Iron–sulfur cluster proteins play a role in plant innate immunity by regulating plant defense-related genes.
Abstract
Iron–sulfur (Fe–S) clusters are inorganic cofactors that are present in all kingdoms of life as part of a large number of proteins involved in several cellular processes, including DNA replication and metabolism. In this work, we demonstrate an additional role for two Fe–S cluster genes in biotic stress responses in plants. Eleven Fe–S cluster genes, including the NITROGEN FIXATION S-LIKE1 (NFS1) and its interactor FRATAXIN (FH), when silenced in Nicotiana benthamiana, compromised nonhost resistance to Pseudomonas syringae pv. tomato T1. NbNFS1 expression was induced by pathogens and salicylic acid. Arabidopsis (Arabidopsis thaliana) atnfs and atfh mutants, with reduced AtNFS1 or AtFH gene expression, respectively, showed increased susceptibility to both host and nonhost pathogen infection. Arabidopsis AtNFS1 and AtFH overexpressor lines displayed decreased susceptibility to infection by host pathogen P. syringae pv. tomato DC3000. The AtNFS1 overexpression line exhibited constitutive upregulation of several defense-related genes and enrichment of gene ontology terms related to immunity and salicylic acid responses. Our results demonstrate that NFS1 and its interactor FH are involved not only in nonhost resistance but also in basal resistance, suggesting a new role of the Fe–S cluster pathway in plant immunity.
Iron–sulfur (Fe–S) clusters are versatile cofactors formed by iron atoms and inorganic sulfide. Several plant metabolic pathways and proteins require Fe–S clusters for proper functioning (Johnson et al., 2005; Balk and Schaedler, 2014). Biosynthesis of Fe–S clusters requires a complex machinery. Until now, around 46 genes involved in Fe–S cluster biosynthesis have been identified in Arabidopsis (Arabidopsis thaliana; Balk and Pilon, 2011). Fe–S clusters are generally linked to the polypeptide chain via the interaction of the iron atoms with Cys residues (Balk and Lobréaux, 2005). Three types of Fe–S cluster biosynthetic systems have been described so far: (1) the nitrogen fixation system from azothropic bacteria; (2) the sulfur mobilization system found in plastids; and (3) the iron–sulfur cluster system, commonly found in bacteria and mitochondria of eukaryotic cells (Lill, 2009; Balk and Pilon, 2011). There are common steps to all systems: first, the release of sulfur from Cys to form Ala; second, the transfer of sulfur to scaffold proteins where Fe–S clusters are assembled; and the third and last step is transfer of the mature Fe–S cluster to an apoprotein. Each of these three steps are mechanistically conserved by the requirement for a Cys desulfurase (Johnson et al., 2005).
The main function of Fe–S proteins is electron transfer through the different oxidation states of iron that can generate redox potential (Balk and Lobréaux, 2005). Several Fe–S cluster proteins contribute to electron transfer in the respiratory electron transport chain (Lill, 2009; Couturier et al., 2013). They are also involved in other cellular processes, including redox and catalytic activity through their enzymes (e.g. aconitase, which can convert citrate to isocitrate; Tong and Rouault, 2007) and regulatory proteins (Fe–S clusters are oxygen stress sensors in bacteria; Kiley and Beinert, 2003). Several transcription factors in bacteria, such as the iron–sulfur cluster regulator and the nitrite-sensitive transcription repressor, have Fe–S cluster cofactors that sense changes in superoxide, Fe–S clusters, and oxygen contents (Rouault, 2006). Defects in Fe–S cluster protein biogenesis have a drastic impact in several metabolic pathways for all organisms, including amino acid biosynthesis, gene expression, DNA replication, DNA repair, protein translation, amino acid metabolism, photosynthesis, respiration, and development (Beinert, 2000, Rouault, 2006; Lill and Mühlenhoff, 2008; Lill, 2009; Rouault, 2012; Stehling and Lill, 2013; Balk and Schaedler, 2014). It has been shown in mammals that Thio modification of transfer RNAs (T-RNAs) require the mitochondrial Cys desulfurase, NITROGEN FIXATION S-LIKE1 (NFS1), as a sulfur donor (Mueller, 2006; Nakai et al., 2017; Braymer and Winge, 2018).
In Arabidopsis, there are two NFS genes coding for Cys desulfurases, a mitochondrial (AtNFS1) and a chloroplastic one (AtNFS2), which share only 18% amino acid identity among them. AtNFS1 and AtNFS2 both have Cys desulfurase activity required for the mobilization of sulfur from Cys that is vital for the biosynthesis of Fe–S clusters in their respective subcellular compartments (Léon et al., 2002; Frazzon et al., 2007; Turowski et al., 2012). However, only the mitochondrial AtNFS1 was shown to be required for cytosolic Fe–S enzyme activity (Bernard et al., 2013). The function of many Fe–S proteins, including AtNFS1, is essential for normal plant development in Arabidopsis. T-DNA knockout mutants of AtNFS1 are embryo-lethal, thus making it challenging for genetic analysis of AtNFS1 (Frazzon et al., 2007; Couturier et al., 2013; Balk and Schaedler, 2014). AtNFS1 is thought to provide sulfur for the biogenesis of Fe–S clusters in plants (Balk and Lobréaux, 2005). It can catalyze the conversion of Cys to Ala, forming sulfane sulfur as a by-product (Balk and Lobréaux, 2005; Balk and Pilon, 2011). Arabidopsis AtNFS1 overexpression lines have increased levels of Fe and S while mutant plants had lower amounts of S, further reinforcing the role of AtNFS1 in Fe and S homeostasis (Armas et al., 2019). The Arabidopsis AtNFS1 protein has been expressed and purified from Escherichia coli and is shown to catalyze the release of sulfide from Cys (Frazzon et al., 2007). Another study confirmed a Cys desulfurase activity for AtNFS1 by showing that it can catalyze l-Ala production in increasing l-Cys concentrations (Turowski et al., 2012). The same study also showed that AtNFS1 binds to an Arabidopsis FRATAXIN homolog (AtFH) using in vitro pull-down, and that addition of AtFH protein increases the catalytic activity of AtNFS1 by 3-fold. FH is a structurally well-conserved protein across different kingdoms of life (Gibson et al., 1996). AtFH showed dual localization in mitochondria and chloroplasts in plants and is known to bind to iron molecules and to be involved in iron homeostasis, Fe–S cluster biosynthesis, and mitochondrial energy metabolism (Turowski et al., 2015). In yeast, FH is associated with mitochondrial membranes and is involved in mitochondrial iron homeostasis. The yeast fh mutant has growth defects, accumulates mitochondrial iron, exhibits a higher sensitivity to oxidative stress, and has reduced mitochondrial respiration (Ristow et al., 2000). In humans, deficiency in FH function due to a point mutation in the FH gene has been related to a degenerative disease known as Friedreich’s ataxia that leads to premature death (Babcock et al., 1997). In Arabidopsis, atfh knockout mutants were shown to be lethal, but a few AtFH knockdown lines are viable and showed retarded growth, reduced fresh weight, and seed number (Busi et al., 2006; Balk and Schaedler, 2014).
In plants, the biological role of Fe–S proteins in genotoxic stress and abiotic stress is gradually emerging (Liang et al., 2014; Iñigo et al., 2016). There are no reports thus far that directly show Fe–S cluster genes’ involvement in plant biotic stress resistance/tolerance. Identification of genes and pathways involved in plant disease resistance is important to develop disease-resistant crops either by breeding or biotechnological approaches. One of the problems of using classical resistance (R) genes in crops to confer disease resistance is their durability because R genes are often effective against a specific pathogen strain. Nonhost resistance (NHR), on the other hand, can be used to confer broad and durable disease resistance (Mysore and Ryu, 2004; Niks and Marcel, 2009; Gill et al., 2015; Lee et al., 2016, 2017). NHR is a type of resistance shown by all plant species against most potential pathogens (nonhost pathogens) and usually results in compromise in, or a failure in, establishing virulence (Heath, 2000; Mysore and Ryu, 2004). Several genes involved in NHR have also been used to confer resistance against economically important diseases in crop plants (for review, see Fonseca and Mysore, 2019).
In this study, we demonstrate the role of Fe–S cluster genes in NHR and plant immunity. We identified 11 Fe–S cluster genes to play a role in NHR by virus-induced gene silencing (VIGS) in Nicotiana benthamiana. We further characterized the role of the mitochondrial AtNFS1 and its interactor AtFH in NHR in Arabidopsis using mutants and overexpression lines that exhibited enhanced susceptibility and increased disease resistance, respectively, against host and nonhost pathogens. This is the first report showing the role of Fe–S cluster genes in combating plant biotic stresses.
RESULTS
VIGS of the NbNFS1 Gene Results in Increased Susceptibility to Nonhost Bacterial Pathogens in N. benthamiana
A forward genetics approach using the Tobacco rattle virus (TRV)-based VIGS system to silence genes induced by a mixed complementary DNA (cDNA) elicitor library (del Pozo et al., 2004; Anand et al., 2007; Senthil-Kumar and Mysore, 2014; Senthil-Kumar et al., 2018) has been used to successfully identify several genes involved not only in NHR but also in host resistance such as gene-for-gene and pathogen associated molecular patterns–mediated immunity (Rojas et al., 2012; Senthil-Kumar et al., 2013; Nagaraj et al., 2016; Kaundal et al., 2017). One of the cDNA clones identified in this screening, NbME26D10 (https://vigs.noble.org), displayed homology to the AtNFS1 gene from Arabidopsis. AtNFS1 is annotated to encode a long (AtNFS1-1) and a short (AtNFS1-2) isoform protein, which likely originated due to alternate splicing. The AtNFS1 amino acid sequence is well conserved across plants, animals, and even prokaryotes, as shown by multiple alignment (Supplemental Fig. S1).
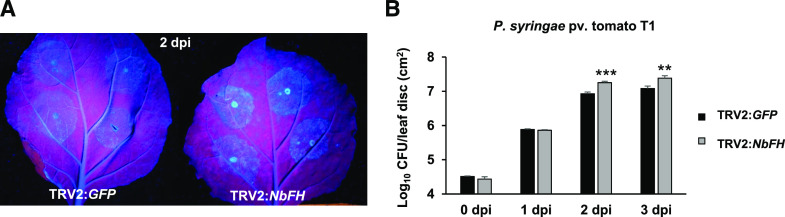
To further confirm that NFS1 plays a role in NHR, two VIGS constructs were generated with distinct regions of the NbNFS1 coding sequence (NbNFS1-a; 522 to 822 bp and NbNFS1-b; 120 to 420 bp), for the downregulation of NbNFS1 in N. benthamiana (Supplemental Fig. S2A). The TRV2:GFP construct was used as a control (GFP has no sequence similarity to plant genomic DNA and thus will not cause gene silencing) to rule out the possibility of TRV or Agrobacterium tumefaciens infection alone causing the observed phenotype. NbNFS1 transcript levels were significantly downregulated by VIGS (Supplemental Fig. S2B). TRV2:PHYTOENE DESATURASE (PDS) was used as a positive control for VIGS (Supplemental Fig. S3). Three weeks after silencing, fully expanded N. benthamiana leaves above the TRV inoculation site were infiltrated with nonhost pathogen Pseudomonas syringae pv. tomato T1 expressing GFPuv (encodes GFP that is optimized for maximal fluorescence when excited by UV light; Wang et al., 2007) using a needleless syringe. We monitored multiplication of P. syringae pv. tomato T1 under UV light. Multiplication of GFPuv-labeled bacteria was more pronounced in NbNFS1-silenced plants compared to control plants 2 d after pathogen inoculation (Fig. 1A). Bacterial multiplication was quantified at 0, 1, 2, and 3 d after infection. NbNFS1-silenced plants were significantly more susceptible compared to the unsilenced control plants upon P. syringae pv. tomato T1 infection at 1, 2, and 3 d after pathogen infection (Fig. 1B). NbNFS1-silenced plants, when compared to the control, were also significantly more susceptible to another nonhost pathogen, P. syringae pv. glycinea at 1, 2, and 3 d after pathogen infection (Supplemental Fig. S3C). To investigate whether the NbNFS1 transcript levels were induced upon pathogen infection, we monitored transcript levels in wild-type N. benthamiana plants at 0 h, 8 h, 1 d, and 3 d after spray inoculation with the nonhost bacterial pathogen P. syringae pv. tomato T1 and the host pathogen P. syringae pv. tabaci. NbNFS1 expression was significantly induced after infection with both host and nonhost pathogens at 3 d post infection (dpi; Fig. 1, C and D). NbNFS1 transcripts were quantified in unsilenced (TRV2:GFP) and NbNFS1-silenced plants after treatment with salicylic acid (SA), a key phytohormone involved in plant defense. Interestingly, NbNFS1 transcripts were significantly upregulated upon 1 mm of SA treatment at 12-h post infection in control plants while, in silenced plants, NbNFS1 transcripts remained unaltered. This indicates that NbNFS1 could be involved in the SA-mediated plant defense pathway (Fig. 1E). We also measured the expression of the SA-mediated defense marker gene, NbPATHOGENESIS RELATED1 (NbPR1), between control and silenced NbNFS1 plants and found it to be significantly reduced in NbNFS1-silenced plants at basal levels without any pathogen treatment (Fig. 1F). Collectively, the above results suggest a role for NbNFS1 in nonhost defense responses in plants and the SA signaling pathway. Because NbNFS1-silenced plants showed disease susceptibility to bacterial nonhost pathogens, we hypothesized that other Fe–S pathway genes may also play a role in NHR.
