Hyp controls the fate of the CLE40 precursor, resulting in subtilase-mediated release of the bioactive peptide or CLE40 degradation for the modified and unmodified forms, respectively.
Abstract
Small posttranslationally modified signaling peptides are proteolytically derived from larger precursor proteins and subject to several additional steps of modification, including Pro hydroxylation, Hyp glycosylation, and/or Tyr sulfation. The processing proteases and the relevance of posttranslational modifications for peptide biogenesis and activity are largely unknown. In this study these questions were addressed for the Clavata3/Endosperm Surrounding Region (CLE) peptide CLE40, a peptide regulator of stem cell differentiation in the Arabidopsis (Arabidopsis thaliana) root meristem. We identify three subtilases (SBT1.4, SBT1.7, and SBT4.13) that cleave the CLE40 precursor redundantly at two sites. C-terminal processing releases the mature peptide from its precursor and is thus required for signal biogenesis. SBT-mediated cleavage at a second site within the mature peptide attenuates the signal. The second cleavage is prevented by Pro hydroxylation, resulting in the formation of mature and bioactive CLE40 in planta. Our data reveal a role for posttranslational modification by Pro hydroxylation in the regulation of CLE40 formation and activity.
The size of the stem cell population in shoot and root apical meristems is negatively regulated by Clavata3/Endosperm Surrounding Region (CLE) peptides. In the shoot meristem, CLAVATA3 (CLV3) is secreted from stem cells to inhibit the expression of the homeobox transcription factor WUSCHEL (WUS) in the organizing center. WUS promotes stem cell identity including the expression of CLV3, resulting in a negative feedback loop controlling stem cell proliferation (Brand et al., 2000; Schoof et al., 2000). CLE40 is the closest homolog of CLV3 and controls stem cell maintenance in the root (Stahl et al., 2009). CLE40 expression in the differentiation zone of the root stele and in differentiated columella cells negatively regulates WUS-related Homeobox5 (WOX5) expression in the quiescent center, which is required to maintain stem cell identity in adjacent cells (Sarkar et al., 2007; Stahl et al., 2009). In cle40 loss-of-function roots, differentiation of columella stem cells is delayed, resulting in additional stem cells in the distal meristem (Stahl et al., 2009). Treatment with CLE40 peptides, on the other hand, reduces the number of stem cells, resulting in shorter roots (Fiers et al., 2005; Stahl et al., 2013). Similarly, overexpression of CLE40 or external application of CLE40 peptide arrest shoot and flower development, indicating that CLE40 can substitute for CLV3 function in the shoot (Hobe et al., 2003).
CLV3 and CLE40 belong to the large family of posttranslationally modified CLE peptides. They are synthesized as preproproteins with an N-terminal signal peptide, a central variable domain, and a conserved 14-amino acid CLE domain close to the C terminus. Proteolytic processing is required to release 12- or 13-amino acid peptides from the C-terminal region (Yamaguchi et al., 2016; Stührwohldt and Schaller, 2019). Some CLE peptides have been purified from plants and structurally characterized, revealing additional posttranslational modifications, including Pro hydroxylation at P4 and/or P7 and arabinosylation of the second Hyp residue. Ohyama et al. (2009) identified CLV3 as triarabinosylated, 13-amino acid glycopeptide and found glycosylation of Hyp-7 to be important for bioactivity and high-affinity binding to the CLV1 receptor. Likewise, a CLE peptide from Lotus japonicus (CLE-RS) was identified as a 13-mer arabinosylated at Hyp-7, and glycosylation was shown to be important for its function as a negative regulator of nodulation (Okamoto et al., 2013). CLE40 from soybean (Glycine max) was shown to depend on glycosylation at P4 for full activity (Corcilius et al., 2017). The 12-amino acid Tracheary Differentiation Factor (TDIF; CLE41/44), on the other hand, lacks glycosylation, and its activity does not depend on Pro hydroxylation (Ito et al., 2006; Ohyama et al., 2008). CLE40 from Arabidopsis (Arabidopsis thaliana) is 12 or 13 amino acids long and hydroxylated at P4 (RQVHypTGSDPLHH and RQVHypTGSDPLHHK, respectively). Both peptides were shown to be active (Stahl et al., 2013; Czyzewicz et al., 2015), and the relevance of Pro hydroxylation is unknown.
As compared with our understanding of peptide perception and signal transduction mechanisms (Hohmann et al., 2017; Song et al., 2017), we know relatively little about how plant signaling peptides are produced. This is particularly true for the large group of peptides that depend on a cascade of posttranslational modifications for maturation and activity (Stührwohldt and Schaller, 2019). All posttranslationally modified peptides require proteolytic cleavage to release the active peptide from its precursor protein. The identification of the processing proteases is a challenge due to the high number of potential candidates (888 genes in Arabidopsis [MEROPS database, release 12.1]; Rawlings et al., 2016), and it is hindered by frequently low expression levels, by functional redundancy, and by the lack of clear, conserved processing sites. Functional redundancy in the large family of subtilisin-like Ser proteases (subtilases [SBTs]; Schaller et al., 2018) has recently been overcome by the use of an inhibitor-based loss-of-function approach. Tissue-specific expression of subtilase inhibitors in the abscission zones of Arabidopsis flowers phenocopied the abscission defect of the inflorescence deficient in abscission (ida) mutant, revealing a role for SBT4.12, SBT4.13, and SBT5.2 in the maturation of the IDA peptide (Schardon et al., 2016). The same approach was used to demonstrate that SBTs are involved in the maturation of Root Meristem Growth Factor6 (alias CLEL6 or Golven1; Stührwohldt et al., 2020) and in the activation of the tomato (Solanum lycopersicum) immune protease Required for Cladosporium Resistance3 (Paulus et al., 2020).
By using the inhibitor-based loss-of-function approach, we show here that SBTs are required for the formation of the CLE40 peptide in Arabidopsis. We identify three SBTs that cleave the CLE40 precursor (proCLE40) redundantly at two sites. C-terminal processing is required for the liberation of mature CLE40 peptide (matCLE40) and signal formation. In contrast, cleavage at a second site within matCLE40 attenuates the CLE40 signal. The second cleavage is prevented by Pro hydroxylation, resulting in the formation of mature and bioactive CLE40 in planta. Our data reveal a role for posttranslational Pro hydroxylation in the regulation of CLE40 formation and activity.
RESULTS
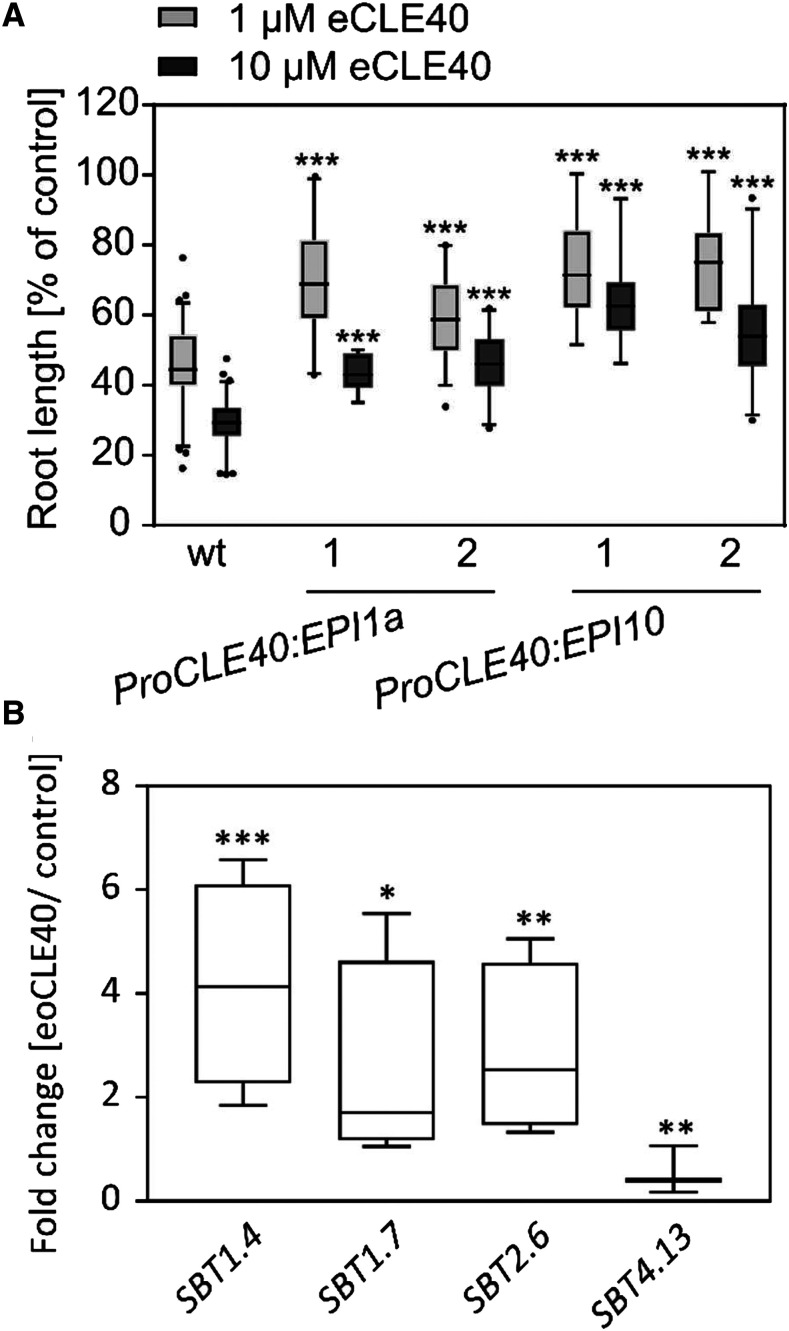
Overexpression of proCLE40 or treatment of Arabidopsis seedlings with the matCLE40 12-mer or 13-mer peptide [RQVHypTGSDPLHH(K)] impairs root growth (Fiers et al., 2005; Stahl et al., 2009, 2013). Similarly, we observed a severe reduction in root length when seedlings were treated with N- and C-terminally extended CLE40 (eCLE40 [ANEVEERQVPTGSDPLHHKHIPFTP]; extensions underlined) carrying six amino acids of the precursor at either end of the nonmodified CLE40 13-mer (Fig. 1A). The activity of eCLE40 in this bioassay indicates that the peptide is taken up by the seedlings and activated in planta, entailing proteolytic processing and, possibly, Pro hydroxylation. The enzymes required for these modifications are unknown. To test the potential involvement of SBTs in CLE40 maturation, we employed the SBT-specific Extracellular Proteinase Inhibitors 1a (EPI1a) and EPI10 to target SBT activity where it is needed for precursor processing (Schardon et al., 2016; Stührwohldt et al., 2017, 2020). Assuming that the processing proteases are needed in tissues where CLE40 is expressed, we used the CLE40 promoter to drive the expression of the EPIs in transgenic Arabidopsis plants. Consistent with a role for SBTs in CLE40 maturation, seedlings expressing EPI1a or EPI10 were significantly less responsive to the eCLE40 precursor peptide than the wild type (Fig. 1A).
