In Brachypodium distachyon, the GRAS family transcriptional regulator RAM1 is partially required for arbuscule development and, when overexpressed, increases arbuscule density.
Abstract
Arbuscular mycorrhizal (AM) symbiosis is a mutually beneficial association of plants and fungi of the subphylum Glomeromycotina. Endosymbiotic AM fungi colonize the inner cortical cells of the roots, where they form branched hyphae called arbuscules that function in nutrient exchange with the plant. To support arbuscule development and subsequent bidirectional nutrient exchange, the root cortical cells undergo substantial transcriptional reprogramming. REDUCED ARBUSCULAR MYCORRHIZA1 (RAM1), previously studied in several dicot plant species, is a major regulator of this cortical cell transcriptional program. Here, we generated ram1 mutants and RAM1 overexpressors in a monocot, Brachypodium distachyon. The AM phenotypes of two ram1 lines revealed that RAM1 is only partly required to enable arbuscule development in B. distachyon. Transgenic lines constitutively overexpressing BdRAM1 showed constitutive expression of AM-inducible genes even in the shoots. Following inoculation with AM fungi, BdRAM1-overexpressing plants showed higher arbuscule densities relative to controls, indicating the potential to manipulate the relative proportion of symbiotic interfaces via modulation of RAM1. However, the overexpressors also show altered expression of hormone biosynthesis genes and aberrant growth patterns, including stunted bushy shoots and poor seed set. While these phenotypes possibly provide additional clues about the scope of influence of BdRAM1, they also indicate that directed approaches to increase the density of symbiotic interfaces will require a more focused, potentially cell type specific manipulation of transcription factor gene expression.
The GRAS (for GA3 INSENSITIVE [GAI], REPRESSOR OF GAI [RGA], and SCARECROW [SCR]) transcription factor REDUCED ARBUSCULAR MYCORRHIZA (RAM1) has been characterized in three dicot plant species where it is a major regulator of arbuscular mycorrhizal (AM) symbiosis. In Medicago truncatula, Lotus japonicus, and Petunia hybrida ram1 mutants, AM fungi display limited arbuscule branching and reduced hyphal colonization of the root, which results in a nonfunctional symbiosis (Gobbato et al., 2013; Park et al., 2015; Rich et al., 2015; Xue et al., 2015; Pimprikar et al., 2016). RAM1 expression is induced in colonized cortical cells and is regulated by CYCLOPS, a transcription factor of the common symbiosis signaling pathway (Pimprikar et al., 2016) and also by DELLA proteins (Park et al., 2015; Pimprikar et al., 2016), negative regulators of GA signaling (Davière and Achard, 2013). RNA sequencing of ram1 mutants (Luginbuehl et al., 2017), as well as smaller scale gene expression analyses of roots overexpressing RAM1 (Park et al., 2015; Jiang et al., 2017), indicate that RAM1 either directly or indirectly regulates expression of several symbiosis-associated transcription factors, including the GRAS transcription factor RAD1, and three AP2-domain transcription factors of the WRINKLED5 (WRI5) family. RAM1 also either directly or indirectly regulates expression of genes involved in the production and transfer of lipids to the fungal symbiont (e.g. FatM, RAM2, and STR) and the phosphate transporter PT4 (Gobbato et al., 2012; Park et al., 2015; Pimprikar et al., 2016; Luginbuehl et al., 2017; Jiang et al., 2018). However, to date, only one lipid biosynthesis gene, RAM2, has been established as a direct target of RAM1 (Gobbato et al., 2012). Regulation of the other lipid biosynthesis and transport genes likely occurs indirectly through the action of WRI5 family genes (Luginbuehl et al., 2017; Jiang et al., 2018).
RAD1, a GRAS transcription factor very closely related to RAM1 (Supplemental Fig. S1; Park et al., 2015; Xue et al., 2015) is also required for AM symbiosis. In L. japonicus rad1 mutants, AM fungi display defective arbuscule branching phenotypes reminiscent of those seen in ram1 (Xue et al., 2015); however, in M. truncatula rad1, AM fungi show normal arbuscule branching but reduced colonization levels (Park et al., 2015). In line with this observation, several predicted RAM1 target genes were induced in colonized L. japonicus ram1 mutants, but induction was completely abolished in M. truncatula and P. hybrida ram1 (Park et al., 2015; Pimprikar et al., 2016; Luginbuehl et al., 2017; Rich et al., 2017). Thus, there are slight differences in regulation of AM symbiosis genes even between relatively closely related plant species (Pimprikar and Gutjahr, 2018).
Several other GRAS proteins are essential for AM symbiosis, including DELLA/SLR1, a negative regulator of GA signaling (Floss et al., 2013, 2017; Foo et al., 2013; Yu et al., 2014). In della mutants, AM fungi show a severely reduced ability to enter cortical cells, and as a result almost no arbuscules are formed (Floss et al., 2013; Foo et al., 2013; Yu et al., 2014). Arbuscules are ephemeral structures, and the few arbuscules that are formed in della mutants display an increased lifespan, indicating that DELLA not only regulates arbuscule formation but also their degradation (Floss et al., 2017). Two other GRAS transcription factors critical for hormone signaling and AM symbiosis are NSP1 and NSP2. These transcription factors regulate phosphate-dependent strigolactone (SL) biosynthesis in M. truncatula and rice (Oryza sativa; Liu et al., 2011). SLs serve as direct plant communication molecules with AM fungi at the onset of the symbiosis. Mutants impaired in NSP or enzymes required for SL biosynthesis show a reduction in fungal entry into the root and consequently reduced colonization (Gomez-Roldan et al., 2008; Liu et al., 2011; Kobae et al., 2018). Thus, there are several examples of GRAS factors that connect hormone signaling and AM symbiosis.
Many GRAS factors operate in complexes with other GRAS proteins, and emerging evidence suggests that this is also true of those involved in AM symbiosis. M. truncatula and L. japonicus RAM1 were reported to interact with RAD1 and NSP2 (which also interact with each other), but not NSP1 (Gobbato et al., 2012; Park et al., 2015; Xue et al., 2015; Heck et al., 2016). In addition, rice RAM1 interacts with the GRAS transcription factor DIP1, which in turn interacts with DELLA (Yu et al., 2014). M. truncatula DELLA proteins were found to interact with numerous other GRAS transcription factors, including RAD1, MIG1, NSP1, and NSP2 (Floss et al., 2016; Fonouni-Farde et al., 2016; Heck et al., 2016; Jin et al., 2016). While their functional significance for symbiosis remains to be determined, the interactions suggest the existence of interconnected transcriptional modules regulated by multiple GRAS transcription factors.
