blocks auxin-dependent ARF5 activation, which is required for hypophysis specification and root initiation (Schlereth et al., 2010). Similarly, venation patterning defects are also reported in mp and bdl mutants (Berleth et al., 2000). Reduced vein complexity in tni cotyledons could be due either to reduced auxin transport or reduced signaling. However, we did not see any significant difference in expression of the auxin transporter PIN1 between Col-0 and tni (Supplemental Fig. S10). Another major auxin-related defect in tni is fewer lateral roots, a phenotype also seen in plants with stabilized AUX/IAAs such as IAA3, IAA14, IAA18, IAA19, and IAA28 (Tian and Reed, 1999; Fukaki et al., 2002; Uehara et al., 2008). Gain-of-function mutation in these repressors produces either no or very few lateral roots. ARF7 and ARF19 act downstream of IAA14/SLR to promote lateral root initiation (Okushima et al., 2005, 2007). Genetic interaction of tni with arf7-1 showed an additive effect in lateral root emergence, which can be interpreted as both these genes working in parallel pathways. However, it should be noted that ARF19 compensates for the loss of ARF7 function in arf7-1, which shows a much weaker phenotype compared to the arf7-1 arf19-1 double mutant. Moreover, the hypomorphic nature of the tni allele may contribute to the additive phenotype in the arf7-1 tni double mutant, and perhaps in tir1-1 tni and aux1-7 tni mutants as well. Besides, tir1-1 and aux1-7 are also weak alleles, and combination of two weaker alleles is expected to show an additive phenotype, even if they work in the same pathway.
One possible mechanism of reduced auxin response in tni could be the stabilization of AUX/IAAs, including IAA17 and IAA18 (Fig. 7). The Arabidopsis genome encodes 29 AUX/IAAs with a conserved DII, which interacts with TIR1 and undergoes polyubiquitination and degradation by the 26S proteasome with varying kinetics (Gray et al., 2001; Reed, 2001). Increased DII:VENUS signal in tni is in agreement with the stabilization of multiple AUX/IAAs. However, our study falls short of providing direct evidence that the tni phenotype is mediated by AUX/IAA stabilization. It also does not resolve whether AUX/IAAs in tni are stabilized in the free form or in the polyubiquitinated form. It is possible that accumulation of Lys-48-linked free polyubiquitin chains creates a roadblock for protein degradation by the 26S proteasome, as reported in yeast and humans (Amerik et al., 1997; Dayal, et al., 2009), leading to an increase in polyubiquitinated target proteins. Direct measurement of polyubiquitinated AUX/IAAs in wild-type and tni plants would be required to determine whether they are part of this protein pool.
Since degradation of negative regulators by the ubiquitin-proteasome pathway is a general theme of signaling for several plant hormones, it is likely that the response pathways of other hormones would also be affected in tni. For example, the DELLA proteins that suppress GA signaling are also degraded by the 26S proteasome pathway (Wang and Deng, 2011; Davière and Achard 2013) and hence are likely to be stabilized in the tni mutant. Whereas this is expected to cause reduced GA response in the mutant, we had earlier noticed an elevated GA-related phenotype in tni, which was rescued by inhibiting GA synthesis (Karidas et al., 2015). It is possible that stabilized DELLA proteins in the mutant suppress GA signaling to an extent that increased GA biosynthesis is triggered as a feedback response (Nelson and Steber, 2016). Ubiquitin-dependent degradation of hormone repressors may not be the only mechanism of action for TNI. It has been recently shown that UBP14 interacts with the ULTRAVIOLET-B INSENSITIVE4 protein in repressing endoreduplication and organ growth in Arabidopsis (Xu et al., 2016). Interestingly, both GA signaling and low-auxin response promotes entry into the endocycle (Gendreau et al., 1999; Ishida et al., 2010). Thus, the collective phenotypic defects in tni could be a cumulative effect of multiple pathways.