Figure 1.
SBTs are involved in CLE40 propeptide processing. A, The activity of eCLE40 is reduced in plants expressing EPI1a and EPI10 SBT inhibitors compared with wild-type (wt) plants. Two independent transgenic pCLE40:EPI1a and pCLE40:EPI10 lines (1 and 2) and wild-type plants were grown for 5 d on plates containing 0 (control), 1 µm, or 10 µm eCLE40. Root length is shown in percentage of the no-peptide control (means ± se). Asterisks indicate significant differences between eCLE40-treated transgenic and wild-type plants using two-tailed unpaired Student’s t test (***P < 0.001; n ≥ 12). B, Expression of four SBT genes is regulated by eCLE40 treatment. SBT expression was analyzed in 5-mm root tips by quantitative real-time PCR (qPCR). Relative expression levels are shown as fold change in eCLE40-treated plants compared with untreated controls. Asterisks indicate significant differences from the control using Student’s t test (* P < 0.05, **P < 0.01, and ***P < 0.001; n ≥ 3).
Candidate SBTs for proCLE40 Processing
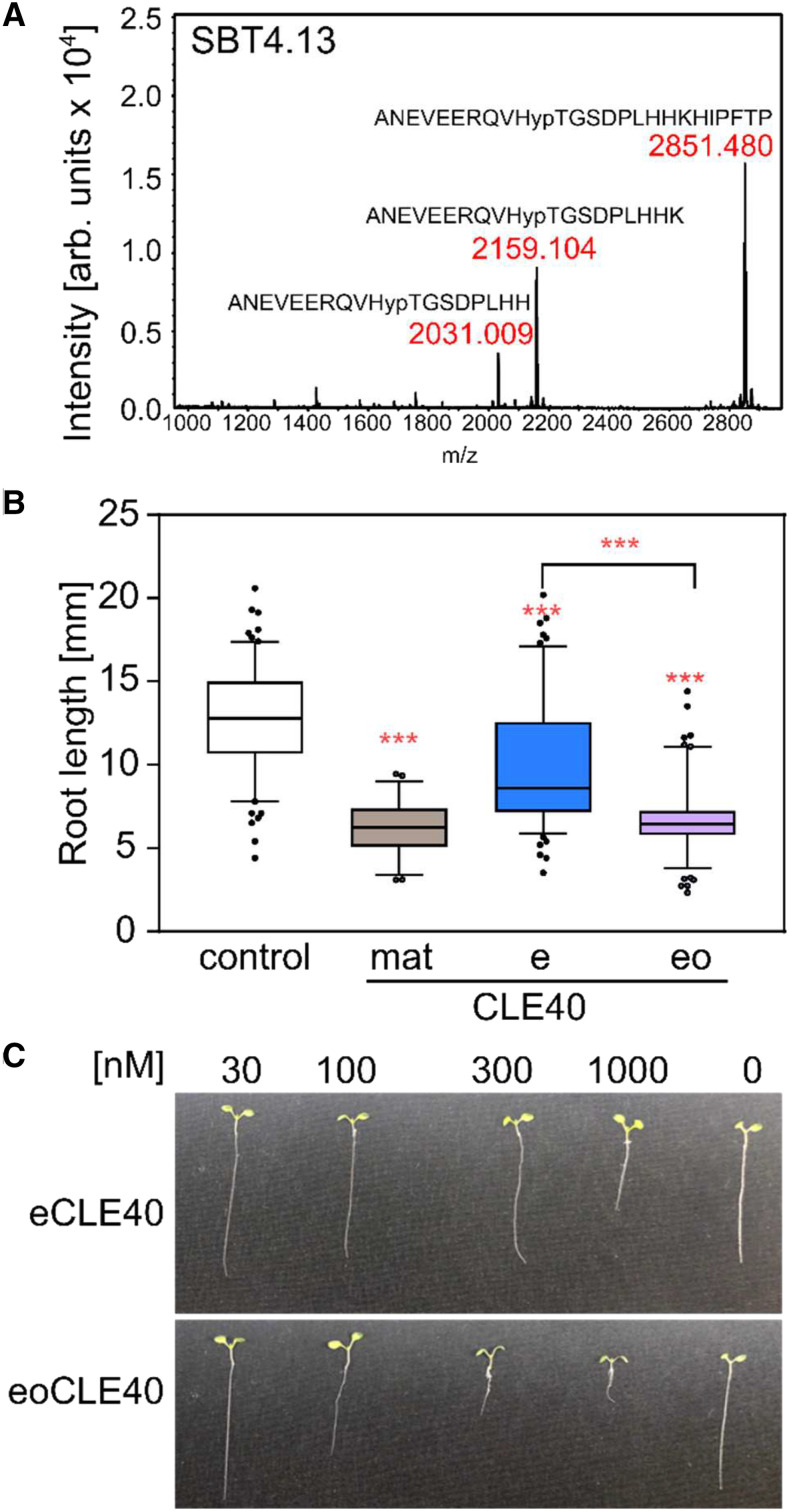
In order to single out individual SBTs as candidates for CLE40 processing, we analyzed the expression of SBT genes in the 5-mm root tip of 6-d-old Arabidopsis seedlings, where CLE40 is known to be expressed (Stahl et al., 2009). Fifty-three SBTs were found to be expressed, although most of them at very low levels relative to the tubulin reference gene (Supplemental Fig. S1). To focus on specific candidates, we tested whether any of these SBT genes respond to treatment with eoCLE40 (eCLE40 hydroxylated at P4; numbering refers to the position in matCLE40). We identified three SBTs that were upregulated in response to eoCLE40 (SBT1.4, SBT1.7, and SBT2.6; Fig. 1B). SBT4.13, for which a precursor-processing function has previously been shown (Schardon et al., 2016), was down-regulated about twofold (Fig. 1B). These four SBTs were considered as candidates for CLE40 precursor processing.