Brachypodium distachyon is a monocot model species capable of forming AM symbiosis (Hong et al., 2012) and amenable to genetic manipulation (Bragg et al., 2015). A recent study identified 48 GRAS transcription factors in the genome of B. distachyon (Niu et al., 2019). Here, we report functional analyses of the GRAS transcription factor RAM1 in a monocot and assess the potential to alter the levels of symbiotic interfaces by manipulating RAM1 expression.
RESULTS AND DISCUSSION
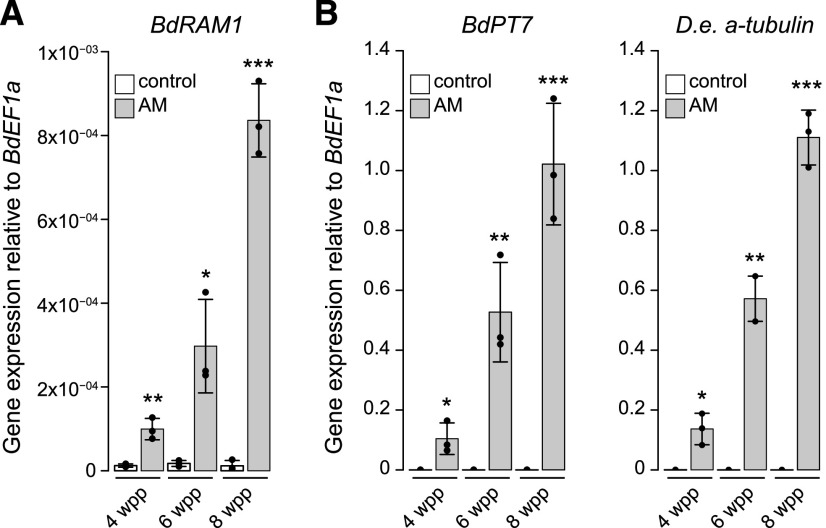
We identified Bradi4g18390 as the single B. distachyon homolog of the GRAS transcription factor RAM1 (Supplemental Fig. S1), a gene that is conserved in AM host plants and missing from nonhosts (Bravo et al., 2016). Similar to orthologous RAM1 genes of M. truncatula (Gobbato et al., 2013; Park et al., 2015), L. japonicus (Xue et al., 2015; Pimprikar et al., 2016), and P. hybrida (Rich et al., 2015), B. distachyon RAM1 expression is induced in mycorrhizal roots. Following inoculation with the AM fungus Diversispora epigaea (formerly Glomus versiforme), BdRAM1 transcripts increased over time in parallel with increasing colonization of the root system as reported by D. epigaea α-tubulin transcripts and the phosphate transporter gene BdPT7 (Hong et al., 2012), a plant gene marker of AM symbiosis (Fig. 1). However, while the transcriptional patterns mirror the marker genes, it is noticeable that BdRAM1 transcript levels are low.
Figure 1.
The B. distachyon ortholog of RAM1 is expressed in roots colonized by AM fungi. A, BdRAM1 gene expression is induced in roots colonized by the AM fungus D. epigaea (gray bars) relative to nonmycorrhizal, mock-inoculated roots (white bars). Plants were harvested 4, 6, and 8 weeks postplanting (wpp). AM-induced BdRAM1 gene expression increases over time. B, Gene expression of the AM marker genes BdPT7 and D. epigaea (D.e.) α-tubulin in D. epigaea-colonized and mock-inoculated control roots over time. A and B, Gene expression was measured by reverse transcription quantitative PCR (RT-qPCR) and normalized to the B. distachyon elongation factor BdEF1α. Bar graphs show the mean, with error bars representing sd. Single points represent individual measurements. Pairwise comparisons of gene expression in AM and control roots were analyzed separately for each time point using Student’s t test (***P < 0.001, **P < 0.01, and *P < 0.05).
The role of RAM1 in AM has been established in at least three dicot host plants (Gobbato et al., 2013; Park et al., 2015; Rich et al., 2015; Xue et al., 2015; Pimprikar et al., 2016), where it is essential to support arbuscule development and appears to act in the upper tier of a transcription factor hierarchy (Luginbuehl et al., 2017); when ectopically overexpressed in roots, RAM1 is sufficient to induce expression of several AM-induced genes in the absence of symbiosis (Park et al., 2015; Pimprikar et al., 2016). Given its AM-inducible expression and pivotal regulatory role, we hypothesized that constitutive, high-level expression of RAM1 might increase the occurrence of arbuscules and possibly overall colonization levels, and this might provide an opportunity to evaluate the functional consequences of modifying colonization patterns. To test this hypothesis, we transformed B. distachyon with an overexpression construct, BdRAM1, under the control of two copies of the constitutively active CaMV 35S promoter (35S:BdRAM1). In addition, we generated B. distachyon ram1 loss-of-function mutants via CRISPR/Cas9 editing.
Arbuscule Development in B. distachyon ram1 Mutants Is Partly Impaired
Five independent transgenic lines carrying a two-guide CRISPR/CAS9 construct targeting BdRAM1 were generated, and two lines in which BdRAM1 had been edited were chosen for subsequent analysis. In both transgenic lines, the genome had been edited by both guides (Supplemental Fig. S2); editing by the upstream-most guide resulted in premature stop codons and created a truncated protein of 16 amino acids in the first line, designated ram1-1. The second line, designated ram1-2, was biallelic, with edits resulting in premature stop codons that generated truncated protein products of 16 and 42 amino acids. Both ram1 lines appeared as wild type with respect to root and shoot morphology.