CONCLUSIONS
We show that TNI encodes the UBP14 enzyme, whose activity is partly compromised in the tni mutant because the mutation causes inefficient splicing of the TNI transcript. This results in an aberrant protein that lacks catalytic activity, leading to an accumulation of polyubiquitin chains and excess polyubiquitinated proteins. This is accompanied by widespread auxin-deficient phenotypes and stabilization of certain AUX/IAA repressors.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) ecotypes Col-0 and Ler were used as wild-type plants. The mutant/transgenic lines DR5::GUS (Ulmasov et al., 1997), DR5::nYFP (Mähönen et al., 2015), IAA2::GUS (Marchant, et al., 2002), HS::AXR3-NT:GUS (N9571), HS:axr3-1-NT:GUS (N9572; Gray et al., 2001), DII-VENUS (N799173), mDII-VENUS (N799174; Brunoud et al., 2012), IAA18:GUS (Ploense et al., 2009), arf7-1 (CS24607; Okushima et al., 2005), aux1-7 (CS3074; Swarup et al., 2004), tir1-1 (CS3798; Ruegger et al., 1998), and ttn6-4 (CS16079; Tzafrir et al., 2002) have been reported earlier. Most of these lines were obtained from the Nottingham Arabidopsis Stock Center or the Arabidopsis Biological Resource Center.
Plant Growth Conditions and Treatments
Seeds were surface sterilized and stratified in darkness for 2 d at 4°C, after which they were transferred to the growth chamber and maintained under long-day conditions with 16 h light (120 μmol m−2 s−1) and 8 h dark at 22°C. For 1-NAA (Sigma-Aldrich) and NPA (Calbiochem) sensitivity assays, seedlings were grown for 4 d in NAA/NPA-free one-half strength Murashige and Skoog medium supplemented with 1% (w/v) Suc (Sigma-Aldrich) and 0.8% (w/v) agar (Hi Media), transferred to 1-NAA- or NPA-containing MS plates, and placed vertically in a growth chamber for an additional 3 d (NAA) or 5 d (NPA). Photographs were taken at 7 or 9 DAG and lateral roots were counted using a differential interference contrast (DIC) microscope (Olympus). Seven-day-old seedlings were treated with MG132 (Sigma-Aldrich) for 16 h in liquid medium containing one-half strength MS salt.
Gravitropism Assay
A gravitropic response assay was performed according to a protocol described earlier (Hobbie et al., 2000). Briefly, one-half strength MS plates containing the seeds were kept vertically in a growth chamber and rotated clockwise by 90° at 4 DAG. ImageJ software (rsbweb.nih.gov/ij/) was used to measure the angle of curvature after 3 d of gravistimulation.
Tissue Clearing
Seedlings were kept in 70% (v/v) ethanol for 24 h, incubated in lactic acid for 30 min, and then mounted in lactic acid on a transparent glass slide and observed under a DIC microscope.
Embryo Dissection
Fertilized ovules were scooped out from siliques and placed on a glass slide containing Hoyer’s medium and chloralhydrate:glycerol:water (8:1:2). Images were acquired using a Zeiss Axio Imager M1 microscope with DIC settings.
Map-Based Cloning
tni in the Col-0 background was crossed to Ler, and the F2 individuals resembling tni were selected. Genomic DNA was extracted from the inflorescence of these individuals using a Nucleon Phytopure kit (GE HealthCare). Polymorph (www.polymorph.weigelworld.org) and dCAPSFinder (http://helix.wustl.edu/dcaps/dcaps.html) software were used to design CAPS and dCAPS markers (Supplemental Table S1). The tni locus was mapped to a 65-kb genomic region with the help of these markers and 509 tni mapping individuals. Among 21 genes in this interval, the candidate genes were amplified and cloned into the pGEM-T-Easy vector (Promega), followed by Sanger DNA sequencing.
Generation of Constructs and Transgenic Lines
The TNI coding sequence was amplified by PCR from complementary DNA (cDNA) using primers P1888 and P1889 (Supplemental Table S2). To generate the TNI overexpression construct 35S::TNI, the coding sequence was cloned downstream of the 35S CaMV promoter in pCAMBIA1302. TNIintron of amplicon size 800 bp was amplified from tni cDNA with primers P1888 and P1774 harboring BglII and BsmI restriction sites and cloned into pGEM-T-Easy-TNI to make pGEM-T-Easy-TNIintron. With BglII and BsmI restriction enzymes, the first 414 bp of TNI in 35S::TNI (pCAMBIA1302) was replaced with TNIintron from pGEM-T-Easy-TNIintron to generate 35S::TNIintron (pCAMBIA1302). The 35S promoter in 35S::TNIintron was replaced with the RPS5a promoter, which was amplified with primers P2230 and P2231 harboring BamHI and BglII sites to obtain RPS5a::TNIintron.