proCLE40 Is Processed by SBTs to Release matCLE40 in Planta
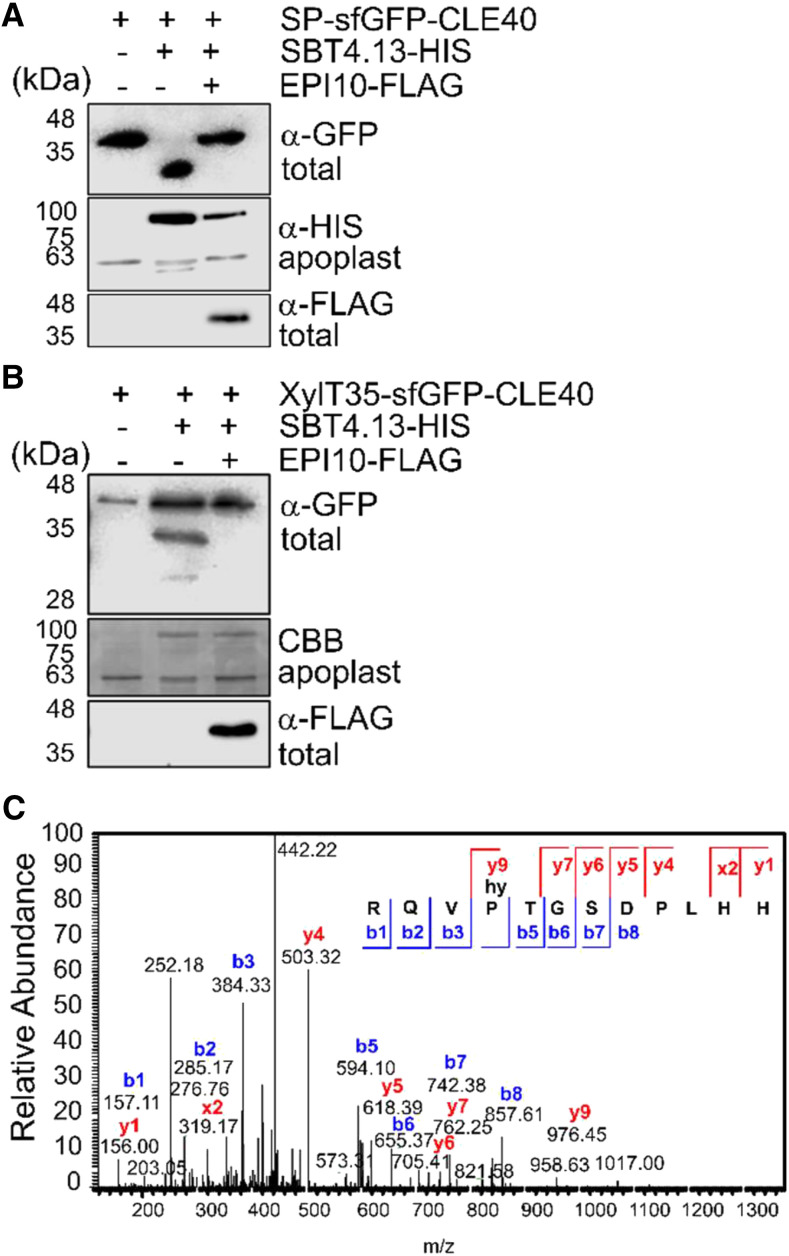
The precursor-processing activity of candidate SBTs was analyzed by coexpression with the superfolder GFP (sfGFP)-tagged CLE40 precursor in Nicotiana benthamiana. In the absence of added SBT-encoding constructs, a single band was detected on anti-GFP immunoblots of total leaf extracts (Fig. 2A), which has been confirmed as the full-length sfGFP-proCLE40 fusion protein by mass spectrometry (MS). Upon coexpression of SBT4.13, a smaller band appeared, indicating processing of the precursor. Coexpression of the EPI10 inhibitor abolished processing, confirming that precursor cleavage is SBT dependent (Fig. 2A). Cleavage of sfGFP-proCLE40 was also observed upon coexpression of SBT1.4 and SBT1.7, resulting in two bands corresponding to the processed and unprocessed forms of the precursor, respectively. Cleavage by SBT1.4 and SBT1.7 was also sensitive to EPI10 inhibition (Supplemental Fig. S2). Addressing the question of whether any of these SBTs are required for proCLE40 processing, we tested whether the sensitivity of the sbt1.4, sbt1.7, and sbt4.13 single-gene loss-of-function mutants to eCLE40 treatment is reduced as compared with the wild type (Supplemental Fig. S3). At 1 µm eCLE40, sensitivity of the mutants was not affected. At 10 µm, we observed a small reduction in sensitivity for sbt1.4 and sbt1.7 (Supplemental Fig. S3). These data suggest a functional redundancy of SBT1.4, SBT1.7, and SBT4.13 with respect to proCLE40 processing.
Figure 2.
The CLE40 precursor is processed by SBTs to release matCLE40. A and B, proCLE40 is cleaved by SBT4.13 in the Golgi. A, The CLE40 precursor equipped with an N-terminal signal peptide (SP) and sfGFP tag was transiently expressed in N. benthamiana together with hexa-His-tagged (HIS) SBT4.13 or the empty vector control (SBT1.4 and SBT1.7 in Supplemental Fig. S2). SBT4.13-mediated cleavage of sfGFP-proCLE40 in total leaf extracts is inhibited by coexpression of FLAG-tagged EPI10 targeted to the endoplasmic reticulum. B, EPI-sensitive processing also occurs when sfGFP-CLE40 is retained in the Golgi by the 35-amino acid membrane anchor of Golgi-resident XylT (XylT35). sfGFP-CLE40 and its cleavage products were detected with α-GFP and EPI10-FLAG with α-FLAG antibodies on western blots of total leaf extracts. Expression of SBT4.13 was confirmed by α-His immunoblotting (A) or Coomassie Brilliant Blue (CBB) staining (B) of cell wall extracts. Proteins were separated by SDS-PAGE using the Laemmli buffer system. Notice that the apparent masses of SP-sfGFP-CLE40 and XylT35-sfGFP-CLE40 fusion proteins (approximately 35 and 40 kD, respectively) are higher than the calculated masses (33.2 and 37.4 kD, respectively). MS analysis confirmed the identity of SP-sfGFP-CLE40 and identified the processing site in the cleavage product at, or in close proximity of, the PTG motif. The apparent mass of the SP-sfGFP-CLE40 cleavage product is lower than the calculated mass of 31.6 kD. The large difference in apparent masses of the full-length and processed forms may be caused by posttranslational modifications of the precursor that are missing from the cleavage product. The difference in expected (65 kD) and apparent (80 kD) mass of SBT4.13 is due to glycosylation. C, proCLE40 is processed in planta to release matCLE40 for secretion into the apoplast. The peptide fraction of cell wall extracts from N. benthamiana leaves expressing CLE40-sfGFP was isolated by ultrafiltration (less than 5-kD MMCO) and solid-phase extraction and analyzed by liquid chromatography-electrospray ionization-MS/MS. The fragmentation spectrum is shown for the Mr = 453.896 precursor ion corresponding to P4-hydroxylated matCLE40. The identity of the peptide is confirmed by the ions that were identified in the y (blue) and b (red) series. m/z, Mass-to-charge ratio.
To address the question of whether the CLE40 precursor is processed within the secretory pathway or only after secretion to the apoplast, the N-terminal signal peptide of the sfGFP-proCLE40 construct was replaced with the membrane anchor of β-1,2-xylosyltransferase (XylT), which is sufficient to target reporter proteins to the medial Golgi (Pagny et al., 2003). The XylT membrane anchor has previously been used as a tool to analyze the subcellular site of processing of Clavata3/Endosperm Surrounding Region-Like (CLEL) peptides (Stührwohldt et al., 2020). When XylT35-sfGFP-proCLE40 was expressed by itself, only the expected full-length protein was detected (Fig. 2B). Processing was observed upon coexpression with SBT4.13, which was inhibited in the presence of EPI10 (Fig. 2B). The data indicate that proCLE40 can be cleaved by SBT4.13 within the secretory pathway. This does not preclude the possibility of extracellular processing if the two proteins are expressed in different cells and only meet in the apoplast.
To identify any peptides released by proteolytic cleavage and secreted into the apoplast, cell wall extracts were prepared from plants expressing C-terminally sfGFP-tagged proCLE40 or wild-type N. benthamiana leaves infiltrated with an empty vector. Peptides were purified by reverse-phase C18 solid-phase extraction and analyzed by MS. matCLE40 (RQVHypTGSDPLHH) was identified in extracts from plants overexpressing proCLE40 (Fig. 2C), along with two peptides that lacked one and two amino acids from the C terminus, respectively. These shorter peptides are likely generated by carboxypeptidase activity, which is abundant in the cell wall of N. benthamiana (Grosse-Holz et al., 2018). We thus consider these truncated peptides as an artifact of the N. benthamiana expression system. All peptides were hydroxylated at P4, which had been reported for other CLE peptides isolated from Arabidopsis and predicted for CLE40 (Ohyama et al., 2008). The identification of matCLE40 in cell wall extracts suggests that proCLE40 is processed by N. benthamiana proteases at two sites that mark the N and C termini of the mature, P4-hydroxylated peptide.
proCLE40 Is Cleaved within the Mature Peptide Sequence in Vitro
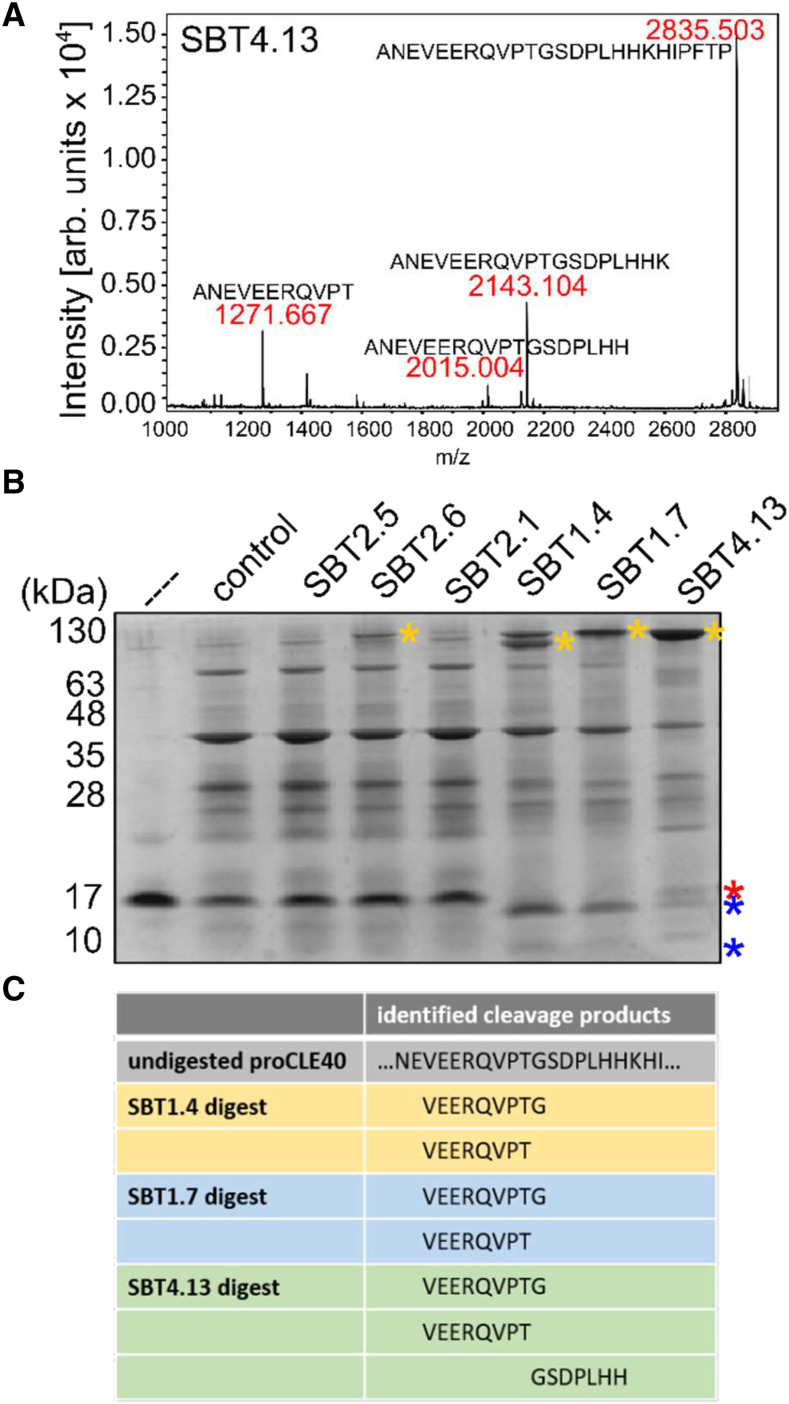
To further characterize SBT-mediated processing, we used the synthetic eCLE40 peptide as a substrate for His-tagged SBT4.13 purified from cell wall extracts by affinity chromatography on Ni-NTA agarose and a fraction mock-purified from empty vector-infiltrated plants as a control. No cleavage was detected for the control treatment, indicating that any background proteases present in N. benthamiana had been removed (Supplemental Fig. S4A). SBT4.13 cleaved eCLE40 to produce peptides corresponding to the C terminus of the bioactive CLE40 12-mer (…HH) and 13-mer (…HHK), respectively (Fig. 3A). Additional cleavage was observed at the PTG motif within matCLE40 (Fig. 3A), in contrast to N-terminal cleavage that was observed upon expression in planta (Fig. 2C).