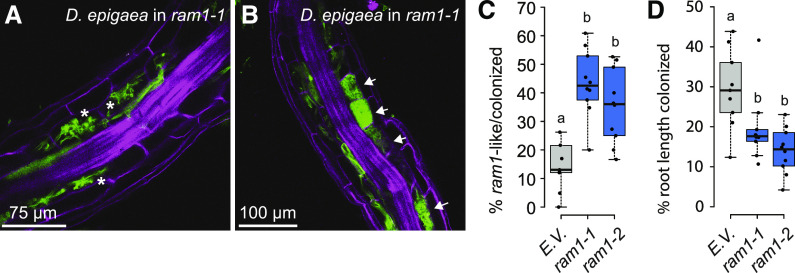
ram1-1 and ram1-2 were inoculated with D. epigaea, and the fungal colonization patterns were examined after 4 weeks. Some ram1 roots showed aberrant infection units, reminiscent of the typical dicot ram1 phenotype, with intraradical hyphae and only small, sparsely branched arbuscules but no fully developed arbuscules (Fig. 2A). However, infection units in other roots of the same plant, or even in other parts of the same root, showed an apparent wild-type morphology with some large, well-branched arbuscules (Fig. 2B). In comparison with the empty vector control, the frequency of aberrant infections in the B. distachyon ram1 mutants was 2.5- to 3-fold higher, and their overall root colonization levels were 34 to 52% lower. Similar results were obtained in experiments across several generations (Supplemental Fig. S2). These results indicate that BdRAM1 is required to enable wild-type levels of arbuscule development similar to its orthologs in dicots; however, the B. distachyon ram1 phenotype is clearly milder than that observed in dicot ram1 mutants. The finding that B. distachyon ram1 can support some full arbuscule development suggests that other proteins or pathways have the potential to compensate for loss of BdRAM1 function. One possible candidate is the GRAS protein RAD1, which is closely related to RAM1 and induced in roots highly colonized by AM fungi (Supplemental Figs. S1 and S3). In legumes, there is evidence of a species-specific micro-diversification of RAM1 and RAD1, with the relative contributions of the two transcription factors to arbuscule development and symbiotic gene expression varying depending on the host species (Park et al., 2015; Xue et al., 2015; Pimprikar et al., 2016; Pimprikar and Gutjahr, 2018). It is therefore conceivable that some diversification of GRAS factor functions has occurred during the evolution of monocots, which might explain the milder arbuscule development phenotype of B. distachyon ram1 mutants relative to M. truncatula ram1. Interestingly, there are other GRAS factor examples where the converse is true, for example the DELLA proteins, where rice slr (Yu et al., 2014) shows a stronger phenotype than the M. truncatula della double or triple mutants (Floss et al., 2013, 2017). However, in the absence of other monocot ram1 mutants for comparison, it is also possible that the milder ram1 phenotype observed here is a feature specific to B. distachyon.
Figure 2.
Arbuscule formation is impaired in B. distachyon ram1 mutants. A, A root piece with ram1-like D. epigaea arbuscules in CRISPR ram1 mutants. ram1-like infections contain solely arbuscules that are not fully developed and show only sparse branching, reminiscent of the ram1 mutant phenotype described previously in dicots (indicated by asterisks). B, A root piece with wild type-like D. epigaea arbuscules in CRISPR ram1 mutants. Wild type-like infections contain fully developed arbuscules (indicated by arrows). D. epigaea fungal structures visualized using WGA-Alexa Fluor 488 (green), with plant cell walls counterstained using propidium iodide (pink). C, Quantification of ram1-like infections relative to the total number of infections in ram1 CRISPR plants and B. distachyon plants transformed with the empty vector (E.V.). The proportion of aberrant infections is increased in two ram1 alleles (ANOVA, P = 2.38 × 10−5). D, Quantification of total D. epigaea root-length colonization in CRISPR ram1 plants relative to E.V. controls. Root length colonization is significantly decreased in two ram1 mutant alleles (ANOVA, P = 0.0014). Pairwise comparisons in C and D were performed using Tukey’s HSD post-hoc test. Different lowercase letters denote significant differences. Box and whisker plots show lower and upper quartiles and minimum and maximum values. The horizontal bar represents the median and the points individual measurements. All results presented in this figure were obtained from the T3 generation.
Overexpression of RAM1 Alters Plant Morphology and Results in Constitutive Expression of AM Marker Genes
The generation of transgenic B. distachyon plants overexpressing RAM1 was surprisingly challenging; from two full-scale independent transformation experiments, only three viable independent transgenic 35S:BdRAM1-overexpressing lines (35S:BdRAM1ox) were obtained, and the seed production from these lines was exceedingly poor. In addition, we obtained two lines, which carried the 35S:BdRAM1 T-DNA but displayed wild type-like BdRAM1 transcript levels (35S:BdRAM1WT). By contrast, transgenic plants carrying 35S:NLS-GFP-GUS (hereto referred to simply as 35S:NLS-GFP), were generated without difficulty. Seed production from the latter two genotypes was not impaired.
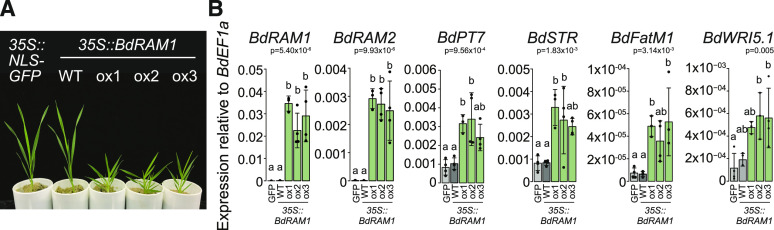
In addition to poor seed production and viability, the shoot and root phenotypes of the three lines with transcriptional up-regulation of BdRAM1 (35S:BdRAM1ox) differed from the vector controls and from the 35S:BdRAM1WT plants. The 35S:BdRAM1ox plants were characterized by a stunted bushy shoot with increased tiller formation and increased leaf angles, as well as a decreased number of node roots (Supplemental Fig. S4). Thus, constitutive overexpression of BdRAM1 clearly influences plant development. While the cause is unknown, it might be the result of ectopic expression of BdRAM1 target genes and/or perhaps an interference of RAM1 with other GRAS transcriptional networks, many of which regulate development (for review, see Cenci and Rouard, 2017). Either way, the aberrant developmental phenotypes likely explain the difficulties in regenerating transgenic lines and their fecundity.