Artificial microRNA against TNI was designed according to the protocol described earlier (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi). Briefly, primers P1918, P1919, P1920, and P1921 were used to clone the amiR fragment and subsequently cloned into pGEM-T-Easy vector. Primers P2887 and P2888 were used to amplify amiR-TNI and cloned into pCAMBIA1302. The 35S CaMV promoter of pCAMBIA1302 was replaced with the RPS5a promoter to generate RPS5a::amiR-TNI (amiR-TNI). The RPS5a promoter sequence was amplified with P2231 and P2238. For pPLT7::TNIintron, the 35SCaMV promoter was replaced with the 1.5-kb pPLT7 in pCAMBIA1302 and pCAMBIA1390 using the primers P2801 and P2802. The coding sequence of TNIintron was cloned downstream of the pPLT7 in pCAMBIA1302 to make the construct pPLT7::TNIintron. The PLT7::amiR-TNI construct was generated similarly. To make the transcriptional fusion of TNI, a 1.9-kb region upstream of the TNI locus was amplified using primers P2759 and P2760 and cloned into pDONR221. The promoter sequence was cloned into the pMDC162 destination vector to make the pTNI::GUS construct.
Agrobacterium tumefaciens GV3101 was transformed with these constructs by electroporation. Flowering Arabidopsis plants were transformed with A. tumefaciens harboring individual constructs through the floral dip method (Clough and Bent, 1998).
TNI CDS was cloned into pGEX-4T-1 using primers P1912 and P1913 with engineered BamH1 and SalI restriction sites, respectively, to create the pGEX4-T-1-TNI construct. pGEX4T-1-TNIwas replaced with TNIintron using SacI and HindIII restriction enzymes to make the pGEX4T-1-TNIintron construct. The TNIC317S mutant was generated using the Q5 site-directed mutagenesis kit (New England Biolabs). pGEM-T-Easy-TNI served as a template for making the site-directed mutant using primers P2346 and P2347. The TNI fragment in pGEM-T-Easy-TNI was replaced with TNIC317S using BglII and SpeI to create pGEM-T-Easy-TNIC317S. From this vector, the TNIC317S coding sequence was moved to pGEX4-T-1 using BamH1 and SalI restriction enzymes for protein expression. The pGEX-4T-1-TNI was used as the template to generate truncated proteins ZnF, ΔZnF, ΔUBA, ΔUBA2, and UBA domains using primer pairs P1912 and P2405, P2403 and P1913, P1912 and P2549, P1912 and P2806, and P2827 and P2808, respectively, with engineered BamH1 and Sal1 restriction sites. All these deletion constructs were subsequently cloned into pGEX-4T-1.
Purification of GST-Tagged Recombinant Proteins from Escherichia coli
Recombinant protein expression and purification was done according to a protocol previously described (Harper and Speicher, 2011), with some modifications. Briefly, transformed E. coli(BL21) cells were induced with 0.5 mm isopropylthio-β-galactoside (Sigma-Aldrich) at midlog phase, incubated at 16°C for 12 h, harvested by centrifugation at 4,000g for 5 min at 4°C, and suspended in lysis buffer (50 mm Tris-HCl [pH 7.4], 150 mm KCl, 0.5 mm EDTA, 10% [v/v] glycerol, 0.5% [v/v] NP-40, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail), followed by sonication until it turned clear. Clear supernatant was collected after centrifugation at 12,000g for 20 min at 4°C and incubated with glutathione beads (Novagen) for 2 h at 4°C with constant shaking. Beads were washed five times with ice-cold cell suspension buffer, and bound protein eluted with 2 mm glutathione. SDS-PAGE was used to analyze the proteins.
In Vitro and In Vivo Deubiquitination Assays
Deubiquitination assays were performed as described earlier (Rao-Naik et al., 2000). Briefly, a 50-μL assay reaction comprising purified protein, 50 mm Tris-HCL (pH 8), 150 mm NaCl, 10 mm β-mercaptoethanol, 0.5 mg mL−1 BSA, and 2 μg of 2- to 7-mer of polyubiquitin (Boston Biochemical) was incubated at 37°C for 3 h. Laemmli buffer (200 mm Tris-HCl [pH 6.8], 10% [w/v] SDS, 30% [v/v] glycerol, 0.05% [v/v] bromophenol blue, and β-mercaptoethanol) was added to stop the reaction. The assay mixture was fractionated by SDS-PAGE and immunoblot analysis was performed using anti-ubiquitin antibody (Novus Biologicals). For in vivo assays, E. coli was cotransformed with constructs along with the p8190-UBQ10 and pACYC184-Ub4 constructs (Rao-Naik et al., 2000), induced with 0.5 mm isopropylthio-β-galactoside, and incubated for 3 h at 37°C. Equal cell aliquots were pelleted down, suspended into lysis buffer, and sonicated until clear. Supernatant was boiled in Laemmli buffer and analyzed by SDS-PAGE. Immunoblotting was done using anti-ubiquitin and anti-His antibodies (Sigma-Aldrich).