Figure 1.
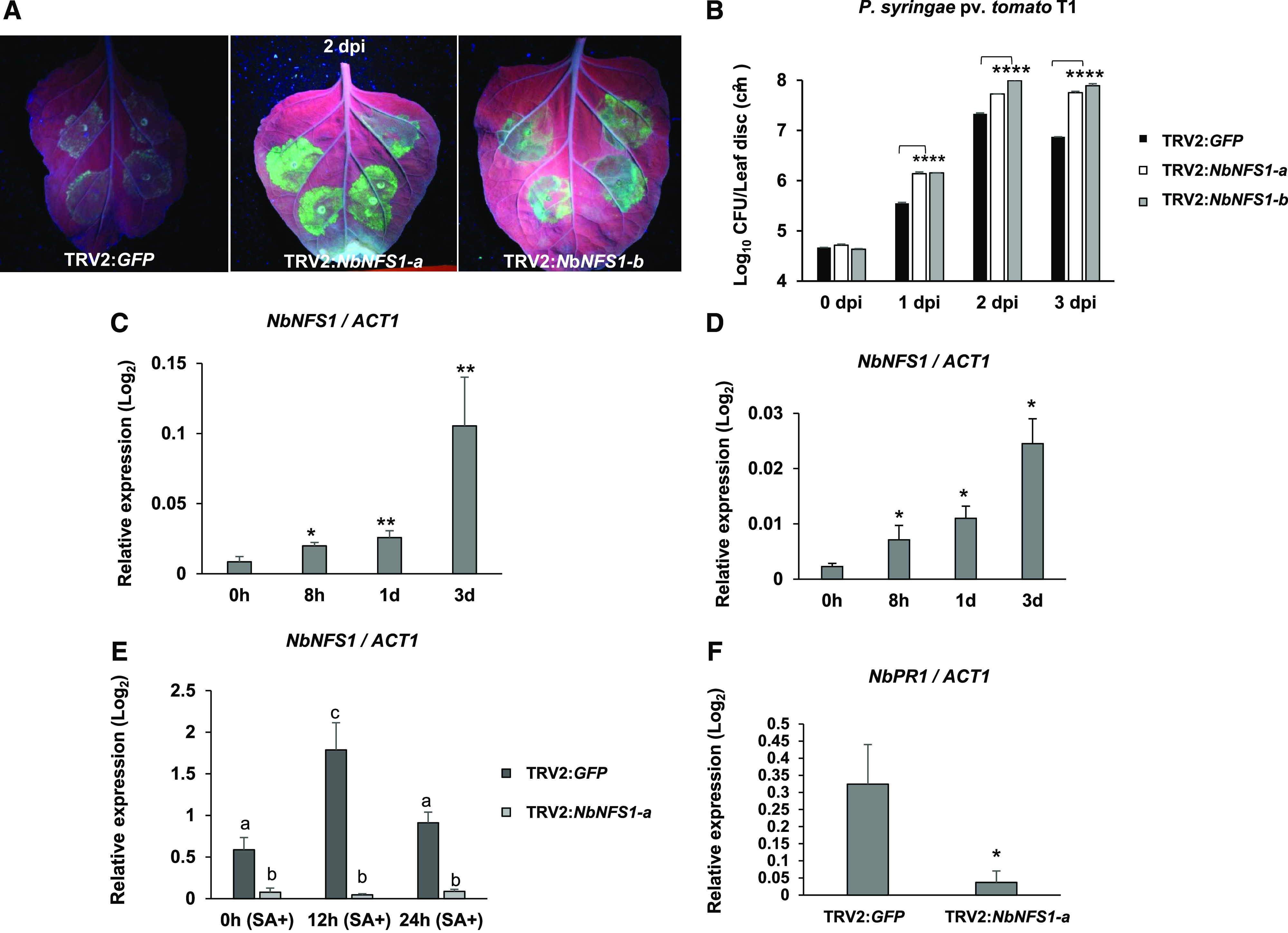
NbNFS1-silenced plants are more susceptible to infection by the nonhost pathogen P. syringae pv. tomato T1 than the control. A, GFPuv-labeled P. syringae pv. tomato T1 accumulation in TRV2:NFS1-a and TRV2:NFS1-b leaves compared to the unsilenced control TRV2:GFP leaf under UV light. A needleless syringe was used to infiltrate the abaxial side of leaves from 6-week-old N. benthamiana plants with P. syringae pv. tomato T1 suspension at 8 × 105 CFU mL−1. Photographs were taken 2 d after infection. B, Quantification of nonhost pathogen multiplication demonstrates a significant increase in bacterial multiplication for NbNFS1-a- and NbNFS1-b-silenced N. benthamiana plants compared to the control. The abaxial side of 6-week-old N. benthamiana leaves was infiltrated with a needleless syringe containing bacterial solution at 8 × 105 CFU/mL−1. The infiltrated region of the leaves was collected at various time points for bacterial quantification. Histograms represent means of four biological replicates. Error bars indicate se. Asterisks indicate significant differences according to one-way ANOVA (****P < 0.0001). All experiments were repeated two times with similar results. C and D, Relative expression of NbNFS1 by RT-qPCR in wild-type N. benthamiana in response to infection by the nonhost bacterial pathogen P. syringae pv. tomato T1 (C) or the host pathogen P. syringae pv. tabaci (D). Three-week-old N. benthamiana plants were sprayed with a bacterial suspension of P. syringae pv. tomato T1 at 8 × 105 CFU mL−1 or P. syringae pv. tabaci at 1.6 × 105 CFU mL−1. Leaf samples were collected at 8 h, 1 d, and 3 d after infection; total RNA was isolated and subjected to RT-qPCR. The expression level was normalized to the ACT1 gene. Histograms represent means of three biological replicates. Error bars represent se. Asterisks represent statistical significance according to Student’s t test (*P < 0.05 and **P < 0.01). All experiments were repeated two times with similar results. E, NbNFS1 expression in NbNFS1-silenced N. benthamiana in response to SA. Six-week–old plants of NbNFS1-silenced (TRV:NbNFS1-a) and control (TRV:GFP) plants were sprayed with 1 mm of SA and leaf samples were collected at various time points after treatment. Total RNA was isolated and subjected to RT-qPCR. The expression level was normalized to the ACT1 gene. Histograms represent means of three biological replicates. Error bars represent se. Lowercase letters represent statistically significant differences according to one-way ANOVA (P value < 0.05). All experiments were repeated two times with similar results. F, NbPR1 expression by RT-qPCR in NbNFS1-silenced N. benthamiana. Total RNA was isolated from 6-week–old NbNFS1-silenced N. benthamiana and nonsilenced control (TRV2:GFP) plants. Histograms represent means of four biological replicates. The expression level was normalized to the ACT1 gene. Error bars represent se. Asterisk represents statistical significance according to Student’s t test (*P < 0.05). All experiments were repeated two times with similar results.
Silencing of Fe–S Cluster Genes in N. benthamiana Compromises Nonhost Resistance
One of the Fe–S genes, NbNFS1, when silenced, resulted in enhanced susceptibility to nonhost pathogens. Therefore, we sought to investigate if other Fe–S cluster pathway genes also play a role in plant defense responses. To do this, we obtained gene sequences for all curated Fe–S cluster genes available in Arabidopsis (Balk and Pilon, 2011; Balk and Lobréaux, 2014). We identified N. benthamiana homologs using a cutoff E value of 1e−10 for these genes using the software BlastX and the N. benthamiana Genome v1.0.1 predicted protein database from the Sol Genomics Network Web site (https://solgenomics.net/; Supplemental Table S1). We then used the Sol Genomics Network VIGS Tool (Fernandez-Pozo et al., 2015) to select a fragment length of ∼300 bp for each N. benthamiana Fe–S cluster homolog and designed primers for Gateway cloning into the TRV2-VIGS vector (Supplemental Table S1).
Twenty-three Fe–S cluster genes were cloned into the TRV2-VIGS binary vector and transformed into A. tumefaciens strain GV2260. VIGS was performed individually for each of the 23 genes in N. benthamiana. Three weeks later, silenced plants were inoculated with a nonhost pathogen as described above for NbNFS1-silenced plants. Eleven out of 23 Fe–S cluster genes, when silenced, compromised NHR (Table 1; Supplemental Fig. S4). Interestingly, a gene that encodes a known interactor of NFS1, NbFH (Turowski et al., 2012), when silenced, also compromised NHR against P. syringae pv. tomato T1 at 2 and 3 d after pathogen infection (Fig. 2). Some of the other Fe–S cluster genes that, when silenced, compromised NHR at 1, 2, and 3 d after pathogen infection, include two mitochondrial ferredoxin genes that display high sequence identity (87%) between them: MITOCHONDRIAL FERREDOXIN1 (MFDX1) and MFDX2. A third mitochondrial ferredoxin gene, PYRIDINE NUCLEOTIDE-DISULFIDE OXIDOREDUCTASE, also displayed compromised NHR when silenced. All three mitochondrial ferredoxins are thought to be involved in electron transfer reactions (Takubo et al., 2003). Other Fe–S cluster-silenced genes that result in increased susceptibility to a nonhost pathogen are GLUTAREDOXIN16 (GRX16), which encodes a plastidial Fe–S cluster assembly machinery enzyme with oxidoreductase activity that can potentially mediate the reduction of disulfide bonds (Garg et al., 2010), and CYTOSOLIC IRON-SULFUR PROTEIN ASSEMBLY1, which encodes a cytosolic Fe–S cluster protein (Supplemental Fig. S4; Supplemental Table S1; Balk et al., 2005; Luo et al., 2012). These results suggest that Fe–S cluster proteins play a role in plant defense, especially for NHR.
Table 1. Fe–S cluster genes silenced in N. benthamiana.
| Gene Name | Proposed Molecular Function | Subcellular Localization | NHR Compromised |
|---|---|---|---|
| NITROGEN FIXATION S-LIKE1 (NFS1) | Cys desulfurase | Mitochondria | Yes |
| FRATAXIN (FH) | Iron donor? | Mitochondria | Yes |
| PYRIDINE NUCLEOTIDE-DISULFIDE OXIDOREDUCTASE (ADXR) | Electron transfer | Mitochondria | Yes |
| MITOCHONDRIAL FERREDOXIN1 (MFDX1) | Electron transfer | Mitochondria | Yes |
| MITOCHONDRIAL FERREDOXIN2 (MFDX2) | Electron transfer | Mitochondria | Yes |
| CYTOSOLIC IRON-SULFUR PROTEIN ASSEMBLY1 (CIA1) | WD40 protein | Cytosolic | Yes |
| NUCLEOTIDE BINDING PROTEIN35 (NBP35) | Scaffold | Cytosolic | Yes |
| DUF59 (CIA3) | DUF59 domain | Cytosolic | Yes |
| SULFUR E1 (SUFE1) | Sulfur mobilization | Plastid | Yes |
| GLUTAREDOXIN16 (GRX16) | Cluster transfer | Plastid | Yes |
| NFU DOMAIN PROTEIN3 (NFU3) | Scaffold | Plastid | Yes |
| ENHANCER7 (CIA2) | DUF59 domain | Cytosolic | No |
| SULFUR E2 (SUFE2) | Activator of NFS2 | Plastid | No |
| SULFUR E3 (SUFE3) | Activator of NFS2 | Plastid | No |
| NFU DOMAIN PROTEIN1 (NFU1) | Scaffold | Plastid | No |
| CHLOROPLAST-LOCALIZED ISCA-LIKE PROTEIN (SUFA) | Cluster transfer | Plastid | No |
| MITOCHONDRIAL COG0354 PROTEIN (IBA57) | Aminomethyltransferase | Mitochondria | No |
| EMBRYO DEFECTIVE 3106 (ERV1) | Sulfhydryl oxidase | Mitochondria | No |
| GOLLUM (NAR1) | Hydrogenase-like | Cytosolic | No |
| ISCU-LIKE3 (ISU3) | Scaffold | Mitochondria | No |
| IRON-SULFUR CLUSTER BIOSYNTHESIS FAMILY PROTEIN (ISA) | Cluster transfer | Mitochondria | No |
| HOMOLOG OF YEAST DRE2 (DRE2) | Ribosomal export | Mitochondria | No |
| HIGH CHLOROPHYLL FLUORESCENCE101 (HCF101) | Chlorophyll-related | Plastid | No |
Figure 2.