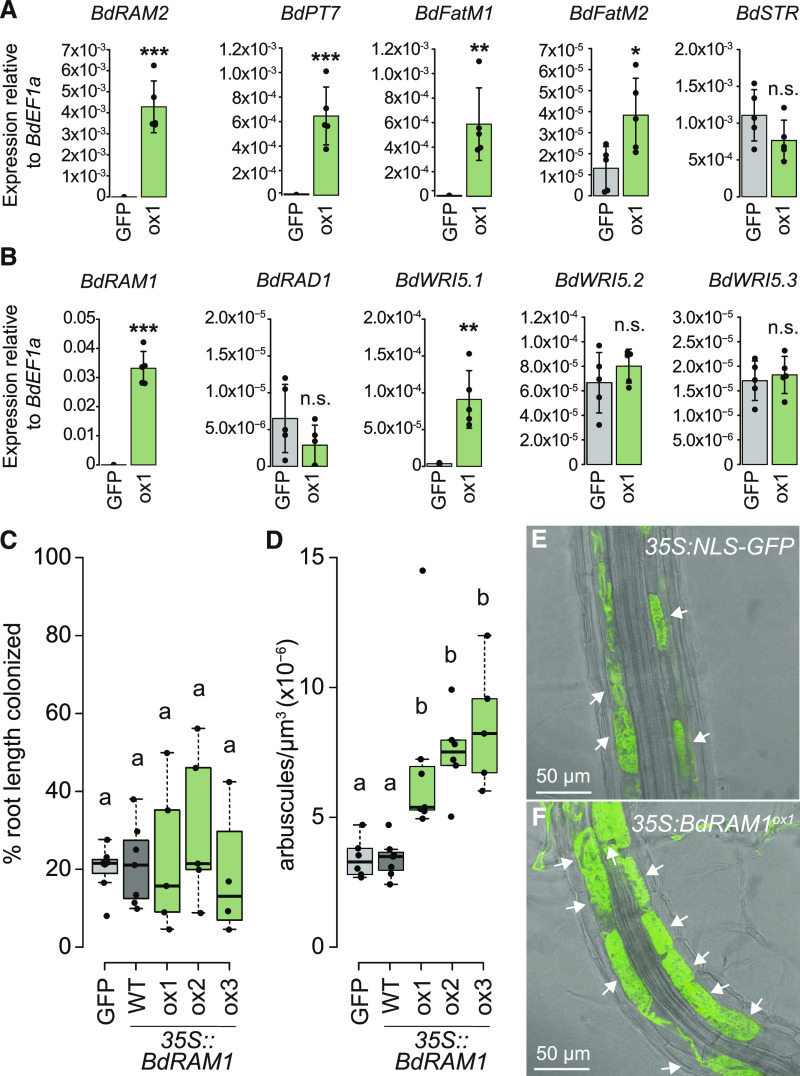
The 35S:BdRAM1ox plants showed constitutive expression of B. distachyon orthologs of RAM2, STR, PT4, and FatM (Fig. 3A); elevated expression of these genes would normally occur only in response to colonization by AM fungi (for example, Harrison et al., 2002; Paszkowski et al., 2002; Gutjahr et al., 2008, 2012; Zhang et al., 2010; Gobbato et al., 2012; Hong et al., 2012; Bravo et al., 2017), and we observed a similar expression pattern in B. distachyon mycorrhizal roots (Fig. 1B; Supplemental Fig. S3). Given the prior knowledge from dicots, we had anticipated that 35S:BdRAM1ox would increase expression of these genes in roots, but it was surprising to see that expression of these genes was also induced in shoots (Fig. 4). There were some exceptions; expression of BdSTR increased in 35S:BdRAM1ox shoots, but not in roots, while BdFatM2 showed the opposite expression pattern (Figs. 3A and 4B; Supplemental Fig. S5). Overall, these data indicate that BdRAM1 alone is sufficient to drive increased expression of these genes in the absence of AM fungi and that transcription cofactors, if required for BdRAM1 function, must be present in all tissues.
Figure 3.
Ectopic overexpression of BdRAM1 promotes arbuscule formation and expression of AM marker genes. A, Gene expression levels of B. distachyon orthologs of MtRAM1 target genes in noncolonized 35S:NLS-GFP (denoted as GFP) and 35S:BdRAM1ox line 1 (denoted as ox1) roots. 35S:BdRAM1ox roots display induced expression of BdRAM1, as well as BdRAM2, BdPT7, BdFatM1, and BdFatM2 in the absence of symbiosis relative to 35S:NLS-GFP control roots. BdSTR gene expression is not affected in these roots. B, Gene expression of B. distachyon RAD1 and WRI5 orthologs. Only expression of BdWRI5.1 is induced in noncolonized 35S:BdRAM1ox roots relative to 35S:NLS-GFP control roots. A and B, Bar graphs show the mean, with error bars representing sd. Single points represent individual measurements. Pairwise comparisons were estimated using the Student’s t test (***P < 0.001, **P < 0.01, and *P < 0.05).; n.s., Not significant. C, Quantification of total root colonization in independent lines transformed with 35S:BdRAM1. There is no difference in overall root colonization between three lines ectopically overexpressing 35S:BdRAM1 (35S:BdRAM1ox, denoted as ox1, ox2, and ox3) and control plants (35S:BdRAM1WT, which does not overexpress BdRAM1 and was therefore denoted as WT [wild type]; and 35S:NLS-GFP, labeled as GFP). ANOVA (P = 0.71). Root-length colonization was quantified using the grid-line method (McGonigle et al., 1990). D, Quantification of D. epigaea arbuscules in a defined area at the fungal hyphopodium. Roots of three independent transgenic 35S:BdRAM1ox lines (denoted as ox1, ox2, and ox3) contain more arbuscules than roots transformed with the control construct 35S:NLS-GFP (GFP) or roots that contain 35S:BdRAM1 but do not overexpress the gene (WT). Arbuscule number was normalized to the volume of the confocal stack. Kruskal-Wallis test (P = 1.32 × 10−4), pairwise comparisons were conducted using the Dunn’s post-hoc test. Different lowercase letters denote significant differences. Box and whisker plots show lower and upper quartiles and minimum and maximum values. The horizontal bar represents the median and the points individual measurements. E, Representative image of a 35S:NLS-GFP root colonized by D. epigaea. F, Representative image of a RAM1-overexpressing 35S:BdRAM1ox (line 1) root colonized by D. epigaea. In E and F, arbuscules are highlighted with arrows.
Figure 4.
RAM1 overexpressors show altered shoot development and constitutively express root AM marker genes in their shoots. A, Photograph of 4.5-week-old B. distachyon plants transformed with 35S:NLS-GFP or 35S:BdRAM1. The three independent transformant lines overexpressing BdRAM1 (ox1, ox2, and ox3) display a bushy stature, whereas the 35S:BdRAM1-transformant line not overexpressing BdRAM1 (wild type [WT]) resembles the 35S:NLS-GFP control plant. B, Gene expression of BdRAM1 and of several root AM marker genes in shoots. All tested genes are strongly induced in 4.5-week-old shoots of three 35S:BdRAM1ox lines (ox1, ox2, and ox3) relative to control plants transformed with 35S:NLS-GFP (GFP) or the 35S:BdRAM1-transformant line not overexpressing BdRAM1 (wild type). Gene expression was measured by RT-qPCR. Bar graphs show the mean, with error bars representing sd. Single points represent individual measurements. Significance values (ANOVA) for each gene are indicated in the figure. Pairwise comparisons were conducted using Tukey’s HSD post-hoc test. Different lowercase letters denote significant differences.
In dicots, RAM1 regulates expression of a second tier of transcription factors including RAD1 and three members of the WRINKLED family (WRI5a–WRI5c); the latter directly regulate expression of lipid biosynthesis genes (Park et al., 2015; Luginbuehl et al., 2017; Jiang et al., 2018). We found that BdRAD1 and the three B. distachyon AP2 family transcription factors most closely related to MtWRI5a to MtWRI5c (further denoted as BdWRI5.1, BdWRI5.2, and BdWRI5.3) were strongly induced in wild-type roots colonized with D. epigaea relative to mock-inoculated controls (Supplemental Fig. S3), but interestingly, only BdWRI5.1 was induced in noncolonized 35S:BdRAM1ox roots (Fig. 3B). A similar pattern was observed in 35S:BdRAM1ox shoots (Fig. 4B; Supplemental Fig. S5). Thus, in contrast to M. truncatula, a RAM1-independent pathway likely leads to up-regulation of BdRAD1, BdWRI5.2, and BdWRI5.3 in mycorrhizal roots. This points to functional diversification of the regulatory cascade responsible for the transcriptional reprogramming of roots during AM symbiosis in B. distachyon. In addition, it may provide an explanation for the relatively mild ram1 mutant phenotype we observed (Fig. 2). Future research in other monocot species is required to determine if such a functional diversification is unique to B. distachyon or a monocot-specific phenomenon.