In Vitro Substrate Binding Assay
GST-tagged recombinant proteins were purified using glutathione sepharose beads, as mentioned in the previous section. Five to 50 μL bead-bound protein was incubated with 1 μg Lys-48-linked tetra-ubiquitin (Boston Biochemicals) in sodium phosphate (pH 7.4) in the presence of protease inhibitor cocktail (Roche). The reaction mixture was incubated at 4°C with shaking for 2 h, then stopped by the addition of Laemmli buffer and by boiling the sample for 10 min. The reaction mixture was fractionated by SDS-PAGE, and immunoblotting was performed using anti-ubiquitin antibody.
GUS Staining and MUG Assay
GUS staining was performed as described earlier (Karidas et al., 2015). Seedlings were harvested in ice-cold 90% (v/v) acetone and incubated on ice for 20 min and then in staining buffer (0.5 m sodium phosphate buffer [pH 7.2], 10% [v/v] Triton-X, 100 mm potassium ferrocyanide, and 100 mm potassium ferricyanide) for 20 min at 25°C. Then, 2 mm X-Gluc (Thermo Scientific) was added to the fresh staining buffer containing seedlings and incubated at 37°C for up to 12 h, followed by washing with 70% (v/v) ethanol for 30 to 60 min at 25°C until chlorophyll was removed, mounting on a glass slide in lactic acid, and observation under a DIC microscope.
The 4-Methylumbelliferyl-b-D-glucuronide (MUG) assays were performed according to the previously described protocol (Weigel and Glazebrook, 2002). Briefly, protein was extracted from seedlings using extraction buffer (50 mm sodium phosphate [pH 7.0], 10 mm EDTA, 0.1% [w/v] SDS, 0.1% ]v/v] TritonX-100, 1 mm PMSF, and protease inhibitor cocktail) and centrifuged at 12,000g at 4°C for 15 min. The clear supernatant was then collected, and an equal concentration of proteins was taken for the MUG assay. The MUG assay buffer composition is that of the extraction buffer supplemented with 1 mm MUG (Sigma-Aldrich). Reactions were incubated at 37°C for 20 min and stopped using 0.2 m sodium carbonate. A TECAN fluorimeter was used to measure the fluorescence at 365-nm excitation and 455-nm emission wavelengths.
Antibody Generation
Anti-TNI polyclonal antibody was raised against a synthetic peptide (USV) corresponding to residues 156 to 174 of TNI/UBP14, which was identified as a potent immunogen using the software available at http://tools.iedb.org/main/bcell/. Five milligrams of synthetic peptide with >85% purity was conjugated to the Keyhole limpet hemocyanin carrier. Rabbits were immunized with 1 mg conjugated peptide, followed by three booster immunizations each with500 μg conjugated peptide. Postimmunization antiserum was collected from the rabbits.
Immunoblot Analysis
Proteins were isolated from 7- to 8-d-old seedlings using protein extraction buffer (50 mm Tris-HCl [pH 7.4], 300 mm KCl, 0.5 mm EDTA, 10% [v/v] glycerol, and 0.5% [v/v] NP-40) along with 1 mm PMSF, 50 μm MG132, and complete protease inhibitor cocktail (Roche). Extract was cleared by centrifugation at 12,000g for 15 min at 4°C. Equal concentrations of protein were analyzed in 15% or 10% (v/v) SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore), and immunoblotted using anti-GFP (Roche), antiubiquitin, anti-Lys-48 ubiquitin (Cell Signaling Technology), and anti-Lys-63 (Enzo Life Sciences) antibodies. Enhanced chemiluminescent (Millipore) was used to develop the blots.
Confocal Microscopy
Roots were stained with 10 μg mL−1 propidium iodide (Sigma-Aldrich) for 5 min, mounted on a glass slide, and observed under a laser confocal microscope (LSM 710, Zeiss).
RNA Isolation and cDNA Synthesis
Total RNA was extracted from 7-d-old seedlings using Trizol (Sigma-Aldrich), treated with DNase (Fermentus) for 2 h at 37°C, and precipitated by sodium acetate. Then, 2 μg RNA was used as a template for cDNA preparation using Revert Aid M-MuLV reverse transcriptase (Fermentus), after which 20-μL reverse transcription reactions were set up and the amplified products were visualized using ethidium bromide staining of 1% (w/v) agarose gels.