NbFH-silenced plants are more susceptible to infection by the nonhost pathogen P. syringae pv. tomato T1 than the control. A, GFPuv-labeled P. syringae pv. tomato T1 accumulation in TRV2:FH leaves compared to unsilenced control TRV2:GFP leaves under UV light. A needleless syringe was used to infiltrate the abaxial side of leaves from 6-week-old N. benthamiana plants with GFPuv-expressing P. syringae pv. tomato T1 at 8 × 105 CFU mL−1. Photographs were taken 2 d after infection. B, Quantification of nonhost pathogen multiplication demonstrates a significant increase in bacterial multiplication for NbFH-silenced plants compared to the control. The undersides of leaves were infiltrated with a needleless syringe containing bacterial solution at 8 × 105 CFU mL−1. Histograms represent means of four independent biological replicates. Error bars indicate se. Asterisks indicate significant differences according to one-way ANOVA (**P < 0.01 and ***P < 0.001).
The Fe–S Cluster Biosynthesis Genes AtNFS1 and AtFH Contribute to Arabidopsis Resistance against Host and Nonhost Pathogens
To determine if the role of Fe–S cluster genes in NHR is conserved in other plant species, we obtained Arabidopsis mutants for AtNFS1 and AtFH. We obtained an AtNFS1 heterozygous mutant from the GABI-KAT collection with a T-DNA insertion located on exon 2 (GABI-KAT 211c08), referred to here as atnfs1-7, and a homozygous SALK line with an insertion located in the 5′ untranslated region (UTR; SALK_083681C), referred to as atnfs1-2 (Supplemental Fig. S5). Because complete loss of NFS1 leads to lethality or severe developmental defects with no viable seed production, we could not obtain a homozygous atnfs1 complete knockout line (Couturier et al., 2013; Balk and Schaedler, 2014). The atnfs1-7 heterozygous mutant showed reduced AtNFS1 expression (Supplemental Fig. S5) and shorter primary root length when compared to wild-type Col-0 plants (Supplemental Fig. S6B). The homozygous atnfs1-2 mutant had reduced AtNFS1 expression (Supplemental Fig. S5). Both atnfs1-7 and atnfs1-2 mutants were inoculated with a host pathogen P. syringae pv. tomato DC3000 and the nonhost pathogens Pseudomonas syringae pv. tabaci and Pseudomonas syringae pv. phaseolicola by flood inoculation (Ishiga et al., 2011, 2017), and samples were collected at 0 h and 3 d after infection for bacterial quantification. In agreement with our previous N. benthamiana NbNFS1-silenced plants (Fig. 1), both atnfs1 mutants were also susceptible to a nonhost pathogen 3 d after pathogen infection (Fig. 3, E and F). Both mutants were also susceptible to the host pathogen P. syringae pv. tomato DC3000 3 d after pathogen infection, when compared to the wild-type plants (Fig. 3, B and C). The observed difference in Log10 colony-forming units (CFUs) between the atnfs1-7 mutant and the wild-type (Fig. 3, B and E), for example, is equivalent to a 3-fold change increase for the atnfs1-7 mutant. The Arabidopsis atnfs1-7 mutant also showed disease-related symptoms such as necrosis and wilting 3 dpi compared to wild-type plants (Fig. 3, A and D). Similar to NbNFS1 (Fig. 1), the AtNFS1 transcript was also significantly induced by the nonhost pathogens and 1 mm of SA in wild-type Col-0 (Supplemental Fig. S7).
Figure 3.
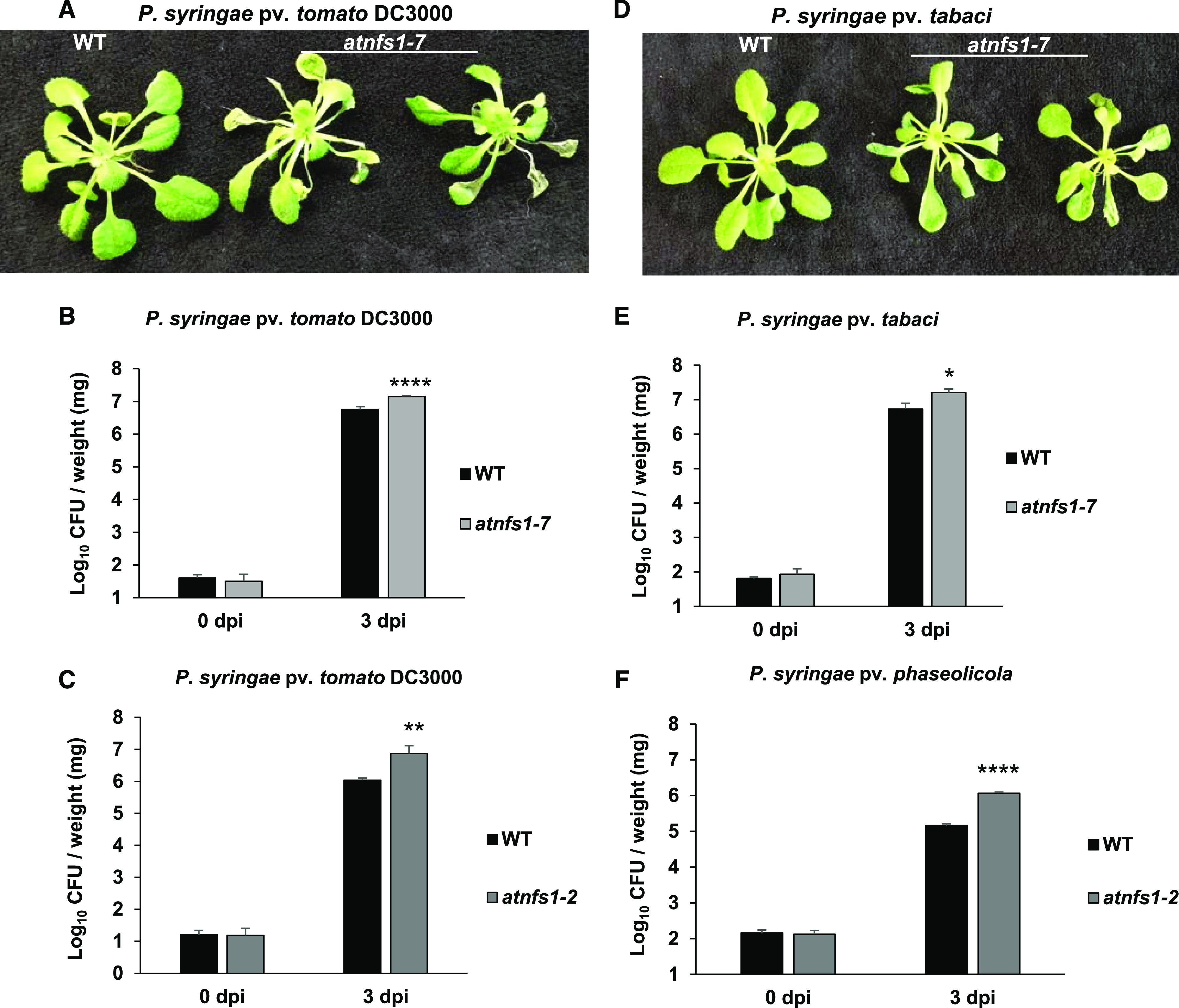
Arabidopsis atnfs mutant lines are more susceptible to host and nonhost pathogens. A, Disease symptoms of the atnfs1-7 mutants upon inoculation with the host pathogen P. syringae pv. tomato DC3000. Three-week-old Col-0 (wild type [WT]) and atnfs1-7 Arabidopsis lines grown on petri plates were flood-inoculated with P. syringae pv. tomato DC3000 at 8 × 104 CFU mL−1. Photographs were taken 3 dpi. B and C, Quantification of host pathogen multiplication in the atnfs1-7 and atnfs1-2 mutant lines. Three-week-old wild-type, atnfs1-7, and atnfs1-2 plants were flood-inoculated with P. syringae pv. tomato DC3000 at 8 × 104 CFU mL−1. Samples (rosettes) were collected at 0 h and 3 dpi for bacterial quantification. Histograms represent means of four biological replicates. Error bars indicate se. Asterisks indicate significant differences according to Student’s t test (**P < 0.01 and ****P < 0.0001). All experiments were repeated two times with similar results. D, Disease symptoms of atnfs1-7 mutants upon inoculation with the nonhost pathogen P. syringae pv. tabaci. Three-week-old wild-type and atnfs1-7 Arabidopsis lines grown on petri plates were flood-inoculated with P. syringae pv. tabaci at 8 × 105 CFU mL−1. Photographs were taken 3 dpi. E, Quantification of nonhost pathogen multiplication in the atnfs1-7 mutant. Three-week–old wild-type and atnfs1-7 mutant plants were flood-inoculated with P. syringae pv. tabaci at 8 × 105 CFU mL−1. Samples (rosettes) were collected at 0 h and 3 dpi for bacterial quantification. Histograms represent means of four biological replicates. Error bars indicate se. Asterisk indicates a significant difference according to Student’s t test (*P < 0.05). All experiments were repeated two times with similar results. F, Quantification of nonhost pathogen multiplication in the atnfs1-2 mutant. Three-week-old wild-type and atnfs1-2 mutant plants were flood-inoculated with P. syringae pv. phaseolicola at 8 × 105 CFU mL−1. Samples (rosettes) were collected at 0 h and 3 dpi for bacterial quantification. Histograms represent means of four biological replicates. Error bars indicate se. Asterisks indicate a significant difference according to Student’s t test (****P < 0.0001). All experiments were repeated two times with similar results.
The susceptibility of two atfh mutants to host and nonhost pathogens was also tested. The homozygous mutant line atfh1 (SALK_054434C), with a T-DNA insertion located in the 3′UTR, and atfh2 (SALK_021263C, 5′UTR), were identified and obtained (Supplemental Fig. S8). Similar to AtNFS1, complete loss of AtFH function in Arabidopsis leads to plant lethality (Busi et al., 2006; Couturier et al., 2013; Balk and Schaedler, 2014). Reverse transcription quantitative PCR (RT-qPCR) analysis showed that expression of AtFH in the atfh1 and atfh2 mutant lines is significantly reduced compared to the wild-type control (Supplemental Fig. S7). We performed a pathogen flood-inoculation assay using both athf1 and atfh2 mutants and wild-type control plants upon infection with the nonhost pathogen P. syringae pv. phaseolicola and the host pathogen P. syringae pv. tomato DC3000. Similar to NbFH VIGS-silenced plants (Fig. 2), the Arabidopsis atfh1 and atfh2 mutant lines were highly susceptible to nonhost pathogens 3 d after pathogen infection when compared to wild-type Col-0 (Fig. 4, A and B). In addition, both atfh1 and atfh2 mutants also supported more host bacterial multiplication when compared to wild-type Col-0 at 3 d after pathogen infection (Fig. 4C). Differences in Log10 CFU observed between the atfh mutants and the wild type is equivalent to a 2-fold change increase for the atfh mutant. (Fig. 4C). Similar to the atnfs1-7 mutant, atfh1 mutant seedlings (10-d-old) also showed slight developmental defects, such as shorter roots at the seedling stage, when compared to wild-type plants (Supplemental Fig. S6A). No significant difference in shoot biomass could be observed for the atfh1 and atnfs1-7 mutants and the wild type in adult plants (23-d-old; Supplemental Fig. S9). These results suggest that both AtNFS1 and AtFH play a role in NHR and basal resistance in Arabidopsis.
Figure 4.