Arbuscule Density Is Higher in RAM1 Overexpressors Relative to Controls
The initial goal of this study was to test the hypothesis that constitutive overexpression of RAM1 would increase arbuscule density and/or colonization and then to use the plants to address secondary hypotheses about symbiotic performance.
To test the first hypothesis, we grew 35S:BdRAM1ox, 35S:BdRAM1WT, and 35S:NLS-GFP control plants in substrate containing D. epigaea spores and evaluated colonization levels and arbuscule morphology. Colonization levels in 35S:BdRAM1ox and control plants did not differ significantly, although the variation was much greater in the 35S:BdRAM1ox plants (Fig. 3C). Arbuscules in 35S:BdRAM1ox plants showed a wild-type morphology, but the number of arbuscules, which we assessed within a defined root volume below the hyphopodium, was on average 2-fold greater in the 35S:BdRAMox plants relative to controls (Fig. 3, D–F; Supplemental Fig. S5). Thus, 35S:BdRAM1ox plants have a higher capacity to establish and/or to maintain arbuscules relative to the control plants. As RAM1 regulates the expression of several other transcription factors, as well as genes involved in lipid biosynthesis and nutrient transport, the increased arbuscule density in the 35S:BdRAM1ox plants may result from a combination of factors including arbuscule initiation and/or regulation of arbuscule lifespan.
Unfortunately, the severe shoot growth and branching phenotype of the BdRAM1 overexpressors prevented a fair evaluation of symbiotic performance (Supplemental Fig. S5). While colonized 35S:BdRAM1ox plants and controls both showed an increase in shoot fresh weight and tiller number relative to their respective mock-inoculated controls, the differences in the developmental architecture of these lines precluded direct physiological comparisons. Consequently, it was not possible to determine whether the increased arbuscule density influenced symbiotic performance.
Hormone Biosynthetic and Regulatory Gene Expression Is Altered in BdRAM1 Overexpressors
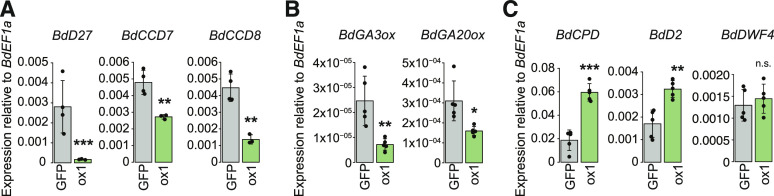
The shoot architecture phenotype of the 35S:BdRAM1ox plants is reminiscent of the phenotypes of several monocot hormone mutants. For example, rice and B. distachyon mutants defective in GA, SL, and brassinosteroid (BR) biosynthesis or signaling display dwarf phenotypes with increased tillering (e.g. Spielmeyer et al., 2002; Ishikawa et al., 2005; Asano et al., 2009; Lin et al., 2009; Thole et al., 2012). To obtain further clues about the BdRAM1 overexpression phenotype, we evaluated the expression of several genes associated with SL, GA, and BR signaling. B. distachyon orthologs of genes involved in SL biosynthesis (BdD27, BdCCD7, and BdCCD8; Seto and Yamaguchi, 2014) and GA biosynthesis (potential orthologs of Arabidopsis [Arabidopsis thaliana] GA3ox1 and GA20ox1; Kakei et al., 2015) were down-regulated, while key BR biosynthesis genes (BdCPD and BdD2/CYP90D) but not BdDWF4 (Kakei et al., 2015) were elevated in noncolonized 35S:BdRAM1ox roots relative to the controls (Fig. 5). The GA receptor GID1 and the GA regulator DELLA/SLR1 (Davière and Achard, 2013) as well as the regulators of SL signaling D3 and D53 (Seto and Yamaguchi, 2014), and the B. distachyon BR receptor BdBRI1 and the BR-responsive transcription factor BdBZR1 (Corvalán and Choe, 2017) were differentially regulated in 35S:BdRAM1ox roots relative to controls (Supplemental Fig. S6). In addition, altered expression of some BR biosynthesis genes and GA biosynthesis and signaling genes was observed in 35S:BdRAM1ox shoots (Supplemental Table S1). Thus, the transcript data indicate a disturbance in hormone biosynthetic and regulatory gene expression likely contributing to the altered shoot architecture. Because of substantial cross talk between hormone signaling pathways (Itoh et al., 2001; Umehara et al., 2008; Unterholzner et al., 2015; Corvalán and Choe, 2017), it is not possible to predict the initial cause. As GA, SL, and BR hormone pathways each involve regulation via GRAS transcription factors (Tong et al., 2009; Liu et al., 2011; Chen et al., 2013), it is possible that ectopic overexpression of BdRAM1 disturbs GRAS-factor complexes, leading to misregulation of these pathways. Alternatively, one of the native functions of BdRAM1 may be to regulate aspects of hormone signaling. For example, in rice, RAM1 interacts with a DELLA-interacting protein (DIP), and therefore it is possible that one of RAM1’s native functions is to influence GA signaling and that this is exacerbated in the 35S:BdRAM1ox line, leading to further downstream effects on other pathways. SL promotes initiation of AM symbiosis, influencing both primary and secondary infections, and BR is a positive regulator of AM symbiosis in some plant species (for review, see Kobae et al., 2018; Liao et al., 2018; Müller and Harrison, 2019); however, direct effects on arbuscule development have not been reported for these hormones. By contrast, it is well established that GA is involved in arbuscule formation and regulation of arbuscule lifespan (Floss et al., 2013, 2017; Yu et al., 2014). If misregulation of GA biosynthesis gene expression translates to disturbed GA homeostasis in 35S:BdRAM1ox roots, an imbalance in GA-regulated arbuscule formation and degradation could result (Floss et al., 2013, 2017). Such a scenario might explain the increased arbuscule numbers in 35S:BdRAM1ox roots as well as a dwarf shoot phenotype.
Figure 5.