Figure 3.
SBTs cleave the CLE40 precursor in vitro. A, Cleavage of the extended CLE40 precursor peptide by SBT4.13. eCLE40 was digested with affinity-purified SBT4.13 (control reaction in Supplemental Fig. S3A). Reaction products were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). The mass spectrum shows ion intensities (arbitrary units) and masses for the substrate peptide (m/z = 2,835.503) and three cleavage products. B, Recombinant proCLE40 is processed by SBT1.4, SBT1.7, and SBT4.13. Recombinant proCLE40 (first lane marked with the dashed line) was incubated with cell wall extracts from mock-infiltrated plants (control) and from plants expressing the indicated SBTs. The protein fraction (greater than 10 kD) was analyzed by Tris-Tricine-PAGE. Notice that on Tris-Tricine gels the apparent mass of SBTs (100–130 kD; yellow asterisks) is higher compared with SDS-PAGE with the Laemmli buffer system (compare with Fig. 2). proCLE40 and its cleavage products are indicated by red and blue asterisks, respectively. C, SBTs cleave proCLE40 at the C terminus and at an internal site of the mature peptide. The peptide fraction (less than 10 kD) of the digest in B was analyzed by liquid chromatography-electrospray ionization-MS/MS. Precursor-derived cleavage products are shown for SBT1.4, SBT1.7, and the SBT4.13 digest.
To confirm this unexpected result, and to exclude the possibility that the synthetic eCLE40 peptide is processed differently compared with the full-length precursor, His-tagged proCLE40 was expressed in Escherichia coli and purified from bacterial extracts. Recombinant proCLE40 was incubated in vitro with SBTs extracted from agroinfiltrated N. benthamiana leaves. Reaction products were separated by ultrafiltration (10-kD molecular mass cuttoff [MMCO]) into protein and peptide fractions. The protein fraction was analyzed by Tris-Tricine-PAGE, revealing processing of the recombinant precursor by SBT1.4, SBT1.7, and SBT4.13 (Fig. 3B, blue asterisks) but not by SBT2.1, SBT2.5, or SBT2.6. The peptide fraction was analyzed by MS. Two peptides were identified in the SBT1.4, SBT1.7, and SBT4.13 digests (VEERQVPT and VEERQVPTG) resulting from cleavage at T5 and G6 of matCLE40, respectively (Fig. 3C). Cleavage at the N terminus of matCLE40 (RQV….) was not observed. The corresponding C-terminal cleavage product (GSDPLHH) was identified in the SBT4.13 digest (Fig. 3C). The data obtained for the E. coli-produced recombinant precursor (Fig. 3, B and C) are fully consistent with those for synthetic eCLE40 (Fig. 3A). This indicates that SBTs cleave the CLE40 precursor to mark the C terminus of matCLE40 and at a second site, the PTG motif, within the mature CLE40 sequence (Fig. 4A).
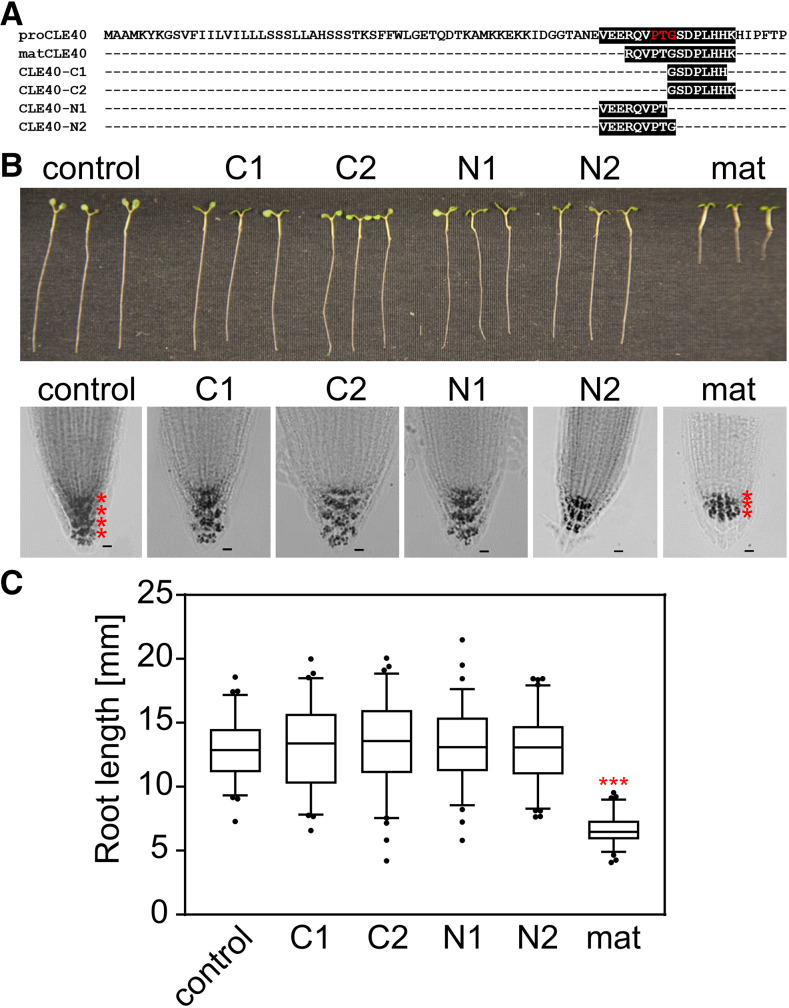
Figure 4.
CLE40 fragments generated by SBTs from recombinant proCLE40 are inactive. A, Peptides (C1, C2, N1, and N2) generated by SBTs from recombinant CLE40 precursor (compare with Fig. 3C) aligned with the precursor sequence. The internal PTG motif is highlighted in red. B and C, CLE40 fragments resulting from cleavage at the PTG motif are inactive. Root tips of 5-d-old seedlings were treated with 1 µm matCLE40 and CLE40 fragments C1, C2, N1, and N2. Starch staining reveals a reduction in columella cell layers, as indicated by red asterisks. Bars = 10 µm. C, Root growth is inhibited in matCLE40-treated seedlings as compared with N1-, N2-, C1-, and C2-treated seedlings and controls. Data show means ± se of n ≥ 73 seedlings of one representative experiment out of three independent experiments. Asterisks indicate significant differences from the untreated control using Student’s t test (P < 0.001).
Root bioassays indicated that the C-terminal (C1 and C2) and N-terminal (N1 and N2) peptides resulting from cleavage at the internal PTG motif are inactive. As compared with matCLE40 treatment, which affected columella cell differentiation and root length of Arabidopsis seedlings, we did not see any effect of the cleavage products on the integrity of the root meristem, the differentiation of columella cells, or root growth (Fig. 4, B and C). These results thus suggest that SBTs are not involved in maturation but rather in the inactivation of the CLE40 signaling peptide.
SBT Cleavage Site Selection Depends on P4 and T5
Site-directed mutants with Ala substitutions for P4 and T5 were generated to assess the relevance of the PTG motif for internal processing. Precursor mutants were expressed in E. coli, purified, and digested with SBTs extracted from N. benthamiana leaves, and the resulting peptide fraction was analyzed as above. Cleavage at the PTG motif was indeed suppressed as a result of the P4A and T5A substitutions. However, very much to our surprise, we now detected cleavage of the precursor at the genuine N and C termini of matCLE40, resulting in the peptides RQVATGGSDPLHH and RQVPAGGSDPLHH (Fig. 5, A and B).