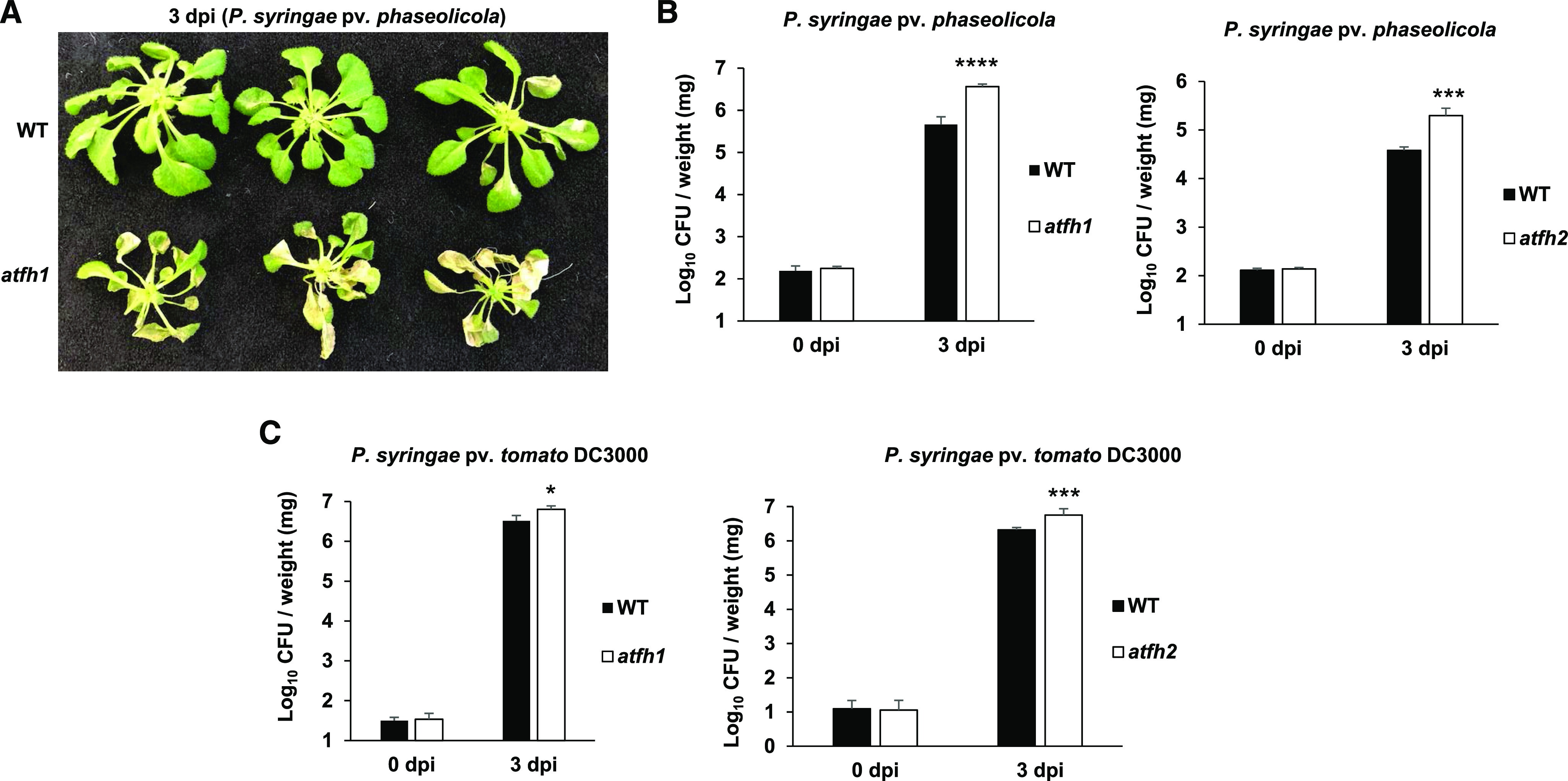
Arabidopsis atfh mutant lines are more susceptible to host and nonhost pathogens P. syringae pv. tomato DC3000 and P. syringae pv. phaseolicola, respectively. A, Disease symptoms of the atfh1 mutant upon inoculation with the nonhost pathogen P. syringae pv. tabaci. Three-week-old wild-type (WT) and atfh1 Arabidopsis lines grown on petri plates were flood-inoculated with P. syringae pv. phaseolicola at 8 × 105 CFU mL−1. Photographs were taken 3 dpi. B, Quantification of nonhost pathogen multiplication in the atfh1 and atfh2 mutant lines. Three-week-old wild-type, atfh1, and atfh2 plants were flood-inoculated with P. syringae pv. phaseolicola at 8 × 105 CFU mL−1. C, Quantification of host pathogen multiplication in the atfh1 and atfh2 mutant lines. Three-week-old wild-type, atfh1, and atfh2 plants were flood-inoculated with P. syringae pv. tomato DC3000 at 8 × 104 CFU mL−1. Samples (rosettes) were collected at 0 h and 3 dpi for bacterial quantification. Histograms represent means of four biological replicates. Error bars indicate se. Asterisks indicate a significant difference compared to the wild type according to Student’s t test (*P < 0.05 and ***P < 0.001). All experiments were repeated two times with similar results.
AtNFS1 and AtFH Overexpression Lines Showed Enhanced Resistance to Infection with the Host Pathogen P. syringae pv. tomato DC3000
The recent annotation of the Arabidopsis genome (Araport11) shows three possible variant coding sequences for AtNFS1: two long isoforms AtNFS1.1 and AtNFS1.3 with 1,362 bp that show identical sequences to each other, and a short isoform AtNFS1.2 with 978 bp and missing 83 amino acid residues from its N-terminal region compared to AtNFS1.1 (Supplemental Fig. S10). AtNFS1.3 is possibly a redundant form and therefore was excluded from this study. Both the long and short AtNFS1 isoforms were amplified from cDNA (Supplemental Fig. S10).
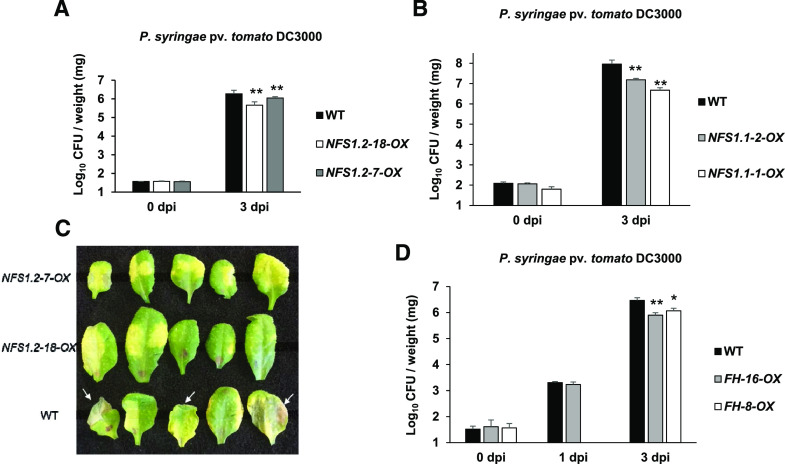
To further confirm AtNFS1’s role in disease resistance, Arabidopsis lines overexpressing either the long AtNFS1.1 isoform or the short AtNFS1.2 isoform were generated. The expression of both AtNFS1.1 and AtNFS1.2 was significantly upregulated by ∼10-fold in their respective overexpression lines (Supplemental Fig. S11). Three- to 4-week-old AtNFS1.1 (NFS1.1-1-OX and NFS1.1-2-OX) and AtNFS1.2 (NFS1.2-7-OX and NFS1.2-18-OX) overexpression lines and the wild type were flood-inoculated (Ishiga et al., 2011) with the host pathogen P. syringae pv. tomato DC3000. Both AtNFS1.1 and AtNFS1.2 overexpression lines were less susceptible to host pathogen infection 3 d after infection in relation to the wild-type control plants, indicating that both the AtNFS1 short and long isoforms have a role in disease resistance (Fig. 5, A and B). Additionally, the overexpression lines also showed fewer disease symptoms, such as wilting and water-soaked lesion formation, compared to wild-type plants upon leaf infiltration using a needleless syringe with P. syringae pv. tomato DC3000 (CFU = 8 × 106) 3 dpi (Fig. 5C). We also tested the growth of the host virulent pathogen Pseudomonas syringae pv. maculicola ES4326 in the short isoform overexpressor lines (NFS1.2-7-OX and NFS1.2-18-OX) and wild-type plants using the flood-inoculation method. As shown in Supplemental Fig. S12, the AtNFS1 overexpression lines exhibited enhanced resistance to P. syringae pv. maculicola ES4326 3 d after pathogen infection (Supplemental Fig. S12) in a similar manner to the resistance observed against P. syringae pv. tomato DC3000 shown above.
Figure 5.
Arabidopsis AtNFS1 and AtFH overexpressor lines are more resistant to the host pathogen P. syringae pv. tomato DC3000. A, Quantification of host pathogen multiplication in AtNFS1 short isoform overexpressor lines. Three-week-old wild-type (WT) and overexpressor of AtNFS1 short isoform (NFS1.2-18-OX and NFS1.2-7-OX) plants were flood-inoculated with P. syringae pv. tomato DC3000 at 8 × 104 CFU mL−1. B, Quantification of host pathogen multiplication in AtNFS1 long isoform overexpressor lines. Three-week-old wild type and overexpressor of AtNFS1 long isoform (NFS1.1-1-OX and NFS1.2-2-OX) plants were flood-inoculated with P. syringae pv. tomato DC3000 at 8 × 104 CFU mL−1. Samples (rosettes) were collected at 0 h and 3 dpi for bacterial quantification. For A and B, histograms represent means of four biological replicates. Error bars indicate se. Asterisks indicate significant differences according to Student’s t test (**P < 0.01). All experiments were repeated two times with similar results. C, Disease symptoms observed in the AtNFS1 overexpressor lines and wild-type plants upon inoculation with a host pathogen. Full-grown leaves of 4-week-old, soil-grown wild-type and AtNFS1 overexpressor (NFS1.2-18-OX and NFS1.2-7-OX) lines were inoculated with P. syringae pv. tomato DC3000 at 8 × 106 CFU mL−1 using a needleless syringe. Photographs were taken 3 d after inoculation. White arrows denote disease symptoms such as wilting and necrosis. D, Quantification of host pathogen multiplication in AtFH overexpressor lines. Four-week-old wild-type and AtFH overexpressor lines (FH-16-OX and FH-8-OX) were flood-inoculated with P. syringae pv. tomato DC3000 at 8 × 104 CFU mL−1. Samples (rosettes) were collected at 0 h and 3 dpi for bacterial quantification. Histograms represent means of four biological replicates. Error bars indicate se. Asterisks indicate significant differences compared to the wild type according to Student’s t test (**P < 0.01 and *P < 0.05).
In addition to AtNFS1 overexpression lines, we also generated two Arabidopsis AtFH overexpression lines (FH-16-OX and FH-8-OX). Upon flood inoculation of both FH-OX lines with the host pathogen P. syringae pv. tomato DC3000, similar to AtNFS1-OX lines both AtFH-OX lines showed reduced disease symptoms and accumulated less bacteria when compared to wild-type Col-0 3 d after pathogen infection (Fig. 5D). We did not observe any obvious phenotypic developmental changes for the AtNFS1 and AtFH overexpression lines. These results indicate that both isoforms of AtNFS1 and AtFH are involved in basal resistance against P. syringae pv. tomato DC3000 in Arabidopsis.
AtNFS1 and AtFH Overexpression or Knockdown Does Not Significantly Impair Shoot Respiration or Biomass in Adult Arabidopsis Plants
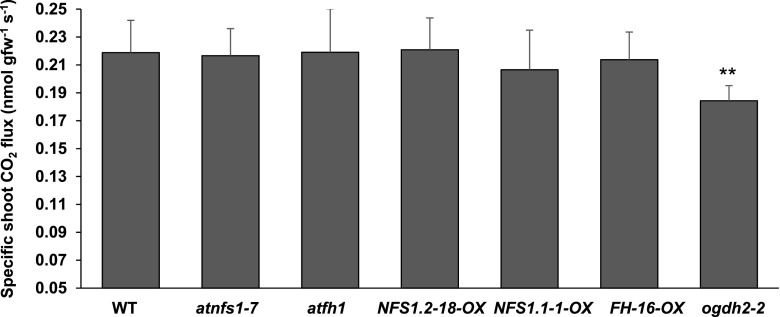
Both AtNFS1 and AtFH localize in the mitochondria, and full knockout lines have impaired growth causing embryo lethality possibly by affecting plant respiration. Knockdown mutants with reduced AtNFS1 or AtFH gene expression, however, are viable, and generate healthy plants. To test a possible effect of the AtNFS1 and AtFH mutations or overexpression on mitochondrial respiration rate, dark respiration of the entire shoot (hereafter shoot respiration) was chosen as the most relevant metric to exclude nonmitochondrial photorespiration (Graham, 1980). We measured shoot biomass and CO2 flux in the wild-type, AtNFS1, or AtFH knockdown lines, AtNFS1 or AtFH overexpression lines, and a E1-2-OXOGLUTARATE DEHYDROGENASE knockout mutant (ogdh2-2, SALK_055824C) as a control (Supplemental Fig. S13), which has been previously shown to have a reduced respiration rate (Condori-Apfata et al., 2019). No significant differences were observed in the shoot respiration rate or biomass among all genotypes tested compared to wild-type plants, with the exception of the control ogdh2-2 mutant (Fig. 6). This suggests that the disease phenotypes observed in the AtNFS1 or AtFH knockdown and overexpression lines are not due to an indirect effect from changes in respiration rates or plant growth.
Figure 6.
Overexpression or knockdown of AtNFS1 and AtFH does not affect shoot dark respiration of 4-week-old Arabidopsis plants. Arabidopsis wild-type (WT; Col-0), atfh1, AtFH-16-OX, atnfs1-7, AtNFS1.2-18-OX, AtNFS1.1-1-OX, and atogdh2-2 seeds were germinated and seedlings were grown in solid one-half strength MS-agar media. The ogdh2-2 mutant was used as a positive control of respiration because it has been shown to reduce dark respiration. After 5 d of growth, evenly germinated seedlings were transferred to a Falcon 6-well culture plate containing liquid one-half strength MS media. Ten plants were pooled together per well to make a biological replicate with a total of five biological replicates per genotype. After another 18 d of growth in liquid media, the plants were transferred to darkness 1 h before CO2 flux measurements using a LI-850 CO2/water gas analyzer. Error bars represent se of five biological replicates from two different experiments. Asterisks denote statistical significance according to Student’s t test (**P < 0.01). Experiments were repeated two times with similar results.