Expression of hormone biosynthesis genes is altered in roots of RAM1 overexpressors. A, B. distachyon orthologs of three genes involved in the SL biosynthesis pathway (BdD27, BdCCD7, and BdCCD8) are down-regulated in noncolonized roots ectopically overexpressing BdRAM1 (35S:BdRAM1ox line 1, denoted as ox1) relative to 35S:NLS-GFP (“GFP”) control roots. B, Two genes with a putative function in GA biosynthesis (BdGA3ox1 and BdGA20ox1) are down-regulated in 35S:BdRAM1ox roots. C, Two B. distachyon genes orthologous to known brassinosteroid biosynthesis genes (BdCPD and BdD2/BdCYP91D) are induced in 35S:BdRAM1ox roots. A third gene, BdDWF4, is not affected. Gene expression was measured by RT-qPCR. Bar graphs show the mean, with error bars representing sd. Single points represent individual measurements. Pairwise comparisons were estimated using the Student’s t test (***P < 0.001, **P < 0.01, and *P < 0.05). n.s., Not significant.
CONCLUSION
In conclusion, BdRAM1, similar to its orthologs in dicots, regulates arbuscule development and transcriptional regulation of several AM symbiosis-induced genes, although it is likely that there is some functional redundancy with other GRAS or WRI5 transcription factors. Constitutive overexpression of 35S:BdRAM1 increased arbuscule density relative to control plants; although the plants were unsuitable for experiments to assess the functional consequences of increasing the symbiotic interfaces, the data nevertheless indicate that it is possible to manipulate arbuscule density through expression of RAM1. Whether this is a direct effect via elevated expression of known BdRAM1 target genes or an indirect effect through phytohormones that influence plant architecture as well as arbuscule initiation, development, or lifespan is currently unknown.
Future research should focus on increasing RAM1 gene expression specifically in the root cortex. We predict such a strategy would increase arbuscule density without the accompanying developmental defects and would enable evaluation of the consequences of increasing the density of symbiotic interfaces and the effects on nutrient exchange during AM symbiosis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Brachypodium distachyon plants were grown in a growth chamber under a 12-h light (24°C)/12-h dark (22°C) regime. For all experiments that were conducted in the absence of an AM fungal symbiont, B. distachyon plants were grown in 20.5-cm-long cones filled with sterile Terragreen (Oli-Dri) and play sand (Quikrete) in a ratio of 1:1. For all experiments involving AM symbiosis, B. distachyon plants were grown in cones filled with a sand-gravel mix and were inoculated with 250 Diversispora epigaea spores (formerly Glomus versiforme) as previously described (Müller et al., 2019). For mock-inoculated controls, we added an appropriate volume of filtered spore wash solution instead of the spores. Unless otherwise stated, B. distachyon plants were fertilized once per week with one-fourth-strength Hoagland’s fertilizer containing 20 µm Pi and harvested 4 to 5 weeks after transplanting to cones.
To monitor AM growth responses, seedlings were planted into pots (three seedlings per 11-cm-diameter pot and eight pots per genotype) containing a 1:20 mixture of autoclaved N7/N8 soil (Watts-Williams et al., 2019) to sand/gravel mix. The sand/gravel mix is a 2:2:1 mixture of play sand, fine black sand, and gravel as described in Floss et al. (2017). Two hundred fifty surface-sterilized D. epigaea spores were placed below each plant. Beginning at 3 weeks postplanting, the pots were fertilized weekly with 50 mL of one-fourth-strength Hoagland’s solution lacking phosphate and 9 mL of 0.5 mm Ca3(PO4)2. Plants were harvested at 9 weeks postplanting. The growth chamber conditions were as described above.
Plasmid Generation
To clone the CRISPR/Cas9 construct targeting BdRAM1, we used the vector and cloning system described previously (Xie et al., 2015). To design the primers (shown in Supplemental Table S2), gene-specific guide RNA sequences targeting Bradi4g18390 were identified using CRISPR-P (Lei et al., 2014) and CRISPR-PLANT (Xie et al., 2015) and selected based on their location in the coding sequence and low number of off-target sites. We generated a two-guide CRISPR/Cas9 construct that targeted Bradi4g18390 at positions 32 to 54 bp (guide RNA1) and 280 to 302 bp (guide RNA2) downstream of the transcription start site (Supplemental Fig. S2). As a negative control, we used the empty vector pRGEB32 (Xie et al., 2015).
To clone 35S:BdRAM1 overexpression constructs, the coding sequence of Bradi4g18390 was amplified using gene-specific primers flanked by attB1 and attB2 recombination sites (Supplemental Table S2) and cloned into pDONR221, resulting in the pENTR1-2 BdRAM1 entry clone. pENTR1-2 clones containing the coding sequence of NLS-GFP-GUS, as well as pENTR4-1 entry clones containing the CaMV35S promoter and pENTR2-3 containing the CaMV35S terminator were cloned previously (Ivanov and Harrison, 2014; Floss et al., 2017). To assemble the binary vectors for B. distachyon transformation, four vectors (pENTR4-1 containing the double CaMV35S promoter, pENTR1-2 containing BdRAM1 or NLS-GFP-GUS, pENTR2-3 containing the CaMV35S terminator, and pHb7m34GW; Karimi et al., 2005) were combined to generate 35S:BdRAM1 or 35S:NLS-GFP using the multisite gateway cloning system (Invitrogen). All vector sequences were confirmed by Sanger sequencing.
Generation of B. distachyon Transformants
The CRISPR/Cas9 constructs targeting BdRAM1 as well as the 35S:BdRAM1 and 35S:NLS-GFP constructs were transformed into B. distachyon (accession Bd21-3) following a previously established protocol (Bragg et al., 2015). Plantlets emerging from transformed calli (selectable marker, hygromycin) were transplanted into Metro-Mix 350 and genotyped to test for the presence of the construct (see Supplemental Table S2 for primer sequences). In addition, in the case of the CRISPR/Cas9 constructs, the CRISPR/Cas9 target loci were amplified using flanking primers, and purified PCR products were Sanger sequenced in order to identify gene edits.
Visualization and Quantification of Fungal Root Colonization
Fungal colonization of B. distachyon roots was visualized by staining with wheat-germ agglutinin coupled to Alexa Fluor 488 as previously described (Hong et al., 2012). Roots were observed using a Leica M205 stereomicroscope, and root colonization was quantified using the gridline-intersect method (McGonigle et al., 1990). To quantify the ram1 phenotype, roots intersecting the gridlines were scored into one of three categories: (1) not colonized; (2) colonized with wild type-like arbuscules; and (3) colonized with aberrant (sparsely branched or collapsed arbuscules) or no arbuscules. The ratio of category 3 over the overall number of intersections of colonized roots (category 2 + 3) × 100 was used to determine the percentage of intersections without arbuscules/total colonization. Total root-length colonization was calculated as the percentage of category 2 + 3 over the total number of intersections counted × 100. To study arbuscule morphology, wheat-germ agglutinin-Alexa Fluor 488-stained roots were counterstained with propidium iodide to visualize plant cell walls and observed with a Leica SP5 confocal microscope. To quantify arbuscule numbers in 35S:BdRAM1ox roots, confocal stacks from highly colonized roots were taken so that the fungal hyphopodium was in the center of the image to ensure we captured infections of similar developmental stages. The total number of arbuscules per stack was assessed manually using the Fiji image analysis package (Schindelin et al., 2012). Stack depth (z plane) was chosen to encompass the whole infection, and arbuscule numbers were normalized against the stack volume (length of x × y × z planes). To avoid potentially confounding effects caused by different B. distachyon root types, we selected only thin lateral roots with a single layer of cortical cells for analysis.