Figure 5.
SBT-mediated processing of recombinant CLE40 depends on P4 and T5. A and B, Site-directed P4A and T5A precursor mutants were expressed in E. coli and digested with cell wall extracts from N. benthamiana leaves expressing SBT1.7 or SBT4.13. The peptide fraction was isolated by ultrafiltration (10-kD MMCO). Peptides identified by MS are shown; matCLE40 is highlighted in red. C to E, proCLE40 is processed by Arabidopsis SBTs in a P4-dependent manner. C, Erlenmeyer flask with Arabidopsis seedlings grown in submerged culture for the collection of cell wall exudates. D, Processing of proCLE40 by Arabidopsis proteases is sensitive to EPI1a inhibition. Recombinant proCLE40 was digested with cell wall exudates of Arabidopsis wild type (wt) and of seedlings expressing EPI1a under the control of the indicated promoters. The red and blue asterisks show full-length proCLE40 and a processed form after SDS-PAGE and Coomassie blue staining, respectively. The dashed line indicates the untreated proCLE40. E, MS analysis of peptides generated from recombinant proCLE40 and the site-directed P4A mutant by Arabidopsis cell wall exudates. The analysis was performed as for A and B. matCLE40 is shown in red.
To confirm these results in Arabidopsis, seedlings were grown in submerged culture (Fig. 5C) to collect apoplastic exudates from the Columbia-0 wild type and from transgenic lines expressing the SBT inhibitor EPI1a under the control of several different promoters (ProEPF1, Cauliflower Mosaic Virus [CaMV] Pro-35S, or ProCLE44). Recombinant proCLE40 was cleaved upon incubation with wild-type exudate. No cleavage was observed with exudates from EPI1a-expressing lines, confirming that cleavage is mediated by SBTs (Fig. 5D). Identification of the cleavage products generated from the CLE40 precursor by wild-type exudates revealed processing within the PTG motif, as peptides of the sequence VEERQVPT, VEERQVPTG, and PTGSDPLHH were produced (Fig. 5E). In contrast, peptides correctly processed at the N and C termini of the CLE40 dodecamer were generated by wild-type exudates from the site-directed proCLE40 P4A mutant (e.g. RQVATGGSDPLHH), and cleavage at the internal PTG motif was no longer observed (Fig. 5E).
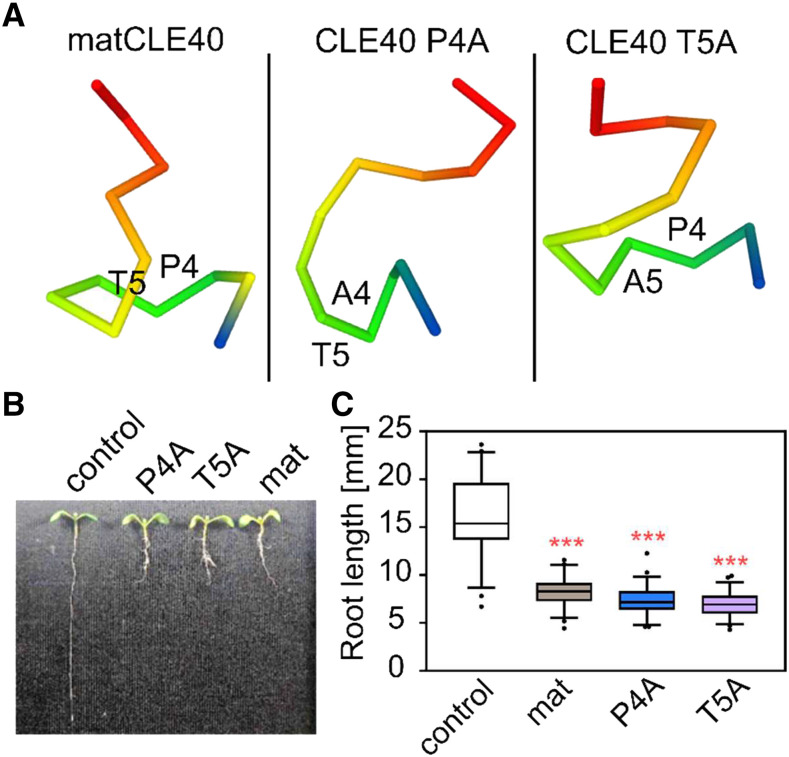
These data indicate that the CLE40 precursor can be cleaved by SBTs at two sites, at the genuine C terminus and at an internal site of matCLE40, and that cleavage at the internal site depends on P4 and T5. In an attempt to explain the strong effect of P4A and T5A substitutions on internal processing, we modeled the peptide structures. The results suggest that Ala substitutions at P4 or T5 may have a considerable impact on peptide conformation (Fig. 6A). The predicted structural differences may explain why SBT-mediated cleavage at the internal site is impaired in P4A and T5A precursor mutants.
Figure 6.
Impact of P4A and T5A substitutions on CLE40 structure and activity. A, Peptide structure predictions suggest that Ala substitutions for P4 and T5 may have an impact on peptide conformation. Peptides are shown from the N terminus (blue) to the C terminus (red); amino acids in positions 4 and 5 are indicated. B and C, Bioactivity (inhibition of root growth) is not affected by P4A and T5A substitutions. B, Representative 6-d-old Arabidopsis seedlings grown on medium containing 1 µm matCLE40 (mat), the P4A and T5A substituted peptides, or no peptide as a control. C, Root length of peptide-treated as compared with control seedlings (means ± se; n ≥ 50). Asterisks indicate statistically significant differences from the control using (***P < 0.001).
Differential Processing Is Controlled by Hydroxylation of P4
To reconcile the relevance of P4 and T5 for cleavage site recognition with the observations that (1) proCLE40 produced in E. coli is cleaved internally and at the mature C terminus (Figs. 3B and 5E) while (2) proCLE40 expressed in planta is not cleaved internally but rather at the genuine N and C termini of matCLE40 (Fig. 2C), we hypothesized that cleavage site selection may be controlled by posttranslational modification of P4. Indeed, peptides generated in planta were found to be hydroxylated at P4 (Fig. 2C), while proCLE40 produced in E. coli lacks this posttranslational modification. To assess the impact of Pro hydroxylation on cleavage site selection, we digested synthetic eoCLE40 with purified SBT4.13 (Fig. 7A) and compared the reaction products with those obtained for the unmodified eCLE40 digest (Fig. 3A). In contrast to the unmodified peptide, which was cleaved at both the internal PTG motif and the C terminus (Fig. 3A), eoCLE40 was only cleaved C terminally to selectively produce peptides with the genuine C termini of matCLE40 (Fig. 7A). Cleavage at the N terminus of matCLE40 was not observed for either of the two peptides, suggesting that this site is processed by unidentified extracellular proteases of Arabidopsis (Fig. 5E) or N. benthamiana (Figs. 2C and 5, A and B). These data confirm the impact of Pro hydroxylation on cleavage site selection and suggest that the CLE40 precursor is differentially processed in planta depending on the extent of posttranslational modification.
Figure 7.
Relevance of Pro hydroxylation for CLE40 precursor peptide processing and activity. A, Cleavage of the extended CLE40 peptide by SBT4.13 (compare with Fig. 3A) is prevented by hydroxylation of P4. eoCLE40 was digested with affinity-purified SBT4.13 (control reaction in Supplemental Fig. S3B). Reaction products were analyzed by MALDI-TOF MS. The mass spectrum shows ion intensities (arbitrary units) and masses for the substrate peptide (m/z = 2,851.480) and two C-terminally processed cleavage products. B and C, Hydroxylation at P4 is required for full activity of the extended CLE40 peptide. Bioactivity (root growth inhibition) was assayed in seedlings grown for 5 d on plates containing 1 µm matCLE40, eCLE40, eoCLE40, or no peptide as a control. B, Root length of peptide-treated as compared with control seedlings (means ± se; n ≥ 49). Asterisks indicate statistically significant differences of peptide-treated roots and the control, and between eCLE40- and eoCLE40-treated roots using Student’s t test (***P < 0.001). C, Representative seedlings treated with the indicated concentrations of eCLE40 or eoCLE40 as described for B.
Impact of Pro Hydroxylation on Bioactivity of CLE40
Pro hydroxylation may be important for the activity of plant signaling peptides if it is required for receptor binding and activation. For example, receptor binding and activation assays indicated that hydroxylation of P9 is required for maximum activity of IDA (Butenko et al., 2014), and structural analysis confirmed that Hyp in position 9 (Hyp-9) establishes specific hydrogen bonds within a binding pocket of the IDA receptor (Santiago et al., 2016). This does not seem to be the case for CLE40. We did not detect any difference in activity for matCLE40 and its P4A and T5A mutants in bioassays indicating that Hyp in position 4 is not required for bioactivity. Treatment with 1 µm of the Ala-substituted derivatives affected root length to the same extent as matCLE40 (Fig. 6, B and C). A comparison of dose-response curves also failed to reveal any reduction in bioactivity for the P4A and T5A mutants as compared with the wild-type peptide (Supplemental Fig. S5, A and B). Since bioactivity is not affected, the residues in positions 4 and 5 do not seem to be relevant for receptor binding and activation. This conclusion is consistent with crystal structure analysis of the TDIF (alias CLE41/44)/receptor complex, showing that the solvent-exposed Hyp-4 does not contribute to peptide-receptor interaction (Morita et al., 2016; Li et al., 2017).