Accession Numbers
Sequence data for this article can be found in the Arabidopsis Genome Initiative (www.arabidopsis.org) under accession numbers: At3G23030 (IAA2), At1G04250 (AXR3), At1G51950 (IAA18), At5G20730 (ARF7), At2G38120 (AUX1), At3G62980 (TIR1), At1G73590 (PIN1), and At3G20630 (TTN6).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Proembryo and cotyledon phenotype in tni.
Supplemental Figure S2. Differentially expressed auxin-related genes in tni.
Supplemental Figure S3. Cloning of the tni locus.
Supplemental Figure S4. Tissue-specific expression of the TNI promoter.
Supplemental Figure S5. Effects of TNI misexpression on lateral roots.
Supplemental Figure S6. TNIintron is catalytically inactive.
Supplemental Figure S7. Accumulation of polyubiquitinated proteins in the tni mutant.
Supplemental Figure S8. In vitro disassembly of ubiquitin conjugates from tni plant extract.
Supplemental Figure S9. Expression of recombinant TNI protein in E. coli.
Supplemental Figure S10. PIN1::GUS reporter assay in tni.
Supplemental Table S1. List of markers used for fine mapping of the TNI locus, including their nature and position in the genome.
Supplemental Table S2. List of primers used for generating constructs.
Acknowledgments
We thank B. Vijaya Lakshmi Vadde (Indian Institute of Science) for the anti-TNI antibody, M. Sowmya Spandana (Indian Institute of Science) for help in making clones, Jason Reed (University of North Carolina) for IAA18:GUS, Judy Callis (University of California, Davis) for the ScUBP14, Ub6, and His-Ub4 constructs, Ben Scheres (Wageningen University) for the DR5::GUS and DR5::nYFP lines, Kalika Prasad (IISER Thiruvananthapuram) for the pPLT7 construct, and P. Ajit Kumar (Indian Institute of Science) for access to a Zeiss microscope.
Footnotes
This work was supported by the Ministry of Human Resource Development, Government of India (fellowships to P.M. and P.K.), the Department of Science and Technology-Fund for Improvement of Science and Technology Infrastructure, University Grants Commission Centre for Advanced Study, and the Department of Biotechnology-India Institute of Science Partnership Program Phase-II (grant no. BT/PR27952/INF/22/212/2018 to U.N.).
References
- Amerik Ayu, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M(1997) In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J 16: 4826–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Z, Liu Y, Ou Y, Li J, Zhang B, Sun D, Sun Y, Tang W(2018) Regulation of the stability of RGF1 receptor by the ubiquitin-specific proteases UBP12/UBP13 is critical for root meristem maintenance. Proc Natl Acad Sci USA 115: 1123–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR(1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J 8: 505–520 [Google Scholar]
- Berleth T, Mattsson J, Hardtke CS(2000) Vascular continuity and auxin signaling. Trends Plant Sci 5: 387–393 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B(2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Boscá S, Knauer S, Laux T(2011) Embryonic development in Arabidopsis thaliana: From the zygote division to the shoot meristem. Front Plant Sci 2: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Callis J.(2014) The ubiquitination machinery of the ubiquitin system. The Arabidopsis Book 12: e0174, doi:10.1199/tab.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, et al. (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davière JM, Achard P(2013) Gibberellin signaling in plants. Development 140: 1147–1151 [DOI] [PubMed] [Google Scholar]
- Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK(2009) Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem 284: 5030–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I.(2012) Lateral root initiation: One step at a time. New Phytol 193: 867–873 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD(2001) The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J 27: 393–405 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M(2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Gendreau E, Orbovic V, Höfte H, Traas J(1999) Gibberellin and ethylene control endoreduplication levels in the Arabidopsis thaliana hypocotyl. Planta 209: 513–516 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M(2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G(2002) The Arabidopsis Bodenlos gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G(1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Harper S, Speicher DW(2011) Purification of proteins fused to glutathione S-transferase. Methods Mol Biol 681: 259–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Liu NY, Bandyopadhyay A, Estelle M(2000) The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127: 23–32 [DOI] [PubMed] [Google Scholar]
- Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K(2010) Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137: 63–71 [DOI] [PubMed] [Google Scholar]
- Isono E, Nagel MK(2014) Deubiquitylating enzymes and their emerging role in plant biology. Front Plant Sci 5: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko MG, Napsucialy-Mendivil S, Dubrovsky JG(2010) Auxin-induced inhibition of lateral root initiation contributes to root system shaping in Arabidopsis thaliana. Plant J 64: 740–752 [DOI] [PubMed] [Google Scholar]