RNA Isolation, cDNA Synthesis, and RT-qPCR
RNA isolation, cDNA synthesis, and RT-qPCR were performed as previously described (Müller et al., 2019). Primers used to quantify expression of target genes are shown in Supplemental Table S2. Ct values of the tested genes were normalized against BdEF1α (resulting in ΔCt), and relative expression levels were calculated with the equation 2−ΔCt.
Assessment of Plant Morphology
Plants were grown in the absence of AM fungi, and whole plants were harvested 2, 4, and 6 weeks after planting. Tiller and node root numbers were counted, and maximal root system and shoot length were measured. The angle between individual leaves and the stem was measured on images of the same plants using the Fiji image analysis package (Schindelin et al., 2012).
Phylogenetic Analyses
B. distachyon orthologs of Medicago truncatula RAM1, RAD1, RAM2, PT4, FatM, STR, and WRI5a to WRI5c as well as Oryza sativa D27, D17/CCD7, D10/CCD8, D3, D53, D14, D14L, and SLR1 were identified using phylogenetic approaches described previously (Supplemental Figs. S1 and S7; Bravo et al., 2016). B. distachyon genes putatively involved in BR and GA biosynthesis and signaling were identified previously (Kakei et al., 2015; Corvalán and Choe, 2017; Niu et al., 2019).
Statistical Analyses and Data Representation
All experiments were performed using three to 10 biological replicates. All experiments were repeated at least two times. The distribution of residuals was tested for normality using the Shapiro-Wilk test. If normality assumption was met, pairwise comparisons were analyzed using a two-sided Student’s t test. For multiple comparisons, the raw data were subjected to a one-way ANOVA followed by Tukey’s post-hoc test. If normality assumption was not met, data were analyzed using the Kruskal-Wallis test followed by Dunn’s post-hoc test (P values adjusted after Benjamini-Hochberg). All statistical analyses were performed using R software. Quantification data for n > 5 biological replicates are represented as box and whisker plots, which show the lower and upper quartiles as well as the minimum and maximum values. The horizontal line in the box plots represents the median. Points represent single measurements. For datasets with less than five measurements per genotype, bar plots were chosen. Bars represent the mean and error bars the sd. Points represent single measurements.
Accession Numbers
B. distachyon gene identifiers for all genes referred to in this manuscript can be found in Supplemental Table S2.
Supplemental Data
The following materials are available.
Supplemental Figure S1. Phylogenetic tree showing GRAS transcription factors related to BdRAM1 in the AM host species (B. distachyon, O. sativa, Hordeum vulgare, Zea mays, M. truncatula) and the nonhost species Arabidopsis.
Supplemental Figure S2. B. distachyon CRISPR/Cas9 edited ram1 mutants.
Supplemental Figure S3. Gene expression of selected AM marker genes in colonized roots.
Supplemental Figure S4. Developmental phenotypes caused by ectopic overexpression of BdRAM1 measured at 2, 4, and 6 weeks postplanting.
Supplemental Figure S5. Growth response experiment and analysis of arbuscule numbers and gene expression in 35S:BdRAM1ox plants.
Supplemental Figure S6. Ectopic overexpression of BdRAM1 influences expression of genes associated with SL, GA, and BR signaling.
Supplemental Figure S7. Phylogenetic trees used to identify B. distachyon orthologs of AM marker genes.
Supplemental Table S1. Gene expression fold change of selected hormone marker genes in shoots overexpressing BdRAM1.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We thank Sophia Cotraccia, Cassandra Proctor, and Stephanie Roh for technical assistance and the Boyce Thompson Institute Biotechnology Center for generating some of the B. distachyon transgenic lines.
Footnotes
This work was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (grant no. DE–SC0012460), the TRIAD Foundation, the Swiss National Science Foundation (Early Postdoc.Mobility fellowship to L.M.M.), the German Research Foundation (Postdoctoral Fellowship to L.M.M.), and a Marie Curie Fellowship (grant no. FP7–PEOPLE–2013–IOF–624739 to L.C.-S.).
Articles can be viewed without a subscription.
References
- Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim RB, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M(2009) Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol Genet Genomics 281: 223–231 [DOI] [PubMed] [Google Scholar]
- Bragg JN, Anderton A, Nieu R, Vogel JP(2015) Brachypodium distachyon. Methods Mol Biol 1223: 17–33 [DOI] [PubMed] [Google Scholar]
- Bravo A, Brands M, Wewer V, Dörmann P, Harrison MJ(2017) Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol 214: 1631–1645 [DOI] [PubMed] [Google Scholar]
- Bravo A, York T, Pumplin N, Mueller LA, Harrison MJ(2016) Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat Plants 2: 15208. [DOI] [PubMed] [Google Scholar]
- Cenci A, Rouard M(2017) Evolutionary analyses of GRAS transcription factors in angiosperms. Front Plant Sci 8: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Xiong G, Cui X, Yan M, Xu T, Qian Q, Xue Y, Li J, Wang Y(2013) OsGRAS19 may be a novel component involved in the brassinosteroid signaling pathway in rice. Mol Plant 6: 988–991 [DOI] [PubMed] [Google Scholar]
- Corvalán C, Choe S(2017) Identification of brassinosteroid genes in Brachypodium distachyon. BMC Plant Biol 17: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière JM, Achard P(2013) Gibberellin signaling in plants. Development 140: 1147–1151 [DOI] [PubMed] [Google Scholar]
- Floss DS, Gomez SK, Park HJ, MacLean AM, Müller LM, Bhattarai KK, Lévesque-Tremblay V, Maldonado-Mendoza IE, Harrison MJ(2017) A transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Curr Biol 27: 1206–1212 [DOI] [PubMed] [Google Scholar]
- Floss DS, Lévesque-Tremblay V, Park HJ, Harrison MJ(2016) DELLA proteins regulate expression of a subset of AM symbiosis-induced genes in Medicago truncatula. Plant Signal Behav 11: e1162369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss DS, Levy JG, Levesque-Tremblay V, Pumplin N, Harrison MJ(2013) DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 110: E5025–E5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonouni-Farde C, Tan S, Baudin M, Brault M, Wen J, Mysore KS, Niebel A, Frugier F, Diet A(2016) DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat Commun 7: 12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Ross JJ, Jones WT, Reid JB(2013) Plant hormones in arbuscular mycorrhizal symbioses: An emerging role for gibberellins. Ann Bot 111: 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbato E, Marsh JF, Vernié T, Wang E, Maillet F, Kim J, Miller JB, Sun J, Bano SA, Ratet P, et al. (2012) A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr Biol 22: 2236–2241 [DOI] [PubMed] [Google Scholar]