Alternatively, Pro hydroxylation may affect bioactivity if it allows for selective degradation of either the hydroxylated or the nonmodified form of the peptide precursor. Supporting this scenario, only inactive CLE40 fragments were produced by SBTs from the nonmodified (E. coli-produced) precursor and from the eCLE40 peptide, whereas cleavage at the internal, inactivating site was not observed with hydroxylated eoCLE40. To test whether differential processing is relevant for the regulation of CLE40 activity in vivo, the root length bioassay was conducted to compare the activity of matCLE40 with eCLE40 and eoCLE40, which both require processing for activation. Root growth was inhibited by treatment with 1 µm eoCLE40 as efficiently as with matCLE40, while eCLE40 was much less active (Fig. 7, B and C). The difference in activity cannot be explained by the presence or absence of Hyp in the mature peptide, since amino acid substitution in this position did not affect bioactivity (Fig. 6, B and C). We conclude that the difference in activity is caused by the presence or absence of Hyp in the precursor, resulting in differential processing for the production of bioactive CLE40 only from the hydroxylated precursor peptide.
DISCUSSION
Using an established, inhibitor-based loss-of-function approach (Schardon et al., 2016), we show here that SBT activity is required for the formation of bioactive CLE40 peptides. Transgenic plants expressing SBT inhibitors were less responsive to extended CLE40 precursor peptide that depends on processing for full activity (Fig. 1). Furthermore, cell wall exudates of transgenic Arabidopsis seedlings expressing SBT inhibitors, unlike wild-type exudates, were not capable of cleaving the CLE40 precursor in vitro (Fig. 5). The SBTs involved in proCLE40 processing were identified as SBT1.4, SBT1.7, and SBT4.13, which cleave the precursor at the C terminus of the CLE40 peptide. Two different C termini were identified corresponding to the CLE40 12-mer (RQVHypTGSDPLHH) and 13-mer (RQVHypTGSDPLHHK). Both peptides have previously been shown to be bioactive (Stahl et al., 2013; Czyzewicz et al., 2015).
In addition to C-terminal processing, SBTs were found to cleave CLE40 at an internal site that we refer to as the PTG motif. The relevance of P4 and T5 for SBT-mediated precursor processing was confirmed by site-directed mutagenesis (Fig. 5). Using synthetic peptides as substrates for affinity-purified SBT4.13, we further showed that cleavage at the PTG motif is prevented by hydroxylation of P4 (Fig. 7). The PT/G cleavage site (with the slash indicating the cleaved bond) matches the substrate selectivity of SBT4.13 (Schardon et al., 2016). SBT4.13 was previously shown to cleave the IDA precursor at PK/G to mark the N terminus of the IDA peptide. P in position −2 relative to the cleavage site was reported to be particularly important for processing (Schardon et al., 2016). We hypothesize that Hyp in this position, similar to Ala substitution of P4 or T5, results in a conformational change of the CLE40 peptide (Fig. 6A) that prevents recognition by the processing protease. Similarly, substitution of G6 in CLV3, CLE8, CLE19, and CLE22 was hypothesized to affect conformational flexibility, resulting in altered peptide receptor interaction and antagonistic peptides (Song et al., 2013; Xu et al., 2015). In CLE40, however, this effect was not observed (Czyzewicz et al., 2015).
The bioactivity of P4A- and T5A-substituted CLE40 did not differ significantly from that of the hydroxylated wild-type peptide (Fig. 6), suggesting that P4 hydroxylation is not essential for peptide receptor interaction or downstream signaling. Similarly, in TDIF (CLE41/44), hydroxylation of P4 is not required and substitution of S5 does not affect bioactivity (Ito et al., 2006). In contrast, in extended CLE40 precursor peptides, hydroxylation of P4 had a strong impact on activity. Hydroxylated eoCLE40 was as active in bioassays as matCLE40, while the activity of nonhydroxylated eCLE40 was clearly reduced (Fig. 7). We attribute this difference in activity to differential processing of the hydroxylated and nonhydroxylated forms of the precursor. Pro hydroxylation protects the precursor from SBT-mediated cleavage at the internal processing site, giving rise to the bioactive peptide (Figs. 2 and 7).
Our study reveals a novel role for posttranslational modification of plant signaling peptides. For some peptides, posttranslational modification was shown to be important for receptor binding and, hence, for bioactivity. In IDA, for example, hydroxylation of P9 is required for maximum activity (Butenko et al., 2014), and the hydroxyl group specifically interacts with the binding pocket of the IDA receptor (Santiago et al., 2016). Likewise, Tyr sulfation was found to be crucial for the interaction of phytosulfokine, Casparian strip integrity factors, and Twisted Seed1 with their respective receptors (Wang et al., 2015; Doll et al., 2020; Okuda et al., 2020). Also in CLE peptides, P7 hydroxylation and subsequent glycosylation were shown to be important for bioactivity and receptor binding (Ohyama et al., 2009; Okamoto et al., 2013). For many other signaling peptides, the function of posttranslational modification remained obscure, including P4 hydroxylation of CLE peptides. Indeed, as mentioned above for TDIF (CLE41/44), P4 hydroxylation is not required for activity, and in the receptor complex Hyp-4 is solvent exposed and does not make specific receptor contacts (Ito et al., 2006; Morita et al., 2016; Zhang et al., 2016). We show here that this modification controls differential processing and the formation of the bioactive CLE40 peptide.
How widespread this phenomenon is, whether it extends to other members of the CLE family and other types of posttranslational modifications like additional glycosylation or Tyr sulfation, remains to be seen. Pro hydroxylation is frequent also in other peptide families, including root meristem growth factors, plant peptide containing sulfated Tyr residues, and C-terminally encoded peptides (Amano et al., 2007; Ohyama et al., 2008; Matsuzaki et al., 2010; Zhang et al., 2016), where it may play a similar role in controlling peptide processing and stability.
Another open question relates to the enzymes responsible for Pro hydroxylation and where they are expressed. In Arabidopsis, there are 13 known genes for prolyl-4-hydroxylases (P4Hs), and the resulting enzymes are located to the endoplasmic reticulum and Golgi membranes within the secretory pathway (Yuasa et al., 2005; Velasquez et al., 2015). Which of these enzymes contribute to the posttranslational modification of signaling peptides is currently unknown. Our data indicated that SBT4.13 can cleave the CLE40 precursor in the secretory pathway before the precursor reaches the trans-Golgi network (Fig. 2). Therefore, for the formation of bioactive matCLE40, the respective P4H would be required in the same cell to allow for hydroxylation of P4 before the precursor encounters the processing protease. In this model, the limiting step for matCLE40 formation is the availability of P4Hs within the secretory pathway of cells that express the precursor and the processing protease. Alternatively, if proCLE40 and processing SBTs are produced by different cells or cell types, they would meet first in the apoplast, and the activity of as yet unknown extracellular P4Hs may control differential processing and peptide signal formation. Hence, we hypothesize that the limiting factor for CLE40 maturation is the coexpression of a modifying P4H that modifies the precursor before it is finally activated by SBTs. If no P4H is coexpressed, the peptide is inactivated at the identified internal processing site.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown on potting compost with 3.6% (v/v) sand and 7.2% (v/v) perlite. To minimize the risk of thrips infection, seeds were incubated overnight at −75°C. They were resuspended in 0.1% (w/v) agar, stratified for 48 h at 4°C in the dark, and grown under short-day conditions (12-h photoperiod) at 22°C and 100 to 120 µE white light. For sterile growth experiments, Arabidopsis seeds were surface sterilized in 70% (v/v) ethanol for 15 min, washed five times in water, and laid out on square plates containing 0.5× Murashige and Skoog medium (Murashige and Skoog, 1962), 1% (w/v) Suc, and 0.38% (w/v) Gelrite. Seeds were stratified for 2 d at 4°C and grown as above for the time indicated. Root length was measured using ImageJ (http://rsbweb.nih.gov/ij/). Loss-of-function mutants sbt1.4 (SALK-054778), sbt1.7 (Gabi Kat 140B02), and sbt4.13 (SAIL_228_A04) were previously described (Rautengarten et al., 2005). All experiments were carried out at least three times with similar results. Media were supplemented with peptides as indicated: CLE40-C1 (GSDPLHH), CLE40-C2 (GSDPLHHK), CLE40-N1 (VEERQVPT), CLE40-N2 (VEERQVPTG), CLE40-P4A (RQVATGSDPLHHK), CLE40-T5A (RQVPAGSDPLHHK), matCLE40 (RQVHypTGSDPLHHK), eCLE40 (ANEVEERQVPTGSDPLHHKHIPFTP), or eoCLE40 (ANEVEERQVHypTGSDPLHHKHIPFTP). All peptides used in this study were synthesized by PepMIC and obtained at greater than 95% purity. Lyophilized peptides were reconstituted in water. Peptide concentration was determined by a modified Lowry procedure using the DC Protein Assay (Bio-Rad) according to the manufacturer’s instructions. Nicotiana benthamiana seeds were provided by Agroscience and were grown at a daylength of 16 h at 28°C. N. benthamiana plants were fertilized once per week with universal fertilizer (Wuxal; 2 mL L−1) and used for experiments at the age of 4 to 6 weeks. For starch staining, roots were incubated for 1 min in Lugol solution (6 mm iodine, 43 mm potassium iodide, and 0.2 n HCl) and destained in distilled water.