AtNFS1 Is Ubiquitously Expressed in Several Organs in Arabidopsis
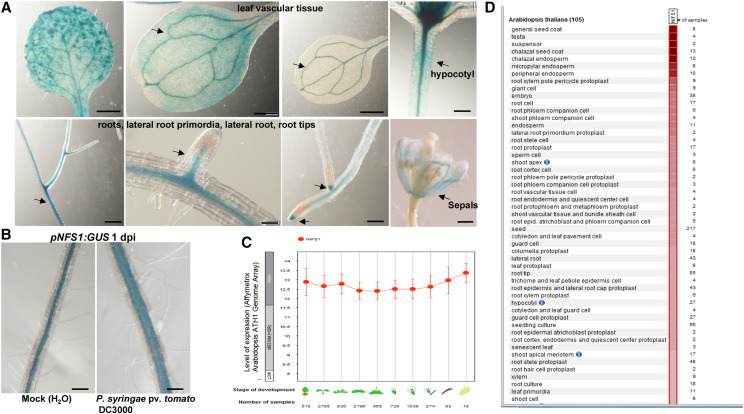
To study the expression pattern of AtNFS1 in various Arabidopsis tissues, a 906-bp promoter fragment (from +7 coding sequence [CDS] to −991 bp) of AtNFS1 was amplified by PCR, and cloned into a binary vector for generation of a GUS reporter construct, pNFS1:GUS. The expression pattern of the GUS gene under the control of the AtNFS1 promoter was determined by GUS histochemical staining of 2-week-old homozygous stable transgenic plants expressing pNFS1:GUS. AtNFS1 is highly expressed in all plant tissues according to publicly available gene expression data from the tool Genevestigator (Fig. 7, C and D). Consistent with the gene expression data, we observed GUS expression in most plant organs including leaf vasculature, leaf veins, the hypocotyl, primary and lateral roots, emerging lateral roots, and at the root tips (Fig. 7A). We further confirmed whether AtNFS1 expression is induced by a host pathogen by infecting pNFS1:GUS expressing plants with P. syringae pv. tomato DC3000. Surprisingly, we did not observe any difference in GUS staining in the leaves, but we did observe increased GUS staining in the roots of plants drenched with the bacterial pathogen suspension (Fig. 7B).
Figure 7.
Tissue-specific expression of AtNFS1 in Arabidopsis. A, Histochemical staining of different organs (arrows) of transgenic Arabidopsis seedlings expressing pNFS1:GUS. One-month–old pNFS1:GUS-expressing Arabidopsis plants were stained with X-gluc. Photographs of leaf vasculature, hypocotyl, roots, lateral roots, root tips, and flower bud sepals were taken 24 h after staining using a stereo microscope. Scale bars = 200 μm for roots and hypocotyl, 500 μm for sepals, and 1 mm for leaves. B, GUS expression in roots of pNFS1:GUS-expressing transgenic plants in response to host pathogen infection. Three-week-old wild-type (Col-0) and pNFS1:GUS-expressing plants were drenched with a 30-mL bacterial suspension of host pathogen P. syringae pv. tomato DC3000 at 8 × 106 CFU mL−1 and plants were stained with X-gluc. Photographs were taken under a stereo microscope 24 h after staining. Scale bars = 200 μm. C and D, Expression levels of the AtNFS1 gene from publicly available gene expression data at different developmental stages and organs in Arabidopsis using the software Genevestigator.
AtNFS1 Localizes to Mitochondria
Sequence prediction analysis of AtNFS1 indicated a mitochondrial localization (score of 0.83), and also a possible chloroplastic localization, albeit with a low score (0.16) according to the program Multiloc2 (Blum et al., 2009). AtNFS1 was previously shown to localize to mitochondria although no mitochondrial marker was used (Frazzon et al., 2007). To confirm the subcellular localization of AtNFS1, this time using a mitochondrial colocalization protein marker, and to rule out a possible chloroplast localization, we fused the CDS of AtNFS1 (AtNFS1.1) with GFP and cloned this into an A. tumefaciens binary vector. AtNFS1:GFP was transiently expressed in N. benthamiana along with a plant mitochondrial marker, mCherry (Nelson et al., 2007). Two days after the coinfiltration, leaves were observed under a confocal microscope and colocalization of AtNFS1:GFP and mCherry could be observed specifically in the mitochondria but not in the chloroplast (Supplemental Fig. S14).
Overexpression of AtNFS1 Results in Upregulation of Several Defense-Related Genes
To understand the mechanism of reduced susceptibility to host pathogens in AtNFS1 overexpression lines, we used the NFS1.2-18-OX line as a representative line to perform RNA sequencing (RNA-seq) upon P. syringae pv. tomato DC3000 infection. The aerial part of 5-week–old Arabidopsis seedlings was collected from three plants (n = 3) each for wild-type and NFS1.2-18-OX line, at different time points (0, 3, and 5 dpi) upon flood-inoculation, with P. syringae pv. tomato DC3000 (CFU = 1.6 × 105). Total RNA was isolated from these samples. Individual RNA-seq libraries were prepared, and uniquely indexed for Illumina sequencing. Differentially expressed genes (DEGs) were identified using the criteria: Log2 fold change > 1 and < −1; false discovery rate (FDR) < 0.05, for the NFS1.2-18-OX line and the wild type upon P. syringae pv. tomato DC3000 infection. Based on these criteria, a total of 129 DEGs were identified for all time points between the NFS1.2-18-OX line and the wild type (Fig. 8A; Supplemental Table S2). A total of 85 upregulated DEGs for all time points were identified in the NFS1.2-18-OX line compared to the wild type (Fig. 8A; Supplemental Table S3). Subsequently, these 85 upregulated DEGs were functionally classified according to gene ontology (GO) terms using the program AgriGO2 (Tian et al., 2017; Supplemental Table S4). To identify the overrepresented functional categories, a singular enrichment analysis (SEA) was performed. SEA compares each annotated gene to all annotated expressed genes. In total, 46 GO terms displayed overrepresentation for biological processes (FDR < 0.001; Supplemental Fig. S15A; Supplemental Table S4). Interestingly, 12 overrepresented GO terms belong to defense-related processes such as immune response, innate immune response, response to biotic stimulus, systemic acquired resistance, and SA-mediated signaling pathway. We also found 29 GO terms overrepresented (FDR < 0.05) for the same dataset of 85 upregulated DEGs using the software PANTHER (Fig. 8B; Supplemental Table S5; Mi et al., 2019). Similar to AgriGO2, the same defense-related GO pathways were found to be enriched using PANTHER such as response to SA, systemic acquired resistance, and immune responses (Fig. 8B). In addition to defense-related processes, we also observed overrepresentation of GO terms related to Fe–S clusters such as metal ion binding, cofactor binding, and adenyl nucleotide binding (Supplemental Fig. S15B; Supplemental Table S4). Additionally, using AgriGO2 and PANTHER, we also identified overrepresented GO terms from other biological processes that may also play a role in plant defense such as response to oxygen and response to stress (Supplemental Tables S4 and S5).
Figure 8.
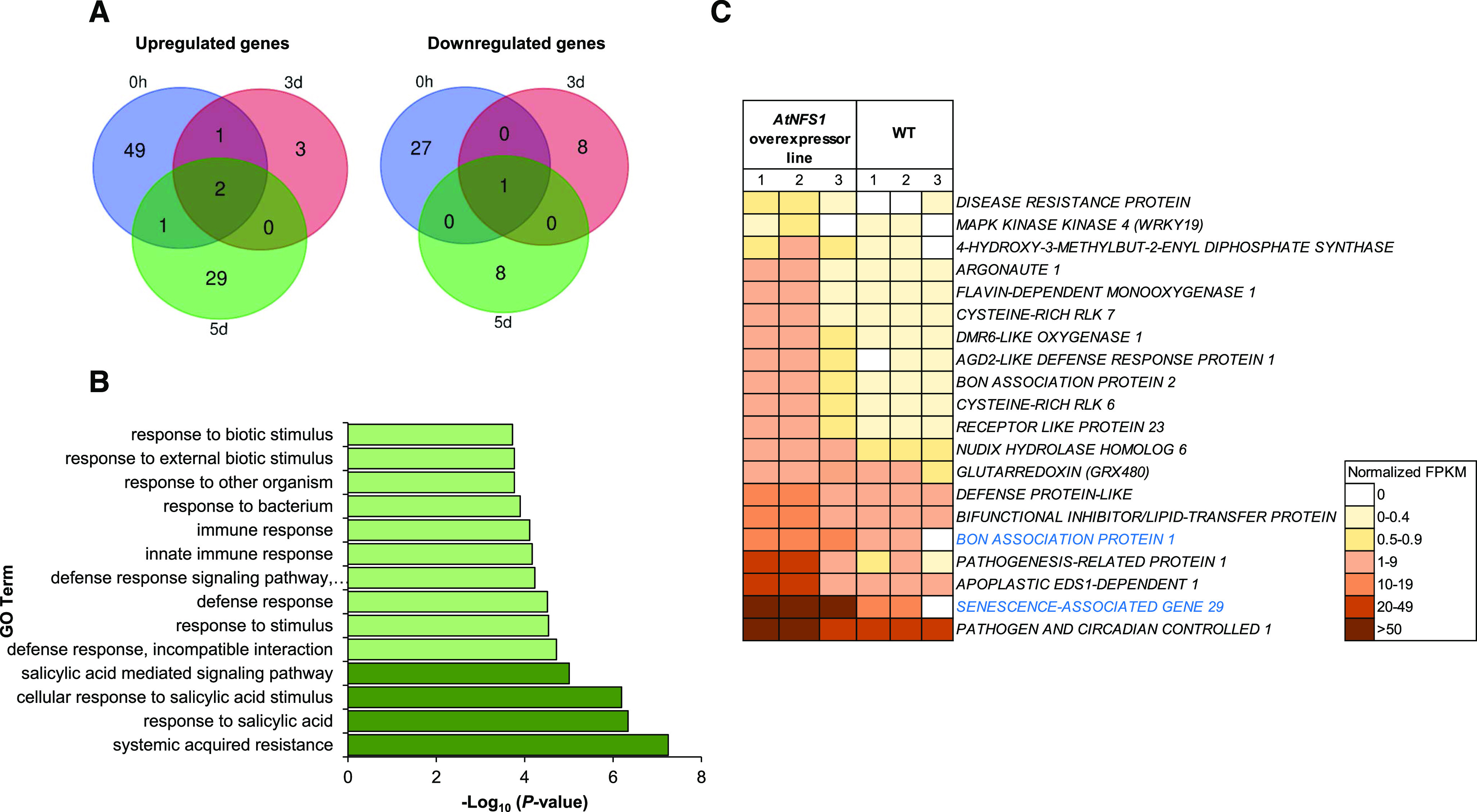
RNA-seq analysis showing upregulation of defense- and SA-responsive genes between the NFS1 overexpressor line and wild-type (WT) plants upon P. syringae pv. tomato DC3000 infection. A, The aerial part (leaf and stem) of 5-week-old Arabidopsis seedlings was collected from three plants (n = 3) for two genotypes (wild type and NFS1.2-18-OX), at different time points (0, 3, and 5 d) upon flood-inoculation with the host pathogen P. syringae pv. tomato DC3000 (1.6 × 105 CFU mL−1). RNA was isolated from the samples and subjected to RNA-seq. The Venn diagram depicts DEGs (Log2 fold change > 1, < −1; FDR < 0.05) identified between a NFS1 overexpressor line and the wild type for each time point. B, GO enrichment analysis of DEGs from RNA-seq data. The bar chart shows all the significantly enriched defense-related GO terms in the NFS1 overexpressor line compared to wild-type plants with the P-value cutoff as 0.05. SA-related GO terms appear in dark green. C, Heatmap showing raw, normalized FPKM values from upregulated defense-related genes identified in the AtNFS1 overexpressor line in relation to the wild type. Heatmap of 20 upregulated genes color-coded from white to brown according to increasing FPKM values. Eighteen defense-related genes belong to the 0-h time point (gene names shown in black) while only two genes belong to the 5-d time point (gene names highlighted in blue). The color legend represents raw FPKM-normalized values.
Strikingly, a total of 53 upregulated DEGs were identified at the 0-h basal control (fold change > 1; FDR < 0.05) in the NFS1.2-18-OX line compared to the wild type (Supplemental Table S6). Interestingly, most of the defense-related DEGs upregulated at all time points in the NFS1.2-18-OX line compared to the wild type belonged to the 0-h time point (18 out of 20; Fig. 8C). Additionally, a total of 28 downregulated DEGs were identified at the 0-h time point between the NFS1.2-18-OX line and the wild type (Fig. 8A; Supplemental Table S7), but no molecular functions related to defense responses were found; other GO terms like hydrolase activity and binding were identified from these DEGs.