- Jeong JS, Jung C, Seo JS, Kim JK, Chua NH(2017) The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 29: 1406–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karidas P. (2014) Map-based cloning and characterization of TARANI, a global regulator of Arabidopsis development. PhD thesis. Indian Institute of Science, Bangalore, India
- Karidas P, Challa KR, Nath U(2015) The tarani mutation alters surface curvature in Arabidopsis leaves by perturbing the patterns of surface expansion and cell division. J Exp Bot 66: 2107–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD(2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Křeček P, Skůpa P, Libus J, Naramoto S, Tejos R, Friml J, Zažímalová E(2009) The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M(1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10: 403–413 [DOI] [PubMed] [Google Scholar]
- Leyser O.(2018) Auxin signaling. Plant Physiol 176: 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang F, Zhang H, He H, Ma L, Deng XW(2008) Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J 55: 844–856 [DOI] [PubMed] [Google Scholar]
- Luo M, Luo MZ, Buzas D, Finnegan J, Helliwell C, Dennis ES, Peacock WJ, Chaudhury A(2008) UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 180: 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B(2014) PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar P, Nath U(2020) De-ubiquitinases on the move: An emerging field in plant biology. Plant Biol (Stuttg) 22: 563–572 [DOI] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G(2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T(2003) Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol 131: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen TET, Komander D(2017) Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem 86: 159–192 [DOI] [PubMed] [Google Scholar]
- Nelson SK, Steber CM (2016) Gibberellin hormone signal perception: Down-regulating DELLA repressors of plant growth and development. In Hedden P, Thomas SG, eds, The Giberellins, Annual Plant Reviews, 49 John Wiley and Sons, Hoboken, NJ, pp 153–188 [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M(2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CW, Ryu KY(2014) Cellular ubiquitin pool dynamics and homeostasis. BMB Rep 47: 475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploense SE, Wu MF, Nagpal P, Reed JW(2009) A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Grigg SP, Barkoulas M, Yadav RK, Sanchez-Perez GF, Pinon V, Blilou I, Hofhuis H, Dhonukshe P, Galinha C, et al. (2011) Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr Biol 21: 1123–1128 [DOI] [PubMed] [Google Scholar]
- Rao-Naik C, Chandler JS, McArdle B, Callis J(2000) Ubiquitin-specific proteases from Arabidopsis thaliana: Cloning of AtUBP5 and analysis of substrate specificity of AtUBP3, AtUBP4, and AtUBP5 using Escherichia coli in vivo and in vitro assays. Arch Biochem Biophys 379: 198–208 [DOI] [PubMed] [Google Scholar]
- Reed JW.(2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M(1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P(1994) Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120: 2475–2487 [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D(2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ(2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261 [DOI] [PubMed] [Google Scholar]
- Sieburth LE.(1999) Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol 121: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, Bressan RA, Zhu JK(2007) Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738 [DOI] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, et al. (2004) Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HMO, Haseloff J, Beemster GTS, Bhalerao R, Bennett MJ(2005) Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7: 1057–1065 [DOI] [PubMed] [Google Scholar]
- ten Hove CA, Lu KJ, Weijers D(2015) Building a plant: Cell fate specification in the early Arabidopsis embryo. Development 142: 420–430 [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed JW(1999) Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, McElver JA, Liu Cm CM, Yang LJ, Wu JQ, Martinez A, Patton DA, Meinke DW(2002) Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol 128: 38–51 [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H(2008) Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol 49: 1025–1038 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ(1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Deng XW(2011) Plant ubiquitin-proteasome pathway and its role in gibberellin signaling. Cell Res 21: 1286–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J(2002) Arabidopsis, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Woodbury, NY [Google Scholar]
- Xu Y, Jin W, Li N, Zhang W, Liu C, Li C, Li Y(2016) UBIQUITIN-SPECIFIC PROTEASE 14 interacts with ULTRAVIOLET-B INSENSITIVE 4 to regulate endoreduplication and cell and organ growth in Arabidopsis. Plant Cell 28: 1200–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter CM, Wagner D(2014) LEAFY and polar auxin transport coordinately regulate Arabidopsis flower development. Plants (Basel) 3: 251–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD(2000) The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol 124: 1828–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]