Generation of Expression Constructs
Open reading frames (ORFs) were amplified by PCR using the primers listed in Supplemental Table S1. All PCR products containing flanking restriction sites were cloned into Topo2.1 (Life Technologies) and verified by sequencing (Macrogen). Inserts were cloned into corresponding restriction sites of pART7 (Gleave, 1992) under the control of the constitutive CaMV 35S promoter and terminator. The entire expression cassette was transferred into pART27 (Gleave, 1992) and transformed into Agrobacterium tumefaciens strain C58C1 or GV3101 for transient expression experiments. SBT4.13 and SBT4.13-6H constructs have been described before (Schardon et al., 2016; Hohl et al., 2017). For sfGFP-CLE40 constructs, overlapping PCR was used to fuse the CLEL6 signal peptide or the first 35 amino acids of XylT (At5g55500) to the 5′ (N-terminal) end of sfGFP (Pagny et al., 2003; Pédelacq et al., 2006; Boulaflous et al., 2009). The EcoRI site upstream of the sfGFP ORF was used to generate CLE40-sfGFP constructs. Orientation was tested by PCR and sequencing. The EPI10-FLAG-KDEL construct has been described before (Stührwohldt et al., 2020).
For constructs expressing EPI inhibitors under the control of the CLE40 promoter, the EPI1a ORF was amplified from clones described by Schardon et al. (2016). Overlapping PCR was used to fuse the signal peptide and the FLAG tag to the EPI1a ORF. Terminal EcoRI and XhoI restriction sites were included with the PCR primers and used for cloning into pGreen0029 upstream of the neomycin phosphotransferase II terminator. EPI10 constructs were described previously (Schardon et al., 2016). CLE40 was PCR amplified including NotI and EcoRI restriction sites and ligated into pGreen229 using corresponding restriction sites upstream of the EPI1a or EPI10 construct. EPF1, CaMV 35S, and CLE44 constructs were generated accordingly. Plasmids were transformed into GV3101 containing the pSOUP helper plasmid and transformed into Arabidopsis Columbia-0 by floral dip (Clough and Bent, 1998). Transgenic lines were selected on glufosinate, and homozygous lines in the T3 or T4 generation were used in further experiments.
For Escherichia coli expression constructs, the CLE40 ORF was amplified from cDNA by PCR, cloned into Topo2.1 as described above, and transferred into pET21a (Novagen) using the indicated restriction sites. Mutations were introduced by primers and were confirmed by sequencing.
Transient Expression in N. benthamiana and Protein Extraction
A. tumefaciens strains C58C1 and GV3101 were used for transient expression experiments in N. benthamiana. Bacteria were grown on plates containing appropriate antibiotics (rifampicin, tetracycline, and spectinomycin for C58C1; gentamycin and spectinomycin for GV3101) at 28°C and were washed off the plates in 10 mm MES, pH 5.6 containing 10 mm MgCl2. Acetosyringone (150 µm) was added prior to infiltration of the cell suspension into N. benthamiana leaves with a blunt syringe. Two to 3 d after infiltration, the leaves were harvested. For total protein extraction, leaves were shock frozen in liquid nitrogen and ground to a fine powder. The powder was thawed and resuspended in 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 10 mm β-mercaptoethanol containing 0.5% (w/v) Triton X-100 and supplemented with proteinase inhibitor cocktail (SERVA Electrophoresis). Cell debris was removed by centrifugation (16,000g, 4°C, 10 min). Extracts were kept at 4°C until use on the same day or stored at −20°C for later analysis. For apoplastic fractions, the harvested leaf material was washed three times with cold distilled, deionized water and vacuum infiltrated twice with ice-cold 50 mm sodium phosphate buffer, pH 7, and 300 mm NaCl at 70 mbar for 1 min. Cell wall extracts were obtained by centrifugation through glass wool for 10 min and 4°C at 1,500g and subsequent centrifugation at 12,000g for 10 min to remove any insoluble material.
Expression and Purification of Recombinant Proteins
E. coli cells were grown in Luria-Bertani medium with appropriate antibiotics at 220 rpm and 37°C to an A600 of 0.6. Protein expression was induced with 1 mm isopropylthio-β-galactoside for 2 to 4 h at 30°C. Cells were harvested by centrifugation (3,500g, 4°C, 20 min) and resuspended in extraction buffer (50 mm sodium phosphate buffer, pH 7.4, and 300 mm NaCl including 5 mm phenylmethylsulfonyl fluoride, 0.1 mg mL−1 lysozyme, and 800 units mL−1 DNaseI). Cells were lysed by sonication (SONOPULS HB2070 with MS72 sonic needle; Bandelin Electronics) at 4°C for 3 min. The solution was centrifuged (15,000g, 4°C, 20 min), and the cleared supernatant was subjected to affinity chromatography on Ni-NTA agarose (Qiagen). Recombinant proteins were bound to the column for 2 h, the column was washed extensively in buffer containing 20 mm imidazole, and recombinant proteins were eluted in buffer containing 400 mm imidazole.
Cleavage of Peptide Precursors by SBTs and Identification of Cleavage Products
For in vitro digests of recombinant CLE40 precursors (Fig. 3, B and C), SBTs were expressed by agroinfiltration in N. benthamiana leaves and apoplastic fractions were isolated as described above. Cell wall extracts were concentrated by ultrafiltration (Vivaspin, Sartorius; 30-kD MMCO). Ten micrograms of recombinant propeptide and at least 4 µg of protein in enriched apoplastic fractions including the SBT of interest were incubated for 15 min. The peptide fraction was isolated by ultrafiltration (10-kD MMCO). Peptides were purified from the filtrate using C18 reverse-phase stage tips (Agilent). C18 discs were conditioned in 80% (v/v) acetonitrile and equilibrated in 0.5% (v/v) acetic acid by centrifugation at 5,000 rpm for 1 min each. Peptides were applied to the tips by centrifugation at 3,000 rpm for 1 min, washed with 0.5% (v/v) acetic acid for 1 min, eluted in 80% (v/v) acetonitrile, and dried by vacuum centrifugation for subsequent MS analysis. Proteins remaining in the filtride were analyzed by Tris-Tricine-PAGE (Schägger, 2006).
Immunoblotting
After SDS-PAGE, proteins were transferred to nitrocellulose membranes using a semidry blotting apparatus according to the manufacturer’s recommendations (Bio-Rad Laboratories). Skim milk powder (6% [w/v]) in Tris-buffered saline was used as a blocking solution, except for 6xHis-tagged proteins, where 1% (w/v) BSA was used. GFP-coupled proteins were detected by α-GFP antibody (Thermo Fisher Scientific), SBTs by a mixture of polyclonal antisera directed against fragments of tomato (Solanum lycopersicum) SBT1, SBT2, SBT3, and SBT4a, 6xHis-tagged proteins by a monoclonal mouse α-6xHis antibody from Dianova, and FLAG-tagged proteins by α-FLAG antibody directly coupled to horseradish peroxidase (Sigma-Aldrich), used at the indicated concentrations and according to the manufacturers’ instructions. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG was used as secondary antibody. Blots were developed by enhanced chemiluminescence detection with an Odyssey Fc imager (LI-COR Biotechnology) or by x-ray film.
MS
Peptides generated from transiently expressed proCLE40-sfGFP (Fig. 2C) or by in vitro digests of recombinant precursors with SBTs obtained from cell wall extracts (Figs. 3 and 5) were analyzed via liquid chromatography-MS/MS using a nanoflow EASY-nLC 1000 System (Thermo Fisher Scientific) for HPLC separation and an Orbitrap hybrid mass spectrometer (Q-Exactive, Thermo Scientific) for mass analysis. Peptides were eluted from a 75-μm analytical C18 column (PepMan, Thermo Fisher Scientific) on a linear gradient from 4% to 64% (v/v) acetonitrile over 135 min and sprayed directly into the Q-Exactive Plus mass spectrometer. Proteins were identified via MS/MS on the basis of the fragmentation spectra of multiple charged peptides. Up to 12 data-dependent MS/MS spectra were acquired in the Orbitrap for each full-scan spectrum. Full-scan spectra were acquired at 70,000 full-width half-maximum resolution and fragment spectra at a resolution of 35,000 full-width half-maximum.
Peptides generated by purified SBT4.13 from eCLE40 or eoCLE40 substrates (Figs. 3A and 7A) were analyzed by MALDI-TOF MS. Two micromolar eCLEL40/eoCLE40 was incubated with SBT4.13 or the mock-purified control fraction in 50 mm potassium phosphate buffer and 10 mm sodium chloride, pH 5.5, at room temperature. The reaction was stopped after 30 min by the addition of 1% (v/v) trifluoroacetic acid. Sample aliquots (1.5 µL) were mixed with an equal volume of the crystallization matrix (5 mg mL−1 α-cyano-4-hydroxy-transcinnamic acid in 50% [v/v] acetonitrile and 0.1% [v/v] trifluoroacetic acid), and mass spectra were recorded with an AutoflexIII mass spectrometer (Bruker Daltonics) in the reflector mode with external calibration (Peptide Calibration Standard II, Bruker Daltonics). Flex Analysis 3.0 was used for data analysis with a 50-ppm ion mass tolerance.
Real-Time PCR
RNA was isolated from 5-mm root tips of about 100 seedlings as previously described with minor modifications (Kutschmar et al., 2009). cDNA was synthesized using 1 µg of total RNA, oligo(dT) primers, and RevertAid Reverse Transcriptase (Thermo Fisher Scientific). Primers for qPCR are shown in Supplemental Table S2. PCR (25-µL total volume) was performed in biological triplicates with two technical repeats on the obtained cDNAs using a CFX96 Real-Time PCR Detection System (Bio-Rad). Primer efficiencies and optimal primer concentrations were determined experimentally. qPCR was performed with Taq polymerase expressed in and purified from E. coli and SYBR-Green (Cambrex Bio Science). Relative SBT mRNA levels were determined after normalization to three reference genes (Actin2, Elongation Factor, and Tubulin) as described (Pfaffl, 2001). For SBT expression analysis in root tips, SBT transcript levels are shown relative to the expression of the housekeeping gene Tubulin.