We validated the differential expression of seven upregulated defense-related genes (0-h time point) from our RNA-seq dataset by RT-qPCR (Supplemental Fig. S16) and confirmed their increased expression in the NFS1.2-18-OX line compared to the wild type in the absence of pathogen infection. Overall, these results suggest that the overexpression of AtNFS1 constitutively upregulates several defense-related genes. Even though several defense-response–related genes were constitutively upregulated in the NFS1 overexpressor line compared to the wild type, we did not see any developmental defects like stunting or yield-penalty phenotypes associated with it.
DISCUSSION
Fe–S proteins carry out the synthesis and assembly of Fe–S clusters mainly in plastids, mitochondria, and in the cytosol of plants. Fe–S clusters are protein cofactors involved in catalysis, electron transport, and sensing of ambient conditions required for several plant physiological processes such as amino acid metabolism, DNA replication, and development (Couturier et al., 2013). In this study, we show a new role for the Fe–S cluster pathway in plant innate immunity. Using a VIGS-mediated forward genetics screening in N. benthamiana, we identified the CYS DESULFURASE coding gene, NbNFS1, which, when silenced, compromised NHR against the bacterial pathogens tested. Expression of NbNFS1 was induced by the host pathogen, nonhost pathogens, and SA. NbPR1 expression was decreased at 0 h after infection in the NbNFS1-silenced plants compared to the unsilenced control. Both atnfs1-7 and atnfs1-2 mutants from Arabidopsis also showed increased susceptibility to host and nonhost pathogen infection compared to wild-type plants. Arabidopsis AtNFS1 overexpressors exhibited resistance against the host pathogen P. syringae pv. tomato DC3000. AtNFS1 transcript was significantly induced upon nonhost pathogen infection and SA treatment in wild-type plants similarly to N. benthamiana NbNFS1. This result is consistent with an earlier report showing induction of soybean (Glycine max) GmNFS1 expression in roots upon treatment with SA (Heis et al., 2011). Furthermore, we showed that the nuclear-encoded AtNFS1:GFP colocalized in mitochondria along with the mitochondria tracker mCherry. This rules out a possible plastidial localization for AtNFS1, and further confirms previous studies (Frazzon et al., 2007).
In addition to NbFS1, we show that 11 out of 23 other Fe–S cluster genes tested also play a role in NHR in N. benthamiana. Along with NFS1, we also further characterized its interactor, FH (Frazzon et al., 2007; Turowski et al., 2015), for its role in basal and NHR in N. benthamiana and Arabidopsis. Among the 11 Fe–S cluster genes found to be involved in NHR, here we highlight a few for their possible roles in plant defense. Mitochondrial ferredoxin1 (NbMFDX1) and NbMFDX2 and a ferredoxin reductase (NbMFDXR) are involved in a variety of redox reactions in the cell, electron transfer, and reduction of cytochrome c in the presence of NADP (NADPH; Takubo et al., 2003). These cellular processes have been shown to be important for plant defense responses such as hypersensitive response (Dayakar et al., 2003). We also identified a member of the glutaredoxin family, GLUTARREDOXIN16 (NbGRX16), which resulted in increased susceptibility to nonhost pathogens when silenced. Glutaredoxins are known to mediate redox regulation of proteins because of their capacity to catalyze disulfide bonds. GRX480 (encoded by At1g28480) has been shown to interact with TGACG SEQUENCE-SPECIFIC BINDING PROTEIN (TGA) transcription factors in a yeast two-hybrid screening, using TGA as bait (Ndamukong et al., 2007). TGA transcription factors are well-known regulators of PR genes because they are known to bind the defense nonexpresser of PR gene1 protein (NPR1; Niggeweg et al., 2000; Kesarwani et al., 2007). Further, it was shown that GRX480 transcripts were significantly induced by SA (Ndamukong et al., 2007). We also investigated the role of the AtNFS1 interactor protein, AtFH, in disease resistance. Reduced expression of FH in N. benthamiana and Arabidopsis made the plants more susceptible to both host and nonhost pathogens. Overexpression of AtFH in Arabidopsis reduced bacterial accumulation of the host pathogen P. syringae pv. tomato DC3000 when compared to wild-type Col-0. FHs in general are known to bind iron molecules and are involved in iron homeostasis, respiration, Fe–S cluster biogenesis, and redox control (Turowski et al., 2012).
Transcriptome analysis of the AtNFS1 overexpressor line compared to the wild type identified several upregulated DEGs involved in plant defense responses in the AtNFS1 overexpressor line compared to the wild-type control. Several GO terms belonging to plant immunity were found to be significantly overrepresented in the upregulated gene list (Supplemental Table S8). For example, the pathogenesis-related gene PR1, PATHOGEN AND CIRCADIAN CONTROLLED1, which is upregulated in response to both virulent and avirulent strains of P. syringae pv. tomato (Mir et al., 2013), and the systemic acquired resistance-related MONOOXYGENASE1, were upregulated in the AtNFS1 overexpressor line compared to the wild type (Fig. 8C; Supplemental Table S3). Among the top five upregulated DEGs with the highest fold change, we found the genes encoding Cys-rich receptor-like kinases 6 and 7, which are involved in pathogen-induced activation of defense responses against host pathogen infection (Yeh et al., 2015). Most defense-related upregulated DEGs identified here belong to the 0-h time point, suggesting that AtNFS1 has a role in basal disease resistance. We did not observe any significant differences in terms of developmental phenotype or size between the AtNFS1 overexpressor lines and wild-type plants. This is probably because the upregulation of defense genes was moderate (<5-fold) in contrast to the constitutive expresser of PR genes mutants, for example, which have constitutive expression of PR genes causing severe stunting (Bowling et al., 1994; Clarke et al., 1998). This moderate upregulation of defense-related genes may suggest that NFS1 overexpression would be a good potential strategy for generating disease-resistant crops.
To address a possible pleiotropic effect in AtNFS1 or AtFH overexpressor or mutant lines resulting in disease-resistant or -susceptible phenotypes, we tested whether knockdown mutants and overexpressors of AtFH and AtNFS1 differ in shoot respiration or growth. We found no significant differences in shoot respiration or fresh shoot biomass in adult plants among all the lines tested. This suggests that the observed disease phenotypes in AtNFS1 or AtFH overexpressor or mutant lines are not an indirect effect due to changes in respiration rates from mitochondrial defects, but likely stem from immunity regulation changes. The lack of difference observed in respiration rates and fresh biomass for adult plants between all genotypes is in agreement with previous shoot biomass data collected for the AtNFS1 overexpressor or knockdown lines in Arabidopsis (Armas et al., 2019).
In agreement with the upregulation of NFS1 transcripts both in Arabidopsis and N. benthamiana after SA treatment, we also found several genes involved in SA signaling to be upregulated in the AtNFS1 overexpression line compared to the wild type (Supplemental Fig. S15A; Supplemental Tables S3 and S7). The gene AGD2-LIKE DEFENSE RESPONSE PROTEIN1 was induced by 5.5-fold at the 0-h time point compared to the wild type in the AtNFS1 overexpressor line. AGD2-LIKE DEFENSE RESPONSE PROTEIN1 has been shown to be required for the accumulation of SA besides affecting basal plant defense responses (Cecchini et al., 2015). The above-mentioned PATHOGEN AND CIRCADIAN CONTROLLED1 (Mir et al., 2013) and the chloroplastic Fe–S cluster binding ferredoxin 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE SYNTHASE are SA-responsive genes and the later has been shown to modulate SA-mediated disease resistance (Supplemental Tables S3 and S8; Gil et al., 2005). These data suggest that AtNFS1 may play a role in SA-mediated plant defense signaling.
In conclusion, the genetic and transcriptomic data, in combination with disease phenotype assays presented here, suggest that the Fe–S cluster interactor proteins NFS1 and FH are involved not only in NHR but also in basal resistance. We propose a new role for Fe–S cluster genes in plant immunity responses that has been overlooked until now. Our results also suggest that NFS1, its interactor FH, and possibly other Fe–S cluster genes, could be new targets for crop breeding and genetic engineering to generate durable disease-resistant varieties.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) plants were grown on one-half strength Murashige and Skoog (MS) media in a controlled chamber maintained at a 22°C constant temperature and 50% relative humidity with a 12-h/12-h day/night cycle. For experiments involving soil-grown plants, agar-germinated plants were transferred to pots containing Metro-Mix 360 (Sun Gro Horticulture) at 2 weeks after germination and maintained at the same conditions as described above in a growth chamber. Nicotiana benthamiana plants were grown in pots with Metro-Mix 360 in the greenhouse under a 16-h/8-h day/night photoperiod. Mutants characterized in this study are as follows: SALK_054434C (atfh1), SALK_021263C (atfh2), Gabi-Kat_211c08 (atnfs1-7), SALK_083681C (atnfs1-2), and SALK_055824C (ogdh2-2). Seeds were obtained from Arabidopsis Biological Resource Center stock (https://abrc.osu.edu/) and belong to the SALK T-DNA collection (http://signal.salk.edu/cgi-bin/tdnaexpress; Alonso et al., 2003). The Arabidopsis atnfs1-7 mutant was obtained from the Gabi-Kat collection (https://www.Gabi-Kat .de/). All SALK and Gabi-Kat lines were grown in one-half strength MS media plates containing kanamycin (50 μg mL−1) and sulfadiazine (8 μg mL−1), respectively, and genotyped using PCR to confirm homozygosity (Supplemental Table S9).
Cloning and Arabidopsis Transformation
The CDS of AtNFS1 and AtFH were amplified by PCR from Arabidopsis cDNA (1 μg) with Phusion High-Fidelity DNA Polymerase (New England Biolabs), purified using a QIAquick PCR Purification Kit (Qiagen) to remove any primer dimers, and incubated with BP clonase (Invitrogen) to generate an entry vector (pDONR207) by transformation of TOP10-competent (Invitrogen) cells. The product from BP reaction was introduced into the following binary vectors containing the Cauliflower mosaic virus promoter (CaMV 35S) that drives the gene construct: pMDC32 for overexpression lines (NFS1-OX) and pMDC83 (AtNFS1:GFP) via LR reaction (LR clonase; Invitrogen), and then transformed into Agrobacterium tumefaciens strain GV3101 for stable transformation in Arabidopsis using the floral-dip method (Clough and Bent, 1998). All cloned genes were checked using Sanger sequencing. The 907-bp promoter region of AtNFS1 was amplified by PCR from genomic DNA (50 ng) and cloned into the GUS reporter binary vector pMDC162 by GATEWAY cloning and stably-transformed into Arabidopsis in the same way as described above for GUS histochemical studies. All primers used in this work can be found in Supplemental Table S1.
VIGS
Fragments from Fe–S cluster genes (∼300 bp) were amplified by PCR from N. benthamiana cDNA and cloned (GATEWAY) into entry vector pMDC207 followed by LR cloning into the VIGS TRV2 vector in the same way as described in the above section. Plasmids were transformed into A. tumefaciens strain GV2260 for VIGS. A normalized library obtained from mixed elicitors-treated N. benthamiana leaves (Anand et al., 2007; Senthil-Kumar et al., 2014) was used to screen genes involved in NHR.
VIGS of Fe–S cluster genes was performed in N. benthamiana as described in Senthil-Kumar and Mysore (2014). Briefly, A. tumefaciens strain GV2260 carrying TRV2 with the gene of interest (GOI) to be silenced (TRV2:GOI), TRV2:GFP, TRV2:PDS were grown overnight in LB media with appropriate antibiotics. Cultures were harvested by centrifugation and resuspended in infiltration buffer (10 mm of MES and 150 μM of acetosyringone at pH 5.6). Samples were incubated for 3 h, and resuspended in 10 mm of MES buffer before infiltration. Suspensions of TRV2:GOI and TRV1 were mixed in a 1:1 ratio to a final optical density (OD600nm) of 0.4 before infiltration into 3-week-old N. benthamiana plants and allowed another 3 weeks for silencing to occur. TRV2:PDS was used as an overall positive control for every VIGS-silencing experiment, and TRV2:GFP was used as the unsilenced control.
Pathogen Infection Assays
We used the flood-inoculation method that mimics natural infection for our disease assays in Arabidopsis (Uppalapati et al., 2008; Ishiga et al., 2011, 2017). In brief, 4-week-old plants grown in one-half strength MS media were incubated for 5 min with 40 mL of a bacterial suspension with 0.015% (v/v) Silwet l-77, containing either one of the following strains: Pseudomonas syringae pv. tomato DC3000 (host), P. syringae pv. tabaci (nonhost), and P. syringae pv. phaseolicola (nonhost) at the concentrations of 8 × 104, 1.6 × 106, and 8 × 105 CFUs, respectively. Disease symptoms were observed at 3 or 5 d after infection. We collected infected tissues at 0 h, 1 d, and 3 d after the infection and weighed each plant for normalization of CFU. Four plants per time point (n = 4) per genotype were used. Whole seedlings were placed in tubes containing grinding beads and 200 μL of sterile water and ground for 1 min to obtain a homogeneous suspension. Suspensions were 10-fold serially diluted for plating. Aliquots of 10 μL were then plated on KB medium with the appropriate selection antibiotic and incubated at 28°C for 2 d after which CFUs were counted (Durrant et al., 2007; Fonseca and Dong, 2014). For disease symptom visualization, we used a CFU of 8 × 106 and infiltrated fully grown leaves of 4-week-old soil-grown Arabidopsis plants using a needleless syringe. Disease symptoms were observed and recorded 24 h to 48 h after infiltration.