- Gobbato E, Wang E, Higgins G, Bano SA, Henry C, Schultze M, Oldroyd GED(2013) RAM1 and RAM2 function and expression during arbuscular mycorrhizal symbiosis and Aphanomyces euteiches colonization. Plant Signal Behav 8: e26049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot J-P, Letisse F, Matusova R, Danoun S, Portais J-C, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Banba M, Croset V, An K, Miyao A, An G, Hirochika H, Imaizumi-Anraku H, Paszkowski U(2008) Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20: 2989–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Radovanovic D, Geoffroy J, Zhang Q, Siegler H, Chiapello M, Casieri L, An K, An G, Guiderdoni E, et al. (2012) The half-size ABC transporters STR1 and STR2 are indispensable for mycorrhizal arbuscule formation in rice. Plant J 69: 906–920 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J(2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck C, Kuhn H, Heidt S, Walter S, Rieger N, Requena N(2016) Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr Biol 26: 2770–2778 [DOI] [PubMed] [Google Scholar]
- Hong JJ, Park YS, Bravo A, Bhattarai KK, Daniels DA, Harrison MJ(2012) Diversity of morphology and function in arbuscular mycorrhizal symbioses in Brachypodium distachyon. Planta 236: 851–865 [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J(2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46: 79–86 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M(2001) Cloning and functional analysis of two gibberellin 3 beta-hydroxylase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci USA 98: 8909–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Harrison MJ(2014) A set of fluorescent protein-based markers expressed from constitutive and arbuscular mycorrhiza-inducible promoters to label organelles, membranes and cytoskeletal elements in Medicago truncatula. Plant J 80: 1151–1163 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wang W, Xie Q, Liu N, Liu L, Wang D, Zhang X, Yang C, Chen X, Tang D, et al. (2017) Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xie Q, Wang W, Yang J, Zhang X, Yu N, Zhou Y, Wang E(2018) Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol Plant 11: 1344–1359 [DOI] [PubMed] [Google Scholar]
- Jin Y, Liu H, Luo D, Yu N, Dong W, Wang C, Zhang X, Dai H, Yang J, Wang E(2016) DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat Commun 7: 12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakei Y, Mochida K, Sakurai T, Yoshida T, Shinozaki K, Shimada Y(2015) Transcriptome analysis of hormone-induced gene expression in Brachypodium distachyon. Sci Rep 5: 14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P(2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kobae Y, Kameoka H, Sugimura Y, Saito K, Ohtomo R, Fujiwara T, Kyozuka J(2018) Strigolactone biosynthesis genes of rice are required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant Cell Physiol 59: 544–553 [DOI] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu H-Y, Li S, Xing F, Chen L-L(2014) CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7: 1494–1496 [DOI] [PubMed] [Google Scholar]
- Liao D, Wang S, Cui M, Liu J, Chen A, Xu G(2018) Phytohormones regulate the development of arbuscular mycorrhizal symbiosis. Int J Mol Sci 19: 3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, et al. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, et al. (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23: 3853–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, Oldroyd GED, Eastmond PJ(2017) Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356: 1175–1178 [DOI] [PubMed] [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA(1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115: 495–501 [DOI] [PubMed] [Google Scholar]
- Müller LM, Flokova K, Schnabel E, Sun X, Fei Z, Frugoli J, Bouwmeester HJ, Harrison MJ(2019) A CLE-SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nat Plants 5: 933–939 [DOI] [PubMed] [Google Scholar]
- Müller LM, Harrison MJ(2019) Phytohormones, miRNAs, and peptide signals integrate plant phosphorus status with arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 50: 132–139 [DOI] [PubMed] [Google Scholar]
- Niu X, Chen S, Li J, Liu Y, Ji W, Li H(2019) Genome-wide identification of GRAS genes in Brachypodium distachyon and functional characterization of BdSLR1 and BdSLRL1. BMC Genomics 20: 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Floss DS, Levesque-Tremblay V, Bravo A, Harrison MJ(2015) Hyphal branching during arbuscule development requires Reduced Arbuscular Mycorrhiza1. Plant Physiol 169: 2774–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP(2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimprikar P, Carbonnel S, Paries M, Katzer K, Klingl V, Bohmer MJ, Karl L, Floss DS, Harrison MJ, Parniske M, et al. (2016) A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr Biol 26: 987–998 [DOI] [PubMed] [Google Scholar]
- Pimprikar P, Gutjahr C(2018) Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol 59: 673–690 [DOI] [PubMed] [Google Scholar]
- Rich MK, Courty PE, Roux C, Reinhardt D(2017) Role of the GRAS transcription factor ATA/RAM1 in the transcriptional reprogramming of arbuscular mycorrhiza in Petunia hybrida. BMC Genomics 18: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich MK, Schorderet M, Bapaume L, Falquet L, Morel P, Vandenbussche M, Reinhardt D(2015) The Petunia GRAS transcription factor ATA/RAM1 regulates symbiotic gene expression and fungal morphogenesis in arbuscular mycorrhiza. Plant Physiol 168: 788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y, Yamaguchi S(2014) Strigolactone biosynthesis and perception. Curr Opin Plant Biol 21: 1–6 [DOI] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM(2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA 99: 9043–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole V, Peraldi A, Worland B, Nicholson P, Doonan JH, Vain P(2012) T-DNA mutagenesis in Brachypodium distachyon. J Exp Bot 63: 567–576 [DOI] [PubMed] [Google Scholar]
- Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C(2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58: 803–816 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Unterholzner SJ, Rozhon W, Papacek M, Ciomas J, Lange T, Kugler KG, Mayer KF, Sieberer T, Poppenberger B(2015) Brassinosteroids are master regulators of gibberellin biosynthesis in Arabidopsis. Plant Cell 27: 2261–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts-Williams SJ, Emmett BD, Levesque-Tremblay V, MacLean AM, Sun X, Satterlee JW, Fei Z, Harrison MJ(2019) Diverse Sorghum bicolor accessions show marked variation in growth and transcriptional responses to arbuscular mycorrhizal fungi. Plant Cell Environ 42: 1758–1774 [DOI] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y(2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Cui H, Buer B, Vijayakumar V, Delaux P-M, Junkermann S, Bucher M(2015) Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol 167: 854–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Luo D, Zhang X, Liu J, Wang W, Jin Y, Dong W, Liu J, Liu H, Yang W, et al. (2014) A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res 24: 130–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Blaylock LA, Harrison MJ(2010) Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 22: 1483–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]