Statistics
All statistical tests were performed in GraphPad Prism v. 7.03 (GraphPad Software). The unpaired, two-tailed Student’s t test was used in all experiments, mostly compared with the wild type or the untreated controls. Statistically significant differences are shown with asterisks: ***P < 0.001, **P < 0.01, or *P < 0.05. Data are represented as boxes with medians and whiskers with 5th and 95th percentiles.
Accession Numbers
Sequence data can be found in The Arabidopsis Information Resourceunder the following accession numbers: CLE40 (At5g12990), XylT (At5g55500), SBT1.4 (At3g14067), SBT1.7 (At5g67360), and SBT4.13 (At5g59120).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The majority of SBTs are expressed in Arabidopsis root tips.
Supplemental Figure S2. The CLE40 precursor is cleaved by SBT1.4 and SBT1.7 in planta.
Supplemental Figure S3. sbt single-gene loss-of-function mutants respond to eCLE40 treatment similar to the wild type.
Supplemental Figure S4. Control for Figures 3A and 7A.
Supplemental Figure S5. Bioactivity of matCLE40 (inhibition of root growth) is not affected by P4A and T5A substitutions.
Supplemental Table S1. Primers used for this study.
Supplemental Table S2. Primers used for qPCR analysis.
Acknowledgments
We thank Lhana Stein (Physiology and Biochemistry, Institute of Biology, University of Hohenheim) for excellent technical support. We also thank Jens Pfannstiel and Berit Würtz (University’s Core Facility Service Unit Mass Spectrometry) for mass spectral analyses.
References
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y(2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA 104: 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulaflous A, Saint-Jore-Dupas C, Herranz-Gordo MC, Pagny-Salehabadi S, Plasson C, Garidou F, Kiefer-Meyer MC, Ritzenthaler C, Faye L, Gomord V(2009) Cytosolic N-terminal arginine-based signals together with a luminal signal target a type II membrane protein to the plant ER. BMC Plant Biol 9: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R(2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Butenko MA, Wildhagen M, Albert M, Jehle A, Kalbacher H, Aalen RB, Felix G(2014) Tools and strategies to match peptide-ligand receptor pairs. Plant Cell 26: 1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corcilius L, Hastwell AH, Zhang M, Williams J, Mackay JP, Gresshoff PM, Ferguson BJ, Payne RJ(2017) Arabinosylation modulates the growth-regulating activity of the peptide hormone CLE40a from soybean. Cell Chem Biol 24: 1347–1355.e7 [DOI] [PubMed] [Google Scholar]
- Czyzewicz N, Wildhagen M, Cattaneo P, Stahl Y, Pinto KG, Aalen RB, Butenko MA, Simon R, Hardtke CS, De Smet I(2015) Antagonistic peptide technology for functional dissection of CLE peptides revisited. J Exp Bot 66: 5367–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll NM, Royek S, Fujita S, Okuda S, Chamot S, Stintzi A, Widiez T, Hothorn M, Schaller A, Geldner N, et al. (2020) A two-way molecular dialogue between embryo and endosperm is required for seed development. Science 367: 431–435 [DOI] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM(2005) The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP.(1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Grosse-Holz F, Kelly S, Blaskowski S, Kaschani F, Kaiser M, van der Hoorn RAL(2018) The transcriptome, extracellular proteome and active secretome of agroinfiltrated Nicotiana benthamiana uncover a large, diverse protease repertoire. Plant Biotechnol J 16: 1068–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobe M, Müller R, Grünewald M, Brand U, Simon R(2003) Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Genes Evol 213: 371–381 [DOI] [PubMed] [Google Scholar]
- Hohl M, Stintzi A, Schaller A(2017) A novel subtilase inhibitor in plants shows structural and functional similarities to protease propeptides. J Biol Chem 292: 6389–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann U, Lau K, Hothorn M(2017) The structural basis of ligand perception and signal activation by receptor kinases. Annu Rev Plant Biol 68: 109–137 [DOI] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H(2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Kutschmar A, Rzewuski G, Stührwohldt N, Beemster GTS, Inzé D, Sauter M(2009) PSK-α promotes root growth in Arabidopsis. New Phytol 181: 820–831 [DOI] [PubMed] [Google Scholar]
- Li Z, Chakraborty S, Xu G(2017) Differential CLE peptide perception by plant receptors implicated from structural and functional analyses of TDIF-TDR interactions. PLoS ONE 12: e0175317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y(2010) Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067 [DOI] [PubMed] [Google Scholar]
- Morita J, Kato K, Nakane T, Kondo Y, Fukuda H, Nishimasu H, Ishitani R, Nureki O(2016) Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide. Nat Commun 7: 12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F(1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y(2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J 55: 152–160 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y(2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M(2013) Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat Commun 4: 2191. [DOI] [PubMed] [Google Scholar]
- Okuda S, Fujita S, Moretti A, Hohmann U, Doblas VG, Ma Y, Pfister A, Brandt B, Geldner N, Hothorn M(2020) Molecular mechanism for the recognition of sequence-divergent CIF peptides by the plant receptor kinases GSO1/SGN3 and GSO2. Proc Natl Acad Sci USA 117: 2693–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagny S, Bouissonnie F, Sarkar M, Follet-Gueye ML, Driouich A, Schachter H, Faye L, Gomord V(2003) Structural requirements for Arabidopsis beta1,2-xylosyltransferase activity and targeting to the Golgi. Plant J 33: 189–203 [DOI] [PubMed] [Google Scholar]
- Paulus JK, Kourelis J, Ramasubramanian S, Homma F, Godson A, Hörger AC, Hong TN, Krahn D, Ossorio Carballo L, Wang S, et al. (2020) Extracellular proteolytic cascade in tomato activates immune protease Rcr3. Proc Natl Acad Sci USA 117: 17409–17417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS(2006) Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24: 79–88 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW.(2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Steinhauser D, Büssis D, Stintzi A, Schaller A, Kopka J, Altmann T(2005) Inferring hypotheses on functional relationships of genes: Analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput Biol 1: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Finn R(2016) Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 44: D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Brandt B, Wildhagen M, Hohmann U, Hothorn LA, Butenko MA, Hothorn M(2016) Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5: e15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T(2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Schägger H.(2006) Tricine-SDS-PAGE. Nat Protoc 1: 16–22 [DOI] [PubMed] [Google Scholar]
- Schaller A, Stintzi A, Rivas S, Serrano I, Chichkova NV, Vartapetian AB, Martínez D, Guiamét JJ, Sueldo DJ, van der Hoorn RAL, et al. (2018) From structure to function: A family portrait of plant subtilases. New Phytol 218: 901–915 [DOI] [PubMed] [Google Scholar]
- Schardon K, Hohl M, Graff L, Pfannstiel J, Schulze W, Stintzi A, Schaller A(2016) Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354: 1594–1597 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T(2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Song W, Han Z, Wang J, Lin G, Chai J(2017) Structural insights into ligand recognition and activation of plant receptor kinases. Curr Opin Struct Biol 43: 18–27 [DOI] [PubMed] [Google Scholar]
- Song XF, Guo P, Ren SC, Xu TT, Liu CM(2013) Antagonistic peptide technology for functional dissection of CLV3/ESR genes in Arabidopsis. Plant Physiol 161: 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, Kühnemuth R, Weidtkamp-Peters S, Pinto KG, Kirschner GK, Schmid JB, Wink RH, Hülsewede A, et al. (2013) Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol 23: 362–371 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R(2009) A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Stührwohldt N, Schaller A(2019) Regulation of plant peptide hormones and growth factors by post-translational modification. Plant Biol (Stuttg) 21(Suppl 1): 49–63 [DOI] [PubMed] [Google Scholar]
- Stührwohldt N, Schardon K, Stintzi A, Schaller A(2017) A toolbox for the analysis of peptide signal biogenesis. Mol Plant 10: 1023–1025 [DOI] [PubMed] [Google Scholar]
- Stührwohldt N, Scholl S, Lang L, Katzenberger J, Schumacher K, Schaller A(2020) The biogenesis of CLEL peptides involves several processing events in consecutive compartments of the secretory pathway. eLife 9: e55580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Poulsen CP, Oikawa A, Dilokpimol A, Halim A, Mangano S, Denita Juarez SP, Marzol E, Salgado Salter JD, et al. (2015) Complex regulation of prolyl-4-hydroxylases impacts root hair expansion. Mol Plant 8: 734–746 [DOI] [PubMed] [Google Scholar]
- Wang J, Li H, Han Z, Zhang H, Wang T, Lin G, Chang J, Yang W, Chai J(2015) Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525: 265–268 [DOI] [PubMed] [Google Scholar]
- Xu TT, Ren SC, Song XF, Liu CM(2015) CLE19 expressed in the embryo regulates both cotyledon establishment and endosperm development in Arabidopsis. J Exp Bot 66: 5217–5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi YL, Ishida T, Sawa S(2016) CLE peptides and their signaling pathways in plant development. J Exp Bot 67: 4813–4826 [DOI] [PubMed] [Google Scholar]
- Yuasa K, Toyooka K, Fukuda H, Matsuoka K(2005) Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J 41: 81–94 [DOI] [PubMed] [Google Scholar]
- Zhang H, Lin X, Han Z, Qu LJ, Chai J(2016) Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res 26: 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]