For pathogen infection assays in N. benthamiana, VIGS-silenced and control plants were infiltrated with a needleless syringe at the abaxial part of the leaf with a bacterial suspension containing the following nonhost pathogens: P. syringae pv. tomato T1 and Pseudomonas syringae pv. glycinea at 8 × 105 CFU. We collected tissues at 0, 1, 2, and 3 d after infection. Two leaf-disks of 10-mm diameter were collected per plant from at least five plants (n = 5) using a 0.5-cm2 borer. Leaf discs were placed in tubes containing grinding beads and 500 μL of sterile distilled water, and ground to obtain a homogeneous suspension. Suspensions of 20 μL were transferred to a plate containing 180 μL of sterile distilled water and 10× serial dilutions were made. Aliquots of 10 μL were then plated on KB medium with the appropriately selected antibiotic and incubated at 30°C for 2 d, after which CFUs were counted. We monitored bacterial growth using GFPuv-labeled P. syringae strains 2 to 3 dpi in N. benthamiana. Statistical analyses were done using Student’s t test of the differences between two means of log-transformed data or by one-way ANOVA using the software GraphPad Prism (GraphPad Software).
Subcellular Localization
To confirm mitochondrial localization of AtNFS1, we coinfiltrated N. benthamiana leaves with A. tumefaciens strain GV2260 carrying a binary vector pMDC83 that has the CaMV 35S promoter driving AtNFS1-GFP and another A. tumefaciens GV2260 strain with the red fluorescent organelle marker protein mCHERRY fused to a mitochondrial targeting sequence from yeast (Saccharomyces cerevisiae), cytochrome oxidase IV (Nelson et al., 2007). Briefly, A. tumefaciens strains carrying the above-mentioned constructs were grown overnight and centrifuged at 13,000 rpm to pellet bacteria. The pellet was resuspended and incubated in induction buffer (10 mm of MES and 150 µm of acetosyringone at pH 5.6) for 4 h with gentle shaking. Samples were centrifuged and the pellet was resuspended in 10 mm of MES buffer to a final OD600nm = 0.15 each. Both A. tumefaciens strains were mixed at a 1:1 ratio and used to infiltrate leaves of 4-week–old greenhouse-grown N. benthamiana plants using a needleless syringe. Two d after infiltration, the infiltrated area of the leaves were observed with an inverted model no. TCS-SP8-X point scanning confocal laser-scanning microscope (Leica Microsystems) equipped with a 40× water-immersion objective. GFP was excited with the 488-nm line of the white light laser and emission was detected at 493 to 551 nm. mCherry was excited with the 587-nm line of the Argon laser and emission was detected at 604 to 661 nm.
Histochemical GUS Staining
GUS histochemical staining of Arabidopsis transgenic lines expressing pNFS1:GUS was done using the chromogenic substrate X-gluc according to Hull and Devic (1995). Whole Arabidopsis plants or specific tissue were submerged in 2 mm of X-gluc solution in 100 mm of sodium phosphate at pH 7.0, 10 mm of EDTA, 1 mm of K3Fe(CN)6, and 0.1% (v/v) Triton X-100; vacuum was applied for 2 min, then tissues were incubated at 37°C overnight. The following day, the tissues were cleared by successive washes in 70% (v/v) ethanol to remove chlorophyll for visualization. Images of GUS staining were taken with the model no. SZX12 stereo microscope (Olympus) using the transmitted light.
RNA Extraction and RT-qPCR
Total RNA was isolated from 50 mg of leaves using an RNeasy Mini Kit (Qiagen), followed by TURBO DNase treatment (Invitrogen). At least 1 μg of RNA was used for cDNA synthesis with SuperScript III reverse transcriptase (Invitrogen), performed according to the manufacturer’s instructions. Amplicons were amplified from cDNA using gene-specific primers (Supplemental Table S1), and their relative gene expression values were normalized to the AtACTIN2 gene for Arabidopsis samples and the NbACTIN1 (ACT1) gene for N. benthamiana using the comparative CT method (Livak and Schmittgen, 2001). qPCR was performed in a Bio-Rad instrument (Eppendorf) using SYBR Green (Applied Biosystems) to monitor double-stranded DNA synthesis. The program used to perform reactions was as follows: 95°C for 5 mins, followed by 40 cycles of extension at 72°C for 20 s, and annealing at 60°C for 1 min. To assess amplicon quality, the melting curve was generated at 65°C until 95°C with 0.5°C increments for 5 s.
RNA-Seq Experiment
Total RNA from Arabidopsis was isolated from 60 mg of leaf tissue using a Spectrum Plant Total RNA Kit (Sigma-Aldrich), and residual DNA was removed from each RNA sample by DNase treatment using a TURBO DNA-free Kit (Thermo Fisher Scientific) and then purified with an RNeasy MinElute Cleanup Kit (Qiagen). The quality and profile of the RNA samples was checked on a model no. 2100 Bioanalyzer (Agilent Technologies) using an RNA 6000 Nano Kit (Agilent Technologies). The RNA samples were quantified on a Qubit 2.0 Fluorometer with a Qubit RNA BR Assay Kit (Invitrogen). RNA-seq libraries were prepared using a TruSeq Stranded mRNA Sample Prep Kit (Illumina). Briefly, mRNA was purified from 1 μg of total RNA, fragmented, and then converted to double-stranded DNA for sequencing. Individual libraries were uniquely indexed using TruSeq RNA Single Indexes (Illumina) and pooled in an equimolar ratio. The pooled libraries were sequenced on a Hiseq4000 sequencing machine (Illumina).
The sequencing quality of the reads generated were examined by the software FastQC (v0.10.1; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and only high-quality samples were used. The filtered clean reads were mapped to the Arabidopsis genome (Araport11, release 2016 downloaded from https://www.araport.org/) to estimate raw counts and effective read lengths with the tool Salmon v0.7.2 (Patro et al., 2017). All samples displayed a high mapping rate to the Arabidopsis genome (>95%). Raw counts from Salmon were normalized across all samples by median normalization, and differential expression was estimated using the DESeq2 module from the package Bioconductor R (Love et al., 2014) and an in-house Perl script. Gene expression was quantified as fragments per kilobase of exon per million fragments mapped (FPKM) values, and identified DEGs were required to have an FDR < 0.05. The software DESeq2 (Love et al., 2014) was further used to perform differential expression analysis, which included dynamic filtering of low expressed genes to prevent spurious identification of DEGs. Hierarchical tree graphs for biological process and molecular function using overrepresented GO terms from all DEGs were obtained by SEA, using the program AgriGO2 (Tian et al., 2017). GO terms were declared “enriched” when FDR < 0.001. Overrepresented GO terms using the same DEGs dataset was confirmed using PANTHER default FDR < 0.05 (Mi et al., 2019).
Shoot Respiration
Arabidopsis wild-type (Col-0), ogdh2-2, atfh1, FH-16-OX, atnfs1-7, NFS1.2-18-OX, and NFS1.1-1-OX seeds were germinated and seedlings were grown in solid one-half strength MS-agar media. After 5 d of growth, evenly germinated seedlings were transferred to a Falcon 6-well culture plate (Corning) with each well containing liquid one-half strength MS media. Nine plants were pooled together per well to make a biological replicate with a total of five biological replicates per genotype. After another 18 d of growth on liquid media, the plants were transferred to darkness 1 h before CO2 flux measurements by wrapping an entire plate in aluminum foil. Shoots were severed from the roots, then the fresh weight of the shoots was recorded before transferring the shoots into a light- and air-tight chamber having a volume of 7.5 cm3. The respiration measurements were performed using a LI-850 CO2/water gas analyzer (LI-COR Biosciences) in a dark room with a supplemental photosynthetically inactive yellow LED light. The chamber was connected to the LI-850 instrument, buried in a bead bath with the temperature set at room temperature (23°C), and the CO2 flux was measured with an observation duration of 2 min 30 s using the software LI-8x0 v1.02 (Li-Cor Biosciences). Specific shoot respiration rate (nmol CO2 gfw−1 s−1) was calculated from the raw CO2 flux data using R (v.3.6.0; https://www.r-project.org/), where a linear regression was used to calculate the CO2 flux rate with dead band set at 60 s, and then divided by total shoot fresh weight.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative (https://www.arabidopsis.org/) or GenBank (https://www.ncbi.nlm.nih.gov/genbank/) databases under the following accession numbers: AtNFS1 (AT5G65720) and AtFH (AT4G03240). For all accession codes related to Fe–S cluster genes discussed in this article, please refer to Supplemental Table S1.
Supplemental Material
The following supplemental materials are available.
Supplemental Figure S1. Amino acid sequence alignment of AtNFS1.
Supplemental Figure S2. Two distinct VIGS-silenced regions of the NbNFS1 gene show a significant transcript downregulation.
Supplemental Figure S3. Silencing of the NbNFS1 gene in N. benthamiana enhances multiplication of nonhost pathogens.
Supplemental Figure S4. Several VIGS-silenced genes from the Fe–S cluster pathway are more susceptible to P. syringae pv. tomato T1 infection compared to the unsilenced control.
Supplemental Figure S5. The atnfs1-7 and atnfs1-2 mutants have reduced AtNFS1 expression compared to the wild type.
Supplemental Figure S6. The Fe–S cluster mutants atfh and atnfs1-7 affect primary root length in Arabidopsis.
Supplemental Figure S7. AtNFS1 expression is induced by a nonhost pathogen and SA.
Supplemental Figure S8. The atfh1 and atfh2 mutants have decreased expression of AtFH compared to the wild type.
Supplemental Figure S9. The atfh1 and atnfs1-7 mutants do not display significant changes compared to wild-type plants in fresh shoot biomass in adult Arabidopsis plants.
Supplemental Figure S10. Amplicons of the long (AtNFS1.1) and short (AtNFS1.2) isoforms of AtNFS1.
Supplemental Figure S11. Relative expression of the AtNFS1 gene in the wild-type and AtNFS1 overexpression lines.
Supplemental Figure S12. Arabidopsis AtNFS1 short isoform overexpression lines display decreased virulent pathogen growth (P. syringae pv. maculicola ES4326) compared to the wild type.
Supplemental Figure S13. The ogdh2-2 mutant is a complete knockout.
Supplemental Figure S14. Subcellular localization of AtNFS1 protein.
Supplemental Figure S15. Several GO terms related to defense responses are significantly enriched for upregulated DEGs between the NFS1.2-18-OX line and the wild type.
Supplemental Figure S16. Validation of DEGs identified with RNA-seq by RT-qPCR.
Supplemental Table S1. Identification of Fe–S cluster genes in N. benthamiana by sequence similarity to Arabidopsis Fe–S cluster genes.
Supplemental Table S2. All DEGs identified in the NFS1.2-18-OX line compared to the wild type (Log2 fold change > 1, < −1; FDR = 0.05).
Supplemental Table S3. All upregulated DEGs in the NFS1.2-18-OX line compared to the wild type.
Supplemental Table S4. Overrepresented GO terms for all upregulated genes using Agri-GO in the NFS1.2-18-OX line compared to the wild type.
Supplemental Table S5. Overrepresented GO terms for all upregulated genes using PANTHER in the NFS1.2-18-OX line compared to the wild type.
Supplemental Table S6. DEGs upregulated at 0-h basal control in the NFS1.2-18-OX line compared to the wild type.
Supplemental Table S7. All downregulated DEGs in the NFS1.2-18-OX line compared to the wild type.
Supplemental Table S8. DEGs related to SA and defense response overrepresented in the NFS1.2-18-OX line compared to the wild type according to the software PANTHER.
Supplemental Table S9. All primers used in this work.
Acknowledgments
We thank Yuhong Tang, Guifen Li, and Nick Krom for support with omics data, and Sunhee Oh for providing technical support. We also thank Jin Nakashima and Jaydeep Kolape for the support with microscopy and Janie Gallaway for greenhouse support.
Footnotes
This work was supported by Noble Research Institute, LLC.
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anand A, Vaghchhipawala Z, Ryu CM, Kang L, Wang K, del-Pozo O, Martin GB, Mysore KS(2007) Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Mol Plant Microbe Interact 20: 41–52 [DOI] [PubMed] [Google Scholar]
- Armas AM, Balparda M, Turowski VR, Busi MV, Pagani MA, Gomez-Casati DF(2019) Altered levels of mitochondrial NFS1 affect cellular Fe and S contents in plants. Plant Cell Rep 38: 981–990 [DOI] [PubMed] [Google Scholar]
- Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J(1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Balk J, Aguilar Netz DJ, Tepper K, Pierik AJ, Lill R(2005) The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron–sulfur protein assembly. Mol Cell Biol 25: 10833–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Lobréaux S(2005) Biogenesis of iron–sulfur proteins in plants. Trends Plant Sci 10: 324–331 [DOI] [PubMed] [Google Scholar]
- Balk J, Pilon M(2011) Ancient and essential: The assembly of iron–sulfur clusters in plants. Trends Plant Sci 16: 218–226 [DOI] [PubMed] [Google Scholar]
- Balk J, Schaedler TA(2014) Iron cofactor assembly in plants. Annu Rev Plant Biol 65: 125–153 [DOI] [PubMed] [Google Scholar]
- Beinert H.(2000) Iron-sulfur proteins: Ancient structures, still full of surprises. J Biol Inorg Chem 5: 2–15 [DOI] [PubMed] [Google Scholar]
- Bernard DG, Netz DJ, Lagny TJ, Pierik AJ, Balk J(2013) Requirements of the cytosolic iron–sulfur cluster assembly pathway in Arabidopsis. Philos Trans R Soc Lond B Biol Sci 368: 20120259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum T, Briesemeister S, Kohlbacher O(2009) MultiLoc2: Integrating phylogeny and gene ontology terms improves subcellular protein localization prediction. BMC Bioinformatics 10: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X(1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braymer JJ, Winge DR(2018) Sulfur from within: Cytosolic tRNA thiouridinylation. Cell Chem Biol 25: 645–647 [DOI] [PubMed] [Google Scholar]
- Busi MV, Maliandi MV, Valdez H, Clemente M, Zabaleta EJ, Araya A, Gomez-Casati DF(2006) Deficiency of Arabidopsis thaliana frataxin alters activity of mitochondrial Fe–S proteins and induces oxidative stress. Plant J 48: 873–882 [DOI] [PubMed] [Google Scholar]
- Cecchini NM, Jung HW, Engle NL, Tschaplinski TJ, Greenberg JT(2015) ALD1 regulates basal immune components and early inducible defense responses in Arabidopsis. Mol Plant Microbe Interact 28: 455–466 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Liu Y, Klessig DF, Dong X(1998) Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10: 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Condori-Apfata JA, Batista-Silva W, Medeiros DB, Vargas JR, Valente LML, Heyneke E, Pérez-Diaz JL, Fernie AR, Araújo WL, Nunes-Nesi A(2019) The Arabidopsis E1 subunit of the 2-oxoglutarate dehydrogenase complex modulates plant growth and seed production. Plant Mol Biol 101: 183–202 [DOI] [PubMed] [Google Scholar]
- Couturier J, Touraine B, Briat JF, Gaymard F, Rouhier N(2013) The iron–sulfur cluster assembly machineries in plants: Current knowledge and open questions. Front Plant Sci 4: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayakar BV, Lin HJ, Chen CH, Ger MJ, Lee BH, Pai CH, Chow D, Huang HE, Hwang SY, Chung MC, et al. (2003) Ferredoxin from sweet pepper (Capsicum annuum L.) intensifying Harpin(PSS)-mediated hypersensitive response shows an enhanced production of active oxygen species (AOS). Plant Mol Biol 51: 913–924 [DOI] [PubMed] [Google Scholar]
- del Pozo O, Pedley KF, Martin GB(2004) MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Wang S, Dong X(2007) Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc Natl Acad Sci USA 104: 4223–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N, Rosli HG, Martin GB, Mueller LA(2015) The SGN VIGS tool: User-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol Plant 8: 486–488 [DOI] [PubMed] [Google Scholar]
- Fonseca JP, Dong X(2014) Functional characterization of a Nudix hydrolase AtNUDX8 upon pathogen attack indicates a positive role in plant immune responses. PLoS One 9: e114119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca JP, Mysore KS(2019) Genes involved in nonhost disease resistance as a key to engineer durable resistance in crops. Plant Sci 279: 108–116 [DOI] [PubMed] [Google Scholar]
- Frazzon AP, Ramirez MV, Warek U, Balk J, Frazzon J, Dean DR, Winkel BS(2007) Functional analysis of Arabidopsis genes involved in mitochondrial iron-sulfur cluster assembly. Plant Mol Biol 64: 225–240 [DOI] [PubMed] [Google Scholar]
- Garg R, Jhanwar S, Tyagi AK, Jain M(2010) Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res 17: 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson TJ, Koonin EV, Musco G, Pastore A, Bork P(1996) Friedreich’s ataxia protein: Phylogenetic evidence for mitochondrial dysfunction. Trends Neurosci 19: 465–468 [DOI] [PubMed] [Google Scholar]
- Gil MJ, Coego A, Mauch-Mani B, Jordá L, Vera P(2005) The Arabidopsis csb3 mutant reveals a regulatory link between salicylic acid-mediated disease resistance and the methyl-erythritol 4-phosphate pathway. Plant J 44: 155–166 [DOI] [PubMed] [Google Scholar]
- Gill US, Lee S, Mysore KS(2015) Host versus nonhost resistance: Distinct wars with similar arsenals. Phytopathology 105: 580–587 [DOI] [PubMed] [Google Scholar]
- Graham D.(1980) Effects of light on “dark” respiration In Davies DD, ed, The Biochemistry of Plants, Vol 2 Academic Press, New York, pp 525–579 [Google Scholar]
- Heath MC.(2000) Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol 3: 315–319 [DOI] [PubMed] [Google Scholar]
- Heis MD, Ditmer EM, de Oliveira LA, Frazzon AP, Margis R, Frazzon J(2011) Differential expression of cysteine desulfurases in soybean. BMC Plant Biol 11: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull GA, Devic M(1995) The beta-glucuronidase (gus) reporter gene system. Gene fusions, spectrophotometric, fluorometric, and histochemical detection. In H Jones, ed, Methods in Plant Molecular Biology, Vol 49: Plant Gene Transfer and Expression Protocols Humana Press, Totowa, NJ, pp 125–141 [DOI] [PubMed] [Google Scholar]
- Iñigo S, Durand AN, Ritter A, Le Gall S, Termathe M, Klassen R, Tohge T, De Coninck B, Van Leene J, De Clercq R, et al. (2016) Glutaredoxin grxs17 associates with the cytosolic iron–sulfur cluster assembly pathway. Plant Physiol 172: 858–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y, Ishiga T, Uppalapati SR, Mysore KS(2011) Arabidopsis seedling flood-inoculation technique: A rapid and reliable assay for studying plant-bacterial interactions. Plant Methods 7: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y, Watanabe M, Ishiga T, Tohge T, Matsuura T, Ikeda Y, Hoefgen R, Fernie AR, Mysore KS(2017) The SAL-PAP chloroplast retrograde pathway contributes to plant immunity by regulating glucosinolate pathway and phytohormone signaling. Mol Plant Microbe Interact 30: 829–841 [DOI] [PubMed] [Google Scholar]
- Kaundal A, Ramu VS, Oh S, Lee S, Pant B, Lee HK, Rojas CM, Senthil-Kumar M, Mysore KS(2017) GENERAL CONTROL NONREPRESSIBLE4 degrades 14-3-3 and the RIN4 complex to regulate stomatal aperture with implications on nonhost disease resistance and drought tolerance. Plant Cell 29: 2233–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani M, Yoo J, Dong X(2007) Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol 144: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiley PJ, Beinert H(2003) The role of Fe–S proteins in sensing and regulation in bacteria. Curr Opin Microbiol 6: 181–185 [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK(2005) Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem 74: 247–281 [DOI] [PubMed] [Google Scholar]
- Lee HA, Lee HY, Seo E, Lee J, Kim SB, Oh S, Choi E, Choi E, Lee SE, Choi D(2017) Current understandings of plant nonhost resistance. Mol Plant Microbe Interact 30: 5–15 [DOI] [PubMed] [Google Scholar]
- Lee S, Whitaker VM, Hutton SF(2016) Mini review: Potential applications of non-host resistance for crop improvement. Front Plant Sci 7: 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S, Touraine B, Briat JF, Lobréaux S(2002) The AtNFS2 gene from Arabidopsis thaliana encodes a NifS-like plastidial cysteine desulphurase. Biochem J 366: 557–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Qin L, Liu P, Wang M, Ye H(2014) Genes for iron–sulphur cluster assembly are targets of abiotic stress in rice, Oryza sativa. Plant Cell Environ 37: 780–794 [DOI] [PubMed] [Google Scholar]
- Lill R.(2009) Function and biogenesis of iron–sulphur proteins. Nature 460: 831–838 [DOI] [PubMed] [Google Scholar]
- Lill R, Mühlenhoff U(2008) Maturation of iron–sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu Rev Biochem 77: 669–700 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S(2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Bernard DG, Balk J, Hai H, Cui X(2012) The DUF59 family gene AE7 acts in the cytosolic iron–sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis. Plant Cell 24: 4135–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, Thomas PD(2019) Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc 14: 703–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir R, Hernández ML, Abou-Mansour E, Martínez-Rivas JM, Mauch F, Métraux JP, León J(2013) Pathogen and Circadian Controlled 1 (PCC1) regulates polar lipid content, ABA-related responses, and pathogen defence in Arabidopsis thaliana. J Exp Bot 64: 3385–3395 [DOI] [PubMed] [Google Scholar]
- Mueller EG.(2006) Trafficking in persulfides: Delivering sulfur in biosynthetic pathways. Nat Chem Biol 2: 185–194 [DOI] [PubMed] [Google Scholar]
- Mysore KS, Ryu CM(2004) Nonhost resistance: How much do we know? Trends Plant Sci 9: 97–104 [DOI] [PubMed] [Google Scholar]
- Nagaraj S, Senthil-kumar M, Ramu VS, Wang K, Mysore KS(2016) Plant ribosomal proteins, RPL12 and RPL19, play a role in nonhost disease resistance against bacterial pathogens. Front Plant Sci 6: 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Yano T(2017) Sulfur modifications of the wobble U34 in tRNAs and their intracellular localization in eukaryotic cells. Biomolecules 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C(2007) SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J 50: 128–139 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A(2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Niggeweg R, Thurow C, Kegler C, Gatz C(2000) Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J Biol Chem 275: 19897–19905 [DOI] [PubMed] [Google Scholar]
- Niks RE, Marcel TC(2009) Nonhost and basal resistance: How to explain specificity? New Phytol 182: 817–828 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C(2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Pfister MF, Yee AJ, Schubert M, Michael L, Zhang CY, Ueki K, Michael MD II, Lowell BB, Kahn CR(2000) Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. Proc Natl Acad Sci USA 97: 12239–12243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Wang K, Ryu CM, Kaundal A, Mysore KS(2012) Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24: 336–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA.(2012) Biogenesis of iron–sulfur clusters in mammalian cells: New insights and relevance to human disease. Dis Model Mech 5: 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA.(2006) The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol 2: 406–414 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Lee HK, Mysore KS(2013) VIGS-mediated forward genetics screening for identification of genes involved in nonhost resistance. J Vis Exp 78: e51033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS(2014) Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat Protoc 9: 1549–1562 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Wang M, Chang J, Ramegowda V, Del Pozo O, Liu Y, Doraiswamy V, Lee H-K, Ryu C-M, Wang K, et al. (2018) Virus-induced gene silencing database for phenomics and functional genomics in Nicotiana benthamiana. Plant Direct 2: e00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O, Lill R(2013) The role of mitochondria in cellular iron–sulfur protein biogenesis: Mechanisms, connected processes, and diseases. Cold Spring Harb Perspect Biol 5: a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K, Morikawa T, Nonaka Y, Mizutani M, Takenaka S, Takabe K, Takahashi MA, Ohta D(2003) Identification and molecular characterization of mitochondrial ferredoxins and ferredoxin reductase from Arabidopsis. Plant Mol Biol 52: 817–830 [DOI] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z(2017) agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45(W1): W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W-H, Rouault T(2007) Metabolic regulation of citrate and iron by aconitases: Role of iron-sulfur cluster biogenesis. BioMetals 20: 549–564 [DOI] [PubMed] [Google Scholar]
- Turowski VR, Aknin C, Maliandi MV, Buchensky C, Leaden L, Peralta DA, Busi MV, Araya A, Gomez-Casati DF(2015) Frataxin is localized to both the chloroplast and mitochondrion and is involved in chloroplast Fe–S protein function in Arabidopsis. PLoS One 10: e0141443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski VR, Busi MV, Gomez-Casati DF(2012) Structural and functional studies of the mitochondrial cysteine desulfurase from Arabidopsis thaliana. Mol Plant 5: 1001–1010 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Wangdi T, Urbanczyk-Wochniak E, Ishiga T, Mysore KS, Bender CL(2008) Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: phenotypic and gene expression analyses of the virulence function of coronatine. Mol Plant Microbe Interact 21: 383–395 [DOI] [PubMed] [Google Scholar]
- Wang K, Kang L, Anand A, Lazarovits G, Mysore KS(2007) Monitoring in planta bacterial infection at both cellular and whole-plant levels using the green fluorescent protein variant GFPuv. New Phytol 174: 212–223 [DOI] [PubMed] [Google Scholar]
- Yeh YH, Chang YH, Huang PY, Huang JB, Zimmerli L(2015) Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front Plant Sci 6: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]